

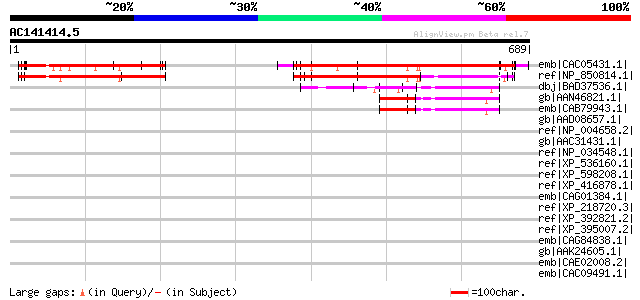
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC141414.5 - phase: 0 /pseudo
(689 letters)
Database: nr
2,540,612 sequences; 863,360,394 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
emb|CAC05431.1| putative protein [Arabidopsis thaliana] gi|15240... 489 e-136
ref|NP_850814.1| expressed protein [Arabidopsis thaliana] 327 1e-87
dbj|BAD37536.1| protein kinase-like [Oryza sativa (japonica cult... 64 2e-08
gb|AAN46821.1| At4g32250/F10M6_110 [Arabidopsis thaliana] gi|214... 58 1e-06
emb|CAB79943.1| putative protein [Arabidopsis thaliana] gi|28646... 58 1e-06
gb|AAD08657.1| HERC2 [Homo sapiens] 46 0.005
ref|NP_004658.2| hect domain and RLD 2 [Homo sapiens] 46 0.005
gb|AAC31431.1| rjs [Mus musculus] 45 0.006
ref|NP_034548.1| hect (homologous to the E6-AP (UBE3A) carboxyl ... 45 0.006
ref|XP_536160.1| PREDICTED: hypothetical protein XP_536160 [Cani... 45 0.008
ref|XP_598208.1| PREDICTED: similar to hect domain and RLD 2, pa... 45 0.008
ref|XP_416878.1| PREDICTED: similar to hect domain and RLD 2 [Ga... 45 0.008
emb|CAG01384.1| unnamed protein product [Tetraodon nigroviridis] 45 0.010
ref|XP_218720.3| PREDICTED: similar to putative E3 ligase [Rattu... 40 0.33
ref|XP_392821.2| PREDICTED: similar to CG17492-PA [Apis mellifera] 38 1.3
ref|XP_395007.2| PREDICTED: similar to hect (homologous to the E... 37 1.6
emb|CAG84838.1| unnamed protein product [Debaryomyces hansenii C... 36 3.7
gb|AAK24605.1| peptidase, M16 family [Caulobacter crescentus CB1... 36 4.8
emb|CAE02008.2| OJ000223_09.10 [Oryza sativa (japonica cultivar-... 35 6.3
emb|CAC09491.1| contains similarity to F12A21.16 [Oryza sativa (... 35 6.3
>emb|CAC05431.1| putative protein [Arabidopsis thaliana]
gi|15240647|ref|NP_196858.1| expressed protein
[Arabidopsis thaliana]
Length = 788
Score = 489 bits (1259), Expect = e-136
Identities = 219/295 (74%), Positives = 261/295 (88%)
Query: 377 SMVTGRQSLWKVSPGDAERLPGXEVGDWVRSKPSLGNRPSYDWNSVGRESLAVVHSVQDS 436
+ VTGRQ+LW+VSPGDAE L G EVGDWVRSKPSLGNRPSYDW++VGRES+AVVHS+Q++
Sbjct: 383 AQVTGRQTLWRVSPGDAELLSGFEVGDWVRSKPSLGNRPSYDWSNVGRESIAVVHSIQET 442
Query: 437 GYLELACCFRKGKWITHYTDVEKVPSFKVGQYVRFRPGLAEPRFGWGGAQPESQGIITNI 496
GYLELACCFRKG+W THYTD+EK+P+ KVGQ+V F+ G+ EPR+GW A+P+S+GIIT +
Sbjct: 443 GYLELACCFRKGRWSTHYTDLEKIPALKVGQFVHFQKGITEPRWGWRAAKPDSRGIITTV 502
Query: 497 HADGEVRVAFFGLSGLWKGDPSDLQAEQIFEVGEWVRLKENVNNWKSIGPGSVGVVQGIG 556
HADGEVRVAFFGL GLW+GDP+DL+ E +FEVGEWVRL+E V+ WKS+GPGSVGVV G+G
Sbjct: 503 HADGEVRVAFFGLPGLWRGDPADLEVEPMFEVGEWVRLREGVSCWKSVGPGSVGVVHGVG 562
Query: 557 YEGGETDRSTFVGFCGEQEKWVGPSSHLERVDKLIVGQKVRVKQNVKQPRFGWSGHTHAS 616
YEG E D +T V FCGEQE+W GP+SHLE+ KL+VGQK RVK VKQPRFGWSGH+H S
Sbjct: 563 YEGDEWDGTTSVSFCGEQERWAGPTSHLEKAKKLVVGQKTRVKLAVKQPRFGWSGHSHGS 622
Query: 617 IGTIQAIDADGKLWIYTPAGSRTWMLDPSEVEVVEEKELCIGDWVRVRASVSTPT 671
+GTI AIDADGKL IYTPAGS+TWMLDPSEVE +EE+EL IGDWVRV+AS++TPT
Sbjct: 623 VGTISAIDADGKLRIYTPAGSKTWMLDPSEVETIEEEELKIGDWVRVKASITTPT 677
Score = 327 bits (837), Expect = 1e-87
Identities = 151/196 (77%), Positives = 175/196 (89%), Gaps = 3/196 (1%)
Query: 12 SPITFDVLDWVKFKRTVTEPKHGWQGAKPNSVGFVQSVPGRDNDDLIVSFCSGEVRVLTS 71
S IT++V DWVKFKR +T P HGWQGAKP SVGFVQ++ + +D+I++FCSGE RVL +
Sbjct: 16 SSITYNVGDWVKFKRGITTPLHGWQGAKPKSVGFVQTI--LEKEDMIIAFCSGEARVLAN 73
Query: 72 EIVKLIPLDRGQHVQLKGDVNEPRFGWRGQSRDSIGTVLCVDPEDGILRVGFPGASRGWK 131
E+VKLIPLDRGQHV+L+ DV EPRFGWRGQSRDS+GTVLCVD EDGILRVGFPGASRGWK
Sbjct: 74 EVVKLIPLDRGQHVRLRADVKEPRFGWRGQSRDSVGTVLCVD-EDGILRVGFPGASRGWK 132
Query: 132 ADPAEMERVEEFKVGDWVRVRPTLTTSKHGLGNVVPGTIGIVYCIRPDSSLLVELSYVQN 191
ADPAEMERVEEFKVGDWVR+R LT++KHG G+VVPG++GIVYC+RPDSSLLVELSY+ N
Sbjct: 133 ADPAEMERVEEFKVGDWVRIRQNLTSAKHGFGSVVPGSMGIVYCVRPDSSLLVELSYLPN 192
Query: 192 PWHCEPEEIEHVPPFR 207
PWHCEPEE+E V PFR
Sbjct: 193 PWHCEPEEVEPVAPFR 208
Score = 171 bits (433), Expect = 7e-41
Identities = 109/355 (30%), Positives = 164/355 (45%), Gaps = 30/355 (8%)
Query: 356 GGQMNHQQLLEK*CELTWMVPSMVTGRQSLWKVSPGDAERLPGX---------------- 399
GG+ +H E ++ + R W+ P D E++
Sbjct: 219 GGETHHSVGKISEIENDGLLIIEIPNRPIPWQADPSDMEKIDDFKHVKSFDSFNTEPLYH 278
Query: 400 -EVGDWVRSKPSLGNRPSYDWNSVGRESLAVVHSVQDSGYLELACCFRKGKWITHYTDVE 458
+VGDWVR K S+ + P Y W + R S+ V+HS+ + G + +A CFR + TDVE
Sbjct: 279 EQVGDWVRVKASVSS-PKYGWEDITRNSIGVMHSLDEDGDVGIAFCFRSKPFSCSVTDVE 337
Query: 459 KVPSFKVGQYVRFRPGLAEPRFGWGGAQPESQGIITNIHADGEVRVAFFGLSGLWKGDPS 518
KV F VGQ + P + +PR GW P + G + I DG + G LW+ P
Sbjct: 338 KVTPFHVGQEIHMTPSITQPRLGWSNETPATIGKVMRIDMDGTLSAQVTGRQTLWRVSPG 397
Query: 519 DLQAEQIFEVGEWVRLKENVNN-----WKSIGPGSVGVVQGIGYEGGETDRSTFVGFCGE 573
D + FEVG+WVR K ++ N W ++G S+ VV I E G + + C
Sbjct: 398 DAELLSGFEVGDWVRSKPSLGNRPSYDWSNVGRESIAVVHSI-QETGYLE----LACCFR 452
Query: 574 QEKWVGPSSHLERVDKLIVGQKVRVKQNVKQPRFGWSGHTHASIGTIQAIDADGKLWIYT 633
+ +W + LE++ L VGQ V ++ + +PR+GW S G I + ADG++ +
Sbjct: 453 KGRWSTHYTDLEKIPALKVGQFVHFQKGITEPRWGWRAAKPDSRGIITTVHADGEVRVAF 512
Query: 634 PAGSRTWMLDPSEVEVVEEKELCIGDWVRVRASVSTPTPPLGGSESFKHRGGASG 688
W DP+++EV E +G+WVR+R VS GS H G G
Sbjct: 513 FGLPGLWRGDPADLEV--EPMFEVGEWVRLREGVSCWKSVGPGSVGVVHGVGYEG 565
Score = 155 bits (393), Expect = 3e-36
Identities = 93/283 (32%), Positives = 142/283 (49%), Gaps = 23/283 (8%)
Query: 381 GRQSLWKVSPGDAERLPGXEVGDWVRSKPSLGNRPSYDWNSVGRESLAVVHSV-----QD 435
G LW+ P D E P EVG+WVR + + W SVG S+ VVH V +
Sbjct: 514 GLPGLWRGDPADLEVEPMFEVGEWVRLREGVSC-----WKSVGPGSVGVVHGVGYEGDEW 568
Query: 436 SGYLELACCFRKGKWITHYTDVEKVPSFKVGQYVRFRPGLAEPRFGWGGAQPESQGIITN 495
G ++ C + +W + +EK VGQ R + + +PRFGW G S G I+
