

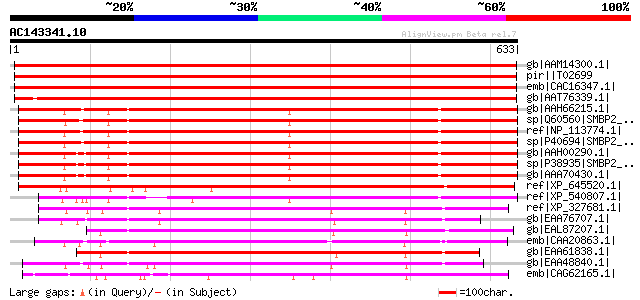
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC143341.10 + phase: 0
(633 letters)
Database: nr
2,540,612 sequences; 863,360,394 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
gb|AAM14300.1| putative helicase [Arabidopsis thaliana] gi|15027... 1022 0.0
pir||T02699 probable helicase At2g03270 [imported] - Arabidopsis... 1022 0.0
emb|CAC16347.1| putative helicase [Arabidopsis thaliana] 1021 0.0
gb|AAT76339.1| putative DNA helicase [Oryza sativa (japonica cul... 951 0.0
gb|AAH66215.1| Ighmbp2 protein [Mus musculus] 528 e-148
sp|Q60560|SMBP2_MESAU DNA-binding protein SMUBP-2 (Immunoglobuli... 526 e-148
ref|NP_113774.1| immunoglobulin mu binding protein 2 [Rattus nor... 526 e-147
sp|P40694|SMBP2_MOUSE DNA-binding protein SMUBP-2 (Immunoglobuli... 524 e-147
gb|AAH00290.1| IGHMBP2 protein [Homo sapiens] 520 e-146
sp|P38935|SMBP2_HUMAN DNA-binding protein SMUBP-2 (Immunoglobuli... 520 e-146
gb|AAA70430.1| DNA helicase [Homo sapiens] 518 e-145
ref|XP_645520.1| hypothetical protein DDB0168508 [Dictyostelium ... 485 e-135
ref|XP_540807.1| PREDICTED: similar to DNA helicase [Canis famil... 463 e-129
ref|XP_327681.1| hypothetical protein [Neurospora crassa] gi|289... 428 e-118
gb|EAA76707.1| hypothetical protein FG06867.1 [Gibberella zeae P... 416 e-114
gb|EAL87207.1| DNA helicase, putative [Aspergillus fumigatus Af293] 412 e-113
emb|CAA20863.1| SPCC737.07c [Schizosaccharomyces pombe] gi|19075... 407 e-112
gb|EAA61838.1| hypothetical protein AN7652.2 [Aspergillus nidula... 395 e-108
gb|EAA48840.1| hypothetical protein MG00498.4 [Magnaporthe grise... 387 e-106
emb|CAG62165.1| unnamed protein product [Candida glabrata CBS138... 379 e-103
>gb|AAM14300.1| putative helicase [Arabidopsis thaliana] gi|15027927|gb|AAK76494.1|
putative helicase [Arabidopsis thaliana]
gi|20197741|gb|AAD17447.2| putative helicase
[Arabidopsis thaliana] gi|20197343|gb|AAM15033.1|
putative helicase [Arabidopsis thaliana]
gi|18395518|ref|NP_565299.1| DNA-binding protein,
putative [Arabidopsis thaliana]
Length = 639
Score = 1022 bits (2643), Expect = 0.0
Identities = 519/628 (82%), Positives = 575/628 (90%), Gaps = 1/628 (0%)
Query: 6 SFMILLFFFHQDAEISSSIATGASRNLDTAQKRGSTILNLKCVDVQTGLMGKSLIELQST 65
S M L ++AEIS S+ +GASRN++TAQK+G+TILNLKCVDVQTGLMGKSLIE QS
Sbjct: 12 STMAPLIDMEKEAEISMSLTSGASRNIETAQKKGTTILNLKCVDVQTGLMGKSLIEFQSN 71
Query: 66 KADVLPAHKFGTHDVVVLKLNKADLGSPALGQGVVYRLKDSSITVAFDDIPEDGLNSPLR 125
K DVLPAHKFG HDVVVLKLNK+DLGS L QGVVYRLKDSSITV FD++PE+GLN+ LR
Sbjct: 72 KGDVLPAHKFGNHDVVVLKLNKSDLGSSPLAQGVVYRLKDSSITVVFDEVPEEGLNTSLR 131
Query: 126 LEKVANEVTYHRMKDALIQLSKGVHKGPASDLIPVLFGERQPTVSKKDVV-FTSINKNLD 184
LEK+ANEVTY RMKD LIQLSKGV +GPASDL+PVLFGERQP+VSKKDV FT NKNLD
Sbjct: 132 LEKLANEVTYRRMKDTLIQLSKGVLRGPASDLVPVLFGERQPSVSKKDVKSFTPFNKNLD 191
Query: 185 YSQKDAISKALSSKNVFLLHGPPGTGKTTTVVEIILQEVKRGSKILACAASNIAVDNIVE 244
SQKDAI+KALSSK+VFLLHGPPGTGKTTTVVEI+LQEVKRGSKILACAASNIAVDNIVE
Sbjct: 192 QSQKDAITKALSSKDVFLLHGPPGTGKTTTVVEIVLQEVKRGSKILACAASNIAVDNIVE 251
Query: 245 RLVPHRVKLVRIGHPARLLPQVVDSALDAQVLRGDNSGLANDIRKEMKVLNGKLLKTKEK 304
RLVPH+VKLVR+GHPARLLPQV+DSALDAQVL+GDNSGLANDIRKEMK LNGKLLK K+K
Sbjct: 252 RLVPHKVKLVRVGHPARLLPQVLDSALDAQVLKGDNSGLANDIRKEMKALNGKLLKAKDK 311
Query: 305 NTRREIQKELRTLSREERKRQQLAVTDVIKTSDVILTTLIGASSKKLGNTSFDLVIIDEA 364
NTRR IQKELRTL +EERKRQQLAV+DVIK +DVILTTL GA ++KL N +FDLVIIDE
Sbjct: 312 NTRRLIQKELRTLGKEERKRQQLAVSDVIKNADVILTTLTGALTRKLDNRTFDLVIIDEG 371
Query: 365 AQALEVACWIPLLKGTRCILAGDHLQLPPTIQSVEAEKKGLGRTLFERLAELYGDEVTSM 424
AQALEVACWI LLKG+RCILAGDHLQLPPTIQS EAE+KGLGRTLFERLA+LYGDE+ SM
Sbjct: 372 AQALEVACWIALLKGSRCILAGDHLQLPPTIQSAEAERKGLGRTLFERLADLYGDEIKSM 431
Query: 425 LTVQYRMHQLIMDWSSKELYNSKVKAHACVASHMLYDLEGVKKTSSTEPTLLLIDTAGCD 484
LTVQYRMH+LIM+WSSKELY++K+ AH+ VASHML+DLE V K+SSTE TLLL+DTAGCD
Sbjct: 432 LTVQYRMHELIMNWSSKELYDNKITAHSSVASHMLFDLENVTKSSSTEATLLLVDTAGCD 491
Query: 485 MEEKKDEEDSTLNEGESEVAMAHAKRLVQSGVLPSDIGIITPYAAQVVLLKMLKNKENSL 544
MEEKKDEE+ST NEGE+EVAMAHAKRL++SGV PSDIGIITPYAAQV+LL++L+ KE L
Sbjct: 492 MEEKKDEEESTYNEGEAEVAMAHAKRLMESGVQPSDIGIITPYAAQVMLLRILRGKEEKL 551
Query: 545 KDIEISTVDGFQGREKEAIIISMVRSNSKKEVGFLSDRRRMNVAVTRARRQCCIVCDTET 604
KD+EISTVDGFQGREKEAIIISMVRSNSKKEVGFL D+RRMNVAVTR+RRQCCIVCDTET
Sbjct: 552 KDMEISTVDGFQGREKEAIIISMVRSNSKKEVGFLKDQRRMNVAVTRSRRQCCIVCDTET 611
Query: 605 VSSDGFLKRLIEYFEEHGEYQSASEYQN 632
VSSD FLKR+IEYFEEHGEY SASEY N
Sbjct: 612 VSSDAFLKRMIEYFEEHGEYLSASEYTN 639
>pir||T02699 probable helicase At2g03270 [imported] - Arabidopsis thaliana
Length = 635
Score = 1022 bits (2643), Expect = 0.0
Identities = 519/628 (82%), Positives = 575/628 (90%), Gaps = 1/628 (0%)
Query: 6 SFMILLFFFHQDAEISSSIATGASRNLDTAQKRGSTILNLKCVDVQTGLMGKSLIELQST 65
S M L ++AEIS S+ +GASRN++TAQK+G+TILNLKCVDVQTGLMGKSLIE QS
Sbjct: 8 STMAPLIDMEKEAEISMSLTSGASRNIETAQKKGTTILNLKCVDVQTGLMGKSLIEFQSN 67
Query: 66 KADVLPAHKFGTHDVVVLKLNKADLGSPALGQGVVYRLKDSSITVAFDDIPEDGLNSPLR 125
K DVLPAHKFG HDVVVLKLNK+DLGS L QGVVYRLKDSSITV FD++PE+GLN+ LR
Sbjct: 68 KGDVLPAHKFGNHDVVVLKLNKSDLGSSPLAQGVVYRLKDSSITVVFDEVPEEGLNTSLR 127
Query: 126 LEKVANEVTYHRMKDALIQLSKGVHKGPASDLIPVLFGERQPTVSKKDVV-FTSINKNLD 184
LEK+ANEVTY RMKD LIQLSKGV +GPASDL+PVLFGERQP+VSKKDV FT NKNLD
Sbjct: 128 LEKLANEVTYRRMKDTLIQLSKGVLRGPASDLVPVLFGERQPSVSKKDVKSFTPFNKNLD 187
Query: 185 YSQKDAISKALSSKNVFLLHGPPGTGKTTTVVEIILQEVKRGSKILACAASNIAVDNIVE 244
SQKDAI+KALSSK+VFLLHGPPGTGKTTTVVEI+LQEVKRGSKILACAASNIAVDNIVE
Sbjct: 188 QSQKDAITKALSSKDVFLLHGPPGTGKTTTVVEIVLQEVKRGSKILACAASNIAVDNIVE 247
Query: 245 RLVPHRVKLVRIGHPARLLPQVVDSALDAQVLRGDNSGLANDIRKEMKVLNGKLLKTKEK 304
RLVPH+VKLVR+GHPARLLPQV+DSALDAQVL+GDNSGLANDIRKEMK LNGKLLK K+K
Sbjct: 248 RLVPHKVKLVRVGHPARLLPQVLDSALDAQVLKGDNSGLANDIRKEMKALNGKLLKAKDK 307
Query: 305 NTRREIQKELRTLSREERKRQQLAVTDVIKTSDVILTTLIGASSKKLGNTSFDLVIIDEA 364
NTRR IQKELRTL +EERKRQQLAV+DVIK +DVILTTL GA ++KL N +FDLVIIDE
Sbjct: 308 NTRRLIQKELRTLGKEERKRQQLAVSDVIKNADVILTTLTGALTRKLDNRTFDLVIIDEG 367
Query: 365 AQALEVACWIPLLKGTRCILAGDHLQLPPTIQSVEAEKKGLGRTLFERLAELYGDEVTSM 424
AQALEVACWI LLKG+RCILAGDHLQLPPTIQS EAE+KGLGRTLFERLA+LYGDE+ SM
Sbjct: 368 AQALEVACWIALLKGSRCILAGDHLQLPPTIQSAEAERKGLGRTLFERLADLYGDEIKSM 427
Query: 425 LTVQYRMHQLIMDWSSKELYNSKVKAHACVASHMLYDLEGVKKTSSTEPTLLLIDTAGCD 484
LTVQYRMH+LIM+WSSKELY++K+ AH+ VASHML+DLE V K+SSTE TLLL+DTAGCD
Sbjct: 428 LTVQYRMHELIMNWSSKELYDNKITAHSSVASHMLFDLENVTKSSSTEATLLLVDTAGCD 487
Query: 485 MEEKKDEEDSTLNEGESEVAMAHAKRLVQSGVLPSDIGIITPYAAQVVLLKMLKNKENSL 544
MEEKKDEE+ST NEGE+EVAMAHAKRL++SGV PSDIGIITPYAAQV+LL++L+ KE L
