

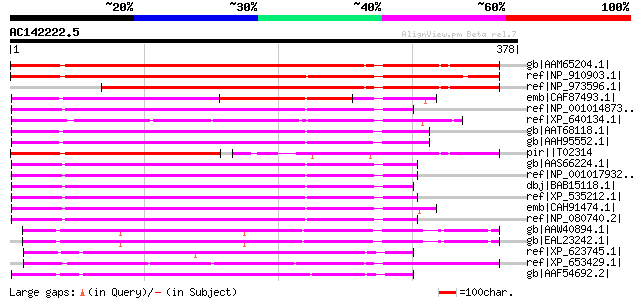
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC142222.5 + phase: 0
(378 letters)
Database: nr
2,540,612 sequences; 863,360,394 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
gb|AAM65204.1| unknown [Arabidopsis thaliana] gi|26450050|dbj|BA... 481 e-134
ref|NP_910903.1| putative transducin / WD-40 repeat protein [Ory... 432 e-120
ref|NP_973596.1| transducin family protein / WD-40 repeat family... 402 e-110
emb|CAF87493.1| unnamed protein product [Tetraodon nigroviridis] 216 1e-54
ref|NP_001014873.1| hypothetical protein LOC508709 [Bos taurus] ... 213 6e-54
ref|XP_640134.1| hypothetical protein DDB0204846 [Dictyostelium ... 213 6e-54
gb|AAT68118.1| AK025355-like [Danio rerio] gi|51467990|ref|NP_00... 213 8e-54
gb|AAH95552.1| Flj20195l protein [Danio rerio] 213 8e-54
pir||T02314 hypothetical protein At2g34260 [imported] - Arabidop... 212 1e-53
gb|AAS66224.1| LRRG00133 [Rattus norvegicus] 211 4e-53
ref|NP_001017932.1| WD repeat domain 55 [Rattus norvegicus] 211 4e-53
dbj|BAB15118.1| unnamed protein product [Homo sapiens] gi|462497... 210 5e-53
ref|XP_535212.1| PREDICTED: similar to hypothetical protein FLJ2... 210 5e-53
emb|CAH91474.1| hypothetical protein [Pongo pygmaeus] 207 3e-52
ref|NP_080740.2| WD repeat domain 55 [Mus musculus] gi|26344866|... 207 4e-52
gb|AAW40894.1| protein monoubiquitination-related protein, putat... 202 2e-50
gb|EAL23242.1| hypothetical protein CNBA3580 [Cryptococcus neofo... 198 3e-49
ref|XP_623745.1| PREDICTED: similar to CG14722-PA [Apis mellifera] 171 3e-41
ref|XP_653429.1| WD repeat protein [Entamoeba histolytica HM-1:I... 166 1e-39
gb|AAF54692.2| CG14722-PA [Drosophila melanogaster] gi|17862296|... 155 1e-36
>gb|AAM65204.1| unknown [Arabidopsis thaliana] gi|26450050|dbj|BAC42145.1| unknown
protein [Arabidopsis thaliana]
gi|28973067|gb|AAO63858.1| putative G-protein
beta-subunit (transducin) [Arabidopsis thaliana]
gi|20197166|gb|AAC27402.2| expressed protein
[Arabidopsis thaliana] gi|18403495|ref|NP_565782.1|
transducin family protein / WD-40 repeat family protein
[Arabidopsis thaliana]
Length = 353
Score = 481 bits (1238), Expect = e-134
Identities = 241/365 (66%), Positives = 291/365 (79%), Gaps = 12/365 (3%)
Query: 1 MEINLGKLAFDIDFHPSDNLVATGLIDGDLHLYRYSSDNTNSDPVRLLEIHAHTESCRAA 60
MEI+LG AF IDFHPS NLVA GLIDG LHLYRY SD S VR ++ AH ESCRA
Sbjct: 1 MEIDLGANAFGIDFHPSTNLVAAGLIDGHLHLYRYDSD---SSLVRERKVRAHKESCRAV 57
Query: 61 RFINGGRALLTGSPDFSILATDVETGSTIARLDNAHEAAVNRLVNLTESTVASGDDDGCI 120
RFI+ G+ ++T S D SILATDVETG+ +A L+NAHE AVN L+N+TE+T+ASGDD GC+
Sbjct: 58 RFIDDGQRIVTASADCSILATDVETGAQVAHLENAHEDAVNTLINVTETTIASGDDKGCV 117
Query: 121 KVWDTRERSCCNSFEAHEDYISDITFASDAMKLLATSGDGTLSVCNLRRNKVQAQSEFSE 180
K+WDTR+RSC + F AHEDYIS +TFASD+MKL+ TSGDGTLSVCNLR +KVQ+QSEFSE
Sbjct: 118 KIWDTRQRSCSHEFNAHEDYISGMTFASDSMKLVVTSGDGTLSVCNLRTSKVQSQSEFSE 177
Query: 181 DELLSVVLMKNGRKVVCGSQTGIMLLYSWGCFKDCSDRFVDLSSNSIDTMLKLDEDRIIT 240
DELLSVV+MKNGRKV+CG+Q G +LLYSWG FKDCSDRFVDL+ NS+D +LKLDEDR+IT
Sbjct: 178 DELLSVVIMKNGRKVICGTQNGTLLLYSWGFFKDCSDRFVDLAPNSVDALLKLDEDRLIT 237
Query: 241 GSENGMINLVGILPNRVIQPVAEHSEYPVERLGN*RFVAFSHDRKFLGSIAHDQMLKLWD 300
G +NG+I+LVGILPNR+IQP+ H +YP+E L A SHD+KFLGS AHD MLKLW+
Sbjct: 238 GCDNGIISLVGILPNRIIQPIGSH-DYPIEDL------ALSHDKKFLGSTAHDSMLKLWN 290
Query: 301 LDNILQGSRNTQRNENGGVDNDVDSDNEDEMDVDNNTSKFTKGNKRKNASKGHAAGDSNN 360
L+ IL+GS N +G + D DSDN D MD+DN+ SK +KG+KRK SK + +NN
Sbjct: 291 LEEILEGSNVNSGNASGAAE-DSDSDN-DGMDLDNDPSKSSKGSKRKTKSKANTLNATNN 348
Query: 361 FFADL 365
FFADL
Sbjct: 349 FFADL 353
>ref|NP_910903.1| putative transducin / WD-40 repeat protein [Oryza sativa (japonica
cultivar-group)] gi|22831203|dbj|BAC16061.1| putative
transducin / WD-40 repeat protein [Oryza sativa
(japonica cultivar-group)]
Length = 353
Score = 432 bits (1111), Expect = e-120
