

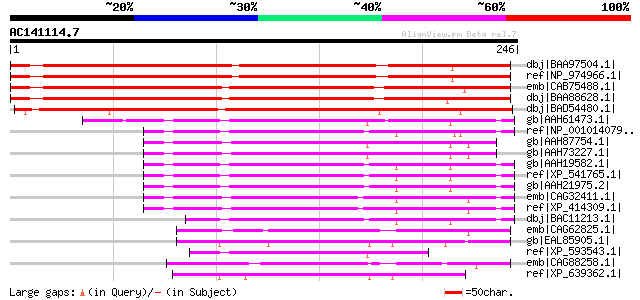
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC141114.7 + phase: 0
(246 letters)
Database: nr
2,540,612 sequences; 863,360,394 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
dbj|BAA97504.1| tRNA intron endonuclease [Arabidopsis thaliana] ... 259 5e-68
ref|NP_974966.1| tRNA-splicing endonuclease, putative [Arabidops... 259 5e-68
emb|CAB75488.1| putative protein [Arabidopsis thaliana] gi|62320... 254 1e-66
dbj|BAA88628.1| tRNA intron endonuclease [Arabidopsis thaliana] ... 253 3e-66
dbj|BAD54480.1| putative tRNA intron endonuclease [Oryza sativa ... 212 9e-54
gb|AAH61473.1| TRNA splicing endonuclease 2 homolog (SEN2, S. ce... 100 5e-20
ref|NP_001014079.1| hypothetical LOC312649 [Rattus norvegicus] g... 100 6e-20
gb|AAH87754.1| LOC496636 protein [Xenopus tropicalis] 96 1e-18
gb|AAH73227.1| MGC80553 protein [Xenopus laevis] 95 2e-18
gb|AAH19582.1| TRNA splicing endonuclease 2 homolog (SEN2, S. ce... 94 4e-18
ref|XP_541765.1| PREDICTED: similar to hypothetical protein MGC2... 94 4e-18
gb|AAH21975.2| TSEN2 protein [Homo sapiens] 94 4e-18
emb|CAG32411.1| hypothetical protein [Gallus gallus] 92 2e-17
ref|XP_414309.1| PREDICTED: similar to hypothetical protein MGC2... 92 2e-17
dbj|BAC11213.1| unnamed protein product [Homo sapiens] 86 1e-15
emb|CAG62825.1| unnamed protein product [Candida glabrata CBS138... 74 3e-12
gb|EAL85905.1| tRNA-splicing endonuclease subunit SEN2,putative ... 72 1e-11
ref|XP_593543.1| PREDICTED: similar to hypothetical protein MGC2... 72 2e-11
emb|CAG88258.1| unnamed protein product [Debaryomyces hansenii C... 71 3e-11
ref|XP_639362.1| hypothetical protein DDB0204993 [Dictyostelium ... 71 3e-11
>dbj|BAA97504.1| tRNA intron endonuclease [Arabidopsis thaliana]
gi|28827218|gb|AAO50453.1| putative tRNA intron
endonuclease [Arabidopsis thaliana]
gi|28393426|gb|AAO42135.1| putative tRNA intron
endonuclease [Arabidopsis thaliana]
gi|15238643|ref|NP_200831.1| tRNA-splicing endonuclease,
putative [Arabidopsis thaliana]
Length = 250
Score = 259 bits (662), Expect = 5e-68
Identities = 138/245 (56%), Positives = 166/245 (67%), Gaps = 16/245 (6%)
Query: 1 MAPRWKGKDAKSKKEAEAEALKEPMSKIISQLQSSLVQSNTCGFLSDNCVHLTVQAEQLD 60
MAPRWK K A EA+AL EP+SK +S+LQ SL ++ + G LS V L V+ EQ +
Sbjct: 1 MAPRWKWKGA------EAKALAEPISKSVSELQLSLAETESSGTLSSCNVLLAVEPEQAE 54
Query: 61 LLDKACFGRPVRIVEKDMYWFQLTVEEAFYLCYSLKCLKIKINVGADTGPLNDEELWHYF 120
LLD+ CFGR V EK W QL+ EEAF+L Y+LKC+KI + N+ + W Y
Sbjct: 55 LLDRCCFGRLVLSAEKIKKWIQLSFEEAFFLHYNLKCIKISLQGRCLE---NEVDTWLYM 111
Query: 121 RSKKETFPYFYKAYSHLRMKNWVVRSGAQYGVDLIVYRHHPARVHSEYGVLVLSHDKDGD 180
+SK+ FP F+KAYSHLR KNWV+RSG QYGVD + YRHHP+ VHSEY VLV S D D
