

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC141414.11 - phase: 0
(857 letters)
Database: nr
2,540,612 sequences; 863,360,394 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
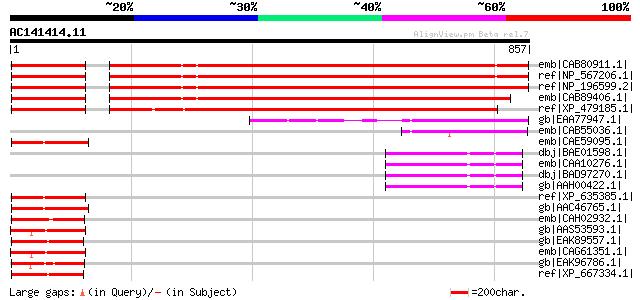
emb|CAB80911.1| putative protein [Arabidopsis thaliana] gi|52621... 684 0.0
ref|NP_567206.1| helicase domain-containing protein / IBR domain... 684 0.0
ref|NP_196599.2| helicase domain-containing protein / IBR domain... 675 0.0
emb|CAB89406.1| putative protein [Arabidopsis thaliana] gi|11357... 629 e-178
ref|XP_479185.1| putative DEAD/H (Asp-Glu-Ala-Asp/His) box polyp... 566 e-160
gb|EAA77947.1| hypothetical protein FG07753.1 [Gibberella zeae P... 177 1e-42
emb|CAB55036.1| Hypothetical protein Y57A10A.31 [Caenorhabditis ... 137 2e-30
emb|CAE59095.1| Hypothetical protein CBG02387 [Caenorhabditis br... 107 2e-21
dbj|BAE01598.1| unnamed protein product [Macaca fascicularis] 107 2e-21
emb|CAA10276.1| Ariadne-2 protein (ARI2) [Homo sapiens] 107 2e-21
dbj|BAD97270.1| ariadne homolog 2 variant [Homo sapiens] 106 3e-21
gb|AAH00422.1| Ariadne homolog 2 [Homo sapiens] gi|3930776|gb|AA... 106 3e-21
ref|XP_635385.1| hypothetical protein DDB0189299 [Dictyostelium ... 106 3e-21
gb|AAC46765.1| Masculinisation of germline protein 5 [Caenorhabd... 106 4e-21
emb|CAH02932.1| unnamed protein product [Kluyveromyces lactis NR... 105 5e-21
gb|AAS53593.1| AFR222Wp [Ashbya gossypii ATCC 10895] gi|45198740... 105 5e-21
gb|EAK89557.1| PRP43 involved in spliceosome disassembly mRNA sp... 105 6e-21
emb|CAG61351.1| unnamed protein product [Candida glabrata CBS138... 105 8e-21
gb|EAK96786.1| hypothetical protein CaO19.10336 [Candida albican... 105 8e-21
ref|XP_667334.1| RNA helicase [Cryptosporidium hominis] gi|54658... 105 8e-21
>emb|CAB80911.1| putative protein [Arabidopsis thaliana] gi|5262156|emb|CAB45785.1|
putative protein [Arabidopsis thaliana]
gi|7486358|pir||T10542 hypothetical protein F3I3.40 -
Arabidopsis thaliana
Length = 2322
Score = 684 bits (1766), Expect = 0.0
Identities = 344/695 (49%), Positives = 466/695 (66%), Gaps = 6/695 (0%)
Query: 165 IDEMELLMFFEKNTSGCICDMQKFTGMVKDVEDKAKWGKITFMTSNAAKRAAELDGEEFC 224
+D+ ELL F EK GCIC + KF +D ++K KWG+ITF+T +A +A E+ +F
Sbjct: 1077 VDDRELLTFLEKKIDGCICSIYKFAANKQDCDEKEKWGRITFLTPESAMKATEIQKFDFK 1136
Query: 225 GSPLKIVHSQSAMGGDTTFS-FPAVEARISWLRRPIKAVGIIKCDKNDVDFIIRDFENLI 283
GS LK+ S S GG F +V A+I W R+ G +KC D+ I+ D +L
Sbjct: 1137 GSVLKVFPSLSTGGGIFKMPYFSSVTAKIRWPRKESSGRGCLKCPSGDIHSILGDITSLE 1196
Query: 284 VDGRRYVRCAPSDKYLDNILITGLDKEVPETKILDILRSATSRRILDFF-FKRGDAVENP 342
+ G YV D+ILI+GL ++ E ++LD+L T RR L+FF F++ +V+ P
Sbjct: 1197 I-GTNYVHIQRDQLSNDSILISGLG-DLSEAEVLDVLEFRTQRRDLNFFIFRKKYSVQCP 1254
Query: 343 PCSMIAETILKEISPLMPKKKPHISSCRVQVFPPKPKDYSMNALIHFDGRLHLEAAKALE 402
+ E + K I M K P + +VQVF PK +Y M ALI FDGRLHLEAAKAL+
Sbjct: 1255 SPTACEEELHKRIFARMSAKNPEPNCVQVQVFEPKEDNYFMRALIKFDGRLHLEAAKALQ 1314
Query: 403 KIDGKVLPGFHSWQKIKTQRLFHSTLIFSPPVYHVIKGQLEKVLARFNNLEGLEWKLDIT 462
+++G+VLPG WQKIK ++LF S++I S +Y+ +K QL +LARF +G E L+ T
Sbjct: 1315 ELNGEVLPGCLPWQKIKCEQLFQSSIICSASIYNTVKRQLNVLLARFERQKGGECCLEPT 1374
Query: 463 PNGSHRVKITANATKTVAEGRRLLEELWRGKVIVHDNLTPATLQPILSKDGSSLTSSIQK 522
NG++RVKITA AT+ VAE RR LEEL RGK I H TP +Q ++S+DG +L IQ+
Sbjct: 1375 HNGAYRVKITAYATRPVAEMRRELEELLRGKPINHPGFTPRVVQHLMSRDGINLMRKIQQ 1434
Query: 523 ATSTYIQFDRRNMKLRIFGSPDKIALAEKKLIQSLLSLHDEKQSVICLSGRDLPSDFMKQ 582
T TYI DR N+ +RI G+ +KIA AE++L+QSL+ H+ KQ I L G ++ D MK+
Sbjct: 1435 ETETYILLDRHNLTVRICGTSEKIAKAEQELVQSLMDYHESKQLEIHLRGPEIRPDLMKE 1494
Query: 583 VVKNFGPDLHGLKEKVPGADLRLNTRNRTILCHGNSELKSRVEEITFEIARLSNPSSERF 642
VVK FGP+L G+KEKV G DL+LNTR I HG+ E++ V+++ E+AR + E+
Sbjct: 1495 VVKRFGPELQGIKEKVHGVDLKLNTRYHVIQVHGSKEMRQEVQKMVNELAREKSALGEKP 1554
Query: 643 D-TGPSCPICLCEVEDGYQLEGCGHLFCQSCMVEQCESAIKNQGSFPIRCAHQGCGNHIL 701
D CPICL EV+DGY LEGC HLFC++C++EQ E++++N +FPI C+H CG I+
Sbjct: 1555 DEIELECPICLSEVDDGYSLEGCSHLFCKACLLEQFEASMRNFDAFPILCSHIDCGAPIV 1614
Query: 702 LVDFRTLLSNDKLEELFRASLGAFVASSSGTYRFCPSPDCPSIYRVADPDTASAPFVCGA 761
+ D R LLS +KL+EL ASL AFV SS G RFC +PDCPSIYRVA P + PF+CGA