Identities = 219/366 (59%), Positives = 284/366 (76%), Gaps = 14/366 (3%)
Query: 1 MEINLGKLAFDIDFHPSDNLVATGLIDGDLHLYRYSSDNTNSDPVRLLEIHAHTESCRAA 60
ME ++ FD+ FHPS LVAT LI G+L+L+RY+++ S P RL +H ESCRA
Sbjct: 1 MEALHEEMPFDLAFHPSSPLVATSLITGELYLFRYAAE---SQPERLFAAKSHKESCRAV 57
Query: 61 RFINGGRALLTGSPDFSILATDVETGSTIARLDNAHEAAVNRLVNLTESTVASGDDDGCI 120
RF+ G +LTGS D SILA+DVETG IARL++AHE +NRLV LTE+TVASGDD+GCI
Sbjct: 58 RFVESGNVILTGSADCSILASDVETGKPIARLEDAHENGINRLVCLTETTVASGDDEGCI 117
Query: 121 KVWDTRERSCCNSFEAHEDYISDITFASDAMKLLATSGDGTLSVCNLRRNKVQAQSEFSE 180
KVWDTRERSCCN+F HEDYISD+T+ SD+ ++LATSGDGTLSV NLRRNKV++QSEFSE
Sbjct: 118 KVWDTRERSCCNTFHCHEDYISDMTYVSDSTQILATSGDGTLSVSNLRRNKVKSQSEFSE 177
Query: 181 DELLSVVLMKNGRKVVCGSQTGIMLLYSWGCFKDCSDRFVDLSSNSIDTMLKLDEDRIIT 240
DELLSVV+MKNG+KVVCG+ +G +LLYSWG FKDCSDRF+ + S+DTMLKLDE+ +I+
Sbjct: 178 DELLSVVVMKNGKKVVCGTPSGALLLYSWGFFKDCSDRFLG-HAQSVDTMLKLDEETLIS 236
Query: 241 GSENGMINLVGILPNRVIQPVAEHSEYPVERLGN*RFVAFSHDRKFLGSIAHDQMLKLWD 300
G+ +G+I LVGILPNR+IQP+AEHSEYP+E L AFS+DR +LGSI+HD+MLKLWD
Sbjct: 237 GASDGVIRLVGILPNRIIQPLAEHSEYPIEAL------AFSNDRNYLGSISHDKMLKLWD 290
Query: 301 LDNILQGSRNTQRNENGGVDNDVDSDNEDEMDVDNNTSKFTKGNKRKNASKGHAAG-DSN 359
L ++L + Q ++ G D+D D+ ++D+D N+S KG++ SKG ++ ++
Sbjct: 291 LQDLLNRQQLVQDDKLGEQDSDDSDDDGMDVDMDPNSS---KGSRSTKTSKGQSSDRPTS 347
Query: 360 NFFADL 365
+FFADL
Sbjct: 348 DFFADL 353
>ref|NP_973596.1| transducin family protein / WD-40 repeat family protein
[Arabidopsis thaliana]
Length = 296
Score = 402 bits (1032), Expect = e-110
Identities = 197/297 (66%), Positives = 242/297 (81%), Gaps = 9/297 (3%)
Query: 69 LLTGSPDFSILATDVETGSTIARLDNAHEAAVNRLVNLTESTVASGDDDGCIKVWDTRER 128
++T S D SILATDVETG+ +A L+NAHE AVN L+N+TE+T+ASGDD GC+K+WDTR+R
Sbjct: 9 IVTASADCSILATDVETGAQVAHLENAHEDAVNTLINVTETTIASGDDKGCVKIWDTRQR 68
Query: 129 SCCNSFEAHEDYISDITFASDAMKLLATSGDGTLSVCNLRRNKVQAQSEFSEDELLSVVL 188
SC + F AHEDYIS +TFASD+MKL+ TSGDGTLSVCNLR +KVQ+QSEFSEDELLSVV+
Sbjct: 69 SCSHEFNAHEDYISGMTFASDSMKLVVTSGDGTLSVCNLRTSKVQSQSEFSEDELLSVVI 128
Query: 189 MKNGRKVVCGSQTGIMLLYSWGCFKDCSDRFVDLSSNSIDTMLKLDEDRIITGSENGMIN 248
MKNGRKV+CG+Q G +LLYSWG FKDCSDRFVDL+ NS+D +LKLDEDR+ITG +NG+I+
Sbjct: 129 MKNGRKVICGTQNGTLLLYSWGFFKDCSDRFVDLAPNSVDALLKLDEDRLITGCDNGIIS 188
Query: 249 LVGILPNRVIQPVAEHSEYPVERLGN*RFVAFSHDRKFLGSIAHDQMLKLWDLDNILQGS 308
LVGILPNR+IQP+ H +YP+E L A SHD+KFLGS AHD MLKLW+L+ IL+GS
Sbjct: 189 LVGILPNRIIQPIGSH-DYPIEDL------ALSHDKKFLGSTAHDSMLKLWNLEEILEGS 241
Query: 309 RNTQRNENGGVDNDVDSDNEDEMDVDNNTSKFTKGNKRKNASKGHAAGDSNNFFADL 365
N +G + D DSDN D MD+DN+ SK +KG+KRK SK + +NNFFADL
Sbjct: 242 NVNSGNASGAAE-DSDSDN-DGMDLDNDPSKSSKGSKRKTKSKANTLNATNNFFADL 296
>emb|CAF87493.1| unnamed protein product [Tetraodon nigroviridis]
Length = 615
Score = 216 bits (549), Expect = 1e-54
Identities = 121/319 (37%), Positives = 182/319 (56%), Gaps = 11/319 (3%)
Query: 2 EINLGKLAFDIDFHPSDNLVATGLIDGDLHLYRYSSDNTNSDPVRLLEIHAHTESCRAAR 61
+I L +A + HPS +++A G +DGD+++Y YS T D L H +SCR R
Sbjct: 19 DIRLEAIANSVALHPSRDILALGDVDGDVYVYSYSC--TEGDNRELWSSGHHVKSCRQVR 76
Query: 62 FINGGRALLTGSPDFSILATDVETGSTIARLDNAHEAAVNRLVNLTESTVASGDDDGCIK 121
F G+ L + S D ++ DVE G ++R+ AH+AA+N L+ + E+ +A+GDD G +K
Sbjct: 77 FSADGQKLYSVSRDKAVHLLDVERGQLLSRIRGAHDAAINSLLLVDENVLATGDDVGGLK 136
Query: 122 VWDTRERSCCNSFEAHEDYISDITFASDAMKLLATSGDGTLSVCNLRRNKVQAQSEFSED 181
VWD R+ + H+DYISD+ LL SGDG + V N++R + + SE
Sbjct: 137 VWDMRKGTAVMDERQHQDYISDMAVDQAKRTLLTASGDGCMGVFNIKRRRWELLSEDQAG 196
Query: 182 ELLSVVLMKNGRKVVCGSQTGIMLLYSWGCFKDCSDRFVDLSSNSIDTMLKLDEDRIITG 241
+L SVVLMK GRKV CGS G + L++W F SDRF L + S++ ++ + + + T
Sbjct: 197 DLTSVVLMKKGRKVACGSSEGTVYLFNWSGFGATSDRFA-LGAESVECIVPVSDSVMCTA 255
Query: 242 SENGMINLVGILPNRVIQPVAEHSEYPVERLGN*RFVAFSHDRKFLGSIAHDQMLKLWDL 301
S +G I V +LPNRV+ + +H+ PVE L A S D + L S AHDQ++K WD+
Sbjct: 256 SIDGYIRAVNLLPNRVLGCIGQHAGEPVEEL------AKSWDARLLASSAHDQLVKFWDI 309
