

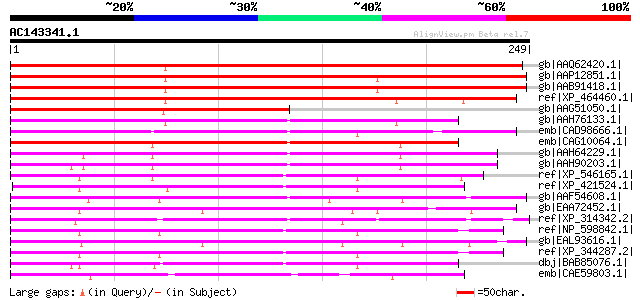
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC143341.1 + phase: 0 /partial
(249 letters)
Database: nr
2,540,612 sequences; 863,360,394 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
gb|AAQ62420.1| At3g12170 [Arabidopsis thaliana] gi|9294116|dbj|B... 365 e-100
gb|AAP12851.1| At5g06910 [Arabidopsis thaliana] gi|9759547|dbj|B... 354 1e-96
gb|AAB91418.1| DnaJ homologue [Arabidopsis thaliana] 352 5e-96
ref|XP_464460.1| putative DnaJ homolog, subfamily C, member 9 [O... 346 4e-94
gb|AAG51050.1| DnaJ protein, putative, 3' partial; 1110-1 [Arabi... 218 2e-55
gb|AAH76133.1| Zgc:92648 [Danio rerio] gi|50539934|ref|NP_001002... 147 3e-34
emb|CAD98666.1| DNAJ protein-like, possible [Cryptosporidium par... 146 6e-34
emb|CAG10064.1| unnamed protein product [Tetraodon nigroviridis] 145 8e-34
gb|AAH64229.1| Hypothetical protein MGC76175 [Xenopus tropicalis... 144 3e-33
gb|AAH90203.1| Unknown (protein for MGC:85182) [Xenopus laevis] 141 1e-32
ref|XP_546165.1| PREDICTED: similar to DnaJ homolog, subfamily C... 140 2e-32
ref|XP_421524.1| PREDICTED: similar to DnaJ homolog, subfamily C... 140 2e-32
gb|AAF54608.1| CG6693-PA [Drosophila melanogaster] gi|17862026|g... 137 4e-31
gb|EAA72452.1| hypothetical protein FG08755.1 [Gibberella zeae P... 135 1e-30
ref|XP_314342.2| ENSANGP00000013114 [Anopheles gambiae str. PEST... 134 2e-30
ref|NP_598842.1| DnaJ homolog, subfamily C, member 9 [Mus muscul... 133 4e-30
gb|EAL93616.1| DnaJ domain protein [Aspergillus fumigatus Af293] 133 4e-30
ref|XP_344287.2| PREDICTED: similar to J domain of DnaJ-like-pro... 130 3e-29
dbj|BAB85076.1| unnamed protein product [Homo sapiens] gi|180282... 129 6e-29
emb|CAE59803.1| Hypothetical protein CBG03265 [Caenorhabditis br... 129 7e-29
>gb|AAQ62420.1| At3g12170 [Arabidopsis thaliana] gi|9294116|dbj|BAB01967.1| dnaJ
protein-like [Arabidopsis thaliana]
gi|12322048|gb|AAG51071.1| DnaJ protein, putative;
5702-7336 [Arabidopsis thaliana]
gi|15230424|ref|NP_187824.1| DNAJ heat shock N-terminal
domain-containing protein [Arabidopsis thaliana]
gi|51968762|dbj|BAD43073.1| hypothetical protein
[Arabidopsis thaliana]
Length = 262
Score = 365 bits (936), Expect = e-100
Identities = 181/247 (73%), Positives = 215/247 (86%), Gaps = 1/247 (0%)
Query: 1 VLGLEKTASQQEIKKAYYKLALRLHPDKNPGDEEAKEKFQQLQKVISILGDEEKRALYDQ 60
VLG+E TAS QEI+KAY+KLALRLHPDKN DE+AKEKFQQLQKVISILGDEEKRA+YDQ
Sbjct: 15 VLGVEATASPQEIRKAYHKLALRLHPDKNKDDEDAKEKFQQLQKVISILGDEEKRAVYDQ 74
Query: 61 TGCIDDDDLAGDV-QNLHAFFKTMYKKVTEADIEEFEANYRGSDSEKNDLIELYKKYKGN 119
TG +DD DL+GDV NL FFK MYKKVTE DIEEFEANYRGS+SEKNDLIELY K+KG
Sbjct: 75 TGSVDDADLSGDVVDNLRDFFKAMYKKVTEEDIEEFEANYRGSESEKNDLIELYNKFKGK 134
Query: 120 MKRLFCSMLCSDAKLDSHRFKDILDEAIAAGELKATKAYQKWAKEVSETKPPTSPLKRKA 179
M RLFCSMLCS+ KLDSHRFKDI+DEAIAAGE+K+TKAY+KWAKE++E +PPT+P K +
Sbjct: 135 MSRLFCSMLCSNPKLDSHRFKDIIDEAIAAGEVKSTKAYKKWAKEIAEMEPPTNPQKMRR 194
Query: 180 KSNKQSETDLCAIISQRRNERKGQFDSMFASLVSKYGGSDMPEPSEEEFEAAQKKLEKGR 239
K+ K ++ DL A++SQR +ERK +FDSMF+SLVS+YG + EP+EEEFEAAQ+K+E R
Sbjct: 195 KAAKAADKDLYAVMSQRGDERKEKFDSMFSSLVSRYGSNADSEPNEEEFEAAQRKVESRR 254
Query: 240 SSKKSKQ 246
