

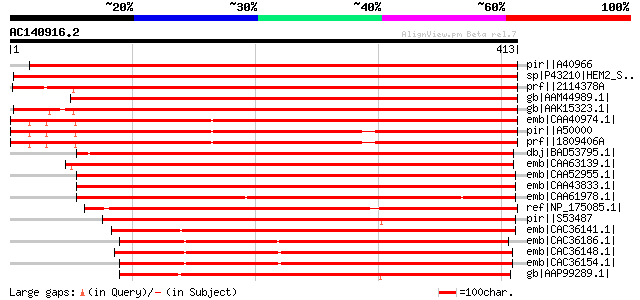
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC140916.2 + phase: 0
(413 letters)
Database: nr
2,540,612 sequences; 863,360,394 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
pir||A40966 porphobilinogen synthase (EC 4.2.1.24) ALAD [validat... 750 0.0
sp|P43210|HEM2_SOYBN Delta-aminolevulinic acid dehydratase, chlo... 716 0.0
prf||2114378A aminolevulinate dehydratase 680 0.0
gb|AAM44989.1| putative aminolevulinate dehydratase [Arabidopsis... 662 0.0
gb|AAK15323.1| aminolevulinate dehydratase [Raphanus sativus] 661 0.0
emb|CAA40974.1| porphobilinogen synthase [Spinacia oleracea] gi|... 642 0.0
pir||A50000 porphobilinogen synthase (EC 4.2.1.24) precursor - s... 614 e-174
prf||1809406A aminolevulinate dehydratase 614 e-174
dbj|BAD53795.1| putative aminolevulinate dehydratase [Oryza sati... 599 e-170
emb|CAA63139.1| aminolevulinate dehydratase [Hordeum vulgare sub... 585 e-165
emb|CAA52955.1| 5-aminolevulinic acid dehydratase [Selaginella m... 573 e-162
emb|CAA43833.1| delta-aminolevulinic acid dehydratase; porphobil... 569 e-161
emb|CAA61978.1| porphobilinogen synthase [Physcomitrella patens]... 562 e-159
ref|NP_175085.1| porphobilinogen synthase, putative / delta-amin... 494 e-138
pir||S53487 porphobilinogen synthase (EC 4.2.1.24) precursor - C... 434 e-120
emb|CAC36141.1| ALA dehydratase [Cyanophora paradoxa] 433 e-120
emb|CAC36186.1| ALA dehydratase [Odontella sinensis] 410 e-113
emb|CAC36148.1| ALA dehydratase [Fucus vesiculosus] 402 e-111
emb|CAC36154.1| ALA dehydratase [Laminaria digitata] 392 e-108
gb|AAP99289.1| Delta-aminolevulinic acid dehydratase [Prochloroc... 387 e-106
>pir||A40966 porphobilinogen synthase (EC 4.2.1.24) ALAD [validated] - garden
pea (fragment) gi|232244|sp|P30124|HEM2_PEA
Delta-aminolevulinic acid dehydratase, chloroplast
precursor (Porphobilinogen synthase) (ALADH)
gi|169031|gb|AAA33640.1| aminolevulinic acid dehydratase
Length = 398
Score = 750 bits (1936), Expect = 0.0
Identities = 375/398 (94%), Positives = 384/398 (96%), Gaps = 1/398 (0%)
Query: 17 HTYVDLKPALPLKNYLSFSSSKRR-PPCLFTVRASDADFEAAVVAGNVPDAPPVPPTPAA 75
HT+VDLK L NYLSFSSSKRR PP LFTVRASD+DFEAAVVAG VP+APPVPPTPA+
Sbjct: 1 HTFVDLKSPFTLSNYLSFSSSKRRQPPSLFTVRASDSDFEAAVVAGKVPEAPPVPPTPAS 60
Query: 76 PAGTPVVTSLPIQRRPRRNRRSATHRSAFQETTLSPANFVYPLFIHEGEEDTPIGAMPGC 135
PAGTPVV SLPIQRRPRRNRRS RSAFQETTLSPANFVYPLFIHEGEEDTPIGAMPGC
Sbjct: 61 PAGTPVVPSLPIQRRPRRNRRSPALRSAFQETTLSPANFVYPLFIHEGEEDTPIGAMPGC 120
Query: 136 YRLGWRHGLIEEVAKARDVGVNSVVLFPKIPDALKTPTGDEAYNENGLVPRTIRLLKDKY 195
YRLGWRHGL+EEVAKARDVGVNSVVLFPKIPDALKTPTGDEAYNE+GLVPR+IRLLKDKY
Sbjct: 121 YRLGWRHGLLEEVAKARDVGVNSVVLFPKIPDALKTPTGDEAYNEDGLVPRSIRLLKDKY 180
Query: 196 PDLVIYTDVALDPYSSDGHDGIVREDGVIMNDETVHQLCKQAVAQARAGADVVSPSDMMD 255
PDL+IYTDVALDPYSSDGHDGIVREDGVIMNDETVHQLCKQAVAQARAGADVVSPSDMMD
Sbjct: 181 PDLIIYTDVALDPYSSDGHDGIVREDGVIMNDETVHQLCKQAVAQARAGADVVSPSDMMD 240
Query: 256 GRVGAMRAALDAEGFQHVSIMSYTAKYASSFYGPFREALDSNPRFGDKKTYQMNPANYRE 315
GRVGAMR ALDAEGFQHVSIMSYTAKYASSFYGPFREALDSNPRFGDKKTYQMNPANYRE
Sbjct: 241 GRVGAMRVALDAEGFQHVSIMSYTAKYASSFYGPFREALDSNPRFGDKKTYQMNPANYRE 300
Query: 316 ALTEMREDETEGADILLVKPGLPYLDIIRLLRDNSPLPIAAYQVSGEYSMIKAGGALKMI 375
ALTEMREDE+EGADILLVKPGLPYLDIIRLLRDNSPLPIAAYQVSGEYSMIKAGGALKMI
Sbjct: 301 ALTEMREDESEGADILLVKPGLPYLDIIRLLRDNSPLPIAAYQVSGEYSMIKAGGALKMI 360
Query: 376 DEEKVMMESLLCLRRAGADIILTYFALQAARCLCGEKR 413
DEEKVMMESLLCLRRAGADIILTYFALQAAR LCGEKR
Sbjct: 361 DEEKVMMESLLCLRRAGADIILTYFALQAARTLCGEKR 398
>sp|P43210|HEM2_SOYBN Delta-aminolevulinic acid dehydratase, chloroplast precursor
(Porphobilinogen synthase) (ALADH)
gi|7437030|pir||T06351 porphobilinogen synthase (EC
4.2.1.24) - soybean gi|468000|gb|AAA18342.1|
delta-aminolevulinic acid dehydratase
Length = 412
Score = 716 bits (1847), Expect = 0.0
Identities = 359/411 (87%), Positives = 379/411 (91%), Gaps = 1/411 (0%)
Query: 4 SSTIPNTPLTLNSHTYVDLKPALPLKNYLSFSSSK-RRPPCLFTVRASDADFEAAVVAGN 62
+S+IPN P NS +YV L+ L N+ S ++K R LF VRASD++FEAAVVAG
Sbjct: 2 ASSIPNAPSAFNSQSYVGLRAPLRTFNFSSPQAAKIPRSQRLFVVRASDSEFEAAVVAGK 61
Query: 63 VPDAPPVPPTPAAPAGTPVVTSLPIQRRPRRNRRSATHRSAFQETTLSPANFVYPLFIHE 122
VP APPV P PAAP GTPVV SLP+ RRPRRNR+S RSAFQET++SPANFVYPLFIHE
Sbjct: 62 VPPAPPVRPRPAAPVGTPVVPSLPLHRRPRRNRKSPALRSAFQETSISPANFVYPLFIHE 121
Query: 123 GEEDTPIGAMPGCYRLGWRHGLIEEVAKARDVGVNSVVLFPKIPDALKTPTGDEAYNENG 182
GEEDTPIGAMPGCYRLGWRHGL+EEVAKARDVGVNSVVLFPKIPDALK+PTGDEAYNENG
