

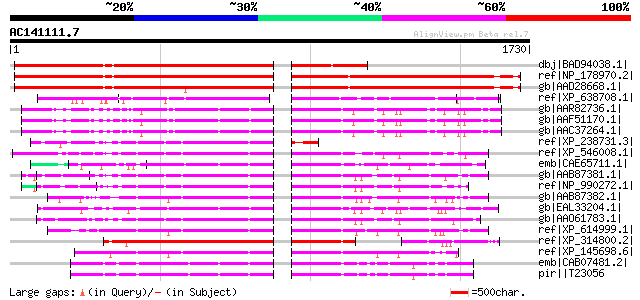
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC141111.7 - phase: 0 /pseudo
(1730 letters)
Database: nr
2,540,612 sequences; 863,360,394 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
dbj|BAD94038.1| pseudogene [Arabidopsis thaliana] 1092 0.0
ref|NP_178970.2| chromodomain-helicase-DNA-binding family protei... 1066 0.0
gb|AAD28668.1| putative chromodomain-helicase-DNA-binding protei... 1058 0.0
ref|XP_638708.1| hypothetical protein DDB0220640 [Dictyostelium ... 478 e-133
gb|AAR82736.1| SD21488p [Drosophila melanogaster] 428 e-118
gb|AAF51170.1| CG3733-PA [Drosophila melanogaster] gi|17137266|r... 428 e-118
gb|AAC37264.1| chromodomain-helicase-DNA-binding protein gi|7511... 427 e-117
ref|XP_238731.3| PREDICTED: similar to Chromodomain-helicase-DNA... 422 e-116
ref|XP_546008.1| PREDICTED: similar to Chromodomain-helicase-DNA... 421 e-115
emb|CAE65711.1| Hypothetical protein CBG10790 [Caenorhabditis br... 419 e-115
gb|AAB87381.1| CHD1 [Homo sapiens] gi|3182949|sp|O14646|CHD1_HUM... 419 e-115
ref|NP_990272.1| chromo-helicase-DNA-binding on the Z chromosome... 418 e-115
gb|AAB87382.1| CHD2 [Homo sapiens] gi|3182950|sp|O14647|CHD2_HUM... 416 e-114
gb|EAL33204.1| GA17649-PA [Drosophila pseudoobscura] 416 e-114
gb|AAO61783.1| chromo-helicase DNA-binding protein [Taeniopygia ... 416 e-114
ref|XP_614999.1| PREDICTED: similar to Chromodomain-helicase-DNA... 414 e-113
ref|XP_314800.2| ENSANGP00000011045 [Anopheles gambiae str. PEST... 413 e-113
ref|XP_145698.6| PREDICTED: similar to Chromodomain-helicase-DNA... 412 e-113
emb|CAB07481.2| Hypothetical protein H06O01.2 [Caenorhabditis el... 410 e-112
pir||T23056 chromodomain helicase H06O01.2 - Caenorhabditis elegans 410 e-112
>dbj|BAD94038.1| pseudogene [Arabidopsis thaliana]
Length = 1221
Score = 1092 bits (2824), Expect = 0.0
Identities = 566/880 (64%), Positives = 679/880 (76%), Gaps = 24/880 (2%)
Query: 15 VGQNADRTRSEKEFYMNMEAQYESDAEPDDASGKQNEAAAVDRLSTRESNVETTSRNPSA 74
+ ++ D T SE+ F MNM+ QY+SD EP + + NE A + +S+ +++++
Sbjct: 34 LNEDVDGTYSERGFDMNMDVQYQSDPEPGCSIRQPNETAVDNVADPVDSHYQSSTKRLGV 93
Query: 75 SERWGSSYLKDCQPMSPQNGSESGDDSKSGSDYRNEDEFEDNSSEGRGEKLGSEDEDGQK 134
+ RWGS++ KDCQPM + GS+ DS+SG Y+ EDN S R EKL SE+E+ +
Sbjct: 94 TGRWGSTFWKDCQPMGQREGSDPAKDSQSG--YKEAYHSEDNHSNDRSEKLDSENENDNE 151
Query: 135 DS------GKGQRGDSDVPAEEMLSDDSYGQDGEEQGESVHSRGFRPSTGSNSCLQPTS- 187
+ K Q G +DVPA+EMLSD+ Y QD + Q + VH +G+ T S S + S
Sbjct: 152 NEEEDNEMNKHQSGQADVPADEMLSDEYYEQDEDNQSDHVHYKGYSNPTNSRSLPKAGSA 211
Query: 188 ----TNVNRRVHRKSRILDDAEDDDDDADYE-EDEPDEDDPDDADFEP--ATSGRGANKY 240
+ +R +H+ D D + DAD + E+E DEDDP+DADFEP A GA+K
Sbjct: 212 VHSNSRTSRAIHKNIHYSDSNHDHNGDADMDYEEEEDEDDPEDADFEPYDAADDGGASKK 271
Query: 241 KDWEGEDSDEVDDSDEDIDVSDNDDLYFDKKAKGRQRGKFGPSVRSTRDCKAFTASSRQR 300
+ SDE +SDE+ID+SD +D Y KK K RQ+ K + + K+F SSRQ+
Sbjct: 272 YGQGWDVSDEDPESDEEIDLSDYEDDYGTKKPKVRQQSKGFRKSSAGLERKSFHVSSRQK 331
Query: 301 RVKSSFEDEDENSTAEDSDSESDEDFKSLKKRGVRVRKNNGRSSAATSFSRPSNEVRSSS 360
R K+S++D+D EDS++++DE F+SL +RG +R+NNGRS T+ S+EVRSS+
Sbjct: 332 R-KTSYQDDDSE---EDSENDNDEGFRSLARRGTTLRQNNGRS---TNTIGQSSEVRSST 384
Query: 361 RTIRKVSYVESDESEGADEG-TKKSQKEEIEVDDGDSVEKVLWHQPKGMAAEAQRNNQSM 419
R++RKVSYVES++SE D+G +K+QK++IE +D D +EKVLWHQ KGM + Q NN+S
Sbjct: 385 RSVRKVSYVESEDSEDIDDGKNRKNQKDDIEEEDADVIEKVLWHQLKGMGEDVQTNNKST 444
Query: 420 EPVLMSHLFDSEPDWNNMEFLIKWKGQSHLHCQWKSFVDLQNLSGFKKVLNYTKRVTEEI 479
PVL+S LFD+EPDWN MEFLIKWKGQSHLHCQWK+ DLQNLSGFKKVLNYTK+VTEEI
Sbjct: 445 VPVLVSQLFDTEPDWNEMEFLIKWKGQSHLHCQWKTLSDLQNLSGFKKVLNYTKKVTEEI 504
Query: 480 RNRMGISREEIEVNDVSKEMDIDIIKQNSQVERIIADRISNDNSGNVFPEYLVKWQGLSY 539
R R +SREEIEVNDVSKEMD+DIIKQNSQVERIIADRIS D G+V PEYLVKWQGLSY
Sbjct: 505 RYRTALSREEIEVNDVSKEMDLDIIKQNSQVERIIADRISKDGLGDVVPEYLVKWQGLSY 564
Query: 540 AEVTWEKDIDIAFAQHTIDEYKTREAAMSVQGKMVDFQRRQSKGSLRKLDEQPEWLKGGK 599
AE TWEKD+DIAFAQ IDEYK RE +++VQGKMV+ QR + K SLRKLDEQPEWL GG
Sbjct: 565 AEATWEKDVDIAFAQVAIDEYKAREVSIAVQGKMVEQQRTKGKASLRKLDEQPEWLIGGT 624
Query: 600 LRDYQLEGLNFLVNSWKNDTNVVLADEMGLGKTVQSVSMLGFLQNAQQIHGPFLVVVPLS 659
LRDYQLEGLNFLVNSW NDTNV+LADEMGLGKTVQSVSMLGFLQN QQI GPFLVVVPLS
Sbjct: 625 LRDYQLEGLNFLVNSWLNDTNVILADEMGLGKTVQSVSMLGFLQNTQQIPGPFLVVVPLS 684
Query: 660 TLSNWAKEFRKWLPDLNVIVYVGTRSSREVCQQYEFCNEKKAGKQIKFNALLTTYEVVLK 719
TL+NWAKEFRKWLP +N+IVYVGTR+SREVCQQYEF NEKK G+ IKFNALLTTYEVVLK
Sbjct: 685 TLANWAKEFRKWLPGMNIIVYVGTRASREVCQQYEFYNEKKVGRPIKFNALLTTYEVVLK 744
Query: 720 DKAVLSKIKWNYLMVDEAHRLKNSEAQLYTALSEFNTKNKLLITGTPLQNSVEELWALLH 779
DKAVLSKIKW YLMVDEAHRLKNSEAQLYTAL EF+TKNKLLITGTPLQNSVEELWALLH
Sbjct: 745 DKAVLSKIKWIYLMVDEAHRLKNSEAQLYTALLEFSTKNKLLITGTPLQNSVEELWALLH 804
Query: 780 FLDSDKFKSKDEFAQNYKNLSSFNENELSNLHMELRPHMLRRVIKDVEKSLPPKIERILR 839
FLD KFK+KDEF +NYKNL+SFNE+EL+NLH+ELRPH+LRRVIKDVEKSLPPKIERILR
Sbjct: 805 FLDPGKFKNKDEFVENYKNLNSFNESELANLHLELRPHILRRVIKDVEKSLPPKIERILR 864
Query: 840 VDMSPLQKQYYKWILERNFRDLNKGVRGNQSKYCSRIEEV 879
V+MSPLQKQYYKWILERNF DLNKGVRGNQ + + E+
Sbjct: 865 VEMSPLQKQYYKWILERNFHDLNKGVRGNQVSLLNIVVEL 904
Score = 426 bits (1096), Expect = e-117
Identities = 217/254 (85%), Positives = 232/254 (90%), Gaps = 2/254 (0%)
Query: 938 DILAQYMSLRGFQFQRLDGSTKSELRQQAMDHFNAPGSDDFCFLLSTRAGGLGINLATAD 997
DILA+Y+SLRGFQFQRLDGSTK+ELRQQAMDHFNAP SDDFCFLLSTRAGGLGINLATAD
Sbjct: 969 DILAEYLSLRGFQFQRLDGSTKAELRQQAMDHFNAPASDDFCFLLSTRAGGLGINLATAD 1028
Query: 998 TVIIFDSDWNPQNDLQAMSRAHRIGQREVVNIYRFVTSKSVEEDILERAKKKMVLDHLVI 1057
TV+IFDSDWNPQNDLQAMSRAHRIGQ+EVVNIYRFVTSKSVEE+ILERAK+KMVLDHLVI
Sbjct: 1029 TVVIFDSDWNPQNDLQAMSRAHRIGQQEVVNIYRFVTSKSVEEEILERAKRKMVLDHLVI 1088
Query: 1058 QKLNAEGKLEKKEAKKGGSFFDKNELSAILRFGAEELFKEERNDEESKKRLLSMDIDEIL 1117
QKLNAEG+LEK+E KK GS FDKNELSAILRFGAEELFKE++NDEESKKRLLSMDIDEIL
Sbjct: 1089 QKLNAEGRLEKRETKK-GSNFDKNELSAILRFGAEELFKEDKNDEESKKRLLSMDIDEIL 1147
Query: 1118 ERAEKVEEKENGGEQAHELLSAFKVANFCNDEDDGSFWSRWIKADSVAQAENALAPRAAR 1177
ERAE+VEEK E HELL AFKVANFCN EDDGSFWSRWIK DSV AE ALAPRAAR
Sbjct: 1148 ERAEQVEEKHT-DETEHELLGAFKVANFCNAEDDGSFWSRWIKPDSVVTAEEALAPRAAR 1206
Query: 1178 NIKSYAEADQSERS 1191
N KSY + +R+
Sbjct: 1207 NTKSYVDPSHPDRT 1220
>ref|NP_178970.2| chromodomain-helicase-DNA-binding family protein / CHD family
protein [Arabidopsis thaliana]
Length = 1722
Score = 1066 bits (2758), Expect = 0.0
Identities = 558/880 (63%), Positives = 672/880 (75%), Gaps = 26/880 (2%)
Query: 15 VGQNADRTRSEKEFYMNMEAQYESDAEPDDASGKQNEAAAVDRLSTRESNVETTSRNPSA 74
+ ++ D T SE+ F MNM+ QY+SD EP + + NE A + +S+ +++++
Sbjct: 34 LNEDVDGTYSERGFDMNMDVQYQSDPEPGCSIRQPNETAVDNVADPVDSHYQSSTKRLGV 93
Query: 75 SERWGSSYLKDCQPMSPQNGSESGDDSKSGSDYRNEDEFEDNSSEGRGEKLGSEDEDGQK 134
+ RWGS++ KDCQPM + GS+ DS+SG Y+ EDN S R EKL SE+E+ +
Sbjct: 94 TGRWGSTFWKDCQPMGQREGSDPAKDSQSG--YKEAYHSEDNHSNDRSEKLDSENENDNE 151
Query: 135 DS------GKGQRGDSDVPAEEMLSDDSYGQDGEEQGESVHSRGFRPSTGSNSCLQPTS- 187
+ K Q G +DVPA+EMLSD+ Y QD + Q + VH +G+ T S S + S
Sbjct: 152 NEEEDNEMNKHQSGQADVPADEMLSDEYYEQDEDNQSDHVHYKGYSNPTNSRSLPKAGSA 211
Query: 188 ----TNVNRRVHRKSRILDDAEDDDDDADYE-EDEPDEDDPDDADFEP--ATSGRGANKY 240
+ +R +H+ D D + DAD + E+E DEDDP+DADFEP A GA+K
Sbjct: 212 VHSNSRTSRAIHKNIHYSDSNHDHNGDADMDYEEEEDEDDPEDADFEPYDAADDGGASKK 271
Query: 241 KDWEGEDSDEVDDSDEDIDVSDNDDLYFDKKAKGRQRGKFGPSVRSTRDCKAFTASSRQR 300
+ SDE +SDE+ID+SD +D Y KK K RQ+ K + + K+F SSRQ+
Sbjct: 272 HGQGWDVSDEDPESDEEIDLSDYEDDYGTKKPKVRQQSKGFRKSSAGLERKSFHVSSRQK 331
Query: 301 RVKSSFEDEDENSTAEDSDSESDEDFKSLKKRGVRVRKNNGRSSAATSFSRPSNEVRSSS 360
R K+S++D+D EDS++++DE F+SL +RG +R+NNGRS T+ S+EVRSS+
Sbjct: 332 R-KTSYQDDDSE---EDSENDNDEGFRSLARRGTTLRQNNGRS---TNTIGQSSEVRSST 384
Query: 361 RTIRKVSYVESDESEGADEG-TKKSQKEEIEVDDGDSVEKVLWHQPKGMAAEAQRNNQSM 419
R++RKVSYVES++SE D+G +K+QK++IE +D D +EKVLWHQ KGM + Q NN+S
Sbjct: 385 RSVRKVSYVESEDSEDIDDGKNRKNQKDDIEEEDADVIEKVLWHQLKGMGEDVQTNNKST 444
Query: 420 EPVLMSHLFDSEPDWNNMEFLIKWKGQSHLHCQWKSFVDLQNLSGFKKVLNYTKRVTEEI 479
PVL+S LFD+EPDWN MEFLIKWKGQSHLHCQWK+ DLQNLSGFKKVLNYTK+VTEEI
Sbjct: 445 VPVLVSQLFDTEPDWNEMEFLIKWKGQSHLHCQWKTLSDLQNLSGFKKVLNYTKKVTEEI 504
Query: 480 RNRMGISREEIEVNDVSKEMDIDIIKQNSQVERIIADRISNDNSGNVFPEYLVKWQGLSY 539
R R +SREEIEVNDVSKEMD+DIIKQNSQVERIIADRIS D G+V PEYLVKWQGLSY
Sbjct: 505 RYRTALSREEIEVNDVSKEMDLDIIKQNSQVERIIADRISKDGLGDVVPEYLVKWQGLSY 564
Query: 540 AEVTWEKDIDIAFAQHTIDEYKTREAAMSVQGKMVDFQRRQSKGSLRKLDEQPEWLKGGK 599
AE TWEKD+DIAFAQ IDEYK RE +++VQGKMV+ QR + K SLRKLDEQPEWL GG
Sbjct: 565 AEATWEKDVDIAFAQVAIDEYKAREVSIAVQGKMVEQQRTKGKASLRKLDEQPEWLIGGT 624
Query: 600 LRDYQLEGLNFLVNSWKNDTNVVLADEMGLGKTVQSVSMLGFLQNAQQIHGPFLVVVPLS 659
LRDYQLEGLNFLVNSW NDTNV+LADEMGLGKTVQSVSMLGFLQN QQI GPFLVVVPLS
Sbjct: 625 LRDYQLEGLNFLVNSWLNDTNVILADEMGLGKTVQSVSMLGFLQNTQQIPGPFLVVVPLS 684
Query: 660 TLSNWAKEFRKWLPDLNVIVYVGTRSSREVCQQYEFCNEKKAGKQIKFNALLTTYEVVLK 719
TL+NWAKEFRKWLP +N+IVYVGTR+SREV + + K G+ IKFNALLTTYEVVLK
Sbjct: 685 TLANWAKEFRKWLPGMNIIVYVGTRASREVRNKTN--DVHKVGRPIKFNALLTTYEVVLK 742
Query: 720 DKAVLSKIKWNYLMVDEAHRLKNSEAQLYTALSEFNTKNKLLITGTPLQNSVEELWALLH 779
DKAVLSKIKW YLMVDEAHRLKNSEAQLYTAL EF+TKNKLLITGTPLQNSVEELWALLH
Sbjct: 743 DKAVLSKIKWIYLMVDEAHRLKNSEAQLYTALLEFSTKNKLLITGTPLQNSVEELWALLH 802
Query: 780 FLDSDKFKSKDEFAQNYKNLSSFNENELSNLHMELRPHMLRRVIKDVEKSLPPKIERILR 839
FLD KFK+KDEF +NYKNLSSFNE+EL+NLH+ELRPH+LRRVIKDVEKSLPPKIERILR
Sbjct: 803 FLDPGKFKNKDEFVENYKNLSSFNESELANLHLELRPHILRRVIKDVEKSLPPKIERILR 862
Query: 840 VDMSPLQKQYYKWILERNFRDLNKGVRGNQSKYCSRIEEV 879
V+MSPLQKQYYKWILERNF DLNKGVRGNQ + + E+
Sbjct: 863 VEMSPLQKQYYKWILERNFHDLNKGVRGNQVSLLNIVVEL 902
Score = 1005 bits (2598), Expect = 0.0
Identities = 529/773 (68%), Positives = 610/773 (78%), Gaps = 39/773 (5%)
Query: 938 DILAQYMSLRGFQFQRLDGSTKSELRQQAMDHFNAPGSDDFCFLLSTRAGGLGINLATAD 997
DILA+Y+SLRGFQFQRLDGSTK+ELRQQAMDHFNAP SDDFCFLLSTRAGGLGINLATAD
Sbjct: 967 DILAEYLSLRGFQFQRLDGSTKAELRQQAMDHFNAPASDDFCFLLSTRAGGLGINLATAD 1026
Query: 998 TVIIFDSDWNPQNDLQAMSRAHRIGQREVVNIYRFVTSKSVEEDILERAKKKMVLDHLVI 1057
TV+IFDSDWNPQNDLQAMSRAHRIGQ+EVVNIYRFVTSKSVEE+ILERAK+KMVLDHLVI
Sbjct: 1027 TVVIFDSDWNPQNDLQAMSRAHRIGQQEVVNIYRFVTSKSVEEEILERAKRKMVLDHLVI 1086
Query: 1058 QKLNAEGKLEKKEAKKGGSFFDKNELSAILRFGAEELFKEERNDEESKKRLLSMDIDEIL 1117
QKLNAEG+LEK+E KKG +F DKNELSAILRFGAEELFKE++NDEESKKRLLSMDIDEIL
Sbjct: 1087 QKLNAEGRLEKRETKKGSNF-DKNELSAILRFGAEELFKEDKNDEESKKRLLSMDIDEIL 1145
Query: 1118 ERAEKVEEKENGGEQAHELLSAFKVANFCNDEDDGSFWSRWIKADSVAQAENALAPRAAR 1177
ERAE+VEEK E HELL AFKVANFCN EDDGSFWSRWIK DSV AE ALAPRAAR
Sbjct: 1146 ERAEQVEEKHTD-ETEHELLGAFKVANFCNAEDDGSFWSRWIKPDSVVTAEEALAPRAAR 1204
Query: 1178 NIKSYAEADQSERSKKRKKKENEP---TERIPKRRKADYSAHVISMIDGASAQVRSWSYG 1234
N KSY + +R+ KRKKK +EP TER KRRK +Y +++G SAQVR WSYG
Sbjct: 1205 NTKSYVDPSHPDRTSKRKKKGSEPPEHTERSQKRRKTEYFVPSTPLLEGTSAQVRGWSYG 1264
Query: 1235 NLSKRDALRFSRSVMKFGNESQINLIVAEVGGAIEAAPLKAQVELFNALIDGCREAVEVG 1294
NL KRDA RF R+VMKFGN +Q+ I EVGG +EAAP +AQVELF+ALIDGC+E+VE G
Sbjct: 1265 NLPKRDAQRFYRTVMKFGNHNQMACIAEEVGGVVEAAPEEAQVELFDALIDGCKESVETG 1324
Query: 1295 SLDLKGPLLDFYGVPMKANELLIRVQELQLLAKRISRYEDPIAQFRVLTYLKPSNWSKGC 1354
+ + KGP+LDF+GVP+KANELL RVQ LQLL+KRISRY DPI+QFRVL+YLKPSNWSKGC
Sbjct: 1325 NFEPKGPVLDFFGVPVKANELLKRVQGLQLLSKRISRYNDPISQFRVLSYLKPSNWSKGC 1384
Query: 1355 GWNQIDDARLLLGVHYHGYGNWEVIRLDERLGLTKKIAPVELQHHETFLPRAPNLRDRAN 1414
GWNQIDDARLLLG+ YHG+GNWE IRLDE LGLTKKIAPVELQHHETFLPRAPNL++RA
Sbjct: 1385 GWNQIDDARLLLGILYHGFGNWEKIRLDESLGLTKKIAPVELQHHETFLPRAPNLKERAT 1444
Query: 1415 ALLEQELAVLGVKNASSKVGRKTSKKEREEREHLVDISLSRGQEKKKNIGSSKVNVQMRK 1474
ALLE ELA G KN ++K RK SKK +++L++ + ++++ G + V++ K
Sbjct: 1445 ALLEMELAAAGGKNTNAKASRKNSKK---VKDNLINQFKAPARDRRGKSGPANVSLLSTK 1501
Query: 1475 DRLQKPLNVEPIVKEEGEMSDDDDVYEQFKEGKWKEWCQDLMVEEMKTLKRLHRLQTTSA 1534
D +K EP+VKEEGEMSDD +VYEQFKE KW EWC+D++ +E+KTL RL RLQTTSA
Sbjct: 1502 DGPRKTQKAEPLVKEEGEMSDDGEVYEQFKEQKWMEWCEDVLADEIKTLGRLQRLQTTSA 1561
Query: 1535 SLPKEKVLSKIRNYLQLLGRRIDQIVSEQEDEPHKQDRMTTRLWKYVSTFSHLSGERLHQ 1594
LPKEKVL KIR YL++LGRRID IV E E++ +KQDRMT RLW YVSTFS+LSG+RL+Q
Sbjct: 1562 DLPKEKVLFKIRRYLEILGRRIDAIVLEHEEDLYKQDRMTMRLWNYVSTFSNLSGDRLNQ 1621
Query: 1595 IYSKLKLE-QNAVGVGSSLPNGSVSGPFSRNGNPNSSFPRPMERQTRFQNVTAHPMREQT 1653
IYSKLK E + GVG S NGS R +RQ +F+ +Q
Sbjct: 1622 IYSKLKQEKEEEEGVGPSHLNGS----------------RNFQRQQKFKTAGNSQGSQQV 1665
Query: 1654 Y---DTGMSEAWKRRRRAENDGCFQGQPPPQRITSNGIRPLDPNSLGILGAGP 1703
+ DT EAWKRRRR END Q + P IT++ NSLGILG GP
Sbjct: 1666 HKGIDTAKFEAWKRRRRTEND--VQTERP--TITNS-------NSLGILGPGP 1707
>gb|AAD28668.1| putative chromodomain-helicase-DNA-binding protein [Arabidopsis
thaliana] gi|25411492|pir||C84507 hypothetical protein
At2g13370 [imported] - Arabidopsis thaliana
Length = 1738
Score = 1058 bits (2737), Expect = 0.0
Identities = 559/896 (62%), Positives = 673/896 (74%), Gaps = 42/896 (4%)
Query: 15 VGQNADRTRSEKEFYMNMEAQYESDAEPDDASGKQNEAAAVDRLSTRESNVETTSRNPSA 74
+ ++ D T SE+ F MNM+ QY+SD EP + + NE A + +S+ +++++
Sbjct: 34 LNEDVDGTYSERGFDMNMDVQYQSDPEPGCSIRQPNETAVDNVADPVDSHYQSSTKRLGV 93
Query: 75 SERWGSSYLKDCQPMSPQNGSESGDDSKSGSDYRNEDEFEDNSSEGRGEKLGSEDEDGQK 134
+ RWGS++ KDCQPM + GS+ DS+SG Y+ EDN S R EKL SE+E+ +
Sbjct: 94 TGRWGSTFWKDCQPMGQREGSDPAKDSQSG--YKEAYHSEDNHSNDRSEKLDSENENDNE 151
Query: 135 DS------GKGQRGDSDVPAEEMLSDDSYGQDGEEQGESVHSRGFRPSTGSNSCLQPTS- 187
+ K Q G +DVPA+EMLSD+ Y QD + Q + VH +G+ T S S + S
Sbjct: 152 NEEEDNEMNKHQSGQADVPADEMLSDEYYEQDEDNQSDHVHYKGYSNPTNSRSLPKAGSA 211
Query: 188 ----TNVNRRVHRKSRILDDAEDDDDDADYE-EDEPDEDDPDDADFEP--ATSGRGANKY 240
+ +R +H+ D D + DAD + E+E DEDDP+DADFEP A GA+K
Sbjct: 212 VHSNSRTSRAIHKNIHYSDSNHDHNGDADMDYEEEEDEDDPEDADFEPYDAADDGGASKK 271
Query: 241 KDWEGEDSDEVDDSDEDIDVSDNDDLYFDKKAKGRQRGKFGPSVRSTRDCKAFTASSRQR 300
+ SDE +SDE+ID+SD +D Y KK K RQ+ K + + K+F SSRQ+
Sbjct: 272 HGQGWDVSDEDPESDEEIDLSDYEDDYGTKKPKVRQQSKGFRKSSAGLERKSFHVSSRQK 331
Query: 301 RVKSSFEDEDENSTAEDSDSESDEDFKSLKKRGVRVRKNNGRSSAATSFSRPSNEVRSSS 360
R K+S++D+D EDS++++DE F+SL +RG +R+NNGRS T+ S+EVRSS+
Sbjct: 332 R-KTSYQDDDSE---EDSENDNDEGFRSLARRGTTLRQNNGRS---TNTIGQSSEVRSST 384
Query: 361 RTIRKVSYVESDESEGADEG-TKKSQKEEIEVDDGDSVEKVLWHQPKGMAAEAQRNNQSM 419
R++RKVSYVES++SE D+G +K+QK++IE +D D +EKVLWHQ KGM + Q NN+S
Sbjct: 385 RSVRKVSYVESEDSEDIDDGKNRKNQKDDIEEEDADVIEKVLWHQLKGMGEDVQTNNKST 444
Query: 420 EPVLMSHLFDSEPDWNNMEFLIKWKGQSHLHCQWKSFVDLQNLSGFKKVLNYTKRVTEEI 479
PVL+S LFD+EPDWN MEFLIKWKGQSHLHCQWK+ DLQNLSGFKKVLNYTK+VTEEI
Sbjct: 445 VPVLVSQLFDTEPDWNEMEFLIKWKGQSHLHCQWKTLSDLQNLSGFKKVLNYTKKVTEEI 504
Query: 480 RNRMGISREEIEVNDVSKEMDIDIIKQNSQVERIIADRISNDNSGNVFPEYLVKWQGLSY 539
R R +SREEIEVNDVSKEMD+DIIKQNSQVERIIADRIS D G+V PEYLVKWQGLSY
Sbjct: 505 RYRTALSREEIEVNDVSKEMDLDIIKQNSQVERIIADRISKDGLGDVVPEYLVKWQGLSY 564
Query: 540 AEVTWEKDIDIAFAQHTIDEYKTREAAMSVQGKMVDFQRRQSKG---------------- 583
AE TWEKD+DIAFAQ IDEYK RE +++VQGKMV+ QR + KG
Sbjct: 565 AEATWEKDVDIAFAQVAIDEYKAREVSIAVQGKMVEQQRTKGKGENSFSNAELWLLFSVA 624
Query: 584 SLRKLDEQPEWLKGGKLRDYQLEGLNFLVNSWKNDTNVVLADEMGLGKTVQSVSMLGFLQ 643
SLRKLDEQPEWL GG LRDYQLEGLNFLVNSW NDTNV+LADEMGLGKTVQSVSMLGFLQ
Sbjct: 625 SLRKLDEQPEWLIGGTLRDYQLEGLNFLVNSWLNDTNVILADEMGLGKTVQSVSMLGFLQ 684
Query: 644 NAQQIHGPFLVVVPLSTLSNWAKEFRKWLPDLNVIVYVGTRSSREVCQQYEFCNEKKAGK 703
N QQI GPFLVVVPLSTL+NWAKEFRKWLP +N+IVYVGTR+SREV + + K G+
Sbjct: 685 NTQQIPGPFLVVVPLSTLANWAKEFRKWLPGMNIIVYVGTRASREVRNKTN--DVHKVGR 742
Query: 704 QIKFNALLTTYEVVLKDKAVLSKIKWNYLMVDEAHRLKNSEAQLYTALSEFNTKNKLLIT 763
IKFNALLTTYEVVLKDKAVLSKIKW YLMVDEAHRLKNSEAQLYTAL EF+TKNKLLIT
Sbjct: 743 PIKFNALLTTYEVVLKDKAVLSKIKWIYLMVDEAHRLKNSEAQLYTALLEFSTKNKLLIT 802
Query: 764 GTPLQNSVEELWALLHFLDSDKFKSKDEFAQNYKNLSSFNENELSNLHMELRPHMLRRVI 823
GTPLQNSVEELWALLHFLD KFK+KDEF +NYKNLSSFNE+EL+NLH+ELRPH+LRRVI