Sbjct: 112 KSKRPNFPMFFKAYSHLRSKNWVLRSGLQYGVDFVAYRHHPSLVHSEYSVLVQSGDSD-- 169
Query: 181 LNGRLRVWSDVHCTTRLLGGVAKTLLVLYVNKN--GNNDESLLCLTNYTVEERTISRWSP 238
RLRVWSD+HC RL G VAKTLL LYVN N G + L+CL N+TVEE+TISRWSP
Sbjct: 170 ---RLRVWSDIHCAVRLSGSVAKTLLTLYVNGNFKGEDVNLLVCLENFTVEEQTISRWSP 226
Query: 239 EQCRE 243
E RE
Sbjct: 227 ELSRE 231
>ref|NP_974966.1| tRNA-splicing endonuclease, putative [Arabidopsis thaliana]
Length = 255
Score = 259 bits (662), Expect = 5e-68
Identities = 138/245 (56%), Positives = 166/245 (67%), Gaps = 16/245 (6%)
Query: 1 MAPRWKGKDAKSKKEAEAEALKEPMSKIISQLQSSLVQSNTCGFLSDNCVHLTVQAEQLD 60
MAPRWK K A EA+AL EP+SK +S+LQ SL ++ + G LS V L V+ EQ +
Sbjct: 6 MAPRWKWKGA------EAKALAEPISKSVSELQLSLAETESSGTLSSCNVLLAVEPEQAE 59
Query: 61 LLDKACFGRPVRIVEKDMYWFQLTVEEAFYLCYSLKCLKIKINVGADTGPLNDEELWHYF 120
LLD+ CFGR V EK W QL+ EEAF+L Y+LKC+KI + N+ + W Y
Sbjct: 60 LLDRCCFGRLVLSAEKIKKWIQLSFEEAFFLHYNLKCIKISLQGRCLE---NEVDTWLYM 116
Query: 121 RSKKETFPYFYKAYSHLRMKNWVVRSGAQYGVDLIVYRHHPARVHSEYGVLVLSHDKDGD 180
+SK+ FP F+KAYSHLR KNWV+RSG QYGVD + YRHHP+ VHSEY VLV S D D
Sbjct: 117 KSKRPNFPMFFKAYSHLRSKNWVLRSGLQYGVDFVAYRHHPSLVHSEYSVLVQSGDSD-- 174
Query: 181 LNGRLRVWSDVHCTTRLLGGVAKTLLVLYVNKN--GNNDESLLCLTNYTVEERTISRWSP 238
RLRVWSD+HC RL G VAKTLL LYVN N G + L+CL N+TVEE+TISRWSP
Sbjct: 175 ---RLRVWSDIHCAVRLSGSVAKTLLTLYVNGNFKGEDVNLLVCLENFTVEEQTISRWSP 231
Query: 239 EQCRE 243
E RE
Sbjct: 232 ELSRE 236
>emb|CAB75488.1| putative protein [Arabidopsis thaliana] gi|62320332|dbj|BAD94688.1|
putative protein [Arabidopsis thaliana]
gi|15231186|ref|NP_190145.1| tRNA-splicing endonuclease,
putative [Arabidopsis thaliana] gi|11290094|pir||T47499
hypothetical protein F9K21.170 - Arabidopsis thaliana
Length = 237
Score = 254 bits (650), Expect = 1e-66
Identities = 136/245 (55%), Positives = 168/245 (68%), Gaps = 15/245 (6%)
Query: 1 MAPRWKGKDAKSKKEAEAEALKEPMSKIISQLQSSLVQSNTCGFLSDNCVHLTVQAEQLD 60
MAPRWK K A EA+AL EP+SK +S+L+SSL Q+ GFLS V L+V++E+ +
Sbjct: 1 MAPRWKWKGA------EAKALAEPVSKTVSELRSSLTQTEALGFLSSCNVLLSVESEEAE 54
Query: 61 LLDKACFGRPVRIVEKDMYWFQLTVEEAFYLCYSLKCLKIKINVGADTGPLNDEELWHYF 120
LLD+ CFGR V EKD W QL+ EEAF+L Y LKC+KI ++ N+ +LW
Sbjct: 55 LLDRCCFGRLVVGAEKDKRWIQLSFEEAFFLFYKLKCIKICLH---GRSLENEVDLWRSM 111
Query: 121 RSKKETFPYFYKAYSHLRMKNWVVRSGAQYGVDLIVYRHHPARVHSEYGVLVLSHDKDGD 180
S K+ F YKAYSHLR KNW+VRSG QYGVD +VYRHHP+ VHSEY VLV S
Sbjct: 112 SSFKQDFAILYKAYSHLRSKNWIVRSGLQYGVDFVVYRHHPSLVHSEYAVLVQSIGG--- 168
Query: 181 LNGRLRVWSDVHCTTRLLGGVAKTLLVLYVNKNGNNDES--LLCLTNYTVEERTISRWSP 238
N RL+VWSD+HC+ RL G VAK+LLVLYVN+ N ++ LCL +YTVEE+TI RWSP
Sbjct: 169 -NDRLKVWSDIHCSVRLTGSVAKSLLVLYVNRKVNTEKMNLPLCLEDYTVEEQTIRRWSP 227
Query: 239 EQCRE 243
E RE
Sbjct: 228 ELSRE 232