Sbjct: 569 DGTTSVSFCGEQERWAGPTSHLEKAKKLVVGQKTRVKLAVKQPRFGWSGHSHGSVGTISA 628
Query: 496 IHADGEVRVAFFGLSGLWKGDPSDLQA--EQIFEVGEWVRLKENVNN----WKSIGPGSV 549
I ADG++R+ S W DPS+++ E+ ++G+WVR+K ++ W + P S
Sbjct: 629 IDADGKLRIYTPAGSKTWMLDPSEVETIEEEELKIGDWVRVKASITTPTYQWGEVNPSST 688
Query: 550 GVVQGIGYEGGETDRSTFVGFCGEQEKWVGPSSHLERVDKLIVGQKVRVKQNVKQPRFGW 609
GVV + D V FC W+ + LER+ +G +V++K + PR+GW
Sbjct: 689 GVVHRM------EDGDLCVSFCFLDRLWLCKAGELERIRPFRIGDRVKIKDGLVTPRWGW 742
Query: 610 SGHTHASIGTIQAIDADGKLWI-YTPAGSRTWMLDPSEVEVVE 651
THAS G + +DA+GKL I + R W+ DP+++ + E
Sbjct: 743 GMETHASKGHVVGVDANGKLRIKFLWREGRPWIGDPADIVLDE 785
Score = 149 bits (376), Expect = 3e-34
Identities = 91/289 (31%), Positives = 146/289 (50%), Gaps = 35/289 (12%)
Query: 401 VGDWVRSKPSLGNRPSYDWNSVGRESLAVVHSVQDSGYLELACCFRKGKWITHYTDVEKV 460
VGDWV+ K + P + W +S+ V ++ + + +A C + + + + +V K+
Sbjct: 22 VGDWVKFKRGI-TTPLHGWQGAKPKSVGFVQTILEKEDMIIAFCSGEARVLAN--EVVKL 78
Query: 461 PSFKVGQYVRFRPGLAEPRFGWGGAQPESQGIITNIHADGEVRVAFFGLSGLWKGDPSDL 520
GQ+VR R + EPRFGW G +S G + + DG +RV F G S WK DP+++
Sbjct: 79 IPLDRGQHVRLRADVKEPRFGWRGQSRDSVGTVLCVDEDGILRVGFPGASRGWKADPAEM 138
Query: 521 QAEQIFEVGEWVRLKENVNNWK----SIGPGSVGVVQGIGYEGGETDRSTFVGFCGEQEK 576
+ + F+VG+WVR+++N+ + K S+ PGS+G+V + D S V
Sbjct: 139 ERVEEFKVGDWVRIRQNLTSAKHGFGSVVPGSMGIVYCV-----RPDSSLLVELSYLPNP 193
Query: 577 WVGPSSHLERVDKLIVGQKVRVKQNVKQPRFGWSGHTHASIGTIQAIDADGKLWIYTPAG 636
W +E V +++V +PR+ W G TH S+G I I+ DG L I P
Sbjct: 194 WHCEPEEVEPV--------APFRRSVAEPRYAWGGETHHSVGKISEIENDGLLIIEIPNR 245
Query: 637 SRTWMLDPSEVEVVEEKELC---------------IGDWVRVRASVSTP 670
W DPS++E +++ + +GDWVRV+ASVS+P
Sbjct: 246 PIPWQADPSDMEKIDDFKHVKSFDSFNTEPLYHEQVGDWVRVKASVSSP 294
Score = 139 bits (350), Expect = 3e-31
Identities = 88/303 (29%), Positives = 141/303 (46%), Gaps = 37/303 (12%)
Query: 386 WKVSPGDAERLPGXEVGDWVRSKPSLGNRPSYDWNSVGRESLAVVHSVQDSGYLELACCF 445
WK P + ER+ +VGDWVR + +L + + SV S+ +V+ V+ L + +
Sbjct: 131 WKADPAEMERVEEFKVGDWVRIRQNL-TSAKHGFGSVVPGSMGIVYCVRPDSSLLVELSY 189
Query: 446 RKGKWITHYTDVEKVPSFKVGQYVRFRPGLAEPRFGWGGAQPESQGIITNIHADGEVRVA 505
W +VE V FR +AEPR+ WGG S G I+ I DG + +
Sbjct: 190 LPNPWHCEPEEVEPV--------APFRRSVAEPRYAWGGETHHSVGKISEIENDGLLIIE 241
Query: 506 FFGLSGLWKGDPSDLQAEQIF-----------------EVGEWVRLKENVNN----WKSI 544
W+ DPSD++ F +VG+WVR+K +V++ W+ I
Sbjct: 242 IPNRPIPWQADPSDMEKIDDFKHVKSFDSFNTEPLYHEQVGDWVRVKASVSSPKYGWEDI 301
Query: 545 GPGSVGVVQGIGYEGGETDRSTFVGFCGEQEKWVGPSSHLERVDKLIVGQKVRVKQNVKQ 604
S+GV+ + + D + FC + + + +E+V VGQ++ + ++ Q
Sbjct: 302 TRNSIGVMHSL-----DEDGDVGIAFCFRSKPFSCSVTDVEKVTPFHVGQEIHMTPSITQ 356
Query: 605 PRFGWSGHTHASIGTIQAIDADGKLWIYTPAGSRTWMLDPSEVEVVEEKELCIGDWVRVR 664
PR GWS T A+IG + ID DG L W + P + E++ E +GDWVR +
Sbjct: 357 PRLGWSNETPATIGKVMRIDMDGTLSAQVTGRQTLWRVSPGDAELLSGFE--VGDWVRSK 414
Query: 665 ASV 667
S+
Sbjct: 415 PSL 417
Score = 125 bits (315), Expect = 4e-27
Identities = 75/200 (37%), Positives = 110/200 (54%), Gaps = 15/200 (7%)
Query: 16 FDVLDWVKFKRTVTEPKHGWQGAKPNSVGFVQSVPGRDNDD----LIVSFCSGEVRVL-- 69
F+V +WV+ + V+ W+ P SVG V V G + D+ VSFC + R
Sbjct: 532 FEVGEWVRLREGVS----CWKSVGPGSVGVVHGV-GYEGDEWDGTTSVSFCGEQERWAGP 586
Query: 70 TSEIVKLIPLDRGQHVQLKGDVNEPRFGWRGQSRDSIGTVLCVDPEDGILRVGFPGASRG 129
TS + K L GQ ++K V +PRFGW G S S+GT+ +D DG LR+ P S+
Sbjct: 587 TSHLEKAKKLVVGQKTRVKLAVKQPRFGWSGHSHGSVGTISAIDA-DGKLRIYTPAGSKT 645
Query: 130 WKADPAEMERVEE--FKVGDWVRVRPTLTTSKHGLGNVVPGTIGIVYCIRPDSSLLVELS 187
W DP+E+E +EE K+GDWVRV+ ++TT + G V P + G+V+ + D L V
Sbjct: 646 WMLDPSEVETIEEEELKIGDWVRVKASITTPTYQWGEVNPSSTGVVHRME-DGDLCVSFC 704
Query: 188 YVQNPWHCEPEEIEHVPPFR 207
++ W C+ E+E + PFR
Sbjct: 705 FLDRLWLCKAGELERIRPFR 724
Score = 104 bits (260), Expect = 8e-21
Identities = 59/210 (28%), Positives = 105/210 (49%), Gaps = 30/210 (14%)
Query: 16 FDVLDWVKFKRTVTEPKHGWQGAKPNSVGFVQSVPGRDNDDLIV--SFCSGEVRVLTSEI 73
F V DWV+ ++ +T KHG+ P S+G V V R + L+V S+ E+
Sbjct: 144 FKVGDWVRIRQNLTSAKHGFGSVVPGSMGIVYCV--RPDSSLLVELSYLPNPWHCEPEEV 201
Query: 74 VKLIPLDRGQHVQLKGDVNEPRFGWRGQSRDSIGTVLCVDPEDGILRVGFPGASRGWKAD 133
+ P R V EPR+ W G++ S+G + ++ DG+L + P W+AD
Sbjct: 202 EPVAPFRRS--------VAEPRYAWGGETHHSVGKISEIE-NDGLLIIEIPNRPIPWQAD 252
Query: 134 PAEMERVEEFK-----------------VGDWVRVRPTLTTSKHGLGNVVPGTIGIVYCI 176
P++ME++++FK VGDWVRV+ ++++ K+G ++ +IG+++ +
Sbjct: 253 PSDMEKIDDFKHVKSFDSFNTEPLYHEQVGDWVRVKASVSSPKYGWEDITRNSIGVMHSL 312
Query: 177 RPDSSLLVELSYVQNPWHCEPEEIEHVPPF 206
D + + + P+ C ++E V PF
Sbjct: 313 DEDGDVGIAFCFRSKPFSCSVTDVEKVTPF 342
Score = 102 bits (254), Expect = 4e-20
Identities = 54/191 (28%), Positives = 98/191 (51%), Gaps = 6/191 (3%)
Query: 20 DWVKFKRTVTEPKHGWQGAKPNSVGFVQSVPGRDNDDLIVSFC--SGEVRVLTSEIVKLI 77
DWV+ K +V+ PK+GW+ NS+G + S+ ++ D+ ++FC S +++ K+
Sbjct: 283 DWVRVKASVSSPKYGWEDITRNSIGVMHSLD--EDGDVGIAFCFRSKPFSCSVTDVEKVT 340
Query: 78 PLDRGQHVQLKGDVNEPRFGWRGQSRDSIGTVLCVDPEDGILRVGFPGASRGWKADPAEM 137
P GQ + + + +PR GW ++ +IG V+ +D DG L G W+ P +
Sbjct: 341 PFHVGQEIHMTPSITQPRLGWSNETPATIGKVMRID-MDGTLSAQVTGRQTLWRVSPGDA 399
Query: 138 ERVEEFKVGDWVRVRPTL-TTSKHGLGNVVPGTIGIVYCIRPDSSLLVELSYVQNPWHCE 196
E + F+VGDWVR +P+L + NV +I +V+ I+ L + + + W
Sbjct: 400 ELLSGFEVGDWVRSKPSLGNRPSYDWSNVGRESIAVVHSIQETGYLELACCFRKGRWSTH 459
Query: 197 PEEIEHVPPFR 207
++E +P +
Sbjct: 460 YTDLEKIPALK 470
Score = 89.7 bits (221), Expect = 3e-16
Identities = 53/160 (33%), Positives = 83/160 (51%), Gaps = 6/160 (3%)
Query: 16 FDVLDWVKFKRTV-TEPKHGWQGAKPNSVGFVQSVPGRDNDDLIVSFCSGEVRVLTSEIV 74
F+V DWV+ K ++ P + W S+ V S+ +L F G +++
Sbjct: 405 FEVGDWVRSKPSLGNRPSYDWSNVGRESIAVVHSIQETGYLELACCFRKGRWSTHYTDLE 464