Sbjct: 803 GTPLQNSVEELWALLHFLDPGKFKNKDEFVENYKNLSSFNESELANLHLELRPHILRRVI 862
Query: 824 KDVEKSLPPKIERILRVDMSPLQKQYYKWILERNFRDLNKGVRGNQSKYCSRIEEV 879
KDVEKSLPPKIERILRV+MSPLQKQYYKWILERNF DLNKGVRGNQ + + E+
Sbjct: 863 KDVEKSLPPKIERILRVEMSPLQKQYYKWILERNFHDLNKGVRGNQVSLLNIVVEL 918
Score = 1005 bits (2598), Expect = 0.0
Identities = 529/773 (68%), Positives = 610/773 (78%), Gaps = 39/773 (5%)
Query: 938 DILAQYMSLRGFQFQRLDGSTKSELRQQAMDHFNAPGSDDFCFLLSTRAGGLGINLATAD 997
DILA+Y+SLRGFQFQRLDGSTK+ELRQQAMDHFNAP SDDFCFLLSTRAGGLGINLATAD
Sbjct: 983 DILAEYLSLRGFQFQRLDGSTKAELRQQAMDHFNAPASDDFCFLLSTRAGGLGINLATAD 1042
Query: 998 TVIIFDSDWNPQNDLQAMSRAHRIGQREVVNIYRFVTSKSVEEDILERAKKKMVLDHLVI 1057
TV+IFDSDWNPQNDLQAMSRAHRIGQ+EVVNIYRFVTSKSVEE+ILERAK+KMVLDHLVI
Sbjct: 1043 TVVIFDSDWNPQNDLQAMSRAHRIGQQEVVNIYRFVTSKSVEEEILERAKRKMVLDHLVI 1102
Query: 1058 QKLNAEGKLEKKEAKKGGSFFDKNELSAILRFGAEELFKEERNDEESKKRLLSMDIDEIL 1117
QKLNAEG+LEK+E KKG +F DKNELSAILRFGAEELFKE++NDEESKKRLLSMDIDEIL
Sbjct: 1103 QKLNAEGRLEKRETKKGSNF-DKNELSAILRFGAEELFKEDKNDEESKKRLLSMDIDEIL 1161
Query: 1118 ERAEKVEEKENGGEQAHELLSAFKVANFCNDEDDGSFWSRWIKADSVAQAENALAPRAAR 1177
ERAE+VEEK E HELL AFKVANFCN EDDGSFWSRWIK DSV AE ALAPRAAR
Sbjct: 1162 ERAEQVEEKHTD-ETEHELLGAFKVANFCNAEDDGSFWSRWIKPDSVVTAEEALAPRAAR 1220
Query: 1178 NIKSYAEADQSERSKKRKKKENEP---TERIPKRRKADYSAHVISMIDGASAQVRSWSYG 1234
N KSY + +R+ KRKKK +EP TER KRRK +Y +++G SAQVR WSYG
Sbjct: 1221 NTKSYVDPSHPDRTSKRKKKGSEPPEHTERSQKRRKTEYFVPSTPLLEGTSAQVRGWSYG 1280
Query: 1235 NLSKRDALRFSRSVMKFGNESQINLIVAEVGGAIEAAPLKAQVELFNALIDGCREAVEVG 1294
NL KRDA RF R+VMKFGN +Q+ I EVGG +EAAP +AQVELF+ALIDGC+E+VE G
Sbjct: 1281 NLPKRDAQRFYRTVMKFGNHNQMACIAEEVGGVVEAAPEEAQVELFDALIDGCKESVETG 1340
Query: 1295 SLDLKGPLLDFYGVPMKANELLIRVQELQLLAKRISRYEDPIAQFRVLTYLKPSNWSKGC 1354
+ + KGP+LDF+GVP+KANELL RVQ LQLL+KRISRY DPI+QFRVL+YLKPSNWSKGC
Sbjct: 1341 NFEPKGPVLDFFGVPVKANELLKRVQGLQLLSKRISRYNDPISQFRVLSYLKPSNWSKGC 1400
Query: 1355 GWNQIDDARLLLGVHYHGYGNWEVIRLDERLGLTKKIAPVELQHHETFLPRAPNLRDRAN 1414
GWNQIDDARLLLG+ YHG+GNWE IRLDE LGLTKKIAPVELQHHETFLPRAPNL++RA
Sbjct: 1401 GWNQIDDARLLLGILYHGFGNWEKIRLDESLGLTKKIAPVELQHHETFLPRAPNLKERAT 1460
Query: 1415 ALLEQELAVLGVKNASSKVGRKTSKKEREEREHLVDISLSRGQEKKKNIGSSKVNVQMRK 1474
ALLE ELA G KN ++K RK SKK +++L++ + ++++ G + V++ K
Sbjct: 1461 ALLEMELAAAGGKNTNAKASRKNSKK---VKDNLINQFKAPARDRRGKSGPANVSLLSTK 1517
Query: 1475 DRLQKPLNVEPIVKEEGEMSDDDDVYEQFKEGKWKEWCQDLMVEEMKTLKRLHRLQTTSA 1534
D +K EP+VKEEGEMSDD +VYEQFKE KW EWC+D++ +E+KTL RL RLQTTSA
Sbjct: 1518 DGPRKTQKAEPLVKEEGEMSDDGEVYEQFKEQKWMEWCEDVLADEIKTLGRLQRLQTTSA 1577
Query: 1535 SLPKEKVLSKIRNYLQLLGRRIDQIVSEQEDEPHKQDRMTTRLWKYVSTFSHLSGERLHQ 1594
LPKEKVL KIR YL++LGRRID IV E E++ +KQDRMT RLW YVSTFS+LSG+RL+Q
Sbjct: 1578 DLPKEKVLFKIRRYLEILGRRIDAIVLEHEEDLYKQDRMTMRLWNYVSTFSNLSGDRLNQ 1637
Query: 1595 IYSKLKLE-QNAVGVGSSLPNGSVSGPFSRNGNPNSSFPRPMERQTRFQNVTAHPMREQT 1653
IYSKLK E + GVG S NGS R +RQ +F+ +Q
Sbjct: 1638 IYSKLKQEKEEEEGVGPSHLNGS----------------RNFQRQQKFKTAGNSQGSQQV 1681
Query: 1654 Y---DTGMSEAWKRRRRAENDGCFQGQPPPQRITSNGIRPLDPNSLGILGAGP 1703
+ DT EAWKRRRR END Q + P IT++ NSLGILG GP
Sbjct: 1682 HKGIDTAKFEAWKRRRRTEND--VQTERP--TITNS-------NSLGILGPGP 1723
>ref|XP_638708.1| hypothetical protein DDB0220640 [Dictyostelium discoideum]
gi|60467321|gb|EAL65353.1| CHD gene family protein
containing chromodomain, helicase domain, and DNA-binding
domain [Dictyostelium discoideum]
Length = 1917
Score = 478 bits (1230), Expect = e-133
Identities = 305/863 (35%), Positives = 451/863 (51%), Gaps = 102/863 (11%)
Query: 93 NGSESGDDSKSGSDYRN---EDEFEDNSSEGRGEKLGSEDEDGQKDSGKGQRGDSDVPAE 149
N S SG+++ + ++ N + +NS+ + + +G K +G+ +E
Sbjct: 167 NNSNSGNNNNNNNNNNNIFNQTIPNNNSNNNNNNNNINNNNNGLKKDISNFKGNESSSSE 226
Query: 150 EMLSDDSYGQDGEEQGESVHSRGFRPSTG-----SNSCLQPTSTNVNRRVHRKSRILDDA 204
S ++ E Q ++ + + + T S LQP + + V + +
Sbjct: 227 SSSESSSSEEESESQEDTPNKQRNKVLTQQLASESEEELQPVNKKKSTMVKKVDSDFESE 286
Query: 205 ED-------DDDDADYEEDEPDE--------DDPDDADFEPATSGRGANKYK-------D 242
ED D+D+DYE E +E D DD + G G K
Sbjct: 287 EDVEEEQSGKDNDSDYEPSESEEQSEEDYESDTDDDDVYTKKKRGGGGRKQTASTTTTTT 346
Query: 243 WEGEDSDEVDDSDEDIDVSDNDDLYFDKKAKGRQRGKFGPSVRSTRDCKAFTASSRQRRV 302
G + + S+ D + S++D Y K K +Q + + ++R RR
Sbjct: 347 RSGRQAKRITYSESDSEESESDSDYEPNKKKKKQTNGRKRKGNNFQKENWTIPATRARRG 406
Query: 303 KSSFEDEDENSTAE---------DSDSESDEDF----KSLKKRGVRVRKNNGRSSAATSF 349
K++ + E+E++ ++ S S SDED+ K KK RKN S ++
Sbjct: 407 KAAIDSEEESNGSDGDQFVNNVFSSSSSSDEDYSDNDKKNKKSKRTKRKNKTESDEESTI 466
Query: 350 SRPSNEVRSSSRTIRKVSYVESDESEGAD---------EGTKKSQKEEIEVDDGDSVEKV 400
+ + R S S SD+ EG D G S+ + K
Sbjct: 467 THLNLRTRGSGNI--DFSIFNSDDEEGEDGSGGGGGSGSGLSTSETTNSSTAAATASSKP 524
Query: 401 LWHQPKGMAAEAQRNNQSMEPVLMSHLFDSEPDWNNME---FLIKWKGQSHLHCQWKSFV 457
L ++P A+ + ++ L ++EP+ N+E FL+KWKG +++H W ++
Sbjct: 525 LIYEP----ADEDVIEKILDHRLKKDCSENEPEIFNVEDYEFLVKWKGWAYIHNTWDAYE 580
Query: 458 DLQNLSGFKKVLNYTKRVTEEIRNRMGISREEIEVNDVSKEMDIDIIKQNSQVERIIADR 517
L N G KK+ NY K + + I+ R S+E+IE D++KE+ I+ + VER+IA R
Sbjct: 581 SLNNFKGNKKLQNYVKGILDNIQWRQEASKEDIEQADITKELMKQEIQDYTIVERVIAQR 640
Query: 518 --------------------------------ISNDNSGNVFPEYLVKWQGLSYAEVTWE 545
S++ N+ +YLVKW+ L+Y+EVTWE
Sbjct: 641 EVAQSRDQIVNSTTAIDPSTGESTTTTTTTTASSSEYKSNI--QYLVKWKVLAYSEVTWE 698
Query: 546 KDIDIAFAQHTIDEYKTREAAMS---VQGKMVDFQRRQSKGSLRKLDEQPEWLKGGKLRD 602
DIA Q ID + R+ ++ + ++R +G KLD QP W+ G LRD
Sbjct: 699 YPEDIAEFQTEIDTFLIRQQNNQNAPMKANTISAKKRLDQG-FTKLDTQPSWISAGTLRD 757
Query: 603 YQLEGLNFLVNSWKNDTNVVLADEMGLGKTVQSVSMLGFLQNAQQIHGPFLVVVPLSTLS 662
YQ+EGLN+LV+SW N+TNV+LADEMGLGKT+Q++S L +L N Q I GPFLVVVPLST+
Sbjct: 758 YQMEGLNWLVHSWMNNTNVILADEMGLGKTIQTISFLSYLFNEQDIKGPFLVVVPLSTIE 817
Query: 663 NWAKEFRKWLPDLNVIVYVGTRSSREVCQQYEFCNEKKAG-KQIKFNALLTTYEVVLKDK 721
NW +EF KW P +NVIVY GT SR++ + YEF + G K++ FN LLTTY+ +LKDK
Sbjct: 818 NWQREFAKWAPAMNVIVYTGTGQSRDIIRLYEFYTTNRLGKKKLNFNVLLTTYDFILKDK 877
Query: 722 AVLSKIKWNYLMVDEAHRLKNSEAQLYTALSEFNTKNKLLITGTPLQNSVEELWALLHFL 781
L IKW +L VDEAHRLKNSE+ L+ L +NT N+LL+TGTPLQNS++ELW LL+FL
Sbjct: 878 NTLGTIKWEFLAVDEAHRLKNSESVLHEVLKLYNTTNRLLVTGTPLQNSLKELWNLLNFL 937
Query: 782 DSDKFKSKDEFAQNYKNLSSFNENELSNLHMELRPHMLRRVIKDVEKSLPPKIERILRVD 841
+KF S +F Y +L ++++ LH L+PH+LRR+ KDVEKSLPPK ERILRVD
Sbjct: 938 MPNKFTSLKDFQDQYSDLK--ENDQIAQLHSVLKPHLLRRIKKDVEKSLPPKTERILRVD 995
Query: 842 MSPLQKQYYKWILERNFRDLNKG 864
+S +QK+YYKWIL +NF++LNKG
Sbjct: 996 LSNVQKKYYKWILTKNFQELNKG 1018
Score = 311 bits (797), Expect = 1e-82
Identities = 250/716 (34%), Positives = 345/716 (47%), Gaps = 64/716 (8%)
Query: 938 DILAQYMSLRGFQFQRLDGSTKSELRQQAMDHFNAPGSDDFCFLLSTRAGGLGINLATAD 997
DILA Y+ R FQFQRLDGS E R QAMD FNA S DFCFLLST+AGGLGINL+TAD
Sbjct: 1095 DILADYLKGRSFQFQRLDGSMSREKRSQAMDRFNAVDSPDFCFLLSTKAGGLGINLSTAD 1154
Query: 998 TVIIFDSDWNPQNDLQAMSRAHRIGQREVVNIYRFVTSKSVEEDILERAKKKMVLDHLVI 1057
TVIIFDSDWNPQNDLQA +RAHRIGQ+ VNIYR V+ SVEEDILERAK+KMVLDHLVI
Sbjct: 1155 TVIIFDSDWNPQNDLQAEARAHRIGQKNHVNIYRLVSKSSVEEDILERAKQKMVLDHLVI 1214
Query: 1058 QKLNAEGKLEKKEAKKGGSFFDKNELSAILRFGAEELFKEERNDEESKKRLLSMDIDEIL 1117
Q + + + K + F+K EL AIL+FGAEELFKE E + MDIDEIL
Sbjct: 1215 QTME-KSQTAKSNTPNNSNVFNKEELEAILKFGAEELFKE---TGEEANPIEEMDIDEIL 1270
Query: 1118 ERAEKVEEKENGGEQAHELLSAFKVANFCNDEDDGSFWSRWIKADSVAQAENALAPRAAR 1177
RAE E + ELL++F+VANF + + W + P A R
Sbjct: 1271 SRAE-TREASDSTTAGEELLNSFRVANFSTTSKEEQQNATW----------EDIIPDADR 1319
Query: 1178 NIKSYAEADQSERSKKRKKKENEPTERIPKRRKADYSAHVISMIDGASAQVRSWSYGNLS 1237
E DQ R++ E + P R +D +A+++ +
Sbjct: 1320 QKAVQEEKDQLLLLGPRRRTETS-SSTAPTRNISDPVV--------ITARLKKDINQIYN 1370
Query: 1238 KRDALRFSRSVMKFGNESQINLIVAEVGGAIEAAPLKAQVELFNALIDGCREAVEVGSLD 1297
K++ RS+ KFG + I+A+ + PL+ E+ +ID +AV + D
Sbjct: 1371 KKEIKILIRSLKKFGTHKRCREILADAD--FQNKPLRPTEEICKEIIDAAEKAVR-DNQD 1427
Query: 1298 LKGPLLDFYGVPMKANELLIRVQELQLLAKRISRYEDPIAQFRVLTYLKPSNWSKGCGWN 1357
L + G + A E + R+Q++++LA+ + QFRV ++P +W+ W
Sbjct: 1428 SDKIHLVYNGTDINATEFVQRIQDMEVLAELCQPWIKDYQQFRVTFPIRPVSWT--IKWG 1485
Query: 1358 QIDDARLLLGVHYHGYGNWEVIRLDERLGLTKKI-----APVELQHHETFLP----RAPN 1408
+DA LL+G++ HG NWE I+ D LG I AP+ L + +P +
Sbjct: 1486 PKEDAMLLMGIYKHGVSNWESIQKDTSLGFGDIIEIDNTAPL-LDANGVPIPSNKLKGAA 1544
Query: 1409 LRDRANALLEQELAVLGVKNASSKVGRKTSKKEREEREHLVDISLSRGQEKK--KNIGSS 1466
L+ R ++LL+ K+ S +K+SK+ +E+ E I RG++ K K + ++
Sbjct: 1545 LQRRVDSLLK------SAKDISKTTNKKSSKQTKEKEEGEFKIPKKRGRKSKEIKEVSNN 1598
Query: 1467 KVNVQMRKDRLQKPLNVEPIVKE---------EGEMSDDDDVYEQFKEGKWKEWCQDLMV 1517
N + K+ N + SDDDD + K+ E +
Sbjct: 1599 NHNNKETKENNNGDNNNNNNNNNSSNNRKRTTNNKYSDDDDDDDNDKDNDNDEDYMEEEE 1658
Query: 1518 EEMKTLKRLHRLQTTSASLPKEKVLSKIRNYLQLLGRRIDQIVSEQEDEPHKQDRMTTRL 1577
++ KT + + S L KE + K ++ GR E P K TT
Sbjct: 1659 KKEKTKNTRTKKRIKSEELQKEILSPKKTSF----GRLSVPPNKMGERSPSKSTTTTTTT 1714
Query: 1578 WKYVSTFS----HLSGERLHQIYSKLKLEQNAVGVGSSLPNGSVSGPFSRNGNPNS 1629
S S H S + K G G S + S S SRN N S
Sbjct: 1715 TTSSSKSSRNSKHHSDDDDDDYRDKDSNSGRGRGRGRSSNSSSSSSSSSRNKNHES 1770
Score = 52.8 bits (125), Expect = 1e-04
Identities = 34/147 (23%), Positives = 68/147 (46%), Gaps = 4/147 (2%)
Query: 1488 KEEGEMSDDDDVYEQFKEGKWKEWCQDLMVEEMKTLKRLHRLQTTSASLPKEKVLSKIRN 1547
+ + SD++D +++ + CQ + + L+ L + + +E+ +SK +
Sbjct: 1764 RNKNHESDEED--DKYYDKNLLRKCQQYLSPIKELLENFRSLSDSKVQISREEKVSKTKK 1821
Query: 1548 YLQLLGRRIDQIVSEQEDEPHKQDRMTTRLWKYVSTFSHLSGERLHQIYSKLKLEQNAVG 1607
YL LG+ I I+ E++ K + + LW S ++ G+ L +Y K+K ++
Sbjct: 1822 YLLSLGKEISIILDEKKSANIKD--LDSHLWYSASKYTASGGKELKSLYDKMKKSSSSNN 1879
Query: 1608 VGSSLPNGSVSGPFSRNGNPNSSFPRP 1634
SS + S S N + +SS +P
Sbjct: 1880 DSSSSSSSRSSNSSSSNNSNSSSSSKP 1906
Score = 49.7 bits (117), Expect = 9e-04
Identities = 65/307 (21%), Positives = 122/307 (39%), Gaps = 68/307 (22%)
Query: 93 NGSESGDDSKSGSDYRNEDEFEDNSSEGRGEKLGSEDEDGQKDSGKGQRGDSDVPAEEML 152
N S + + + Y ++D+ +DN + ++ E+E+ +K+ K R + +EE+
Sbjct: 1620 NNSSNNRKRTTNNKYSDDDDDDDNDKDNDNDEDYMEEEE-KKEKTKNTRTKKRIKSEELQ 1678
Query: 153 SD------DSYGQ---DGEEQGESVHSRGFRPSTGSNSCLQPTSTNVNRRVHRKSRILDD 203
+ S+G+ + GE S+ +T T+T + + R S+
Sbjct: 1679 KEILSPKKTSFGRLSVPPNKMGERSPSKSTTTTT--------TTTTSSSKSSRNSK---- 1726
Query: 204 AEDDDDDADYEEDEPDEDDPDDADFEPATSGRGANKYKDWEGEDSDEVDDSDEDIDVSDN 263
DDDD DY + + + SGRG + + S +++ + +
Sbjct: 1727 HHSDDDDDDYRDKD-------------SNSGRGRGRGRSSNSSSSSSSSSRNKNHESDEE 1773
Query: 264 DDLYFDKKAKGRQRGKFGP------SVRSTRDCKAFTASSRQRRVK-------------S 304
DD Y+DK + + P + RS D K SR+ +V S
Sbjct: 1774 DDKYYDKNLLRKCQQYLSPIKELLENFRSLSDSK--VQISREEKVSKTKKYLLSLGKEIS 1831
Query: 305 SFEDEDENSTAEDSDS-----------ESDEDFKSLKKRGVRVRKNNGRSSAATSFSRPS 353
DE +++ +D DS ++ KSL + + +N SS+++S SR S
Sbjct: 1832 IILDEKKSANIKDLDSHLWYSASKYTASGGKELKSLYDKMKKSSSSNNDSSSSSS-SRSS 1890
Query: 354 NEVRSSS 360
N S++
Sbjct: 1891 NSSSSNN 1897
Score = 41.6 bits (96), Expect = 0.24
Identities = 50/235 (21%), Positives = 95/235 (40%), Gaps = 31/235 (13%)
Query: 121 RGEKLGSEDEDGQKDSGKGQRGDSDVPAEEMLSDDSYGQDGEEQGESVHSRGFRPSTGSN 180
+ K E E+G+ K +RG +E+ +++ ++ +E + G N
Sbjct: 1566 KSSKQTKEKEEGEFKIPK-KRGRKSKEIKEVSNNNHNNKETKEN-----------NNGDN 1613
Query: 181 SCLQPTSTNVNRRVHRKSRILDDAEDDDDDADYEEDEPDEDDPDDADFEPATSGRGANKY 240
+ + N N +RK + DDDDD D ++D +++D + + + + K
Sbjct: 1614 N---NNNNNNNSSNNRKRTTNNKYSDDDDDDDNDKDNDNDEDYMEEEEKKEKTKNTRTKK 1670
Query: 241 KDWEGEDSDEVDDSDEDIDVSDNDDLYFDKKAKGRQRGKFGPSVRSTRDCKAFTASSRQR 300
+ E E+ +S + + G+ PS +T T+SS+
Sbjct: 1671 RIKSEELQKEI--------LSPKKTSFGRLSVPPNKMGERSPSKSTTTTTTTTTSSSKSS 1722
Query: 301 RVKSSFEDEDENSTAEDSDSESDEDFKSLKKRGVRVRKNNGRSSAATSFSRPSNE 355
R D+D+ D D+D S + RG R R +N SS+++S ++E
Sbjct: 1723 RNSKHHSDDDD-------DDYRDKDSNSGRGRG-RGRSSNSSSSSSSSSRNKNHE 1769
Score = 37.7 bits (86), Expect = 3.5
Identities = 45/231 (19%), Positives = 88/231 (37%), Gaps = 31/231 (13%)
Query: 16 GQNADRTRSEKEFYMNMEAQYESDAEPDDASGKQNEAAAVDRLSTRESNVETTSRNPSAS 75
G++ D E E YESD + DD K+ + + S TT+R+ +
Sbjct: 295 GKDNDSDYEPSESEEQSEEDYESDTDDDDVYTKKKRGGGGRKQTA--STTTTTTRSGRQA 352
Query: 76 ERWGSSYLKDCQPMSPQNGSESGDDSKSGSDYRNEDEFEDNSSEGRGEKLGSEDEDGQKD 135
+R S ++S+S SDY + + ++ + + + E+
Sbjct: 353 KRITYS-------------ESDSEESESDSDYEPNKKKKKQTNGRKRKGNNFQKENWTIP 399
Query: 136 SGKGQRGDSDVPAEEMLSDDSYGQDGEEQGESVHSRGFRPSTGSNSCLQPTSTNVNRRVH 195
+ + +RG + + +EE +S G DG++ +V S S+ + ++R
Sbjct: 400 ATRARRGKAAIDSEE----ESNGSDGDQFVNNVFSSS--SSSDEDYSDNDKKNKKSKRTK 453
Query: 196 RKSRILDDAED----------DDDDADYEEDEPDEDDPDDADFEPATSGRG 236
RK++ D E + D+ D+++ +D SG G
Sbjct: 454 RKNKTESDEESTITHLNLRTRGSGNIDFSIFNSDDEEGEDGSGGGGGSGSG 504
>gb|AAR82736.1| SD21488p [Drosophila melanogaster]
Length = 1645
Score = 428 bits (1101), Expect = e-118
Identities = 286/862 (33%), Positives = 451/862 (52%), Gaps = 120/862 (13%)
Query: 38 SDAEPDDASGKQNEAAAVDRLSTRESNVE----TTSRNPSASERWGSSYLKDCQPMSPQN 93
SD++ D +SG ++ + + +V T + + S+ G + ++ +
Sbjct: 41 SDSDSDSSSGNSSDGRSSPEPEDKSLSVAGFPPTAAAAQADSKTNGFTDDQEDSSSDGSS 100
Query: 94 GSESGDDSKSGSDYRNEDEFEDNSSEGRGEKLGSEDEDGQKDSGKGQRGDSDVPAEEMLS 153
GS+S D++ SD RN+ N+S + +E+ED + ++G+ Q PA + +
Sbjct: 101 GSDSDSDAEGPSDQRNQSINNANTSSSLPKPEQNEEEDNETEAGQQQ------PASDASA 154
Query: 154 DDSYGQDGEEQGESVHSRGFRPSTGSNSCLQPTSTNVNRRVHRKSRILDDAEDDDDDADY 213
D+S + S++ + PTS++ +E++++D
Sbjct: 155 DES--------------------SDSSANVSPTSSS------------SSSEEEEEDYRP 182
Query: 214 EEDEPDEDDPDDADFEPATSGRGANKYKDWEGEDSDEVDDSDEDIDVSDNDDLYFDKKAK 273
+ P A+ + + NK K W+ ++SDE +DSD+++ + K A
Sbjct: 183 KRTRQARKPPTAAE-KSKKAPAPKNKKKTWDSDESDESEDSDDEVSTAQKR-----KPAA 236
Query: 274 GRQRGKFGPSVRSTRDCKAFTASSRQRRVKSSFEDEDENSTAEDSDSESDEDFKSLKKRG 333
R K A +QRR F ED SD+D S K+
Sbjct: 237 TTSRSKL--------------AQQQQRRRVKPFSSED-----------SDDDDAS-KRCA 270
Query: 334 VRVRKNNGRSSAATSFSRPSNEVRSSSRTIRKVSYVESDESEGADEGTKKSQKEEIEVDD 393
R R AA S+ S + + S + + Y DES+ A ++E+ E
Sbjct: 271 TR------RKGAAVSYKEASEDEATDSEDLLEFEY---DESQAATTAATAEEEEKCE--- 318
Query: 394 GDSVEKVLWHQPKGMAAEAQRNNQSMEPVLMSHLFDSEPDWNNME---------FLIKWK 444
++E++L + NQ+ + + FD ++ + FLIKWK
Sbjct: 319 --TIERILAQRA---GKRGCTGNQTTIYAIEENGFDPHAGFDEKQTSDAETEAQFLIKWK 373
Query: 445 GQSHLHCQWKSFVDLQNLS--GFKKVLNYTKRVTEEIRNRMGISREEIEVNDVSKEMDID 502
G S++H W+S L+++ G KK+ N+ K+ E+ R E+I+ + E+ +
Sbjct: 374 GWSYIHNTWESEATLRDMKAKGMKKLDNFIKKEKEQAYWRRYAGPEDIDYFECQLELQHE 433
Query: 503 IIKQNSQVERIIADRISNDNSGNVFPEYLVKWQGLSYAEVTWEKDIDIAFA----QHTID 558
++K + V+RIIA D+ EYL KWQ L YAE TWE D A Q +
Sbjct: 434 LLKSYNNVDRIIAKGSKPDDGTE---EYLCKWQSLPYAESTWE---DAALVLRKWQRCAE 487
Query: 559 EYKTREAAMSVQGKMVDFQRRQSKGSLRKLDEQPEWLKGG-KLRDYQLEGLNFLVNSWKN 617
++ RE++ + + + K S ++ QPE+L G LRDYQ++GLN+L++SW
Sbjct: 488 QFNDRESSKCTPSRHCRVIKYRPKFS--RIKNQPEFLSSGLTLRDYQMDGLNWLLHSWCK 545
Query: 618 DTNVVLADEMGLGKTVQSVSMLGFLQNAQQIHGPFLVVVPLSTLSNWAKEFRKWLPDLNV 677
+ +V+LADEMGLGKT+Q++ L L ++GPFL VVPLST++ W +EF W PD+NV
Sbjct: 546 ENSVILADEMGLGKTIQTICFLYSLFKIHHLYGPFLCVVPLSTMTAWQREFDLWAPDMNV 605
Query: 678 IVYVGTRSSREVCQQYEFCNEKKAGKQIKFNALLTTYEVVLKDKAVLSKIKWNYLMVDEA 737
+ Y+G SRE+ QQYE+ + ++ K++KFN +LTTYE+VLKDK L ++W L+VDEA
Sbjct: 606 VTYLGDIKSRELIQQYEW--QFESSKRLKFNCILTTYEIVLKDKQFLGTLQWAALLVDEA 663
Query: 738 HRLKNSEAQLYTALSEFNTKNKLLITGTPLQNSVEELWALLHFLDSDKFKSKDEFAQNYK 797
HRLKN ++ LY +L EF+T ++LLITGTPLQNS++ELWALLHF+ DKF + + F +
Sbjct: 664 HRLKNDDSLLYKSLKEFDTNHRLLITGTPLQNSLKELWALLHFIMPDKFDTWENFEVQHG 723
Query: 798 NLSSFNENELSNLHMELRPHMLRRVIKDVEKSLPPKIERILRVDMSPLQKQYYKWILERN 857
N + + LH +L P++LRRV KDVEKSLP K+E+ILRV+M+ LQKQYYKWIL +N
Sbjct: 724 NA---EDKGYTRLHQQLEPYILRRVKKDVEKSLPAKVEQILRVEMTSLQKQYYKWILTKN 780
Query: 858 FRDLNKGVRGNQSKYCSRIEEV 879
F L KG RG+ S + + + E+
Sbjct: 781 FDALRKGKRGSTSTFLNIVIEL 802
