

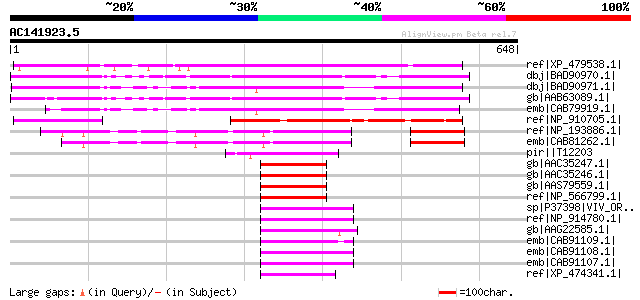
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC141923.5 - phase: 0 /pseudo
(648 letters)
Database: nr
2,540,612 sequences; 863,360,394 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
ref|XP_479538.1| VP1/ABI3 family regulatory protein-like [Oryza ... 461 e-128
dbj|BAD90970.1| transcription factor B3-EAR motif [Arabidopsis t... 422 e-116
dbj|BAD90971.1| transcription factor B3-EAR motif family [Arabid... 417 e-115
gb|AAB63089.1| putative VP1/ABI3 family regulatory protein [Arab... 365 3e-99
emb|CAB79919.1| predicted protein [Arabidopsis thaliana] gi|2827... 315 2e-84
ref|NP_910705.1| VP1/ABI3 family regulatory protein-like protein... 264 8e-69
ref|NP_193886.1| transcriptional factor B3 family protein [Arabi... 188 4e-46
emb|CAB81262.1| VP1 like protein [Arabidopsis thaliana] gi|30803... 160 2e-37
pir||T12203 transcription factor Vp1 - common ice plant gi|32191... 85 6e-15
gb|AAC35247.1| FUSCA3 [Arabidopsis thaliana] 84 1e-14
gb|AAC35246.1| FUSCA3 [Arabidopsis thaliana] 84 1e-14
gb|AAS79559.1| transcriptional regulator [Arabidopsis thaliana] ... 84 1e-14
ref|NP_566799.1| transcriptional regulator (FUSCA3) [Arabidopsis... 84 1e-14
sp|P37398|VIV_ORYSA Viviparous protein homolog gi|391885|dbj|BAA... 84 1e-14
ref|NP_914780.1| transcription activator VP1 - rice [Oryza sativ... 84 1e-14
gb|AAG22585.1| transcription factor viviparous 1 [Picea abies] 84 1e-14
emb|CAB91109.1| VIVIPAROUS1 protein [Triticum aestivum] 84 1e-14
emb|CAB91108.1| VIVIPAROUS1 protein [Triticum aestivum] 83 3e-14
emb|CAB91107.1| VIVIPAROUS1 protein [Triticum aestivum] 83 3e-14
ref|XP_474341.1| OSJNBa0064G10.6 [Oryza sativa (japonica cultiva... 82 5e-14
>ref|XP_479538.1| VP1/ABI3 family regulatory protein-like [Oryza sativa (japonica
cultivar-group)] gi|34394741|dbj|BAC84102.1| VP1/ABI3
family regulatory protein-like [Oryza sativa (japonica
cultivar-group)]
Length = 949
Score = 461 bits (1187), Expect = e-128
Identities = 269/614 (43%), Positives = 357/614 (57%), Gaps = 57/614 (9%)
Query: 6 CMNNYC----KEGSTGEWKKGWSLISGGFAKLCNKCGLAYENSLFCDKFHRHETGWRKCS 61
CMN C + GEW+KGW L SGGFA LC+KCGLAYE +FCD FH+ E+GWR CS
Sbjct: 17 CMNAACGAPAPSPAGGEWRKGWPLRSGGFAVLCDKCGLAYEQLVFCDIFHQKESGWRDCS 76
Query: 62 NCSKPIHSGCIVSKSSFEYLDFGGITCVTCVKPSQL----------CLNTENHNRFSQTT 111
C K +H GCI SK+SF+ LD GG+ CVTC+K S + +++N+ R +
Sbjct: 77 FCGKRLHCGCIASKNSFDLLDSGGVQCVTCIKNSAVQSVPSPVVPKLFSSQNNQRLFGKS 136
Query: 112 KNNASDQYGEHIDGRLLVEQA--------GKGNLMQLCRIGEVGESSRWPQAQRDAMVSC 163
+ S G ++ L+ A K NL + + E G+SS + +
Sbjct: 137 DDLLS---GRPLETSSLMVDARNDDLTIIAKNNLPFMVKNVEAGQSSNILRQKE------ 187
Query: 164 IGPKTEEVKCQF---NKEDTRFLNVMKHSSHLTAFTTLENNRPSWETKSIDETLS---LK 217
+ ++K + + D + + S EN P+ E+ + E+LS L
Sbjct: 188 LENGARQIKWELPTLSIGDMGRIPFLTRSQSALESRRDENKDPTTESTT-SESLSEACLN 246
Query: 218 MALGTSSRNS-----------VLPLATEIGEGKLEGKASSHFQQGQTSQSILAQLSKTGI 266
M+LG +S + +L T EG+ A S FQ Q ++ L + + G
Sbjct: 247 MSLGIASNGNKLEATSTVERPMLSPTTGFPEGRELTTALSPFQHAQRARHFLTRPPRVGE 306
Query: 267 AMNLETNKGMISH-PPRRPPADVKGKNQLLSRYWPRITDQELEKLSGDLKSTVVPLFEKV 325
+ K M+ H RPPA+ +G+NQLL RYWPRITDQEL+++SGD ST+VPLFEKV
Sbjct: 307 GAVFDPTKDMLPHLRVARPPAEGRGRNQLLPRYWPRITDQELQQISGDSNSTIVPLFEKV 366
Query: 326 LSPSDAGRIGRLVLPKACAEAFLPRILQSEGVPLQFQDIMGNEWTFQFRFWPNNNSRMYV 385
LS SDAGRIGRLVLPKACAEA+ P I Q EG PL QD G EW FQFRFWPNNNSRMYV
Sbjct: 367 LSASDAGRIGRLVLPKACAEAYFPPISQPEGRPLTIQDAKGKEWHFQFRFWPNNNSRMYV 426