Sbjct: 488 MEEKKDEEESTYNEGEAEVAMAHAKRLMESGVQPSDIGIITPYAAQVMLLRILRGKEEKL 547
Query: 545 KDIEISTVDGFQGREKEAIIISMVRSNSKKEVGFLSDRRRMNVAVTRARRQCCIVCDTET 604
KD+EISTVDGFQGREKEAIIISMVRSNSKKEVGFL D+RRMNVAVTR+RRQCCIVCDTET
Sbjct: 548 KDMEISTVDGFQGREKEAIIISMVRSNSKKEVGFLKDQRRMNVAVTRSRRQCCIVCDTET 607
Query: 605 VSSDGFLKRLIEYFEEHGEYQSASEYQN 632
VSSD FLKR+IEYFEEHGEY SASEY N
Sbjct: 608 VSSDAFLKRMIEYFEEHGEYLSASEYTN 635
>emb|CAC16347.1| putative helicase [Arabidopsis thaliana]
Length = 635
Score = 1021 bits (2639), Expect = 0.0
Identities = 518/628 (82%), Positives = 574/628 (90%), Gaps = 1/628 (0%)
Query: 6 SFMILLFFFHQDAEISSSIATGASRNLDTAQKRGSTILNLKCVDVQTGLMGKSLIELQST 65
S M L ++AEIS S+ +GASRN++TAQK+G+TILNLKCVDVQTGLMGKSLIE QS
Sbjct: 8 STMAPLIDMEKEAEISMSLTSGASRNIETAQKKGTTILNLKCVDVQTGLMGKSLIEFQSN 67
Query: 66 KADVLPAHKFGTHDVVVLKLNKADLGSPALGQGVVYRLKDSSITVAFDDIPEDGLNSPLR 125
K D LPAHKFG HDVVVLKLNK+DLGS L QGVVYRLKDSSITV FD++PE+GLN+ LR
Sbjct: 68 KGDALPAHKFGNHDVVVLKLNKSDLGSSPLAQGVVYRLKDSSITVVFDEVPEEGLNTSLR 127
Query: 126 LEKVANEVTYHRMKDALIQLSKGVHKGPASDLIPVLFGERQPTVSKKDVV-FTSINKNLD 184
LEK+ANEVTY RMKD LIQLSKGV +GPASDL+PVLFGERQP+VSKKDV FT NKNLD
Sbjct: 128 LEKLANEVTYRRMKDTLIQLSKGVLRGPASDLVPVLFGERQPSVSKKDVKSFTPFNKNLD 187
Query: 185 YSQKDAISKALSSKNVFLLHGPPGTGKTTTVVEIILQEVKRGSKILACAASNIAVDNIVE 244
SQKDAI+KALSSK+VFLLHGPPGTGKTTTVVEI+LQEVKRGSKILACAASNIAVDNIVE
Sbjct: 188 QSQKDAITKALSSKDVFLLHGPPGTGKTTTVVEIVLQEVKRGSKILACAASNIAVDNIVE 247
Query: 245 RLVPHRVKLVRIGHPARLLPQVVDSALDAQVLRGDNSGLANDIRKEMKVLNGKLLKTKEK 304
RLVPH+VKLVR+GHPARLLPQV+DSALDAQVL+GDNSGLANDIRKEMK LNGKLLK K+K
Sbjct: 248 RLVPHKVKLVRVGHPARLLPQVLDSALDAQVLKGDNSGLANDIRKEMKALNGKLLKAKDK 307
Query: 305 NTRREIQKELRTLSREERKRQQLAVTDVIKTSDVILTTLIGASSKKLGNTSFDLVIIDEA 364
NTRR IQKELRTL +EERKRQQLAV+DVIK +DVILTTL GA ++KL N +FDLVIIDE
Sbjct: 308 NTRRLIQKELRTLGKEERKRQQLAVSDVIKNADVILTTLTGALTRKLDNRTFDLVIIDEG 367
Query: 365 AQALEVACWIPLLKGTRCILAGDHLQLPPTIQSVEAEKKGLGRTLFERLAELYGDEVTSM 424
AQALEVACWI LLKG+RCILAGDHLQLPPTIQS EAE+KGLGRTLFERLA+LYGDE+ SM
Sbjct: 368 AQALEVACWIALLKGSRCILAGDHLQLPPTIQSAEAERKGLGRTLFERLADLYGDEIKSM 427
Query: 425 LTVQYRMHQLIMDWSSKELYNSKVKAHACVASHMLYDLEGVKKTSSTEPTLLLIDTAGCD 484
LTVQYRMH+LIM+WSSKELY++K+ AH+ VASHML+DLE V K+SSTE TLLL+DTAGCD
Sbjct: 428 LTVQYRMHELIMNWSSKELYDNKITAHSSVASHMLFDLENVTKSSSTEATLLLVDTAGCD 487
Query: 485 MEEKKDEEDSTLNEGESEVAMAHAKRLVQSGVLPSDIGIITPYAAQVVLLKMLKNKENSL 544
MEEKKDEE+ST NEGE+EVAMAHAKRL++SGV PSDIGIITPYAAQV+LL++L+ KE L
Sbjct: 488 MEEKKDEEESTYNEGEAEVAMAHAKRLMESGVQPSDIGIITPYAAQVMLLRILRGKEEKL 547
Query: 545 KDIEISTVDGFQGREKEAIIISMVRSNSKKEVGFLSDRRRMNVAVTRARRQCCIVCDTET 604
KD+EISTVDGFQGREKEAIIISMVRSNSKKEVGFL D+RRMNVAVTR+RRQCCIVCDTET
Sbjct: 548 KDMEISTVDGFQGREKEAIIISMVRSNSKKEVGFLKDQRRMNVAVTRSRRQCCIVCDTET 607
Query: 605 VSSDGFLKRLIEYFEEHGEYQSASEYQN 632
VSSD FLKR+IEYFEEHGEY SASEY N
Sbjct: 608 VSSDAFLKRMIEYFEEHGEYLSASEYTN 635
>gb|AAT76339.1| putative DNA helicase [Oryza sativa (japonica cultivar-group)]
Length = 651
Score = 951 bits (2459), Expect = 0.0
Identities = 481/627 (76%), Positives = 545/627 (86%), Gaps = 4/627 (0%)
Query: 6 SFMILLFFFHQDAEISSSIATGASRNLDTAQKRGSTILNLKCVDVQTGLMGKSLIELQST 65
S M L + AEIS+ AT + T ++RG + NLKC D QTGLMGK+L+E Q
Sbjct: 29 SSMAPLIDLEKAAEISAESATSSK----TLERRGCVMANLKCTDAQTGLMGKTLLEFQPN 84
Query: 66 KADVLPAHKFGTHDVVVLKLNKADLGSPALGQGVVYRLKDSSITVAFDDIPEDGLNSPLR 125
K DVLP HKFGTHDVV LK NKAD GS ALGQGVVYRLKDSSITVAFDDIPEDGLNSPLR
Sbjct: 85 KGDVLPPHKFGTHDVVALKPNKADAGSAALGQGVVYRLKDSSITVAFDDIPEDGLNSPLR 144
Query: 126 LEKVANEVTYHRMKDALIQLSKGVHKGPASDLIPVLFGERQPTVSKKDVVFTSINKNLDY 185
LEK+ANEVTY RMKDAL+QLSKG+ GP+++LIPVLFGE P SK F+ NKNLD
Sbjct: 145 LEKLANEVTYRRMKDALVQLSKGIQTGPSANLIPVLFGENPPMSSKDVAKFSPFNKNLDE 204
Query: 186 SQKDAISKALSSKNVFLLHGPPGTGKTTTVVEIILQEVKRGSKILACAASNIAVDNIVER 245
SQKDAISKAL S++VFLLHGPPGTGKTTT++EIILQEVKRGSKILACAASNIAVDNIVER
Sbjct: 205 SQKDAISKALRSRDVFLLHGPPGTGKTTTIIEIILQEVKRGSKILACAASNIAVDNIVER 264
Query: 246 LVPHRVKLVRIGHPARLLPQVVDSALDAQVLRGDNSGLANDIRKEMKVLNGKLLKTKEKN 305
L +R KLVR+GHPARLLPQV+DSALDAQVLR DNS LA DIRKEMKVLN KLLK K+KN
Sbjct: 265 LSRYRTKLVRLGHPARLLPQVLDSALDAQVLRADNSSLAGDIRKEMKVLNSKLLKAKDKN 324
Query: 306 TRREIQKELRTLSREERKRQQLAVTDVIKTSDVILTTLIGASSKKLGNTSFDLVIIDEAA 365
T+R+I+KELRTL++EERKRQQLAV DVIK +DV+L+TL GASSKKL +FDLVIIDEAA
Sbjct: 325 TKRDIRKELRTLAKEERKRQQLAVADVIKNADVVLSTLTGASSKKLDGITFDLVIIDEAA 384
Query: 366 QALEVACWIPLLKGTRCILAGDHLQLPPTIQSVEAEKKGLGRTLFERLAELYGDEVTSML 425
QALE+ACWI LLKG RC+LAGDHLQLPPTIQS EAEKKG+G+TLFERL E YGD++TSML
Sbjct: 385 QALEMACWIALLKGPRCVLAGDHLQLPPTIQSAEAEKKGMGKTLFERLTEAYGDQITSML 444
Query: 426 TVQYRMHQLIMDWSSKELYNSKVKAHACVASHMLYDLEGVKKTSSTEPTLLLIDTAGCDM 485
T+QYRMH+LIM+WSSKELYN+K+KAH+ VA HMLYD+E VK++SSTEPT++LIDT GCDM
Sbjct: 445 TIQYRMHELIMNWSSKELYNNKIKAHSSVADHMLYDIEEVKRSSSTEPTIILIDTTGCDM 504
Query: 486 EEKKDEEDSTLNEGESEVAMAHAKRLVQSGVLPSDIGIITPYAAQVVLLKMLKNKENSLK 545
EE KDEE+ST+NEGE+ V++AHAK LV+SGV SDIGIITPYAAQV LKM++NK+ LK
Sbjct: 505 EEVKDEEESTMNEGEAAVSIAHAKLLVESGVRASDIGIITPYAAQVTCLKMMRNKDTKLK 564
Query: 546 DIEISTVDGFQGREKEAIIISMVRSNSKKEVGFLSDRRRMNVAVTRARRQCCIVCDTETV 605
D+EISTVDGFQGREKEAIIISMVRSNSKKEVGFLSD RRMNVAVTRARRQCC+VCD ETV
Sbjct: 565 DLEISTVDGFQGREKEAIIISMVRSNSKKEVGFLSDHRRMNVAVTRARRQCCLVCDVETV 624
Query: 606 SSDGFLKRLIEYFEEHGEYQSASEYQN 632
S+D FLKRL+EYFEE+GEY SASEYQ+
Sbjct: 625 SNDKFLKRLVEYFEENGEYLSASEYQS 651
>gb|AAH66215.1| Ighmbp2 protein [Mus musculus]
Length = 866
Score = 528 bits (1359), Expect = e-148
Identities = 295/635 (46%), Positives = 408/635 (63%), Gaps = 18/635 (2%)
Query: 11 LFFFHQDAEISSSIATGASRNLDTAQKRGSTILNLKCVDVQTGLMGKSLIELQSTK---A 67
L +DAE+ + +L Q RG +L L+ +TGL G+ L+ + K A
Sbjct: 16 LLELERDAEVEERRSWQEHSSLRELQSRGVCLLKLQVSSQRTGLYGQRLVTFEPRKFGPA 75
Query: 68 DVLPAHKFGTHDVVVLKLNKADLGSPALGQGVVYRLKDSSITVAFDDIPEDGLN----SP 123
VLP++ F + D+V L + L GV+ R+ S+TVAFD+ + LN +
Sbjct: 76 VVLPSNSFTSGDIVGLYDTNEN---SQLATGVLTRITQKSVTVAFDESHDLQLNLDRENT 132
Query: 124 LRLEKVANEVTYHRMKDALIQLSKGVHKGPASDLIPVLFGERQPTVSKKDVVFTSINKNL 183
RL K+AN+VTY R+K AL+ L K H GPAS LI +L G P+ + + + N L
Sbjct: 133 YRLLKLANDVTYKRLKKALMTLKK-YHSGPASSLIDILLGSSTPSPAMEIPPLSFYNTTL 191
Query: 184 DYSQKDAISKALSSKNVFLLHGPPGTGKTTTVVEIILQEVKRGSKILACAASNIAVDNIV 243
D SQK+A+S AL+ K + ++HGPPGTGKTTTVVEIILQ VK+G K+L CA SNIAVDN+V
Sbjct: 192 DLSQKEAVSFALAQKELAIIHGPPGTGKTTTVVEIILQAVKQGLKVLCCAPSNIAVDNLV 251
Query: 244 ERLVPHRVKLVRIGHPARLLPQVVDSALDAQVLRGDNSGLANDIRKEMKVLNGKLLKTKE 303
ERL + +++R+GHPARLL V +LDA + R DN+ + DIR+++ + GK KT++
Sbjct: 252 ERLALCKKRILRLGHPARLLESVQHHSLDAVLARSDNAQIVADIRRDIDQVFGKNKKTQD 311
