

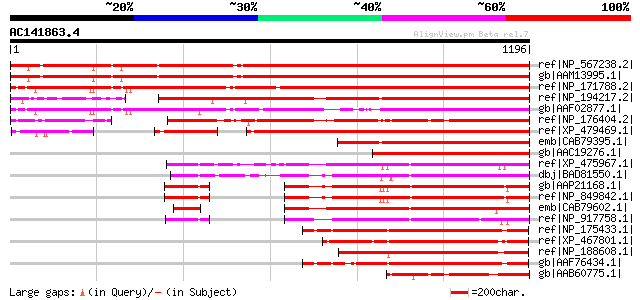
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC141863.4 + phase: 0 /partial
(1196 letters)
Database: nr
2,540,612 sequences; 863,360,394 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
ref|NP_567238.2| AAA-type ATPase family protein [Arabidopsis tha... 1164 0.0
gb|AAM13995.1| unknown protein [Arabidopsis thaliana] 1161 0.0
ref|NP_171788.2| AAA-type ATPase family protein [Arabidopsis tha... 1095 0.0
ref|NP_194217.2| AAA-type ATPase family protein [Arabidopsis tha... 949 0.0
gb|AAF02877.1| Unknown protein [Arabidopsis thaliana] gi|2551840... 868 0.0
ref|NP_176404.2| AAA-type ATPase family protein [Arabidopsis tha... 831 0.0
ref|XP_479469.1| putative MSP1(mitochondrial sorting of proteins... 831 0.0
emb|CAB79395.1| putative protein [Arabidopsis thaliana] gi|46782... 623 e-176
gb|AAC19276.1| T14P8.7 [Arabidopsis thaliana] gi|7269007|emb|CAB... 602 e-170
ref|XP_475967.1| unknown protein [Oryza sativa (japonica cultiva... 536 e-150
dbj|BAD81550.1| unknown protein [Oryza sativa (japonica cultivar... 518 e-145
gb|AAP21168.1| At1g64110/F22C12_22 [Arabidopsis thaliana] gi|184... 479 e-133
ref|NP_849842.1| AAA-type ATPase family protein [Arabidopsis tha... 479 e-133
emb|CAB79602.1| putative protein [Arabidopsis thaliana] gi|44553... 475 e-132
ref|NP_917758.1| P0501G01.20 [Oryza sativa (japonica cultivar-gr... 436 e-120
ref|NP_175433.1| AAA-type ATPase family protein [Arabidopsis tha... 432 e-119
ref|XP_467801.1| transitional endoplasmic reticulum ATPase-like ... 430 e-118
ref|NP_188608.1| AAA-type ATPase family protein [Arabidopsis tha... 422 e-116
gb|AAF76434.1| Contains similarity to p60 katanin from Chlamydom... 421 e-116
gb|AAB60775.1| Similar to Xenopus TER ATPase (gb|X54240). [Arabi... 392 e-107
>ref|NP_567238.2| AAA-type ATPase family protein [Arabidopsis thaliana]
Length = 1265
Score = 1164 bits (3010), Expect = 0.0
Identities = 661/1236 (53%), Positives = 821/1236 (65%), Gaps = 62/1236 (5%)
Query: 1 PDNGAASSEKPPENSNP--EPSADPGK----CAQPDAQ-IDEPVAAAD------DDKADT 47
P +++SE P EN P +P ++ G+ + P A ++PV D + D
Sbjct: 51 PAGSSSASEVPIENQGPASDPGSESGEPELGSSDPQAMDAEKPVVTTDVPVMENSPETDA 110
Query: 48 TPPIADASTPTLVADKPRASFSSWSLYQKQNPNLESSAPWCRLLSQSAQHPNVSICIPNF 107
P + +TPT+ + + S K APW +LLSQ +Q+P+ I P F
Sbjct: 111 NPEVEVLATPTVAGEAVADADKS-----KAAKKRALKAPWAKLLSQYSQNPHRVIRGPVF 165
Query: 108 TIGSSRNCNFHLKDHTISGNLCKIKHTQREGSDVAVLESTGSKGSVIVNGTLVKKSTCCT 167
T+G R C+ ++D + LC++K ++ G VA LE G+ V VNG +KSTC
Sbjct: 166 TVGR-RGCDLSIRDQAMPSTLCELKQSEHGGPSVASLEILGNGVIVHVNGKCYQKSTCVH 224
Query: 168 LNSGDEVVFGLHGNHSYIFQQVNTE--------VAVKGAEVQSGIGKFMQLERRSGD--- 216
L GDEV+F L+G H+YIFQ V E ++ E + K + +E R+GD
Sbjct: 225 LRGGDEVIFSLNGKHAYIFQPVKDENLAAPDRASSLSICEARGAPLKGVHVETRAGDVDG 284
Query: 217 PSAVAGASILASLSNLRQDLTRWKSPSQTASKPHQGADV-----SIHTVLPDGTEIELDG 271
S V GASILASLS LR P A K Q V S + + D + D
Sbjct: 285 ASDVDGASILASLSKLRS--FHLLPPIAKAGKRQQNPAVPVVPSSFNDCISDTDMNDADS 342
Query: 272 LGNSTPSMGTDKAADAEA---SNKNTPMDCDPEDA--GAEPGNVKYSGVNDLLRPFFRIL 326
+ +K A A +N+N +D D A+ GNV +G +RP +L
Sbjct: 343 NNDHAAVASVEKIAAASTPGTANENLNVDGSGLDPFQEADGGNVPAAGYE--IRPIVHLL 400
Query: 327 AGSTTCKLKLSKSICKQVLEERNGAED--TQAASTSGTSVRCAVFKEDAHAAILDGKEQE 384
S++ ++ S S ++L+ER ++ + +S S R FK+ +L+ + +
Sbjct: 401 GESSSFDIRGSIS---RLLDERREVKEFLREFDLSSTISTRRQAFKDSLRGGVLNAQNID 457
Query: 385 VSFDNFPYYLSENTKNVLIAACFIHLKH-KEHAKYTADLPTVNPRILLSGPAGSEIYSEM 443
+SF+NFPYYLS TK VL+ + ++H+ ++A + DL T PRILLSGP+GSEIY EM
Sbjct: 458 ISFENFPYYLSATTKGVLMISMYVHMNGGSKYANFATDLTTACPRILLSGPSGSEIYQEM 517
Query: 444 LVKALAKYFGAKLLIFDSQLLLGGLSSKEAELLKDGFNAEKSCSCPKQSPTATDMAKSTD 503
L KALAK FGAKL+I DS LL GG ++EAE K+G E+ K++ A + +
Sbjct: 518 LAKALAKQFGAKLMIVDSLLLPGGSPAREAESSKEGSRRERLSMLAKRAVQAAQVLQHKK 577
Query: 504 PPAS-ETDTPSSSNVPTPLGLESQAKLETDSVPSTSGTAKNCLFKLGDRVKYSSSSACLY 562
P +S + D S L SQA + + ++ T+K+ FK GDRVK+ SA
Sbjct: 578 PTSSVDADITGGST------LSSQALPKQEV---STATSKSYTFKAGDRVKFVGPSASAI 628
Query: 563 QTSSSRYKGPSNGSRGKVVLIFDDNPLSKIGVRFDKPIPDGVDLGSACEAGQGFFCNITD 622
+ + +GP+ GS+GKV L F+DN SKIG+RFD+P+ DG DLG CE GFFC +
Sbjct: 629 SSLQGQLRGPAIGSQGKVALAFEDNCASKIGIRFDRPVQDGNDLGGLCEEDHGFFCAASS 688
Query: 623 LRLENSGIDELDKSLINTLFEVVTSESRDSPFILFMKEAEKSIVGNGDPYS-FKSKLEKL 681
LRLE S D+ DK +N +FEV SES ILF+K+ EKS+VGN D Y+ KSKLE L
Sbjct: 689 LRLEGSSSDDADKLAVNEIFEVALSESEGGSLILFLKDIEKSLVGNSDVYATLKSKLETL 748
Query: 682 PDNVVVIGSHTHSDSRKEKSHAGGLLFTKFGSNQTALLDLAFPDSFGRLHDRGKEVPKPN 741
P+N+VVI S T DSRKEKSH GG LFTKFG NQTALLDLAFPD+FG+LHDR KE PK
Sbjct: 749 PENIVVIASQTQLDSRKEKSHPGGFLFTKFGGNQTALLDLAFPDNFGKLHDRSKETPKSM 808
Query: 742 KTLTKLFPNKVTIHMPQDEALLASWKQQLDRDVETLKIKGNLHHLRTVLSRSGMESDGLE 801
K +T+LFPNK+ I +PQ+EALL+ WK++LDRD E LK++ N+ + VL+++ ++ L
Sbjct: 809 KQITRLFPNKIAIQLPQEEALLSDWKEKLDRDTEILKVQANITSILAVLAKNKLDCPDLG 868
Query: 802 SLCVKDLTLTNENSEKILGWALSHHLMQNPEADA-DAKLVLSSESIQYGIGIFQAIQNES 860
+LC+KD TL +E+ EK++GWA HHLM E D KLV+S+ESI YG+ IQNE+
Sbjct: 869 TLCIKDQTLPSESVEKVVGWAFGHHLMICTEPIVKDNKLVISAESISYGLQTLHDIQNEN 928
Query: 861 KSLKKSLKDVVTENEFEKRLLGDVIPPNDIGVTFDDIGALENVKDTLKELVMLPLQRPEL 920
KSLKKSLKDVVTENEFEK+LL DVIPP+DIGV+FDDIGALENVK+TLKELVMLPLQRPEL
Sbjct: 929 KSLKKSLKDVVTENEFEKKLLSDVIPPSDIGVSFDDIGALENVKETLKELVMLPLQRPEL 988
Query: 921 FCKGQLTKPCKGILLFGPPGTGKTMLAKAVATDAGANFINISMSSITSKWFGEGEKYVKA 980
F KGQLTKP KGILLFGPPGTGKTMLAKAVAT+AGANFINISMSSITSKWFGEGEKYVKA
Sbjct: 989 FDKGQLTKPTKGILLFGPPGTGKTMLAKAVATEAGANFINISMSSITSKWFGEGEKYVKA 1048
Query: 981 VFSLASKIAPSVIFVDEVDSMLGRRENPGEHEAMRKMKNEFMVNWDGLRTKDTERVIVLA 1040
VFSLASKIAPSVIFVDEVDSMLGRRENPGEHEAMRKMKNEFMVNWDGLRTKD ERV+VLA
Sbjct: 1049 VFSLASKIAPSVIFVDEVDSMLGRRENPGEHEAMRKMKNEFMVNWDGLRTKDRERVLVLA 1108
Query: 1041 ATNRPYDLDEAVIRRLPRRLMVNLPDAPNRAKILKVILAKEDLSSDVDLGAIANMTDGYS 1100
ATNRP+DLDEAVIRRLPRRLMVNLPDA NR+KIL VILAKE+++ DVDL AIANMTDGYS
Sbjct: 1109 ATNRPFDLDEAVIRRLPRRLMVNLPDATNRSKILSVILAKEEIAPDVDLEAIANMTDGYS 1168
Query: 1101 GSDLKNLCVTAAHRPIKEILEKEKKELAAAVAEGRPAPALRGSDDIRSLNMEDFKHAHQQ 1160
GSDLKNLCVTAAH PI+EILEKEKKE AA AE RP P L D+RSL M DFK AH Q
Sbjct: 1169 GSDLKNLCVTAAHFPIREILEKEKKEKTAAQAENRPTPPLYSCTDVRSLTMNDFKAAHDQ 1228
Query: 1161 VCASVSSESVNMTELVQWNELYGEGGSRVKKALSYF 1196
VCASVSS+S NM EL QWNELYGEGGSR K +LSYF
Sbjct: 1229 VCASVSSDSSNMNELQQWNELYGEGGSRKKTSLSYF 1264
>gb|AAM13995.1| unknown protein [Arabidopsis thaliana]
Length = 1265
Score = 1161 bits (3004), Expect = 0.0
Identities = 660/1236 (53%), Positives = 820/1236 (65%), Gaps = 62/1236 (5%)
Query: 1 PDNGAASSEKPPENSNP--EPSADPGK----CAQPDAQ-IDEPVAAAD------DDKADT 47
P +++SE P EN P +P ++ G+ + P A ++PV D + D
Sbjct: 51 PAGSSSASEVPIENQGPASDPGSESGEPELGSSDPQAMDAEKPVVTTDVPVMENSPETDA 110
Query: 48 TPPIADASTPTLVADKPRASFSSWSLYQKQNPNLESSAPWCRLLSQSAQHPNVSICIPNF 107
P + +TPT+ + + S K APW +LLSQ +Q+P+ I P F
Sbjct: 111 NPEVEVLATPTVAGEAVADADKS-----KAAKKRALKAPWAKLLSQYSQNPHRVIRGPVF 165
Query: 108 TIGSSRNCNFHLKDHTISGNLCKIKHTQREGSDVAVLESTGSKGSVIVNGTLVKKSTCCT 167
T+G R C+ ++D + LC++K ++ G VA LE G+ V VNG +KSTC
Sbjct: 166 TVGR-RGCDLSIRDQAMPSTLCELKQSEHGGPSVASLEILGNGVIVHVNGKCYQKSTCVH 224
Query: 168 LNSGDEVVFGLHGNHSYIFQQVNTE--------VAVKGAEVQSGIGKFMQLERRSGD--- 216
L GDEV+F L+G H+YIFQ V E ++ E + K + +E R+GD
Sbjct: 225 LRGGDEVIFSLNGKHAYIFQPVKDENLAAPDRASSLSICEARGAPLKGVHVETRAGDVDG 284
Query: 217 PSAVAGASILASLSNLRQDLTRWKSPSQTASKPHQGADV-----SIHTVLPDGTEIELDG 271
S V GASILASLS LR P A K Q V S + + D + D
Sbjct: 285 ASDVDGASILASLSKLRS--FHLLPPIAKAGKRQQNPAVPVVPSSFNDCISDTDMNDADS 342
Query: 272 LGNSTPSMGTDKAADAEA---SNKNTPMDCDPEDA--GAEPGNVKYSGVNDLLRPFFRIL 326
+ +K A A +N+N +D D A+ GNV +G +RP +L
Sbjct: 343 NNDHAAVASVEKIAAASTPGTANENLNVDGSGLDPFQEADGGNVPAAGYE--IRPIVHLL 400
Query: 327 AGSTTCKLKLSKSICKQVLEERNGAED--TQAASTSGTSVRCAVFKEDAHAAILDGKEQE 384
S++ ++ S S ++L+ER ++ + +S S R FK+ +L+ + +
Sbjct: 401 GESSSFDIRGSIS---RLLDERREVKEFLREFDLSSTISTRRQAFKDSLRGGVLNAQNID 457
Query: 385 VSFDNFPYYLSENTKNVLIAACFIHLKH-KEHAKYTADLPTVNPRILLSGPAGSEIYSEM 443
+SF+NFPYYLS TK VL+ + ++H+ ++A + DL T PRILLSGP+ SEIY EM
Sbjct: 458 ISFENFPYYLSATTKGVLMISMYVHMNGGSKYANFATDLTTACPRILLSGPSSSEIYQEM 517
