

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC143339.10 + phase: 0 /pseudo
(349 letters)
Database: nr
2,540,612 sequences; 863,360,394 total letters
Searching..................................................done
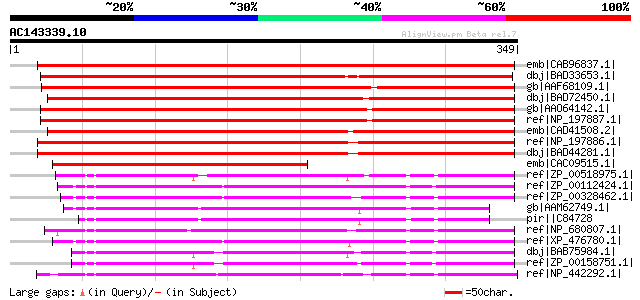
Score E
Sequences producing significant alignments: (bits) Value
emb|CAB96837.1| putative protein [Arabidopsis thaliana] gi|15238... 456 e-127
dbj|BAD33653.1| integral membrane transporter-like protein [Oryz... 420 e-116
gb|AAF68109.1| F20B17.13 [Arabidopsis thaliana] gi|15219976|ref|... 321 2e-86
dbj|BAD72450.1| integral membrane transporter-like protein [Oryz... 316 6e-85
gb|AAO64142.1| putative membrane protein [Arabidopsis thaliana] 308 2e-82
ref|NP_197887.1| integral membrane transporter family protein [A... 308 2e-82
emb|CAD41508.2| OSJNBa0029H02.6 [Oryza sativa (japonica cultivar... 297 3e-79
ref|NP_197886.1| integral membrane transporter family protein [A... 282 9e-75
dbj|BAD44281.1| putative membrane protein [Arabidopsis thaliana] 282 9e-75
emb|CAC09515.1| hypothetical protein [Oryza sativa (indica culti... 179 1e-43
ref|ZP_00518975.1| Biopterin transport-related protein BT1 [Croc... 144 5e-33
ref|ZP_00112424.1| COG0477: Permeases of the major facilitator s... 143 9e-33
ref|ZP_00328462.1| COG0477: Permeases of the major facilitator s... 141 3e-32
gb|AAM62749.1| unknown [Arabidopsis thaliana] gi|28059039|gb|AAO... 137 4e-31
pir||C84728 hypothetical protein At2g32040 [imported] - Arabidop... 136 1e-30
ref|NP_680807.1| hypothetical protein tll0016 [Thermosynechococc... 133 7e-30
ref|XP_476780.1| putative integral membrane protein [Oryza sativ... 133 9e-30
dbj|BAB75984.1| all4285 [Nostoc sp. PCC 7120] gi|25533268|pir||A... 133 9e-30
ref|ZP_00158751.1| COG0477: Permeases of the major facilitator s... 130 6e-29
ref|NP_442292.1| integral membrane protein [Synechocystis sp. PC... 125 1e-27
>emb|CAB96837.1| putative protein [Arabidopsis thaliana]
gi|15238279|ref|NP_196643.1| integral membrane
transporter family protein [Arabidopsis thaliana]
gi|11358302|pir||T50791 hypothetical protein T30N20_90 -
Arabidopsis thaliana
Length = 503
Score = 456 bits (1172), Expect = e-127
Identities = 203/328 (61%), Positives = 267/328 (80%)
Query: 20 LLSILTEPIEWLTMLTNNLNPTFIAGIFLVYGIGQGFTGSLFKLVTDYYWKDVQKLQPST 79
++S+L +P +WL ML++ LN +F+ G+ LVYG+ QGF+GS+FK+VTDYYWKDVQ++QPS
Sbjct: 35 VVSVLIQPFQWLQMLSSRLNLSFVLGVVLVYGVNQGFSGSIFKVVTDYYWKDVQQVQPSV 94
Query: 80 VQLYVGIYFIPWILKPIWGILTDAFPILGYRRRPYFVVAGVVGTVSATIVAVGGEVSVVA 139
VQLY+G+Y+IPW+++PIWG+ TD FPI GY+R+PYFVV+GV+G VSA + V G++
Sbjct: 95 VQLYMGLYYIPWVMRPIWGLFTDVFPIKGYKRKPYFVVSGVLGLVSAIAIVVLGKLPAAL 154
Query: 140 ALMCFVGVSASLAIADVTIDACIARNSIEVPGLAPDLQSLCGFCSGSGALLGYLASGFFV 199
AL C +GVSA++AIADV IDACIA NSI + LAPD+QSLC CS +GAL+GY SG FV
Sbjct: 155 ALSCLLGVSAAMAIADVVIDACIATNSINIRSLAPDIQSLCMVCSSAGALVGYATSGVFV 214
Query: 200 HHLGPQGSLGLMAVFPALTIVLGFVIYEKRTTGLHNEKKKGVVENVGTTIRSMYKTIKYP 259
H LGPQG+LG++A PA ++LGF IYEKR++ + +K K + +G ++ M KT+KYP
Sbjct: 215 HQLGPQGALGVLAFSPATIVILGFFIYEKRSSTVPTQKTKKDTDGLGVAVKGMCKTVKYP 274
Query: 260 QAWKPSLYMFLSLALNVTTHEGHFYWYTDSKVGPAFSQEFVGIIYAIGAVASIIGVLIYH 319
+ WKPSLYMF+SLALN++THEGHFYWYTD GPAFSQEFVGIIYA+GA+AS+ GVLIYH
Sbjct: 275 EVWKPSLYMFISLALNISTHEGHFYWYTDPTAGPAFSQEFVGIIYAVGALASMFGVLIYH 334
Query: 320 KYLKNYTFRNLVFYAQLLYAISGLLDLM 347
K LK Y+FRN++F+AQLLY SG+LDL+
Sbjct: 335 KKLKGYSFRNILFFAQLLYVFSGMLDLV 362
>dbj|BAD33653.1| integral membrane transporter-like protein [Oryza sativa (japonica
cultivar-group)] gi|50725892|dbj|BAD33420.1| integral
membrane transporter-like protein [Oryza sativa
(japonica cultivar-group)]
Length = 504
Score = 420 bits (1080), Expect = e-116
Identities = 196/326 (60%), Positives = 253/326 (77%), Gaps = 3/326 (0%)
