

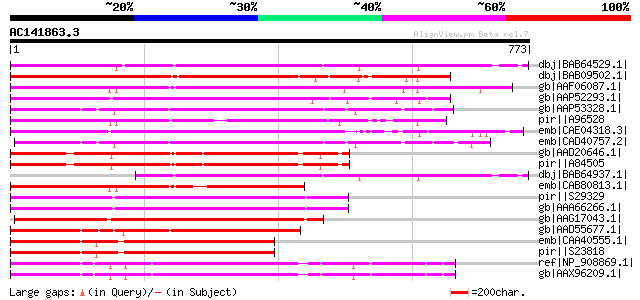
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC141863.3 - phase: 0
(773 letters)
Database: nr
2,540,612 sequences; 863,360,394 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
dbj|BAB64529.1| TdcA1-ORF2 [Daucus carota] 629 e-178
dbj|BAB09502.1| transposon protein-like [Arabidopsis thaliana] 570 e-161
gb|AAF06087.1| Similar to gi|4325351 T25H8.2 TNP2 protein homolo... 561 e-158
gb|AAP52293.1| putative transposable element [Oryza sativa (japo... 485 e-135
gb|AAP53328.1| putative TNP2-like transposon protein [Oryza sati... 476 e-132
pir||A96528 protein F27J15.14 [imported] - Arabidopsis thaliana ... 473 e-131
emb|CAE04318.3| OSJNBb0016D16.9 [Oryza sativa (japonica cultivar... 462 e-128
emb|CAD40757.2| OSJNBa0081G05.10 [Oryza sativa (japonica cultiva... 447 e-124
gb|AAD20646.1| putative TNP2-like transposon protein [Arabidopsi... 427 e-118
pir||A84505 probable TNP2-like transposon protein [imported] - A... 427 e-118
dbj|BAB64937.1| TdcA1-ORF1-ORF2 [Daucus carota] 426 e-117
emb|CAB80813.1| putative transposon protein [Arabidopsis thalian... 414 e-114
pir||S29329 hypothetical protein 1 - maize transposon En-1 gi|22... 401 e-110
gb|AAA66266.1| unknown protein 401 e-110
gb|AAG17043.1| transposase [Zea mays] 392 e-107
gb|AAD55677.1| putative transposase protein [Zea mays] 381 e-104
emb|CAA40555.1| TNP2 [Antirrhinum majus] 372 e-101
pir||S23818 hypothetical protein Tnp2 - garden snapdragon transp... 372 e-101
ref|NP_908869.1| B1151H08.16 [Oryza sativa (japonica cultivar-gr... 362 2e-98
gb|AAX96209.1| transposon protein, putative, CACTA, En/Spm sub-c... 362 3e-98
>dbj|BAB64529.1| TdcA1-ORF2 [Daucus carota]
Length = 812
Score = 629 bits (1621), Expect = e-178
Identities = 345/793 (43%), Positives = 467/793 (58%), Gaps = 40/793 (5%)
Query: 1 MCMTKPYLFLSCIVPGPDNPKAGIDVFLQPLIDDLKRLW-VGELTYDISRKQNFKLRAAL 59
MC + Y+FLS +VPGP NPK IDVFLQPLI +LK LW VG T+D S KQNF++RAAL
Sbjct: 35 MCSKEEYMFLSILVPGPRNPKQKIDVFLQPLISELKMLWEVGVETWDTSLKQNFQMRAAL 94
Query: 60 MWTINDFPAYGMLSGWSTHGRFACPHCMENTKAFTLESGGKSSWFDCHRPFLPHNHPFRR 119
MWTI+DFPAY MLSGW T G ACPHC A+ L+ GGK +WFD HR FLP NHPFR+
Sbjct: 95 MWTISDFPAYSMLSGWKTAGHLACPHCAHEHDAYNLKHGGKPTWFDNHRKFLPANHPFRK 154
Query: 120 LKNGFKKDERVFVGPPPKITSEEVWTRV--CDYPKVTDYG----RAVPIPRYGVDHHWTK 173
KN F K + V PPP T E+V + K+T+ G A I Y W K
Sbjct: 155 NKNWFTKGKVVSEFPPPIRTGEDVLQEIESLGLMKITELGSEEHNAKIIKTYNCG--WKK 212
Query: 174 RSIFWDLPYWKDNLLRHNLDVMHIEKNVFDNIFNTVMDVKGKTKDNEKARKDLQIYCKRK 233
RSIFWDLPYW+ +RHNLDVMHIEKNVF+NIFNT+M ++GKTKDN KAR D+ + C+R
Sbjct: 213 RSIFWDLPYWRTLSIRHNLDVMHIEKNVFENIFNTIMAIEGKTKDNAKARADIALLCRRP 272
Query: 234 DLELKPQPNGKLLKPKANYNLSAQEAKLVCQWLKDLRMPDGYSSNIGRCADVNTGRLHGM 293
+L + PKA Y+L + + VC+WL+DL+ PDGY SN+GRC D+ +L GM
Sbjct: 273 ELAIDESTRKY---PKACYSLDKKGKEAVCKWLQDLKFPDGYVSNMGRCIDMKKYKLFGM 329
Query: 294 KSHDSHIFMENFIPIAFSS-LPKHVLDPLIEISQFFKNLCASTLRVDELEKMEKNMPIII 352
KSHD H+FM+ +PIAF LP +V + + E+S FFK+L ++TL+V ++ ++E+ +P+I+
Sbjct: 330 KSHDCHVFMQRLMPIAFREFLPNNVWEAVTELSLFFKDLTSATLKVSDMCRLEEQIPVIL 389
Query: 353 CQLEQIFPPGFFDSMEHLPVHLAYEAILGGPVQYRWMYPFERFMGDSKRAVKNLAKVEGS 412
C+LE+IFPP FDSMEHL VHL YEA + GPVQYRWMYPFERF+ K +KN A+VEGS
Sbjct: 390 CKLERIFPPALFDSMEHLLVHLPYEARIAGPVQYRWMYPFERFLRHLKNNIKNKARVEGS 449
Query: 413 IVAIYTAKETTYFIGHYFVDRLLTPSN--TRNEVQVNDLRDPSTLSVFNLDGRHAGKVLQ 470
+ Y +E + F HYF + T RN+ S+F+ GR +G
Sbjct: 450 MCNAYLVEEASTFCSHYFEQHVQTKHRKVPRNDSSGGGENFEGNFSIFSHPGRASG-CAN 508
Query: 471 YWMVDQKEMRSAHVHVLINCAEVKPYLEEFIAFYAEFGEGSTVGSIHEY----FPAWFKQ 526
+D +E +AH +VL+NC EV PY E FI E + T I + F WFKQ
Sbjct: 509 VRYLDDREYMAAHNYVLLNCPEVAPYTEIFINMVRENNQSITDAEIDKCLESDFALWFKQ 568
Query: 527 RVYNAEPTDEVIRLRQLSQGPLQCANEFHTYFVNGYKFHTHSWTEGKKTINSGVYVKGVT 586
N I +R ++ GPL+ ++VNGYKFHT + + T NSGV +KG
Sbjct: 569 YAQNPSLVPNEI-VRDIASGPLRSVRSVPIFYVNGYKFHTRKYGANRSTFNSGVCIKGSN 627
Query: 587 -DGGEDDFYGVIKHIYELSYND---NKVVLFYCDWFDPSPR-WTKINKLCNTVDIRVDKK 641
+D++G+I I L Y K LF C+WFDP+P T+++ V++ KK
Sbjct: 628 YSETSNDYFGIIDEILILEYPRLPIKKTTLFKCEWFDPTPSVGTRVHLRFKMVEVNRKKK 687
Query: 642 YKEYDPFIMAHNVRQVYYVPYPPTLPRKRGWSVAIKTKPRGRIETEE--RNEEVAYQADD 699
Y+PFI+A QVY+ YP K W V K K R +E + ++ + YQ +
Sbjct: 688 LSVYEPFILASQAMQVYFCNYPSLRRDKMDWLVVCKIKARPLVELSQASQSHQEPYQDET 747
Query: 700 MTHVNEIIEVDQVTSWQDSQVEGDQVDPSILEMRNEVEDENEDVVDDNNEGDHQDNQVDD 759
++N I D T D++ IL++ + E++ D++ + + +Q D
Sbjct: 748 PENLNRIDIRDIPTHLNDNE--------GILDLDDGEGSPEEEIELDSDSEEGRSSQSD- 798
Query: 760 GDVDEYNYASDNN 772
D +NY SD+N
Sbjct: 799 ---DIHNYGSDSN 808
>dbj|BAB09502.1| transposon protein-like [Arabidopsis thaliana]
Length = 1089
Score = 570 bits (1470), Expect = e-161
Identities = 310/677 (45%), Positives = 415/677 (60%), Gaps = 33/677 (4%)
Query: 1 MCMTKPYLFLSCIVPGPDNPKAGIDVFLQPLIDDLKRLWV-GELTYDISRKQNFKLRAAL 59
MCM + ++FL+ +VPGP++PK +D+FLQPLI++LK LWV G YDIS KQNF L+ L
Sbjct: 369 MCMKQEFMFLTILVPGPNHPKRSLDIFLQPLIEELKDLWVNGVEAYDISTKQNFLLKVVL 428
Query: 60 MWTINDFPAYGMLSGWSTHGRFACPHCMENTKAFTLESGGKSSWFDCHRPFLPHNHPFRR 119
MWTI+DFPAYGMLSGW+THGR ACP+C + T AF L++G K+ WFDCHR FLP NH +R
Sbjct: 429 MWTISDFPAYGMLSGWTTHGRLACPYCSDQTGAFWLKNGRKTCWFDCHRCFLPVNHSYRG 488
Query: 120 LKNGFKKDERVFVGPPPKITSEEVWTRVCDYPKVTDY-GRAVPIPRYGVDHHWTKRSIFW 178
K FKK + V P +T EE++ VC PK D G + YG H+W K+SI W
Sbjct: 489 NKKDFKKGKVVEDSKPEILTGEELYNEVCCLPKTVDCGGNHGRLEGYGKTHNWHKQSILW 548
Query: 179 DLPYWKDNLLRHNLDVMHIEKNVFDNIFNTVMDVKGKTKDNEKARKDLQIYCKRKDLELK 238
+L YWKD LRHNLDVMHIEKNV DN T+++V+GKTKDN K+R DLQ C RKDL L
Sbjct: 549 ELSYWKDLKLRHNLDVMHIEKNVLDNFIKTLLNVQGKTKDNIKSRLDLQENCNRKDLHLT 608
Query: 239 PQPNGKLLKPKANYNLSAQEAKLVCQWL-KDLRMPDGYSSNIGRCADVNTGRLHGMKSHD 297
P+ GK PK + L ++ +WL KD++ DGYSS++ C D+ +L GMKSHD
Sbjct: 609 PE--GKAPIPK--FRLKPDAKEIFLRWLEKDVKFSDGYSSSLANCVDLPGRKLTGMKSHD 664
Query: 298 SHIFMENFIPIAFSSL-PKHVLDPLIEISQFFKNLCASTLRVDELEKMEKNMPIIICQLE 356
H+ M+ PIAF+ L K V D L +S FF++LCA T+ D +E ++KN+ +++C LE
Sbjct: 665 CHVLMQRLFPIAFAELMDKSVHDALSSVSLFFRDLCARTIHKDGIEMLQKNICMVLCNLE 724
Query: 357 QIFPPGFFDSMEHLPVHLAYEAILGGPVQYRWMYPFERFMGDSKRAVKNLAKVEGSIVAI 416
