

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC141436.6 - phase: 0
(1173 letters)
Database: nr
2,540,612 sequences; 863,360,394 total letters
Searching..................................................done
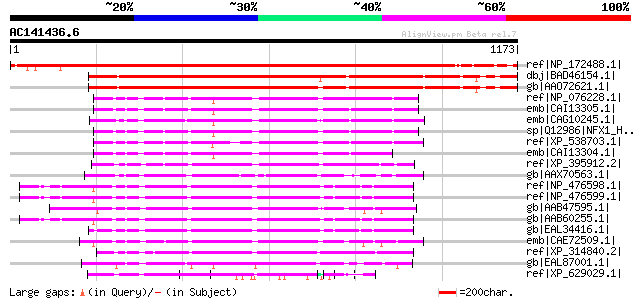
Score E
Sequences producing significant alignments: (bits) Value
ref|NP_172488.1| NF-X1 type zinc finger family protein [Arabidop... 1426 0.0
dbj|BAD46154.1| putative TF-like protein [Oryza sativa (japonica... 1316 0.0
gb|AAO72621.1| TF-like protein [Oryza sativa (japonica cultivar-... 1281 0.0
ref|NP_076228.1| nuclear transcription factor, X-box binding 1 [... 582 e-164
emb|CAI13305.1| nuclear transcription factor, X-box binding 1 [H... 578 e-163
emb|CAG10245.1| unnamed protein product [Tetraodon nigroviridis] 573 e-162
sp|Q12986|NFX1_HUMAN Transcriptional repressor NF-X1 (Nuclear tr... 568 e-160
ref|XP_538703.1| PREDICTED: similar to nuclear transcription fac... 565 e-159
emb|CAI13304.1| nuclear transcription factor, X-box binding 1 [H... 556 e-156
ref|XP_395912.2| PREDICTED: similar to ENSANGP00000015832 [Apis ... 528 e-148
gb|AAX70563.1| hypothetical protein, conserved [Trypanosoma brucei] 525 e-147
ref|NP_476598.1| CG3647-PA, isoform A [Drosophila melanogaster] ... 511 e-143
ref|NP_476599.1| CG3647-PB, isoform B [Drosophila melanogaster] ... 511 e-143
gb|AAB47595.1| Hypothetical protein C16A3.7 [Caenorhabditis eleg... 506 e-141
gb|AAB60255.1| shuttle craft protein [Drosophila melanogaster] 505 e-141
gb|EAL34416.1| GA17585-PA [Drosophila pseudoobscura] 504 e-141
emb|CAE72509.1| Hypothetical protein CBG19688 [Caenorhabditis br... 499 e-139
ref|XP_314840.2| ENSANGP00000015832 [Anopheles gambiae str. PEST... 497 e-139
gb|EAL87001.1| NF-X1 finger transcription factor, putative [Aspe... 496 e-138
ref|XP_629029.1| hypothetical protein DDB0220630 [Dictyostelium ... 479 e-133
>ref|NP_172488.1| NF-X1 type zinc finger family protein [Arabidopsis thaliana]
gi|4914319|gb|AAD32867.1| F14N23.5 [Arabidopsis thaliana]
gi|25402589|pir||D86236 protein F14N23.5 [imported] -
Arabidopsis thaliana
Length = 1188
Score = 1426 bits (3692), Expect = 0.0
Identities = 691/1212 (57%), Positives = 830/1212 (68%), Gaps = 63/1212 (5%)
Query: 1 MSLQQRRERREGSRFPSHRPPRQE-WIPKGAGASSSASTT-----STTTTASTTTAAAAT 54
MS Q RR+R + SHR Q+ WIP+ + SS +T + T A +A+
Sbjct: 1 MSFQVRRDRSDDR---SHRFNHQQTWIPRNSSTSSVVVNEPLLPPNTDRNSETLDAGSAS 57
Query: 55 TTV--------DQPTSLHSHEKNTNSDGGSSNQGAVVAPTFARHRSNHHVAHRVEREHVA 106
V P S + H++++N NQ +H+ + ++ +H
Sbjct: 58 RPVYLQRQHNASGPPSYNHHQRSSNIGPPPPNQHRRYNAPDNQHQRSDNIGPPQPNQHRR 117
Query: 107 HRT-DRDHVAH------RVERERVAHRVEREHVAHRMEREHVAHRVEREHVAHRV--ERE 157
+ D H + R R + E +H +R R + + + R+
Sbjct: 118 YNAPDNQHQRSDNSGPPQPYRHRRNNAPENQHQRSDNIGPPPPNRQRRNNASGTLPDNRQ 177
Query: 158 HVAHRVERGRGRSGNMAGRQYGSRDSSLPQLVQEIQEKLTKGTVECMICYDMVRRSAPIW 217
VA R D +LPQLVQE+QEKL K ++ECMICYD V RSA IW
Sbjct: 178 RVASRTRPVNQGKRVAKEENVVLTDPNLPQLVQELQEKLVKSSIECMICYDKVGRSANIW 237
Query: 218 SCSSCYSIFHLNCIKKWARAPTSVDLSAEKNLGFNWRCPGCQSVQHTSSKDIKYACFCGK 277
SCSSCYSIFH+NCIK+WARAPTSVDL AEKN G NWRCPGCQSVQ TSSK+I Y CFCGK
Sbjct: 238 SCSSCYSIFHINCIKRWARAPTSVDLLAEKNQGDNWRCPGCQSVQLTSSKEISYRCFCGK 297
Query: 278 RVDPPSDLYLTPHSCGEPCGKPLEKEVFVTEERKDELCPHACVLQCHPGPCPPCKAFAPP 337
R DPPSD YLTPHSCGEPCGKPLEKE E +++LCPH CVLQCHPGPCPPCKAFAPP
Sbjct: 298 RRDPPSDPYLTPHSCGEPCGKPLEKEFAPAETTEEDLCPHVCVLQCHPGPCPPCKAFAPP 357
Query: 338 RLCPCGKKRIATRCSDRQSDLTCGQRCDKLLDCGRHHCENACHVGPCDPCQVLIEASCFC 397
R CPCGKK + TRCS+R+SDL CGQRCDKLL CGRH CE CHVGPCDPCQVL+ A+CFC
Sbjct: 358 RSCPCGKKMVTTRCSERRSDLVCGQRCDKLLSCGRHQCERTCHVGPCDPCQVLVNATCFC 417
Query: 398 SKMTQVLFCGEMAMKGEFEAEGGVFSCGSNCGNVLGCSNHICREVCHPGSCGECEFLPSR 457
K + + CG+M +KGE +AE GV+SC NCG LGC NH C EVCHPG CG+C+ LPSR
Sbjct: 418 KKKVETVICGDMNVKGELKAEDGVYSCSFNCGKPLGCGNHFCSEVCHPGPCGDCDLLPSR 477
Query: 458 VKACCCGKTKLEDE-RKSCVDPIPTCSKVCSKTLRCGVHACKETCHVGECPPCKVLISQK 516
VK C CG T+LE++ R+SC+DPIP+CS VC K L C +H C E CH G+CPPC V ++QK
Sbjct: 478 VKTCYCGNTRLEEQIRQSCLDPIPSCSNVCRKLLPCRLHTCNEMCHAGDCPPCLVQVNQK 537
Query: 517 CRCGSTSRTVECYKTT--ENQKFTCQKPCGAKKNCGRHRCSEKCCPLSGPNNGLTTPDWD 574
CRCGSTSR VECY TT E +KF C KPCG KKNCGRHRCSE+CCPL + DWD
Sbjct: 538 CRCGSTSRAVECYITTSSEAEKFVCAKPCGRKKNCGRHRCSERCCPLLNGKKNDLSGDWD 597
Query: 575 PHFCSMLCGKKLRCGQHVCETLCHSGHCPPCLETIFTDLACACGMTSIPPPLPCGTMPPL 634
PH C + C KKLRCGQH CE+LCHSGHCPPCLE IFTDL CACG TSIPPPL CGT P
Sbjct: 598 PHVCQIPCQKKLRCGQHSCESLCHSGHCPPCLEMIFTDLTCACGRTSIPPPLSCGTPVPS 657
Query: 635 CQLPCSVPQPCGHSGSHSCHFGDCPPCSVPVSKECVGGHVILRNIPCGSNNIKCNNPCGR 694
CQLPC +PQPCGHS +H CHFGDCPPCS PV K+CVGGHV+LRNIPCG +I+C CG+
Sbjct: 658 CQLPCPIPQPCGHSDTHGCHFGDCPPCSTPVEKKCVGGHVVLRNIPCGLKDIRCTKICGK 717
Query: 695 TRQCGLHACGRSCHSPPCDILPGIVKGLRAACGQTCGAPRSGCRHMCMALCHPGCPCPDA 754
TR+CG+HAC R+CH PCD G+R C Q CGAPR+ CRH C ALCHP PCPD
Sbjct: 718 TRRCGMHACARTCHPEPCDSFNESEAGMRVTCRQKCGAPRTDCRHTCAALCHPSAPCPDL 777
Query: 755 RCEFPVTITCSCGRISANVPCDVGG---NNSNYNADAIFEASIIQKLPMPLQPVDANGQK 811
RCEF VTITCSCGRI+A VPCD GG N SN A EAS++QKLP PLQPV+++G +
Sbjct: 778 RCEFSVTITCSCGRITATVPCDAGGRSANGSNVYCAAYDEASVLQKLPAPLQPVESSGNR 837
Query: 812 VPLGQRKLMCDEECAKLERKRVLADAFDIT-PSLDALHFGENSSY-ELLSDTFRRDPKWV 869
+PLGQRKL CD+ECAKLERKRVL DAFDIT P+L+ALHF ENS+ E++SD +RRDPKWV
Sbjct: 838 IPLGQRKLSCDDECAKLERKRVLQDAFDITPPNLEALHFSENSAMTEIISDLYRRDPKWV 897
Query: 870 LAIEERCKILVLGKSKGTTHGLKVHVFCPMIKDKRDAVRMIAERWKLAVNAAGWEPKRFI 929
LA+EERCK LVLGK++G+T LKVH+FCPM KDKRD VR+IAERWKL V+ AGWEPKRF
Sbjct: 898 LAVEERCKFLVLGKARGSTSALKVHIFCPMQKDKRDTVRLIAERWKLGVSNAGWEPKRFT 957
Query: 930 VISATQKSKAPARVLGVK-GTTTLNAPLPTAFDPLVDMDPRLVVSFPDLPRDADISALVL 988
V+ T KSK P R++G + G ++ P P +D LVDMDP LVVSF DLPR+A+ISALVL
Sbjct: 958 VVHVTAKSKPPTRIIGARGGAISIGGPHPPFYDSLVDMDPGLVVSFLDLPREANISALVL 1017
Query: 989 RFGGECELVWLNDRNALAVFHDPARAATAMRRLDHGTVYQGAVSFVQNAGASAASSVTSA 1048
RFGGECELVWLND+NALAVFHD ARAATAMRRL+HG+VY GAV VQ+ G S S+ +
Sbjct: 1018 RFGGECELVWLNDKNALAVFHDHARAATAMRRLEHGSVYHGAV-VVQSGGQS--PSLNNV 1074
Query: 1049 WGGTKEGAL----RSNPWKKAAVLDPGWKEDSWGDEQWTTAGDSANIQPSAL---KKEAP 1101
WG + + NPW++A + + +DSWG E G S + Q SAL K +P
Sbjct: 1075 WGKLPGSSAWDVDKGNPWRRAVIQE---SDDSWGAEDSPIGGSSTDAQASALRSAKSNSP 1131
Query: 1102 IPASLNPWNVLNHESSSSSSPATVIRSVASGKQTESGNVSTKVEPSAGGADGGNSDATEA 1161
I S+N W+VL + +S+S+ + +A +++ S + K +P G +
Sbjct: 1132 IVTSVNRWSVLEPKKASTST----LEPIAQIEESSSSKTTGK-QPVEGSGE--------- 1177
Query: 1162 AEVVDDWEKAFE 1173
EVVDDWEK E
Sbjct: 1178 -EVVDDWEKVCE 1188
>dbj|BAD46154.1| putative TF-like protein [Oryza sativa (japonica cultivar-group)]
Length = 1117
Score = 1316 bits (3407), Expect = 0.0
Identities = 611/1014 (60%), Positives = 724/1014 (71%), Gaps = 53/1014 (5%)
Query: 182 DSSLPQLVQEIQEKLTKGTVECMICYDMVRRSAPIWSCSSCYSIFHLNCIKKWARAPTSV 241
D ++PQLVQEIQ+KL +G VECMICYDMVRRSAP+WSC SC+SIFHL CI+KWAR+P S
Sbjct: 135 DGAVPQLVQEIQDKLARGAVECMICYDMVRRSAPVWSCGSCFSIFHLPCIRKWARSPASA 194
Query: 242 DLSAEKNLGFNWRCPGCQSVQHTSSKDIKYACFCGKRVDPPSDLYLTPHSCGEPCGKPLE 301
+++ + +WRCPGCQSV ++++ Y CFCG+R +PP+DL+LTPHSCGEPC KPLE
Sbjct: 195 ADASDPDS--SWRCPGCQSVHAVPARELAYTCFCGRRREPPNDLFLTPHSCGEPCSKPLE 252
Query: 302 KE--VFVTEERKDELCPHACVLQCHPGPCPPCKAFAPPRLCPCGKKRIATRCSDRQSDLT 359
K ++ CPH CVLQCHPGPCPPCKAFAP RLCPCGK+ I RC+DR + +T
Sbjct: 253 KADPAVKADDAAATRCPHVCVLQCHPGPCPPCKAFAPDRLCPCGKQTIVRRCADRTTPVT 312
Query: 360 CGQRCDKLLDCGRHHCENACHVGPCDPCQVLIEASCFCSKMTQVLFCGEMAMKGEFEAEG 419
CGQRCD+LL C RH CE CH GPC C VLI A CFC K T+ L CGEM +KG +
Sbjct: 313 CGQRCDRLLPCRRHRCEKVCHTGPCGDCNVLISARCFCGKKTETLLCGEMELKGNLSEKD 372
Query: 420 GVFSCGSNCGNVLGCSNHICREVCHPGSCGECEFLPSRVKACCCGKTKLEDERKSCVDPI 479
GVFSC C ++L C NH C+++CHPG CGECE +P +V AC CGKT+L ++R SC+DPI
Sbjct: 373 GVFSCSEACSHMLSCGNHACQDICHPGPCGECELMPGKVTACHCGKTRLLEKRASCLDPI 432
Query: 480 PTCSKVCSKTLRCGVHACKETCHVGECPPCKVLISQKCRCGSTSRTVECYKTTENQKFTC 539
PTC KVC K L CGVH CK TCH G+CPPC V + Q+CRCGS+ + VECYK E ++F C
Sbjct: 433 PTCDKVCDKKLPCGVHRCKVTCHEGDCPPCVVRVEQRCRCGSSGQMVECYKVLE-EEFRC 491
Query: 540 QKPCGAKKNCGRHRCSEKCCPLSGPNNGLTTPDWDPHFCSMLCGKKLRCGQHVCETLCHS 599
KPCG KKNCGRHRCSE CCPLS P L +WDPH C + CGKKLRCGQH C+ LCHS
Sbjct: 492 NKPCGRKKNCGRHRCSECCCPLSKPLARLEGGNWDPHLCQIPCGKKLRCGQHGCQLLCHS 551
Query: 600 GHCPPCLETIFTDLACACGMTSIPPPLPCGTMPPLCQLPCSVPQPCGHSGSHSCHFGDCP 659
GHCPPCLETIF DL CACG TSIPPPLPCGT P C C VPQPCGH +H CHFGDCP
Sbjct: 552 GHCPPCLETIFNDLTCACGRTSIPPPLPCGTPTPSCPHQCLVPQPCGHPATHQCHFGDCP 611
Query: 660 PCSVPVSKECVGGHVILRNIPCGSNNIKCNNPCGRTRQCGLHACGRSCHSPPCDILP--- 716
PC VPV +EC+GGHV+LRNIPCGS +I+CN PCG+ RQCG+HAC RSCH PCD P
Sbjct: 612 PCVVPVMRECIGGHVVLRNIPCGSKDIRCNQPCGKNRQCGMHACNRSCHPSPCDPPPANG 671
Query: 717 --GIVKGLRAACGQTCGAPRSGCRHMCMALCHPGCPCPDARCEFPVTITCSCGRISANVP 774
G RA+CGQ CGAPR C+H C A CHP PCPD RCEFP+TI CSCGRI+A VP
Sbjct: 672 DASSSTGGRASCGQVCGAPRRECKHTCTAPCHPSSPCPDLRCEFPMTIACSCGRITATVP 731
Query: 775 CDVGGNNSNYNADAIFEASIIQKLPMPLQPVDANGQKVPLGQRKLMCDEECAKLERKRVL 834