Query: 386 LEGVTPCIQSLQLNAGDTVTFSRIDPGEKFLFGFRRSLTSIVTQDASTSSHSNGILIKDT 445
LEGVTPCIQSLQL AGDTVTFSRI+PG K + GFR++ ++ D+ S+ +NG ++ DT
Sbjct: 427 LEGVTPCIQSLQLQAGDTVTFSRIEPGGKLVMGFRKATNTVSLPDSQISAIANGSILGDT 486
Query: 446 NFSGAPQNLNSLSSFSNLLQSMKGNGEPYLNG-HSEHLRLGNGTADWLKTANSEEEMNNG 504
FS +NL +S +S LQS+KG + + + + H+ +G WLKT + G
Sbjct: 487 LFSSTNENLAIVSGYSGFLQSIKGAADLHTSSIYDHHVNSADGDVSWLKTDKFGSRPDEG 546
Query: 505 PLQRLVSVSEKKRSRNIGTKTKRLHIHSEDAMELRLTWEEAQEFLCPPPSVEPNFVTIED 564
LQ L KR RNIG+K++RL + +E+A EL+L W+E QE L P P+ +P V IED
Sbjct: 547 SLQFL------KRGRNIGSKSRRLSMDAEEAWELKLYWDEVQELLRPAPTAKPTVVMIED 600
Query: 565 QVFEEYDEPPVFGK 578
EEYDEPPVF K
Sbjct: 601 YEIEEYDEPPVFAK 614
>dbj|BAD90970.1| transcription factor B3-EAR motif [Arabidopsis thaliana]
gi|17064832|gb|AAL32570.1| putative VP1/ABI3 family
regulatory protein [Arabidopsis thaliana]
gi|30684597|ref|NP_850146.1| transcriptional factor B3
family protein [Arabidopsis thaliana]
Length = 790
Score = 422 bits (1086), Expect = e-116
Identities = 256/591 (43%), Positives = 335/591 (56%), Gaps = 61/591 (10%)
Query: 1 MGVEVCMNNYCKEGSTGEWKKGWSLISGGFAKLCNKCGLAYENSLFCDKFHRHETGWRKC 60
MG ++CMN C ST EWKKGW L SG A LC +CG AYE+SLFC++FH+ ++GWR+C
Sbjct: 6 MGSKMCMNASCGTTSTVEWKKGWPLRSGLLADLCYRCGSAYESSLFCEQFHKDQSGWREC 65
Query: 61 SNCSKPIHSGCIVSKSSFEYLDFGGITCVTCVKPSQLCLNT--ENHNRFSQTTKNNASDQ 118
CSK +H GCI SK + E +D+GG+ C TC QL LNT EN FS+ +D+
Sbjct: 66 YLCSKRLHCGCIASKVTIELMDYGGVGCSTCACCHQLNLNTRGENPGVFSRLPMKTLADR 125
Query: 119 YGEHIDGRLLVEQAGKGNLMQLCRIGEVGESSRWPQAQRDAMVSCIGPKTEEVKCQFNKE 178
+H++G E G+ G+ P G K EE
Sbjct: 126 --QHVNG----ESGGRNE----------GDLFSQPLVMG-------GDKREEFM-----P 157
Query: 179 DTRFLNVMKHSSHLTAFTTLENNRPSWETKSIDETLSLKMALGTSSRNSVLPLATEIGEG 238
F +M S T L+ E+ + +L++ +A+ S + ATE EG
Sbjct: 158 HRGFGKLMSPESTTTGHR-LDAAGEMHESSPLQPSLNMGLAVNPFSPS----FATEAVEG 212
Query: 239 KLEGKASSHFQQGQTSQSILAQLSKTGIAMNLETNKGMISHPPRRPPADVKGKNQLLSRY 298
S ++ +IL + S+ I+ +K + R PP + +G+ LL RY
Sbjct: 213 MKHISPSQSNMVHCSASNILQKPSRPAISTPPVASKSAQARIGR-PPVEGRGRGHLLPRY 271
Query: 299 WPRITDQELEKLSGDLKSTVVPLFEKVLSPSDAGRIGRLVLPKACAEAFLPRILQSEGVP 358
WP+ TD+E++++SG+L +VPLFEK LS SDAGRIGRLVLPKACAEA+ P I QSEG+P
Sbjct: 272 WPKYTDKEVQQISGNLNLNIVPLFEKTLSASDAGRIGRLVLPKACAEAYFPPISQSEGIP 331
Query: 359 LQFQDIMGNEWTFQFRFWPNNNSRMYVLEGVTPCIQSLQLNAGDTVTFSRIDPGEKFLFG 418
L+ QD+ G EWTFQFR+WPNNNSRMYVLEGVTPCIQS+ L AGDTVTFSR+DPG K + G
Sbjct: 332 LKIQDVRGREWTFQFRYWPNNNSRMYVLEGVTPCIQSMMLQAGDTVTFSRVDPGGKLIMG 391
Query: 419 FRRSLTSIVTQDASTSSHSNGILIKDTNFSGAPQNLNSLSSFSNLLQSMKGNGEPYLNGH 478
R++ + D +NG +DT+ SG +N S++ S + K LNG
Sbjct: 392 SRKAANA---GDMQGCGLTNGTSTEDTSSSGVTENPPSINGSSCISLIPK-----ELNGM 443
Query: 479 SEHLRLGNGTADWLKTANSEEEMNNGPL-QRLVSVSEKKRSRNIGTKTKRLHIHSEDAME 537
E+L E N G + V EKKR+R IG K KRL +HSE++ME
Sbjct: 444 PENL---------------NSETNGGRIGDDPTRVKEKKRTRTIGAKNKRLLLHSEESME 488
Query: 538 LRLTWEEAQEFLCPPPSVEPNFVTIEDQVFEEYDEPPVFGK-IKTNASPSG 587
LRLTWEEAQ+ L P PSV+P V IE+Q EEYDEPPVFGK PSG
Sbjct: 489 LRLTWEEAQDLLRPSPSVKPTIVVIEEQEIEEYDEPPVFGKRTIVTTKPSG 539
>dbj|BAD90971.1| transcription factor B3-EAR motif family [Arabidopsis thaliana]
gi|42567319|ref|NP_194929.2| transcriptional factor B3
family protein [Arabidopsis thaliana]
Length = 780
Score = 417 bits (1071), Expect = e-115
Identities = 247/583 (42%), Positives = 323/583 (55%), Gaps = 87/583 (14%)