+IFPP FFD MEHL +HL +EA LGGPVQ+RWMY FER+MG K+ VKN AKVEGSIV
Sbjct: 725 KIFPPSFFDVMEHLLIHLPHEASLGGPVQFRWMYCFERYMGHLKKKVKNKAKVEGSIVEQ 784
Query: 417 YTAKETTYFIGHYFVDRLLTPSNTRNEVQVNDLRDPST------LSVFNLDGRHAGKVLQ 470
Y +E + F +YF DR + N + + T +F+ GR +GK +
Sbjct: 785 YINEEISTFCTYYF-DRHIKTKNRAGDRHYDGGNQEDTHEFDGVPDIFSQAGRDSGKESE 843
Query: 471 YWMVDQKEMRSAHVHVLINCAEVKPYLEEFIAFYAEFGEGSTVGSIHE----YFPAWFKQ 526
W+ D K+ AH ++L NC +++P+ F G + ++E + +W K
Sbjct: 844 IWLQD-KDYHIAHRYILRNCDQLRPFERLFDESLIAANPGISEKDLNELREKQYSSWLK- 901
Query: 527 RVYNAEPTDEVIRLRQLSQGPLQCANEFHTYFVNGYKFHTHSWTEGKKTINSGVYVKGVT 586
+ N+ P L + GP+ + YF GY FHT+ + KK N GV VKG T
Sbjct: 902 KYDNSYPE----WLLSIVHGPMVKVTCWPMYFCRGYIFHTYDHGKNKKNANYGVCVKGTT 957
Query: 587 DGG---EDDFYGVIKHIYELSYN---DNKVVLFYCDWFDPS-PRWTKINKLCNTVDIRVD 639
E DFYG+++ IYEL Y D KVV+F CDW+D R + NK +DI
Sbjct: 958 SSSSNEEADFYGILREIYELHYPGHVDLKVVVFKCDWYDSKVGRGIRRNK-SGIIDINAK 1016
Query: 640 KKYKEYDPFIMAHNVRQ 656
+ Y+EYDPF++A Q
Sbjct: 1017 RHYEEYDPFVLASQADQ 1033
>gb|AAF06087.1| Similar to gi|4325351 T25H8.2 TNP2 protein homolog from Arabidopsis
thaliana BAC gb|AF128394 gi|25518667|pir||F86485
hypothetical protein F28J9.11 [imported] - Arabidopsis
thaliana
Length = 1121
Score = 561 bits (1447), Expect = e-158
Identities = 320/778 (41%), Positives = 439/778 (56%), Gaps = 35/778 (4%)
Query: 1 MCMTKPYLFLSCIVPGPDNPKAGIDVFLQPLIDDLKRLW-VGELTYDISRKQNFKLRAAL 59
MCM YLFL+ + GP++P+A +DVFLQPLI++LK W G YD+S QNF L+A L
Sbjct: 348 MCMNTEYLFLTILNSGPNHPRASLDVFLQPLIEELKEFWSTGVDAYDVSLSQNFNLKAVL 407
Query: 60 MWTINDFPAYGMLSGWSTHGRFACPHCMENTKAFTLESGGKSSWFDCHRPFLPHNHPFRR 119
+WTI+DFPAY ML GW+THG+ +CP CME+TK+F L +G K+ WFDCHR FLPH HP RR
Sbjct: 408 LWTISDFPAYNMLLGWTTHGKLSCPVCMESTKSFYLPNGRKTCWFDCHRRFLPHGHPSRR 467
Query: 120 LKNGFKKDERVFVG-PPPKITSEEVWTR---VCDYPKVTDYG---RAVPIPRYGVDHHWT 172
+ F K PP +T E+V+ + PK D G + YG +H+W
Sbjct: 468 NRKDFLKGRDASSEYPPESLTGEQVYYERLASVNPPKTKDVGGNGHEKKMRGYGKEHNWH 527
Query: 173 KRSIFWDLPYWKDNLLRHNLDVMHIEKNVFDNIFNTVMDVKGKTKDNEKARKDLQIYCKR 232
K SI W+L YWKD LRHN+DVMH EKN DNI NT+M VKGK+KDN +R D++ +C R
Sbjct: 528 KESILWELSYWKDLNLRHNIDVMHTEKNFLDNIMNTLMRVKGKSKDNIMSRLDIEKFCSR 587
Query: 233 KDLELKPQPNGKLLKPKANYNLSAQEAKLVCQWLK-DLRMPDGYSSNIGRCADVNTGRLH 291
L + +GK P Y L+ + + + Q +K D+R PDGYSS++ C D++ G+
Sbjct: 588 PGLHI--DSSGKA--PFPAYTLTEEAKQSLLQCVKYDIRFPDGYSSDLASCVDLDNGKFS 643
Query: 292 GMKSHDSHIFMENFIPIAFSSL-PKHVLDPLIEISQFFKNLCASTLRVDELEKMEKNMPI 350
GMKSHD H+FME +P F+ L ++V L I FF++LC+ TL+ ++ +++N+ +
Sbjct: 644 GMKSHDCHVFMERLLPFIFAELLDRNVHLALSGIGAFFRDLCSRTLQTSRVQILKQNIVL 703
Query: 351 IICQLEQIFPPGFFDSMEHLPVHLAYEAILGGPVQYRWMYPFERFMGDSKRAVKNLAKVE 410
IIC E+IFPP FFD MEHLP+HL YEA LGGPVQYRWMYPFERF K KN
Sbjct: 704 IICNFEKIFPPSFFDVMEHLPIHLPYEAELGGPVQYRWMYPFERFFKKLKGKAKNKRYAA 763
Query: 411 GSIVAIYTAKETTYFIGHYFVDRLLTPSN-TR-NEVQVNDLRDPSTLSVFNLDGRHAGKV 468
GSIV Y E YF HYF D + T S TR NE +V ++F GR +G++
Sbjct: 764 GSIVESYINDEIAYFSEHYFADHIQTKSRLTRFNEGEVPVYHVSGVPNIFMHVGRPSGEM 823
Query: 469 LQYWMVDQKEMRSAHVHVLINCAEVKPY---LEEFI-AFYAEFGEGSTVGSIHEYFPAWF 524
W++ +K+ +SAH +VL NC KP+ E+++ A Y E E + W
Sbjct: 824 HVDWLL-EKDYQSAHAYVLRNCDYFKPFESMFEDYLSAKYPCLPEKELYARRAEEYHLWV 882
Query: 525 KQRVYNAEPTDEV-IRLRQLSQGPLQCANEFHTYFVNGYKFHTHSWTEGKKTINSGVYVK 583
K+ V T ++++ QGPL + YF GY FHT + G+KT N G+ VK
Sbjct: 883 KEYVTYWNTTSPFPTWVQEIVQGPLNKVKTWPMYFTRGYLFHTQNHGAGRKTCNYGICVK 942
Query: 584 G---VTDGGEDDFYGVIKHIYELSYN---DNKVVLFYCDWFDPSPRWTKINKLCNTVDIR 637
G E DFYG + I EL Y + ++ LF C W+DP VDI
Sbjct: 943 GENYADSSDEADFYGTLTDIIELEYEGIVNLRITLFKCKWYDPKIGRGTRRSHSGVVDIL 1002
Query: 638 VDKKYKEYDPFIMAHNVRQVYYVPYPPTLPRKRGWSVAIKTKPRGRIETE-ERNEEVAYQ 696
+KY +Y+PFI+ QV Y+PYP T K W +K PRG I E E N+ Q
Sbjct: 1003 STRKYNKYEPFILGSQADQVCYIPYPYTKKPKNIWLNVLKVNPRGNISGEYENNDPTLLQ 1062
Query: 697 ADD-----MTHVNEIIEVDQVTSWQDSQVEGDQVDPSIL-EMRNEVEDENEDVVDDNN 748
++ +T + +++ V + ++ D D E R + +ED V+D +
Sbjct: 1063 TENDDDVLLTTIEDLVLETPVANLNPIILDYDVGDAEPEDEFRCNLSSLDEDEVEDED 1120
>gb|AAP52293.1| putative transposable element [Oryza sativa (japonica
cultivar-group)] gi|37531408|ref|NP_920006.1| putative
transposable element [Oryza sativa (japonica
cultivar-group)] gi|22655757|gb|AAN04174.1| Putative
transposable element [Oryza sativa (japonica
cultivar-group)] gi|21672010|gb|AAM74372.1| Putative
transposable element [Oryza sativa (japonica
cultivar-group)]
Length = 992
Score = 485 bits (1249), Expect = e-135
Identities = 258/672 (38%), Positives = 394/672 (58%), Gaps = 22/672 (3%)
Query: 1 MCMTKPYLFLSCIVPGPDNPKAGIDVFLQPLIDDLKRLWV-GELTYDISRKQNFKLRAAL 59
MC + LS ++PGP++P ++VF++PL+D+L LW+ G +TYD KQNF ++AA+
Sbjct: 265 MCTKDNNILLSLVIPGPEHPGKNLNVFMEPLVDELLDLWMNGAVTYDRFLKQNFNMKAAV 324
Query: 60 MWTINDFPAYGMLSGWSTHGRFACPHCMENTKAFTLESGGKSSWFDCHRPFLPHNHPFRR 119
+WTI+DFPAYG++S WSTHG+ ACP C TKAF L+ G K+ WFDCHR FLP NH FRR
Sbjct: 325 VWTIHDFPAYGLVSCWSTHGKLACPICGGGTKAFQLKHGHKACWFDCHRRFLPSNHEFRR 384
Query: 120 LKNGFKKDERVFVGPPPKITSEEVWTRVCDYPKVTDYGRAVPIPRYGVDHHWTKRSIFWD 179
F+K +V PP ++T +E++ ++ V D +G H+WT S W
Sbjct: 385 SLKCFRKKTKVLDPPPERLTGDEIYKQLMSL--VPDRTGKHTFEGFGETHNWTGISGLWK 442
Query: 180 LPYWKDNLLRHNLDVMHIEKNVFDNIFNTVMDVKGKTKDNEKARKDLQIYCKRKDLELKP 239
LPY+K +LRHN+DVMH EKNV + + +T +D++ KTKDN KAR D+ C R L +
Sbjct: 443 LPYFKKLMLRHNIDVMHNEKNVAEAVLSTCLDIQDKTKDNVKARLDMADICDRPTLNMTK 502
Query: 240 QPNGKLLKPKANYNLSAQEAKLVCQWLKDLRMPDGYSSNIGRCADVNTGRLHGMKSHDSH 299
NG+ KP+A Y ++ ++ + +W KDL+ PD +++N+ + +++ + G+KSHD H
Sbjct: 503 STNGRWQKPRAKYCVTKEDKLTIMKWFKDLKFPDRFAANLKKAVNLSQCKFIGLKSHDFH 562
Query: 300 IFMENFIPIAFSS-LPKHVLDPLIEISQFFKNLCASTLRVDELEKMEKNMPIIICQLEQI 358
IF+E +P+A +P+ L E+S F++ LCA + +++ ++EK + +++C+LE++
Sbjct: 563 IFIERLLPVALRGFIPEKEWKDLSELSFFYRQLCAKEIDKEQMHRLEKEIVVLLCKLEKM 622
Query: 359 FPPGFFDSMEHLPVHLAYEAILGGPVQYRWMYPFERFMGDSKRAVKNLAKVEGSIVAIYT 418
FP GFF+SM+HL +HL YEA +GGPVQ+RWMYPFER M + V+N A+VEGSIV Y
Sbjct: 623 FPLGFFNSMQHLILHLPYEARVGGPVQFRWMYPFERCMCKLRLKVRNKARVEGSIVEAYI 682
Query: 419 AKETTYFIGHYFVDRLLTPSNTRNEVQVNDL---RDPSTLSVFNLDGRHAGKVLQYWMVD 475
+ETT F+ YF + T N N D TL VF G G+ + +
Sbjct: 683 VEETTNFMSIYFNKDIRTTWNKPNRYNNGGTVVQNDGCTLDVFQEQGMLHGRGVPRDLSP 742