Query: 75 KLIPLDRGQHVQLKGDVNEPRFGWRGQSRDSIGTVLCVDPEDGILRVGFPGASRGWKADP 134
K+ L GQ V + + EPR+GWR DS G + V DG +RV F G W+ DP
Sbjct: 465 KIPALKVGQFVHFQKGITEPRWGWRAAKPDSRGIITTVH-ADGEVRVAFFGLPGLWRGDP 523
Query: 135 AEMERVEEFKVGDWVRVRPTLTTSKHGLGNVVPGTIGIVY 174
A++E F+VG+WVR+R ++ K +V PG++G+V+
Sbjct: 524 ADLEVEPMFEVGEWVRLREGVSCWK----SVGPGSVGVVH 559
Score = 82.4 bits (202), Expect = 4e-14
Identities = 54/193 (27%), Positives = 91/193 (46%), Gaps = 11/193 (5%)
Query: 462 SFKVGQYVRFRPGLAEPRFGWGGAQPESQGIITNIHADGEVRVAFFGLSGLWKGDPSDLQ 521
++ VG +V+F+ G+ P GW GA+P+S G + I ++ +AF SG + +++
Sbjct: 19 TYNVGDWVKFKRGITTPLHGWQGAKPKSVGFVQTILEKEDMIIAF--CSGEARVLANEVV 76
Query: 522 AEQIFEVGEWVRLKENVN----NWKSIGPGSVGVVQGIGYEGGETDRSTFVGFCGEQEKW 577
+ G+ VRL+ +V W+ SVG V + + D VGF G W
Sbjct: 77 KLIPLDRGQHVRLRADVKEPRFGWRGQSRDSVGTVLCV-----DEDGILRVGFPGASRGW 131
Query: 578 VGPSSHLERVDKLIVGQKVRVKQNVKQPRFGWSGHTHASIGTIQAIDADGKLWIYTPAGS 637
+ +ERV++ VG VR++QN+ + G+ S+G + + D L +
Sbjct: 132 KADPAEMERVEEFKVGDWVRIRQNLTSAKHGFGSVVPGSMGIVYCVRPDSSLLVELSYLP 191
Query: 638 RTWMLDPSEVEVV 650
W +P EVE V
Sbjct: 192 NPWHCEPEEVEPV 204
Score = 66.6 bits (161), Expect = 3e-09
Identities = 49/182 (26%), Positives = 82/182 (44%), Gaps = 7/182 (3%)
Query: 23 KFKRTVTEPKHGWQGAKPNSVGFVQSVPGRDNDDLIVSFCSGEVRVLTSEIVKLIP---L 79
+ K V +P+ GW G SVG + ++ D I + + +L V+ I L
Sbjct: 603 RVKLAVKQPRFGWSGHSHGSVGTISAIDA-DGKLRIYTPAGSKTWMLDPSEVETIEEEEL 661
Query: 80 DRGQHVQLKGDVNEPRFGWRGQSRDSIGTVLCVDPEDGILRVGFPGASRGWKADPAEMER 139
G V++K + P + W + S G V + EDG L V F R W E+ER
Sbjct: 662 KIGDWVRVKASITTPTYQWGEVNPSSTGVVHRM--EDGDLCVSFCFLDRLWLCKAGELER 719
Query: 140 VEEFKVGDWVRVRPTLTTSKHGLGNVVPGTIGIVYCIRPDSSLLVELSYVQ-NPWHCEPE 198
+ F++GD V+++ L T + G G + G V + + L ++ + + PW +P
Sbjct: 720 IRPFRIGDRVKIKDGLVTPRWGWGMETHASKGHVVGVDANGKLRIKFLWREGRPWIGDPA 779
Query: 199 EI 200
+I
Sbjct: 780 DI 781
Score = 65.9 bits (159), Expect = 4e-09
Identities = 46/187 (24%), Positives = 81/187 (42%), Gaps = 8/187 (4%)
Query: 21 WVKFKRTVTEPKHGWQGAKPNSVGFVQSVPGRDNDDLIVSFCSGEVRVLTSEIVKLIPLD 80
+V F++ +TEP+ GW+ AKP+S G + +V + G R +++ +
Sbjct: 474 FVHFQKGITEPRWGWRAAKPDSRGIITTVHADGEVRVAFFGLPGLWRGDPADLEVEPMFE 533
Query: 81 RGQHVQLKGDVNEPRFGWRGQSRDSIGTVLCV----DPEDGILRVGFPGASRGWKADPAE 136
G+ V+L+ V+ W+ S+G V V D DG V F G W +
Sbjct: 534 VGEWVRLREGVS----CWKSVGPGSVGVVHGVGYEGDEWDGTTSVSFCGEQERWAGPTSH 589
Query: 137 MERVEEFKVGDWVRVRPTLTTSKHGLGNVVPGTIGIVYCIRPDSSLLVELSYVQNPWHCE 196
+E+ ++ VG RV+ + + G G++G + I D L + W +
Sbjct: 590 LEKAKKLVVGQKTRVKLAVKQPRFGWSGHSHGSVGTISAIDADGKLRIYTPAGSKTWMLD 649
Query: 197 PEEIEHV 203
P E+E +
Sbjct: 650 PSEVETI 656
Score = 62.8 bits (151), Expect = 4e-08
Identities = 41/121 (33%), Positives = 64/121 (52%), Gaps = 7/121 (5%)
Query: 20 DWVKFKRTVTEPKHGWQGAKPNSVGFVQSVPGRDNDDLIVSFCSGEVRVL--TSEIVKLI 77
DWV+ K ++T P + W P+S G V + ++ DL VSFC + L E+ ++
Sbjct: 665 DWVRVKASITTPTYQWGEVNPSSTGVVHRM---EDGDLCVSFCFLDRLWLCKAGELERIR 721
Query: 78 PLDRGQHVQLKGDVNEPRFGWRGQSRDSIGTVLCVDPEDGILRVGFP-GASRGWKADPAE 136
P G V++K + PR+GW ++ S G V+ VD +G LR+ F R W DPA+
Sbjct: 722 PFRIGDRVKIKDGLVTPRWGWGMETHASKGHVVGVD-ANGKLRIKFLWREGRPWIGDPAD 780
Query: 137 M 137
+
Sbjct: 781 I 781
Score = 62.0 bits (149), Expect = 6e-08
Identities = 46/195 (23%), Positives = 81/195 (40%), Gaps = 8/195 (4%)
Query: 16 FDVLDWVKFKRTVTEPKHGWQGAKPNSVGFVQSVPGRDNDDLIVSFCSGEV---RVLTSE 72
F V + ++T+P+ GW P ++G V + D D + + +G RV +
Sbjct: 342 FHVGQEIHMTPSITQPRLGWSNETPATIGKVMRI---DMDGTLSAQVTGRQTLWRVSPGD 398
Query: 73 IVKLIPLDRGQHVQLKGDV-NEPRFGWRGQSRDSIGTVLCVDPEDGILRVGFPGASRGWK 131
L + G V+ K + N P + W R+SI V + E G L + W
Sbjct: 399 AELLSGFEVGDWVRSKPSLGNRPSYDWSNVGRESIAVVHSIQ-ETGYLELACCFRKGRWS 457
Query: 132 ADPAEMERVEEFKVGDWVRVRPTLTTSKHGLGNVVPGTIGIVYCIRPDSSLLVELSYVQN 191
++E++ KVG +V + +T + G P + GI+ + D + V +
Sbjct: 458 THYTDLEKIPALKVGQFVHFQKGITEPRWGWRAAKPDSRGIITTVHADGEVRVAFFGLPG 517
Query: 192 PWHCEPEEIEHVPPF 206
W +P ++E P F
Sbjct: 518 LWRGDPADLEVEPMF 532
Score = 47.8 bits (112), Expect = 0.001
Identities = 31/138 (22%), Positives = 60/138 (43%), Gaps = 3/138 (2%)
Query: 82 GQHVQLKGDVNEPRFGWRGQSRDSIGTVLCVDPEDGILRVGFPGASRGWKADPAEMERVE 141
G V+ K + P GW+G S+G V + ++ ++ G +R + ++ ++
Sbjct: 23 GDWVKFKRGITTPLHGWQGAKPKSVGFVQTILEKEDMIIAFCSGEARVLANEVVKLIPLD 82
Query: 142 EFKVGDWVRVRPTLTTSKHGLGNVVPGTIGIVYCIRPDSSLLVELSYVQNPWHCEPEEIE 201
G VR+R + + G ++G V C+ D L V W +P E+E
Sbjct: 83 R---GQHVRLRADVKEPRFGWRGQSRDSVGTVLCVDEDGILRVGFPGASRGWKADPAEME 139
Query: 202 HVPPFRNLDMLGVVRHIT 219
V F+ D + + +++T
Sbjct: 140 RVEEFKVGDWVRIRQNLT 157
>ref|NP_850814.1| expressed protein [Arabidopsis thaliana]
Length = 558
Score = 327 bits (837), Expect = 1e-87
Identities = 151/196 (77%), Positives = 175/196 (89%), Gaps = 3/196 (1%)
Query: 12 SPITFDVLDWVKFKRTVTEPKHGWQGAKPNSVGFVQSVPGRDNDDLIVSFCSGEVRVLTS 71
S IT++V DWVKFKR +T P HGWQGAKP SVGFVQ++ + +D+I++FCSGE RVL +
Sbjct: 16 SSITYNVGDWVKFKRGITTPLHGWQGAKPKSVGFVQTI--LEKEDMIIAFCSGEARVLAN 73
Query: 72 EIVKLIPLDRGQHVQLKGDVNEPRFGWRGQSRDSIGTVLCVDPEDGILRVGFPGASRGWK 131
E+VKLIPLDRGQHV+L+ DV EPRFGWRGQSRDS+GTVLCVD EDGILRVGFPGASRGWK
Sbjct: 74 EVVKLIPLDRGQHVRLRADVKEPRFGWRGQSRDSVGTVLCVD-EDGILRVGFPGASRGWK 132
Query: 132 ADPAEMERVEEFKVGDWVRVRPTLTTSKHGLGNVVPGTIGIVYCIRPDSSLLVELSYVQN 191
ADPAEMERVEEFKVGDWVR+R LT++KHG G+VVPG++GIVYC+RPDSSLLVELSY+ N
Sbjct: 133 ADPAEMERVEEFKVGDWVRIRQNLTSAKHGFGSVVPGSMGIVYCVRPDSSLLVELSYLPN 192
Query: 192 PWHCEPEEIEHVPPFR 207
PWHCEPEE+E V PFR
Sbjct: 193 PWHCEPEEVEPVAPFR 208
Score = 269 bits (688), Expect = 2e-70
Identities = 118/171 (69%), Positives = 149/171 (87%), Gaps = 2/171 (1%)
Query: 377 SMVTGRQSLWKVSPGDAERLPGXEVGDWVRSKPSLGNRPSYDWNSVGRESLAVVHSVQDS 436