Query: 302 DNILQGS--RNTQRNENGG 318
+ S + +R + GG
Sbjct: 310 STLASVSVDEHRKRKKKGG 328
Score = 79.3 bits (194), Expect = 2e-13
Identities = 41/99 (41%), Positives = 60/99 (60%), Gaps = 1/99 (1%)
Query: 157 SGDGTLSVCNLRRNKVQAQSEFSEDELLSVVLMKNGRKVVCGSQTGIMLLYSWGCFKDCS 216
SGDG + V N++R + + SE +L SVVLMK GRKV CGS G + L++W F S
Sbjct: 401 SGDGCMGVFNIKRRRWELLSEDQAGDLTSVVLMKKGRKVACGSSEGTVYLFNWSGFGATS 460
Query: 217 DRFVDLSSNSIDTMLKLDEDRIITGSENGMINLVGILPN 255
DRF L + S++ ++ + + + T S +G I V +LPN
Sbjct: 461 DRFA-LGAESVECIVPVSDSVMCTASIDGYIRAVNLLPN 498
>ref|NP_001014873.1| hypothetical protein LOC508709 [Bos taurus]
gi|61553137|gb|AAX46356.1| hypothetical protein FLJ20195
[Bos taurus]
Length = 382
Score = 213 bits (543), Expect = 6e-54
Identities = 123/300 (41%), Positives = 168/300 (56%), Gaps = 9/300 (3%)
Query: 2 EINLGKLAFDIDFHPSDNLVATGLIDGDLHLYRYSSDNTNSDPVRLLEIHAHTESCRAAR 61
+I L A + FHP+ +L+A G +DGD+ ++ YS + L H +SCRA
Sbjct: 34 DIVLEAPASGLAFHPARDLLAAGDVDGDVFVFSYSCQE--GETKELWSSGHHLKSCRAVV 91
Query: 62 FINGGRALLTGSPDFSILATDVETGSTIARLDNAHEAAVNRLVNLTESTVASGDDDGCIK 121
F G+ L+T S D +I DVE G R+ AH A +N L+ + E+ +A+GDD G I+
Sbjct: 92 FSEDGQKLVTVSKDKAIHFLDVELGRLERRISKAHGAPINSLLLVDENVLATGDDTGGIR 151
Query: 122 VWDTRERSCCNSFEAHEDYISDITFASDAMKLLATSGDGTLSVCNLRRNKVQAQSEFSED 181
+WD R+ HE+YI+D+ D LL SGDG L V N++R + + SE
Sbjct: 152 LWDQRKEGPLMDMRQHEEYIADMALDPDKKLLLTASGDGCLGVFNIKRRRFELLSEPQSG 211
Query: 182 ELLSVVLMKNGRKVVCGSQTGIMLLYSWGCFKDCSDRFVDLSSNSIDTMLKLDEDRIITG 241
+L SV LMK GRKV CGS G + L++W F SDRF L + SID M+ + E + G
Sbjct: 212 DLTSVTLMKYGRKVACGSSEGTIYLFNWDGFGATSDRFA-LRAESIDCMVPVTESLLCAG 270
Query: 242 SENGMINLVGILPNRVIQPVAEHSEYPVERLGN*RFVAFSHDRKFLGSIAHDQMLKLWDL 301
S +G+I V ILPNRV+ V +H+E PVE L A SH FL S HDQ LK WD+
Sbjct: 271 STDGVIRAVNILPNRVVGSVGQHAEEPVENL------ALSHCGCFLASSGHDQRLKFWDM 324
>ref|XP_640134.1| hypothetical protein DDB0204846 [Dictyostelium discoideum]
gi|60468136|gb|EAL66146.1| hypothetical protein
DDB0204846 [Dictyostelium discoideum]
Length = 408
Score = 213 bits (543), Expect = 6e-54
Identities = 122/343 (35%), Positives = 195/343 (56%), Gaps = 22/343 (6%)
Query: 2 EINLGKLAFDIDFHPSDNLVATGLIDGDLHLYRYSSDNTNSDPVRLLEIHAHTESCRAAR 61
+I+ + F ++FHP+++L+ +G L L++YS D L + H CR A
Sbjct: 15 DISFNTIPFSLNFHPTEDLLVVSDAEGRLKLFKYSLDENEVK----LSLRPHQSGCRQAN 70
Query: 62 FINGGRALLTGSPDFSILATDVETGSTIARLDNAHEAAVNRLVNLTESTVASGDDDGCIK 121
F + G+ + T S D S+ D+ TGS + + AH+ +N LV+ E V +GDD+G IK
Sbjct: 71 FSSDGKYIFTASSDCSMKVIDINTGSILYTREEAHDYPINCLVS-KEFMVFTGDDEGTIK 129
Query: 122 VWDTRERSCCNSFEAHEDYISDITFASDAMKLLATSGDGTLSVCNLRRNKVQAQSEFSED 181
VWD R+++ F+ H D+ISDIT D + ATSGDG +S+ N R + SE S++
Sbjct: 130 VWDMRQQNIVCEFQEHGDFISDITTIDDR-HIAATSGDGGVSIYNFVRKSMDDISEKSDN 188
Query: 182 ELLSVVLMKNGRKVVCGSQTGIMLLYSWGCFKDCSDRFVDLSSNSIDTMLKLDEDRIITG 241
ELLS + + NG+K+VCGSQ G +L+Y ++ +F S+D ++K++ + +G
Sbjct: 189 ELLSCLSLDNGQKLVCGSQDGSILIYDRNNLENVK-KFAG-HPQSVDALVKVNNNTFFSG 246
Query: 242 SENGMINLVGILPNRVIQPVAEHSEYPVERLGN*RFVAFSHDRKFLGSIAHDQMLKLWDL 301
S +G+I +G+ P +++ V EHS +P+ER+ A S D ++LGSI+HD LK W++
Sbjct: 247 SSDGIIRFIGLRPKKLLGVVGEHSTFPIERM------AISRDNRYLGSISHDFSLKFWNV 300
Query: 302 DNILQ-------GSRNTQRNENGGVDNDVDSDNEDEMDVDNNT 337
+ + S N NE D+D D D+E E D++N T
Sbjct: 301 ASFYEEDESEDADSANANANEIENDDDDDDDDDEIE-DIENAT 342
>gb|AAT68118.1| AK025355-like [Danio rerio] gi|51467990|ref|NP_001003871.1|
hypothetical protein FLJ20195-like [Danio rerio]
Length = 386
Score = 213 bits (542), Expect = 8e-54
Identities = 114/312 (36%), Positives = 174/312 (55%), Gaps = 9/312 (2%)
Query: 2 EINLGKLAFDIDFHPSDNLVATGLIDGDLHLYRYSSDNTNSDPVRLLEIHAHTESCRAAR 61
+I L + I FHP +++A G IDGD++L+ YS T + L H +SCR
Sbjct: 31 DIKLEAIVNTIAFHPKQDILAAGDIDGDIYLFSYSC--TEGENKELWSSGHHLKSCRKVL 88
Query: 62 FINGGRALLTGSPDFSILATDVETGSTIARLDNAHEAAVNRLVNLTESTVASGDDDGCIK 121
F + G+ L + S D +I DVE G R+ AH+ +N ++ + E+ A+GDD+G +K