Query: 3 VEVCMNNYCKEGST-GEWKKGWSLISGGFAKLCNKCGLAYENSLFCDKFHRHETGWRKCS 61
++VCMN C ST GEWKKGW + SG A LC+KCG AYE S+FC+ FH E+GWR+C+
Sbjct: 4 IKVCMNALCGAASTSGEWKKGWPMRSGDLASLCDKCGCAYEQSIFCEVFHAKESGWRECN 63
Query: 62 NCSKPIHSGCIVSKSSFEYLDFGGITCVTCVKPSQLCLNTENHNRFSQTTKNNASDQYGE 121
+C K +H GCI S+ E L+ GG+TC++C K S L +H + + AS E
Sbjct: 64 SCDKRLHCGCIASRFMMELLENGGVTCISCAKKSGLISMNVSHESNGKDFPSFAS---AE 120
Query: 122 HIDGRLLVEQAGKGNLMQLCRIGEVGESSRWPQAQRDAMVSCIGPKTEEVKCQFNKEDTR 181
H+ ++E+ +L+ RI D S + K EE +
Sbjct: 121 HVGS--VLERTNLKHLLHFQRI--------------DPTHSSLQMKQEESLLPSS----- 159
Query: 182 FLNVMKHSSHLTAFTTLENNRPSWETKSIDETLSLKMALGTSSRNSVLPLATEIGEGKLE 241
L+ ++H + E K + +L ++LG + S P + +
Sbjct: 160 -LDALRHKT---------------ERKELSAQPNLSISLGPTLMTS--PFHDAAVDDR-- 199
Query: 242 GKASSHFQQGQTSQSILAQLSKTG-IAMNLETNKGMISH-PPRRPPADVKGKNQLLSRYW 299
K +S FQ S+ +L + + + IA +E + ++S RPP + +GK QLL RYW
Sbjct: 200 SKTNSIFQLAPRSRQLLPKPANSAPIAAGMEPSGSLVSQIHVARPPPEGRGKTQLLPRYW 259
Query: 300 PRITDQELEKLSGDL----KSTVVPLFEKVLSPSDAGRIGRLVLPKACAEAFLPRILQSE 355
PRITDQEL +LSG S ++PLFEKVLS SDAGRIGRLVLPKACAEA+ P I E
Sbjct: 260 PRITDQELLQLSGQYPHLSNSKIIPLFEKVLSASDAGRIGRLVLPKACAEAYFPPISLPE 319
Query: 356 GVPLQFQDIMGNEWTFQFRFWPNNNSRMYVLEGVTPCIQSLQLNAGDTVTFSRIDPGEKF 415
G+PL+ QDI G EW FQFRFWPNNNSRMYVLEGVTPCIQS+QL AGDTVTFSR +P K
Sbjct: 320 GLPLKIQDIKGKEWVFQFRFWPNNNSRMYVLEGVTPCIQSMQLQAGDTVTFSRTEPEGKL 379
Query: 416 LFGFRRSLTSIVTQDASTSSHSNGILIKDTNFSGAPQNLNSLSSFSNLLQSMKGNGEPYL 475
+ G+R++ S T Q KG+ EP L
Sbjct: 380 VMGYRKATNSTAT------------------------------------QMFKGSSEPNL 403
Query: 476 NGHSEHLRLGNGTADWLKTANSEEEMNNGPLQRLVSVSEKKRSRNIGTKTKRLHIHSEDA 535
N S L G G +W K SE+ + + S +KR RNIGTK+KRL I S D
Sbjct: 404 NMFSNSLNPGCGDINWSKLEKSEDMAKDNLFLQSSLTSARKRVRNIGTKSKRLLIDSVDV 463
Query: 536 MELRLTWEEAQEFLCPPPSVEPNFVTIEDQVFEEYDEPPVFGK 578
+EL++TWEEAQE L PP S +P+ T+E+Q FEEYDEPPVFGK
Sbjct: 464 LELKITWEEAQELLRPPQSTKPSIFTLENQDFEEYDEPPVFGK 506
>gb|AAB63089.1| putative VP1/ABI3 family regulatory protein [Arabidopsis thaliana]
gi|25408077|pir||G84708 probable VP1/ABI3 family
regulatory protein [imported] - Arabidopsis thaliana
Length = 780
Score = 365 bits (936), Expect = 3e-99
Identities = 242/591 (40%), Positives = 316/591 (52%), Gaps = 66/591 (11%)
Query: 1 MGVEVCMNNYCKEGSTGEWKKGWSLISGGFAKLCNKCGLAYENSLFCDKFHRHETGWRKC 60
MG ++CMN C ST EWKKGW L SG A LC + S+ + F R
Sbjct: 1 MGSKMCMNASCGTTSTVEWKKGWPLRSGLLADLCYHLRMRVLYSV--NNFIRTNLVG--- 55
Query: 61 SNCSKPIHSGCIVSKSSFEYLDFGGITCVTCVKPSQLCLNT--ENHNRFSQTTKNNASDQ 118
N +H GCI SK + E +D+GG+ C TC QL LNT EN FS+ +D+
Sbjct: 56 GNAICRLHCGCIASKVTIELMDYGGVGCSTCACCHQLNLNTRGENPGVFSRLPMKTLADR 115
Query: 119 YGEHIDGRLLVEQAGKGNLMQLCRIGEVGESSRWPQAQRDAMVSCIGPKTEEVKCQFNKE 178
+H++G E G+ G+ P G K EE
Sbjct: 116 --QHVNG----ESGGRNE----------GDLFSQPLVMG-------GDKREEFM-----P 147
Query: 179 DTRFLNVMKHSSHLTAFTTLENNRPSWETKSIDETLSLKMALGTSSRNSVLPLATEIGEG 238
F +M S T L+ E+ + +L++ +A+ S + ATE EG
Sbjct: 148 HRGFGKLMSPESTTTGHR-LDAAGEMHESSPLQPSLNMGLAVNPFSPS----FATEAVEG 202
Query: 239 KLEGKASSHFQQGQTSQSILAQLSKTGIAMNLETNKGMISHPPRRPPADVKGKNQLLSRY 298
S ++ +IL + S+ I+ +K + R PP + +G+ LL RY
Sbjct: 203 MKHISPSQSNMVHCSASNILQKPSRPAISTPPVASKSAQARIGR-PPVEGRGRGHLLPRY 261
Query: 299 WPRITDQELEKLSGDLKSTVVPLFEKVLSPSDAGRIGRLVLPKACAEAFLPRILQSEGVP 358
WP+ TD+E++++SG+L +VPLFEK LS SDAGRIGRLVLPKACAEA+ P I QSEG+P
Sbjct: 262 WPKYTDKEVQQISGNLNLNIVPLFEKTLSASDAGRIGRLVLPKACAEAYFPPISQSEGIP 321