SSKKS++
Sbjct: 255 SSKKSRK 261
>gb|AAP12851.1| At5g06910 [Arabidopsis thaliana] gi|9759547|dbj|BAB11149.1| DnaJ
homologue [Arabidopsis thaliana]
gi|15240212|ref|NP_196308.1| DNAJ heat shock protein,
putative (J6) [Arabidopsis thaliana]
gi|66774117|sp|Q9FL54|DNAJ6_ARATH Chaperone protein dnaJ
6 (AtJ6) (AtDjC6)
Length = 284
Score = 354 bits (908), Expect = 1e-96
Identities = 173/252 (68%), Positives = 211/252 (83%), Gaps = 4/252 (1%)
Query: 1 VLGLEKTASQQEIKKAYYKLALRLHPDKNPGDEEAKEKFQQLQKVISILGDEEKRALYDQ 60
VLG+E+ A+ QEI+KAY+KLAL+LHPDKN D+EAK+KFQQLQKVISILGDEEKRA+YDQ
Sbjct: 33 VLGVERRATSQEIRKAYHKLALKLHPDKNQDDKEAKDKFQQLQKVISILGDEEKRAVYDQ 92
Query: 61 TGCIDDDDLAGDV-QNLHAFFKTMYKKVTEADIEEFEANYRGSDSEKNDLIELYKKYKGN 119
TG IDD D+ GD +NL FF+ MYKKV EADIEEFEANYRGS+SEK DL+EL+ K+KG
Sbjct: 93 TGSIDDADIPGDAFENLRDFFRDMYKKVNEADIEEFEANYRGSESEKKDLLELFNKFKGK 152
Query: 120 MKRLFCSMLCSDAKLDSHRFKDILDEAIAAGELKATKAYQKWAKEVSETKPPTSPL---K 176
M RLFCSMLCSD KLDSHRFKD+LDEAIAAGE+K++KAY+KWA ++SETKPPTSPL K
Sbjct: 153 MNRLFCSMLCSDPKLDSHRFKDMLDEAIAAGEVKSSKAYEKWANKISETKPPTSPLRKRK 212
Query: 177 RKAKSNKQSETDLCAIISQRRNERKGQFDSMFASLVSKYGGSDMPEPSEEEFEAAQKKLE 236
+K + K SETDLC +I++R+ ERKG+ DSMF+SL+S+YGG EP+EEEFEAAQ+++E
Sbjct: 213 KKKSAAKDSETDLCLMIAKRQEERKGKVDSMFSSLISRYGGDAEAEPTEEEFEAAQRRIE 272
Query: 237 KGRSSKKSKQSK 248
R K + K
Sbjct: 273 SKRKPSKKSRGK 284
>gb|AAB91418.1| DnaJ homologue [Arabidopsis thaliana]
Length = 284
Score = 352 bits (903), Expect = 5e-96
Identities = 172/252 (68%), Positives = 211/252 (83%), Gaps = 4/252 (1%)
Query: 1 VLGLEKTASQQEIKKAYYKLALRLHPDKNPGDEEAKEKFQQLQKVISILGDEEKRALYDQ 60
VLG+E+ A+ QEI+KAY+KLAL+LHPDKN D+EAK+KFQQLQKVISILGDEEKRA+YDQ
Sbjct: 33 VLGVERRATSQEIRKAYHKLALKLHPDKNQDDKEAKDKFQQLQKVISILGDEEKRAVYDQ 92
Query: 61 TGCIDDDDLAGDV-QNLHAFFKTMYKKVTEADIEEFEANYRGSDSEKNDLIELYKKYKGN 119
TG IDD D+ GD +NL FF+ MYKKVTEADIEEFEA YRGS+SEK DL+EL+ K+KG
Sbjct: 93 TGSIDDADIPGDAFENLRDFFRDMYKKVTEADIEEFEATYRGSESEKKDLLELFNKFKGK 152
Query: 120 MKRLFCSMLCSDAKLDSHRFKDILDEAIAAGELKATKAYQKWAKEVSETKPPTSPL---K 176
M RLFCSMLCSD KLDSHRFKD+LDEAIAAGE+K++KAY+KWA ++SETKPPTSPL K
Sbjct: 153 MNRLFCSMLCSDPKLDSHRFKDMLDEAIAAGEVKSSKAYEKWANKISETKPPTSPLRKRK 212
Query: 177 RKAKSNKQSETDLCAIISQRRNERKGQFDSMFASLVSKYGGSDMPEPSEEEFEAAQKKLE 236
+K + K SETDLC +I++R+ ERKG+ DSMF+SL+S+YGG EP+EEEFEA+Q+++E
Sbjct: 213 KKKSAPKDSETDLCLMIAKRQEERKGKVDSMFSSLISRYGGDAEAEPTEEEFEASQRRIE 272
Query: 237 KGRSSKKSKQSK 248
R K + K
Sbjct: 273 TQRKPSKKSRGK 284
>ref|XP_464460.1| putative DnaJ homolog, subfamily C, member 9 [Oryza sativa
(japonica cultivar-group)] gi|49388138|dbj|BAD25266.1|
putative DnaJ homolog, subfamily C, member 9 [Oryza
sativa (japonica cultivar-group)]
gi|49388122|dbj|BAD25253.1| putative DnaJ homolog,
subfamily C, member 9 [Oryza sativa (japonica
cultivar-group)]
Length = 282
Score = 346 bits (887), Expect = 4e-94
Identities = 174/248 (70%), Positives = 206/248 (82%), Gaps = 5/248 (2%)
Query: 1 VLGLEKTASQQEIKKAYYKLALRLHPDKNPGDEEAKEKFQQLQKVISILGDEEKRALYDQ 60
+LG+E+TASQQEIKKAY+KLALRLHPDKNPGDEEAKEKFQQLQKVISILGDEEKRALYD+
Sbjct: 34 ILGVERTASQQEIKKAYHKLALRLHPDKNPGDEEAKEKFQQLQKVISILGDEEKRALYDE 93
Query: 61 TGCIDDDDLAGDV-QNLHAFFKTMYKKVTEADIEEFEANYRGSDSEKNDLIELYKKYKGN 119