Query: 476 QKEMRSAHVHVLINCAEVKPYLEEFI----AFYAEFGEGSTVGSIHEYFPAWFKQRVYNA 531
Q E+ A ++VL NC+ V + E+F A + + E F WFK N
Sbjct: 743 Q-ELNVAMLYVLTNCSMVDKFQEKFSEEKHAMHNNLSVEGLDQLMREEFVDWFKNACRND 801
Query: 532 EPTDEVIRLRQLSQGPLQCANEFHTYFVNGYKFHTHSWTEGKK---TINSGVYVKGVT-D 587
D+ L L+ G +++Y +NGY+F + + + ++ T+NSGV + T D
Sbjct: 802 PSADD--DLWNLATGCKPRVLSYNSYDINGYRFRSEKYEKSRRRLTTVNSGVCLSSFTSD 859
Query: 588 GGEDDFYGVIKHIYELSYNDNK---VVLFYCDWFDPSPRWTKINKLCNTVDIRVDKKYKE 644
+ D+YGVI+ I +L+++ + +VL C WFDP R + ++I+ K
Sbjct: 860 DQQLDYYGVIEDIIKLTFDAGRKIEIVLLQCRWFDPI-RGKRSTPTLGMIEIKSTSKLTN 918
Query: 645 YDPFIMAHNVRQ 656
++PF MAH Q
Sbjct: 919 FEPFAMAHQATQ 930
>gb|AAP53328.1| putative TNP2-like transposon protein [Oryza sativa (japonica
cultivar-group)] gi|37533478|ref|NP_921041.1| putative
TNP2-like transposon protein [Oryza sativa (japonica
cultivar-group)] gi|20279447|gb|AAM18727.1| putative
TNP2-like transposon protein [Oryza sativa (japonica
cultivar-group)]
Length = 1571
Score = 476 bits (1224), Expect = e-132
Identities = 272/677 (40%), Positives = 401/677 (59%), Gaps = 33/677 (4%)
Query: 1 MCMTKPYLFLSCIVPGPDNPKAGIDVFLQPLIDDLKRLWV-GELTYDISRKQNFKLRAAL 59
MC+ + + LS ++PGPD P IDV+LQPLI++L LW G TYD S K NFKLRAAL
Sbjct: 370 MCIKQQNVMLSMLLPGPDGPGDAIDVYLQPLIEELIELWEDGVDTYDSSTKTNFKLRAAL 429
Query: 60 MWTINDFPAYGMLSGWSTHGRFACPHCMENTKAFTLESGGKSSWFDCHRPFLPHNHPFRR 119
+WT++DFPAYG +SG ST G+ ACP C + T + L+ G K + HR FL +H +R
Sbjct: 430 LWTVHDFPAYGNISGHSTKGKLACPVCHKETNSMWLKKGQKFCYMG-HRCFLRRSHRWRN 488
Query: 120 LKNGFK--KDERVFVGPPPKITSEEVWTRVCDYPKVT---DYGRAVPIPRYGVDHHWTKR 174
+ F K+ R GPP +++ ++V +V D + D + + R +W KR
Sbjct: 489 DRTSFDGTKESR---GPPMELSGDDVLKQVQDLQGIILSKDMSKKTKVSRSDRGDNWKKR 545
Query: 175 SIFWDLPYWKDNLLRHNLDVMHIEKNVFDNIFNTVMDVKGKTKDNEKARKDLQIYCKRKD 234
SIF++LPY+K LLRHNLDVMHIEKN+ D+I T+M+VKGKTKDN R DL++ R D
Sbjct: 546 SIFFELPYFKTLLLRHNLDVMHIEKNICDSIIGTLMNVKGKTKDNINTRSDLKLMNLRPD 605
Query: 235 LELKPQPNGKLLKPKANYNLSAQEAKLVCQWLKDLRMPDGYSSNIGRCADVNTGRLHGMK 294
L GK++ P A Y LS + K + + K+L++PDG+SSNI RC ++ ++ G+K
Sbjct: 606 LH-PVDDGGKVVLPPAPYTLSPDQKKALLSFFKELKVPDGFSSNISRCVNMKERKMSGLK 664
Query: 295 SHDSHIFMENFIPIAFSSL-PKHVLDPLIEISQFFKNLCASTLRVDELEKMEKNMPIIIC 353
SHD H+ + + +P+A L P+++ +PL+E+SQFFK L + L V++LE+M+ +P+I+C
Sbjct: 665 SHDCHVILNHILPLALRGLLPENIYEPLVELSQFFKKLNSKALSVEQLEEMDAQIPVILC 724
Query: 354 QLEQIFPPGFFDSMEHLPVHLAYEAILGGPVQYRWMYPFERFMGDSKRAVKNLAKVEGSI 413
+LE+ FPP FFD M HLPVHLA EA++GGP YRWMY FER + K V+N+A+ EGSI
Sbjct: 725 KLEKEFPPSFFDVMMHLPVHLAREALIGGPTMYRWMYLFERQIHYLKSLVRNMARPEGSI 784
Query: 414 VAIYTAKETTYFIGHYFVDRLLTPSNTRNEVQVNDLRDPSTLSVFNLDGRHAGKVLQYWM 473
Y A E Y +D + T N + + + LS+F+ GR G +
Sbjct: 785 AEGYIADEFMTLSSRY-LDDVETKHNRPGRIHDTSIGNKFNLSIFSCAGRPIGS-RKTRD 842
Query: 474 VDQKEMRSAHVHVLINCAEVKPYLEEFIAFYAEFGEGSTV----GSIHEYFPAWFKQRVY 529
+D E AH+++L NC EV+ Y+ E Y EGS++ + ++ F WFK+++Y
Sbjct: 843 LDMLESEQAHIYILRNCDEVQAYISE----YCNSTEGSSMVQFTSTWNKKFINWFKEKIY 898
Query: 530 NAEPTD--EVIR-LRQLSQGPLQCANEFHTYFVNGYKFHTHSWTEGKKTINSGVYVKGVT 586
D EV+ L LS+GPL+ F Y +NG+++ T + T NSGV V
Sbjct: 899 QQHQHDKSEVMEDLLSLSRGPLKNITCFTGYDMNGFRYRTERRDSRRCTQNSGVMVL--- 955
Query: 587 DGGEDDFYGVIKHIYELSY-NDNKVVLFYCDWFD--PSPRWTKINKLCNTVDIRVDKKYK 643
G + ++YGV+ I E+ Y +V LF C+W D R +I+K + + + +
Sbjct: 956 -GDDVEYYGVLTEILEIQYLGGRRVPLFRCNWVDVFNKDRGIRIDKY-GLTSVNLQRLLQ 1013
Query: 644 EYDPFIMAHNVRQVYYV 660
+PF++A QV++V
Sbjct: 1014 TDEPFVLASQAAQVFFV 1030
>pir||A96528 protein F27J15.14 [imported] - Arabidopsis thaliana
gi|7770347|gb|AAF69717.1| F27J15.14 [Arabidopsis
thaliana]
Length = 1526
Score = 473 bits (1217), Expect = e-131
Identities = 268/672 (39%), Positives = 379/672 (55%), Gaps = 62/672 (9%)
Query: 1 MCMTKPYLFLSCIVPGPDNPKAGIDVFLQPLIDDLKRLW-VGELTYDISRKQNFKLRAAL 59
MCM YLFL+ + GP++P+ +DVFLQPLI++LK LW G YD+S QNF L+A L
Sbjct: 366 MCMNIEYLFLTILNSGPNHPRGSLDVFLQPLIEELKELWSTGIDAYDVSLNQNFNLKAVL 425
Query: 60 MWTINDFPAYGMLSGWSTHGRFACPHCMENTKAFTLESGGKSSWFDCHRPFLPHNHPFRR 119
+WTI+DFPAY MLSGW+THG+ +CP CME+T +F L +G K+ WFDCHR FL H HP R+
Sbjct: 426 LWTISDFPAYSMLSGWTTHGKLSCPICMESTNSFYLPNGRKTCWFDCHRRFLSHGHPLRK 485
Query: 120 LKNGFKKDERVFVG-PPPKITSEEVWTRVC---DYPKVTDYG---RAVPIPRYGVDHHWT 172
K F+K + PP +T E+++ + P+ D G +P YG +H+W
Sbjct: 486 NKKDFRKGKDASTEYPPESLTGEQIYYERLSGVNPPRTKDVGGNGHEKKMPGYGKEHNWH 545
Query: 173 KRSIFWDLPYWKDNLLRHNLDVMHIEKNVFDNIFNTVMDVKGKTKDNEKARKDLQIYCKR 232
K SI W+LPYWKD LRH +DVMH EKN DNI NT+M VKGK+KDN AR D++ +C R
Sbjct: 546 KESILWELPYWKDLNLRHCIDVMHTEKNFLDNIMNTLMSVKGKSKDNIMARMDIERFCSR 605
Query: 233 KDLELKPQPNGKLLKPKANYNLSAQEAKLVCQWLKD-LRMPDGYSSNIGRCADVNTGRLH 291
DL + + K P Y L+ + + Q +K ++ PDGYSS++ C D+ G+L
Sbjct: 606 PDLHI----DSKRKAPFPAYTLTNEAKMSLLQCVKHAIKFPDGYSSDLSSCVDMENGKLS 661
Query: 292 GMKSHDSHIFMENFIPIAFSSLPKHVLDPLIEISQFFKNLCASTLRVDELEKMEKNMPII 351
+ + H+ + + FF++LC+ TL+ ++ +++N+ +I
Sbjct: 662 ELLDRNVHLALSG-------------------VGAFFRDLCSRTLQKSHVQILKQNIVLI 702
Query: 352 ICQLEQIFPPGFFDSMEHLPVHLAYEAILGGPVQYRWMYPFERFMGDSKRAVKNLAKVEG 411
IC LE+IFPP FFD MEHLP+HL YEA LGGPVQYRWMYPFERF K KN G
Sbjct: 703 ICNLEKIFPPSFFDVMEHLPIHLPYEAELGGPVQYRWMYPFERFFKKLKGKAKNKRYAAG 762
Query: 412 SIVAIYTAKETTYFIGHYFVDRLLTPSN-TR-NEVQVNDLRDPSTLSVFNLDGRHAGKVL 469
SIV Y E +YF HYF D + T S TR NE +V P ++F+ GR +G++
Sbjct: 763 SIVESYINDEISYFSEHYFADDIQTKSRLTRFNEGEVPVYHVPGVPTIFSSVGRPSGEIR 822
Query: 470 QYWMVDQKEMRSAHVHVLINC---AEVKPYLEEFIAF-YAEFGEGSTVGSIHEYFPAWFK 525
+ W+ +++ + AH +V+ NC ++ E+F++ Y E +E + W
Sbjct: 823 EVWL-SEEDYQCAHGYVIRNCDYFQVIESMFEDFLSIKYPGLNEKELFVKRNEEYHVWVT 881
Query: 526 Q-RVYNAEPTDEVIRLRQLSQGPLQCANEFHTYFVNGYKFHTHSWTEGKKTINSGVYVKG 584
N PT ++++ GPL ++ T+ ++G G+KT N GV VKG
Sbjct: 882 YWNSSNPFPT----WVQEIVNGPL---HKVKTWPIHG---------AGRKTCNYGVCVKG 925
Query: 585 --VTDGGE-DDFYGVIKHIYELSYN---DNKVVLFYCDWFDPSPRWTKINKLCNTVDIRV 638
TD + DFYG + I EL Y K+ LF C W+DP VD+
Sbjct: 926 ENYTDASDAADFYGNLTDIIELEYEGVVSLKITLFKCSWYDPKLGRGTRRSNSGVVDVLS 985
Query: 639 DKKYKEYDPFIM 650
+KY +Y+PFI+
Sbjct: 986 SRKYNKYEPFIL 997
>emb|CAE04318.3| OSJNBb0016D16.9 [Oryza sativa (japonica cultivar-group)]