Query: 22 SILTEPIEWLTMLTNNLNPTFIAGIFLVYGIGQGFTGSLFKLVTDYYWKDVQKLQPSTVQ 81
S +P+ WL ML L TF+AG+ LVYG+ QGF GS F++ +DYYWKDVQ++QP+TVQ
Sbjct: 21 STALQPVRWLRMLCRELGATFVAGVVLVYGLSQGFAGSFFRVASDYYWKDVQRVQPATVQ 80
Query: 82 LYVGIYFIPWILKPIWGILTDAFPILGYRRRPYFVVAGVVGTVSATIVAVGGEVSVVAAL 141
L ++FIPW+LKP+WGI+TD FP+ GYRRRPYF+ AGV+GT SA IV + + + +A+
Sbjct: 81 LLSAVFFIPWVLKPLWGIMTDVFPVRGYRRRPYFLFAGVLGTASAAIVTMVNGLPMTSAI 140
Query: 142 MCFVGVSASLAIADVTIDACIARNSIEVPGLAPDLQSLCGFCSGSGALLGYLASGFFVHH 201
+ FVG+S ++AIADVTIDACIA+N I+ P L PD+QSLC F S GAL+GY SG FVHH
Sbjct: 141 LSFVGISTAVAIADVTIDACIAKNGIDKPSLVPDMQSLCAFSSSLGALIGYATSGMFVHH 200
Query: 202 LGPQGSLGLMAVFPALTIVLGFVIYEKRTTGLHNEKKKGVVENVGTTIRSMYKTIKYPQA 261
LG QG+LG+MA+ PA + LGF IYE + HN K+K V+ V ++ M +TIKYP
Sbjct: 201 LGAQGALGVMALPPATLVFLGFFIYELKMY-QHNVKEK-VLNKVHMAVKGMAQTIKYPVV 258
Query: 262 WKPSLYMFLSLALNVTTHEGHFYWYTDSK-VGPAFSQEFVGIIYAIGAVASIIGVLIYHK 320
WKPSLYMFLSLAL+++THEG FYWYT + P FSQEFVG+++AIGAVAS++GVL+YHK
Sbjct: 259 WKPSLYMFLSLALSISTHEGQFYWYTSKEPPNPGFSQEFVGMVHAIGAVASMVGVLVYHK 318
Query: 321 YLKNYTFRNLVFYAQLLYAISGLLDL 346
YLK+Y FR+++FYAQLLY +SGLLDL
Sbjct: 319 YLKDYPFRSILFYAQLLYGVSGLLDL 344
>gb|AAF68109.1| F20B17.13 [Arabidopsis thaliana] gi|15219976|ref|NP_178089.1|
integral membrane transporter family protein
[Arabidopsis thaliana] gi|12324591|gb|AAG52252.1|
hypothetical protein; 109043-107192 [Arabidopsis
thaliana] gi|25406597|pir||A96828 hypothetical protein
F19K16.31 [imported] - Arabidopsis thaliana
Length = 497
Score = 321 bits (823), Expect = 2e-86
Identities = 153/325 (47%), Positives = 217/325 (66%), Gaps = 3/325 (0%)
Query: 23 ILTEPIEWLTMLTNNLNPTFIAGIFLVYGIGQGFTGSLFKLVTDYYWKDVQKLQPSTVQL 82
+L P+ WL ML L+ +F G+ +VYG+ QG L K+ T YY+KD QK+QPS Q+
Sbjct: 30 VLKRPLRWLRMLIEELHWSFFFGVIIVYGVSQGLGKGLSKVSTQYYFKDEQKIQPSQAQI 89
Query: 83 YVGIYFIPWILKPIWGILTDAFPILGYRRRPYFVVAGVVGTVSATIVAVGGEVSVVAALM 142
YVG+ IPWI+KP+WG+LTD P+LGYRRRPYF++AG + +S ++ + + + AL
Sbjct: 90 YVGLIQIPWIIKPLWGLLTDVVPVLGYRRRPYFILAGFLAMISMMVLWLHTNLHLALALS 149
Query: 143 CFVGVSASLAIADVTIDACIARNSIEVPGLAPDLQSLCGFCSGSGALLGYLASGFFVHHL 202
C V SA +AIADVTIDAC+ + SI P LA D+QSLCG CS G+L+G+ SG VH +
Sbjct: 150 CLVAGSAGVAIADVTIDACVTQCSISHPTLAADMQSLCGLCSSIGSLVGFSLSGVLVHLV 209
Query: 203 GPQGSLGLMAVFPALTIVLGFVIYEKRTTGLHNEKKKGVVENVGTTIRSMYKTIKYPQAW 262
G +G GL+ V L +V+G V+ E + L + + G+ I +KT +Y + W
Sbjct: 210 GSKGVYGLLGVTAGLLVVVGMVLKESPSRSLGRKHVNDKFLDAGSAI---WKTFQYGEVW 266
Query: 263 KPSLYMFLSLALNVTTHEGHFYWYTDSKVGPAFSQEFVGIIYAIGAVASIIGVLIYHKYL 322
+P L+M LS A+++ HEG FYWYTDSK GP+FS+E VG I + GA+ S++G+L+Y +L
Sbjct: 267 RPCLFMLLSAAVSLHIHEGMFYWYTDSKDGPSFSKEAVGSIMSFGAIGSLVGILLYQNFL 326
Query: 323 KNYTFRNLVFYAQLLYAISGLLDLM 347
KN+ FRN+VF+A L +SG LDL+
Sbjct: 327 KNFPFRNVVFWALSLSVLSGFLDLI 351
>dbj|BAD72450.1| integral membrane transporter-like protein [Oryza sativa (japonica
cultivar-group)]
Length = 539
Score = 316 bits (810), Expect = 6e-85
Identities = 150/322 (46%), Positives = 221/322 (68%), Gaps = 5/322 (1%)
Query: 27 PIEWLTMLTNNLNPTFIAGIFLVYGIGQGFTGSLFKLVTDYYWKDVQKLQPSTVQLYVGI 86
P+ W ML L+ +F+AG+ YG QG G + ++ +DYYWKDVQ++QPS Q+Y G+
Sbjct: 46 PVGWFRMLGRELHWSFVAGVVATYGASQGLGGGVMRVASDYYWKDVQRVQPSAAQVYQGV 105
Query: 87 YFIPWILKPIWGILTDAFPILGYRRRPYFVVAGVVGTVSATIVAVGGEVSVVAALMCFVG 146
IPW++KP+WG+LTD PI GYRRRPYFV+AG +G V+ ++++ ++ V+ AL+ +
Sbjct: 106 TSIPWMVKPLWGLLTDVLPIAGYRRRPYFVIAGFMGVVAMLVLSLHSKLHVLFALLALMA 165
Query: 147 VSASLAIADVTIDACIARNSIEVPGLAPDLQSLCGFCSGSGALLGYLASGFFVHHLGPQG 206
SAS+AIADVTIDAC+A NSI P LA D+ SL GFC+ G L+G+ SGF VH +G QG
Sbjct: 166 GSASVAIADVTIDACVAENSIVHPHLAADMISLNGFCASVGGLIGFSISGFLVHAIGSQG 225
Query: 207 SLGLMAVFPALTIVLGFVIYEKRTTGLHNE-KKKGVVENVGTTIRSMYKTIKYPQAWKPS 265
+LG++A+ AL I+ G +I + E VE R+M T+K P+ W+P
Sbjct: 226 ALGMLAIPSALVILAGMMIKDVHMPNFPYELAHMKFVE----ASRTMMATLKCPEVWRPC 281