Query: 444 LVKALAKYFGAKLLIFDSQLLLGGLSSKEAELLKDGFNAEKSCSCPKQSPTATDMAKSTD 503
L KALAK FGAKL+I DS LL GG ++EAE K+G E+ K++ A + +
Sbjct: 518 LAKALAKQFGAKLMIVDSLLLPGGSPAREAESSKEGSRRERLSMLAKRAVQAAQVLQHKK 577
Query: 504 PPAS-ETDTPSSSNVPTPLGLESQAKLETDSVPSTSGTAKNCLFKLGDRVKYSSSSACLY 562
P +S + D S L SQA + + ++ T+K+ FK GDRVK+ SA
Sbjct: 578 PTSSVDADITGGST------LSSQALPKQEV---STATSKSYTFKAGDRVKFVGPSASAI 628
Query: 563 QTSSSRYKGPSNGSRGKVVLIFDDNPLSKIGVRFDKPIPDGVDLGSACEAGQGFFCNITD 622
+ + +GP+ GS+GKV L F+DN SKIG+RFD+P+ DG DLG CE GFFC +
Sbjct: 629 SSLQGQLRGPAIGSQGKVALAFEDNCASKIGIRFDRPVQDGNDLGGLCEEDHGFFCAASS 688
Query: 623 LRLENSGIDELDKSLINTLFEVVTSESRDSPFILFMKEAEKSIVGNGDPYS-FKSKLEKL 681
LRLE S D+ DK +N +FEV SES ILF+K+ EKS+VGN D Y+ KSKLE L
Sbjct: 689 LRLEGSSSDDADKLAVNEIFEVALSESEGGSLILFLKDIEKSLVGNSDVYATLKSKLETL 748
Query: 682 PDNVVVIGSHTHSDSRKEKSHAGGLLFTKFGSNQTALLDLAFPDSFGRLHDRGKEVPKPN 741
P+N+VVI S T DSRKEKSH GG LFTKFG NQTALLDLAFPD+FG+LHDR KE PK
Sbjct: 749 PENIVVIASQTQLDSRKEKSHPGGFLFTKFGGNQTALLDLAFPDNFGKLHDRSKETPKSM 808
Query: 742 KTLTKLFPNKVTIHMPQDEALLASWKQQLDRDVETLKIKGNLHHLRTVLSRSGMESDGLE 801
K +T+LFPNK+ I +PQ+EALL+ WK++LDRD E LK++ N+ + VL+++ ++ L
Sbjct: 809 KQITRLFPNKIAIQLPQEEALLSDWKEKLDRDTEILKVQANITSILAVLAKNKLDCPDLG 868
Query: 802 SLCVKDLTLTNENSEKILGWALSHHLMQNPEADA-DAKLVLSSESIQYGIGIFQAIQNES 860
+LC+KD TL +E+ EK++GWA HHLM E D KLV+S+ESI YG+ IQNE+
Sbjct: 869 TLCIKDQTLPSESVEKVVGWAFGHHLMICTEPIVKDNKLVISAESISYGLQTLHDIQNEN 928
Query: 861 KSLKKSLKDVVTENEFEKRLLGDVIPPNDIGVTFDDIGALENVKDTLKELVMLPLQRPEL 920
KSLKKSLKDVVTENEFEK+LL DVIPP+DIGV+FDDIGALENVK+TLKELVMLPLQRPEL
Sbjct: 929 KSLKKSLKDVVTENEFEKKLLSDVIPPSDIGVSFDDIGALENVKETLKELVMLPLQRPEL 988
Query: 921 FCKGQLTKPCKGILLFGPPGTGKTMLAKAVATDAGANFINISMSSITSKWFGEGEKYVKA 980
F KGQLTKP KGILLFGPPGTGKTMLAKAVAT+AGANFINISMSSITSKWFGEGEKYVKA
Sbjct: 989 FDKGQLTKPTKGILLFGPPGTGKTMLAKAVATEAGANFINISMSSITSKWFGEGEKYVKA 1048
Query: 981 VFSLASKIAPSVIFVDEVDSMLGRRENPGEHEAMRKMKNEFMVNWDGLRTKDTERVIVLA 1040
VFSLASKIAPSVIFVDEVDSMLGRRENPGEHEAMRKMKNEFMVNWDGLRTKD ERV+VLA
Sbjct: 1049 VFSLASKIAPSVIFVDEVDSMLGRRENPGEHEAMRKMKNEFMVNWDGLRTKDRERVLVLA 1108
Query: 1041 ATNRPYDLDEAVIRRLPRRLMVNLPDAPNRAKILKVILAKEDLSSDVDLGAIANMTDGYS 1100
ATNRP+DLDEAVIRRLPRRLMVNLPDA NR+KIL VILAKE+++ DVDL AIANMTDGYS
Sbjct: 1109 ATNRPFDLDEAVIRRLPRRLMVNLPDATNRSKILSVILAKEEIAPDVDLEAIANMTDGYS 1168
Query: 1101 GSDLKNLCVTAAHRPIKEILEKEKKELAAAVAEGRPAPALRGSDDIRSLNMEDFKHAHQQ 1160
GSDLKNLCVTAAH PI+EILEKEKKE AA AE RP P L D+RSL M DFK AH Q
Sbjct: 1169 GSDLKNLCVTAAHFPIREILEKEKKEKTAAQAENRPTPPLYSCTDVRSLTMNDFKAAHDQ 1228
Query: 1161 VCASVSSESVNMTELVQWNELYGEGGSRVKKALSYF 1196
VCASVSS+S NM EL QWNELYGEGGSR K +LSYF
Sbjct: 1229 VCASVSSDSSNMNELQQWNELYGEGGSRKKTSLSYF 1264
>ref|NP_171788.2| AAA-type ATPase family protein [Arabidopsis thaliana]
Length = 1252
Score = 1095 bits (2832), Expect = 0.0
Identities = 640/1244 (51%), Positives = 819/1244 (65%), Gaps = 74/1244 (5%)
Query: 1 PDNGAASSEKPPENSNPEPSADPGKCA-QPDAQIDEPVAAADDDKADTTPPIADASTPT- 58
P + +++SE P +N P +DPG + P+ + +P + D ++ TT + T T
Sbjct: 34 PASSSSASEVPIDNQ--APVSDPGSISGDPELRTSDP-QSNDAERPVTTTDVPAMETDTN 90
Query: 59 -----LVADKPRASFSSWSLYQKQNPNLESSAPWCRLLSQSAQHPNVSICIPNFTIGSSR 113
LV P + K + + APW +LLSQ Q+P++ + FT+G R
Sbjct: 91 PELEGLVTPTPAGEVVVEAEKSKSSKKRIAKAPWAKLLSQFPQNPHLVMRGSVFTVGR-R 149
Query: 114 NCNFHLKDHTISGNLCKIKHTQREGSDVAVLESTGSKGSVIVNGTLVKKSTCCTLNSGDE 173
C+ ++DH++ LC+++ ++ G VA LE G+ V VNG + ++STC L GDE
Sbjct: 150 ACDLCIRDHSMPNVLCELRQSEHGGPSVASLEIIGNGVLVQVNGKIYQRSTCVHLRGGDE 209
Query: 174 VVFGLHGNHSY-----------IFQQVNTE--------VAVKGAEVQSGIGKFMQLERRS 214
++F G H+Y IFQ + E ++ E QS K + +E R+
Sbjct: 210 IIFTTPGKHAYVSFYKFFENVQIFQPLKDENLAAPDRTSSLSLFEAQSAPLKGLHVETRA 269
Query: 215 GDPSAVAG-ASILASLSNLRQDLTRWKSPSQTASKPHQGADVSIHTVLPDGTE-----IE 268
D S+V G AS+LAS+S L+ + P+ + K Q ++V VLP + ++
Sbjct: 270 RDSSSVDGTASLLASISKLQN--VPFLPPTAKSVKRQQNSEVP---VLPSSCDDFILDVD 324
Query: 269 LDGLGNSTP-----SMGTDKAADAEASNKNTPMDCDPEDAGAEP--GNVKYSGVNDLLRP 321
L+ ++ SM A+ + A+N + D + D EP GN+ +RP
Sbjct: 325 LNDADSNNDHAAIASMEKTVASTSCAANDDHDADGNGMDPFQEPEAGNIPDPAYE--IRP 382
Query: 322 FFRILAGSTTCKLK--LSKSICKQVLEERNGAEDTQAASTSGTSVRCAVFKEDAHAAILD 379
+L + L+ +SK + + E R ++ + S S + R A K+ IL+
Sbjct: 383 ILSLLGDPSEFDLRGSISKILVDERREVREMPKEYERPSASVLTRRQA-HKDSLRGGILN 441
Query: 380 GKEQEVSFDNFPYYLSENTKNVLIAACFIHLKH-KEHAKYTADLPTVNPRILLSGPAGSE 438
++ EVSF+NFPY+LS TK+VL+ + + H+K+ KE+A+Y +DLPT PRILLSGP+GSE
Sbjct: 442 PQDIEVSFENFPYFLSGTTKDVLMISTYAHIKYGKEYAEYASDLPTACPRILLSGPSGSE 501
Query: 439 IYSEMLVKALAKYFGAKLLIFDSQLLLGGLSSKEAELLKDGFNAEKSCSCPKQSPTATDM 498
IY EML KALAK GAKL+I DS LL GG + KEA+ K+ E+ K++ A
Sbjct: 502 IYQEMLAKALAKQCGAKLMIVDSLLLPGGSTPKEADTTKESSRRERLSVLAKRAVQAAQA 561
Query: 499 AKSTDPPASETDTPSSSNVPTPLGLESQAKLETDSVPS---TSGTAKNCLFKLGDRVKYS 555
A + P SS G+ + L + +V ++ T+K+ FK GDRV++
Sbjct: 562 A------VLQHKKPISS---VEAGITGGSTLSSQAVRRQEVSTATSKSYTFKAGDRVRFL 612
Query: 556 SSSACLYQTSSSRYKGPSNGSRGKVVLIFDDNPLSKIGVRFDKPIPDGVDLGSACEAGQG 615
S + + +GP+ G +GKV+L F+ N SKIGVRFD+ IPDG DLG CE
Sbjct: 613 GPSTSSLASLRAPPRGPATGFQGKVLLAFEGNGSSKIGVRFDRSIPDGNDLGGLCEEDHA 672
Query: 616 FFCNITDLRLENSGIDELDKSLINTLFEVVTSESRDSPFILFMKEAEKSIVGNGDPY-SF 674
+ LRLE+S D+ DK IN +FEV +ES ILF+K+ EKS+ GN D Y +
Sbjct: 673 -----SSLRLESSSSDDADKLAINEIFEVAFNESERGSLILFLKDIEKSVSGNTDVYITL 727
Query: 675 KSKLEKLPDNVVVIGSHTHSDSRKEKSHAGGLLFTKFGSNQTALLDLAFPDSFG-RLHDR 733
KSKLE LP+N+VVI S T D+RKEKSH GG LFTKFGSNQTALLDLAFPD+FG RL DR
Sbjct: 728 KSKLENLPENIVVIASQTQLDNRKEKSHPGGFLFTKFGSNQTALLDLAFPDTFGGRLQDR 787
Query: 734 GKEVPKPNKTLTKLFPNKVTIHMPQDEALLASWKQQLDRDVETLKIKGNLHHLRTVLSRS 793
E+PK K +T+LFPNKVTI +P+DEA L WK +L+RD E LK + N+ +R VLS++
Sbjct: 788 NTEMPKAVKQITRLFPNKVTIQLPEDEASLVDWKDKLERDTEILKAQANITSIRAVLSKN 847
Query: 794 GMESDGLESLCVKDLTLTNENSEKILGWALSHHLMQNPEADA-DAKLVLSSESIQYGIGI 852
+ +E LC+KD TL +++ EK++G+A +HHLM E D KL++S+ESI YG+ +
Sbjct: 848 QLVCPDIEILCIKDQTLPSDSVEKVVGFAFNHHLMNCSEPTVKDNKLIISAESITYGLQL 907
Query: 853 FQAIQNESKSLKKSLKDVVTENEFEKRLLGDVIPPNDIGVTFDDIGALENVKDTLKELVM 912
IQNE+KS KKSLKDVVTENEFEK+LL DVIPP+DIGV+F DIGALENVKDTLKELVM
Sbjct: 908 LHEIQNENKSTKKSLKDVVTENEFEKKLLSDVIPPSDIGVSFSDIGALENVKDTLKELVM 967
Query: 913 LPLQRPELFCKGQLTKPCKGILLFGPPGTGKTMLAKAVATDAGANFINISMSSITSKWFG 972
LPLQRPELF KGQLTKP KGILLFGPPGTGKTMLAKAVAT+AGANFINISMSSITSKWFG
Sbjct: 968 LPLQRPELFGKGQLTKPTKGILLFGPPGTGKTMLAKAVATEAGANFINISMSSITSKWFG 1027
Query: 973 EGEKYVKAVFSLASKIAPSVIFVDEVDSMLGRRENPGEHEAMRKMKNEFMVNWDGLRTKD 1032
EGEKYVKAVFSLASKIAPSVIFVDEVDSMLGRRENPGEHEAMRKMKNEFM+NWDGLRTKD
Sbjct: 1028 EGEKYVKAVFSLASKIAPSVIFVDEVDSMLGRRENPGEHEAMRKMKNEFMINWDGLRTKD 1087
Query: 1033 TERVIVLAATNRPYDLDEAVIRRLPRRLMVNLPDAPNRAKILKVILAKEDLSSDVDLGAI 1092
ERV+VLAATNRP+DLDEAVIRRLPRRLMVNLPD+ NR+KIL VILAKE+++ DVDL AI
Sbjct: 1088 KERVLVLAATNRPFDLDEAVIRRLPRRLMVNLPDSANRSKILSVILAKEEMAEDVDLEAI 1147
Query: 1093 ANMTDGYSGSDLKNLCVTAAHRPIKEILEKEKKELAAAVAEGRPAPALRGSDDIRSLNME 1152
ANMTDGYSGSDLKNLCVTAAH PI+EILEKEKKE + A AE R P L S D+R LNM
Sbjct: 1148 ANMTDGYSGSDLKNLCVTAAHLPIREILEKEKKERSVAQAENRAMPQLYSSTDVRPLNMN 1207
Query: 1153 DFKHAHQQVCASVSSESVNMTELVQWNELYGEGGSRVKKALSYF 1196
DFK AH QVCASV+S+S NM EL QWNELYGEGGSR K +LSYF
Sbjct: 1208 DFKTAHDQVCASVASDSSNMNELQQWNELYGEGGSRKKTSLSYF 1251
>ref|NP_194217.2| AAA-type ATPase family protein [Arabidopsis thaliana]
Length = 1122
Score = 949 bits (2454), Expect = 0.0
Identities = 514/891 (57%), Positives = 633/891 (70%), Gaps = 67/891 (7%)
Query: 343 QVLEERNGA-EDTQAASTSGTSVRCAVFKEDAHAAILDGKEQEVSFDNFPYYLSENTKNV 401
++L+E+N ++Q ASTSG ++ A+F+E A + G+ EVSF NFPYYLSE TK
Sbjct: 261 EILDEKNEVTSNSQQASTSGNGLQSAIFREAIQAGFVRGENMEVSFKNFPYYLSEYTKAA 320
Query: 402 LIAACFIHLKHKEHAKYTADLPTVNPRILLSGPAGSEIYSEMLVKALAKYFGAKLLIFDS 461
L+ A +IHLK KE+ ++ +D+ +NPRILLSGPAGSEIY E L KALA+ AKLLIFDS
Sbjct: 321 LLYASYIHLKKKEYVQFVSDMTPMNPRILLSGPAGSEIYQETLAKALARDLEAKLLIFDS 380
Query: 462 QLLLG-----------------------GLSSKEAELLKDGFNAEKSCSCPKQSPTATDM 498
+LG L++KE E L+DG + KSC P QS D
Sbjct: 381 YPILGFTRGKFLHLHLFVYFPDYGYEITALTAKEVESLRDGLASNKSCKLPNQSIELIDQ 440
Query: 499 AKSTDPPASETDTPSSSNVPTPLGLESQAKLETDSVP-STSGTAK------NCLF-KLGD 550