Sbjct: 122 GEEDTPIGAMPGCYRLGWRHGLVEEVAKARDVGVNSVVLFPKIPDALKSPTGDEAYNENG 181
Query: 183 LVPRTIRLLKDKYPDLVIYTDVALDPYSSDGHDGIVREDGVIMNDETVHQLCKQAVAQAR 242
LVPRTIRLLKDKYPDLVIYTDVALDPYSSDGHDGIVREDGVIMNDETVHQLCKQAVAQA+
Sbjct: 182 LVPRTIRLLKDKYPDLVIYTDVALDPYSSDGHDGIVREDGVIMNDETVHQLCKQAVAQAQ 241
Query: 243 AGADVVSPSDMMDGRVGAMRAALDAEGFQHVSIMSYTAKYASSFYGPFREALDSNPRFGD 302
AGADVVSPSDMMDGRVGA+RAALDAEGFQHVSIMSYTAKYASSFYGPFREALDSNPRFGD
Sbjct: 242 AGADVVSPSDMMDGRVGALRAALDAEGFQHVSIMSYTAKYASSFYGPFREALDSNPRFGD 301
Query: 303 KKTYQMNPANYREALTEMREDETEGADILLVKPGLPYLDIIRLLRDNSPLPIAAYQVSGE 362
KKTYQMNPANYREALTEMREDE+EGADILLVKPGLPYLDIIRLLRDNSPLPIAAYQVSGE
Sbjct: 302 KKTYQMNPANYREALTEMREDESEGADILLVKPGLPYLDIIRLLRDNSPLPIAAYQVSGE 361
Query: 363 YSMIKAGGALKMIDEEKVMMESLLCLRRAGADIILTYFALQAARCLCGEKR 413
Y+MIKA GALKMIDEEKVMMESL+CLRRAGADIILTY ALQAARCLCGEKR
Sbjct: 362 YAMIKAAGALKMIDEEKVMMESLMCLRRAGADIILTYSALQAARCLCGEKR 412
>prf||2114378A aminolevulinate dehydratase
Length = 430
Score = 680 bits (1755), Expect = 0.0
Identities = 346/431 (80%), Positives = 373/431 (86%), Gaps = 22/431 (5%)
Query: 3 SSSTIPNTPLTLNSHTY-VDLKPALPLKNYLSFSSSKRRPPCLFTVRAS----------- 50
+S+ + N P + + + V LKP+ L + + S K + T+RAS
Sbjct: 2 ASAAMLNAPCNIGAVKFEVKLKPSPNL--FCARPSVKLNQRRVLTIRASKEGHDNGSSSG 59
Query: 51 --------DADFEAAVVAGNVPDAPPVPPTPAAPAGTPVVTSLPIQRRPRRNRRSATHRS 102
D + EAAVVAGNVP+APPVPP PAAP GTP+V+SLPI RRPRRNRRS+ R+
Sbjct: 60 PLRKMGLTDEECEAAVVAGNVPEAPPVPPKPAAPDGTPIVSSLPINRRPRRNRRSSAARA 119
Query: 103 AFQETTLSPANFVYPLFIHEGEEDTPIGAMPGCYRLGWRHGLIEEVAKARDVGVNSVVLF 162
AFQET +SPAN VYPLFIHEGEEDTPIGAMPGCYRLGWRHGL+EEVAKARDVGVNS+VLF
Sbjct: 120 AFQETNISPANLVYPLFIHEGEEDTPIGAMPGCYRLGWRHGLVEEVAKARDVGVNSIVLF 179
Query: 163 PKIPDALKTPTGDEAYNENGLVPRTIRLLKDKYPDLVIYTDVALDPYSSDGHDGIVREDG 222
PK+PDALKT TGDEAYN+NGLVPRTIRLLKDKYPDLVIYTDVALDPYSSDGHDGIVREDG
Sbjct: 180 PKVPDALKTSTGDEAYNDNGLVPRTIRLLKDKYPDLVIYTDVALDPYSSDGHDGIVREDG 239
Query: 223 VIMNDETVHQLCKQAVAQARAGADVVSPSDMMDGRVGAMRAALDAEGFQHVSIMSYTAKY 282
VIMNDETVHQLCKQAVAQARAGADVVSPSDMMDGRVGA+RAALDAEGFQHVSIMSYTAKY
Sbjct: 240 VIMNDETVHQLCKQAVAQARAGADVVSPSDMMDGRVGAIRAALDAEGFQHVSIMSYTAKY 299
Query: 283 ASSFYGPFREALDSNPRFGDKKTYQMNPANYREALTEMREDETEGADILLVKPGLPYLDI 342
ASSFYGPFREALDSNPRFGDKKTYQMNPANYREAL EM+ DE+EGADILLVKPGLPYLDI
Sbjct: 300 ASSFYGPFREALDSNPRFGDKKTYQMNPANYREALVEMQADESEGADILLVKPGLPYLDI 359
Query: 343 IRLLRDNSPLPIAAYQVSGEYSMIKAGGALKMIDEEKVMMESLLCLRRAGADIILTYFAL 402
IRLLRD SPLPIAAYQVSGEYSMIKAGG LKMIDEE+VMMESL+CLRRAGADIILTYFAL
Sbjct: 360 IRLLRDKSPLPIAAYQVSGEYSMIKAGGVLKMIDEERVMMESLMCLRRAGADIILTYFAL 419
Query: 403 QAARCLCGEKR 413
QA RCLCGEKR
Sbjct: 420 QAGRCLCGEKR 430
>gb|AAM44989.1| putative aminolevulinate dehydratase [Arabidopsis thaliana]
gi|11935205|gb|AAG42018.1| putative aminolevulinate
dehydratase [Arabidopsis thaliana]
gi|22136066|gb|AAM91111.1| putative aminolevulinate
dehydratase [Arabidopsis thaliana]
gi|21539421|gb|AAM53263.1| putative aminolevulinate
dehydratase [Arabidopsis thaliana]
gi|19698815|gb|AAL91143.1| putative aminolevulinate
dehydratase [Arabidopsis thaliana]
gi|13605645|gb|AAK32816.1| At1g69740/T6C23_6
[Arabidopsis thaliana] gi|15222443|ref|NP_177132.1|
porphobilinogen synthase, putative /
delta-aminolevulinic acid dehydratase, putative
[Arabidopsis thaliana] gi|16323332|gb|AAL15379.1|
At1g69740/T6C23_6 [Arabidopsis thaliana]
gi|12325198|gb|AAG52549.1| putative aminolevulinate
dehydratase; 38705-36189 [Arabidopsis thaliana]
gi|25291926|pir||D96719 hypothetical protein T6C23.6
[imported] - Arabidopsis thaliana
gi|12229846|sp|Q9SFH9|HEM2_ARATH Delta-aminolevulinic
acid dehydratase, chloroplast precursor (Porphobilinogen
synthase) (ALADH)
Length = 430
Score = 662 bits (1707), Expect = 0.0
Identities = 323/364 (88%), Positives = 342/364 (93%)
Query: 50 SDADFEAAVVAGNVPDAPPVPPTPAAPAGTPVVTSLPIQRRPRRNRRSATHRSAFQETTL 109
SDA+ EAAV AGNVP+APPVPP PAAP GTP++ L + RRPRRNR S R+AFQET +
Sbjct: 67 SDAECEAAVAAGNVPEAPPVPPKPAAPVGTPIIKPLNLSRRPRRNRASPVTRAAFQETDI 126
Query: 110 SPANFVYPLFIHEGEEDTPIGAMPGCYRLGWRHGLIEEVAKARDVGVNSVVLFPKIPDAL 169
SPANFVYPLFIHEGEEDTPIGAMPGCYRLGWRHGL++EVAKAR VGVNS+VLFPK+P+AL
Sbjct: 127 SPANFVYPLFIHEGEEDTPIGAMPGCYRLGWRHGLVQEVAKARAVGVNSIVLFPKVPEAL 186
Query: 170 KTPTGDEAYNENGLVPRTIRLLKDKYPDLVIYTDVALDPYSSDGHDGIVREDGVIMNDET 229
K TGDEAYN+NGLVPRTIRLLKDKYPDL+IYTDVALDPYSSDGHDGIVREDGVIMNDET
Sbjct: 187 KNSTGDEAYNDNGLVPRTIRLLKDKYPDLIIYTDVALDPYSSDGHDGIVREDGVIMNDET 246
Query: 230 VHQLCKQAVAQARAGADVVSPSDMMDGRVGAMRAALDAEGFQHVSIMSYTAKYASSFYGP 289