Query: 359 LQFQDIMGNEWTFQFRFWPNNNSRMYVLEGVTPCIQSLQLNAGDTVTFSRIDPGEKFLFG 418
L+ QD+ G EWTFQFR+WPNNNSRMYVLEGVTPCIQS+ L AGDTVTFSR+DPG K + G
Sbjct: 322 LKIQDVRGREWTFQFRYWPNNNSRMYVLEGVTPCIQSMMLQAGDTVTFSRVDPGGKLIMG 381
Query: 419 FRRSLTSIVTQDASTSSHSNGILIKDTNFSGAPQNLNSLSSFSNLLQSMKGNGEPYLNGH 478
R++ + D +NG +DT+ SG +N S++ S + K LNG
Sbjct: 382 SRKAANA---GDMQGCGLTNGTSTEDTSSSGVTENPPSINGSSCISLIPK-----ELNGM 433
Query: 479 SEHLRLGNGTADWLKTANSEEEMNNGPL-QRLVSVSEKKRSRNIGTKTKRLHIHSEDAME 537
E+L E N G + V EKKR+R IG K KRL +HSE++ME
Sbjct: 434 PENL---------------NSETNGGRIGDDPTRVKEKKRTRTIGAKNKRLLLHSEESME 478
Query: 538 LRLTWEEAQEFLCPPPSVEPNFVTIEDQVFEEYDEPPVFGK-IKTNASPSG 587
LRLTWEEAQ+ L P PSV+P V IE+Q EEYDEPPVFGK PSG
Sbjct: 479 LRLTWEEAQDLLRPSPSVKPTIVVIEEQEIEEYDEPPVFGKRTIVTTKPSG 529
>emb|CAB79919.1| predicted protein [Arabidopsis thaliana] gi|2827635|emb|CAA16588.1|
predicted protein [Arabidopsis thaliana]
gi|7485366|pir||T04644 hypothetical protein F10N7.180 -
Arabidopsis thaliana
Length = 675
Score = 315 bits (808), Expect = 2e-84
Identities = 210/545 (38%), Positives = 277/545 (50%), Gaps = 108/545 (19%)
Query: 46 FCDKFHRHETGWRKCSNCSKPIHSGCIVSKSSFEYLDFGGITCVTCVKPSQLCLNTENHN 105
F DKF R K +H GCI S+ E L+ GG+TC++C K S L +H
Sbjct: 9 FLDKFLR------------KRLHCGCIASRFMMELLENGGVTCISCAKKSGLISMNVSHE 56
Query: 106 RFSQTTKNNASDQYGEHIDGRLLVEQAGKGNLMQLCRIGEVGESSRWPQAQRDAMVSCIG 165
+ + AS EH+ ++E+ +L+ RI D S +
Sbjct: 57 SNGKDFPSFAS---AEHVGS--VLERTNLKHLLHFQRI--------------DPTHSSLQ 97
Query: 166 PKTEEVKCQFNKEDTRFLNVMKHSSHLTAFTTLENNRPSWETKSIDETLSLKMALGTSSR 225
K EE + L+ ++H + E K + +L ++LG +
Sbjct: 98 MKQEESLLPSS------LDALRHKT---------------ERKELSAQPNLSISLGPTLM 136
Query: 226 NSVLPLATEIGEGKLEGKASSHFQQGQTSQSILAQLSKTG-IAMNLETNKGMISH-PPRR 283
S P + + K +S FQ S+ +L + + + IA +E + ++S R
Sbjct: 137 TS--PFHDAAVDDR--SKTNSIFQLAPRSRQLLPKPANSAPIAAGMEPSGSLVSQIHVAR 192
Query: 284 PPADVKGKNQLLSRYWPRITDQELEKLSGDL--------------KSTVVPLFEKVLSPS 329
PP + +GK QLL RYWPRITDQEL +LSG S ++PLFEKVLS S
Sbjct: 193 PPPEGRGKTQLLPRYWPRITDQELLQLSGQYPHLYESLTVYFPSSNSKIIPLFEKVLSAS 252
Query: 330 DAGRIGRLVLPKACAEAFLPRILQSEGVPLQFQDIMGNEWTFQFRFWPNNNSRMYVLEGV 389
DAGRIGRLVLPKACAEA+ P I EG+PL+ QDI G EW FQFRFWPNNNSRMYVLEGV
Sbjct: 253 DAGRIGRLVLPKACAEAYFPPISLPEGLPLKIQDIKGKEWVFQFRFWPNNNSRMYVLEGV 312
Query: 390 TPCIQSLQLNAGDTVTFSRIDPGEKFLFGFRRSLTSIVTQDASTSSHSNGILIKDTNFSG 449
TPCIQS+QL AGDTVTFSR +P K + G+R++ S T
Sbjct: 313 TPCIQSMQLQAGDTVTFSRTEPEGKLVMGYRKATNSTAT--------------------- 351
Query: 450 APQNLNSLSSFSNLLQSMKGNGEPYLNGHSEHLRLGNGTADWLKTANSEEEMNNGPLQRL 509
Q KG+ EP LN S L G G +W K SE+ + +
Sbjct: 352 ---------------QMFKGSSEPNLNMFSNSLNPGCGDINWSKLEKSEDMAKDNLFLQS 396
Query: 510 VSVSEKKRSRNIGTKTKRLHIHSEDAMELRLTWEEAQEFLCPPPSVEPNFVTIEDQVFEE 569
S +KR RNIGTK+KRL I S D +EL++TWEEAQE L PP S +P+ T+E+Q FEE
Sbjct: 397 SLTSARKRVRNIGTKSKRLLIDSVDVLELKITWEEAQELLRPPQSTKPSIFTLENQDFEE 456
Query: 570 YDEPP 574
YD+ P
Sbjct: 457 YDKLP 461
>ref|NP_910705.1| VP1/ABI3 family regulatory protein-like protein [Oryza sativa
(japonica cultivar-group)] gi|23617202|dbj|BAC20873.1|
VP1/ABI3 family regulatory protein-like protein [Oryza
sativa (japonica cultivar-group)]
Length = 947
Score = 264 bits (674), Expect = 8e-69
Identities = 147/298 (49%), Positives = 185/298 (61%), Gaps = 22/298 (7%)
Query: 283 RPPADVKGKNQLLSRYWPRITDQELEKLSGDLKSTVVPLFEKVLSPSDAGRIGRLVLPKA 342
RP D K ++QLL RYWPRITDQEL+ LSGD S + PLFEK+LS SDAGRIGRLVLPK