gi|50928325|ref|XP_473690.1| OSJNBb0016D16.9 [Oryza
sativa (japonica cultivar-group)]
Length = 1097
Score = 462 bits (1190), Expect = e-128
Identities = 281/787 (35%), Positives = 412/787 (51%), Gaps = 74/787 (9%)
Query: 1 MCMTKPYLFLSCIVPGPDNPKAGIDVFLQPLIDDLKRLWV-GELTYDISRKQNFKLRAAL 59
+CM K +FLS I+PGPD P I ++++PL+DDL W G TYD + KQNF +R +
Sbjct: 362 VCMQKHNMFLSLIIPGPDYPGKKISMYMEPLVDDLLHAWEHGVQTYDRATKQNFNMRVSY 421
Query: 60 MWTINDFPAYGMLSGWSTHGRFACPHCMENTKAFTLESGGKSSWFDCHRPFLPHNHPFRR 119
+++ +D PAYG+ GW HG+ CP CME K L+ GGK S+FDCHR FLPH H FR
Sbjct: 422 LFSFHDLPAYGIFCGWCVHGKMPCPVCMEVLKGRRLKFGGKYSFFDCHRQFLPHGHIFRN 481
Query: 120 LKNGFKKDERVFVGPPPKITSEEVWTRVCDYPKVTDYGRAVPIPRYGVDHHWTKRSIFWD 179
N F + V PP + +EEV R+ ++ YG DH+WT W
Sbjct: 482 DPNSFLANTTVTTEPPHRFKTEEVHVRL---QRLQPAANGEGFEGYGEDHNWTHIPGLWR 538
Query: 180 LPYWKDNLLRHNLDVMHIEKNVFDNIFNTVMDVKGKTKDNEKARKDLQIYCKRKDLEL-K 238
LPY+ +L HN+DVMH EKNV + IFNT D+ KTKDN KAR D I C R DL + +
Sbjct: 539 LPYFHKLVLPHNIDVMHNEKNVAEAIFNTCFDIPEKTKDNVKARLDQAILCNRPDLNMIR 598
Query: 239 PQPNGKLLKPKANYNLSAQEAKLVCQWLKDLRMPDGYSSNIGRCADVNTGRLHGMKSHDS 298
Q +G+ LKP+A++ L+ + K + +W + L+ PDGY SN+ R + R++G+KSHD
Sbjct: 599 RQTSGQWLKPRADFCLNRTQKKEILEWFQTLKFPDGYGSNLRRGVNFKKMRINGLKSHDY 658
Query: 299 HIFMENFIPIAFSS-LPKHVLDPLIEISQFFKNLCASTLRVDELEKMEKNMPIIICQLEQ 357
HI ME +P+ F ++ + + E+S F++ LCA + ELE ME +P+++C+LE
Sbjct: 659 HIMMERLLPVMFRGYFQPYLWEVIAELSFFYRKLCAKEVDPIELESMELQVPVLLCKLEM 718
Query: 358 IFPPGFFDSMEHLPVHLAYEAILGGPVQYRWMYPFERFMGDSKRAVKNLAKVEGSIVAIY 417
IFP GFF+ M+HL +HL YEA +GGPVQYRWMYP ER D K VKN A+VE SI Y
Sbjct: 719 IFPSGFFNPMQHLILHLPYEARMGGPVQYRWMYPGERDQKDLKSKVKNRARVEASIAEAY 778
Query: 418 TAKETTYFIGHYFVDRLLTPSNTRNEVQVNDLRDPSTLSVFNLDGRHAGKVLQYWMVDQK 477
E F YF D++ T N V +LS+F++ G + + + +
Sbjct: 779 ILDEIANFTTIYFADQVYTVHNPVPRYNVTVESRECSLSLFSIKGDSTSRGVTRHLT-EV 837
Query: 478 EMRSAHVHVLINCAEVKPYLEEFIAFYAEFGEGSTVGSIHEYFPAWFKQRVYNAEPTDEV 537
E +A ++VL N EV Y+ +F +HE W ++ EPT +
Sbjct: 838 EWEAAMLYVLTNLPEVDDYVGKF---------------LHE---EWSRR----GEPTRQQ 875
Query: 538 IRLRQLSQGPLQCANEFHTYFVNGYKFHTHSWTEGKKTINSGVYVKGVTDGGEDDFYGVI 597
+ L G F T+F + + TI G + D+YG I
Sbjct: 876 -KENLLRNGARNGCPNFVTWF----------YQQCLVTIGQG------ENSDITDYYGYI 918
Query: 598 KHIYELSYNDNK---VVLFYCDWFDPSPRWTKINKLCNTVDIRVDKKYKEYDPFIMAHNV 654
K I ELS++ + +VLF C WFDP+ T+ V++ Y+PF++AH
Sbjct: 919 KEIIELSFHGDSELTLVLFNCHWFDPAQ--TRYTPQYGLVEVAHSSTLAVYEPFVVAHQA 976
Query: 655 RQVYYVPYP-PTLPRKRGWSVAIKTKPRGRIETE--------ERNEEVAYQADD---MTH 702
QVYY+PYP ++P W V K +P G+I N+ V Y +D T
Sbjct: 977 TQVYYIPYPCKSVPALIDWWVVYKVQPTGKIAAPVDKDYDFLPNNDNVHYFQEDGLQGTF 1036
Query: 703 VNEIIE----VDQVTSWQDSQVEGDQVDPSILEMRNEVEDENEDVVDDNNEGDHQDNQVD 758
V +++E V + T+ + D+ D +L N ++ D+++ + +++ +
Sbjct: 1037 VVDLMEGLHNVSEATTRDAEGIVNDK-DLQLLNGSNVLQQ------DESDSSEFDEDEEE 1089
Query: 759 DGDVDEY 765
+DEY
Sbjct: 1090 GPIIDEY 1096
>emb|CAD40757.2| OSJNBa0081G05.10 [Oryza sativa (japonica cultivar-group)]
gi|50923505|ref|XP_472113.1| OSJNBa0081G05.10 [Oryza
sativa (japonica cultivar-group)]
Length = 1490
Score = 447 bits (1150), Expect = e-124
Identities = 274/732 (37%), Positives = 409/732 (55%), Gaps = 44/732 (6%)
Query: 8 LFLSCIVPGPDNPKAGIDVFLQPLIDDLKRLWV-GELTYDISRKQNFKLRAALMWTINDF 66
+ LS ++PGPD P IDV+LQPLI++L LW G TYD S K NFKL AAL+WT++DF
Sbjct: 339 VMLSMLLPGPDGPGDAIDVYLQPLIEELIELWEDGVDTYDSSTKTNFKLHAALLWTVHDF 398
Query: 67 PAYGMLSGWSTHGRFACPHCMENTKAFTLESGGKSSWFDCHRPFLPHNHPFRRLKNGFK- 125
PAYG +SG ST G+ ACP C + T + L+ G K + HR FL +H +R + F
Sbjct: 399 PAYGNISGHSTKGKLACPVCHKETNSMWLKKGRKFCYMG-HRRFLRKSHRWRNDRTSFDG 457
Query: 126 -KDERVFVGPPPKITSEEVWTRVCDYPKVT---DYGRAVPIPRYGVDHHWTKRSIFWDLP 181
K+ R GP +++ ++V +V D + D + + R +W KRSIF++LP
Sbjct: 458 TKESR---GPSKELSGDDVLKQVQDLQGIILSKDMSKKTKVSRSDRGDNWKKRSIFFELP 514
Query: 182 YWKDNLLRHNLDVMHIEKNVFDNIFNTVMDVKGKTKDNEKARKDLQIYCKRKDLELKPQP 241
Y+K LLRHNLDVMHI KN+ D+I T+M+VKGKTKDN R DL++ R DL
Sbjct: 515 YFKTLLLRHNLDVMHIGKNICDSIIGTLMNVKGKTKDNINTRSDLKLMNLRPDLH-PVDD 573
Query: 242 NGKLLKPKANYNLSAQEAKLVCQWLKDLRMPDGYSSNIGRCADVNTGRLHGMKSHDSHIF 301
GK++ P A Y L + K + + K+L++ DG+SSNI RC ++ ++ G+KSHD H+
Sbjct: 574 GGKVVLPPAPYTLFPDQKKALLSFFKELKVLDGFSSNISRCVNMKERKMSGLKSHDCHVI 633
Query: 302 MENFIPIAFSSL-PKHVLDPLIEISQFFKNLCASTLRVDELEKMEKNMPIIICQLEQIFP 360
+ + +P+A L P+++ +P +E+SQFFK L + L V++LE+M+ +P+I+C+LE+ FP
Sbjct: 634 LNHILPLALRGLLPENIYEPHVELSQFFKKLNSKALSVEQLEEMDAQIPVILCKLEKEFP 693
Query: 361 PGFFDSMEHLPVHLAYEAILGGPVQYRWMYPFERFMGDSKRAVKNLAKVEGSIVAIYTAK 420
P FFD M HLPVHLA EA++GGP YRWMY FER + K V+N+A+ EGSI Y A
Sbjct: 694 PSFFDVMMHLPVHLAREALIGGPTIYRWMYLFERQIHCLKSLVRNMARPEGSIAEGYIAD 753
Query: 421 ETTYFIGHYFVDRLLTPSNTRNEVQVNDLRDPSTLSVFNLDGRHAGKVLQYWMVDQKEMR 480
E Y +D + T N + + + LS+F+ GR G + +D E
Sbjct: 754 EFMTLNSRY-LDDVETKHNRPGRIHDTSIGNKFNLSIFSCAGRPIGS-RKTRDLDMLESE 811
Query: 481 SAHVHVLINCAEVKPYLEEFIAFYAEFGEGSTV----GSIHEYFPAWFKQRVYNAEPTD- 535
AH+++L NC EV+ Y+ E Y EGS++ + ++ F WFK+++Y D
Sbjct: 812 KAHIYILRNCDEVQAYISE----YCNSTEGSSMVQFTSTWNKKFINWFKEKIYQQHQHDK 867
Query: 536 -EVIR-LRQLSQGPLQCANEFHTYFVNGYKFHTHSWTEGKKTINSGVYVKGVTDGGEDDF 593
EV+ L LS+G L+ F Y +NG+++ T + T NSGV V G + ++
Sbjct: 868 SEVMEDLLSLSRGLLKNITCFTGYDMNGFRYRTERRDSHRCTQNSGVMVL----GDDVEY 923
Query: 594 YGVIKHIYELSY-NDNKVVLFYCDWFD--PSPRWTKINKLCNTVDIRVDKKYKEYDPFIM 650
YGV+ I E+ Y +V LF C+W D R I+K + + + + + F++
Sbjct: 924 YGVLTEILEIQYLGGRRVPLFRCNWVDVFNKDRGILIDKY-GLTSVNLQRLLQTDELFVL 982
Query: 651 AHNVRQVYYVPYPPTLPRKRGWSVAIKTKPRGRIET-------EERNEEVAYQADDMTHV 703
A QV +V + +GW + K +PR + RN A Q ++ +
Sbjct: 983 ASQAAQVSFVKD----NQIKGWHLIEKMQPRDTYQVVVMISGRYGRNRVSALQNNNSKFI 1038
Query: 704 NEIIEVDQVTSW 715
+ + QV W
Sbjct: 1039 SPGLLQQQVARW 1050
>gb|AAD20646.1| putative TNP2-like transposon protein [Arabidopsis thaliana]
Length = 1040
Score = 427 bits (1099), Expect = e-118
Identities = 227/524 (43%), Positives = 320/524 (60%), Gaps = 44/524 (8%)
Query: 1 MCMTKPYLFLSCIVPGPDNPKAGIDVFLQPLIDDLKRLWV-GELTYDISRKQNFKLRAAL 59
MCM ++FLS +VPGP +PK +DVFLQPLI +LK+LW G T+D S KQNF +RA L
Sbjct: 541 MCMQSEFMFLSILVPGPKHPKRALDVFLQPLIHELKKLWYEGVHTFDYSSKQNFNMRAML 600
