

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC142394.6 - phase: 0
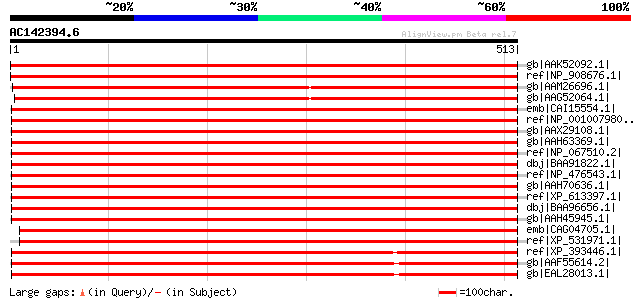
(513 letters)
Database: nr
2,540,612 sequences; 863,360,394 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
gb|AAK52092.1| WD-40 repeat protein [Lycopersicon esculentum] 926 0.0
ref|NP_908676.1| P0426D06.17 [Oryza sativa (japonica cultivar-gr... 920 0.0
gb|AAM26696.1| At1g73720/F25P22_14 [Arabidopsis thaliana] gi|180... 908 0.0
gb|AAG52064.1| unknown protein; 53481-57666 [Arabidopsis thalian... 897 0.0
emb|CAI15554.1| RP11-54K16.3 [Homo sapiens] gi|39652282|dbj|BAD0... 708 0.0
ref|NP_001007980.1| similar to Smu-1 suppressor of mec-8 and unc... 708 0.0
gb|AAX29108.1| smu-1 suppressor of mec-8 and unc-52-like [synthe... 708 0.0
gb|AAH63369.1| Hypothetical protein MGC75979 [Xenopus tropicalis... 706 0.0
ref|NP_067510.2| smu-1 suppressor of mec-8 and unc-52 homolog [M... 705 0.0
dbj|BAA91822.1| unnamed protein product [Homo sapiens] gi|892267... 705 0.0
ref|NP_476543.1| smu-1 suppressor of mec-8 and unc-52 homolog [R... 704 0.0
gb|AAH70636.1| MGC81475 protein [Xenopus laevis] 704 0.0
ref|XP_613397.1| PREDICTED: similar to Smu-1 suppressor of mec-8... 703 0.0
dbj|BAA96656.1| unnamed protein product [Mus musculus] 703 0.0
gb|AAH45945.1| Smu1 protein [Danio rerio] gi|41055638|ref|NP_956... 699 0.0
emb|CAG04705.1| unnamed protein product [Tetraodon nigroviridis] 699 0.0
ref|XP_531971.1| PREDICTED: similar to Smu-1 suppressor of mec-8... 698 0.0
ref|XP_393446.1| PREDICTED: similar to ENSANGP00000015224 [Apis ... 677 0.0
gb|AAF55614.2| CG5451-PA [Drosophila melanogaster] gi|15292169|g... 671 0.0
gb|EAL28013.1| GA18890-PA [Drosophila pseudoobscura] 669 0.0
>gb|AAK52092.1| WD-40 repeat protein [Lycopersicon esculentum]
Length = 514
Score = 926 bits (2393), Expect = 0.0
Identities = 444/512 (86%), Positives = 484/512 (93%)
Query: 2 STLEIEARDVIKTVLQFCKENSLHQTFQTLQNECQVSLNTVDSIETFVADINGGRWDAIL 61
STLEIEARDVIK VLQFCKENSLHQ+FQTLQNECQVSLNTVDS+ETF+ADIN GRWDA+L
Sbjct: 3 STLEIEARDVIKIVLQFCKENSLHQSFQTLQNECQVSLNTVDSLETFIADINSGRWDAVL 62
Query: 62 PQVSQLKLPRNTLEDLYEQIVLEMIELRELDTARAILRQTQVMGVMKQEQPERYLRLEHL 121
PQV+QLKLPR LEDLYEQIVLEMIELRE+DTARAILRQT VMGVMKQEQPERYLRLEHL
Sbjct: 63 PQVAQLKLPRKILEDLYEQIVLEMIELREMDTARAILRQTNVMGVMKQEQPERYLRLEHL 122
Query: 122 LVRTYFDPNEAYQESTKEKRRAQIAQAIAAEVTVVPPSRLMALIGQALKWQQHQGLLPPG 181
LVRTYFDP+EAY +STKEKRRAQI+QA++AEV+VVPPSRLMALIGQALKWQQHQGLLPPG
Sbjct: 123 LVRTYFDPHEAYNDSTKEKRRAQISQALSAEVSVVPPSRLMALIGQALKWQQHQGLLPPG 182
Query: 182 TQFDLFRGTAAMKQDVDDMYPTNLSHTIKFGAKSHAECARFSPDGQYLVSCSIDGFIEVW 241
TQFDLFRGTAAMKQD +DMYPT L HTIKF KSH EC+RFSPDGQ+LVSCS+DGFIEVW
Sbjct: 183 TQFDLFRGTAAMKQDEEDMYPTTLGHTIKFPKKSHPECSRFSPDGQFLVSCSVDGFIEVW 242
Query: 242 DYISGKLKKDLQYQAEETFMMHDEPVLCVDFSRDSEMIASGSTDGKIKVWRIRTAQCLRR 301
D+ISGKLKKDLQYQA+ETFMMHD+ VLCVDFSRDSEM+ASGS DGKIKVWRIRT QCLRR
Sbjct: 243 DHISGKLKKDLQYQADETFMMHDDAVLCVDFSRDSEMLASGSQDGKIKVWRIRTGQCLRR 302
Query: 302 LEHAHSQGVTSVSFSRDGSQLLSTSFDSTARIHGLKSGKMLKEFRGHTSYVNDATFTNDG 361
LE AHSQGVTS+ FSRDG+Q+LSTSFDSTARIHGLKSGK+LKEFRGH+SYVNDA FT DG
Sbjct: 303 LERAHSQGVTSLVFSRDGTQILSTSFDSTARIHGLKSGKLLKEFRGHSSYVNDAIFTFDG 362
Query: 362 TRVITASSDCTVKVWDLKTTDCLQTFKPPPPLRGGDASVNSVFIFPKNTEHIVVCNKTSS 421
+RVITASSD +VKVWD KTT+CLQTF+PPPPLRGGDASVNSV +FPKN EHI+VCNKTSS
Sbjct: 363 SRVITASSDGSVKVWDAKTTECLQTFRPPPPLRGGDASVNSVHLFPKNNEHIIVCNKTSS 422
Query: 422 IYIMTLQGQVVKSFSSGKREGGDFVAACISPKGEWIYCVGEDRNMYCFSYQSGKLEHLMT 481
IY+MTLQGQVVKSFSSGKREGGDFVAAC+SPKGEWIYCVGEDRN+YCFSYQS KLEHLM
Sbjct: 423 IYLMTLQGQVVKSFSSGKREGGDFVAACLSPKGEWIYCVGEDRNLYCFSYQSNKLEHLMK 482
Query: 482 VHDKEIIGVTHHPHRNLVATFSDDHTMKLWKP 513
VH+K++IG+THHPH NLVAT+ +D TMKLWKP
Sbjct: 483 VHEKDVIGITHHPHINLVATYGEDCTMKLWKP 514
>ref|NP_908676.1| P0426D06.17 [Oryza sativa (japonica cultivar-group)]
gi|12328578|dbj|BAB21237.1| putative WD-40 repeat
protein [Oryza sativa (japonica cultivar-group)]
Length = 517
Score = 920 bits (2377), Expect = 0.0
Identities = 441/512 (86%), Positives = 481/512 (93%)
Query: 2 STLEIEARDVIKTVLQFCKENSLHQTFQTLQNECQVSLNTVDSIETFVADINGGRWDAIL 61
STLEIEARDV+K VLQFCKENSL QTFQTLQNECQVSLNTVDSI+TF+ADIN GRWDA+L
Sbjct: 6 STLEIEARDVVKIVLQFCKENSLQQTFQTLQNECQVSLNTVDSIDTFIADINAGRWDAVL 65
Query: 62 PQVSQLKLPRNTLEDLYEQIVLEMIELRELDTARAILRQTQVMGVMKQEQPERYLRLEHL 121
PQV+QLKLPR LEDLYEQIVLEM ELRELDTARAILRQTQVMGVMKQEQPERYLRLEHL
Sbjct: 66 PQVAQLKLPRKKLEDLYEQIVLEMAELRELDTARAILRQTQVMGVMKQEQPERYLRLEHL 125
Query: 122 LVRTYFDPNEAYQESTKEKRRAQIAQAIAAEVTVVPPSRLMALIGQALKWQQHQGLLPPG 181
LVRTYFDPNEAYQESTKEKRRAQI+QAIA+EV+VVPPSRLMALIGQALKWQQHQGLLPPG
Sbjct: 126 LVRTYFDPNEAYQESTKEKRRAQISQAIASEVSVVPPSRLMALIGQALKWQQHQGLLPPG 185
Query: 182 TQFDLFRGTAAMKQDVDDMYPTNLSHTIKFGAKSHAECARFSPDGQYLVSCSIDGFIEVW 241
TQFDLFRGTAAMKQD ++ YPT LSH KFG K+H ECARFSPDGQYLVSCS+DG IEVW
Sbjct: 186 TQFDLFRGTAAMKQDEEETYPTTLSHQTKFGKKTHPECARFSPDGQYLVSCSVDGIIEVW 245
Query: 242 DYISGKLKKDLQYQAEETFMMHDEPVLCVDFSRDSEMIASGSTDGKIKVWRIRTAQCLRR 301
DYISGKLKKDLQYQA+E+FMMHD+ VL VDFSRDSEM+ASGS DGKIKVWRIRT QCLRR
Sbjct: 246 DYISGKLKKDLQYQADESFMMHDDAVLSVDFSRDSEMLASGSQDGKIKVWRIRTGQCLRR 305
Query: 302 LEHAHSQGVTSVSFSRDGSQLLSTSFDSTARIHGLKSGKMLKEFRGHTSYVNDATFTNDG 361
LE AH++GVTSV+FSRDG+Q+LS+SFD+TAR+HGLKSGKMLKEFRGH SYVN A F+ DG
Sbjct: 306 LERAHAKGVTSVTFSRDGTQILSSSFDTTARVHGLKSGKMLKEFRGHNSYVNCAIFSTDG 365
Query: 362 TRVITASSDCTVKVWDLKTTDCLQTFKPPPPLRGGDASVNSVFIFPKNTEHIVVCNKTSS 421
+RVITASSDCTVKVWD KTTDCLQTFKPPPPLRGGDA+VNSV + PKN++HI+VCNKTSS
Sbjct: 366 SRVITASSDCTVKVWDTKTTDCLQTFKPPPPLRGGDATVNSVHLSPKNSDHIIVCNKTSS 425