Query: 266 LYMFLSLALNVTTHEGHFYWYTDSKVGPAFSQEFVGIIYAIGAVASIIGVLIYHKYLKNY 325
+YM++SLAL+V EG FYWYTD G +FS+ +G I+A+G+V S+IGV++Y LK++
Sbjct: 282 VYMYMSLALSVDIQEGMFYWYTDRNAGLSFSEGLIGFIFAVGSVGSLIGVILYQNILKDH 341
Query: 326 TFRNLVFYAQLLYAISGLLDLM 347
+FR+++ +QLL ++SG+LDL+
Sbjct: 342 SFRSVLCLSQLLLSLSGMLDLI 363
>gb|AAO64142.1| putative membrane protein [Arabidopsis thaliana]
Length = 499
Score = 308 bits (788), Expect = 2e-82
Identities = 152/326 (46%), Positives = 213/326 (64%), Gaps = 3/326 (0%)
Query: 22 SILTEPIEWLTMLTNNLNPTFIAGIFLVYGIGQGFTGSLFKLVTDYYWKDVQKLQPSTVQ 81
+++ P+ WL ML++ L+ +F+ G+ +YGI QG GSL ++ T+YY KDVQK+QPS Q
Sbjct: 25 NVVCGPVRWLKMLSSELHWSFVFGVVSLYGINQGLGGSLGRVATEYYMKDVQKVQPSESQ 84
Query: 82 LYVGIYFIPWILKPIWGILTDAFPILGYRRRPYFVVAGVVGTVSATIVAVGGEVSVVAAL 141
I IPWI+KP+WGILTD PI G+ RRPYF++AGV+G VS +++ + + AL
Sbjct: 85 ALTAITKIPWIIKPLWGILTDVLPIFGFHRRPYFILAGVLGVVSLLFISLHSNLHLYLAL 144
Query: 142 MCFVGVSASLAIADVTIDACIARNSIEVPGLAPDLQSLCGFCSGSGALLGYLASGFFVHH 201
SA++AIADVTIDAC A NSI+ P LA D+QSLC S GALLG+ SG VH
Sbjct: 145 FWMTISSAAMAIADVTIDACTAYNSIKHPSLASDMQSLCSLSSSIGALLGFFMSGILVHL 204
Query: 202 LGPQGSLGLMAVFPALTIVLGFVIYEKRTTGLHNEKKKGVVENVGTTIRSMYKTIKYPQA 261
+G +G GL+ AL V+G V E G ++ + G ++M++T+K
Sbjct: 205 VGSKGVFGLLTFPFALVSVVGIVFSEPHVPGFSYKQVNQKFTDAG---KAMWRTMKCSDV 261
Query: 262 WKPSLYMFLSLALNVTTHEGHFYWYTDSKVGPAFSQEFVGIIYAIGAVASIIGVLIYHKY 321
W+PSLYM++SL L + HEG FYW+TDSK GP F+QE VG I +IG++ SI+ +Y
Sbjct: 262 WRPSLYMYISLTLGLNIHEGLFYWFTDSKDGPLFAQETVGFILSIGSIGSILAATLYQLV 321
Query: 322 LKNYTFRNLVFYAQLLYAISGLLDLM 347
LK++ FR L + QLL+A+SG+LDL+
Sbjct: 322 LKDHPFRGLCLWTQLLFALSGMLDLI 347
>ref|NP_197887.1| integral membrane transporter family protein [Arabidopsis thaliana]
gi|58331763|gb|AAW70379.1| At5g25050 [Arabidopsis
thaliana]
Length = 499
Score = 308 bits (788), Expect = 2e-82
Identities = 152/326 (46%), Positives = 213/326 (64%), Gaps = 3/326 (0%)
Query: 22 SILTEPIEWLTMLTNNLNPTFIAGIFLVYGIGQGFTGSLFKLVTDYYWKDVQKLQPSTVQ 81
+++ P+ WL ML++ L+ +F+ G+ +YGI QG GSL ++ T+YY KDVQK+QPS Q
Sbjct: 25 NVVCGPVRWLKMLSSELHWSFVFGVVSLYGINQGLGGSLGRVATEYYMKDVQKVQPSESQ 84
Query: 82 LYVGIYFIPWILKPIWGILTDAFPILGYRRRPYFVVAGVVGTVSATIVAVGGEVSVVAAL 141
I IPWI+KP+WGILTD PI G+ RRPYF++AGV+G VS +++ + + AL
Sbjct: 85 ALTAITKIPWIIKPLWGILTDVLPIFGFHRRPYFILAGVLGVVSLLFISLHSNLHLYLAL 144
Query: 142 MCFVGVSASLAIADVTIDACIARNSIEVPGLAPDLQSLCGFCSGSGALLGYLASGFFVHH 201
SA++AIADVTIDAC A NSI+ P LA D+QSLC S GALLG+ SG VH
Sbjct: 145 FWMTISSAAMAIADVTIDACTAYNSIKHPSLASDMQSLCSLSSSIGALLGFFMSGILVHL 204
Query: 202 LGPQGSLGLMAVFPALTIVLGFVIYEKRTTGLHNEKKKGVVENVGTTIRSMYKTIKYPQA 261
+G +G GL+ AL V+G V E G ++ + G ++M++T+K
Sbjct: 205 VGSKGVFGLLTFPFALVSVVGIVFSEPHVPGFSYKQVNQKFTDAG---KAMWRTMKCSDV 261
Query: 262 WKPSLYMFLSLALNVTTHEGHFYWYTDSKVGPAFSQEFVGIIYAIGAVASIIGVLIYHKY 321
W+PSLYM++SL L + HEG FYW+TDSK GP F+QE VG I +IG++ SI+ +Y
Sbjct: 262 WRPSLYMYISLTLGLNIHEGLFYWFTDSKDGPLFAQETVGFILSIGSIGSILAATLYQLV 321
Query: 322 LKNYTFRNLVFYAQLLYAISGLLDLM 347
LK++ FR L + QLL+A+SG+LDL+
Sbjct: 322 LKDHPFRGLCLWTQLLFALSGMLDLI 347
>emb|CAD41508.2| OSJNBa0029H02.6 [Oryza sativa (japonica cultivar-group)]
gi|50926062|ref|XP_473050.1| OSJNBa0029H02.6 [Oryza
sativa (japonica cultivar-group)]
Length = 527
Score = 297 bits (761), Expect = 3e-79
Identities = 140/321 (43%), Positives = 212/321 (65%), Gaps = 3/321 (0%)
Query: 27 PIEWLTMLTNNLNPTFIAGIFLVYGIGQGFTGSLFKLVTDYYWKDVQKLQPSTVQLYVGI 86
P WL L+ L+ +F+ + VYG QG ++ + YYWKDVQ++QPS Q Y G
Sbjct: 23 PGGWLRRLSRELHWSFVLAVVAVYGACQGVGDAVGGVAAGYYWKDVQRVQPSAAQFYQGF 82
Query: 87 YFIPWILKPIWGILTDAFPILGYRRRPYFVVAGVVGTVSATIVAVGGEVSVVAALMCFVG 146
PW++KPIWG+LTD P+ GYRRRPYF++AGV+G S +++ ++ ++ A++
Sbjct: 83 VSAPWVVKPIWGLLTDVVPVAGYRRRPYFLLAGVIGASSMLTLSLHRKLGIMPAILALTA 142