KS+D A SS + +SQ +LE +++P S + T K +CL K+
Sbjct: 441 GKSSDLSAGG-GVASSLSPAASSDSDSQLQLEPETLPRSVNHTLKKGMPPLHCLQQKILL 499
Query: 551 RVKYSSSSACLYQTSSSRYKGPSNGSRGKVVLIFDDNPLSKIGVRFDKPIPDGVDLGSAC 610
+ + S L+ + +GP NG+ GKV+L+FD+NP +K+GVRFDKPIPDGVDLG C
Sbjct: 500 QSSWISGLRILHLEEKNTCRGPPNGTTGKVILVFDENPSAKVGVRFDKPIPDGVDLGELC 559
Query: 611 EAGQGFFCNITDLRLENSGIDELDKSLINTLFEVVTSESRDSPFILFMKEAEKSIVGNGD 670
E+G GFFC TDL ++S +L + L+NTLFEVV SESR PFILF+K+AEKS+ GN D
Sbjct: 560 ESGHGFFCKATDLPFKSSSFKDLVRLLVNTLFEVVHSESRTCPFILFLKDAEKSVAGNFD 619
Query: 671 PYS-FKSKLEKLPDNVVVIGSHTHSDSRKEKSHAGGLLFTKFGSNQTALLDLAFPDSFGR 729
YS F+ +LE LP+NV+VI S THSD K K GR
Sbjct: 620 LYSAFQIRLEYLPENVIVICSQTHSDHLKVKD-------------------------IGR 654
Query: 730 LHDRGKEVPKPNKTLTKLFPNKVTIHMPQDEALLASWKQQLDRDVETLKIKGNLHHLRTV 789
+GKEVP + L +LF NK+TI MPQDE L WK Q+DRD ET K+K N +HLR V
Sbjct: 655 QKKQGKEVPHATELLAELFENKITIQMPQDEKRLTLWKHQMDRDAETSKVKSNFNHLRMV 714
Query: 790 LSRSGMESDGLES----LCVKDLTLTNENSEKILGWALSHHLMQNPEADADAKLVLSSES 845
L R G+ +GLE+ +C+KDLTL ++ EKI+GWA +H+ +NP+ D AK+ LS ES
Sbjct: 715 LRRRGLGCEGLETTWSRMCLKDLTLQRDSVEKIIGWAFGNHISKNPDTDP-AKVTLSRES 773
Query: 846 IQYGIGIFQAIQNESKSLKKSLKDVVTENEFEKRLLGDVIPPNDIGVTFDDIGALENVKD 905
I++GIG+ +QN+ K S KD+V EN FEKRLL DVI P+DI VTFDDIGALE VKD
Sbjct: 774 IEFGIGL---LQNDLKGSTSSKKDIVVENVFEKRLLSDVILPSDIDVTFDDIGALEKVKD 830
Query: 906 TLKELVMLPLQRPELFCKGQLTKPCKGILLFGPPGTGKTMLAKAVATDAGANFINISMSS 965
LKELVMLPLQRPELFCKG+LTKPCKGILLFGPPGTGKTMLAKAVA +A ANFINISMSS
Sbjct: 831 ILKELVMLPLQRPELFCKGELTKPCKGILLFGPPGTGKTMLAKAVAKEADANFINISMSS 890
Query: 966 ITSKWFGEGEKYVKAVFSLASKIAPSVIFVDEVDSMLGRRENPGEHEAMRKMKNEFMVNW 1025
ITSKWFGEGEKYVKAVFSLASK++PSVIFVDEVDSMLGRRE+P EHEA RK+KNEFM++W
Sbjct: 891 ITSKWFGEGEKYVKAVFSLASKMSPSVIFVDEVDSMLGRREHPREHEASRKIKNEFMMHW 950
Query: 1026 DGLRTKDTERVIVLAATNRPYDLDEAVIRRLPRRLMVNLPDAPNRAKILKVILAKEDLSS 1085
DGL T++ ERV+VLAATNRP+DLDEAVIRRLPRRLMV LPD NRA ILKVILAKEDLS
Sbjct: 951 DGLTTQERERVLVLAATNRPFDLDEAVIRRLPRRLMVGLPDTSNRAFILKVILAKEDLSP 1010
Query: 1086 DVDLGAIANMTDGYSGSDLKNLCVTAAHRPIKEILEKEKKELAAAVAEGRPAPALRGSDD 1145
D+D+G IA+MT+GYSGSDLKNLCVTAAHRPIKEILEKEK+E AA+A+G+ P L GS D
Sbjct: 1011 DLDIGEIASMTNGYSGSDLKNLCVTAAHRPIKEILEKEKRERDAALAQGKVPPPLSGSSD 1070
Query: 1146 IRSLNMEDFKHAHQQVCASVSSESVNMTELVQWNELYGEGGSRVKKALSYF 1196
+R+LN+EDF+ AH+ V ASVSSES MT L QWN+L+GEGGS +++ S++
Sbjct: 1071 LRALNVEDFRDAHKWVSASVSSESATMTALQQWNKLHGEGGSGKQQSFSFY 1121
Score = 97.4 bits (241), Expect = 2e-18
Identities = 84/272 (30%), Positives = 119/272 (42%), Gaps = 62/272 (22%)
Query: 2 DNGAASSEKPPENSNPEPSADPGKCA-------QPDAQIDEPVAAADDDKADTTPPIADA 54
D + +K + + P +D KC D+QID AAA + PP+A A
Sbjct: 32 DKSPSKRQKLEDGGDTLPPSDSSKCVLGDTTPTSGDSQIDASAAAATTSQP---PPVAQA 88
Query: 55 STPTLVADKPRASFSSWSLYQKQNPNLESSAPWCRLLSQSAQHPNVSICIPNFTIGSSRN 114
+ +ASF W+ + N PWCRLLSQSAQ+P+++I + F
Sbjct: 89 IL------QEKASFERWTYVHSRFEN-----PWCRLLSQSAQYPSINIFLSVFKF----- 132
Query: 115 CNFHLKDHTISGNLCKIKHTQREGSDVAVLESTGSKGSVIVNGTLVKKSTCCTLNSGDEV 174
D +S KI QR+G+ +AVLE+ G+ G + +NG + + LNSGDEV
Sbjct: 133 -----LDGELSSYSFKITRIQRKGNVLAVLETMGNNGHMWINGNYAEGNINHVLNSGDEV 187
Query: 175 VFGLHGNHSYIFQQVNTEVAVKGAEVQSGIGKFMQLERRSGDPSAVAGASILASLSNLRQ 234
V+ Q VA K VQ GKF+ LER + G SI++SL L
Sbjct: 188 VY-----------QQMPIVAAKPGSVQVPAGKFLDLER-------MTGHSIISSLERL-- 227
Query: 235 DLTRWKSPSQTASKPHQGADVSIHTVLPDGTE 266
+SK HQ + + DG E
Sbjct: 228 --------IHASSKSHQAPESMVQV---DGME 248
>gb|AAF02877.1| Unknown protein [Arabidopsis thaliana] gi|25518408|pir||C86159
hypothetical protein F22D16.11 - Arabidopsis thaliana
Length = 1217
Score = 868 bits (2243), Expect = 0.0
Identities = 568/1274 (44%), Positives = 733/1274 (56%), Gaps = 175/1274 (13%)
Query: 1 PDNGAASSEKPPENSNPEPSADPGKCA-QPDAQIDEPVAAADDDKADTTPPIADASTPT- 58
P + +++SE P +N P +DPG + P+ + +P + D ++ TT + T T
Sbjct: 40 PASSSSASEVPIDNQ--APVSDPGSISGDPELRTSDP-QSNDAERPVTTTDVPAMETDTN 96
Query: 59 -----LVADKPRASFSSWSLYQKQNPNLESSAPWCRLLSQSAQHPNVSICIPNFTIGSSR 113
LV P + K + + APW +LLSQ Q+P++ + FT+G R
Sbjct: 97 PELEGLVTPTPAGEVVVEAEKSKSSKKRIAKAPWAKLLSQFPQNPHLVMRGSVFTVGR-R 155
Query: 114 NCNFHLKDHTISGNLCKIKHTQREGSDVAVLESTGSKGSVIVNGTLVKKSTCCTLNSGDE 173
C+ ++DH++ LC+++ ++ G VA LE G+ V VNG + ++STC L GDE
Sbjct: 156 ACDLCIRDHSMPNVLCELRQSEHGGPSVASLEIIGNGVLVQVNGKIYQRSTCVHLRGGDE 215
Query: 174 VVFGLHGNHSY--------------IFQQVNTE--------VAVKGAEVQSGIGKFMQLE 211
++F G H+Y IFQ + E ++ E QS K + +E
Sbjct: 216 IIFTTPGKHAYVSFYKFFEKLSTVQIFQPLKDENLAAPDRTSSLSLFEAQSAPLKGLHVE 275
Query: 212 RRSGDPSAVAG-ASILASLSNLRQDLTRWKSPSQTASKPHQGADVSIHTVLPDGTE---- 266
R+ D S+V G AS+LAS+S L+ + P+ + K Q ++V VLP +
Sbjct: 276 TRARDSSSVDGTASLLASISKLQN--VPFLPPTAKSVKRQQNSEVP---VLPSSCDDFIL 330
Query: 267 -IELDGLGNSTP-----SMGTDKAADAEASNKNTPMDCDPEDAGAEP--GNVKYSGVNDL 318
++L+ ++ SM A+ + A+N + D + D EP GN+
Sbjct: 331 DVDLNDADSNNDHAAIASMEKTVASTSCAANDDHDADGNGMDPFQEPEAGNIPDPAYE-- 388
Query: 319 LRPFFRILAGSTTCKLK--LSKSICKQVLEERNGAEDTQAASTSGTSVRCAVFKEDAHAA 376
+RP +L + L+ +SK + + E R ++ + S S + R A K+
Sbjct: 389 IRPILSLLGDPSEFDLRGSISKILVDERREVREMPKEYERPSASVLTRRQA-HKDSLRGG 447
Query: 377 ILDGKEQEVSFDNFPYYLSENTKNVLIAACFIHLKH-KEHAKYTADLPTVNPRILLSGPA 435
IL+ ++ EVSF+NFPY+LS TK+VL+ + + H+K+ KE+A+Y +DLPT PRILLSGP+
Sbjct: 448 ILNPQDIEVSFENFPYFLSGTTKDVLMISTYAHIKYGKEYAEYASDLPTACPRILLSGPS 507
Query: 436 G----------------------------SEIYSEMLVKALAKYFGAKLLIFDSQLLLGG 467
G SEIY EML KALAK GAKL+I DS LL GG
Sbjct: 508 GKLWTSIVYESFVSHFHFPNKFSYGIFEGSEIYQEMLAKALAKQCGAKLMIVDSLLLPGG 567
Query: 468 LSSKEAELLKDGFNAEKSCSCPKQSPTATDMAKSTDPPASETDTPSSSNVPTPLGLESQA 527
+ KEA+ K+ E+ K++ A A + P SS G+ +
Sbjct: 568 STPKEADTTKESSRRERLSVLAKRAVQAAQAA------VLQHKKPISS---VEAGITGGS 618
Query: 528 KLETDSVPS---TSGTAKNCLFKLGDRVKYSSSSACLYQTSSSRYKGPSNGSRGKVVLIF 584
L + +V ++ T+K+ FK GDR + L + K + +
Sbjct: 619 TLSSQAVRRQEVSTATSKSYTFKAGDRQQVFKEKYSLRLKAMDLQKSGLD--------LI 670
Query: 585 DDNPLSKIGVRF-DKPIPDGVDLGSACEAGQGFFCNITDLRLENSGIDELDKSLINTLFE 643
D ++ I V + K + V + + + LRLE+S D+ DK IN +FE
Sbjct: 671 DRYQMAMILVVYAKKTMVFFVLVIFVLISTTHLRLKASSLRLESSSSDDADKLAINEIFE 730
Query: 644 VVTSESRDSPFILFMKEAEKSIVGNGDPY-SFKSKLEKLPDNVVVIGSHTHSDSRKEKSH 702
V +ES ILF+K+ EKS+ GN D Y + KSKLE LP+N+VVI S T D+RKEKSH
Sbjct: 731 VAFNESERGSLILFLKDIEKSVSGNTDVYITLKSKLENLPENIVVIASQTQLDNRKEKSH 790
Query: 703 AGGLLFTKFGSNQTALLDLAFPDSFGRLHDRGKEVPKPNKTLTKLFPNKVTIHMPQDEAL 762
GG LFTKFGSNQTALLDLAFPD EA
Sbjct: 791 PGGFLFTKFGSNQTALLDLAFPD----------------------------------EAS 816
Query: 763 LASWKQQLDRDVETLKIKGNLHHLRTVLSRSGMESDGLESLCVKDLTLTNENSEKILGWA 822
L WK +L+RD E LK + N+ +R L +C+ + + N E GW
Sbjct: 817 LVDWKDKLERDTEILKAQANITSIRAHLV-----------ICLIENHMINRCGES--GWL 863
Query: 823 LSHHLMQNPEADADAKLVLSSESIQYGIGIFQAIQNESKSLKKSLKDVVTENEFEKRLLG 882
Q+P + + + ++ KDVVTENEFEK+LL
Sbjct: 864 C----FQSPSYE-----------------LLRTYSQGQQAYHLGRKDVVTENEFEKKLLS 902
Query: 883 DVIPPNDIGVTFDDIGALENVKDTLKELVMLPLQRPELFCKGQLTKPCKGILLFGPPGTG 942
DVIPP+DIGV+F DIGALENVKDTLKELVMLPLQRPELF KGQLTKP KGILLFGPPGTG
Sbjct: 903 DVIPPSDIGVSFSDIGALENVKDTLKELVMLPLQRPELFGKGQLTKPTKGILLFGPPGTG 962
Query: 943 KTMLAKAVATDAGANFINISMSSITSKWFGEGEKYVKAVFSLASKIAPSVIFVDEVDSML 1002
KTMLAKAVAT+AGANFINISMSSITSKWFGEGEKYVKAVFSLASKIAPSVIFVDEVDSML
Sbjct: 963 KTMLAKAVATEAGANFINISMSSITSKWFGEGEKYVKAVFSLASKIAPSVIFVDEVDSML 1022
Query: 1003 GRRENPGEHEAMRKMKNEFMVNWDGLRTKDTERVIVLAATNRPYDLDEAVIRRLPRRLMV 1062
GRRENPGEHEAMRKMKNEFM+NWDGLRTKD ERV+VLAATNRP+DLDEAVIRRLPRRLMV
Sbjct: 1023 GRRENPGEHEAMRKMKNEFMINWDGLRTKDKERVLVLAATNRPFDLDEAVIRRLPRRLMV 1082
Query: 1063 NLPDAPNRAKILKVILAKEDLSSDVDLGAIANMTDGYSGSDLKNLCVTAAHRPIKEILEK 1122
NLPD+ NR+KIL VILAKE+++ DVDL AIANMTDGYSGSDLKNLCVTAAH PI+EILEK
Sbjct: 1083 NLPDSANRSKILSVILAKEEMAEDVDLEAIANMTDGYSGSDLKNLCVTAAHLPIREILEK 1142
Query: 1123 EKKELAAAVAEGRPAPALRGSDDIRSLNMEDFKHAHQQVCASVSSESVNMTELVQWNELY 1182
EKKE + A AE R P L S D+R LNM DFK AH QVCASV+S+S NM EL QWNELY
Sbjct: 1143 EKKERSVAQAENRAMPQLYSSTDVRPLNMNDFKTAHDQVCASVASDSSNMNELQQWNELY 1202
Query: 1183 GEGGSRVKKALSYF 1196
GEGGSR K +LSYF
Sbjct: 1203 GEGGSRKKTSLSYF 1216