VHQLCKQAV+QARAGADVVSPSDMMDGRVGA+R+ALDAEGFQ+VSIMSYTAKYASSFYGP
Sbjct: 247 VHQLCKQAVSQARAGADVVSPSDMMDGRVGAIRSALDAEGFQNVSIMSYTAKYASSFYGP 306
Query: 290 FREALDSNPRFGDKKTYQMNPANYREALTEMREDETEGADILLVKPGLPYLDIIRLLRDN 349
FREALDSNPRFGDKKTYQMNPANYREAL E REDE EGADILLVKPGLPYLDIIRLLRD
Sbjct: 307 FREALDSNPRFGDKKTYQMNPANYREALIEAREDEAEGADILLVKPGLPYLDIIRLLRDK 366
Query: 350 SPLPIAAYQVSGEYSMIKAGGALKMIDEEKVMMESLLCLRRAGADIILTYFALQAARCLC 409
SPLPIAAYQVSGEYSMIKAGG LKMIDEEKVMMESL+CLRRAGADIILTYFALQAA CLC
Sbjct: 367 SPLPIAAYQVSGEYSMIKAGGVLKMIDEEKVMMESLMCLRRAGADIILTYFALQAATCLC 426
Query: 410 GEKR 413
GEKR
Sbjct: 427 GEKR 430
>gb|AAK15323.1| aminolevulinate dehydratase [Raphanus sativus]
Length = 426
Score = 661 bits (1705), Expect = 0.0
Identities = 338/428 (78%), Positives = 367/428 (84%), Gaps = 21/428 (4%)
Query: 4 SSTIPNTPLTLNSHTYVDLKPALPLKNY--------LSFSSSKRRPPCLFTVRAS----- 50
++T+ N + S +D K + L++ L F++S+RR VRAS
Sbjct: 2 ATTLFNASCSFPSIKVIDCKSYVGLRSNANQVRVASLPFATSQRRS---LVVRASNGHAK 58
Query: 51 -----DADFEAAVVAGNVPDAPPVPPTPAAPAGTPVVTSLPIQRRPRRNRRSATHRSAFQ 105
DA+ EA V AGNVP+APPVPP PAAPAGTPV+ L + RRPRRNR S T R+AFQ
Sbjct: 59 KLGRSDAECEADVAAGNVPEAPPVPPKPAAPAGTPVIQPLNLNRRPRRNRASPTVRAAFQ 118
Query: 106 ETTLSPANFVYPLFIHEGEEDTPIGAMPGCYRLGWRHGLIEEVAKARDVGVNSVVLFPKI 165
ET +SPANFVYPLFIHEGEEDTPIGAMPGCYRLGWRHGL++EVAKAR VGVNS+VLFPK+
Sbjct: 119 ETDISPANFVYPLFIHEGEEDTPIGAMPGCYRLGWRHGLLQEVAKARAVGVNSIVLFPKV 178
Query: 166 PDALKTPTGDEAYNENGLVPRTIRLLKDKYPDLVIYTDVALDPYSSDGHDGIVREDGVIM 225
P+ALK PTGDEAYN+NGLVPRTIRLLKDKYPDL+IYTDVALDPYSSDGHDGIVREDGVIM
Sbjct: 179 PEALKNPTGDEAYNDNGLVPRTIRLLKDKYPDLIIYTDVALDPYSSDGHDGIVREDGVIM 238
Query: 226 NDETVHQLCKQAVAQARAGADVVSPSDMMDGRVGAMRAALDAEGFQHVSIMSYTAKYASS 285
NDETVHQLCKQAV+QARAGADVVSPSDMMDGRVGA+RAALDAEGFQ+VSIMSYTAKYASS
Sbjct: 239 NDETVHQLCKQAVSQARAGADVVSPSDMMDGRVGAIRAALDAEGFQNVSIMSYTAKYASS 298
Query: 286 FYGPFREALDSNPRFGDKKTYQMNPANYREALTEMREDETEGADILLVKPGLPYLDIIRL 345
FYGPFREALDSNPRFGDKKTYQMNPANYREAL E REDE EGADILLVKPGLPYLDIIRL
Sbjct: 299 FYGPFREALDSNPRFGDKKTYQMNPANYREALIEAREDEAEGADILLVKPGLPYLDIIRL 358
Query: 346 LRDNSPLPIAAYQVSGEYSMIKAGGALKMIDEEKVMMESLLCLRRAGADIILTYFALQAA 405
LRD SPLPIAAYQVSGEYSMIKAGG LKMIDEEKVMMESL+CLRRAGADIILTYFALQAA
Sbjct: 359 LRDKSPLPIAAYQVSGEYSMIKAGGVLKMIDEEKVMMESLMCLRRAGADIILTYFALQAA 418
Query: 406 RCLCGEKR 413
LC +KR
Sbjct: 419 TYLCNQKR 426
>emb|CAA40974.1| porphobilinogen synthase [Spinacia oleracea]
gi|1170215|sp|P24493|HEM2_SPIOL Delta-aminolevulinic
acid dehydratase, chloroplast precursor (Porphobilinogen
synthase) (ALADH) (ALAD)
Length = 433
Score = 642 bits (1657), Expect = 0.0
Identities = 335/434 (77%), Positives = 359/434 (82%), Gaps = 22/434 (5%)
Query: 1 MASSSTIPNTPLTL----NSHTYVDLKPALPL---KNYLSFSSSKRRPPCLFTVRASDA- 52
MAS+ IP T+ NS + L + K S R P VRAS+
Sbjct: 1 MASTFNIPCNAGTIKNFNNSQRNLGFSSNLGINFAKTRFSNCGDSGRIPSQLVVRASERR 60
Query: 53 -------------DFEAAVVAGNVPDAPPVPPTPAAPAGTPVVTSLPIQRRPRRNRRSAT 99
+ EAAVVAGN P APPVPPTP AP+GTP V+ L + RRPRRNR S
Sbjct: 61 DNLTQQKTGLSIEECEAAVVAGNAPSAPPVPPTPKAPSGTPSVSPLSLGRRPRRNRTSPV 120
Query: 100 HRSAFQETTLSPANFVYPLFIHEGEEDTPIGAMPGCYRLGWRHGLIEEVAKARDVGVNSV 159
R+AFQETTLSPAN VYPLFIHEGEEDTPIGAMPGCYRLGWRHGL+EEVAKARDV VNS+
Sbjct: 121 FRAAFQETTLSPANVVYPLFIHEGEEDTPIGAMPGCYRLGWRHGLVEEVAKARDVVVNSI 180
Query: 160 VLFPKIPDALKTPTGDEAYNENGLVPRTIRLLKDKYPDLVIYTDVALDPYSSDGHDGIVR 219
V+FPK PDALK+PTGDEAYNENGLVPRTIR+LKDK+PDL+IYTDVALDPY DGHDGIV
Sbjct: 181 VVFPK-PDALKSPTGDEAYNENGLVPRTIRMLKDKFPDLIIYTDVALDPYYYDGHDGIVT 239
Query: 220 EDGVIMNDETVHQLCKQAVAQARAGADVVSPSDMMDGRVGAMRAALDAEGFQHVSIMSYT 279
+ GVIMNDETVHQLCKQAVAQARAGADVVSPSDMMDGRVGA+RAALDAEG+ +VSIMSYT
Sbjct: 240 QHGVIMNDETVHQLCKQAVAQARAGADVVSPSDMMDGRVGAIRAALDAEGYSNVSIMSYT 299
Query: 280 AKYASSFYGPFREALDSNPRFGDKKTYQMNPANYREALTEMREDETEGADILLVKPGLPY 339
AKYASSFYGPFREALDSNPRFGDKKTYQMNPANYREAL E +EDE+EGADILLVKPGLPY
Sbjct: 300 AKYASSFYGPFREALDSNPRFGDKKTYQMNPANYREALIETQEDESEGADILLVKPGLPY 359
Query: 340 LDIIRLLRDNSPLPIAAYQVSGEYSMIKAGGALKMIDEEKVMMESLLCLRRAGADIILTY 399
LDIIRLLRDNS LPIAAYQVSGEYSMIKAGG LKMIDEEKVM+ESLLCLRRAGADIILTY
Sbjct: 360 LDIIRLLRDNSDLPIAAYQVSGEYSMIKAGGVLKMIDEEKVMLESLLCLRRAGADIILTY 419
Query: 400 FALQAARCLCGEKR 413
FALQAARCLCGEKR
Sbjct: 420 FALQAARCLCGEKR 433
>pir||A50000 porphobilinogen synthase (EC 4.2.1.24) precursor - spinach
Length = 424
Score = 614 bits (1583), Expect = e-174
Identities = 325/434 (74%), Positives = 349/434 (79%), Gaps = 32/434 (7%)
Query: 1 MASSSTIPNTPLTL----NSHTYVDLKPALPL---KNYLSFSSSKRRPPCLFTVRASDA- 52