Score = 291 bits (746), Expect = 1e-76
Identities = 252/793 (31%), Positives = 367/793 (45%), Gaps = 117/793 (14%)
Query: 938 DILAQYMSLRGFQFQRLDGSTKSELRQQAMDHFNAPGSDDFCFLLSTRAGGLGINLATAD 997
D+LA Y+ R F FQRLDGS K E+R+QA+DHFNA GS DFCFLLSTRAGGLGINLATAD
Sbjct: 866 DVLADYLQKRHFPFQRLDGSIKGEMRRQALDHFNAEGSQDFCFLLSTRAGGLGINLATAD 925
Query: 998 TVIIFDSDWNPQNDLQAMSRAHRIGQREVVNIYRFVTSKSVEEDILERAKKKMVLDHLVI 1057
TVIIFDSDWNPQNDLQA +RAHRIGQ+ VNIYR VT++SVEE I+ERAK+KMVLDHLVI
Sbjct: 926 TVIIFDSDWNPQNDLQAQARAHRIGQKNQVNIYRLVTARSVEEQIVERAKQKMVLDHLVI 985
Query: 1058 QKLNAEGKLEKKEAKKGGSF----FDKNELSAILRFGAEELFKEERNDEESKKRLLSMDI 1113
Q+++ G+ ++ G S F+K++LSAIL+FGAEELFK+E+ ++ L DI
Sbjct: 986 QRMDTTGRTVLDKSGNGHSSNSNPFNKDDLSAILKFGAEELFKDEQEHDDD----LVCDI 1041
Query: 1114 DEILERAEKVEEKENGGEQAHELLSAFKVA------------------NFCNDEDDGSFW 1155
DEIL RAE E+ A +LLSAFKVA N +EDD W
Sbjct: 1042 DEILRRAE--TRNEDPEMPADDLLSAFKVASIAAFEEEPSDSVSKQDQNAAGEEDDSKDW 1099
Query: 1156 SRWIKADSVAQAENALAPRAARNI----KSYAEADQSERSKKRKKKENEPTERIPKRRKA 1211
I ++ + ++ + A+Q+E + K + +
Sbjct: 1100 DDIIPEGFRKAIDDQERAKEMEDLYLPPRRKTAANQNEGKRGAGKGGKGKQQADDSGGDS 1159
Query: 1212 DYSAHVISMIDGASAQVRSWSYGNLSKRDAL---------RFSRSVMKFGNESQINLIVA 1262
DY DG+ R G + ++ + RF RS KF +A
Sbjct: 1160 DYELG----SDGSGDDGRPRKRGRPTMKEKITGFTDAELRRFIRSYKKFPAPLHRMEAIA 1215
Query: 1263 EVGGAIEAAPLKAQVELFNALIDGC---------REAVEVGSLDLKGP---------LLD 1304
++ PL L L D C E+ + + G +
Sbjct: 1216 -CDAELQEKPLAELKRLGEMLHDRCVQFLHEHKEEESKTAATDETPGAKQRRARATFSVK 1274
Query: 1305 FYGVPMKANELLIRVQELQLLAKRISRYEDPIAQFRVLTYLKPSNWSKGCGWNQIDDARL 1364
GV A +LL QELQ L + + + Q+ +K W +D +L
Sbjct: 1275 LGGVSFNAKKLLACEQELQPLNEIMPSMPEERQQWSF--NIKTRAPVFDVDWGIEEDTKL 1332
Query: 1365 LLGVHYHGYGNWEVIRLDERLGLTKKIAPVELQHHETFLPRAPNLRDRANALLEQELAVL 1424
L G++ +G G+WE ++LD L LT KI ++T P+A L+ RA LL+ ++
Sbjct: 1333 LCGIYQYGIGSWEQMKLDPTLKLTDKIL-----LNDTRKPQAKQLQTRAEYLLK----II 1383
Query: 1425 GVKNASSKVGRKTSKKEREEREH-----------LVDISLSRGQEKKKNIGSSKVNVQMR 1473
+K G++ ++ R R + +D G++ + + + +
Sbjct: 1384 KKNVELTKGGQRRQRRPRASRANDAKAASQSASSTIDAKPHDGEDAAGDARTVAESSNSQ 1443
Query: 1474 KDRLQKPLNVEPIVKEEGE----------------MSDDDD----VYEQFKEGKWKEWCQ 1513
D + P ++ G+ SD++ + E + E
Sbjct: 1444 VDPSNASPHNAPATEQHGDPAKKAKKSKARSKKTSASDNNGNKPMHFTANNEPRALEVLG 1503
Query: 1514 DL-------MVEEMKTLKR-LHRLQTTSASLPKEKVLSKIRNYLQLLGRRIDQIVSEQED 1565
DL E+M+ +K+ L L SL + L R+ L +G++ID ++ +
Sbjct: 1504 DLDPSIFNECKEKMRPVKKALKALDQPDVSLSDQDQLQHTRDCLLQIGKQIDVCLNPYAE 1563
Query: 1566 EPHKQDRMTTRLWKYVSTFSHLSGERLHQIYSKLKLEQNAVGVGSSLPNGSVSGPFSRNG 1625
K+ R + LW +VS F+ L +RL +IY K L+Q A G G + S
Sbjct: 1564 TEKKEWR--SNLWYFVSKFTELDAKRLFKIY-KHALKQKAGGDGEAKGKDKGSSGSPAKS 1620
Query: 1626 NPNSSFPRPMERQ 1638
PN E++
Sbjct: 1621 KPNGVTTEEKEKE 1633
>gb|AAF51170.1| CG3733-PA [Drosophila melanogaster] gi|17137266|ref|NP_477197.1|
CG3733-PA [Drosophila melanogaster]
Length = 1883
Score = 428 bits (1101), Expect = e-118
Identities = 286/862 (33%), Positives = 451/862 (52%), Gaps = 120/862 (13%)
Query: 38 SDAEPDDASGKQNEAAAVDRLSTRESNVE----TTSRNPSASERWGSSYLKDCQPMSPQN 93
SD++ D +SG ++ + + +V T + + S+ G + ++ +
Sbjct: 41 SDSDSDSSSGNSSDGRSSPEPEDKSLSVAGFPPTAAAAQADSKTNGFTDDQEDSSSDGSS 100
Query: 94 GSESGDDSKSGSDYRNEDEFEDNSSEGRGEKLGSEDEDGQKDSGKGQRGDSDVPAEEMLS 153
GS+S D++ SD RN+ N+S + +E+ED + ++G+ Q PA + +
Sbjct: 101 GSDSDSDAEGPSDQRNQSINNANTSSSLPKPEQNEEEDNETEAGQQQ------PASDASA 154
Query: 154 DDSYGQDGEEQGESVHSRGFRPSTGSNSCLQPTSTNVNRRVHRKSRILDDAEDDDDDADY 213
D+S + S++ + PTS++ +E++++D
Sbjct: 155 DES--------------------SDSSANVSPTSSS------------SSSEEEEEDYRP 182
Query: 214 EEDEPDEDDPDDADFEPATSGRGANKYKDWEGEDSDEVDDSDEDIDVSDNDDLYFDKKAK 273
+ P A+ + + NK K W+ ++SDE +DSD+++ + K A
Sbjct: 183 KRTRQARKPPTAAE-KSKKAPAPKNKKKTWDSDESDESEDSDDEVSTAQKR-----KPAA 236
Query: 274 GRQRGKFGPSVRSTRDCKAFTASSRQRRVKSSFEDEDENSTAEDSDSESDEDFKSLKKRG 333
R K A +QRR F ED SD+D S K+
Sbjct: 237 TTSRSKL--------------AQQQQRRRVKPFSSED-----------SDDDDAS-KRCA 270
Query: 334 VRVRKNNGRSSAATSFSRPSNEVRSSSRTIRKVSYVESDESEGADEGTKKSQKEEIEVDD 393
R R AA S+ S + + S + + Y DES+ A ++E+ E
Sbjct: 271 TR------RKGAAVSYKEASEDEATDSEDLLEFEY---DESQAATTAATAEEEEKCE--- 318
Query: 394 GDSVEKVLWHQPKGMAAEAQRNNQSMEPVLMSHLFDSEPDWNNME---------FLIKWK 444
++E++L + NQ+ + + FD ++ + FLIKWK
Sbjct: 319 --TIERILAQRA---GKRGCTGNQTTIYAIEENGFDPHAGFDEKQTPDAETEAQFLIKWK 373
Query: 445 GQSHLHCQWKSFVDLQNLS--GFKKVLNYTKRVTEEIRNRMGISREEIEVNDVSKEMDID 502
G S++H W+S L+++ G KK+ N+ K+ E+ R E+I+ + E+ +
Sbjct: 374 GWSYIHNTWESEATLRDMKAKGMKKLDNFIKKEKEQAYWRRYAGPEDIDYFECQLELQHE 433
Query: 503 IIKQNSQVERIIADRISNDNSGNVFPEYLVKWQGLSYAEVTWEKDIDIAFA----QHTID 558
++K + V+RIIA D+ EYL KWQ L YAE TWE D A Q +
Sbjct: 434 LLKSYNNVDRIIAKGSKPDDGTE---EYLCKWQSLPYAESTWE---DAALVLRKWQRCAE 487
Query: 559 EYKTREAAMSVQGKMVDFQRRQSKGSLRKLDEQPEWLKGG-KLRDYQLEGLNFLVNSWKN 617
++ RE++ + + + K S ++ QPE+L G LRDYQ++GLN+L++SW
Sbjct: 488 QFNDRESSKCTPSRHCRVIKYRPKFS--RIKNQPEFLSSGLTLRDYQMDGLNWLLHSWCK 545
Query: 618 DTNVVLADEMGLGKTVQSVSMLGFLQNAQQIHGPFLVVVPLSTLSNWAKEFRKWLPDLNV 677
+ +V+LADEMGLGKT+Q++ L L ++GPFL VVPLST++ W +EF W PD+NV
Sbjct: 546 ENSVILADEMGLGKTIQTICFLYSLFKIHHLYGPFLCVVPLSTMTAWQREFDLWAPDMNV 605
Query: 678 IVYVGTRSSREVCQQYEFCNEKKAGKQIKFNALLTTYEVVLKDKAVLSKIKWNYLMVDEA 737
+ Y+G SRE+ QQYE+ + ++ K++KFN +LTTYE+VLKDK L ++W L+VDEA
Sbjct: 606 VTYLGDIKSRELIQQYEW--QFESSKRLKFNCILTTYEIVLKDKQFLGTLQWAALLVDEA 663
Query: 738 HRLKNSEAQLYTALSEFNTKNKLLITGTPLQNSVEELWALLHFLDSDKFKSKDEFAQNYK 797
HRLKN ++ LY +L EF+T ++LLITGTPLQNS++ELWALLHF+ DKF + + F +
Sbjct: 664 HRLKNDDSLLYKSLKEFDTNHRLLITGTPLQNSLKELWALLHFIMPDKFDTWENFEVQHG 723
Query: 798 NLSSFNENELSNLHMELRPHMLRRVIKDVEKSLPPKIERILRVDMSPLQKQYYKWILERN 857
N + + LH +L P++LRRV KDVEKSLP K+E+ILRV+M+ LQKQYYKWIL +N
Sbjct: 724 NA---EDKGYTRLHQQLEPYILRRVKKDVEKSLPAKVEQILRVEMTSLQKQYYKWILTKN 780
Query: 858 FRDLNKGVRGNQSKYCSRIEEV 879
F L KG RG+ S + + + E+
Sbjct: 781 FDALRKGKRGSTSTFLNIVIEL 802
Score = 291 bits (745), Expect = 1e-76
Identities = 252/793 (31%), Positives = 367/793 (45%), Gaps = 117/793 (14%)
Query: 938 DILAQYMSLRGFQFQRLDGSTKSELRQQAMDHFNAPGSDDFCFLLSTRAGGLGINLATAD 997
D+LA Y+ R F FQRLDGS K E+R+QA+DHFNA GS DFCFLLSTRAGGLGINLATAD
Sbjct: 866 DVLADYLQKRHFPFQRLDGSIKGEMRRQALDHFNAEGSQDFCFLLSTRAGGLGINLATAD 925
Query: 998 TVIIFDSDWNPQNDLQAMSRAHRIGQREVVNIYRFVTSKSVEEDILERAKKKMVLDHLVI 1057
TVIIFDSDWNPQNDLQA +RAHRIGQ+ VNIYR VT++SVEE I+ERAK+KMVLDHLVI
Sbjct: 926 TVIIFDSDWNPQNDLQAQARAHRIGQKNQVNIYRLVTARSVEEQIVERAKQKMVLDHLVI 985
Query: 1058 QKLNAEGKLEKKEAKKGGSF----FDKNELSAILRFGAEELFKEERNDEESKKRLLSMDI 1113
Q+++ G+ ++ G S F+K++LSAIL+FGAEELFK+E+ ++ L DI
Sbjct: 986 QRMDTTGRTVLDKSGNGHSSNSNPFNKDDLSAILKFGAEELFKDEQEHDDD----LVCDI 1041
Query: 1114 DEILERAEKVEEKENGGEQAHELLSAFKVA------------------NFCNDEDDGSFW 1155
DEIL RAE E+ A +LLSAFKVA N +EDD W
Sbjct: 1042 DEILRRAE--TRNEDPEMPADDLLSAFKVASIAAFEEEPSDSVSKQDQNAAGEEDDSKDW 1099
Query: 1156 SRWIKADSVAQAENALAPRAARNI----KSYAEADQSERSKKRKKKENEPTERIPKRRKA 1211
I ++ + ++ + A+Q+E + K + +
Sbjct: 1100 DDIIPEGFRKAIDDQERAKEMEDLYLPPRRKTAANQNEGKRGAGKGGKGKQQADDSGGDS 1159
Query: 1212 DYSAHVISMIDGASAQVRSWSYGNLSKRDAL---------RFSRSVMKFGNESQINLIVA 1262
DY DG+ R G + ++ + RF RS KF +A
Sbjct: 1160 DYELG----SDGSGDDGRPRKRGRPTMKEKITGFTDAELRRFIRSYKKFPAPLHRMEAIA 1215
Query: 1263 EVGGAIEAAPLKAQVELFNALIDGC---------REAVEVGSLDLKGP---------LLD 1304
++ PL L L D C E+ + + G +
Sbjct: 1216 -CDAELQEKPLAELKRLGEMLHDRCVQFLHEHKEEESKTAATDETPGAKQRRARATFSVK 1274
Query: 1305 FYGVPMKANELLIRVQELQLLAKRISRYEDPIAQFRVLTYLKPSNWSKGCGWNQIDDARL 1364
GV A +LL QELQ L + + + Q+ +K W +D +L
Sbjct: 1275 LGGVSFNAKKLLACEQELQPLNEIMPSMPEERQQWSF--NIKTRAPVFDVDWGIEEDTKL 1332
Query: 1365 LLGVHYHGYGNWEVIRLDERLGLTKKIAPVELQHHETFLPRAPNLRDRANALLEQELAVL 1424
L G++ +G G+WE ++LD L LT KI ++T P+A L+ RA LL+ ++
Sbjct: 1333 LCGIYQYGIGSWEQMKLDPTLKLTDKIL-----LNDTRKPQAKQLQTRAEYLLK----II 1383
Query: 1425 GVKNASSKVGRKTSKKEREEREH-----------LVDISLSRGQEKKKNIGSSKVNVQMR 1473
+K G++ ++ R R + +D G++ + + + +
Sbjct: 1384 KKNVELTKGGQRRQRRPRASRANDAKAASQSASSTIDAKPHDGEDAAGDARTVAESSNSQ 1443
Query: 1474 KDRLQKPLNVEPIVKEEGE----------------MSDDDD----VYEQFKEGKWKEWCQ 1513
D + P ++ G+ SD++ + E + E
Sbjct: 1444 VDPSTASPHNAPATEQHGDPAKKAKKSKARSKKTSASDNNGNKPMHFTANNEPRALEVLG 1503
Query: 1514 DL-------MVEEMKTLKR-LHRLQTTSASLPKEKVLSKIRNYLQLLGRRIDQIVSEQED 1565
DL E+M+ +K+ L L SL + L R+ L +G++ID ++ +
Sbjct: 1504 DLDPSIFNECKEKMRPVKKALKALDQPDVSLSDQDQLQHTRDCLLQIGKQIDVCLNPYAE 1563
Query: 1566 EPHKQDRMTTRLWKYVSTFSHLSGERLHQIYSKLKLEQNAVGVGSSLPNGSVSGPFSRNG 1625
K+ R + LW +VS F+ L +RL +IY K L+Q A G G + S
Sbjct: 1564 TEKKEWR--SNLWYFVSKFTELDAKRLFKIY-KHALKQKAGGDGEAKGKDKGSSGSPAKS 1620
Query: 1626 NPNSSFPRPMERQ 1638
PN E++
Sbjct: 1621 KPNGVTTEEKEKE 1633
>gb|AAC37264.1| chromodomain-helicase-DNA-binding protein gi|7511822|pir||T13944
chromodomain helicase - fruit fly (Drosophila
melanogaster)
Length = 1883
Score = 427 bits (1099), Expect = e-117
Identities = 286/862 (33%), Positives = 451/862 (52%), Gaps = 120/862 (13%)
Query: 38 SDAEPDDASGKQNEAAAVDRLSTRESNVE----TTSRNPSASERWGSSYLKDCQPMSPQN 93
SD++ D +SG ++ + + +V T + + S+ G + ++ +
Sbjct: 41 SDSDSDSSSGNSSDGRSSPEPEDKSLSVAGFPPTAAAAQADSKTNGFTDDQEDSSSDGSS 100
Query: 94 GSESGDDSKSGSDYRNEDEFEDNSSEGRGEKLGSEDEDGQKDSGKGQRGDSDVPAEEMLS 153
GS+S D++ SD RN+ N+S + +E+ED + ++G+ Q PA + +
Sbjct: 101 GSDSDSDAEGPSDQRNQSINNANTSSSLPKPEQNEEEDNETEAGQQQ------PASDASA 154
Query: 154 DDSYGQDGEEQGESVHSRGFRPSTGSNSCLQPTSTNVNRRVHRKSRILDDAEDDDDDADY 213
D+S + S++ + PTS++ +E++++D
Sbjct: 155 DES--------------------SDSSANVSPTSSS------------SSSEEEEEDYRP 182
Query: 214 EEDEPDEDDPDDADFEPATSGRGANKYKDWEGEDSDEVDDSDEDIDVSDNDDLYFDKKAK 273
+ P A+ + + NK K W+ ++SDE +DSD+++ + K A
Sbjct: 183 KRTRQARKPPTAAE-KSKKAPAPKNKKKTWDSDESDESEDSDDEVSTAQKR-----KPAA 236
Query: 274 GRQRGKFGPSVRSTRDCKAFTASSRQRRVKSSFEDEDENSTAEDSDSESDEDFKSLKKRG 333
R K A +QRR F ED SD+D S K+
Sbjct: 237 TTSRSKL--------------AQQQQRRRVKPFSSED-----------SDDDDAS-KRCA 270
Query: 334 VRVRKNNGRSSAATSFSRPSNEVRSSSRTIRKVSYVESDESEGADEGTKKSQKEEIEVDD 393
R R AA S+ S + + S + + Y DES+ A ++E+ E
Sbjct: 271 TR------RKGAAVSYKEASEDEATDSEDLLEFEY---DESQAATTAATAEEEEKCE--- 318
Query: 394 GDSVEKVLWHQPKGMAAEAQRNNQSMEPVLMSHLFDSEPDWNNME---------FLIKWK 444
++E++L + NQ+ + + FD ++ + FLIKWK
Sbjct: 319 --TIERILAQRA---GKRGCTGNQTTIYAIEENGFDPHAGFDEKQTPDAETEAQFLIKWK 373
Query: 445 GQSHLHCQWKSFVDLQNLS--GFKKVLNYTKRVTEEIRNRMGISREEIEVNDVSKEMDID 502
G S++H W+S L+++ G KK+ N+ K+ E+ R E+I+ + E+ +
Sbjct: 374 GWSYIHNTWESEATLRDMKAKGMKKLDNFIKKEKEQAYWRRYAGPEDIDYFECQLELQHE 433
Query: 503 IIKQNSQVERIIADRISNDNSGNVFPEYLVKWQGLSYAEVTWEKDIDIAFA----QHTID 558
++K + V+RIIA D+ EYL KWQ L YAE TWE D A Q +
Sbjct: 434 LLKSYNNVDRIIAKGSKPDDGTE---EYLCKWQSLPYAESTWE---DAALVLRKWQRCAE 487
Query: 559 EYKTREAAMSVQGKMVDFQRRQSKGSLRKLDEQPEWLKGG-KLRDYQLEGLNFLVNSWKN 617
++ RE++ + + + K S ++ QPE+L G LRDYQ++GLN+L++SW
Sbjct: 488 QFNDRESSKCTPSRHCRVIKYRPKFS--RIKNQPEFLSSGLTLRDYQMDGLNWLLHSWCK 545
Query: 618 DTNVVLADEMGLGKTVQSVSMLGFLQNAQQIHGPFLVVVPLSTLSNWAKEFRKWLPDLNV 677
+ +V+LADEMGLGKT+Q++ L L ++GPFL VVPLST++ W +EF W PD+NV
Sbjct: 546 ENSVILADEMGLGKTIQTICFLYSLFKIHHLYGPFLCVVPLSTMTAWQREFDLWAPDMNV 605
Query: 678 IVYVGTRSSREVCQQYEFCNEKKAGKQIKFNALLTTYEVVLKDKAVLSKIKWNYLMVDEA 737
+ Y+G SRE+ QQYE+ + ++ K++KFN +LTTYE+VLKDK L ++W L+VDEA
Sbjct: 606 VTYLGDIKSRELIQQYEW--QFESSKRLKFNCILTTYEIVLKDKQFLGTLQWAALLVDEA 663
Query: 738 HRLKNSEAQLYTALSEFNTKNKLLITGTPLQNSVEELWALLHFLDSDKFKSKDEFAQNYK 797
HRLKN ++ LY +L EF+T ++LLITGTPLQNS++ELWALLHF+ DKF + + F +
Sbjct: 664 HRLKNDDSLLYKSLKEFDTNHRLLITGTPLQNSLKELWALLHFIMPDKFDTWENFEVQHG 723
Query: 798 NLSSFNENELSNLHMELRPHMLRRVIKDVEKSLPPKIERILRVDMSPLQKQYYKWILERN 857
N + + LH +L P++LRRV KDVEKSLP K+E+ILRV+M+ LQKQYYKWIL +N
Sbjct: 724 NA---EDKGHTRLHQQLEPYILRRVKKDVEKSLPAKVEQILRVEMTSLQKQYYKWILTKN 780
Query: 858 FRDLNKGVRGNQSKYCSRIEEV 879
F L KG RG+ S + + + E+
Sbjct: 781 FDALRKGKRGSTSTFLNIVIEL 802
Score = 291 bits (746), Expect = 1e-76
Identities = 252/793 (31%), Positives = 367/793 (45%), Gaps = 117/793 (14%)
Query: 938 DILAQYMSLRGFQFQRLDGSTKSELRQQAMDHFNAPGSDDFCFLLSTRAGGLGINLATAD 997
D+LA Y+ R F FQRLDGS K E+R+QA+DHFNA GS DFCFLLSTRAGGLGINLATAD
Sbjct: 866 DVLADYLQKRHFPFQRLDGSIKGEMRRQALDHFNAEGSQDFCFLLSTRAGGLGINLATAD 925
Query: 998 TVIIFDSDWNPQNDLQAMSRAHRIGQREVVNIYRFVTSKSVEEDILERAKKKMVLDHLVI 1057
TVIIFDSDWNPQNDLQA +RAHRIGQ+ VNIYR VT++SVEE I+ERAK+KMVLDHLVI
Sbjct: 926 TVIIFDSDWNPQNDLQAQARAHRIGQKNQVNIYRLVTARSVEEQIVERAKQKMVLDHLVI 985
Query: 1058 QKLNAEGKLEKKEAKKGGSF----FDKNELSAILRFGAEELFKEERNDEESKKRLLSMDI 1113
Q+++ G+ ++ G S F+K++LSAIL+FGAEELFK+E+ ++ L DI
Sbjct: 986 QRMDTTGRTVLDKSGNGHSSNSNPFNKDDLSAILKFGAEELFKDEQEHDDD----LVCDI 1041
Query: 1114 DEILERAEKVEEKENGGEQAHELLSAFKVA------------------NFCNDEDDGSFW 1155
DEIL RAE E+ A +LLSAFKVA N +EDD W
Sbjct: 1042 DEILRRAE--TRNEDPEMPADDLLSAFKVASIAAFEEEPSDSVSKQDQNAAGEEDDSKDW 1099
Query: 1156 SRWIKADSVAQAENALAPRAARNI----KSYAEADQSERSKKRKKKENEPTERIPKRRKA 1211
I ++ + ++ + A+Q+E + K + +
Sbjct: 1100 DDIIPEGFRKAIDDQERAKEMEDLYLPPRRKTAANQNEGKRGAGKGGKGKQQADDSGGDS 1159
Query: 1212 DYSAHVISMIDGASAQVRSWSYGNLSKRDAL---------RFSRSVMKFGNESQINLIVA 1262
DY DG+ R G + ++ + RF RS KF +A
Sbjct: 1160 DYELG----SDGSGDDGRPRKRGRPTMKEKITGFTDAELRRFIRSYKKFPAPLHRMEAIA 1215
Query: 1263 EVGGAIEAAPLKAQVELFNALIDGC---------REAVEVGSLDLKGP---------LLD 1304
++ PL L L D C E+ + + G +
Sbjct: 1216 -CDAELQEKPLAELKRLGEMLHDRCVQFLHEHKEEESKTAATDETPGAKQRRARATFSVK 1274
Query: 1305 FYGVPMKANELLIRVQELQLLAKRISRYEDPIAQFRVLTYLKPSNWSKGCGWNQIDDARL 1364
GV A +LL QELQ L + + + Q+ +K W +D +L
Sbjct: 1275 LGGVSFNAKKLLACEQELQPLNEIMPSMPEERQQWSF--NIKTRAPVFDVDWGIEEDTKL 1332
Query: 1365 LLGVHYHGYGNWEVIRLDERLGLTKKIAPVELQHHETFLPRAPNLRDRANALLEQELAVL 1424
L G++ +G G+WE ++LD L LT KI ++T P+A L+ RA LL+ ++
Sbjct: 1333 LCGIYQYGIGSWEQMKLDPTLKLTDKIL-----LNDTRKPQAKQLQTRAEYLLK----II 1383
Query: 1425 GVKNASSKVGRKTSKKEREEREH-----------LVDISLSRGQEKKKNIGSSKVNVQMR 1473
+K G++ ++ R R + +D G++ + + + +
Sbjct: 1384 KKNVELTKGGQRRQRRPRASRANDAKAASQSASSTIDAKPHDGEDAAGDARTVAESSNSQ 1443
Query: 1474 KDRLQKPLNVEPIVKEEGE----------------MSDDDD----VYEQFKEGKWKEWCQ 1513
D + P ++ G+ SD++ + E + E
Sbjct: 1444 VDPSNASPHNAPATEQHGDPAKKAKKSKARSKKTSASDNNGNKPMHFTANNEPRALEVLG 1503
Query: 1514 DL-------MVEEMKTLKR-LHRLQTTSASLPKEKVLSKIRNYLQLLGRRIDQIVSEQED 1565
DL E+M+ +K+ L L SL + L R+ L +G++ID ++ +
Sbjct: 1504 DLDPSIFNECKEKMRPVKKALKALDQPDVSLSDQDQLQHTRDCLLQIGKQIDVCLNPYAE 1563
Query: 1566 EPHKQDRMTTRLWKYVSTFSHLSGERLHQIYSKLKLEQNAVGVGSSLPNGSVSGPFSRNG 1625
K+ R + LW +VS F+ L +RL +IY K L+Q A G G + S
Sbjct: 1564 TEKKEWR--SNLWYFVSKFTELDAKRLFKIY-KHALKQKAGGDGEAKGKDKGSSGSPAKS 1620
Query: 1626 NPNSSFPRPMERQ 1638
PN E++
Sbjct: 1621 KPNGVTTEEKEKE 1633
>ref|XP_238731.3| PREDICTED: similar to Chromodomain-helicase-DNA-binding protein 1
(CHD-1) [Rattus norvegicus]
Length = 1103
Score = 422 bits (1085), Expect = e-116
Identities = 280/827 (33%), Positives = 426/827 (50%), Gaps = 87/827 (10%)
Query: 70 RNPSASE----RWGSSYLKDCQPMSPQNGSESGDDSKSGSDYRNEDEFEDNSSEGRGEKL 125
R P+ S+ W S ++ +C +GS SG S S S +SS+G +
Sbjct: 218 RGPALSQGSIYEWFSFFIFNCSQSEDDSGSASGSGSGSSSG---------SSSDGSSSQS 268
Query: 126 GSEDEDGQKDSGKGQRGDSDVPAEEMLSDDSYGQDGEEQGES-------VHSRGFRPSTG 178
GS D D DSG +SD E + DG E +S S R T
Sbjct: 269 GSSDSDSGSDSGSQSESESDTSRENKVQAKPPKVDGAEFWKSNPSILAVQRSAMLRKQTQ 328
Query: 179 SNSCLQPTSTNVNRRVHRKSRILDDAEDDDDDADYEEDEPDEDDPDDADFEPATSGRGAN 238
+P S+N ++ +D+D + +D D++ + SG +
Sbjct: 329 QPQQQRPASSNSGS---------EEDSSSSEDSDASSSDVKRKKHNDEDWQMSGSGSPSQ 379
Query: 239 KYKDWEGEDSDEVDDSDEDIDVSDNDDLYFDKKAKGRQRGKFGPSVRSTRDCKAFTASSR 298
D E ED + D + + +K + R + K G + +