Sbjct: 89 FSSDGQKLFSVSKDKAIHIMDVEAGKLETRIPKAHKVPINAMLLIDENIFATGDDEGTLK 148
Query: 122 VWDTRERSCCNSFEAHEDYISDITFASDAMKLLATSGDGTLSVCNLRRNKVQAQSEFSED 181
VWD R+ + + HEDYISDIT LL +SGDGTL V N++R + + SE
Sbjct: 149 VWDMRKGTSFMDLKHHEDYISDITIDQAKRTLLTSSGDGTLGVFNIKRRRFELLSEIQNG 208
Query: 182 ELLSVVLMKNGRKVVCGSQTGIMLLYSWGCFKDCSDRFVDLSSNSIDTMLKLDEDRIITG 241
+L SV +MK GRKVVCGS G + +++W F SDRF + + S+D ++ + + +
Sbjct: 209 DLTSVSIMKRGRKVVCGSGEGTIYIFNWNGFGATSDRFA-VQAESVDCIVPITDSILCAA 267
Query: 242 SENGMINLVGILPNRVIQPVAEHSEYPVERLGN*RFVAFSHDRKFLGSIAHDQMLKLWDL 301
S +G+I + ILPNRV+ + +H +E +A D FL S AHD+++K WD+
Sbjct: 268 STDGVIRAINILPNRVVGSIGQHVGEAIEE------IARCRDTHFLASCAHDELIKFWDI 321
Query: 302 DNILQGSRNTQR 313
++ N R
Sbjct: 322 SSLPDEKVNDYR 333
>gb|AAH95552.1| Flj20195l protein [Danio rerio]
Length = 387
Score = 213 bits (542), Expect = 8e-54
Identities = 114/312 (36%), Positives = 174/312 (55%), Gaps = 9/312 (2%)
Query: 2 EINLGKLAFDIDFHPSDNLVATGLIDGDLHLYRYSSDNTNSDPVRLLEIHAHTESCRAAR 61
+I L + I FHP +++A G IDGD++L+ YS T + L H +SCR
Sbjct: 31 DIKLEAIVNTIAFHPKQDILAAGDIDGDIYLFSYSC--TEGENKELWSSGHHLKSCRKVL 88
Query: 62 FINGGRALLTGSPDFSILATDVETGSTIARLDNAHEAAVNRLVNLTESTVASGDDDGCIK 121
F + G+ L + S D +I DVE G R+ AH+ +N ++ + E+ A+GDD+G +K
Sbjct: 89 FSSDGQKLFSVSKDKAIHIMDVEAGKLETRIPKAHKVPINAMLLIDENIFATGDDEGTLK 148
Query: 122 VWDTRERSCCNSFEAHEDYISDITFASDAMKLLATSGDGTLSVCNLRRNKVQAQSEFSED 181
VWD R+ + + HEDYISDIT LL +SGDGTL V N++R + + SE
Sbjct: 149 VWDMRKGTSFMDLKHHEDYISDITIDQAKRTLLTSSGDGTLGVFNIKRRRFELLSEIQNG 208
Query: 182 ELLSVVLMKNGRKVVCGSQTGIMLLYSWGCFKDCSDRFVDLSSNSIDTMLKLDEDRIITG 241
+L SV +MK GRKVVCGS G + +++W F SDRF + + S+D ++ + + +
Sbjct: 209 DLTSVSIMKRGRKVVCGSGEGTIYIFNWNGFGATSDRFA-VQAESVDCIVPITDSILCAA 267
Query: 242 SENGMINLVGILPNRVIQPVAEHSEYPVERLGN*RFVAFSHDRKFLGSIAHDQMLKLWDL 301
S +G+I + ILPNRV+ + +H +E +A D FL S AHD+++K WD+
Sbjct: 268 STDGVIRAINILPNRVVGSIGQHVGEAIEE------IARCRDTHFLASCAHDELIKFWDI 321
Query: 302 DNILQGSRNTQR 313
++ N R
Sbjct: 322 SSLPDEKVNDYR 333
>pir||T02314 hypothetical protein At2g34260 [imported] - Arabidopsis thaliana
Length = 236
Score = 212 bits (540), Expect = 1e-53
Identities = 104/157 (66%), Positives = 125/157 (79%), Gaps = 3/157 (1%)
Query: 1 MEINLGKLAFDIDFHPSDNLVATGLIDGDLHLYRYSSDNTNSDPVRLLEIHAHTESCRAA 60
MEI+LG AF IDFHPS NLVA GLIDG LHLYRY SD S VR ++ AH ESCRA
Sbjct: 1 MEIDLGANAFGIDFHPSTNLVAAGLIDGHLHLYRYDSD---SSLVRERKVRAHKESCRAV 57
Query: 61 RFINGGRALLTGSPDFSILATDVETGSTIARLDNAHEAAVNRLVNLTESTVASGDDDGCI 120
RFI+ G+ ++T S D SILATDVETG+ +A L+NAHE AVN L+N+TE+T+ASGDD GC+
Sbjct: 58 RFIDDGQRIVTASADCSILATDVETGAQVAHLENAHEDAVNTLINVTETTIASGDDKGCV 117
Query: 121 KVWDTRERSCCNSFEAHEDYISDITFASDAMKLLATS 157
K+WDTR+RSC + F AHEDYIS +TFASD+MKL+ TS
Sbjct: 118 KIWDTRQRSCSHEFNAHEDYISGMTFASDSMKLVVTS 154
Score = 76.6 bits (187), Expect = 1e-12
Identities = 61/214 (28%), Positives = 98/214 (45%), Gaps = 34/214 (15%)
Query: 167 LRRNKVQAQSEFSEDELLSVVLMKNGRKVVCGSQTGIMLLYSWGCFKDCSDRFVDLSS-- 224
+R KV+A E +V + +G+++V S DCS D+ +
Sbjct: 42 VRERKVRAHKESCR----AVRFIDDGQRIVTASA-------------DCSILATDVETGA 84
Query: 225 ----------NSIDTMLKLDEDRIITGSENGMINLVGILPNRVIQPVAEHSEY---PVER 271
++++T++ + E I +G + G + + H +Y
Sbjct: 85 QVAHLENAHEDAVNTLINVTETTIASGDDKGCVKIWDTRQRSCSHEFNAHEDYISGMTFA 144
Query: 272 LGN*RFVAFSHDRKFLGSIAHDQMLKLWDLDNILQGSRNTQRNENGGVDNDVDSDNEDEM 331
+ + V S L + D +LKLW+L+ IL+GS N +G + D DSDN D M
Sbjct: 145 SDSMKLVVTSDRFVDLAPNSVDALLKLWNLEEILEGSNVNSGNASGAAE-DSDSDN-DGM 202
Query: 332 DVDNNTSKFTKGNKRKNASKGHAAGDSNNFFADL 365
D+DN+ SK +KG+KRK SK + +NNFFADL
Sbjct: 203 DLDNDPSKSSKGSKRKTKSKANTLNATNNFFADL 236
>gb|AAS66224.1| LRRG00133 [Rattus norvegicus]
Length = 872
Score = 211 bits (536), Expect = 4e-53
Identities = 120/303 (39%), Positives = 168/303 (54%), Gaps = 9/303 (2%)
Query: 2 EINLGKLAFDIDFHPSDNLVATGLIDGDLHLYRYSSDNTNSDPVRLLEIHAHTESCRAAR 61
+I L A + FHP+ +L+A G +DGD+ ++ YS + L H +SCRA