>ref|NP_176404.2| AAA-type ATPase family protein [Arabidopsis thaliana]
Length = 1025
Score = 831 bits (2147), Expect = 0.0
Identities = 467/842 (55%), Positives = 573/842 (67%), Gaps = 101/842 (11%)
Query: 365 RCAVFKEDAHAAILDGKEQEVSFDNFPYYLSENTKNVLIAACFIHLKHKE--HAKYTADL 422
+ A F+E A I+DGK E SF+NFPYYLSE+TK VL+A +HL +A Y +DL
Sbjct: 274 QAAKFREYIRAGIVDGKRLEFSFENFPYYLSEHTKYVLLAVSDMHLNKMNIGYAPYASDL 333
Query: 423 PTVNPRILLSGPAGSEIYSEMLVKALAKYFGAKLLIFDSQLLLGGLSSKEAELLKDGFNA 482
+NPRILLSGPAGSEIY E+L KALA F AKLLIFDS +LG +++KE E L +G
Sbjct: 334 TILNPRILLSGPAGSEIYQEILAKALANSFNAKLLIFDSNPILGVMTAKEFESLMNG--- 390
Query: 483 EKSCSCPKQSPTATDMAKSTDPPASETDTPSSSNVPTPLGLESQAKLETDSVPSTSGTAK 542
P D KS D + + D S++P+P + P + GT
Sbjct: 391 ----------PALIDRGKSLDLSSGQGD----SSIPSPA-----------TSPRSFGTPI 425
Query: 543 NCLFKL-------GDRVKYSSSSACLYQTSSSRYKGPSNGSRGKVVLIFDDNPLSKIGVR 595
+ L L GDRV++ C +S +GP G GKV+L+FD+NP +K+GVR
Sbjct: 426 SGLLILHWGKTLAGDRVRFFGDELCPGLPTS---RGPPYGFIGKVLLVFDENPSAKVGVR 482
Query: 596 FDKPIPDGVDLGSACEAGQGFFCNITDLRLENSGIDELDKSLINTLFEVVTSESRDSPFI 655
F+ P+PDGVDLG CE G GFFC+ TDL+ E+S D+L++ L+ LFEV +SR P I
Sbjct: 483 FENPVPDGVDLGQLCEMGHGFFCSATDLQFESSASDDLNELLVTKLFEVAHDQSRTCPVI 542
Query: 656 LFMKEAEKSIVGNGDPYS-FKSKLEKLPDNVVVIGSHTHSDSRKEKSHAGGLLFTKFGSN 714
+F+K+AEK VGN S FKSKLE + DN++VI S THSD+ KEK
Sbjct: 543 IFLKDAEKYFVGNSHFCSAFKSKLEVISDNLIVICSQTHSDNPKEKG------------- 589
Query: 715 QTALLDLAFPDSFGRLHDRGKEVPKPNKTLTKLFPNKVTIHMPQDEALLASWKQQLDRDV 774
GRL D LF NKVTI+MPQ E LL SWK LDRD
Sbjct: 590 ------------IGRLTD--------------LFVNKVTIYMPQGEELLKSWKYHLDRDA 623
Query: 775 ETLKIKGNLHHLRTVLSRSGMESDGLESLCVKDLTLTNENSEKILGWALSHHLMQNPEAD 834
ETLK+K N +HLR VL R G+E +G+E+LC+KDLTL +++EKI+GWALSHH+ NP AD
Sbjct: 624 ETLKMKANYNHLRMVLGRCGIECEGIETLCMKDLTLRRDSAEKIIGWALSHHIKSNPGAD 683
Query: 835 ADAKLVLSSESIQYGIGIFQAIQNESKSLKKSLKDVVTENEFEKRLLGDVIPPNDIGVTF 894
D +++LS ES++ GI + ++ ESK KSLKD+VTEN FE + D+IPP++IGVTF
Sbjct: 684 PDVRVILSLESLKCGI---ELLEIESK---KSLKDIVTENTFE---ISDIIPPSEIGVTF 734
Query: 895 DDIGALENVKDTLKELVMLPLQRPELFCKGQLTKPCKGILLFGPPGTGKTMLAKAVATDA 954
DDIGALENVKDTLKELVMLP Q PELFCKGQLTKPC GILLFGP GTGKTMLAKAVAT+A
Sbjct: 735 DDIGALENVKDTLKELVMLPFQWPELFCKGQLTKPCNGILLFGPSGTGKTMLAKAVATEA 794
Query: 955 GANFINISMSSITSKWFGEGEKYVKAVFSLASKIAPSVIFVDEVDSMLGRRENPGEHEAM 1014
GAN IN+SMS +WF EGEKYVKAVFSLASKI+PS+IF+DEV+SML H
Sbjct: 795 GANLINMSMS----RWFSEGEKYVKAVFSLASKISPSIIFLDEVESML--------HRYR 842
Query: 1015 RKMKNEFMVNWDGLRTKDTERVIVLAATNRPYDLDEAVIRRLPRRLMVNLPDAPNRAKIL 1074
K KNEF++NWDGLRT + ERV+VLAATNRP+DLDEAVIRRLP RLMV LPDA +R+KIL
Sbjct: 843 LKTKNEFIINWDGLRTNEKERVLVLAATNRPFDLDEAVIRRLPHRLMVGLPDARSRSKIL 902
Query: 1075 KVILAKEDLSSDVDLGAIANMTDGYSGSDLKNLCVTAAHRPIKEILEKEKKELAAAVAEG 1134
KVIL+KEDLS D D+ +A+MT+GYSG+DLKNLCVTAA R I EI+EKEK E AAVAEG
Sbjct: 903 KVILSKEDLSPDFDIDEVASMTNGYSGNDLKNLCVTAARRRIIEIVEKEKSERDAAVAEG 962
Query: 1135 RPAPALRGSDDIRSLNMEDFKHAHQQVCASVSSESVNMTELVQWNELYGEGGSRVKKALS 1194
R PA G D+R L MEDF++A + V S+SS+SVNMT L QWNE YGEGGSR ++ S
Sbjct: 963 RVPPAGSGGSDLRVLKMEDFRNALELVSMSISSKSVNMTALRQWNEDYGEGGSRRNESFS 1022
Query: 1195 YF 1196
+
Sbjct: 1023 QY 1024
Score = 107 bits (266), Expect = 3e-21
Identities = 80/235 (34%), Positives = 116/235 (49%), Gaps = 33/235 (14%)
Query: 3 NGAASSEKPPENSNPEPSADPGKCAQPDAQIDEPVAAADDDKADTTPP--IADASTPTLV 60
+G++ PP + PS D ++ D A+ D +A T+ IA+ TPTL
Sbjct: 20 DGSSGKRIPPSS----PSGDKSPSSKRSKLGDGSGASTDSSEAPTSEDAKIAEGLTPTL- 74
Query: 61 ADKPRASFSSWSLYQKQNPNLESSAPWCRLLSQSAQHPNVSICIPNFTIGSSRNCNFHLK 120
P +SFS W+ + PWC+LLSQSA+ N+ + + T GS +F L
Sbjct: 75 ---PDSSFSGWTYR-----HCTFKTPWCKLLSQSAKQQNLCLYESSCTFGSCLTSDFTLH 126
Query: 121 DHTISGNLCKIKHTQREGSDVAVLESTGSKGSVIVNGTLVKKSTCCTLNSGDEVVFGLHG 180
D + LCKI QR G+ VAVL+ TG+ G + +N V K+ L+SGDE++
Sbjct: 127 DRNLGAFLCKITRIQRNGNVVAVLDITGTGGPLRINKAFVIKNVSHELHSGDELM----- 181
Query: 181 NHSYIFQQVNTEVAVKGAEVQSGIGKFMQLERRSGDPSAVAGASILASLSNLRQD 235
++V V Q GKF+QLER + DPS V S+LASL R++
Sbjct: 182 ----------SKVTVISGGEQVPAGKFLQLEREARDPSRV---SMLASLEISREN 223
>ref|XP_479469.1| putative MSP1(mitochondrial sorting of proteins) protein [Oryza
sativa (japonica cultivar-group)]
gi|33146850|dbj|BAC79845.1| putative MSP1(mitochondrial
sorting of proteins) protein [Oryza sativa (japonica
cultivar-group)]
Length = 1081
Score = 831 bits (2146), Expect = 0.0
Identities = 428/659 (64%), Positives = 514/659 (77%), Gaps = 13/659 (1%)
Query: 546 FKLGDRVKYSSSSACLYQTSSSRYKG---PSNGSRGKVVLIFDDNPLSKIGVRFDKPIPD 602
+K GDRV+Y S Q++ +G P GS+G+V L F++N SK+GVRFDK IP
Sbjct: 427 YKKGDRVRYIGS----VQSTGIILEGQRAPDYGSQGEVRLPFEENESSKVGVRFDKKIPG 482
Query: 603 GVDLGSACEAGQGFFCNITDLRLENSGIDELDKSLINTLFEVVTSESRDSPFILFMKEAE 662
G+DLG CE +GFFC + L L+ G ++ K + ++E + ES+ P ILF+K+ E
Sbjct: 483 GIDLGGNCEVDRGFFCPVDSLCLDGPGWEDRAKHPFDVIYEFASEESQHGPLILFLKDVE 542
Query: 663 KSIVGNGDPYSFKSKLEKLPDNVVVIGSHTHSDSRKEKSHAGGLLFTKFGSNQTALLDLA 722
K + + K+K+E P V ++GS H+DSRK+KS++G +KF +Q A+LDL
Sbjct: 543 KMCGNSYSYHGLKNKIESFPAGVFIVGSQIHTDSRKDKSNSGSPFLSKFPYSQ-AILDLT 601
Query: 723 FPDSFGRLHDRGKEVPKPNKTLTKLFPNKVTIHMPQDEALLASWKQQLDRDVETLKIKGN 782
F DSFGR++D+ KE K K LTKLFPNKVTI PQDE L+ WKQ LDRDVE LK K N
Sbjct: 602 FQDSFGRVNDKNKEALKIAKHLTKLFPNKVTIQTPQDELELSQWKQLLDRDVEILKAKAN 661
Query: 783 LHHLRTVLSRSGMESDGLE-SLCVKDLTLTNENSEKILGWALSHHLMQN--PEADADAKL 839
+++ L+R+G+E +E S CVKD LTNE +K++G+ALSH + P + D L
Sbjct: 662 TSKIQSFLTRNGLECADIETSACVKDRILTNECVDKVVGYALSHQFKHSTIPTRENDGLL 721
Query: 840 VLSSESIQYGIGIFQAIQNE--SKSLKKSLKDVVTENEFEKRLLGDVIPPNDIGVTFDDI 897
LS ES+++G+ + ++Q++ KS KKSLKDV TENEFEKRLLGDVIPP++IGVTF+DI
Sbjct: 722 ALSGESLKHGVELLDSMQSDPKKKSTKKSLKDVTTENEFEKRLLGDVIPPDEIGVTFEDI 781
Query: 898 GALENVKDTLKELVMLPLQRPELFCKGQLTKPCKGILLFGPPGTGKTMLAKAVATDAGAN 957
GALENVK+TLKELVMLPLQRPELF KGQL KPCKGILLFGPPGTGKTMLAKAVAT+AGAN
Sbjct: 782 GALENVKETLKELVMLPLQRPELFSKGQLMKPCKGILLFGPPGTGKTMLAKAVATEAGAN 841
Query: 958 FINISMSSITSKWFGEGEKYVKAVFSLASKIAPSVIFVDEVDSMLGRRENPGEHEAMRKM 1017
FINISMSSI SKWFGEGEKYVKAVFSLASKIAPSVIFVDEVD MLGRRENPGEHEAMRKM
Sbjct: 842 FINISMSSIASKWFGEGEKYVKAVFSLASKIAPSVIFVDEVDGMLGRRENPGEHEAMRKM 901
Query: 1018 KNEFMVNWDGLRTKDTERVIVLAATNRPYDLDEAVIRRLPRRLMVNLPDAPNRAKILKVI 1077
KNEFMVNWDGLRTKD ERV+VLAATNRP+DLDEAV+RRLPRRLMVNLPDA NR KIL VI
Sbjct: 902 KNEFMVNWDGLRTKDKERVLVLAATNRPFDLDEAVVRRLPRRLMVNLPDASNRKKILSVI 961
Query: 1078 LAKEDLSSDVDLGAIANMTDGYSGSDLKNLCVTAAHRPIKEILEKEKKELAAAVAEGRPA 1137
LAKEDL+ DVDL A+AN+TDGYSGSD+KNLCVTAAH PI+EILE+EKKE A+A AE +P
Sbjct: 962 LAKEDLADDVDLEALANLTDGYSGSDMKNLCVTAAHCPIREILEREKKERASAEAENKPL 1021
Query: 1138 PALRGSDDIRSLNMEDFKHAHQQVCASVSSESVNMTELVQWNELYGEGGSRVKKALSYF 1196
P R S D+RSL M DFKHAH+QVCAS++S+S NMTEL+QWN+LYGEGGSR K +LSYF
Sbjct: 1022 PPPRSSSDVRSLRMNDFKHAHEQVCASITSDSRNMTELIQWNDLYGEGGSRKKTSLSYF 1080
Score = 117 bits (294), Expect = 2e-24
Identities = 68/148 (45%), Positives = 94/148 (62%), Gaps = 9/148 (6%)
Query: 335 KLSKSICKQVLEERNGAEDTQAASTSGTSV-RCAVFKEDAHAAILDGKEQEVSFDNFPYY 393
++ S+CK + E+ +Q AS V + + KED +++ + SFD+FPYY
Sbjct: 289 EIVSSLCKTMEEQ------SQLASEENLQVAQHQLLKEDLKKVVVNASDISDSFDSFPYY 342
Query: 394 LSENTKNVLIAACFIHLKHKEHAKYTADLPTVNPRILLSGPAGSEIYSEMLVKALAKYFG 453
LSENTKN L+++ +++L KE K+T + ++ R+LLSGPAGSEIY E LVKAL K+FG
Sbjct: 343 LSENTKNALLSSAYVNLCCKESIKWTKHISSLCQRVLLSGPAGSEIYQESLVKALTKHFG 402
Query: 454 AKLLIFDSQLLLGGLS--SKEAELLKDG 479
AKLLI D LL G S SKE+E K G
Sbjct: 403 AKLLIIDPSLLASGQSSKSKESESYKKG 430
Score = 76.3 bits (186), Expect = 6e-12
Identities = 69/217 (31%), Positives = 97/217 (43%), Gaps = 41/217 (18%)
Query: 4 GAASSEKPPENSNPEPSADPGKCAQPDAQIDEPVAAAD--DDKADTTPPIADASTPTL-- 59
GAA S P S GK A+ +A + P A A D A + D+S L
Sbjct: 32 GAAGSSTPRRRS--------GKRAKAEATVGTPAAKAGGADATAAAAIDVIDSSVENLHG 83
Query: 60 ----VADKPRASFSSWSLYQKQN------PNLES--------------SAPWCRLLSQSA 95
P +S S S +K+ P+ E + W RL+SQS+
Sbjct: 84 VARPTGAVPASSTVSNSGVKKKRTKYINVPSAEELSLWKARQAVANGRAEAWGRLISQSS 143
Query: 96 QHPNVSICIPNFTIGSSRNCNFHLKDHTISGNLCKIKHTQREGSDVAVLESTGSKGSVIV 155
+ P+V I +FT+G N + L + +CK+KH +R A LE SK +V V
Sbjct: 144 ESPSVPIYTTHFTVGHGGNYDLRLTESFPGSLICKLKHVKRG----AALEIYVSK-AVHV 198
Query: 156 NGTLVKKSTCCTLNSGDEVVFGLHGNHSYIFQQVNTE 192