+ VTGRQ+LW+VSPGDAE L G EVGDWVRSKPSLGNRPSYDW++VGRES+AVVHS+Q++
Sbjct: 387 AQVTGRQTLWRVSPGDAELLSGFEVGDWVRSKPSLGNRPSYDWSNVGRESIAVVHSIQET 446
Query: 437 GYLELACCFRKGKWITHYTDVEKVPSFKVGQYVRFRPGLAEPRFGWGGAQPESQGIITNI 496
GYLELACCFRKG+W THYTD+EK+P+ KVGQ+V F+ G+ EPR+GW A+P+S+GIIT +
Sbjct: 447 GYLELACCFRKGRWSTHYTDLEKIPALKVGQFVHFQKGITEPRWGWRAAKPDSRGIITTV 506
Query: 497 HADGEVRVAFFGLSGLWKGDPSDLQAEQIFEVGEW--VRLKENVNNWKSIG 545
HADGEVRVAFFGL GLW+GDP+DL+ E +FEVGEW +++K+ + W +G
Sbjct: 507 HADGEVRVAFFGLPGLWRGDPADLEVEPMFEVGEWRPMQVKDMLWEWMQMG 557
Score = 160 bits (406), Expect = 1e-37
Identities = 92/274 (33%), Positives = 139/274 (50%), Gaps = 13/274 (4%)
Query: 392 DAERLPGXEVGDWVRSKPSLGNRPSYDWNSVGRESLAVVHSVQDSGYLELACCFRKGKWI 451
+ E L +VGDWVR K S+ + P Y W + R S+ V+HS+ + G + +A CFR +
Sbjct: 276 NTEPLYHEQVGDWVRVKASVSS-PKYGWEDITRNSIGVMHSLDEDGDVGIAFCFRSKPFS 334
Query: 452 THYTDVEKVPSFKVGQYVRFRPGLAEPRFGWGGAQPESQGIITNIHADGEVRVAFFGLSG 511
TDVEKV F VGQ + P + +PR GW P + G + I DG + G
Sbjct: 335 CSVTDVEKVTPFHVGQEIHMTPSITQPRLGWSNETPATIGKVMRIDMDGTLSAQVTGRQT 394
Query: 512 LWKGDPSDLQAEQIFEVGEWVRLKENVNN-----WKSIGPGSVGVVQGIGYEGGETDRST 566
LW+ P D + FEVG+WVR K ++ N W ++G S+ VV I E G +
Sbjct: 395 LWRVSPGDAELLSGFEVGDWVRSKPSLGNRPSYDWSNVGRESIAVVHSI-QETGYLE--- 450
Query: 567 FVGFCGEQEKWVGPSSHLERVDKLIVGQKVRVKQNVKQPRFGWSGHTHASIGTIQAIDAD 626
+ C + +W + LE++ L VGQ V ++ + +PR+GW S G I + AD
Sbjct: 451 -LACCFRKGRWSTHYTDLEKIPALKVGQFVHFQKGITEPRWGWRAAKPDSRGIITTVHAD 509
Query: 627 GKLWIYTPAGSRTWMLDPSEVEVVEEKELCIGDW 660
G++ + W DP+++EV E +G+W
Sbjct: 510 GEVRVAFFGLPGLWRGDPADLEV--EPMFEVGEW 541
Score = 149 bits (377), Expect = 2e-34
Identities = 92/293 (31%), Positives = 147/293 (49%), Gaps = 39/293 (13%)
Query: 401 VGDWVRSKPSLGNRPSYDWNSVGRESLAVVHSVQDSGYLELACCFRKGKWITHYTDVEKV 460
VGDWV+ K + P + W +S+ V ++ + + +A C + + + + +V K+
Sbjct: 22 VGDWVKFKRGI-TTPLHGWQGAKPKSVGFVQTILEKEDMIIAFCSGEARVLAN--EVVKL 78
Query: 461 PSFKVGQYVRFRPGLAEPRFGWGGAQPESQGIITNIHADGEVRVAFFGLSGLWKGDPSDL 520
GQ+VR R + EPRFGW G +S G + + DG +RV F G S WK DP+++
Sbjct: 79 IPLDRGQHVRLRADVKEPRFGWRGQSRDSVGTVLCVDEDGILRVGFPGASRGWKADPAEM 138
Query: 521 QAEQIFEVGEWVRLKENVNNWK----SIGPGSVGVVQGIGYEGGETDRSTFVGFCGEQEK 576
+ + F+VG+WVR+++N+ + K S+ PGS+G+V + D S V
Sbjct: 139 ERVEEFKVGDWVRIRQNLTSAKHGFGSVVPGSMGIVYCV-----RPDSSLLVELSYLPNP 193
Query: 577 WVGPSSHLERVDKLIVGQKVRVKQNVKQPRFGWSGHTHASIGTIQAIDADGKLWIYTPAG 636
W +E V +++V +PR+ W G TH S+G I I+ DG L I P
Sbjct: 194 WHCEPEEVEPV--------APFRRSVAEPRYAWGGETHHSVGKISEIENDGLLIIEIPNR 245
Query: 637 SRTWMLDPSEVEVVEEKEL-------------------CIGDWVRVRASVSTP 670
W DPS++E +++ +L +GDWVRV+ASVS+P
Sbjct: 246 PIPWQADPSDMEKIDDFKLDVQHVKSFDSFNTEPLYHEQVGDWVRVKASVSSP 298
Score = 137 bits (346), Expect = 9e-31
Identities = 88/307 (28%), Positives = 141/307 (45%), Gaps = 41/307 (13%)
Query: 386 WKVSPGDAERLPGXEVGDWVRSKPSLGNRPSYDWNSVGRESLAVVHSVQDSGYLELACCF 445
WK P + ER+ +VGDWVR + +L + + SV S+ +V+ V+ L + +
Sbjct: 131 WKADPAEMERVEEFKVGDWVRIRQNL-TSAKHGFGSVVPGSMGIVYCVRPDSSLLVELSY 189
Query: 446 RKGKWITHYTDVEKVPSFKVGQYVRFRPGLAEPRFGWGGAQPESQGIITNIHADGEVRVA 505
W +VE V FR +AEPR+ WGG S G I+ I DG + +
Sbjct: 190 LPNPWHCEPEEVEPV--------APFRRSVAEPRYAWGGETHHSVGKISEIENDGLLIIE 241
Query: 506 FFGLSGLWKGDPSDLQAEQIF---------------------EVGEWVRLKENVNN---- 540
W+ DPSD++ F +VG+WVR+K +V++
Sbjct: 242 IPNRPIPWQADPSDMEKIDDFKLDVQHVKSFDSFNTEPLYHEQVGDWVRVKASVSSPKYG 301
Query: 541 WKSIGPGSVGVVQGIGYEGGETDRSTFVGFCGEQEKWVGPSSHLERVDKLIVGQKVRVKQ 600
W+ I S+GV+ + + D + FC + + + +E+V VGQ++ +
Sbjct: 302 WEDITRNSIGVMHSL-----DEDGDVGIAFCFRSKPFSCSVTDVEKVTPFHVGQEIHMTP 356
Query: 601 NVKQPRFGWSGHTHASIGTIQAIDADGKLWIYTPAGSRTWMLDPSEVEVVEEKELCIGDW 660
++ QPR GWS T A+IG + ID DG L W + P + E++ E +GDW
Sbjct: 357 SITQPRLGWSNETPATIGKVMRIDMDGTLSAQVTGRQTLWRVSPGDAELLSGFE--VGDW 414
Query: 661 VRVRASV 667
VR + S+
Sbjct: 415 VRSKPSL 421
Score = 103 bits (256), Expect = 2e-20
Identities = 59/214 (27%), Positives = 105/214 (48%), Gaps = 34/214 (15%)
Query: 16 FDVLDWVKFKRTVTEPKHGWQGAKPNSVGFVQSVPGRDNDDLIV--SFCSGEVRVLTSEI 73
F V DWV+ ++ +T KHG+ P S+G V V R + L+V S+ E+
Sbjct: 144 FKVGDWVRIRQNLTSAKHGFGSVVPGSMGIVYCV--RPDSSLLVELSYLPNPWHCEPEEV 201
Query: 74 VKLIPLDRGQHVQLKGDVNEPRFGWRGQSRDSIGTVLCVDPEDGILRVGFPGASRGWKAD 133
+ P R V EPR+ W G++ S+G + ++ DG+L + P W+AD
Sbjct: 202 EPVAPFRRS--------VAEPRYAWGGETHHSVGKISEIE-NDGLLIIEIPNRPIPWQAD 252
Query: 134 PAEMERVEEFK---------------------VGDWVRVRPTLTTSKHGLGNVVPGTIGI 172
P++ME++++FK VGDWVRV+ ++++ K+G ++ +IG+
Sbjct: 253 PSDMEKIDDFKLDVQHVKSFDSFNTEPLYHEQVGDWVRVKASVSSPKYGWEDITRNSIGV 312
Query: 173 VYCIRPDSSLLVELSYVQNPWHCEPEEIEHVPPF 206
++ + D + + + P+ C ++E V PF
Sbjct: 313 MHSLDEDGDVGIAFCFRSKPFSCSVTDVEKVTPF 346
Score = 102 bits (254), Expect = 4e-20
Identities = 54/191 (28%), Positives = 98/191 (51%), Gaps = 6/191 (3%)
Query: 20 DWVKFKRTVTEPKHGWQGAKPNSVGFVQSVPGRDNDDLIVSFC--SGEVRVLTSEIVKLI 77
DWV+ K +V+ PK+GW+ NS+G + S+ ++ D+ ++FC S +++ K+
Sbjct: 287 DWVRVKASVSSPKYGWEDITRNSIGVMHSLD--EDGDVGIAFCFRSKPFSCSVTDVEKVT 344
Query: 78 PLDRGQHVQLKGDVNEPRFGWRGQSRDSIGTVLCVDPEDGILRVGFPGASRGWKADPAEM 137
P GQ + + + +PR GW ++ +IG V+ +D DG L G W+ P +
Sbjct: 345 PFHVGQEIHMTPSITQPRLGWSNETPATIGKVMRID-MDGTLSAQVTGRQTLWRVSPGDA 403
Query: 138 ERVEEFKVGDWVRVRPTL-TTSKHGLGNVVPGTIGIVYCIRPDSSLLVELSYVQNPWHCE 196
E + F+VGDWVR +P+L + NV +I +V+ I+ L + + + W
Sbjct: 404 ELLSGFEVGDWVRSKPSLGNRPSYDWSNVGRESIAVVHSIQETGYLELACCFRKGRWSTH 463
Query: 197 PEEIEHVPPFR 207
++E +P +
Sbjct: 464 YTDLEKIPALK 474
Score = 82.4 bits (202), Expect = 4e-14
Identities = 54/193 (27%), Positives = 91/193 (46%), Gaps = 11/193 (5%)
