

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC141414.1 - phase: 0
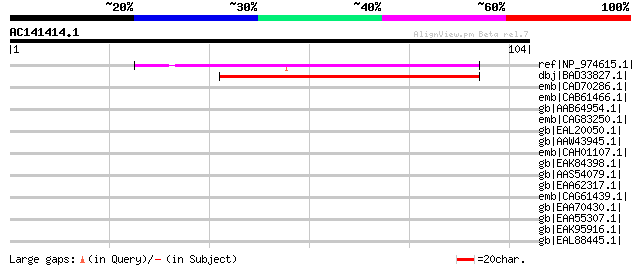
(104 letters)
Database: nr
2,540,612 sequences; 863,360,394 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
ref|NP_974615.1| Expressed protein [Arabidopsis thaliana] 76 2e-13
dbj|BAD33827.1| hypothetical protein [Oryza sativa (japonica cul... 73 1e-12
emb|CAD70286.1| conserved hypothetical protein [Neurospora crass... 42 0.004
emb|CAB61466.1| SPAC227.17c [Schizosaccharomyces pombe] gi|19113... 41 0.008
gb|AAB64954.1| Ydr512cp; CAI: 0.08 [Saccharomyces cerevisiae] gi... 39 0.031
emb|CAG83250.1| unnamed protein product [Yarrowia lipolytica CLI... 39 0.031
gb|EAL20050.1| hypothetical protein CNBF3760 [Cryptococcus neofo... 37 0.12
gb|AAW43945.1| hypothetical protein CNF00930 [Cryptococcus neofo... 37 0.12
emb|CAH01107.1| unnamed protein product [Kluyveromyces lactis NR... 37 0.15
gb|EAK84398.1| hypothetical protein UM03168.1 [Ustilago maydis 5... 35 0.34
gb|AAS54079.1| AFR707Cp [Ashbya gossypii ATCC 10895] gi|45199226... 35 0.45
gb|EAA62317.1| hypothetical protein AN5136.2 [Aspergillus nidula... 35 0.59
emb|CAG61439.1| unnamed protein product [Candida glabrata CBS138... 33 1.7
gb|EAA70430.1| hypothetical protein FG00837.1 [Gibberella zeae P... 32 2.9
gb|EAA55307.1| hypothetical protein MG06964.4 [Magnaporthe grise... 32 2.9
gb|EAK95916.1| hypothetical protein CaO19.11017 [Candida albican... 32 3.8
gb|EAL88445.1| conserved hypothetical protein [Aspergillus fumig... 31 8.5
>ref|NP_974615.1| Expressed protein [Arabidopsis thaliana]
Length = 127
Score = 75.9 bits (185), Expect = 2e-13
Identities = 41/88 (46%), Positives = 53/88 (59%), Gaps = 20/88 (22%)
Query: 26 MSSEKEELSSTVKRKLSCFEHFDALWFCY-------------------SPVYQMKQYYRL 66
MSSE E+ SS+V+R++SC + FDALWFCY +P YQM+QYYR+
Sbjct: 1 MSSENED-SSSVRRRVSCTKCFDALWFCYWMSRWSNITSNGDDTGNDLAPFYQMQQYYRV 59
Query: 67 GTLDNCRGKWKAWTDCLMLKTKPKSQVE 94
G LD+C K+ DCL LKTK S+ E
Sbjct: 60 GKLDDCTKKFSDLFDCLSLKTKRASEAE 87
>dbj|BAD33827.1| hypothetical protein [Oryza sativa (japonica cultivar-group)]
Length = 169
Score = 73.2 bits (178), Expect = 1e-12
Identities = 30/52 (57%), Positives = 36/52 (68%)
Query: 43 CFEHFDALWFCYSPVYQMKQYYRLGTLDNCRGKWKAWTDCLMLKTKPKSQVE 94
C + FDALWFCYSP YQ++ YYR G DNC GKW DCL LKT+ ++ E
Sbjct: 23 CVKCFDALWFCYSPYYQLQNYYRHGEFDNCFGKWGDLVDCLWLKTRRAAEAE 74
>emb|CAD70286.1| conserved hypothetical protein [Neurospora crassa]
gi|32419611|ref|XP_330249.1| predicted protein
[Neurospora crassa] gi|28924843|gb|EAA33941.1| predicted