Query: 304 KNTRREIQKELRTLSREERKRQQLAVTDVIKTSDVILTTLIGASS----KKLGNTSFDLV 359
K + + E++ L RE ++R++ A+ + +DV+L T GASS K L FD+V
Sbjct: 312 KREKGNFRSEIKLLRRELKEREEAAIVQSLTAADVVLATNTGASSDGPLKLLPEDYFDVV 371
Query: 360 IIDEAAQALEVACWIPLLKGTRCILAGDHLQLPPTIQSVEAEKKGLGRTLFERLAELYGD 419
++DE AQALE +CWIPLLK +CILAGDH QLPPT S A GL R+L ERLAE +G
Sbjct: 372 VVDECAQALEASCWIPLLKAPKCILAGDHRQLPPTTASHRAALAGLSRSLMERLAEKHGA 431
Query: 420 EVTSMLTVQYRMHQLIMDWSSKELYNSKVKAHACVASHMLYDLEGVKKTSSTEPTLLLID 479
V MLTVQYRMHQ IM W+S+ +Y+ ++ +H VA H+L DL GV T T LLLID
Sbjct: 432 GVVRMLTVQYRMHQAIMCWASEAMYHGQLTSHPSVAGHLLKDLPGVTDTEETRVPLLLID 491
Query: 480 TAGCDMEEKKDEE-DSTLNEGESEVAMAHAKRLVQSGVLPSDIGIITPYAAQVVLLKMLK 538
TAGC + E ++E+ S N GE + H + LV +GV DI +I PY QV LL+ +
Sbjct: 492 TAGCGLLELEEEDSQSKGNPGEVRLVTLHIQALVDAGVQAGDIAVIAPYNLQVDLLR--Q 549
Query: 539 NKENSLKDIEISTVDGFQGREKEAIIISMVRSNSKKEVGFLSDRRRMNVAVTRARRQCCI 598
+ N ++EI +VDGFQGREKEA++++ VRSN K EVGFL++ RR+NVAVTRARR +
Sbjct: 550 SLSNKHPELEIKSVDGFQGREKEAVLLTFVRSNRKGEVGFLAEDRRINVAVTRARRHVAV 609
Query: 599 VCDTETVSSDGFLKRLIEYFEEHGEYQSASEYQNE 633
+CD+ TV++ FLK L++YF EHGE ++A EY ++
Sbjct: 610 ICDSHTVNNHAFLKTLVDYFTEHGEVRTAFEYLDD 644
>sp|Q60560|SMBP2_MESAU DNA-binding protein SMUBP-2 (Immunoglobulin mu binding protein 2)
(SMUBP-2) (Insulin II gene enhancer-binding protein)
(RIPE3B-binding complex 3B2 p110 subunit) (RIP-1)
gi|290919|gb|AAB00104.1| insulin II gene
enhancer-binding protein
Length = 989
Score = 526 bits (1356), Expect = e-148
Identities = 301/637 (47%), Positives = 406/637 (63%), Gaps = 22/637 (3%)
Query: 11 LFFFHQDAEISSSIATGASRNLDTAQKRGSTILNLKCVDVQTGLMGKSLIELQSTKAD-- 68
L +DAE+ + +L Q RG +L L+ TGL G+ L+ + K
Sbjct: 16 LLELERDAEVEERRSWQEHSSLKELQSRGVCLLKLQVSSQCTGLYGQRLVTFEPRKLGPV 75
Query: 69 -VLPAHKFGTHDVVVL-KLNKADLGSPALGQGVVYRLKDSSITVAFDDIPEDGLN----S 122
VLP++ F + D+V L N+ S L GV+ R+ S+TVAFD+ + LN +
Sbjct: 76 VVLPSNSFTSGDIVGLYDANE----SSQLATGVLTRITQKSVTVAFDESHDFQLNLDREN 131
Query: 123 PLRLEKVANEVTYHRMKDALIQLSKGVHKGPASDLIPVLFGERQPTVSKKDVVFTSINKN 182
RL K+AN+VTY R+K AL+ L K H GPAS LI VL G P+ + + FT N
Sbjct: 132 TYRLLKLANDVTYKRLKKALMTLKK-YHSGPASSLIDVLLGGSSPSPTTEIPPFTFYNTA 190
Query: 183 LDYSQKDAISKALSSKNVFLLHGPPGTGKTTTVVEIILQEVKRGSKILACAASNIAVDNI 242
LD SQK+A+S AL+ K V ++HGPPGTGKTTTVVEIILQ VK+G KIL CA SN+AVDN+
Sbjct: 191 LDPSQKEAVSFALAQKEVAIIHGPPGTGKTTTVVEIILQAVKQGLKILCCAPSNVAVDNL 250
Query: 243 VERLVPHRVKLVRIGHPARLLPQVVDSALDAQVLRGDNSGLANDIRKEMKVLNGKLLKTK 302
VERL + +++R+GHPARLL +LDA + R DN+ + DIRK++ + GK KT+
Sbjct: 251 VERLALCKKRILRLGHPARLLESAQQHSLDAVLARSDNAQIVADIRKDIDQVFGKNKKTQ 310
Query: 303 EKNTRREIQKELRTLSREERKRQQLAVTDVIKTSDVILTTLIGASS----KKLGNTSFDL 358
+K + + E++ L +E ++R++ A+ + +DV+L T GASS K L FD+
Sbjct: 311 DKREKSNFRNEIKLLRKELKEREEAAIVQSLTAADVVLATNTGASSDGPLKLLPENHFDV 370
Query: 359 VIIDEAAQALEVACWIPLLKGTRCILAGDHLQLPPTIQSVEAEKKGLGRTLFERLAELYG 418
V++DE AQALE +CWIPLLK +CILAGDH QLPPT S +A GL R+L ERL E +G
Sbjct: 371 VVVDECAQALEASCWIPLLKAPKCILAGDHRQLPPTTISHKAALAGLSRSLMERLVEKHG 430
Query: 419 DEVTSMLTVQYRMHQLIMDWSSKELYNSKVKAHACVASHMLYDLEGVKKTSSTEPTLLLI 478
MLTVQYRMHQ I W+S+ +Y+ ++ AH VA H+L DL GV T T LLLI
Sbjct: 431 AGAVRMLTVQYRMHQAITRWASEAMYHGQLTAHPSVAGHLLKDLPGVADTEETSVPLLLI 490
Query: 479 DTAGCDMEEKKDEEDSTL--NEGESEVAMAHAKRLVQSGVLPSDIGIITPYAAQVVLLKM 536
DTAGC + E DEEDS N GE + H + LV +GV DI +I PY QV LL+
Sbjct: 491 DTAGCGLLE-LDEEDSQSKGNPGEVRLVTLHIQALVDAGVHAGDIAVIAPYNLQVDLLR- 548
Query: 537 LKNKENSLKDIEISTVDGFQGREKEAIIISMVRSNSKKEVGFLSDRRRMNVAVTRARRQC 596
++ N ++EI +VDGFQGREKEA+I++ VRSN K EVGFL++ RR+NVAVTRARR
Sbjct: 549 -QSLSNKHPELEIKSVDGFQGREKEAVILTFVRSNRKGEVGFLAEDRRINVAVTRARRHV 607
Query: 597 CIVCDTETVSSDGFLKRLIEYFEEHGEYQSASEYQNE 633
++CD+ TV++ FLK L++YF EHGE ++A EY ++
Sbjct: 608 AVICDSRTVNNHAFLKTLVDYFTEHGEVRTAFEYLDD 644
>ref|NP_113774.1| immunoglobulin mu binding protein 2 [Rattus norvegicus]
gi|11066349|gb|AAG28561.1| antifreeze-enhancer binding
protein AEP [Rattus norvegicus]
Length = 988
Score = 526 bits (1354), Expect = e-147
Identities = 296/635 (46%), Positives = 406/635 (63%), Gaps = 18/635 (2%)
Query: 11 LFFFHQDAEISSSIATGASRNLDTAQKRGSTILNLKCVDVQTGLMGKSLIELQSTK---A 67
L +DAE+ + +L Q RG +L L+ +TGL G+ L+ + K A
Sbjct: 16 LLELERDAEVEERRSWQEHSSLKELQSRGVCLLKLQVSGQRTGLYGQRLVTFEPRKFGPA 75
Query: 68 DVLPAHKFGTHDVVVLKLNKADLGSPALGQGVVYRLKDSSITVAFDDIPEDGLN----SP 123
VLP++ F + D+V L S L GV+ R+ S+ VAFD+ + LN +
Sbjct: 76 VVLPSNSFTSGDIVGLYDTNE---SSQLATGVLTRITQKSVIVAFDESHDFQLNLDRENT 132
Query: 124 LRLEKVANEVTYHRMKDALIQLSKGVHKGPASDLIPVLFGERQPTVSKKDVVFTSINKNL 183
RL K+AN+VTY R+K AL+ L K H GPAS LI VL G P+ + + T N L
Sbjct: 133 YRLLKLANDVTYKRLKKALLTLKK-YHSGPASSLIDVLLGGSTPSPATEIPPLTFYNTTL 191
Query: 184 DYSQKDAISKALSSKNVFLLHGPPGTGKTTTVVEIILQEVKRGSKILACAASNIAVDNIV 243
D SQK+A+S AL+ K V ++HGPPGTGKTTTVVEIILQ VK+G K+L CA SNIAVDN+V
Sbjct: 192 DPSQKEAVSFALAQKEVAIIHGPPGTGKTTTVVEIILQAVKQGLKVLCCAPSNIAVDNLV 251
Query: 244 ERLVPHRVKLVRIGHPARLLPQVVDSALDAQVLRGDNSGLANDIRKEMKVLNGKLLKTKE 303
ERL + +++R+GHPARLL V +LDA + R DN+ + DIR+++ + GK KT++
Sbjct: 252 ERLALCKKQILRLGHPARLLESVQQHSLDAVLARSDNAQIVADIRRDIDQVFGKNKKTQD 311
Query: 304 KNTRREIQKELRTLSREERKRQQLAVTDVIKTSDVILTTLIGASS----KKLGNTSFDLV 359
K + + E++ L +E ++R++ A+ + +DV+L T GAS+ K L FD+V
Sbjct: 312 KREKSNFRNEIKLLRKELKEREEAAIVQSLSAADVVLATNTGASTDGPLKLLPEDYFDVV 371
Query: 360 IIDEAAQALEVACWIPLLKGTRCILAGDHLQLPPTIQSVEAEKKGLGRTLFERLAELYGD 419
++DE AQALE +CWIPLLK +CILAGDH QLPPT S +A GL R+L ERLAE +G
Sbjct: 372 VVDECAQALEASCWIPLLKAPKCILAGDHKQLPPTTVSHKAALAGLSRSLMERLAEKHGA 431
Query: 420 EVTSMLTVQYRMHQLIMDWSSKELYNSKVKAHACVASHMLYDLEGVKKTSSTEPTLLLID 479
V ML VQYRMHQ I W+S+ +Y+ ++ AH VA H+L DL GV T T LLLID
Sbjct: 432 AVVRMLAVQYRMHQAITRWASEAMYHGQLTAHPSVAGHLLKDLPGVADTEETSVPLLLID 491
Query: 480 TAGCDMEEKKDEE-DSTLNEGESEVAMAHAKRLVQSGVLPSDIGIITPYAAQVVLLKMLK 538
TAGC + E ++E+ S N GE + H + LV +GV DI +I PY QV LL+ +
Sbjct: 492 TAGCGLLELEEEDSQSKGNPGEVRLVTLHIQALVDAGVQAGDIAVIAPYNLQVDLLR--Q 549
Query: 539 NKENSLKDIEISTVDGFQGREKEAIIISMVRSNSKKEVGFLSDRRRMNVAVTRARRQCCI 598
+ N ++EI +VDGFQGREKEA+I++ VRSN K EVGFL++ RR+NVAVTRARR +
Sbjct: 550 SLSNKHPELEIKSVDGFQGREKEAVILTFVRSNRKGEVGFLAEDRRINVAVTRARRHVAV 609
Query: 599 VCDTETVSSDGFLKRLIEYFEEHGEYQSASEYQNE 633
+CD+ TV++ FLK L++YF EHGE ++A EY ++
Sbjct: 610 ICDSHTVNNHAFLKTLVDYFTEHGEVRTAFEYLDD 644
>sp|P40694|SMBP2_MOUSE DNA-binding protein SMUBP-2 (Immunoglobulin mu binding protein 2)
(SMUBP-2) (Cardiac transcription factor 1) (CATF1)
gi|6678029|ref|NP_033238.1| immunoglobulin mu binding
protein 2 [Mus musculus] gi|293806|gb|AAA40143.1|
DNA-binding protein
Length = 993