Sbjct: 380 SGSDSESEDERDKSSCDGTESDYEPKNKVKSRKPQNRSKSKSGKKI----------LGQK 429
Query: 299 QRRVKSSFEDEDENSTAEDSDSESDEDFKSLKKRGVRVRKNNGRSSAATSFSRPSNEVRS 358
+R++ SS EDED+ ED D++ K + R A + S +E
Sbjct: 430 KRQIDSS-EDEDD----EDYDND----------------KRSSRRQATVNVSYKEDEEMK 468
Query: 359 SSRTIRKVSYVESDESEGADEGTKKSQKEEIEVDDGDSVEKVLWHQPKGMAAEAQRNNQS 418
+ D E E +++ EE E + +V G
Sbjct: 469 TD---------SDDLLEVCGEDVPQTEDEEFETIERFMDCRVGRKGATGATTTIYAIEAD 519
Query: 419 MEPVLMSHLFDSEPDWNNMEFLIKWKGQSHLHCQWKSFVDL--QNLSGFKKVLNYTKRVT 476
+P + F+ + ++++LIKWKG SH+H W++ L QN+ G KK+ NY K+
Sbjct: 520 GDP---NAGFEKSKEPGDVQYLIKWKGWSHIHNTWETEETLKQQNVRGMKKLDNYKKKDQ 576
Query: 477 EEIRNRMGISREEIEVNDVSKEMDIDIIKQNSQVERIIADRISNDNSGNVFPEYLVKWQG 536
E R S E++E + +E+ D+ KQ VERIIA SN S P+Y KWQG
Sbjct: 577 ETKRWLKNASPEDVEYYNCQQELTDDLHKQYQIVERIIAH--SNQKSAAGLPDYYCKWQG 634
Query: 537 LSYAEVTWEKDIDIAFAQHT-IDEYKTREAAMSVQGKMVDFQRRQSKGSLRKLDEQPEWL 595
L Y+E +WE I+ T IDEY +R + + K D + + + L +QP ++
Sbjct: 635 LPYSECSWEDGALISKKFQTCIDEYFSRNQSKTTPFK--DCKVLKQRPRFVALKKQPSYI 692
Query: 596 ---KGGKLRDYQLEGLNFLVNSWKNDTNVVLADEMGLGKTVQSVSMLGFLQNAQQIHGPF 652
+G +LRDYQL GLN+L +SW + +LADEMGLGKT+Q++S L +L + Q++GPF
Sbjct: 693 GGHEGLELRDYQLNGLNWLAHSWCKGNSCILADEMGLGKTIQTISFLNYLFHEHQLYGPF 752
Query: 653 LVVVPLSTLSNWAKEFRKWLPDLNVIVYVGTRSSREVCQQYEFCNEKKAGKQIKFNALLT 712
L+VVPLSTL++W +E + W +N +VY+G +SR + + +E+ + + K++KFN LLT
Sbjct: 753 LLVVPLSTLTSWQREIQTWASQMNAVVYLGDINSRNMIRTHEWMHPQT--KRLKFNILLT 810
Query: 713 TYEVVLKDKAVLSKIKWNYLMVDEAHRLKNSEAQLYTALSEFNTKNKLLITGTPLQNSVE 772
TYE++LKDKA L + W ++ VDEAHRLKN ++ LY L +F + ++LLITGTPLQNS++
Sbjct: 811 TYEILLKDKAFLGGLNWAFIGVDEAHRLKNDDSLLYKTLIDFKSNHRLLITGTPLQNSLK 870
Query: 773 ELWALLHFLDSDKFKSKDEFAQNYKNLSSFNENELSNLHMELRPHMLRRVIKDVEKSLPP 832
ELW+LLHF+ +KF S ++F + + E ++LH EL P +LRRV KDVEKSLP
Sbjct: 871 ELWSLLHFIMPEKFSSWEDFEEEH---GKGREYGYASLHKELEPFLLRRVKKDVEKSLPA 927
Query: 833 KIERILRVDMSPLQKQYYKWILERNFRDLNKGVRGNQSKYCSRIEEV 879
K+E+ILR++MS LQKQYYKWIL RN++ L+KG +G+ S + + + E+
Sbjct: 928 KVEQILRMEMSALQKQYYKWILTRNYKALSKGSKGSTSGFLNIMMEL 974
Score = 101 bits (251), Expect = 3e-19
Identities = 53/90 (58%), Positives = 60/90 (65%), Gaps = 24/90 (26%)
Query: 938 DILAQYMSLRGFQFQRLDGSTKSELRQQAMDHFNAPGSDDFCFLLSTRAGGLGINLATAD 997
DILA+Y+ R F FQ DFCFLLSTRAGGLGINLA+AD
Sbjct: 1037 DILAEYLKYRQFPFQ------------------------DFCFLLSTRAGGLGINLASAD 1072
Query: 998 TVIIFDSDWNPQNDLQAMSRAHRIGQREVV 1027
TV+IFDSDWNPQNDLQA +RAHRIGQ++ V
Sbjct: 1073 TVVIFDSDWNPQNDLQAQARAHRIGQKKQV 1102
Score = 45.8 bits (107), Expect = 0.013
Identities = 61/253 (24%), Positives = 100/253 (39%), Gaps = 38/253 (15%)
Query: 16 GQNADRTRSEKEFYMNMEAQYESDAEPDDASGKQNEAAAVDRLSTRESNVETTSRNPSAS 75
G ++ S+ + + +Q ES+++ + Q + VD +SN + SA
Sbjct: 263 GSSSQSGSSDSDSGSDSGSQSESESDTSRENKVQAKPPKVDGAEFWKSNPSILAVQRSAM 322
Query: 76 ERWGSSYLKDCQPMSPQNGSESGDDSKSGSDYRNEDEFEDNSSEGRGEKLGSEDEDGQKD 135
R + + +P S +GSE S SD + D ++ DED Q
Sbjct: 323 LRKQTQQPQQQRPASSNSGSEEDSSSSEDSDASSSDV----------KRKKHNDEDWQM- 371
Query: 136 SGKGQRGDSDVPAEEMLSDDSYGQDGEEQG----ESVHSRGFRPSTGSNSCLQPTSTNVN 191
SG G S +E D DG E V SR +P S S +
Sbjct: 372 SGSGSPSQSGSDSESEDERDKSSCDGTESDYEPKNKVKSR--KPQNRSK------SKSGK 423
Query: 192 RRVHRKSRILDDAEDDDDDADYEEDEPDEDDPDDADFEPATSGRGANKYKDWEGEDSDEV 251
+ + +K R +D +ED+DD+ DY+ D+ +S R A ++ ED +
Sbjct: 424 KILGQKKRQIDSSEDEDDE-DYDNDK-------------RSSRRQATVNVSYK-EDEEMK 468
Query: 252 DDSDEDIDVSDND 264
DSD+ ++V D
Sbjct: 469 TDSDDLLEVCGED 481
>ref|XP_546008.1| PREDICTED: similar to Chromodomain-helicase-DNA-binding protein 1
(CHD-1) [Canis familiaris]
Length = 1790
Score = 421 bits (1082), Expect = e-115
Identities = 289/888 (32%), Positives = 452/888 (50%), Gaps = 77/888 (8%)
Query: 8 SWKIKARVGQNADRTRSEKEFYMNMEAQYESDAEPDDAS---GKQNEAAAVDRLSTRESN 64
+W RV ++T E +F +N +++ + +S G QN + S ++
Sbjct: 26 AWSKPLRVVWQRNQTEREPDFTVNTDSRCDCVGVNQCSSVLAGCQNPVRGLPYFSHFQAQ 85
Query: 65 VETTSRNPSASERWGSSYLKDCQPMSPQNGSESGDDSKSGSDYRNEDEFEDNSSEGRGEK 124
+ + ++ + L +P P G DD + +SS+G +
Sbjct: 86 ALRSDVSDYTLQKGKEASLL-IKPQKPILGHPQSDDDSGSASGSGSGSSSGSSSDGSSSQ 144
Query: 125 LGSEDEDGQKDSGKGQRGDSDVPAEEMLSDDSYGQDGEEQGESVHSRGFRPSTGSNSCLQ 184
GS D D +SG +SD E + DG E F S+ S +Q
Sbjct: 145 SGSSDSDSGSESGSQSESESDTSRENKVQAKPPKVDGAE---------FWKSSPSILAVQ 195
Query: 185 PTSTNVNRRVHRKSRILDDAEDDDDDADYEEDEPDEDDPDDADFEPATSGRGANKYKDWE 244
++ + +K + ++ EED +D DD+ E R +K +DW+
Sbjct: 196 RSA------MLKKQQQQQQQHPASSNSGSEEDSSSSEDSDDSSNEVK---RKKHKDEDWQ 246
Query: 245 ----GEDSDEVDDSD---EDIDVSDNDDLYFDKKAKGRQRGKFGPSVRSTRDCKAFTASS 297
G S DS+ E+ D S D+ D + K + + + P RS
Sbjct: 247 MSGSGSPSQSGSDSESEEEERDKSSCDETESDYEPKNKVKSR-KPQNRSKAKNGKKILGQ 305
Query: 298 RQRRVKSSFEDEDENSTAEDSDSESDEDFKSLKKRGVRVRKNNGRSSAATSFSRPSNEVR 357
++R++ SS E++DE ED D++ K + R A + S +E
Sbjct: 306 KKRQIDSSEEEDDE----EDYDND----------------KRSSRRQATVNVSYKEDEEM 345
Query: 358 SSSRTIRKVSYVESDESEGADEGTKKSQKEEIEVDDGDSVEKVLWHQPKGMAAEAQRNNQ 417
+ D E E + ++EE E + ++ G
Sbjct: 346 KTD---------SDDLLEVCGEDVPQPEEEEFETIERFMDCRIGRKGATGATTTIYAVEA 396
Query: 418 SMEPVLMSHLFDSEPDWNNMEFLIKWKGQSHLHCQWKSFVDL--QNLSGFKKVLNYTKRV 475
+P + F+ + +++LIKWKG SH+H W++ L QN+ G KK+ NY K+
Sbjct: 397 DGDP---NAGFEKNKEPGEIQYLIKWKGWSHIHNTWETEETLKQQNVRGMKKLDNYKKKD 453
Query: 476 TEEIRNRMGISREEIEVNDVSKEMDIDIIKQNSQVERIIADRISNDNSGNVFPEYLVKWQ 535
E R S E++E + +E+ D+ KQ VERIIA SN S +P+Y KWQ
Sbjct: 454 QETKRWLKNASPEDVEYYNCQQELTDDLHKQYQIVERIIAH--SNQKSAAGYPDYYCKWQ 511
Query: 536 GLSYAEVTWEKDIDIAFA-QHTIDEYKTREAAMSVQGKMVDFQRRQSKGSLRKLDEQPEW 594
GL Y+E +WE I+ Q IDEY +R + + K D + + + L +QP +
Sbjct: 512 GLPYSECSWEDGALISKKFQACIDEYFSRNQSKTTPFK--DCKVLKQRPRFVALKKQPSY 569
Query: 595 LKGGK---LRDYQLEGLNFLVNSWKNDTNVVLADEMGLGKTVQSVSMLGFLQNAQQIHGP 651
+ G + LRDYQL GLN+L +SW + +LADEMGLGKT+Q++S L +L + Q++GP
Sbjct: 570 IGGHESLELRDYQLNGLNWLAHSWCKGNSCILADEMGLGKTIQTISFLNYLFHEHQLYGP 629
Query: 652 FLVVVPLSTLSNWAKEFRKWLPDLNVIVYVGTRSSREVCQQYEFCNEKKAGKQIKFNALL 711
FL+VVPLSTL++W +E + W +N +VY+G +SR + + +E+ + + K++KFN LL
Sbjct: 630 FLLVVPLSTLTSWQREIQTWASQMNAVVYLGDINSRNMIRTHEWMHPQT--KRLKFNILL 687
Query: 712 TTYEVVLKDKAVLSKIKWNYLMVDEAHRLKNSEAQLYTALSEFNTKNKLLITGTPLQNSV 771
TTYE++LKDKA L + W ++ VDEAHRLKN ++ LY L +F + ++LLITGTPLQNS+
Sbjct: 688 TTYEILLKDKAFLGGLNWAFIGVDEAHRLKNDDSLLYKTLIDFKSNHRLLITGTPLQNSL 747
Query: 772 EELWALLHFLDSDKFKSKDEFAQNYKNLSSFNENELSNLHMELRPHMLRRVIKDVEKSLP 831
+ELW+LLHF+ +KF S ++F + + E ++LH EL P +LRRV KDVEKSLP
Sbjct: 748 KELWSLLHFIMPEKFSSWEDFEEEH---GKGREYGYASLHKELEPFLLRRVKKDVEKSLP 804
Query: 832 PKIERILRVDMSPLQKQYYKWILERNFRDLNKGVRGNQSKYCSRIEEV 879
K+E+ILR++MS LQKQYYKWIL RN++ L+KG +G+ S + + + E+
Sbjct: 805 AKVEQILRMEMSALQKQYYKWILTRNYKALSKGSKGSTSGFLNIMMEL 852
Score = 348 bits (893), Expect = 9e-94
Identities = 259/714 (36%), Positives = 361/714 (50%), Gaps = 106/714 (14%)
Query: 938 DILAQYMSLRGFQFQRLDGSTKSELRQQAMDHFNAPGSDDFCFLLSTRAGGLGINLATAD 997
DILA+Y+ R F FQRLDGS K ELR+QA+DHFNA GS+DFCFLLSTRAGGLGINLA+AD
Sbjct: 915 DILAEYLKYRQFPFQRLDGSIKGELRKQALDHFNAEGSEDFCFLLSTRAGGLGINLASAD 974
Query: 998 TVIIFDSDWNPQNDLQAMSRAHRIGQREVVNIYRFVTSKSVEEDILERAKKKMVLDHLVI 1057
TV+IFDSDWNPQNDLQA +RAHRIGQ++ VNIYR VT SVEEDILERAKKKMVLDHLVI
Sbjct: 975 TVVIFDSDWNPQNDLQAQARAHRIGQKKQVNIYRLVTKGSVEEDILERAKKKMVLDHLVI 1034
Query: 1058 QKLNAEGK--LEKKEAKKGGSFFDKNELSAILRFGAEELFKEERNDEESKKRLLSMDIDE 1115
Q+++ GK L A + F+K ELSAIL+FGAEELFKE +E+ + MDIDE
Sbjct: 1035 QRMDTTGKTVLHTGSAPSSSTPFNKEELSAILKFGAEELFKEPEGEEQEPQ---EMDIDE 1091
Query: 1116 ILERAEKVEEKENGGEQAHELLSAFKVANFCN-DEDDGSFWSRWIKADSVAQAENALAPR 1174
IL+RAE E + ELLS FKVANF N DEDD I+ + R
Sbjct: 1092 ILKRAETHENEPGPLTVGDELLSQFKVANFSNMDEDD-------IELE---------PER 1135
Query: 1175 AARNIKSYAEADQSER-SKKRKKKENEPTERIPKRRKADYSAHVISMIDGASAQVRSWSY 1233
++N + DQ R ++ ++KE E +P+ R A IS + RS Y
Sbjct: 1136 NSKNWEEIIPEDQRRRLEEEERQKELEEIYMLPRMRNC---AKQISFNGSEGRRSRSRRY 1192
Query: 1234 G-----NLSKRDALR-------FSRSVMKFGNESQINLIVAEVGGAIEAAPLKAQVELFN 1281
++S+R + R +K ++++I + A A + +
Sbjct: 1193 SGSDSDSISERKRPKKRGRPRTIPRENIKGFSDAEIRRLDAIARDAELVDKSETDLRRLG 1252
Query: 1282 ALI-DGCREAVEVGSL----------DLKGPLLDFYGVPMKANELLIRVQELQLLAKRIS 1330
L+ +GC +A++ S +KGP GV + A ++ +EL L K I
Sbjct: 1253 ELVHNGCIKALKDNSSGTERTGGRLGKVKGPTFRISGVQVNAKLVISHEEELIPLHKSIP 1312
Query: 1331 RYEDPIAQFRVLTYLKPSNWSKGCGWNQIDDARLLLGVHYHGYGNWEVIRLDERLGLTKK 1390
+ Q+ + + K +++ W + DD+ LL+G++ +GYG+WE+I++D L LT K
Sbjct: 1313 SDPEERKQYTIPCHTKAAHFD--IDWGKEDDSNLLIGIYEYGYGSWEMIKMDPDLSLTHK 1370
Query: 1391 IAPVELQHHETFLPRAPNLRDRANALLEQELAVLGVKNASSKVGRKTSKKEREEREHLVD 1450
I P + P+A L+ RA+ L++ L K A G SK+ +
Sbjct: 1371 ILPDDPDKK----PQAKQLQTRADYLIKLLSRDLAKKEAQRLSGAGGSKRRKAR------ 1420
Query: 1451 ISLSRGQEKKKNIGSSKVNVQMRKDRLQKPLNVEPIVKEEGEMSDDDDVYEQFKEGKWKE 1510
+K K + S KV +M+ D P E SD+DD + KE K
Sbjct: 1421 ------AKKNKAMKSIKVKEEMKSDSSPLP----------SEKSDEDDDKSESKERSKKS 1464
Query: 1511 WCQDLMV---------------------------EEMKTLK-RLHRLQTTSASLPKEKVL 1542
D V E M+ +K L +L L + + L
Sbjct: 1465 SVSDAPVHITASGEPVPISEESEELDQKTFSICKERMRPVKAALKQLDRPEKGLSEREQL 1524
Query: 1543 SKIRNYLQLLGRRIDQIVSEQEDEPHKQDRMTTRLWKYVSTFSHLSGERLHQIY 1596
R L +G I + + E + P + + LW +VS F+ +LH++Y
Sbjct: 1525 EHTRQCLIKIGDHITECLKEYTN-PEQIKQWRKNLWIFVSKFTEFDARKLHKLY 1577
>emb|CAE65711.1| Hypothetical protein CBG10790 [Caenorhabditis briggsae]
Length = 1463
Score = 419 bits (1077), Expect = e-115
Identities = 260/712 (36%), Positives = 393/712 (54%), Gaps = 71/712 (9%)
Query: 196 RKSRILDDAEDDDDDADYEEDEPDEDDPDDADFEPATSGRGANKYKDWEGEDSDEVDDSD 255
RK++++++ + E++E D D ++A A K D E SD+ ++S
Sbjct: 24 RKTKVIEEGVSSGSGSSEEKEEEDSDYENEAK---------AEKSPDSTSEQSDDSEESS 74
Query: 256 EDIDVSDNDDLYFDKKAKGRQRGKFGPSVRSTRDCKAFTASSRQRRVKSSFEDEDENSTA 315
D S+ D+ KK RG + + + F S R +R + S ++ S +
Sbjct: 75 GDSSNSEADEPA-PKKKVNNSRGMDAETRKLLEEENYFRRSQRSKRTEESVSEKSGESES 133
Query: 316 EDSDSESDEDFKSLKKRGVRVRKNNGRSSAATSFSRPSNEVRSSSRTIRKVSYVESDESE 375
D D D K R V+K + + R S + VSYVE + E
Sbjct: 134 SDDDEWDDGGRKKKSSRRSAVKKTVVKKVVQSVIKRKSVK--------GNVSYVEKNSDE 185
Query: 376 GADEGTKKSQKEEIEVDDG------------DSVEKVL-WHQPKGMAAE--------AQR 414
D+ + +E D+G ++VE+V+ W A A++
Sbjct: 186 DIDDD------DVLEWDEGPAVPVEGPAVLTETVERVIKWRIGVPGATGSGTTCYNVAEK 239
Query: 415 NNQSMEPVLMSHLFDSEPDWNNMEFLIKWKGQSHLHCQWKS--FVDLQNLSGFKKVLNYT 472
+ + +PV + +F +KW G SHLH W+S + + + G KKV NY
Sbjct: 240 GDPNEQPV----------EKTEKQFFVKWTGWSHLHNTWESENSLTIMHAKGIKKVQNYV 289
Query: 473 KRVTEEIRNRMGISREEIEVNDVSKEMDIDIIKQNSQVERIIADRISNDNS--GNVFPEY 530
K+ E + +E IE + ++M ++ ++ +VER++A + S D + G+ E+
Sbjct: 290 KKQKEVEMWKRSADKEYIEFYECEQQMAEELCEEYKKVERVVAHQTSRDRAPDGSFATEF 349
Query: 531 LVKWQGLSYAEVTWEKDIDIAFAQHTIDEYKTREAAMSVQGKMVDFQRRQSKGSLRKLDE 590
L+KW GL YA+ TWE + +A Q I Y R + K R++ K KL+
Sbjct: 350 LIKWSGLPYADCTWEDEKMVAPDQ--IQGYYHRVENLKPPNKNATVLRKRPK--FEKLES 405
Query: 591 QPEWLKGG---KLRDYQLEGLNFLVNSWKNDTNVVLADEMGLGKTVQSVSMLGFLQNAQQ 647
QP++LK KLRDYQLEGLN+++ +W + +LADEMGLGKT+QS+S+L L +
Sbjct: 406 QPDYLKTNGDHKLRDYQLEGLNWMIYAWCKGNSSILADEMGLGKTIQSISLLASLFHRYD 465
Query: 648 IHGPFLVVVPLSTLSNWAKEFRKWLPDLNVIVYVGTRSSREVCQQYEFCNEKKAGKQIKF 707
+ GP+LVVVPLST++ W KEF +W PD+N+I+Y+G SR++ +QYE+ K++K
Sbjct: 466 LAGPYLVVVPLSTMAAWQKEFAQWAPDINLIIYMGDVVSRDMIRQYEWF--VGGTKKMKV 523
Query: 708 NALLTTYEVVLKDKAVLSKIKWNYLMVDEAHRLKNSEAQLYTALSEFNTKNKLLITGTPL 767
NA+LTTYE++LKDKA LS + W L+VDEAHRLKN E+ LY L +F +KLLITGTPL
Sbjct: 524 NAILTTYEILLKDKAFLSSVDWAALLVDEAHRLKNDESLLYKCLIQFRFNHKLLITGTPL 583
Query: 768 QNSVEELWALLHFLDSDKFKSKDEFAQNYKNLSSFNENELSNLHMELRPHMLRRVIKDVE 827
QNS++ELWALLHF+ +KF +EF + + N +S LH +L P +LRRV KDVE
Sbjct: 584 QNSLKELWALLHFIMPEKFDCWEEFETAH---NESNHKGISALHKKLEPFLLRRVKKDVE 640
Query: 828 KSLPPKIERILRVDMSPLQKQYYKWILERNFRDLNKGVRGNQSKYCSRIEEV 879
KSLPPK E+ILRVDM+ QKQ+YKWIL +N+R+L+KGV+G+ + + + + E+
Sbjct: 641 KSLPPKTEQILRVDMTAHQKQFYKWILTKNYRELSKGVKGSINGFVNLVMEL 692
Score = 292 bits (748), Expect = 6e-77
Identities = 238/689 (34%), Positives = 339/689 (48%), Gaps = 93/689 (13%)
Query: 938 DILAQYMSLRGFQFQRLDGSTKSELRQQAMDHFNAPGSDDFCFLLSTRAGGLGINLATAD 997
DIL +Y+ LR F QRLDGS +++LR+QA+DH+NAPGS DF FLLSTRAGGLGINLATAD
Sbjct: 754 DILQEYLQLRRFPSQRLDGSMRADLRKQALDHYNAPGSTDFAFLLSTRAGGLGINLATAD 813
Query: 998 TVIIFDSDWNPQNDLQAMSRAHRIGQREVVNIYRFVTSKSVEEDILERAKKKMVLDHLVI 1057
TVIIFDSDWNPQNDLQAMSRAHRIGQ + VNIYR VT SVEE+I+ERAK+K+VLDHLVI
Sbjct: 814 TVIIFDSDWNPQNDLQAMSRAHRIGQTKTVNIYRLVTKGSVEEEIVERAKRKLVLDHLVI 873
Query: 1058 QKLNAEGK--LEKKEAKKGGSFFDKNELSAILRFGAEELFKEERNDEESKKRLLSMDIDE 1115
Q+++ GK L K G FDK ELSAIL+FGA ELFKE+ +E+ + +DID
Sbjct: 874 QRMDTTGKTVLSKNATASGSVPFDKQELSAILKFGAVELFKEKEGEEQEPE----VDIDR 929
Query: 1116 ILERAEKVEEKENGGEQAHELLSAFKVANFCNDEDDGSFWSRWIKADSVAQAENALAPRA 1175
IL AE E +E ++ +ELLS+FK ANF DE+ +A A + A
Sbjct: 930 ILMGAETREAEEEVLKE-NELLSSFKYANFAIDEE-----------KDIAAATDEWA--- 974
Query: 1176 ARNIKSYAEADQSERSKKRKKKENEPTERIPKRRKADYSAHVISMIDG--------ASAQ 1227
E D++ ++ + KE P++RK V DG +
Sbjct: 975 ----AIIPEEDRNRILEEERMKELAEMNLAPRQRKQPMPQVVEDGEDGDDDDDEEDGGKK 1030
Query: 1228 VRSWSYGNLSKRDALRFSRSVMKFGNESQINLIVAEVGGAIEAAPLKAQVELFNALIDGC 1287
G+ + + RF R+ KF + +A+ +E + ++L +L + C
Sbjct: 1031 KSKKKIGSFTVAEIKRFIRAFRKFSMPLERLEEIAQ-DAELEEHSTEEVMKLVESLTEAC 1089
Query: 1288 REAV-EVGSLDLKGPLL--DFYGVPMKANEL--LIRVQELQLLAKRISR--------YED 1334
++A E S + G G K + + Q + K+I R +E
Sbjct: 1090 KKAADEFDSAEKNGRAASSSAEGADKKEKDADRKFKFQTCDVNLKQIERSHAELKPLHEA 1149
Query: 1335 PIAQFRVLTYLKPSNWSKGCGWN----QIDDARLLLGVHYHGYGNWEVIRLDERLGLTKK 1390
+ + ++ P N GW+ DD LL GV +GYG+WE I++D LGL K
Sbjct: 1150 LKSVDKKTSFKPPMNAKPQKGWDVEWKWDDDGALLWGVWKYGYGSWEAIKMDPSLGLADK 1209
Query: 1391 IAPVELQHHETFLPRAPNLRDRANALLEQELAVLGVKNASSKVGRKTSKK--EREEREHL 1448
I +T P+ NL+ R + LL K+ K KK E+++R+
Sbjct: 1210 I----FIKDKTKKPQGKNLQVRVDYLL--------------KLMAKDKKKGPEKKDRKRK 1251
Query: 1449 VDISLSRGQEKKK----NIGSSKVNVQMRKDRLQKPLNVEPIVKEEGEMSDDDDVYEQFK 1504
D + S +KKK N+ + + K + + + + ++ D +Y
Sbjct: 1252 ADDTPSGAPDKKKKHTNNVAPDADHKKKEKKEKSEDKSRSSLKDQLSVLTIDKSLYGGVL 1311
Query: 1505 EGKWKE---WCQDLMVEEMKTLKRLHRLQTTSASLPKEKVLSKIRNYLQLLGRRIDQIVS 1561
E + C L + K +K+L Q T + K YL LG D ++
Sbjct: 1312 EDSSAKPFLECVKLCMPVHKYMKKLKEAQETKNKEDETK-------YLTRLG---DSFLA 1361
Query: 1562 EQEDEPHKQDRMTTR-----LWKYVSTFS 1585
E K+ + R LW ++S F+
Sbjct: 1362 NLETLLKKKPKTNIRKWYNYLWIFLSKFT 1390
Score = 60.8 bits (146), Expect = 4e-07
Identities = 98/413 (23%), Positives = 160/413 (38%), Gaps = 78/413 (18%)
Query: 69 SRNPSASERWGSSYLKDCQPMSPQNGSESGDDSKSGSDYRNEDEFEDNSSEGRGEK---- 124
+++ S+S S K +PM E G S SGS E+E D +E + EK
Sbjct: 3 NQSESSSGSDADSDEKSDEPMRKTKVIEEGVSSGSGSSEEKEEEDSDYENEAKAEKSPDS 62
Query: 125 LGSEDEDGQKDSGKGQRGDSDVPAEEMLSDDSYGQDGEEQGESVHSRGFRPSTGSNSCLQ 184
+ +D ++ SG ++D PA + ++S G D E + L+
Sbjct: 63 TSEQSDDSEESSGDSSNSEADEPAPKKKVNNSRGMDAE----------------TRKLLE 106
Query: 185 PTSTNVNRRVHRKSRILDDAEDDDDDADYEEDEPDEDDPDDADFEPATSGRGA------- 237
N RR R R E+ + E + D+D+ DD + +S R A
Sbjct: 107 --EENYFRRSQRSKR----TEESVSEKSGESESSDDDEWDDGGRKKKSSRRSAVKKTVVK 160
Query: 238 ------NKYKDWEGEDSDEVDDSDEDIDVSDNDDLYFDKKAKGRQRGKFGPSVRSTRDCK 291
K K +G S +SDEDID D+D L +D+ GP+V +
Sbjct: 161 KVVQSVIKRKSVKGNVSYVEKNSDEDID--DDDVLEWDE----------GPAV--PVEGP 206
Query: 292 AFTASSRQRRVK-------SSFEDEDENSTAEDSDSESDEDFKSLKKRGVRVRKNNGRSS 344
A + +R +K ++ + AE D K+ K+ V K G S
Sbjct: 207 AVLTETVERVIKWRIGVPGATGSGTTCYNVAEKGDPNEQPVEKTEKQFFV---KWTGWSH 263
Query: 345 AATSFSRPSNEVRSSSRTIRKV-SYVESDESEGADEGTKKSQKEEIEVDDGDSVEKVLWH 403
++ ++ ++ I+KV +YV+ + + + + KE IE +
Sbjct: 264 LHNTWESENSLTIMHAKGIKKVQNYVKKQKE--VEMWKRSADKEYIE----------FYE 311
Query: 404 QPKGMAAEAQRNNQSMEPVLMSHLF-DSEPDWN-NMEFLIKWKGQSHLHCQWK 454
