

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC140913.6 - phase: 0
(831 letters)
Database: nr
2,540,612 sequences; 863,360,394 total letters
Searching..................................................done
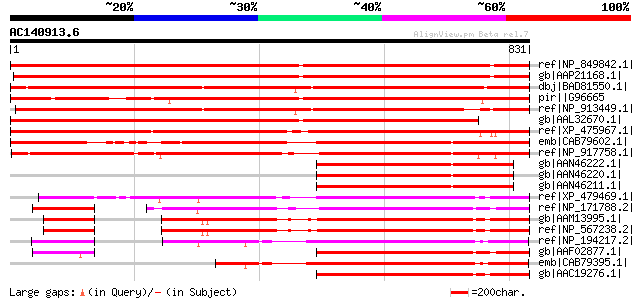
Score E
Sequences producing significant alignments: (bits) Value
ref|NP_849842.1| AAA-type ATPase family protein [Arabidopsis tha... 1111 0.0
gb|AAP21168.1| At1g64110/F22C12_22 [Arabidopsis thaliana] gi|184... 1107 0.0
dbj|BAD81550.1| unknown protein [Oryza sativa (japonica cultivar... 1058 0.0
pir||G96665 protein F22C12.12 [imported] - Arabidopsis thaliana ... 1030 0.0
ref|NP_913449.1| P0492F05.26 [Oryza sativa (japonica cultivar-gr... 1008 0.0
gb|AAL32670.1| similar to homeobox protein [Arabidopsis thaliana] 1005 0.0
ref|XP_475967.1| unknown protein [Oryza sativa (japonica cultiva... 1003 0.0
emb|CAB79602.1| putative protein [Arabidopsis thaliana] gi|44553... 933 0.0
ref|NP_917758.1| P0501G01.20 [Oryza sativa (japonica cultivar-gr... 743 0.0
gb|AAN46222.1| unknown protein [Arabidopsis lyrata] gi|28188591|... 521 e-146
gb|AAN46220.1| unknown protein [Arabidopsis thaliana] gi|2818858... 521 e-146
gb|AAN46211.1| unknown protein [Arabidopsis thaliana] 518 e-145
ref|XP_479469.1| putative MSP1(mitochondrial sorting of proteins... 513 e-143
ref|NP_171788.2| AAA-type ATPase family protein [Arabidopsis tha... 486 e-135
gb|AAM13995.1| unknown protein [Arabidopsis thaliana] 476 e-132
ref|NP_567238.2| AAA-type ATPase family protein [Arabidopsis tha... 476 e-132
ref|NP_194217.2| AAA-type ATPase family protein [Arabidopsis tha... 449 e-124
gb|AAF02877.1| Unknown protein [Arabidopsis thaliana] gi|2551840... 417 e-115
emb|CAB79395.1| putative protein [Arabidopsis thaliana] gi|46782... 410 e-113
gb|AAC19276.1| T14P8.7 [Arabidopsis thaliana] gi|7269007|emb|CAB... 410 e-112
>ref|NP_849842.1| AAA-type ATPase family protein [Arabidopsis thaliana]
Length = 829
Score = 1111 bits (2873), Expect = 0.0
Identities = 589/839 (70%), Positives = 678/839 (80%), Gaps = 18/839 (2%)
Query: 1 MEQKGMLISA-ALSVGVGVGLGLASGQTMFK-PNTYSSSSNALTPDKIENEMLRLVVDGR 58
M+ K ML+SA + VGVGVGLGLASGQ + K SSS+NA+T DK+E E+LR VVDGR
Sbjct: 1 MDSKQMLLSALGVGVGVGVGLGLASGQAVGKWAGGNSSSNNAVTADKMEKEILRQVVDGR 60