Sbjct: 522 DIVLEAPASGLAFHPTRDLLAAGDVDGDVFVFAYSCQE--GETKELWSSGHHLKSCRAVV 579
Query: 62 FINGGRALLTGSPDFSILATDVETGSTIARLDNAHEAAVNRLVNLTESTVASGDDDGCIK 121
F G+ L+T S D +I DVE G R+ AH A +N L+ + E+ + +GDD G I+
Sbjct: 580 FSEDGQKLVTVSKDKAIHVLDVEQGQLERRISKAHSAPINSLLLVDENVLVTGDDTGGIR 639
Query: 122 VWDTRERSCCNSFEAHEDYISDITFASDAMKLLATSGDGTLSVCNLRRNKVQAQSEFSED 181
+WD R+ HE+YI+D+ LL SGDG L V N++R + + SE
Sbjct: 640 LWDQRKEGPLMDMRQHEEYIADMALDPAKKLLLTASGDGCLGVFNIKRRRFELLSEPQSG 699
Query: 182 ELLSVVLMKNGRKVVCGSQTGIMLLYSWGCFKDCSDRFVDLSSNSIDTMLKLDEDRIITG 241
+L SV LMK G+KV CGS G + L++W F SDRF L + SID M+ + E+ + TG
Sbjct: 700 DLTSVALMKYGKKVACGSSEGTIYLFNWNGFGATSDRFA-LRAESIDCMVPVTENLLCTG 758
Query: 242 SENGMINLVGILPNRVIQPVAEHSEYPVERLGN*RFVAFSHDRKFLGSIAHDQMLKLWDL 301
S +G+I V ILPNRV+ V +H+ PVE L A SH FL S HDQ LK WD+
Sbjct: 759 STDGIIRAVNILPNRVVGTVGQHAGEPVEEL------ALSHCGHFLASSGHDQRLKFWDM 812
Query: 302 DNI 304
+
Sbjct: 813 TQL 815
>ref|NP_001017932.1| WD repeat domain 55 [Rattus norvegicus]
Length = 384
Score = 211 bits (536), Expect = 4e-53
Identities = 120/303 (39%), Positives = 168/303 (54%), Gaps = 9/303 (2%)
Query: 2 EINLGKLAFDIDFHPSDNLVATGLIDGDLHLYRYSSDNTNSDPVRLLEIHAHTESCRAAR 61
+I L A + FHP+ +L+A G +DGD+ ++ YS + L H +SCRA
Sbjct: 34 DIVLEAPASGLAFHPTRDLLAAGDVDGDVFVFAYSCQE--GETKELWSSGHHLKSCRAVV 91
Query: 62 FINGGRALLTGSPDFSILATDVETGSTIARLDNAHEAAVNRLVNLTESTVASGDDDGCIK 121
F G+ L+T S D +I DVE G R+ AH A +N L+ + E+ + +GDD G I+
Sbjct: 92 FSEDGQKLVTVSKDKAIHVLDVEQGQLERRISKAHSAPINSLLLVDENVLVTGDDTGGIR 151
Query: 122 VWDTRERSCCNSFEAHEDYISDITFASDAMKLLATSGDGTLSVCNLRRNKVQAQSEFSED 181
+WD R+ HE+YI+D+ LL SGDG L V N++R + + SE
Sbjct: 152 LWDQRKEGPLMDMRQHEEYIADMALDPAKKLLLTASGDGCLGVFNIKRRRFELLSEPQSG 211
Query: 182 ELLSVVLMKNGRKVVCGSQTGIMLLYSWGCFKDCSDRFVDLSSNSIDTMLKLDEDRIITG 241
+L SV LMK G+KV CGS G + L++W F SDRF L + SID M+ + E+ + TG
Sbjct: 212 DLTSVALMKYGKKVACGSSEGTIYLFNWNGFGATSDRFA-LRAESIDCMVPVTENLLCTG 270
Query: 242 SENGMINLVGILPNRVIQPVAEHSEYPVERLGN*RFVAFSHDRKFLGSIAHDQMLKLWDL 301
S +G+I V ILPNRV+ V +H+ PVE L A SH FL S HDQ LK WD+
Sbjct: 271 STDGIIRAVNILPNRVVGTVGQHAGEPVEEL------ALSHCGHFLASSGHDQRLKFWDM 324
Query: 302 DNI 304
+
Sbjct: 325 TQL 327
>dbj|BAB15118.1| unnamed protein product [Homo sapiens] gi|46249769|gb|AAH68485.1|
WD repeat domain 55 [Homo sapiens]
gi|38327642|ref|NP_060176.2| WD repeat domain 55 [Homo
sapiens]
Length = 383
Score = 210 bits (535), Expect = 5e-53
Identities = 119/300 (39%), Positives = 168/300 (55%), Gaps = 9/300 (3%)
Query: 2 EINLGKLAFDIDFHPSDNLVATGLIDGDLHLYRYSSDNTNSDPVRLLEIHAHTESCRAAR 61
+I L A + FHP+ +L+A G +DGD+ ++ YS + L H ++CRA
Sbjct: 34 DIVLEAPASGLAFHPARDLLAAGDVDGDVFVFSYSCQE--GETKELWSSGHHLKACRAVA 91
Query: 62 FINGGRALLTGSPDFSILATDVETGSTIARLDNAHEAAVNRLVNLTESTVASGDDDGCIK 121
F G+ L+T S D +I DVE G R+ AH A +N L+ + E+ +A+GDD G I+
Sbjct: 92 FSEDGQKLITVSKDKAIHVLDVEQGQLERRVSKAHGAPINSLLLVDENVLATGDDTGGIR 151
Query: 122 VWDTRERSCCNSFEAHEDYISDITFASDAMKLLATSGDGTLSVCNLRRNKVQAQSEFSED 181
+WD R+ HE+YI+D+ LL SGDG L + N++R + + SE
Sbjct: 152 LWDQRKEGPLMDMRQHEEYIADMALDPAKKLLLTASGDGCLGIFNIKRRRFELLSEPQSG 211
Query: 182 ELLSVVLMKNGRKVVCGSQTGIMLLYSWGCFKDCSDRFVDLSSNSIDTMLKLDEDRIITG 241
+L SV LMK G+KV CGS G + L++W F SDRF L + SID M+ + E + TG
Sbjct: 212 DLTSVTLMKWGKKVACGSSEGTIYLFNWNGFGATSDRFA-LRAESIDCMVPVTESLLCTG 270
Query: 242 SENGMINLVGILPNRVIQPVAEHSEYPVERLGN*RFVAFSHDRKFLGSIAHDQMLKLWDL 301
S +G+I V ILPNRV+ V +H+ PVE L A SH +FL S HDQ LK WD+
Sbjct: 271 STDGVIRAVNILPNRVVGSVGQHTGEPVEEL------ALSHCGRFLASSGHDQRLKFWDM 324
>ref|XP_535212.1| PREDICTED: similar to hypothetical protein FLJ20195 [Canis
familiaris]
Length = 953
Score = 210 bits (535), Expect = 5e-53
Identities = 119/303 (39%), Positives = 168/303 (55%), Gaps = 9/303 (2%)
Query: 2 EINLGKLAFDIDFHPSDNLVATGLIDGDLHLYRYSSDNTNSDPVRLLEIHAHTESCRAAR 61
+I L A + FHP+ +L+A G +DGD+ ++ YS + L H +SCRA