NG ++ K+ TL GDEV+F G H+YIFQQ+ E
Sbjct: 199 NGKVLDKTAKVTLVGGDEVIFSSLGRHAYIFQQLPEE 235
>emb|CAB79395.1| putative protein [Arabidopsis thaliana] gi|4678264|emb|CAB41125.1|
putative protein [Arabidopsis thaliana]
gi|23296350|gb|AAN13049.1| unknown protein [Arabidopsis
thaliana] gi|7486530|pir||T06669 hypothetical protein
F6I7.60 - Arabidopsis thaliana
Length = 442
Score = 623 bits (1607), Expect = e-176
Identities = 316/445 (71%), Positives = 371/445 (83%), Gaps = 8/445 (1%)
Query: 756 MPQDEALLASWKQQLDRDVETLKIKGNLHHLRTVLSRSGMESDGLES----LCVKDLTLT 811
MPQDE L WK Q+DRD ET K+K N +HLR VL R G+ +GLE+ +C+KDLTL
Sbjct: 1 MPQDEKRLTLWKHQMDRDAETSKVKSNFNHLRMVLRRRGLGCEGLETTWSRMCLKDLTLQ 60
Query: 812 NENSEKILGWALSHHLMQNPEADADAKLVLSSESIQYGIGIFQAIQNESKSLKKSLKDVV 871
++ EKI+GWA +H+ +NP+ D AK+ LS ESI++GIG+ +QN+ K S KD+V
Sbjct: 61 RDSVEKIIGWAFGNHISKNPDTDP-AKVTLSRESIEFGIGL---LQNDLKGSTSSKKDIV 116
Query: 872 TENEFEKRLLGDVIPPNDIGVTFDDIGALENVKDTLKELVMLPLQRPELFCKGQLTKPCK 931
EN FEKRLL DVI P+DI VTFDDIGALE VKD LKELVMLPLQRPELFCKG+LTKPCK
Sbjct: 117 VENVFEKRLLSDVILPSDIDVTFDDIGALEKVKDILKELVMLPLQRPELFCKGELTKPCK 176
Query: 932 GILLFGPPGTGKTMLAKAVATDAGANFINISMSSITSKWFGEGEKYVKAVFSLASKIAPS 991
GILLFGPPGTGKTMLAKAVA +A ANFINISMSSITSKWFGEGEKYVKAVFSLASK++PS
Sbjct: 177 GILLFGPPGTGKTMLAKAVAKEADANFINISMSSITSKWFGEGEKYVKAVFSLASKMSPS 236
Query: 992 VIFVDEVDSMLGRRENPGEHEAMRKMKNEFMVNWDGLRTKDTERVIVLAATNRPYDLDEA 1051
VIFVDEVDSMLGRRE+P EHEA RK+KNEFM++WDGL T++ ERV+VLAATNRP+DLDEA
Sbjct: 237 VIFVDEVDSMLGRREHPREHEASRKIKNEFMMHWDGLTTQERERVLVLAATNRPFDLDEA 296
Query: 1052 VIRRLPRRLMVNLPDAPNRAKILKVILAKEDLSSDVDLGAIANMTDGYSGSDLKNLCVTA 1111
VIRRLPRRLMV LPD NRA ILKVILAKEDLS D+D+G IA+MT+GYSGSDLKNLCVTA
Sbjct: 297 VIRRLPRRLMVGLPDTSNRAFILKVILAKEDLSPDLDIGEIASMTNGYSGSDLKNLCVTA 356
Query: 1112 AHRPIKEILEKEKKELAAAVAEGRPAPALRGSDDIRSLNMEDFKHAHQQVCASVSSESVN 1171
AHRPIKEILEKEK+E AA+A+G+ P L GS D+R+LN+EDF+ AH+ V ASVSSES
Sbjct: 357 AHRPIKEILEKEKRERDAALAQGKVPPPLSGSSDLRALNVEDFRDAHKWVSASVSSESAT 416
Query: 1172 MTELVQWNELYGEGGSRVKKALSYF 1196
MT L QWN+L+GEGGS +++ S++
Sbjct: 417 MTALQQWNKLHGEGGSGKQQSFSFY 441
>gb|AAC19276.1| T14P8.7 [Arabidopsis thaliana] gi|7269007|emb|CAB80740.1| AT4g02470
[Arabidopsis thaliana] gi|7487016|pir||T01303
hypothetical protein T14P8.7 - Arabidopsis thaliana
Length = 371
Score = 602 bits (1551), Expect = e-170
Identities = 307/361 (85%), Positives = 326/361 (90%)
Query: 836 DAKLVLSSESIQYGIGIFQAIQNESKSLKKSLKDVVTENEFEKRLLGDVIPPNDIGVTFD 895
D KLV+S+ESI YG+ IQNE+KSLKKSLKDVVTENEFEK+LL DVIPP+DIGV+FD
Sbjct: 10 DNKLVISAESISYGLQTLHDIQNENKSLKKSLKDVVTENEFEKKLLSDVIPPSDIGVSFD 69
Query: 896 DIGALENVKDTLKELVMLPLQRPELFCKGQLTKPCKGILLFGPPGTGKTMLAKAVATDAG 955
DIGALENVK+TLKELVMLPLQRPELF KGQLTKP KGILLFGPPGTGKTMLAKAVAT+AG
Sbjct: 70 DIGALENVKETLKELVMLPLQRPELFDKGQLTKPTKGILLFGPPGTGKTMLAKAVATEAG 129
Query: 956 ANFINISMSSITSKWFGEGEKYVKAVFSLASKIAPSVIFVDEVDSMLGRRENPGEHEAMR 1015
ANFINISMSSITSKWFGEGEKYVKAVFSLASKIAPSVIFVDEVDSMLGRRENPGEHEAMR
Sbjct: 130 ANFINISMSSITSKWFGEGEKYVKAVFSLASKIAPSVIFVDEVDSMLGRRENPGEHEAMR 189
Query: 1016 KMKNEFMVNWDGLRTKDTERVIVLAATNRPYDLDEAVIRRLPRRLMVNLPDAPNRAKILK 1075
KMKNEFMVNWDGLRTKD ERV+VLAATNRP+DLDEAVIRRLPRRLMVNLPDA NR+KIL
Sbjct: 190 KMKNEFMVNWDGLRTKDRERVLVLAATNRPFDLDEAVIRRLPRRLMVNLPDATNRSKILS 249
Query: 1076 VILAKEDLSSDVDLGAIANMTDGYSGSDLKNLCVTAAHRPIKEILEKEKKELAAAVAEGR 1135
VILAKE+++ DVDL AIANMTDGYSGSDLKNLCVTAAH PI+EILEKEKKE AA AE R
Sbjct: 250 VILAKEEIAPDVDLEAIANMTDGYSGSDLKNLCVTAAHFPIREILEKEKKEKTAAQAENR 309
Query: 1136 PAPALRGSDDIRSLNMEDFKHAHQQVCASVSSESVNMTELVQWNELYGEGGSRVKKALSY 1195
P P L D+RSL M DFK AH QVCASVSS+S NM EL QWNELYGEGGSR K +LSY
Sbjct: 310 PTPPLYSCTDVRSLTMNDFKAAHDQVCASVSSDSSNMNELQQWNELYGEGGSRKKTSLSY 369
Query: 1196 F 1196
F
Sbjct: 370 F 370
>ref|XP_475967.1| unknown protein [Oryza sativa (japonica cultivar-group)]
gi|48843801|gb|AAT47060.1| unknown protein [Oryza sativa
(japonica cultivar-group)]
Length = 855
Score = 536 bits (1380), Expect = e-150
Identities = 345/923 (37%), Positives = 499/923 (53%), Gaps = 192/923 (20%)
Query: 361 GTSVRCAVFKEDAHAAILDGKEQEVSFDNFPYYLSENTKNVLIAACFIHLKHKEHAKYTA 420
G + A + + ++DG+E +V+FD F YYLSE TK VLI+A F+HLK + +K+
Sbjct: 37 GAGIGVAELEAELRRLVVDGREGDVTFDEFRYYLSERTKEVLISAAFVHLKQADLSKHIR 96
Query: 421 DLPTVNPRILLSGPAGSEIYSEMLVKALAKYFGAKLLIFDSQLLLGGLSSKEAELLKDGF 480
+L + ILLSGP +E Y + L +AL+ Y+ A+LLI D + SK +
Sbjct: 97 NLCAASRAILLSGP--TEPYLQSLARALSHYYKAQLLILDVTDFSLRIQSK--------Y 146
Query: 481 NAEKSCSCPKQSPTATDMAKSTDPPASETDTPSSSNVPTPLGLESQAKLETDSVPSTSGT 540
+ QS + T + +D S T P S+ E + L+
Sbjct: 147 GSSSKGLAQSQSISETTFGRMSDLIGSFTIFPKSA--------EPRESLQR--------- 189
Query: 541 AKNCLFKLGDRVKYSSSSACLYQTSSSRYKGPSNGSRGKVVLIFDDNPLSKIGVRFDKPI 600
QTSS+ + + + + + +S
Sbjct: 190 ----------------------QTSSADVRSRGSEASSNAPPLRKNASMSS--------- 218
Query: 601 PDGVDLGSACEAGQGFFCNITDLRLENSGIDELDKSLINTLFEVVTSESRDSPFILFMKE 660
D D+ S C A +++ R + DE K LI +L++V+ S + ++P IL++++
Sbjct: 219 -DISDVSSQCSAH-----SVSARRTSSWCFDE--KVLIQSLYKVMVSVAENNPVILYIRD 270
Query: 661 AEKSIVGNGDPYS-FKSKLEKLPDNVVVIGSHTHSDSRKEKSHAGGLLFTKFGSNQTALL 719
++ + + YS F+ L KL V+++GS
Sbjct: 271 VDQLLHRSQRTYSLFQKMLAKLTGQVLILGS----------------------------- 301
Query: 720 DLAFPDSFGRLHDRGKEVPKPNKTLTKLFPNKVTIHMPQDEALLASWKQQLDRDVETLKI 779
RL D + ++ ++ LFP V I P++E L SWK Q++ D + ++I
Sbjct: 302 ---------RLLDSDSDHTDVDERVSSLFPFHVDIKPPEEETHLDSWKTQMEEDTKKIQI 352
Query: 780 KGNLHHLRTVLSRSGMESDGLESLCVKDLTLTNENSEKILGWALSHHLMQNPEAD-ADAK 838
+ N +H+ VLS + ++ D L S+C D + + E+I+ A+S+H++ N + + + K
Sbjct: 353 QDNRNHIIEVLSANDLDCDDLSSICQADTMVLSNYIEEIIVSAVSYHMIHNKDPEYKNGK 412
Query: 839 LVLSSESIQYGIGIFQAI----------------------QNESKSLKKSLKD------- 869
LVLSS+S+ +G+ IFQ KS LKD
Sbjct: 413 LVLSSKSLSHGLSIFQESGFGGKETLKLEDDLKGATGPKKSETEKSATVPLKDGDGPLPP 472
Query: 870 ---VVTENEFEKRLLGDVIPPNDIGVTFDDIGALENVKDTLKELVMLPLQRPELFCKGQL 926
+ +NEFEKR+ +VIP ++IGVTFDDIGAL ++K++L+ELVMLPL+RP+LF KG L
Sbjct: 473 PKPEIPDNEFEKRIRPEVIPASEIGVTFDDIGALADIKESLQELVMLPLRRPDLF-KGGL 531
Query: 927 TKPCKGILLFGPPGTGKTMLAKAVATDAGANFINISMSSITSKWFGEGEKYVKAVFSLAS 986
KPC+GILLFGPPGTGKTMLAKA+A DAGA+FIN+SMS+ITSKWFGE EK V+A+FSLA+
Sbjct: 532 LKPCRGILLFGPPGTGKTMLAKAIANDAGASFINVSMSTITSKWFGEDEKNVRALFSLAA 591
Query: 987 KIAPSVIFVDEVDSMLGRRENPGEHEAMRKMKNEFMVNWDGLRTKDTERVIVLAATNRPY 1046
K+AP++IFVDEVDSMLG+R GEHEAMRK+KNEFM +WDGL +K ER++VLAATNRP+
Sbjct: 592 KVAPTIIFVDEVDSMLGQRARCGEHEAMRKIKNEFMSHWDGLLSKSGERILVLAATNRPF 651
Query: 1047 DLDEAVIRRLPRRLMVNLPDAPNRAKILKVILAKEDLSSDVDLGAIANMTDGYSGSDLKN 1106
DLDEA+IRR RR+MV LP +R IL+ +L+KE ++ D+D +A MT+GYSGSDLKN
Sbjct: 652 DLDEAIIRRFERRIMVGLPTLDSRELILRTLLSKEKVAEDIDYKELATMTEGYSGSDLKN 711
Query: 1107 LCVTAAHRPIKEILEKEK-KEL---------AAAVAEGRPAP------------------ 1138
LCVTAA+RP++E+L++E+ KE+ AA AE +P
Sbjct: 712 LCVTAAYRPVRELLKREREKEMERRANEAKEKAATAENSESPESKKEKENSENPESKEKE 771
Query: 1139 -------------------------ALRGSDDIRSLNMEDFKHAHQQVCASVSSESVNMT 1173
G+ D+R L MED + A QV AS ++E M
Sbjct: 772 KERKENSENKEEKTENKQDNSKAEGGTEGTIDLRPLTMEDLRQAKNQVAASFATEGAVMN 831
Query: 1174 ELVQWNELYGEGGSRVKKALSYF 1196
EL QWN+LYGEGGSR K+ L+YF
Sbjct: 832 ELKQWNDLYGEGGSRKKQQLTYF 854
>dbj|BAD81550.1| unknown protein [Oryza sativa (japonica cultivar-group)]
gi|56784122|dbj|BAD81507.1| unknown protein [Oryza sativa
(japonica cultivar-group)]
Length = 840
Score = 518 bits (1334), Expect = e-145
Identities = 335/899 (37%), Positives = 494/899 (54%), Gaps = 177/899 (19%)
Query: 370 KEDAHAAILDGKEQEVSFDNFPYYLSENTKNVLIAACFIHLKHKEHAKYTADLPTVNPRI 429
+++ ++DG + V+FD FPYYLSE T+ +L +A ++HLK + ++YT +L + I
Sbjct: 46 EQELRRLVVDGADSRVTFDGFPYYLSEQTRVLLTSAAYVHLKQADISQYTRNLAPASRAI 105
Query: 430 LLSGPAGSEIYSEMLVKALAKYFGAKLLIFDSQLLLGGLSSKEAELLKDGFNAEKSCSCP 489
LLSGPA E+Y +ML KALA YF AKLL+ D L + SK + S
Sbjct: 106 LLSGPA--ELYQQMLAKALAHYFEAKLLLLDPTDFLIKIHSKYG-------GGSSTDSSF 156
Query: 490 KQSPTATDMAKSTDPPASETDTPSSSNVPTPLGLE-SQAKLETDSVPSTSGTAKNCLFKL 548
K+S + T + K + S + P + + S ++ S STS K
Sbjct: 157 KRSISETTLEKVSGLLGSLSILPQKEKPKGTIRRQSSMTDMKLRSSESTSSFPK------ 210
Query: 549 GDRVKYSSSSACLYQTSSSRYKGPSNGSRGKVVLIFDDNPLSKIGVRFDKPIPDGVDLGS 608