TG DDD L G+ NL +F+ +YKKVTEADIEEFEA YRGSDSEK DL +LY K+KGN
Sbjct: 94 TGIADDDALVGEAADNLQEYFRAVYKKVTEADIEEFEAKYRGSDSEKKDLKDLYTKFKGN 153
Query: 120 MKRLFCSMLCSDAKLDSHRFKDILDEAIAAGELKATKAYQKWAKEVSETKPPTSPLKRKA 179
M RLFCSM+CSD KLDSHRFKDI+DEAIA GELK+TKAY KWAK++SE +PPT+PL+R+
Sbjct: 154 MNRLFCSMICSDPKLDSHRFKDIIDEAIAEGELKSTKAYDKWAKKISEIEPPTNPLERRV 213
Query: 180 KSNKQ--SETDLCAIISQRRNERKGQFDSMFASLVSKYG--GSDMPEPSEEEFEAAQKKL 235
K NK+ E DL ISQRR +RK +FDS+ +S++SK GS EP+EEEFE A+++L
Sbjct: 214 KKNKKKSEENDLILAISQRRAQRKDRFDSVLSSIMSKCDPKGSSSSEPTEEEFEQARQRL 273
Query: 236 EKGRSSKK 243
EK RS +
Sbjct: 274 EKKRSKNR 281
>gb|AAG51050.1| DnaJ protein, putative, 3' partial; 1110-1 [Arabidopsis thaliana]
Length = 149
Score = 218 bits (554), Expect = 2e-55
Identities = 109/135 (80%), Positives = 119/135 (87%), Gaps = 1/135 (0%)
Query: 1 VLGLEKTASQQEIKKAYYKLALRLHPDKNPGDEEAKEKFQQLQKVISILGDEEKRALYDQ 60
VLG+E TAS QEI+KAY+KLALRLHPDKN DE+AKEKFQQLQKVISILGDEEKRA+YDQ
Sbjct: 15 VLGVEATASPQEIRKAYHKLALRLHPDKNKDDEDAKEKFQQLQKVISILGDEEKRAVYDQ 74
Query: 61 TGCIDDDDLAGD-VQNLHAFFKTMYKKVTEADIEEFEANYRGSDSEKNDLIELYKKYKGN 119
TG +DD DL+GD V NL FFK MYKKVTE DIEEFEANYRGS+SEKNDLIELY K+KG
Sbjct: 75 TGSVDDADLSGDVVDNLRDFFKAMYKKVTEEDIEEFEANYRGSESEKNDLIELYNKFKGK 134
Query: 120 MKRLFCSMLCSDAKL 134
M RLFCSMLCS+ KL
Sbjct: 135 MSRLFCSMLCSNPKL 149
>gb|AAH76133.1| Zgc:92648 [Danio rerio] gi|50539934|ref|NP_001002433.1| zgc:92648
[Danio rerio]
Length = 252
Score = 147 bits (371), Expect = 3e-34
Identities = 82/222 (36%), Positives = 133/222 (58%), Gaps = 8/222 (3%)
Query: 1 VLGLEKTASQQEIKKAYYKLALRLHPDKNPGDEEAKEKFQQLQKVISILGDEEKRALYDQ 60
VLG+ K A EI++ YYK++L++HPD+ PGD+ A KFQ L KV ++L D+E+RA+YD+
Sbjct: 19 VLGVCKEAPDSEIRRGYYKVSLQVHPDRAPGDQSATTKFQVLGKVYAVLADKEQRAVYDE 78
Query: 61 TGCIDDDDLAGDV-QNLHAFFKTMYKKVTEADIEEFEANYRGSDSEKNDLIELYKKYKGN 119
G +D++ ++ D +N ++ ++ K+T DI +FE Y+GSD E DL +Y +++G+
Sbjct: 79 QGIVDEESVSLDQDRNWEEHWRRLFPKITLQDILDFEKQYKGSDEEVEDLKRVYLQHEGD 138
Query: 120 MKRLFCSMLCSDAKLDSHRFKDILDEAIAAGELKATKAYQKWAKEVSETKPPTSPLKRKA 179
M ++ S LC + D R KDIL AI A E+ A KA+ + + + + +RK
Sbjct: 139 MDQIMESALCCSYE-DEPRVKDILQRAIDAEEVPAYKAFTHESVKKKNIRKRKAEKERKE 197
Query: 180 KSNKQ------SETDLCAIISQRRNERKGQFDSMFASLVSKY 215
Q SE L A+I QR+ ++ F+S+ + L +KY
Sbjct: 198 AEEMQEEMGLNSEDSLVAMIKQRQKAKENGFNSLISDLEAKY 239
>emb|CAD98666.1| DNAJ protein-like, possible [Cryptosporidium parvum]
Length = 247
Score = 146 bits (368), Expect = 6e-34
Identities = 85/244 (34%), Positives = 135/244 (54%), Gaps = 7/244 (2%)
Query: 1 VLGLEKTASQQEIKKAYYKLALRLHPDKNPGDEEAKEKFQQLQKVISILGDEEKRALYDQ 60
++G+ A EIKK Y AL LHPDKN DE++KE+FQ+LQK IL +EE R LYD+
Sbjct: 10 IIGVSPNAGAAEIKKEYRLRALALHPDKNQNDEKSKERFQELQKAYEILRNEESRKLYDE 69
Query: 61 TGCIDDDDLAGDVQNLHAFFKTMYKKVTEADIEEFEANYRGSDSEKNDLIELYKKYKGNM 120
TG I+ ++ ++ +FK KK++E DI+E++ YRGSD E DL Y ++ GN
Sbjct: 70 TGIIEGEE-GKKFDDIINYFKQFTKKISEKDIQEYKERYRGSDDEWEDLSNFYLRFNGNC 128
Query: 121 KRLFCSMLCSDAKLDSHRFKDILDEAIAAGELKATKAYQKWAKEV-SETKPPTSPLKRKA 179
K L + S+ + D + + ++++AI G L K ++ KE+ ++ K + +KR+
Sbjct: 129 KLLLEYIPFSEPE-DINYYVSMIEDAIKDGRLPQKKEFKGSIKELHAQGKKWKAKMKREK 187
Query: 180 KSNKQSETDLCAIISQRRNERKGQFDSMFASLVSKYGGSDMPEPSEEEFEAAQKKLEKGR 239