Query: 422 IYIMTLQGQVVKSFSSGKREGGDFVAACISPKGEWIYCVGEDRNMYCFSYQSGKLEHLMT 481
IYIMTLQGQVVKSFSSGKREGGDF+AA +SPKGEWIYCVGED NMYCFSYQSGKLEHLM
Sbjct: 426 IYIMTLQGQVVKSFSSGKREGGDFLAASVSPKGEWIYCVGEDMNMYCFSYQSGKLEHLMK 485
Query: 482 VHDKEIIGVTHHPHRNLVATFSDDHTMKLWKP 513
VHDK++IG+THHPHRNL+AT+++D TMK WKP
Sbjct: 486 VHDKDVIGITHHPHRNLIATYAEDCTMKTWKP 517
>gb|AAM26696.1| At1g73720/F25P22_14 [Arabidopsis thaliana]
gi|18087515|gb|AAL58892.1| At1g73720/F25P22_14
[Arabidopsis thaliana] gi|22330602|ref|NP_177513.2|
transducin family protein / WD-40 repeat family protein
[Arabidopsis thaliana]
Length = 511
Score = 908 bits (2346), Expect = 0.0
Identities = 433/510 (84%), Positives = 481/510 (93%), Gaps = 1/510 (0%)
Query: 4 LEIEARDVIKTVLQFCKENSLHQTFQTLQNECQVSLNTVDSIETFVADINGGRWDAILPQ 63
LEIEARDVIK +LQFCKENSL+QTFQTLQ+ECQVSLNTVDS+ETF++DIN GRWD++LPQ
Sbjct: 3 LEIEARDVIKIMLQFCKENSLNQTFQTLQSECQVSLNTVDSVETFISDINSGRWDSVLPQ 62
Query: 64 VSQLKLPRNTLEDLYEQIVLEMIELRELDTARAILRQTQVMGVMKQEQPERYLRLEHLLV 123
VSQLKLPRN LEDLYEQIVLEMIELRELDTARAILRQTQVMGVMKQEQ ERYLR+EHLLV
Sbjct: 63 VSQLKLPRNKLEDLYEQIVLEMIELRELDTARAILRQTQVMGVMKQEQAERYLRMEHLLV 122
Query: 124 RTYFDPNEAYQESTKEKRRAQIAQAIAAEVTVVPPSRLMALIGQALKWQQHQGLLPPGTQ 183
R+YFDP+EAY +STKE++RAQIAQA+AAEVTVVPPSRLMALIGQALKWQQHQGLLPPGTQ
Sbjct: 123 RSYFDPHEAYGDSTKERKRAQIAQAVAAEVTVVPPSRLMALIGQALKWQQHQGLLPPGTQ 182
Query: 184 FDLFRGTAAMKQDVDDMYPTNLSHTIKFGAKSHAECARFSPDGQYLVSCSIDGFIEVWDY 243
FDLFRGTAAMKQDV+D +P L+HTIKFG KSHAECARFSPDGQ+L S S+DGFIEVWDY
Sbjct: 183 FDLFRGTAAMKQDVEDTHPNVLTHTIKFGKKSHAECARFSPDGQFLASSSVDGFIEVWDY 242
Query: 244 ISGKLKKDLQYQAEETFMMHDEPVLCVDFSRDSEMIASGSTDGKIKVWRIRTAQCLRRLE 303
ISGKLKKDLQYQA+E+FMMHD+PVLC+DFSRDSEM+ASGS DGKIK+WRIRT C+RR +
Sbjct: 243 ISGKLKKDLQYQADESFMMHDDPVLCIDFSRDSEMLASGSQDGKIKIWRIRTGVCIRRFD 302
Query: 304 HAHSQGVTSVSFSRDGSQLLSTSFDSTARIHGLKSGKMLKEFRGHTSYVNDATFTNDGTR 363
AHSQGVTS+SFSRDGSQLLSTSFD TARIHGLKSGK+LKEFRGHTSYVN A FT+DG+R
Sbjct: 303 -AHSQGVTSLSFSRDGSQLLSTSFDQTARIHGLKSGKLLKEFRGHTSYVNHAIFTSDGSR 361
Query: 364 VITASSDCTVKVWDLKTTDCLQTFKPPPPLRGGDASVNSVFIFPKNTEHIVVCNKTSSIY 423
+ITASSDCTVKVWD KTTDCLQTFKPPPPLRG DASVNS+ +FPKNTEHIVVCNKTSSIY
Sbjct: 362 IITASSDCTVKVWDSKTTDCLQTFKPPPPLRGTDASVNSIHLFPKNTEHIVVCNKTSSIY 421
Query: 424 IMTLQGQVVKSFSSGKREGGDFVAACISPKGEWIYCVGEDRNMYCFSYQSGKLEHLMTVH 483
IMTLQGQVVKSFSSG REGGDFVAAC+S KG+WIYC+GED+ +YCF+YQSG LEH M VH
Sbjct: 422 IMTLQGQVVKSFSSGNREGGDFVAACVSTKGDWIYCIGEDKKLYCFNYQSGGLEHFMMVH 481
Query: 484 DKEIIGVTHHPHRNLVATFSDDHTMKLWKP 513
+K++IG+THHPHRNL+AT+S+D TMKLWKP
Sbjct: 482 EKDVIGITHHPHRNLLATYSEDCTMKLWKP 511
>gb|AAG52064.1| unknown protein; 53481-57666 [Arabidopsis thaliana]
gi|25406289|pir||D96764 unknown protein F25P22.14
[imported] - Arabidopsis thaliana
Length = 522
Score = 897 bits (2319), Expect = 0.0
Identities = 427/508 (84%), Positives = 476/508 (93%), Gaps = 1/508 (0%)
Query: 6 IEARDVIKTVLQFCKENSLHQTFQTLQNECQVSLNTVDSIETFVADINGGRWDAILPQVS 65
+E VIK +LQFCKENSL+QTFQTLQ+ECQVSLNTVDS+ETF++DIN GRWD++LPQVS
Sbjct: 16 VECLSVIKIMLQFCKENSLNQTFQTLQSECQVSLNTVDSVETFISDINSGRWDSVLPQVS 75
Query: 66 QLKLPRNTLEDLYEQIVLEMIELRELDTARAILRQTQVMGVMKQEQPERYLRLEHLLVRT 125
QLKLPRN LEDLYEQIVLEMIELRELDTARAILRQTQVMGVMKQEQ ERYLR+EHLLVR+
Sbjct: 76 QLKLPRNKLEDLYEQIVLEMIELRELDTARAILRQTQVMGVMKQEQAERYLRMEHLLVRS 135
Query: 126 YFDPNEAYQESTKEKRRAQIAQAIAAEVTVVPPSRLMALIGQALKWQQHQGLLPPGTQFD 185
YFDP+EAY +STKE++RAQIAQA+AAEVTVVPPSRLMALIGQALKWQQHQGLLPPGTQFD
Sbjct: 136 YFDPHEAYGDSTKERKRAQIAQAVAAEVTVVPPSRLMALIGQALKWQQHQGLLPPGTQFD 195
Query: 186 LFRGTAAMKQDVDDMYPTNLSHTIKFGAKSHAECARFSPDGQYLVSCSIDGFIEVWDYIS 245
LFRGTAAMKQDV+D +P L+HTIKFG KSHAECARFSPDGQ+L S S+DGFIEVWDYIS
Sbjct: 196 LFRGTAAMKQDVEDTHPNVLTHTIKFGKKSHAECARFSPDGQFLASSSVDGFIEVWDYIS 255
Query: 246 GKLKKDLQYQAEETFMMHDEPVLCVDFSRDSEMIASGSTDGKIKVWRIRTAQCLRRLEHA 305
GKLKKDLQYQA+E+FMMHD+PVLC+DFSRDSEM+ASGS DGKIK+WRIRT C+RR + A
Sbjct: 256 GKLKKDLQYQADESFMMHDDPVLCIDFSRDSEMLASGSQDGKIKIWRIRTGVCIRRFD-A 314
Query: 306 HSQGVTSVSFSRDGSQLLSTSFDSTARIHGLKSGKMLKEFRGHTSYVNDATFTNDGTRVI 365
HSQGVTS+SFSRDGSQLLSTSFD TARIHGLKSGK+LKEFRGHTSYVN A FT+DG+R+I
Sbjct: 315 HSQGVTSLSFSRDGSQLLSTSFDQTARIHGLKSGKLLKEFRGHTSYVNHAIFTSDGSRII 374
Query: 366 TASSDCTVKVWDLKTTDCLQTFKPPPPLRGGDASVNSVFIFPKNTEHIVVCNKTSSIYIM 425
TASSDCTVKVWD KTTDCLQTFKPPPPLRG DASVNS+ +FPKNTEHIVVCNKTSSIYIM
Sbjct: 375 TASSDCTVKVWDSKTTDCLQTFKPPPPLRGTDASVNSIHLFPKNTEHIVVCNKTSSIYIM 434
Query: 426 TLQGQVVKSFSSGKREGGDFVAACISPKGEWIYCVGEDRNMYCFSYQSGKLEHLMTVHDK 485
TLQGQVVKSFSSG REGGDFVAAC+S KG+WIYC+GED+ +YCF+YQSG LEH M VH+K
Sbjct: 435 TLQGQVVKSFSSGNREGGDFVAACVSTKGDWIYCIGEDKKLYCFNYQSGGLEHFMMVHEK 494
Query: 486 EIIGVTHHPHRNLVATFSDDHTMKLWKP 513
++IG+THHPHRNL+AT+S+D TMKLWKP
Sbjct: 495 DVIGITHHPHRNLLATYSEDCTMKLWKP 522
>emb|CAI15554.1| RP11-54K16.3 [Homo sapiens] gi|39652282|dbj|BAD04854.1| SMU-1
[Cricetulus griseus] gi|35505294|gb|AAH57446.1| Smu-1
suppressor of mec-8 and unc-52 homolog [Mus musculus]
gi|12804047|gb|AAH02876.1| Smu-1 suppressor of mec-8 and
unc-52 homolog [Homo sapiens]
Length = 513
Score = 708 bits (1827), Expect = 0.0
Identities = 334/512 (65%), Positives = 419/512 (81%), Gaps = 1/512 (0%)
Query: 3 TLEIEARDVIKTVLQFCKENSLHQTFQTLQNECQVSLNTVDSIETFVADINGGRWDAILP 62
++EIE+ DVI+ ++Q+ KENSLH+ TLQ E VSLNTVDSIE+FVADIN G WD +L
Sbjct: 2 SIEIESSDVIRLIMQYLKENSLHRALATLQEETTVSLNTVDSIESFVADINSGHWDTVLQ 61
Query: 63 QVSQLKLPRNTLEDLYEQIVLEMIELRELDTARAILRQTQVMGVMKQEQPERYLRLEHLL 122
+ LKLP TL DLYEQ+VLE+IELREL AR++LRQT M ++KQ QPERY+ LE+LL
Sbjct: 62 AIQSLKLPDKTLIDLYEQVVLELIELRELGAARSLLRQTDPMIMLKQTQPERYIHLENLL 121