>dbj|BAA88628.1| tRNA intron endonuclease [Arabidopsis thaliana]
gi|6635256|dbj|BAA88627.1| tRNA intron endonuclease
[Arabidopsis thaliana]
Length = 237
Score = 253 bits (647), Expect = 3e-66
Identities = 138/245 (56%), Positives = 163/245 (66%), Gaps = 15/245 (6%)
Query: 1 MAPRWKGKDAKSKKEAEAEALKEPMSKIISQLQSSLVQSNTCGFLSDNCVHLTVQAEQLD 60
MAPRWK K A EA+AL EP+SK + +LQSSL Q+ GFLS V L+V++EQ +
Sbjct: 1 MAPRWKWKGA------EAKALAEPVSKTVLELQSSLTQTEALGFLSSCNVLLSVESEQAE 54
Query: 61 LLDKACFGRPVRIVEKDMYWFQLTVEEAFYLCYSLKCLKIKINVGADTGPLNDEELWHYF 120
LLD+ CFGR V EKD W QL+ EEAF+L Y LKC+KI ++ N+ +LW
Sbjct: 55 LLDRCCFGRLVVSAEKDKRWIQLSFEEAFFLFYKLKCIKICLH---GRSLENEVDLWRSM 111
Query: 121 RSKKETFPYFYKAYSHLRMKNWVVRSGAQYGVDLIVYRHHPARVHSEYGVLVLSHDKDGD 180
S K+ F YKAYSHLR KNW+VRSG QYGVD +VYRHHP+ VHSEY VLV S
Sbjct: 112 SSFKQDFAILYKAYSHLRSKNWIVRSGLQYGVDFVVYRHHPSLVHSEYAVLVQSISG--- 168
Query: 181 LNGRLRVWSDVHCTTRLLGGVAKTLLVLYVN--KNGNNDESLLCLTNYTVEERTISRWSP 238
N RL+VWSD+HC+ RL G VAKTLLVLYVN N LCL YTVEE+TI RWSP
Sbjct: 169 -NDRLKVWSDIHCSVRLTGSVAKTLLVLYVNGQVKTENMNLPLCLDEYTVEEQTIRRWSP 227
Query: 239 EQCRE 243
E RE
Sbjct: 228 ELSRE 232
>dbj|BAD54480.1| putative tRNA intron endonuclease [Oryza sativa (japonica
cultivar-group)] gi|53791958|dbj|BAD54220.1| putative
tRNA intron endonuclease [Oryza sativa (japonica
cultivar-group)]
Length = 293
Score = 212 bits (539), Expect = 9e-54
Identities = 114/247 (46%), Positives = 152/247 (61%), Gaps = 11/247 (4%)
Query: 3 PRWK-GKDAKSKKEAEAEALKEPMSKIISQLQSSLVQSNTCGFLSD--NCVHLTVQAEQL 59
PRWK GKD K + + A PMS I+S+L++S + S LS L V EQ
Sbjct: 6 PRWKKGKDGK---DFASLAAANPMSAIVSELKASFISSKPVAILSGPGGSAVLGVGPEQA 62
Query: 60 DLLDKACFGRPVRIVEKDMYWFQLTVEEAFYLCYSLKCLKIKINVGADTGPLNDEELWHY 119
+L++A FG + +WFQL+ EE FYLC++L C+++ D +++ ELW Y
Sbjct: 63 VILNRAAFGHAIENATAQKHWFQLSPEEVFYLCHALNCIRVD---SLDNKQMSEIELWDY 119
Query: 120 FRSKKETFPYFYKAYSHLRMKNWVVRSGAQYGVDLIVYRHHPARVHSEYGVLVLSHDKD- 178
FRS E+FP YKAY+HLR+KNWVVRSG QYG D + YRHHPA VHSE+ V+V+ +
Sbjct: 120 FRSGSESFPEMYKAYAHLRLKNWVVRSGLQYGADFVAYRHHPALVHSEFAVVVVPEGAEF 179
Query: 179 GDLNGRLRVWSDVHCTTRLLGGVAKTLLVLYVNKNGNND-ESLLCLTNYTVEERTISRWS 237
G+ GRL VWSD+ C R G VAKTLLVL ++ + + S CL V ERTI+RW
Sbjct: 180 GNRCGRLEVWSDLLCALRASGSVAKTLLVLTISSSSKCELSSPDCLEQLVVHERTITRWI 239
Query: 238 PEQCRER 244
+QCRE+
Sbjct: 240 LQQCREQ 246
>gb|AAH61473.1| TRNA splicing endonuclease 2 homolog (SEN2, S. cerevisiae) [Mus
musculus] gi|39841065|ref|NP_950198.1| tRNA splicing
endonuclease 2 homolog (SEN2, S. cerevisiae) [Mus
musculus] gi|50428912|sp|Q6P7W5|SEN2_MOUSE tRNA-splicing
endonuclease subunit Sen2 (tRNA-intron endonuclease
Sen2)
Length = 460
Score = 100 bits (248), Expect = 5e-20
Identities = 69/216 (31%), Positives = 106/216 (48%), Gaps = 18/216 (8%)