MAS+ IP T+ NS + L + K S R P VRAS+
Sbjct: 2 MASTFNIPCNAGTIKNFNNSQRNLGFSSNLGINFAKTRFSNCGDSGRIPSQLVVRASERR 61
Query: 53 -------------DFEAAVVAGNVPDAPPVPPTPAAPAGTPVVTSLPIQRRPRRNRRSAT 99
+ EAAVVAGN P APPVPPTP AP+GTP V+ L + RRPRRNR S
Sbjct: 62 DNLTQQKTGLSIEECEAAVVAGNAPSAPPVPPTPKAPSGTPSVSPLSLGRRPRRNRTSPV 121
Query: 100 HRSAFQETTLSPANFVYPLFIHEGEEDTPIGAMPGCYRLGWRHGLIEEVAKARDVGVNSV 159
R+AFQETTLSPAN VYPLFIHEGEEDTPIGAMPGCYRLGWRHGL+EEVAKARDV VNS+
Sbjct: 122 FRAAFQETTLSPANVVYPLFIHEGEEDTPIGAMPGCYRLGWRHGLVEEVAKARDVVVNSI 181
Query: 160 VLFPKIPDALKTPTGDEAYNENGLVPRTIRLLKDKYPDLVIYTDVALDPYSSDGHDGIVR 219
V+FPK PDALK+PTGDEAYNENGLVPRTIR+LKDK+PDL+IYTDVALDPY DGHDGIV
Sbjct: 182 VVFPK-PDALKSPTGDEAYNENGLVPRTIRMLKDKFPDLIIYTDVALDPYYYDGHDGIVT 240
Query: 220 EDGVIMNDETVHQLCKQAVAQARAGADVVSPSDMMDGRVGAMRAALDAEGFQHVSIMSYT 279
+ GVIMNDETVHQLCKQAVAQARAGADVVSPSDMMDGRVGA+RAALDAEG+ +VSIMSYT
Sbjct: 241 QHGVIMNDETVHQLCKQAVAQARAGADVVSPSDMMDGRVGAIRAALDAEGYSNVSIMSYT 300
Query: 280 AKYASSFYGPFREALDSNPRFGDKKTYQMNPANYREALTEMREDETEGADILLVKPGLPY 339
AKYASSFY PRFGDKKTYQMNPANYREAL E +EDE+EGADILLVKPGLPY
Sbjct: 301 AKYASSFY----------PRFGDKKTYQMNPANYREALIETQEDESEGADILLVKPGLPY 350
Query: 340 LDIIRLLRDNSPLPIAAYQVSGEYSMIKAGGALKMIDEEKVMMESLLCLRRAGADIILTY 399
LDIIRLLRDNS LPIAAYQVSGEYSMIKAGG LKMIDEEKVM+ESLLCLRRAGADIILTY
Sbjct: 351 LDIIRLLRDNSDLPIAAYQVSGEYSMIKAGGVLKMIDEEKVMLESLLCLRRAGADIILTY 410
Query: 400 FALQAARCLCGEKR 413
FALQAARCLCGEKR
Sbjct: 411 FALQAARCLCGEKR 424
>prf||1809406A aminolevulinate dehydratase
Length = 423
Score = 614 bits (1583), Expect = e-174
Identities = 325/434 (74%), Positives = 349/434 (79%), Gaps = 32/434 (7%)
Query: 1 MASSSTIPNTPLTL----NSHTYVDLKPALPL---KNYLSFSSSKRRPPCLFTVRASDA- 52
MAS+ IP T+ NS + L + K S R P VRAS+
Sbjct: 1 MASTFNIPCNAGTIKNFNNSQRNLGFSSNLGINFAKTRFSNCGDSGRIPSQLVVRASERR 60
Query: 53 -------------DFEAAVVAGNVPDAPPVPPTPAAPAGTPVVTSLPIQRRPRRNRRSAT 99
+ EAAVVAGN P APPVPPTP AP+GTP V+ L + RRPRRNR S
Sbjct: 61 DNLTQQKTGLSIEECEAAVVAGNAPSAPPVPPTPKAPSGTPSVSPLSLGRRPRRNRTSPV 120
Query: 100 HRSAFQETTLSPANFVYPLFIHEGEEDTPIGAMPGCYRLGWRHGLIEEVAKARDVGVNSV 159
R+AFQETTLSPAN VYPLFIHEGEEDTPIGAMPGCYRLGWRHGL+EEVAKARDV VNS+
Sbjct: 121 FRAAFQETTLSPANVVYPLFIHEGEEDTPIGAMPGCYRLGWRHGLVEEVAKARDVVVNSI 180
Query: 160 VLFPKIPDALKTPTGDEAYNENGLVPRTIRLLKDKYPDLVIYTDVALDPYSSDGHDGIVR 219
V+FPK PDALK+PTGDEAYNENGLVPRTIR+LKDK+PDL+IYTDVALDPY DGHDGIV
Sbjct: 181 VVFPK-PDALKSPTGDEAYNENGLVPRTIRMLKDKFPDLIIYTDVALDPYYYDGHDGIVT 239
Query: 220 EDGVIMNDETVHQLCKQAVAQARAGADVVSPSDMMDGRVGAMRAALDAEGFQHVSIMSYT 279
+ GVIMNDETVHQLCKQAVAQARAGADVVSPSDMMDGRVGA+RAALDAEG+ +VSIMSYT
Sbjct: 240 QHGVIMNDETVHQLCKQAVAQARAGADVVSPSDMMDGRVGAIRAALDAEGYSNVSIMSYT 299
Query: 280 AKYASSFYGPFREALDSNPRFGDKKTYQMNPANYREALTEMREDETEGADILLVKPGLPY 339
AKYASSFY PRFGDKKTYQMNPANYREAL E +EDE+EGADILLVKPGLPY
Sbjct: 300 AKYASSFY----------PRFGDKKTYQMNPANYREALIETQEDESEGADILLVKPGLPY 349
Query: 340 LDIIRLLRDNSPLPIAAYQVSGEYSMIKAGGALKMIDEEKVMMESLLCLRRAGADIILTY 399
LDIIRLLRDNS LPIAAYQVSGEYSMIKAGG LKMIDEEKVM+ESLLCLRRAGADIILTY
Sbjct: 350 LDIIRLLRDNSDLPIAAYQVSGEYSMIKAGGVLKMIDEEKVMLESLLCLRRAGADIILTY 409
Query: 400 FALQAARCLCGEKR 413
FALQAARCLCGEKR
Sbjct: 410 FALQAARCLCGEKR 423
>dbj|BAD53795.1| putative aminolevulinate dehydratase [Oryza sativa (japonica
cultivar-group)]
Length = 426
Score = 599 bits (1545), Expect = e-170
Identities = 298/356 (83%), Positives = 319/356 (88%), Gaps = 1/356 (0%)
Query: 55 EAAVVAGNVPDAPPVPPTPAAPAGTPVVTSLPIQRRPRRNRRSATHRSAFQETTLSPANF 114
EA VAG P APP P AP GTP + L + +RPRRNRRS R+AFQETT+SPAN
Sbjct: 68 EADAVAGRFP-APPPLVRPKAPEGTPQIRPLDLTKRPRRNRRSPALRAAFQETTISPANL 126
Query: 115 VYPLFIHEGEEDTPIGAMPGCYRLGWRHGLIEEVAKARDVGVNSVVLFPKIPDALKTPTG 174
V PLFIHEGE+D PIGAMPGCYRLGWRHGL++EV K+RDVGVNS VLFPK+PDALK+ +G
Sbjct: 127 VLPLFIHEGEDDAPIGAMPGCYRLGWRHGLLDEVYKSRDVGVNSFVLFPKVPDALKSQSG 186
Query: 175 DEAYNENGLVPRTIRLLKDKYPDLVIYTDVALDPYSSDGHDGIVREDGVIMNDETVHQLC 234
DEAYN+NGLVPRTIRLLKDK+PD+V+YTDVALDPYSSDGHDGIVREDGVIMNDETV+QLC
Sbjct: 187 DEAYNDNGLVPRTIRLLKDKFPDIVVYTDVALDPYSSDGHDGIVREDGVIMNDETVYQLC 246
Query: 235 KQAVAQARAGADVVSPSDMMDGRVGAMRAALDAEGFQHVSIMSYTAKYASSFYGPFREAL 294
KQAV+QARAGADVVSPSDMMDGRVGA+RAALDAEGF VSIMSYTAKYASSFYGPFREAL
Sbjct: 247 KQAVSQARAGADVVSPSDMMDGRVGAIRAALDAEGFHDVSIMSYTAKYASSFYGPFREAL 306
Query: 295 DSNPRFGDKKTYQMNPANYREALTEMREDETEGADILLVKPGLPYLDIIRLLRDNSPLPI 354
DSNPRFGDKKTYQMNPANYREAL E DE EGADILLVKPGLPYLD+IRLLRDNS LPI
Sbjct: 307 DSNPRFGDKKTYQMNPANYREALLETAADEAEGADILLVKPGLPYLDVIRLLRDNSALPI 366
Query: 355 AAYQVSGEYSMIKAGGALKMIDEEKVMMESLLCLRRAGADIILTYFALQAARCLCG 410