Sbjct: 414 RPRMDAKARSQLLPRYWPRITDQELQHLSGDSNSVITPLFEKMLSASDAGRIGRLVLPKK 473
Query: 343 CAEAFLPRILQSEGVPLQFQDIMGNEWTFQFRFWPNNNSRMYVLEGVTPCIQSLQLNAGD 402
CAEA EG+PL+ QD G EW FQFRFWPNNNSRMYVLEGVTPCIQS+QL AGD
Sbjct: 474 CAEA--------EGLPLKVQDATGKEWVFQFRFWPNNNSRMYVLEGVTPCIQSMQLQAGD 525
Query: 403 TVTFSRIDPGEKFLFGFRRSLTSIVTQDASTSSHSNGIL--IKDTNFSGAPQNLNSLSSF 460
TVTFSRIDP K + GFR++ QD T +NG+L + N P + S
Sbjct: 526 TVTFSRIDPEGKLVMGFRKATNLSAEQDQPTKP-ANGVLPPPEANNKVVVPDS----SPN 580
Query: 461 SNLLQSMKGNGEPYLNGHSEHLRLGNGTADWLKTANSEEEMNNGPLQRLVSVSEKKRSRN 520
+ + + +K N E + E L ++ PL K+++ +
Sbjct: 581 AAVPRPIKVNTESKSSSPVEQATACKIDKGALPQKEGPGTSSSSPL------PVKRKATS 634
Query: 521 IGTKTKRLHIHSEDAMELRLTWEEAQEFLCPPPSVEPNFVTIEDQVFEEYDEPPVFGK 578
+G K KR H+ SE++MEL++TWEEAQE L PPP P+ V ++ FEEY+EPP+ G+
Sbjct: 635 VGPKIKRFHMDSEESMELKITWEEAQELLRPPPKA-PSIVVVDGHEFEEYEEPPILGR 691
Score = 100 bits (248), Expect = 2e-19
Identities = 44/115 (38%), Positives = 66/115 (57%), Gaps = 1/115 (0%)
Query: 5 VCMNNYCKEG-STGEWKKGWSLISGGFAKLCNKCGLAYENSLFCDKFHRHETGWRKCSNC 63
+C N +CK+ S G ++GW L +G FA+LC++C ++E+ FC+ FH GWR C +C
Sbjct: 82 ICFNAHCKDPKSDGPRRRGWRLRNGDFAELCDRCYHSFEHGGFCETFHLEVAGWRNCESC 141
Query: 64 SKPIHSGCIVSKSSFEYLDFGGITCVTCVKPSQLCLNTENHNRFSQTTKNNASDQ 118
K +H GCIVS +F +LD GG+ CV C + S + S N +D+
Sbjct: 142 GKRLHCGCIVSVHAFVHLDAGGVECVMCARKSHAAMAPSQIWSSSMHMAQNVADR 196
>ref|NP_193886.1| transcriptional factor B3 family protein [Arabidopsis thaliana]
Length = 721
Score = 188 bits (478), Expect = 4e-46
Identities = 135/418 (32%), Positives = 200/418 (47%), Gaps = 51/418 (12%)
Query: 40 AYENSLFCDKFHRHETGWRKCSNCSKP----IHSGCIVSKSSFEYLDFGGITCVTCVK-- 93
AYE FCD FH+ +GWR C +C K IH GCI S S++ +D GGI C+ C +
Sbjct: 31 AYEQGKFCDVFHQRASGWRCCESCGKHFVQRIHCGCIASASAYTLMDAGGIECLACARKK 90
Query: 94 ----PSQLCLNTENHNRFSQTTKNNASDQYGEHIDGRLLVEQAGKGNLMQLCRIGEVGES 149
P + N S T N S Q +D +L Q G +
Sbjct: 91 FALSPISEKFKDLSINWSSSTRSNQISYQPPSCLDPSVL----------QFDFRNRGGNN 140
Query: 150 SRWPQAQRDAMVSCIGPKTEEVKCQFNKEDTRFLNVMKHSSHLTAFTTLENNRPSWETKS 209
A ++ + +C T E K N + ++ ++ F + P
Sbjct: 141 EFSQPASKERVTAC----TMEKKRGMNDMIGKLMSENSKHYRVSPFPNVNVYHP------ 190
Query: 210 IDETLSLKMALGTSSRNSVLPLATEI---GEGKLEGKASSHFQQGQTSQSILAQLSKTGI 266
+SLK + +P+ T I G +L+G H + + L+
Sbjct: 191 ---LISLKEGPCGTQLAFPVPITTPIEKTGHSRLDGSNLWHTRNSSPLSRLHNDLNGGAD 247
Query: 267 AMNLETNKGMISHPPRRPPADVKGKNQLLSRYWPRIT--DQELEKLSGDLKSTVVPLFE- 323
+ ++ +++H + GK Q++ R+WP+++ +Q L+ S + S+++
Sbjct: 248 SPFESKSRNVMAH------LETPGKYQVVPRFWPKVSYKNQVLQNQSKEYPSSLIDTTLE 301
Query: 324 ---KVLSPSDAGRIGRLVLPKACAEAFLPRILQSEGVPLQFQDIMGNEWTFQFRFWPNNN 380
K+LS +D G+ RLVLPK AEAFLP++ ++GVPL QD MG EW FQFRFWP++
Sbjct: 302 YNFKILSATDTGK--RLVLPKKYAEAFLPQLSHTKGVPLTVQDPMGKEWRFQFRFWPSSK 359
Query: 381 SRMYVLEGVTPCIQSLQLNAGDTVTFSRIDPGEKFLFGFRR-SLTSIVTQDASTSSHS 437
R+YVLEGVTP IQ+LQL AGDTV FSR+DP K + GFR+ S+T Q HS
Sbjct: 360 GRIYVLEGVTPFIQTLQLQAGDTVIFSRLDPERKLILGFRKASITQSSDQADPADMHS 417
Score = 63.5 bits (153), Expect = 2e-08
Identities = 31/69 (44%), Positives = 45/69 (64%)
Query: 513 SEKKRSRNIGTKTKRLHIHSEDAMELRLTWEEAQEFLCPPPSVEPNFVTIEDQVFEEYDE 572
S KK+S + T++KR + D L+LTWEEAQ FL PPP++ P+ V IED FEEY++
Sbjct: 438 SGKKKSSMMITRSKRQKVEKGDDNLLKLTWEEAQGFLLPPPNLTPSRVVIEDYEFEEYED 497
Query: 573 PPVFGKIKT 581
V ++ +
Sbjct: 498 LAVVSQVSS 506
>emb|CAB81262.1| VP1 like protein [Arabidopsis thaliana] gi|3080399|emb|CAA18719.1|
VP1 like protein [Arabidopsis thaliana]