Score = 524 bits (1349), Expect = e-147
Identities = 293/635 (46%), Positives = 407/635 (63%), Gaps = 18/635 (2%)
Query: 11 LFFFHQDAEISSSIATGASRNLDTAQKRGSTILNLKCVDVQTGLMGKSLIELQSTK---A 67
L +DAE+ + +L Q RG +L L+ +TGL G+ L+ + K A
Sbjct: 16 LLELERDAEVEERRSWQEHSSLRELQSRGVCLLKLQVSSQRTGLYGQRLVTFEPRKFGPA 75
Query: 68 DVLPAHKFGTHDVVVLKLNKADLGSPALGQGVVYRLKDSSITVAFDDIPEDGLN----SP 123
VLP++ F + D+V L + L GV+ R+ S+TVAFD+ + LN +
Sbjct: 76 VVLPSNSFTSGDIVGLYDTNEN---SQLATGVLTRITQKSVTVAFDESHDLQLNLDRENT 132
Query: 124 LRLEKVANEVTYHRMKDALIQLSKGVHKGPASDLIPVLFGERQPTVSKKDVVFTSINKNL 183
RL K+AN+VTY R+K AL+ L K H GPAS LI +L G P+ + + + N L
Sbjct: 133 YRLLKLANDVTYKRLKKALMTLKK-YHSGPASSLIDILLGSSTPSPAMEIPPLSFYNTTL 191
Query: 184 DYSQKDAISKALSSKNVFLLHGPPGTGKTTTVVEIILQEVKRGSKILACAASNIAVDNIV 243
D SQK+A+S AL+ K + ++HGPPGTGKTTTVVEIILQ VK+G K+L CA SNIAVDN+V
Sbjct: 192 DLSQKEAVSFALAQKELAIIHGPPGTGKTTTVVEIILQAVKQGLKVLCCAPSNIAVDNLV 251
Query: 244 ERLVPHRVKLVRIGHPARLLPQVVDSALDAQVLRGDNSGLANDIRKEMKVLNGKLLKTKE 303
ERL + +++R+GHPARLL V +LDA + R DN+ + DIR+++ + GK KT++
Sbjct: 252 ERLALCKKRILRLGHPARLLESVQHHSLDAVLARSDNAQIVADIRRDIDQVFGKNKKTQD 311
Query: 304 KNTRREIQKELRTLSREERKRQQLAVTDVIKTSDVILTTLIGASS----KKLGNTSFDLV 359
K + + E++ L +E ++R++ A+ + +DV+L T GASS K L FD+V
Sbjct: 312 KREKGNFRSEIKLLRKELKEREEAAIVQSLTAADVVLATNTGASSDGPLKLLPEDYFDVV 371
Query: 360 IIDEAAQALEVACWIPLLKGTRCILAGDHLQLPPTIQSVEAEKKGLGRTLFERLAELYGD 419
++DE AQALE +CWIPLLK +CILAGDH QLPPT S A GL R+L ERLAE +G
Sbjct: 372 VVDECAQALEASCWIPLLKAPKCILAGDHRQLPPTTVSHRAALAGLSRSLMERLAEKHGA 431
Query: 420 EVTSMLTVQYRMHQLIMDWSSKELYNSKVKAHACVASHMLYDLEGVKKTSSTEPTLLLID 479
V MLTVQYRMHQ IM W+S+ +Y+ + +H VA H+L DL GV T T LLLID
Sbjct: 432 GVVRMLTVQYRMHQAIMCWASEAMYHGQFTSHPSVAGHLLKDLPGVTDTEETRVPLLLID 491
Query: 480 TAGCDMEEKKDEE-DSTLNEGESEVAMAHAKRLVQSGVLPSDIGIITPYAAQVVLLKMLK 538
TAGC + E ++E+ S N GE + H + LV +GV DI +I PY QV LL+ +
Sbjct: 492 TAGCGLLELEEEDSQSKGNPGEVRLVTLHIQALVDAGVQAGDIAVIAPYNLQVDLLR--Q 549
Query: 539 NKENSLKDIEISTVDGFQGREKEAIIISMVRSNSKKEVGFLSDRRRMNVAVTRARRQCCI 598
+ N ++EI +VDGFQGREKEA++++ VRSN K EVGFL++ RR+NVAVTRARR +
Sbjct: 550 SLSNKHPELEIKSVDGFQGREKEAVLLTFVRSNRKGEVGFLAEDRRINVAVTRARRHVAV 609
Query: 599 VCDTETVSSDGFLKRLIEYFEEHGEYQSASEYQNE 633
+CD+ TV++ FL+ L++YF EHGE ++A EY ++
Sbjct: 610 ICDSHTVNNHAFLETLVDYFTEHGEVRTAFEYLDD 644
>gb|AAH00290.1| IGHMBP2 protein [Homo sapiens]
Length = 868
Score = 520 bits (1340), Expect = e-146
Identities = 294/635 (46%), Positives = 415/635 (65%), Gaps = 17/635 (2%)
Query: 11 LFFFHQDAEISSSIATGASRNLDTAQKRGSTILNLKCVDVQTGLMGKSLIELQSTK---A 67
L +DAE+ + + +L Q RG +L L+ +TGL G+ L+ + + A
Sbjct: 16 LLELERDAEVEERRSWQENISLKELQSRGVCLLKLQVSSQRTGLYGRLLVTFEPRRYGSA 75
Query: 68 DVLPAHKFGTHDVVVLKLNKADLGSPALGQGVVYRLKDSSITVAFDDIPEDGLN----SP 123
LP++ F + D+V L + A+ GS L G++ R+ S+TVAFD+ + L+ +
Sbjct: 76 AALPSNSFTSGDIVGL-YDAANEGSQ-LATGILTRVTQKSVTVAFDESHDFQLSLDRENS 133
Query: 124 LRLEKVANEVTYHRMKDALIQLSKGVHKGPASDLIPVLFGERQPTVSKKDVVFTSINKNL 183
RL K+AN+VTY R+K ALI L K H GPAS LI VLFG P+ + + T N L
Sbjct: 134 YRLLKLANDVTYRRLKKALIALKK-YHSGPASSLIEVLFGRSAPSPASEIHPLTFFNTCL 192
Query: 184 DYSQKDAISKALSSKNVFLLHGPPGTGKTTTVVEIILQEVKRGSKILACAASNIAVDNIV 243
D SQK+A+S ALS K + ++HGPPGTGKTTTVVEIILQ VK+G K+L CA SNIAVDN+V
Sbjct: 193 DTSQKEAVSFALSQKELAIIHGPPGTGKTTTVVEIILQAVKQGLKVLCCAPSNIAVDNLV 252
Query: 244 ERLVPHRVKLVRIGHPARLLPQVVDSALDAQVLRGDNSGLANDIRKEMKVLNGKLLKTKE 303
ERL + +++R+GHPARLL + +LDA + R D++ + DIRK++ + K KT++
Sbjct: 253 ERLALCKQRILRLGHPARLLESIQQHSLDAVLARSDSAQIVADIRKDIDQVFVKNKKTQD 312
Query: 304 KNTRREIQKELRTLSREERKRQQLAVTDVIKTSDVILTTLIGASS----KKLGNTSFDLV 359
K + + E++ L +E ++R++ A+ + + +++V+L T GAS+ K L + FD+V
Sbjct: 313 KREKSNFRNEIKLLRKELKEREEAAMLESLTSANVVLATNTGASADGPLKLLPESYFDVV 372
Query: 360 IIDEAAQALEVACWIPLLKGTRCILAGDHLQLPPTIQSVEAEKKGLGRTLFERLAELYGD 419
+IDE AQALE +CWIPLLK +CILAGDH QLPPT S +A GL +L ERLAE YG
Sbjct: 373 VIDECAQALEASCWIPLLKARKCILAGDHKQLPPTTVSHKAALAGLSLSLMERLAEEYGA 432
Query: 420 EVTSMLTVQYRMHQLIMDWSSKELYNSKVKAHACVASHMLYDLEGVKKTSSTEPTLLLID 479
V LTVQYRMHQ IM W+S +Y ++ AH+ VA H+L DL GV T T LLL+D
Sbjct: 433 RVVRTLTVQYRMHQAIMRWASDTMYLGQLTAHSSVARHLLRDLPGVAATEETGVPLLLVD 492
Query: 480 TAGCDM-EEKKDEEDSTLNEGESEVAMAHAKRLVQSGVLPSDIGIITPYAAQVVLLKMLK 538
TAGC + E ++++E S N GE + H + LV +GV DI +++PY QV LL+ +
Sbjct: 493 TAGCGLFELEEEDEQSKGNPGEVRLVSLHIQALVDAGVPARDIAVVSPYNLQVDLLR--Q 550
Query: 539 NKENSLKDIEISTVDGFQGREKEAIIISMVRSNSKKEVGFLSDRRRMNVAVTRARRQCCI 598
+ + ++EI +VDGFQGREKEA+I+S VRSN K EVGFL++ RR+NVAVTRARR +
Sbjct: 551 SLVHRHPELEIKSVDGFQGREKEAVILSFVRSNRKGEVGFLAEDRRINVAVTRARRHVAV 610
Query: 599 VCDTETVSSDGFLKRLIEYFEEHGEYQSASEYQNE 633
+CD+ TV++ FLK L+EYF +HGE ++A EY ++
Sbjct: 611 ICDSRTVNNHAFLKTLVEYFTQHGEVRTAFEYLDD 645
>sp|P38935|SMBP2_HUMAN DNA-binding protein SMUBP-2 (Immunoglobulin mu binding protein 2)
(SMUBP-2) (Glial factor-1) (GF-1)
gi|4504623|ref|NP_002171.1| immunoglobulin mu binding
protein 2 [Homo sapiens] gi|401776|gb|AAA53082.1|
DNA-binding protein
Length = 993
Score = 520 bits (1339), Expect = e-146
Identities = 296/635 (46%), Positives = 414/635 (64%), Gaps = 17/635 (2%)
Query: 11 LFFFHQDAEISSSIATGASRNLDTAQKRGSTILNLKCVDVQTGLMGKSLIELQSTK---A 67
L +DAE+ + + +L Q RG +L L+ +TGL G+ L+ + + A
Sbjct: 16 LLELERDAEVEERRSWQENISLKELQSRGVCLLKLQVSSQRTGLYGRLLVTFEPRRYGSA 75
Query: 68 DVLPAHKFGTHDVVVLKLNKADLGSPALGQGVVYRLKDSSITVAFDDIPEDGLN----SP 123
LP++ F + D+V L + A+ GS L G++ R+ S+TVAFD+ + L+ +
Sbjct: 76 AALPSNSFTSGDIVGL-YDAANEGSQ-LATGILTRVTQKSVTVAFDESHDFQLSLDRENS 133
Query: 124 LRLEKVANEVTYHRMKDALIQLSKGVHKGPASDLIPVLFGERQPTVSKKDVVFTSINKNL 183
RL K+AN+VTY R+K ALI L K H GPAS LI VLFG P+ + + T N L
Sbjct: 134 YRLLKLANDVTYRRLKKALIALKK-YHSGPASSLIEVLFGRSAPSPASEIHPLTFFNTCL 192
Query: 184 DYSQKDAISKALSSKNVFLLHGPPGTGKTTTVVEIILQEVKRGSKILACAASNIAVDNIV 243
D SQK+A+S ALS K + ++HGPPGTGKTTTVVEIILQ VK+G K+L CA SNIAVDN+V
Sbjct: 193 DTSQKEAVSFALSQKELAIIHGPPGTGKTTTVVEIILQAVKQGLKVLCCAPSNIAVDNLV 252
Query: 244 ERLVPHRVKLVRIGHPARLLPQVVDSALDAQVLRGDNSGLANDIRKEMKVLNGKLLKTKE 303
ERL + +++R+GHPARLL V +LDA + R D++ DIRK++ + K KT++
Sbjct: 253 ERLALCKQRILRLGHPARLLESVQQHSLDAVLARSDSAQNVADIRKDIDQVFVKNKKTQD 312
Query: 304 KNTRREIQKELRTLSREERKRQQLAVTDVIKTSDVILTTLIGASS----KKLGNTSFDLV 359
K + + E++ L +E ++R++ A+ + + +++V+L T GAS+ K L + FD+V
Sbjct: 313 KREKSNFRNEIKLLRKELKEREEAAMLESLTSANVVLATNTGASADGPLKLLPESYFDVV 372
Query: 360 IIDEAAQALEVACWIPLLKGTRCILAGDHLQLPPTIQSVEAEKKGLGRTLFERLAELYGD 419
+IDE AQALE +CWIPLLK +CILAGDH QLPPT S +A GL +L ERLAE YG
Sbjct: 373 VIDECAQALEASCWIPLLKARKCILAGDHKQLPPTTVSHKAALAGLSLSLMERLAEEYGA 432