Query: 123 VRTYFDPNEAYQE-STKEKRRAQIAQAIAAEVTVVPPSRLMALIGQALKWQQHQGLLPPG 181
R+YFDP EAY + S+KEKRRA IAQA+A EV+VVPPSRLMAL+GQALKWQQHQGLLPPG
Sbjct: 122 ARSYFDPREAYPDGSSKEKRRAAIAQALAGEVSVVPPSRLMALLGQALKWQQHQGLLPPG 181
Query: 182 TQFDLFRGTAAMKQDVDDMYPTNLSHTIKFGAKSHAECARFSPDGQYLVSCSIDGFIEVW 241
DLFRG AA+K ++ +PT LS IKFG KSH ECARFSPDGQYLV+ S+DGFIEVW
Sbjct: 182 MTIDLFRGKAAVKDVEEEKFPTQLSRHIKFGQKSHVECARFSPDGQYLVTGSVDGFIEVW 241
Query: 242 DYISGKLKKDLQYQAEETFMMHDEPVLCVDFSRDSEMIASGSTDGKIKVWRIRTAQCLRR 301
++ +GK++KDL+YQA++ FMM D+ VLC+ FSRD+EM+A+G+ DGKIKVW+I++ QCLRR
Sbjct: 242 NFTTGKIRKDLKYQAQDNFMMMDDAVLCMCFSRDTEMLATGAQDGKIKVWKIQSGQCLRR 301
Query: 302 LEHAHSQGVTSVSFSRDGSQLLSTSFDSTARIHGLKSGKMLKEFRGHTSYVNDATFTNDG 361
E AHS+GVT +SFS+D SQ+LS SFD T RIHGLKSGK LKEFRGH+S+VN+ATFT DG
Sbjct: 302 FERAHSKGVTCLSFSKDSSQILSASFDQTIRIHGLKSGKTLKEFRGHSSFVNEATFTQDG 361
Query: 362 TRVITASSDCTVKVWDLKTTDCLQTFKPPPPLRGGDASVNSVFIFPKNTEHIVVCNKTSS 421
+I+ASSD TVK+W++KTT+C TFK G D +VNSV + PKN EH VVCN++++
Sbjct: 362 HYIISASSDGTVKIWNMKTTECSNTFKSLGSTAGTDITVNSVILLPKNPEHFVVCNRSNT 421
Query: 422 IYIMTLQGQVVKSFSSGKREGGDFVAACISPKGEWIYCVGEDRNMYCFSYQSGKLEHLMT 481
+ IM +QGQ+V+SFSSGKREGGDFV +SP+GEWIYCVGED +YCFS +GKLE +T
Sbjct: 422 VVIMNMQGQIVRSFSSGKREGGDFVCCALSPRGEWIYCVGEDFVLYCFSTVTGKLERTLT 481
Query: 482 VHDKEIIGVTHHPHRNLVATFSDDHTMKLWKP 513
VH+K++IG+ HHPH+NL+AT+S+D +KLWKP
Sbjct: 482 VHEKDVIGIAHHPHQNLIATYSEDGLLKLWKP 513
>ref|NP_001007980.1| similar to Smu-1 suppressor of mec-8 and unc-52 homolog [Gallus
gallus] gi|53127424|emb|CAG31095.1| hypothetical protein
[Gallus gallus]
Length = 513
Score = 708 bits (1827), Expect = 0.0
Identities = 335/512 (65%), Positives = 419/512 (81%), Gaps = 1/512 (0%)
Query: 3 TLEIEARDVIKTVLQFCKENSLHQTFQTLQNECQVSLNTVDSIETFVADINGGRWDAILP 62
++EIE+ DVI+ ++Q+ KENSLH+ TLQ E VSLNTVDSIE+FVADIN G WD +L
Sbjct: 2 SIEIESSDVIRLIMQYLKENSLHRALATLQEETTVSLNTVDSIESFVADINSGHWDTVLQ 61
Query: 63 QVSQLKLPRNTLEDLYEQIVLEMIELRELDTARAILRQTQVMGVMKQEQPERYLRLEHLL 122
+ LKLP TL DLYEQ+VLE+IELREL AR++LRQT M ++KQ QPERY+ LE+LL
Sbjct: 62 AIQSLKLPDKTLIDLYEQVVLELIELRELGAARSLLRQTDPMIMLKQTQPERYIHLENLL 121
Query: 123 VRTYFDPNEAYQE-STKEKRRAQIAQAIAAEVTVVPPSRLMALIGQALKWQQHQGLLPPG 181
R+YFDP EAY + S+KEKRRA IAQA+A EV+VVPPSRLMAL+GQALKWQQHQGLLPPG
Sbjct: 122 ARSYFDPREAYPDGSSKEKRRAAIAQALAGEVSVVPPSRLMALLGQALKWQQHQGLLPPG 181
Query: 182 TQFDLFRGTAAMKQDVDDMYPTNLSHTIKFGAKSHAECARFSPDGQYLVSCSIDGFIEVW 241
DLFRG AA+K ++ +PT LS IKFG KSH ECARFSPDGQYLV+ S+DGFIEVW
Sbjct: 182 MTIDLFRGKAAVKDVEEEKFPTQLSRHIKFGQKSHVECARFSPDGQYLVTGSVDGFIEVW 241
Query: 242 DYISGKLKKDLQYQAEETFMMHDEPVLCVDFSRDSEMIASGSTDGKIKVWRIRTAQCLRR 301
++ +GK++KDL+YQA++ FMM D+ VLC+ FSRD+EM+A+G+ DGKIKVW+I++ QCLRR
Sbjct: 242 NFTTGKIRKDLKYQAQDNFMMMDDAVLCMCFSRDTEMLATGAQDGKIKVWKIQSGQCLRR 301
Query: 302 LEHAHSQGVTSVSFSRDGSQLLSTSFDSTARIHGLKSGKMLKEFRGHTSYVNDATFTNDG 361
E AHS+GVT +SFS+D SQ+LS SFD T RIHGLKSGK LKEFRGH+S+VN+ATFT DG
Sbjct: 302 FERAHSKGVTCLSFSKDSSQILSASFDQTIRIHGLKSGKTLKEFRGHSSFVNEATFTQDG 361
Query: 362 TRVITASSDCTVKVWDLKTTDCLQTFKPPPPLRGGDASVNSVFIFPKNTEHIVVCNKTSS 421
+I+ASSD TVKVW++KTT+C TFK G D +VNSV + PKN EH VVCN++++
Sbjct: 362 HYIISASSDGTVKVWNVKTTECSNTFKSLGSTAGTDITVNSVILLPKNPEHFVVCNRSNT 421
Query: 422 IYIMTLQGQVVKSFSSGKREGGDFVAACISPKGEWIYCVGEDRNMYCFSYQSGKLEHLMT 481
+ IM +QGQ+V+SFSSGKREGGDFV +SP+GEWIYCVGED +YCFS +GKLE +T
Sbjct: 422 VVIMNMQGQIVRSFSSGKREGGDFVCCALSPRGEWIYCVGEDFVLYCFSTVTGKLERTLT 481
Query: 482 VHDKEIIGVTHHPHRNLVATFSDDHTMKLWKP 513
VH+K++IG+ HHPH+NL+AT+S+D +KLWKP
Sbjct: 482 VHEKDVIGIAHHPHQNLIATYSEDGLLKLWKP 513
>gb|AAX29108.1| smu-1 suppressor of mec-8 and unc-52-like [synthetic construct]
Length = 514
Score = 708 bits (1827), Expect = 0.0
Identities = 334/512 (65%), Positives = 419/512 (81%), Gaps = 1/512 (0%)
Query: 3 TLEIEARDVIKTVLQFCKENSLHQTFQTLQNECQVSLNTVDSIETFVADINGGRWDAILP 62
++EIE+ DVI+ ++Q+ KENSLH+ TLQ E VSLNTVDSIE+FVADIN G WD +L
Sbjct: 2 SIEIESSDVIRLIMQYLKENSLHRALATLQEETTVSLNTVDSIESFVADINSGHWDTVLQ 61
Query: 63 QVSQLKLPRNTLEDLYEQIVLEMIELRELDTARAILRQTQVMGVMKQEQPERYLRLEHLL 122
+ LKLP TL DLYEQ+VLE+IELREL AR++LRQT M ++KQ QPERY+ LE+LL
Sbjct: 62 AIQSLKLPDKTLIDLYEQVVLELIELRELGAARSLLRQTDPMIMLKQTQPERYIHLENLL 121
Query: 123 VRTYFDPNEAYQE-STKEKRRAQIAQAIAAEVTVVPPSRLMALIGQALKWQQHQGLLPPG 181
R+YFDP EAY + S+KEKRRA IAQA+A EV+VVPPSRLMAL+GQALKWQQHQGLLPPG
Sbjct: 122 ARSYFDPREAYPDGSSKEKRRAAIAQALAGEVSVVPPSRLMALLGQALKWQQHQGLLPPG 181
Query: 182 TQFDLFRGTAAMKQDVDDMYPTNLSHTIKFGAKSHAECARFSPDGQYLVSCSIDGFIEVW 241
DLFRG AA+K ++ +PT LS IKFG KSH ECARFSPDGQYLV+ S+DGFIEVW
Sbjct: 182 MTIDLFRGKAAVKDVEEEKFPTQLSRHIKFGQKSHVECARFSPDGQYLVTGSVDGFIEVW 241
Query: 242 DYISGKLKKDLQYQAEETFMMHDEPVLCVDFSRDSEMIASGSTDGKIKVWRIRTAQCLRR 301
++ +GK++KDL+YQA++ FMM D+ VLC+ FSRD+EM+A+G+ DGKIKVW+I++ QCLRR
Sbjct: 242 NFTTGKIRKDLKYQAQDNFMMMDDAVLCMCFSRDTEMLATGAQDGKIKVWKIQSGQCLRR 301
Query: 302 LEHAHSQGVTSVSFSRDGSQLLSTSFDSTARIHGLKSGKMLKEFRGHTSYVNDATFTNDG 361
E AHS+GVT +SFS+D SQ+LS SFD T RIHGLKSGK LKEFRGH+S+VN+ATFT DG
Sbjct: 302 FERAHSKGVTCLSFSKDSSQILSASFDQTIRIHGLKSGKTLKEFRGHSSFVNEATFTQDG 361
Query: 362 TRVITASSDCTVKVWDLKTTDCLQTFKPPPPLRGGDASVNSVFIFPKNTEHIVVCNKTSS 421
+I+ASSD TVK+W++KTT+C TFK G D +VNSV + PKN EH VVCN++++
Sbjct: 362 HYIISASSDGTVKIWNMKTTECSNTFKSLGSTAGTDITVNSVILLPKNPEHFVVCNRSNT 421
Query: 422 IYIMTLQGQVVKSFSSGKREGGDFVAACISPKGEWIYCVGEDRNMYCFSYQSGKLEHLMT 481
+ IM +QGQ+V+SFSSGKREGGDFV +SP+GEWIYCVGED +YCFS +GKLE +T
Sbjct: 422 VVIMNMQGQIVRSFSSGKREGGDFVCCALSPRGEWIYCVGEDFVLYCFSTVTGKLERTLT 481
Query: 482 VHDKEIIGVTHHPHRNLVATFSDDHTMKLWKP 513
VH+K++IG+ HHPH+NL+AT+S+D +KLWKP
Sbjct: 482 VHEKDVIGIAHHPHQNLIATYSEDGLLKLWKP 513