Sbjct: 1615 VADMRALLSQEKLDELISASLSAFVTSSDGKLRFCSTPDCPSIYRVAGPQESGEPFICGA 1674
Query: 762 CYSETCTRCHIEYHPYVSCERYRQFKDDPDSSLRDWCKGKEQVKNCPACGHVIEKVDGCN 821
C+SETCTRCH+EYHP ++CERY++FK++PD SL+DW KGK+ VK CP C IEK DGCN
Sbjct: 1675 CHSETCTRCHLEYHPLITCERYKKFKENPDLSLKDWAKGKD-VKECPICKSTIEKTDGCN 1733
Query: 822 HIECKCGKHICWVCLEFFTTSGECYSHMDTIHLGV 856
H++C+CGKHICW CL+ FT + CY+H+ TIH G+
Sbjct: 1734 HLQCRCGKHICWTCLDVFTQAEPCYAHLRTIHGGI 1768
Score = 184 bits (467), Expect = 1e-44
Identities = 85/121 (70%), Positives = 107/121 (88%)
Query: 4 QITVLIGETGSGKSTQLVQFLADSGVGANESIVCTQPRRIAAKSLAERVREESGGCYEDS 63
QI VLIGETGSGKSTQLVQFLADSGV A+ESIVCTQPR+IAA +L +RVREES GCYE++
Sbjct: 320 QIMVLIGETGSGKSTQLVQFLADSGVAASESIVCTQPRKIAAMTLTDRVREESSGCYEEN 379
Query: 64 SIKCYSSFSSWNKFDSRIIFMTDHCLLQHYMSDKNFTGISCIIVDEAHERSINTDLLLAL 123
++ C +FSS + S++++MTD+CLLQHYM D++ +GISC+I+DEAHERS+NTDLLLAL
Sbjct: 380 TVSCTPTFSSTEEISSKVVYMTDNCLLQHYMKDRSLSGISCVIIDEAHERSLNTDLLLAL 439
Query: 124 V 124
+
Sbjct: 440 L 440
>ref|NP_567206.1| helicase domain-containing protein / IBR domain-containing protein /
zinc finger protein-related [Arabidopsis thaliana]
Length = 1787
Score = 684 bits (1766), Expect = 0.0
Identities = 344/695 (49%), Positives = 466/695 (66%), Gaps = 6/695 (0%)
Query: 165 IDEMELLMFFEKNTSGCICDMQKFTGMVKDVEDKAKWGKITFMTSNAAKRAAELDGEEFC 224
+D+ ELL F EK GCIC + KF +D ++K KWG+ITF+T +A +A E+ +F
Sbjct: 1077 VDDRELLTFLEKKIDGCICSIYKFAANKQDCDEKEKWGRITFLTPESAMKATEIQKFDFK 1136
Query: 225 GSPLKIVHSQSAMGGDTTFS-FPAVEARISWLRRPIKAVGIIKCDKNDVDFIIRDFENLI 283
GS LK+ S S GG F +V A+I W R+ G +KC D+ I+ D +L
Sbjct: 1137 GSVLKVFPSLSTGGGIFKMPYFSSVTAKIRWPRKESSGRGCLKCPSGDIHSILGDITSLE 1196
Query: 284 VDGRRYVRCAPSDKYLDNILITGLDKEVPETKILDILRSATSRRILDFF-FKRGDAVENP 342
+ G YV D+ILI+GL ++ E ++LD+L T RR L+FF F++ +V+ P
Sbjct: 1197 I-GTNYVHIQRDQLSNDSILISGLG-DLSEAEVLDVLEFRTQRRDLNFFIFRKKYSVQCP 1254
Query: 343 PCSMIAETILKEISPLMPKKKPHISSCRVQVFPPKPKDYSMNALIHFDGRLHLEAAKALE 402
+ E + K I M K P + +VQVF PK +Y M ALI FDGRLHLEAAKAL+
Sbjct: 1255 SPTACEEELHKRIFARMSAKNPEPNCVQVQVFEPKEDNYFMRALIKFDGRLHLEAAKALQ 1314
Query: 403 KIDGKVLPGFHSWQKIKTQRLFHSTLIFSPPVYHVIKGQLEKVLARFNNLEGLEWKLDIT 462
+++G+VLPG WQKIK ++LF S++I S +Y+ +K QL +LARF +G E L+ T
Sbjct: 1315 ELNGEVLPGCLPWQKIKCEQLFQSSIICSASIYNTVKRQLNVLLARFERQKGGECCLEPT 1374
Query: 463 PNGSHRVKITANATKTVAEGRRLLEELWRGKVIVHDNLTPATLQPILSKDGSSLTSSIQK 522
NG++RVKITA AT+ VAE RR LEEL RGK I H TP +Q ++S+DG +L IQ+
Sbjct: 1375 HNGAYRVKITAYATRPVAEMRRELEELLRGKPINHPGFTPRVVQHLMSRDGINLMRKIQQ 1434
Query: 523 ATSTYIQFDRRNMKLRIFGSPDKIALAEKKLIQSLLSLHDEKQSVICLSGRDLPSDFMKQ 582
T TYI DR N+ +RI G+ +KIA AE++L+QSL+ H+ KQ I L G ++ D MK+
Sbjct: 1435 ETETYILLDRHNLTVRICGTSEKIAKAEQELVQSLMDYHESKQLEIHLRGPEIRPDLMKE 1494
Query: 583 VVKNFGPDLHGLKEKVPGADLRLNTRNRTILCHGNSELKSRVEEITFEIARLSNPSSERF 642
VVK FGP+L G+KEKV G DL+LNTR I HG+ E++ V+++ E+AR + E+
Sbjct: 1495 VVKRFGPELQGIKEKVHGVDLKLNTRYHVIQVHGSKEMRQEVQKMVNELAREKSALGEKP 1554
Query: 643 D-TGPSCPICLCEVEDGYQLEGCGHLFCQSCMVEQCESAIKNQGSFPIRCAHQGCGNHIL 701
D CPICL EV+DGY LEGC HLFC++C++EQ E++++N +FPI C+H CG I+
Sbjct: 1555 DEIELECPICLSEVDDGYSLEGCSHLFCKACLLEQFEASMRNFDAFPILCSHIDCGAPIV 1614
Query: 702 LVDFRTLLSNDKLEELFRASLGAFVASSSGTYRFCPSPDCPSIYRVADPDTASAPFVCGA 761
+ D R LLS +KL+EL ASL AFV SS G RFC +PDCPSIYRVA P + PF+CGA
Sbjct: 1615 VADMRALLSQEKLDELISASLSAFVTSSDGKLRFCSTPDCPSIYRVAGPQESGEPFICGA 1674
Query: 762 CYSETCTRCHIEYHPYVSCERYRQFKDDPDSSLRDWCKGKEQVKNCPACGHVIEKVDGCN 821
C+SETCTRCH+EYHP ++CERY++FK++PD SL+DW KGK+ VK CP C IEK DGCN
Sbjct: 1675 CHSETCTRCHLEYHPLITCERYKKFKENPDLSLKDWAKGKD-VKECPICKSTIEKTDGCN 1733
Query: 822 HIECKCGKHICWVCLEFFTTSGECYSHMDTIHLGV 856
H++C+CGKHICW CL+ FT + CY+H+ TIH G+
Sbjct: 1734 HLQCRCGKHICWTCLDVFTQAEPCYAHLRTIHGGI 1768
Score = 184 bits (467), Expect = 1e-44
Identities = 85/121 (70%), Positives = 107/121 (88%)
Query: 4 QITVLIGETGSGKSTQLVQFLADSGVGANESIVCTQPRRIAAKSLAERVREESGGCYEDS 63
QI VLIGETGSGKSTQLVQFLADSGV A+ESIVCTQPR+IAA +L +RVREES GCYE++
Sbjct: 320 QIMVLIGETGSGKSTQLVQFLADSGVAASESIVCTQPRKIAAMTLTDRVREESSGCYEEN 379
Query: 64 SIKCYSSFSSWNKFDSRIIFMTDHCLLQHYMSDKNFTGISCIIVDEAHERSINTDLLLAL 123