Query: 420 EVTSMLTVQYRMHQLIMDWSSKELYNSKVKAHACVASHMLYDLEGVKKTSSTEPTLLLID 479
V LTVQYRMHQ IM W+S +Y +V AH+ VA H+L DL GV T T LLL+D
Sbjct: 433 RVVRTLTVQYRMHQAIMRWASDTMYLGQVTAHSSVARHLLRDLPGVAATEETGVPLLLVD 492
Query: 480 TAGCDM-EEKKDEEDSTLNEGESEVAMAHAKRLVQSGVLPSDIGIITPYAAQVVLLKMLK 538
TAGC + E ++++E S N GE + H + LV +GV DI +++PY QV LL+ +
Sbjct: 493 TAGCGLFELEEEDEQSKGNPGEVRLVSLHIQALVDAGVPARDIAVVSPYNLQVDLLR--Q 550
Query: 539 NKENSLKDIEISTVDGFQGREKEAIIISMVRSNSKKEVGFLSDRRRMNVAVTRARRQCCI 598
+ + ++EI +VDGFQGREKEA+I+S VRSN K EVGFL++ RR+NVAVTRARR +
Sbjct: 551 SLVHRHPELEIKSVDGFQGREKEAVILSFVRSNRKGEVGFLAEDRRINVAVTRARRHVAV 610
Query: 599 VCDTETVSSDGFLKRLIEYFEEHGEYQSASEYQNE 633
+CD+ TV++ FLK L+EYF +HGE ++A EY ++
Sbjct: 611 ICDSRTVNNHAFLKTLVEYFTQHGEVRTAFEYLDD 645
>gb|AAA70430.1| DNA helicase [Homo sapiens]
Length = 993
Score = 518 bits (1334), Expect = e-145
Identities = 293/635 (46%), Positives = 414/635 (65%), Gaps = 17/635 (2%)
Query: 11 LFFFHQDAEISSSIATGASRNLDTAQKRGSTILNLKCVDVQTGLMGKSLIELQSTK---A 67
L +DAE+ + + +L Q RG +L L+ +TGL G+ L+ + + A
Sbjct: 16 LLELERDAEVEERRSWQENISLKELQSRGVCLLKLQVSSQRTGLYGRLLVTFEPRRYGSA 75
Query: 68 DVLPAHKFGTHDVVVLKLNKADLGSPALGQGVVYRLKDSSITVAFDDIPEDGLN----SP 123
LP++ F + D+V L + A+ GS L G++ R+ S+TVAFD+ + L+ +
Sbjct: 76 AALPSNSFTSGDIVGL-YDAANEGSQ-LATGILTRVTQKSVTVAFDESHDFQLSLDRENS 133
Query: 124 LRLEKVANEVTYHRMKDALIQLSKGVHKGPASDLIPVLFGERQPTVSKKDVVFTSINKNL 183
RL K+AN+VTY R+K ALI L K H GPAS LI VLFG P+ + + T N L
Sbjct: 134 YRLLKLANDVTYRRLKKALIALKK-YHSGPASSLIEVLFGRSAPSPASEIHPLTFFNTCL 192
Query: 184 DYSQKDAISKALSSKNVFLLHGPPGTGKTTTVVEIILQEVKRGSKILACAASNIAVDNIV 243
D SQK+A+ ALS K + ++HGPPGTGKTTTVVEIILQ VK+G K+L CA SNIAVDN+V
Sbjct: 193 DTSQKEAVLFALSQKELAIIHGPPGTGKTTTVVEIILQAVKQGLKVLCCAPSNIAVDNLV 252
Query: 244 ERLVPHRVKLVRIGHPARLLPQVVDSALDAQVLRGDNSGLANDIRKEMKVLNGKLLKTKE 303
ERL + +++R+GHPARLL + +LDA + R D++ + DIRK++ + K KT++
Sbjct: 253 ERLALCKQRILRLGHPARLLESIQQHSLDAVLARSDSAQIVADIRKDIDQVFVKNKKTQD 312
Query: 304 KNTRREIQKELRTLSREERKRQQLAVTDVIKTSDVILTTLIGASS----KKLGNTSFDLV 359
K + + E++ L +E ++R++ A+ + + +++V+L T GAS+ K L + FD+V
Sbjct: 313 KREKSNFRNEIKLLRKELKEREEAAMLESLTSANVVLATNTGASADGPLKLLPESYFDVV 372
Query: 360 IIDEAAQALEVACWIPLLKGTRCILAGDHLQLPPTIQSVEAEKKGLGRTLFERLAELYGD 419
+IDE AQALE +CWIPLLK +CILAGDH QLPPT S +A GL +L ERLAE YG
Sbjct: 373 VIDECAQALEASCWIPLLKARKCILAGDHKQLPPTTVSHKAALAGLSLSLMERLAEEYGA 432
Query: 420 EVTSMLTVQYRMHQLIMDWSSKELYNSKVKAHACVASHMLYDLEGVKKTSSTEPTLLLID 479
V LTVQYRMHQ IM W+S +Y ++ AH+ VA H+L DL GV T T LLL+D
Sbjct: 433 RVVRTLTVQYRMHQAIMRWASDTMYLGQLTAHSSVARHLLRDLPGVAATEETGVPLLLVD 492
Query: 480 TAGCDM-EEKKDEEDSTLNEGESEVAMAHAKRLVQSGVLPSDIGIITPYAAQVVLLKMLK 538
TAGC + E ++++E S N GE + H + LV +GV DI +++PY QV LL+ +
Sbjct: 493 TAGCGLFELEEEDEQSKGNPGEVRLVSLHIQALVDAGVPARDIAVVSPYNLQVDLLR--Q 550
Query: 539 NKENSLKDIEISTVDGFQGREKEAIIISMVRSNSKKEVGFLSDRRRMNVAVTRARRQCCI 598
+ + ++EI +VDGFQGREKEA+I+S VRSN K EVGFL++ RR+NVAVTRARR +
Sbjct: 551 SLVHRHPELEIKSVDGFQGREKEAVILSFVRSNRKGEVGFLAEDRRINVAVTRARRHVAV 610
Query: 599 VCDTETVSSDGFLKRLIEYFEEHGEYQSASEYQNE 633
+CD+ TV++ FLK L+EYF +HGE ++A EY ++
Sbjct: 611 ICDSRTVNNHAFLKTLVEYFTQHGEVRTAFEYLDD 645
>ref|XP_645520.1| hypothetical protein DDB0168508 [Dictyostelium discoideum]
gi|42734059|gb|AAS38931.1| similar to Rattus norvegicus
(Rat). Antifreeze-enhancer binding protein AEP
[Dictyostelium discoideum] gi|60473615|gb|EAL71556.1|
hypothetical protein DDB0168508 [Dictyostelium
discoideum]
Length = 1024
Score = 485 bits (1249), Expect = e-135
Identities = 288/687 (41%), Positives = 421/687 (60%), Gaps = 69/687 (10%)
Query: 11 LFFFHQDAEISSSIATGASRNLDTAQKRGSTILNLKCVDVQTGLMGKSLIEL-------- 62
L +++E++ +I A+ + + +G TI +K ++ TGL G+ LI+L
Sbjct: 11 LLDIEKESEVNEAIENFATLSNKELELKGVTINKVKILNFSTGLSGRILIKLARASFVDN 70
Query: 63 -----------QSTKADV----------LPAHKFGTHDVVVLKLNKADLGSPALGQGVVY 101
Q+ D LP HK D+V ++ +K+ G+ +GVVY
Sbjct: 71 GGSNNNNNNKKQNNDKDSDGDDEDDNFDLPPHKISNGDIVGIRPSKSKPGTNHYFKGVVY 130
Query: 102 RLKDSSITVAFDDIPEDG--LNSPL---------RLEKVANEVTYHRMKDALIQLSKGVH 150
++ I +AFDD ED N P+ ++K+AN+VTY +++++L +L V+
Sbjct: 131 KVDSRKIVIAFDDNYEDSDPNNRPMLDEYFQTLYSIDKLANDVTYKKIRESLDKLKLNVN 190
Query: 151 K---GPASDLIPVLFGER-QPT--------VSKKDVVFTSINKNLDYSQKDAISKALSSK 198
K + LI +L + QP+ ++K+ INK L+ SQK+AI +LSS
Sbjct: 191 KRTGNSENSLINLLLNDGYQPSNNNSYFQQINKEKFEQQLINKGLNQSQKEAILFSLSSN 250
Query: 199 NVFLLHGPPGTGKTTTVVEIILQEVKRGSKILACAASNIAVDNIVERLVPHR-------- 250
+V +HGPPGTGKTTTVVE I+Q +K G K+LAC SN++VDN++E+L+ +
Sbjct: 251 DVACIHGPPGTGKTTTVVEFIVQLIKSGKKVLACGPSNLSVDNMLEKLLEYSNSSSCNGF 310
Query: 251 -VKLVRIGHPARLLPQVVDSALDAQVLRGDNSGLANDIRKEMKVLNGKLLKTKEKNTRRE 309
+ RIGHP R+LPQ++ LD + + + I+ E+K L+ +LLK K+ + RR
Sbjct: 311 LINATRIGHPTRILPQLLKHTLDHKTKNSEGGQIIKGIKDEIKSLSKQLLKVKQHSERRV 370
Query: 310 IQKELRTLSREERKRQQLAVTDVIKTSDVILTTLIGASSKKL-GNTSFDLVIIDEAAQAL 368
IQ ++ L + + R++ + VI S+VIL+T GAS L G +FD V+IDE AQAL
Sbjct: 371 IQSSIKELRIDLKNREKSLIQQVINDSNVILSTNTGASDSSLKGIDNFDWVVIDECAQAL 430
Query: 369 EVACWIPLLKGTRCILAGDHLQLPPTIQSVEAEKKGLGRTLFERLAELYGDEVTSMLTVQ 428
E +CWIP+ KG + +LAGDH QLPPTI S+EA K GL TLFER+ + YGD+V+ +L VQ
Sbjct: 431 EASCWIPIQKGNKLLLAGDHQQLPPTIHSMEAAKMGLSITLFERIIKQYGDQVSRLLNVQ 490
Query: 429 YRMHQLIMDWSSKELYNSKVKAHACVASHMLY--DLEGVKKTSSTEPTLLLIDTAGCDME 486
YRM+ IMDWSS E YNSK+ A V++H+L D ++ T +T LL+IDT+GCDME
Sbjct: 491 YRMNHKIMDWSSMEFYNSKMIADKSVSNHLLVTGDSPKIRNTLTTTCPLLMIDTSGCDME 550
Query: 487 EKKDEE-DSTLNEGESEVAMAHAKRLVQSGVLPSDIGIITPYAAQVVLLKMLKNKENSLK 545
E +D+E +S N GE V H ++L++ GV P+DIG+ITPY QV LLK +K+ S
Sbjct: 551 ESQDDEGESKFNNGEVIVVKRHIEKLIECGVKPNDIGVITPYNGQVKLLKSYLSKKYS-- 608
Query: 546 DIEISTVDGFQGREKEAIIISMVRSN--SKKEVGFLSDRRRMNVAVTRARRQCCIVCDTE 603
+EI TVDGFQGREK+ IIISMVRSN + +VGFL++ RR NVA+TRAR+ +VCDT+
Sbjct: 609 SMEIGTVDGFQGREKDVIIISMVRSNTDAPHKVGFLTEDRRTNVAITRARKHVVVVCDTD 668
Query: 604 TVSSDGFLKRLIEYFEEHGEYQSASEY 630
T+SS LKR+++YF+ +G ++SA EY
Sbjct: 669 TISSHEPLKRMVDYFKLNGLFRSALEY 695
>ref|XP_540807.1| PREDICTED: similar to DNA helicase [Canis familiaris]
Length = 1068
Score = 463 bits (1192), Expect = e-129
Identities = 283/664 (42%), Positives = 392/664 (58%), Gaps = 94/664 (14%)
Query: 36 QKRGSTILNLKCVDVQTGLMGKSLIELQS---TKADVLPAHKFGTHDVVV---LKLNKAD 89
Q RG +L L+ +TGL G+ L+ L+ T A VLP++ F + + LK +A+
Sbjct: 139 QSRGVCLLKLQVSSQRTGLYGRLLVTLEPRRCTSAAVLPSNSFTSGLCICDEYLKYTRAN 198
Query: 90 -------LGSPA-----------------LGQGVVYRLKDSSITVAFDDIPEDGLN---- 121
LG A L G++ R+ S+TVAFD + L+
Sbjct: 199 PAGEDEQLGEAAEPPTKGDIVGLYDEGNQLATGILTRITQRSVTVAFDASHDFQLSLDRD 258
Query: 122 --SPLRLEKVANEVTYHRMKDALIQLSKGVHKGPASDLIPVLFGERQPTVSKKDVVFTSI 179