Query: 60 MWTINDFPAYGMLSGWSTHGRFACPHCMENTKAFTLESGGKSSWFDCHRPFLPHNHPFRR 119
MWTI+DFPAY MLSGW+ HGR P K WF+CHR FLP +HP+RR
Sbjct: 601 MWTISDFPAYRMLSGWTMHGRLYVP-----------TKDRKPCWFNCHRRFLPLHHPYRR 649
Query: 120 LKNGFKKDERVFVGPPPKITSEEVWTRVCDY------PKVTDYGRAVPIP-RYGVDHHWT 172
K F+K++ V + PP ++ +++ ++ Y + ++ +P YG H+W
Sbjct: 650 NKKLFRKNKIVRLLPPSYVSGTDLFEQIDYYGAQETCKRGGNWHTTANMPDEYGSTHNWY 709
Query: 173 KRSIFWDLPYWKDNLLRHNLDVMHIEKNVFDNIFNTVMDVKGKTKDNEKARKDLQIYCKR 232
K+SIFW LPYWKD LLRHNL VMHIEKN FDNI NT+++++GKTKD K+R DL C +
Sbjct: 710 KQSIFWQLPYWKDLLLRHNLYVMHIEKNFFDNIMNTLLNLQGKTKDTLKSRLDLAEICSK 769
Query: 233 KDLELKPQPNGKLLKPKANYNLSAQEAKLVCQWL-KDLRMPDGYSSNIGRCADVNTGRLH 291
+L + +GKL P + + LS + K + +W+ +++ PDGY S RC + +
Sbjct: 770 PELHVTR--DGKL--PVSKFRLSNEAKKALFEWVVAEVKFPDGYVSKFSRCVEQGQ-KFS 824
Query: 292 GMKSHDSHIFMENFIP-IAFSSLPKHVLDPLIEISQFFKNLCASTLRVDELEKMEKNMPI 350
GMKSHD H+FM+ +P + LP ++ + + I FFK+LC TL D +E+++KN+P+
Sbjct: 825 GMKSHDCHVFMQRLLPFVLVELLPTNIHEAIAGIGVFFKDLCTRTLTTDTIEQLDKNIPV 884
Query: 351 IICQLEQIFPPGFFDSMEHLPVHLAYEAILGGPVQYRWMYPFERFMGDSKRAVKNLAKVE 410
++C LE++F P FFD MEHLP+HL +E LGGPVQ+RWMYPFERFM K KNLA+VE
Sbjct: 885 LLCNLEKLFSPAFFDVMEHLPIHLPHEVALGGPVQFRWMYPFERFMKSLKGKAKNLARVE 944
Query: 411 GSIVAIYTAKETTYFIGHYFVDRLLTPSNTRNEVQVNDLRDPSTLSVFNLD--------- 461
GSIVA +ET++F +YF +PS + + D +N+
Sbjct: 945 GSIVAGSLTEETSHFTSYYF-----SPSVRTKKTRPRCYDDGGVAPTYNVPDVPDTFAQI 999
Query: 462 GRHAGKVLQYWMVDQKEMRSAHVHVLINCAEVKPYLEEFIAFYA 505
GR AGK+ + W D R+AH ++L N Y+++F + A
Sbjct: 1000 GRLAGKLKEVWWDDHTLSRAAHNYILRNV----DYIQKFERYKA 1039
>pir||A84505 probable TNP2-like transposon protein [imported] - Arabidopsis
thaliana
Length = 1040
Score = 427 bits (1099), Expect = e-118
Identities = 227/524 (43%), Positives = 320/524 (60%), Gaps = 44/524 (8%)
Query: 1 MCMTKPYLFLSCIVPGPDNPKAGIDVFLQPLIDDLKRLWV-GELTYDISRKQNFKLRAAL 59
MCM ++FLS +VPGP +PK +DVFLQPLI +LK+LW G T+D S KQNF +RA L
Sbjct: 541 MCMQSEFMFLSILVPGPKHPKRALDVFLQPLIHELKKLWYEGVHTFDYSSKQNFNMRAML 600
Query: 60 MWTINDFPAYGMLSGWSTHGRFACPHCMENTKAFTLESGGKSSWFDCHRPFLPHNHPFRR 119
MWTI+DFPAY MLSGW+ HGR P K WF+CHR FLP +HP+RR
Sbjct: 601 MWTISDFPAYRMLSGWTMHGRLYVP-----------TKDRKPCWFNCHRRFLPLHHPYRR 649
Query: 120 LKNGFKKDERVFVGPPPKITSEEVWTRVCDY------PKVTDYGRAVPIP-RYGVDHHWT 172
K F+K++ V + PP ++ +++ ++ Y + ++ +P YG H+W
Sbjct: 650 NKKLFRKNKIVRLLPPSYVSGTDLFEQIDYYGAQETCKRGGNWHTTANMPDEYGSTHNWY 709
Query: 173 KRSIFWDLPYWKDNLLRHNLDVMHIEKNVFDNIFNTVMDVKGKTKDNEKARKDLQIYCKR 232
K+SIFW LPYWKD LLRHNL VMHIEKN FDNI NT+++++GKTKD K+R DL C +
Sbjct: 710 KQSIFWQLPYWKDLLLRHNLYVMHIEKNFFDNIMNTLLNLQGKTKDTLKSRLDLAEICSK 769
Query: 233 KDLELKPQPNGKLLKPKANYNLSAQEAKLVCQWL-KDLRMPDGYSSNIGRCADVNTGRLH 291
+L + +GKL P + + LS + K + +W+ +++ PDGY S RC + +
Sbjct: 770 PELHVTR--DGKL--PVSKFRLSNEAKKALFEWVVAEVKFPDGYVSKFSRCVEQGQ-KFS 824
Query: 292 GMKSHDSHIFMENFIP-IAFSSLPKHVLDPLIEISQFFKNLCASTLRVDELEKMEKNMPI 350
GMKSHD H+FM+ +P + LP ++ + + I FFK+LC TL D +E+++KN+P+
Sbjct: 825 GMKSHDCHVFMQRLLPFVLVELLPTNIHEAIAGIGVFFKDLCTRTLTTDTIEQLDKNIPV 884
Query: 351 IICQLEQIFPPGFFDSMEHLPVHLAYEAILGGPVQYRWMYPFERFMGDSKRAVKNLAKVE 410
++C LE++F P FFD MEHLP+HL +E LGGPVQ+RWMYPFERFM K KNLA+VE
Sbjct: 885 LLCNLEKLFSPAFFDVMEHLPIHLPHEVALGGPVQFRWMYPFERFMKSLKGKAKNLARVE 944
Query: 411 GSIVAIYTAKETTYFIGHYFVDRLLTPSNTRNEVQVNDLRDPSTLSVFNLD--------- 461
GSIVA +ET++F +YF +PS + + D +N+
Sbjct: 945 GSIVAGSLTEETSHFTSYYF-----SPSVRTKKTRPRCYDDGGVAPTYNVPDVPDTFAQI 999
Query: 462 GRHAGKVLQYWMVDQKEMRSAHVHVLINCAEVKPYLEEFIAFYA 505
GR AGK+ + W D R+AH ++L N Y+++F + A
Sbjct: 1000 GRLAGKLKEVWWDDHTLSRAAHNYILRNV----DYIQKFERYKA 1039
>dbj|BAB64937.1| TdcA1-ORF1-ORF2 [Daucus carota]
Length = 988
Score = 426 bits (1096), Expect = e-117
Identities = 238/599 (39%), Positives = 342/599 (56%), Gaps = 31/599 (5%)
Query: 188 LRHNLDVMHIEKNVFDNIFNTVMDVKGKTKDNEKARKDLQIYCKRKDLELKPQPNGKLLK 247
+RHNLDVMHIEKNVF+NIFNT+M ++GKTKDN KAR D+ + C+R +L +
Sbjct: 3 IRHNLDVMHIEKNVFENIFNTIMAIEGKTKDNAKARADIALLCRRPELAIDESTRKY--- 59
Query: 248 PKANYNLSAQEAKLVCQWLKDLRMPDGYSSNIGRCADVNTGRLHGMKSHDSHIFMENFIP 307
PKA Y+L + + VC+WL+DL+ PDGY SN+GRC D+ +L GMKSHD H+FM+ +P
Sbjct: 60 PKACYSLDKKGKEAVCKWLQDLKFPDGYVSNMGRCIDMKKYKLFGMKSHDCHVFMQRLMP 119
Query: 308 IAFSS-LPKHVLDPLIEISQFFKNLCASTLRVDELEKMEKNMPIIICQLEQIFPPGFFDS 366
IAF LP +V + + E+S FFK+L ++TL+V ++ ++E+ +P+I+C+LE+IFPP FDS
Sbjct: 120 IAFREFLPNNVWEAVTELSLFFKDLTSATLKVSDMCRLEEQIPVILCKLERIFPPALFDS 179
Query: 367 MEHLPVHLAYEAILGGPVQYRWMYPFERFMGDSKRAVKNLAKVEGSIVAIYTAKETTYFI 426
MEHL VHL YEA + GPVQYRWMYPFERF+ K +KN A+VEGS+ Y +E + F
Sbjct: 180 MEHLLVHLPYEARIAGPVQYRWMYPFERFLRHLKNNIKNKARVEGSMCNAYLVEEASTFC 239
Query: 427 GHYFVDRLLTPSN--TRNEVQVNDLRDPSTLSVFNLDGRHAGKVLQYWMVDQKEMRSAHV 484
HYF + T RN+ S+F+ GR +G +D +E +AH
Sbjct: 240 SHYFEQHVQTKHRKVPRNDSSGGGENFEGNFSIFSHPGRASG-CANVRYLDDREYMAAHN 298
Query: 485 HVLINCAEVKPYLEEFIAFYAEFGEGSTVGSIHEY----FPAWFKQRVYNAEPTDEVIRL 540
+VL+NC EV PY E FI E + T I + F WFKQ N I +
Sbjct: 299 YVLLNCPEVAPYTEIFINMVRENNQSITDAEIDKCLESDFALWFKQYAQNPSLVPNEI-V 357
Query: 541 RQLSQGPLQCANEFHTYFVNGYKFHTHSWTEGKKTINSGVYVKGVT-DGGEDDFYGVIKH 599
R ++ GPL+ ++VNGYKFHT + + T NSGV +KG +D++G+I
Sbjct: 358 RDIASGPLRSVRSVPIFYVNGYKFHTRKYGANRSTFNSGVCIKGSNYSETSNDYFGIIDE 417
Query: 600 IYELSYND---NKVVLFYCDWFDPSPR-WTKINKLCNTVDIRVDKKYKEYDPFIMAHNVR 655
I L Y K LF C+WFDP+P T+++ V++ KK Y+PFI+A
Sbjct: 418 ILILEYPRLPIKKTTLFKCEWFDPTPSVGTRVHLRFKMVEVNRKKKLSVYEPFILASQAM 477
Query: 656 QVYYVPYPPTLPRKRGWSVAIKTKPRGRIETEE--RNEEVAYQADDMTHVNEIIEVDQVT 713
QVY+ YP K W V K K R +E + ++ + YQ + ++N I D T
Sbjct: 478 QVYFCNYPSLRRDKMDWLVVCKIKARPLVELSQASQSHQEPYQDETPENLNRIDIRDIPT 537
Query: 714 SWQDSQVEGDQVDPSILEMRNEVEDENEDVVDDNNEGDHQDNQVDDGDVDEYNYASDNN 772
D++ IL++ + E++ D++ + + +Q D D +NY SD+N
Sbjct: 538 HLNDNE--------GILDLDDGEGSPEEEIELDSDSEEGRSSQSD----DIHNYGSDSN 584
>emb|CAB80813.1| putative transposon protein [Arabidopsis thaliana]
gi|4325351|gb|AAD17349.1| similar to Antirrhinum majus
(garden snapdragon) TNP2 protein (GB:X57297)
[Arabidopsis thaliana] gi|25407250|pir||D85049 probable
transposon protein [imported] - Arabidopsis thaliana
Length = 817