++ C +FSS + S++++MTD+CLLQHYM D++ +GISC+I+DEAHERS+NTDLLLAL
Sbjct: 380 TVSCTPTFSSTEEISSKVVYMTDNCLLQHYMKDRSLSGISCVIIDEAHERSLNTDLLLAL 439
Query: 124 V 124
+
Sbjct: 440 L 440
>ref|NP_196599.2| helicase domain-containing protein / IBR domain-containing protein /
zinc finger protein-related [Arabidopsis thaliana]
Length = 1775
Score = 675 bits (1741), Expect = 0.0
Identities = 341/695 (49%), Positives = 462/695 (66%), Gaps = 6/695 (0%)
Query: 165 IDEMELLMFFEKNTSGCICDMQKFTGMVKDVEDKAKWGKITFMTSNAAKRAAELDGEEFC 224
+D+ ELL F EK G IC + KF +D ++K KWG+ITF+T +A +A E+ F
Sbjct: 1080 VDDRELLTFLEKKIDGSICSIYKFAANKQDCDEKEKWGRITFLTPESAMKATEIQKFYFK 1139
Query: 225 GSPLKIVHSQSAMGGDTTFS-FPAVEARISWLRRPIKAVGIIKCDKNDVDFIIRDFENLI 283
GS LK+ S S GG F +V A+I W RR G +KC D+ I+ D +L
Sbjct: 1140 GSVLKLFPSLSTGGGIFKMPYFSSVTAKIRWPRRESSGRGCLKCPSGDIHRILGDISSLE 1199
Query: 284 VDGRRYVRCAPSDKYLDNILITGLDKEVPETKILDILRSATSRRILDFF-FKRGDAVENP 342
+ G YV + D+ILI+GL ++ E ++LD+L T RR L+FF F++ +V+ P
Sbjct: 1200 I-GTNYVHIQRDQQSNDSILISGLG-DLSEAEVLDVLEFRTQRRDLNFFIFRKKYSVQCP 1257
Query: 343 PCSMIAETILKEISPLMPKKKPHISSCRVQVFPPKPKDYSMNALIHFDGRLHLEAAKALE 402
+ E + K I M K P + +VQVF PK +Y M ALI FDGRLH EAAKAL+
Sbjct: 1258 SPTACEEELHKRIFARMSAKNPEPNCVQVQVFEPKEDNYFMRALIKFDGRLHFEAAKALQ 1317
Query: 403 KIDGKVLPGFHSWQKIKTQRLFHSTLIFSPPVYHVIKGQLEKVLARFNNLEGLEWKLDIT 462
+++G+VLPG WQKIK ++LF S++I S +Y+ +K QL +LARF +G E L+ T
Sbjct: 1318 ELNGEVLPGCLPWQKIKCEQLFQSSIICSASIYNTVKRQLNVLLARFERQKGGECCLEPT 1377
Query: 463 PNGSHRVKITANATKTVAEGRRLLEELWRGKVIVHDNLTPATLQPILSKDGSSLTSSIQK 522
NG++RVKITA AT+ VAE RR LEEL RG+ I H T LQ ++S+DG +L IQ+
Sbjct: 1378 HNGAYRVKITAYATRPVAEMRRELEELLRGRPINHPGFTRRVLQHLMSRDGINLMRKIQQ 1437
Query: 523 ATSTYIQFDRRNMKLRIFGSPDKIALAEKKLIQSLLSLHDEKQSVICLSGRDLPSDFMKQ 582
T TYI DR N+ +RI G+ +KIA AE++LIQ+L+ H+ KQ I L G ++ D MK+
Sbjct: 1438 ETETYILLDRHNLTVRICGTSEKIAKAEQELIQALMDYHESKQLEIHLRGPEIRPDLMKE 1497
Query: 583 VVKNFGPDLHGLKEKVPGADLRLNTRNRTILCHGNSELKSRVEEITFEIARLSNPSSERF 642
VVK FGP+L G+KEKV G DL+LNTR I HG+ E++ V+++ E+AR + E+
Sbjct: 1498 VVKRFGPELQGIKEKVHGVDLKLNTRYHVIQVHGSKEMRQEVQKMVNELAREKSALGEKP 1557
Query: 643 D-TGPSCPICLCEVEDGYQLEGCGHLFCQSCMVEQCESAIKNQGSFPIRCAHQGCGNHIL 701
D CPICL EV+DGY LEGC HLFC++C++EQ E++++N +FPI C+H CG I+
Sbjct: 1558 DEIEVECPICLSEVDDGYSLEGCSHLFCKACLLEQFEASMRNFDAFPILCSHIDCGAPIV 1617
Query: 702 LVDFRTLLSNDKLEELFRASLGAFVASSSGTYRFCPSPDCPSIYRVADPDTASAPFVCGA 761
L D R LLS +KL+ELF ASL +FV SS G +RFC +PDCPS+YRVA P + PF+CGA
Sbjct: 1618 LADMRALLSQEKLDELFSASLSSFVTSSDGKFRFCSTPDCPSVYRVAGPQESGEPFICGA 1677
Query: 762 CYSETCTRCHIEYHPYVSCERYRQFKDDPDSSLRDWCKGKEQVKNCPACGHVIEKVDGCN 821
C+SE CTRCH+EYHP ++CERY++FK++PD SL+DW KGK VK CP C IEK DGCN
Sbjct: 1678 CHSEICTRCHLEYHPLITCERYKKFKENPDLSLKDWAKGK-NVKECPICKSTIEKTDGCN 1736
Query: 822 HIECKCGKHICWVCLEFFTTSGECYSHMDTIHLGV 856
H++C+CGKHICW CL+ FT CY+H+ TIH G+
Sbjct: 1737 HMKCRCGKHICWTCLDVFTQEEPCYAHLRTIHGGI 1771
Score = 186 bits (471), Expect = 4e-45
Identities = 86/121 (71%), Positives = 108/121 (89%)
Query: 4 QITVLIGETGSGKSTQLVQFLADSGVGANESIVCTQPRRIAAKSLAERVREESGGCYEDS 63
QI VLIGETGSGKSTQLVQFLADSGV A+ESIVCTQPR+IAA +LA+RVREES GCYE++
Sbjct: 323 QIMVLIGETGSGKSTQLVQFLADSGVAASESIVCTQPRKIAAMTLADRVREESSGCYEEN 382
Query: 64 SIKCYSSFSSWNKFDSRIIFMTDHCLLQHYMSDKNFTGISCIIVDEAHERSINTDLLLAL 123
++ C +FSS + S++++MTD+CLLQHYM D++ +GISC+I+DEAHERS+NTDLLLAL
Sbjct: 383 TVSCTPTFSSTEEISSKVVYMTDNCLLQHYMKDRSLSGISCVIIDEAHERSLNTDLLLAL 442
Query: 124 V 124
+
Sbjct: 443 L 443
>emb|CAB89406.1| putative protein [Arabidopsis thaliana] gi|11357363|pir||T50002
hypothetical protein F12B17.280 - Arabidopsis thaliana
Length = 1751
Score = 629 bits (1621), Expect = e-178
Identities = 323/665 (48%), Positives = 440/665 (65%), Gaps = 6/665 (0%)
Query: 165 IDEMELLMFFEKNTSGCICDMQKFTGMVKDVEDKAKWGKITFMTSNAAKRAAELDGEEFC 224
+D+ ELL F EK G IC + KF +D ++K KWG+ITF+T +A +A E+ F
Sbjct: 1080 VDDRELLTFLEKKIDGSICSIYKFAANKQDCDEKEKWGRITFLTPESAMKATEIQKFYFK 1139
Query: 225 GSPLKIVHSQSAMGGDTTFS-FPAVEARISWLRRPIKAVGIIKCDKNDVDFIIRDFENLI 283
GS LK+ S S GG F +V A+I W RR G +KC D+ I+ D +L
Sbjct: 1140 GSVLKLFPSLSTGGGIFKMPYFSSVTAKIRWPRRESSGRGCLKCPSGDIHRILGDISSLE 1199