AAYQVSGEYSMIKAGGAL MIDEEKVMMESL+CLRRAGADIILTYFA QAA LCG
Sbjct: 367 AAYQVSGEYSMIKAGGALNMIDEEKVMMESLMCLRRAGADIILTYFARQAANVLCG 422
>emb|CAA63139.1| aminolevulinate dehydratase [Hordeum vulgare subsp. vulgare]
gi|7437032|pir||T04472 probable porphobilinogen synthase
(EC 4.2.1.24) - barley gi|2495171|sp|Q42836|HEM2_HORVU
Delta-aminolevulinic acid dehydratase, chloroplast
precursor (Porphobilinogen synthase) (ALADH)
Length = 428
Score = 585 bits (1507), Expect = e-165
Identities = 294/369 (79%), Positives = 320/369 (86%), Gaps = 4/369 (1%)
Query: 46 TVRA----SDADFEAAVVAGNVPDAPPVPPTPAAPAGTPVVTSLPIQRRPRRNRRSATHR 101
TVRA S + EA VAG P V TP AP GTP + L + +RPRRNRRS R
Sbjct: 55 TVRAPSGRSIEECEADAVAGRFPAPSCVCQTPKAPDGTPEIRPLDMAKRPRRNRRSPALR 114
Query: 102 SAFQETTLSPANFVYPLFIHEGEEDTPIGAMPGCYRLGWRHGLIEEVAKARDVGVNSVVL 161
+AFQET++SPAN V PLFIHEGEED PIGAMPGC+RLGW+HGL+ EV KARDVGVNS VL
Sbjct: 115 AAFQETSISPANLVLPLFIHEGEEDAPIGAMPGCFRLGWQHGLLAEVYKARDVGVNSFVL 174
Query: 162 FPKIPDALKTPTGDEAYNENGLVPRTIRLLKDKYPDLVIYTDVALDPYSSDGHDGIVRED 221
FPK+PDALK+PTG EAYN+NGLVPRTIRLLKDK+PD+++YTDVALDPYSSDGHDGIVR+D
Sbjct: 175 FPKVPDALKSPTGVEAYNDNGLVPRTIRLLKDKFPDIIVYTDVALDPYSSDGHDGIVRKD 234
Query: 222 GVIMNDETVHQLCKQAVAQARAGADVVSPSDMMDGRVGAMRAALDAEGFQHVSIMSYTAK 281
GVI+NDETV+QLCKQAV+QARAGADVVSPS+MMDGRVGA+R+ALDAEGF VSIMSYTAK
Sbjct: 235 GVILNDETVYQLCKQAVSQARAGADVVSPSNMMDGRVGAIRSALDAEGFNDVSIMSYTAK 294
Query: 282 YASSFYGPFREALDSNPRFGDKKTYQMNPANYREALTEMREDETEGADILLVKPGLPYLD 341
YASSFYGPFREALDSNPRFGDKKTYQMNPANYREAL E DE EGADILLVKPGLPYLD
Sbjct: 295 YASSFYGPFREALDSNPRFGDKKTYQMNPANYREALLETAADEAEGADILLVKPGLPYLD 354
Query: 342 IIRLLRDNSPLPIAAYQVSGEYSMIKAGGALKMIDEEKVMMESLLCLRRAGADIILTYFA 401
IIRL RDNS LPIAAYQVSGEYSMIKAGGAL MIDEEKVMMESL+CLRRAGAD+ILTYFA
Sbjct: 355 IIRLSRDNSALPIAAYQVSGEYSMIKAGGALNMIDEEKVMMESLMCLRRAGADVILTYFA 414
Query: 402 LQAARCLCG 410
Q LCG
Sbjct: 415 RQPPAVLCG 423
>emb|CAA52955.1| 5-aminolevulinic acid dehydratase [Selaginella martensii]
gi|1170213|sp|P45623|HEM2_SELMA Delta-aminolevulinic
acid dehydratase, chloroplast precursor (Porphobilinogen
synthase) (ALADH) (ALAD)
Length = 417
Score = 573 bits (1477), Expect = e-162
Identities = 283/358 (79%), Positives = 311/358 (86%)
Query: 55 EAAVVAGNVPDAPPVPPTPAAPAGTPVVTSLPIQRRPRRNRRSATHRSAFQETTLSPANF 114
EA VV+GN P AP P AP GTPVV L + RPRRNR+SA R AFQETTL+PANF
Sbjct: 60 EADVVSGNPPAAPAAPAKAKAPPGTPVVKPLRLTSRPRRNRKSAALRDAFQETTLTPANF 119
Query: 115 VYPLFIHEGEEDTPIGAMPGCYRLGWRHGLIEEVAKARDVGVNSVVLFPKIPDALKTPTG 174
+ PLFIHEGEED+PIGAMPGC RLGWRHGLI+EV KARDVGVNSVVLFPKIPDALK+ TG
Sbjct: 120 ILPLFIHEGEEDSPIGAMPGCSRLGWRHGLIDEVYKARDVGVNSVVLFPKIPDALKSSTG 179
Query: 175 DEAYNENGLVPRTIRLLKDKYPDLVIYTDVALDPYSSDGHDGIVREDGVIMNDETVHQLC 234
DEAYN +GLVPR IR LKDK+PDLVIYTDVALDPYSSDGHDGIVREDGVIMNDETVHQLC
Sbjct: 180 DEAYNPDGLVPRAIRTLKDKFPDLVIYTDVALDPYSSDGHDGIVREDGVIMNDETVHQLC 239
Query: 235 KQAVAQARAGADVVSPSDMMDGRVGAMRAALDAEGFQHVSIMSYTAKYASSFYGPFREAL 294
KQAVAQA AGADVVSPSDMMDGRVGA+R ALD G+ HVSIM+YTAKYAS+FY PFRE L
Sbjct: 240 KQAVAQAEAGADVVSPSDMMDGRVGAIRTALDEAGYYHVSIMAYTAKYASAFYEPFREEL 299
Query: 295 DSNPRFGDKKTYQMNPANYREALTEMREDETEGADILLVKPGLPYLDIIRLLRDNSPLPI 354
DSNPRFGDKKTYQMNP NYREAL E+ DE+EGADIL+VKP +PYL +IRLLRD S LPI
Sbjct: 300 DSNPRFGDKKTYQMNPENYREALLEVHADESEGADILMVKPAMPYLHVIRLLRDTSALPI 359
Query: 355 AAYQVSGEYSMIKAGGALKMIDEEKVMMESLLCLRRAGADIILTYFALQAARCLCGEK 412
+AYQVSGEYSMIKA + M+DE+K ++ESLLC++RAGAD+ILTY ALQAAR LCGEK
Sbjct: 360 SAYQVSGEYSMIKAAASQGMLDEKKAILESLLCIKRAGADVILTYAALQAARWLCGEK 417
>emb|CAA43833.1| delta-aminolevulinic acid dehydratase; porphobilinogen synthase
[Selaginella martensii] gi|99459|pir||S16738
porphobilinogen synthase (EC 4.2.1.24) precursor -
Martens's spike moss (fragment)
Length = 401
Score = 569 bits (1466), Expect = e-161
Identities = 283/359 (78%), Positives = 312/359 (86%), Gaps = 1/359 (0%)
Query: 55 EAAVVAGNVPDAPPVPPTPAAPAGTPVVTSLPIQRRPRRNRRSATHRSAFQETTLSPANF 114
EA VV+GN P AP P AP+GTPVV L + RPRRNR+SA R AFQETTL+PANF
Sbjct: 43 EADVVSGNPPAAPAAPAKAKAPSGTPVVKPLRLTSRPRRNRKSAALRDAFQETTLTPANF 102
Query: 115 VYPLFIHEGEEDTPIGAMPGCYRLGWRHGLIEEVAKARDVGVNSVVLFPKIPDALKTPTG 174
+ PLFIHEGEED+PIGAMPGC RLGWRHGLI+EV KARDVGVNSVVLFPKIPDALK+ TG
Sbjct: 103 ILPLFIHEGEEDSPIGAMPGCSRLGWRHGLIDEVYKARDVGVNSVVLFPKIPDALKSSTG 162
Query: 175 DEAYNENGLVPRTIRLLKDKYPDLVIYTDVALDPYSSDGHDGIVREDGVIMNDETVHQLC 234
DEAYN NGLVPR IR LKDK+PDLVIYTDVALDPYSSDGHDGIVREDGVIMNDETVHQLC
Sbjct: 163 DEAYNPNGLVPRAIRTLKDKFPDLVIYTDVALDPYSSDGHDGIVREDGVIMNDETVHQLC 222