Query: 147 VSASLAIADVTIDACIARNSIEVPGLAPDLQSLCGFCSGSGALLGYLASGFFVHHLGPQG 206
SA A+ADVT+DA +A+NSI LA D+QSLCGF S GALLG+ SG VH +G QG
Sbjct: 143 QSAGAAVADVTVDALVAQNSITHQPLAADMQSLCGFSSSVGALLGFSISGLLVHSMGSQG 202
Query: 207 SLGLMAVFPALTIVLGFVIYEKRTTGLHNEKKKGVVENVGTTIRSMYKTIKYPQAWKPSL 266
+LGL+++ AL + G ++ E+R T K V I+SM T+K + W+P +
Sbjct: 203 ALGLLSIPSALVFLAGVLLKERRVTDF---DYKQVHRKFYKAIQSMGATLKCAEVWRPCV 259
Query: 267 YMFLSLALNVTTHEGHFYWYTDSKVGPAFSQEFVGIIYAIGAVASIIGVLIYHKYLKNYT 326
YM++SL+L++ G FYWYTD VGP FS+EF+G++Y++G++ S++GVL+Y LK+
Sbjct: 260 YMYVSLSLSLDIQAGMFYWYTDPIVGPGFSEEFIGLVYSVGSIGSLLGVLLYQISLKDCP 319
Query: 327 FRNLVFYAQLLYAISGLLDLM 347
FR ++F+ Q+L +++G+LDL+
Sbjct: 320 FRGVLFWGQVLSSLAGMLDLI 340
>ref|NP_197886.1| integral membrane transporter family protein [Arabidopsis thaliana]
Length = 492
Score = 282 bits (722), Expect = 9e-75
Identities = 148/329 (44%), Positives = 206/329 (61%), Gaps = 7/329 (2%)
Query: 20 LLSILTEPIEWLTMLTNNLNPTFIAGIFLVYGIGQGFTGSLFKLVTDYYWKDVQKLQPST 79
L S++ P+ WL ML + L+ +++ + GI QGF SL + TDYY KDVQK+QPS
Sbjct: 23 LRSVVCGPVRWLKMLASKLHWSYVFAVVSNCGINQGFGYSLGHVATDYYMKDVQKVQPSQ 82
Query: 80 VQLYVGIYFIPWIL-KPIWGILTDAFPILGYRRRPYFVVAGVVGTVSATIVAVGGEVSVV 138
Q I I WI+ KP++GILTD PI G+ RRPYF++AGV+G VS +++ + +
Sbjct: 83 YQALSAITRISWIIFKPLFGILTDVLPIFGFHRRPYFILAGVIGVVSLLFISLQSNLHLY 142
Query: 139 AALMCFVGVSASLAIADVTIDACIARNSIEVPGLAPDLQSLCGFCSGSGALLGYLASGFF 198
AL SA++AIADVTIDAC NS + P LA D+QSLC G LLG+ SG
Sbjct: 143 LALFWMTISSAAMAIADVTIDACTTYNSNKHPSLASDMQSLCSLSYSIGELLGFFMSGIL 202
Query: 199 VHHLGPQGSLGLMAVFPALTIVLGFVIYEKRTTGLHNEKKKGVVENVGTTIRSMYKTIKY 258
VH +G +G GL+ AL V+G + E R G +N +++M++TIK
Sbjct: 203 VHLVGSKGVFGLLTFTFALVSVVGVLFSEPRVHGF------SFKQNFTNAMKAMWRTIKC 256
Query: 259 PQAWKPSLYMFLSLALNVTTHEGHFYWYTDSKVGPAFSQEFVGIIYAIGAVASIIGVLIY 318
W+PSLYMF++ AL + EG FYW+TDSK GP F+QE VG I +IG++ SI+GVL+Y
Sbjct: 257 SDVWQPSLYMFITRALGLDIKEGLFYWFTDSKDGPLFAQETVGFILSIGSIGSILGVLLY 316
Query: 319 HKYLKNYTFRNLVFYAQLLYAISGLLDLM 347
+ LK++ FR L + QLL+A+SG+ DL+
Sbjct: 317 NLRLKDHPFRKLFLWTQLLFALSGMFDLI 345
>dbj|BAD44281.1| putative membrane protein [Arabidopsis thaliana]
Length = 434
Score = 282 bits (722), Expect = 9e-75
Identities = 148/329 (44%), Positives = 206/329 (61%), Gaps = 7/329 (2%)
Query: 20 LLSILTEPIEWLTMLTNNLNPTFIAGIFLVYGIGQGFTGSLFKLVTDYYWKDVQKLQPST 79
L S++ P+ WL ML + L+ +++ + GI QGF SL + TDYY KDVQK+QPS
Sbjct: 23 LRSVVCGPVRWLKMLASKLHWSYVFAVVSNCGINQGFGYSLGHVATDYYMKDVQKVQPSQ 82
Query: 80 VQLYVGIYFIPWIL-KPIWGILTDAFPILGYRRRPYFVVAGVVGTVSATIVAVGGEVSVV 138
Q I I WI+ KP++GILTD PI G+ RRPYF++AGV+G VS +++ + +
Sbjct: 83 YQALSAITRISWIIFKPLFGILTDVLPIFGFHRRPYFILAGVIGVVSLLFISLQSNLHLY 142
Query: 139 AALMCFVGVSASLAIADVTIDACIARNSIEVPGLAPDLQSLCGFCSGSGALLGYLASGFF 198
AL SA++AIADVTIDAC NS + P LA D+QSLC G LLG+ SG
Sbjct: 143 LALFWMTISSAAMAIADVTIDACTTYNSNKHPSLASDMQSLCSLSYSIGELLGFFMSGIL 202
Query: 199 VHHLGPQGSLGLMAVFPALTIVLGFVIYEKRTTGLHNEKKKGVVENVGTTIRSMYKTIKY 258
VH +G +G GL+ AL V+G + E R G +N +++M++TIK
Sbjct: 203 VHLVGSKGVFGLLTFTFALVSVVGVLFSEPRVHGF------SFKQNFTNAMKAMWRTIKC 256
Query: 259 PQAWKPSLYMFLSLALNVTTHEGHFYWYTDSKVGPAFSQEFVGIIYAIGAVASIIGVLIY 318
W+PSLYMF++ AL + EG FYW+TDSK GP F+QE VG I +IG++ SI+GVL+Y
Sbjct: 257 SDVWQPSLYMFITRALGLDIKEGLFYWFTDSKDGPLFAQETVGFILSIGSIGSILGVLLY 316
Query: 319 HKYLKNYTFRNLVFYAQLLYAISGLLDLM 347
+ LK++ FR L + QLL+A+SG+ DL+
Sbjct: 317 NLRLKDHPFRKLFLWTQLLFALSGMFDLI 345
>emb|CAC09515.1| hypothetical protein [Oryza sativa (indica cultivar-group)]
Length = 402
Score = 179 bits (453), Expect = 1e-43
Identities = 82/176 (46%), Positives = 115/176 (64%)
Query: 30 WLTMLTNNLNPTFIAGIFLVYGIGQGFTGSLFKLVTDYYWKDVQKLQPSTVQLYVGIYFI 89
WL L+ L+ +F+ + VYG QG ++ + YYWKDVQ++QPS Q Y G
Sbjct: 35 WLRRLSRELHWSFVLAVVAVYGACQGVGDAVGGVAAGYYWKDVQRVQPSAAQFYQGFVSA 94