+ + DL I +R G FAS++SK+ E E +F+ Q+ L K +
Sbjct: 188 SKHSSNMEDLVQAIQSSSKKRMGG----FASIISKFSDDSFEEVDESKFQEIQENLLKNK 243
Query: 240 SSKK 243
K+
Sbjct: 244 KKKR 247
>emb|CAG10064.1| unnamed protein product [Tetraodon nigroviridis]
Length = 245
Score = 145 bits (367), Expect = 8e-34
Identities = 75/223 (33%), Positives = 138/223 (61%), Gaps = 9/223 (4%)
Query: 1 VLGLEKTASQQEIKKAYYKLALRLHPDKNPGDEEAKEKFQQLQKVISILGDEEKRALYDQ 60
+LG+EK A++ +++++YYK++L++HPD+ P D A EKFQ L +V ++L D E+RA+YD+
Sbjct: 19 ILGIEKDAAEGDVRRSYYKVSLKVHPDRAPEDPLATEKFQVLGQVYAVLSDTEQRAVYDE 78
Query: 61 TGCIDDD-DLAGDVQNLHAFFKTMYKKVTEADIEEFEANYRGSDSEKNDLIELYKKYKGN 119
G +D+D D+ + +++ ++ K+T DI +FE Y+GS+ EK D+I+LY ++KG+
Sbjct: 79 QGLVDEDSDILRQDRCWEDYWRLLFPKITLQDILDFEKTYKGSEEEKRDVIQLYLQHKGD 138
Query: 120 MKRLFCSMLCSDAKLDSHRFKDILDEAIAAGELKATKAYQKWAKEVSETKPPTSPLKRKA 179
M + S+LCS+ + D R I+ AI G +KA A+ + +++ + + + +++
Sbjct: 139 MDAITASVLCSNQE-DEPRICSIIQAAIDDGAVKAFAAFTRESEKKKKARRRKADREQRE 197
Query: 180 KSNKQSE-------TDLCAIISQRRNERKGQFDSMFASLVSKY 215
+Q E L ++ QR+ R+ QF+S + L +KY
Sbjct: 198 AEERQKEMGLGQEDDSLVMMLQQRQKSREKQFNSFLSDLEAKY 240
>gb|AAH64229.1| Hypothetical protein MGC76175 [Xenopus tropicalis]
gi|45361439|ref|NP_989296.1| hypothetical protein
MGC76175 [Xenopus tropicalis]
Length = 262
Score = 144 bits (362), Expect = 3e-33
Identities = 81/244 (33%), Positives = 143/244 (58%), Gaps = 11/244 (4%)
Query: 1 VLGLEKTASQQEIKKAYYKLALRLHPDKNPGDEE--AKEKFQQLQKVISILGDEEKRALY 58
VLG+ K A + EI++ Y++++L++HPD+ +E+ A +FQ L KV ++L D+E+RALY
Sbjct: 20 VLGVRKEAGEGEIRRGYHRVSLKVHPDRVQQEEKENATAEFQILGKVYAVLSDKEQRALY 79
Query: 59 DQTGCIDDD-DLAGDVQNLHAFFKTMYKKVTEADIEEFEANYRGSDSEKNDLIELYKKYK 117
D+ G +D++ D +N +++ ++KK+T DI+ +E Y+GS+ EKND+I Y ++
Sbjct: 80 DEQGIVDEETDTLSQDKNWEEYWRLLFKKITVEDIKAYEEKYKGSEEEKNDIISAYMDFE 139
Query: 118 GNMKRLFCSMLCSDAKLDSHRFKDILDEAIAAGELKATKAYQKWAKEVSETKPPTSPLKR 177
GNM + S+ C+D + D R + I+ +AI + E+ + A+ K +K+ E + + +
Sbjct: 140 GNMDGIMESVPCADFE-DEPRIRHIIQKAIKSKEIPSYNAFVKESKKKREQRNKRAHEEA 198
Query: 178 KAKSNKQSE-------TDLCAIISQRRNERKGQFDSMFASLVSKYGGSDMPEPSEEEFEA 230
K + E DL A+I +R+N+RK + DS F L +KY + S+ +
Sbjct: 199 KEAEEIKKEMGLGDDSDDLKALIQRRQNDRKKEMDSFFDQLEAKYCNTSKKATSKSKAPK 258
Query: 231 AQKK 234
KK
Sbjct: 259 KGKK 262
>gb|AAH90203.1| Unknown (protein for MGC:85182) [Xenopus laevis]
Length = 262
Score = 141 bits (356), Expect = 1e-32
Identities = 79/244 (32%), Positives = 142/244 (57%), Gaps = 11/244 (4%)
Query: 1 VLGLEKTASQQEIKKAYYKLALRLHPDK-NPGDEE-AKEKFQQLQKVISILGDEEKRALY 58
VLG+ K A + EI++ Y++++L++HPD+ G++E A +FQ L KV ++L D+E+RALY
Sbjct: 20 VLGVRKEAGEGEIRRGYHRVSLKVHPDRVQDGEKETATAEFQILGKVYAVLSDKEQRALY 79
Query: 59 DQTGCIDDD-DLAGDVQNLHAFFKTMYKKVTEADIEEFEANYRGSDSEKNDLIELYKKYK 117
D+ G +D++ D +N +++ ++KK+T DI+ +E Y+GS+ EKND+I Y ++
Sbjct: 80 DEQGIVDEETDTLSQDRNWEEYWRLLFKKITVEDIKAYEEKYKGSEEEKNDIISAYMDFE 139
Query: 118 GNMKRLFCSMLCSDAKLDSHRFKDILDEAIAAGELKATKAYQKWAKEVSETKPPTSPLKR 177
G++ + S+ C+D + D R + I+ +AI + E+ + + K K+ E + + +
Sbjct: 140 GDLDGIMESVPCADFE-DEPRIRQIIQKAIKSKEIPSYDTFVKETKKKREQRKKRAHEEA 198
Query: 178 KAKSNKQSE-------TDLCAIISQRRNERKGQFDSMFASLVSKYGGSDMPEPSEEEFEA 230
K + E DL A+I +R+N+RK + D F L +KY + PS+ +