C GG N D +FE SIIQKLPMPLQPV+++G++VPLGQRKL CDE+CAK+ERKRVL
Sbjct: 732 CSAGG---TANGDNMFEVSIIQKLPMPLQPVESDGRRVPLGQRKLSCDEDCAKMERKRVL 788
Query: 835 ADAFDIT-PSLDALHFGENS-SYELLSDTFRRDPKWVLAIEERCKILVLGKSKGTTHG-L 891
A+AFDIT P+LDALHFGENS + +LLSD FRR+PKWV+AIEERCK LVLGK++G + G L
Sbjct: 789 AEAFDITPPNLDALHFGENSNASDLLSDLFRREPKWVMAIEERCKFLVLGKTRGNSSGNL 848
Query: 892 KVHVFCPMIKDKRDAVRMIAERWKLAVNAAGWEPKRFIVISATQKSKAPARVLGVKGTTT 951
KVHVFC M KDKRDA+R+IA+RWKL+V AAGWEPKRFI I T KSKAPAR+LG K
Sbjct: 849 KVHVFCHMTKDKRDAIRVIADRWKLSVQAAGWEPKRFITIHPTPKSKAPARILGSKPGVF 908
Query: 952 LNAPLPTAFDPLVDMDPRLVVSFPDLPRDADISALVLRFGGECELVWLNDRNALAVFHDP 1011
+ A P FDPLVDMDPRLVV+ DLPRDAD+SALVLRFGGECELVWLND+NA+AVF+DP
Sbjct: 909 VAASHP-FFDPLVDMDPRLVVAMLDLPRDADVSALVLRFGGECELVWLNDKNAVAVFNDP 967
Query: 1012 ARAATAMRRLDHGTVYQGAVSFVQNAGASAASSVTSAW-GGTKEGAL----RSNPWKKAA 1066
ARAATA+RRLD+G+ YQGA F+ ++ A + W G K+G +NPWKKA
Sbjct: 968 ARAATALRRLDYGSAYQGAAVFLPSSSAQPG----NVWVAGQKDGVAATKSSANPWKKAT 1023
Query: 1067 VLDPGWKEDSW-------GDEQWTTAGDSANIQPSALKKEAPIPASLNPWNVLNHESSSS 1119
+P W W GD+ A + + N WN L ++++S
Sbjct: 1024 ASEPDPSSGDWTGVLGQAPGSVWRRGGDTV----------AQVMGTSNRWNALESDAATS 1073
Query: 1120 SSPATVIRSVASGKQTESGNVSTKVEPSAGGADGGNSDATEAAEVVDDWEKAFE 1173
S P + T SAG + + VDDWE+A E
Sbjct: 1074 SRPV----------EESKPAPRTDAVSSAGPSTAPPVSKMQPEVEVDDWEEACE 1117
>gb|AAO72621.1| TF-like protein [Oryza sativa (japonica cultivar-group)]
Length = 1017
Score = 1281 bits (3316), Expect = 0.0
Identities = 600/1010 (59%), Positives = 713/1010 (70%), Gaps = 60/1010 (5%)
Query: 182 DSSLPQLVQEIQEKLTKGTVECMICYDMVRRSAPIWSCSSCYSIFHLNCIKKWARAPTSV 241
+ ++PQLVQEIQ+KL +G VECMICYDMVRRSAP+WSC SC+SIFHL CI+KWAR+P S
Sbjct: 50 NXAVPQLVQEIQDKLARGAVECMICYDMVRRSAPVWSCGSCFSIFHLPCIRKWARSPASA 109
Query: 242 DLSAEKNLGFNWRCPGCQSVQHTSSKDIKYACFCGKRVDPPSDLYLTPHSCGEPCGKPLE 301
+++ + +WRCPGCQSV ++++ Y CFCG+R +PP+DL+L PHSCGEPC KPLE
Sbjct: 110 ADASDPDS--SWRCPGCQSVHAVPARELAYTCFCGRRREPPNDLFLXPHSCGEPCSKPLE 167
Query: 302 KE--VFVTEERKDELCPHACVLQCHPGPCPPCKAFAPPRLCPCGKKRIATRCSDRQSDLT 359
K ++ CPH CVLQCHPGPCPPCKAFAP RLCPCGK+ I RC+DR + +T
Sbjct: 168 KADPAVKADDAAATRCPHVCVLQCHPGPCPPCKAFAPDRLCPCGKQTIVRRCADRTTPVT 227
Query: 360 CGQRCDKLLDCGRHHCENACHVGPCDPCQVLIEASCFCSKMTQVLFCGEMAMKGEFEAEG 419
CGQRCD+LL C RH CE CH GPC C VLI A CFC K T+ L CGEM +KG +
Sbjct: 228 CGQRCDRLLPCRRHRCEKVCHTGPCGDCNVLISARCFCGKKTETLLCGEMELKGNLSEKD 287
Query: 420 GVFSCGSNCGNVLGCSNHICREVCHPGSCGECEFLPSRVKACCCGKTKLEDERKSCVDPI 479
GVFSC C ++L C NH C+++CHPG CGECE +P +V AC CGKT+L ++R SC+DPI
Sbjct: 288 GVFSCSEACSHMLSCGNHACQDICHPGPCGECELMPGKVTACHCGKTRLLEKRASCLDPI 347
Query: 480 PTCSKVCSKTLRCGVHACKETCHVGECPPCKVLISQKCRCGSTSRTVECYKTTENQKFTC 539
PTC KVC K L CGVH CK TCH G+CPPC V + Q+CRCGS+ + VECYK E ++F C
Sbjct: 348 PTCDKVCDKKLPCGVHRCKVTCHEGDCPPCVVRVEQRCRCGSSGQMVECYKVLE-EEFRC 406
Query: 540 QKPCGAKKNCGRHRCSEKCCPLSGPNNGLTTPDWDPHFCSMLCGKKLRCGQHVCETLCHS 599
KPCG KKNCGRHRCSE CCPLS P L +WDPH C + CGKKLRCGQH C+ LCHS
Sbjct: 407 NKPCGRKKNCGRHRCSECCCPLSKPLARLEGGNWDPHLCQIPCGKKLRCGQHGCQLLCHS 466
Query: 600 GHCPPCLETIFTDLACACGMTSIPPPLPCGTMPPLCQLPCSVPQPCGHSGSHSCHFGDCP 659
GHCPPCLETIF DL CACG TSIPPPLPCGT P C C VPQPCGH +H CHFGDCP
Sbjct: 467 GHCPPCLETIFNDLTCACGRTSIPPPLPCGTPTPSCPHQCLVPQPCGHPATHQCHFGDCP 526
Query: 660 PCSVPVSKECVGGHVILRNIPCGSNNIKCNNPCGRTRQCGLHACGRSCHSPPCDILPGIV 719
PC VPV +EC+GGHV+LRNIPCGS +I+CN PCG+ RQCG+HAC RSCH PCD
Sbjct: 527 PCVVPVMRECIGGHVVLRNIPCGSKDIRCNQPCGKNRQCGMHACNRSCHPSPCD------ 580
Query: 720 KGLRAACGQTCGAPR-SGCRHMCMALCHPGCPCPDARCEFPVTITCSCGRISANVPCDVG 778
CGQ CGAPR G + P PCPD RCEFP+TI CSCGRI+A VPC G
Sbjct: 581 -----PCGQVCGAPRXZGGXXTFDSSMPPILPCPDLRCEFPMTIACSCGRITATVPCSAG 635
Query: 779 GNNSNYNADAIFEASIIQKLPMPLQPVDANGQKVPLGQRKLMCDEECAKLERKRVLADAF 838
G N D +FE SIIQKLPMPLQPV+++G++VPLGQRKL CDE+CAK+ERKRVLA+AF
Sbjct: 636 G---TANGDNMFEVSIIQKLPMPLQPVESDGRRVPLGQRKLSCDEDCAKMERKRVLAEAF 692
Query: 839 DIT-PSLDALHFGENS-SYELLSDTFRRDPKWVLAIEERCKILVLGKSKGTTHG-LKVHV 895
DIT P+LDALHFGENS + +LLSD FRR+PKWV+AIEERCK LVLGK++G + G LKVHV
Sbjct: 693 DITPPNLDALHFGENSNASDLLSDLFRREPKWVMAIEERCKFLVLGKTRGNSSGNLKVHV 752
Query: 896 FCPMIKDKRDAVRMIAERWKLAVNAAGWEPKRFIVISATQKSKAPARVLGVKGTTTLNAP 955
FC M KDKRDA+R+IA+RWKL+V AAGWEPKRFI I T KSKAPAR+LG K + A
Sbjct: 753 FCHMTKDKRDAIRVIADRWKLSVQAAGWEPKRFITIHPTPKSKAPARILGSKPGVFVAAS 812
Query: 956 LPTAFDPLVDMDPRLVVSFPDLPRDADISALVLRFGGECELVWLNDRNALAVFHDPARAA 1015
P FDPLVDMDPRLVV+ DLPRDAD+SALVLRFGGECELVWLND+NA+AVF+DPARAA
Sbjct: 813 HP-FFDPLVDMDPRLVVAMLDLPRDADVSALVLRFGGECELVWLNDKNAVAVFNDPARAA 871
Query: 1016 TAMRRLDHGTVYQGAVSFVQNAGASAASSVTSAW-GGTKEGAL----RSNPWKKAAVLDP 1070
TA+RRLD+G+ YQGA F+ ++ A + W G K+G +NPWKKA +P
Sbjct: 872 TALRRLDYGSAYQGAAVFLPSSSAQPG----NVWVAGQKDGVAATKSSANPWKKATASEP 927
Query: 1071 GWKEDSW-------GDEQWTTAGDSANIQPSALKKEAPIPASLNPWNVLNHESSSSSSPA 1123
W W GD+ A + + N WN L ++++SS P
Sbjct: 928 DPSSGDWTGVLGQAPGSVWRRGGDTV----------AQVMGTSNRWNALESDAATSSRPV 977
Query: 1124 TVIRSVASGKQTESGNVSTKVEPSAGGADGGNSDATEAAEVVDDWEKAFE 1173
+ T SAG + + VDDWE+A E
Sbjct: 978 ----------EESKPAPRTDAVSSAGPSTAPPVSKMQPEVEVDDWEEACE 1017
>ref|NP_076228.1| nuclear transcription factor, X-box binding 1 [Mus musculus]
gi|6980018|gb|AAF34700.1| cysteine-rich protein NFX-1
[Mus musculus]
Length = 1114
Score = 582 bits (1501), Expect = e-164
Identities = 303/769 (39%), Positives = 407/769 (52%), Gaps = 63/769 (8%)
Query: 194 EKLTKGTVECMICYDMVRRSAPIWSCSSCYSIFHLNCIKKWARAPTSVDLSAEKNLGFNW 253
E+LT ECM+C ++V+ +AP+WSC SC+ +FHLNCIKKWAR+P S A+ G W
Sbjct: 343 EQLTTEKYECMVCCELVQVTAPVWSCQSCFHVFHLNCIKKWARSPAS---HADGQSG--W 397
Query: 254 RCPGCQSVQ-HTSSKDIKYACFCGKRVDPPSDLYLTPHSCGEPCGKPLEKEVFVTEERKD 312
RCP CQ+V H + Y CFCGK +P PHSCGE C K ++
Sbjct: 398 RCPACQNVSAHVPNT---YTCFCGKVKNPEWSRNEIPHSCGEVCRK----------KQPG 444
Query: 313 ELCPHACVLQCHPGPCPPCKAFAPPRLCPCGKKRIATRCSDRQSDLTCGQRCDKLLDCGR 372
+ CPH+C L CHPGPCPPC AF + C CG+ R RC S + C C+ +L+CG+
Sbjct: 445 QDCPHSCNLLCHPGPCPPCPAFTT-KTCECGRTRHTVRCGQPVS-VHCSNACENILNCGQ 502
Query: 373 HHCENACHVGPCDPCQVLIEASCFCSKMTQVLFCGEMAMKGEFEAEGGVFSCGSNCGNVL 432
HHC CH G C PC++++ C+C ++ + CG K + G FSC CG L
Sbjct: 503 HHCAELCHGGQCQPCRIILNQVCYCGSTSRDVLCGTDVGKSD---GFGDFSCLKICGKDL 559
Query: 433 GCSNHICREVCHPGSCGECEFLPSRVKACCCGKTKLE-------DERKSCVDPIPTCSKV 485
C +H C +VCHP C C LP V+ C CG+T L + RK+C+DP+P+C KV
Sbjct: 560 KCGSHTCSQVCHPQPCQPCPRLPHLVRYCPCGQTPLSQLLEHGSNARKTCMDPVPSCGKV 619
Query: 486 CSKTLRCG----VHACKETCHVGECPPCKVLISQKCRCGSTSRTVECYKT-TENQKFTCQ 540
C K L CG +H C++ CH G+C PC CRC ++ + C +E+ F C
Sbjct: 620 CGKPLACGSSDFIHTCEKLCHEGDCGPCSRTSVISCRCSFRTKELPCTSLKSEDATFMCD 679
Query: 541 KPCGAKKNCGRHRCSEKCCPLSGPNNGLTTPDWDPHFCSMLCGKKLRCGQHVCETLCHSG 600
K C K+ CGRH+C+E CC H C ++CG+KLRCG H CE CH G
Sbjct: 680 KRCNKKRLCGRHKCNEICCVDK------------EHKCPLICGRKLRCGLHRCEEPCHRG 727
Query: 601 HCPPCLETIFTDLACACGMTSIPPPLPCGTMPPLCQLPCSVPQPCGHSGSHSCHFGD-CP 659
+C C + F +L C CG + I PP+PCGT PP C C+ C H HSCH + CP
Sbjct: 728 NCQTCWQASFDELTCHCGASVIYPPVPCGTRPPECTQTCARIHECDHPVYHSCHSEEKCP 787
Query: 660 PCSVPVSKECVGGHVILRNIPCGSNNIKCNNPCGRTRQCGLHACGRSCHSPPCDILPGIV 719
PC+ K C+G H + NIPC +I C PC CG+H C R CH C +
Sbjct: 788 PCTFLTQKWCMGKHELRSNIPCHLVDISCGLPCSAMLPCGMHKCQRLCHKGECLV----- 842
Query: 720 KGLRAACGQTCGAPRSGCRHMCMALCHPGCPCPDARCEFPVTITCSCGRISANVPCDVGG 779
AC Q C PR C H CMA CHP PCP C+ V + C CGR V C
Sbjct: 843 ---DEACKQPCTTPRGDCGHPCMAPCHPSLPCPVTACKAKVELQCECGRRKEMVICS-EA 898
Query: 780 NNSNYNADAIFEASIIQKLPM-PLQPVDANGQKVPLGQRKLMCDEECAKLERKRVLADAF 838
+ + AI AS I + + + K + Q +L CDEECA LER++ LA+AF
Sbjct: 899 SGTYQRIVAISMASKITDMQLGDSVEISKLITKKEVQQARLQCDEECAALERRKRLAEAF 958
Query: 839 DITPSLDALHFGENSS--YELLSDTFRRDPKWVLAIEERCKILVLGKSKGTTHGLKVHVF 896
DIT D + ++S + L D R+D K+V +E+ + LV +KG + K H F
Sbjct: 959 DITDDSDPFNVRSSASKFSDSLKDDARKDLKFVSDVEKEMETLVEAVNKG-KNNKKSHCF 1017
Query: 897 CPMIKDKRDAVRMIAERWKLAVNAAGWEPKRFIVISATQ-KSKAPARVL 944
PM +D R + +A+ + L + EPKR +V++A + KS P L
Sbjct: 1018 PPMNRDHRRIIHDLAQVYGLESISYDSEPKRNVVVTAVRGKSVCPPTTL 1066
>emb|CAI13305.1| nuclear transcription factor, X-box binding 1 [Homo sapiens]
gi|55663201|emb|CAH72746.1| nuclear transcription factor,
X-box binding 1 [Homo sapiens] gi|15082473|gb|AAH12151.1|
Nuclear transcription factor, X-box binding 1, isoform 1
[Homo sapiens] gi|21361137|ref|NP_002495.2| nuclear
transcription factor, X-box binding 1 isoform 1 [Homo
sapiens]
Length = 1120
Score = 578 bits (1491), Expect = e-163
Identities = 303/769 (39%), Positives = 403/769 (52%), Gaps = 63/769 (8%)
Query: 194 EKLTKGTVECMICYDMVRRSAPIWSCSSCYSIFHLNCIKKWARAPTSVDLSAEKNLGFNW 253
E+LT ECM+C ++VR +AP+WSC SCY +FHLNCIKKWAR+P S A+ G W
Sbjct: 349 EQLTTEKYECMVCCELVRVTAPVWSCQSCYHVFHLNCIKKWARSPAS---QADGQSG--W 403
Query: 254 RCPGCQSVQ-HTSSKDIKYACFCGKRVDPPSDLYLTPHSCGEPCGKPLEKEVFVTEERKD 312
RCP CQ+V H + Y CFCGK +P PHSCGE C K ++
Sbjct: 404 RCPACQNVSAHVPNT---YTCFCGKVKNPEWSRNEIPHSCGEVCRK----------KQPG 450
Query: 313 ELCPHACVLQCHPGPCPPCKAFAPPRLCPCGKKRIATRCSDRQSDLTCGQRCDKLLDCGR 372