>gb|AAH63369.1| Hypothetical protein MGC75979 [Xenopus tropicalis]
gi|45361221|ref|NP_989188.1| hypothetical protein
LOC394796 [Xenopus tropicalis]
Length = 513
Score = 706 bits (1822), Expect = 0.0
Identities = 333/512 (65%), Positives = 419/512 (81%), Gaps = 1/512 (0%)
Query: 3 TLEIEARDVIKTVLQFCKENSLHQTFQTLQNECQVSLNTVDSIETFVADINGGRWDAILP 62
++EIE+ DVI+ ++Q+ KENSLH+T TLQ E VSLNTVDSIE+FVADIN G WD +L
Sbjct: 2 SIEIESSDVIRLIMQYLKENSLHRTLATLQEETTVSLNTVDSIESFVADINSGHWDTVLQ 61
Query: 63 QVSQLKLPRNTLEDLYEQIVLEMIELRELDTARAILRQTQVMGVMKQEQPERYLRLEHLL 122
+ LKLP TL DLYEQ+VLE+IELREL AR++LRQT M ++KQ QPERY+ LE+LL
Sbjct: 62 AIQSLKLPDKTLIDLYEQVVLELIELRELGAARSLLRQTDPMIMLKQNQPERYIHLENLL 121
Query: 123 VRTYFDPNEAYQE-STKEKRRAQIAQAIAAEVTVVPPSRLMALIGQALKWQQHQGLLPPG 181
R+YFDP EAY + S+KEKRRA IAQA+A EV+VVPPSRLMAL+GQALKWQQHQGLLPPG
Sbjct: 122 ARSYFDPREAYPDGSSKEKRRAAIAQALAGEVSVVPPSRLMALLGQALKWQQHQGLLPPG 181
Query: 182 TQFDLFRGTAAMKQDVDDMYPTNLSHTIKFGAKSHAECARFSPDGQYLVSCSIDGFIEVW 241
DLFRG AA+K ++ +PT LS IKFG KSH ECARFSPDGQYLV+ S+DGFIEVW
Sbjct: 182 MIIDLFRGKAAVKDVEEEKFPTQLSRHIKFGQKSHVECARFSPDGQYLVTGSVDGFIEVW 241
Query: 242 DYISGKLKKDLQYQAEETFMMHDEPVLCVDFSRDSEMIASGSTDGKIKVWRIRTAQCLRR 301
++ +GK++KDL+YQA++ FMM D+ VLC+ FSRD+EM+A+G+ DGKIKVW+I++ QCLRR
Sbjct: 242 NFTTGKIRKDLKYQAQDNFMMMDDAVLCMCFSRDTEMLATGAQDGKIKVWKIQSGQCLRR 301
Query: 302 LEHAHSQGVTSVSFSRDGSQLLSTSFDSTARIHGLKSGKMLKEFRGHTSYVNDATFTNDG 361
E AHS+GVT +SFS+D SQ+LS SFD T R+HGLKSGK LKEFRGH+S+VN+ATFT DG
Sbjct: 302 FERAHSKGVTCLSFSKDCSQILSASFDQTIRVHGLKSGKTLKEFRGHSSFVNEATFTQDG 361
Query: 362 TRVITASSDCTVKVWDLKTTDCLQTFKPPPPLRGGDASVNSVFIFPKNTEHIVVCNKTSS 421
+I+ASSD TVK+W++KTT+C TFK G D +VNSV + PKN EH VVCN++++
Sbjct: 362 HYIISASSDGTVKIWNMKTTECSNTFKSLGSTAGTDITVNSVILLPKNPEHFVVCNRSNT 421
Query: 422 IYIMTLQGQVVKSFSSGKREGGDFVAACISPKGEWIYCVGEDRNMYCFSYQSGKLEHLMT 481
+ IM +QGQ+V+SFSSGKREGGDFV +SP+GEWIYCVGED +YCFS +GKLE +T
Sbjct: 422 VVIMNMQGQIVRSFSSGKREGGDFVCCTLSPRGEWIYCVGEDFVLYCFSTVTGKLERTLT 481
Query: 482 VHDKEIIGVTHHPHRNLVATFSDDHTMKLWKP 513
VH+K++IG+ HHPH+NL+ T+S+D +KLWKP
Sbjct: 482 VHEKDVIGIAHHPHQNLIGTYSEDGLLKLWKP 513
>ref|NP_067510.2| smu-1 suppressor of mec-8 and unc-52 homolog [Mus musculus]
gi|12834195|dbj|BAB22820.1| unnamed protein product [Mus
musculus]
Length = 513
Score = 705 bits (1820), Expect = 0.0
Identities = 333/512 (65%), Positives = 418/512 (81%), Gaps = 1/512 (0%)
Query: 3 TLEIEARDVIKTVLQFCKENSLHQTFQTLQNECQVSLNTVDSIETFVADINGGRWDAILP 62
++EIE+ DVI+ ++Q+ KENSLH+ TLQ E VSLNTVDSIE+FVADIN G WD +L
Sbjct: 2 SIEIESSDVIRLIMQYLKENSLHRALATLQEETTVSLNTVDSIESFVADINSGHWDTVLQ 61
Query: 63 QVSQLKLPRNTLEDLYEQIVLEMIELRELDTARAILRQTQVMGVMKQEQPERYLRLEHLL 122
+ LKLP TL DLYEQ+VLE+IELREL AR++LRQT M ++KQ QPERY+ LE+LL
Sbjct: 62 AIQSLKLPDKTLIDLYEQVVLELIELRELGAARSLLRQTDPMIMLKQTQPERYIHLENLL 121
Query: 123 VRTYFDPNEAYQE-STKEKRRAQIAQAIAAEVTVVPPSRLMALIGQALKWQQHQGLLPPG 181
R+YFDP EAY + S+KEKRRA IAQA+A EV+VVPPSRLMAL+GQALKWQQHQGLLPPG
Sbjct: 122 ARSYFDPREAYPDGSSKEKRRAAIAQALAGEVSVVPPSRLMALLGQALKWQQHQGLLPPG 181
Query: 182 TQFDLFRGTAAMKQDVDDMYPTNLSHTIKFGAKSHAECARFSPDGQYLVSCSIDGFIEVW 241
DLFRG AA+K ++ +PT LS IKFG KSH ECARFSPDGQYLV+ S+DGFIEVW
Sbjct: 182 MTIDLFRGKAAVKDVEEEKFPTQLSRHIKFGQKSHVECARFSPDGQYLVTGSVDGFIEVW 241
Query: 242 DYISGKLKKDLQYQAEETFMMHDEPVLCVDFSRDSEMIASGSTDGKIKVWRIRTAQCLRR 301
++ +GK++KDL+YQA++ FMM D+ VLC+ FSRD+EM+A+G+ DGKIKVW+I++ QCLRR
Sbjct: 242 NFTTGKIRKDLKYQAQDNFMMMDDAVLCMCFSRDTEMLATGAQDGKIKVWKIQSGQCLRR 301
Query: 302 LEHAHSQGVTSVSFSRDGSQLLSTSFDSTARIHGLKSGKMLKEFRGHTSYVNDATFTNDG 361
E AHS+GVT +SFS+D SQ+LS SFD T RIHGLKSGK LKEF GH+S+VN+ATFT DG
Sbjct: 302 FERAHSKGVTCLSFSKDSSQILSASFDQTIRIHGLKSGKTLKEFLGHSSFVNEATFTQDG 361
Query: 362 TRVITASSDCTVKVWDLKTTDCLQTFKPPPPLRGGDASVNSVFIFPKNTEHIVVCNKTSS 421
+I+ASSD TVK+W++KTT+C TFK G D +VNSV + PKN EH VVCN++++
Sbjct: 362 HYIISASSDGTVKIWNMKTTECSNTFKSLGSTAGTDITVNSVILLPKNPEHFVVCNRSNT 421
Query: 422 IYIMTLQGQVVKSFSSGKREGGDFVAACISPKGEWIYCVGEDRNMYCFSYQSGKLEHLMT 481
+ IM +QGQ+V+SFSSGKREGGDFV +SP+GEWIYCVGED +YCFS +GKLE +T
Sbjct: 422 VVIMNMQGQIVRSFSSGKREGGDFVCCALSPRGEWIYCVGEDFVLYCFSTVTGKLERTLT 481
Query: 482 VHDKEIIGVTHHPHRNLVATFSDDHTMKLWKP 513
VH+K++IG+ HHPH+NL+AT+S+D +KLWKP
Sbjct: 482 VHEKDVIGIAHHPHQNLIATYSEDGLLKLWKP 513
>dbj|BAA91822.1| unnamed protein product [Homo sapiens] gi|8922679|ref|NP_060695.1|
smu-1 suppressor of mec-8 and unc-52 homolog [Homo
sapiens]
Length = 513
Score = 705 bits (1820), Expect = 0.0
Identities = 333/512 (65%), Positives = 418/512 (81%), Gaps = 1/512 (0%)
Query: 3 TLEIEARDVIKTVLQFCKENSLHQTFQTLQNECQVSLNTVDSIETFVADINGGRWDAILP 62
++EIE+ DVI+ ++Q+ KENSLH+ TLQ E VSLNTVDSIE+FVADIN G WD +L
Sbjct: 2 SIEIESSDVIRLIMQYLKENSLHRALATLQEETTVSLNTVDSIESFVADINSGHWDTVLQ 61
Query: 63 QVSQLKLPRNTLEDLYEQIVLEMIELRELDTARAILRQTQVMGVMKQEQPERYLRLEHLL 122
+ LKLP TL DLYEQ+VLE+IELREL AR++LRQT M ++KQ QPERY+ LE+LL
Sbjct: 62 AIQSLKLPDKTLIDLYEQVVLELIELRELGAARSLLRQTDPMIMLKQTQPERYIHLENLL 121
Query: 123 VRTYFDPNEAYQE-STKEKRRAQIAQAIAAEVTVVPPSRLMALIGQALKWQQHQGLLPPG 181
R+YFDP EAY + S+KEKRRA IAQA+A EV+VVPPSRLMAL+GQALKWQQHQGLLPPG
Sbjct: 122 ARSYFDPREAYPDGSSKEKRRAAIAQALAGEVSVVPPSRLMALLGQALKWQQHQGLLPPG 181
Query: 182 TQFDLFRGTAAMKQDVDDMYPTNLSHTIKFGAKSHAECARFSPDGQYLVSCSIDGFIEVW 241
DLFRG AA+K ++ +PT LS IKFG KSH ECARFSPDGQYLV+ S+DGFIEVW
Sbjct: 182 MTIDLFRGKAAVKDVEEEKFPTQLSRHIKFGQKSHVECARFSPDGQYLVTGSVDGFIEVW 241
Query: 242 DYISGKLKKDLQYQAEETFMMHDEPVLCVDFSRDSEMIASGSTDGKIKVWRIRTAQCLRR 301
++ +GK++KDL+YQA++ FMM D+ VLC+ FSRD+EM+A+G+ DGKIKVW+I++ QCLRR
Sbjct: 242 NFTTGKIRKDLKYQAQDNFMMMDDAVLCMCFSRDTEMLATGAQDGKIKVWKIQSGQCLRR 301