+K ++S++ SS +GP N NP S
Sbjct: 211 ---LKRNASTSS--DMSSLASQGPPN------------NPAS------------------ 235
Query: 609 ACEAGQGFFCNITDLRLENSGIDELDKSLINTLFEVVTSESRDSPFILFMKEAEKSIVGN 668
LR +S + +K L+ +++V+ S S+ +P +L++++ EK + +
Sbjct: 236 --------------LRRASSWTFD-EKILVQAVYKVLHSVSKKNPIVLYIRDVEKFLHKS 280
Query: 669 GDPY-SFKSKLEKLPDNVVVIGSHTHSDSRKEKSHAGGLLFTKFGSNQTALLDLAFPDSF 727
Y F+ L KL V+V+GS ++D+ F +
Sbjct: 281 KKMYVMFEKLLNKLEGPVLVLGSR--------------------------IVDMDFDEEL 314
Query: 728 GRLHDRGKEVPKPNKTLTKLFPNKVTIHMPQDEALLASWKQQLDRDVETLKIKGNLHHLR 787
++ LT LFP + I P++E L SW QL+ D++ ++ + N +H+
Sbjct: 315 -------------DERLTALFPYNIEIKPPENENHLVSWNSQLEEDMKMIQFQDNRNHIT 361
Query: 788 TVLSRSGMESDGLESLCVKDLTLTNENSEKILGWALSHHLMQNPEAD-ADAKLVLSSESI 846
VL+ + +E D L S+C+ D + E+I+ A+S+HLM + + + KL+LS++S+
Sbjct: 362 EVLAENDLECDDLGSICLSDTMVLGRYIEEIVVSAVSYHLMNKKDPEYRNGKLLLSAKSL 421
Query: 847 QYGIGIFQ-------------AIQNESKSLKKSLKDVVTENE------------------ 875
+ + IFQ A ++ SK + + ++E
Sbjct: 422 SHALEIFQENKMYDKDSMKLEAKRDASKVADRGIAPFAAKSETKPATLLPPVPPTAAAAP 481
Query: 876 ----------FEK---------------------RLLGDVIPPNDIGVTFDDIGALENVK 904
FEK R+ +VIP N+IGVTFDDIGAL ++K
Sbjct: 482 PVESKAEPEKFEKKDNPSPAAKAPEMPPDNEFEKRIRPEVIPANEIGVTFDDIGALSDIK 541
Query: 905 DTLKELVMLPLQRPELFCKGQLTKPCKGILLFGPPGTGKTMLAKAVATDAGANFINISMS 964
++L+ELVMLPL+RP+LF KG L KPC+GILLFGPPGTGKTMLAKA+A +A A+FIN+SMS
Sbjct: 542 ESLQELVMLPLRRPDLF-KGGLLKPCRGILLFGPPGTGKTMLAKAIANEAQASFINVSMS 600
Query: 965 SITSKWFGEGEKYVKAVFSLASKIAPSVIFVDEVDSMLGRRENPGEHEAMRKMKNEFMVN 1024
+ITSKWFGE EK V+A+F+LA+K++P++IFVDEVDSMLG+R GEHEAMRK+KNEFM +
Sbjct: 601 TITSKWFGEDEKNVRALFTLAAKVSPTIIFVDEVDSMLGQRNRAGEHEAMRKIKNEFMTH 660
Query: 1025 WDGLRTKDTERVIVLAATNRPYDLDEAVIRRLPRRLMVNLPDAPNRAKILKVILAKEDLS 1084
WDGL ++ ++++VLAATNRP+DLDEA+IRR RR+MV LP +R IL+ +L+KE +
Sbjct: 661 WDGLLSRPDQKILVLAATNRPFDLDEAIIRRFERRIMVGLPSLESRELILRSLLSKEKVD 720
Query: 1085 SDVDLGAIANMTDGYSGSDLKNLCVTAAHRPIKEILEKE-KKELAAAVAEG---RPAPAL 1140
+D +A MT+GYSGSDLKNLC TAA+RP++E+++KE KKEL +G A +
Sbjct: 721 GGLDYKELATMTEGYSGSDLKNLCTTAAYRPVRELIQKERKKELEKKREQGGNASDASKM 780
Query: 1141 RGSDD---IRSLNMEDFKHAHQQVCASVSSESVNMTELVQWNELYGEGGSRVKKALSYF 1196
+ D+ +R LNM+D K A QV AS ++E M EL QWNELYGEGGSR K+ L+YF
Sbjct: 781 KEKDETIILRPLNMKDLKEAKNQVAASFAAEGTIMGELKQWNELYGEGGSRKKQQLTYF 839
>gb|AAP21168.1| At1g64110/F22C12_22 [Arabidopsis thaliana]
gi|18407974|ref|NP_564824.1| AAA-type ATPase family
protein [Arabidopsis thaliana] gi|15810167|gb|AAL06985.1|
At1g64110/F22C12_22 [Arabidopsis thaliana]
Length = 824
Score = 479 bits (1234), Expect = e-133
Identities = 271/619 (43%), Positives = 388/619 (61%), Gaps = 96/619 (15%)
Query: 634 DKSLINTLFEVVTSESRDSPFILFMKEAEKSIVGNGDPYS-FKSKLEKLPDNVVVIGSHT 692
+K L+ +L++V+ S+ +P +L++++ E + + Y+ F+ L+KL V+++GS
Sbjct: 245 EKLLVQSLYKVLAYVSKANPIVLYLRDVENFLFRSQRTYNLFQKLLQKLSGPVLILGSR- 303
Query: 693 HSDSRKEKSHAGGLLFTKFGSNQTALLDLAFPDSFGRLHDRGKEVPKPNKTLTKLFPNKV 752
++DL+ D+ +E+ ++ L+ +FP +
Sbjct: 304 -------------------------IVDLSSEDA--------QEI---DEKLSAVFPYNI 327
Query: 753 TIHMPQDEALLASWKQQLDRDVETLKIKGNLHHLRTVLSRSGMESDGLESLCVKDLTLTN 812
I P+DE L SWK QL+RD+ ++ + N +H+ VLS + + D LES+ +D + +
Sbjct: 328 DIRPPEDETHLVSWKSQLERDMNMIQTQDNRNHIMEVLSENDLICDDLESISFEDTKVLS 387
Query: 813 ENSEKILGWALSHHLMQNPEAD-ADAKLVLSSESIQYGIGIF------------QAIQNE 859
E+I+ ALS+HLM N + + + KLV+SS S+ +G +F Q + E
Sbjct: 388 NYIEEIVVSALSYHLMNNKDPEYRNGKLVISSISLSHGFSLFREGKAGGREKLKQKTKEE 447
Query: 860 S------KSLKKSLK---------------------------DVVTENEFEKRLLGDVIP 886
S +S+K K +V +NEFEKR+ +VIP
Sbjct: 448 SSKEVKAESIKPETKTESVTTVSSKEEPEKEAKAEKVTPKAPEVAPDNEFEKRIRPEVIP 507
Query: 887 PNDIGVTFDDIGALENVKDTLKELVMLPLQRPELFCKGQLTKPCKGILLFGPPGTGKTML 946
+I VTF DIGAL+ +K++L+ELVMLPL+RP+LF G L KPC+GILLFGPPGTGKTML
Sbjct: 508 AEEINVTFKDIGALDEIKESLQELVMLPLRRPDLFTGG-LLKPCRGILLFGPPGTGKTML 566
Query: 947 AKAVATDAGANFINISMSSITSKWFGEGEKYVKAVFSLASKIAPSVIFVDEVDSMLGRRE 1006
AKA+A +AGA+FIN+SMS+ITSKWFGE EK V+A+F+LASK++P++IFVDEVDSMLG+R
Sbjct: 567 AKAIAKEAGASFINVSMSTITSKWFGEDEKNVRALFTLASKVSPTIIFVDEVDSMLGQRT 626
Query: 1007 NPGEHEAMRKMKNEFMVNWDGLRTKDTERVIVLAATNRPYDLDEAVIRRLPRRLMVNLPD 1066
GEHEAMRK+KNEFM +WDGL TK ER++VLAATNRP+DLDEA+IRR RR+MV LP
Sbjct: 627 RVGEHEAMRKIKNEFMSHWDGLMTKPGERILVLAATNRPFDLDEAIIRRFERRIMVGLPA 686
Query: 1067 APNRAKILKVILAKEDLSSDVDLGAIANMTDGYSGSDLKNLCVTAAHRPIKEILEKEKKE 1126
NR KIL+ +LAKE + ++D +A MT+GY+GSDLKNLC TAA+RP++E++++E+ +
Sbjct: 687 VENREKILRTLLAKEKVDENLDYKELAMMTEGYTGSDLKNLCTTAAYRPVRELIQQERIK 746
Query: 1127 LAAAVAEGRPAPALRGSDD---------IRSLNMEDFKHAHQQVCASVSSESVNMTELVQ 1177
+ P A G +D +R LN +DFK A QV AS ++E M EL Q
Sbjct: 747 DTEKKKQREPTKA--GEEDEGKEERVITLRPLNRQDFKEAKNQVAASFAAEGAGMGELKQ 804
Query: 1178 WNELYGEGGSRVKKALSYF 1196
WNELYGEGGSR K+ L+YF
Sbjct: 805 WNELYGEGGSRKKEQLTYF 823
Score = 97.1 bits (240), Expect = 3e-18
Identities = 47/103 (45%), Positives = 71/103 (68%), Gaps = 2/103 (1%)
Query: 358 STSGTSVRCAVFKEDAHAAILDGKEQEVSFDNFPYYLSENTKNVLIAACFIHLKHKEHAK 417
S+S +V +++ ++DG+E +++FD FPYYLSE T+ +L +A ++HLKH + +K
Sbjct: 32 SSSNNAVTADKMEKEILRQVVDGRESKITFDEFPYYLSEQTRVLLTSAAYVHLKHFDASK 91
Query: 418 YTADLPTVNPRILLSGPAGSEIYSEMLVKALAKYFGAKLLIFD 460
YT +L + ILLSGPA E+Y +ML KALA +F AKLL+ D
Sbjct: 92 YTRNLSPASRAILLSGPA--ELYQQMLAKALAHFFDAKLLLLD 132
>ref|NP_849842.1| AAA-type ATPase family protein [Arabidopsis thaliana]
Length = 829
Score = 479 bits (1234), Expect = e-133
Identities = 271/619 (43%), Positives = 388/619 (61%), Gaps = 96/619 (15%)
Query: 634 DKSLINTLFEVVTSESRDSPFILFMKEAEKSIVGNGDPYS-FKSKLEKLPDNVVVIGSHT 692
+K L+ +L++V+ S+ +P +L++++ E + + Y+ F+ L+KL V+++GS
Sbjct: 250 EKLLVQSLYKVLAYVSKANPIVLYLRDVENFLFRSQRTYNLFQKLLQKLSGPVLILGSR- 308
Query: 693 HSDSRKEKSHAGGLLFTKFGSNQTALLDLAFPDSFGRLHDRGKEVPKPNKTLTKLFPNKV 752
++DL+ D+ +E+ ++ L+ +FP +
Sbjct: 309 -------------------------IVDLSSEDA--------QEI---DEKLSAVFPYNI 332
Query: 753 TIHMPQDEALLASWKQQLDRDVETLKIKGNLHHLRTVLSRSGMESDGLESLCVKDLTLTN 812
I P+DE L SWK QL+RD+ ++ + N +H+ VLS + + D LES+ +D + +
Sbjct: 333 DIRPPEDETHLVSWKSQLERDMNMIQTQDNRNHIMEVLSENDLICDDLESISFEDTKVLS 392
Query: 813 ENSEKILGWALSHHLMQNPEAD-ADAKLVLSSESIQYGIGIF------------QAIQNE 859
E+I+ ALS+HLM N + + + KLV+SS S+ +G +F Q + E
Sbjct: 393 NYIEEIVVSALSYHLMNNKDPEYRNGKLVISSISLSHGFSLFREGKAGGREKLKQKTKEE 452
Query: 860 S------KSLKKSLK---------------------------DVVTENEFEKRLLGDVIP 886
S +S+K K +V +NEFEKR+ +VIP
Sbjct: 453 SSKEVKAESIKPETKTESVTTVSSKEEPEKEAKAEKVTPKAPEVAPDNEFEKRIRPEVIP 512
Query: 887 PNDIGVTFDDIGALENVKDTLKELVMLPLQRPELFCKGQLTKPCKGILLFGPPGTGKTML 946
+I VTF DIGAL+ +K++L+ELVMLPL+RP+LF G L KPC+GILLFGPPGTGKTML
Sbjct: 513 AEEINVTFKDIGALDEIKESLQELVMLPLRRPDLFTGG-LLKPCRGILLFGPPGTGKTML 571
Query: 947 AKAVATDAGANFINISMSSITSKWFGEGEKYVKAVFSLASKIAPSVIFVDEVDSMLGRRE 1006
AKA+A +AGA+FIN+SMS+ITSKWFGE EK V+A+F+LASK++P++IFVDEVDSMLG+R
Sbjct: 572 AKAIAKEAGASFINVSMSTITSKWFGEDEKNVRALFTLASKVSPTIIFVDEVDSMLGQRT 631
Query: 1007 NPGEHEAMRKMKNEFMVNWDGLRTKDTERVIVLAATNRPYDLDEAVIRRLPRRLMVNLPD 1066
GEHEAMRK+KNEFM +WDGL TK ER++VLAATNRP+DLDEA+IRR RR+MV LP
Sbjct: 632 RVGEHEAMRKIKNEFMSHWDGLMTKPGERILVLAATNRPFDLDEAIIRRFERRIMVGLPA 691
Query: 1067 APNRAKILKVILAKEDLSSDVDLGAIANMTDGYSGSDLKNLCVTAAHRPIKEILEKEKKE 1126
NR KIL+ +LAKE + ++D +A MT+GY+GSDLKNLC TAA+RP++E++++E+ +
Sbjct: 692 VENREKILRTLLAKEKVDENLDYKELAMMTEGYTGSDLKNLCTTAAYRPVRELIQQERIK 751
Query: 1127 LAAAVAEGRPAPALRGSDD---------IRSLNMEDFKHAHQQVCASVSSESVNMTELVQ 1177
+ P A G +D +R LN +DFK A QV AS ++E M EL Q
Sbjct: 752 DTEKKKQREPTKA--GEEDEGKEERVITLRPLNRQDFKEAKNQVAASFAAEGAGMGELKQ 809
Query: 1178 WNELYGEGGSRVKKALSYF 1196
WNELYGEGGSR K+ L+YF
Sbjct: 810 WNELYGEGGSRKKEQLTYF 828
Score = 97.1 bits (240), Expect = 3e-18
Identities = 47/103 (45%), Positives = 71/103 (68%), Gaps = 2/103 (1%)
Query: 358 STSGTSVRCAVFKEDAHAAILDGKEQEVSFDNFPYYLSENTKNVLIAACFIHLKHKEHAK 417
S+S +V +++ ++DG+E +++FD FPYYLSE T+ +L +A ++HLKH + +K
Sbjct: 37 SSSNNAVTADKMEKEILRQVVDGRESKITFDEFPYYLSEQTRVLLTSAAYVHLKHFDASK 96
Query: 418 YTADLPTVNPRILLSGPAGSEIYSEMLVKALAKYFGAKLLIFD 460
YT +L + ILLSGPA E+Y +ML KALA +F AKLL+ D