Query: 284 VDGRRYVRCAPSDKYLDNILITGLDKEVPETKILDILRSATSRRILDFF-FKRGDAVENP 342
+ G YV + D+ILI+GL ++ E ++LD+L T RR L+FF F++ +V+ P
Sbjct: 1200 I-GTNYVHIQRDQQSNDSILISGLG-DLSEAEVLDVLEFRTQRRDLNFFIFRKKYSVQCP 1257
Query: 343 PCSMIAETILKEISPLMPKKKPHISSCRVQVFPPKPKDYSMNALIHFDGRLHLEAAKALE 402
+ E + K I M K P + +VQVF PK +Y M ALI FDGRLH EAAKAL+
Sbjct: 1258 SPTACEEELHKRIFARMSAKNPEPNCVQVQVFEPKEDNYFMRALIKFDGRLHFEAAKALQ 1317
Query: 403 KIDGKVLPGFHSWQKIKTQRLFHSTLIFSPPVYHVIKGQLEKVLARFNNLEGLEWKLDIT 462
+++G+VLPG WQKIK ++LF S++I S +Y+ +K QL +LARF +G E L+ T
Sbjct: 1318 ELNGEVLPGCLPWQKIKCEQLFQSSIICSASIYNTVKRQLNVLLARFERQKGGECCLEPT 1377
Query: 463 PNGSHRVKITANATKTVAEGRRLLEELWRGKVIVHDNLTPATLQPILSKDGSSLTSSIQK 522
NG++RVKITA AT+ VAE RR LEEL RG+ I H T LQ ++S+DG +L IQ+
Sbjct: 1378 HNGAYRVKITAYATRPVAEMRRELEELLRGRPINHPGFTRRVLQHLMSRDGINLMRKIQQ 1437
Query: 523 ATSTYIQFDRRNMKLRIFGSPDKIALAEKKLIQSLLSLHDEKQSVICLSGRDLPSDFMKQ 582
T TYI DR N+ +RI G+ +KIA AE++LIQ+L+ H+ KQ I L G ++ D MK+
Sbjct: 1438 ETETYILLDRHNLTVRICGTSEKIAKAEQELIQALMDYHESKQLEIHLRGPEIRPDLMKE 1497
Query: 583 VVKNFGPDLHGLKEKVPGADLRLNTRNRTILCHGNSELKSRVEEITFEIARLSNPSSERF 642
VVK FGP+L G+KEKV G DL+LNTR I HG+ E++ V+++ E+AR + E+
Sbjct: 1498 VVKRFGPELQGIKEKVHGVDLKLNTRYHVIQVHGSKEMRQEVQKMVNELAREKSALGEKP 1557
Query: 643 D-TGPSCPICLCEVEDGYQLEGCGHLFCQSCMVEQCESAIKNQGSFPIRCAHQGCGNHIL 701
D CPICL EV+DGY LEGC HLFC++C++EQ E++++N +FPI C+H CG I+
Sbjct: 1558 DEIEVECPICLSEVDDGYSLEGCSHLFCKACLLEQFEASMRNFDAFPILCSHIDCGAPIV 1617
Query: 702 LVDFRTLLSNDKLEELFRASLGAFVASSSGTYRFCPSPDCPSIYRVADPDTASAPFVCGA 761
L D R LLS +KL+ELF ASL +FV SS G +RFC +PDCPS+YRVA P + PF+CGA
Sbjct: 1618 LADMRALLSQEKLDELFSASLSSFVTSSDGKFRFCSTPDCPSVYRVAGPQESGEPFICGA 1677
Query: 762 CYSETCTRCHIEYHPYVSCERYRQFKDDPDSSLRDWCKGKEQVKNCPACGHVIEKVDGCN 821
C+SE CTRCH+EYHP ++CERY++FK++PD SL+DW KGK VK CP C IEK DGCN
Sbjct: 1678 CHSEICTRCHLEYHPLITCERYKKFKENPDLSLKDWAKGK-NVKECPICKSTIEKTDGCN 1736
Query: 822 HIECK 826
H++C+
Sbjct: 1737 HMKCR 1741
Score = 186 bits (471), Expect = 4e-45
Identities = 86/121 (71%), Positives = 108/121 (89%)
Query: 4 QITVLIGETGSGKSTQLVQFLADSGVGANESIVCTQPRRIAAKSLAERVREESGGCYEDS 63
QI VLIGETGSGKSTQLVQFLADSGV A+ESIVCTQPR+IAA +LA+RVREES GCYE++
Sbjct: 323 QIMVLIGETGSGKSTQLVQFLADSGVAASESIVCTQPRKIAAMTLADRVREESSGCYEEN 382
Query: 64 SIKCYSSFSSWNKFDSRIIFMTDHCLLQHYMSDKNFTGISCIIVDEAHERSINTDLLLAL 123
++ C +FSS + S++++MTD+CLLQHYM D++ +GISC+I+DEAHERS+NTDLLLAL
Sbjct: 383 TVSCTPTFSSTEEISSKVVYMTDNCLLQHYMKDRSLSGISCVIIDEAHERSLNTDLLLAL 442
Query: 124 V 124
+
Sbjct: 443 L 443
>ref|XP_479185.1| putative DEAD/H (Asp-Glu-Ala-Asp/His) box polypeptide 8 (RNA
helicase) [Oryza sativa (japonica cultivar-group)]
gi|33146632|dbj|BAC79920.1| putative DEAD/H
(Asp-Glu-Ala-Asp/His) box polypeptide 8 (RNA helicase)
[Oryza sativa (japonica cultivar-group)]
Length = 1686
Score = 567 bits (1460), Expect = e-160
Identities = 287/643 (44%), Positives = 419/643 (64%), Gaps = 6/643 (0%)
Query: 165 IDEMELLMFFEKNTSGCICDMQKFTGMVKDVEDKAKWGKITFMTSNAAKRA-AELDGEEF 223
+++ EL+ + SG + + K G ++ D+ KWGK TF+ A+ A ++L+G EF
Sbjct: 1043 LNDKELICLVDTLISG-VANFYKLYGNLQVASDETKWGKFTFLNPEYAEDAVSKLNGMEF 1101
Query: 224 CGSPLKIVHSQSAMGGDTTFSFPAVEARISWLRRPIKAVGIIKCDKNDVDFIIRDFENLI 283
GSPLK+V S+ + FPAV A++SW + + + ++ C + +F+++D L
Sbjct: 1102 HGSPLKVVPVCSS--SNRGLPFPAVRAKVSWPLKQSRGLALVTCASGEAEFVVKDCFALG 1159
Query: 284 VDGRRYVRCAPSDKYLDNILITGLDKEVPETKILDILRSATSRRILDFFFKRGDAVENPP 343
V GR Y+ C S ++ + I + G+ V E ++ D RS T+R+I+D RG + P
Sbjct: 1160 VGGR-YINCEVSTRHENCIFVRGIPMHVTEPELYDAFRSTTTRKIVDVHLLRGTPIAAPS 1218
Query: 344 CSMIAETILKEISPLMPKKKPHISSCRVQVFPPKPKDYSMNALIHFDGRLHLEAAKALEK 403
S+ AE + +EIS MPKK + RV+V P+ D M A I FDG LH EAA+ALE
Sbjct: 1219 ASLCAEALNREISSFMPKKIFPAQNFRVEVLTPEENDSVMRATITFDGSLHREAARALEH 1278
Query: 404 IDGKVLPGFHSWQKIKTQRLFHSTLIFSPPVYHVIKGQLEKVLARFNNLEGLEWKLDITP 463
+ G VLP WQ I+ Q +FHST+ VY+VI + +L F + +G+ + L+
Sbjct: 1279 LQGSVLPCCLPWQTIQCQHVFHSTVSCQVRVYNVISQAVASLLESFRSQKGVSYNLEKNE 1338
Query: 464 NGSHRVKITANATKTVAEGRRLLEELWRGKVIVHDNLTPATLQPILSKDGSSLTSSIQKA 523
G RVK+TANATKT+A+ RR LE L +GK I H +LT +T+Q ++S+DG + S+++