protein [Neurospora crassa]
Length = 282
Score = 42.0 bits (97), Expect = 0.004
Identities = 17/47 (36%), Positives = 24/47 (50%)
Query: 41 LSCFEHFDALWFCYSPVYQMKQYYRLGTLDNCRGKWKAWTDCLMLKT 87
+SC + FD W C +P Q YR GT+ NC W + C+ K+
Sbjct: 155 MSCRDAFDYAWHCQTPGAQFNAVYRYGTMKNCSELWDDFWFCMRTKS 201
>emb|CAB61466.1| SPAC227.17c [Schizosaccharomyces pombe]
gi|19113883|ref|NP_592971.1| hypothetical protein
SPAC227.17c [Schizosaccharomyces pombe 972h-]
gi|11289169|pir||T50173 hypothetical protein
SPAC227.17c [imported] - fission yeast
(Schizosaccharomyces pombe)
gi|50401807|sp|Q9UTC2|YIDH_SCHPO Hypothetical protein
C227.17c in chromosome I
Length = 121
Score = 40.8 bits (94), Expect = 0.008
Identities = 17/51 (33%), Positives = 27/51 (52%)
Query: 46 HFDALWFCYSPVYQMKQYYRLGTLDNCRGKWKAWTDCLMLKTKPKSQVEVG 96
+FD + C + QM YYR G + C+G +K + CL K+K + + G
Sbjct: 39 YFDQMAMCLTVFSQMNHYYRYGEYNRCKGNFKDFKWCLSTKSKAHEEAQEG 89
>gb|AAB64954.1| Ydr512cp; CAI: 0.08 [Saccharomyces cerevisiae]
gi|6320721|ref|NP_010800.1| Non-essential protein of
unknown function required for transcriptional induction
of the early meiotic-specific transcription factor IME1,
also required for sporulation; Emi1p [Saccharomyces
cerevisiae] gi|2131539|pir||S69569 hypothetical protein
YDR512c - yeast (Saccharomyces cerevisiae)
Length = 187
Score = 38.9 bits (89), Expect = 0.031
Identities = 19/64 (29%), Positives = 31/64 (47%)
Query: 26 MSSEKEELSSTVKRKLSCFEHFDALWFCYSPVYQMKQYYRLGTLDNCRGKWKAWTDCLML 85
+ ++E +S+ +SC E FD L CYS Q + YYR G +C + + C++
Sbjct: 90 LRKKQERMSTKYPTTMSCREAFDQLTSCYSIGGQFRSYYRYGDFTSCDKQVSKFKFCIIH 149
Query: 86 KTKP 89
P
Sbjct: 150 GNDP 153
>emb|CAG83250.1| unnamed protein product [Yarrowia lipolytica CLIB99]
gi|50547055|ref|XP_500997.1| hypothetical protein
[Yarrowia lipolytica]
Length = 181
Score = 38.9 bits (89), Expect = 0.031
Identities = 17/51 (33%), Positives = 29/51 (56%)
Query: 40 KLSCFEHFDALWFCYSPVYQMKQYYRLGTLDNCRGKWKAWTDCLMLKTKPK 90
++SC+E F+ + CYS Q++ YR G+L+ C W C+ +K P+
Sbjct: 100 EMSCWEAFNEVVACYSVGGQVRNLYRFGSLNYCDKPKDKWKFCMGVKLDPE 150
>gb|EAL20050.1| hypothetical protein CNBF3760 [Cryptococcus neoformans var.
neoformans B-3501A]
Length = 163
Score = 37.0 bits (84), Expect = 0.12
Identities = 16/46 (34%), Positives = 25/46 (53%)
Query: 43 CFEHFDALWFCYSPVYQMKQYYRLGTLDNCRGKWKAWTDCLMLKTK 88
C D CY+ V Q++ YR G L NC K++ + C+ LK++
Sbjct: 64 CMRLLDEFLMCYALVPQLRNIYRHGELHNCSWKFQDFKYCMSLKSE 109
>gb|AAW43945.1| hypothetical protein CNF00930 [Cryptococcus neoformans var.