Score = 414 bits (1064), Expect = e-114
Identities = 217/448 (48%), Positives = 277/448 (61%), Gaps = 32/448 (7%)
Query: 1 MCMTKPYLFLSCIVPGPDNPKAGIDVFLQPLIDDLKRLW-VGELTYDISRKQNFKLRAAL 59
MCM YLFL+ + GP++P+A +DVFLQPLI++LK LW G YD+S QNF L+A L
Sbjct: 391 MCMNTEYLFLTILNSGPNHPRASLDVFLQPLIEELKELWCTGVDAYDVSLSQNFNLKAVL 450
Query: 60 MWTINDFPAYGMLSGWSTHGRFACPHCMENTKAFTLESGGKSSWFDCHRPFLPHNHPFRR 119
+WTI+DFPAY MLSGW+THG+ +CP CME+TK+F L +G K+ WFDCHR FLPH HP RR
Sbjct: 451 LWTISDFPAYSMLSGWTTHGKLSCPVCMESTKSFYLPNGRKTCWFDCHRRFLPHGHPSRR 510
Query: 120 LKNGFKKDERVFVG-PPPKITSEEVWTR---VCDYPKVTDYG---RAVPIPRYGVDHHWT 172
K F K PP +T E+V+ + PK D G + YG +H+W
Sbjct: 511 NKKDFLKGRDASSEYPPESLTGEQVYYERLASVNPPKTKDVGGNGHEKKMRGYGKEHNWH 570
Query: 173 KRSIFWDLPYWKDNLLRHNLDVMHIEKNVFDNIFNTVMDVKGKTKDNEKARKDLQIYCKR 232
K SI W+L YWKD LRHN+DVMH EKN DNI NT++ VKGK+KDN +R D++ YC R
Sbjct: 571 KESILWELSYWKDLNLRHNIDVMHTEKNFLDNIMNTLLGVKGKSKDNIMSRLDIEKYCSR 630
Query: 233 KDLELKPQPNGKLLKPKANYNLSAQEAKLVCQWLK-DLRMPDGYSSNIGRCADVNTGRLH 291
L + GK P Y L+ + + + Q +K D+R PDG
Sbjct: 631 PGLHI--DSTGKA--PFPPYKLTEEAKQSLFQCVKHDVRFPDG----------------- 669
Query: 292 GMKSHDSHIFMENFIPIAFSSL-PKHVLDPLIEISQFFKNLCASTLRVDELEKMEKNMPI 350
MKSHD H+FME +P F+ L ++V L I FF++LC+ TL+ ++ +++N+ +
Sbjct: 670 -MKSHDCHVFMERLLPFIFAELLDRNVHLALSGIRAFFRDLCSRTLQTSRVQILKQNIVL 728
Query: 351 IICQLEQIFPPGFFDSMEHLPVHLAYEAILGGPVQYRWMYPFERFMGDSKRAVKNLAKVE 410
IIC LE+IFPP FFD MEHLP+HL YEA LGGPVQYRWMYPFERF K KN
Sbjct: 729 IICNLEKIFPPSFFDVMEHLPIHLPYEAELGGPVQYRWMYPFERFFKKLKGKAKNKRYAA 788
Query: 411 GSIVAIYTAKETTYFIGHYFVDRLLTPS 438
GSIV Y E YF HYF D + T S
Sbjct: 789 GSIVESYINDEIAYFSEHYFADHIQTKS 816
>pir||S29329 hypothetical protein 1 - maize transposon En-1
gi|225005|prf||1206239A ORF 1
Length = 905
Score = 401 bits (1031), Expect = e-110
Identities = 206/504 (40%), Positives = 297/504 (58%), Gaps = 7/504 (1%)
Query: 2 CMTKPYLFLSCIVPGPDNPKAGIDVFLQPLIDDLKRLWVGELTYDISRKQNFKLRAALMW 61
CM + +FL+ IVPGP +P I+VF++PLI++LK LW G YD K F LRAA +W
Sbjct: 359 CMKEEVMFLALIVPGPKDPVTKINVFMEPLIEELKMLWQGVEAYDSHLKCCFTLRAAYLW 418
Query: 62 TINDFPAYGMLSGWSTHGRFACPHCMENTKAFTLESGGKSSWFDCHRPFLPHNHPFRRLK 121
+I+D AYG+ SGW HG CP CM +++A+ LE G K ++FD HR LP+NHPFR+
Sbjct: 419 SIHDLLAYGIFSGWCVHGILRCPICMGDSQAYRLEHGKKETFFDVHRRLLPYNHPFRKDT 478
Query: 122 NGFKKDERVFVGPPPKITSEEVWTRVCDYPKVTDYGRAVPIPRYGVDHHWTKRSIFWDLP 181
F+K +RV GPP + T E + + D G YG +H+WT S W+LP
Sbjct: 479 KSFRKGKRVRDGPPKRQTGENIMRQHRDLKP----GVGGRFQGYGKEHNWTHISFIWELP 534
Query: 182 YWKDNLLRHNLDVMHIEKNVFDNIFNTVMDVKGKTKDNEKARKDLQIYCKRKDLELKPQP 241
Y K LL HN+D+MH E+NV ++I + D G+TKDN AR+DL C R LEL+ P
Sbjct: 535 YTKALLLPHNIDLMHQERNVAESIISMCFDFTGQTKDNMNARRDLAELCDRPHLELRKNP 594
Query: 242 NGKLLKPKANYNLSAQEAKLVCQWLKDLRMPDGYSSNIGRCADVNTGRLHGMKSHDSHIF 301
+G +P+A Y L QE + + QWLK LR PD Y++NI R +++TG+L G+KSHD HI
Sbjct: 595 SGSESRPQAPYCLKRQEREEIFQWLKKLRFPDRYAANIKRAVNLDTGKLVGLKSHDYHIL 654
Query: 302 MENFIPIAFSS-LPKHVLDPLIEISQFFKNLCASTLRVDELEKMEKNMPIIICQLEQIFP 360
+E +P+ F V E+S F+K +CA + + + EK + +++C++E++FP
Sbjct: 655 IERLVPVMFRGYFSPDVWKIFAELSYFYKQICAKEISKKLMLRFEKEIVVLVCKMEKVFP 714
Query: 361 PGFFDSMEHLPVHLAYEAILGGPVQYRWMYPFERFMGDSKRAVKNLAKVEGSIVAIYTAK 420
PGFF+ M+HL VHL +EA++GGP Q+RWMY ER + + V+N A+VEG I + A+
Sbjct: 715 PGFFNCMQHLLVHLPWEALVGGPAQFRWMYSQERELKKLRGMVRNKARVEGCIAEAFAAR 774
Query: 421 ETTYFIGHYFVDRLLTPSNTRNEVQVNDLRDPSTLSVFNLDGRHAGKVLQYWMVDQKEMR 480
E T F YF D + T V + + LS F DG+ G + +V E
Sbjct: 775 EITLFSSKYFSDTNNVNAQT-TRYHVAEQAPITDLSAFKWDGKGVGAYTSH-LVGTIERN 832
Query: 481 SAHVHVLINCAEVKPYLEEFIAFY 504
+ + +N E+ PY + F + Y
Sbjct: 833 KTLLFLYVNMPELHPYFQIFDSIY 856
>gb|AAA66266.1| unknown protein
Length = 897
Score = 401 bits (1031), Expect = e-110
Identities = 206/504 (40%), Positives = 297/504 (58%), Gaps = 7/504 (1%)
Query: 2 CMTKPYLFLSCIVPGPDNPKAGIDVFLQPLIDDLKRLWVGELTYDISRKQNFKLRAALMW 61
CM + +FL+ IVPGP +P I+VF++PLI++LK LW G YD K F LRAA +W
Sbjct: 351 CMKEEVMFLALIVPGPKDPVTKINVFMEPLIEELKMLWQGVEAYDSHLKCCFTLRAAYLW 410
Query: 62 TINDFPAYGMLSGWSTHGRFACPHCMENTKAFTLESGGKSSWFDCHRPFLPHNHPFRRLK 121
+I+D AYG+ SGW HG CP CM +++A+ LE G K ++FD HR LP+NHPFR+
Sbjct: 411 SIHDLLAYGIFSGWCVHGILRCPICMGDSQAYRLEHGKKETFFDVHRRLLPYNHPFRKDT 470
Query: 122 NGFKKDERVFVGPPPKITSEEVWTRVCDYPKVTDYGRAVPIPRYGVDHHWTKRSIFWDLP 181
F+K +RV GPP + T E + + D G YG +H+WT S W+LP
Sbjct: 471 KSFRKGKRVRDGPPKRQTGENIMRQHRDLKP----GVGGRFQGYGKEHNWTHISFIWELP 526
Query: 182 YWKDNLLRHNLDVMHIEKNVFDNIFNTVMDVKGKTKDNEKARKDLQIYCKRKDLELKPQP 241
Y K LL HN+D+MH E+NV ++I + D G+TKDN AR+DL C R LEL+ P
Sbjct: 527 YTKALLLPHNIDLMHQERNVAESIISMCFDFTGQTKDNMNARRDLAELCDRPHLELRKNP 586
Query: 242 NGKLLKPKANYNLSAQEAKLVCQWLKDLRMPDGYSSNIGRCADVNTGRLHGMKSHDSHIF 301
+G +P+A Y L QE + + QWLK LR PD Y++NI R +++TG+L G+KSHD HI
Sbjct: 587 SGSESRPQAPYCLKRQEREEIFQWLKKLRFPDRYAANIKRAVNLDTGKLVGLKSHDYHIL 646
Query: 302 MENFIPIAFSS-LPKHVLDPLIEISQFFKNLCASTLRVDELEKMEKNMPIIICQLEQIFP 360
+E +P+ F V E+S F+K +CA + + + EK + +++C++E++FP
Sbjct: 647 IERLVPVMFRGYFSPDVWKIFAELSYFYKQICAKEISKKLMLRFEKEIVVLVCKMEKVFP 706
Query: 361 PGFFDSMEHLPVHLAYEAILGGPVQYRWMYPFERFMGDSKRAVKNLAKVEGSIVAIYTAK 420
PGFF+ M+HL VHL +EA++GGP Q+RWMY ER + + V+N A+VEG I + A+
Sbjct: 707 PGFFNCMQHLLVHLPWEALVGGPAQFRWMYSQERELKKLRGMVRNKARVEGCIAEAFAAR 766
Query: 421 ETTYFIGHYFVDRLLTPSNTRNEVQVNDLRDPSTLSVFNLDGRHAGKVLQYWMVDQKEMR 480
E T F YF D + T V + + LS F DG+ G + +V E
Sbjct: 767 EITLFSSKYFSDTNNVNAQT-TRYHVAEQAPITDLSAFKWDGKGVGAYTSH-LVGTIERN 824
Query: 481 SAHVHVLINCAEVKPYLEEFIAFY 504
+ + +N E+ PY + F + Y
Sbjct: 825 KTLLFLYVNMPELHPYFQIFDSIY 848
>gb|AAG17043.1| transposase [Zea mays]
Length = 854
Score = 392 bits (1006), Expect = e-107
Identities = 193/464 (41%), Positives = 286/464 (61%), Gaps = 10/464 (2%)
Query: 7 YLFLSCIVPGPDNPKAGIDVFLQPLIDDLKRLWVGELTYDISRKQNFKLRAALMWTINDF 66
++FL+ ++PGP++P +++F++PLI++LK+LW G YD ++ F +RAA +W+++D
Sbjct: 368 FMFLALVIPGPEHPGPKLNMFVRPLIEELKQLWRGVKAYDSHTEKEFTMRAAYLWSVHDL 427
Query: 67 PAYGMLSGWSTHGRFACPHCMENTKAFTLESGGKSSWFDCHRPFLPHNHPFRRLKNGFKK 126
AYG SGW HGR CP CM +T AF L+ GGK S+FD HR + P H FR F+
Sbjct: 428 LAYGDWSGWCVHGRLCCPICMNDTDAFRLKHGGKVSFFDAHRRWTPFKHDFRNSLTAFRG 487