gi|25407580|pir||H85245 VP1 like protein [imported] -
Arabidopsis thaliana gi|7485714|pir||T05163 hypothetical
protein F18E5.170 - Arabidopsis thaliana (fragment)
Length = 739
Score = 160 bits (404), Expect = 2e-37
Identities = 121/387 (31%), Positives = 183/387 (47%), Gaps = 47/387 (12%)
Query: 67 IHSGCIVSKSSFEYLDFGGITCVTCVK------PSQLCLNTENHNRFSQTTKNNASDQYG 120
IH GCI S S++ +D GGI C+ C + P + N S T N S Q
Sbjct: 80 IHCGCIASASAYTLMDAGGIECLACARKKFALSPISEKFKDLSINWSSSTRSNQISYQPP 139
Query: 121 EHIDGRLLVEQAGKGNLMQLCRIGEVGESSRWPQAQRDAMVSCIGPKTEEVKCQFNKEDT 180
+D +L Q G + A ++ + +C T E K N
Sbjct: 140 SCLDPSVL----------QFDFRNRGGNNEFSQPASKERVTAC----TMEKKRGMNDMIG 185
Query: 181 RFLNVMKHSSHLTAFTTLENNRPSWETKSIDETLSLKMALGTSSRNSVLPLATEI---GE 237
+ ++ ++ F + P +SLK + +P+ T I G
Sbjct: 186 KLMSENSKHYRVSPFPNVNVYHP---------LISLKEGPCGTQLAFPVPITTPIEKTGH 236
Query: 238 GKLEGKASSHFQQGQTSQSILAQLSKTGIAMNLETNKGMISHPPRRPPADVKGKNQLLSR 297
+L+G H + + L+ + ++ +++H + GK Q++ R
Sbjct: 237 SRLDGSNLWHTRNSSPLSRLHNDLNGGADSPFESKSRNVMAH------LETPGKYQVVPR 290
Query: 298 YWPRIT--DQELEKLSGDLKSTVVPLFE----KVLSPSDAGRIGRLVLPKACAEAFLPRI 351
+WP+++ +Q L+ S + S+++ K+LS +D G+ RLVLPK AEAFLP++
Sbjct: 291 FWPKVSYKNQVLQNQSKEYPSSLIDTTLEYNFKILSATDTGK--RLVLPKKYAEAFLPQL 348
Query: 352 LQSEGVPLQFQDIMGNEWTFQFRFWPNNNSRMYVLEGVTPCIQSLQLNAGDTVTFSRIDP 411
++GVPL QD MG EW FQFRFWP++ R+YVLEGVTP IQ+LQL AGDTV FSR+DP
Sbjct: 349 SHTKGVPLTVQDPMGKEWRFQFRFWPSSKGRIYVLEGVTPFIQTLQLQAGDTVIFSRLDP 408
Query: 412 GEKFLFGFRR-SLTSIVTQDASTSSHS 437
K + GFR+ S+T Q HS
Sbjct: 409 ERKLILGFRKASITQSSDQADPADMHS 435
Score = 63.5 bits (153), Expect = 2e-08
Identities = 31/69 (44%), Positives = 45/69 (64%)
Query: 513 SEKKRSRNIGTKTKRLHIHSEDAMELRLTWEEAQEFLCPPPSVEPNFVTIEDQVFEEYDE 572
S KK+S + T++KR + D L+LTWEEAQ FL PPP++ P+ V IED FEEY++
Sbjct: 456 SGKKKSSMMITRSKRQKVEKGDDNLLKLTWEEAQGFLLPPPNLTPSRVVIEDYEFEEYED 515
Query: 573 PPVFGKIKT 581
V ++ +
Sbjct: 516 LAVVSQVSS 524
>pir||T12203 transcription factor Vp1 - common ice plant
gi|3219155|dbj|BAA28779.1| transcription factor Vp1
[Mesembryanthemum crystallinum]
Length = 790
Score = 85.1 bits (209), Expect = 6e-15
Identities = 56/151 (37%), Positives = 80/151 (52%), Gaps = 8/151 (5%)
Query: 277 ISHPPRRPPADVKGKNQLLSRYWPRITDQE----LEKLSG-DLKSTVVPLFEKVLSPSDA 331
+SHP PP V G+ Q L R P + +EK G + + L +KVL SD
Sbjct: 596 VSHPVGGPPPMV-GQMQGLERAAPSGNGFQRQGGVEKKQGWKSEKNLRFLLQKVLKQSDV 654
Query: 332 GRIGRLVLPKACAEAFLPRILQSEGVPLQFQDI-MGNEWTFQFRFWPNNNSRMYVLEGVT 390
G +GR+VLPK AE LP + +G+P+ +DI W ++RFWPNN SRMY+LE
Sbjct: 655 GNLGRIVLPKKEAETHLPELEARDGIPIAMEDIGTSRVWNMRYRFWPNNKSRMYLLENTG 714
Query: 391 PCIQSLQLNAGD-TVTFSRIDPGEKFLFGFR 420
++S L GD V +S + G+ + G +
Sbjct: 715 DFVRSNGLQEGDFIVIYSDVKCGKYMIRGVK 745
>gb|AAC35247.1| FUSCA3 [Arabidopsis thaliana]
Length = 312
Score = 84.3 bits (207), Expect = 1e-14
Identities = 39/85 (45%), Positives = 56/85 (65%), Gaps = 1/85 (1%)
Query: 321 LFEKVLSPSDAGRIGRLVLPKACAEAFLPRILQSEGVPLQFQDIMG-NEWTFQFRFWPNN 379
LF+K L SD + R++LPK AEA LP + EG+P++ +D+ G + WTF++R+WPNN
Sbjct: 90 LFQKELKNSDVSSLRRMILPKKAAEAHLPALECKEGIPIRMEDLDGFHVWTFKYRYWPNN 149
Query: 380 NSRMYVLEGVTPCIQSLQLNAGDTV 404
NSRMYVLE + + L GD +
Sbjct: 150 NSRMYVLENTGDFVNAHGLQLGDFI 174
>gb|AAC35246.1| FUSCA3 [Arabidopsis thaliana]
Length = 310
Score = 84.3 bits (207), Expect = 1e-14
Identities = 39/85 (45%), Positives = 56/85 (65%), Gaps = 1/85 (1%)
Query: 321 LFEKVLSPSDAGRIGRLVLPKACAEAFLPRILQSEGVPLQFQDIMG-NEWTFQFRFWPNN 379