Sbjct: 199 KEAEEMKKEMGLGDDNDDLKALIQKRQNDRKKEMDGFFDQLEAKYCSNSKKAPSKSKAPK 258
Query: 231 AQKK 234
KK
Sbjct: 259 RGKK 262
>ref|XP_546165.1| PREDICTED: similar to DnaJ homolog, subfamily C, member 9 [Canis
familiaris]
Length = 259
Score = 140 bits (354), Expect = 2e-32
Identities = 80/242 (33%), Positives = 143/242 (59%), Gaps = 16/242 (6%)
Query: 1 VLGLEKTASQQEIKKAYYKLALRLHPDKNPGD--EEAKEKFQQLQKVISILGDEEKRALY 58
VLG+ + AS E+++ Y+K++L++HPD+ D E+A +FQ L KV S+L D+E+RALY
Sbjct: 19 VLGVRREASDGEVRRGYHKVSLQVHPDRVGDDDKEDATRRFQILGKVYSVLSDKEQRALY 78
Query: 59 DQTGCIDDD-DLAGDVQNLHAFFKTMYKKVTEADIEEFEANYRGSDSEKNDLIELYKKYK 117
D+ G +D+D D+ ++ A+++ ++KK++ DI+ FE Y+GS+ E D+ + Y +K
Sbjct: 79 DEQGTVDEDSDVLNQDRDWEAYWRLLFKKISLEDIQAFEKTYKGSEEELADIKQAYLDFK 138
Query: 118 GNMKRLFCSMLCSDAKLDSHRFKDILDEAIAAGELKATKAYQKWAKEV-------SETKP 170
G+M ++ S+LC + R ++I+ EAI AGE+ + A+ K +K+ ++ +
Sbjct: 139 GDMDQIMESVLCVQ-YTEEPRIRNIIQEAIDAGEIPSYNAFVKESKQKMNARKRRAQEEA 197
Query: 171 PTSPLKRKAKSNKQSETDLCAIISQRRNERKGQFDSMFASLVSKY-----GGSDMPEPSE 225
+ + RK + +L AII R+ +R+ + DS A + +KY GG P E
Sbjct: 198 KEAEISRKELGLDEGVDNLKAIIQSRQKDRQKEMDSFLAQMEAKYCKPKRGGKKTPLKKE 257
Query: 226 EE 227
++
Sbjct: 258 KK 259
>ref|XP_421524.1| PREDICTED: similar to DnaJ homolog, subfamily C, member 9; DnaJ
protein SB73 [Gallus gallus]
Length = 260
Score = 140 bits (354), Expect = 2e-32
Identities = 81/227 (35%), Positives = 132/227 (57%), Gaps = 11/227 (4%)
Query: 2 LGLEKTASQQEIKKAYYKLALRLHPDKNPGD--EEAKEKFQQLQKVISILGDEEKRALYD 59
LG+ + AS +EI++AY++ +LR+HPD+ D EEA FQ L K ++L D E+RA+YD
Sbjct: 20 LGVRREASPEEIRRAYHRASLRVHPDRAEPDAKEEATRLFQILGKAYAVLSDAEQRAVYD 79
Query: 60 QTGCIDDDDLAGDVQ-NLHAFFKTMYKKVTEADIEEFEANYRGSDSEKNDLIELYKKYKG 118
+ G +D++ A + + +++ ++KK+T DI++FE NY+ S+ E D+ Y ++G
Sbjct: 80 EQGTVDEEGEALRAERDWQEYWRLLFKKITIKDIQDFEKNYKDSEQELADIKSAYMDFEG 139
Query: 119 NMKRLFCSMLCSDAKLDSHRFKDILDEAIAAGELKATKAYQKWAKEV-------SETKPP 171
+M R+ S+LC D D R K I++ AI AGEL + KA+ K +K+ +E +
Sbjct: 140 DMDRIMESVLCVD-YTDEPRVKKIIERAIDAGELPSYKAFVKESKQKMTARKRRAEKEAK 198
Query: 172 TSPLKRKAKSNKQSETDLCAIISQRRNERKGQFDSMFASLVSKYGGS 218
+ RK E DL A+I R +RK + D A + +KYG +
Sbjct: 199 EAEESRKELGLGDGEDDLKALIQSRNKDRKKEMDDFLAQMEAKYGSN 245
>gb|AAF54608.1| CG6693-PA [Drosophila melanogaster] gi|17862026|gb|AAL39490.1|
LD05521p [Drosophila melanogaster]
gi|21356411|ref|NP_650052.1| CG6693-PA [Drosophila
melanogaster]
Length = 299
Score = 137 bits (344), Expect = 4e-31
Identities = 88/268 (32%), Positives = 144/268 (52%), Gaps = 23/268 (8%)
Query: 1 VLGLEKTASQQEIKKAYYKLALRLHPDKNPGDEEAK--EKFQQLQKVISILGDEEKRALY 58
++ L + A ++E+KKAY+KL+L +HPD+ P +++A+ EKF+ L K+ +L D +KRALY
Sbjct: 19 LMELARGAGEKEVKKAYHKLSLLVHPDRVPEEQKAESTEKFKVLSKLYQVLTDTQKRALY 78
Query: 59 DQTGCIDDDDLA-GDVQNLHAFFKTMYKKVTEADIEEFEANYRGSDSEKNDLIELYKKYK 117
D+ G IDDDD + + + + ++K +TE DI +E Y S+ E+ DL + Y K
Sbjct: 79 DEQGVIDDDDESESKLSSWLELWSKIFKPITEEDINNYEKEYVESELERTDLKKAYLGGK 138
Query: 118 GNMKRLFCSMLCSDAKLDSHRFKDILDEAIAAGEL------------KATKAYQKWAKEV 165
G + L + + D R + I+ + IA+GE+ K K +QK+A+E
Sbjct: 139 GCINYLMNHVPFMKVE-DEPRIQKIVQDMIASGEVPEYKIFTEEPAAKRKKRHQKYAREF 197
Query: 166 SETKPPTSPLKRKAKSNKQSET-----DLCAIISQRRNERKGQFDSMFASLVSKYGGSDM 220