Sbjct: 1339 YGIFRVKLTANATKTIADLRRPLEILMKGKTINHPDLTLSTVQLLMSRDGVADLKSVEQE 1398
Query: 524 TSTYIQFDRRNMKLRIFGSPDKIALAEKKLIQSLLSLHDEKQSVICLSGRDLPSDFMKQV 583
T TYI +DR+++ +++FG D++A AE+KLI +LL L D+K I L GR+LP + MK++
Sbjct: 1399 TGTYILYDRQSLNIKVFGLQDQVAAAEEKLIHALLQLRDKKPLDIRLRGRNLPPNLMKEM 1458
Query: 584 VKNFGPDLHGLKEKVPGADLRLNTRNRTILCHGNSELKSRVEEITFEIARLSNPSS-ERF 642
+K FG DL GLK +VP +LRLN R T+ G+ E K RVEE+ E+ + + +
Sbjct: 1459 LKKFGADLEGLKREVPAVELRLNLRQHTLYVRGSKEDKQRVEEMISELVNSTKYNGLLQL 1518
Query: 643 DTGPSCPICLCEVEDGYQLEGCGHLFCQSCMVEQCESAIKNQGSFPIRCAHQGCGNHILL 702
+CPICLCEVED ++LE CGH+FC +C+V+QCESA+K+ FP+ C GC +L+
Sbjct: 1519 PLENACPICLCEVEDPFKLESCGHVFCLTCLVDQCESALKSHDGFPLCCLKNGCKKQLLV 1578
Query: 703 VDFRTLLSNDKLEELFRASLGAFVASSSGTYRFCPSPDCPSIYRVADPDTASAPFVCGAC 762
VD R+LLS++KLEELFRASL AFVAS++G YRFCP+PDCPSIY+VA D S PFVCGAC
Sbjct: 1579 VDLRSLLSSEKLEELFRASLRAFVASNAGKYRFCPTPDCPSIYQVAAADAESKPFVCGAC 1638
Query: 763 YSETCTRCHIEYHPYVSCERYRQFKDDPDSSLRDWCKGKEQVK 805
+ E C +CH+EYHP++SCE Y+++K+DPD++L +W KGKE VK
Sbjct: 1639 FVEICNKCHLEYHPFISCEAYKEYKEDPDATLLEWRKGKENVK 1681
Score = 155 bits (393), Expect = 4e-36
Identities = 81/121 (66%), Positives = 95/121 (77%), Gaps = 1/121 (0%)
Query: 4 QITVLIGETGSGKSTQLVQFLADSGVGANESIVCTQPRRIAAKSLAERVREESGGCYEDS 63
Q+ +LIGETGSGKSTQLVQ+LADSG+ AN SIVCTQPR+IAA SLA RV EES GCY D+
Sbjct: 280 QVMILIGETGSGKSTQLVQYLADSGLAANGSIVCTQPRKIAAISLAHRVVEESNGCYGDN 339
Query: 64 SIKCYSSFSSWNKFDSRIIFMTDHCLLQHYMSDKNFTGISCIIVDEAHERSINTDLLLAL 123
+ S+F F S+II+ TD+CLL H M+D GIS IIVDEAHERS+NTDLLLAL
Sbjct: 340 FV-LNSTFLDHQDFSSKIIYTTDNCLLHHCMNDMGLDGISYIIVDEAHERSLNTDLLLAL 398
Query: 124 V 124
+
Sbjct: 399 I 399
>gb|EAA77947.1| hypothetical protein FG07753.1 [Gibberella zeae PH-1]
gi|46126751|ref|XP_387929.1| hypothetical protein
FG07753.1 [Gibberella zeae PH-1]
Length = 781
Score = 177 bits (450), Expect = 1e-42
Identities = 127/467 (27%), Positives = 203/467 (43%), Gaps = 81/467 (17%)
Query: 397 AAKALEKIDGKVLPGFHSWQKIKTQRLFHSTLIFSPPVYHVIKGQLEKVLARFNNLEGLE 456
A A+ ++ LP F+S K+ Q L S VY+ +K ++ ++++ + +
Sbjct: 388 AQAAVSSLNETELP-FNSTGKLFVQLLTSVKFKVSVRVYNAVKKSIDAHKSQWSR-QFVH 445
Query: 457 WKLDITPNGSHRV-KITANATKTVAEGRRLLEELWRGKVIVHD--NLTPATLQPILSKDG 513
+ + G +R+ KI + VA+ +R LE + G V+ D N+ + L+ +SK+
Sbjct: 446 YSA-LPERGFNRILKIEGEDRQLVAQAKRTLENIITGTVMTMDGKNIWYSNLK--ISKNA 502
Query: 514 SSLTSSIQKATSTYIQFDRRNMKLRIFGSPDKIALAEKKLIQSLLSLHDEKQSVICLSGR 573
+++ I D R R FG D++A A + L
Sbjct: 503 YKKLQKLEQDFEVVIIRDIRASNFRAFGPEDRLAEAAEAL-------------------- 542
Query: 574 DLPSDFMKQVVKNFGPDLHGLKEK-VPGADLRLNTRNRTILCHGNSELKSRVEEITFEIA 632
+Q+V + D H +KE VP L T
Sbjct: 543 -------QQLVDDLKTDGHAVKETAVPVKTKNLET------------------------- 570
Query: 633 RLSNPSSERFDTGPSCPICLCEVEDGYQLEGCGHLFCQSCMVEQCESAIKNQGSFPIRC- 691
C +C E E+ + CGH++C C V C+S + G F I+C
Sbjct: 571 --------------DCSVCFGEAEESLETS-CGHIYCNICFVNMCQSGESSSGDFSIKCV 615
Query: 692 -AHQGCGNHILLVDFRTLLSNDKLEELFRASLGAFVASSSGTYRFCPSPDCPSIYRVADP 750
A C + + + +TLL ++ LE + AS +F+ S +R+CP+PDC +YRV+ P
Sbjct: 616 GASDACKKILPISEIQTLLLSETLENILDASFASFIRSHPTEFRYCPTPDCDQVYRVSSP 675
Query: 751 DTASAPFVCGACYSETCTRCHIEYHPYVSCERYRQFKDDPDSSLRDWCKGKEQVKNCPAC 810
+ F C C++ TCT C+ HP +SC + + D L K VK+CP C
Sbjct: 676 EKVPFMFTCSRCFTSTCTACNAS-HPGISCSKNKGSNSDDIEKLMK-AKKDLGVKDCPKC 733
Query: 811 GHVIEKVDGCNHIEC-KCGKHICWVCLEFFTTSGECYSHMDTIHLGV 856
I+K +GCNH+ C C HICWVCL F +CY+HM +H G+
Sbjct: 734 TTAIQKSEGCNHMTCFACRTHICWVCLATFAQDTDCYAHMRRLHGGI 780
>emb|CAB55036.1| Hypothetical protein Y57A10A.31 [Caenorhabditis elegans]
gi|17537627|ref|NP_496609.1| zn-finger, cysteine-rich
C6HC (2M589) [Caenorhabditis elegans]
gi|7510231|pir||T31653 hypothetical protein Y57A10A.ff -
Caenorhabditis elegans
Length = 1048
Score = 137 bits (345), Expect = 2e-30
Identities = 76/220 (34%), Positives = 106/220 (47%), Gaps = 14/220 (6%)
Query: 648 CPICLCEV---EDGYQLEGCGHLFCQSCMVEQCESAIKNQGSFPIRCAHQGCGNHILLVD 704
CPIC E D Y+LE CGH+ C++C+ Q S I Q F IRC GC + +
Sbjct: 804 CPICTAETGISNDFYRLE-CGHVMCRTCINTQIRSRIDEQ-QFNIRCEIDGCHRVLTPAE 861
Query: 705 FRTLL--SNDKLEELFRASLGAF-------VASSSGTYRFCPSPDCPSIYRVADPDTASA 755