+ MA E + +E V+ D PD + EFLIKW G + C W+
Sbjct: 312 CEQQMAEELCEEYKKVERVVAHQTSRDRAPDGSFATEFLIKWSGLPYADCTWE 364
>gb|AAB87381.1| CHD1 [Homo sapiens] gi|3182949|sp|O14646|CHD1_HUMAN
Chromodomain-helicase-DNA-binding protein 1 (CHD-1)
gi|4557447|ref|NP_001261.1| chromodomain helicase DNA
binding protein 1 [Homo sapiens]
Length = 1709
Score = 419 bits (1077), Expect = e-115
Identities = 274/798 (34%), Positives = 420/798 (52%), Gaps = 67/798 (8%)
Query: 90 SPQNGSESGDDSKSGSDYRNEDEFEDNSSEGRGEKLGSEDEDGQKDSGKGQRGDSDVPAE 149
S Q+ +SG S SGS + +SS+G + GS D D +SG +SD E
Sbjct: 17 SSQSDDDSGSASGSGSGSSS-----GSSSDGSSSQSGSSDSDSGSESGSQSESESDTSRE 71
Query: 150 EMLSDDSYGQDGEEQGESVHSRGFRPSTGSNSCLQPTSTNVNRRVHRKSRILDDAEDDD- 208
+ DG E +S S + ++ L+ ++ H+ S ED
Sbjct: 72 NKVQAKPPKVDGAEFWKSSPSI---LAVQRSAILKKQQQQQQQQQHQASSNSGSEEDSSS 128
Query: 209 -DDADYEEDEPDEDDPDDADFEPATSGRGANKYKDWEGEDSDEVDDSDEDIDVSDNDDLY 267
+D+D E D D++ + SG + D E E+ E DE + +
Sbjct: 129 SEDSDDSSSEVKRKKHKDEDWQMSGSGSPSQSGSDSESEEEREKSSCDETESDYEPKNKV 188
Query: 268 FDKKAKGRQRGKFGPSVRSTRDCKAFTASSRQRRVKSSFEDEDENSTAEDSDSESDEDFK 327
+K + R + K G + ++R++ SS ED+DE ED D++
Sbjct: 189 KSRKPQNRSKSKNGKKI----------LGQKKRQIDSSEEDDDE----EDYDND------ 228
Query: 328 SLKKRGVRVRKNNGRSSAATSFSRPSNEVRSSSRTIRKVSYVESDESEGADEGTKKSQKE 387
K + R A + S +E + D E E + ++E
Sbjct: 229 ----------KRSSRRQATVNVSYKEDEEMKTD---------SDDLLEVCGEDVPQPEEE 269
Query: 388 EIEVDDGDSVEKVLWHQPKGMAAEAQRNNQSMEPVLMSHLFDSEPDWNNMEFLIKWKGQS 447
E E + ++ G +P + F+ + +++LIKWKG S
Sbjct: 270 EFETIERFMDCRIGRKGATGATTTIYAVEADGDP---NAGFEKNKEPGEIQYLIKWKGWS 326
Query: 448 HLHCQWKSFVDL--QNLSGFKKVLNYTKRVTEEIRNRMGISREEIEVNDVSKEMDIDIIK 505
H+H W++ L QN+ G KK+ NY K+ E R S E++E + +E+ D+ K
Sbjct: 327 HIHNTWETEETLKQQNVRGMKKLDNYKKKDQETKRWLKNASPEDVEYYNCQQELTDDLHK 386
Query: 506 QNSQVERIIADRISNDNSGNVFPEYLVKWQGLSYAEVTWEKDIDIAFA-QHTIDEYKTRE 564
Q V RIIA SN S +P+Y KWQGL Y+E +WE I+ Q IDEY +R
Sbjct: 387 QYQIVGRIIAH--SNQKSAAGYPDYYCKWQGLPYSECSWEDGALISKKFQACIDEYFSRN 444
Query: 565 AAMSVQGKMVDFQRRQSKGSLRKLDEQPEWL---KGGKLRDYQLEGLNFLVNSWKNDTNV 621
+ + K D + + + L +QP ++ +G +LRDYQL GLN+L +SW +
Sbjct: 445 QSKTTPFK--DCKVLKQRPRFVALKKQPSYIGGHEGLELRDYQLNGLNWLAHSWCKGNSC 502
Query: 622 VLADEMGLGKTVQSVSMLGFLQNAQQIHGPFLVVVPLSTLSNWAKEFRKWLPDLNVIVYV 681
+LADEMGLGKT+Q++S L +L + Q++GPFL+VVPLSTL++W +E + W +N +VY+
Sbjct: 503 ILADEMGLGKTIQTISFLNYLFHEHQLYGPFLLVVPLSTLTSWQREIQTWASQMNAVVYL 562
Query: 682 GTRSSREVCQQYEFCNEKKAGKQIKFNALLTTYEVVLKDKAVLSKIKWNYLMVDEAHRLK 741
G +SR + + +E+ + + K++KFN LLTTYE++LKDKA L + W ++ VDEAHRLK
Sbjct: 563 GDINSRNMIRTHEWTHHQT--KRLKFNILLTTYEILLKDKAFLGGLNWAFIGVDEAHRLK 620
Query: 742 NSEAQLYTALSEFNTKNKLLITGTPLQNSVEELWALLHFLDSDKFKSKDEFAQNYKNLSS 801
N ++ LY L +F + ++LLITGTPLQNS++ELW+LLHF+ +KF S ++F + +
Sbjct: 621 NDDSLLYKTLIDFKSNHRLLITGTPLQNSLKELWSLLHFIMPEKFSSWEDFEEEH---GK 677
Query: 802 FNENELSNLHMELRPHMLRRVIKDVEKSLPPKIERILRVDMSPLQKQYYKWILERNFRDL 861
E ++LH EL P +LRRV KDVEKSLP K+E+ILR++MS LQKQYYKWIL RN++ L
Sbjct: 678 GREYGYASLHKELEPFLLRRVKKDVEKSLPAKVEQILRMEMSALQKQYYKWILTRNYKAL 737
Query: 862 NKGVRGNQSKYCSRIEEV 879
+KG +G+ S + + + E+
Sbjct: 738 SKGSKGSTSGFLNIMMEL 755
Score = 345 bits (885), Expect = 8e-93
Identities = 253/717 (35%), Positives = 358/717 (49%), Gaps = 95/717 (13%)
Query: 938 DILAQYMSLRGFQFQRLDGSTKSELRQQAMDHFNAPGSDDFCFLLSTRAGGLGINLATAD 997
DILA+Y+ R F FQRLDGS K ELR+QA+DHFNA GS+DFCFLLSTRAGGLGINLA+AD
Sbjct: 818 DILAEYLKYRQFPFQRLDGSIKGELRKQALDHFNAEGSEDFCFLLSTRAGGLGINLASAD 877
Query: 998 TVIIFDSDWNPQNDLQAMSRAHRIGQREVVNIYRFVTSKSVEEDILERAKKKMVLDHLVI 1057
TV+IFDSDWNPQNDLQA +RAHRIGQ++ VNIYR VT SVEEDILERAKKKMVLDHLVI
Sbjct: 878 TVVIFDSDWNPQNDLQAQARAHRIGQKKQVNIYRLVTKGSVEEDILERAKKKMVLDHLVI 937
Query: 1058 QKLNAEGK--LEKKEAKKGGSFFDKNELSAILRFGAEELFKEERNDEESKKRLLSMDIDE 1115
Q+++ GK L A + F+K ELSAIL+FGAEELFKE +E+ + MDIDE
Sbjct: 938 QRMDTTGKTVLHTGSAPSSSTPFNKEELSAILKFGAEELFKEPEGEEQEPQ---EMDIDE 994
Query: 1116 ILERAEKVEEKENGGEQAHELLSAFKVANFCN-DEDD--------GSFWSRWIKADSVAQ 1166
IL+RAE E + ELLS FKVANF N DEDD W I D +
Sbjct: 995 ILKRAETHENEPGPLTVGDELLSQFKVANFSNMDEDDIELEPERNSKNWEEIIPEDQRRR 1054
Query: 1167 AENA----------LAPRAARNIKSYA-EADQSERSKKRKKKENEPTE----RIPKRRKA 1211
E + PR K + + RS+ R+ ++ + PK+R
Sbjct: 1055 LEEEERQKELEEIYMLPRMRNCAKQISFNGSEGRRSRSRRYSGSDSDSISEGKRPKKR-- 1112
Query: 1212 DYSAHVISMIDGASAQVRSWSYGNLSKRDALRFSRSVMKFGNESQINLIVAEVGGAIEAA 1271
G + + S + RF +S KFG + +A ++ +
Sbjct: 1113 -----------GRPRTIPRENIKGFSDAEIRRFIKSYKKFGGPLERLDAIARDAELVDKS 1161
Query: 1272 PLKAQVELFNALIDGCREAVEVGSL----------DLKGPLLDFYGVPMKANELLIRVQE 1321
+ L + +GC +A++ S +KGP GV + A ++ +E
Sbjct: 1162 ETDLR-RLGELVHNGCIKALKDSSSGTERTGGRLGKVKGPTFRISGVQVNAKLVISHEEE 1220
Query: 1322 LQLLAKRISRYEDPIAQFRVLTYLKPSNWSKGCGWNQIDDARLLLGVHYHGYGNWEVIRL 1381
L L K I + Q+ + + K +++ W + DD+ LL+G++ +GYG+WE+I++
Sbjct: 1221 LIPLHKSIPSDPEERKQYTIPCHTKAAHFD--IDWGKEDDSNLLIGIYEYGYGSWEMIKM 1278
Query: 1382 DERLGLTKKIAPVELQHHETFLPRAPNLRDRANALLEQELAVLGVKNASSKVGRKTSKKE 1441
D L LT KI P + P+A L+ RA+ L++ L K A S G +K
Sbjct: 1279 DPDLSLTHKILPDDPDKK----PQAKQLQTRADYLIKLLSRDLAKKEALSGAGSSKRRKA 1334
Query: 1442 REEREHLV--------------DISLSRGQEKKKNIGSSKVNVQMRKDR---LQKPLNV- 1483
R ++ + + + E + SK + + R + P+++
Sbjct: 1335 RAKKNKAMKSIKVKEEIKSDSSPLPSEKSDEDDDKLSESKSDGRERSKKSSVSDAPVHIT 1394
Query: 1484 ---EPI-VKEEGEMSDDDDVYEQFKEGKWKEWCQDLMVEEMKTLKRLHRLQTTSASLPKE 1539
EP+ + EE E D K C++ M LK+L R L +
Sbjct: 1395 ASGEPVPISEESEELDQ----------KTFSICKERMRPVKAALKQLDR---PEKGLSER 1441
Query: 1540 KVLSKIRNYLQLLGRRIDQIVSEQEDEPHKQDRMTTRLWKYVSTFSHLSGERLHQIY 1596
+ L R L +G I + + E + P + + LW +VS F+ +LH++Y
Sbjct: 1442 EQLEHTRQCLIKIGDHITECLKEYTN-PEQIKQWRKNLWIFVSKFTEFDARKLHKLY 1497
Score = 61.6 bits (148), Expect = 2e-07
Identities = 60/238 (25%), Positives = 103/238 (43%), Gaps = 36/238 (15%)
Query: 38 SDAEPDDASGKQNEAAAVDRLSTRESNVETTSRNPSASERWGSS----------YLKDCQ 87
SD++ SG Q+E+ + ++RE+ V+ +E W SS LK Q
Sbjct: 50 SDSDSGSESGSQSESESD---TSRENKVQAKPPKVDGAEFWKSSPSILAVQRSAILKKQQ 106
Query: 88 PMSPQNGSESGDDSKSGSDYRNEDEFEDNSSEGRGEKLGSEDEDGQ-KDSGKGQRGDSDV 146
Q ++ +S S D + ++ +D+SSE + +K +DED Q SG + SD
Sbjct: 107 QQQQQQQHQASSNSGSEEDSSSSEDSDDSSSEVKRKK--HKDEDWQMSGSGSPSQSGSDS 164
Query: 147 PAEEMLSDDSYGQDGEEQGESVHSRGFRPSTGSNSCLQPTSTNVNRRVHRKSRILDDAED 206
+EE S + + + +P S S N + + +K R +D +E+
Sbjct: 165 ESEEEREKSSCDETESDYEPKNKVKSRKPQNRSK------SKNGKKILGQKKRQIDSSEE 218
Query: 207 DDDDADYEEDEPDEDDPDDADFEPATSGRGANKYKDWEGEDSDEVDDSDEDIDVSDND 264
DDD+ DY+ D+ +S R A ++ ED + DSD+ ++V D
Sbjct: 219 DDDEEDYDNDK-------------RSSRRQATVNVSYK-EDEEMKTDSDDLLEVCGED 262
>ref|NP_990272.1| chromo-helicase-DNA-binding on the Z chromosome protein [Gallus
gallus] gi|2501846|gb|AAC60282.1|
chromo-helicase-DNA-binding on the Z chromosome protein
[Gallus gallus]
Length = 1808
Score = 418 bits (1074), Expect = e-115
Identities = 277/803 (34%), Positives = 422/803 (52%), Gaps = 76/803 (9%)
Query: 90 SPQNGSESGDDSKSGSDYRNEDEFEDNSSEGRGEKLGSEDEDGQKDSGKGQRGDSDVPAE 149
S S S DDS S S +SS+G + GS D + +SG +SD E
Sbjct: 13 SSGESSRSDDDSGSASG-SGSGSSSGSSSDGSSSQSGSSDSESGSESGSQSESESDTSRE 71
Query: 150 E-MLSDDSYGQDGEEQGESVHSRGFRPSTGSNSCLQPTSTNVNRRVHRKSRILDDAEDDD 208
+ + DG E +S P+ V R K +
Sbjct: 72 KKQVQAKPPKADGSEFWKS----------------SPSILAVQRSAVLKKQQQQQKAASS 115
Query: 209 DDADYEEDEPDEDDPDDADFEPATSGRGANKYKDWEGEDSDEVD------DSDEDIDVSD 262
D E+ ED DD+ E + + +K +DW+ S V +S ED D S
Sbjct: 116 DSGSEEDSSSSEDSADDSSSE---TKKKKHKDEDWQMSGSGSVSGTGSDSESAEDGDKSS 172
Query: 263 NDDLYFDKKAKGRQRGKFGPSVRSTRDCKAFTASSRQRRVKSSFEDEDENSTAEDSDSES 322
++ D + K + + + PS + K T ++R++ SS E+ED+
Sbjct: 173 CEESESDYEPKNKVKSRKPPSRIKPKSGKKSTGQ-KKRQLDSSEEEEDD----------- 220
Query: 323 DEDFKSLKKRGVRVRKNNGRSSAATSFSRPSNEVRSSSRTIRKVSYVESDESEGADEGTK 382
DED+ KRG R R + + + E ++ S D E E
Sbjct: 221 DEDYD---KRGSR------RQATVNVSYKEAEETKTDS----------DDLLEVCGEDVP 261
Query: 383 KSQKEEIEVDDGDSVEKVLWHQPKGMAAEAQRNNQSMEPVLMSHLFDSEPDWNNMEFLIK 442
+++++E E + ++ G + +P + F+ + +++LIK
Sbjct: 262 QTEEDEFETIEKFMDSRIGRKGATGASTTIYAVEADGDP---NAGFEKSKELGEIQYLIK 318
Query: 443 WKGQSHLHCQWKSFVDL--QNLSGFKKVLNYTKRVTEEIRNRMGISREEIEVNDVSKEMD 500
WKG SH+H W++ L QN+ G K+ NY K+ E R S E++E + +E+
Sbjct: 319 WKGWSHIHNTWETEETLKQQNVKGMNKLDNYKKKDQETKRWLKNASPEDVEYYNCQQELT 378
Query: 501 IDIIKQNSQVERIIADRISNDNSGNVFPEYLVKWQGLSYAEVTWEKDIDIAFA-QHTIDE 559
D+ KQ VERIIA SN S +P+Y KWQGL Y+E +WE IA Q IDE
Sbjct: 379 DDLHKQYQIVERIIAH--SNQKSAAGYPDYYCKWQGLPYSECSWEDGALIAKKFQARIDE 436
Query: 560 YKTREAAMSVQGKMVDFQRRQSKGSLRKLDEQPEWLKGGK---LRDYQLEGLNFLVNSWK 616
Y +R + + K D + + + L +QP ++ G + LRDYQL GLN+L +SW
Sbjct: 437 YFSRNQSKTTPFK--DCKVLKQRPRFVALKKQPSYIGGHESLELRDYQLNGLNWLAHSWC 494
Query: 617 NDTNVVLADEMGLGKTVQSVSMLGFLQNAQQIHGPFLVVVPLSTLSNWAKEFRKWLPDLN 676
+ +LADEMGLGKT+Q++S L +L + Q++GPFL+ VPLSTL++W +E + W P +N
Sbjct: 495 KGNSCILADEMGLGKTIQTISFLNYLFHEHQLYGPFLLRVPLSTLTSWQREIQTWAPQMN 554
Query: 677 VIVYVGTRSSREVCQQYEFCNEKKAGKQIKFNALLTTYEVVLKDKAVLSKIKWNYLMVDE 736
+VY+G +SR + + +E+ + + K++KFN LLTTYE++LKDK+ L + W ++ VDE
Sbjct: 555 AVVYLGDITSRNMIRTHEWMHPQT--KRLKFNILLTTYEILLKDKSFLGGLNWAFIGVDE 612
Query: 737 AHRLKNSEAQLYTALSEFNTKNKLLITGTPLQNSVEELWALLHFLDSDKFKSKDEFAQNY 796
AHRLKN ++ LY L +F + ++LLITGTPLQNS++ELW+LLHF+ +KF S ++F + +
Sbjct: 613 AHRLKNDDSLLYRTLIDFKSNHRLLITGTPLQNSLKELWSLLHFIMPEKFSSWEDFEEEH 672
Query: 797 KNLSSFNENELSNLHMELRPHMLRRVIKDVEKSLPPKIERILRVDMSPLQKQYYKWILER 856
E ++LH EL P +LRRV KDVEKSLP K+E+ILR++MS LQKQYYKWIL R
Sbjct: 673 ---GKGREYGYASLHKELEPFLLRRVKKDVEKSLPAKVEQILRMEMSALQKQYYKWILTR 729
Query: 857 NFRDLNKGVRGNQSKYCSRIEEV 879
N++ L+KG +G+ S + + + E+
Sbjct: 730 NYKALSKGSKGSTSGFLNIMMEL 752
Score = 333 bits (855), Expect = 2e-89
Identities = 230/622 (36%), Positives = 322/622 (50%), Gaps = 85/622 (13%)
Query: 938 DILAQYMSLRGFQFQRLDGSTKSELRQQAMDHFNAPGSDDFCFLLSTRAGGLGINLATAD 997
DILA+Y+ R F FQRLDGS K ELR+QA+DHFNA GS+DFCFLLSTRAGGLGINLA+AD
Sbjct: 815 DILAEYLKYRQFPFQRLDGSIKGELRKQALDHFNAEGSEDFCFLLSTRAGGLGINLASAD 874
Query: 998 TVIIFDSDWNPQNDLQAMSRAHRIGQREVVNIYRFVTSKSVEEDILERAKKKMVLDHLVI 1057
TV+IFDSDWNPQNDLQA +RAHRIGQ++ VNIYR VT SVEEDILERAKKKMVLDHLVI
Sbjct: 875 TVVIFDSDWNPQNDLQAQARAHRIGQKKQVNIYRLVTKGSVEEDILERAKKKMVLDHLVI 934
Query: 1058 QKLNAEGK--LEKKEAKKGGSFFDKNELSAILRFGAEELFKEERNDEESKKRLLSMDIDE 1115
Q+++ GK L + F+K ELSAIL+FGAEELFKE +E+ + MDIDE
Sbjct: 935 QRMDTTGKTVLHTGSTPSSSTPFNKEELSAILKFGAEELFKEPEGEEQEPQ---EMDIDE 991
Query: 1116 ILERAEKVEEKENGGEQAHELLSAFKVANFCN-DEDD--------GSFWSRWIKADSVAQ 1166
IL+RAE E + ELLS FKVANF N DEDD W I +
Sbjct: 992 ILKRAETRENEPGPLTVGDELLSQFKVANFSNMDEDDIELEPERNSRNWEEIIPESQRRR 1051
Query: 1167 AENA----------LAPRAARNIKSYAEADQSERSKKRKKKENEPTERIPKRRKADYSAH 1216
E + PR K + R + ++ ++ I +R++
Sbjct: 1052 IEEEERQKELEEIYMLPRMRNCAKQISFNGSEGRRSRSRRYSGSDSDSITERKRPKKR-- 1109
Query: 1217 VISMIDGASAQVRSWSYGNLSKRDALRFSRSVMKFGNESQINLIVAEVGGAIEAAPLKAQ 1276
G + + S + RF +S KFG + VA ++ + +
Sbjct: 1110 ------GRPRTIPRENIKGFSDAEIRRFIKSYKKFGGPLERLDAVARDAELVDKSETDLR 1163
Query: 1277 VELFNALIDGCREAVEVGSL----------DLKGPLLDFYGVPMKANELLIRVQELQLLA 1326
L + +GC +A++ S +KGP GV + A ++ +EL L
Sbjct: 1164 -RLGELVHNGCIKALKDNSSGQERAGGRLGKVKGPTFRISGVQVNAKLVISHEEELAPLH 1222
Query: 1327 KRISRYEDPIAQFRVLTYLKPSNWSKGCGWNQIDDARLLLGVHYHGYGNWEVIRLDERLG 1386
K I + ++ + + K +++ W + DD+ LL+G++ +GYG+WE+I++D L
Sbjct: 1223 KSIPSDPEERKRYVIPCHTKAAHFD--IDWGKEDDSNLLVGIYEYGYGSWEMIKMDPDLS 1280
Query: 1387 LTKKIAPVELQHHETFLPRAPNLRDRANALLEQELAVLGVKNASSKVGRKTSKKEREERE 1446
LT+KI P + P+A L+ RA+ L++ L K A G SK+ +
Sbjct: 1281 LTQKILPDDPDKK----PQAKQLQTRADYLIKLLNKDLARKEAQRLAGAGNSKRRKT--- 1333
Query: 1447 HLVDISLSRGQEKKKNIGSSKVNVQMRKDRLQKPLNVEPIVKEEGEMSDDDDVYEQFKEG 1506
+ KK + +SK+ +++ D +P E SD+DD E K
Sbjct: 1334 ----------RNKKNKMKASKIKEEIKSDSSPQP----------SEKSDEDDEEEDNK-- 1371
Query: 1507 KWKEWCQDLMVEEMKTLKRLHR 1528
+E+ ++K LH+
Sbjct: 1372 -----------DEIVSVKHLHK 1382
Score = 57.4 bits (137), Expect = 4e-06
Identities = 64/263 (24%), Positives = 106/263 (39%), Gaps = 36/263 (13%)
Query: 38 SDAEPDDASGKQNEAAAVDRLSTRESNVETTSRNPSASERWGSS----------YLKDCQ 87
SD+E SG Q+E+ + S + V+ SE W SS LK Q
Sbjct: 50 SDSESGSESGSQSESES--DTSREKKQVQAKPPKADGSEFWKSSPSILAVQRSAVLKKQQ 107
Query: 88 PMSPQNGSESGDDSKSGSDYRNEDEFEDNSSEGRGEKLGSEDEDGQKDSGKGQRGDSDVP 147
S+SG + S S +ED +D+SSE + +K +DED Q SG G +
Sbjct: 108 QQQKAASSDSGSEEDSSS---SEDSADDSSSETKKKK--HKDEDWQM-SGSGSVSGTG-- 159
Query: 148 AEEMLSDDSYGQDGEEQGESVHSRGFRPSTGSNSCLQPT---STNVNRRVHRKSRILDDA 204
SD +DG++ + P S P+ + + +K R LD +
Sbjct: 160 -----SDSESAEDGDKSSCEESESDYEPKNKVKSRKPPSRIKPKSGKKSTGQKKRQLDSS 214
Query: 205 EDDDDDADYEEDEPDEDDPDDADFEPATSGRGANKYKDWEGEDSDEVDDSDEDIDVSDND 264
E+++DD DED AT + ++ + + D ++ ED+ ++ D
Sbjct: 215 EEEEDD--------DEDYDKRGSRRQATVNVSYKEAEETKTDSDDLLEVCGEDVPQTEED 266
Query: 265 DLYFDKKAKGRQRGKFGPSVRST 287
+ +K + G+ G + ST
Sbjct: 267 EFETIEKFMDSRIGRKGATGAST 289
Score = 42.4 bits (98), Expect = 0.14
Identities = 42/189 (22%), Positives = 80/189 (42%), Gaps = 28/189 (14%)
Query: 1427 KNASSKVGRKTSKKEREEREHLVDISLSRGQEKKKN-----------IGSSKVNVQMRKD 1475
+ K ++ K+E++E+E ++ +EK++N + KVN +M+ +
Sbjct: 1408 RETKEKENKRELKREKKEKEDKKELKEKDNKEKRENKVKESTQKEKEVKEEKVN-EMKSE 1466
Query: 1476 RLQK----PLNVEPI----VKEEGEMSDDDDVYEQFKEGKWKEWCQDLMVEEMKTLKRLH 1527
+K PL P+ E +S++ + Q K C++ M LK+L
Sbjct: 1467 NKEKSKKIPLLDTPVHITATSEPVPISEESEELHQ----KTFSVCKERMRPVKAALKQLD 1522
Query: 1528 RLQTTSASLPKEKVLSKIRNYLQLLGRRIDQIVSEQEDEPHKQDRMTTRLWKYVSTFSHL 1587
R L + + L R L +G I + + E + P + + LW +VS F+
Sbjct: 1523 R---PEKGLSEREQLEHTRQCLIKIGDHITECLKEYTN-PEQIKQWRKNLWIFVSKFTEF 1578
Query: 1588 SGERLHQIY 1596
+LH++Y
Sbjct: 1579 DARKLHKLY 1587
>gb|AAB87382.1| CHD2 [Homo sapiens] gi|3182950|sp|O14647|CHD2_HUMAN
Chromodomain-helicase-DNA-binding protein 2 (CHD-2)
gi|4557449|ref|NP_001262.1| chromodomain helicase DNA
binding protein 2 [Homo sapiens]
Length = 1739
Score = 416 bits (1070), Expect = e-114
Identities = 280/810 (34%), Positives = 426/810 (52%), Gaps = 112/810 (13%)
Query: 127 SEDEDGQKDSGKGQRGDSDVPAEEMLSDDSYGQDGEEQG---------------ESVHSR 171
++D+ ++DS S +EE DS Q EQG ES S+
Sbjct: 4 NKDKSQEEDSSLHSNASSHSASEEASGSDSGSQSESEQGSDPGSGHGSESNSSSESSESQ 63
Query: 172 GFRPSTGSNSCLQPTSTNVNRR-VHRKSRILDDA---EDDDDDADYEEDEPDEDDPDDAD 227
S + S QP + +K RI D E+ D +P +
Sbjct: 64 SESESESAGSKSQPVLPEAKEKPASKKERIADVKKMWEEYPDVYGVRRSNRSRQEPSRFN 123
Query: 228 F-EPATSG-----------RGANKYKDWEGEDSDEVDDSDEDIDVSDNDDLYFDKKAKGR 275
E A+SG R K + W+ E S+ D+ ++ + KK K R
Sbjct: 124 IKEEASSGSESGSPKRRGQRQLKKQEKWKQEPSE--DEQEQGTSAESEPE---QKKVKAR 178
Query: 276 QRGKFGPSVRSTRDCKAFTASSRQRRVKSSFEDEDENSTAEDSDSESDEDFKSLKKRGVR 335
+ P R T + RVK + + +DS E D+D ++ K++ R
Sbjct: 179 R-----PVPRRTVP---------KPRVKKQPKTQRGKRKKQDSSDEDDDDDEAPKRQTRR 224
Query: 336 VRKNNGRSSAATSFSRPSNEVRSSSRTIRKVSYVESDESEGADEGTKKSQKEEIEV--DD 393
R + VSY E D+ E + + E ++ D+
Sbjct: 225 -------------------------RAAKNVSYKEDDDFETDSDDLIEMTGEGVDEQQDN 259
Query: 394 GDSVEKVLWHQ-----PKGMAAEAQRNNQSMEPVLMSHLFDSEPDWNNMEFLIKWKGQSH 448
+++EKVL + G + + +P S FD+E D +++LIKWKG S+
Sbjct: 260 SETIEKVLDSRLGKKGATGASTTVYAIEANGDP---SGDFDTEKDEGEIQYLIKWKGWSY 316
Query: 449 LHCQWKSFVDLQN--LSGFKKVLNYTKRVTEEIRNRMG-ISREEIEVNDVSKEMDIDIIK 505
+H W+S LQ + G KK+ N+ K+ +EI+ +G +S E++E + +E+ ++ K
Sbjct: 317 IHSTWESEESLQQQKVKGLKKLENFKKK-EDEIKQWLGKVSPEDVEYFNCQQELASELNK 375
Query: 506 QNSQVERIIADRISNDNSGNVF-------------PEYLVKWQGLSYAEVTWEKDIDIAF 552
Q VER+IA + S G PEYL KW GL Y+E +WE + I
Sbjct: 376 QYQIVERVIAVKTSKSTLGQTDFPAHSRKPAPSNEPEYLCKWMGLPYSECSWEDEALIGK 435
Query: 553 A-QHTIDEYKTREAAMSVQGKMVDFQRRQSKGSLRKLDEQPEWLKGG--KLRDYQLEGLN 609
Q+ ID + +R + ++ + + + + + L +QP +L G +LRDYQLEGLN