Query: 235 KQAVAQARAGADVVSPSDMMDGRVGAMRAALDAEGFQHVSIMSYTAKYASSFYGPFREAL 294
KQAVAQA AGADVVSPSD+MDGRVGA+R ALD G+ HV IM+YTAKYAS+ Y PFREAL
Sbjct: 223 KQAVAQAEAGADVVSPSDIMDGRVGAIRTALDEAGYYHVWIMAYTAKYASALYEPFREAL 282
Query: 295 DSNPRFGDKKTYQMNPANYREALTEMREDETEGADILLVKPGLPYLDIIRLLRDNSPLPI 354
DSNPRFGDKKTYQMNPANYREAL E+ DE+EGADIL+VKP +PYLD+IRLL D S LPI
Sbjct: 283 DSNPRFGDKKTYQMNPANYREALLEVHADESEGADILMVKPAMPYLDVIRLLPDTSALPI 342
Query: 355 AAYQVSGEYSMIKAGGALKM-IDEEKVMMESLLCLRRAGADIILTYFALQAARCLCGEK 412
+AYQVSGEYSMIKA + M +DE+K ++ESLLC++RAGAD+ILTY ALQAAR LCGEK
Sbjct: 343 SAYQVSGEYSMIKAAASQGMLVDEKKAILESLLCIKRAGADVILTYAALQAARWLCGEK 401
>emb|CAA61978.1| porphobilinogen synthase [Physcomitrella patens]
gi|2118311|pir||S58169 porphobilinogen synthase (EC
4.2.1.24) - moss (Physcomitrella patens)
gi|2495172|sp|Q43058|HEM2_PHYPA Delta-aminolevulinic
acid dehydratase, chloroplast precursor (Porphobilinogen
synthase) (ALADH)
Length = 430
Score = 562 bits (1449), Expect = e-159
Identities = 279/358 (77%), Positives = 314/358 (86%), Gaps = 2/358 (0%)
Query: 55 EAAVVAGNVPDAPPVPPTPAAPAGTPVVTSLPIQRRPRRNRRSATHRSAFQETTLSPANF 114
EA VVAGN P APPVP P+AP GTP ++ L + RPRRNRRS R+AFQETT+SPANF
Sbjct: 74 EANVVAGNAPAAPPVPAKPSAPEGTPAISPLVMPARPRRNRRSPALRAAFQETTISPANF 133
Query: 115 VYPLFIHEGEEDTPIGAMPGCYRLGWRHGLIEEVAKARDVGVNSVVLFPKIPDALKTPTG 174
+ PLF+HEGE++ PIGAMPGC RLGWRHGLI+EV KARDVGVNSVVLFPK+PDALK+ TG
Sbjct: 134 ILPLFVHEGEQNAPIGAMPGCQRLGWRHGLIDEVYKARDVGVNSVVLFPKVPDALKSSTG 193
Query: 175 DEAYNENGLVPRTIRLLKDKYPDLVIYTDVALDPYSSDGHDGIVREDGVIMNDETVHQLC 234
DEAYN +GLVPR IRLLK PDLVIYTDVALDPYSSDGHDGIVREDG+IMNDETVHQLC
Sbjct: 194 DEAYNPDGLVPRCIRLLK-AIPDLVIYTDVALDPYSSDGHDGIVREDGLIMNDETVHQLC 252
Query: 235 KQAVAQARAGADVVSPSDMMDGRVGAMRAALDAEGFQHVSIMSYTAKYASSFYGPFREAL 294
KQAVAQA+AGADVVSPSDMMDGRVGA+R ALD G Q VSI++YTAKYAS+FYGP REAL
Sbjct: 253 KQAVAQAQAGADVVSPSDMMDGRVGAIRKALDLAGHQDVSIIAYTAKYASAFYGPSREAL 312
Query: 295 DSNPRFGDKKTYQMNPANYREALTEMREDETEGADILLVKPGLPYLDIIRLLRDNSPLPI 354
DSNPRFGDKKTYQMNPANYREAL E R DE EGADIL+VKP +PYLD+IRLLRDN+ LPI
Sbjct: 313 DSNPRFGDKKTYQMNPANYREALIETRMDEAEGADILMVKPAMPYLDVIRLLRDNTALPI 372
Query: 355 AAYQVSGEYSMIKAGGALKMIDEEKVMMESLLCLRRAGADIILTYFALQAARCLCGEK 412
+AYQVSGEYSMI+A G M+DE+K ++ESLL +RRAGAD+ILTYFA+QAA+ LC E+
Sbjct: 373 SAYQVSGEYSMIRA-GCRGMLDEKKAVLESLLSIRRAGADVILTYFAIQAAQWLCAER 429
>ref|NP_175085.1| porphobilinogen synthase, putative / delta-aminolevulinic acid
dehydratase, putative [Arabidopsis thaliana]
gi|13876503|gb|AAK43479.1| delta-aminolevulinic acid
dehydratase (Alad), putative [Arabidopsis thaliana]
Length = 406
Score = 494 bits (1272), Expect = e-138
Identities = 251/352 (71%), Positives = 288/352 (81%), Gaps = 9/352 (2%)
Query: 62 NVPDAPPVPPTPAAPAGTPVVTSLPIQRRPRRNRRSATHRSAFQETTLSPANFVYPLFIH 121
N +APPVP PA P + L + RR RRNR+ T R+AFQET +SPANF+YPLFIH
Sbjct: 59 NALEAPPVPSKPATPL---IDQPLQLSRRARRNRKCPTQRAAFQETNISPANFIYPLFIH 115
Query: 122 EGEEDTPIGAMPGCYRLGWRHGLIEEVAKARDVGVNSVVLFPKIPDALKTPTGDEAYNEN 181
EGE D PI +MPG Y LGWRHGLIEEVA+A DVGVNSV L+PK+P+ALK+PTG+EA+N+N
Sbjct: 116 EGEVDIPITSMPGRYMLGWRHGLIEEVARALDVGVNSVKLYPKVPEALKSPTGEEAFNDN 175
Query: 182 GLVPRTIRLLKDKYPDLVIYTDVALDPYSSDGHDGIVREDGVIMNDETVHQLCKQAVAQA 241
GL+PRT+RLLKD++PDLVIYTDV D YS+ GH GIV EDGVI+NDET+HQL KQAV+QA
Sbjct: 176 GLIPRTVRLLKDRFPDLVIYTDVNFDEYSTTGHGGIVGEDGVILNDETIHQLRKQAVSQA 235
Query: 242 RAGADVVSPSDMMDGRVGAMRAALDAEGFQHVSIMSYTAKYASSFYGPFREALDSNPRFG 301
RAGADVV S+M+DGRVGA+RAALDAEGFQ VSIMSY+ KY SS YG FR+
Sbjct: 236 RAGADVVCTSEMLDGRVGAVRAALDAEGFQDVSIMSYSVKYTSSLYGRFRKVQ------L 289
Query: 302 DKKTYQMNPANYREALTEMREDETEGADILLVKPGLPYLDIIRLLRDNSPLPIAAYQVSG 361
DKKTYQ+NPAN REAL E REDE EGADIL+VKP LP LDIIRLL++ + LPI A QVSG
Sbjct: 290 DKKTYQINPANSREALLEAREDEAEGADILMVKPALPSLDIIRLLKNQTLLPIGACQVSG 349
Query: 362 EYSMIKAGGALKMIDEEKVMMESLLCLRRAGADIILTYFALQAARCLCGEKR 413
EYSMIKA G LKMIDEEKVMMESLLC+RRAGAD+ILTYFALQAA LCGE +
Sbjct: 350 EYSMIKAAGLLKMIDEEKVMMESLLCIRRAGADLILTYFALQAATKLCGENK 401
>pir||S53487 porphobilinogen synthase (EC 4.2.1.24) precursor - Chlamydomonas
reinhardtii gi|642579|gb|AAA79515.1|
delta-aminolevulinic acid dehydratase precursor
gi|2495170|sp|Q42682|HEM2_CHLRE Delta-aminolevulinic
acid dehydratase, chloroplast precursor (Porphobilinogen
synthase) (ALADH)
Length = 390
Score = 434 bits (1117), Expect = e-120
Identities = 210/346 (60%), Positives = 270/346 (77%), Gaps = 9/346 (2%)
Query: 76 PAGTPVVTSLPIQRRPRRNRRSATHRSAFQETTLSPANFVYPLFIHE-GEEDTPIGAMPG 134