Query: 36 LVQSNTCGFLSDNCVHLTVQAEQLDLLDKACFGRPVRIVEKDMYWFQLTVEEAFYLCYSL 95
L++ CG + + + + L C P RI E + QL++EEAF+L Y+L
Sbjct: 252 LIEEELCGAQEEEAAAASDE-KLLKRKKLVCRRNPYRIFE----YLQLSLEEAFFLAYAL 306
Query: 96 KCLKIKINVGADTGPLNDEELWHYFRSKKETFPYFYKAYSHLRMKNWVVRSGAQYGVDLI 155
CL I + PL +LW F + + TF Y AY + R K WV + G +YG DL+
Sbjct: 307 GCLSIYY----EKEPLTIVKLWQAFTAVQPTFRTTYMAYHYFRSKGWVPKVGLKYGTDLL 362
Query: 156 VYRHHPARVHSEYGVLV--LSHDKDGDLNGRLRVWSDVHCTTRLLGGVAKTLLVLYVNK- 212
+YR P H+ Y V++ L + +G L R W + +R+ G V+K L++ Y+ K
Sbjct: 363 LYRKGPPFYHASYSVIIELLDDNYEGSLR-RPFSWKSLAALSRVSGNVSKELMLCYLIKP 421
Query: 213 ---NGNNDESLLCLTNYTVEERTISRWSPEQCRERS 245
+ E+ C+ V+E +SRW RERS
Sbjct: 422 STMTAEDMETPECMKRIQVQEVILSRW--VSSRERS 455
>ref|NP_001014079.1| hypothetical LOC312649 [Rattus norvegicus]
gi|56585031|gb|AAH87631.1| Hypothetical LOC312649
[Rattus norvegicus]
Length = 463
Score = 99.8 bits (247), Expect = 6e-20
Identities = 67/187 (35%), Positives = 96/187 (50%), Gaps = 19/187 (10%)
Query: 66 CFGRPVRIVEKDMYWFQLTVEEAFYLCYSLKCLKIKINVGADTGPLNDEELWHYFRSKKE 125
C P RI E + QL++EEAF+L Y+L CL I + PL +LW F + +
Sbjct: 284 CRRNPYRIFE----YLQLSLEEAFFLAYALGCLSIYY----EKEPLTIVKLWQAFTAVQP 335
Query: 126 TFPYFYKAYSHLRMKNWVVRSGAQYGVDLIVYRHHPARVHSEYGVLVLSHDKDGDLNGRL 185
TF Y AY + R K WV + G +YG DL++YR P H+ Y V++ D + G L
Sbjct: 336 TFRTTYMAYHYFRSKGWVPKVGLKYGTDLLLYRKGPPFYHASYSVII--ELVDDNFEGSL 393
Query: 186 R---VWSDVHCTTRLLGGVAKTLLVLYVNKNG---NND-ESLLCLTNYTVEERTISRWSP 238
R W + +R+ G V+K L++ Y+ K N D E+ C+ V+E +SRW
Sbjct: 394 RRPFSWKSLAALSRVSGNVSKELMLCYLIKPSTMTNEDMETPECMRRIQVQEVILSRW-- 451
Query: 239 EQCRERS 245
RERS
Sbjct: 452 VSSRERS 458
>gb|AAH87754.1| LOC496636 protein [Xenopus tropicalis]
Length = 493
Score = 95.5 bits (236), Expect = 1e-18
Identities = 62/176 (35%), Positives = 87/176 (49%), Gaps = 13/176 (7%)
Query: 66 CFGRPVRIVEKDMYWFQLTVEEAFYLCYSLKCLKIKINVGADTGPLNDEELWHYFRSKKE 125
C P R+ E + QL+ EEAF+L ++L CL + N PL +LW F + +
Sbjct: 314 CRRNPFRVFE----YLQLSREEAFFLVFALGCLTVSYN----KEPLTILKLWEVFSAAQA 365
Query: 126 TFPYFYKAYSHLRMKNWVVRSGAQYGVDLIVYRHHPARVHSEYGVLV-LSHDKDGDLNGR 184
+F Y AY H R K WV + G +YG DL++YR P H+ Y V+V L +DK R
Sbjct: 366 SFSTTYMAYHHFRSKGWVPKVGLKYGTDLLLYRKGPPFYHASYSVIVELVNDKCEGAPLR 425
Query: 185 LRVWSDVHCTTRLLGGVAKTLLVLYVNK-NGNNDESLL---CLTNYTVEERTISRW 236
W + R V+K LL Y+ K G D L+ C+ V+E +SRW
Sbjct: 426 PLSWRSLAGLHRTTANVSKELLFCYLIKPAGFTDTDLMSPACINRIKVQELIVSRW 481
>gb|AAH73227.1| MGC80553 protein [Xenopus laevis]
Length = 460
Score = 94.7 bits (234), Expect = 2e-18
Identities = 63/176 (35%), Positives = 86/176 (48%), Gaps = 13/176 (7%)
Query: 66 CFGRPVRIVEKDMYWFQLTVEEAFYLCYSLKCLKIKINVGADTGPLNDEELWHYFRSKKE 125
C P RI E + QL+ EEAF+L Y+L CL I PL ++LW F + +