+ CPH+C L CHPGPCPPC AF + C CG+ R RC S + C C+ +L+CG+
Sbjct: 451 QDCPHSCNLLCHPGPCPPCPAFMT-KTCECGRTRHTVRCGQAVS-VHCSNPCENILNCGQ 508
Query: 373 HHCENACHVGPCDPCQVLIEASCFCSKMTQVLFCGEMAMKGEFEAEGGVFSCGSNCGNVL 432
H C CH G C PCQ+++ C+C ++ + CG K + G FSC CG L
Sbjct: 509 HQCAELCHGGQCQPCQIILNQVCYCGSTSRDVLCGTDVGKSD---GFGDFSCLKICGKDL 565
Query: 433 GCSNHICREVCHPGSCGECEFLPSRVKACCCGKTKLED-------ERKSCVDPIPTCSKV 485
C NH C +VCHP C +C LP V+ C CG+T L RK+C+DP+P+C KV
Sbjct: 566 KCGNHTCSQVCHPQPCQQCPRLPQLVRCCPCGQTPLSQLLELGSSSRKTCMDPVPSCGKV 625
Query: 486 CSKTLRCG----VHACKETCHVGECPPCKVLISQKCRCGSTSRTVECYKT-TENQKFTCQ 540
C K L CG +H C++ CH G+C PC CRC ++ + C +E+ F C
Sbjct: 626 CGKPLPCGSLDFIHTCEKLCHEGDCGPCSRTSVISCRCSFRTKELPCTSLKSEDATFMCD 685
Query: 541 KPCGAKKNCGRHRCSEKCCPLSGPNNGLTTPDWDPHFCSMLCGKKLRCGQHVCETLCHSG 600
K C K+ CGRH+C+E CC H C ++CG+KLRCG H CE CH G
Sbjct: 686 KRCNKKRLCGRHKCNEICCVDK------------EHKCPLICGRKLRCGLHRCEEPCHRG 733
Query: 601 HCPPCLETIFTDLACACGMTSIPPPLPCGTMPPLCQLPCSVPQPCGHSGSHSCHFGD-CP 659
+C C + F +L C CG + I PP+PCGT PP C C+ C H HSCH + CP
Sbjct: 734 NCQTCWQASFDELTCHCGASVIYPPVPCGTRPPECTQTCARVHECDHPVYHSCHSEEKCP 793
Query: 660 PCSVPVSKECVGGHVILRNIPCGSNNIKCNNPCGRTRQCGLHACGRSCHSPPCDILPGIV 719
PC+ K C+G H NIPC +I C PC T CG+H C R CH C +
Sbjct: 794 PCTFLTQKWCMGKHEFRSNIPCHLVDISCGLPCSATLPCGMHKCQRLCHKGECLV----- 848
Query: 720 KGLRAACGQTCGAPRSGCRHMCMALCHPGCPCPDARCEFPVTITCSCGRISANVPCDVGG 779
C Q C PR+ C H CMA CH PCP C+ V + C CGR V C
Sbjct: 849 ---DEPCKQPCTTPRADCGHPCMAPCHTSSPCPVTACKAKVELQCECGRRKEMVICSEAS 905
Query: 780 NNSNYNADAIFEASIIQKLPM-PLQPVDANGQKVPLGQRKLMCDEECAKLERKRVLADAF 838
+ A AI AS I + + + K + Q +L CDEEC+ LERK+ LA+AF
Sbjct: 906 STYQRIA-AISMASKITDMQLGGSVEISKLITKKEVHQARLECDEECSALERKKRLAEAF 964
Query: 839 DITPSLDALHFGENSS--YELLSDTFRRDPKWVLAIEERCKILVLGKSKGTTHGLKVHVF 896
I+ D + + S + L + R+D K+V +E+ + LV +KG + K H F
Sbjct: 965 HISEDSDPFNIRSSGSKFSDSLKEDARKDLKFVSDVEKEMETLVEAVNKG-KNSKKSHSF 1023
Query: 897 CPMIKDKRDAVRMIAERWKLAVNAAGWEPKRFIVISATQ-KSKAPARVL 944
PM +D R + +A+ + L + EPKR +V++A + KS P L
Sbjct: 1024 PPMNRDHRRIIHDLAQVYGLESVSYDSEPKRNVVVTAIRGKSVCPPTTL 1072
>emb|CAG10245.1| unnamed protein product [Tetraodon nigroviridis]
Length = 990
Score = 573 bits (1478), Expect = e-162
Identities = 314/796 (39%), Positives = 411/796 (51%), Gaps = 68/796 (8%)
Query: 186 PQLVQEIQEKLTKGTVECMICYDMVRRSAPIWSCSSCYSIFHLNCIKKWARAPTS-VDLS 244
P L + E+L++ ECM+C D++R +AP+WSC SC+ +FHLNCIKKWAR+P S D S
Sbjct: 209 PPLAGCLIEQLSEEKYECMVCCDIIRPAAPVWSCQSCFHVFHLNCIKKWARSPASQADDS 268
Query: 245 AEKNLGFNWRCPGCQSV--QHTSSKDIKYACFCGKRVDPPSDLYLTPHSCGEPCGKPLEK 302
AE WRCP CQ+V +H +S Y CFCGK +P PHSCG CGK
Sbjct: 269 AE-----GWRCPACQNVALKHPTS----YTCFCGKVTNPEWQRRRIPHSCGNMCGK---- 315
Query: 303 EVFVTEERKDELCPHACVLQCHPGPCPPCKAFAPPRLCPCGKKRIATRCSDRQSDLTCGQ 362
+R C H C + CHPGPCP C AF + C CGK RC + + L C +
Sbjct: 316 ------KRSAGDCNHPCNILCHPGPCPQCPAFVT-KSCVCGKISQPMRCG-QGTLLRCQE 367
Query: 363 RCDKLLDCGRHHCENACHVGPCDPCQVLIEASCFCSKMTQVLFCGEMAMKGEFEAEGGVF 422
C LL+CG H C CH G C PCQ+ ++ C+C + CG A K F+ G F
Sbjct: 368 ACSALLNCGEHTCPQVCHPGACQPCQIQVQQVCYCGVSRRGSLCG--ADKEGFDGSGH-F 424
Query: 423 SCGSNCGNVLGCSNHICREVCHPGSCGECEFLPSRVKACCCGKTKLED-------ERKSC 475
SCG C +L C H C++ CHPG C C PS V+ C CG+T L ER++C
Sbjct: 425 SCGKQCEMMLKCGFHRCQQQCHPGPCQPCPLSPSLVETCPCGQTPLAKLLELGYPERENC 484
Query: 476 VDPIPTCSKVCSKTLRCG----VHACKETCHVGECPPCKVLISQKCRCGSTSRTVEC--Y 529
DPIP+C K C++ L CG VH C++ CH G C PC + + +CRCGS +R V C
Sbjct: 485 TDPIPSCGKTCNRPLACGSSDSVHLCEKLCHEGRCGPCSLTSALRCRCGSKTREVPCETI 544
Query: 530 KTTENQKFTCQKPCGAKKNCGRHRCSEKCCPLSGPNNGLTTPDWDPHFCSMLCGKKLRCG 589
+T E FTC+K C K++CGRH+C E CC + H C +CG KL CG
Sbjct: 545 QTEEGLVFTCEKRCSKKRSCGRHKCGELCCVIV------------EHKCLQICGYKLNCG 592
Query: 590 QHVCETLCHSGHCPPCLETIFTDLACACGMTSIPPPLPCGTMPPLCQLPCSVPQPCGHSG 649
H C+ CH G+C PC ++ F +L C CG+T + PP+ CGT PP C+ C+ C H
Sbjct: 593 LHRCQEPCHRGNCDPCWQSSFDELTCHCGLTVLYPPIACGTKPPECKNMCTRRHECEHPV 652
Query: 650 SHSCHFGD-CPPCSVPVSKECVGGHVILRNIPCGSNNIKCNNPCGRTRQCGLHACGRSCH 708
H+CH + CPPC+ K C+G H NIPC +I C C + C H C R CH
Sbjct: 653 FHNCHSEEKCPPCTFLTEKWCMGKHEKRSNIPCHLQDISCGLVCNKALPCESHRCRRICH 712
Query: 709 SPPCDILPGIVKGLRAACGQTCGAPRSGCRHMCMALCHPGCPCPDARCEFPVTITCSCGR 768
C + G C Q C PR C H C A CH G CP C VT+ C CGR
Sbjct: 713 RGEC-----LADG---GCLQPCSLPRPDCGHPCSAPCHRGADCPGTPCTSKVTLQCGCGR 764
Query: 769 ISANVPCDVGGNNSNYNADAIFEASIIQKLPM-PLQPVDANGQKVPLGQRKLMCDEECAK 827
V C ++ A AI AS + + + + K + Q +L CD+EC
Sbjct: 765 RKELVLCTEAASSYQRYA-AIAMASKLSDMQLGDSMDIGLLLTKKEVNQARLECDQECIT 823
Query: 828 LERKRVLADAFDITPSLDALHFGENSS--YELLSDTFRRDPKWVLAIEERCKILVLGKSK 885
LER R LA+A I PS + G ++S L D R+D K+V +EE K LV +K
Sbjct: 824 LERNRRLAEALQIDPSSEPFISGRSTSAFSSSLKDDARKDLKFVAEVEEEMKNLVELTNK 883
Query: 886 GTTHGLKVHVFCPMIKDKRDAVRMIAERWKLAVNAAGWEPKRFIVISATQKSKAPAR--V 943
G + H F PM ++ R V +AE + L + EPKR +VI+A + A R +
Sbjct: 884 G-KQPKRSHCFPPMNREHRKMVHELAEAYALESVSYDSEPKRNVVITAHKGKAACPRWSL 942
Query: 944 LGVKGTTTLNAPLPTA 959
V T AP P A
Sbjct: 943 TSVIERETARAPPPIA 958
>sp|Q12986|NFX1_HUMAN Transcriptional repressor NF-X1 (Nuclear transcription factor, X
box-binding, 1) gi|563217|gb|AAA69517.1| NFX1
Length = 1104
Score = 568 bits (1464), Expect = e-160
Identities = 300/769 (39%), Positives = 401/769 (52%), Gaps = 63/769 (8%)
Query: 194 EKLTKGTVECMICYDMVRRSAPIWSCSSCYSIFHLNCIKKWARAPTSVDLSAEKNLGFNW 253
E+LT ECM+C ++VR +AP+WSC SCY +FHLNCIKKWAR+P S A+ G W
Sbjct: 333 EQLTTEKYECMVCCELVRVTAPVWSCQSCYHVFHLNCIKKWARSPAS---QADGQSG--W 387
Query: 254 RCPGCQSVQ-HTSSKDIKYACFCGKRVDPPSDLYLTPHSCGEPCGKPLEKEVFVTEERKD 312
RCP CQ+V H + ++CFCGK +P PHSCGE C K ++
Sbjct: 388 RCPACQNVSAHVPNT---FSCFCGKVKNPEWSRNEIPHSCGEVCRK----------KQPG 434
Query: 313 ELCPHACVLQCHPGPCPPCKAFAPPRLCPCGKKRIATRCSDRQSDLTCGQRCDKLLDCGR 372
+ CPH+C L CHPGPCPPC AF + C CG+ R RC S + C C+ +L+CG+
Sbjct: 435 QDCPHSCNLLCHPGPCPPCPAFMT-KTCECGRTRHTVRCGQAVS-VHCSNPCENILNCGQ 492
Query: 373 HHCENACHVGPCDPCQVLIEASCFCSKMTQVLFCGEMAMKGEFEAEGGVFSCGSNCGNVL 432
H C CH G C PCQ+++ C+C ++ + CG K + G FSC CG L
Sbjct: 493 HQCAELCHGGQCQPCQIILNQVCYCGSTSRDVLCGTDVGKSD---GFGDFSCLKTCGKDL 549
Query: 433 GCSNHICREVCHPGSCGECEFLPSRVKACCCGKTKLED-------ERKSCVDPIPTCSKV 485
C NH C +VCHP C +C LP V+ C CG+T L RK+C+DP+P+C KV
Sbjct: 550 KCGNHTCSQVCHPQPCQQCPRLPQLVRCCPCGQTPLSQLLELGSSSRKTCMDPVPSCGKV 609
Query: 486 CSKTLRCG----VHACKETCHVGECPPCKVLISQKCRCGSTSRTVECYKT-TENQKFTCQ 540
C K L CG +H C++ CH G+C P CRC ++ + C +E+ F C
Sbjct: 610 CGKPLPCGSLDFIHTCEKLCHEGDCGPVSRTSVISCRCSFRTKELPCTSLKSEDATFMCD 669
Query: 541 KPCGAKKNCGRHRCSEKCCPLSGPNNGLTTPDWDPHFCSMLCGKKLRCGQHVCETLCHSG 600
K C K+ CGRH+C+E CC H C + CG+KLRCG H CE CH G
Sbjct: 670 KRCNKKRLCGRHKCNEICCVDK------------EHKCPLNCGRKLRCGLHRCEEPCHRG 717
Query: 601 HCPPCLETIFTDLACACGMTSIPPPLPCGTMPPLCQLPCSVPQPCGHSGSHSCHFGD-CP 659
+C C + F +L C CG + I PP+PCGT PP C C+ C H HS H + CP
Sbjct: 718 NCQTCWQASFDELTCHCGASVIYPPVPCGTRPPECTQTCARVHECDHPVYHSGHSEEKCP 777
Query: 660 PCSVPVSKECVGGHVILRNIPCGSNNIKCNNPCGRTRQCGLHACGRSCHSPPCDILPGIV 719
PC+ K C+G H NIPC +I C PC T CG+H C R CH C +
Sbjct: 778 PCTFLTQKWCMGKHEFRSNIPCHLVDISCGLPCSATLPCGMHKCQRLCHKGECLV----- 832
Query: 720 KGLRAACGQTCGAPRSGCRHMCMALCHPGCPCPDARCEFPVTITCSCGRISANVPCDVGG 779
C Q C PR+ C H CMA CH PCP C+ V + C CGR V C
Sbjct: 833 ---DEPCKQPCTTPRADCGHPCMAPCHTSSPCPVTACKAKVELQCECGRRKEMVICSEAS 889
Query: 780 NNSNYNADAIFEASIIQKLPM-PLQPVDANGQKVPLGQRKLMCDEECAKLERKRVLADAF 838
+ A AI AS I + + + K + Q +L CDEEC+ LERK+ LA+AF
Sbjct: 890 STYQRIA-AISMASKITDMQLGGSVEISKLITKKEVHQARLECDEECSALERKKRLAEAF 948
Query: 839 DITPSLDALHFGENSS--YELLSDTFRRDPKWVLAIEERCKILVLGKSKGTTHGLKVHVF 896
I+ D + + S + L + R+D K+V +E+ + LV +KG + K H F
Sbjct: 949 HISEDSDPFNIRSSGSKFSDSLKEDARKDLKFVSDVEKEMETLVEAVNKG-KNSKKSHSF 1007
Query: 897 CPMIKDKRDAVRMIAERWKLAVNAAGWEPKRFIVISATQ-KSKAPARVL 944
PM +D R + +A+ + L + EPKR +V++A + KS P L
Sbjct: 1008 PPMNRDHRRIIHDLAQVYGLESVSYDSEPKRNVVVTAIRGKSVCPPTTL 1056
>ref|XP_538703.1| PREDICTED: similar to nuclear transcription factor, X-box binding 1
isoform 1 [Canis familiaris]
Length = 1223
Score = 565 bits (1456), Expect = e-159
Identities = 300/782 (38%), Positives = 399/782 (50%), Gaps = 85/782 (10%)
Query: 194 EKLTKGTVECMICYDMVRRSAPIWSCSSCYSIFHLNCIKKWARAPTSVDLSAEKNLGFNW 253
E+LT ECM+C ++VR +AP+WSC SCY +FHLNCIKKWAR+P S A+ G W
Sbjct: 498 EQLTTEKYECMVCCELVRVTAPVWSCQSCYHVFHLNCIKKWARSPAS---QADGQSG--W 552
Query: 254 RCPGCQSVQ-HTSSKDIKYACFCGKRVDPPSDLYLTPHSCGEPCGKPLEKEVFVTEERKD 312
RCP CQ+V H + Y CFCGK +P PHSCGE C K ++
Sbjct: 553 RCPACQNVSAHVPNT---YTCFCGKVKNPEWSRNEIPHSCGEVCRK----------KQPG 599
Query: 313 ELCPHACVLQCHPGPCPPCKAFAPPRLCPCGKKRIATRCSDRQSDLTCGQRCDKLLDCGR 372
+ CPH+C L CHPGPCPPC AF + C CG+ R RC S + C C+ LL+CG+
Sbjct: 600 QDCPHSCNLLCHPGPCPPCPAFMT-KTCECGRTRHTVRCGQAVS-VHCSNPCENLLNCGQ 657
Query: 373 HHCENACHVGPCDPCQVLIEASCFCSKMTQVLFCGEMAMKGEFEAEGGVFSCGSNCGNVL 432
HHC CH G C PC++++ C+C ++ + CG K + G F C CG L