++ +D+++EL L + ++ CPS DC I +D S
Sbjct: 862 IMNIILGGSDRMKELDTQKLRRLTDRAKQLLIQTNEDLAACPSADCVGILSKSDDGLISE 921
Query: 756 PFVCGACYSETCTRCHIEYHPYVSCERYRQFKDDPDSSLRDWCKGKEQVKNCPACGHVIE 815
C AC C +C E HP +CE + + + DS R +VK CP C ++E
Sbjct: 922 FKKCEACDRSYCRKCLAEPHPDDTCEEFARIRRPEDSIARYQKDMGSRVKKCPKCAVLVE 981
Query: 816 KVDGCNHIECKCGKHICWVCLEFFTTSGECYSHMDTIHLG 855
K +GCNH++C CG H CW CL +SG+CY HM H G
Sbjct: 982 KREGCNHMQCGCGTHYCWTCLYVAESSGDCYRHMQAEHGG 1021
>emb|CAE59095.1| Hypothetical protein CBG02387 [Caenorhabditis briggsae]
Length = 1205
Score = 107 bits (267), Expect = 2e-21
Identities = 57/126 (45%), Positives = 83/126 (65%), Gaps = 1/126 (0%)
Query: 4 QITVLIGETGSGKSTQLVQFLADSGVGANESIVCTQPRRIAAKSLAERVREESGGCYEDS 63
QI V++GETGSGK+TQ+ Q+ ++G+G I CTQPRR+AA S+A+RV EE GC S
Sbjct: 562 QILVVVGETGSGKTTQMTQYAIEAGLGRRGKIGCTQPRRVAAMSVAKRVAEEY-GCKLGS 620
Query: 64 SIKCYSSFSSWNKFDSRIIFMTDHCLLQHYMSDKNFTGISCIIVDEAHERSINTDLLLAL 123
+ F D+ I +MTD LL+ + D + +G S I++DEAHER+I+TD+L L
Sbjct: 621 DVGYTIRFEDCTSQDTVIKYMTDGMLLRECLIDPDLSGYSLIMLDEAHERTIHTDVLFGL 680
Query: 124 VPSSNR 129
+ ++ R
Sbjct: 681 LKAAAR 686
>dbj|BAE01598.1| unnamed protein product [Macaca fascicularis]
Length = 421
Score = 107 bits (266), Expect = 2e-21
Identities = 71/232 (30%), Positives = 96/232 (40%), Gaps = 10/232 (4%)
Query: 621 KSRVEEITFEIARLSNPSSERFDTGPS--CPICLCEVEDGYQLE-GCGHLFCQSCMVEQC 677
KS ++ E NPS + P C +C+ V L C H FC+SC + C
Sbjct: 38 KSNSAQLLVEARVQPNPSKHVPTSHPPHHCAVCMQFVRKENLLSLACQHQFCRSCWEQHC 97
Query: 678 ESAIKNQGSFPIRCAHQGCGNHILLVDFRTLLSNDKLEELFRASLGAFVASSSGTYRFCP 737
IK+ + C Q C LL N++L E +R L S + CP
Sbjct: 98 SVLIKDGVGVGVSCMAQDCPLRTPEDFVFPLLPNEELREKYRRYLFRDYVESHYQLQLCP 157
Query: 738 SPDCPSIYRVADPDTASAPFVCGACYSETCTRCHIEYHPYVSCERYRQF--KDDPDSSLR 795
DCP + RV +P C C C +C YH C R++ K DS
Sbjct: 158 GADCPMVIRVQEPRARRVQ--CNRCNEVFCFKCRQMYHAPTDCATIRKWLTKCADDSETA 215
Query: 796 DWCKGKEQVKNCPACGHVIEKVDGCNHIEC-KCGKHICWVCLEFFTTSGECY 846
++ K+CP C IEK GCNH++C KC CW+CL + T G Y
Sbjct: 216 NYISA--HTKDCPKCNICIEKNGGCNHMQCSKCKHDFCWMCLGDWKTHGSEY 265
>emb|CAA10276.1| Ariadne-2 protein (ARI2) [Homo sapiens]
Length = 493
Score = 107 bits (266), Expect = 2e-21
Identities = 70/232 (30%), Positives = 96/232 (41%), Gaps = 10/232 (4%)
Query: 621 KSRVEEITFEIARLSNPSSERFDTGPS--CPICLCEVEDGYQLE-GCGHLFCQSCMVEQC 677
KS ++ E NPS + P C +C+ V L C H FC+SC + C
Sbjct: 110 KSNSAQLLVEARVQPNPSKHVPTSHPPHHCAVCMQFVRKENLLSLACQHQFCRSCWEQHC 169
Query: 678 ESAIKNQGSFPIRCAHQGCGNHILLVDFRTLLSNDKLEELFRASLGAFVASSSGTYRFCP 737
+K+ + C Q C LL N++L E +R L S + CP
Sbjct: 170 SVLVKDGVGVGVSCMAQDCPLRTPEDFVFPLLPNEELREKYRRYLFRDYVESHYQLQLCP 229
Query: 738 SPDCPSIYRVADPDTASAPFVCGACYSETCTRCHIEYHPYVSCERYRQF--KDDPDSSLR 795
DCP + RV +P C C C +C YH C R++ K DS
Sbjct: 230 GADCPMVIRVQEPRARRVQ--CNRCNEVFCFKCRQMYHAPTDCATIRKWLTKLQDDSETA 287
Query: 796 DWCKGKEQVKNCPACGHVIEKVDGCNHIEC-KCGKHICWVCLEFFTTSGECY 846
++ K+CP C IEK GCNH++C KC CW+CL + T G Y
Sbjct: 288 NYISA--HTKDCPKCNICIEKNGGCNHMQCSKCKHDFCWMCLGDWKTHGSEY 337
>dbj|BAD97270.1| ariadne homolog 2 variant [Homo sapiens]
Length = 493
Score = 106 bits (265), Expect = 3e-21
Identities = 70/232 (30%), Positives = 96/232 (41%), Gaps = 10/232 (4%)
Query: 621 KSRVEEITFEIARLSNPSSERFDTGPS--CPICLCEVEDGYQLE-GCGHLFCQSCMVEQC 677
KS ++ E NPS + P C +C+ V L C H FC+SC + C
Sbjct: 110 KSNSAQLLVEARVQPNPSKHVPTSHPPHHCAVCMQFVRKENLLSLACQHQFCRSCWEQHC 169
Query: 678 ESAIKNQGSFPIRCAHQGCGNHILLVDFRTLLSNDKLEELFRASLGAFVASSSGTYRFCP 737
+K+ + C Q C LL N++L E +R L S + CP
Sbjct: 170 SVLVKDGVGVGVSCMAQDCPLRTPEDFVFPLLPNEELREKYRRYLFRDYVESHYQLQLCP 229
Query: 738 SPDCPSIYRVADPDTASAPFVCGACYSETCTRCHIEYHPYVSCERYRQF--KDDPDSSLR 795
DCP + RV +P C C C +C YH C R++ K DS
Sbjct: 230 GADCPMVIRVQEPRARRVQ--CNRCNEVFCFKCRQMYHAPTDCATIRKWLTKCADDSETA 287
Query: 796 DWCKGKEQVKNCPACGHVIEKVDGCNHIEC-KCGKHICWVCLEFFTTSGECY 846
++ K+CP C IEK GCNH++C KC CW+CL + T G Y
Sbjct: 288 NYISA--HTKDCPKCNICIEKNGGCNHMQCSKCKHDFCWMCLGDWKTHGSEY 337
>gb|AAH00422.1| Ariadne homolog 2 [Homo sapiens] gi|3930776|gb|AAC82469.1| TRIAD1
type I [Homo sapiens] gi|18202259|sp|O95376|ARI2_HUMAN