neoformans JEC21] gi|58268192|ref|XP_571252.1|
hypothetical protein CNF00930 [Cryptococcus neoformans
var. neoformans JEC21]
Length = 163
Score = 37.0 bits (84), Expect = 0.12
Identities = 16/46 (34%), Positives = 25/46 (53%)
Query: 43 CFEHFDALWFCYSPVYQMKQYYRLGTLDNCRGKWKAWTDCLMLKTK 88
C D CY+ V Q++ YR G L NC K++ + C+ LK++
Sbjct: 64 CMRLLDEFLMCYALVPQLRNIYRHGELHNCSWKFQDFKYCMSLKSE 109
>emb|CAH01107.1| unnamed protein product [Kluyveromyces lactis NRRL Y-1140]
gi|50304601|ref|XP_452256.1| unnamed protein product
[Kluyveromyces lactis]
Length = 86
Score = 36.6 bits (83), Expect = 0.15
Identities = 17/57 (29%), Positives = 30/57 (51%)
Query: 33 LSSTVKRKLSCFEHFDALWFCYSPVYQMKQYYRLGTLDNCRGKWKAWTDCLMLKTKP 89
+SS +SC + FD L C+S Q + YYR G ++ C +++ + C++ P
Sbjct: 1 MSSNYPTTMSCTDAFDQLTGCFSLGGQFRNYYRHGHMNECLKEFEKFKFCVIHSKDP 57
>gb|EAK84398.1| hypothetical protein UM03168.1 [Ustilago maydis 521]
gi|49073076|ref|XP_400783.1| hypothetical protein
UM03168.1 [Ustilago maydis 521]
Length = 160
Score = 35.4 bits (80), Expect = 0.34
Identities = 18/53 (33%), Positives = 26/53 (48%)
Query: 34 SSTVKRKLSCFEHFDALWFCYSPVYQMKQYYRLGTLDNCRGKWKAWTDCLMLK 86
S+ V SC FD C++ Q+K YR G L +C+ K + CL +K
Sbjct: 28 STPVSEMPSCTNMFDKWAACFALGPQLKAVYRYGGLQDCKAKLDDFKHCLTMK 80
>gb|AAS54079.1| AFR707Cp [Ashbya gossypii ATCC 10895]
gi|45199226|ref|NP_986255.1| AFR707Cp [Eremothecium
gossypii]
Length = 97
Score = 35.0 bits (79), Expect = 0.45
Identities = 17/61 (27%), Positives = 31/61 (49%)
Query: 29 EKEELSSTVKRKLSCFEHFDALWFCYSPVYQMKQYYRLGTLDNCRGKWKAWTDCLMLKTK 88
E ++ + ++SC E FD L CYS ++ YYR G + C + + + C++ +
Sbjct: 8 EPQQPAEQYPSQISCTEAFDRLTDCYSIGGLVRHYYRHGEFNGCSKEIERFKFCILHGSD 67
Query: 89 P 89
P
Sbjct: 68 P 68
>gb|EAA62317.1| hypothetical protein AN5136.2 [Aspergillus nidulans FGSC A4]
gi|67537932|ref|XP_662740.1| hypothetical protein
AN5136_2 [Aspergillus nidulans FGSC A4]
gi|49095624|ref|XP_409273.1| hypothetical protein
AN5136.2 [Aspergillus nidulans FGSC A4]
Length = 221
Score = 34.7 bits (78), Expect = 0.59
Identities = 16/47 (34%), Positives = 24/47 (51%)
Query: 41 LSCFEHFDALWFCYSPVYQMKQYYRLGTLDNCRGKWKAWTDCLMLKT 87
+SC + FD +FC S Q YR G L +C W+ + C+ +T
Sbjct: 105 MSCRDAFDYAFFCQSLGGQFVNVYRYGELRSCSEHWENFWLCMKTRT 151
>emb|CAG61439.1| unnamed protein product [Candida glabrata CBS138]
gi|50292091|ref|XP_448478.1| unnamed protein product
[Candida glabrata]
Length = 88
Score = 33.1 bits (74), Expect = 1.7
Identities = 20/64 (31%), Positives = 28/64 (43%), Gaps = 4/64 (6%)
Query: 26 MSSEKEELSSTVKRKLSCFEHFDALWFCYSPVYQMKQYYRLGTLDNCRGKWKAWTDCLML 85
MS E + +T +SC E FD L C S Q + YYR G C + + C++
Sbjct: 1 MSQENVKYPTT----MSCREAFDQLTTCLSIGGQFRNYYRYGEFSACEKQVNKFKFCILN 56
Query: 86 KTKP 89