Query: 127 DERVFVGPPPKITSEEVWTRVCDYPKVTDYGRAVPIPRYGVDHHWTKRSIFWDLPYWKDN 186
++ GPP + T+ ++ + G YG DH+WT S W+LPY K
Sbjct: 488 GAKIRNGPPKRQTAPQIMA----WHACLKQGENDRFQGYGEDHNWTHISSIWELPYAKAL 543
Query: 187 LLRHNLDVMHIEKNVFDNIFNTVMDVKGKTKDNEKARKDLQIYCKRKDLELKPQPNGKLL 246
++ HN+D+MH E+NV ++I +T DV KTKDN KARKD+ CKR LELK G
Sbjct: 544 IMPHNIDLMHQERNVAESIISTCFDVTDKTKDNMKARKDMAEICKRPMLELKVSDKGHES 603
Query: 247 KPKANYNLSAQEAKLVCQWLKDLRMPDGYSSNIGRCADVNTGRLHGMKSHDSHIFMENFI 306
+P+A+Y L E K + +WLK+L+ PD Y++N+ R ++ TG+L G+KSHD HI ME +
Sbjct: 604 RPRADYCLKPDERKEIFKWLKNLKFPDRYAANLKRSVNLKTGKLIGLKSHDYHIIMERLM 663
Query: 307 PIAFSSLPKHVLDPLI-EISQFFKNLCASTLRVDELEKMEKNMPIIICQLEQIFPPGFFD 365
P+ F K L + E+S F++ +CA T+ ++K EK +PI+IC+ E++FPPGFF+
Sbjct: 664 PVMFRGYFKDELWSIFAELSYFYREVCAKTVSKRLMQKFEKEIPILICKFEKVFPPGFFN 723
Query: 366 SMEHLPVHLAYEAILGGPVQYRWMYPFERFMGDSKRAVKNLAKVEGSIVAIYTAKETTYF 425
M+HL VHL +EA++GGPVQ+RWMYP ER + + +V+N A+VEG I + KE + F
Sbjct: 724 VMQHLIVHLPWEALVGGPVQFRWMYPIERALKKLRASVRNKARVEGCIAEAFALKEISQF 783
Query: 426 IGHYF--VDRLLTPSNTRNEVQVNDLRDPSTLSVFNLDGRHAGK 467
YF + + PS + V + STL +F G+ GK
Sbjct: 784 STRYFARANNVFAPS---VRLHVENESPQSTLQIFANPGKAVGK 824
>gb|AAD55677.1| putative transposase protein [Zea mays]
Length = 793
Score = 381 bits (978), Expect = e-104
Identities = 196/438 (44%), Positives = 281/438 (63%), Gaps = 10/438 (2%)
Query: 1 MCMTKPYLFLSCIVPGPDNPKAGIDVFLQPLIDDLKRLWV-GELTYDISRKQNFKLRAAL 59
+C+ +P+ LS ++PG ++P IDV+LQPLID+LK LW G T+D K+NF LRA L
Sbjct: 355 LCLKQPFWMLSMLIPGKNSPGNNIDVYLQPLIDELKDLWEKGVETWDAKEKKNFNLRAIL 414
Query: 60 MWTINDFPAYGMLSGWSTHGRFACPHCMENTKAFTLESGGKSSWFDCHRPFLPHNHPFRR 119
+WTINDFPAYGMLSGWST G+FACP+C ++T L+ G K + HR FLP +H +R+
Sbjct: 415 LWTINDFPAYGMLSGWSTKGKFACPYCHKDTDFLWLKCGSKHCYMG-HRRFLPMDHRWRK 473
Query: 120 LKNGFKKDERVFVGPPPKITSEEVWTRVCDYPKVTDYGRAVPIPRYGVD----HHWTKRS 175
K F PP ++ E V + + ++T +G++ + + H+W K+S
Sbjct: 474 NKASFNNTVETRE-PPVPLSGEGVLQQYESFEQIT-FGKSTRKRKQREEETRWHNWRKKS 531
Query: 176 IFWDLPYWKDNLLRHNLDVMHIEKNVFDNIFNTVMDVKGKTKDNEKARKDLQIYCKRKDL 235
IF++L YW+ L+RHNLDVMHIEKN+ D+I T++ ++GK+KD EKAR D++ RKD
Sbjct: 532 IFFELQYWEKLLVRHNLDVMHIEKNICDSILGTLLGMEGKSKDGEKARLDMEHLGIRKDQ 591
Query: 236 ELKPQPNGKLLKPKANYNLSAQEAKLVCQWLKDLRMPDGYSSNIGRCADVNTGRLHGMKS 295
+ NGK P A Y L E +CQ+L+ ++MPDGY+SNI R DVN ++ G+K+
Sbjct: 592 HIV-LGNGKYTLPSALYTLGKDEMIWLCQFLEGVKMPDGYASNIKRSVDVNKCKVSGLKT 650
Query: 296 HDSHIFMENFIPIAFSS-LPKHVLDPLIEISQFFKNLCASTLRVDELEKMEKNMPIIICQ 354
HD H+ + +PIA + LP++V PLI++S++F +C+ L ++LE++ ++P +C+
Sbjct: 651 HDCHVIFQKLLPIAVRNVLPENVALPLIQLSRYFNAICSKELSAEDLEQLSTSIPETLCR 710
Query: 355 LEQIFPPGFFDSMEHLPVHLAYEAILGGPVQYRWMYPFERFMGDSKRAVKNLAKVEGSIV 414
LE IFPP FFD M HL +HLA EA LGGPV YRWMYP ER++ K V+N A EGSI
Sbjct: 711 LEMIFPPAFFDIMIHLSIHLAEEAKLGGPVCYRWMYPIERYLRTLKAYVRNKAHPEGSIA 770
Query: 415 AIYTAKETTYFIGHYFVD 432
Y +E F + D
Sbjct: 771 EGYILEECMTFCSRFLQD 788
>emb|CAA40555.1| TNP2 [Antirrhinum majus]
Length = 752
Score = 372 bits (956), Expect = e-101
Identities = 186/398 (46%), Positives = 258/398 (64%), Gaps = 12/398 (3%)
Query: 1 MCMTKPYLFLSCIVPGPDNPKAGIDVFLQPLIDDLKRLWVGELTYDISRKQNFKLRAALM 60
MCM +P+ FL+ ++PGP P IDV++QPLI++L+ LW G TYD S K+NF +RAAL+
Sbjct: 363 MCMKEPFFFLTLLIPGPSPPGNNIDVYMQPLIEELQELWGGVNTYDASAKENFNVRAALL 422
Query: 61 WTINDFPAYGMLSGWSTHGRFACPHCMENTKAFTLESGGKSSWFDCHRPFLPHNHPFRRL 120
WTIND+PAY LSGWST G ACP C ++T + L+ K + HR FL HP+R+
Sbjct: 423 WTINDYPAYANLSGWSTKGELACPSCHKDTCSKYLQKSHKYCYMG-HRRFLNRYHPYRKD 481
Query: 121 KNGFKKDE---RVFVGPPPKITSEEVWTRVCDYPKVTDYGRAVPIPRYGVDHHWTKRSIF 177
F +E R + + SEE+ + K D +P+ +W KRSIF
Sbjct: 482 TKSFDGNEEYRRAPIALTGDMVSEEITGFNIKFGKKVDDNPTLPL-------NWKKRSIF 534
Query: 178 WDLPYWKDNLLRHNLDVMHIEKNVFDNIFNTVMDVKGKTKDNEKARKDLQIYCKRKDLEL 237
+DLPYWKD+LLRHN DVMHIEKNV ++I T+++++G+TKD+E +R DL+ R +L
Sbjct: 535 FDLPYWKDSLLRHNFDVMHIEKNVCESIIGTLLNLEGRTKDHENSRLDLKDMGIRSELHP 594
Query: 238 KPQPNGKLLKPKANYNLSAQEAKLVCQWLKDLRMPDGYSSNIGRCADVNTGRLHGMKSHD 297
+GK P A Y++S +E ++V + LK +++PDGY+SNI R + ++ G+KSHD
Sbjct: 595 ISLESGKHYLPAACYSMSKKEKEIVFEILKTVKVPDGYASNISRRVQLKPNKISGLKSHD 654
Query: 298 SHIFMENFIPIAFSS-LPKHVLDPLIEISQFFKNLCASTLRVDELEKMEKNMPIIICQLE 356
HI M+ +PIA LPKHV PLI++ FF+ LC+ L +L +M K++ +C LE
Sbjct: 655 HHILMQQLLPIALRKVLPKHVRTPLIKLCTFFRELCSKVLNPQDLVRMGKDIAKTLCDLE 714
Query: 357 QIFPPGFFDSMEHLPVHLAYEAILGGPVQYRWMYPFER 394
+IFPP FFD M HLP+HLAYEA + GPVQYRWMYP ER
Sbjct: 715 KIFPPSFFDIMMHLPIHLAYEAQIAGPVQYRWMYPIER 752
>pir||S23818 hypothetical protein Tnp2 - garden snapdragon transposable element
Tam1
Length = 752
Score = 372 bits (956), Expect = e-101
Identities = 186/398 (46%), Positives = 258/398 (64%), Gaps = 12/398 (3%)
Query: 1 MCMTKPYLFLSCIVPGPDNPKAGIDVFLQPLIDDLKRLWVGELTYDISRKQNFKLRAALM 60
MCM +P+ FL+ ++PGP P IDV++QPLI++L+ LW G TYD S K+NF +RAAL+
Sbjct: 363 MCMKEPFFFLTLLIPGPSPPGNNIDVYMQPLIEELQELWGGVNTYDASAKENFNVRAALL 422
Query: 61 WTINDFPAYGMLSGWSTHGRFACPHCMENTKAFTLESGGKSSWFDCHRPFLPHNHPFRRL 120
WTIND+PAY LSGWST G ACP C ++T + L+ K + HR FL HP+R+
Sbjct: 423 WTINDYPAYANLSGWSTKGELACPSCHKDTCSKYLQKSHKYCYMG-HRRFLNRYHPYRKD 481
Query: 121 KNGFKKDE---RVFVGPPPKITSEEVWTRVCDYPKVTDYGRAVPIPRYGVDHHWTKRSIF 177
F +E R + + SEE+ + K D +P+ +W KRSIF
Sbjct: 482 TKSFDGNEEYRRAPIALTGDMVSEEITGFNIKFGKKVDDNPTLPL-------NWKKRSIF 534
Query: 178 WDLPYWKDNLLRHNLDVMHIEKNVFDNIFNTVMDVKGKTKDNEKARKDLQIYCKRKDLEL 237
+DLPYWKD+LLRHN DVMHIEKNV ++I T+++++G+TKD+E +R DL+ R +L
Sbjct: 535 FDLPYWKDSLLRHNFDVMHIEKNVCESIIGTLLNLEGRTKDHENSRLDLKDMGIRSELHP 594
Query: 238 KPQPNGKLLKPKANYNLSAQEAKLVCQWLKDLRMPDGYSSNIGRCADVNTGRLHGMKSHD 297
+GK P A Y++S +E ++V + LK +++PDGY+SNI R + ++ G+KSHD
Sbjct: 595 ISLESGKHYLPAACYSMSKKEKEIVFEILKTVKVPDGYASNISRRVQLKPNKISGLKSHD 654
Query: 298 SHIFMENFIPIAFSS-LPKHVLDPLIEISQFFKNLCASTLRVDELEKMEKNMPIIICQLE 356
HI M+ +PIA LPKHV PLI++ FF+ LC+ L +L +M K++ +C LE
Sbjct: 655 HHILMQQLLPIALRKVLPKHVRTPLIKLCTFFRELCSKVLNPQDLVRMGKDIAKTLCDLE 714
Query: 357 QIFPPGFFDSMEHLPVHLAYEAILGGPVQYRWMYPFER 394
+IFPP FFD M HLP+HLAYEA + GPVQYRWMYP ER
Sbjct: 715 KIFPPSFFDIMMHLPIHLAYEAQIAGPVQYRWMYPIER 752
>ref|NP_908869.1| B1151H08.16 [Oryza sativa (japonica cultivar-group)]