Sbjct: 281 CRRNPFRIFE----YLQLSREEAFFLVYALGCLTISYK----KEPLTIQKLWEVFCAAQP 332
Query: 126 TFPYFYKAYSHLRMKNWVVRSGAQYGVDLIVYRHHPARVHSEYGVLV-LSHDKDGDLNGR 184
+F Y AY H R K WV + G +YG DL++YR P H+ Y V+V L +DK R
Sbjct: 333 SFSTKYLAYHHFRSKGWVPKVGLKYGTDLLLYRKGPPFYHASYSVIVELVNDKGEGTPLR 392
Query: 185 LRVWSDVHCTTRLLGGVAKTLLVLYVNK-NGNNDESLL---CLTNYTVEERTISRW 236
W + R V+K LL Y+ K G D + C+ V+E +SRW
Sbjct: 393 PLSWRSLAGLHRTTANVSKELLFCYLIKPAGFTDSDFISPACIHRIKVQELIVSRW 448
>gb|AAH19582.1| TRNA splicing endonuclease 2 homolog (SEN2, S. cerevisiae) [Homo
sapiens] gi|13278906|gb|AAH04211.1| TSEN2 protein [Homo
sapiens] gi|13278819|gb|AAH04178.1| TSEN2 protein [Homo
sapiens] gi|13376882|ref|NP_079541.1| tRNA splicing
endonuclease 2 homolog (SEN2, S. cerevisiae) [Homo
sapiens] gi|50428914|sp|Q8NCE0|SEN2_HUMAN tRNA-splicing
endonuclease subunit SEN2 (tRNA-intron endonuclease
SEN2) (HsSen2 protein)
Length = 465
Score = 93.6 bits (231), Expect = 4e-18
Identities = 65/187 (34%), Positives = 91/187 (47%), Gaps = 19/187 (10%)
Query: 66 CFGRPVRIVEKDMYWFQLTVEEAFYLCYSLKCLKIKINVGADTGPLNDEELWHYFRSKKE 125
C P RI E + QL++EEAF+L Y+L CL I + PL +LW F +
Sbjct: 286 CRRNPYRIFE----YLQLSLEEAFFLVYALGCLSIYY----EKEPLTIVKLWKAFTVVQP 337
Query: 126 TFPYFYKAYSHLRMKNWVVRSGAQYGVDLIVYRHHPARVHSEYGVLVLSHDKDGDLNGRL 185
TF Y AY + R K WV + G +YG DL++YR P H+ Y V++ D G L
Sbjct: 338 TFRTTYMAYHYFRSKGWVPKVGLKYGTDLLLYRKGPPFYHASYSVII--ELVDDHFEGSL 395
Query: 186 R---VWSDVHCTTRLLGGVAKTLLVLYVNK----NGNNDESLLCLTNYTVEERTISRWSP 238
R W + +R+ V+K L++ Y+ K ES C+ V+E +SRW
Sbjct: 396 RRPLSWKSLAALSRVSVNVSKELMLCYLIKPSTMTDKEMESPECMKRIKVQEVILSRW-- 453
Query: 239 EQCRERS 245
RERS
Sbjct: 454 VSSRERS 460
>ref|XP_541765.1| PREDICTED: similar to hypothetical protein MGC2776 [Canis
familiaris]
Length = 470
Score = 93.6 bits (231), Expect = 4e-18
Identities = 65/187 (34%), Positives = 91/187 (47%), Gaps = 19/187 (10%)
Query: 66 CFGRPVRIVEKDMYWFQLTVEEAFYLCYSLKCLKIKINVGADTGPLNDEELWHYFRSKKE 125
C P RI+E + QL++EEAF+L Y+L CL I + PL +LW F +
Sbjct: 291 CRRNPYRILE----YLQLSLEEAFFLVYALGCLSIYY----EKEPLTIMKLWKAFTVVQP 342
Query: 126 TFPYFYKAYSHLRMKNWVVRSGAQYGVDLIVYRHHPARVHSEYGVLVLSHDKDGDLNGRL 185
TF Y AY + R K WV + G +YG DL++YR P H+ Y V++ D G L
Sbjct: 343 TFRTTYMAYHYFRSKGWVPKVGLKYGTDLLLYRKGPPFYHASYSVII--ELVDDHFEGSL 400
Query: 186 R---VWSDVHCTTRLLGGVAKTLLVLYVNK----NGNNDESLLCLTNYTVEERTISRWSP 238
R W + R+ V+K L++ Y+ K ES C+ V+E +SRW
Sbjct: 401 RRPFSWRSLAALNRVSVNVSKELMLCYLIKPSTMTDKEMESPECMKQIKVQEVILSRW-- 458
Query: 239 EQCRERS 245
RERS
Sbjct: 459 VSSRERS 465
>gb|AAH21975.2| TSEN2 protein [Homo sapiens]
Length = 255
Score = 93.6 bits (231), Expect = 4e-18
Identities = 65/187 (34%), Positives = 91/187 (47%), Gaps = 19/187 (10%)