Sbjct: 97 YTRNLSPASRAILLSGPA--ELYQQMLAKALAHFFDAKLLLLD 137
>emb|CAB79602.1| putative protein [Arabidopsis thaliana] gi|4455359|emb|CAB36769.1|
putative protein [Arabidopsis thaliana]
gi|15234347|ref|NP_194529.1| AAA-type ATPase family
protein [Arabidopsis thaliana] gi|7487959|pir||T02901
MSP1 protein homolog T13J8.110 - Arabidopsis thaliana
Length = 726
Score = 475 bits (1223), Expect = e-132
Identities = 261/581 (44%), Positives = 375/581 (63%), Gaps = 58/581 (9%)
Query: 634 DKSLINTLFEVVTSESRDSPFILFMKEAEKSIVGNGDPYSFKSKLEKLPDNVVVIGSHTH 693
++ + +L++V+ S S +P I+++++ EK F+ L KL V+V+GS
Sbjct: 185 ERLFLQSLYKVLVSISETNPIIIYLRDVEKLCQSERFYKLFQRLLTKLSGPVLVLGS--- 241
Query: 694 SDSRKEKSHAGGLLFTKFGSNQTALLDLAFPDSFGRLHDRGKEVPKPNKTLTKLFPNKVT 753
RL + + + + ++ LFP +
Sbjct: 242 -----------------------------------RLLEPEDDCQEVGEGISALFPYNIE 266
Query: 754 IHMPQDEALLASWKQQLDRDVETLKIKGNLHHLRTVLSRSGMESDGLESLCVKDLTLTNE 813
I P+DE L SWK + + D++ ++ + N +H+ VL+ + +E D L S+C D +
Sbjct: 267 IRPPEDENQLMSWKTRFEDDMKVIQFQDNKNHIAEVLAANDLECDDLGSICHADTMFLSS 326
Query: 814 NSEKILGWALSHHLMQNPEAD-ADAKLVLSSESIQYGIGIFQAIQN---ESKSLKKSL-- 867
+ E+I+ A+S+HLM N E + + +LV+SS S+ +G+ I Q Q +S L ++
Sbjct: 327 HIEEIVVSAISYHLMNNKEPEYKNGRLVISSNSLSHGLNILQEGQGCFEDSLKLDTNIDS 386
Query: 868 KDVVTENEFEKRLLGDVIPPNDIGVTFDDIGALENVKDTLKELVMLPLQRPELFCKGQLT 927
K+V +NEFEKR+ +VIP N+IGVTF DIG+L+ K++L+ELVMLPL+RP+LF KG L
Sbjct: 387 KEVAPDNEFEKRIRPEVIPANEIGVTFADIGSLDETKESLQELVMLPLRRPDLF-KGGLL 445
Query: 928 KPCKGILLFGPPGTGKTMLAKAVATDAGANFINISMSSITSKWFGEGEKYVKAVFSLASK 987
KPC+GILLFGPPGTGKTM+AKA+A +AGA+FIN+SMS+ITSKWFGE EK V+A+F+LA+K
Sbjct: 446 KPCRGILLFGPPGTGKTMMAKAIANEAGASFINVSMSTITSKWFGEDEKNVRALFTLAAK 505
Query: 988 IAPSVIFVDEVDSMLGRRENPGEHEAMRKMKNEFMVNWDGLRTKDTERVIVLAATNRPYD 1047
++P++IFVDEVDSMLG+R GEHEAMRK+KNEFM +WDGL + +R++VLAATNRP+D
Sbjct: 506 VSPTIIFVDEVDSMLGQRTRVGEHEAMRKIKNEFMTHWDGLMSNAGDRILVLAATNRPFD 565
Query: 1048 LDEAVIRRLPRRLMVNLPDAPNRAKILKVILAKEDLSSDVDLGAIANMTDGYSGSDLKNL 1107
LDEA+IRR RR+MV LP +R KIL+ +L+KE + ++D +A MTDGYSGSDLKN
Sbjct: 566 LDEAIIRRFERRIMVGLPSVESREKILRTLLSKEK-TENLDFQELAQMTDGYSGSDLKNF 624
Query: 1108 CVTAAHRPIKEIL--------EKEKKELAAAVAEGRPAPALRGSDD----IRSLNMEDFK 1155
C TAA+RP++E++ E+ K+E A +E S++ +R L+MED K
Sbjct: 625 CTTAAYRPVRELIKQECLKDQERRKREEAEKNSEEGSEAKEEVSEERGITLRPLSMEDMK 684
Query: 1156 HAHQQVCASVSSESVNMTELVQWNELYGEGGSRVKKALSYF 1196
A QV AS ++E M EL QWN+LYGEGGSR K+ LSYF
Sbjct: 685 VAKSQVAASFAAEGAGMNELKQWNDLYGEGGSRKKEQLSYF 725
Score = 68.2 bits (165), Expect = 2e-09
Identities = 31/62 (50%), Positives = 45/62 (72%)
Query: 377 ILDGKEQEVSFDNFPYYLSENTKNVLIAACFIHLKHKEHAKYTADLPTVNPRILLSGPAG 436
I+DG+E V+FD FPYYLSE T+ +L +A ++HLK + +K+T +L + ILLSGPA
Sbjct: 54 IVDGRESSVTFDEFPYYLSEKTRLLLTSAAYVHLKQSDISKHTRNLAPGSKAILLSGPAD 113
Query: 437 SE 438
+E
Sbjct: 114 TE 115
>ref|NP_917758.1| P0501G01.20 [Oryza sativa (japonica cultivar-group)]
gi|12313686|dbj|BAB21091.1| cell division cycle gene
CDC48-like [Oryza sativa (japonica cultivar-group)]
Length = 812
Score = 436 bits (1122), Expect = e-120
Identities = 251/612 (41%), Positives = 364/612 (59%), Gaps = 89/612 (14%)
Query: 634 DKSLINTLFEVVTSESRDSPFILFMKEAEKSIVGNGDPYS-FKSKLEKLPDNVVVIGSHT 692
+K LI +L++++ S S SP IL++++ + + + Y F+ L+KL V+VIGS
Sbjct: 240 EKVLIQSLYKIIVSASEISPVILYIRDVDDLLGSSEKAYCMFQKMLKKLSGRVIVIGSQF 299
Query: 693 HSDSRKEKSHAGGLLFTKFGSNQTALLDLAFPDSFGRLHDRGKEVPKPNKTLTKLFPNKV 752
D + +++ LFP +
Sbjct: 300 LDDDEDREDI--------------------------------------EESVCALFPCIL 321
Query: 753 TIHMPQDEALLASWKQQLDRDVETLKIKGNLHHLRTVLSRSGMESDGLESLCVKD-LTLT 811
P+D+ LL WK Q++ D + +++ VL+ + +E + L S+ D +
Sbjct: 322 ETKPPKDKVLLEKWKTQMEEDSNNNNNQVVQNYIAEVLAENNLECEDLSSINADDDCKII 381
Query: 812 NENSEKILGWALSHHLMQNPEAD-ADAKLVLSSESIQYGIGIFQAIQNESKSL---KKSL 867
E+I+ ++S+HLM N + LV+SSES+ +G+ IFQ + K K
Sbjct: 382 VAYLEEIITPSVSYHLMNNKNPKYRNGNLVISSESLSHGLRIFQESNDLGKDTVEAKDET 441
Query: 868 KDVVTENEFEKRLLGDVIPPNDIGVTFDDIGALENVKDTLKELVMLPLQRPELFCKGQLT 927
+ VV +NE+EK++ VIP N+IGVTFDDIGAL ++K+ L ELVMLPLQRP+ F KG L
Sbjct: 442 EMVVPDNEYEKKIRPTVIPANEIGVTFDDIGALADIKECLHELVMLPLQRPDFF-KGGLL 500
Query: 928 KPCKGILLFGPPGTGKTMLAKAVATDAGANFINISMSSITSKWFGEGEKYVKAVFSLASK 987
KPCKG+LLFGPPGTGKTMLAKA+A AGA+F+NISM+S+TSKW+GE EK ++A+FSLA+K
Sbjct: 501 KPCKGVLLFGPPGTGKTMLAKALANAAGASFLNISMASMTSKWYGESEKCIQALFSLAAK 560
Query: 988 IAPSVIFVDEVDSMLGRRENPGEHEAMRKMKNEFMVNWDGLRTKDTERVIVLAATNRPYD 1047
+AP++IF+DEVDSMLG+R+N E+EA R++KNEFM +WDGL +K ER++VLAATNRP+D
Sbjct: 561 LAPAIIFIDEVDSMLGKRDNHSENEASRRVKNEFMAHWDGLLSKSNERILVLAATNRPFD 620
Query: 1048 LDEAVIRRLPRRLMVNLPDAPNRAKILKVILAKEDLSSDVDLGAIANMTDGYSGSDLKNL 1107
LD+AVIRR R+MV LP +R ILK +L+KE + ++D +A MT+GY+ SDLKN+
Sbjct: 621 LDDAVIRRFEHRIMVGLPTLESRELILKTLLSKETV-ENIDFKELAKMTEGYTSSDLKNI 679
Query: 1108 CVTAAHRPIKEILEKEKKELAAAVA----------------------------------- 1132
CVTAA+ P++E+L+KEK ++ A
Sbjct: 680 CVTAAYHPVRELLQKEKNKVKKETAPETKQEPKEKTKIQENGTKSSDSKTEKDKLDNKEG 739
Query: 1133 -EGRPAPALRGSD-------DIRSLNMEDFKHAHQQVCASVSSESVNMTELVQWNELYGE 1184
+ +PA SD +R LNMED + A +V AS +SE V M ++ +WNELYG+
Sbjct: 740 KKDKPADKKDKSDKGDAGETTLRPLNMEDLRKAKDEVAASFASEGVVMNQIKEWNELYGK 799
Query: 1185 GGSRVKKALSYF 1196
GGSR ++ L+YF
Sbjct: 800 GGSRKREQLTYF 811
Score = 69.7 bits (169), Expect = 6e-10
Identities = 39/101 (38%), Positives = 55/101 (53%), Gaps = 2/101 (1%)
Query: 360 SGTSVRCAVFKEDAHAAILDGKEQEVSFDNFPYYLSENTKNVLIAACFIHLKHKEHAKYT 419
+G A + + ++DG++ VSFD+FPYYLSE +K L + F+HL +
Sbjct: 33 TGGGATAAEVEAELRCLVVDGRDVGVSFDDFPYYLSEQSKLALTSTAFVHLSPTILPNHI 92
Query: 420 ADLPTVNPRILLSGPAGSEIYSEMLVKALAKYFGAKLLIFD 460
L + ILL GP SE Y + L KALA F A+LL+ D
Sbjct: 93 RVLSASSRTILLCGP--SEAYLQSLAKALANQFSARLLLLD 131
>ref|NP_175433.1| AAA-type ATPase family protein [Arabidopsis thaliana]
Length = 640
Score = 432 bits (1111), Expect = e-119
Identities = 231/523 (44%), Positives = 336/523 (64%), Gaps = 20/523 (3%)
Query: 675 KSKLEKLPDNVVVI-GSHTHSDSRKEKSHAGGLLFTKFGSNQTALLDLAFPDSFGRLHDR 733
K +KL +V+I G + KE+ FT N + ++ L P
Sbjct: 136 KEMFDKLSGPIVMICGQNKIETGSKEREK-----FTMVLPNLSRVVKLPLPLKGLTEGFT 190
Query: 734 GKEVPKPNKTLTKLFPNKVTIHMPQDEALLASWKQQLDRDVETLKIKGNLHHLRTVLSRS 793
G+ + N+ + KLF N + +H P++E L +K+QL D + + N++ L L
Sbjct: 191 GRGKSEENE-IYKLFTNVMRLHPPKEEDTLRLFKKQLGEDRRIVISRSNINELLKALEEH 249
Query: 794 GMESDGLESLCVKDLTLTNENSEKILGWALSHHLMQNP-EADADAKLVLSSESIQYGIGI 852
+ L + + LT + +EK +GWA +H+L P +L L ES++ I
Sbjct: 250 ELLCTDLYQVNTDGVILTKQKAEKAIGWAKNHYLASCPVPLVKGGRLSLPRESLEISIAR 309
Query: 853 FQAIQNESKSLKKSLKDVVTENEFEKRLLGDVIPPNDIGVTFDDIGALENVKDTLKELVM 912
+ +++ S ++LK++ ++E+E+ + V+ P +IGV F+DIGALE+VK L ELV+
Sbjct: 310 LRKLEDNSLKPSQNLKNIA-KDEYERNFVSAVVAPGEIGVKFEDIGALEDVKKALNELVI 368
Query: 913 LPLQRPELFCKGQLTKPCKGILLFGPPGTGKTMLAKAVATDAGANFINISMSSITSKWFG 972
LP++RPELF +G L +PCKGILLFGPPGTGKT+LAKA+AT+AGANFI+I+ S++TSKWFG
Sbjct: 369 LPMRRPELFARGNLLRPCKGILLFGPPGTGKTLLAKALATEAGANFISITGSTLTSKWFG 428
Query: 973 EGEKYVKAVFSLASKIAPSVIFVDEVDSMLGRRENPGEHEAMRKMKNEFMVNWDGLRTKD 1032
+ EK KA+FS A+K+AP +IFVDE+DS+LG R EHEA R+M+NEFM WDGLR+KD
Sbjct: 429 DAEKLTKALFSFATKLAPVIIFVDEIDSLLGARGGSSEHEATRRMRNEFMAAWDGLRSKD 488
Query: 1033 TERVIVLAATNRPYDLDEAVIRRLPRRLMVNLPDAPNRAKILKVILAKEDLSSDVDLGAI 1092
++R+++L ATNRP+DLD+AVIRRLPRR+ V+LPDA NR KILK+ L E+L SD +
Sbjct: 489 SQRILILGATNRPFDLDDAVIRRLPRRIYVDLPDAENRLKILKIFLTPENLESDFQFEKL 548
Query: 1093 ANMTDGYSGSDLKNLCVTAAHRPIKEILEKEKKELAAAVAEGRPAPALRGSDDIRSLNME 1152
A T+GYSGSDLKNLC+ AA+RP++E+L++E+K A + G +RSL+++
Sbjct: 549 AKETEGYSGSDLKNLCIAAAYRPVQELLQEEQKGARAEASPG-----------LRSLSLD 597
Query: 1153 DFKHAHQQVCASVSSESVNMTELVQWNELYGEGGSRVKKALSY 1195
DF + +V SV+ ++ M EL +WNE YGEGGSR K +
Sbjct: 598 DFIQSKAKVSPSVAYDATTMNELRKWNEQYGEGGSRTKSPFGF 640
>ref|XP_467801.1| transitional endoplasmic reticulum ATPase-like [Oryza sativa
(japonica cultivar-group)] gi|46390947|dbj|BAD16461.1|
transitional endoplasmic reticulum ATPase-like [Oryza
sativa (japonica cultivar-group)]
Length = 473
Score = 430 bits (1106), Expect = e-118
Identities = 225/476 (47%), Positives = 322/476 (67%), Gaps = 16/476 (3%)