P
Sbjct: 57 AKDP 60
>gb|EAA70430.1| hypothetical protein FG00837.1 [Gibberella zeae PH-1]
gi|46107908|ref|XP_381013.1| hypothetical protein
FG00837.1 [Gibberella zeae PH-1]
Length = 210
Score = 32.3 bits (72), Expect = 2.9
Identities = 14/46 (30%), Positives = 21/46 (45%)
Query: 41 LSCFEHFDALWFCYSPVYQMKQYYRLGTLDNCRGKWKAWTDCLMLK 86
+SC + FD W C Q YR G + +C W + C+ +K
Sbjct: 112 MSCRQAFDQAWSCNGMGGQFIAVYRYGEMRSCSEHWDDFWFCMRVK 157
>gb|EAA55307.1| hypothetical protein MG06964.4 [Magnaporthe grisea 70-15]
gi|39978159|ref|XP_370467.1| hypothetical protein
MG06964.4 [Magnaporthe grisea 70-15]
Length = 274
Score = 32.3 bits (72), Expect = 2.9
Identities = 16/59 (27%), Positives = 29/59 (49%)
Query: 28 SEKEELSSTVKRKLSCFEHFDALWFCYSPVYQMKQYYRLGTLDNCRGKWKAWTDCLMLK 86
+++E + S + +SC + FD + C S Q YR G + +C W + C+ +K
Sbjct: 127 AKEEIVQSLMPTDMSCRDAFDYAYGCNSIRGQFNAVYRYGEVRSCSELWDNFWFCMRVK 185
>gb|EAK95916.1| hypothetical protein CaO19.11017 [Candida albicans SC5314]
gi|46436491|gb|EAK95852.1| hypothetical protein
CaO19.3533 [Candida albicans SC5314]
Length = 149
Score = 32.0 bits (71), Expect = 3.8
Identities = 18/64 (28%), Positives = 33/64 (51%), Gaps = 10/64 (15%)
Query: 26 MSSEKEELSSTVKRKLSCFEH---------FDALWFCYSPVYQMKQYYRLGTLDNC-RGK 75
+ ++E++ ++++ S E+ D L C++ Q+K YYR GT D+C R +
Sbjct: 22 VQKRRKEITKSIQKDTSLNEYPGSVSMMTALDELIACFALGGQVKNYYRYGTYDSCTRQR 81
Query: 76 WKAW 79
K W
Sbjct: 82 EKFW 85
>gb|EAL88445.1| conserved hypothetical protein [Aspergillus fumigatus Af293]
Length = 227
Score = 30.8 bits (68), Expect = 8.5
Identities = 15/43 (34%), Positives = 20/43 (45%)
Query: 41 LSCFEHFDALWFCYSPVYQMKQYYRLGTLDNCRGKWKAWTDCL 83
+SC FD +FC S Q YR G L +C W + C+
Sbjct: 115 MSCRSAFDYAFFCQSFGGQFVNVYRYGELRSCSEHWDNFWLCM 157
Database: nr
Posted date: Jul 5, 2005 12:34 AM
Number of letters in database: 863,360,394
Number of sequences in database: 2,540,612
Lambda K H
0.326 0.137 0.445
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 168,996,350
Number of Sequences: 2540612
Number of extensions: 5627075
Number of successful extensions: 12398
Number of sequences better than 10.0: 17
Number of HSP's better than 10.0 without gapping: 16
Number of HSP's successfully gapped in prelim test: 1
Number of HSP's that attempted gapping in prelim test: 12376
Number of HSP's gapped (non-prelim): 17
length of query: 104
length of database: 863,360,394
effective HSP length: 80
effective length of query: 24
effective length of database: 660,111,434
effective search space: 15842674416
effective search space used: 15842674416
T: 11
A: 40
X1: 15 ( 7.1 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 40 (21.7 bits)
S2: 68 (30.8 bits)
Medicago: description of AC141414.1