Query: 302 LEHAHSQGVTSVSFSRDGSQLLSTSFDSTARIHGLKSGKMLKEFRGHTSYVNDATFTNDG 361
E AHS+GVT +SFS+D SQ+LS SFD T RIHGLKSGK LKEFRGH+S+VN+ATFT DG
Sbjct: 302 FERAHSKGVTCLSFSKDSSQILSASFDQTIRIHGLKSGKTLKEFRGHSSFVNEATFTQDG 361
Query: 362 TRVITASSDCTVKVWDLKTTDCLQTFKPPPPLRGGDASVNSVFIFPKNTEHIVVCNKTSS 421
+I+ASSD TVK+W++KTT+C TFK G D +VNSV + PKN EH VVCN++++
Sbjct: 362 HYIISASSDGTVKIWNMKTTECSNTFKSLGSTAGTDITVNSVILLPKNPEHFVVCNRSNT 421
Query: 422 IYIMTLQGQVVKSFSSGKREGGDFVAACISPKGEWIYCVGEDRNMYCFSYQSGKLEHLMT 481
+ IM +QGQ+V+SFSSGKREGGDFV +SP+GEWIYCVGED +YCFS +GKLE +T
Sbjct: 422 VVIMNMQGQIVRSFSSGKREGGDFVCCALSPRGEWIYCVGEDFVLYCFSTVTGKLERTLT 481
Query: 482 VHDKEIIGVTHHPHRNLVATFSDDHTMKLWKP 513
VH+K++IG+ HHPH+N +AT+S+D +KLWKP
Sbjct: 482 VHEKDVIGIAHHPHQNPIATYSEDGLLKLWKP 513
>ref|NP_476543.1| smu-1 suppressor of mec-8 and unc-52 homolog [Rattus norvegicus]
gi|13660779|gb|AAK33013.1| brain-enriched WD-repeat
protein [Rattus norvegicus]
Length = 513
Score = 704 bits (1817), Expect = 0.0
Identities = 332/512 (64%), Positives = 417/512 (80%), Gaps = 1/512 (0%)
Query: 3 TLEIEARDVIKTVLQFCKENSLHQTFQTLQNECQVSLNTVDSIETFVADINGGRWDAILP 62
++EIE+ DVI+ ++Q+ KENSLH+ T E VSLNTVDSIE+FVADIN G WD +L
Sbjct: 2 SIEIESSDVIRLIMQYLKENSLHRALATFTEETTVSLNTVDSIESFVADINSGHWDTVLQ 61
Query: 63 QVSQLKLPRNTLEDLYEQIVLEMIELRELDTARAILRQTQVMGVMKQEQPERYLRLEHLL 122
+ LKLP TL DLYEQ+VLE+IELREL AR++LRQT M ++KQ QPERY+ LE+LL
Sbjct: 62 AIQSLKLPDKTLIDLYEQVVLELIELRELGAARSLLRQTDPMIMLKQTQPERYIHLENLL 121
Query: 123 VRTYFDPNEAYQE-STKEKRRAQIAQAIAAEVTVVPPSRLMALIGQALKWQQHQGLLPPG 181
R+YFDP EAY + S+KEKRRA IAQA+A EV+VVPPSRLMAL+GQALKWQQHQGLLPPG
Sbjct: 122 ARSYFDPREAYPDGSSKEKRRAAIAQALAGEVSVVPPSRLMALLGQALKWQQHQGLLPPG 181
Query: 182 TQFDLFRGTAAMKQDVDDMYPTNLSHTIKFGAKSHAECARFSPDGQYLVSCSIDGFIEVW 241
DLFRG AA+K ++ +PT LS IKFG KSH ECARFSPDGQYLV+ S+DGFIEVW
Sbjct: 182 MTIDLFRGKAAVKDVEEEKFPTQLSRHIKFGQKSHVECARFSPDGQYLVTGSVDGFIEVW 241
Query: 242 DYISGKLKKDLQYQAEETFMMHDEPVLCVDFSRDSEMIASGSTDGKIKVWRIRTAQCLRR 301
++ +GK++KDL+YQA++ FMM D+ VLC+ FSRD+EM+A+G+ DGKIKVW+I++ QCLRR
Sbjct: 242 NFTTGKIRKDLKYQAQDNFMMMDDAVLCMCFSRDTEMLATGAQDGKIKVWKIQSGQCLRR 301
Query: 302 LEHAHSQGVTSVSFSRDGSQLLSTSFDSTARIHGLKSGKMLKEFRGHTSYVNDATFTNDG 361
E AHS+GVT +SFS+D SQ+LS SFD T RIHGLKSGK LKEFRGH+S+VN+ATFT DG
Sbjct: 302 FERAHSKGVTCLSFSKDSSQILSASFDQTIRIHGLKSGKTLKEFRGHSSFVNEATFTQDG 361
Query: 362 TRVITASSDCTVKVWDLKTTDCLQTFKPPPPLRGGDASVNSVFIFPKNTEHIVVCNKTSS 421
+I+ASSD TVK+W++KTT+C TFK G D +VNSV + PKN EH VVCN++++
Sbjct: 362 HYIISASSDGTVKIWNMKTTECSNTFKSLGSTAGTDITVNSVILLPKNPEHFVVCNRSNT 421
Query: 422 IYIMTLQGQVVKSFSSGKREGGDFVAACISPKGEWIYCVGEDRNMYCFSYQSGKLEHLMT 481
+ IM +QGQ+V+SFSSGKREGGDFV +SP+GEWIYCVGED +YCFS +GKLE +T
Sbjct: 422 VVIMNMQGQIVRSFSSGKREGGDFVCCALSPRGEWIYCVGEDFVLYCFSTVTGKLERTLT 481
Query: 482 VHDKEIIGVTHHPHRNLVATFSDDHTMKLWKP 513
VH+K++IG+ HHPH+NL+AT+S+D +KLWKP
Sbjct: 482 VHEKDVIGIAHHPHQNLIATYSEDGLLKLWKP 513
>gb|AAH70636.1| MGC81475 protein [Xenopus laevis]
Length = 513
Score = 704 bits (1816), Expect = 0.0
Identities = 332/512 (64%), Positives = 417/512 (80%), Gaps = 1/512 (0%)
Query: 3 TLEIEARDVIKTVLQFCKENSLHQTFQTLQNECQVSLNTVDSIETFVADINGGRWDAILP 62
++EIE+ DVI+ ++Q+ KENSLH+T TLQ E VSLNTVDSIE+FVADIN G WD +L
Sbjct: 2 SIEIESSDVIRLIMQYLKENSLHRTLATLQEETTVSLNTVDSIESFVADINSGHWDTVLQ 61
Query: 63 QVSQLKLPRNTLEDLYEQIVLEMIELRELDTARAILRQTQVMGVMKQEQPERYLRLEHLL 122
+ LKLP TL DLYEQ+VLE+IELREL AR++LRQT M ++KQ Q ERY+ LE+LL
Sbjct: 62 AIQSLKLPDKTLIDLYEQVVLELIELRELGAARSLLRQTDPMIMLKQNQSERYIHLENLL 121
Query: 123 VRTYFDPNEAYQE-STKEKRRAQIAQAIAAEVTVVPPSRLMALIGQALKWQQHQGLLPPG 181
R+YFDP EAY + S+KEKRR IAQA+A EV+VVPPSRLMAL+GQALKWQQHQGLLPPG
Sbjct: 122 ARSYFDPREAYPDGSSKEKRRTAIAQALAGEVSVVPPSRLMALLGQALKWQQHQGLLPPG 181
Query: 182 TQFDLFRGTAAMKQDVDDMYPTNLSHTIKFGAKSHAECARFSPDGQYLVSCSIDGFIEVW 241
DLFRG AA+K ++ +PT LS IKFG KSH ECARFSPDGQYLV+ S+DGFIEVW
Sbjct: 182 MTIDLFRGKAAVKDVEEEKFPTQLSRHIKFGQKSHVECARFSPDGQYLVTGSVDGFIEVW 241
Query: 242 DYISGKLKKDLQYQAEETFMMHDEPVLCVDFSRDSEMIASGSTDGKIKVWRIRTAQCLRR 301
++ +GK++KDL+YQA++ FMM D+ VLC+ FSRD+EM+A+G+ DGKIKVW+I++ QCLRR
Sbjct: 242 NFTTGKIRKDLKYQAQDNFMMMDDAVLCMCFSRDTEMLATGAQDGKIKVWKIQSGQCLRR 301
Query: 302 LEHAHSQGVTSVSFSRDGSQLLSTSFDSTARIHGLKSGKMLKEFRGHTSYVNDATFTNDG 361
E AHS+GVT +SFS+D SQ+LS SFD T RIHGLKSGK LKEFRGH+S+VN+ATFT DG
Sbjct: 302 FERAHSKGVTCLSFSKDSSQILSASFDQTIRIHGLKSGKTLKEFRGHSSFVNEATFTQDG 361
Query: 362 TRVITASSDCTVKVWDLKTTDCLQTFKPPPPLRGGDASVNSVFIFPKNTEHIVVCNKTSS 421
+I+ASSD TVK+W++KTT+C TFK G D +VNSV + PKN EH VVCN++++
Sbjct: 362 HYIISASSDGTVKIWNMKTTECSNTFKSLGSTAGTDITVNSVILLPKNPEHFVVCNRSNT 421
Query: 422 IYIMTLQGQVVKSFSSGKREGGDFVAACISPKGEWIYCVGEDRNMYCFSYQSGKLEHLMT 481
+ IM +QGQ+V+SFSSGKREGGDFV +SP+GEWIYCVGED +YCFS +GKLE +T
Sbjct: 422 VVIMNMQGQIVRSFSSGKREGGDFVCCTLSPRGEWIYCVGEDFVLYCFSTVTGKLERTLT 481
Query: 482 VHDKEIIGVTHHPHRNLVATFSDDHTMKLWKP 513
VH+K++IG+ HHPH+NL+ T+S+D +KLWKP
Sbjct: 482 VHEKDVIGIAHHPHQNLIGTYSEDGLLKLWKP 513
>ref|XP_613397.1| PREDICTED: similar to Smu-1 suppressor of mec-8 and unc-52 homolog
[Bos taurus]
Length = 515
Score = 703 bits (1815), Expect = 0.0
Identities = 335/514 (65%), Positives = 419/514 (81%), Gaps = 3/514 (0%)
Query: 3 TLEIEARDVIKTVLQFCKENSLHQTFQTLQNECQVSLNTVDSIETFVADINGGRWDAILP 62
++EIE+ DVI+ ++Q+ KENSLH+ TLQ E VSLNTVDSIE+FVADIN G WD +L
Sbjct: 2 SIEIESSDVIRLIMQYLKENSLHRALATLQEETTVSLNTVDSIESFVADINSGHWDTVLQ 61
Query: 63 QVSQLKLPRNTLEDLYEQIVLEMIELRELDTARAILRQTQVMGVMKQEQPERYLRLEHLL 122
+ LKLP TL DLYEQ+VLE+IELREL AR++LRQT M ++KQ QPERY+ LE+LL
Sbjct: 62 AIQSLKLPDKTLIDLYEQVVLELIELRELGAARSLLRQTDPMIMLKQTQPERYIHLENLL 121
Query: 123 VRTYFDPNEAYQE-STKEKRRAQIAQAIAAEVTVVPPSRLMALIGQALKWQQHQGLLPPG 181