Length = 1095
Score = 362 bits (930), Expect = 2e-98
Identities = 237/690 (34%), Positives = 352/690 (50%), Gaps = 51/690 (7%)
Query: 1 MCMTKPYLFLSCIVPGPDNPKAGIDVFLQPLIDDLKRLW-VGELTYDISRKQNFKLRAAL 59
+C + Y+ L+ ++ GP P IDVFL+P+IDD +RLW G T+D ++ F L A L
Sbjct: 368 LCFKRKYIMLAMLIQGPRQPGNDIDVFLEPIIDDFERLWNEGTRTWDAYAQEYFNLHAML 427
Query: 60 MWTINDFPAYGMLSGWSTHGRFACPHCMENTKAFTLESGGKSSWFDCHRPFLPHNHPFRR 119
TIND+PA G LSG + G++AC CME T++ L+ K+ + HR FLP HP+R
Sbjct: 428 FCTINDYPALGNLSGQTVKGKWACSECMEETRSKWLKHSHKTVYMG-HRRFLPRYHPYRN 486
Query: 120 LKNGFKKDERVFVGPPPKITSEEVWTRVC----DYPKVTDYGRAVP--IPRYGVDHH--- 170
++ F R GPP ++T EV V ++ K G+ + + H
Sbjct: 487 MRKNFN-GHRDTAGPPTELTGTEVHNLVMGITNEFGKKRKVGKRKEKSTSKEKTEEHVEK 545
Query: 171 --------WTKRSIFWDLPYWKDNLLRHNLDVMHIEKNVFDNIFNTVMDVKGKTKDNEKA 222
W K+SIFW LPYWKD +RH +D+MH+EKNV +++ +++ G TKD A
Sbjct: 546 QKTKERSMWKKKSIFWRLPYWKDLEVRHCIDLMHVEKNVCESLMGLLLN-PGTTKDGLNA 604
Query: 223 RKDLQIYCKRKDLELKPQPNGKLLKPKANYNLSAQEAKLVCQWLKDLRMPDGYSSNIGRC 282
R+DL+ R +L +G++ P A Y LS +E + L +++P GYSS I R
Sbjct: 605 RRDLEDMGVRSELHPITTESGRVYLPPACYTLSKEEKIDLLTCLSGIKVPSGYSSRISRL 664
Query: 283 ADVNTGRLHGMKSHDSHIFMENFIPIAFSS-LPKHVLDPLIEISQFFKNLCASTLRVDEL 341
+ +L GMKSHD H+ + +P+A + LP V + + FF + + + L
Sbjct: 665 VSLQDLKLVGMKSHDCHVLITQLLPVAIRNILPPKVRHTIQRLCSFFHAIGQKIIDPEGL 724
Query: 342 EKMEKNMPIIICQLEQIFPPGFFDSMEHLPVHLAYEAILGGPVQYRWMYPFERFMGDSKR 401
++++ + +C LE FPP FFD MEHLPVHL + GP MYP ER++G K
Sbjct: 725 DELQAELVKTLCHLEMYFPPTFFDIMEHLPVHLVRQTKCCGPAFMTQMYPCERYLGILKG 784
Query: 402 AVKNLAKVEGSIVAIYTAKETTYFIGHYFVDRLLTPSNTRNEVQVNDLRDPSTLSVFNLD 461
V+N + EGSI+ YT +E F Y + + + P + LD
Sbjct: 785 YVRNRSHPEGSIIESYTTEEAIEFCVDYM-------------SETSSIGLPRSHHEGRLD 831
Query: 462 GRHAGKVLQYWM-VDQKEMRSAHVHVLINCAEVKPYLEEFIAFYAEFGEG---STVGSIH 517
G G V + + +D+K AH VL + EV PY++E +A + G S V + H
Sbjct: 832 G--VGTVGRKTIRLDRKVYDKAHFTVLQHMTEVVPYVDEHLAVIRQENPGRSESWVRNKH 889
Query: 518 -EYFPAWFKQRVYNAE--PTDEVIRLRQLSQGPLQCANEFHTYFVNGYKFHTHSWTEGKK 574
F W K R+ + P++ L+ LSQGP A + Y +NGY FHT
Sbjct: 890 MSSFNEWLKNRIARLQNLPSE---TLQWLSQGPEWSATTWQGYDINGYTFHTVKQDSKCT 946
Query: 575 TINSGVYVKGVTDGG-EDDFYGVIKHIYELSYNDNKVVLFYCDWFDPSPRWTKINKLCNT 633
NSG+ ++ +DGG D +YG ++ I EL Y KV LF C W D R K++ T
Sbjct: 947 VQNSGLCIEAASDGGRRDQYYGRVEQILELDYLKFKVPLFRCRWVD--LRNVKVDNEAFT 1004
Query: 634 VDIRVDKKYKEYDPFIMAHNVRQVYYVPYP 663
+ YK+ +PF++A V QV+Y+ P
Sbjct: 1005 TVNLANNAYKD-EPFVLAKQVVQVFYIVDP 1033
>gb|AAX96209.1| transposon protein, putative, CACTA, En/Spm sub-class [Oryza sativa
(japonica cultivar-group)] gi|62732737|gb|AAX94856.1|
transposon protein, putative, CACTA, En/Spm sub-class
[Oryza sativa (japonica cultivar-group)]
Length = 1100
Score = 362 bits (929), Expect = 3e-98
Identities = 237/688 (34%), Positives = 349/688 (50%), Gaps = 47/688 (6%)
Query: 1 MCMTKPYLFLSCIVPGPDNPKAGIDVFLQPLIDDLKRLW-VGELTYDISRKQNFKLRAAL 59
+C + Y+ L+ ++ GP P IDVFL+P+IDD +RLW G T+D ++ F L A L
Sbjct: 362 LCFKRKYIMLAMLIQGPRQPGNDIDVFLEPIIDDFERLWDEGTRTWDAYAQEYFNLHAIL 421
Query: 60 MWTINDFPAYGMLSGWSTHGRFACPHCMENTKAFTLESGGKSSWFDCHRPFLPHNHPFRR 119
TIND+PA G LSG + G++AC CME T++ L+ K+ + HR FLP HP+R
Sbjct: 422 FCTINDYPALGNLSGQTVKGKWACSECMEETRSKWLKHSHKTVYMG-HRRFLPRYHPYRN 480
Query: 120 LKNGFKKDERVFVGPPPKITSEEVWTRVC----DYPKVTDYGRAVP--IPRYGVDHH--- 170
++ F R GPP ++T EV V ++ K G+ + + H
Sbjct: 481 MRKNFN-GHRDTAGPPAELTGTEVHNLVMGITNEFGKKRKVGKRKEKSTSKEKTEEHVEK 539
Query: 171 --------WTKRSIFWDLPYWKDNLLRHNLDVMHIEKNVFDNIFNTVMDVKGKTKDNEKA 222
W K+SIFW LPYWKD +RH +D+MH+EKNV +++ +++ G TKD A
Sbjct: 540 QKTKERSMWKKKSIFWRLPYWKDLEVRHCIDLMHVEKNVCESLMGLLLN-PGTTKDGLNA 598
Query: 223 RKDLQIYCKRKDLELKPQPNGKLLKPKANYNLSAQEAKLVCQWLKDLRMPDGYSSNIGRC 282
R+DL+ R +L +G++ P A Y LS +E + L +++P GYSS I R
Sbjct: 599 RRDLEDMGVRSELHPITTESGRVYLPPACYTLSKEEKIDLLTCLSGIKVPSGYSSRISRL 658
Query: 283 ADVNTGRLHGMKSHDSHIFMENFIPIAFSS-LPKHVLDPLIEISQFFKNLCASTLRVDEL 341
+ +L GMKSHD H+ + +PIA + LP V + + FF + + + L
Sbjct: 659 VSLQDLKLVGMKSHDCHVLITQLLPIAIRNILPPKVRHTIQRLCSFFHAIGQKIIDPEGL 718
Query: 342 EKMEKNMPIIICQLEQIFPPGFFDSMEHLPVHLAYEAILGGPVQYRWMYPFERFMGDSKR 401
++++ + +C LE FPP FFD MEHLPVHL + GP MYP ER++G K
Sbjct: 719 DELQAELVRTLCHLEMYFPPTFFDIMEHLPVHLVRQTKCCGPAFMTQMYPCERYLGILKG 778
Query: 402 AVKNLAKVEGSIVAIYTAKETTYFIGHYFVDRLLTPSNTRNEVQVNDLRDPSTLSVFNLD 461
V+N + EGSI+ YT +E F Y + + + P + LD
Sbjct: 779 YVRNRSHPEGSIIESYTTEEAIEFCVDYM-------------SETSSIGLPRSHHEGRLD 825
Query: 462 GRHAGKVLQYWM-VDQKEMRSAHVHVLINCAEVKPYLEEFIAFYAEFGEG---STVGSIH 517
G G V + + +D+K AH VL + EV PY++E +A + G S V + H
Sbjct: 826 G--VGTVGRKTIRLDRKVYDKAHFTVLQHMTEVVPYVDEHLAVIRQENPGRSESWVRNKH 883
Query: 518 -EYFPAWFKQRVYNAEPTDEVIRLRQLSQGPLQCANEFHTYFVNGYKFHTHSWTEGKKTI 576
F W K R+ + L+ LSQGP A + Y +NGY FHT
Sbjct: 884 MSSFNKWLKNRIARLQNLSSE-TLQWLSQGPEWSATTWQGYDINGYTFHTVKQDSKCTVQ 942
Query: 577 NSGVYVKGVTDGG-EDDFYGVIKHIYELSYNDNKVVLFYCDWFDPSPRWTKINKLCNTVD 635
NSG+ ++ +DGG D +YG ++ I EL Y KV LF C W D R K++ T
Sbjct: 943 NSGLRIEAASDGGRRDQYYGKVEQILELDYLKFKVPLFRCRWVD--LRNVKVDNEGFTTV 1000
Query: 636 IRVDKKYKEYDPFIMAHNVRQVYYVPYP 663
+ YK+ +PF++A V QV+Y+ P
Sbjct: 1001 NLANNAYKD-EPFVLAKQVVQVFYIVDP 1027
Database: nr
Posted date: Jul 5, 2005 12:34 AM
Number of letters in database: 863,360,394
Number of sequences in database: 2,540,612
Lambda K H
0.320 0.138 0.438
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 1,509,770,459
Number of Sequences: 2540612
Number of extensions: 69790591
Number of successful extensions: 223291
Number of sequences better than 10.0: 985
Number of HSP's better than 10.0 without gapping: 641
Number of HSP's successfully gapped in prelim test: 355
Number of HSP's that attempted gapping in prelim test: 211000
Number of HSP's gapped (non-prelim): 4982
length of query: 773
length of database: 863,360,394
effective HSP length: 136
effective length of query: 637
effective length of database: 517,837,162
effective search space: 329862272194
effective search space used: 329862272194
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 79 (35.0 bits)
Medicago: description of AC141863.3