Query: 90 PWILKPIWGILTDAFPILGYRRRPYFVVAGVVGTVSATIVAVGGEVSVVAALMCFVGVSA 149
PW++KP+WG+LTD P+ GYRRRPYF++AGV+G S +A+ ++ ++ A++ SA
Sbjct: 95 PWVVKPVWGLLTDVVPVAGYRRRPYFLLAGVIGASSMLTLALHRKLGIMPAILALTAQSA 154
Query: 150 SLAIADVTIDACIARNSIEVPGLAPDLQSLCGFCSGSGALLGYLASGFFVHHLGPQ 205
A+ADVT+DA +A+NSI LA D+QSLCGF S GALLG+ SG VH +G Q
Sbjct: 155 GAAVADVTVDALVAQNSITHQPLAADMQSLCGFSSSVGALLGFSISGLLVHSMGSQ 210
>ref|ZP_00518975.1| Biopterin transport-related protein BT1 [Crocosphaera watsonii WH
8501] gi|67852502|gb|EAM47942.1| Biopterin
transport-related protein BT1 [Crocosphaera watsonii WH
8501]
Length = 484
Score = 144 bits (362), Expect = 5e-33
Identities = 98/323 (30%), Positives = 176/323 (54%), Gaps = 21/323 (6%)
Query: 32 TMLTNNLNPTFIAGIFLVYGIGQGFTGSLFKLVTDYYWKDVQKLQPSTVQLYVGIYFIPW 91
T+L N + I VY + QG G L +L ++ KD L P+ + GI IPW
Sbjct: 18 TVLFGNKPSAELFAILSVYFV-QGILG-LARLAVSFFLKDDLGLTPAQMGALTGIAAIPW 75
Query: 92 ILKPIWGILTDAFPILGYRRRPYFVVAGVVGTVS----ATIVAVGGEVSVVAALMCFVGV 147
++KP++G ++D PI YRRRPY V++G++G++S AT+V+ +V A + +
Sbjct: 76 VIKPLFGFISDGLPIFSYRRRPYLVISGLLGSLSWILLATVVS-----NVWLATVTLLLT 130
Query: 148 SASLAIADVTIDACIA-RNSIEVPGLAPDLQSLCGFCSGSGALLGYLASGFFVHHLGPQG 206
S S+AI+DV +D+ + R E A LQSL S G L+ SG + Q
Sbjct: 131 SLSVAISDVIVDSLVVERARKESLSQAGSLQSLTWGVSAFGGLITAYLSGLLLERFSNQI 190
Query: 207 SLGLMAVFPALTIVLGFVIYEKRTT--GLHNEKKKGVVENVGTTIRSMYKTIKYPQAWKP 264
+ A+FP + + ++I E+ + L +++ + +V+ I+ ++K IK + P
Sbjct: 191 VFEITAIFPFIVCLAAWLIAEEPVSKNELLSQQDQSLVKQ---QIKQVWKAIKQKSIFLP 247
Query: 265 SLYMFLSLALNVTTHEGHFYWYTDSKVGPAFSQEFVGIIYAIGAVASIIGVLIYHKYLKN 324
+ ++FL A + F+++ +++G F EF+G + + +VA+++GVLIY K+LKN
Sbjct: 248 TAFVFLWQA--TPNGDSAFFYFATNELG--FEPEFLGRVRLVTSVAALLGVLIYQKFLKN 303
Query: 325 YTFRNLVFYAQLLYAISGLLDLM 347
+FRN++ ++ ++ + G+ L+
Sbjct: 304 VSFRNILGWSVVISSAFGMTTLL 326
>ref|ZP_00112424.1| COG0477: Permeases of the major facilitator superfamily [Nostoc
punctiforme PCC 73102]
Length = 471
Score = 143 bits (360), Expect = 9e-33
Identities = 94/315 (29%), Positives = 165/315 (51%), Gaps = 9/315 (2%)
Query: 34 LTNNLNPTFIAGIFLVYGIGQGFTGSLFKLVTDYYWKDVQKLQPSTVQLYVGIYFIPWIL 93
L N N IA I VY + QG G L +L ++ KD L P V +GI F+PW++
Sbjct: 21 LGNEPNAELIA-ILTVYFV-QGILG-LSRLAVSFFLKDELLLSPVQVSALLGIVFLPWMI 77
Query: 94 KPIWGILTDAFPILGYRRRPYFVVAGVVGTVSATIVAVGGEVSVVAALMCFVGVSASLAI 153
KP++G ++D PI GYRRRPY +++G++GT S +A S A L +G S S+A+
Sbjct: 78 KPVFGFISDGLPIFGYRRRPYLILSGILGTASWMSLATIVHTSWAATLAIAIG-SLSVAM 136
Query: 154 ADVTIDACIA-RNSIEVPGLAPDLQSLCGFCSGSGALLGYLASGFFVHHLGPQGSLGLMA 212
+DV +D+ + R E A LQS+C S G L+ SG + + + G+ A
Sbjct: 137 SDVIVDSLVVERARGESQAKAGSLQSVCWGASAIGGLITAYFSGLLLQYFTTRTVFGITA 196
Query: 213 VFPALTIVLGFVIYEKRTTGLHNEKKKGVVENVGTTIRSMYKTIKYPQAWKPSLYMFLSL 272
+FP + + ++I E + NE + + ++ + + I W P+ ++F+
Sbjct: 197 LFPLIVSGVAWLIAESPVSKGTNESNQTNPLPIKHQLKQLRQAITQKTIWLPTAFVFIWQ 256
Query: 273 ALNVTTHEGHFYWYTDSKVGPAFSQEFVGIIYAIGAVASIIGVLIYHKYLKNYTFRNLVF 332
A E F++++ +++ F EF+G + + + AS+ GV I+ ++LK+ FR +
Sbjct: 257 A--TPNAESAFFYFSTNEL--HFEPEFLGRVNLVTSFASLAGVWIFQRFLKSIPFRVIFA 312
Query: 333 YAQLLYAISGLLDLM 347
++ +L ++ G+ L+
Sbjct: 313 WSTVLSSMLGMTMLL 327
>ref|ZP_00328462.1| COG0477: Permeases of the major facilitator superfamily
[Trichodesmium erythraeum IMS101]
Length = 458
Score = 141 bits (356), Expect = 3e-32
Identities = 93/313 (29%), Positives = 162/313 (51%), Gaps = 15/313 (4%)
Query: 36 NNLNPTFIAGIFLVYGIGQGFTGSLFKLVTDYYWKDVQKLQPSTVQLYVGIYFIPWILKP 95
N P IA I LVY + QG G L L ++ KD + P+ V +GI IPW++KP
Sbjct: 23 NEPTPELIA-ILLVYFV-QGILG-LGSLAISFFLKDELAMTPAEVSAMLGIVIIPWMIKP 79
Query: 96 IWGILTDAFPILGYRRRPYFVVAGVVGTVSATIVAVGGEVSVVAALMCFVGVSASLAIAD 155
++G +TD PILGYRR PY + +G++G S +A + + A + + S S+A++D
Sbjct: 80 LFGFITDGVPILGYRRGPYIIFSGILGAFSWVALATWVDTPLTATVAIAMS-SLSVAMSD 138