Ariadne-2 protein homolog (ARI-2) (Triad1 protein)
(HT005) gi|48145687|emb|CAG33066.1| ARIH2 [Homo sapiens]
gi|5453557|ref|NP_006312.1| ariadne homolog 2 [Homo
sapiens]
Length = 493
Score = 106 bits (265), Expect = 3e-21
Identities = 70/232 (30%), Positives = 96/232 (41%), Gaps = 10/232 (4%)
Query: 621 KSRVEEITFEIARLSNPSSERFDTGPS--CPICLCEVEDGYQLE-GCGHLFCQSCMVEQC 677
KS ++ E NPS + P C +C+ V L C H FC+SC + C
Sbjct: 110 KSNSAQLLVEARVQPNPSKHVPTSHPPHHCAVCMQFVRKENLLSLACQHQFCRSCWEQHC 169
Query: 678 ESAIKNQGSFPIRCAHQGCGNHILLVDFRTLLSNDKLEELFRASLGAFVASSSGTYRFCP 737
+K+ + C Q C LL N++L E +R L S + CP
Sbjct: 170 SVLVKDGVGVGVSCMAQDCPLRTPEDFVFPLLPNEELREKYRRYLFRDYVESHYQLQLCP 229
Query: 738 SPDCPSIYRVADPDTASAPFVCGACYSETCTRCHIEYHPYVSCERYRQF--KDDPDSSLR 795
DCP + RV +P C C C +C YH C R++ K DS
Sbjct: 230 GADCPMVIRVQEPRARRVQ--CNRCNEVFCFKCRQMYHAPTDCATIRKWLTKCADDSETA 287
Query: 796 DWCKGKEQVKNCPACGHVIEKVDGCNHIEC-KCGKHICWVCLEFFTTSGECY 846
++ K+CP C IEK GCNH++C KC CW+CL + T G Y
Sbjct: 288 NYISA--HTKDCPKCNICIEKNGGCNHMQCSKCKHDFCWMCLGDWKTHGSEY 337
>ref|XP_635385.1| hypothetical protein DDB0189299 [Dictyostelium discoideum]
gi|60463693|gb|EAL61875.1| hypothetical protein
DDB0189299 [Dictyostelium discoideum]
Length = 1160
Score = 106 bits (265), Expect = 3e-21
Identities = 57/121 (47%), Positives = 79/121 (65%), Gaps = 1/121 (0%)
Query: 4 QITVLIGETGSGKSTQLVQFLADSGVGANESIVCTQPRRIAAKSLAERVREESGGCYEDS 63
Q+ V+IGETGSGK+TQ+ Q+LA++G G I CTQPRR+AA S+++RV EE GC
Sbjct: 525 QLLVVIGETGSGKTTQMAQYLAEAGYGTRGKIGCTQPRRVAAMSVSKRVAEEF-GCQLGQ 583
Query: 64 SIKCYSSFSSWNKFDSRIIFMTDHCLLQHYMSDKNFTGISCIIVDEAHERSINTDLLLAL 123
+ F ++ I FMTD LL+ + D N + S II+DEAHER+I+TD+L L
Sbjct: 584 EVGYAIRFEDCTSPETIIKFMTDGILLRECLLDPNLSAYSVIILDEAHERTISTDVLFGL 643
Query: 124 V 124
+
Sbjct: 644 L 644
>gb|AAC46765.1| Masculinisation of germline protein 5 [Caenorhabditis elegans]
gi|2500543|sp|Q09530|MOG5_CAEEL Probable pre-mRNA
splicing factor ATP-dependent RNA helicase mog-5 (Sex
determination protein mog-5) (Masculinization of germ
line protein 5) gi|17535281|ref|NP_495019.1| sex
determination DEAH box protein, posttranscriptional
regulatory factor similar to pre-mRNA splicing factor
ATP-dependent RNA helicase., Masculinisation Of Germline
MOG-5 (135.8 kD) (mog-5) [Caenorhabditis elegans]
gi|9864170|gb|AAG01332.1| sex determining protein MOG-5
[Caenorhabditis elegans]
Length = 1200
Score = 106 bits (264), Expect = 4e-21
Identities = 56/126 (44%), Positives = 83/126 (65%), Gaps = 1/126 (0%)
Query: 4 QITVLIGETGSGKSTQLVQFLADSGVGANESIVCTQPRRIAAKSLAERVREESGGCYEDS 63
QI V++GETGSGK+TQ+ Q+ ++G+G I CTQPRR+AA S+A+RV EE GC +
Sbjct: 557 QILVVVGETGSGKTTQMTQYAIEAGLGRRGKIGCTQPRRVAAMSVAKRVAEEY-GCKLGT 615
Query: 64 SIKCYSSFSSWNKFDSRIIFMTDHCLLQHYMSDKNFTGISCIIVDEAHERSINTDLLLAL 123
+ F D+ I +MTD LL+ + D + +G S I++DEAHER+I+TD+L L
Sbjct: 616 DVGYTIRFEDCTSQDTIIKYMTDGMLLRECLIDPDLSGYSLIMLDEAHERTIHTDVLFGL 675
Query: 124 VPSSNR 129
+ ++ R
Sbjct: 676 LKAAAR 681
>emb|CAH02932.1| unnamed protein product [Kluyveromyces lactis NRRL Y-1140]
gi|50302815|ref|XP_451344.1| unnamed protein product
[Kluyveromyces lactis]
Length = 1064
Score = 105 bits (263), Expect = 5e-21
Identities = 64/124 (51%), Positives = 80/124 (63%), Gaps = 8/124 (6%)
Query: 4 QITVLIGETGSGKSTQLVQFLADSGVGANESIV-CTQPRRIAAKSLAERVREESG---GC 59
QITV+IGETGSGK+TQL QFL + G AN ++ CTQPRR+AA S+AERV +E G
Sbjct: 364 QITVVIGETGSGKTTQLAQFLYEDGFTANGRMIGCTQPRRVAAVSVAERVSKEMDTPIGV 423
Query: 60 YEDSSIKCYSSFSSWNKFDSRIIFMTDHCLLQHYMSDKNFTGISCIIVDEAHERSINTDL 119
SI+ F ++I FMTD LL+ M D SCII+DEAHERS+NTD+
Sbjct: 424 KVGYSIR----FEDQTSDSTKIKFMTDGILLREAMIDPLLEKYSCIIMDEAHERSLNTDV 479
Query: 120 LLAL 123
LL +
Sbjct: 480 LLGI 483
>gb|AAS53593.1| AFR222Wp [Ashbya gossypii ATCC 10895] gi|45198740|ref|NP_985769.1|
AFR222Wp [Eremothecium gossypii]
Length = 1234
Score = 105 bits (263), Expect = 5e-21
Identities = 58/129 (44%), Positives = 80/129 (61%), Gaps = 8/129 (6%)
Query: 2 HVQITVLIGETGSGKSTQLVQFLADSGVGANES------IVCTQPRRIAAKSLAERVREE 55
H + ++ G TGSGK+TQ+ QFL +SG G ++ I TQPRR+AA S+A+RV E
Sbjct: 398 HNDVVIICGATGSGKTTQVPQFLYESGYGHPDAPDTPGMIAITQPRRVAAVSMAKRVGSE 457