P GTP+VT + RPRRNRRS + R++ +E +SPANF+ P+FIHE ++ PI +MPG
Sbjct: 45 PEGTPIVTPQDLPSRPRRNRRSESFRASVREVNVSPANFILPIFIHEESNQNVPIASMPG 104
Query: 135 CYRLGWRHGLIEEVAKARDVGVNSVVLFPKIPDALKTPTGDEAYNENGLVPRTIRLLKDK 194
RL + +I+ VA+ R GVN VV+FPK PD LKT T +EA+N+NGL RTIRLLKD
Sbjct: 105 INRLAYGKNVIDYVAEPRSYGVNQVVVFPKTPDHLKTQTAEEAFNKNGLSQRTIRLLKDS 164
Query: 195 YPDLVIYTDVALDPYSSDGHDGIVREDGVIMNDETVHQLCKQAVAQARAGADVVSPSDMM 254
+PDL +YTDVALDPY+SDGHDGIV + GVI+NDET+ LC+QAV+QA AGADVVSPSDMM
Sbjct: 165 FPDLEVYTDVALDPYNSDGHDGIVSDAGVILNDETIEYLCRQAVSQAEAGADVVSPSDMM 224
Query: 255 DGRVGAMRAALDAEGFQHVSIMSYTAKYASSFYGPFREALDSNPRFG--------DKKTY 306
DGRVGA+R ALD EGF +VSIMSYTAKYAS++YGPFR+AL S P+ G +KKTY
Sbjct: 225 DGRVGAIRRALDREGFTNVSIMSYTAKYASAYYGPFRDALASAPKPGQAHRRIPPNKKTY 284
Query: 307 QMNPANYREALTEMREDETEGADILLVKPGLPYLDIIRLLRDNSPLPIAAYQVSGEYSMI 366
QM+PANYREA+ E + DE EGADI++VKPG+PYLD++RLLR+ SPLP+A Y VSGEY+M+
Sbjct: 285 QMDPANYREAIREAKADEAEGADIMMVKPGMPYLDVVRLLRETSPLPVAVYHVSGEYAML 344
Query: 367 KAGGALKMIDEEKVMMESLLCLRRAGADIILTYFALQAARCLCGEK 412
KA ++E+ ++E++ C RRAG D+ILTY+ ++A++ L GEK
Sbjct: 345 KAAAERGWLNEKDAVLEAMTCFRRAGGDLILTYYGIEASKWLAGEK 390
>emb|CAC36141.1| ALA dehydratase [Cyanophora paradoxa]
Length = 419
Score = 433 bits (1114), Expect = e-120
Identities = 210/330 (63%), Positives = 267/330 (80%), Gaps = 2/330 (0%)
Query: 84 SLPIQRRPRRNRRSATHRSAFQETTLSPANFVYPLFIHEGEEDTPIGAMPGCYRLGWRHG 143
+L ++RRPRRNR+ A R ++ETT+ P NF+ PLFIH+G+++ PI AMPGC+RL R G
Sbjct: 85 ALAMRRRPRRNRKGAAMRGMYRETTVGPENFILPLFIHDGDDNQPISAMPGCFRLS-REG 143
Query: 144 LIEEVAKARDVGVNSVVLFPKIPDALKTPTGDEAYNENGLVPRTIRLLKDKYPDLVIYTD 203
L++EV A G+ +VVLFPK+PD LKT G E +N +G+VPRT+R+LK+K+P+L++ TD
Sbjct: 144 LLKEVEGAYSEGIRAVVLFPKVPDNLKTSDGAECFNPDGIVPRTVRMLKEKFPELLVITD 203
Query: 204 VALDPYSSDGHDGIV-REDGVIMNDETVHQLCKQAVAQARAGADVVSPSDMMDGRVGAMR 262
+ALDPY+SDGHDGIV + +G I+NDETV QLCKQA+ A AGAD+VSPSDMMDGR+GA+R
Sbjct: 204 IALDPYNSDGHDGIVDKTNGKIINDETVEQLCKQALCHAAAGADIVSPSDMMDGRIGAIR 263
Query: 263 AALDAEGFQHVSIMSYTAKYASSFYGPFREALDSNPRFGDKKTYQMNPANYREALTEMRE 322
ALD G+ VSI+SY AKYAS++YGPFR ALDS+PRFGDK TYQM+PAN REAL E
Sbjct: 264 DALDGAGYTDVSILSYCAKYASAYYGPFRTALDSDPRFGDKTTYQMDPANVREALIEAEL 323
Query: 323 DETEGADILLVKPGLPYLDIIRLLRDNSPLPIAAYQVSGEYSMIKAGGALKMIDEEKVMM 382
D EGAD+++VKPGLPYLD+I+ LR+NSPLPIAAYQVSGEY+MIKA A +DE KV+M
Sbjct: 324 DLEEGADMIMVKPGLPYLDVIKTLRENSPLPIAAYQVSGEYAMIKAAVANGWLDERKVVM 383
Query: 383 ESLLCLRRAGADIILTYFALQAARCLCGEK 412
ESL+ LRRAGAD ILTYFA A+R + ++
Sbjct: 384 ESLIALRRAGADFILTYFARPASRWIAEDR 413
>emb|CAC36186.1| ALA dehydratase [Odontella sinensis]
Length = 412
Score = 410 bits (1053), Expect = e-113
Identities = 204/317 (64%), Positives = 256/317 (80%), Gaps = 2/317 (0%)
Query: 90 RPRRNRRSATHRSAFQETTLSPANFVYPLFIHEGEEDTPIGAMPGCYRLGWRHGLIEEVA 149
RPRRNR+SA R +E ++PANF+YPLFIHE + +T I +MPGC R + +++EV
Sbjct: 90 RPRRNRKSAAVRGMVRENIVTPANFIYPLFIHEEDFNTEIPSMPGCERHSLPN-MLKEVG 148
Query: 150 KARDVGVNSVVLFPKIPDALKTPTGDEAYNENGLVPRTIRLLKDKYPDLVIYTDVALDPY 209
+A D+GV + VLFPK+PD LKT G EAYN +G+V R IR++K+ YPD V+ TDVALDPY
Sbjct: 149 EAYDLGVKTFVLFPKVPDELKTNLGVEAYNPDGIVHRAIRMIKEAYPDSVVCTDVALDPY 208
Query: 210 SSDGHDGIVREDGVIMNDETVHQLCKQAVAQARAGADVVSPSDMMDGRVGAMRAALDAEG 269
S GHDG+V EDG I+ND T++QLCKQAV+QARAGADVV+PSDMMDGRVGA+R ALDAEG
Sbjct: 209 SDQGHDGVV-EDGKILNDVTINQLCKQAVSQARAGADVVAPSDMMDGRVGAIRDALDAEG 267
Query: 270 FQHVSIMSYTAKYASSFYGPFREALDSNPRFGDKKTYQMNPANYREALTEMREDETEGAD 329
F VSI++YTAKYAS++YGPFR+ALDS+P FGDKKTYQ +PAN REAL E D EGAD
Sbjct: 268 FTDVSILAYTAKYASAYYGPFRDALDSHPGFGDKKTYQQDPANGREALIEAALDAAEGAD 327
Query: 330 ILLVKPGLPYLDIIRLLRDNSPLPIAAYQVSGEYSMIKAGGALKMIDEEKVMMESLLCLR 389
+L+VKPG+PYLDIIR L+D +PLP+AAY VSGEY+MIKA ++E+ V++E+L C +
Sbjct: 328 MLMVKPGMPYLDIIRRLKDATPLPVAAYHVSGEYAMIKAACEKGWLEEKDVVLETLTCFK 387
Query: 390 RAGADIILTYFALQAAR 406
RAGADIILTY+A QAA+
Sbjct: 388 RAGADIILTYYAKQAAQ 404
>emb|CAC36148.1| ALA dehydratase [Fucus vesiculosus]
Length = 425
Score = 402 bits (1034), Expect = e-111
Identities = 197/324 (60%), Positives = 252/324 (76%), Gaps = 2/324 (0%)
Query: 86 PIQRRPRRNRRSATHRSAFQETTLSPANFVYPLFIHEGEEDTPIGAMPGCYRLGWRHGLI 145
P + RPRRNR+S RS +E + PANF+YPLFIH+ + + I +MPGC R H ++
Sbjct: 99 PQKVRPRRNRKSEAMRSMVRENIVRPANFIYPLFIHDKDFEVEIQSMPGCLRHSLPH-ML 157
Query: 146 EEVAKARDVGVNSVVLFPKIPDALKTPTGDEAYNENGLVPRTIRLLKDKYPDLVIYTDVA 205
EEV +A GV + VLFP++PD LKT G+E YN G+VPR + ++K+K+PD ++ TDVA
Sbjct: 158 EEVEEAWKFGVRTFVLFPQVPDHLKTNYGEECYNPEGIVPRAVSMIKEKFPDSIVCTDVA 217
Query: 206 LDPYSSDGHDGIVREDGVIMNDETVHQLCKQAVAQARAGADVVSPSDMMDGRVGAMRAAL 265