E K LKR+ K + DL +I RRN+R+ F S+ L+ KYG D
Sbjct: 198 KEAKVIKERLKRRQKEKDDQDLADNGGDLQQMILARRNQRESNFGSLMDRLMEKYGNED- 256
Query: 221 PEPSEEEFEAAQKKLEKGRSSKKSKQSK 248
+ +F A +KK +K + +++K
Sbjct: 257 -DSDTVDFSAFEKKKKKSKKPAAKQETK 283
>gb|EAA72452.1| hypothetical protein FG08755.1 [Gibberella zeae PH-1]
gi|46128755|ref|XP_388931.1| hypothetical protein
FG08755.1 [Gibberella zeae PH-1]
Length = 297
Score = 135 bits (339), Expect = 1e-30
Identities = 91/281 (32%), Positives = 140/281 (49%), Gaps = 41/281 (14%)
Query: 1 VLGLEKTASQQEIKKAYYKLALRLHPDKNPGD--EEAKEKFQQLQKVISILGDEEKRALY 58
VL LEK A+ +IK+AY K AL+ HPDK D E A E FQ + +IL D +R Y
Sbjct: 20 VLNLEKIATGDQIKQAYRKAALKHHPDKVAQDQKETAHETFQAIAFAYAILSDPTRRKRY 79
Query: 59 DQTGCIDDDDLAGDVQNLHAFFKTMYKKVTEAD-IEEFEANYRGSDSEKNDLIELYKKYK 117
D+TG + + + N +++ +K+ D IE+F Y+GSD EK D+++ Y+ +
Sbjct: 80 DETGSTSESIVDSEGFNWSDYYREQFKESVSGDAIEKFAKKYKGSDEEKGDVLDAYEDCE 139
Query: 118 GNMKRLFCSMLCSDAKLDSHRFKDILDEAIAAGELKATKAYQKWAK------------EV 165
G+M L+ ++ SD D RF+DI+++AI + ++ + AY K K E
Sbjct: 140 GDMDALYERVILSDVLEDDERFRDIINKAIKSKKVSSFPAYTKETKKKREGRMKMAREEA 199
Query: 166 SETKPPTSPL------------KRKAKSNKQSETDLCAIISQRRNERKGQFDSMFASLVS 213
+E + L K+K K SE DL A+I +R+ +R +S L
Sbjct: 200 TEAEDYAKELGVHDKLFGDKKGKKKGKGKGSSEDDLAALIQKRQKDRS---ESFLDHLTE 256
Query: 214 KYGGSDM-----------PEPSEEEFEAAQKKLEKGRSSKK 243
KYG + EPSEE F+AA +L+ + SK+
Sbjct: 257 KYGAKESKGKKGKKRPVEDEPSEEAFQAAASRLKGSKRSKR 297
>ref|XP_314342.2| ENSANGP00000013114 [Anopheles gambiae str. PEST]
gi|55240289|gb|EAA09682.2| ENSANGP00000013114 [Anopheles
gambiae str. PEST]
Length = 280
Score = 134 bits (337), Expect = 2e-30
Identities = 86/253 (33%), Positives = 144/253 (55%), Gaps = 14/253 (5%)
Query: 1 VLGLEKTASQQEIKKAYYKLALRLHPDKNP--GDEEAKEKFQQLQKVISILGDEEKRALY 58
+ G+EK+AS QEIKKAYY+L+L+ HPD+ P +EA EKF+ L K+ +IL +++ RA+Y
Sbjct: 19 LFGVEKSASDQEIKKAYYRLSLQTHPDRVPESDKQEATEKFKVLSKLYNILSNKDSRAIY 78
Query: 59 DQTGCIDDDDLAGDVQNLHAFFKTMYKKVTEADIEEFEANYRGSDSEKNDLIELYKKYKG 118
D+ G +DDDD A N A ++ +K +T DI+ ++ +Y GS++E+ND+ Y + KG
Sbjct: 79 DERGTVDDDDNAS--TNWLARWQQFFKPLTTEDIDNYQKSYVGSETERNDIKRAYLRGKG 136
Query: 119 NMKRLFCSMLCSDAKLDSHRFKDILDEAIAAGELKATKAY--QKWAKEVSETKPPTSPLK 176
+ C++ + D R I+ E I + E+ K + + AK K K
Sbjct: 137 CKNSMMCTVPFMQCE-DEPRIAAIVQEMIDSKEVPEYKIFTNEPEAKRKQRHKKYAREAK 195
Query: 177 RKAKSNKQSETDLCAIISQRRNERKGQFDSMFASLVSKYGGSDMPEPSEEEFEAAQKKLE 236
A K+ + D ++ Q +RK F S+ SL +KYG +D + S+E +++ ++ +
Sbjct: 196 -MASQMKKKQDDTSSLEQQIALKRKSAFSSLIESLEAKYGNND--DDSDELYQSEEEPPK 252
Query: 237 KGRSSKKSKQSKR 249
K +K +Q+KR
Sbjct: 253 K----RKMQQAKR 261
>ref|NP_598842.1| DnaJ homolog, subfamily C, member 9 [Mus musculus]
gi|23271492|gb|AAH23787.1| DnaJ homolog, subfamily C,
member 9 [Mus musculus] gi|20073291|gb|AAH27012.1| DnaJ
homolog, subfamily C, member 9 [Mus musculus]
gi|37515263|gb|AAH14686.2| DnaJ homolog, subfamily C,
member 9 [Mus musculus]
gi|52782788|sp|Q91WN1|DNJC9_MOUSE DnaJ homolog subfamily
C member 9 gi|26346198|dbj|BAC36750.1| unnamed protein
product [Mus musculus] gi|26344886|dbj|BAC36092.1|
unnamed protein product [Mus musculus]
Length = 259
Score = 133 bits (335), Expect = 4e-30