Query: 462 SFKVGQYVRFRPGLAEPRFGWGGAQPESQGIITNIHADGEVRVAFFGLSGLWKGDPSDLQ 521
++ VG +V+F+ G+ P GW GA+P+S G + I ++ +AF SG + +++
Sbjct: 19 TYNVGDWVKFKRGITTPLHGWQGAKPKSVGFVQTILEKEDMIIAF--CSGEARVLANEVV 76
Query: 522 AEQIFEVGEWVRLKENVN----NWKSIGPGSVGVVQGIGYEGGETDRSTFVGFCGEQEKW 577
+ G+ VRL+ +V W+ SVG V + + D VGF G W
Sbjct: 77 KLIPLDRGQHVRLRADVKEPRFGWRGQSRDSVGTVLCV-----DEDGILRVGFPGASRGW 131
Query: 578 VGPSSHLERVDKLIVGQKVRVKQNVKQPRFGWSGHTHASIGTIQAIDADGKLWIYTPAGS 637
+ +ERV++ VG VR++QN+ + G+ S+G + + D L +
Sbjct: 132 KADPAEMERVEEFKVGDWVRIRQNLTSAKHGFGSVVPGSMGIVYCVRPDSSLLVELSYLP 191
Query: 638 RTWMLDPSEVEVV 650
W +P EVE V
Sbjct: 192 NPWHCEPEEVEPV 204
Score = 76.6 bits (187), Expect = 2e-12
Identities = 44/134 (32%), Positives = 66/134 (48%), Gaps = 2/134 (1%)
Query: 16 FDVLDWVKFKRTV-TEPKHGWQGAKPNSVGFVQSVPGRDNDDLIVSFCSGEVRVLTSEIV 74
F+V DWV+ K ++ P + W S+ V S+ +L F G +++
Sbjct: 409 FEVGDWVRSKPSLGNRPSYDWSNVGRESIAVVHSIQETGYLELACCFRKGRWSTHYTDLE 468
Query: 75 KLIPLDRGQHVQLKGDVNEPRFGWRGQSRDSIGTVLCVDPEDGILRVGFPGASRGWKADP 134
K+ L GQ V + + EPR+GWR DS G + V DG +RV F G W+ DP
Sbjct: 469 KIPALKVGQFVHFQKGITEPRWGWRAAKPDSRGIITTVH-ADGEVRVAFFGLPGLWRGDP 527
Query: 135 AEMERVEEFKVGDW 148
A++E F+VG+W
Sbjct: 528 ADLEVEPMFEVGEW 541
Score = 62.0 bits (149), Expect = 6e-08
Identities = 46/195 (23%), Positives = 81/195 (40%), Gaps = 8/195 (4%)
Query: 16 FDVLDWVKFKRTVTEPKHGWQGAKPNSVGFVQSVPGRDNDDLIVSFCSGEV---RVLTSE 72
F V + ++T+P+ GW P ++G V + D D + + +G RV +
Sbjct: 346 FHVGQEIHMTPSITQPRLGWSNETPATIGKVMRI---DMDGTLSAQVTGRQTLWRVSPGD 402
Query: 73 IVKLIPLDRGQHVQLKGDV-NEPRFGWRGQSRDSIGTVLCVDPEDGILRVGFPGASRGWK 131
L + G V+ K + N P + W R+SI V + E G L + W
Sbjct: 403 AELLSGFEVGDWVRSKPSLGNRPSYDWSNVGRESIAVVHSIQ-ETGYLELACCFRKGRWS 461
Query: 132 ADPAEMERVEEFKVGDWVRVRPTLTTSKHGLGNVVPGTIGIVYCIRPDSSLLVELSYVQN 191
++E++ KVG +V + +T + G P + GI+ + D + V +
Sbjct: 462 THYTDLEKIPALKVGQFVHFQKGITEPRWGWRAAKPDSRGIITTVHADGEVRVAFFGLPG 521
Query: 192 PWHCEPEEIEHVPPF 206
W +P ++E P F
Sbjct: 522 LWRGDPADLEVEPMF 536
Score = 47.8 bits (112), Expect = 0.001
Identities = 31/138 (22%), Positives = 60/138 (43%), Gaps = 3/138 (2%)
Query: 82 GQHVQLKGDVNEPRFGWRGQSRDSIGTVLCVDPEDGILRVGFPGASRGWKADPAEMERVE 141
G V+ K + P GW+G S+G V + ++ ++ G +R + ++ ++
Sbjct: 23 GDWVKFKRGITTPLHGWQGAKPKSVGFVQTILEKEDMIIAFCSGEARVLANEVVKLIPLD 82
Query: 142 EFKVGDWVRVRPTLTTSKHGLGNVVPGTIGIVYCIRPDSSLLVELSYVQNPWHCEPEEIE 201
G VR+R + + G ++G V C+ D L V W +P E+E
Sbjct: 83 R---GQHVRLRADVKEPRFGWRGQSRDSVGTVLCVDEDGILRVGFPGASRGWKADPAEME 139
Query: 202 HVPPFRNLDMLGVVRHIT 219
V F+ D + + +++T
Sbjct: 140 RVEEFKVGDWVRIRQNLT 157
>dbj|BAD37536.1| protein kinase-like [Oryza sativa (japonica cultivar-group)]
gi|51536357|dbj|BAD37488.1| protein kinase-like [Oryza
sativa (japonica cultivar-group)]
Length = 630
Score = 63.9 bits (154), Expect = 2e-08
Identities = 52/168 (30%), Positives = 73/168 (42%), Gaps = 18/168 (10%)
Query: 491 GIITNIHADGEVRVAFFGLSGLWKGDPSDLQAEQI-FEVGEWVRLKENVNNWKSIGPGSV 549
G+ N DG + V GL K S ++ F G+WVRL+E+ V
Sbjct: 377 GMEDNGERDGYILVRIHGLHDPLKVRSSTVERVTYGFAAGDWVRLREDEKK-----RSQV 431
Query: 550 GVVQGIGYEGGETDRSTFVGFCGEQEKWVGPSSHLERVDKLIVGQKVRVKQNVKQPRFGW 609
G++ I G + +VG G W G S L+ + VGQ VR+K N+ P+F W
Sbjct: 432 GILHSIDRSG-----TVYVGLIGVDTLWKGEYSDLQMAEAYCVGQFVRLKANISSPQFEW 486
Query: 610 SGHTHASI--GTIQAIDADGKLWIYTPAGSR-----TWMLDPSEVEVV 650
+ G I I +G L+I P + + DPSEVEVV
Sbjct: 487 QRKRGGGLATGRISQILPNGCLFIKFPGKFNLGEVCSCLADPSEVEVV 534
Score = 48.9 bits (115), Expect = 5e-04
Identities = 38/145 (26%), Positives = 66/145 (45%), Gaps = 22/145 (15%)
Query: 387 KVSPGDAERLP-GXEVGDWVRSKPSLGNRPSYDWNSVGRESLAVVHSVQDSGYLELACCF 445
KV ER+ G GDWVR + + R + ++HS+ SG + +
Sbjct: 400 KVRSSTVERVTYGFAAGDWVRLRE----------DEKKRSQVGILHSIDRSGTVYVGLIG 449
Query: 446 RKGKWITHYTDVEKVPSFKVGQYVRFRPGLAEPRFGW----GGAQPESQGIITNIHADGE 501
W Y+D++ ++ VGQ+VR + ++ P+F W GG + G I+ I +G
Sbjct: 450 VDTLWKGEYSDLQMAEAYCVGQFVRLKANISSPQFEWQRKRGGGL--ATGRISQILPNGC 507
Query: 502 VRVAFFGLSGLWK-----GDPSDLQ 521
+ + F G L + DPS+++
Sbjct: 508 LFIKFPGKFNLGEVCSCLADPSEVE 532
Score = 48.5 bits (114), Expect = 7e-04
Identities = 32/86 (37%), Positives = 46/86 (53%), Gaps = 12/86 (13%)
Query: 457 VEKVP-SFKVGQYVRFRPGLAEPRFGWGGAQPESQ-GIITNIHADGEVRVAFFGLSGLWK 514
VE+V F G +VR R + SQ GI+ +I G V V G+ LWK
Sbjct: 406 VERVTYGFAAGDWVRLRED----------EKKRSQVGILHSIDRSGTVYVGLIGVDTLWK 455
Query: 515 GDPSDLQAEQIFEVGEWVRLKENVNN 540
G+ SDLQ + + VG++VRLK N+++
Sbjct: 456 GEYSDLQMAEAYCVGQFVRLKANISS 481
Score = 42.0 bits (97), Expect = 0.067
Identities = 38/148 (25%), Positives = 72/148 (47%), Gaps = 18/148 (12%)
Query: 527 EVGEWVRLKENVNNWKSIGPGSVGVVQG--IGYE-GGETDRSTFVGFCGEQEKWVGPSSH 583
+VG+ VR ++ N + P ++ V G +G E GE D V G + SS
Sbjct: 349 QVGDKVRSRKLKN---TCSPTTMEVPDGTIVGMEDNGERDGYILVRIHGLHDPLKVRSST 405
Query: 584 LERVDK-LIVGQKVRVKQNVKQPRFGWSGHTHASIGTIQAIDADGKLWIYTPAGSRTWML 642
+ERV G VR++++ K+ + +G + +ID G +++ W
Sbjct: 406 VERVTYGFAAGDWVRLREDEKK---------RSQVGILHSIDRSGTVYVGLIGVDTLWKG 456
Query: 643 DPSEVEVVEEKELCIGDWVRVRASVSTP 670
+ S++++ E C+G +VR++A++S+P
Sbjct: 457 EYSDLQMAEA--YCVGQFVRLKANISSP 482
>gb|AAN46821.1| At4g32250/F10M6_110 [Arabidopsis thaliana]
gi|21436453|gb|AAM51427.1| unknown protein [Arabidopsis
thaliana] gi|20259492|gb|AAM13866.1| unknown protein
[Arabidopsis thaliana] gi|21703136|gb|AAM74508.1|
AT4g32250/F10M6_110 [Arabidopsis thaliana]
gi|30689316|ref|NP_849560.1| protein kinase family
protein [Arabidopsis thaliana]
gi|22329080|ref|NP_194952.2| protein kinase family
protein [Arabidopsis thaliana]
Length = 611
Score = 57.8 bits (138), Expect = 1e-06