Sbjct: 436 KFQNCIDSFHSRNNSKTIPTR--ECKALKQRPRFVALKKQPAYLGGENLELRDYQLEGLN 493
Query: 610 FLVNSWKNDTNVVLADEMGLGKTVQSVSMLGFLQNAQQIHGPFLVVVPLSTLSNWAKEFR 669
+L +SW + +V+LADEMGLGKT+Q++S L +L + Q++GPFL+VVPLSTL++W +EF
Sbjct: 494 WLAHSWCKNNSVILADEMGLGKTIQTISFLSYLFHQHQLYGPFLIVVPLSTLTSWQREFE 553
Query: 670 KWLPDLNVIVYVGTRSSREVCQQYEFCNEKKAGKQIKFNALLTTYEVVLKDKAVLSKIKW 729
W P++NV+VY+G SR ++YE+ + + K++KFNAL+TTYE++LKDK VL I W
Sbjct: 554 IWAPEINVVVYIGDLMSRNTIREYEWIHSQT--KRLKFNALITTYEILLKDKTVLGSINW 611
Query: 730 NYLMVDEAHRLKNSEAQLYTALSEFNTKNKLLITGTPLQNSVEELWALLHFLDSDKFKSK 789
+L VDEAHRLKN ++ LY L +F + ++LLITGTPLQNS++ELW+LLHF+ +KF+
Sbjct: 612 AFLGVDEAHRLKNDDSLLYKTLIDFKSNHRLLITGTPLQNSLKELWSLLHFIMPEKFEFW 671
Query: 790 DEFAQNYKNLSSFNENELSNLHMELRPHMLRRVIKDVEKSLPPKIERILRVDMSPLQKQY 849
++F +++ EN +LH L P +LRRV KDVEKSLP K+E+ILRV+MS LQKQY
Sbjct: 672 EDFEEDH---GKGRENGYQSLHKVLEPFLLRRVKKDVEKSLPAKVEQILRVEMSALQKQY 728
Query: 850 YKWILERNFRDLNKGVRGNQSKYCSRIEEV 879
YKWIL RN++ L KG RG+ S + + + E+
Sbjct: 729 YKWILTRNYKALAKGTRGSTSGFLNIVMEL 758
Score = 347 bits (891), Expect = 2e-93
Identities = 263/767 (34%), Positives = 377/767 (48%), Gaps = 143/767 (18%)
Query: 938 DILAQYMSLRGFQFQRLDGSTKSELRQQAMDHFNAPGSDDFCFLLSTRAGGLGINLATAD 997
DILA+Y++++ + FQRLDGS K E+R+QA+DHFNA GS+DFCFLLSTRAGGLGINLA+AD
Sbjct: 821 DILAEYLTIKHYPFQRLDGSIKGEIRKQALDHFNADGSEDFCFLLSTRAGGLGINLASAD 880
Query: 998 TVIIFDSDWNPQNDLQAMSRAHRIGQREVVNIYRFVTSKSVEEDILERAKKKMVLDHLVI 1057
TV+IFDSDWNPQNDLQA +RAHRIGQ++ VNIYR VT +VEE+I+ERAKKKMVLDHLVI
Sbjct: 881 TVVIFDSDWNPQNDLQAQARAHRIGQKKQVNIYRLVTKGTVEEEIIERAKKKMVLDHLVI 940
Query: 1058 QKLNAEGK--LEKKEAKKGGSFFDKNELSAILRFGAEELFKEERNDEESKKRLLSMDIDE 1115
Q+++ G+ LE + + F+K EL+AIL+FGAE+LFKE +E + MDIDE
Sbjct: 941 QRMDTTGRTILENNSGRSNSNPFNKEELTAILKFGAEDLFKELEGEESEPQ---EMDIDE 997
Query: 1116 ILERAEKVEEKENGGEQAHELLSAFKVANFCNDEDDGSF-------WSRWIKADSVAQAE 1168
IL AE E E ELLS FKVANF ED+ W I + + E
Sbjct: 998 ILRLAE-TRENEVSTSATDELLSQFKVANFATMEDEEELEERPHKDWDEIIPEEQRKKVE 1056
Query: 1169 NA----------LAPRAARNIKSYA--EADQSERSKKRKKK--------ENEPTERIPKR 1208
+ PR + K ++D SK++ ++ E+ ++ PKR
Sbjct: 1057 EEERQKELEEIYMLPRIRSSTKKAQTNDSDSDTESKRQAQRSSASESETEDSDDDKKPKR 1116
Query: 1209 RKADYSAHVISMIDGASAQVRSWSYGNLSKRDALRFSRSVMKFGNESQINLIVAEVGGAI 1268
R G VR + + RF ++ KFG + +A +
Sbjct: 1117 R-------------GRPRSVRKDLVEGFTDAEIRRFIKAYKKFGLPLERLECLARDAELV 1163
Query: 1269 E--AAPLKAQVELF-NALIDGCREAVE---------VGSLDLKGPLLDFYGVPMKANELL 1316
+ A LK EL N+ + +E E G +GP + GV + ++
Sbjct: 1164 DKSVADLKRLGELIHNSCVSAMQEYEEQLKENASEGKGPGKRRGPTIKISGVQVNVKSII 1223
Query: 1317 IRVQELQLLAKRISRYEDPIAQFRVLTYLKPSNWSKGCGWNQIDDARLLLGVHYHGYGNW 1376
+E ++L K I + ++ + +K +++ W DD+RLLLG++ HGYGNW
Sbjct: 1224 QHEEEFEMLHKSIPVDPEEKKKYCLTCRVKAAHFD--VEWGVEDDSRLLLGIYEHGYGNW 1281
Query: 1377 EVIRLDERLGLTKKIAPVELQHHETFLPRAPNLRDRANAL-------LEQELAVLGVKNA 1429
E+I+ D L LT KI PVE P+ L+ RA+ L LE++ AV G + A
Sbjct: 1282 ELIKTDPELKLTDKILPVETDKK----PQGKQLQTRADYLLKLLRKGLEKKGAVTGGEEA 1337
Query: 1430 SSKVGRKTSKKE----REEREHLVDIS-----------------------LSRGQEKKKN 1462
K + KKE R + EH +++S + + Q+KK+N
Sbjct: 1338 KLKKRKPRVKKENKVPRLKEEHGIELSSPRHSDNPSEEGEVKDDGLEKSPMKKKQKKKEN 1397
Query: 1463 IGSSKVNVQMRKD-----------------------------RLQKPLNV----EPIVKE 1489
+ + + RKD R Q P+++ EP+
Sbjct: 1398 KENKEKQMSSRKDKEGDKERKKSKDKKEKPKSGDAKSSSKSKRSQGPVHITAGSEPV--P 1455
Query: 1490 EGEMSDDDDVYEQFKEGKWKEWCQDLMVEEMKTLKRLHRLQTTSASLPKEKVLSKIRNYL 1549
GE DDD E F C++ M K LK+L + L ++ L RN L
Sbjct: 1456 IGEDEDDDLDQETF------SICKERMRPVKKALKQLDK---PDKGLNVQEQLEHTRNCL 1506
Query: 1550 QLLGRRIDQIVSEQEDEPHKQDRMTTRLWKYVSTFSHLSGERLHQIY 1596
+G RI + + D+ H + LW +VS F+ +LH++Y
Sbjct: 1507 LKIGDRIAECLKAYSDQEHIK-LWRRNLWIFVSKFTEFDARKLHKLY 1552
>gb|EAL33204.1| GA17649-PA [Drosophila pseudoobscura]
Length = 1908
Score = 416 bits (1068), Expect = e-114
Identities = 279/832 (33%), Positives = 422/832 (50%), Gaps = 103/832 (12%)
Query: 93 NGSESGDDSKSGSDYRNEDEFEDNSSEGRGE-----KLGSEDEDGQKDSGKGQRGDSDVP 147
+GS SG S SGSD + D NSS+GR K S G + + DS
Sbjct: 27 SGSGSGSGS-SGSD-SDSDSSSGNSSDGRSSPEPNVKPSSTSVAGFPPTAAAAQADSKTN 84
Query: 148 A-----EEMLSDDSYGQDGEEQGESVHSRGFRPSTG----SNSCLQPTSTNVNRRVHRKS 198
EE S S G D + E + +TG +N+ T+++ ++
Sbjct: 85 GFTDDQEESSSGGSSGSDSDSDPEETRTGADHTATGGKLSANTSTSTTNSSSKPAADSEA 144
Query: 199 RILDDAEDDDDDADYEEDEPDEDDPDDADFEPATSGRGAN-----------------KYK 241
DDA D +A +D ++ D+ P + + K K
Sbjct: 145 SADDDASDSSPNASPTSSSSSSEDEEE-DYRPKRTRQARKPPTAAPDKSKRAPPHKKKKK 203
Query: 242 DWEGEDSDEVDDSDEDIDVSDNDDLYFDKKAKGRQRGKFGPSVRSTRDCKAFTASSRQRR 301
W+ ++SDE D+SDE+ K P+ +T K ++RR
Sbjct: 204 TWDSDESDESDESDEESSAP-----------------KRKPAPATTSRNKPVPQQQQRRR 246
Query: 302 VKSSFEDEDENSTAEDSDSESDEDFKSLKKRGVRVRKNNGRSSAATSFSRPSNEVRSSSR 361
VKS D DS+ D+ K R R+ AA S+ S + + S
Sbjct: 247 VKSFSSD----------DSDDDDASKRCATR---------RTGAAVSYKEASEDEATDSE 287
Query: 362 TIRKVSYVESDESEGADEGTKKSQKEEIEVD----------DGDSVEKVLWHQPKGMAAE 411
+ + Y E + A + + E IE G+ P E
Sbjct: 288 DLLECEYDEIQAAASAAAAVEDEKCETIERILAQRMGRRGCTGNKTTIYAIEDPHAKVEE 347
Query: 412 AQRNNQSMEPVLMSHLFDSEPDWNNMEFLIKWKGQSHLHCQWKSFVDLQNLS--GFKKVL 469
++ ++ E + P ++LIKWKG S++H W+S L+++ G KK+
Sbjct: 348 VEKVAEAEEDL---------PKEEEEQYLIKWKGWSYIHNTWESERTLRDMKAKGMKKLD 398
Query: 470 NYTKRVTEEIRNRMGISREEIEVNDVSKEMDIDIIKQNSQVERIIADRISNDNSGNVFPE 529
N+ K+ E R E+I+ + +E+ +++K + V+RIIA D + E
Sbjct: 399 NFIKKEQETAYWRRYAGPEDIDYFECQQELQHELLKSYNNVDRIIAKGSKPDVGTD---E 455
Query: 530 YLVKWQGLSYAEVTWEK-DIDIAFAQHTIDEYKTREAAMSVQGKMVDFQRRQSKGSLRKL 588
+L KWQ L YAE TWE + + Q +++ RE++ + + + K S ++
Sbjct: 456 FLCKWQSLPYAEATWEDATLVLRKWQRCAEQFHDRESSKCTPSRHCRVLKYRPKFS--RI 513
Query: 589 DEQPEWLKGGK-LRDYQLEGLNFLVNSWKNDTNVVLADEMGLGKTVQSVSMLGFLQNAQQ 647
QP++L G LRDYQ++GLN+L++SW + +V+LADEMGLGKT+Q++ L L
Sbjct: 514 KNQPDYLVAGLVLRDYQMDGLNWLLHSWCKENSVILADEMGLGKTIQTICFLYSLFKVHH 573
Query: 648 IHGPFLVVVPLSTLSNWAKEFRKWLPDLNVIVYVGTRSSREVCQQYEFCNEKKAGKQIKF 707
++GPFL VVPLST++ W +EF W PD+NV+ Y+G SRE+ QQYE+ + + K++KF
Sbjct: 574 LYGPFLCVVPLSTMTAWQREFDLWAPDMNVVTYLGDIKSREMIQQYEW--QFEGSKRLKF 631
Query: 708 NALLTTYEVVLKDKAVLSKIKWNYLMVDEAHRLKNSEAQLYTALSEFNTKNKLLITGTPL 767
N +LTTYE+VLKDK L ++W L+VDEAHRLKN ++ LY +L EF+T ++LLITGTPL
Sbjct: 632 NCILTTYEIVLKDKQFLGTLQWAALLVDEAHRLKNDDSLLYKSLKEFDTNHRLLITGTPL 691
Query: 768 QNSVEELWALLHFLDSDKFKSKDEFAQNYKNLSSFNENELSNLHMELRPHMLRRVIKDVE 827
QNS++ELWALLHF+ +KF + D F + N + + LH +L P++LRRV KDVE
Sbjct: 692 QNSLKELWALLHFIMPEKFDTWDNFELQHGNA---EDKGYTRLHQQLEPYILRRVKKDVE 748
Query: 828 KSLPPKIERILRVDMSPLQKQYYKWILERNFRDLNKGVRGNQSKYCSRIEEV 879
KSLP K+E+ILRV+M+ LQKQYYKWIL +NF L KG +G+ S + + + E+
Sbjct: 749 KSLPAKVEQILRVEMTSLQKQYYKWILTKNFDALRKGKKGSTSTFLNIVIEL 800
Score = 290 bits (741), Expect = 4e-76
Identities = 267/820 (32%), Positives = 372/820 (44%), Gaps = 174/820 (21%)
Query: 938 DILAQYMSLRGFQFQRLDGSTKSELRQQAMDHFNAPGSDDFCFLLSTRAGGLGINLATAD 997
D+LA Y+ R F FQRLDGS K E+R+QA+DHFNA GS DFCFLLSTRAGGLGINLATAD
Sbjct: 864 DVLADYLQKRHFPFQRLDGSIKGEMRRQALDHFNAEGSQDFCFLLSTRAGGLGINLATAD 923
Query: 998 TVIIFDSDWNPQNDLQAMSRAHRIGQREVVNIYRFVTSKSVEEDILERAKKKMVLDHLVI 1057
TVIIFDSDWNPQNDLQA +RAHRIGQ+ VNIYR VT++SVEE I+ERAK+KMVLDHLVI
Sbjct: 924 TVIIFDSDWNPQNDLQAQARAHRIGQKNQVNIYRLVTARSVEEQIVERAKQKMVLDHLVI 983
Query: 1058 QKLNAEGKLEKKEAKKGGSF----FDKNELSAILRFGAEELFKEERNDEESKKRLLSMDI 1113
Q+++ G+ ++ G S F+K++LSAIL+FGAEELFK+E +E + L DI
Sbjct: 984 QRMDTTGRTVLDKSGNGHSSNSNPFNKDDLSAILKFGAEELFKDEPEHDEHE---LVCDI 1040
Query: 1114 DEILERAE-KVEEKENGGEQAHELLSAFKVANFC------------------NDEDDGSF 1154
DEIL RAE + ++ E G+ +LLSAFKVA+ + EDD
Sbjct: 1041 DEILRRAETRNDDPEMPGD---DLLSAFKVASIAAFEEEPSESNKQDVDAGDDAEDDSKD 1097
Query: 1155 WSRWIK---ADSVAQAENA-------LAPRAARNIKSYAEADQSERSKKRKKKENEPTER 1204
W I + E A L PR + +E+ + K + K+ + +
Sbjct: 1098 WDDIIPEGFRKVIEDQEKAKEIEDLYLPPRRKTAAANQSESKRGAGGKAGRSKQADDS-- 1155
Query: 1205 IPKRRKADYSAHVISMIDGASAQVRSWSYGNLSKR-------DALRFSRSVMKFGNESQI 1257
D + S G + R + ++ + RF RS KF
Sbjct: 1156 ------GDSEYELGSDASGDEGRPRKRGRPTMKEKITGFTDAELRRFIRSYKKFPAPLH- 1208
Query: 1258 NLIVAEVGGAIEAAPLKAQVELFNALIDGC--------REAVEVGSLDLKGP-------- 1301
L ++ PL L L D C E ++ D G
Sbjct: 1209 RLEAIACDAELQEKPLTELKRLGEMLHDRCVQFLDEHKEEEIKTVQDDTPGAKQRRARAT 1268
Query: 1302 -LLDFYGVPMKANELLIRVQELQLLAKRISRYEDPIAQFRVLTY-LKPSNWSKGCGWNQI 1359
+ GV A +LL QELQ L++ I A+ + T+ +K W
Sbjct: 1269 YSVKLGGVSFNAKKLLTSEQELQPLSEII---PSGAAERQQWTFNIKTRAPLFDVEWGNE 1325
Query: 1360 DDARLLLGVHYHGYGNWEVIRLDERLGLTKKIAPVELQHHETFLPRAPNLRDRANALLE- 1418
+D +LL G++ +G G+WE ++LD L LT KI ++T P+A L+ RA LL+
Sbjct: 1326 EDTKLLCGIYQYGIGSWEQMKLDPTLKLTDKIL-----LNDTRKPQAKQLQTRAEYLLKI 1380
Query: 1419 ----QELAVLGVK-----------NASSKVGRKTS-----------KKEREEREHLVD-- 1450
EL G + NA SK G ++ K E E VD
Sbjct: 1381 IKKNVELTKKGGQKRQRRPRAARINAESKAGNNSNNHSATSGAMAESKTLGEAEETVDGG 1440
Query: 1451 -----------------------------------ISLSRGQEKKKNIGSSKVNVQMRKD 1475
S+ + KK + + N M
Sbjct: 1441 RGALDNSSSHLDPSSTASPHLTPASEVSGGGTAQRTKKSKARSKKTSASDNNGNKPMHFT 1500
Query: 1476 RLQKPLNVEPIVKEEGEMSDDDDVYEQFKEGKWKEWCQDLMVEEMKTLKR-LHRLQTTSA 1534
+P +E + G++ D ++ + K E+M+ +K+ L L
Sbjct: 1501 ANNEPRALEVL----GDL--DPSIFNECK-------------EKMRPVKKALKALDQPDL 1541
Query: 1535 SLPKEKVLSKIRNYLQLLGRRIDQIVSEQEDEPHKQDRMTTRLWKYVSTFSHLSGERLHQ 1594
SL + L R+ L +GR+ID + + K+ R + LW +VS F+ L + L +
Sbjct: 1542 SLSDQDQLQHTRDCLLQIGRQIDVCLQPYGESEKKEWR--SNLWYFVSKFTELDAKHLFK 1599
Query: 1595 IYS---KLKLEQNAVGVGSSLPNG---SVS-GPFSRNGNP 1627
IY K +L + + G G S SVS RNG P
Sbjct: 1600 IYKHALKQQLSRESGGKGKSRAGSKEDSVSPSKIRRNGLP 1639
Score = 40.8 bits (94), Expect = 0.41
Identities = 54/221 (24%), Positives = 85/221 (38%), Gaps = 35/221 (15%)
Query: 37 ESDAEPDDASGKQNEAAAVDRLSTRESNVETTSRNPSASERWGSSYLKDCQPMSPQNGSE 96
+SD++P++ + A +LS S T S + A++ S+ D SP +
Sbjct: 102 DSDSDPEETRTGADHTATGGKLSANTSTSTTNSSSKPAADSEASAD-DDASDSSP---NA 157
Query: 97 SGDDSKSGSDYRNEDEFEDNSSEGRGEKLGSEDEDGQKDSGKGQRGDSDVPAEEMLSDDS 156
S S S S+ ED + + R + D+ + K ++ D SD+S
Sbjct: 158 SPTSSSSSSEDEEEDYRPKRTRQARKPPTAAPDKSKRAPPHKKKKKTWD-------SDES 210
Query: 157 YGQDGEEQGESVHSRGFRPSTGSNSCLQPTSTNVNRRVHRKSRILDDAEDDDDDADYEED 216
D ++ S R P+T S + +P RR R+ + DD DD
Sbjct: 211 DESDESDEESSAPKRKPAPATTSRN--KPVPQQQQRR-----RVKSFSSDDSDD------ 257
Query: 217 EPDEDDPDDADFEPATSGRGAN-KYKDWEGEDSDEVDDSDE 256
DDA AT GA YK+ DE DS++
Sbjct: 258 -------DDASKRCATRRTGAAVSYKE---ASEDEATDSED 288
>gb|AAO61783.1| chromo-helicase DNA-binding protein [Taeniopygia guttata]
Length = 1806
Score = 416 bits (1068), Expect = e-114
Identities = 277/801 (34%), Positives = 426/801 (52%), Gaps = 68/801 (8%)
Query: 90 SPQNGSESGDDSKSGSDYRNEDEFEDNSSEGRGEKLGSEDEDGQKDSGKGQRGDSDVPAE 149
S + S S DDS S S +SS+G + GS D D SG +SD E
Sbjct: 13 SSRESSRSDDDSGSASA-SGSGSSSGSSSDGSSSQSGSSDSDCGSGSGTQSESESDTSRE 71
Query: 150 EMLSDDSYGQDGEEQGESVHSRGFRPSTGSNSCLQPTSTNVNRRVHRKSRILDDAEDDD- 208
+ + + + V F S+ S +Q ++ + ++ +K+ D ++D
Sbjct: 72 K--------KQIQSKPPKVDGSEFWKSSPSILAVQRSAV-LKKQQQQKAASSDSVSEEDS 122
Query: 209 ----DDADYEEDEPDEDDPDDADFEPATSGRGANKYKDWEGEDSDEVDDSDEDIDVSDND 264
D AD E + D D++ + SG + D E E+ E +E+
Sbjct: 123 SSSEDSADDSSSETKKKTHKDEDWQMSGSGSMSGTGSDSESEEDREKSSCEEN-----ES 177
Query: 265 DLYFDKKAKGRQRGKFGPSVRSTRDCKAFTASSRQRRVKSSFEDEDENSTAEDSDSESDE 324
D K K R+ P +R +A ++R++ SS +D+D D + E D+
Sbjct: 178 DYEPKNKVKSRK-----PPIRIKPKSGKKSAGPKKRQLDSSEDDDD------DEEEEEDD 226
Query: 325 DFKSLKKRGVRVRKNNGRSSAATSFSRPSNEVRSSSRTIRKVSYVESDESEGADEGTKKS 384
D+ KRG R R + + + E ++ S D E E +
Sbjct: 227 DYD---KRGSR------RQATVNISYKEAEETKTDS----------DDLLEVCGEDVPQP 267
Query: 385 QKEEIEVDDGDSVEKVLWHQPKGMAAEAQRNNQSMEPVLMSHLFDSEPDWNNMEFLIKWK 444
+++E E + +V G A +P + F+ + +++LIKWK
Sbjct: 268 EEDEFETIEKFMDSRVGRKGATGAATTIYAVEADGDP---NAGFEKSKEPAEVQYLIKWK 324
Query: 445 GQSHLHCQWKSFVDL--QNLSGFKKVLNYTKRVTEEIRNRMGISREEIEVNDVSKEMDID 502
G SH+H W++ L QN+ G KK+ NY K+ E R S E++E + +E+ D
Sbjct: 325 GWSHIHNTWETEETLKQQNVKGMKKLDNYKKKDQETKRWLKNASPEDVEYYNCQQELTDD 384
Query: 503 IIKQNSQVERIIADRISNDNSGNVFPEYLVKWQGLSYAEVTWEKDIDIAFA-QHTIDEYK 561
+ KQ VERIIA SN S +P+Y KWQGL Y+E +WE IA Q IDEY
Sbjct: 385 LHKQYQIVERIIAH--SNQKSAAGYPDYYCKWQGLPYSECSWEDGALIAKKFQARIDEYF 442
Query: 562 TREAAMSVQGKMVDFQRRQSKGSLRKLDEQPEWLKGGK---LRDYQLEGLNFLVNSWKND 618
+R + + K D + + + L +QP ++ G + LRDYQL GLN+L +SW
Sbjct: 443 SRNQSKTTPFK--DCKVLKQRPRFVALKKQPSYIGGHESLGLRDYQLNGLNWLAHSWCKG 500
Query: 619 TNVVLADEMGLGKTVQSVSMLGFLQNAQQIHGPFLVVVPLSTLSNWAKEFRKWLPDLNVI 678
+ +LADEMGLGKT+Q++S L +L + Q++GPFL+VVPLSTL++W +E + P +N +
Sbjct: 501 NSCILADEMGLGKTIQTISFLNYLFHEHQLYGPFLLVVPLSTLTSWQREIQTRAPQMNAV 560
Query: 679 VYVGTRSSREVCQQYEFCNEKKAGKQIKFNALLTTYEVVLKDKAVLSKIKWNYLMVDEAH 738
VY+G +SR + + +E+ + + K++KFN LLTTYE++LKDK+ L + W ++ VDEAH
Sbjct: 561 VYLGDITSRNMIRTHEWMHPQT--KRLKFNILLTTYEILLKDKSFLGGLNWAFIGVDEAH 618
Query: 739 RLKNSEAQLYTALSEFNTKNKLLITGTPLQNSVEELWALLHFLDSDKFKSKDEFAQNYKN 798
RLKN ++ LY L +F + ++LLITGTPLQNS++ELW+LLHF+ +KF S ++F + +
Sbjct: 619 RLKNDDSLLYKTLIDFKSNHRLLITGTPLQNSLKELWSLLHFIMPEKFSSWEDFEEEH-- 676
Query: 799 LSSFNENELSNLHMELRPHMLRRVIKDVEKSLPPKIERILRVDMSPLQKQYYKWILERNF 858
E ++LH EL P +LRRV KDVEKSLP K+E+ILR++MS LQKQYYKWIL N+
Sbjct: 677 -GKGREYGYASLHKELEPFLLRRVKKDVEKSLPAKVEQILRMEMSALQKQYYKWILTGNY 735
Query: 859 RDLNKGVRGNQSKYCSRIEEV 879
+ L+KG +G+ S + + + E+
Sbjct: 736 KALSKGSKGSTSGFLNIMMEL 756
Score = 328 bits (841), Expect = 1e-87
Identities = 242/682 (35%), Positives = 350/682 (50%), Gaps = 78/682 (11%)
Query: 938 DILAQYMSLRGFQFQRLDGSTKSELRQQAMDHFNAPGSDDFCFLLSTRAGGLGINLATAD 997
DILA+Y+ R F FQRLDGS K ELR+QA+DHFNA GS+DFCFLLSTRAGGLGINLA+AD
Sbjct: 819 DILAEYLKYRQFPFQRLDGSIKGELRKQALDHFNAEGSEDFCFLLSTRAGGLGINLASAD 878
Query: 998 TVIIFDSDWNPQNDLQAMSRAHRIGQREVVNIYRFVTSKSVEEDILERAKKKMVLDHLVI 1057
TV+IFDSDWNPQNDLQA +RAHRIGQ++ VNIYR VT SVEEDILERAKKKMVLDHLVI
Sbjct: 879 TVVIFDSDWNPQNDLQAQARAHRIGQKKQVNIYRLVTKGSVEEDILERAKKKMVLDHLVI 938
Query: 1058 QKLNAEGK--LEKKEAKKGGSFFDKNELSAILRFGAEELFKEERNDEESKKRLLSMDIDE 1115
Q+++ GK L + F+K ELSAIL+FGAEELFKE +E+ + MDIDE
Sbjct: 939 QRMDTTGKTVLHTGSTPSSSTPFNKEELSAILKFGAEELFKEPEGEEQEPQ---EMDIDE 995
Query: 1116 ILERAEKVEEKENGGEQAHELLSAFKVANFCN-DEDD--------GSFWSRWIKADSVAQ 1166
IL+RAE E + ELLS FKVANF N DEDD W I +
Sbjct: 996 ILKRAETRENEPGPLTVGDELLSQFKVANFSNMDEDDIELEPERNSRNWEEIIPEVQRRR 1055
Query: 1167 AENA----------LAPRAARNIKSYAEADQSERSKKRKKKENEPTERIPKRRKADYSAH 1216
E + PR K + R + ++ ++ I +R++
Sbjct: 1056 IEEEERQKELEEIYMLPRMRNCAKQISFNGSEGRRSRSRRYSGSDSDSISERKRPKKR-- 1113
Query: 1217 VISMIDGASAQVRSWSYGNLSKRDALRFSRSVMKFGNESQINLIVAEVGGAIEAAPLKAQ 1276
G + + S + RF +S KFG + VA ++ + +
Sbjct: 1114 ------GRPRTIPRENIKGFSDAEIRRFIKSYKKFGGPLERLDAVARDAELVDKSETDLR 1167
Query: 1277 VELFNALIDGCREAVEVGSL----------DLKGPLLDFYGVPMKANELLIRVQELQLLA 1326
L + +GC +A++ S +KGP GV + A ++ +EL L
Sbjct: 1168 -RLGELVHNGCIKALKDNSSGQERAGGRLGKVKGPTFRISGVQVNAKLVISHEEELAPLH 1226
Query: 1327 KRISRYEDPIAQFRVLTYLKPSNWSKGCGWNQIDDARLLLGVHYHGYGNWEVIRLDERLG 1386
K I + ++ + + K +++ W + DD+ LL+G++ +GYG+WE+I++D L
Sbjct: 1227 KSIPSDPEERKRYVIPCHTKAAHFD--IDWGKEDDSNLLIGIYEYGYGSWEMIKMDPDLS 1284
Query: 1387 LTKKIAPVELQHHETFLPRAPNLRDRANALLEQELAVLGVKNASSKVGRKTSKKEREERE 1446
LT+KI P + P+A L+ RA+ L++ L K A G SK+ + +
Sbjct: 1285 LTQKILPDDPDKK----PQAKQLQTRADYLIKLLNKDLARKEAQRLAGAGNSKRRKTRTK 1340
Query: 1447 HLVDISLSRGQEKKKNIGSSKVNVQMRKDRLQ---------KPLNVEPIVKEEGEMSDDD 1497
+ ++ +E+ K+ S + + + +D + K L+ + ++E E +
Sbjct: 1341 KNKVLKAAKLKEEIKSDSSPQPSEKSDEDDDENNKDEIVSVKHLHKKFKAEKENEEKPEP 1400
Query: 1498 DV-----YEQFKEGKWKEWCQDLM-----VEEMKTLKRLHRLQTTSASLPKEKVLSKIRN 1547
D E+ +E K KE +DL E+ K LK R S KEK + +