Query: 56 SGGCYEDSSIKCYSSFSSWNKFDSRIIFMTDHCLLQHYMSDKNFTGISCIIVDEAHERSI 115
G + F S K D+R+ FMTD LL+ M+D T SCII+DEAHER+I
Sbjct: 458 LGN--HGDKVAYQIRFDSTTKEDTRVKFMTDGVLLREMMNDFRLTKYSCIIIDEAHERNI 515
Query: 116 NTDLLLALV 124
NTD+L+ ++
Sbjct: 516 NTDILIGML 524
>gb|EAK89557.1| PRP43 involved in spliceosome disassembly mRNA splicing
[Cryptosporidium parvum] gi|66361342|ref|XP_627301.1|
PRP43 involved in spliceosome disassembly mRNA splicing
[Cryptosporidium parvum]
Length = 714
Score = 105 bits (262), Expect = 6e-21
Identities = 55/119 (46%), Positives = 80/119 (67%), Gaps = 3/119 (2%)
Query: 4 QITVLIGETGSGKSTQLVQFLADSGVGANESIVCTQPRRIAAKSLAERVREESGGCYEDS 63
Q+ +L+G+TGSGK+TQ QF+ +SG+G N I CTQPRR+AA S+A+RV EE C D
Sbjct: 63 QVVILVGDTGSGKTTQCPQFILESGLGGNLKIACTQPRRVAAMSVAQRVSEEMDVCLGD- 121
Query: 64 SIKCYS-SFSSWNKFDSRIIFMTDHCLLQHYMSDKNFTGISCIIVDEAHERSINTDLLL 121
I Y+ F ++R+ ++TD LL+ M D + + II+DEAHER+I+TD+L+
Sbjct: 122 -IVGYTIRFEDKTNENTRLKYVTDGMLLREAMYDNDLSQYGVIIIDEAHERTISTDILM 179
>emb|CAG61351.1| unnamed protein product [Candida glabrata CBS138]
gi|50291915|ref|XP_448390.1| unnamed protein product
[Candida glabrata]
Length = 1295
Score = 105 bits (261), Expect = 8e-21
Identities = 59/129 (45%), Positives = 81/129 (62%), Gaps = 8/129 (6%)
Query: 2 HVQITVLIGETGSGKSTQLVQFLADSGVGANES------IVCTQPRRIAAKSLAERVREE 55
H + ++ GETGSGK+TQ+ QFL +SG G+ +S I TQPRR+AA S+AERV E
Sbjct: 445 HNDVVIICGETGSGKTTQVPQFLYESGYGSPDSTEHGGMIGITQPRRVAAVSMAERVSNE 504
Query: 56 SGGCYEDSSIKCYSSFSSWNKFDSRIIFMTDHCLLQHYMSDKNFTGISCIIVDEAHERSI 115
G + F S +K D+R+ FMTD LL+ M D + S II+DEAHER+I
Sbjct: 505 LGN--HGDKVAYQIRFDSTSKEDTRVKFMTDGVLLRELMEDFLLSKYSAIIIDEAHERNI 562
Query: 116 NTDLLLALV 124
NTD+L+ ++
Sbjct: 563 NTDILIGML 571
>gb|EAK96786.1| hypothetical protein CaO19.10336 [Candida albicans SC5314]
gi|46437388|gb|EAK96735.1| hypothetical protein
CaO19.2818 [Candida albicans SC5314]
Length = 1070
Score = 105 bits (261), Expect = 8e-21
Identities = 62/130 (47%), Positives = 82/130 (62%), Gaps = 13/130 (10%)
Query: 4 QITVLIGETGSGKSTQLVQFLADSGVGAN-------ESIVCTQPRRIAAKSLAERVREES 56
Q+T++IGETGSGK+TQL QFL + G GAN I CTQPRR+AA S+A+RV EE
Sbjct: 375 QVTIVIGETGSGKTTQLTQFLYEDGFGANIDKNGEKRIIACTQPRRVAAMSVAKRVSEEM 434
Query: 57 GGCYEDSSIKCYSSFSSWNKFDSR---IIFMTDHCLLQHYMSDKNFTGISCIIVDEAHER 113
C + F +K D++ I +MT+ LL+ ++D SCII+DEAHER
Sbjct: 435 N-CKLGEEVGYSIRFE--DKTDNKKTVIKYMTEGILLREILADPMLANYSCIIMDEAHER 491
Query: 114 SINTDLLLAL 123
S+NTD+LL L
Sbjct: 492 SLNTDILLGL 501
>ref|XP_667334.1| RNA helicase [Cryptosporidium hominis] gi|54658451|gb|EAL37096.1|
RNA helicase [Cryptosporidium hominis]
Length = 714
Score = 105 bits (261), Expect = 8e-21
Identities = 53/118 (44%), Positives = 78/118 (65%), Gaps = 1/118 (0%)
Query: 4 QITVLIGETGSGKSTQLVQFLADSGVGANESIVCTQPRRIAAKSLAERVREESGGCYEDS 63
Q+ +L+G+TGSGK+TQ QF+ +SG+G N I CTQPRR+AA S+A+RV EE C D
Sbjct: 63 QVVILVGDTGSGKTTQCPQFILESGLGGNLKIACTQPRRVAAMSVAQRVSEEMDVCLGD- 121
Query: 64 SIKCYSSFSSWNKFDSRIIFMTDHCLLQHYMSDKNFTGISCIIVDEAHERSINTDLLL 121
+ F ++R+ ++TD LL+ M D + + II+DEAHER+I+TD+L+
Sbjct: 122 VVGYTIRFEDKTNENTRLKYVTDGMLLREAMYDNDLSQYGVIIIDEAHERTISTDILM 179
Database: nr
Posted date: Jul 5, 2005 12:34 AM
Number of letters in database: 863,360,394
Number of sequences in database: 2,540,612
Lambda K H
0.321 0.137 0.422
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 1,436,065,545
Number of Sequences: 2540612
Number of extensions: 60565715
Number of successful extensions: 163966
Number of sequences better than 10.0: 1516
Number of HSP's better than 10.0 without gapping: 853
Number of HSP's successfully gapped in prelim test: 665
Number of HSP's that attempted gapping in prelim test: 160410
Number of HSP's gapped (non-prelim): 1976
length of query: 857
length of database: 863,360,394
effective HSP length: 137
effective length of query: 720
effective length of database: 515,296,550
effective search space: 371013516000
effective search space used: 371013516000
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.9 bits)
S2: 80 (35.4 bits)
Medicago: description of AC141414.11