RL K+AN++TY R+K ALI L K H GPAS LI VLFG P+ +
Sbjct: 259 RERAYRLLKLANDITYKRLKKALITLKK-YHSGPASSLIEVLFGGSAPSPA--------- 308
Query: 180 NKNLDYSQKDAISKALSSKNVFLLHGPPGTGKTTTVVEIILQEVKRG------------- 226
S+ + ++HGPPGTGKTTTVVEIILQ V++G
Sbjct: 309 ----------------SNTELAIIHGPPGTGKTTTVVEIILQAVRQGLKVSYGCCCPVRM 352
Query: 227 ------------SKILACAASNIAVDNIVERLVPHRVKLVRIGHPARLLPQVVDSALDAQ 274
S++L CA SNIAVDN+VERL + K++R+GHPARLL + +LDA
Sbjct: 353 DVLLVLGSQELGSQVLCCAPSNIAVDNLVERLARCKQKILRLGHPARLLESIQQHSLDAV 412
Query: 275 VLRGDNSGLANDIRKEMKVLNGKLLKTKEKNTRREIQKELRTLSREERKRQQLAVTDVIK 334
+ R DN+ + DIRK++ +T+++ + + E++ L +E ++R++ A+ + +
Sbjct: 413 LARSDNAQIVADIRKDIDQAFVNNRQTQDRREKSSVWNEVKLLRKELKEREEAAMLESLT 472
Query: 335 TSDVILTTLIGASS----KKLGNTSFDLVIIDEAAQALEVACWIPLLKGTRCILAGDHLQ 390
++ V+L T GASS K L +T FD+V+IDE AQALE +CWIPLLK +CILAGDH Q
Sbjct: 473 SAAVVLATNTGASSDGPLKLLPDTHFDVVVIDECAQALEASCWIPLLKARKCILAGDHKQ 532
Query: 391 LPPTIQSVEAEKKGLGRTLFERLAELYGDEVTSMLTVQYRMHQLIMDWSSKELYNSKVKA 450
LPPT S +A GL +L ERLAE +G V LTVQYRMHQ IM W+S+ LY+ ++ A
Sbjct: 533 LPPTTVSHKAALAGLSLSLMERLAEEHGARVVRTLTVQYRMHQAIMRWASEALYHGQLTA 592
Query: 451 HACVASHMLYDLEGVKKTSSTEPTLLLIDTAGCDMEEKKDEED-STLNEGESEVAMAHAK 509
H VA H+L DL GV T T LLL+DTAGC + E +D++D S N GE + H +
Sbjct: 593 HPSVAGHLLRDLPGVAATEETGIPLLLVDTAGCGLFELEDDDDQSKGNPGEVRLVSLHIQ 652
Query: 510 RLVQSGVLPSDIGIITPYAAQVVLLKMLKNKENSLKDIEISTVDGFQGREKEAIIISMVR 569
LV +GV SDI +ITPY QV LL+ ++ + ++EI +VDGFQGREKEA+++S VR
Sbjct: 653 ALVDAGVQASDIAVITPYNLQVDLLR--QSLAHRHPELEIKSVDGFQGREKEAVVLSFVR 710
Query: 570 SNSKKEVGFLSDRRRMNVAVTRARRQCCIVCDTETVSSDGFLKRLIEYFEEHGEYQSASE 629
SN K EVGFL++ RR+NVAVTRARR +VCD+ TVS+ FLK L+++F EHGE ++A E
Sbjct: 711 SNRKGEVGFLAEDRRINVAVTRARRHVAVVCDSRTVSNHAFLKTLVDHFTEHGEVRTAFE 770
Query: 630 YQNE 633
Y ++
Sbjct: 771 YLDD 774
>ref|XP_327681.1| hypothetical protein [Neurospora crassa] gi|28919845|gb|EAA29243.1|
hypothetical protein [Neurospora crassa]
Length = 716
Score = 428 bits (1100), Expect = e-118
Identities = 265/664 (39%), Positives = 373/664 (55%), Gaps = 80/664 (12%)
Query: 36 QKRGSTILNLKCVDVQTGLMGKSLIELQSTKADV----------------------LPAH 73
Q+ G + NL +TG GK+++EL A V LP H
Sbjct: 44 QRAGLALTNLVVNAQRTGYGGKTVLELGPDSATVTITSSSSSSSSGQKGGGYDGPELPEH 103
Query: 74 KFGTHDVVVLKLNKADLGSPAL--------GQGVVYRLKDSSITVAFDD-------IPED 118
T D+VV+ A +GVV R+ + VA D+ +
Sbjct: 104 GIRTGDIVVVSEQPAGSAKKREVRELEKKGAKGVVTRVGRGVVGVALDEDKGEENVVGTG 163
Query: 119 GLNSPLR--LEKVANEVTYHRMKDALIQLSKGVHKGPASDLIPVLFGERQPTVSKKDVVF 176
GL R + K+A++VTY RM + +L K + + S I VLFG+ P+ D+
Sbjct: 164 GLGGSGRVWMVKLADDVTYRRMNGTMEKLQK-MSESEHSMFIRVLFGQSSPSPVPADLTN 222
Query: 177 TSINKNLDY-------SQKDAISKALSSKNVFLLHGPPGTGKTTTVVEIILQEVKRGSKI 229
N+D+ SQKDAI AL+S+ + L+HGPPGTGKT T++E+ILQ ++R +I
Sbjct: 223 DPEVGNIDWVDPTLNDSQKDAIRFALASREIALIHGPPGTGKTHTLIELILQLLRRDLRI 282
Query: 230 LACAASNIAVDNIVERLVPHRVKLVRIGHPARLLPQVVDSALDAQVLRGDNSGLANDIRK 289
L C SNI+VDNIVERL PH++ ++R+GHPARLLP V+ +LD D + D+R
Sbjct: 283 LVCGPSNISVDNIVERLSPHKLPILRLGHPARLLPSVLSHSLDILTTTSDAGAIVKDVRA 342
Query: 290 EMKVLNGKLLKTKEKNTRREIQKELRTLSREERKRQQLAVTDVIKTSDVILTTLIGASSK 349
EM + KT+ RR I EL+ L +E R+R++ V+D++ S V+L TL GA
Sbjct: 343 EMDAKQASIRKTRNGRERRAIYAELKELRKEYRERERRCVSDLVGRSKVVLATLHGAGGY 402
Query: 350 KLGNTSFDLVIIDEAAQALEVACWIPLLKGTRCILAGDHLQLPPTIQSVEA--------- 400
+L N FD+VIIDEA+QALE CW+PLL + +LAGDHLQLPPTI+S+ +
Sbjct: 403 QLKNEEFDVVIIDEASQALEAQCWVPLLWAKKAVLAGDHLQLPPTIKSLNSRAATTKEKK 462
Query: 401 EKKGLGRTLFERLAELYGDEVTSMLTVQYRMHQLIMDWSSKELYNSKVKAHACVASHMLY 460
E + L TLF+RL L+G + MLT QYRMH+ IM + S ELY SK+ A V +L
Sbjct: 463 EGQTLETTLFDRLLTLHGPSIKRMLTTQYRMHEKIMRFPSDELYESKLVAAEHVRERLLK 522
Query: 461 DL--EGVKKTSSTEPTLLLIDTAGCDMEEKKDEE------------------DSTLNEGE 500
DL EGV+ T L+ IDT G D EK +EE +S NE E
Sbjct: 523 DLPYEGVEANDDTTEPLIFIDTQGGDFPEKNEEELDGNGSTDKKKVIRSLHGESKSNEME 582
Query: 501 SEVAMAHAKRLVQSGVLPSDIGIITPYAAQVVLLKMLKNKENSLKDIEISTVDGFQGREK 560
+ + H + L+++GV P DI ++TPY AQ+ +L LK++ IE+ +VDGFQGREK
Sbjct: 583 AALVRQHVRGLIKAGVRPDDIAVVTPYNAQLSILAPLKDE---FPGIELGSVDGFQGREK 639
Query: 561 EAIIISMVRSNSKKEVGFLSDRRRMNVAVTRARRQCCIVCDTETVSSDG-FLKRLIEYFE 619
EA+I+S+VRSN + EVGFL ++RR+NVA+TR +R ++ D+ETV FLKR +E+ E
Sbjct: 640 EAVIVSLVRSNDEGEVGFLGEKRRLNVAMTRPKRSLTVIGDSETVKKGSTFLKRWMEFLE 699
Query: 620 EHGE 623
E+ +
Sbjct: 700 ENAD 703
>gb|EAA76707.1| hypothetical protein FG06867.1 [Gibberella zeae PH-1]
gi|46124979|ref|XP_387043.1| hypothetical protein
FG06867.1 [Gibberella zeae PH-1]
Length = 685
Score = 416 bits (1068), Expect = e-114
Identities = 253/601 (42%), Positives = 350/601 (58%), Gaps = 54/601 (8%)
Query: 36 QKRGSTILNLKCVDVQTGLMGKSLIELQ----STKADVLPAHKFGTHDVVVL-------- 83
Q+ G + NL +TGL G++++EL + AD LP H T D+V++
Sbjct: 45 QRAGIALTNLVVSGQRTGLGGRTVVELSPDAATGSADELPEHGLRTGDIVLVAEQPAGSA 104
Query: 84 -KLNKADLGSPALGQGVVYRLKDSSITVAFDDIPED-GLNSPLRLEKVANEVTYHRMKDA 141
K DL +GVV +++ ++VA D+ E+ + K+A+EVTY RM
Sbjct: 105 KKREVKDLEKKG-ARGVVTKVRREWVSVAIDEGKEEVAFTGRVWAVKLADEVTYKRMNWT 163
Query: 142 LIQLSKGVHKGPASDLIPVLFGERQPTVSKKDVVFTSINKNLDY-------SQKDAISKA 194
+ +L+K + + S L VLFG P+ D+ NLD+ SQKDAI A
Sbjct: 164 MEKLNK-MDESEYSSLTRVLFGLSSPSPVATDLTKDESVGNLDWFDPTLNDSQKDAIRFA 222
Query: 195 LSSKNVFLLHGPPGTGKTTTVVEIILQEVKRGSKILACAASNIAVDNIVERLVPHRVKLV 254
L+S+ V L+HGPPGTGKT T++E+ILQ +K +IL C SNI+VDN+VERL PH+V ++
Sbjct: 223 LASREVALIHGPPGTGKTHTLIELILQMIKLEQRILVCGPSNISVDNVVERLAPHKVPIL 282
Query: 255 RIGHPARLLPQVVDSALDAQVLRGDNSGLANDIRKEMKVLNGKLLKTKEKNTRREIQKEL 314
R+GHPARLLP VVD +LD + + DIR EM + KTK R+ I +L
Sbjct: 283 RLGHPARLLPSVVDHSLDVLTQTSEAGTIVKDIRSEMDTKQASIKKTKSGKERKAIYNDL 342
Query: 315 RTLSREERKRQQLAVTDVIKTSDVILTTLIGASSKKLGNTSFDLVIIDEAAQALEVACWI 374
+ L +E R+R++ V+ +I S V+L TL GA +L N FD+VIIDEA+QALE CW+
Sbjct: 343 KELRKEFRERERRCVSTLIGGSKVVLATLHGAGGYQLRNEEFDVVIIDEASQALEAQCWV 402
Query: 375 PLLKGTRCILAGDHLQLPPTIQSVEAEKKG-------------LGRTLFERLAELYGDEV 421
PL+ + + AGDHLQLPPTI+S + K L TLF+RL L+G +
Sbjct: 403 PLVSAKKVVCAGDHLQLPPTIKSTNTKVKAPVKEGVKITKGATLEVTLFDRLLALHGPSI 462
Query: 422 TSMLTVQYRMHQLIMDWSSKELYNSKVKAHACVASHMLYDLE-GVKKTSSTEPTLLLIDT 480
MLT QYRMH+ IM + S ELY+SK+ A V +L DLE V+ T ++ IDT
Sbjct: 463 KRMLTTQYRMHESIMRFPSDELYDSKLIAADAVKHRLLKDLEYEVEDNEDTSEPVIFIDT 522
Query: 481 AGCDMEEKKDEE--------------DSTLNEGESEVAMAHAKRLVQSGVLPSDIGIITP 526
G D EK +E+ DS NE E+ + H K+LV +GV P DI ++TP
Sbjct: 523 QGGDFPEKNEEDDKDTPKKGKASLHGDSKSNEMEAALVQQHVKQLVAAGVRPEDIAVVTP 582
Query: 527 YAAQVVLLKMLKNKENSLKDIEISTVDGFQGREKEAIIISMVRSNSKKEVGFLSDRRRMN 586
Y AQ+ +L LK+K IE+ +VDGFQGREKEA+I+S+VRSNS+ EVGFL +RRR+N