Sbjct: 1401 DTGIKKEAEEKRETKEKENKRDLKREKKEKEDKKELKE-KRENKVKESTQKEKEVKE--- 1456
Query: 1548 YLQLLGRRIDQIVSEQEDEPHK 1569
++++I SE +++ K
Sbjct: 1457 ------EKVNEIKSENKEKTKK 1472
Score = 44.3 bits (103), Expect = 0.037
Identities = 46/186 (24%), Positives = 81/186 (42%), Gaps = 30/186 (16%)
Query: 1427 KNASSKVGRKTSKKEREEREHLVDISLSRGQE---KKKNIGSSKVNVQMRKDR------- 1476
K +K K KKE+E+++ L + ++ +E K+K + KVN +++
Sbjct: 1415 KEKENKRDLKREKKEKEDKKELKEKRENKVKESTQKEKEVKEEKVNEIKSENKEKTKKIP 1474
Query: 1477 -LQKPLNV----EPI-VKEEGEMSDDDDVYEQFKEGKWKEWCQDLMVEEMKTLKRLHRLQ 1530
L P+++ EPI + EE E D K C++ M LK+L R +
Sbjct: 1475 LLDTPVHITASSEPIPISEESEELDQ----------KTFSVCKERMRPVKAALKQLDRPE 1524
Query: 1531 TTSASLPKEKVLSKIRNYLQLLGRRIDQIVSEQEDEPHKQDRMTTRLWKYVSTFSHLSGE 1590
L + + L R L +G I + + E + P + + LW +VS F+
Sbjct: 1525 K---GLSEREQLEHTRQCLIKIGDHITECLKEYTN-PEQIKQWRKNLWIFVSKFTEFDAR 1580
Query: 1591 RLHQIY 1596
+LH++Y
Sbjct: 1581 KLHKLY 1586
>ref|XP_614999.1| PREDICTED: similar to Chromodomain-helicase-DNA-binding protein 2
(CHD-2), partial [Bos taurus]
Length = 1816
Score = 414 bits (1064), Expect = e-113
Identities = 278/795 (34%), Positives = 432/795 (53%), Gaps = 83/795 (10%)
Query: 127 SEDEDGQKDSGKGQRGDSDVPAEEMLSDDSYGQDGEEQGESVHSRGFRPSTGSNSCLQPT 186
++D+ ++DS S +EE DS Q EQG S S S+ +
Sbjct: 4 NKDKSQEEDSSLHSNASSHSASEEASGSDSGSQSESEQGSDPGSGHGSESNSSSESSESQ 63
Query: 187 STNVNRRVHRKSR-ILDDAEDDDDDADYEEDEPDEDDPDDADFEPATSG-RGANKYKDWE 244
S + + KS+ +L +A++ +PA+ R A+ K WE
Sbjct: 64 SESESESAGSKSQPVLPEAKE----------------------KPASKKERIADVKKMWE 101
Query: 245 G-EDSDEVDDSDEDIDVSDNDDLYFDKKAKGRQRGKFG-PSVRSTRDCKAFTASSRQRRV 302
D V S+ S + F+ K + + G P R R K +R+
Sbjct: 102 EYPDVYGVRRSNR----SRQEPSRFNIKEEASSGSESGSPKRRGQRQLK----KQEKRKW 153
Query: 303 KSSFEDEDENSTAEDSDSESDEDFKSLKKRGV---RVRKN----NGRSSAATSFSRPSNE 355
+ S +++++ S+AE ++ + + + +R V RV+K G+ S +
Sbjct: 154 ELSDDEQEQGSSAESEPEQTVKARRPVPRRTVPKPRVKKQPKTQRGKRKKQDSSDDEDED 213
Query: 356 VRSSSRTIRK-----VSYVESDESEGADEGTKKSQKEEIEV--DDGDSVEKVLWHQ---- 404
+ R R+ VSY E D+ E + + E ++ D+ +++EKVL +
Sbjct: 214 DEAPKRQTRRRAAKNVSYKEDDDFETDSDDLIEMTGEGVDEQQDNSETIEKVLDSRLGKK 273
Query: 405 -PKGMAAEAQRNNQSMEPVLMSHLFDSEPDWNNMEFLIKWKGQSHLHCQWKSFVDLQN-- 461
G + + +P S FD E D +++LIKWKG S++H W+S LQ
Sbjct: 274 GATGASTTVYAVEANGDP---SSDFDPEKDEGEVQYLIKWKGWSYIHSTWESEESLQQQK 330
Query: 462 LSGFKKVLNYTKRVTEEIRNRMG-ISREEIEVNDVSKEMDIDIIKQNSQVERIIADRISN 520
+ G KK+ N+ K+ +EI+ +G +S E++E + +E+ ++ KQ VER+IA + S
Sbjct: 331 VKGLKKLENFKKK-EDEIKQWLGKVSPEDVEYFNCQQELASELNKQYQIVERVIAVKTSK 389
Query: 521 DNSGNVF-------------PEYLVKWQGLSYAEVTWEKDIDIAFA-QHTIDEYKTREAA 566
G PEYL KW GL Y+E +WE + I Q ID + +R +
Sbjct: 390 STLGQTDFPAHSRKPAPSNEPEYLCKWMGLPYSECSWEDEALIGKKFQSCIDSFHSRNNS 449
Query: 567 MSVQGKMVDFQRRQSKGSLRKLDEQPEWLKGG--KLRDYQLEGLNFLVNSWKNDTNVVLA 624
++ + + + + + L +QP +L G +LRDYQLEGLN+L +SW +V+LA
Sbjct: 450 KTIPTR--ECKALKQRPRFVALKKQPAYLGGENLELRDYQLEGLNWLAHSWCKSNSVILA 507
Query: 625 DEMGLGKTVQSVSMLGFLQNAQQIHGPFLVVVPLSTLSNWAKEFRKWLPDLNVIVYVGTR 684
DEMGLGKT+Q++S L +L + Q++GPFL+VVPLSTL++W +EF W P++NV+VY+G
Sbjct: 508 DEMGLGKTIQTISFLSYLFHQHQLYGPFLIVVPLSTLTSWQREFEIWAPEINVVVYIGDL 567
Query: 685 SSREVCQQYEFCNEKKAGKQIKFNALLTTYEVVLKDKAVLSKIKWNYLMVDEAHRLKNSE 744
SR ++YE+ + + K++KFNAL+TTYE++LKDK VL I W +L VDEAHRLKN +
Sbjct: 568 MSRNTIREYEWIHSQT--KRLKFNALITTYEILLKDKTVLGSINWAFLGVDEAHRLKNDD 625
Query: 745 AQLYTALSEFNTKNKLLITGTPLQNSVEELWALLHFLDSDKFKSKDEFAQNYKNLSSFNE 804
+ LY L +F + ++LLITGTPLQNS++ELW+LLHF+ +KF+ ++F +++ E
Sbjct: 626 SLLYKTLIDFKSNHRLLITGTPLQNSLKELWSLLHFIMPEKFEFWEDFEEDH---GKGRE 682
Query: 805 NELSNLHMELRPHMLRRVIKDVEKSLPPKIERILRVDMSPLQKQYYKWILERNFRDLNKG 864
N +LH L P +LRRV KDVEKSLP K+E+ILRV+MS LQKQYYKWIL RN++ L KG
Sbjct: 683 NGYQSLHKVLEPFLLRRVKKDVEKSLPAKVEQILRVEMSALQKQYYKWILTRNYKALAKG 742
Query: 865 VRGNQSKYCSRIEEV 879
RG+ S + + + E+
Sbjct: 743 TRGSTSGFLNIVMEL 757
Score = 353 bits (905), Expect = 4e-95
Identities = 256/726 (35%), Positives = 363/726 (49%), Gaps = 93/726 (12%)
Query: 938 DILAQYMSLRGFQFQRLDGSTKSELRQQAMDHFNAPGSDDFCFLLSTRAGGLGINLATAD 997
DILA+Y++++ + FQRLDGS K E+R+QA+DHFNA GS+DFCFLLSTRAGGLGINLA+AD
Sbjct: 820 DILAEYLTIKHYPFQRLDGSIKGEIRKQALDHFNADGSEDFCFLLSTRAGGLGINLASAD 879
Query: 998 TVIIFDSDWNPQNDLQAMSRAHRIGQREVVNIYRFVTSKSVEEDILERAKKKMVLDHLVI 1057
TV+IFDSDWNPQNDLQA +RAHRIGQ++ VNIYR VT +VEE+I+ERAKKKMVLDHLVI
Sbjct: 880 TVVIFDSDWNPQNDLQAQARAHRIGQKKQVNIYRLVTKGTVEEEIIERAKKKMVLDHLVI 939
Query: 1058 QKLNAEGK--LEKKEAKKGGSFFDKNELSAILRFGAEELFKEERNDEESKKRLLSMDIDE 1115
Q+++ G+ LE + + F+K EL+AIL+FGAE+LFKE +E + MDIDE
Sbjct: 940 QRMDTTGRTVLENNSGRSNSNPFNKEELTAILKFGAEDLFKELEGEESEPQ---EMDIDE 996
Query: 1116 ILERAEKVEEKENGGEQAHELLSAFKVANFCNDEDDGSF-------WSRWIKADSVAQAE 1168
IL AE E E ELLS FKVANF ED+ W I + + E
Sbjct: 997 ILRLAE-TRENEVSTSATDELLSQFKVANFATMEDEEELDERPHKDWDEIIPEEQRKKVE 1055
Query: 1169 NALAPRAARNIKSYAEADQSERSKKRKKKENEPTERIPKRRKADYSAHVISMID------ 1222
+ I RS +K + N+ +R+A S+ S D
Sbjct: 1056 EEERQKELEEIYMLPRI----RSSTKKAQTNDSDSDTESKRQAQRSSASESETDDSDDDK 1111
Query: 1223 -----GASAQVRSWSYGNLSKRDALRFSRSVMKFGNESQINLIVAEVGGAIE--AAPLKA 1275
G VR + + RF ++ KFG + +A ++ A LK
Sbjct: 1112 KPKRRGRPRSVRKDLVEGFTDAEIRRFIKAYKKFGLPLERLECIARDAELVDKSVADLKR 1171
Query: 1276 QVELF-NALIDGCREAVE---------VGSLDLKGPLLDFYGVPMKANELLIRVQELQLL 1325
EL N+ + +E E G +GP + GV + ++ +E ++L
Sbjct: 1172 LGELIHNSCVSAMQEYEEQLKENASEGKGPGKKRGPTIKISGVQVNVKSIIQHEEEFEML 1231
Query: 1326 AKRISRYEDPIAQFRVLTYLKPSNWSKGCGWNQIDDARLLLGVHYHGYGNWEVIRLDERL 1385
K I + ++ + +K +++ W DD+RLLLG++ HGYGNWE+I+ D L
Sbjct: 1232 HKSIPVDPEEKKKYCLSCRVKAAHFD--VEWGVEDDSRLLLGIYEHGYGNWELIKTDPEL 1289
Query: 1386 GLTKKIAPVELQHHETFLPRAPNLRDRANAL-------LEQELAVLGVKNASSKVG---- 1434
LT KI PVE P+ L+ RA+ L LE+E AV G + S+ G
Sbjct: 1290 KLTDKILPVETDKK----PQGKQLQTRADYLLKLLRKSLEKEGAVTGGEEVSAAAGLASP 1345
Query: 1435 -----------RKTSKKEREE---------REHLVDISLSRGQEKKKNIGSSKVNVQMRK 1474
+K KKE +E ++ D R ++KK+ +
Sbjct: 1346 GDDGLEKSPTKKKQKKKENKENKEKQMSSRKDKEGDKERKRSKDKKEKPKGGDAKSSSKS 1405
Query: 1475 DRLQKPLNV----EPIVKEEGEMSDDDDVYEQFKEGKWKEWCQDLMVEEMKTLKRLHRLQ 1530
R Q P+++ EP+ E E D D E F C++ M K LK+L +
Sbjct: 1406 KRSQGPVHITAGSEPVPIGEDEEEDLDQ--ETF------SICKERMRPVKKALKQLDK-- 1455
Query: 1531 TTSASLPKEKVLSKIRNYLQLLGRRIDQIVSEQEDEPHKQDRMTTRLWKYVSTFSHLSGE 1590
L ++ L R+ L +G RI + + D+ H + LW +VS F+
Sbjct: 1456 -PDKGLSVQEQLEHTRSCLLKIGDRIAECLKAYSDQEHTK-LWRRNLWIFVSKFTEFDAR 1513
Query: 1591 RLHQIY 1596
+LH++Y
Sbjct: 1514 KLHKLY 1519
>ref|XP_314800.2| ENSANGP00000011045 [Anopheles gambiae str. PEST]
gi|55239891|gb|EAA10171.2| ENSANGP00000011045 [Anopheles
gambiae str. PEST]
Length = 1436
Score = 413 bits (1061), Expect = e-113
Identities = 248/594 (41%), Positives = 362/594 (60%), Gaps = 38/594 (6%)
Query: 311 ENSTAEDSDSESDEDFKSLKKRGVRVRKNNGRSSAATSFSRPSNEVRSSSRTIRK---VS 367
+ STA ++ S++ + KK+ K+N SS S S + RSS+ T RK VS
Sbjct: 6 KRSTASNAKSKAAAAAAASKKK-----KSNAYSSDDGSNSDDDRK-RSSAATRRKNNTVS 59
Query: 368 YVES--DESEGADEGTKKSQKEE-------IEVDDGDSVEKVLWHQP-----KGMAAEAQ 413
Y E DE +D+ + Q+ E E + +++E+++ + G
Sbjct: 60 YKEESGDEPTDSDDLLEVDQEAEEAAAALAAEEEKAETIERIIGRRKGRRGATGAVTTVY 119
Query: 414 RNNQSMEP-VLMSHLFDSEPDWNNMEFLIKWKGQSHLHCQWKSFVDL--QNLSGFKKVLN 470
++ +P VL E + +FLIKW G S+LHC W+S L Q + G KK+ N
Sbjct: 120 AIEENGDPNVLGGEGVGREQEEEEEQFLIKWTGWSYLHCTWESEETLREQKVKGMKKLEN 179
Query: 471 YTKRVTE-EIRNRMGISREEIEVNDVSKEMDIDIIKQNSQVERIIADRISNDNSGNVFPE 529
Y KR + E + E+I+ + +E+ D++K VERIIA + + G+ E
Sbjct: 180 YIKREQQLEYWRKYQAGPEDIDYYECQQELQQDLLKSYYHVERIIA-QANKAEEGDSGLE 238
Query: 530 YLVKWQGLSYAEVTWEKDIDIAFA-QHTIDEYKTREAAMSVQGKMVDFQRRQSKGSLRKL 588
YL KW+ L Y++ TWE I Q I E+ RE + K R + + + L
Sbjct: 239 YLCKWESLPYSDSTWEDAGLIRRKWQQKIVEFHEREESRRTPSKHCKAIRYRP--NFKHL 296
Query: 589 DEQPEWL---KGGKLRDYQLEGLNFLVNSWKNDTNVVLADEMGLGKTVQSVSMLGFLQNA 645
+QPE+L +G KLRDYQ++GLN+L+ +W D +V+LADEMGLGKT+Q++ L +L +
Sbjct: 297 KQQPEYLGEERGLKLRDYQMDGLNWLILTWCKDNSVILADEMGLGKTIQTICFLYYLFKS 356
Query: 646 QQIHGPFLVVVPLSTLSNWAKEFRKWLPDLNVIVYVGTRSSREVCQQYEFCNEKKAGKQI 705
QQ++GPFL VVPLST+ W +EF W P+LNV+ Y+G SRE+ +QYE+C E K++
Sbjct: 357 QQLYGPFLCVVPLSTMPAWQREFGIWAPELNVVTYLGDVQSREIIRQYEWCYEST--KKL 414
Query: 706 KFNALLTTYEVVLKDKAVLSKIKWNYLMVDEAHRLKNSEAQLYTALSEFNTKNKLLITGT 765
KFNA+LTTYE++LKDK L I W L+VDEAHRLKN ++ LY AL EF+T ++LLITGT
Sbjct: 415 KFNAILTTYEILLKDKTFLGSIGWASLLVDEAHRLKNDDSLLYKALKEFDTNHRLLITGT 474
Query: 766 PLQNSVEELWALLHFLDSDKFKSKDEFAQNYKNLSSFNENELSNLHMELRPHMLRRVIKD 825
PLQNS++ELWALLHF+ ++F+S D+F +NY N + N+ + LH EL P++LRRV KD
Sbjct: 475 PLQNSLKELWALLHFIMPERFESWDDFERNYGNTT--NDKSYTKLHKELEPYILRRVKKD 532
Query: 826 VEKSLPPKIERILRVDMSPLQKQYYKWILERNFRDLNKGVRGNQSKYCSRIEEV 879
VEKSLP K+E+ILRV+M+ +Q+QYYKWIL +NF L KG++G+ + + + E+
Sbjct: 533 VEKSLPAKVEQILRVEMTSIQRQYYKWILSKNFDALRKGMKGSVGTFLNIVIEL 586
Score = 264 bits (675), Expect = 2e-68
Identities = 143/215 (66%), Positives = 165/215 (76%), Gaps = 8/215 (3%)
Query: 938 DILAQYMSLRGFQFQRLDGSTKSELRQQAMDHFNAPGSDDFCFLLSTRAGGLGINLATAD 997
DILA+Y+ R F FQRLDGS K ELR+QA+DHFNA GS DFCFLLSTRAGGLGINLATAD
Sbjct: 650 DILAEYLQKRHFSFQRLDGSIKGELRKQALDHFNAEGSTDFCFLLSTRAGGLGINLATAD 709
Query: 998 TVIIFDSDWNPQNDLQAMSRAHRIGQREVVNIYRFVTSKSVEEDILERAKKKMVLDHLVI 1057
TVIIFDSDWNPQNDLQA +RAHRIGQ+ VNIYR VT+ SVEE+I+ERAK+KMVLDHLVI
Sbjct: 710 TVIIFDSDWNPQNDLQAQARAHRIGQKNQVNIYRLVTAHSVEENIVERAKQKMVLDHLVI 769
Query: 1058 QKLNAEGK--LEKKEAKKGGSFFDKNELSAILRFGAEELFKEERNDEESKKRLLSMDIDE 1115
Q+++ G+ L+K + F+K+ELSAIL+FGAEELFKEE + +E L DIDE
Sbjct: 770 QRMDTTGRTVLDKNGGSNTSNPFNKDELSAILKFGAEELFKEEEDGDEE----LVCDIDE 825
Query: 1116 ILERAEKVEEKENGGEQAHELLSAFKVANFCNDED 1150
IL RAE +E G ELLSAF V F DED
Sbjct: 826 ILRRAETRDEAP--GMPGDELLSAFNVTTFDFDED 858
Score = 70.9 bits (172), Expect = 4e-10
Identities = 90/366 (24%), Positives = 152/366 (40%), Gaps = 56/366 (15%)
Query: 1307 GVPMKANELLIRVQELQLLAKRISRYEDPIAQFRVLTYLKPSNWSKGCGWNQIDDARLLL 1366
G ++ V+ELQ L + I A++ + +P N+ W DD+RLL
Sbjct: 1084 GASFNVKTMMQCVEELQPLDEVIPSDAAERARWTLNIKTRPPNFD--VDWGGEDDSRLLR 1141
Query: 1367 GVHYHGYGNWEVIRLDERLGLTKKIAPVELQHHETFLPRAPNLRDRANALLEQELAVLGV 1426
G++ G G+WE +++D LGL KI L + + P+ +L+ RA LL+ L +
Sbjct: 1142 GIYQFGIGSWEAMKMDPSLGLADKI----LSNDASRKPQGKHLQSRAEYLLKVLRKTLEL 1197
Query: 1427 KNASSKVGRKTSKKE--------------------REEREHLVDISL----------SRG 1456
K SK ++ KE R+ + S+ SR
Sbjct: 1198 KRGPSKPRKQRKPKEMKSALGGAGMAPVPVVSSPLRDGGDASFSCSISATIAAHSNGSRA 1257
Query: 1457 QEKKKNIGSS---------KVNVQMRKDRLQKPLNVEPIVKEEGEM--SDDDDVYEQFKE 1505
++ +GSS K + +KDR QK + K G M + +++
Sbjct: 1258 ADEHAAVGSSNHATPAPDKKDKKKSKKDR-QKDAKKDKKNKNTGPMHFTANNEPCALNIL 1316
Query: 1506 GKWKEWCQDLMVEEMKTLKR-LHRLQTTSASLPKEKVLSKIRNYLQLLGRRIDQIVSEQE 1564
G + E+M+ +K+ L L SL +E+ L + R L +G +I+ ++E
Sbjct: 1317 GDLDPTVFNECKEKMRPVKKALKTLDNPDESLSQEEQLRQTRACLLSIGDQINLCLAEYR 1376
Query: 1565 DEPHKQDRMTTRLWKYVSTFSHLSGERLHQIYSKLKLEQNAVGVGSSLPNGSVSGPFSRN 1624
D P K + LW +VS F+ +L ++Y K L+++ GS+ G G
Sbjct: 1377 D-PEKAKEWRSNLWYFVSKFTEFDARKLFKLY-KHALKRSE---GSATSGGEAGG--GGG 1429
Query: 1625 GNPNSS 1630
G NS+
Sbjct: 1430 GTTNST 1435
>ref|XP_145698.6| PREDICTED: similar to Chromodomain-helicase-DNA-binding protein 2
(CHD-2) [Mus musculus]
Length = 1847
Score = 412 bits (1059), Expect = e-113
Identities = 254/708 (35%), Positives = 398/708 (55%), Gaps = 54/708 (7%)
Query: 215 EDEPDEDDPDDADFEPATS-GRGANKYKDWEGEDSDEVDDSDEDIDVSDNDDLYFDKKAK 273
E+ D ++ E + G G + E S+ +S+ + S + + + K K
Sbjct: 32 EEVSGSDSGSQSESEQGSEPGSGHGSESNSSSESSESQSESESESAGSKSQPVLPEAKEK 91
Query: 274 GRQRGKFGPSVRST----RDCKAFTASSRQRRVKSSFEDEDENSTAEDSDSESDEDFKSL 329
+ + V+ D S+R R+ S F ++E S+ +S S + L
Sbjct: 92 PASKKERIADVKKMWEEYPDVYGVRRSNRSRQEPSRFNVKEEASSGSESGSPKRRGQRQL 151
Query: 330 KKRGV------------RVRKNNGRSSAATSFSRPSNEVRSSSRTIRKVSYVESDESEGA 377
KK+ V R ++ SS + + ++ R + VSY E D+ E
Sbjct: 152 KKQTVPKPQVKKQPKIQRGKRKKQESSDDDDDDDEAPKRQTRRRAAKNVSYKEDDDFETD 211
Query: 378 DEGTKKSQKE--EIEVDDGDSVEKVLWHQ-----PKGMAAEAQRNNQSMEPVLMSHLFDS 430
+ + E + + D+ +++EKVL + G + + +P S FD+
Sbjct: 212 SDDLIEMTGEGGDEQQDNSETIEKVLDSRLGKKGATGASTTVYAVEANGDP---SDDFDT 268
Query: 431 EPDWNNMEFLIKWKGQSHLHCQWKSFVDLQN--LSGFKKVLNYTKRVTEEIRNRMG-ISR 487
E + +++LIKWKG S++H W+S LQ + G KK+ N+ K+ +E++ +G +S
Sbjct: 269 EREEGEVQYLIKWKGWSYIHSTWESEDSLQQQKVKGLKKLENFKKK-EDEVKQWLGKVSP 327
Query: 488 EEIEVNDVSKEMDIDIIKQNSQVERIIADRISNDNSGNVF-------------PEYLVKW 534
E++E +E+ ++ KQ VER+IA + S G PEYL KW
Sbjct: 328 EDVEYFSCQQELASELNKQYQIVERVIAVKTSKSTLGQTDFPAHSRKPAPSNEPEYLCKW 387
Query: 535 QGLSYAEVTWEKDIDIAFA-QHTIDEYKTREAAMSVQGKMVDFQRRQSKGSLRKLDEQPE 593
GL Y+E +WE + I Q+ ID + +R + ++ + + + + + L +QP
Sbjct: 388 MGLPYSECSWEDEALIGKKFQNCIDSFHSRNNSKTIPTR--ECKALKQRPRFVALKKQPA 445
Query: 594 WLKGG--KLRDYQLEGLNFLVNSWKNDTNVVLADEMGLGKTVQSVSMLGFLQNAQQIHGP 651
+L G +LRDYQLEGLN+L +SW +V+LADEMGLGKT+Q++S L +L + Q++GP
Sbjct: 446 YLGGESLELRDYQLEGLNWLAHSWCKSNSVILADEMGLGKTIQTISFLSYLFHQHQLYGP 505
Query: 652 FLVVVPLSTLSNWAKEFRKWLPDLNVIVYVGTRSSREVCQQYEFCNEKKAGKQIKFNALL 711
FL+VVPLSTL++W +EF W P++NV+VY+G SR ++YE+ + + K++KFNAL+
Sbjct: 506 FLIVVPLSTLTSWQREFEIWAPEINVVVYIGDLMSRNTIREYEWIHSQT--KRLKFNALI 563
Query: 712 TTYEVVLKDKAVLSKIKWNYLMVDEAHRLKNSEAQLYTALSEFNTKNKLLITGTPLQNSV 771
TTYE++LKDK VL I W +L VDEAHRLKN ++ LY L +F + ++LLITGTPLQNS+
Sbjct: 564 TTYEILLKDKTVLGSINWAFLGVDEAHRLKNDDSLLYKTLIDFKSNHRLLITGTPLQNSL 623
Query: 772 EELWALLHFLDSDKFKSKDEFAQNYKNLSSFNENELSNLHMELRPHMLRRVIKDVEKSLP 831
+ELW+LLHF+ +KF+ ++F +++ EN +LH L P +LRRV KDVEKSLP
Sbjct: 624 KELWSLLHFIMPEKFEFWEDFEEDH---GKGRENGYQSLHKVLEPFLLRRVKKDVEKSLP 680
Query: 832 PKIERILRVDMSPLQKQYYKWILERNFRDLNKGVRGNQSKYCSRIEEV 879
K+E+ILRV+MS LQKQYYKWIL RN++ L KG RG+ S + + + E+
Sbjct: 681 AKVEQILRVEMSALQKQYYKWILTRNYKALAKGTRGSTSGFLNIVMEL 728
Score = 324 bits (830), Expect = 2e-86
Identities = 225/594 (37%), Positives = 311/594 (51%), Gaps = 68/594 (11%)
Query: 938 DILAQYMSLRGFQFQRLDGSTKSELRQQAMDHFNAPGSDDFCFLLSTRAGGLGINLATAD 997
DILA+Y++++ + FQRLDGS K E+R+QA+DHFNA GS+DFCFLLSTRAGGLGINLA+AD
Sbjct: 791 DILAEYLTIKHYPFQRLDGSIKGEIRKQALDHFNADGSEDFCFLLSTRAGGLGINLASAD 850
Query: 998 TVIIFDSDWNPQNDLQAMSRAHRIGQREVVNIYRFVTSKSVEEDILERAKKKMVLDHLVI 1057
TV+IFDSDWNPQNDLQA +RAHRIGQ++ VNIYR VT +VEE+I+ERAKKKMVLDHLVI
Sbjct: 851 TVVIFDSDWNPQNDLQAQARAHRIGQKKQVNIYRLVTKGTVEEEIIERAKKKMVLDHLVI 910
Query: 1058 QKLNAEGK--LEKKEAKKGGSFFDKNELSAILRFGAEELFKEERNDEESKKRLLSMDIDE 1115
Q+++ G+ LE + + F+K EL+AIL+FGAE+LFKE +E + MDIDE
Sbjct: 911 QRMDTTGRTVLENNSGRSNSNPFNKEELTAILKFGAEDLFKEIEGEESEPQ---EMDIDE 967
Query: 1116 ILERAEKVEEKENGGEQAHELLSAFKVANFCNDEDDGSFWSRWIKA-DSVAQAENALAPR 1174
IL AE E E ELLS FKVANF ED+ R K D + E
Sbjct: 968 ILRLAE-TRENEVSTSATDELLSQFKVANFATMEDEEELEERPHKDWDEIIPEEQRKKVE 1026
Query: 1175 AARNIKSYAEADQSE--RSKKRKKKENEPTERIPKRRKADYSAHVISMID---------- 1222
K E RS +K + N+ +R+A S+ S D
Sbjct: 1027 EEERQKELEEIYMLPRIRSSTKKAQTNDSDSDTESKRQAQRSSASESETDDSDDDKKPKR 1086
Query: 1223 -GASAQVRSWSYGNLSKRDALRFSRSVMKFGNESQINLIVAEVGGAIE--AAPLKAQVEL 1279
G VR + + RF ++ KFG + +A ++ A LK EL
Sbjct: 1087 RGRPRSVRKDLVEGFTDAEIRRFIKAYKKFGLPLERLECIARDAELVDKSVADLKRLGEL 1146
Query: 1280 F-NALIDGCREAVE---------VGSLDLKGPLLDFYGVPMKANELLIRVQELQLLAKRI 1329
N+ + +E E G +GP + GV + ++ +E ++L K I
Sbjct: 1147 IHNSCVSAMQEYEEQLKESTSEGKGPGKRRGPTIKISGVQVNVKSIIQHEEEFEMLHKSI 1206
Query: 1330 SRYEDPIAQFRVLTYLKPSNWSKGCGWNQIDDARLLLGVHYHGYGNWEVIRLDERLGLTK 1389
+ ++ + +K +++ W DD+RLLLG++ HGYGNWE+I+ D L LT
Sbjct: 1207 PVDPEEKKKYCLTCRVKAAHFD--VEWGVEDDSRLLLGIYEHGYGNWELIKTDPELKLTD 1264
Query: 1390 KIAPVELQHHETFLPRAPNLRDRANALLEQELAVLGVKNASSKVGRKTSKKEREEREHLV 1449
KI PVE P+ L+ R + LL K+ RK +K+
Sbjct: 1265 KILPVETDKK----PQGKQLQTRVDYLL--------------KLLRKGLEKKG------- 1299