Sbjct: 658 HHCAELCHGGQCQPCRIILNQVCYCGSTSRDVLCGTDVGKSD---GFGDFGCLKICGKDL 714
Query: 433 GCSNHICREVCHPGSCGECEFLPSRVKACCCGKTKL-------EDERKSCVDPIPTCSKV 485
C NH C +VCHP C C LP V+ C CG+T L RK+C+DP+P+C KV
Sbjct: 715 KCGNHTCSQVCHPQPCQPCPRLPQLVRYCPCGQTPLCQLLELGSSTRKTCMDPVPSCGKV 774
Query: 486 CSKTLRCG----VHACKETCHVGECPPCKVLISQKCRCGSTSRTVECYKTTENQKFTCQK 541
C K L CG +H C++ CH G+C PC + + F C K
Sbjct: 775 CGKPLPCGSLDFIHTCEKLCHEGDCGPC----------------------SPDATFMCDK 812
Query: 542 PCGAKKNCGRHRCSEKCCPLSGPNNGLTTPDWDPHFCSMLCGKKLRCGQHVCETLCHSGH 601
C K+ CGRH+C+E CC H C ++CG+KLRCG H CE CH G+
Sbjct: 813 RCNKKRLCGRHKCNEICCVDK------------EHKCPLICGRKLRCGLHRCEEPCHRGN 860
Query: 602 CPPCLETIFTDLACACGMTSIPPPLPCGTMPPLCQLPCSVPQPCGHSGSHSCHFGD-CPP 660
C C + F +L C CG + I PP+PCGT PP C C+ C H HSCH + CPP
Sbjct: 861 CQSCWQASFDELTCHCGASVIYPPVPCGTRPPECTQTCARVHECDHPVYHSCHSEEKCPP 920
Query: 661 CSVPVSKECVGGHVILRNIPCGSNNIKCNNPCGRTRQCGLHACGRSCHSPPCDILPGIVK 720
C+ K C+G H + NIPC +I C PC T CG+H C R CH C +
Sbjct: 921 CTFLTQKWCMGKHELRSNIPCHLADISCGLPCSATLPCGMHKCQRLCHKGECLV------ 974
Query: 721 GLRAACGQTCGAPRSGCRHMCMALCHPGCPCPDARCEFPVTITCSCGRISANVPCDVGGN 780
C Q C PR+ C H CMA CHP PCP C+ + + C CGR + C +
Sbjct: 975 --DEPCKQPCTTPRADCGHPCMAPCHPSSPCPMTACKAKIELQCECGRRKEMMICSEASS 1032
Query: 781 NSNYNADAIFEASIIQKLPM-PLQPVDANGQKVPLGQRKLMCDEECAKLERKRVLADAFD 839
A AI AS I + + + K + Q +L CDEEC LERK+ LA+AF
Sbjct: 1033 TYQRIA-AISMASKITDMQLGDSVEISKLITKKEIHQARLECDEECLALERKKRLAEAFH 1091
Query: 840 ITPSLDALHFGENSS--YELLSDTFRRDPKWVLAIEERCKILVLGKSKGTTHGLKVHVFC 897
I+ D + + S + L D R+D K+V IE+ + LV +KG + K H F
Sbjct: 1092 ISEDSDPFNVRSSGSKFSDSLKDDARKDLKFVSDIEKEMEALVEAVNKG-KNSKKSHCFP 1150
Query: 898 PMIKDKRDAVRMIAERWKLAVNAAGWEPKRFIVISATQKSK--APARVLGVKGTTTLNAP 955
PM +D R + +A+ + L + EPKR +V++A + P ++GV + P
Sbjct: 1151 PMNRDHRRIIHDLAQVYGLESVSYDSEPKRNVVVTAIRGKSICPPTTLIGVLEREMQSRP 1210
Query: 956 LP 957
P
Sbjct: 1211 PP 1212
>emb|CAI13304.1| nuclear transcription factor, X-box binding 1 [Homo sapiens]
gi|55663200|emb|CAH72745.1| nuclear transcription factor,
X-box binding 1 [Homo sapiens] gi|13242069|gb|AAK16545.1|
nuclear transcription factor NFX2 [Homo sapiens]
gi|22212925|ref|NP_667344.1| nuclear transcription
factor, X-box binding 1 isoform 2 [Homo sapiens]
Length = 1024
Score = 556 bits (1433), Expect = e-156
Identities = 283/709 (39%), Positives = 372/709 (51%), Gaps = 61/709 (8%)
Query: 194 EKLTKGTVECMICYDMVRRSAPIWSCSSCYSIFHLNCIKKWARAPTSVDLSAEKNLGFNW 253
E+LT ECM+C ++VR +AP+WSC SCY +FHLNCIKKWAR+P S A+ G W
Sbjct: 349 EQLTTEKYECMVCCELVRVTAPVWSCQSCYHVFHLNCIKKWARSPAS---QADGQSG--W 403
Query: 254 RCPGCQSVQ-HTSSKDIKYACFCGKRVDPPSDLYLTPHSCGEPCGKPLEKEVFVTEERKD 312
RCP CQ+V H + Y CFCGK +P PHSCGE C K ++
Sbjct: 404 RCPACQNVSAHVPNT---YTCFCGKVKNPEWSRNEIPHSCGEVCRK----------KQPG 450
Query: 313 ELCPHACVLQCHPGPCPPCKAFAPPRLCPCGKKRIATRCSDRQSDLTCGQRCDKLLDCGR 372
+ CPH+C L CHPGPCPPC AF + C CG+ R RC S + C C+ +L+CG+
Sbjct: 451 QDCPHSCNLLCHPGPCPPCPAFMT-KTCECGRTRHTVRCGQAVS-VHCSNPCENILNCGQ 508
Query: 373 HHCENACHVGPCDPCQVLIEASCFCSKMTQVLFCGEMAMKGEFEAEGGVFSCGSNCGNVL 432
H C CH G C PCQ+++ C+C ++ + CG K + G FSC CG L
Sbjct: 509 HQCAELCHGGQCQPCQIILNQVCYCGSTSRDVLCGTDVGKSD---GFGDFSCLKICGKDL 565
Query: 433 GCSNHICREVCHPGSCGECEFLPSRVKACCCGKTKLED-------ERKSCVDPIPTCSKV 485
C NH C +VCHP C +C LP V+ C CG+T L RK+C+DP+P+C KV
Sbjct: 566 KCGNHTCSQVCHPQPCQQCPRLPQLVRCCPCGQTPLSQLLELGSSSRKTCMDPVPSCGKV 625
Query: 486 CSKTLRCG----VHACKETCHVGECPPCKVLISQKCRCGSTSRTVECYKT-TENQKFTCQ 540
C K L CG +H C++ CH G+C PC CRC ++ + C +E+ F C
Sbjct: 626 CGKPLPCGSLDFIHTCEKLCHEGDCGPCSRTSVISCRCSFRTKELPCTSLKSEDATFMCD 685
Query: 541 KPCGAKKNCGRHRCSEKCCPLSGPNNGLTTPDWDPHFCSMLCGKKLRCGQHVCETLCHSG 600
K C K+ CGRH+C+E CC H C ++CG+KLRCG H CE CH G
Sbjct: 686 KRCNKKRLCGRHKCNEICCVDK------------EHKCPLICGRKLRCGLHRCEEPCHRG 733
Query: 601 HCPPCLETIFTDLACACGMTSIPPPLPCGTMPPLCQLPCSVPQPCGHSGSHSCHFGD-CP 659
+C C + F +L C CG + I PP+PCGT PP C C+ C H HSCH + CP
Sbjct: 734 NCQTCWQASFDELTCHCGASVIYPPVPCGTRPPECTQTCARVHECDHPVYHSCHSEEKCP 793
Query: 660 PCSVPVSKECVGGHVILRNIPCGSNNIKCNNPCGRTRQCGLHACGRSCHSPPCDILPGIV 719
PC+ K C+G H NIPC +I C PC T CG+H C R CH C +
Sbjct: 794 PCTFLTQKWCMGKHEFRSNIPCHLVDISCGLPCSATLPCGMHKCQRLCHKGECLV----- 848
Query: 720 KGLRAACGQTCGAPRSGCRHMCMALCHPGCPCPDARCEFPVTITCSCGRISANVPCDVGG 779
C Q C PR+ C H CMA CH PCP C+ V + C CGR V C
Sbjct: 849 ---DEPCKQPCTTPRADCGHPCMAPCHTSSPCPVTACKAKVELQCECGRRKEMVICSEAS 905
Query: 780 NNSNYNADAIFEASIIQKLPM-PLQPVDANGQKVPLGQRKLMCDEECAKLERKRVLADAF 838
+ A AI AS I + + + K + Q +L CDEEC+ LERK+ LA+AF
Sbjct: 906 STYQRIA-AISMASKITDMQLGGSVEISKLITKKEVHQARLECDEECSALERKKRLAEAF 964
Query: 839 DITPSLDALHFGENSS--YELLSDTFRRDPKWVLAIEERCKILVLGKSK 885
I+ D + + S + L + R+D K+V +E+ + LV +K
Sbjct: 965 HISEDSDPFNIRSSGSKFSDSLKEDARKDLKFVSDVEKEMETLVEAVNK 1013
>ref|XP_395912.2| PREDICTED: similar to ENSANGP00000015832 [Apis mellifera]
Length = 994
Score = 528 bits (1359), Expect = e-148
Identities = 273/760 (35%), Positives = 375/760 (48%), Gaps = 72/760 (9%)
Query: 190 QEIQEKLTKGTVECMICYDMVRRSAPIWSCSSCYSIFHLNCIKKWARAPTSVDLSAEKNL 249
+ + E+L +G +EC++C + ++++ IWSCS+CY + HL CIKKWA++ + +
Sbjct: 222 ERLTEQLNRGQLECLVCCEYIKQNDYIWSCSNCYHVLHLKCIKKWAKSSQAEN------- 274
Query: 250 GFNWRCPGCQSVQHTSSKDIKYACFCGKRVDPPSDLYLTPHSCGEPCGKPLEKEVFVTEE 309
WRCP CQ+V +D Y CFCGK P + HSCGE CG+ L K
Sbjct: 275 --GWRCPACQNVNLIIPED--YFCFCGKTKAPEWNRRDVAHSCGEICGRMLSKNN----- 325
Query: 310 RKDELCPHACVLQCHPGPCPPCKAFAPPRLCPCGKKRIATRCSDRQSDLTCGQRCDKLLD 369
CPH C L CHPG CP C A + C CGK +CS + L C CDK L+
Sbjct: 326 -----CPHKCTLLCHPGSCPQCTAMVA-KYCGCGKTLQTVQCSVHKL-LLCDSICDKNLN 378
Query: 370 CGRHHCENACHVGPCDPCQVLIEASCFCSKMTQVLFCGEMAMKGEFEAEGGVFSCGSNCG 429
CGRH C+ CH G C C+ I CFC K T+ + C +SC + CG
Sbjct: 379 CGRHKCQKKCHHGECANCEETITQKCFCGKNTREITCENNISL--------TYSCETVCG 430
Query: 430 NVLGCSNHICREVCHPGSCGECEFLPSRVKACCCGKTKLEDERKSCVDPIPTCSKVCSKT 489
+L C NH C ++CH +C C +P ++ CCCG+T L D+R+SC+DPIP C K+CSK
Sbjct: 431 KLLECGNHTCTKLCHADACESCSLIPEKITTCCCGQTPLTDKRQSCLDPIPVCEKICSKN 490
Query: 490 LRCGV----HACKETCHVGECPPCKVLISQKCRCGSTSRTVECYKT-TENQKFTCQKPCG 544
L+CG H CK CH G+CP C + KCRCG+ R + C ++ C+K C
Sbjct: 491 LKCGQPNNPHKCKARCHEGDCPVCDLSTDVKCRCGNMDREIACKDLISKADDARCEKKCI 550
Query: 545 AKKNCGRHRCSEKCCPLSGPNNGLTTPDWDPHFCSMLCGKKLRCGQHVCETLCHSGHCPP 604
K++CG+H+C++ CC H C ++C K L CG+H CE CH G C P
Sbjct: 551 KKRSCGKHKCNQLCCIEI------------EHICPLICSKTLSCGRHKCEQRCHKGRCQP 598
Query: 605 CLETIFTDLACACGMTSIPPPLPCGTMPPLCQLPCSVPQPCGHSGSHSCHFG-DCPPCSV 663
C + F +L C CG + I PP+PCGT P+C PCS CGH H+CH CPPC+V
Sbjct: 599 CWRSSFDELYCECGSSVIYPPVPCGTRRPVCDKPCSRQHSCGHEVLHNCHSEITCPPCTV 658
Query: 664 PVSKECVGGHVILRNIPCGSNNIKCNNPCGRTRQCGLHACGRSCHSPPCDILPGIVKGLR 723
+ C G H + + +PC N I C C + CG H C CH PC+ PG
Sbjct: 659 LTQRWCHGKHELRKAVPCHVNEISCGLSCNKPISCGRHKCIIICHPGPCE-KPG------ 711
Query: 724 AACGQTCGAPRSGCRHMCMALCHPGCPCPDARCEFPVTITCSCGRISANVPCDVGGNNSN 783
C Q C R C H+C A CH G CPD C+ V +TC CG + C
Sbjct: 712 QQCSQPCTTLREMCGHICAAPCHEG-KCPDTPCKEMVKVTCQCGHRTMFRVCAENSKEYQ 770
Query: 784 YNADAIFEASIIQKL---PMPLQPVDANGQKVPLGQRKLMCDEECAKLERKRVLADAFD- 839
A +I + + + L+ V G K + L C++EC +ER R LA
Sbjct: 771 RIASSILASKMADMQLGHSINLEEVFGQGVKKQNQLKTLECNDECKIIERNRRLALGLQI 830
Query: 840 ITPSLDALHFGENSSYELLSDTFRRDPKWVLAIEERCKILV----LGKSKGTTHGLKVHV 895
I P L S + + ++DP + + E+ LV + K K ++
Sbjct: 831 INPDLSGKLMPRYSDF--MKQWAKKDPHFCQMVHEKLTELVQLAKISKQKSRSYS----- 883
Query: 896 FCPMIKDKRDAVRMIAERWKLAVNAAGWEPKRFIVISATQ 935
F M KDKR + E + A EPKR +V +A +
Sbjct: 884 FDSMNKDKRHFIHESCEHFGCESQAYDQEPKRNVVATAVK 923
>gb|AAX70563.1| hypothetical protein, conserved [Trypanosoma brucei]
Length = 807
Score = 525 bits (1353), Expect = e-147
Identities = 287/786 (36%), Positives = 365/786 (45%), Gaps = 98/786 (12%)
Query: 174 AGRQYGSRDSSLPQLVQEIQEKLTKGTVECMICYDMVRRSAPIWSCSSCYSIFHLNCIKK 233
AG Q GS ++ Q + I E+L EC C + +R ++SC CY IFHL CIK+
Sbjct: 6 AGDQDGSSYDAVVQRINGIAEQLKSLRYECANCLNKIRHDKAVFSCRDCYCIFHLYCIKR 65
Query: 234 WARAPTSVDLSAEKNLGFNWRCPGCQSVQHTSSKDIKYACFCGKRVDPPSDLYLTPHSCG 293
WA+ S D +A +RCP CQ VQ + D+KY CFCGK DP D ++TPHSCG
Sbjct: 66 WAKEDASGDGNA-------FRCPHCQVVQRS---DLKYYCFCGKVRDPKYDPHITPHSCG 115
Query: 294 EPCGKPLEKEVFVTEERKDELCPHACVLQCHPGPCPPCKAFAPPRLCPCGKKRIATRCSD 353
+ CGK + CPH+C L CHPGPC PC A + P C CG RC
Sbjct: 116 DTCGKA-----------RPNNCPHSCPLPCHPGPCVPCSAVSGPGSCGCGSTSYTWRCGQ 164
Query: 354 RQSDLTCGQRCDKLLDCGRHHCENACHVGPCDPCQVLIEASCFCSKMTQVLFCGEMAMKG 413
TC + C K+L CG+H C + CH G C C SC C + + CG
Sbjct: 165 PDPLKTCDRICGKMLSCGKHLCPDKCHHGACRGCSKQSTTSCHCGSEVRPMRCGST---- 220
Query: 414 EFEAEGGVFSCGSNCGNVLGCSNHICREVCHPGSCGECEFLPSRVKACCCGKTKLEDERK 473
VFSCG CG L C H C +CH G+C C+ PS V C CG L R
Sbjct: 221 -------VFSCGKVCGRELKCGVHHCERMCHEGTCPPCKTDPSVVATCPCGSVPLVVVRT 273
Query: 474 SCVDPIPTCSKVCSKTLRCGVHACKETCHVGECPPCKVLISQKCRCGSTSRTVECYKTTE 533
C DP+P C VC + L CG H C CH G C PC+ ++ C CG T T+ C
Sbjct: 274 RCSDPVPVCGNVCGRWLDCGEHKCDAKCHEGGCEPCQQQVAATCSCGKTQATLPCLM--- 330