Sbjct: 583 YNAQLAVLAPLKDK---FPGIELGSVDGFQGREKEAVIVSLVRSNSEGEVGFLGERRRLN 639
Query: 587 V 587
V
Sbjct: 640 V 640
>gb|EAL87207.1| DNA helicase, putative [Aspergillus fumigatus Af293]
Length = 730
Score = 412 bits (1058), Expect = e-113
Identities = 246/586 (41%), Positives = 347/586 (58%), Gaps = 57/586 (9%)
Query: 97 QGVVYRLKDSSITVAF------------DDIPEDGLNSPLRLEKVANEVTYHRMKDALIQ 144
+GVV R+ + SI +AF D+ E+ L L K+AN+VT+ RM + +
Sbjct: 147 EGVVTRVGERSIWIAFGQRGGSGRSKEDDEAIEELWGKKLWLIKLANDVTFRRMNQTMEK 206
Query: 145 LSKGVHKGPASDLIPVLFGERQPTVSKKDVV--FTSINKNLDYSQKDAISKALSSKNVFL 202
++K + S + V FG P + ++ L+ SQK+AI AL++++V L
Sbjct: 207 MAK-MTDSEYSHFMRVAFGHTAPMQPDYGAIGPLEFVDPTLNDSQKEAIRFALAARDVAL 265
Query: 203 LHGPPGTGKTTTVVEIILQEVKRGSKILACAASNIAVDNIVERLVPHRVKLVRIGHPARL 262
+HGPPGTGKT T++E+ILQ VKR ++L C SNI+VDNIVERL P+ V +VRIGHPARL
Sbjct: 266 IHGPPGTGKTHTLIELILQMVKRKLRVLVCGPSNISVDNIVERLAPNGVPVVRIGHPARL 325
Query: 263 LPQVVDSALDAQVLRGDNSGLANDIRKEMKVLNGKLLKTKEKNTRREIQKELRTLSREER 322
LP V++ +L+ + + + D+RKE+ + KT+ RR I +L+ L RE R
Sbjct: 326 LPSVLEHSLEVLTHTSEAAAIIKDVRKEIDEKQASIRKTRTGRERRAIYDDLKELRREFR 385
Query: 323 KRQQLAVTDVIKTSDVILTTLIGASSKKLGNTSFDLVIIDEAAQALEVACWIPLLKGTRC 382
+R+ V ++++ S+V+L TL GA +L N FD+V+IDEA+QALE CWIPLL ++
Sbjct: 386 ERESRCVDNLVRESNVVLATLHGAGGHQLKNQKFDVVVIDEASQALEAQCWIPLLSASKV 445
Query: 383 ILAGDHLQLPPTIQSVEAEKK------------------------GLGRTLFERLAELYG 418
+LAGDHLQLPPT++S + K L TLF+RL L+G
Sbjct: 446 VLAGDHLQLPPTVKSSVDKLKNTKKKEKSDTKDASANSAEIIGDFSLETTLFDRLLSLHG 505
Query: 419 DEVTSMLTVQYRMHQLIMDWSSKELYNSKVKAHACVASHMLYDLE-GVKKTSSTEPTLLL 477
+ MLT QYRMH+ IM + S ELY SK+ A V S +L DL V++T T ++
Sbjct: 506 PGIKRMLTTQYRMHEKIMQFPSDELYESKLVAAETVKSRLLKDLPYEVEETDDTREPIVF 565
Query: 478 IDTAGCDMEEKKDEED----------STLNEGESEVAMAHAKRLVQSGVLPSDIGIITPY 527
DT G D EK +++D S NE E+ V H + L+Q+GV P DI ITPY
Sbjct: 566 WDTQGGDFPEKTEDDDVGKKEALLGESKSNEMEALVVAKHVQNLIQAGVKPEDIACITPY 625
Query: 528 AAQVVLL-KMLKNKENSLKDIEISTVDGFQGREKEAIIISMVRSNSKKEVGFLSDRRRMN 586
Q+ LL ML+ K L E+ +VDGFQGREKEA+++S+VRSNS+ EVGFL ++RR+N
Sbjct: 626 NGQLSLLSSMLREKYPGL---ELGSVDGFQGREKEAVVVSLVRSNSEHEVGFLGEKRRLN 682
Query: 587 VAVTRARRQCCIVCDTETVS-SDGFLKRLIEYFEEHGE--YQSASE 629
VA+TR +R CI D+ET+S FLKR + + EEH + Y A E
Sbjct: 683 VAMTRPKRHLCICGDSETISKGSAFLKRWMAFLEEHADLRYPDAGE 728
>emb|CAA20863.1| SPCC737.07c [Schizosaccharomyces pombe]
gi|19075869|ref|NP_588369.1| hypothetical protein
SPCC737.07c [Schizosaccharomyces pombe 972h-]
gi|7492541|pir||T41580 probable dna-binding protein -
fission yeast (Schizosaccharomyces pombe)
Length = 660
Score = 407 bits (1045), Expect = e-112
Identities = 244/617 (39%), Positives = 362/617 (58%), Gaps = 41/617 (6%)
Query: 32 LDTAQKRGSTILNLKCVDVQTGLMGKSLIELQSTKA----DVLPAHKFGTHDVVVLKL-- 85
L Q++G ++NL+ V+TG GK++I+ + A + LPA+ F DVV ++
Sbjct: 46 LSVLQRKGLALINLRIGVVKTGFGGKTIIDFEKDPAFSNGEELPANSFSPGDVVSIRQDF 105
Query: 86 --------NKADLGSPALGQGVVYRLKDSSITVAF---DDIPEDGLNSPLRLEKVANEVT 134
N+ D+ +GVV R+ + I+VA +DIP L + K+ N VT
Sbjct: 106 QSSKKKRPNETDISV----EGVVTRVHERHISVALKSEEDIPSSVTR--LSVVKLVNRVT 159
Query: 135 YHRMKDALIQLSKGVHKGPASDLIPVLFGERQPTVSKKDVVFTSI---NKNLDYSQKDAI 191
Y RM+ +++ + + + S L L G ++ VS + I NK L+ SQK A+
Sbjct: 160 YERMRHTMLEFKRSIPEYRNS-LFYTLIGRKKADVSIDQKLIGDIKYFNKELNASQKKAV 218
Query: 192 SKALSSKNVFLLHGPPGTGKTTTVVEIILQEVKRGSKILACAASNIAVDNIVERLVPHRV 251
+++ K + L+HGPPGTGKT T+VEII Q V R +IL C ASN+AVDNIV+RL +
Sbjct: 219 KFSIAVKELSLIHGPPGTGKTHTLVEIIQQLVLRNKRILVCGASNLAVDNIVDRLSSSGI 278
Query: 252 KLVRIGHPARLLPQVVDSALDAQVLRGDNSGLANDIRKEMKVLNGKLLKTKEKNTRREIQ 311
+VR+GHPARLLP ++D +LD GDN + I +++ V K+ KTK RREI
Sbjct: 279 PMVRLGHPARLLPSILDHSLDVLSRTGDNGDVIRGISEDIDVCLSKITKTKNGRERREIY 338
Query: 312 KELRTLSREERKRQQLAVTDVIKTSDVILTTLIGASSKKLGNTSFDLVIIDEAAQALEVA 371
K +R L ++ RK + V +++ S V+ TL GA S++L FD VIIDEA+QALE
Sbjct: 339 KNIRELRKDYRKYEAKTVANIVSASKVVFCTLHGAGSRQLKGQRFDAVIIDEASQALEPQ 398
Query: 372 CWIPLLKGTRCILAGDHLQLPPTIQSVEAEKKGLGRTLFERLAELYGDEVTSMLTVQYRM 431
CWIPLL + ILAGDH+QL P +QS K ++FERL + GD V L +QYRM
Sbjct: 399 CWIPLLGMNKVILAGDHMQLSPNVQS-----KRPYISMFERLVKSQGDLVKCFLNIQYRM 453
Query: 432 HQLIMDWSSKELYNSKVKAHACVASHMLYDLEGVKKTSSTEPTLLLIDTAGCDMEEKKDE 491
H+LI + S Y+SK+ V +L DLE V++T T+ + DT G E+ + E
Sbjct: 454 HELISKFPSDTFYDSKLVPAEEVKKRLLMDLENVEETELTDSPIYFYDTLGNYQEDDRSE 513
Query: 492 ------EDSTLNEGESEVAMAHAKRLVQSGVLPSDIGIITPYAAQVVLLKMLKNKENSLK 545
+DS N E+++ H L+++G+ DI ++TPY AQV L++ L KE +
Sbjct: 514 DMQNFYQDSKSNHWEAQIVSYHISGLLEAGLEAKDIAVVTPYNAQVALIRQLL-KEKGI- 571
Query: 546 DIEISTVDGFQGREKEAIIISMVRSNSKKEVGFLSDRRRMNVAVTRARRQCCIVCDTETV 605
++E+ +VD QGREKEAII S+VRSN +EVGFL+++RR+NVA+TR +R C++ D+ TV
Sbjct: 572 EVEMGSVDKVQGREKEAIIFSLVRSNDVREVGFLAEKRRLNVAITRPKRHLCVIGDSNTV 631
Query: 606 S-SDGFLKRLIEYFEEH 621
+ F + +++ EE+
Sbjct: 632 KWASEFFHQWVDFLEEN 648
>gb|EAA61838.1| hypothetical protein AN7652.2 [Aspergillus nidulans FGSC A4]
gi|67901330|ref|XP_680921.1| hypothetical protein
AN7652_2 [Aspergillus nidulans FGSC A4]
gi|49111123|ref|XP_411789.1| hypothetical protein
AN7652.2 [Aspergillus nidulans FGSC A4]
Length = 686
Score = 395 bits (1016), Expect = e-108
Identities = 231/544 (42%), Positives = 329/544 (60%), Gaps = 45/544 (8%)
Query: 84 KLNKADLGSPALGQGVVYRLKDSSITVAF-------------DDIPEDGLNSPLRLEKVA 130
K K D S +GVV R+ +SS+ VAF +++ E+ L L K+A
Sbjct: 135 KDKKKDGDSAKGPEGVVTRVGESSVWVAFGQSGGGGRSKEEDEEVIEELYGKKLWLIKLA 194
Query: 131 NEVTYHRMKDALIQLSKGVHKGPASDLIPVLFGERQPTVSKKDVV--FTSINKNLDYSQK 188
N+VT+ RM + +++K + + + + V FG P + I+ L+ SQK
Sbjct: 195 NDVTFRRMNQTMEKMAK-MSESDYTHFVRVAFGHTTPVQPDYEAAGPVEFIDPTLNDSQK 253
Query: 189 DAISKALSSKNVFLLHGPPGTGKTTTVVEIILQEVKRGSKILACAASNIAVDNIVERLVP 248
+AI AL+S+++ L+HGPPGTGKT T++E+I+Q VKR ++L C SNI+VDNIVERL P
Sbjct: 254 EAIQFALASRDIALIHGPPGTGKTHTLIELIIQMVKRNLRVLVCGPSNISVDNIVERLAP 313
Query: 249 HRVKLVRIGHPARLLPQVVDSALDAQVLRGDNSGLANDIRKEMKVLNGKLLKTKEKNTRR 308
++ +VRIGHPARLLP V+D +L+ D + + D+RKE+ + + KT+ +R
Sbjct: 314 SKIPVVRIGHPARLLPSVLDHSLEVLTQTSDAAAIVRDVRKEIDEKHASIRKTRFGREKR 373
Query: 309 EIQKELRTLSREERKRQQLAVTDVIKTSDVILTTLIGASSKKLGNTSFDLVIIDEAAQAL 368
I +++R L RE R+R+ V ++++ S V+L TL GA +L N FD+VIIDEA+QAL
Sbjct: 374 AIYQDIRELRREFRERESKCVDNLVRGSSVVLATLHGAGGHQLKNQKFDVVIIDEASQAL 433
Query: 369 EVACWIPLLKGTRCILAGDHLQLPPTIQSVEAEKKGLG--------------RTLFERLA 414
E CWIPLL + +LAGDHLQLPPT++S + K G +TLF+RL
Sbjct: 434 EAQCWIPLLSAPKVVLAGDHLQLPPTVKSTPHKTKEAGEDGEQDANGSFSLEKTLFDRLL 493
Query: 415 ELYGDEVTSMLTVQYRMHQLIMDWSSKELYNSKVKAHACVASHMLYDLE-GVKKTSSTEP 473
L+G + MLT QYRMH+ IM + S ELY SK+ A V S +L DL V +T T+