Query: 156 VTIDACIA-RNSIEVPGLAPDLQSLCGFCSGSGALLGYLASGFFVHHLGPQGSLGLMAVF 214
V ID+ + R E LQSLC S G L+ SG+ + H PQ + A F
Sbjct: 139 VIIDSLVVERVQKESVSNTGSLQSLCWSASALGGLITAYFSGYLLEHFSPQTVFLITATF 198
Query: 215 PALTIVLGFVIYEKRTTGLHNEKKKGVVENVGTTIRSMYKTIKYPQAWKPSLYMFLSLAL 274
P + ++ +I EK+ + N +NV I+ + K + P+ ++ + A
Sbjct: 199 PLIVSLVPLLIEEKKVSIRIN------FDNVKQQIKQVKKAVIKKSILLPTAFLVIWQA- 251
Query: 275 NVTTHEGHFYWYTDSKVGPAFSQEFVGIIYAIGAVASIIGVLIYHKYLKNYTFRNLVFYA 334
E F++++ +++G F EF+G I + ++A++ GV ++ ++ K+ FR + ++
Sbjct: 252 -TPNSESAFFFFSTNELG--FQAEFLGRIRLVTSIAALFGVWLFQRFFKSVPFRKIFGWS 308
Query: 335 QLLYAISGLLDLM 347
++ A+ G+ L+
Sbjct: 309 TVISALLGMTTLL 321
>gb|AAM62749.1| unknown [Arabidopsis thaliana] gi|28059039|gb|AAO29981.1| unknown
protein [Arabidopsis thaliana]
gi|20197619|gb|AAD15400.2| expressed protein
[Arabidopsis thaliana] gi|19698919|gb|AAL91195.1|
unknown protein [Arabidopsis thaliana]
gi|18402856|ref|NP_565734.1| integral membrane
transporter family protein [Arabidopsis thaliana]
Length = 560
Score = 137 bits (346), Expect = 4e-31
Identities = 90/302 (29%), Positives = 165/302 (53%), Gaps = 18/302 (5%)
Query: 38 LNPTFIAGIFLVYGIGQGFTGSLFKLVTDYYWKDVQKLQPSTVQLYVGIYFIPWILKPIW 97
L+P +A + +VY + QG G L +L +Y KD L P+ + G+ +PW++KP++
Sbjct: 123 LSPDNVA-VAMVYFV-QGVLG-LARLAVSFYLKDDLHLDPAETAVITGLSSLPWLVKPLY 179
Query: 98 GILTDAFPILGYRRRPYFVVAGVVGTVSATIVAVGGEVSVVAALMCFVGVSASLAIADVT 157
G ++D+ P+ GYRRR Y V++G++G S +++A G S +A C + S S+A +DV
Sbjct: 180 GFISDSVPLFGYRRRSYLVLSGLLGAFSWSLMA-GFVDSKYSAAFCILLGSLSVAFSDVV 238
Query: 158 IDACIA-RNSIEVPGLAPDLQSLCGFCSGSGALLGYLASGFFVHHLGPQGSLGLMAVFPA 216
+D+ + R E ++ LQSLC S G ++ SG V G + G+ A+ P
Sbjct: 239 VDSMVVERARGESQSVSGSLQSLCWGSSAFGGIVSSYFSGSLVESYGVRFVFGVTALLPL 298
Query: 217 LTIVLGFVIYEKRTTGLHNEKKK-------GVVENVGTTIRSMYKTIKYPQAWKPSLYMF 269
+T + ++ E+R + +K+ G ++ + ++ IK P + P+L++F
Sbjct: 299 ITSAVAVLVNEQRVVRPASGQKENITLLSPGFLQTSKQNMIQLWGAIKQPNVFLPTLFIF 358
Query: 270 LSLALNVTTH-EGHFYWYTDSKVGPAFSQEFVGIIYAIGAVASIIGVLIYHKYLKNYTFR 328
L A T H + +++T +K+G F+ EF+G + + ++AS++GV +Y+ +LK R
Sbjct: 359 LWQA---TPHSDSAMFYFTTNKLG--FTPEFLGRVKLVTSIASLLGVGLYNGFLKTVPLR 413
Query: 329 NL 330
+
Sbjct: 414 KI 415
>pir||C84728 hypothetical protein At2g32040 [imported] - Arabidopsis thaliana
Length = 429
Score = 136 bits (342), Expect = 1e-30
Identities = 87/292 (29%), Positives = 159/292 (53%), Gaps = 17/292 (5%)
Query: 48 LVYGIGQGFTGSLFKLVTDYYWKDVQKLQPSTVQLYVGIYFIPWILKPIWGILTDAFPIL 107
+VY + QG G L +L +Y KD L P+ + G+ +PW++KP++G ++D+ P+
Sbjct: 1 MVYFV-QGVLG-LARLAVSFYLKDDLHLDPAETAVITGLSSLPWLVKPLYGFISDSVPLF 58
Query: 108 GYRRRPYFVVAGVVGTVSATIVAVGGEVSVVAALMCFVGVSASLAIADVTIDACIA-RNS 166
GYRRR Y V++G++G S +++A G S +A C + S S+A +DV +D+ + R
Sbjct: 59 GYRRRSYLVLSGLLGAFSWSLMA-GFVDSKYSAAFCILLGSLSVAFSDVVVDSMVVERAR 117
Query: 167 IEVPGLAPDLQSLCGFCSGSGALLGYLASGFFVHHLGPQGSLGLMAVFPALTIVLGFVIY 226
E ++ LQSLC S G ++ SG V G + G+ A+ P +T + ++
Sbjct: 118 GESQSVSGSLQSLCWGSSAFGGIVSSYFSGSLVESYGVRFVFGVTALLPLITSAVAVLVN 177
Query: 227 EKRTTGLHNEKKK-------GVVENVGTTIRSMYKTIKYPQAWKPSLYMFLSLALNVTTH 279
E+R + +K+ G ++ + ++ IK P + P+L++FL A T H
Sbjct: 178 EQRVVRPASGQKENITLLSPGFLQTSKQNMIQLWGAIKQPNVFLPTLFIFLWQA---TPH 234
Query: 280 -EGHFYWYTDSKVGPAFSQEFVGIIYAIGAVASIIGVLIYHKYLKNYTFRNL 330
+ +++T +K+G F+ EF+G + + ++AS++GV +Y+ +LK R +
Sbjct: 235 SDSAMFYFTTNKLG--FTPEFLGRVKLVTSIASLLGVGLYNGFLKTVPLRKI 284
>ref|NP_680807.1| hypothetical protein tll0016 [Thermosynechococcus elongatus BP-1]
gi|22293737|dbj|BAC07569.1| tll0016 [Thermosynechococcus
elongatus BP-1]
Length = 451
Score = 133 bits (335), Expect = 7e-30
Identities = 93/330 (28%), Positives = 167/330 (50%), Gaps = 21/330 (6%)
Query: 25 TEPIEWL-----TMLTNNLNPTFIAGIFLVYGIGQGFTGSLFKLVTDYYWKDVQKLQPST 79
++P WL T+L + I LVY + QG G L +L +++KD L P+