Query: 66 CFGRPVRIVEKDMYWFQLTVEEAFYLCYSLKCLKIKINVGADTGPLNDEELWHYFRSKKE 125
C P RI E + QL++EEAF+L Y+L CL I + PL +LW F +
Sbjct: 76 CRRNPYRIFE----YLQLSLEEAFFLVYALGCLSIYY----EKEPLTIVKLWKAFTVVQP 127
Query: 126 TFPYFYKAYSHLRMKNWVVRSGAQYGVDLIVYRHHPARVHSEYGVLVLSHDKDGDLNGRL 185
TF Y AY + R K WV + G +YG DL++YR P H+ Y V++ D G L
Sbjct: 128 TFRTTYMAYHYFRSKGWVPKVGLKYGTDLLLYRKGPPFYHASYSVII--ELVDDHFEGSL 185
Query: 186 R---VWSDVHCTTRLLGGVAKTLLVLYVNK----NGNNDESLLCLTNYTVEERTISRWSP 238
R W + +R+ V+K L++ Y+ K ES C+ V+E +SRW
Sbjct: 186 RRPLSWKSLAALSRVSVNVSKELMLCYLIKPSTMTDKEMESPECMKRIKVQEVILSRW-- 243
Query: 239 EQCRERS 245
RERS
Sbjct: 244 VSSRERS 250
>emb|CAG32411.1| hypothetical protein [Gallus gallus]
Length = 461
Score = 91.7 bits (226), Expect = 2e-17
Identities = 63/187 (33%), Positives = 89/187 (46%), Gaps = 19/187 (10%)
Query: 66 CFGRPVRIVEKDMYWFQLTVEEAFYLCYSLKCLKIKINVGADTGPLNDEELWHYFRSKKE 125
C P RI E + QL++EEAF+L Y+L CL + PL +LW F K
Sbjct: 282 CRRNPFRIFE----YLQLSLEEAFFLVYALGCLTVYYG----EEPLTILKLWEIFSEVKP 333
Query: 126 TFPYFYKAYSHLRMKNWVVRSGAQYGVDLIVYRHHPARVHSEYGVLVLSHDKDGDLNGRL 185
+F Y AY + R K WV + G +YG DL++YR P H+ Y V++ D + G L
Sbjct: 334 SFKTTYMAYHYFRSKGWVPKVGLKYGTDLLLYRKGPPFYHASYS--VIAELVDDNFEGSL 391
Query: 186 R---VWSDVHCTTRLLGGVAKTLLVLYVNKNGNNDESLL----CLTNYTVEERTISRWSP 238
R W + R +K LL+ Y+ K + E + CL V+E ++RW
Sbjct: 392 RRPLSWMSLSGLNRTTVNASKELLLCYLIKPSDMTEEEMATPECLKRIKVQELIVTRW-- 449
Query: 239 EQCRERS 245
RERS
Sbjct: 450 VSSRERS 456
>ref|XP_414309.1| PREDICTED: similar to hypothetical protein MGC2776 [Gallus gallus]
Length = 636
Score = 91.7 bits (226), Expect = 2e-17
Identities = 63/187 (33%), Positives = 89/187 (46%), Gaps = 19/187 (10%)
Query: 66 CFGRPVRIVEKDMYWFQLTVEEAFYLCYSLKCLKIKINVGADTGPLNDEELWHYFRSKKE 125
C P RI E + QL++EEAF+L Y+L CL + PL +LW F K
Sbjct: 457 CRRNPFRIFE----YLQLSLEEAFFLVYALGCLTVYYG----EEPLTILKLWEIFSEVKP 508
Query: 126 TFPYFYKAYSHLRMKNWVVRSGAQYGVDLIVYRHHPARVHSEYGVLVLSHDKDGDLNGRL 185
+F Y AY + R K WV + G +YG DL++YR P H+ Y V++ D + G L
Sbjct: 509 SFKTTYMAYHYFRSKGWVPKVGLKYGTDLLLYRKGPPFYHASYS--VIAELVDDNFEGSL 566
Query: 186 R---VWSDVHCTTRLLGGVAKTLLVLYVNKNGNNDESLL----CLTNYTVEERTISRWSP 238
R W + R +K LL+ Y+ K + E + CL V+E ++RW
Sbjct: 567 RRPLSWMSLSGLNRTTVNASKELLLCYLIKPSDMTEEEMATPECLKRIKVQELIVTRW-- 624
Query: 239 EQCRERS 245
RERS
Sbjct: 625 VSSRERS 631
>dbj|BAC11213.1| unnamed protein product [Homo sapiens]
Length = 439
Score = 85.5 bits (210), Expect = 1e-15
Identities = 58/167 (34%), Positives = 81/167 (47%), Gaps = 15/167 (8%)
Query: 86 EEAFYLCYSLKCLKIKINVGADTGPLNDEELWHYFRSKKETFPYFYKAYSHLRMKNWVVR 145
EEAF+L Y+L CL I + PL +LW F + TF Y AY + R K WV +
Sbjct: 276 EEAFFLVYALGCLSIYY----EKEPLTIVKLWKAFTVVQPTFRTTYMAYHYFRSKGWVPK 331