Identities = 77/247 (31%), Positives = 145/247 (58%), Gaps = 16/247 (6%)
Query: 1 VLGLEKTASQQEIKKAYYKLALRLHPDKNPGD--EEAKEKFQQLQKVISILGDEEKRALY 58
VLG+ + AS E+++ Y+K++L++HPD+ D E+A +FQ L +V ++L D+E++A+Y
Sbjct: 19 VLGVRREASDGEVRRGYHKVSLQVHPDRVEEDQKEDATRRFQILGRVYAVLSDKEQKAVY 78
Query: 59 DQTGCIDDDDLA-GDVQNLHAFFKTMYKKVTEADIEEFEANYRGSDSEKNDLIELYKKYK 117
D+ G +D+D ++ A+++ ++KK++ DI+ FE Y+GS+ E ND+ + Y +K
Sbjct: 79 DEQGTVDEDSAGLNQDRDWDAYWRLLFKKISLEDIQAFEKTYKGSEEELNDIKQAYLDFK 138
Query: 118 GNMKRLFCSMLCSDAKLDSHRFKDILDEAIAAGELKATKAYQKWAKEV-------SETKP 170
G+M ++ S+LC D R ++I+ +AI + E+ A A+ K +K+ ++ +
Sbjct: 139 GDMDQIMESVLCVQ-YTDEPRIRNIIQKAIESKEIPAYSAFVKESKQKMNARKRRAQEEA 197
Query: 171 PTSPLKRKAKSNKQSETDLCAIISQRRNERKGQFDSMFASLVSKYGGSDMPEPSEEEFEA 230
+ L RK ++ +L A+I R+ +R+ + DS A + +KY +PS+
Sbjct: 198 KEAELSRKELGLEEGVDNLKALIQSRQKDRQKEMDSFLAQMEAKY-----CKPSKGGKRT 252
Query: 231 AQKKLEK 237
A KK +K
Sbjct: 253 ALKKEKK 259
>gb|EAL93616.1| DnaJ domain protein [Aspergillus fumigatus Af293]
Length = 323
Score = 133 bits (335), Expect = 4e-30
Identities = 88/289 (30%), Positives = 151/289 (51%), Gaps = 45/289 (15%)
Query: 1 VLGLEKTASQQEIKKAYYKLALRLHPDKNPGDE--EAKEKFQQLQKVISILGDEEKRALY 58
VLG+++ A+Q EIK AY KLAL+ HPDK P D+ +A KFQQ+ +IL DE++R +
Sbjct: 38 VLGVKEDATQDEIKSAYRKLALKHHPDKAPADQKDQAHSKFQQIAFAYAILSDEKRRRRF 97
Query: 59 DQTGCIDDDDLAGDVQNLHAFFKTMYKKVTEAD-IEEFEANYRGSDSEKNDLIELYKKYK 117
D+TG + + + F++ +Y + + I++ + Y+GS E+ D++E + +++
Sbjct: 98 DRTGSTAEAAAGDEDFDWTEFYRDLYSNSVDTEAIDKLKKEYQGSAEEEKDILEAFDRHR 157
Query: 118 GNMKRLFCSMLCSDAKLDSHRFKDILDEAIAAGELKATKAY------------------- 158
G+M R++ S++ S+ D RF+ +D+AIA G+++A K Y
Sbjct: 158 GDMDRVYESVMLSNVLDDDERFRATIDKAIAEGKVEAYKKYTDEPAKKRQARIKRAHQEA 217
Query: 159 ---QKWAKEVSETKPPTSPLKRKAKSNKQSET-------DLCAIISQRRNERKGQFDSMF 208
++ AKE+ E K P + +K KA + K++++ DL A I QR+ R +
Sbjct: 218 KEAEELAKELEEKKRPKTEVKEKAPTKKKAKSKNISDLGDLAAAIKQRQANRMESLMEKW 277
Query: 209 ASLVSKYGGSD---------MPEPSEEEFEAAQKKLEKGRSSKKSKQSK 248
+ G + EP EE F A + ++SKK+K+SK
Sbjct: 278 QTEADTLEGGNGKRTKRKTLFEEPPEEAFAATAAR----QASKKTKKSK 322
>ref|XP_344287.2| PREDICTED: similar to J domain of DnaJ-like-protein 1 - rat [Rattus
norvegicus] gi|25513803|pir||JC7707 J domain of
DnaJ-like-protein 1 - rat
Length = 259
Score = 130 bits (327), Expect = 3e-29
Identities = 77/247 (31%), Positives = 144/247 (58%), Gaps = 16/247 (6%)
Query: 1 VLGLEKTASQQEIKKAYYKLALRLHPDKNPGD--EEAKEKFQQLQKVISILGDEEKRALY 58
VLG+ + AS E+++ Y+K++L++HPD+ D E+A +FQ L +V ++L D+E++A+Y
Sbjct: 19 VLGVRREASDGEVRRGYHKVSLQVHPDRVKEDQKEDATRRFQILGRVYAVLSDKEQKAVY 78
Query: 59 DQTGCIDDDDLA-GDVQNLHAFFKTMYKKVTEADIEEFEANYRGSDSEKNDLIELYKKYK 117
D+ G +D+D ++ A+++ ++KK++ DI+ FE Y+GS+ E ND+ + Y +K
Sbjct: 79 DEQGTVDEDSAGLHQDRDWDAYWRLLFKKISLEDIQAFEKTYKGSEEELNDIKQAYLDFK 138
Query: 118 GNMKRLFCSMLCSDAKLDSHRFKDILDEAIAAGELKATKAYQKWAKEV-------SETKP 170
G+M ++ S+LC D R ++I+ +AI A E+ + A+ K +K+ ++ +
Sbjct: 139 GDMDQIMESVLCVQ-YTDEPRIRNIIQKAIDAKEVPSYNAFVKESKQKMNARKRRAQEEA 197
Query: 171 PTSPLKRKAKSNKQSETDLCAIISQRRNERKGQFDSMFASLVSKYGGSDMPEPSEEEFEA 230
+ L RK + L A+I R+ +R+ + DS A + +KY +PS+ A
Sbjct: 198 KEAELSRKELGLEGGVDSLKALIQSRQKDRQKEMDSFLAQMEAKY-----CKPSKGGKRA 252