Sbjct: 6 SQPQRWLRGLKETLLFGQEPSGELLAILLVYFV-QGVLG-LARLAISFFFKDELGLSPAE 63
Query: 80 VQLYVGIYFIPWILKPIWGILTDAFPILGYRRRPYFVVAGVVGTVSATIVAVGGEVSVV- 138
V +G+ +PW++KP++G+++D+ P+LG+RRR Y V++G++G +++G V+
Sbjct: 64 VAALMGVAALPWVIKPVFGLVSDSLPLLGFRRRSYLVLSGLLG--CGAWLSLGTWVATPW 121
Query: 139 AALMCFVGVSASLAIADVTIDACIA-RNSIEVPGLAPDLQSLCGFCSGSGALLGYLASGF 197
A +G S ++A++DV +D+ + R E LQSL + G +L SG
Sbjct: 122 QATAAILGASIAIALSDVIVDSLVVERARQESATEMGTLQSLSWAATAVGGILTAYFSGQ 181
Query: 198 FVHHLGPQGSLGLMAVFPALTIVLGFVIYEKRTTGLHNEKKKGVVENVGTTIRSMYKTIK 257
+ P+ + AVFP L + I E+R + + I + + +
Sbjct: 182 LLQWFSPRTVFQITAVFPLLVSAVAGAIAEERVMA------RPQLSQTWEHINHVRQAMM 235
Query: 258 YPQAWKPSLYMFLSLALNVTTHEGHFYWYTDSKVGPAFSQEFVGIIYAIGAVASIIGVLI 317
+ W P ++FL A E F+++T +++G F EF+G + + ++AS++GV I
Sbjct: 236 QKRIWLPVAFLFLWQA--TPGSESAFFFFTTNELG--FEAEFLGRVRLVTSIASLLGVWI 291
Query: 318 YHKYLKNYTFRNLVFYAQLLYAISGLLDLM 347
+ +YLK R + F++ +L A GL L+
Sbjct: 292 FQRYLKTLPLRTIFFWSTILSAFLGLSSLI 321
>ref|XP_476780.1| putative integral membrane protein [Oryza sativa (japonica
cultivar-group)] gi|34394451|dbj|BAC83625.1| putative
integral membrane protein [Oryza sativa (japonica
cultivar-group)]
Length = 490
Score = 133 bits (334), Expect = 9e-30
Identities = 88/322 (27%), Positives = 165/322 (50%), Gaps = 12/322 (3%)
Query: 30 WLTMLTNNLNPTFIAGIFLVYGIGQGFTGSLFKLVTDYYWKDVQKLQPSTVQLYVGIYFI 89
+ T+ L+P +A + VY + QG G L +L +Y KD L P+ + G +
Sbjct: 54 YFTISGVELSPDNMA-VATVYFV-QGVLG-LARLAVSFYLKDDLHLDPAETAVISGFSSL 110
Query: 90 PWILKPIWGILTDAFPILGYRRRPYFVVAGVVGTVSATIVAVGGEVSVVAALMCFVGVSA 149
PW++KP++G ++D+ P+ GYRRR Y +++G +G +S ++A + AA +G S
Sbjct: 111 PWLIKPLYGFISDSIPLFGYRRRSYLILSGFLGALSWGLMATLVDSKYSAAFSILLG-SL 169
Query: 150 SLAIADVTIDACIA-RNSIEVPGLAPDLQSLCGFCSGSGALLGYLASGFFVHHLGPQGSL 208
S+A +DV +D+ + R E + LQSLC S G ++ SG V G +
Sbjct: 170 SVAFSDVVVDSMVVERARGESQSTSGSLQSLCWGSSAFGGIVSAYFSGSLVDTYGVRFVF 229
Query: 209 GLMAVFPALTIVLGFVIYEKRTTG---LHNEKKKGVVENVGTTIRSMYKTIKYPQAWKPS 265
G+ A P +T + ++ E R + + G +E IR ++ ++K P + P+
Sbjct: 230 GVTAFLPLMTSAVAVLVNEHRLSSGERAMSHSGSGFIETSKQHIRQLWTSVKQPNIFLPT 289
Query: 266 LYMFLSLALNVTTHEGHFYWYTDSKVGPAFSQEFVGIIYAIGAVASIIGVLIYHKYLKNY 325
L++FL A + +++ +K+G F+ EF+G + + ++AS++GV +Y+ +LK
Sbjct: 290 LFIFLWQA--TPKSDSAMFFFITNKLG--FTPEFLGRVKLVTSIASLLGVGLYNYFLKAV 345
Query: 326 TFRNLVFYAQLLYAISGLLDLM 347
R + ++ + G+ ++
Sbjct: 346 PLRKIFLATTIIGSALGMTQVL 367
>dbj|BAB75984.1| all4285 [Nostoc sp. PCC 7120] gi|25533268|pir||AF2341 hypothetical
protein all4285 [imported] - Nostoc sp. (strain PCC
7120) gi|17231777|ref|NP_488325.1| hypothetical protein
all4285 [Nostoc sp. PCC 7120]
Length = 473
Score = 133 bits (334), Expect = 9e-30
Identities = 91/312 (29%), Positives = 163/312 (52%), Gaps = 22/312 (7%)
Query: 43 IAGIFLVYGIGQGFTGSLFKLVTDYYWKDVQKLQPSTVQLYVGIYFIPWILKPIWGILTD 102
+ I VY + QG G L +L ++ KD L P V +GI +PW++KP++G ++D
Sbjct: 29 LIAILSVYFV-QGILG-LARLAVSFFLKDELLLSPVQVSALMGIVALPWMVKPLFGFVSD 86
Query: 103 AFPILGYRRRPYFVVAGVVGTVS----ATIVAVGGEVSVVAALMCFVGVSASLAIADVTI 158
PI GYRRRPY V++G++G +S ATIV +V AL S S+A++DV +
Sbjct: 87 GLPIFGYRRRPYLVLSGILGAISWISLATIVHTSWAATVAIAL-----GSLSVAVSDVIV 141
Query: 159 DACIA-RNSIEVPGLAPDLQSLCGFCSGSGALLGYLASGFFVHHLGPQGSLGLMAVFPAL 217
D+ + R E LQ+LC S G L+ SG + H + G+ A FP +
Sbjct: 142 DSLVVERAREESVAHVGSLQALCWGASAFGGLITAYFSGLLLEHFTTRTVFGITASFPLI 201
Query: 218 TIVLGFVIYEKRTT--GLHNEKKKGVVENVGTTIRSMYKTIKYPQAWKPSLYMFLSLALN 275
+ ++I E + G N + +V ++ + + W P+ ++F+ A
Sbjct: 202 VSGVAWLIAESPVSKDGSGNTN----LLSVKHQLQQLRQAFTQKSIWLPTAFVFIWQA-- 255
Query: 276 VTTHEGHFYWYTDSKVGPAFSQEFVGIIYAIGAVASIIGVLIYHKYLKNYTFRNLVFYAQ 335
T + F++++ +++ F EF+G + + ++AS+IGV I+ ++LK+ +FR + ++
Sbjct: 256 TPTADSAFFFFSTNEL--HFEPEFLGRVRLVTSLASLIGVWIFQRFLKSVSFRLIFGWST 313
Query: 336 LLYAISGLLDLM 347
++ A+ G+ L+