Query: 146 SGAQYGVDLIVYRHHPARVHSEYGVLVLSHDKDGDLNGRLR---VWSDVHCTTRLLGGVA 202
G +YG DL++YR P H+ Y V++ D G LR W + +R+ V+
Sbjct: 332 VGLKYGTDLLLYRKGPPFYHASYSVII--ELVDDHFEGSLRRPLSWKSLAALSRVSVNVS 389
Query: 203 KTLLVLYVNK----NGNNDESLLCLTNYTVEERTISRWSPEQCRERS 245
K L++ Y+ K ES C+ V+E +SRW RERS
Sbjct: 390 KELMLCYLIKPSTMTDKEMESPECMKRIKVQEVILSRW--VSSRERS 434
>emb|CAG62825.1| unnamed protein product [Candida glabrata CBS138]
gi|50294868|ref|XP_449845.1| unnamed protein product
[Candida glabrata]
Length = 368
Score = 74.3 bits (181), Expect = 3e-12
Identities = 51/172 (29%), Positives = 79/172 (45%), Gaps = 24/172 (13%)
Query: 82 QLTVEEAFYLCYSLKCLKIKINVGADTGPLNDEELWHYFRSKKETFPYFYKAYSHLRMKN 141
+L EA +L ++L L IKIN + E H K Y AY H R
Sbjct: 211 ELMPVEAIFLTFALPVLDIKIN-----DLIKHFEFQHIEELK--VLVRKYAAYHHYRSLK 263
Query: 142 WVVRSGAQYGVDLIVYRHHPARVHSEYGVLVLSHDKDGDLNGRLRVWSDVHCTTRLLGGV 201
W VRSG ++G D ++Y+ P H+E+ ++VL H + D ++ R+ G
Sbjct: 264 WCVRSGIKFGCDYLLYKRGPPFQHAEFAIMVLDHTESHD-------YTWYSSVARVASGA 316
Query: 202 AKTLLVLYVNKNGNNDESLL----------CLTNYTVEERTISRWSPEQCRE 243
KTL++ Y++ G +E+LL L ++ V E T RW P + R+
Sbjct: 317 KKTLILCYIDDQGLTNETLLDLWHQGNLVKLLQHFKVAEVTYKRWIPGKNRD 368
>gb|EAL85905.1| tRNA-splicing endonuclease subunit SEN2,putative [Aspergillus
fumigatus Af293]
Length = 484
Score = 72.0 bits (175), Expect = 1e-11
Identities = 58/194 (29%), Positives = 86/194 (43%), Gaps = 33/194 (17%)
Query: 82 QLTVEEAFYLCYSLKCLKI----KINVGADTGPLNDEELWHYFRSKK--------ETFPY 129
QL+ EEAF+L Y L L + + + T L+ Y+ + + F
Sbjct: 292 QLSNEEAFFLAYGLGVLHVYDHQQKTIIPHTSLLSLFCHNSYYPPRDPSTDLKPDDPFMI 351
Query: 130 FYKAYSHLRMKNWVVRSGAQYGVDLIVYRHHPARVHSEYGVLVL---SHDKDGDLNGR-- 184
Y Y H R WVVRSG ++GVD ++Y P H+E+ V+V+ H + + R
Sbjct: 352 SYIVYHHFRSLGWVVRSGVKFGVDYLLYNRGPVFSHAEFAVVVIPAYEHSYWSETSDRRS 411
Query: 185 ------LRVWSDVHCTTRLLGGVAKTLLVLYV---------NKNGNNDESLLCLTNYTVE 229
R W +HC R+ V K+L+V YV +K + D L L+ Y V
Sbjct: 412 YCATKQARSWWWLHCVNRVQAQVKKSLVVCYVEVPPPRSHDSKTHSEDIGAL-LSRYRVR 470
Query: 230 ERTISRWSPEQCRE 243
E I RW P + R+
Sbjct: 471 EMLIRRWVPNRTRD 484
>ref|XP_593543.1| PREDICTED: similar to hypothetical protein MGC2776, partial [Bos
taurus]
Length = 113
Score = 71.6 bits (174), Expect = 2e-11
Identities = 42/117 (35%), Positives = 60/117 (50%), Gaps = 5/117 (4%)
Query: 88 AFYLCYSLKCLKIKINVGADTGPLNDEELWHYFRSKKETFPYFYKAYSHLRMKNWVVRSG 147
AF+L Y+L CL I + PL +LW+ F + + TF Y AY H R K WV + G
Sbjct: 1 AFFLVYALGCLSIYY----EKEPLTIMKLWNAFSTVQPTFRTTYMAYHHFRSKGWVPKPG 56
Query: 148 AQYGVDLIVYRHHPARVHSEYGVLV-LSHDKDGDLNGRLRVWSDVHCTTRLLGGVAK 203
+YG DL++YR P H+ Y V+V L D+ R W + +R+ V+K
Sbjct: 57 LKYGTDLLLYRKGPPFYHASYSVIVELVDDRFQGAPRRPLSWRSLAALSRVSVSVSK 113