Identities = 48/130 (36%), Positives = 63/130 (47%), Gaps = 17/130 (13%)
Query: 529 GEWVRLKENVNNWKSIGPGSVGVVQGIGYEGGETDRSTFVGFCGEQEKWVGPSSHLERVD 588
G+WVRLK V K P VGV+ I EG + VGF G W G SS L+
Sbjct: 411 GDWVRLK--VRKDKRHSP--VGVLHSIDREG-----NVAVGFIGLPTLWKGTSSQLQMAK 461
Query: 589 KLIVGQKVRVKQNVKQPRFGW--SGHTHASIGTIQAIDADGKLWI----YTPAGSR--TW 640
VGQ V++K NV PRF W G + G I + +G L + P G ++
Sbjct: 462 VYSVGQFVKLKANVVIPRFKWMRKGRGIWATGRISQVLPNGCLEVDFPGMLPFGEEHGSY 521
Query: 641 MLDPSEVEVV 650
+ DP+EVE+V
Sbjct: 522 LADPAEVEIV 531
Score = 50.4 bits (119), Expect = 2e-04
Identities = 22/48 (45%), Positives = 32/48 (65%)
Query: 491 GIITNIHADGEVRVAFFGLSGLWKGDPSDLQAEQIFEVGEWVRLKENV 538
G++ +I +G V V F GL LWKG S LQ +++ VG++V+LK NV
Sbjct: 428 GVLHSIDREGNVAVGFIGLPTLWKGTSSQLQMAKVYSVGQFVKLKANV 475
Score = 40.4 bits (93), Expect = 0.19
Identities = 32/132 (24%), Positives = 56/132 (42%), Gaps = 17/132 (12%)
Query: 398 GXEVGDWVRSKPSLGNRPSYDWNSVGRESLAVVHSVQDSGYLELACCFRKGKWITHYTDV 457
G GDWVR K R S + V+HS+ G + + W + +
Sbjct: 407 GLASGDWVRLKVRKDKRHS---------PVGVLHSIDREGNVAVGFIGLPTLWKGTSSQL 457
Query: 458 EKVPSFKVGQYVRFRPGLAEPRFGW--GGAQPESQGIITNIHADGEVRVAFFGL------ 509
+ + VGQ+V+ + + PRF W G + G I+ + +G + V F G+
Sbjct: 458 QMAKVYSVGQFVKLKANVVIPRFKWMRKGRGIWATGRISQVLPNGCLEVDFPGMLPFGEE 517
Query: 510 SGLWKGDPSDLQ 521
G + DP++++
Sbjct: 518 HGSYLADPAEVE 529
>emb|CAB79943.1| putative protein [Arabidopsis thaliana] gi|2864618|emb|CAA16965.1|
putative protein [Arabidopsis thaliana]
gi|7485349|pir||T05403 hypothetical protein F10M6.110 -
Arabidopsis thaliana
Length = 593
Score = 57.8 bits (138), Expect = 1e-06
Identities = 48/130 (36%), Positives = 63/130 (47%), Gaps = 17/130 (13%)
Query: 529 GEWVRLKENVNNWKSIGPGSVGVVQGIGYEGGETDRSTFVGFCGEQEKWVGPSSHLERVD 588
G+WVRLK V K P VGV+ I EG + VGF G W G SS L+
Sbjct: 393 GDWVRLK--VRKDKRHSP--VGVLHSIDREG-----NVAVGFIGLPTLWKGTSSQLQMAK 443
Query: 589 KLIVGQKVRVKQNVKQPRFGW--SGHTHASIGTIQAIDADGKLWI----YTPAGSR--TW 640
VGQ V++K NV PRF W G + G I + +G L + P G ++
Sbjct: 444 VYSVGQFVKLKANVVIPRFKWMRKGRGIWATGRISQVLPNGCLEVDFPGMLPFGEEHGSY 503
Query: 641 MLDPSEVEVV 650
+ DP+EVE+V
Sbjct: 504 LADPAEVEIV 513
Score = 50.4 bits (119), Expect = 2e-04
Identities = 22/48 (45%), Positives = 32/48 (65%)
Query: 491 GIITNIHADGEVRVAFFGLSGLWKGDPSDLQAEQIFEVGEWVRLKENV 538
G++ +I +G V V F GL LWKG S LQ +++ VG++V+LK NV
Sbjct: 410 GVLHSIDREGNVAVGFIGLPTLWKGTSSQLQMAKVYSVGQFVKLKANV 457
Score = 40.4 bits (93), Expect = 0.19
Identities = 32/132 (24%), Positives = 56/132 (42%), Gaps = 17/132 (12%)
Query: 398 GXEVGDWVRSKPSLGNRPSYDWNSVGRESLAVVHSVQDSGYLELACCFRKGKWITHYTDV 457
G GDWVR K R S + V+HS+ G + + W + +
Sbjct: 389 GLASGDWVRLKVRKDKRHS---------PVGVLHSIDREGNVAVGFIGLPTLWKGTSSQL 439
Query: 458 EKVPSFKVGQYVRFRPGLAEPRFGW--GGAQPESQGIITNIHADGEVRVAFFGL------ 509
+ + VGQ+V+ + + PRF W G + G I+ + +G + V F G+
Sbjct: 440 QMAKVYSVGQFVKLKANVVIPRFKWMRKGRGIWATGRISQVLPNGCLEVDFPGMLPFGEE 499
Query: 510 SGLWKGDPSDLQ 521
G + DP++++
Sbjct: 500 HGSYLADPAEVE 511
>gb|AAD08657.1| HERC2 [Homo sapiens]
Length = 4834
Score = 45.8 bits (107), Expect = 0.005
Identities = 26/70 (37%), Positives = 38/70 (54%), Gaps = 6/70 (8%)
Query: 581 SSHLERVDKLIVGQKVRVKQNVKQPRFGWSGHTHASIGTIQAIDADGKLWIYTPAGSRTW 640
SSH++ +G KVRVK +V P++ W TH S+G ++A A+GK I W
Sbjct: 2636 SSHIK------IGDKVRVKASVTTPKYKWGSVTHQSVGVVKAFSANGKDIIVDFPQQSHW 2689
Query: 641 MLDPSEVEVV 650
SE+E+V
Sbjct: 2690 TGLLSEMELV 2699
Score = 35.8 bits (81), Expect = 4.8
Identities = 15/58 (25%), Positives = 29/58 (49%)
Query: 464 KVGQYVRFRPGLAEPRFGWGGAQPESQGIITNIHADGEVRVAFFGLSGLWKGDPSDLQ 521
K+G VR + + P++ WG +S G++ A+G+ + F W G S+++
Sbjct: 2640 KIGDKVRVKASVTTPKYKWGSVTHQSVGVVKAFSANGKDIIVDFPQQSHWTGLLSEME 2697
Score = 35.0 bits (79), Expect = 8.2
Identities = 21/67 (31%), Positives = 34/67 (50%), Gaps = 1/67 (1%)
Query: 400 EVGDWVRSKPSLGNRPSYDWNSVGRESLAVVHSVQDSGYLELACCFRKGKWITHYTDVEK 459
++GD VR K S+ P Y W SV +S+ VV + +G + ++ W +++E
Sbjct: 2640 KIGDKVRVKASV-TTPKYKWGSVTHQSVGVVKAFSANGKDIIVDFPQQSHWTGLLSEMEL 2698
Query: 460 VPSFKVG 466
VPS G
Sbjct: 2699 VPSIHPG 2705
>ref|NP_004658.2| hect domain and RLD 2 [Homo sapiens]
Length = 4834
Score = 45.8 bits (107), Expect = 0.005
Identities = 26/70 (37%), Positives = 38/70 (54%), Gaps = 6/70 (8%)
Query: 581 SSHLERVDKLIVGQKVRVKQNVKQPRFGWSGHTHASIGTIQAIDADGKLWIYTPAGSRTW 640
SSH++ +G KVRVK +V P++ W TH S+G ++A A+GK I W
Sbjct: 2636 SSHIK------IGDKVRVKASVTTPKYKWGSVTHQSVGVVKAFSANGKDIIVDFPQQSHW 2689
Query: 641 MLDPSEVEVV 650
SE+E+V
Sbjct: 2690 TGLLSEMELV 2699
Score = 35.8 bits (81), Expect = 4.8
Identities = 15/58 (25%), Positives = 29/58 (49%)
Query: 464 KVGQYVRFRPGLAEPRFGWGGAQPESQGIITNIHADGEVRVAFFGLSGLWKGDPSDLQ 521
K+G VR + + P++ WG +S G++ A+G+ + F W G S+++
Sbjct: 2640 KIGDKVRVKASVTTPKYKWGSVTHQSVGVVKAFSANGKDIIVDFPQQSHWTGLLSEME 2697
Score = 35.0 bits (79), Expect = 8.2
Identities = 21/67 (31%), Positives = 34/67 (50%), Gaps = 1/67 (1%)
Query: 400 EVGDWVRSKPSLGNRPSYDWNSVGRESLAVVHSVQDSGYLELACCFRKGKWITHYTDVEK 459
++GD VR K S+ P Y W SV +S+ VV + +G + ++ W +++E
Sbjct: 2640 KIGDKVRVKASV-TTPKYKWGSVTHQSVGVVKAFSANGKDIIVDFPQQSHWTGLLSEMEL 2698
Query: 460 VPSFKVG 466
VPS G
Sbjct: 2699 VPSIHPG 2705
>gb|AAC31431.1| rjs [Mus musculus]
Length = 4836
Score = 45.4 bits (106), Expect = 0.006
Identities = 26/70 (37%), Positives = 38/70 (54%), Gaps = 6/70 (8%)
Query: 581 SSHLERVDKLIVGQKVRVKQNVKQPRFGWSGHTHASIGTIQAIDADGKLWIYTPAGSRTW 640
SSH++ +G KVRVK +V P++ W TH S+G ++A A+GK I W
Sbjct: 2637 SSHIK------IGDKVRVKASVTTPKYKWGSVTHQSVGLVKAFSANGKDIIVDFPQQSHW 2690
Query: 641 MLDPSEVEVV 650
SE+E+V
Sbjct: 2691 TGLLSEMELV 2700