R+YFDP EAY + S+KEKRRA IAQA+A EV+VVPPSRLMAL+GQALKWQQHQGLLPPG
Sbjct: 122 ARSYFDPREAYPDGSSKEKRRAAIAQALAGEVSVVPPSRLMALLGQALKWQQHQGLLPPG 181
Query: 182 TQFDLFRGTAAMKQDVDDMYPTNLSHTIKFGAKSHAECARFSPDGQYLVSCSIDGFIEVW 241
DLFRG AA+K ++ +PT LS IKFG KSH ECARFSPDGQYLV+ S+DGFIEVW
Sbjct: 182 MTIDLFRGKAAVKDVEEEKFPTQLSRHIKFGQKSHVECARFSPDGQYLVTGSVDGFIEVW 241
Query: 242 DYISGKLKKDLQYQAEETFMMHDEPVLCVDFSRDSEMIASGSTDGKIKVWRIRTAQCLRR 301
++ +GK++KDL+YQA++ FMM D+ VLC+ FSRD+EM+A+G+ DGKIKVW+I++ QCLRR
Sbjct: 242 NFTTGKIRKDLKYQAQDNFMMMDDAVLCMCFSRDTEMLATGAQDGKIKVWKIQSGQCLRR 301
Query: 302 LEHAHSQGVTSVSFSRDGSQLLSTSFDSTARIHGLKSGKMLKEFRGHTSYVNDATFTNDG 361
E AHS+GVT +SFS+D SQ+LS SFD T RIHGLKSGK LKEFRGH+S+VN+ATFT DG
Sbjct: 302 FERAHSKGVTCLSFSKDSSQILSASFDQTIRIHGLKSGKTLKEFRGHSSFVNEATFTQDG 361
Query: 362 TRVITASSDCTVKVWDLKTTDCLQTFKPPPPLRGGDASVNSVFIFPKNTEHIVVCNKTSS 421
+I+ASSD TVK+W++KTT+C TFK G D +VNSV + PKN EH VVCN++++
Sbjct: 362 HYIISASSDGTVKIWNMKTTECSNTFKSLGSTAGTDITVNSVILLPKNPEHFVVCNRSNT 421
Query: 422 IYIMTLQGQV--VKSFSSGKREGGDFVAACISPKGEWIYCVGEDRNMYCFSYQSGKLEHL 479
+ IM +QGQV V+SFSSGKREGGDFV +SP+GEWIYCVGED +YCFS +GKLE
Sbjct: 422 VVIMNMQGQVSIVRSFSSGKREGGDFVCCALSPRGEWIYCVGEDFVLYCFSTVTGKLERT 481
Query: 480 MTVHDKEIIGVTHHPHRNLVATFSDDHTMKLWKP 513
+TVH+K++IG+ HHPH+NL+AT+S+D +KLWKP
Sbjct: 482 LTVHEKDVIGIAHHPHQNLIATYSEDGLLKLWKP 515
>dbj|BAA96656.1| unnamed protein product [Mus musculus]
Length = 513
Score = 703 bits (1815), Expect = 0.0
Identities = 333/512 (65%), Positives = 417/512 (81%), Gaps = 1/512 (0%)
Query: 3 TLEIEARDVIKTVLQFCKENSLHQTFQTLQNECQVSLNTVDSIETFVADINGGRWDAILP 62
++EIE+ DVI+ ++Q+ KENSLH+ TLQ E VSLNTVDSIE+FVADIN G WD +L
Sbjct: 2 SIEIESSDVIRLIMQYLKENSLHRALATLQEETTVSLNTVDSIESFVADINSGHWDTVLQ 61
Query: 63 QVSQLKLPRNTLEDLYEQIVLEMIELRELDTARAILRQTQVMGVMKQEQPERYLRLEHLL 122
+ LKLP TL DLYEQ+VLE+IELREL AR++LRQT M ++KQ QPERY+ LE+LL
Sbjct: 62 AIQSLKLPDKTLIDLYEQVVLELIELRELGAARSLLRQTDPMIMLKQTQPERYIHLENLL 121
Query: 123 VRTYFDPNEAYQE-STKEKRRAQIAQAIAAEVTVVPPSRLMALIGQALKWQQHQGLLPPG 181
R+YFDP EAY + S+KEKRRA IAQA+A EV+VVPPSRLMAL+GQALKWQQHQGLLPPG
Sbjct: 122 ARSYFDPREAYPDGSSKEKRRAAIAQALAGEVSVVPPSRLMALLGQALKWQQHQGLLPPG 181
Query: 182 TQFDLFRGTAAMKQDVDDMYPTNLSHTIKFGAKSHAECARFSPDGQYLVSCSIDGFIEVW 241
DLFRG AA+K ++ +PT LS IKFG KSH ECARFSPDGQYLV+ S+DGFIEVW
Sbjct: 182 MTIDLFRGKAAVKDVEEEKFPTQLSRHIKFGQKSHVECARFSPDGQYLVTGSVDGFIEVW 241
Query: 242 DYISGKLKKDLQYQAEETFMMHDEPVLCVDFSRDSEMIASGSTDGKIKVWRIRTAQCLRR 301
++ +GK++KDL+YQA++ FMM D+ VLC+ FSRD+EM+A+G+ DGKIKVW+I++ QCLRR
Sbjct: 242 NFTTGKIRKDLKYQAQDNFMMMDDAVLCMCFSRDTEMLATGAQDGKIKVWKIQSGQCLRR 301
Query: 302 LEHAHSQGVTSVSFSRDGSQLLSTSFDSTARIHGLKSGKMLKEFRGHTSYVNDATFTNDG 361
E AHS+GVT +SFS+D SQ+LS SFD T RIHGLKSGK LKEFRGH+S+VN+ATFT DG
Sbjct: 302 FERAHSKGVTCLSFSKDSSQILSASFDQTIRIHGLKSGKTLKEFRGHSSFVNEATFTQDG 361
Query: 362 TRVITASSDCTVKVWDLKTTDCLQTFKPPPPLRGGDASVNSVFIFPKNTEHIVVCNKTSS 421
+I+ASSD TVK+W++KTT+C TFK G D +VNSV + PKN EH VVCN++++
Sbjct: 362 HYIISASSDGTVKIWNMKTTECSNTFKSLGSTAGTDITVNSVILLPKNPEHFVVCNRSNT 421
Query: 422 IYIMTLQGQVVKSFSSGKREGGDFVAACISPKGEWIYCVGEDRNMYCFSYQSGKLEHLMT 481
+ IM +QGQ+V+SFSSGKREGGDFV +S +GEWIYCVGED YCFS +GKLE +T
Sbjct: 422 VVIMNMQGQIVRSFSSGKREGGDFVCCALSLRGEWIYCVGEDFVFYCFSTVTGKLERTLT 481
Query: 482 VHDKEIIGVTHHPHRNLVATFSDDHTMKLWKP 513
VH+K++IG+ HHPH+NL+AT+S+D +KLWKP
Sbjct: 482 VHEKDVIGIAHHPHQNLIATYSEDGLLKLWKP 513
>gb|AAH45945.1| Smu1 protein [Danio rerio] gi|41055638|ref|NP_956493.1| smu-1
suppressor of mec-8 and unc-52 homolog [Danio rerio]
Length = 513
Score = 699 bits (1804), Expect = 0.0
Identities = 329/512 (64%), Positives = 416/512 (80%), Gaps = 1/512 (0%)
Query: 3 TLEIEARDVIKTVLQFCKENSLHQTFQTLQNECQVSLNTVDSIETFVADINGGRWDAILP 62
++EIE+ DVI+ ++Q+ KENSLH+T LQ E VSLNTVDSIE+FVADIN G WD +L
Sbjct: 2 SIEIESADVIRLIMQYLKENSLHRTLAMLQEETTVSLNTVDSIESFVADINSGHWDTVLQ 61
Query: 63 QVSQLKLPRNTLEDLYEQIVLEMIELRELDTARAILRQTQVMGVMKQEQPERYLRLEHLL 122
+ LKLP TL DLYEQ+VLE+IELREL AR++LRQT M ++KQ QPERY+ LE+LL
Sbjct: 62 AIQSLKLPDKTLIDLYEQVVLELIELRELGAARSLLRQTDPMIMLKQTQPERYIHLENLL 121
Query: 123 VRTYFDPNEAYQE-STKEKRRAQIAQAIAAEVTVVPPSRLMALIGQALKWQQHQGLLPPG 181
R+YFDP EAY + S+KEKRRA IAQA+A EV+VVPPSRLMAL+GQ+LKWQQHQGLLPPG
Sbjct: 122 ARSYFDPREAYPDGSSKEKRRAAIAQALAGEVSVVPPSRLMALLGQSLKWQQHQGLLPPG 181
Query: 182 TQFDLFRGTAAMKQDVDDMYPTNLSHTIKFGAKSHAECARFSPDGQYLVSCSIDGFIEVW 241
DLFRG AA+K ++ +PT L+ IKFG KSH ECARFSPDGQYLV+ S+DGFIEVW
Sbjct: 182 MTIDLFRGKAAVKDVEEERFPTQLARHIKFGQKSHVECARFSPDGQYLVTGSVDGFIEVW 241
Query: 242 DYISGKLKKDLQYQAEETFMMHDEPVLCVDFSRDSEMIASGSTDGKIKVWRIRTAQCLRR 301
++ +GK++KDL+YQA++ FMM D+ VLC+ FSRD+EM+A+G+ DGKIKVW+I++ QCLRR
Sbjct: 242 NFTTGKIRKDLKYQAQDNFMMMDDAVLCMSFSRDTEMLATGAQDGKIKVWKIQSGQCLRR 301
Query: 302 LEHAHSQGVTSVSFSRDGSQLLSTSFDSTARIHGLKSGKMLKEFRGHTSYVNDATFTNDG 361
E AHS+GVT +SFS+D +Q+LS SFD T RIHGLKSGK LKEFRGH+S+VN+AT T DG
Sbjct: 302 YERAHSKGVTCLSFSKDSTQILSASFDQTIRIHGLKSGKCLKEFRGHSSFVNEATLTPDG 361
Query: 362 TRVITASSDCTVKVWDLKTTDCLQTFKPPPPLRGGDASVNSVFIFPKNTEHIVVCNKTSS 421
I+ASSD TVKVW++KTT+C TFK G D +VN+V + PKN EH VVCN++++
Sbjct: 362 HHAISASSDGTVKVWNMKTTECTSTFKSLGTSAGTDITVNNVILLPKNPEHFVVCNRSNT 421
Query: 422 IYIMTLQGQVVKSFSSGKREGGDFVAACISPKGEWIYCVGEDRNMYCFSYQSGKLEHLMT 481
+ IM +QGQ+V+SFSSGKREGGDFV +SP+GEWIYCVGED +YCFS +GKLE +T
Sbjct: 422 VVIMNMQGQIVRSFSSGKREGGDFVCCTLSPRGEWIYCVGEDYVLYCFSTVTGKLERTLT 481
Query: 482 VHDKEIIGVTHHPHRNLVATFSDDHTMKLWKP 513
VH+K++IG+ HHPH+NL+ T+S+D +KLWKP
Sbjct: 482 VHEKDVIGIAHHPHQNLIGTYSEDGLLKLWKP 513
>emb|CAG04705.1| unnamed protein product [Tetraodon nigroviridis]
Length = 504