Sbjct: 314 VISAVLGMTMLL 325
>ref|ZP_00158751.1| COG0477: Permeases of the major facilitator superfamily [Anabaena
variabilis ATCC 29413]
Length = 472
Score = 130 bits (327), Expect = 6e-29
Identities = 88/310 (28%), Positives = 162/310 (51%), Gaps = 18/310 (5%)
Query: 43 IAGIFLVYGIGQGFTGSLFKLVTDYYWKDVQKLQPSTVQLYVGIYFIPWILKPIWGILTD 102
+ I VY + QG G L +L ++ KD L P V +GI +PW++KP++G ++D
Sbjct: 29 LIAILSVYFV-QGILG-LARLAVSFFLKDELLLSPVQVSALMGIVALPWMVKPLFGFVSD 86
Query: 103 AFPILGYRRRPYFVVAGVVGTVS----ATIVAVGGEVSVVAALMCFVGVSASLAIADVTI 158
PI GYRRRPY V++G++G +S ATIV +V AL S S+A++DV +
Sbjct: 87 GLPIFGYRRRPYLVLSGILGAISWISLATIVHTSWAATVAIAL-----GSLSVAVSDVIV 141
Query: 159 DACIA-RNSIEVPGLAPDLQSLCGFCSGSGALLGYLASGFFVHHLGPQGSLGLMAVFPAL 217
D+ + R E LQ+LC S G L+ SG + H + + A FP +
Sbjct: 142 DSLVVERAREESVSHVGSLQALCWGASAFGGLITAYFSGLLLEHFTTRTVFLITASFPLI 201
Query: 218 TIVLGFVIYEKRTTGLHNEKKKGVVENVGTTIRSMYKTIKYPQAWKPSLYMFLSLALNVT 277
+ ++I E + ++ + +V ++ + + W P+ ++F+ A
Sbjct: 202 VSGVAWLIAESPVSKDGSDNTN--LLSVKQQLQQLRQAFTQKSIWLPTAFVFIWQA--TP 257
Query: 278 THEGHFYWYTDSKVGPAFSQEFVGIIYAIGAVASIIGVLIYHKYLKNYTFRNLVFYAQLL 337
T + F++++ +++ F EF+G + + ++AS+IGV I+ ++LK+ +FR + ++ ++
Sbjct: 258 TADSAFFFFSTNEL--HFEPEFLGRVRLVTSLASLIGVWIFQRFLKSVSFRLIFGWSTVI 315
Query: 338 YAISGLLDLM 347
A+ G+ L+
Sbjct: 316 SAVLGMTMLL 325
>ref|NP_442292.1| integral membrane protein [Synechocystis sp. PCC 6803]
gi|1001631|dbj|BAA10362.1| integral membrane protein
[Synechocystis sp. PCC 6803] gi|7470715|pir||S76516
integral membrane protein - Synechocystis sp. (strain
PCC 6803)
Length = 494
Score = 125 bits (315), Expect = 1e-27
Identities = 91/332 (27%), Positives = 166/332 (49%), Gaps = 19/332 (5%)
Query: 19 PLLSILTEPIEWLTMLTNNLNPTFIAGIFLVYGIGQGFTGSLFKLVTDYYWKDVQKLQPS 78
PL L E + L N + I +Y + QG G L +L ++ KD L P+
Sbjct: 17 PLKRFLREKV-----LLGNAPSWELLAILSIYFV-QGVLG-LSRLAVSFFLKDELGLSPA 69
Query: 79 TVQLYVGIYFIPWILKPIWGILTDAFPILGYRRRPYFVVAGVVGTVSATIVAVGGEVSVV 138
+ +G+ PWILKP+ G+++D P+ GYRRR Y ++G++G+ + A
Sbjct: 70 AMGALIGLGAAPWILKPVLGLMSDTVPLFGYRRRSYLWLSGLMGSAGWLLFAAWVSSGTQ 129
Query: 139 AALMCFVGVSASLAIADVTIDACIA-RNSIEVPGLAPDLQSLCGFCSGSGALLGYLASGF 197
A L+ + S S+AI DV +D+ + R E LQSL + G ++ ASG
Sbjct: 130 AGLV-LLFTSLSVAIGDVIVDSLVVERAQRESLAQVGSLQSLTWGAAAVGGIITAYASGA 188
Query: 198 FVHHLGPQGSLGLMAVFPALTIVLGFVIYEKRTTGLHNEKKKGVVENVGTTIRSMYKTIK 257
+ + + A+FP LT+ F+I E +T EK + + I+ +++ ++
Sbjct: 189 LLEWFSTRTVFAITAIFPLLTVGAAFLISEV-STAEEEEKPQPKAQ-----IKLVWQAVR 242
Query: 258 YPQAWKPSLYMFLSLALNVTTHEGHFYWYTDSKVGPAFSQEFVGIIYAIGAVASIIGVLI 317
P+L++F A + E F+++T +++G F +F+G + + +VA +IGV +
Sbjct: 243 QKTILLPTLFIFFWQA--TPSAESAFFYFTTNELG--FEPKFLGRVRLVTSVAGLIGVGL 298
Query: 318 YHKYLKNYTFRNLVFYAQLLYAISGLLDLMCL 349
Y ++LK FR ++ ++ ++ ++ GL L+ +
Sbjct: 299 YQRFLKTLPFRVIMGWSTVISSLLGLTTLILI 330
Database: nr
Posted date: Jul 5, 2005 12:34 AM
Number of letters in database: 863,360,394
Number of sequences in database: 2,540,612
Lambda K H
0.325 0.141 0.433
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 600,868,629
Number of Sequences: 2540612
Number of extensions: 26464453
Number of successful extensions: 82422
Number of sequences better than 10.0: 182
Number of HSP's better than 10.0 without gapping: 58
Number of HSP's successfully gapped in prelim test: 125
Number of HSP's that attempted gapping in prelim test: 82215
Number of HSP's gapped (non-prelim): 224
length of query: 349
length of database: 863,360,394
effective HSP length: 129
effective length of query: 220
effective length of database: 535,621,446
effective search space: 117836718120
effective search space used: 117836718120
T: 11
A: 40
X1: 15 ( 7.0 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 40 (21.6 bits)
S2: 75 (33.5 bits)
Medicago: description of AC143339.10