Sbjct: 604 DIVLEAPASGLAFHPARDLLAAGDVDGDVFVFSYSCQE--GETKELWSSGHHLKSCRAVV 661
Query: 62 FINGGRALLTGSPDFSILATDVETGSTIARLDNAHEAAVNRLVNLTESTVASGDDDGCIK 121
F G+ L+T S D +I DVE G R+ AH A +N L+ + E+ + +GDD G ++
Sbjct: 662 FSEDGQKLVTVSKDKAIHVLDVEQGRLERRISKAHGAPINSLLLVDENVLVTGDDTGGVR 721
Query: 122 VWDTRERSCCNSFEAHEDYISDITFASDAMKLLATSGDGTLSVCNLRRNKVQAQSEFSED 181
+WD R+ HE+YI+D+ LL SGDG L V N+RR + + SE
Sbjct: 722 LWDQRKEGPLMDMRQHEEYIADMALDPSKKLLLTASGDGCLGVFNIRRRRFELLSEPQSG 781
Query: 182 ELLSVVLMKNGRKVVCGSQTGIMLLYSWGCFKDCSDRFVDLSSNSIDTMLKLDEDRIITG 241
+L SV LMK G+KV CGS G + L++W F SDRF L + SID M+ + E + TG
Sbjct: 782 DLTSVTLMKYGKKVACGSSEGTIYLFNWNGFGATSDRFA-LRAESIDCMVPVTESLLCTG 840
Query: 242 SENGMINLVGILPNRVIQPVAEHSEYPVERLGN*RFVAFSHDRKFLGSIAHDQMLKLWDL 301
S +G+I V ILPNRV+ V +H+ PVE+L A SH FL S HDQ LK W++
Sbjct: 841 STDGVIRAVNILPNRVVGSVGQHAGEPVEKL------ALSHCTHFLASSGHDQRLKFWNM 894
Query: 302 DNI 304
+
Sbjct: 895 SQL 897
>emb|CAH91474.1| hypothetical protein [Pongo pygmaeus]
Length = 383
Score = 207 bits (528), Expect = 3e-52
Identities = 123/319 (38%), Positives = 174/319 (53%), Gaps = 11/319 (3%)
Query: 2 EINLGKLAFDIDFHPSDNLVATGLIDGDLHLYRYSSDNTNSDPVRLLEIHAHTESCRAAR 61
+I L A + FHP+ +L+A G +DGD+ ++ YS + L H ++CRA
Sbjct: 34 DIVLEAPASGLAFHPARDLLAAGDVDGDVFVFSYSCQE--GETKELWSSGHHLKACRAVA 91
Query: 62 FINGGRALLTGSPDFSILATDVETGSTIARLDNAHEAAVNRLVNLTESTVASGDDDGCIK 121
F G+ L+T S D +I DVE G R+ AH A +N L+ + E+ +A+GDD G I+
Sbjct: 92 FSEDGQKLVTVSKDKAIHVLDVEQGRLERRISKAHGAPINSLLLVDENVLATGDDMGGIR 151
Query: 122 VWDTRERSCCNSFEAHEDYISDITFASDAMKLLATSGDGTLSVCNLRRNKVQAQSEFSED 181
+WD R+ HE+YI+D+ LL SGDG L V N++R + + SE
Sbjct: 152 LWDQRKEGPLMDMRQHEEYIADMALDPAKKLLLTASGDGCLGVFNIKRRRFELLSEPQSG 211
Query: 182 ELLSVVLMKNGRKVVCGSQTGIMLLYSWGCFKDCSDRFVDLSSNSIDTMLKLDEDRIITG 241
+L SV LMK G+KV CGS G + L++W F SDRF L + SID M+ + E + TG
Sbjct: 212 DLTSVTLMKCGKKVACGSSEGTIYLFNWNGFGATSDRFA-LRAESIDCMVPVTESLLCTG 270
Query: 242 SENGMINLVGILPNRVIQPVAEHSEYPVERLGN*RFVAFSHDRKFLGSIAHDQMLKLWDL 301
S +G+I V ILPNRV+ V +H+ PV L A SH +FL S HDQ LK WD+
Sbjct: 271 STDGVIRAVNILPNRVVGSVGQHTGEPVGEL------ALSHCGRFLASSGHDQRLKFWDM 324
Query: 302 DNI--LQGSRNTQRNENGG 318
+ + QR + GG
Sbjct: 325 AQLRAVVVDDYRQRKKKGG 343
>ref|NP_080740.2| WD repeat domain 55 [Mus musculus] gi|26344866|dbj|BAC36082.1|
unnamed protein product [Mus musculus]
gi|26383897|dbj|BAB31225.2| unnamed protein product [Mus
musculus]
Length = 388
Score = 207 bits (527), Expect = 4e-52
Identities = 118/303 (38%), Positives = 168/303 (54%), Gaps = 9/303 (2%)
Query: 2 EINLGKLAFDIDFHPSDNLVATGLIDGDLHLYRYSSDNTNSDPVRLLEIHAHTESCRAAR 61
+I L A + FHP+ +L+A G +DGD+ ++ YS + L H +SCRA
Sbjct: 35 DIVLEAPASGLAFHPTRDLLAAGDVDGDVFVFAYSCQE--GETKELWSSGHHLKSCRAVV 92
Query: 62 FINGGRALLTGSPDFSILATDVETGSTIARLDNAHEAAVNRLVNLTESTVASGDDDGCIK 121
F G+ L+T S D +I DVE G R+ AH A +N ++ + E+ + +GDD G I+
Sbjct: 93 FSEDGQKLVTVSKDKAIHILDVEQGQLERRISKAHSAPINSVLLVDENALVTGDDTGGIR 152
Query: 122 VWDTRERSCCNSFEAHEDYISDITFASDAMKLLATSGDGTLSVCNLRRNKVQAQSEFSED 181
+WD R+ HE+YI+D+ LL SGDG L V N++R + + SE
Sbjct: 153 LWDQRKEGPLMDMRQHEEYIADMALDPAKKLLLTASGDGCLGVFNIKRRRFELLSEPQSG 212
Query: 182 ELLSVVLMKNGRKVVCGSQTGIMLLYSWGCFKDCSDRFVDLSSNSIDTMLKLDEDRIITG 241
+L SV LMK G+KV CGS G + L++W F SDRF L + SID ++ + E+ + TG
Sbjct: 213 DLTSVALMKYGKKVACGSSEGTIYLFNWNGFGATSDRFA-LRAESIDCIVPVTENLLCTG 271
Query: 242 SENGMINLVGILPNRVIQPVAEHSEYPVERLGN*RFVAFSHDRKFLGSIAHDQMLKLWDL 301
S +G+I V ILPNRV+ V +H+ PVE L A SH FL S HDQ LK WD+
Sbjct: 272 STDGIIRAVNILPNRVVGTVGQHAGEPVEAL------ALSHCGHFLASSGHDQRLKFWDM 325
Query: 302 DNI 304
+
Sbjct: 326 TQL 328
>gb|AAW40894.1| protein monoubiquitination-related protein, putative [Cryptococcus
neoformans var. neoformans JEC21]
gi|58258601|ref|XP_566713.1| protein
monoubiquitination-related protein, putative
[Cryptococcus neoformans var. neoformans JEC21]
Length = 374
Score = 202 bits (513), Expect = 2e-50