LDPYS GHDG+V+ DG I+NDET+ QL KQ V QARAGADVV+PSDMMDGR+GA+R AL
Sbjct: 218 LDPYSDQGHDGVVK-DGQILNDETIMQLTKQVVCQARAGADVVAPSDMMDGRIGAIRDAL 276
Query: 266 DAEGFQHVSIMSYTAKYASSFYGPFREALDSNPRFGDKKTYQMNPANYREALTEMREDET 325
DAEGF +VSIM+YTAKYAS+FYGPFR+ALDS+P FGDKKTYQ +PAN REAL E D
Sbjct: 277 DAEGFTNVSIMAYTAKYASAFYGPFRDALDSHPGFGDKKTYQQDPANGREALLEAALDAE 336
Query: 326 EGADILLVKPGLPYLDIIRLLRDNSPLPIAAYQVSGEYSMIKAGGALKMIDEEKVMMESL 385
EGAD+L+VKPG+PYLD+IR L+++S LPIAAY VSGEY+M+KA +DE + ++E++
Sbjct: 337 EGADMLMVKPGMPYLDVIRRLKESSNLPIAAYHVSGEYAMMKAAVERGWLDEREAVLEAM 396
Query: 386 LCLRRAGADIILTYFALQAARCLC 409
C +RAGAD+ILTY+A AAR LC
Sbjct: 397 TCFKRAGADVILTYYAKDAARWLC 420
>emb|CAC36154.1| ALA dehydratase [Laminaria digitata]
Length = 410
Score = 392 bits (1008), Expect = e-108
Identities = 191/320 (59%), Positives = 246/320 (76%), Gaps = 2/320 (0%)
Query: 90 RPRRNRRSATHRSAFQETTLSPANFVYPLFIHEGEEDTPIGAMPGCYRLGWRHGLIEEVA 149
RPRRNR+S RS +E + PANF+YPLFIH+ + + I MPGC R H +++EV
Sbjct: 88 RPRRNRKSEAMRSMLREAVVRPANFIYPLFIHDEDFEVEIKPMPGCLRHSLSH-MLKEVE 146
Query: 150 KARDVGVNSVVLFPKIPDALKTPTGDEAYNENGLVPRTIRLLKDKYPDLVIYTDVALDPY 209
+A GV + V+FPK+PD LKT DE +N NG+VPR + ++K+++PD ++ TDVALDPY
Sbjct: 147 EAWKFGVRTFVVFPKVPDQLKTNYADECHNPNGIVPRAVSMIKERFPDSIVCTDVALDPY 206
Query: 210 SSDGHDGIVREDGVIMNDETVHQLCKQAVAQARAGADVVSPSDMMDGRVGAMRAALDAEG 269
S GHDG+V+ DG I+NDET+ QL KQ V QARAG+DVV+PSDMMDGRVGA+R ALD+EG
Sbjct: 207 SDQGHDGVVK-DGKILNDETIMQLTKQVVCQARAGSDVVAPSDMMDGRVGAIRDALDSEG 265
Query: 270 FQHVSIMSYTAKYASSFYGPFREALDSNPRFGDKKTYQMNPANYREALTEMREDETEGAD 329
F VSI++YTAKYAS+FYGPFR+ALDS+P FGDKKTYQ +PAN REAL E D EGAD
Sbjct: 266 FTDVSILAYTAKYASAFYGPFRDALDSHPGFGDKKTYQQDPANGREALIEAALDAAEGAD 325
Query: 330 ILLVKPGLPYLDIIRLLRDNSPLPIAAYQVSGEYSMIKAGGALKMIDEEKVMMESLLCLR 389
+L+VKPG+PYLD+I+ LR++S LPI+AY VSGEY+M+KA +DE ++E++ C +
Sbjct: 326 MLMVKPGMPYLDVIQRLRESSNLPISAYHVSGEYAMMKAAVERGWLDERAAVLETMTCFK 385
Query: 390 RAGADIILTYFALQAARCLC 409
RAGAD ILTY+A AAR LC
Sbjct: 386 RAGADAILTYYAKDAARWLC 405
>gb|AAP99289.1| Delta-aminolevulinic acid dehydratase [Prochlorococcus marinus
subsp. marinus str. CCMP1375]
gi|33239695|ref|NP_874637.1| Delta-aminolevulinic acid
dehydratase [Prochlorococcus marinus subsp. marinus str.
CCMP1375]
Length = 334
Score = 387 bits (995), Expect = e-106
Identities = 202/328 (61%), Positives = 244/328 (73%), Gaps = 11/328 (3%)
Query: 90 RPRRNRRSATHRSAFQETTLSPANFVYPLFIHEGEEDTPIGAMPGCYRLGWR-HGLIEEV 148
RPRR RR+ RS QE ++ ++F+YPLFIHEG E PIGAMPG R W ++ EV
Sbjct: 6 RPRRLRRTTALRSMVQEHSVHASDFIYPLFIHEGAEVQPIGAMPGANR--WTLDTVVGEV 63
Query: 149 AKARDVGVNSVVLFPKIPDALKTPTGDEAYNENGLVPRTIRLLKDKYPDLVIYTDVALDP 208
+A ++GV VVLFPKI + LKT G+E +NE GL+PR IR LK + P++ I TDVALDP
Sbjct: 64 TRAWNLGVRCVVLFPKIDEELKTEEGEECFNEKGLIPRAIRRLKTELPEMAIMTDVALDP 123
Query: 209 YSSDGHDGIVREDGVIMNDETVHQLCKQAVAQARAGADVVSPSDMMDGRVGAMRAALDAE 268
YS DGHDGIV +GV++NDETV QLCKQA+ QA AGAD++ PSDMMDGRVGA+R ALD E
Sbjct: 124 YSLDGHDGIVSSEGVVLNDETVSQLCKQAIVQAAAGADLIGPSDMMDGRVGAIREALDEE 183
Query: 269 GFQHVSIMSYTAKYASSFYGPFREALDSNPRF--------GDKKTYQMNPANYREALTEM 320
G+++V I+SYTAKY+S++YGPFREALDS PR DK TYQMNPAN REA+TE
Sbjct: 184 GYENVGIISYTAKYSSAYYGPFREALDSAPRAINNSKPIPKDKNTYQMNPANAREAITEA 243
Query: 321 REDETEGADILLVKPGLPYLDIIRLLRDNSPLPIAAYQVSGEYSMIKAGGALKMIDEEKV 380
+ DE EG+DIL+VKPGL YLDII LR S LPIAAY VSGEYSMIKA IDE+ V
Sbjct: 244 QLDEQEGSDILMVKPGLAYLDIIYRLRQESELPIAAYNVSGEYSMIKAAALQGWIDEKSV 303
Query: 381 MMESLLCLRRAGADIILTYFALQAARCL 408
++E+LL +RAGAD+ILTY A AA L
Sbjct: 304 VLETLLSFKRAGADLILTYHACDAAEWL 331
Database: nr
Posted date: Jul 5, 2005 12:34 AM
Number of letters in database: 863,360,394
Number of sequences in database: 2,540,612
Lambda K H
0.318 0.135 0.397
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 742,191,392
Number of Sequences: 2540612
Number of extensions: 33249473
Number of successful extensions: 150922
Number of sequences better than 10.0: 455
Number of HSP's better than 10.0 without gapping: 375
Number of HSP's successfully gapped in prelim test: 82
Number of HSP's that attempted gapping in prelim test: 149421
Number of HSP's gapped (non-prelim): 587
length of query: 413
length of database: 863,360,394
effective HSP length: 130
effective length of query: 283
effective length of database: 533,080,834
effective search space: 150861876022
effective search space used: 150861876022
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 76 (33.9 bits)
Medicago: description of AC140916.2