LF+K L SD + R++LPK AEA LP + EG+P++ +D+ G + WTF++R+WPNN
Sbjct: 90 LFQKELKNSDVSSLRRMILPKKAAEAHLPALECKEGIPIRMEDLDGFHVWTFKYRYWPNN 149
Query: 380 NSRMYVLEGVTPCIQSLQLNAGDTV 404
NSRMYVLE + + L GD +
Sbjct: 150 NSRMYVLENTGDFVNAHGLQLGDFI 174
>gb|AAS79559.1| transcriptional regulator [Arabidopsis thaliana]
gi|46367488|emb|CAG25870.1| hypothetical protein
[Arabidopsis thaliana]
Length = 310
Score = 84.3 bits (207), Expect = 1e-14
Identities = 39/85 (45%), Positives = 56/85 (65%), Gaps = 1/85 (1%)
Query: 321 LFEKVLSPSDAGRIGRLVLPKACAEAFLPRILQSEGVPLQFQDIMG-NEWTFQFRFWPNN 379
LF+K L SD + R++LPK AEA LP + EG+P++ +D+ G + WTF++R+WPNN
Sbjct: 91 LFQKELKNSDVSSLRRMILPKKAAEAHLPALECKEGIPIRMEDLDGFHVWTFKYRYWPNN 150
Query: 380 NSRMYVLEGVTPCIQSLQLNAGDTV 404
NSRMYVLE + + L GD +
Sbjct: 151 NSRMYVLENTGDFVNAHGLQLGDFI 175
>ref|NP_566799.1| transcriptional regulator (FUSCA3) [Arabidopsis thaliana]
Length = 313
Score = 84.3 bits (207), Expect = 1e-14
Identities = 39/85 (45%), Positives = 56/85 (65%), Gaps = 1/85 (1%)
Query: 321 LFEKVLSPSDAGRIGRLVLPKACAEAFLPRILQSEGVPLQFQDIMG-NEWTFQFRFWPNN 379
LF+K L SD + R++LPK AEA LP + EG+P++ +D+ G + WTF++R+WPNN
Sbjct: 91 LFQKELKNSDVSSLRRMILPKKAAEAHLPALECKEGIPIRMEDLDGFHVWTFKYRYWPNN 150
Query: 380 NSRMYVLEGVTPCIQSLQLNAGDTV 404
NSRMYVLE + + L GD +
Sbjct: 151 NSRMYVLENTGDFVNAHGLQLGDFI 175
>sp|P37398|VIV_ORYSA Viviparous protein homolog gi|391885|dbj|BAA04066.1| VP1 protein
(OSVP1) [Oryza sativa (japonica cultivar-group)]
Length = 728
Score = 84.0 bits (206), Expect = 1e-14
Identities = 47/123 (38%), Positives = 68/123 (55%), Gaps = 4/123 (3%)
Query: 321 LFEKVLSPSDAGRIGRLVLPKACAEAFLPRILQSEGVPLQFQDIMGNE-WTFQFRFWPNN 379
L +KVL SD G +GR+VLPK AE LP + +GV + +DI ++ W ++RFWPNN
Sbjct: 536 LLQKVLKQSDVGSLGRIVLPKKEAEVHLPELKTRDGVSIPMEDIGTSQVWNMRYRFWPNN 595
Query: 380 NSRMYVLEGVTPCIQSLQLNAGD-TVTFSRIDPGEKFLFG--FRRSLTSIVTQDASTSSH 436
SRMY+LE ++S +L GD V +S I G+ + G RR+ + H
Sbjct: 596 KSRMYLLENTGDFVRSNELQEGDFIVIYSDIKSGKYLIRGVKVRRAAQEQGNSSGAVGKH 655
Query: 437 SNG 439
+G
Sbjct: 656 KHG 658
>ref|NP_914780.1| transcription activator VP1 - rice [Oryza sativa (japonica
cultivar-group)] gi|629807|pir||S43768 transcription
activator VP1 - rice
Length = 728
Score = 84.0 bits (206), Expect = 1e-14
Identities = 47/123 (38%), Positives = 68/123 (55%), Gaps = 4/123 (3%)
Query: 321 LFEKVLSPSDAGRIGRLVLPKACAEAFLPRILQSEGVPLQFQDIMGNE-WTFQFRFWPNN 379
L +KVL SD G +GR+VLPK AE LP + +GV + +DI ++ W ++RFWPNN
Sbjct: 536 LLQKVLKQSDVGSLGRIVLPKKEAEVHLPELKTRDGVSIPMEDIGTSQVWNMRYRFWPNN 595
Query: 380 NSRMYVLEGVTPCIQSLQLNAGD-TVTFSRIDPGEKFLFG--FRRSLTSIVTQDASTSSH 436
SRMY+LE ++S +L GD V +S I G+ + G RR+ + H
Sbjct: 596 KSRMYLLENTGDFVRSNELQEGDFIVIYSDIKSGKYLIRGVKVRRAAQEQGNSSGAVGKH 655
Query: 437 SNG 439
+G
Sbjct: 656 KHG 658
>gb|AAG22585.1| transcription factor viviparous 1 [Picea abies]
Length = 828
Score = 84.0 bits (206), Expect = 1e-14
Identities = 49/132 (37%), Positives = 71/132 (53%), Gaps = 8/132 (6%)
Query: 321 LFEKVLSPSDAGRIGRLVLPKACAEAFLPRILQSEGVPLQFQDIMGNE-WTFQFRFWPNN 379
L +KVL SD G +GR+VLPK AE LP + +G+ + +DI+ + W ++RFWPNN
Sbjct: 581 LLQKVLKQSDVGNLGRIVLPKKEAEIHLPELEARDGISIAMEDIVTSRVWNLRYRFWPNN 640
Query: 380 NSRMYVLEGVTPCIQSLQLNAGD-TVTFSRIDPGEKFLFGFR------RSLTSIVTQDAS 432
SRMY+LE ++S L GD V +S G+ + G + S ++ T +
Sbjct: 641 KSRMYLLENTGDFVRSNGLQEGDFIVIYSDTKTGKYMIRGVKVPRSDTTSASAAATPPTT 700
Query: 433 TSSHSNGILIKD 444
T S S LI D
Sbjct: 701 TKSASGSCLIPD 712