Identities = 132/369 (35%), Positives = 194/369 (51%), Gaps = 35/369 (9%)
Query: 10 FDIDFHPSDNLVATGLIDGDLHLYRYSSDNTNSDPVRLLEIHAHTESCRAARFINGGRAL 69
FD+ FHP + +V + L+ G + + Y D+ + + + RA G +
Sbjct: 28 FDVAFHPKEPVVFSSLLTGQVCAWSY--DDATGETSSSWSVRPSKRTARALSIEENGDEI 85
Query: 70 LTGSPDFSILAT---------DVETGSTIARLDNAHEAAVNRLVNLTESTVASGDDDGCI 120
G S+L GS D+AHE +NR+ + + VA+GDDDG I
Sbjct: 86 WMGGKSGSLLCALTPPKVIQLSTRDGSMTRERDSAHECPINRVYCVNRNLVATGDDDGVI 145
Query: 121 KVWDTRERSCCNSFEAHEDYISDITFASDAMKLLATSGDGTLSVCNLRRNKVQ--AQSEF 178
K+WD R+ ++ H DYISD T+ D +L+ATSGDG LSV ++R NK SE
Sbjct: 146 KLWDPRQADSIRTYSQHFDYISDFTYFDDKRQLVATSGDGHLSVIDIRSNKSTPLTVSED 205
Query: 179 SEDELLSVVLMKNGRKVVCGSQTGIMLLYSWGC-FKDCSDRFVDLSSNSIDTMLKLDEDR 237
EDELLS+V +K G+K + GS GI+ +++ + D DR + SID ++ L D
Sbjct: 206 QEDELLSIVPIKGGQKAIVGSGLGILSVWNRQMGWADSVDR-IPGHPASIDAIVALTPDI 264
Query: 238 IITGSENGMINLVGILPNRVIQPVAEHSEYPVERLGN*RFVAFSHDRKFLGSIAHDQMLK 297
I TGSE+GMI ++ +LP++ + VA H E+PVER + + K+LGS++HD+ LK
Sbjct: 265 IATGSEDGMIRVIQVLPHKFLGVVATHEEFPVER------IRLDRNNKWLGSVSHDECLK 318
Query: 298 LWDLDNILQGSRNTQRNENGGVDNDVDSDNEDEMDVDNNTS-KFTKGNKRKNASKGHAAG 356
L D++++ + S D D D EDE D D S K K N K+ S+G A
Sbjct: 319 LTDVEDLFEDS-----------DED-DEMEEDEPDSDEEKSKKKKKDNGMKDMSRGQAEN 366
Query: 357 DSNNFFADL 365
D +FFADL
Sbjct: 367 D-GSFFADL 374
>gb|EAL23242.1| hypothetical protein CNBA3580 [Cryptococcus neoformans var.
neoformans B-3501A]
Length = 384
Score = 198 bits (503), Expect = 3e-49
Identities = 132/379 (34%), Positives = 194/379 (50%), Gaps = 45/379 (11%)
Query: 10 FDIDFHPSDNLVATGLIDGDLHLYRYSSDNTNSDPVRLLEIHAHTESCRAARFINGGRAL 69
FD+ FHP + +V + L+ G + + Y D+ + + + RA G +
Sbjct: 28 FDVAFHPKEPVVFSSLLTGQVCAWSY--DDATGETSSSWSVRPSKRTARALSIEENGDEI 85
Query: 70 LTGSPDFSILAT-------------------DVETGSTIARLDNAHEAAVNRLVNLTEST 110
G S+L GS D+AHE +NR+ + +
Sbjct: 86 WMGGKSGSLLCALTPPKVMCPILLMSNCSQLSTRDGSMTRERDSAHECPINRVYCVNRNL 145
Query: 111 VASGDDDGCIKVWDTRERSCCNSFEAHEDYISDITFASDAMKLLATSGDGTLSVCNLRRN 170
VA+GDDDG IK+WD R+ ++ H DYISD T+ D +L+ATSGDG LSV ++R N
Sbjct: 146 VATGDDDGVIKLWDPRQADSIRTYSQHFDYISDFTYFDDKRQLVATSGDGHLSVIDIRSN 205
Query: 171 KVQ--AQSEFSEDELLSVVLMKNGRKVVCGSQTGIMLLYSWGC-FKDCSDRFVDLSSNSI 227
K SE EDELLS+V +K G+K + GS GI+ +++ + D DR + SI
Sbjct: 206 KSTPLTVSEDQEDELLSIVPIKGGQKAIVGSGLGILSVWNRQMGWADSVDR-IPGHPASI 264
Query: 228 DTMLKLDEDRIITGSENGMINLVGILPNRVIQPVAEHSEYPVERLGN*RFVAFSHDRKFL 287
D ++ L D I TGSE+GMI ++ +LP++ + VA H E+PVER + + K+L
Sbjct: 265 DAIVALTPDIIATGSEDGMIRVIQVLPHKFLGVVATHEEFPVER------IRLDRNNKWL 318
Query: 288 GSIAHDQMLKLWDLDNILQGSRNTQRNENGGVDNDVDSDNEDEMDVDNNTS-KFTKGNKR 346
GS++HD+ LKL D++++ + S D D D EDE D D S K K N
Sbjct: 319 GSVSHDECLKLTDVEDLFEDS-----------DED-DEMEEDEPDSDEEKSKKKKKDNGM 366
Query: 347 KNASKGHAAGDSNNFFADL 365
K+ S+G A D +FFADL
Sbjct: 367 KDMSRGQAEND-GSFFADL 384
>ref|XP_623745.1| PREDICTED: similar to CG14722-PA [Apis mellifera]
Length = 437
Score = 171 bits (433), Expect = 3e-41
Identities = 98/293 (33%), Positives = 162/293 (54%), Gaps = 13/293 (4%)
Query: 11 DIDFHPSDNLVATGLIDGDLHLYRYSSDNTNSDPVRLLEIHAHTESCRAARFINGGRALL 70
DI FHP+ N++A I GD++LY+YS NT + + LE+H ++CR F G L
Sbjct: 116 DICFHPNLNMIAIADIGGDIYLYKYS--NTKVELISTLELHL--KACRDIEFNENGTILF 171
Query: 71 TGSPDFSILATDVETGSTIARLDNAHEAAVNRLVNLTESTVASGDDDGCIKVWDTRERSC 130
+ + D ++ TD+ET I +N HE + + + E A+GDD+G +K+WD R+R
Sbjct: 172 STAKDLCMMLTDLETEKLIRLYENIHEQPIYTMTIIGEHMFATGDDNGVVKMWDLRQRGN 231
Query: 131 CNSFEAH--EDYISDITFASDAMKLLATSGDGTLSVCNLRRNKVQAQSEFSEDELLSVVL 188
F EDY+S IT +A L+ T GDG+L+ N+ K+ QSE ++EL + L
Sbjct: 232 MPVFSLKKMEDYVSAITTNREAKYLVCTCGDGSLTTFNIPGKKLHVQSEEYQEELTCLGL 291
Query: 189 MKNGRKVVCGSQTGIMLLYSWGCFKDCSDRFVDLSSNSIDTMLKLDEDRIITGSENGMIN 248
K+ K++ G+ G M +Y+WG F SD F +L+ +I+ M+ + E+ +ITG E+GM+
Sbjct: 292 FKSETKILVGTNKGKMYVYNWGEFGFHSDEFPNLTKKAINCMIPITENIVITGGEDGMLR 351