Score = 35.4 bits (80), Expect = 6.3
Identities = 15/58 (25%), Positives = 29/58 (49%)
Query: 464 KVGQYVRFRPGLAEPRFGWGGAQPESQGIITNIHADGEVRVAFFGLSGLWKGDPSDLQ 521
K+G VR + + P++ WG +S G++ A+G+ + F W G S+++
Sbjct: 2641 KIGDKVRVKASVTTPKYKWGSVTHQSVGLVKAFSANGKDIIVDFPQQSHWTGLLSEME 2698
>ref|NP_034548.1| hect (homologous to the E6-AP (UBE3A) carboxyl terminus) domain and
RCC1 (CHC1)-like domain (RLD) 2 [Mus musculus]
gi|4079811|gb|AAD08658.1| Herc2 [Mus musculus]
gi|7513677|pir||T14346 herc2 protein - mouse
Length = 4836
Score = 45.4 bits (106), Expect = 0.006
Identities = 26/70 (37%), Positives = 38/70 (54%), Gaps = 6/70 (8%)
Query: 581 SSHLERVDKLIVGQKVRVKQNVKQPRFGWSGHTHASIGTIQAIDADGKLWIYTPAGSRTW 640
SSH++ +G KVRVK +V P++ W TH S+G ++A A+GK I W
Sbjct: 2637 SSHIK------IGDKVRVKASVTTPKYKWGSVTHQSVGLVKAFSANGKDIIVDFPQQSHW 2690
Query: 641 MLDPSEVEVV 650
SE+E+V
Sbjct: 2691 TGLLSEMELV 2700
Score = 35.4 bits (80), Expect = 6.3
Identities = 15/58 (25%), Positives = 29/58 (49%)
Query: 464 KVGQYVRFRPGLAEPRFGWGGAQPESQGIITNIHADGEVRVAFFGLSGLWKGDPSDLQ 521
K+G VR + + P++ WG +S G++ A+G+ + F W G S+++
Sbjct: 2641 KIGDKVRVKASVTTPKYKWGSVTHQSVGLVKAFSANGKDIIVDFPQQSHWTGLLSEME 2698
>ref|XP_536160.1| PREDICTED: hypothetical protein XP_536160 [Canis familiaris]
Length = 4077
Score = 45.1 bits (105), Expect = 0.008
Identities = 23/59 (38%), Positives = 33/59 (54%)
Query: 592 VGQKVRVKQNVKQPRFGWSGHTHASIGTIQAIDADGKLWIYTPAGSRTWMLDPSEVEVV 650
+G KVRVK +V P++ W TH S+G ++A A+GK I W SE+E+V
Sbjct: 1859 IGDKVRVKSSVTTPKYKWGSVTHQSVGIVKAFSANGKDVIVDFPQQSHWTGLLSEMELV 1917
Score = 36.2 bits (82), Expect = 3.7
Identities = 16/58 (27%), Positives = 29/58 (49%)
Query: 464 KVGQYVRFRPGLAEPRFGWGGAQPESQGIITNIHADGEVRVAFFGLSGLWKGDPSDLQ 521
K+G VR + + P++ WG +S GI+ A+G+ + F W G S+++
Sbjct: 1858 KIGDKVRVKSSVTTPKYKWGSVTHQSVGIVKAFSANGKDVIVDFPQQSHWTGLLSEME 1915
>ref|XP_598208.1| PREDICTED: similar to hect domain and RLD 2, partial [Bos taurus]
Length = 479
Score = 45.1 bits (105), Expect = 0.008
Identities = 23/59 (38%), Positives = 33/59 (54%)
Query: 592 VGQKVRVKQNVKQPRFGWSGHTHASIGTIQAIDADGKLWIYTPAGSRTWMLDPSEVEVV 650
+G KVRVK +V P++ W TH S+G ++A A+GK I W SE+E+V
Sbjct: 237 IGDKVRVKASVTTPKYKWGSVTHQSVGVVKAFSANGKDVIVDFPQQSHWTGLLSEMELV 295
Score = 35.8 bits (81), Expect = 4.8
Identities = 15/58 (25%), Positives = 29/58 (49%)
Query: 464 KVGQYVRFRPGLAEPRFGWGGAQPESQGIITNIHADGEVRVAFFGLSGLWKGDPSDLQ 521
K+G VR + + P++ WG +S G++ A+G+ + F W G S+++
Sbjct: 236 KIGDKVRVKASVTTPKYKWGSVTHQSVGVVKAFSANGKDVIVDFPQQSHWTGLLSEME 293
Score = 35.0 bits (79), Expect = 8.2
Identities = 21/67 (31%), Positives = 34/67 (50%), Gaps = 1/67 (1%)
Query: 400 EVGDWVRSKPSLGNRPSYDWNSVGRESLAVVHSVQDSGYLELACCFRKGKWITHYTDVEK 459
++GD VR K S+ P Y W SV +S+ VV + +G + ++ W +++E
Sbjct: 236 KIGDKVRVKASV-TTPKYKWGSVTHQSVGVVKAFSANGKDVIVDFPQQSHWTGLLSEMEL 294
Query: 460 VPSFKVG 466
VPS G
Sbjct: 295 VPSVHPG 301
>ref|XP_416878.1| PREDICTED: similar to hect domain and RLD 2 [Gallus gallus]
Length = 3701
Score = 45.1 bits (105), Expect = 0.008
Identities = 23/59 (38%), Positives = 33/59 (54%)
Query: 592 VGQKVRVKQNVKQPRFGWSGHTHASIGTIQAIDADGKLWIYTPAGSRTWMLDPSEVEVV 650
+G KVRVK +V P++ W TH S+G ++A A+GK I W SE+E+V
Sbjct: 2977 IGDKVRVKTSVTTPKYKWGSVTHRSVGVVKAFSANGKDVIVDFPQQSHWTGLLSEMELV 3035
Score = 35.0 bits (79), Expect = 8.2
Identities = 15/58 (25%), Positives = 28/58 (47%)
Query: 464 KVGQYVRFRPGLAEPRFGWGGAQPESQGIITNIHADGEVRVAFFGLSGLWKGDPSDLQ 521
K+G VR + + P++ WG S G++ A+G+ + F W G S+++
Sbjct: 2976 KIGDKVRVKTSVTTPKYKWGSVTHRSVGVVKAFSANGKDVIVDFPQQSHWTGLLSEME 3033
>emb|CAG01384.1| unnamed protein product [Tetraodon nigroviridis]
Length = 4628
Score = 44.7 bits (104), Expect = 0.010
Identities = 26/71 (36%), Positives = 36/71 (50%), Gaps = 3/71 (4%)
Query: 580 PSSHLERVDKLIVGQKVRVKQNVKQPRFGWSGHTHASIGTIQAIDADGKLWIYTPAGSRT 639
P S L + +G KVRVK V P++ W TH S+G ++A A+GK I
Sbjct: 2395 PQSSLSHIK---IGDKVRVKPAVTTPKYKWGSVTHRSVGVVKAFSANGKDVIVDFPQQSH 2451
Query: 640 WMLDPSEVEVV 650
W SE+E+V
Sbjct: 2452 WTGLLSEMELV 2462
Score = 38.1 bits (87), Expect = 0.96
Identities = 16/58 (27%), Positives = 29/58 (49%)
Query: 464 KVGQYVRFRPGLAEPRFGWGGAQPESQGIITNIHADGEVRVAFFGLSGLWKGDPSDLQ 521
K+G VR +P + P++ WG S G++ A+G+ + F W G S+++
Sbjct: 2403 KIGDKVRVKPAVTTPKYKWGSVTHRSVGVVKAFSANGKDVIVDFPQQSHWTGLLSEME 2460
Score = 37.0 bits (84), Expect = 2.1
Identities = 20/65 (30%), Positives = 34/65 (51%)
Query: 140 VEEFKVGDWVRVRPTLTTSKHGLGNVVPGTIGIVYCIRPDSSLLVELSYVQNPWHCEPEE 199
+ K+GD VRV+P +TT K+ G+V ++G+V + ++ Q+ W E
Sbjct: 2399 LSHIKIGDKVRVKPAVTTPKYKWGSVTHRSVGVVKAFSANGKDVIVDFPQQSHWTGLLSE 2458
Query: 200 IEHVP 204
+E VP
Sbjct: 2459 MELVP 2463
Score = 36.6 bits (83), Expect = 2.8
Identities = 21/67 (31%), Positives = 34/67 (50%), Gaps = 1/67 (1%)
Query: 400 EVGDWVRSKPSLGNRPSYDWNSVGRESLAVVHSVQDSGYLELACCFRKGKWITHYTDVEK 459
++GD VR KP++ P Y W SV S+ VV + +G + ++ W +++E
Sbjct: 2403 KIGDKVRVKPAV-TTPKYKWGSVTHRSVGVVKAFSANGKDVIVDFPQQSHWTGLLSEMEL 2461
Query: 460 VPSFKVG 466
VPS G
Sbjct: 2462 VPSIHPG 2468
>ref|XP_218720.3| PREDICTED: similar to putative E3 ligase [Rattus norvegicus]
Length = 5030
Score = 39.7 bits (91), Expect = 0.33
Identities = 17/45 (37%), Positives = 29/45 (63%), Gaps = 6/45 (13%)
Query: 581 SSHLERVDKLIVGQKVRVKQNVKQPRFGWSGHTHASIGTIQAIDA 625
SSH++ +G KVRVK +V P++ W TH S+G ++A+++
Sbjct: 2733 SSHIK------IGDKVRVKASVTTPKYKWGSVTHQSVGLVKALES 2771
>ref|XP_392821.2| PREDICTED: similar to CG17492-PA [Apis mellifera]
Length = 957
Score = 37.7 bits (86), Expect = 1.3
Identities = 39/137 (28%), Positives = 57/137 (41%), Gaps = 20/137 (14%)