>emb|CAB91109.1| VIVIPAROUS1 protein [Triticum aestivum]
Length = 692
Score = 84.0 bits (206), Expect = 1e-14
Identities = 45/121 (37%), Positives = 68/121 (56%), Gaps = 8/121 (6%)
Query: 321 LFEKVLSPSDAGRIGRLVLPKACAEAFLPRILQSEGVPLQFQDIMGNE-WTFQFRFWPNN 379
L +KVL SD G +GR+VLPK AE LP + +G+ + +DI ++ W+ ++RFWPNN
Sbjct: 534 LLQKVLKQSDVGTLGRIVLPKKEAETHLPELKTGDGISIPIEDIGTSQVWSMRYRFWPNN 593
Query: 380 NSRMYVLEGVTPCIQSLQLNAGD-TVTFSRIDPGEKFLFGFRRSLTSIVTQDASTSSHSN 438
SRMY+LE ++S +L GD V +S + G+ + G + V + H N
Sbjct: 594 KSRMYLLENTGDFVRSNELQEGDFIVLYSDVKSGKYLIRGVK------VRAQQDLAKHKN 647
Query: 439 G 439
G
Sbjct: 648 G 648
>emb|CAB91108.1| VIVIPAROUS1 protein [Triticum aestivum]
Length = 688
Score = 82.8 bits (203), Expect = 3e-14
Identities = 43/121 (35%), Positives = 68/121 (55%), Gaps = 2/121 (1%)
Query: 321 LFEKVLSPSDAGRIGRLVLPKACAEAFLPRILQSEGVPLQFQDIMGNE-WTFQFRFWPNN 379
L +KVL SD G +GR+VLPK AE LP + +G+ + +DI ++ W+ ++RFWPNN
Sbjct: 530 LLQKVLKQSDVGTLGRIVLPKKEAETHLPELKTGDGISIPIEDIGTSQVWSMRYRFWPNN 589
Query: 380 NSRMYVLEGVTPCIQSLQLNAGD-TVTFSRIDPGEKFLFGFRRSLTSIVTQDASTSSHSN 438
SRMY+LE ++S +L GD V +S + G+ + G + + + + S
Sbjct: 590 KSRMYLLENTGDFVRSNELQEGDFIVLYSDVKSGKYLIRGVKVRAQQDLAKHKNASPEKG 649
Query: 439 G 439
G
Sbjct: 650 G 650
>emb|CAB91107.1| VIVIPAROUS1 protein [Triticum aestivum]
Length = 692
Score = 82.8 bits (203), Expect = 3e-14
Identities = 43/121 (35%), Positives = 68/121 (55%), Gaps = 2/121 (1%)
Query: 321 LFEKVLSPSDAGRIGRLVLPKACAEAFLPRILQSEGVPLQFQDIMGNE-WTFQFRFWPNN 379
L +KVL SD G +GR+VLPK AE LP + +G+ + +DI ++ W+ ++RFWPNN
Sbjct: 534 LLQKVLKQSDVGTLGRIVLPKKEAETHLPELKTGDGISIPIEDIGTSQVWSMRYRFWPNN 593
Query: 380 NSRMYVLEGVTPCIQSLQLNAGD-TVTFSRIDPGEKFLFGFRRSLTSIVTQDASTSSHSN 438
SRMY+LE ++S +L GD V +S + G+ + G + + + + S
Sbjct: 594 KSRMYLLENTGDFVRSNELQEGDFIVLYSDVKSGKYLIRGVKVRAQQDLAKHKNASPEKG 653
Query: 439 G 439
G
Sbjct: 654 G 654
>ref|XP_474341.1| OSJNBa0064G10.6 [Oryza sativa (japonica cultivar-group)]
gi|32487511|emb|CAE05755.1| OSJNBa0064G10.6 [Oryza
sativa (japonica cultivar-group)]
Length = 433
Score = 82.0 bits (201), Expect = 5e-14
Identities = 42/97 (43%), Positives = 55/97 (56%), Gaps = 1/97 (1%)
Query: 321 LFEKVLSPSDAGRIGRLVLPKACAEAFLPRILQSEGVPLQFQDI-MGNEWTFQFRFWPNN 379
+ K L+ SD G IGR+V+PK AEA LP + Q EGV L+ D + W F++RFWPNN
Sbjct: 296 ILRKELTNSDVGNIGRIVMPKRDAEAHLPALHQREGVMLKMDDFKLETTWNFKYRFWPNN 355
Query: 380 NSRMYVLEGVTPCIQSLQLNAGDTVTFSRIDPGEKFL 416
SRMYVLE ++ L GD + EK +
Sbjct: 356 KSRMYVLESTGGFVKQHVLQTGDIFIIYKSSESEKLV 392
Database: nr
Posted date: Jul 5, 2005 12:34 AM
Number of letters in database: 863,360,394
Number of sequences in database: 2,540,612
Lambda K H
0.317 0.133 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 1,111,811,931
Number of Sequences: 2540612
Number of extensions: 47574218
Number of successful extensions: 115032
Number of sequences better than 10.0: 220
Number of HSP's better than 10.0 without gapping: 96
Number of HSP's successfully gapped in prelim test: 125
Number of HSP's that attempted gapping in prelim test: 114648
Number of HSP's gapped (non-prelim): 371
length of query: 648
length of database: 863,360,394
effective HSP length: 135
effective length of query: 513
effective length of database: 520,377,774
effective search space: 266953798062
effective search space used: 266953798062
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 78 (34.7 bits)
Medicago: description of AC141923.5