>emb|CAG88258.1| unnamed protein product [Debaryomyces hansenii CBS767]
gi|50422855|ref|XP_460005.1| unnamed protein product
[Debaryomyces hansenii]
Length = 369
Score = 70.9 bits (172), Expect = 3e-11
Identities = 46/183 (25%), Positives = 83/183 (45%), Gaps = 31/183 (16%)
Query: 77 DMYWFQLTVEEAFYLCYSLKCLKIKINVGADTGPLNDEELWHYFRSKKETFPYFYKAYSH 136
D+ + QL E F+L ++L + I +++ + ++ S +F Y Y H
Sbjct: 202 DLEYLQLQKTEVFFLRFALNVINIDLSLSQLFSECCERDI-----SPNNSFILEYVVYHH 256
Query: 137 LRMKNWVVRSGAQYGVDLIVYRHHPARVHSEYGVLVLSHDKDGDLNGRLRVWSDVHCTTR 196
R W VRSG ++G D+++Y+ P H+E+ +L++S D D WS + +R
Sbjct: 257 YRSLGWCVRSGIKFGCDMLLYKRGPPFSHAEHAILIMS-DSHHD-------WSSISSISR 308
Query: 197 LLGGVAKTLLVLYVNKNGNNDESLLCLTN----------------YTVEERTISRWSPEQ 240
++GGV K L++ ++ +G E + N Y + E RW P +
Sbjct: 309 VIGGVKKNLVLTFI--DGPTTEEFDAVVNSSYSCANEKLYNIFKLYKITEILYRRWIPSR 366
Query: 241 CRE 243
R+
Sbjct: 367 NRD 369
>ref|XP_639362.1| hypothetical protein DDB0204993 [Dictyostelium discoideum]
gi|60467996|gb|EAL66007.1| hypothetical protein
DDB0204993 [Dictyostelium discoideum]
Length = 432
Score = 70.9 bits (172), Expect = 3e-11
Identities = 47/181 (25%), Positives = 83/181 (44%), Gaps = 39/181 (21%)
Query: 80 WFQLTVEEAFYLCYSLKCLKI--KINVGADTGPLND-------------EELWHYFRSKK 124
+ QL++ EAFYL YSL CL I + N ND +E W F++ +
Sbjct: 198 YLQLSLCEAFYLIYSLGCLTILRRPNSSDKIIKTNDIDTNKKTMVKMTIQECWKEFQTIE 257
Query: 125 ETFPYFYKAYSHLRMKNWVVRSGAQYGVDLIVYRHHPARVHSEYGVLVL----------- 173
+F Y Y + R W+ RSG +YG D I+Y+ P +H++YG+++
Sbjct: 258 SSFISKYITYHYFRSLGWIPRSGIKYGCDYILYKLKPDLIHAQYGIIIQNRNEQSTNNND 317
Query: 174 -SHDKDGDLNGR------------LRVWSDVHCTTRLLGGVAKTLLVLYVNKNGNNDESL 220
+++ +G++N ++ W ++ R+ VAK ++V + K N+ E
Sbjct: 318 NNNNNNGEINSNQPLFDESSGNRIIQSWDELSGINRVSESVAKGIIVFNIFKPKNHSEKF 377
Query: 221 L 221
+
Sbjct: 378 I 378
Database: nr
Posted date: Jul 5, 2005 12:34 AM
Number of letters in database: 863,360,394
Number of sequences in database: 2,540,612
Lambda K H
0.320 0.136 0.420
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 434,374,292
Number of Sequences: 2540612
Number of extensions: 17994431
Number of successful extensions: 38272
Number of sequences better than 10.0: 65
Number of HSP's better than 10.0 without gapping: 55
Number of HSP's successfully gapped in prelim test: 10
Number of HSP's that attempted gapping in prelim test: 38161
Number of HSP's gapped (non-prelim): 69
length of query: 246
length of database: 863,360,394
effective HSP length: 125
effective length of query: 121
effective length of database: 545,783,894
effective search space: 66039851174
effective search space used: 66039851174
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 73 (32.7 bits)
Medicago: description of AC141114.7