Query: 85 VQLKGDVNEPRFGWRGQSR---DSIGTVLCVDPEDGILRVGFPGASRGWKADPAEMERV- 140
V+ ++ E GW + D IG V + + G +RV F G + W P + +V
Sbjct: 289 VETLKEMQEGHGGWNPRMAEYIDKIGRVHRIT-DKGDIRVQFEGCNNRWTFHPGTLTKVI 347
Query: 141 --EEFKVGDWVRVRPTLTTSK-----HG-----LGNVVPGTIGIVYCIRPDSSLLVELSY 188
+ F +GD VRV+ L K HG + N + G G + I D L V L
Sbjct: 348 SKDTFSLGDIVRVKTDLCAVKLYQQGHGEWIDVMKNAL-GKTGKIIKIYSDGDLRVALD- 405
Query: 189 VQNPWHCEPEEIEHVPP 205
+ W P + VPP
Sbjct: 406 -GHTWTFNPLSVSPVPP 421
>ref|XP_395007.2| PREDICTED: similar to hect (homologous to the E6-AP (UBE3A) carboxyl
terminus) domain and RCC1 (CHC1)-like domain (RLD) 2,
partial [Apis mellifera]
Length = 3746
Score = 37.4 bits (85), Expect = 1.6
Identities = 17/39 (43%), Positives = 23/39 (58%)
Query: 590 LIVGQKVRVKQNVKQPRFGWSGHTHASIGTIQAIDADGK 628
L +G KVRVK +V P++ W H SIG + I +GK
Sbjct: 1533 LKIGDKVRVKASVTVPKYKWGSVNHQSIGIVTGIINNGK 1571
Score = 35.0 bits (79), Expect = 8.2
Identities = 21/64 (32%), Positives = 37/64 (57%), Gaps = 3/64 (4%)
Query: 400 EVGDWVRSKPSLGNRPSYDWNSVGRESLAVVHSVQDSGYLELACCF-RKGKWITHYTDVE 458
++GD VR K S+ P Y W SV +S+ +V + ++G ++A F ++ W ++E
Sbjct: 1534 KIGDKVRVKASV-TVPKYKWGSVNHQSIGIVTGIINNG-KDVAIDFPQQNNWTGCAIEME 1591
Query: 459 KVPS 462
+VPS
Sbjct: 1592 RVPS 1595
Score = 35.0 bits (79), Expect = 8.2
Identities = 16/54 (29%), Positives = 26/54 (47%)
Query: 462 SFKVGQYVRFRPGLAEPRFGWGGAQPESQGIITNIHADGEVRVAFFGLSGLWKG 515
+ K+G VR + + P++ WG +S GI+T I +G+ F W G
Sbjct: 1532 TLKIGDKVRVKASVTVPKYKWGSVNHQSIGIVTGIINNGKDVAIDFPQQNNWTG 1585
>emb|CAG84838.1| unnamed protein product [Debaryomyces hansenii CBS767]
gi|50409334|ref|XP_456863.1| unnamed protein product
[Debaryomyces hansenii]
Length = 545
Score = 36.2 bits (82), Expect = 3.7
Identities = 33/133 (24%), Positives = 58/133 (42%), Gaps = 14/133 (10%)
Query: 373 WMVPSMVTGRQSLWKVSPGDAERLPGXEVGDWVRSKPSLGNRPSYDWNSVGRESLAV--- 429
W+V T Q + G +E L G EV + N+P+Y + V +L +
Sbjct: 69 WLVTQGNTSFQGILNNIGGISEFLEGTEVSEGAVIASPSKNKPNYFYQWVRDAALTIRSL 128
Query: 430 VHSVQDSGYLELACCFRKGKWITHYTDVEKVPSFKVGQY-VRFRPGLAEPRF-------- 480
VH ++D+ C K+I + ++++P+ +G++ R GL EP+F
Sbjct: 129 VHYLEDNNTNNQKICSIIEKYIENNYYLQRLPN-NLGRFDDNTRSGLGEPKFMPDCTAFD 187
Query: 481 -GWGGAQPESQGI 492
WG Q + G+
Sbjct: 188 EHWGRPQNDGPGL 200
>gb|AAK24605.1| peptidase, M16 family [Caulobacter crescentus CB15]
gi|16126873|ref|NP_421437.1| peptidase, M16 family
[Caulobacter crescentus CB15] gi|25290049|pir||A87576
peptidase, M16 family [imported] - Caulobacter
crescentus
Length = 976
Score = 35.8 bits (81), Expect = 4.8
Identities = 40/139 (28%), Positives = 52/139 (36%), Gaps = 11/139 (7%)
Query: 476 AEPRFGWGGAQPESQGIITNIHADGEVRVAFFGLS---GLWKGDPSDLQAEQIFEVGEWV 532
AEP F GGA Q G VRV FG + G W+ + L AEQ + V
Sbjct: 365 AEPPFIAGGAFKGDQF--------GAVRVTTFGATAQPGRWREALTALDAEQRRAIQYGV 416
Query: 533 RLKENVNNWKSIGPGSVGVVQGIGYEGGETDRSTFVGFCGEQEKWVGPSSHLERVDKLIV 592
R E S+ G V G + + + VG G+ E PS +L D +
Sbjct: 417 RQDELDREIASLRAGLVAAAAGEATQRTPSLANQLVGTLGDGEVVTSPSQNLAAFDLFVK 476
Query: 593 GQKVRVKQNVKQPRFGWSG 611
G V + F SG
Sbjct: 477 GLTAERVNAVLKSAFVGSG 495
>emb|CAE02008.2| OJ000223_09.10 [Oryza sativa (japonica cultivar-group)]
gi|38345347|emb|CAE03158.2| OSJNBa0081L15.20 [Oryza
sativa (japonica cultivar-group)]
gi|50925293|ref|XP_472944.1| OSJNBa0081L15.20 [Oryza
sativa (japonica cultivar-group)]
Length = 384
Score = 35.4 bits (80), Expect = 6.3
Identities = 37/136 (27%), Positives = 51/136 (37%), Gaps = 40/136 (29%)
Query: 93 EPRFGWRGQSRDSIGTVLCVDPEDGILRV------GFPGASRGWKADP----AEMERVEE 142
E FGW G S D VL DP+ GIL V G P + W+A P +++ + E
Sbjct: 228 EANFGWLGASPDG---VLGCDPDGGILEVKCPYNKGKPELALPWRAMPYYYMPQVQGLME 284
Query: 143 FKVGDWVRVRPTLTTSKHGLGNVVPGTIGIVYCIRPDSSLLVELSYVQNPWHCEPEEIE- 201
DWV +YC P+ S L + + W E +
Sbjct: 285 IMGRDWVE----------------------LYCWTPNGSSLFRVPRDRGYWELIHEVLRD 322
Query: 202 ----HVPPFRNLDMLG 213
+V P R L +LG
Sbjct: 323 FWWGNVMPARELVLLG 338
>emb|CAC09491.1| contains similarity to F12A21.16 [Oryza sativa (indica
cultivar-group)]
Length = 428
Score = 35.4 bits (80), Expect = 6.3
Identities = 37/136 (27%), Positives = 51/136 (37%), Gaps = 40/136 (29%)
Query: 93 EPRFGWRGQSRDSIGTVLCVDPEDGILRV------GFPGASRGWKADP----AEMERVEE 142
E FGW G S D VL DP+ GIL V G P + W+A P +++ + E
Sbjct: 272 EANFGWLGASPDG---VLGCDPDGGILEVKCPYNKGKPELALPWRAMPYYYMPQVQGLME 328
Query: 143 FKVGDWVRVRPTLTTSKHGLGNVVPGTIGIVYCIRPDSSLLVELSYVQNPWHCEPEEIE- 201
DWV +YC P+ S L + + W E +
Sbjct: 329 IMGRDWVE----------------------LYCWTPNGSSLFRVPRDRGYWELIHEVLRD 366
Query: 202 ----HVPPFRNLDMLG 213
+V P R L +LG
Sbjct: 367 FWWGNVMPARELVLLG 382
Database: nr
Posted date: Jul 5, 2005 12:34 AM
Number of letters in database: 863,360,394
Number of sequences in database: 2,540,612
Lambda K H
0.328 0.144 0.480
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 1,291,184,750
Number of Sequences: 2540612
Number of extensions: 57050364
Number of successful extensions: 131214
Number of sequences better than 10.0: 22
Number of HSP's better than 10.0 without gapping: 15
Number of HSP's successfully gapped in prelim test: 7
Number of HSP's that attempted gapping in prelim test: 130995
Number of HSP's gapped (non-prelim): 132
length of query: 689
length of database: 863,360,394
effective HSP length: 135
effective length of query: 554
effective length of database: 520,377,774
effective search space: 288289286796
effective search space used: 288289286796
T: 11
A: 40
X1: 15 ( 7.1 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 40 (21.7 bits)
S2: 79 (35.0 bits)
Medicago: description of AC141414.5