Query: 722 AFPDSFGRLHDRGKEVPKPNKTLTKLFPNKVTIHMPQDEALLASWKQQLDRDVETLKIKG 781
+ P S RL G+ + ++KLF N + + +P+++ + Q++ D + + +
Sbjct: 10 SLPSSLKRLVG-GRPKYSRSSGISKLFTNSLIVPLPEEDEQRRIFNNQIEEDRKIIISRH 68
Query: 782 NLHHLRTVLSRSGMESDGLESLCVKDLTLTNENSEKILGWALSHHLMQN--PEADADAKL 839
NL L VL + L + + LT + +EK++GWA SH+L P D +L
Sbjct: 69 NLVELHKVLQEHELSCVELLHVKSDGVVLTRQKAEKVVGWARSHYLSSAVLPNIKGD-RL 127
Query: 840 VLSSESIQYGIGIFQAIQNESKSLKKSLKDVVTENEFEKRLLGDVIPPNDIGVTFDDIGA 899
++ ES+ I + ++K +++K++ ++E+E+ + V+PP++IGV FDDIGA
Sbjct: 128 IIPRESLDVAIERLKEQGIKTKRPSQNIKNLA-KDEYERNFISAVVPPDEIGVKFDDIGA 186
Query: 900 LENVKDTLKELVMLPLQRPELFCKGQLTKPCKGILLFGPPGTGKTMLAKAVATDAGANFI 959
LE+VK TL ELV LP++RPELF G L +PCKG+LLFGPPGTGKT+LAKA+AT+AGANFI
Sbjct: 187 LEDVKRTLDELVTLPMRRPELFSHGNLLRPCKGVLLFGPPGTGKTLLAKALATEAGANFI 246
Query: 960 NISMSSITSKWFGEGEKYVKAVFSLASKIAPSVIFVDEVDSMLGRRENPGEHEAMRKMKN 1019
+I+ S++TSKWFG+ EK KA+FS AS++AP +IFVDEVDS+LG R EHEA R+M+N
Sbjct: 247 SITGSTLTSKWFGDAEKLTKALFSFASRLAPVIIFVDEVDSLLGARGGAFEHEATRRMRN 306
Query: 1020 EFMVNWDGLRTKDTERVIVLAATNRPYDLDEAVIRRLPRRLMVNLPDAPNRAKILKVILA 1079
EFM WDGLR+K+++R+++L ATNRP+DLD+AVIRRLPRR+ V+LPDA NR KILK++LA
Sbjct: 307 EFMAAWDGLRSKESQRILILGATNRPFDLDDAVIRRLPRRIYVDLPDAQNRMKILKILLA 366
Query: 1080 KEDLSSDVDLGAIANMTDGYSGSDLKNLCVTAAHRPIKEILEKEKKELAAAVAEGRPAPA 1139
KE+L SD +AN T+GYSGSDLKNLC+ +A+RP+ E+LE+EKK G P
Sbjct: 367 KENLESDFRFDELANSTEGYSGSDLKNLCIASAYRPVHELLEEEKK--------GGPCSQ 418
Query: 1140 LRGSDDIRSLNMEDFKHAHQQVCASVSSESVNMTELVQWNELYGEGGSRVKKALSY 1195
G +R L ++DF A +V SVS ++ +M EL +WNE YGEGGSR + +
Sbjct: 419 NTG---LRPLRLDDFIQAKAKVSPSVSYDATSMNELRKWNEQYGEGGSRTRSPFGF 471
>ref|NP_188608.1| AAA-type ATPase family protein [Arabidopsis thaliana]
Length = 439
Score = 422 bits (1086), Expect = e-116
Identities = 213/445 (47%), Positives = 303/445 (67%), Gaps = 17/445 (3%)
Query: 757 PQDEALLASWKQQLDRDVETLKIKGNLHHLRTVLSRSGMESDGLESLCVKDLTLTNENSE 816
P++E L + +QL D + + NL+ L L + + L + + LT + +E
Sbjct: 6 PKEEENLIVFNKQLGEDRRIVMSRSNLNELLKALEENELLCTDLYQVNTDGVILTKQRAE 65
Query: 817 KILGWALSHHLMQNPEADA-DAKLVLSSESIQYGIGIFQAIQNESKSLKKSLKDV----- 870
K++GWA +H+L P + +L+L ESI+ + +A ++ S+ ++LK
Sbjct: 66 KVIGWARNHYLSSCPSPSIKEGRLILPRESIEISVKRLKAQEDISRKPTQNLKRFLFLQN 125
Query: 871 VTENEFEKRLLGDVIPPNDIGVTFDDIGALENVKDTLKELVMLPLQRPELFCKGQLTKPC 930
+ ++EFE + V+ P +IGV FDDIGALE+VK TL ELV+LP++RPELF +G L +PC
Sbjct: 126 IAKDEFETNFVSAVVAPGEIGVKFDDIGALEHVKKTLNELVILPMRRPELFTRGNLLRPC 185
Query: 931 KGILLFGPPGTGKTMLAKAVATDAGANFINISMSSITSKWFGEGEKYVKAVFSLASKIAP 990
KGILLFGPPGTGKT+LAKA+AT+AGANFI+I+ S++TSKWFG+ EK KA+FS ASK+AP
Sbjct: 186 KGILLFGPPGTGKTLLAKALATEAGANFISITGSTLTSKWFGDAEKLTKALFSFASKLAP 245
Query: 991 SVIFVDEVDSMLGRRENPGEHEAMRKMKNEFMVNWDGLRTKDTERVIVLAATNRPYDLDE 1050
+IFVDEVDS+LG R EHEA R+M+NEFM WDGLR+KD++R+++L ATNRP+DLD+
Sbjct: 246 VIIFVDEVDSLLGARGGAFEHEATRRMRNEFMAAWDGLRSKDSQRILILGATNRPFDLDD 305
Query: 1051 AVIRRLPRRLMVNLPDAPNRAKILKVILAKEDLSSDVDLGAIANMTDGYSGSDLKNLCVT 1110
AVIRRLPRR+ V+LPDA NR KILK+ L E+L + + +A T+GYSGSDLKNLC+
Sbjct: 306 AVIRRLPRRIYVDLPDAENRLKILKIFLTPENLETGFEFDKLAKETEGYSGSDLKNLCIA 365
Query: 1111 AAHRPIKEILEKEKKELAAAVAEGRPAPALRGSDDIRSLNMEDFKHAHQQVCASVSSESV 1170
AA+RP++E+L++E K+ S D+R L+++DF + +V SV+ ++
Sbjct: 366 AAYRPVQELLQEENKD-----------SVTNASPDLRPLSLDDFIQSKAKVSPSVAYDAT 414
Query: 1171 NMTELVQWNELYGEGGSRVKKALSY 1195
M EL +WNE YGEGG+R K +
Sbjct: 415 TMNELRKWNEQYGEGGTRTKSPFGF 439
>gb|AAF76434.1| Contains similarity to p60 katanin from Chlamydomonas reinhardtii
gb|AF205377 and contains an AAA domain PF|00004.
[Arabidopsis thaliana] gi|25405352|pir||G96537
hypothetical protein F2J10.1 [imported] - Arabidopsis
thaliana
Length = 627
Score = 421 bits (1083), Expect = e-116
Identities = 232/523 (44%), Positives = 335/523 (63%), Gaps = 33/523 (6%)
Query: 675 KSKLEKLPDNVVVI-GSHTHSDSRKEKSHAGGLLFTKFGSNQTALLDLAFPDSFGRLHDR 733
K +KL +V+I G + KE+ FT N + ++ L P
Sbjct: 136 KEMFDKLSGPIVMICGQNKIETGSKEREK-----FTMVLPNLSRVVKLPLPLKGLTEGFT 190
Query: 734 GKEVPKPNKTLTKLFPNKVTIHMPQDEALLASWKQQLDRDVETLKIKGNLHHLRTVLSRS 793
G+ + N+ + KLF N + +H P++E L +K+QL D R V+SRS
Sbjct: 191 GRGKSEENE-IYKLFTNVMRLHPPKEEDTLRLFKKQLGED------------RRIVISRS 237
Query: 794 GMESDGLESLCVKDLTLTNENSEKILGWALSHHLMQNP-EADADAKLVLSSESIQYGIGI 852
+ ++ L+ K + +EK +GWA +H+L P +L L ES++ I
Sbjct: 238 NI-NELLKKRPQKLVAFVFAEAEKAIGWAKNHYLASCPVPLVKGGRLSLPRESLEISIAR 296
Query: 853 FQAIQNESKSLKKSLKDVVTENEFEKRLLGDVIPPNDIGVTFDDIGALENVKDTLKELVM 912
+ +++ S ++LK++ ++E+E+ + V+ P +IGV F+DIGALE+VK L ELV+
Sbjct: 297 LRKLEDNSLKPSQNLKNIA-KDEYERNFVSAVVAPGEIGVKFEDIGALEDVKKALNELVI 355
Query: 913 LPLQRPELFCKGQLTKPCKGILLFGPPGTGKTMLAKAVATDAGANFINISMSSITSKWFG 972
LP++RPELF +G L +PCKGILLFGPPGTGKT+LAKA+AT+AGANFI+I+ S++TSKWFG
Sbjct: 356 LPMRRPELFARGNLLRPCKGILLFGPPGTGKTLLAKALATEAGANFISITGSTLTSKWFG 415
Query: 973 EGEKYVKAVFSLASKIAPSVIFVDEVDSMLGRRENPGEHEAMRKMKNEFMVNWDGLRTKD 1032
+ EK KA+FS A+K+AP +IFVDE+DS+LG R EHEA R+M+NEFM WDGLR+KD
Sbjct: 416 DAEKLTKALFSFATKLAPVIIFVDEIDSLLGARGGSSEHEATRRMRNEFMAAWDGLRSKD 475
Query: 1033 TERVIVLAATNRPYDLDEAVIRRLPRRLMVNLPDAPNRAKILKVILAKEDLSSDVDLGAI 1092
++R+++L ATNRP+DLD+AVIRRLPRR+ V+LPDA NR KILK+ L E+L SD +
Sbjct: 476 SQRILILGATNRPFDLDDAVIRRLPRRIYVDLPDAENRLKILKIFLTPENLESDFQFEKL 535
Query: 1093 ANMTDGYSGSDLKNLCVTAAHRPIKEILEKEKKELAAAVAEGRPAPALRGSDDIRSLNME 1152
A T+GYSGSDLKNLC+ AA+RP++E+L++E+K A + G +RSL+++
Sbjct: 536 AKETEGYSGSDLKNLCIAAAYRPVQELLQEEQKGARAEASPG-----------LRSLSLD 584
Query: 1153 DFKHAHQQVCASVSSESVNMTELVQWNELYGEGGSRVKKALSY 1195
DF + +V SV+ ++ M EL +WNE YGEGGSR K +
Sbjct: 585 DFIQSKAKVSPSVAYDATTMNELRKWNEQYGEGGSRTKSPFGF 627
>gb|AAB60775.1| Similar to Xenopus TER ATPase (gb|X54240). [Arabidopsis thaliana]
gi|25289957|pir||G96647 hypothetical protein F19K23.7
[imported] - Arabidopsis thaliana
Length = 330
Score = 392 bits (1007), Expect = e-107
Identities = 216/348 (62%), Positives = 250/348 (71%), Gaps = 55/348 (15%)
Query: 869 DVVTENEFEKRLLGDVIPPNDIGVTFDDIGALENVKDTLKELVMLPLQRPELFCKGQLTK 928
D+VTEN FE + D+IPP++IGVTFDDIGALENVKDTLKELVMLP Q PELFCKGQLTK
Sbjct: 17 DIVTENTFE---ISDIIPPSEIGVTFDDIGALENVKDTLKELVMLPFQWPELFCKGQLTK 73
Query: 929 --------------------PCKGILLFGPPGTGKTMLAKAVATDAGANFINISMSSITS 968
PC GILLFGP GTGKTMLAKAVAT+AGAN IN+SMS
Sbjct: 74 MLTLWIGGFLISLLLYFSTQPCNGILLFGPSGTGKTMLAKAVATEAGANLINMSMS---- 129
Query: 969 KWFGEGEKYVKAVFSLASKIAPSVIFVDEVDSMLGRRENPGEHEAMRKMKNEFMVNWDGL 1028
+WF EGEKYVKAVFSLASKI+PS+IF+DEV+SML H K KNEF++NWDGL
Sbjct: 130 RWFSEGEKYVKAVFSLASKISPSIIFLDEVESML--------HRYRLKTKNEFIINWDGL 181
Query: 1029 RTKDTERVIVLAATNRPYDLDEAVIRRLPRRLMVNLPDAPNRAKILKVILAKEDLSSDVD 1088
RT + ERV+VLAATNRP+DLDEAVIRRLP RLMV LPDA +R+KILKVIL+KEDLS D D
Sbjct: 182 RTNEKERVLVLAATNRPFDLDEAVIRRLPHRLMVGLPDARSRSKILKVILSKEDLSPDFD 241
Query: 1089 LGAIANMTDGYSGSDLKNLCVTAAHRPIKEILEKEKKELAAAVAEGRPAPALRGSDDIRS 1148
+ +A+MT+GYSG+DL KE AAVAEGR PA G D+R
Sbjct: 242 IDEVASMTNGYSGNDL--------------------KERDAAVAEGRVPPAGSGGSDLRV 281
Query: 1149 LNMEDFKHAHQQVCASVSSESVNMTELVQWNELYGEGGSRVKKALSYF 1196
L MEDF++A + V S+SS+SVNMT L QWNE YGEGGSR ++ S +
Sbjct: 282 LKMEDFRNALELVSMSISSKSVNMTALRQWNEDYGEGGSRRNESFSQY 329
Database: nr
Posted date: Jul 5, 2005 12:34 AM
Number of letters in database: 863,360,394
Number of sequences in database: 2,540,612
Lambda K H
0.313 0.131 0.374
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 2,065,823,551
Number of Sequences: 2540612
Number of extensions: 90506936
Number of successful extensions: 251641
Number of sequences better than 10.0: 4411
Number of HSP's better than 10.0 without gapping: 2608
Number of HSP's successfully gapped in prelim test: 1810
Number of HSP's that attempted gapping in prelim test: 240288
Number of HSP's gapped (non-prelim): 6764
length of query: 1196
length of database: 863,360,394
effective HSP length: 140
effective length of query: 1056
effective length of database: 507,674,714
effective search space: 536104497984
effective search space used: 536104497984
T: 11
A: 40
X1: 16 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (21.9 bits)
S2: 81 (35.8 bits)
Medicago: description of AC141863.4