Query: 534 NQKFTCQKPCGAKKNCGRHRCSEKCCPLSGPNNGLTTPDWDPHFCSMLCGKKLRCGQHVC 593
+ F C K C A +CG+H+C +CCP NN D H C +C KKL CG H C
Sbjct: 331 RRSFRCNKVCKAALSCGKHQCRARCCPARKGNNA------DAHCCRQVCNKKLSCG-HNC 383
Query: 594 ETLCHSGHCPPCLETIFTDLACACGMTSIPPPLPCGTMPPLCQLPCSVPQPCGH-SGSHS 652
LCH G CP C + L C CG + PP PCGT PP C PCSVP PCGH +GSH
Sbjct: 384 MELCHRGPCPSCPNVVSERLRCRCGAEELMPPQPCGTKPPQCNRPCSVPLPCGHPTGSHI 443
Query: 653 CHFGDCPPCSVPVSKECVGGHVILRNIPCGSNNIKCNNPCGRTRQCGLHACGRSCHSPPC 712
CHFGDCPPC PV + C GGH I+RN+PC + + C CG+ CG H C R CH PC
Sbjct: 444 CHFGDCPPCMFPVERLCAGGHRIVRNVPCSAESTTCTYKCGKQLSCG-HKCVRFCHGGPC 502
Query: 713 DILPGIVKGLRAACGQTCGAPRSGCRHMCMALCHPGCPCPDARCEFPVTITCSCGRISAN 772
+ CGQ CG + C H C + CH CP C + C C R
Sbjct: 503 -------VDEQRPCGQPCGKIQDVCGHACASKCHGSANCP--VCIVVEELHCECRRRVVQ 553
Query: 773 VPCDVGGNNSNYNADAIFEASIIQKLPMPLQPVDANGQKVPLGQRKLM-CDEECAKLERK 831
V C +IQ +K GQR L+ CD EC ++R
Sbjct: 554 VSC----------------GKLIQ------------FKKEHQGQRYLIKCDAECLAVQRL 585
Query: 832 RVLADAFDITPSLDALHFGENSSYELLSDTFRRDPKWVLAIEERCKILVLGKSKGTTHGL 891
+L+ P + F L + D + + E V+G
Sbjct: 586 EILSKRIKPHPPIKYSVF--------LWEAALTDVEQIRKFERILHTFVVGCE------- 630
Query: 892 KVHVFCPMIKDKRDAVRMIAERWKLAVNAAGWEPKRFIVISATQKSKAPARVLGVK-GTT 950
V M KR V + + + + EP+R +++ T K+K P +L V T
Sbjct: 631 GVMCLGAMAAPKRAIVHNMCHYYHINSESVDPEPRRGCLLTRTPKTKLPDPLLSVAIATP 690
Query: 951 TLNAPL 956
++PL
Sbjct: 691 EQHSPL 696
>ref|NP_476598.1| CG3647-PA, isoform A [Drosophila melanogaster]
gi|22946527|gb|AAN10891.1| CG3647-PA, isoform A
[Drosophila melanogaster]
Length = 1099
Score = 511 bits (1317), Expect = e-143
Identities = 302/967 (31%), Positives = 441/967 (45%), Gaps = 129/967 (13%)
Query: 24 EWIPKGAGASSSASTTSTTTTASTTTAAAATTTVDQPTSLHSHEKNTNSDGGSSNQGAVV 83
E++P A S + + TT ++T + +P +L + E ++ G++NQ
Sbjct: 168 EFVPNFAKLSLEETPAAATTNGNSTASLETAINETRPRTLRAQEP---AERGANNQC--- 221
Query: 84 APTFARHRSNHHVAHRVEREHVAHRT---DRDHVAHRVERERVAHRVEREHVAHRMEREH 140
SNH+ ERE R DRD R +R+R R + ++
Sbjct: 222 --------SNHNYERERERERDRDRDRERDRDRDRDR-DRDRDRDRDRDSRPGNTRQQRR 272
Query: 141 VAHRVEREHVAHRVEREHVAHRVERGRGRSGNMAGRQYGSRD--SSLPQLVQEI------ 192
+R +RE R +R + RS +RD + P+ V ++
Sbjct: 273 SDYRDDREDRYERSDRRRPQKQQRYDNHRSNKRRDDWNRNRDRINGFPRAVDDLDTSNES 332
Query: 193 --------------------------------QEKLTKGT----VECMICYDMVRRSAPI 216
+EKL + +EC++C + ++ P
Sbjct: 333 AHPSPEKQSQLQQISPRRGPPLPPADNEKLSQREKLVRDIEQRRLECLVCVEAIKSHQPT 392
Query: 217 WSCSSCYSIFHLNCIKKWARAPTSVDLSAEKNLGFNWRCPGCQSVQHTSSKDIKYACFCG 276
WSC +CY + HL C WA S++ +G WRCP CQ+V +D Y CFCG
Sbjct: 393 WSCRNCYHMLHLKCTITWAS-------SSKSEVG--WRCPACQNVLQDLPRD--YLCFCG 441
Query: 277 KRVDPPSDLYLTPHSCGEPCGKPLEKEVFVTEERKDELCPHACVLQCHPGPCPPCKAFAP 336
K +PP HSCGE C + E C HAC L CHPGPCPPC+A
Sbjct: 442 KLKNPPVSRTELAHSCGEVCCRI-------------EGCSHACTLLCHPGPCPPCQANVV 488
Query: 337 PRLCPCGKKRIATRCSDRQSDLTCGQRCDKLLDCGRHHCENACHVGPCDPCQVLIEASCF 396
R C CG+ +C+ ++ ++ CG+ CDKLL+CG H C+ CH G C C + C
Sbjct: 489 -RSCGCGRSTKTMQCAMKE-EVLCGEICDKLLNCGEHRCQAECHSGKCAACSEQVVQQCH 546
Query: 397 CSKMTQVLFCGEMAMKGEFEAEGGVFSCGSNCGNVLGCSNHICREVCHPGSCGECEFLPS 456
C K + + C + + +SC +CG L C +H C++ CH GSC C+ P
Sbjct: 547 CGKQERKVPCTRESQ------DKRTYSCKDSCGQPLPCGHHKCKDSCHAGSCRPCKLSPE 600
Query: 457 RVKACCCGKTKLE-DERKSCVDPIPTCSKVCSKTLRCGV----HACKETCHVGECPPCKV 511
++ +C CGK + +R SC+DPIPTC +CS+TLRCG H C CH+G+CPPC
Sbjct: 601 QITSCPCGKMPVPAGQRSSCLDPIPTCEGICSRTLRCGKPAHPHQCGSKCHLGQCPPCPK 660
Query: 512 LISQKCRCGSTSRTVECYKTTEN-QKFTCQKPCGAKKNCGRHRCSEKCCPLSGPNNGLTT 570
KCRCG + ++C + C++ C K++CG+H+C+ +CC
Sbjct: 661 QTGVKCRCGHMDQMIKCRQLCNRADDARCKRRCTKKRSCGKHKCNVECCI---------- 710
Query: 571 PDWDPHFCSMLCGKKLRCGQHVCETLCHSGHCPPCLETIFTDLACACGMTSIPPPLPCGT 630
D D H C + C + L CG+H C+ CH G+CPPC + F +L C CG I PP+PCGT
Sbjct: 711 -DID-HDCPLPCNRTLSCGKHKCDQPCHRGNCPPCYRSSFEELYCECGAEVIYPPVPCGT 768
Query: 631 MPPLCQLPCSVPQPCGHSGSHSCHFG-DCPPCSVPVSKECVGGHVILRNIPCGSNNIKCN 689
P+C+LPCS PC H H+CH G CPPC + +K C G H + + IPC N C
Sbjct: 769 KKPICKLPCSRIHPCDHPPQHNCHSGPTCPPCMIFTTKLCHGNHELRKTIPCSQPNFSCG 828
Query: 690 NPCGRTRQCGLHACGRSCHSPPCDILPGIVKGLRAACGQTCGAPRSGCRHMCMALCHPGC 749
CG+ CG H C + CH PC I C Q+C PR C H C A CH G
Sbjct: 829 MACGKPLPCGGHKCIKPCHEGPCQSAGEI-------CRQSCTKPRPTCGHKCAAACHEGA 881
Query: 750 PCPDARCEFPVTITCSCGRISANVPCDVGGNNSNYNADAIFEASIIQKLP---MPLQPVD 806
CP+ C+ V + C CG N C + A +S+ + M L +
Sbjct: 882 -CPETPCKELVEVQCECGNRKQNRSCQELAREHSRIATIQLASSMAEMSRGNYMELSEIL 940
Query: 807 ANGQKVPLGQRKLMCDEECAKLERKRVLADAFDITPSLDALHFGENSSYELLSDTFRRDP 866
A +K + L C++EC LER R LA A + + D E + +++P
Sbjct: 941 APAKK---SNKTLDCNDECRLLERNRRLAAALS-SGNSDTKQKCLTKYSEFVRGFAKKNP 996
Query: 867 KWVLAIEERCKILVLGKSKGTTHGLKVHVFCPMIKDKRDAVRMIAERWKLAVNAAGWEPK 926
++ E LV +K + + H F M ++KR V + E + + + EP
Sbjct: 997 ALTKSVYETLTDLV-KLAKESKQRSRSHSFPTMNREKRQLVHELCEVFGIESVSYDKEPN 1055
Query: 927 RFIVISA 933
R +V +A
Sbjct: 1056 RNVVATA 1062
>ref|NP_476599.1| CG3647-PB, isoform B [Drosophila melanogaster]
gi|22946526|gb|AAF53441.2| CG3647-PB, isoform B
[Drosophila melanogaster] gi|7287884|gb|AAF44922.1|
symbol=stc; synonym=BG:DS04929.4; match=method:''sim4'',
score:''1000.0'', desc:''GenBank::U09306:stc FBgn0001978
SWISS-PROT:P40798 U09306:224..3544'',
species:''Drosophila melanogaster'';
match=method:''BLASTX'', version:''2.0a19MP-WashU
[05-Feb-1998] [Build sol2.5-ultra 01:47:30
05-Feb-1998]'', score:''877.0'',
desc:''trEMBL::Q18034:SIMILAR TO D. MELANOGASTER SHUTTLE
CRAFT PROTEIN. organism:CAENORHABDITIS ELEGANS.
dbxref:GenBank; U41534; g1109825; -.'',
species:''CAENORHABDITIS ELEGANS
gi|5052648|gb|AAD38654.1| BcDNA.LD22726 [Drosophila
melanogaster] gi|41712398|sp|P40798|STC_DROME Shuttle
craft protein gi|11359837|pir||T44598 hypothetical
protein [imported] - fruit fly (Drosophila melanogaster)
Length = 1106
Score = 511 bits (1317), Expect = e-143
Identities = 302/967 (31%), Positives = 441/967 (45%), Gaps = 129/967 (13%)
Query: 24 EWIPKGAGASSSASTTSTTTTASTTTAAAATTTVDQPTSLHSHEKNTNSDGGSSNQGAVV 83
E++P A S + + TT ++T + +P +L + E ++ G++NQ
Sbjct: 175 EFVPNFAKLSLEETPAAATTNGNSTASLETAINETRPRTLRAQEP---AERGANNQC--- 228
Query: 84 APTFARHRSNHHVAHRVEREHVAHRT---DRDHVAHRVERERVAHRVEREHVAHRMEREH 140
SNH+ ERE R DRD R +R+R R + ++
Sbjct: 229 --------SNHNYERERERERDRDRDRERDRDRDRDR-DRDRDRDRDRDSRPGNTRQQRR 279
Query: 141 VAHRVEREHVAHRVEREHVAHRVERGRGRSGNMAGRQYGSRD--SSLPQLVQEI------ 192
+R +RE R +R + RS +RD + P+ V ++
Sbjct: 280 SDYRDDREDRYERSDRRRPQKQQRYDNHRSNKRRDDWNRNRDRINGFPRAVDDLDTSNES 339
Query: 193 --------------------------------QEKLTKGT----VECMICYDMVRRSAPI 216
+EKL + +EC++C + ++ P
Sbjct: 340 AHPSPEKQSQLQQISPRRGPPLPPADNEKLSQREKLVRDIEQRRLECLVCVEAIKSHQPT 399
Query: 217 WSCSSCYSIFHLNCIKKWARAPTSVDLSAEKNLGFNWRCPGCQSVQHTSSKDIKYACFCG 276
WSC +CY + HL C WA S++ +G WRCP CQ+V +D Y CFCG
Sbjct: 400 WSCRNCYHMLHLKCTITWAS-------SSKSEVG--WRCPACQNVLQDLPRD--YLCFCG 448
Query: 277 KRVDPPSDLYLTPHSCGEPCGKPLEKEVFVTEERKDELCPHACVLQCHPGPCPPCKAFAP 336
K +PP HSCGE C + E C HAC L CHPGPCPPC+A
Sbjct: 449 KLKNPPVSRTELAHSCGEVCCRI-------------EGCSHACTLLCHPGPCPPCQANVV 495
Query: 337 PRLCPCGKKRIATRCSDRQSDLTCGQRCDKLLDCGRHHCENACHVGPCDPCQVLIEASCF 396
R C CG+ +C+ ++ ++ CG+ CDKLL+CG H C+ CH G C C + C
Sbjct: 496 -RSCGCGRSTKTMQCAMKE-EVLCGEICDKLLNCGEHRCQAECHSGKCAACSEQVVQQCH 553
Query: 397 CSKMTQVLFCGEMAMKGEFEAEGGVFSCGSNCGNVLGCSNHICREVCHPGSCGECEFLPS 456
C K + + C + + +SC +CG L C +H C++ CH GSC C+ P
Sbjct: 554 CGKQERKVPCTRESQ------DKRTYSCKDSCGQPLPCGHHKCKDSCHAGSCRPCKLSPE 607
Query: 457 RVKACCCGKTKLE-DERKSCVDPIPTCSKVCSKTLRCGV----HACKETCHVGECPPCKV 511
++ +C CGK + +R SC+DPIPTC +CS+TLRCG H C CH+G+CPPC
Sbjct: 608 QITSCPCGKMPVPAGQRSSCLDPIPTCEGICSRTLRCGKPAHPHQCGSKCHLGQCPPCPK 667
Query: 512 LISQKCRCGSTSRTVECYKTTEN-QKFTCQKPCGAKKNCGRHRCSEKCCPLSGPNNGLTT 570
KCRCG + ++C + C++ C K++CG+H+C+ +CC
Sbjct: 668 QTGVKCRCGHMDQMIKCRQLCNRADDARCKRRCTKKRSCGKHKCNVECCI---------- 717
Query: 571 PDWDPHFCSMLCGKKLRCGQHVCETLCHSGHCPPCLETIFTDLACACGMTSIPPPLPCGT 630
D D H C + C + L CG+H C+ CH G+CPPC + F +L C CG I PP+PCGT
Sbjct: 718 -DID-HDCPLPCNRTLSCGKHKCDQPCHRGNCPPCYRSSFEELYCECGAEVIYPPVPCGT 775
Query: 631 MPPLCQLPCSVPQPCGHSGSHSCHFG-DCPPCSVPVSKECVGGHVILRNIPCGSNNIKCN 689
P+C+LPCS PC H H+CH G CPPC + +K C G H + + IPC N C
Sbjct: 776 KKPICKLPCSRIHPCDHPPQHNCHSGPTCPPCMIFTTKLCHGNHELRKTIPCSQPNFSCG 835
Query: 690 NPCGRTRQCGLHACGRSCHSPPCDILPGIVKGLRAACGQTCGAPRSGCRHMCMALCHPGC 749
CG+ CG H C + CH PC I C Q+C PR C H C A CH G
Sbjct: 836 MACGKPLPCGGHKCIKPCHEGPCQSAGEI-------CRQSCTKPRPTCGHKCAAACHEGA 888
Query: 750 PCPDARCEFPVTITCSCGRISANVPCDVGGNNSNYNADAIFEASIIQKLP---MPLQPVD 806
CP+ C+ V + C CG N C + A +S+ + M L +
Sbjct: 889 -CPETPCKELVEVQCECGNRKQNRSCQELAREHSRIATIQLASSMAEMSRGNYMELSEIL 947
Query: 807 ANGQKVPLGQRKLMCDEECAKLERKRVLADAFDITPSLDALHFGENSSYELLSDTFRRDP 866
A +K + L C++EC LER R LA A + + D E + +++P
Sbjct: 948 APAKK---SNKTLDCNDECRLLERNRRLAAALS-SGNSDTKQKCLTKYSEFVRGFAKKNP 1003