Query: 231 AQKKLEK 237
A +K +K
Sbjct: 253 ALRKEKK 259
>dbj|BAB85076.1| unnamed protein product [Homo sapiens] gi|18028277|gb|AAL56008.1|
DnaJ protein SB73 [Homo sapiens]
gi|52782787|sp|Q8WXX5|DNJC9_HUMAN DnaJ homolog subfamily
C member 9 (DnaJ protein SB73)
gi|27597059|ref|NP_056005.1| DnaJ homolog, subfamily C,
member 9 [Homo sapiens]
Length = 260
Score = 129 bits (325), Expect = 6e-29
Identities = 74/226 (32%), Positives = 135/226 (58%), Gaps = 13/226 (5%)
Query: 1 VLGLEKTASQQEIKKAYYKLALRLHPDK-NPGD-EEAKEKFQQLQKVISILGDEEKRALY 58
VLG+ + AS E+++ Y+K++L++HPD+ GD E+A +FQ L KV S+L D E+RA+Y
Sbjct: 19 VLGVRREASDGEVRRGYHKVSLQVHPDRVGEGDKEDATRRFQILGKVYSVLSDREQRAVY 78
Query: 59 DQTGCIDDDD--LAGDVQNLHAFFKTMYKKVTEADIEEFEANYRGSDSEKNDLIELYKKY 116
D+ G +D+D L D ++ A+++ ++KK++ DI+ FE Y+GS+ E D+ + Y +
Sbjct: 79 DEQGTVDEDSPVLTQD-RDWEAYWRLLFKKISLEDIQAFEKTYKGSEEELADIKQAYLDF 137
Query: 117 KGNMKRLFCSMLCSDAKLDSHRFKDILDEAIAAGELKATKAYQKWAKEV-------SETK 169
KG+M ++ S+LC + R ++I+ +AI AGE+ + A+ K +K+ ++ +
Sbjct: 138 KGDMDQIMESVLCVQ-YTEEPRIRNIIQQAIDAGEVPSYNAFVKESKQKMNARKRRAQEE 196
Query: 170 PPTSPLKRKAKSNKQSETDLCAIISQRRNERKGQFDSMFASLVSKY 215
+ + RK + L A I R+ +R+ + D+ A + +KY
Sbjct: 197 AKEAEMSRKELGLDEGVDSLKAAIQSRQKDRQKEMDNFLAQMEAKY 242
>emb|CAE59803.1| Hypothetical protein CBG03265 [Caenorhabditis briggsae]
Length = 243
Score = 129 bits (324), Expect = 7e-29
Identities = 81/229 (35%), Positives = 128/229 (55%), Gaps = 22/229 (9%)
Query: 1 VLGLEKTASQQEIKKAYYKLALRLHPDKNPGDEEAKE----KFQQLQKVISILGDEEKRA 56
+LG++K +E+KK YY+ ++R HPDK+ EE K+ KFQ L K IL D+E+R
Sbjct: 18 LLGVQKDCDGKELKKGYYRQSMRWHPDKSNLGEEEKQTYTTKFQLLNKAYQILSDDERRK 77
Query: 57 LYDQTGCIDDDDLAGDVQNLHAFFKTMYKKVTEADIEEFEANYRGSDSEKNDLIELYKKY 116
+YD+TG +DD++L DV ++ ++KKVT+ DI+ F Y+GS +K++LI Y K
Sbjct: 78 IYDETGSVDDEELNEDVLKA---WRKIFKKVTKEDIDNFMKTYQGSREQKDELIMHYNKC 134
Query: 117 KGNMKRLFCSMLCSDAKLDSHRFKDILDEAIAAGELKATKAYQKWAKEVSETKPPTSPLK 176
KG++ ++ + ++ D K +D I GE++ TK Y E S T+ K
Sbjct: 135 KGDISKIQEYAIGYESIED---LKAAVDSLIEDGEIEKTKKY-----ETSTTEKKMIAYK 186
Query: 177 RKAKSN-------KQSETDLCAIISQRRNERKGQFDSMFASLVSKYGGS 218
RKA+ S+ DL A+I R+ +R+ + DS +L +KY S
Sbjct: 187 RKAEKEASEAENLTNSDADLMALIRGRQKKREEKNDSFLDALAAKYAPS 235
Database: nr
Posted date: Jul 5, 2005 12:34 AM
Number of letters in database: 863,360,394
Number of sequences in database: 2,540,612
Lambda K H
0.310 0.128 0.349
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 412,351,489
Number of Sequences: 2540612
Number of extensions: 16921918
Number of successful extensions: 60110
Number of sequences better than 10.0: 2743
Number of HSP's better than 10.0 without gapping: 2072
Number of HSP's successfully gapped in prelim test: 675
Number of HSP's that attempted gapping in prelim test: 56421
Number of HSP's gapped (non-prelim): 3143
length of query: 249
length of database: 863,360,394
effective HSP length: 125
effective length of query: 124
effective length of database: 545,783,894
effective search space: 67677202856
effective search space used: 67677202856
T: 11
A: 40
X1: 16 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (21.8 bits)
S2: 73 (32.7 bits)
Medicago: description of AC143341.1