Query: 249 LVGILPNRVIQPVAEHSEYPVERLGN*RFVAFSHDRKFLGSIAHDQMLKLWDL 301
+ P+ + V +H+ +E L + + + S +HD +K W++
Sbjct: 352 ASSLFPHYHLGIVGQHN-LSIETLD------INSNGNLIASSSHDNDIKFWNV 397
>ref|XP_653429.1| WD repeat protein [Entamoeba histolytica HM-1:IMSS]
gi|56470375|gb|EAL48041.1| WD repeat protein [Entamoeba
histolytica HM-1:IMSS]
Length = 363
Score = 166 bits (419), Expect = 1e-39
Identities = 100/355 (28%), Positives = 181/355 (50%), Gaps = 14/355 (3%)
Query: 11 DIDFHPSDNLVATGLIDGDLHLYRYSSDNTNSDPVRLLEIHAHTESCRAARFINGGRALL 70
DI F P++++VA G + G + LY Y + +P + + AH +S R F + G L
Sbjct: 23 DIKFSPTNDVVAVGDMHGLIRLYNYGNIQ---EPNEIFTLKAHEDSTRDVCFNDNGNLLF 79
Query: 71 TGSPDFSILATDVETGSTIARLDNAHEAAVNRLVNLTESTVASGDDDGCIKVWDTRERSC 130
+ S D SI TD+ +A+ D AH+ + L ++ + +GD+ G IKVWD R++ C
Sbjct: 80 STSADGSITITDLNNQKILAKNDKAHKHGIYSL-DVKGNIFVTGDESGRIKVWDMRQQKC 138
Query: 131 CNSFEAHEDYISDITFASDAMKLLATSGDGTLSVCNLRRNKVQAQSEFSEDELLSVVLMK 190
F H DYIS +T + + A GDG +S N+ KV S+ ++LLS+ + K
Sbjct: 139 VVVFNEHHDYISQLTICDNT--IFAAGGDGCMSTWNINSKKVIGISDNMNEDLLSLSITK 196
Query: 191 NGRKVVCGSQTGIMLLYSWGCFKDCSDRFVDLSSNSIDTMLKLDEDRIITGSENGMINLV 250
+VCG Q+G + ++ W + + + SI++++ ++++ IITGS +G+I +V
Sbjct: 197 KDDTLVCGGQSGKLFVWEWDNW-EYPKNALKGHPESIESIIAVNKNTIITGSSDGIIRVV 255
Query: 251 GILPNRVIQPVAEHSEYPVERLGN*RFVAFSHDRKFLGSIAHDQMLKLWDLDNILQGSRN 310
I P++++ V +H ++P+ERL A S D+ L S +H + +K WD+ + +
Sbjct: 256 QINPHKLLGCVGQH-DFPIERL------ALSRDKNILASSSHSRHIKFWDVGYLYNDQDD 308
Query: 311 TQRNENGGVDNDVDSDNEDEMDVDNNTSKFTKGNKRKNASKGHAAGDSNNFFADL 365
+ +++ ++ D + NK+K ++N F+ D+
Sbjct: 309 EESDDSKQKTMVIEKPEADIVTPFQEEKVQNAYNKKKKTKGPQQRNETNPFYKDM 363
>gb|AAF54692.2| CG14722-PA [Drosophila melanogaster] gi|17862296|gb|AAL39625.1|
LD21659p [Drosophila melanogaster]
gi|28571655|ref|NP_650111.3| CG14722-PA [Drosophila
melanogaster]
Length = 498
Score = 155 bits (393), Expect = 1e-36
Identities = 98/300 (32%), Positives = 154/300 (50%), Gaps = 12/300 (4%)
Query: 2 EINLGKLAFDIDFHPSDNLVATGLIDGDLHLYRYSSDNTNSDPVRLLEIHAHTESCRAAR 61
EI L DI FHP +++A I GD+HLY Y DN + +R +E+H+ ++CR
Sbjct: 152 EIKLEDFITDICFHPDRDIIALATIIGDVHLYEY--DNEANKLLRTIEVHS--KACRDVE 207
Query: 62 FINGGRALLTGSPDFSILATDVETGSTIARLDNAHEAAVNRLVNLTESTVASGDDDGCIK 121
F GR LLT S D ++ TD+ET + AH+ A+N L L E+ ASGDD G +K
Sbjct: 208 FTEDGRFLLTCSKDKCVMVTDMETEKLKKLYETAHDDAINTLHVLNENLFASGDDAGTVK 267
Query: 122 VWDTRERSCCNSFEAHEDYISDITFASDAMKLLATSGDGTLSVCNLRRNKVQAQSEFSED 181
+WD R ++ + ED I+ +T + LLATS DG L+ N+ K+ QSE E+
Sbjct: 268 LWDLRTKNAIFELKELEDQITQLTTNEQSKLLLATSADGYLTTFNISARKMYVQSEPYEE 327
Query: 182 ELLSVVLMKNGRKVVCGSQTGIMLLYSWGCFKDCSDRFVDLSSNSIDTMLKLDEDRIITG 241
EL + + + K+V G+ G + Y+WG F D + + S I M+ + +
Sbjct: 328 ELNCMGVYRGDSKLVVGTSKGRLYTYNWGQFGYHCDMYPGIKS-PISLMIPITDRIACVA 386
Query: 242 SENGMINLVGILPNRVIQPVAEHSEYPVERLGN*RFVAFSHDRKFLGSIAHDQMLKLWDL 301
E+G I I P R + V +H+ P+E L + + + S +H+ ++ W++
Sbjct: 387 GEDGNIRACHIAPYRNLGVVGQHN-MPIESLD------VNASGELIASSSHNNDVRFWNV 439
Database: nr
Posted date: Jul 5, 2005 12:34 AM
Number of letters in database: 863,360,394
Number of sequences in database: 2,540,612
Lambda K H
0.319 0.135 0.394
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 621,195,914
Number of Sequences: 2540612
Number of extensions: 26251033
Number of successful extensions: 202679
Number of sequences better than 10.0: 3777
Number of HSP's better than 10.0 without gapping: 1766
Number of HSP's successfully gapped in prelim test: 2061
Number of HSP's that attempted gapping in prelim test: 179288
Number of HSP's gapped (non-prelim): 16071
length of query: 378
length of database: 863,360,394
effective HSP length: 130
effective length of query: 248
effective length of database: 533,080,834
effective search space: 132204046832
effective search space used: 132204046832
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 76 (33.9 bits)
Medicago: description of AC142222.5