Score = 699 bits (1804), Expect = 0.0
Identities = 328/504 (65%), Positives = 414/504 (82%), Gaps = 1/504 (0%)
Query: 11 VIKTVLQFCKENSLHQTFQTLQNECQVSLNTVDSIETFVADINGGRWDAILPQVSQLKLP 70
V++ ++Q+ KEN+L +T TLQ E VSLNTVDSIE+FVADIN G WD +L + LKLP
Sbjct: 1 VVRLIMQYLKENNLQRTLATLQEETTVSLNTVDSIESFVADINSGHWDTVLQAIQSLKLP 60
Query: 71 RNTLEDLYEQIVLEMIELRELDTARAILRQTQVMGVMKQEQPERYLRLEHLLVRTYFDPN 130
TL DLYEQ+VLE+IELREL AR++LRQT M ++KQ QPERY+ LE+LL R+YFDP
Sbjct: 61 DKTLIDLYEQVVLELIELRELGAARSLLRQTDPMIMLKQTQPERYIHLENLLARSYFDPR 120
Query: 131 EAYQE-STKEKRRAQIAQAIAAEVTVVPPSRLMALIGQALKWQQHQGLLPPGTQFDLFRG 189
EAY + S+KEKRRA IAQA+A EV+VVPPSRLMAL+GQ+LKWQQHQGLLPPGT DLFRG
Sbjct: 121 EAYPDGSSKEKRRAAIAQALAGEVSVVPPSRLMALLGQSLKWQQHQGLLPPGTTIDLFRG 180
Query: 190 TAAMKQDVDDMYPTNLSHTIKFGAKSHAECARFSPDGQYLVSCSIDGFIEVWDYISGKLK 249
AA+K ++ +PT LS IKFG KSH EC+RFSPDGQYLV+ S+DGFIEVW++ +GK++
Sbjct: 181 KAAVKDVEEERFPTQLSRQIKFGQKSHVECSRFSPDGQYLVTGSVDGFIEVWNFTTGKIR 240
Query: 250 KDLQYQAEETFMMHDEPVLCVDFSRDSEMIASGSTDGKIKVWRIRTAQCLRRLEHAHSQG 309
KDL+YQA++ FMM D+ VLC+ FSRD+EM+A+G+ DGKIKVW+I++ QCLRR E AHS+G
Sbjct: 241 KDLKYQAQDNFMMMDDAVLCMCFSRDTEMLATGAQDGKIKVWKIQSGQCLRRFERAHSKG 300
Query: 310 VTSVSFSRDGSQLLSTSFDSTARIHGLKSGKMLKEFRGHTSYVNDATFTNDGTRVITASS 369
VT VSF +D SQLLSTSFD T RIHGLKSGK LKEFRGH+S+VN+ATFT DG +I+ASS
Sbjct: 301 VTCVSFCKDSSQLLSTSFDQTIRIHGLKSGKTLKEFRGHSSFVNEATFTPDGHHIISASS 360
Query: 370 DCTVKVWDLKTTDCLQTFKPPPPLRGGDASVNSVFIFPKNTEHIVVCNKTSSIYIMTLQG 429
D TVKVW++KTT+C TFKP G D +VN++ + PKN EH VVCN+++++ IM +QG
Sbjct: 361 DGTVKVWNMKTTECTNTFKPLGTSAGTDITVNNILLLPKNPEHFVVCNRSNTVVIMNMQG 420
Query: 430 QVVKSFSSGKREGGDFVAACISPKGEWIYCVGEDRNMYCFSYQSGKLEHLMTVHDKEIIG 489
Q+V+SFSSGKREGGDFV +SP+GEWIYCVGED +YCFS +GKLE +TVH+K++IG
Sbjct: 421 QIVRSFSSGKREGGDFVCCTLSPRGEWIYCVGEDFVLYCFSTVTGKLERTLTVHEKDVIG 480
Query: 490 VTHHPHRNLVATFSDDHTMKLWKP 513
+ HHPH+NL++T+S+D +KLWKP
Sbjct: 481 IAHHPHQNLISTYSEDGLLKLWKP 504
>ref|XP_531971.1| PREDICTED: similar to Smu-1 suppressor of mec-8 and unc-52 homolog
[Canis familiaris]
Length = 600
Score = 698 bits (1801), Expect = 0.0
Identities = 329/504 (65%), Positives = 412/504 (81%), Gaps = 1/504 (0%)
Query: 11 VIKTVLQFCKENSLHQTFQTLQNECQVSLNTVDSIETFVADINGGRWDAILPQVSQLKLP 70
VI+ ++Q+ KENSLH+ TLQ E VSLNTVDSIE+FVADIN G WD +L + LKLP
Sbjct: 97 VIRLIMQYLKENSLHRALATLQEETTVSLNTVDSIESFVADINSGHWDTVLQAIQSLKLP 156
Query: 71 RNTLEDLYEQIVLEMIELRELDTARAILRQTQVMGVMKQEQPERYLRLEHLLVRTYFDPN 130
TL DLYEQ+VLE+IELREL AR++LRQT M ++KQ QPERY+ LE+LL R+YFDP
Sbjct: 157 DKTLIDLYEQVVLELIELRELGAARSLLRQTDPMIMLKQTQPERYIHLENLLARSYFDPR 216
Query: 131 EAYQE-STKEKRRAQIAQAIAAEVTVVPPSRLMALIGQALKWQQHQGLLPPGTQFDLFRG 189
EAY + S+KEKRRA IAQA+A EV+VVPPSRLMAL+GQALKWQQHQGLLPPG DLFRG
Sbjct: 217 EAYPDGSSKEKRRAAIAQALAGEVSVVPPSRLMALLGQALKWQQHQGLLPPGMTIDLFRG 276
Query: 190 TAAMKQDVDDMYPTNLSHTIKFGAKSHAECARFSPDGQYLVSCSIDGFIEVWDYISGKLK 249
AA+K ++ +PT L+ IKFG KSH ECARFSPDGQYLV+ S+DGFIEVW++ +GK++
Sbjct: 277 KAAVKDVEEEKFPTQLTRHIKFGQKSHVECARFSPDGQYLVTGSVDGFIEVWNFTTGKIR 336
Query: 250 KDLQYQAEETFMMHDEPVLCVDFSRDSEMIASGSTDGKIKVWRIRTAQCLRRLEHAHSQG 309
KDL+YQA++ FMM D+ VLC+ FSRD+EM+A+G+ DGKIKVW+I++ QCLRR E AHS+G
Sbjct: 337 KDLKYQAQDNFMMMDDAVLCMCFSRDTEMLATGAQDGKIKVWKIQSGQCLRRFERAHSKG 396
Query: 310 VTSVSFSRDGSQLLSTSFDSTARIHGLKSGKMLKEFRGHTSYVNDATFTNDGTRVITASS 369
VT +SFS+D SQ+LS SFD T RIHGLKSGK LKEFRGH+S+VN+ATFT DG +I+ASS
Sbjct: 397 VTCLSFSKDSSQILSASFDQTIRIHGLKSGKTLKEFRGHSSFVNEATFTQDGHYIISASS 456
Query: 370 DCTVKVWDLKTTDCLQTFKPPPPLRGGDASVNSVFIFPKNTEHIVVCNKTSSIYIMTLQG 429
D TVK+W++KTT+C TFK G D +VNSV + PKN EH VVCN+++++ IM +QG
Sbjct: 457 DGTVKIWNMKTTECSNTFKSLGSTAGTDITVNSVILLPKNPEHFVVCNRSNTVVIMNMQG 516
Query: 430 QVVKSFSSGKREGGDFVAACISPKGEWIYCVGEDRNMYCFSYQSGKLEHLMTVHDKEIIG 489
Q+V+SFSSGKREGGDFV +SP+GEWIYCVGED +YCFS +GKLE +TVH+K++IG
Sbjct: 517 QIVRSFSSGKREGGDFVCCALSPRGEWIYCVGEDFVLYCFSTVTGKLERTLTVHEKDVIG 576
Query: 490 VTHHPHRNLVATFSDDHTMKLWKP 513
+ HHPH+NL+AT+S+D +KLWKP
Sbjct: 577 IAHHPHQNLIATYSEDGLLKLWKP 600
>ref|XP_393446.1| PREDICTED: similar to ENSANGP00000015224 [Apis mellifera]
Length = 510
Score = 677 bits (1748), Expect = 0.0
Identities = 322/512 (62%), Positives = 409/512 (78%), Gaps = 4/512 (0%)
Query: 3 TLEIEARDVIKTVLQFCKENSLHQTFQTLQNECQVSLNTVDSIETFVADINGGRWDAILP 62
++EIE+ DVI+ + Q+ KE++L +T QTLQ E VSLNTVDS++ FVADIN G WD +L
Sbjct: 2 SIEIESADVIRLIQQYLKESNLVKTLQTLQEETGVSLNTVDSVDGFVADINNGHWDTVLK 61
Query: 63 QVSQLKLPRNTLEDLYEQIVLEMIELRELDTARAILRQTQVMGVMKQEQPERYLRLEHLL 122
+ LKLP L DLYEQ+VLE+IELREL AR++LRQT M +MKQ++PERY+ LE+LL
Sbjct: 62 AIQSLKLPDKKLIDLYEQVVLELIELRELGAARSLLRQTDPMIMMKQQEPERYIHLENLL 121
Query: 123 VRTYFDPNEAYQE-STKEKRRAQIAQAIAAEVTVVPPSRLMALIGQALKWQQHQGLLPPG 181
R+YFDP EAY + STKEKRRA IA A+A EV+VVP SRL+AL+GQALKWQQHQGLLPPG
Sbjct: 122 ARSYFDPREAYPDGSTKEKRRAYIATALAGEVSVVPSSRLLALLGQALKWQQHQGLLPPG 181
Query: 182 TQFDLFRGTAAMKQDVDDMYPTNLSHTIKFGAKSHAECARFSPDGQYLVSCSIDGFIEVW 241
T DLFRG AA++ D+ YPT LS IKFG KSH ECARFSPDGQYLV+ S+DGFIEVW
Sbjct: 182 TTIDLFRGKAAVRDQEDEKYPTQLSKQIKFGQKSHVECARFSPDGQYLVTGSVDGFIEVW 241
Query: 242 DYISGKLKKDLQYQAEETFMMHDEPVLCVDFSRDSEMIASGSTDGKIKVWRIRTAQCLRR 301
++ +GK++KDL+YQA++ FMM ++ VL + FSRDSEM+A G DGKIKVWR+++ QCLRR
Sbjct: 242 NFTTGKIRKDLKYQAQDNFMMMEQAVLSMAFSRDSEMLAGGGQDGKIKVWRVQSGQCLRR 301
Query: 302 LEHAHSQGVTSVSFSRDGSQLLSTSFDSTARIHGLKSGKMLKEFRGHTSYVNDATFTNDG 361
E AHS+GVT + FSRD SQ+LS SFD+T RIHGLKSGK LKEFRGH S+VN+ F DG
Sbjct: 302 FEKAHSKGVTCLQFSRDNSQILSASFDTTIRIHGLKSGKTLKEFRGHASFVNEVVFAPDG 361
Query: 362 TRVITASSDCTVKVWDLKTTDCLQTFKPPPPLRGGDASVNSVFIFPKNTEHIVVCNKTSS 421