Query: 59 ESNVTFDNFPYYLSEQTRVLLTSAAYVHLKHAEVSKYTRNLAPASRTILLSGPAELYQQV 118
ES +TFD FPYYLSEQTRVLLTSAAYVHLKH + SKYTRNL+PASR ILLSGPAELYQQ+
Sbjct: 61 ESKITFDEFPYYLSEQTRVLLTSAAYVHLKHFDASKYTRNLSPASRAILLSGPAELYQQM 120
Query: 119 LAKALTHYFEAKLLLFDVTDFSLKIQSRYGSSNCETS-FTRSTSETALARLSDLFGSFAL 177
LAKAL H+F+AKLLL DV DF+LKIQS+YGS N E+S F RS SE+AL +LS LF SF++
Sbjct: 121 LAKALAHFFDAKLLLLDVNDFALKIQSKYGSGNTESSSFKRSPSESALEQLSGLFSSFSI 180
Query: 178 FPQREENQ--GKIHRQSSGSDLRQMEAEGSYS--KLRRNASASANISSIGLQSNPTNSAP 233
PQREE++ G + RQSSG D++ EGS + KLRRN+SA+ANIS++ SN SAP
Sbjct: 181 LPQREESKAGGTLRRQSSGVDIKSSSMEGSSNPPKLRRNSSAAANISNLASSSNQV-SAP 239
Query: 234 GKHITGWPFDEKILIQTLYKVLLYVSKTYPIVLYMRDADKLLCRSQRIYKLFQTMLTKLS 293
K + W FDEK+L+Q+LYKVL YVSK PIVLY+RD + L RSQR Y LFQ +L KLS
Sbjct: 240 LKRSSSWSFDEKLLVQSLYKVLAYVSKANPIVLYLRDVENFLFRSQRTYNLFQKLLQKLS 299
Query: 294 GPILIIGSRILD-SGNECKRVDEMLTSLFPYNIEIKPPEDESRLVSWKSQFEADMKKIQI 352
GP+LI+GSRI+D S + + +DE L+++FPYNI+I+PPEDE+ LVSWKSQ E DM IQ
Sbjct: 300 GPVLILGSRIVDLSSEDAQEIDEKLSAVFPYNIDIRPPEDETHLVSWKSQLERDMNMIQT 359
Query: 353 QDNKNHIMEVLAANDLDCHDLDSICVADTMVLSNYIEEIIVSAISYHIMKNKEPEYRNGK 412
QDN+NHIMEVL+ NDL C DL+SI DT VLSNYIEEI+VSA+SYH+M NK+PEYRNGK
Sbjct: 360 QDNRNHIMEVLSENDLICDDLESISFEDTKVLSNYIEEIVVSALSYHLMNNKDPEYRNGK 419
Query: 413 LIIPCNSLSHALGIFQAGKFGDRDSLKLEAQAVTSEKKEEGAAVKPEGKTESPAPAVKTE 472
L+I SLSH +F+ GK G R+ LK + + +S K+ + ++KPE KTES V T
Sbjct: 420 LVISSISLSHGFSLFREGKAGGREKLKQKTKEESS-KEVKAESIKPETKTES----VTTV 474
Query: 473 AEIPTSVRKTDGENSVPASKAEVPDNEFEKRIRPEVIPANEIGVTFSDIGALDETKDSLQ 532
+ ++ E P + PDNEFEKRIRPEVIPA EI VTF DIGALDE K+SLQ
Sbjct: 475 SSKEEPEKEAKAEKVTPKAPEVAPDNEFEKRIRPEVIPAEEINVTFKDIGALDEIKESLQ 534
Query: 533 ELVMLPLRRPDLFEGGLLKPCRGILLFGPPGTGKTMLAKAIANEAGASFINVSMSTITSK 592
ELVMLPLRRPDLF GGLLKPCRGILLFGPPGTGKTMLAKAIA EAGASFINVSMSTITSK
Sbjct: 535 ELVMLPLRRPDLFTGGLLKPCRGILLFGPPGTGKTMLAKAIAKEAGASFINVSMSTITSK 594
Query: 593 WFGEDEKNVRALFTLAAKVSPTIIFVDEVDSMLGQRTRVGEHEAMRKIKNEFMSNWDGLT 652
WFGEDEKNVRALFTLA+KVSPTIIFVDEVDSMLGQRTRVGEHEAMRKIKNEFMS+WDGL
Sbjct: 595 WFGEDEKNVRALFTLASKVSPTIIFVDEVDSMLGQRTRVGEHEAMRKIKNEFMSHWDGLM 654
Query: 653 SKSEDRILVLAATNRPFDLDEAIIRRFERRIMVGLPSAENRENILRTLLAKEKVHGGLDF 712
+K +RILVLAATNRPFDLDEAIIRRFERRIMVGLP+ ENRE ILRTLLAKEKV LD+