Query: 867 KWVLAIEERCKILVLGKSKGTTHGLKVHVFCPMIKDKRDAVRMIAERWKLAVNAAGWEPK 926
++ E LV +K + + H F M ++KR V + E + + + EP
Sbjct: 1004 ALTKSVYETLTDLV-KLAKESKQRSRSHSFPTMNREKRQLVHELCEVFGIESVSYDKEPN 1062
Query: 927 RFIVISA 933
R +V +A
Sbjct: 1063 RNVVATA 1069
>gb|AAB47595.1| Hypothetical protein C16A3.7 [Caenorhabditis elegans]
gi|17552206|ref|NP_498394.1| nuclear transcription
factor X-box binding 1 like (125.1 kD) (3H467)
[Caenorhabditis elegans] gi|25395800|pir||A88481 protein
C16A3.6 [imported] - Caenorhabditis elegans
Length = 1119
Score = 506 bits (1303), Expect = e-141
Identities = 292/890 (32%), Positives = 413/890 (45%), Gaps = 102/890 (11%)
Query: 92 SNHHVAHRVEREHVAHRTDRDHVAHRVERERVAHRVEREHVAHRMEREHVAHRVEREHVA 151
S H + ++E +R R + R+ + R++ +++ + +A
Sbjct: 115 STHSNQNSRQQEPSQNRRRRGGQQNTQRRQLEIQEQRGDSHPQNQSRQNNRNQLNVDRIA 174
Query: 152 HRVEREHVAHRVERGRGRSGNMAGRQYGSRDSSLPQLVQEIQEK-LTKGTV--------E 202
++ + G R G A R+ + + + + +K L +G V E
Sbjct: 175 NQQNKSVQNPSRNPGNSRRGQGARRREQKEEPLTEEETKILADKPLREGLVYLLENNKYE 234
Query: 203 CMICYDMVRRSAPIWSCSSCYSIFHLN--CIKKWARAPTSVDLSAEKNLGFNWRCPGCQS 260
C ICY + +WSC +CY IFH++ CI WAR+ S +K WRCP CQ+
Sbjct: 235 CAICYTRITTRQGVWSCKTCYHIFHISTGCITDWARS------SRDKEGANTWRCPTCQT 288
Query: 261 VQHTSSKDIKYACFCGKRVDPPSDLYLTPHSCGEPCGKPLEKEVFVTEERKDELCPHACV 320
T + Y CFCG+ +P + PHSCGE CG + CPH C
Sbjct: 289 ENETMPYN--YYCFCGRMRNPNFRVGEVPHSCGETCGGARKFG-----------CPHPCT 335
Query: 321 LQCHPGPCPPCKAFAPPRLCPCGKKRIATRCSDRQSDLTCGQRCDKLLDCGRHHCENACH 380
CHPGPC CK F + C CGK + + RC Q ++ C C K L CG+H+CE CH
Sbjct: 336 ELCHPGPCIECKLFTT-KSCNCGKTKKSVRCGSDQ-EVMCETVCGKQLSCGQHNCERICH 393
Query: 381 VGPCDPCQVLIEASCFCSKMTQVLFCGEMAMKGEFEAEGGVFSCGSNCGNVLGCSNHICR 440
G C C V++E CFC K + + C A + +SCGS C + C H C
Sbjct: 394 SGDCGECTVILEQDCFCGKTPKEVSCNPCAHEK--------YSCGSECDGMFSCGIHHCT 445
Query: 441 EVCHPGSCGECEFLPSRVKACCCGKTKLED---ERKSCVDPIPTCSKVCSKTLRCGV--- 494
+ CH CGECE +R++ C CG+ L+ RK C D +PTC VC K L CG
Sbjct: 446 KKCHDKECGECETGANRIRTCPCGRNTLQSLGVTRKKCTDMVPTCDSVCDKWLTCGTPGK 505
Query: 495 -HACKETCHVGECPPCKVLISQKCRCGSTSRTVECYKTTENQKFT----CQKPCGAKKNC 549
H C+E CH G CPPC + S CRCG++ + C + + K T C K C KK+C
Sbjct: 506 NHHCREKCHEGPCPPCNLNTSVICRCGTSKGVIPCDEYLQIMKTTGEYLCTKRCRKKKSC 565
Query: 550 GRHRCSEKCCPLSGPNNGLTTPDWDPHFCSMLCGKKLRCGQHVCETLCHSGHCPPCLETI 609
G H+C E CC D HFC +C K+L CG H CE +CH+G C PCL+
Sbjct: 566 GMHKCQEVCCIQ------------DEHFCLQMCNKRLSCGIHTCENVCHAGQCRPCLQAS 613
Query: 610 FTDLACACGMTSIPPPLPCGTMPPLCQLPCSVPQPCGHSGSHSCH-FGDCPPCSVPVSKE 668
F + C CG T PP+PCG P+C PC P C HS SH CH +CPPC+ K
Sbjct: 614 FDEQFCHCGHTVRMPPIPCGARLPVCSQPCVRPHSCDHSVSHKCHGEQNCPPCTQLTEKM 673
Query: 669 CVGGHVILRNIPCGSNNIKCNNPCGRTRQCGLHACGRSCHSPPCDILPGIVKGLRAACGQ 728
C GGH + +NIPC +++ C C + +CG+H C R+CH C+ +G + C +
Sbjct: 674 CYGGHRVRKNIPCHIDSVSCGVVCKKPLKCGVHVCQRTCHGEECE-----KEGEK--CTK 726
Query: 729 TCGAPRSGCRHMCMALCHPGCPCPDARCEFPVTITCSCGRISANVPCDVGGNNSNYNADA 788
C R C H C CH PC + C+ V I+C CGRI + PC D
Sbjct: 727 KCETIRELCEHPCALPCHEDSPCEPSPCKASVRISCECGRIKKDAPC--------CEVDK 778
Query: 789 IFEASIIQKLPMPLQPVDANGQKVPLGQRK----------LMCDEECAKLERKRVLADAF 838
+ S ++K D + V G K + CD+EC KLER R +A+A
Sbjct: 779 MI-LSKLEKEEEEKSESDGEEKVVKEGILKRSTSFSQLNCMKCDDECKKLERNRKVAEAL 837
Query: 839 DITPSLDALHFGENSSYELLS------DTFRRDPKWVLAIEERCKILVLGKSKGTTHGLK 892
++ D +G N +S + R + +V ++E+ LV+ G +
Sbjct: 838 EV----DTDEYGMNKLAPTISFPCYLKEMVRTNIDFVKSVEKILIDLVIQILSGEAYHDT 893
Query: 893 VHVFCP-MIKDKRDAVRMIAERWKLAVNAAGWEPKRFIVISATQ-KSKAP 940
P M +KR V A + +A + PKR IV++A + KS P
Sbjct: 894 FRAHLPAMSIEKRRFVHEYANFFNIASESVDSPPKRSIVLTAVRGKSHQP 943
>gb|AAB60255.1| shuttle craft protein [Drosophila melanogaster]
Length = 1106
Score = 505 bits (1301), Expect = e-141
Identities = 301/967 (31%), Positives = 439/967 (45%), Gaps = 129/967 (13%)
Query: 24 EWIPKGAGASSSASTTSTTTTASTTTAAAATTTVDQPTSLHSHEKNTNSDGGSSNQGAVV 83
E++P A S + + TT ++T + +P +L + E ++ G++NQ
Sbjct: 175 EFVPNFAKLSLEETPAAATTNGNSTASLETAINETRPRTLRAQEP---AERGANNQC--- 228
Query: 84 APTFARHRSNHHVAHRVEREHVAHRT---DRDHVAHRVERERVAHRVEREHVAHRMEREH 140
SNH+ ERE R DRD R +R+R R + ++
Sbjct: 229 --------SNHNYERERERERDRDRDRERDRDRDRDR-DRDRDRDRDRDSRPGNTRQQRR 279
Query: 141 VAHRVEREHVAHRVEREHVAHRVERGRGRSGNMAGRQYGSRD--SSLPQLVQEI------ 192
+R +RE R +R + RS +RD + P+ V ++
Sbjct: 280 SDYRDDREDRYERSDRRRPQKQQRYDNHRSNKRRDDWNRNRDRINGFPRAVDDLDTSNES 339
Query: 193 --------------------------------QEKLTKGT----VECMICYDMVRRSAPI 216
+EKL + +EC++C + ++ P
Sbjct: 340 AHPSPEKQSQLQQISPRRGPPLPPADNEKLSQREKLVRDIEQRRLECLVCVEAIKSHQPT 399
Query: 217 WSCSSCYSIFHLNCIKKWARAPTSVDLSAEKNLGFNWRCPGCQSVQHTSSKDIKYACFCG 276
WSC +CY + HL C WA S++ +G WRCP CQ+V +D Y CFCG
Sbjct: 400 WSCRNCYHMLHLKCTITWAS-------SSKSEVG--WRCPACQNVLQDLPRD--YLCFCG 448
Query: 277 KRVDPPSDLYLTPHSCGEPCGKPLEKEVFVTEERKDELCPHACVLQCHPGPCPPCKAFAP 336
K +PP HSCGE C + E C HAC L CHPGPCPPC+A
Sbjct: 449 KLKNPPVSRTELAHSCGEVCCRI-------------EGCSHACTLLCHPGPCPPCQANVV 495
Query: 337 PRLCPCGKKRIATRCSDRQSDLTCGQRCDKLLDCGRHHCENACHVGPCDPCQVLIEASCF 396
R C CG+ +C+ ++ ++ CG+ CDKLL+CG H C+ CH G C C + C
Sbjct: 496 -RSCGCGRSTKTMQCAMKE-EVLCGEICDKLLNCGEHRCQAECHSGKCAACSEQVVQQCH 553
Query: 397 CSKMTQVLFCGEMAMKGEFEAEGGVFSCGSNCGNVLGCSNHICREVCHPGSCGECEFLPS 456
C K + + C + + +SC +CG L C +H C++ CH GSC C+ P
Sbjct: 554 CGKQERKVPCTRESQ------DKRTYSCKDSCGQPLPCGHHKCKDSCHAGSCRPCKLSPE 607
Query: 457 RVKACCCGKTKLE-DERKSCVDPIPTCSKVCSKTLRCGV----HACKETCHVGECPPCKV 511
++ +C CGK + +R SC+DPIPTC +CS+TLRCG H C CH+G+CPPC
Sbjct: 608 QITSCPCGKIPVPAGQRSSCLDPIPTCEGICSRTLRCGKPAHPHQCGSKCHLGQCPPCPK 667
Query: 512 LISQKCRCGSTSRTVECYKTTEN-QKFTCQKPCGAKKNCGRHRCSEKCCPLSGPNNGLTT 570
KCRCG + ++C + C++ C K++CG+H+C+ +CC
Sbjct: 668 QTGVKCRCGHMDQMIKCRQLCNRADDARCKRRCTKKRSCGKHKCNVECCI---------- 717
Query: 571 PDWDPHFCSMLCGKKLRCGQHVCETLCHSGHCPPCLETIFTDLACACGMTSIPPPLPCGT 630
D D H C + C + L CG+H C+ CH G+CPPC + F +L C CG I PP+PCGT
Sbjct: 718 -DID-HDCPLPCNRTLSCGKHKCDQPCHRGNCPPCYRSSFEELYCECGAEVIYPPVPCGT 775
Query: 631 MPPLCQLPCSVPQPCGHSGSHSCHFG-DCPPCSVPVSKECVGGHVILRNIPCGSNNIKCN 689
P+C+LP S PC H H+CH G CPPC + +K C G H + IPC N C
Sbjct: 776 KKPICKLPSSRIHPCDHPPQHNCHSGPTCPPCMIFTTKLCHGNHDWRKTIPCSQPNFSCG 835
Query: 690 NPCGRTRQCGLHACGRSCHSPPCDILPGIVKGLRAACGQTCGAPRSGCRHMCMALCHPGC 749
CG+ CG H C + CH PC I C Q+C PR C H C A CH G
Sbjct: 836 MACGKPLPCGGHKCIKPCHEGPCQSAGEI-------CRQSCTKPRPTCGHKCAAACHEGA 888
Query: 750 PCPDARCEFPVTITCSCGRISANVPCDVGGNNSNYNADAIFEASIIQKLP---MPLQPVD 806
CP+ C+ V + C CG N C + A +S+ + M L +
Sbjct: 889 -CPETPCKELVEVQCECGNRKQNRSCQELAREHSRIATIQLASSMAEMSRGNYMELSEIL 947
Query: 807 ANGQKVPLGQRKLMCDEECAKLERKRVLADAFDITPSLDALHFGENSSYELLSDTFRRDP 866
A +K + L C++EC LER R LA A + + D E + +++P
Sbjct: 948 APAKK---SNKTLDCNDECRLLERNRRLAAALS-SGNSDTKQKCLTKYSEFVRGFAKKNP 1003
Query: 867 KWVLAIEERCKILVLGKSKGTTHGLKVHVFCPMIKDKRDAVRMIAERWKLAVNAAGWEPK 926
++ E LV +K + + H F M ++KR V + E + + + EP
Sbjct: 1004 ALTKSVYETLTDLV-KLAKESKQRSRSHSFPTMNREKRQLVHELCEVFGIESVSYDKEPN 1062
Query: 927 RFIVISA 933
R +V +A
Sbjct: 1063 RNVVATA 1069
>gb|EAL34416.1| GA17585-PA [Drosophila pseudoobscura]
Length = 1074
Score = 504 bits (1298), Expect = e-141
Identities = 265/761 (34%), Positives = 384/761 (49%), Gaps = 68/761 (8%)
Query: 183 SSLPQLVQEIQEKLTKGTVECMICYDMVRRSAPIWSCSSCYSIFHLNCIKKWARAPTSVD 242
S +L+++I+++ +EC++C + ++ WSC +CY + HL C WA
Sbjct: 335 SQREKLIRDIEQR----RLECLVCVETIKSHHSTWSCQNCYHVIHLKCTITWAS------ 384
Query: 243 LSAEKNLGFNWRCPGCQSVQHTSSKDIKYACFCGKRVDPPSDLYLTPHSCGEPCGKPLEK 302
S++ ++G WRCP CQ+V ++ Y CFCGK +P HSCGE C +
Sbjct: 385 -SSKSDVG--WRCPACQNVLQDLPRE--YLCFCGKLKNPTVSRNELAHSCGEVCCRI--- 436
Query: 303 EVFVTEERKDELCPHACVLQCHPGPCPPCKAFAPPRLCPCGKKRIATRCSDRQSDLTCGQ 362
E C HAC L CHPGPCPPC+A R C CG+ +C+ ++ +L CG+
Sbjct: 437 ----------EGCSHACTLLCHPGPCPPCQANVV-RSCGCGRSSQTMQCAMKE-ELKCGE 484
Query: 363 RCDKLLDCGRHHCENACHVGPCDPCQVLIEASCFCSKMTQVLFCGEMAMKGEFEAEGGVF 422
CDK+L+CG HHC+ CH G C C +E C C K + + C ++ + +
Sbjct: 485 ICDKVLNCGEHHCKEECHSGKCAACPEQVEQHCHCGKQDRQVPCTRESL------DKRNY 538
Query: 423 SCGSNCGNVLGCSNHICREVCHPGSCGECEFLPSRVKACCCGKTKLED-ERKSCVDPIPT 481
SC +CG L C NH C++ CH G+C C+ P ++ +C CGK + +R SC+D IPT
Sbjct: 539 SCKESCGQPLPCGNHKCKDSCHAGNCRPCKLSPEQITSCPCGKMPVPAAQRSSCLDAIPT 598
Query: 482 CSKVCSKTLRCGV----HACKETCHVGECPPCKVLISQKCRCGSTSRTVECYK-TTENQK 536
C +CS+TLRCG H C CH+G+CPPC+ + KCRCG + ++C + +T
Sbjct: 599 CEGICSRTLRCGKPAHPHQCGSKCHLGQCPPCQKQTAVKCRCGHMDQMIKCRQLSTRADD 658
Query: 537 FTCQKPCGAKKNCGRHRCSEKCCPLSGPNNGLTTPDWDPHFCSMLCGKKLRCGQHVCETL 596
C+K C K++CG+H+C+ +CC D D H C + C + L CG+H C+
Sbjct: 659 ARCKKRCIKKRSCGKHKCNAECCI-----------DID-HACPLPCNRTLSCGKHKCDQP 706
Query: 597 CHSGHCPPCLETIFTDLACACGMTSIPPPLPCGTMPPLCQLPCSVPQPCGHSGSHSCH-F 655
CH G+CPPC + F +L C CG I PP+PCGT PLC+ PCS PC HS H+CH
Sbjct: 707 CHRGNCPPCYRSSFEELFCECGAEVIYPPVPCGTKKPLCKRPCSRQHPCDHSPQHNCHSA 766
Query: 656 GDCPPCSVPVSKECVGGHVILRNIPCGSNNIKCNNPCGRTRQCGLHACGRSCHSPPCDIL 715
CPPC + +K C G H + IPC + C CG+ CG H C + CH PC