Sbjct: 494 SLHGPGIKRMLTTQYRMHENIMRFPSDELYESKLIAAESVKSRLLKDLPYNVHETDDTKE 553
Query: 474 TLLLIDTAGCDMEEKKDEE----------DSTLNEGESEVAMAHAKRLVQSGVLPSDIGI 523
++ DT G D EK D+E +S NE E+ V H LVQ+GV P DI +
Sbjct: 554 PVVFWDTQGGDFPEKVDDEEFAKKESLLGESKSNEMEALVVARHVDNLVQAGVRPEDIAV 613
Query: 524 ITPYAAQV-VLLKMLKNKENSLKDIEISTVDGFQGREKEAIIISMVRSNSKKEVGFLSDR 582
ITPY Q+ VL +ML+ K D+E+ +VDGFQGREKEA+++S+VRSNS+ EVGFL ++
Sbjct: 614 ITPYNGQLAVLSQMLREK---YPDLELGSVDGFQGREKEAVVVSLVRSNSEHEVGFLGEK 670
Query: 583 RRMN 586
RR+N
Sbjct: 671 RRLN 674
>gb|EAA48840.1| hypothetical protein MG00498.4 [Magnaporthe grisea 70-15]
gi|39974711|ref|XP_368746.1| hypothetical protein
MG00498.4 [Magnaporthe grisea 70-15]
Length = 1299
Score = 387 bits (995), Expect = e-106
Identities = 253/653 (38%), Positives = 355/653 (53%), Gaps = 83/653 (12%)
Query: 16 QDAEISSSIATGASRNLDTAQKRGSTILNLKCVDVQTGLMGKSLIELQSTKAD---VLPA 72
Q+AE + + ++ Q+ G+ + NL +TG G++++EL S A LP
Sbjct: 408 QEAEAAGAAELISAHTPAALQRAGAALTNLTVQGRRTGAGGRTVVELSSDPAVGGIELPE 467
Query: 73 HKFGTHDVVVLKLNKADLGSPALGQ----------GVVYRLKDSSITVAFD--------- 113
H DVV++ A GS + GVV R++ S + VA D
Sbjct: 468 HGLRVGDVVLVAEQPA--GSAKKREVREMERNGVRGVVTRVRRSDLNVALDAGDDKSGGA 525
Query: 114 DIPEDGLNSPLRLEKVANEVTYHRMKDALIQLSKGVHKGPASDLIPVLFGERQPTVS--- 170
E GL + + ++A++VTY RM + ++ + + S+ I VLFG P+
Sbjct: 526 GKEEKGLAGRVWVVRLADDVTYRRMTWTMEKMER-MADAEHSNFIRVLFGHSSPSPVPSN 584
Query: 171 -KKDVVFTSI---NKNLDYSQKDAISKALSSKNVFLLHGPPGTGKTTTVVEIILQEVKRG 226
K D F + + L+ SQKDAI AL+SK + L+HGPPGTGKT T++E+ILQ +K+G
Sbjct: 585 LKTDPEFGELEWLDPTLNDSQKDAIRFALASKEIALIHGPPGTGKTHTLIELILQMLKQG 644
Query: 227 SKILACAASNIAVDNIVERLVPHRVKLVRIGHPARLLPQVVDSALDAQVLRGDNSGLAND 286
+IL C SNI+VDNIVERL PH+V +VR+GHPARLLP VV +LD + + D
Sbjct: 645 LRILVCGPSNISVDNIVERLAPHKVPIVRLGHPARLLPSVVGHSLDVLTQTSEAGAIVRD 704
Query: 287 IRKEMKVLNGKLLKTKEKNTRREIQKELRTLSREERKRQQLAVTDVIKTSDVILTTLIGA 346
+R EM + KTK R+ I EL L +E R R++ V +++++S V+L TL G+
Sbjct: 705 VRAEMDAKQASIKKTKSGRERKAIYGELHELRKEFRDREKRCVANLLQSSKVVLATLHGS 764
Query: 347 SSKKLGNTSFDLVIIDEAAQALEVACWIPLLKGTRCILAGDHLQLPPTIQSVEA------ 400
+L FD+VIIDEA+QALE CW+ LL + +LAGDHLQLPPTI+S+ +
Sbjct: 765 GGFQLRQEKFDVVIIDEASQALEAQCWVALLSAKKAVLAGDHLQLPPTIKSLNSKAAKTA 824
Query: 401 --------------------EKKG--LGRTLFERLAELYGDEVTSMLTVQYRMHQLIMDW 438
+KKG L TLF+RL +L+G + MLT QYRMH+ IM +
Sbjct: 825 TGGDGSGTADGEESSAGQSRKKKGVTLETTLFDRLLDLHGPAIKRMLTTQYRMHEKIMRF 884
Query: 439 SSKELYNSKVKAHACVASHMLYDLE-GVKKTSSTEPTLLLIDTAGCDMEEKKDEED---- 493
S ELY ++ A V +L DL V+ T T L+ IDT G D E+ DE D
Sbjct: 885 PSDELYGGRLVAAEAVKERLLKDLPYKVEDTDDTSEPLIFIDTQGGDFPERNDEMDNGDA 944
Query: 494 --------------STLNEGESEVAMAHAKRLVQSGVLPSDIGIITPYAAQV-VLLKMLK 538
S NE E+ + H + LV +GV P DI ITPY AQ+ VL +LK
Sbjct: 945 DDKKKTKRMLLHGESKSNEMEAALVAQHVRSLVDAGVKPEDIACITPYNAQLAVLAPLLK 1004
Query: 539 NKENSLKDIEISTVDGFQGREKEAIIISMVRSNSKKEVGFLSDRRRMNVAVTR 591
++ IE+ +VDGFQGREKEA+++S+ RSN EVGFL +RRR+N R
Sbjct: 1005 DR---FPGIELGSVDGFQGREKEAVVVSLCRSNPDGEVGFLGERRRLNAPTRR 1054
>emb|CAG62165.1| unnamed protein product [Candida glabrata CBS138]
gi|50293567|ref|XP_449195.1| unnamed protein product
[Candida glabrata]
Length = 695
Score = 379 bits (974), Expect = e-103
Identities = 259/671 (38%), Positives = 381/671 (56%), Gaps = 69/671 (10%)
Query: 16 QDAEISSSIATGASRNLDTAQKRGSTILNLKCVDVQTGLMGKSLIELQSTKADVLPAHKF 75
QD E+ S+ L + G I NL + +TGL GK +IEL T++ + +
Sbjct: 20 QDIELVESLLKQIP--LKGLSRLGLAISNLTVTNSRTGLAGKLVIELAQTQSTPVSSD-L 76
Query: 76 GTHDVVVLKLNKADLGSPALGQGVVYRLKDS--------SITVAFDDIPED------GLN 121
D+V++K + QGVV ++ ++ S+ +A D+ E+ N
Sbjct: 77 KNGDIVIIKPYSKKEDNDQQCQGVVVKISNAESNSTSPTSLAIAIDESQENIATTMIHSN 136
Query: 122 SPLRLEKVANEVTYHRMKDALIQLSKGVHKGPASDLIPVLF---GERQ-----PTVSKKD 173
+ K N VTY RM L +L + K ++ +I +L G ++ P K +
Sbjct: 137 EKFYVMKTVNTVTYDRMNSTLRKLKEFKEKSISNPIIDILLPTEGVKKLRASIPNAKKSN 196
Query: 174 VVFTSINKNLDYSQKDAISKALSSKNVFLLHGPPGTGKTTTVVEIILQEVKRGS--KILA 231
+ F ++NL+ SQK AI ++ +K + ++HGPPGTGKT TVVE+I Q + + S ++L
Sbjct: 197 IQF--FDENLNDSQKYAIRFSMENK-LSIIHGPPGTGKTYTVVELIRQLISKDSTSRVLV 253
Query: 232 CAASNIAVDNIVERL---VPHRVKLVRIGHPARLLPQVVDSALDAQVLRG-DNSGLANDI 287
CA SNIAVD I+ERL + + L+RIGHPARLL +LD G D++ + NDI
Sbjct: 254 CAPSNIAVDTILERLSKSIKDKRWLLRIGHPARLLKTNWYHSLDILSKEGTDSASIINDI 313
Query: 288 RKEMKVLNGKLLKTKEKNTRREIQKELRTLSREERKRQQLAVTDVIKTSDVILTTLIGAS 347
E+ + K RR+ E++ L +E R R++ + D+IK S V++ TL G+S
Sbjct: 314 SMEITKTIASIKSIKSSGERRKAWAEVKQLRKELRLRERKVINDLIKQSRVVVATLHGSS 373
Query: 348 SKKLGN-----------TSFDLVIIDEAAQALEVACWIPLL-----KGTRCILAGDHLQL 391
S++L N FD VIIDE +Q+LE CWIPL+ K T+ +LAGD QL
Sbjct: 374 SRELCNYYKSIEEGENDNLFDYVIIDEVSQSLEPQCWIPLISHLQSKITKLVLAGDSKQL 433
Query: 392 PPTIQSVEAEK--KGLGRTLFERLAELYGDEVTSMLTVQYRMHQLIMDWSSKELYNSKVK 449
PPTI++ +K K LG TLF+RL +YGDE ++L +QYRM+ IM++SSK +YN ++K
Sbjct: 434 PPTIKTNNNDKVMKVLGTTLFDRLVNMYGDEFKNLLNIQYRMNAKIMEFSSKAMYNGELK 493
Query: 450 AHACVASHMLYDLEGVKKTSSTEPTLLLIDTAGCDMEEKKDEED------------STLN 497
A + V + +L DL GV T+ ++ DT G D E +EED S LN
Sbjct: 494 ADSSVENIVLSDLPGVDSNEETDEPIIWYDTQGDDFPEVDEEEDELKSKSAKFLYSSKLN 553
Query: 498 EGESEVAMAHAKRLVQSGVLPSDIGIITPYAAQVVLLKMLKN-KENS--LKDIEISTVDG 554
E+ + + H ++L++S V IGII+PY AQV LLK L N E+S IEIS+VDG
Sbjct: 554 TNEAYLVLHHVRKLIESNVQQDCIGIISPYNAQVSLLKKLVNGTEDSPVYPLIEISSVDG 613
Query: 555 FQGREKEAIIISMVRSNSKKEVGFLSDRRRMNVAVTRARRQCCIVCDTETV--SSDGFLK 612
FQGREKE II+S+VRSN K EVGFL ++RR+NVA+TR +RQ C++ + ET+ S + FLK
Sbjct: 614 FQGREKECIILSLVRSNDKAEVGFLKEQRRLNVAMTRPKRQLCVIGNIETLQRSGNSFLK 673
Query: 613 RLIEYFEEHGE 623
E+ E++ +
Sbjct: 674 DWTEWSEDNAD 684
Database: nr
Posted date: Jul 5, 2005 12:34 AM
Number of letters in database: 863,360,394
Number of sequences in database: 2,540,612
Lambda K H
0.316 0.133 0.364
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 963,378,360
Number of Sequences: 2540612
Number of extensions: 38870378
Number of successful extensions: 168444
Number of sequences better than 10.0: 1067
Number of HSP's better than 10.0 without gapping: 590
Number of HSP's successfully gapped in prelim test: 477
Number of HSP's that attempted gapping in prelim test: 164748
Number of HSP's gapped (non-prelim): 1985
length of query: 633
length of database: 863,360,394
effective HSP length: 134
effective length of query: 499
effective length of database: 522,918,386
effective search space: 260936274614
effective search space used: 260936274614
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.6 bits)
S2: 78 (34.7 bits)
Medicago: description of AC143341.10