Query: 1450 DISLSRGQEKKKNIGSSKVNVQMRKDRLQKPLNVEPI-------VKEEGEMSDD 1496
+++ G+E K +V + + RL+ +EP EEGE+ DD
Sbjct: 1300 --TVASGEEAKLKKRKPRVKKENKAPRLKDEHGLEPASPRHSDNPSEEGEVKDD 1351
Score = 38.1 bits (87), Expect = 2.7
Identities = 23/80 (28%), Positives = 39/80 (48%), Gaps = 2/80 (2%)
Query: 1518 EEMKTLKR-LHRLQTTSASLPKEKVLSKIRNYLQLLGRRIDQIVSEQEDEPHKQDRMTTR 1576
E M+ +K+ L +L L ++ L RN L +G RI + + D+ H +
Sbjct: 1548 ERMRPVKKALKQLDKPDKGLSVQEQLEHTRNCLLKIGDRIAECLKAYSDQEHIK-LWRRN 1606
Query: 1577 LWKYVSTFSHLSGERLHQIY 1596
LW +VS F+ +LH++Y
Sbjct: 1607 LWIFVSKFTEFDARKLHKLY 1626
>emb|CAB07481.2| Hypothetical protein H06O01.2 [Caenorhabditis elegans]
gi|32563629|ref|NP_491994.2| chromo domain and SNF2
related domain and helicase, C-terminal (1H586)
[Caenorhabditis elegans]
Length = 1461
Score = 410 bits (1055), Expect = e-112
Identities = 260/701 (37%), Positives = 382/701 (54%), Gaps = 50/701 (7%)
Query: 202 DDAEDDDDDADYEEDEPDEDDPDDADFEPATSGRGANKYKDWEGEDSDEVDDSDEDIDVS 261
+ + D + D + DEP ++ + + K D SD+ DDSD+ + S
Sbjct: 6 ESSSGSDAEPDEKTDEPVREEENPEGNSTSEDNDEKEKAADSPESSSDQSDDSDDASENS 65
Query: 262 DND--DLYFDKKAKGRQRGKFGPSVRSTRDCKAFTASSRQRRVKSSFEDEDENSTAEDSD 319
+ D KK RG + + + F S R +R S D E E
Sbjct: 66 SSSEPDEPAPKKKNNNNRGMDAETRKLLENENYFRRSKRSKRQDDSPSDNSE----ESQS 121
Query: 320 SESDEDFKSLKKRGVRVRKNNGRSSAATSFSRPSNEVRSSSRTIRKVSYVESDESEGADE 379
SE D D + +R V + A + R S + V+Y E + + D+
Sbjct: 122 SEDDWDDRRKPRRTVTKKVVKQTKKAVSVVKRKSVK--------GNVNYAEKNSDDDIDD 173
Query: 380 GTKKSQKEEIEVD-DG-----DSVEKVL-WHQ----PKGMAAEAQRNNQSMEPVLMSHLF 428
EE V +G ++VE+V+ W G A +P
Sbjct: 174 DDVLEWDEEPAVPAEGPAILTETVERVIKWRHGVPGATGSATTCYNIADKGDPN------ 227
Query: 429 DSEP-DWNNMEFLIKWKGQSHLHCQWKS--FVDLQNLSGFKKVLNYTKRVTEEIRNRMGI 485
D P D +F +KW G SHLH W+S + L N G KKV NY K+ E +
Sbjct: 228 DQIPGDKTEQQFFVKWTGWSHLHNTWESENSLALMNAKGLKKVQNYVKKQKEVEMWKRSA 287
Query: 486 SREEIEVNDVSKEMDIDIIKQNSQVERIIADRISNDNS--GNVFPEYLVKWQGLSYAEVT 543
+E IE + ++M ++ ++ +VER++A + S D + G++ EYL+KW GL Y++ T
Sbjct: 288 DKEYIEFFECEQQMAEELCEEYKKVERVVAHQTSRDRAADGSMATEYLIKWSGLPYSDCT 347
Query: 544 WEKDIDIAFAQ-----HTIDEYKTREAAMSVQGKMVDFQRRQSKGSLRKLDEQPEWLKGG 598
WE + +A Q H I+ K+ +V K F++ +S K D +
Sbjct: 348 WEDEKMVAPEQIKAYYHRIENLKSPNKNSNVLRKRPKFEKFESMPDFLKTDGESTH---- 403
Query: 599 KLRDYQLEGLNFLVNSWKNDTNVVLADEMGLGKTVQSVSMLGFLQNAQQIHGPFLVVVPL 658
KLRDYQLEGLN++V +W + +LADEMGLGKT+QS+S+L L + + GP+LVVVPL
Sbjct: 404 KLRDYQLEGLNWMVYAWCKGNSSILADEMGLGKTIQSISLLASLFHRYDLAGPYLVVVPL 463
Query: 659 STLSNWAKEFRKWLPDLNVIVYVGTRSSREVCQQYEFCNEKKAGKQIKFNALLTTYEVVL 718
ST++ W KEF +W P++N++VY+G SR++ +QYE+ K++K NA+LTTYE++L
Sbjct: 464 STMAAWQKEFAQWAPEMNLVVYMGDVVSRDMIRQYEWF--VGGTKKMKINAILTTYEILL 521
Query: 719 KDKAVLSKIKWNYLMVDEAHRLKNSEAQLYTALSEFNTKNKLLITGTPLQNSVEELWALL 778
KDKA LS I W L+VDEAHRLKN E+ LY +L++F +KLLITGTPLQNS++ELWALL
Sbjct: 522 KDKAFLSSIDWAALLVDEAHRLKNDESLLYKSLTQFRFNHKLLITGTPLQNSLKELWALL 581
Query: 779 HFLDSDKFKSKDEFAQNYKNLSSFNENELSNLHMELRPHMLRRVIKDVEKSLPPKIERIL 838
HF+ +KF +EF + + N +S LH +L P +LRRV KDVEKSLPPK E+IL
Sbjct: 582 HFIMPEKFDCWEEFETAH---NESNHKGISALHKKLEPFLLRRVKKDVEKSLPPKTEQIL 638
Query: 839 RVDMSPLQKQYYKWILERNFRDLNKGVRGNQSKYCSRIEEV 879
RVDM+ QKQ+YKWIL +N+R+L+KGV+G+ + + + + E+
Sbjct: 639 RVDMTAHQKQFYKWILTKNYRELSKGVKGSINGFVNLVMEL 679
Score = 303 bits (777), Expect = 3e-80
Identities = 224/627 (35%), Positives = 323/627 (50%), Gaps = 54/627 (8%)
Query: 938 DILAQYMSLRGFQFQRLDGSTKSELRQQAMDHFNAPGSDDFCFLLSTRAGGLGINLATAD 997
DIL +Y+ LR F QRLDGS +++LR+QA+DH+NAPGS DF FLLSTRAGGLGINLATAD
Sbjct: 741 DILQEYLQLRRFPSQRLDGSMRADLRKQALDHYNAPGSTDFAFLLSTRAGGLGINLATAD 800
Query: 998 TVIIFDSDWNPQNDLQAMSRAHRIGQREVVNIYRFVTSKSVEEDILERAKKKMVLDHLVI 1057
TVIIFDSDWNPQNDLQAMSRAHRIGQ + VNIYR VT SVEE+I+ERAK+K+VLDHLVI
Sbjct: 801 TVIIFDSDWNPQNDLQAMSRAHRIGQTKTVNIYRLVTKGSVEEEIVERAKRKLVLDHLVI 860
Query: 1058 QKLNAEGK--LEKKEAKKGGSFFDKNELSAILRFGAEELFKEERNDEESKKRLLSMDIDE 1115
Q+++ GK L K G FDK ELSAIL+FGA ELFKE+ +E+ + +DID
Sbjct: 861 QRMDTTGKTVLSKNATASGSVPFDKQELSAILKFGAVELFKEKEGEEQEPE----VDIDR 916
Query: 1116 ILERAEKVEEKENGGEQAHELLSAFKVANFCNDE--DDGSFWSRWIKADSVAQAENALAP 1173
IL AE E +E ++ +ELLS+FK ANF DE D + W ++ E+
Sbjct: 917 ILMGAETREAEEEVMKE-NELLSSFKYANFAIDEEKDIAAATDEWA---AIIPEEDRNRI 972
Query: 1174 RAARNIKSYAEADQSERSKKRKKKENEPTERIPKRRKADYSAHVISMIDGASAQVRSWSY 1233
+K AE + + R +K +P ++ + D D + + +
Sbjct: 973 LEEERMKELAEMNLAPRQRK------QPIPQVVEDDDGDDDEEE----DDTGKKKKKKAV 1022
Query: 1234 GNLSKRDALRFSRSVMKFGNESQINLIVAEVGGAIEAAPLKAQVELFNALIDGCREAVEV 1293
GN + + RF +S KF +A+ +E +L +L + C++A +
Sbjct: 1023 GNFTIPEIKRFIKSFRKFSMPLNRLEEIAQ-DAELEEHSTDEMKKLVESLSEACKKAADE 1081
Query: 1294 GSLDLKGPLLDFYGVPMKANELLIRVQELQLLAKRISRYEDPIAQFRVL-------TYLK 1346
+ K K E + + K+I R + + T K
Sbjct: 1082 FDSNEKNGDAGAAESEKKDIERKFKFHTCDVNLKQIERSHAELKPLHEILKSEETKTSFK 1141
Query: 1347 P---SNWSKG--CGWNQIDDARLLLGVHYHGYGNWEVIRLDERLGLTKKIAPVELQHHET 1401
P + KG W++ DD+ LLLGV +GYG+WE I++D LGL KI +T
Sbjct: 1142 PPANAKLQKGWDVDWSRPDDSALLLGVWKYGYGSWEAIKMDPTLGLADKI----FIKDKT 1197
Query: 1402 FLPRAPNLRDRANALLEQELAVLGVKNASSKVGRKTSKKEREEREHLVDISLSRGQEKKK 1461
P+ NL+ R + LL K S + T KKER+ + V + ++KK+
Sbjct: 1198 KKPQGKNLQVRVDYLL---------KLMSKDKVKTTEKKERKRKADDVPVG---PEKKKR 1245
Query: 1462 NIGSSKVNVQMRKDRLQKPLNVEPIVKEEGEMSDDDDVY-EQFKEGKWKEW--CQDLMVE 1518
+ + + +K ++ N + + +S D +Y ++ K + C L +
Sbjct: 1246 HTNNVPQEGEKKKKEKKEEKNSSSLKDQLALLSIDKSLYGGALEDSSAKPFLECVKLCMP 1305
Query: 1519 EMKTLKRLHRLQTTSASLPKEKVLSKI 1545
K +K+L Q + K L+++
Sbjct: 1306 VHKYMKKLKEAQEAKNQADEAKYLTRL 1332
Score = 39.3 bits (90), Expect = 1.2
Identities = 76/375 (20%), Positives = 140/375 (37%), Gaps = 51/375 (13%)
Query: 100 DSKSGSDYRNEDEFEDNSSEGRGEKLGSEDEDGQKDSGKGQRGDSDVPAEEMLSDDSYGQ 159
+S SGSD +++ ++ E + S ED + +S + D S
Sbjct: 6 ESSSGSDAEPDEKTDEPVREEENPEGNSTSEDNDEKEKAADSPESSSDQSDDSDDASENS 65
Query: 160 DGEEQGESVHSRGFRPSTGSNSCLQPTSTNVN--RRVHRKSRILDDAEDDDDDADYEEDE 217
E E + + G ++ + N N RR R R +DD + EE +
Sbjct: 66 SSSEPDEPAPKKKNNNNRGMDAETRKLLENENYFRRSKRSKR-----QDDSPSDNSEESQ 120
Query: 218 PDEDDPDDADFEPATSGR----------GANKYKDWEGEDSDEVDDSDEDIDVSDNDDLY 267
EDD DD T + K K +G + +SD+DID D+D L
Sbjct: 121 SSEDDWDDRRKPRRTVTKKVVKQTKKAVSVVKRKSVKGNVNYAEKNSDDDID--DDDVLE 178
Query: 268 FDKKAKGRQRGKFGPSVRSTRDCKAFTASSRQRRVKSSFEDEDENSTAEDSDSESDEDFK 327
+D++ P+V + + A + +R +K +A + +D+
Sbjct: 179 WDEE----------PAVPA--EGPAILTETVERVIKWRHGVPGATGSATTCYNIADKGDP 226
Query: 328 SLKKRGVRVR-----KNNGRSSAATSFSRPSNEVRSSSRTIRKV-SYVESDESEGADEGT 381
+ + G + K G S ++ ++ +++ ++KV +YV+ + +
Sbjct: 227 NDQIPGDKTEQQFFVKWTGWSHLHNTWESENSLALMNAKGLKKVQNYVKKQKE--VEMWK 284
Query: 382 KKSQKEEIEVDDGDSVEKVLWHQPKGMAAEAQRNNQSMEPVLMSHLFDSEPDWNNM--EF 439
+ + KE IE + + MA E + +E V+ +M E+
Sbjct: 285 RSADKEYIE----------FFECEQQMAEELCEEYKKVERVVAHQTSRDRAADGSMATEY 334
Query: 440 LIKWKGQSHLHCQWK 454
LIKW G + C W+
Sbjct: 335 LIKWSGLPYSDCTWE 349
Score = 38.5 bits (88), Expect = 2.0
Identities = 47/196 (23%), Positives = 83/196 (41%), Gaps = 31/196 (15%)
Query: 33 EAQYESDAEPDDASGKQNEAAAVDRLSTRESNVETTSRNPSASERWGSSYLKDCQPMSPQ 92
E+ SDAEPD+ + D E N E S + E K+ SP+
Sbjct: 6 ESSSGSDAEPDEKT---------DEPVREEENPEGNSTSEDNDE-------KEKAADSPE 49
Query: 93 NGSESGDDSKSGSDYRNEDEFED-------NSSEGRGEKLGS--EDEDGQKDSGKGQRGD 143
+ S+ DDS S+ + E ++ N++ G + E+E+ + S + +R D
Sbjct: 50 SSSDQSDDSDDASENSSSSEPDEPAPKKKNNNNRGMDAETRKLLENENYFRRSKRSKRQD 109
Query: 144 ---SDVPAEEMLSDDSYGQDGEEQGESVHSRGFRPSTGSNSCLQPTSTNVNRRVHRKSRI 200
SD E S+D + D + +V + + + + S ++ S N V+ +
Sbjct: 110 DSPSDNSEESQSSEDDW-DDRRKPRRTVTKKVVKQTKKAVSVVKRKSVKGN--VNYAEKN 166
Query: 201 LDDAEDDDDDADYEED 216
DD DDDD +++E+
Sbjct: 167 SDDDIDDDDVLEWDEE 182
>pir||T23056 chromodomain helicase H06O01.2 - Caenorhabditis elegans
Length = 1465
Score = 410 bits (1055), Expect = e-112
Identities = 260/701 (37%), Positives = 382/701 (54%), Gaps = 50/701 (7%)
Query: 202 DDAEDDDDDADYEEDEPDEDDPDDADFEPATSGRGANKYKDWEGEDSDEVDDSDEDIDVS 261
+ + D + D + DEP ++ + + K D SD+ DDSD+ + S
Sbjct: 6 ESSSGSDAEPDEKTDEPVREEENPEGNSTSEDNDEKEKAADSPESSSDQSDDSDDASENS 65
Query: 262 DND--DLYFDKKAKGRQRGKFGPSVRSTRDCKAFTASSRQRRVKSSFEDEDENSTAEDSD 319
+ D KK RG + + + F S R +R S D E E
Sbjct: 66 SSSEPDEPAPKKKNNNNRGMDAETRKLLENENYFRRSKRSKRQDDSPSDNSE----ESQS 121
Query: 320 SESDEDFKSLKKRGVRVRKNNGRSSAATSFSRPSNEVRSSSRTIRKVSYVESDESEGADE 379
SE D D + +R V + A + R S + V+Y E + + D+
Sbjct: 122 SEDDWDDRRKPRRTVTKKVVKQTKKAVSVVKRKSVK--------GNVNYAEKNSDDDIDD 173
Query: 380 GTKKSQKEEIEVD-DG-----DSVEKVL-WHQ----PKGMAAEAQRNNQSMEPVLMSHLF 428
EE V +G ++VE+V+ W G A +P
Sbjct: 174 DDVLEWDEEPAVPAEGPAILTETVERVIKWRHGVPGATGSATTCYNIADKGDPN------ 227
Query: 429 DSEP-DWNNMEFLIKWKGQSHLHCQWKS--FVDLQNLSGFKKVLNYTKRVTEEIRNRMGI 485
D P D +F +KW G SHLH W+S + L N G KKV NY K+ E +
Sbjct: 228 DQIPGDKTEQQFFVKWTGWSHLHNTWESENSLALMNAKGLKKVQNYVKKQKEVEMWKRSA 287
Query: 486 SREEIEVNDVSKEMDIDIIKQNSQVERIIADRISNDNS--GNVFPEYLVKWQGLSYAEVT 543
+E IE + ++M ++ ++ +VER++A + S D + G++ EYL+KW GL Y++ T
Sbjct: 288 DKEYIEFFECEQQMAEELCEEYKKVERVVAHQTSRDRAADGSMATEYLIKWSGLPYSDCT 347
Query: 544 WEKDIDIAFAQ-----HTIDEYKTREAAMSVQGKMVDFQRRQSKGSLRKLDEQPEWLKGG 598
WE + +A Q H I+ K+ +V K F++ +S K D +
Sbjct: 348 WEDEKMVAPEQIKAYYHRIENLKSPNKNSNVLRKRPKFEKFESMPDFLKTDGESTH---- 403
Query: 599 KLRDYQLEGLNFLVNSWKNDTNVVLADEMGLGKTVQSVSMLGFLQNAQQIHGPFLVVVPL 658
KLRDYQLEGLN++V +W + +LADEMGLGKT+QS+S+L L + + GP+LVVVPL
Sbjct: 404 KLRDYQLEGLNWMVYAWCKGNSSILADEMGLGKTIQSISLLASLFHRYDLAGPYLVVVPL 463
Query: 659 STLSNWAKEFRKWLPDLNVIVYVGTRSSREVCQQYEFCNEKKAGKQIKFNALLTTYEVVL 718
ST++ W KEF +W P++N++VY+G SR++ +QYE+ K++K NA+LTTYE++L
Sbjct: 464 STMAAWQKEFAQWAPEMNLVVYMGDVVSRDMIRQYEWF--VGGTKKMKINAILTTYEILL 521
Query: 719 KDKAVLSKIKWNYLMVDEAHRLKNSEAQLYTALSEFNTKNKLLITGTPLQNSVEELWALL 778
KDKA LS I W L+VDEAHRLKN E+ LY +L++F +KLLITGTPLQNS++ELWALL
Sbjct: 522 KDKAFLSSIDWAALLVDEAHRLKNDESLLYKSLTQFRFNHKLLITGTPLQNSLKELWALL 581
Query: 779 HFLDSDKFKSKDEFAQNYKNLSSFNENELSNLHMELRPHMLRRVIKDVEKSLPPKIERIL 838
HF+ +KF +EF + + N +S LH +L P +LRRV KDVEKSLPPK E+IL
Sbjct: 582 HFIMPEKFDCWEEFETAH---NESNHKGISALHKKLEPFLLRRVKKDVEKSLPPKTEQIL 638
Query: 839 RVDMSPLQKQYYKWILERNFRDLNKGVRGNQSKYCSRIEEV 879
RVDM+ QKQ+YKWIL +N+R+L+KGV+G+ + + + + E+
Sbjct: 639 RVDMTAHQKQFYKWILTKNYRELSKGVKGSINGFVNLVMEL 679
Score = 303 bits (777), Expect = 3e-80
Identities = 224/627 (35%), Positives = 323/627 (50%), Gaps = 54/627 (8%)
Query: 938 DILAQYMSLRGFQFQRLDGSTKSELRQQAMDHFNAPGSDDFCFLLSTRAGGLGINLATAD 997
DIL +Y+ LR F QRLDGS +++LR+QA+DH+NAPGS DF FLLSTRAGGLGINLATAD
Sbjct: 741 DILQEYLQLRRFPSQRLDGSMRADLRKQALDHYNAPGSTDFAFLLSTRAGGLGINLATAD 800
Query: 998 TVIIFDSDWNPQNDLQAMSRAHRIGQREVVNIYRFVTSKSVEEDILERAKKKMVLDHLVI 1057
TVIIFDSDWNPQNDLQAMSRAHRIGQ + VNIYR VT SVEE+I+ERAK+K+VLDHLVI
Sbjct: 801 TVIIFDSDWNPQNDLQAMSRAHRIGQTKTVNIYRLVTKGSVEEEIVERAKRKLVLDHLVI 860
Query: 1058 QKLNAEGK--LEKKEAKKGGSFFDKNELSAILRFGAEELFKEERNDEESKKRLLSMDIDE 1115
Q+++ GK L K G FDK ELSAIL+FGA ELFKE+ +E+ + +DID
Sbjct: 861 QRMDTTGKTVLSKNATASGSVPFDKQELSAILKFGAVELFKEKEGEEQEPE----VDIDR 916
Query: 1116 ILERAEKVEEKENGGEQAHELLSAFKVANFCNDE--DDGSFWSRWIKADSVAQAENALAP 1173
IL AE E +E ++ +ELLS+FK ANF DE D + W ++ E+
Sbjct: 917 ILMGAETREAEEEVMKE-NELLSSFKYANFAIDEEKDIAAATDEWA---AIIPEEDRNRI 972
Query: 1174 RAARNIKSYAEADQSERSKKRKKKENEPTERIPKRRKADYSAHVISMIDGASAQVRSWSY 1233
+K AE + + R +K +P ++ + D D + + +
Sbjct: 973 LEEERMKELAEMNLAPRQRK------QPIPQVVEDDDGDDDEEE----DDTGKKKKKKAV 1022
Query: 1234 GNLSKRDALRFSRSVMKFGNESQINLIVAEVGGAIEAAPLKAQVELFNALIDGCREAVEV 1293
GN + + RF +S KF +A+ +E +L +L + C++A +
Sbjct: 1023 GNFTIPEIKRFIKSFRKFSMPLNRLEEIAQ-DAELEEHSTDEMKKLVESLSEACKKAADE 1081
Query: 1294 GSLDLKGPLLDFYGVPMKANELLIRVQELQLLAKRISRYEDPIAQFRVL-------TYLK 1346
+ K K E + + K+I R + + T K
Sbjct: 1082 FDSNEKNGDAGAAESEKKDIERKFKFHTCDVNLKQIERSHAELKPLHEILKSEETKTSFK 1141
Query: 1347 P---SNWSKG--CGWNQIDDARLLLGVHYHGYGNWEVIRLDERLGLTKKIAPVELQHHET 1401
P + KG W++ DD+ LLLGV +GYG+WE I++D LGL KI +T
Sbjct: 1142 PPANAKLQKGWDVDWSRPDDSALLLGVWKYGYGSWEAIKMDPTLGLADKI----FIKDKT 1197
Query: 1402 FLPRAPNLRDRANALLEQELAVLGVKNASSKVGRKTSKKEREEREHLVDISLSRGQEKKK 1461
P+ NL+ R + LL K S + T KKER+ + V + ++KK+
Sbjct: 1198 KKPQGKNLQVRVDYLL---------KLMSKDKVKTTEKKERKRKADDVPVG---PEKKKR 1245
Query: 1462 NIGSSKVNVQMRKDRLQKPLNVEPIVKEEGEMSDDDDVY-EQFKEGKWKEW--CQDLMVE 1518
+ + + +K ++ N + + +S D +Y ++ K + C L +
Sbjct: 1246 HTNNVPQEGEKKKKEKKEEKNSSSLKDQLALLSIDKSLYGGALEDSSAKPFLECVKLCMP 1305
Query: 1519 EMKTLKRLHRLQTTSASLPKEKVLSKI 1545
K +K+L Q + K L+++
Sbjct: 1306 VHKYMKKLKEAQEAKNQADEAKYLTRL 1332
Score = 39.3 bits (90), Expect = 1.2
Identities = 76/375 (20%), Positives = 140/375 (37%), Gaps = 51/375 (13%)
Query: 100 DSKSGSDYRNEDEFEDNSSEGRGEKLGSEDEDGQKDSGKGQRGDSDVPAEEMLSDDSYGQ 159
+S SGSD +++ ++ E + S ED + +S + D S
Sbjct: 6 ESSSGSDAEPDEKTDEPVREEENPEGNSTSEDNDEKEKAADSPESSSDQSDDSDDASENS 65
Query: 160 DGEEQGESVHSRGFRPSTGSNSCLQPTSTNVN--RRVHRKSRILDDAEDDDDDADYEEDE 217
E E + + G ++ + N N RR R R +DD + EE +
Sbjct: 66 SSSEPDEPAPKKKNNNNRGMDAETRKLLENENYFRRSKRSKR-----QDDSPSDNSEESQ 120
Query: 218 PDEDDPDDADFEPATSGR----------GANKYKDWEGEDSDEVDDSDEDIDVSDNDDLY 267
EDD DD T + K K +G + +SD+DID D+D L
Sbjct: 121 SSEDDWDDRRKPRRTVTKKVVKQTKKAVSVVKRKSVKGNVNYAEKNSDDDID--DDDVLE 178
Query: 268 FDKKAKGRQRGKFGPSVRSTRDCKAFTASSRQRRVKSSFEDEDENSTAEDSDSESDEDFK 327
+D++ P+V + + A + +R +K +A + +D+
Sbjct: 179 WDEE----------PAVPA--EGPAILTETVERVIKWRHGVPGATGSATTCYNIADKGDP 226
Query: 328 SLKKRGVRVR-----KNNGRSSAATSFSRPSNEVRSSSRTIRKV-SYVESDESEGADEGT 381
+ + G + K G S ++ ++ +++ ++KV +YV+ + +
Sbjct: 227 NDQIPGDKTEQQFFVKWTGWSHLHNTWESENSLALMNAKGLKKVQNYVKKQKE--VEMWK 284
Query: 382 KKSQKEEIEVDDGDSVEKVLWHQPKGMAAEAQRNNQSMEPVLMSHLFDSEPDWNNM--EF 439
+ + KE IE + + MA E + +E V+ +M E+
Sbjct: 285 RSADKEYIE----------FFECEQQMAEELCEEYKKVERVVAHQTSRDRAADGSMATEY 334
Query: 440 LIKWKGQSHLHCQWK 454
LIKW G + C W+
Sbjct: 335 LIKWSGLPYSDCTWE 349
Score = 38.5 bits (88), Expect = 2.0
Identities = 47/196 (23%), Positives = 83/196 (41%), Gaps = 31/196 (15%)
Query: 33 EAQYESDAEPDDASGKQNEAAAVDRLSTRESNVETTSRNPSASERWGSSYLKDCQPMSPQ 92
E+ SDAEPD+ + D E N E S + E K+ SP+
Sbjct: 6 ESSSGSDAEPDEKT---------DEPVREEENPEGNSTSEDNDE-------KEKAADSPE 49
Query: 93 NGSESGDDSKSGSDYRNEDEFED-------NSSEGRGEKLGS--EDEDGQKDSGKGQRGD 143
+ S+ DDS S+ + E ++ N++ G + E+E+ + S + +R D
Sbjct: 50 SSSDQSDDSDDASENSSSSEPDEPAPKKKNNNNRGMDAETRKLLENENYFRRSKRSKRQD 109
Query: 144 ---SDVPAEEMLSDDSYGQDGEEQGESVHSRGFRPSTGSNSCLQPTSTNVNRRVHRKSRI 200
SD E S+D + D + +V + + + + S ++ S N V+ +
Sbjct: 110 DSPSDNSEESQSSEDDW-DDRRKPRRTVTKKVVKQTKKAVSVVKRKSVKGN--VNYAEKN 166
Query: 201 LDDAEDDDDDADYEED 216
DD DDDD +++E+
Sbjct: 167 SDDDIDDDDVLEWDEE 182
Database: nr
Posted date: Jul 5, 2005 12:34 AM
Number of letters in database: 863,360,394
Number of sequences in database: 2,540,612
Lambda K H
0.316 0.133 0.385
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 2,969,596,158
Number of Sequences: 2540612
Number of extensions: 134924921
Number of successful extensions: 925918
Number of sequences better than 10.0: 8890
Number of HSP's better than 10.0 without gapping: 2957
Number of HSP's successfully gapped in prelim test: 6479
Number of HSP's that attempted gapping in prelim test: 677774
Number of HSP's gapped (non-prelim): 76993
length of query: 1730
length of database: 863,360,394
effective HSP length: 142
effective length of query: 1588
effective length of database: 502,593,490
effective search space: 798118462120
effective search space used: 798118462120
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.6 bits)
S2: 83 (36.6 bits)
Medicago: description of AC141111.7