Sbjct: 767 ATCPPCMMFTTKWCHGNHEQRKTIPCSQESFSCGLSCGKPLPCGRHKCIKPCHEGPCQSS 826
Query: 716 PGIVKGLRAACGQTCGAPRSGCRHMCMALCHPGCPCPDARCEFPVTITCSCGRISANVPC 775
P + C Q+C PR+ C H C + CH G CP+ C+ V + C CG + C
Sbjct: 827 PSDI------CRQSCTKPRTLCGHKCASACHEGA-CPETPCKELVDVQCECGNRKLSRSC 879
Query: 776 DVGGNNSNYNADAIFEASIIQKLP---MPLQPVDANGQKVPLGQRKLMCDEECAKLERKR 832
+ A A +S+ + M L + A KV R L C++EC LER R
Sbjct: 880 QELAREHSRIATAQLASSMAEMSRGNYMELSEILAPA-KVNKSNRTLDCNDECRVLERNR 938
Query: 833 VLADAFDITPSLDALHFGENSSYELLSDTFRRDPKWVLAIEERCKILVLGKSKGTTHGLK 892
LA A + + D E L +R+ ++ E LV +K + +
Sbjct: 939 RLAQALQ-SRNPDTQQKSLTKYSEFLRGFAKRNQALTKSVYETLTDLV-KLAKESKQRSR 996
Query: 893 VHVFCPMIKDKRDAVRMIAERWKLAVNAAGWEPKRFIVISA 933
H F M ++KR V + E + + + EP R +V +A
Sbjct: 997 SHSFPTMNREKRQMVHELCEIFGIESVSYDKEPNRNVVATA 1037
>emb|CAE72509.1| Hypothetical protein CBG19688 [Caenorhabditis briggsae]
Length = 1099
Score = 499 bits (1284), Expect = e-139
Identities = 288/846 (34%), Positives = 397/846 (46%), Gaps = 111/846 (13%)
Query: 162 RVERGRGRSGNMA-GRQYGSRDSSLPQLVQE---------IQEKLT----KGTVECMICY 207
R E RG GN G+ R+ QL +E ++E+L EC ICY
Sbjct: 159 REENRRGAPGNYRRGQGARRREQKEEQLTEEETAMLADKPLRERLVYMLENNKYECAICY 218
Query: 208 DMVRRSAPIWSCSSCYSIFHLN--CIKKWARAPTSVDLSAEKNLGFN-WRCPGCQSVQHT 264
+ +WSC +CY IFH++ CI WA++ S +K G N WRCP CQ+ T
Sbjct: 219 TRITTRQGVWSCKTCYHIFHISTGCITDWAKS------SRDKEAGSNTWRCPTCQTENET 272
Query: 265 SSKDIKYACFCGKRVDPPSDLYLTPHSCGEPCGKPLEKEVFVTEERKDELCPHACVLQCH 324
+ Y CFCGK+ +P + PHSCGE CG + CPH C CH
Sbjct: 273 MPYN--YYCFCGKQRNPNFRVGEVPHSCGEICGGTRKFG-----------CPHPCNELCH 319
Query: 325 PGPCPPCKAFAPPRLCPCGKKRIATRCSDRQSDLTCGQRCDKLLDCGRHHCENACHVGPC 384
PGPC CK + + C CGK + + RC QS L C C K L CG+H CE CH G C
Sbjct: 320 PGPCSECKLYVT-KSCNCGKTKKSVRCGSGQSVL-CDTVCGKTLPCGQHTCEKICHSGEC 377
Query: 385 DPCQVLIEASCFCSKMTQVLFCGEMAMKGEFEAEGGVFSCGSNCGNVLGCSNHICREVCH 444
C V++E CFC K + + C A + +SCGS C + C H C + CH
Sbjct: 378 GDCTVVLEQECFCGKKPKEVVCNPTACQK--------YSCGSECDGMFSCGVHRCTKKCH 429
Query: 445 PGSCGECEFLPSRVKACCCGKTKLED---ERKSCVDPIPTCSKVCSKTLRCGV----HAC 497
CG+CE +R++ C CG+ L+ R+ C D +PTC +C K L CG H C
Sbjct: 430 GKECGDCETGVNRIRTCPCGRNSLQSLGIVREKCTDKVPTCDSICDKWLTCGTPGKNHRC 489
Query: 498 KETCHVGECPPCKVLISQKCRCGSTSRTVECYKTTENQKFT----CQKPCGAKKNCGRHR 553
+E CH +CPPC + S CRCGS+ + C + E + T C K C KK+CG H+
Sbjct: 490 REKCHEDQCPPCNLNTSVVCRCGSSKGVILCEEYLEIMRTTGEYLCTKRCRKKKSCGMHK 549
Query: 554 CSEKCCPLSGPNNGLTTPDWDPHFCSMLCGKKLRCGQHVCETLCHSGHCPPCLETIFTDL 613
C E CC D H+C +C K+L CG H CE +CH+G C PCL+ F +
Sbjct: 550 CQEVCCIQ------------DEHYCLQMCNKRLSCGIHTCENVCHAGQCRPCLQASFDEQ 597
Query: 614 ACACGMTSIPPPLPCGTMPPLCQLPCSVPQPCGHSGSHSCH-FGDCPPCSVPVSKECVGG 672
C CG T PP+PCG P+C PC P C H +H CH +CPPC+ K C GG
Sbjct: 598 FCHCGHTVRMPPIPCGARLPVCSQPCVRPHSCDHPVNHKCHGEPNCPPCTHLTDKTCYGG 657
Query: 673 HVILRNIPCGSNNIKCNNPCGRTRQCGLHACGRSCHSPPCDILPGIVKGLRAACGQTCGA 732
H + +NIPC ++ C C + +CG+H C R+CH C+ +G C + C A
Sbjct: 658 HTVRKNIPCHIQSVSCGVICQKPLKCGVHVCQRTCHGDECE-----KEG--EQCTKKCEA 710
Query: 733 PRSGCRHMCMALCHPGCPCPDARCEFPVTITCSCGRISANVPCDVGGNNSNYNADAIFEA 792
R C H C CH PC + C+ V ITC CGR+ PC D + EA
Sbjct: 711 IRERCEHPCGLPCHKDSPCEPSPCKTNVQITCECGRLKKAAPC--------CEVDKLIEA 762
Query: 793 SIIQKLPMPLQPVDANGQKVPLGQ----------RKLMCDEECAKLERKRVLADAFDITP 842
++ +G + G+ + CD++C KLER R +A+A ++
Sbjct: 763 KQEKEETEKSGDSGDSGDQDKPGKLVRSSSFSQLNCMKCDDDCKKLERNRKVAEALEV-- 820
Query: 843 SLDALHFGENSSYELLS------DTFRRDPKWVLAIEERCKILVLGKSKGTTHG--LKVH 894
D +G N +S R + +V +E+ LV+ G + + H
Sbjct: 821 --DTDEYGMNKLTPTISFPCYLKAMVRANIDFVKNVEKVFIDLVIKVLSGEAYHDVFRAH 878
Query: 895 VFCPMIKDKRDAVRMIAERWKLAVNAAGWEPKRFIVISATQ-KSKAPARVLG--VKGTTT 951
+ PM +KR V A + + + PKR IV++A + KS P ++ V
Sbjct: 879 L-PPMSIEKRRFVHEYANFFNIVSESVDSPPKRSIVLTAVRGKSHQPLILISDLVNYKGA 937
Query: 952 LNAPLP 957
L AP P
Sbjct: 938 LKAPGP 943
>ref|XP_314840.2| ENSANGP00000015832 [Anopheles gambiae str. PEST]
gi|55239925|gb|EAA10127.2| ENSANGP00000015832 [Anopheles
gambiae str. PEST]
Length = 744
Score = 497 bits (1280), Expect = e-139
Identities = 267/743 (35%), Positives = 361/743 (47%), Gaps = 62/743 (8%)
Query: 201 VECMICYDMVRRSAPIWSCSSCYSIFHLNCIKKWARAPTSVDLSAEKNLGFNWRCPGCQS 260
+EC++C ++V+ WSC +CY I HL C+ KWA + S D G WRCP CQ
Sbjct: 17 LECLVCCEIVKPMQSTWSCGNCYHILHLGCVTKWATSSQSED-------GSGWRCPACQH 69
Query: 261 VQHTSSKDIKYACFCGKRVDPPSDLYLTPHSCGEPCGKPLEKEVFVTEERKDELCPHACV 320
V ++ Y CFCGK+ +P + HSCGE CG+ ++C HAC
Sbjct: 70 VSKKVPRE--YFCFCGKQKNPAYNREDLAHSCGEVCGRR-------------DVCEHACT 114
Query: 321 LQCHPGPCPPCKAFAPPRLCPCGKKRIATRCSDRQSDLTCGQRCDKLLDCGRHHCENACH 380
L CHPGPCPPC+A + R C CG+ +C ++ +L C C+K L+CG H C CH
Sbjct: 115 LLCHPGPCPPCQA-SVQRRCGCGRLEKTVQCCQKE-ELLCEAVCEKPLNCGLHRCGRQCH 172
Query: 381 VGPCDPCQVLIEASCFCSKMTQVLFCGEMAMKGEFEAEGGVFSCGSNCGNVLGCSNHICR 440
G CD C ++ SC C K + + C + + + CG C LGC NH C
Sbjct: 173 AGSCDTCTEQVQHSCHCGKGERSVVCSVENLAVQR------YGCGKACERPLGCGNHRCA 226
Query: 441 EVCHPGSCGECEFLPSRVKACCCGKTKL-EDERKSCVDPIPTCSKVCSKTLRCGV----H 495
+CH G C C P RV++C CG+ + R+SC+DP+PTCS C + L CG H
Sbjct: 227 RLCHEGECEACADAPDRVRSCPCGREPIVPGSRQSCLDPVPTCSGPCGRKLACGPPGANH 286
Query: 496 ACKETCHVGECPPCKVLISQKCRCGSTSRTVECYK-TTENQKFTCQKPCGAKKNCGRHRC 554
C+ CH G CPPCK + KCRCG + + C + TT C+K C +++C +H+C
Sbjct: 287 CCEAACHRGACPPCKKSTTVKCRCGGMDQPMRCKELTTRADDARCKKRCVRRRSCAKHKC 346
Query: 555 SEKCCPLSGPNNGLTTPDWDPHFCSMLCGKKLRCGQHVCETLCHSGHCPPCLETIFTDLA 614
++ CC D D H C C L CG+H C+ CH G+C PC F +LA
Sbjct: 347 NQVCCI-----------DLD-HICPKTCSLPLSCGRHRCDKPCHKGNCRPCHRVSFDELA 394
Query: 615 CACGMTSIPPPLPCGTMPPLCQLPCSVPQPCGHSGSHSCHF-GDCPPCSVPVSKECVGGH 673
C CG I PP+PCGT P C PC C H H+CH +CPPC V +K C G H
Sbjct: 395 CDCGANVIYPPVPCGTRKPACDRPCGRRHACDHPALHNCHAEAECPPCVVLTTKYCHGRH 454
Query: 674 VILRNIPCGSNNIKCNNPCGRTRQCGLHACGRSCHSPPCDILPGIVKGLRAACGQTCGAP 733
+ IPC + C PC + CG H C R+CH C +G C Q+C
Sbjct: 455 EQRKTIPCYQESFSCGMPCAKPLACGRHKCIRACHEDEC-----APEGAAGVCKQSCTRT 509
Query: 734 RSGCRHMCMALCHPGCPCPDARCEFPVTITCSCGRISANVPCDVGGNNSNYNADAIFEAS 793
R C H C A CH G CPD C+ V +TC CG C A A AS
Sbjct: 510 REACPHPCNAPCHEG-ECPDVPCKIVVEVTCECGNRKQQRTCHDFSKEYRRIATAQL-AS 567
Query: 794 IIQKLPM--PLQPVDANGQKVPLGQRKLMCDEECAKLERKRVLADAFDI-TPSLDALHFG 850
+Q++ ++ D G P + L C+EEC LER R LA A I P L +
Sbjct: 568 SVQEMQRGGSVELSDILGPVRPKNNKTLECNEECRMLERNRRLAIALQIRNPDLPS-KLQ 626
Query: 851 ENSSYELLSDTFRRDPKWVLAIEERCKILVLGKSKGTTHGLKVHVFCPMIKDKRDAVRMI 910
N S E L ++DP V I ++ LV +K + + + F M ++KR V +
Sbjct: 627 PNYS-ETLRAYAKKDPALVQMIHDKLTELV-KLAKESKQKSRSYSFPVMNREKRAIVHEM 684
Query: 911 AERWKLAVNAAGWEPKRFIVISA 933
+ + A EP R +V +A
Sbjct: 685 CHMFGVESVAYDAEPNRNVVATA 707
>gb|EAL87001.1| NF-X1 finger transcription factor, putative [Aspergillus fumigatus
Af293]
Length = 1144
Score = 496 bits (1276), Expect = e-138
Identities = 282/824 (34%), Positives = 376/824 (45%), Gaps = 111/824 (13%)
Query: 166 GRGRSGNMAGRQYG-SRDSSLPQLVQEIQEKLTKGTVECMICYDMVRRSAPIWSCSSCYS 224
G+G+S N+ R + S+ P + I E + EC IC + R + +WSC C++
Sbjct: 169 GKGKSKNIQARPPKVTTKSTAPDIATRIHEDIAHNLYECPICTSELGRRSRVWSCGLCWT 228
Query: 225 IFHLNCIKKWARAPTSVDLSA-------EKNLGFNWRCPGCQSVQHTSSKDIKYACFCGK 277
+FHL+C+KKW++ S A E + WRCPGC Y+C+C K
Sbjct: 229 VFHLSCVKKWSKNEGSAAQDAARRQAEGEPSAPRAWRCPGCNLPHEIFPST--YSCWCEK 286
Query: 278 RVDPPSDLYLTPHSCGEPCGKPLEKEVFVTEERKDELCPHACVLQCHPGPCPPCKAFAPP 337
VDP L PHSCG+ C +P RK CPH C CH GPC PC A P
Sbjct: 287 EVDPRPLPGLPPHSCGQTCSRP----------RKG--CPHPCDATCHAGPCSPCTAMGPT 334
Query: 338 RLCPCGKKRIATRCSDR--QSDLTCGQRCDKLLDCGRHHCENACHVGPCDPCQVLIEASC 395
+ C CG+ RC D ++ +CG+ C LL CG H C CH G C C+V IE C
Sbjct: 335 QDCFCGRNSSTKRCQDTDYENGWSCGEICSDLLPCGEHTCTRPCHEGLCGACEVKIEGRC 394
Query: 396 FCSKMTQVLFCGEMAMKGEFEAEG------------GVFSCGSNCGNVLGCSNHICREVC 443
+C K+ + C + EFE++ G FSCG C C H C + C
Sbjct: 395 YCGKVQTEMLCS--SKDEEFESQMARGDDGTIDEWIGCFSCGDRCSRPFDCGVHFCEKDC 452
Query: 444 HPGSC--GECEFLPSRVKACCCGKTKLED-----ERKSCVDPIPTCSKVCSKTLRCGVHA 496
HP C P V C CGKT L + R SC DPIP CSK C K L CG H+
Sbjct: 453 HPQDAHPAHCPRSPDVVSHCPCGKTPLTEMSDFSPRMSCDDPIPNCSKPCGKMLDCG-HS 511
Query: 497 CKETCHVGECPPCKVLISQKCRCGSTSRTVECYKTTENQKFTCQKPCGAKKNCGRHRCSE 556
C +TCH G C C+ + CRCG T+ C++ + C + C A +CGRH C+E
Sbjct: 512 CDQTCHTGPCGSCRRKLPVSCRCGRTTVVTVCHQGMIEPPW-CFRVCKAGLHCGRHACAE 570
Query: 557 KCCPLSGPNNG-------------LTTPDWD---PHFCSMLCGKKLRCGQHVCETLCHSG 600
+CCP G L D D H C+ +CG+ L+CG+H C +CH G
Sbjct: 571 RCCP--GEQKAIERQAMRRKLKAHLRPSDEDVEAEHICTRVCGRPLKCGRHTCPEICHKG 628
Query: 601 HCPPCLETIFTDLACACGMTSIPPPLPCGTMPPLCQLPCSVPQPCGHSGS-HSCHFGD-- 657
C C E IF D+ C CG T + PPLPCGT PP C PC P+PCGH + H+CH +
Sbjct: 629 PCNTCREAIFEDIPCNCGRTVLSPPLPCGTKPPACSFPCERPKPCGHPQTPHNCHTDEES 688
Query: 658 CPPCSVPVSKECVGGHVILRNIPCGSNNIKCNNPCGRTRQCGLHACGRSCHSPPCDILPG 717
CP C K C+ G +L+N PC +C CG +CG H+C ++CH PG
Sbjct: 689 CPKCPFLTEKACLCGRRVLKNQPCWLAETRCGEVCGEPLKCGSHSCQKTCHR------PG 742
Query: 718 IVKGLRAACGQTCGAPRSGCRHMCMALCHPGCPCPD-ARCEFPVTITCSCGRISANVPCD 776