+I+ASSD TVKVW LKTT+C+ T+K L D +VNS+ + P+N EH VVCN++++
Sbjct: 362 HNLISASSDGTVKVWSLKTTECIGTYK---SLGAADLTVNSIHLLPRNPEHFVVCNRSNT 418
Query: 422 IYIMTLQGQVVKSFSSGKREGGDFVAACISPKGEWIYCVGEDRNMYCFSYQSGKLEHLMT 481
+ IM +QGQ+V+SFSSGKREGGDFV +SP+GE+IYCVGED +YCFS SGKLE +
Sbjct: 419 VVIMNMQGQIVRSFSSGKREGGDFVCTVVSPRGEFIYCVGEDLVLYCFSTSSGKLERTLN 478
Query: 482 VHDKEIIGVTHHPHRNLVATFSDDHTMKLWKP 513
+H+K++IG+THHPH+NL+ ++S+D +KLWKP
Sbjct: 479 IHEKDVIGLTHHPHQNLLCSYSEDGLLKLWKP 510
>gb|AAF55614.2| CG5451-PA [Drosophila melanogaster] gi|15292169|gb|AAK93353.1|
LD41216p [Drosophila melanogaster]
gi|21356791|ref|NP_650766.1| CG5451-PA [Drosophila
melanogaster]
Length = 509
Score = 671 bits (1730), Expect = 0.0
Identities = 316/512 (61%), Positives = 413/512 (79%), Gaps = 5/512 (0%)
Query: 3 TLEIEARDVIKTVLQFCKENSLHQTFQTLQNECQVSLNTVDSIETFVADINGGRWDAILP 62
++EIE+ DVI+ + Q+ KE++L +T QTLQ E VSLNTVDS++ FV DI+ G WD +L
Sbjct: 2 SIEIESADVIRLIQQYLKESNLMKTLQTLQEETGVSLNTVDSVDGFVQDISNGHWDTVLK 61
Query: 63 QVSQLKLPRNTLEDLYEQIVLEMIELRELDTARAILRQTQVMGVMKQEQPERYLRLEHLL 122
LKLP L +LYEQIVLE+IELREL AR++LRQT M ++KQ++PERY+ LE++L
Sbjct: 62 VTQSLKLPDKKLLNLYEQIVLELIELRELGAARSLLRQTDPMTMLKQQEPERYIHLENML 121
Query: 123 VRTYFDPNEAYQE-STKEKRRAQIAQAIAAEVTVVPPSRLMALIGQALKWQQHQGLLPPG 181
R YFDP EAY E S+KEKRR IAQ ++ EV VVP SRL+AL+GQALKWQQHQGLLPPG
Sbjct: 122 QRAYFDPREAYAEGSSKEKRRTVIAQELSGEVHVVPSSRLLALLGQALKWQQHQGLLPPG 181
Query: 182 TQFDLFRGTAAMKQDVDDMYPTNLSHTIKFGAKSHAECARFSPDGQYLVSCSIDGFIEVW 241
T DLFRG AAMK ++MYPT L IKFG KSH ECA+FSPDGQYL++ S+DGF+EVW
Sbjct: 182 TTIDLFRGKAAMKDQEEEMYPTQLFRQIKFGQKSHVECAQFSPDGQYLITGSVDGFLEVW 241
Query: 242 DYISGKLKKDLQYQAEETFMMHDEPVLCVDFSRDSEMIASGSTDGKIKVWRIRTAQCLRR 301
++ +GK++KDL+YQA++ FMM ++ VL ++FSRDSEM+ASG+ DG+IKVWRI T QCLR+
Sbjct: 242 NFTTGKVRKDLKYQAQDQFMMMEQAVLALNFSRDSEMVASGAQDGQIKVWRIITGQCLRK 301
Query: 302 LEHAHSQGVTSVSFSRDGSQLLSTSFDSTARIHGLKSGKMLKEFRGHTSYVNDATFTNDG 361
E AH++G+T + FSRD SQ+LS SFD T R+HGLKSGKMLKEF+GH+S+VN+ATFT DG
Sbjct: 302 FEKAHTKGITCLQFSRDNSQVLSASFDYTVRLHGLKSGKMLKEFKGHSSFVNEATFTPDG 361
Query: 362 TRVITASSDCTVKVWDLKTTDCLQTFKPPPPLRGGDASVNSVFIFPKNTEHIVVCNKTSS 421
V++ASSD TVKVW LKTT+C+ T+KP G + +VN+V I PKN EH +VCN++++
Sbjct: 362 HSVLSASSDGTVKVWSLKTTECVATYKP----LGNELAVNTVLILPKNPEHFIVCNRSNT 417
Query: 422 IYIMTLQGQVVKSFSSGKREGGDFVAACISPKGEWIYCVGEDRNMYCFSYQSGKLEHLMT 481
+ IM +QGQ+V+SFSSGKREGG F++A +SP+GE+IYC GED+ +YCFS SGKLE +
Sbjct: 418 VVIMNMQGQIVRSFSSGKREGGAFISATLSPRGEFIYCAGEDQVLYCFSVSSGKLERTLN 477
Query: 482 VHDKEIIGVTHHPHRNLVATFSDDHTMKLWKP 513
VH+K++IG+THHPH+NL+A++S+D +KLWKP
Sbjct: 478 VHEKDVIGLTHHPHQNLLASYSEDGLLKLWKP 509
>gb|EAL28013.1| GA18890-PA [Drosophila pseudoobscura]
Length = 509
Score = 669 bits (1726), Expect = 0.0
Identities = 314/512 (61%), Positives = 413/512 (80%), Gaps = 5/512 (0%)
Query: 3 TLEIEARDVIKTVLQFCKENSLHQTFQTLQNECQVSLNTVDSIETFVADINGGRWDAILP 62
++EIE+ DVI+ + Q+ KE++L +T QTLQ E VSLNTVDS++ FV DI+ G WD +L
Sbjct: 2 SIEIESADVIRLIQQYLKESNLMKTLQTLQEETGVSLNTVDSVDGFVQDISNGHWDTVLK 61
Query: 63 QVSQLKLPRNTLEDLYEQIVLEMIELRELDTARAILRQTQVMGVMKQEQPERYLRLEHLL 122
LKLP L +LYEQIVLE+IELREL AR++LRQT M ++KQ++PERY+ LE++L
Sbjct: 62 VTQSLKLPDKKLLNLYEQIVLELIELRELGAARSLLRQTDPMSMLKQQEPERYIHLENML 121
Query: 123 VRTYFDPNEAYQE-STKEKRRAQIAQAIAAEVTVVPPSRLMALIGQALKWQQHQGLLPPG 181
R YFDP EAY E S+KEKRR I+Q ++ EV VVP SRL+AL+GQALKWQQHQGLLPPG
Sbjct: 122 QRAYFDPREAYAEGSSKEKRRTAISQELSGEVHVVPSSRLLALLGQALKWQQHQGLLPPG 181
Query: 182 TQFDLFRGTAAMKQDVDDMYPTNLSHTIKFGAKSHAECARFSPDGQYLVSCSIDGFIEVW 241
T DLFRG AAMK ++MYPT L IKFG KSH ECA+FSPDGQYL++ S+DGF+EVW
Sbjct: 182 TTIDLFRGKAAMKDQEEEMYPTQLFRQIKFGQKSHVECAQFSPDGQYLITGSVDGFLEVW 241
Query: 242 DYISGKLKKDLQYQAEETFMMHDEPVLCVDFSRDSEMIASGSTDGKIKVWRIRTAQCLRR 301
++ +GK++KDL+YQA++ FMM ++ VL ++FSRDSEM+ASG+ DG+IKVWRI T QCLR+
Sbjct: 242 NFTTGKVRKDLKYQAQDQFMMMEQAVLALNFSRDSEMVASGAQDGQIKVWRIITGQCLRK 301
Query: 302 LEHAHSQGVTSVSFSRDGSQLLSTSFDSTARIHGLKSGKMLKEFRGHTSYVNDATFTNDG 361
E AH++G+T + FSRD SQ+LS SFD T R+HGLKSGKMLKEF+GH+S+VN+ATFT DG
Sbjct: 302 FEKAHTKGITCLQFSRDNSQVLSASFDYTVRLHGLKSGKMLKEFKGHSSFVNEATFTPDG 361
Query: 362 TRVITASSDCTVKVWDLKTTDCLQTFKPPPPLRGGDASVNSVFIFPKNTEHIVVCNKTSS 421
V++ASSD TVKVW LKTT+C+ T+KP G + +VN+V I PKN EH +VCN++++
Sbjct: 362 HSVLSASSDGTVKVWSLKTTECVATYKP----LGNELAVNTVLILPKNPEHFIVCNRSNT 417
Query: 422 IYIMTLQGQVVKSFSSGKREGGDFVAACISPKGEWIYCVGEDRNMYCFSYQSGKLEHLMT 481
+ IM +QGQ+V+SFSSGKREGG F++A +SP+GE+IYC GED+ +YCFS SGKLE +
Sbjct: 418 VVIMNMQGQIVRSFSSGKREGGAFISATLSPRGEFIYCAGEDQVLYCFSVSSGKLERTLN 477
Query: 482 VHDKEIIGVTHHPHRNLVATFSDDHTMKLWKP 513
VH+K++IG++HHPH+NL+A++S+D +KLWKP
Sbjct: 478 VHEKDVIGLSHHPHQNLLASYSEDGLLKLWKP 509
Database: nr
Posted date: Jul 5, 2005 12:34 AM
Number of letters in database: 863,360,394
Number of sequences in database: 2,540,612
Lambda K H
0.319 0.134 0.398
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 865,596,414
Number of Sequences: 2540612
Number of extensions: 35659815
Number of successful extensions: 150979
Number of sequences better than 10.0: 6242
Number of HSP's better than 10.0 without gapping: 3371
Number of HSP's successfully gapped in prelim test: 2903
Number of HSP's that attempted gapping in prelim test: 92879
Number of HSP's gapped (non-prelim): 29998
length of query: 513
length of database: 863,360,394
effective HSP length: 132
effective length of query: 381
effective length of database: 527,999,610
effective search space: 201167851410
effective search space used: 201167851410
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 77 (34.3 bits)
Medicago: description of AC142394.6