+ C Q CG +S C H C CH CP+ C VT+TC CGR+ + C
Sbjct: 743 ECEDASRPCQQPCGKTKSLCGHPCTEPCHAPYQCPEKTPCTSTVTVTCGCGRLRQSRRC- 801
Query: 777 VGGNNSNYNADAIFEASIIQKLPMP-LQPVDANGQKVPLGQRKLMCDEECAKLERKRVLA 835
NA A + + Q P L P L CD+ECA+LER R LA
Sbjct: 802 --------NAAAASKGPVPQATRGPSLTP--------------LACDDECARLERNRALA 839
Query: 836 DAF--DITPSLDALHFGENSSYELLSDTFRRDPKWVLAIEERCKILVLGKSKGTTHGLKV 893
A +I P+ A S+ ++T + + + L + T H L +
Sbjct: 840 SALGVEINPTTTAASNLSPSTLPYSTETL----DLYVQLSSTSPLSTLQTYESTLHSLAI 895
Query: 894 HV------FCPMIKDKRDAVRMIAERWKLAVNAAGWEPKRFIVI 931
F P R + +A W A + EP R + +
Sbjct: 896 STTQRSVRFQPAKSPLRAFIHSLAADWGFASESFDPEPHRHVFV 939
>ref|XP_629029.1| hypothetical protein DDB0220630 [Dictyostelium discoideum]
gi|60462386|gb|EAL60607.1| NF-X1 type Zn
finger-containing protein [Dictyostelium discoideum]
Length = 1506
Score = 479 bits (1232), Expect = e-133
Identities = 212/546 (38%), Positives = 291/546 (52%), Gaps = 37/546 (6%)
Query: 181 RDSSLPQLVQEIQEKLTKGTVECMICYDMVRRSAPIWSCSSCYSIFHLNCIKKWARAPTS 240
++S L ++ K+ ECM+C++ V ++A IWSCS C+++FH +CIK+W+ +
Sbjct: 344 KNSEYTPLAIQMITKIQANIYECMVCFENVGKNAVIWSCSQCFTMFHSSCIKQWSSKSVT 403
Query: 241 VDLSAEKNLGFNWRCPGCQSVQHTSSKDIKYACFCGKRVDPPSDLYLTPHSCGEPCGKPL 300
+ W+CPGC+ + + ACFCGK DP + Y HSCGE CGK
Sbjct: 404 TE--------GKWKCPGCRYNHEVKPEHFQPACFCGKVKDPQYNPYHLAHSCGEVCGKL- 454
Query: 301 EKEVFVTEERKDELCPHACVLQCHPGPCPPCKAFAPPRLCPCGKKRIATRCSDRQSDLTC 360
R CPH+C + CHPGPC C + + C CGK + C + C
Sbjct: 455 ---------RAKSNCPHSCTMLCHPGPCLNCSSLGDIKDCYCGKTKYRLLCGEVDKGKVC 505
Query: 361 GQRCDKLLDCGRHHCENACHVGPCDPCQVLIEASCFCSK-MTQVLFCGEMAMK-GEFEAE 418
+C+KLL CG H C+ CH G C PC+V+ C+C K + + CG ++ + + +
Sbjct: 506 EGKCEKLLSCGNHRCQQQCHSGSCSPCEVVETQKCYCGKHIDKTKPCGSGSIDTSQSDDD 565
Query: 419 GGVFSCGSNCGNVLGCSNHICREVCHPGSCGECEFLPSRVKACCCGKTKLED---ERKSC 475
FSC C L C NH C+ CH G C C +P RV C C +T LE+ R SC
Sbjct: 566 PRFFSCAEKCDRTLDCGNHKCQRTCHKGDCEPCSLVPDRVDRCNCKQTTLEELGVVRTSC 625
Query: 476 VDPIPTCSKVCSKTLRCGVHACKETCHVGECPPCKVLISQKCRCGSTSRTVECYKTTENQ 535
+DPIP C KVCS L C H+C + CH G C CKV + CRCG T+ +C +N
Sbjct: 626 LDPIPVCKKVCSTLLACKQHSCTDRCHTGPCGSCKVKVRSICRCGKTTENRQCGVIQQNM 685
Query: 536 K------FTCQKPCGAKKNCGRHRCSEKCCPLSGPNNGLTTPDWDPHFCSMLCGKKLRCG 589
FTC C ++C RH C KCCP + ++ + H C+++CGK+L+CG
Sbjct: 686 SSTSAAAFTCGNVCKILRSCKRHECGVKCCPSTSSSD-----EAGNHVCTLVCGKQLKCG 740
Query: 590 QHVCETLCHSGHCPPCLETIFTDLACACGMTSIPPPLPCGTMPPLCQLPCSVPQPCGHSG 649
H C+ LCHSG C C + DLAC CG T I PP+PCGT PP+C PC++ + CGHS
Sbjct: 741 VHKCQQLCHSGKCYNCYINSYDDLACPCGKTVIQPPVPCGTKPPMCNHPCTIKRECGHSN 800
Query: 650 ---SHSCHFGDCPPCSVPVSKECVGGHVILRNIPCGSNNIKCNNPCGRTRQCGLHACGRS 706
H CH G CPPC+ V K C GGH + +++ C + + C CG+ C H C R
Sbjct: 801 EVWEHKCHSGPCPPCTYLVDKMCAGGHQVQKSVKCSTIDASCGRICGKVLSCLSHTCPRI 860
Query: 707 CHSPPC 712
CHS PC
Sbjct: 861 CHSGPC 866
Score = 166 bits (419), Expect = 6e-39
Identities = 126/459 (27%), Positives = 170/459 (36%), Gaps = 116/459 (25%)
Query: 392 EASCFCSKMTQVLFCGEMAMKGEFEAEGGVFSCGSNCGNVLGCSN--HICREVCHPGSCG 449
+ +CFC K+ ++ SCG CG + SN H C +CHPG C
Sbjct: 425 QPACFCGKVKDP----------QYNPYHLAHSCGEVCGKLRAKSNCPHSCTMLCHPGPCL 474
Query: 450 ECEFLPSRVKACCCGKTKLEDERKSC--VDPIPTCSKVCSKTLRCGVHACKETCHVGECP 507
C L +K C CGKTK R C VD C C K L CG H C++ CH G C
Sbjct: 475 NCSSLGD-IKDCYCGKTKY---RLLCGEVDKGKVCEGKCEKLLSCGNHRCQQQCHSGSCS 530
Query: 508 PCKVLISQKCRCGST--------SRTVECYKTTENQKF-TCQKPCGAKKNCGRHRCSEKC 558
PC+V+ +QKC CG S +++ ++ ++ +F +C + C +CG H+C C
Sbjct: 531 PCEVVETQKCYCGKHIDKTKPCGSGSIDTSQSDDDPRFFSCAEKCDRTLDCGNHKCQRTC 590
Query: 559 -------CPLSGPN--------------NGLTTPDWDP-HFCSMLCGKKLRCGQHVCETL 596
C L + T DP C +C L C QH C
Sbjct: 591 HKGDCEPCSLVPDRVDRCNCKQTTLEELGVVRTSCLDPIPVCKKVCSTLLACKQHSCTDR 650
Query: 597 CHSGHCPPCLETIFTDLACACGMTS---------------IPPPLPCGTMPPL------- 634
CH+G C C + + C CG T+ CG + +
Sbjct: 651 CHTGPCGSCKVKVRS--ICRCGKTTENRQCGVIQQNMSSTSAAAFTCGNVCKILRSCKRH 708
Query: 635 --------------------CQLPCSVPQPCG-HSGSHSCHFGDCPPCSVPVSKECV--- 670
C L C CG H CH G C C + +
Sbjct: 709 ECGVKCCPSTSSSDEAGNHVCTLVCGKQLKCGVHKCQQLCHSGKCYNCYINSYDDLACPC 768
Query: 671 GGHVILRNIPCGSNNIKCNNPCGRTRQCGL--HACGRSCHSPPCDILPGIV--------- 719
G VI +PCG+ CN+PC R+CG CHS PC +V
Sbjct: 769 GKTVIQPPVPCGTKPPMCNHPCTIKRECGHSNEVWEHKCHSGPCPPCTYLVDKMCAGGHQ 828
Query: 720 -------KGLRAACGQTCGAPRSGCRHMCMALCHPGCPC 751
+ A+CG+ CG S H C +CH G PC
Sbjct: 829 VQKSVKCSTIDASCGRICGKVLSCLSHTCPRICHSG-PC 866
Score = 113 bits (283), Expect = 3e-23
Identities = 112/392 (28%), Positives = 151/392 (37%), Gaps = 79/392 (20%)
Query: 464 GKTKL----EDERKSCVDP--IPTCSKVCSKTLRCGVHACKETCH----VGECPPCKVLI 513
GKTK DE+ S P I +K+ + C V C E + C C +
Sbjct: 332 GKTKFIFNENDEKNSEYTPLAIQMITKIQANIYECMV--CFENVGKNAVIWSCSQCFTMF 389
Query: 514 SQKCRCGSTSRTVECYKTTENQKFTCQKPCGAKKNCGRHRCSEKCCPLSGPNNGLTTPDW 573
C +S++V TTE K+ C G + N H + + + P +
Sbjct: 390 HSSCIKQWSSKSV----TTEG-KWKCP---GCRYN---HEVKPEHFQPACFCGKVKDPQY 438
Query: 574 DP----HFCSMLCGK---KLRCGQHVCETLCHSGHCPPCLETIFTDLACACGMTSIPPPL 626
+P H C +CGK K C H C LCH G C C ++ C CG T L
Sbjct: 439 NPYHLAHSCGEVCGKLRAKSNC-PHSCTMLCHPGPCLNC-SSLGDIKDCYCGKTKYR--L 494
Query: 627 PCGTMPP--LCQLPCSVPQPCG-HSGSHSCHFGDCPPCSVPVSKECVGGHVILRNIPCGS 683
CG + +C+ C CG H CH G C PC V +++C G I + PCGS
Sbjct: 495 LCGEVDKGKVCEGKCEKLLSCGNHRCQQQCHSGSCSPCEVVETQKCYCGKHIDKTKPCGS 554
Query: 684 NNI------------KCNNPCGRTRQCGLHACGRSCHS---PPCDILPGIVKGLRAACGQ 728
+I C C RT CG H C R+CH PC ++P V R C Q
Sbjct: 555 GSIDTSQSDDDPRFFSCAEKCDRTLDCGNHKCQRTCHKGDCEPCSLVPDRVD--RCNCKQ 612
Query: 729 T----CGAPRSGC------------------RHMCMALCHPGCPCPDARCEFPVTITCSC 766
T G R+ C +H C CH G PC C+ V C C
Sbjct: 613 TTLEELGVVRTSCLDPIPVCKKVCSTLLACKQHSCTDRCHTG-PC--GSCKVKVRSICRC 669
Query: 767 GRISANVPCDVGGNNSNYNADAIFEASIIQKL 798
G+ + N C V N + + A F + K+
Sbjct: 670 GKTTENRQCGVIQQNMSSTSAAAFTCGNVCKI 701
Score = 59.3 bits (142), Expect = 7e-07
Identities = 38/121 (31%), Positives = 49/121 (40%), Gaps = 36/121 (29%)
Query: 726 CGQTCGAPRSGCRHMCMALCHPGCPCP-DARCEFPVTITCSCGRISANVPCDVGGNNSNY 784
CG CG P C H C CHPG PCP + C+ V I C C R S C G +++
Sbjct: 1051 CGYLCGIPLKSCEHTCHQACHPGEPCPTNISCKQKVRIYCKCKRRSVETLC-CGKDDT-- 1107
Query: 785 NADAIFEASIIQKLPMPLQPVDANGQKVPLGQRKLMCDEECAKLERKRVLADAFDITPSL 844
R+L CDE C K ER++ L +AF + +L
Sbjct: 1108 --------------------------------RQLECDEVCEKEEREKQLKEAFYGSHTL 1135
Query: 845 D 845
D
Sbjct: 1136 D 1136
Score = 42.0 bits (97), Expect = 0.12
Identities = 28/89 (31%), Positives = 38/89 (42%), Gaps = 21/89 (23%)
Query: 415 FEAEGGVFSCGSNCGNVLGCSNHICREVCHPG-------SCGE-----CEFLPSRVKACC 462
F++ G +CG CG L H C + CHPG SC + C+ V+ C
Sbjct: 1042 FDSLMGNLTCGYLCGIPLKSCEHTCHQACHPGEPCPTNISCKQKVRIYCKCKRRSVETLC 1101
Query: 463 CGKTKLEDERKSCVDPIPTCSKVCSKTLR 491
CGK +D R+ C +VC K R
Sbjct: 1102 CGK---DDTRQL------ECDEVCEKEER 1121
Score = 41.6 bits (96), Expect = 0.16
Identities = 27/82 (32%), Positives = 34/82 (40%), Gaps = 19/82 (23%)
Query: 291 SCGEPCGKPLEKEVFVTEERKDELCPHACVLQCHPG-PCP---PCKAFAPPRLCPCGKKR 346
+CG CG PL+ C H C CHPG PCP CK C C ++
Sbjct: 1050 TCGYLCGIPLKS------------CEHTCHQACHPGEPCPTNISCKQ-KVRIYCKCKRRS 1096
Query: 347 IATRC--SDRQSDLTCGQRCDK 366
+ T C D L C + C+K
Sbjct: 1097 VETLCCGKDDTRQLECDEVCEK 1118
Score = 41.2 bits (95), Expect = 0.21
Identities = 27/98 (27%), Positives = 39/98 (39%), Gaps = 4/98 (4%)
Query: 450 ECEFLPSRVKACCCGKTKLEDERKSCVDPIPTCSKVCSKTLRCGVHACKETCHVGE-CP- 507
EC R+K + EDE + TC +C L+ H C + CH GE CP
Sbjct: 1019 ECNEEEERIKKEEKEDDEDEDELFDSLMGNLTCGYLCGIPLKSCEHTCHQACHPGEPCPT 1078
Query: 508 --PCKVLISQKCRCGSTSRTVECYKTTENQKFTCQKPC 543
CK + C+C S C + ++ C + C
Sbjct: 1079 NISCKQKVRIYCKCKRRSVETLCCGKDDTRQLECDEVC 1116
Score = 36.2 bits (82), Expect = 6.7
Identities = 20/57 (35%), Positives = 27/57 (47%), Gaps = 5/57 (8%)
Query: 357 DLTCGQRCDKLLDCGRHHCENACHVG-PCD---PCQVLIEASCFCSKMT-QVLFCGE 408
+LTCG C L H C ACH G PC C+ + C C + + + L CG+
Sbjct: 1048 NLTCGYLCGIPLKSCEHTCHQACHPGEPCPTNISCKQKVRIYCKCKRRSVETLCCGK 1104
Database: nr
Posted date: Jul 5, 2005 12:34 AM
Number of letters in database: 863,360,394
Number of sequences in database: 2,540,612
Lambda K H
0.320 0.134 0.445
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 2,184,117,675
Number of Sequences: 2540612
Number of extensions: 101689942
Number of successful extensions: 476678
Number of sequences better than 10.0: 2088
Number of HSP's better than 10.0 without gapping: 470
Number of HSP's successfully gapped in prelim test: 1709
Number of HSP's that attempted gapping in prelim test: 437547
Number of HSP's gapped (non-prelim): 16774
length of query: 1173
length of database: 863,360,394
effective HSP length: 139
effective length of query: 1034
effective length of database: 510,215,326
effective search space: 527562647084
effective search space used: 527562647084
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 81 (35.8 bits)
Medicago: description of AC141436.6