Sbjct: 655 TKPGERILVLAATNRPFDLDEAIIRRFERRIMVGLPAVENREKILRTLLAKEKVDENLDY 714
Query: 713 KELATMTEGYSGSDLKNLCTTAAYRPVRELIQQEIQKDSKKKKDAEGQNSQDAQDAKEEV 772
KELA MTEGY+GSDLKNLCTTAAYRPVRELIQQE KD++KKK E + + + KE
Sbjct: 715 KELAMMTEGYTGSDLKNLCTTAAYRPVRELIQQERIKDTEKKKQREPTKAGEEDEGKE-- 772
Query: 773 EQERVITLRPLNMQDFKMAKSQVAASFAAEGAGMNELRQWNDLYGEGGSRKKEQLSYFL 831
ERVITLRPLN QDFK AK+QVAASFAAEGAGM EL+QWN+LYGEGGSRKKEQL+YFL
Sbjct: 773 --ERVITLRPLNRQDFKEAKNQVAASFAAEGAGMGELKQWNELYGEGGSRKKEQLTYFL 829
>gb|AAP21168.1| At1g64110/F22C12_22 [Arabidopsis thaliana]
gi|18407974|ref|NP_564824.1| AAA-type ATPase family
protein [Arabidopsis thaliana]
gi|15810167|gb|AAL06985.1| At1g64110/F22C12_22
[Arabidopsis thaliana]
Length = 824
Score = 1107 bits (2863), Expect = 0.0
Identities = 587/834 (70%), Positives = 675/834 (80%), Gaps = 18/834 (2%)
Query: 6 MLISA-ALSVGVGVGLGLASGQTMFK-PNTYSSSSNALTPDKIENEMLRLVVDGRESNVT 63
ML+SA + VGVGVGLGLASGQ + K SSS+NA+T DK+E E+LR VVDGRES +T
Sbjct: 1 MLLSALGVGVGVGVGLGLASGQAVGKWAGGNSSSNNAVTADKMEKEILRQVVDGRESKIT 60
Query: 64 FDNFPYYLSEQTRVLLTSAAYVHLKHAEVSKYTRNLAPASRTILLSGPAELYQQVLAKAL 123
FD FPYYLSEQTRVLLTSAAYVHLKH + SKYTRNL+PASR ILLSGPAELYQQ+LAKAL
Sbjct: 61 FDEFPYYLSEQTRVLLTSAAYVHLKHFDASKYTRNLSPASRAILLSGPAELYQQMLAKAL 120
Query: 124 THYFEAKLLLFDVTDFSLKIQSRYGSSNCETS-FTRSTSETALARLSDLFGSFALFPQRE 182
H+F+AKLLL DV DF+LKIQS+YGS N E+S F RS SE+AL +LS LF SF++ PQRE
Sbjct: 121 AHFFDAKLLLLDVNDFALKIQSKYGSGNTESSSFKRSPSESALEQLSGLFSSFSILPQRE 180
Query: 183 ENQ--GKIHRQSSGSDLRQMEAEGSYS--KLRRNASASANISSIGLQSNPTNSAPGKHIT 238
E++ G + RQSSG D++ EGS + KLRRN+SA+ANIS++ SN SAP K +
Sbjct: 181 ESKAGGTLRRQSSGVDIKSSSMEGSSNPPKLRRNSSAAANISNLASSSNQV-SAPLKRSS 239
Query: 239 GWPFDEKILIQTLYKVLLYVSKTYPIVLYMRDADKLLCRSQRIYKLFQTMLTKLSGPILI 298
W FDEK+L+Q+LYKVL YVSK PIVLY+RD + L RSQR Y LFQ +L KLSGP+LI
Sbjct: 240 SWSFDEKLLVQSLYKVLAYVSKANPIVLYLRDVENFLFRSQRTYNLFQKLLQKLSGPVLI 299
Query: 299 IGSRILD-SGNECKRVDEMLTSLFPYNIEIKPPEDESRLVSWKSQFEADMKKIQIQDNKN 357
+GSRI+D S + + +DE L+++FPYNI+I+PPEDE+ LVSWKSQ E DM IQ QDN+N
Sbjct: 300 LGSRIVDLSSEDAQEIDEKLSAVFPYNIDIRPPEDETHLVSWKSQLERDMNMIQTQDNRN 359
Query: 358 HIMEVLAANDLDCHDLDSICVADTMVLSNYIEEIIVSAISYHIMKNKEPEYRNGKLIIPC 417
HIMEVL+ NDL C DL+SI DT VLSNYIEEI+VSA+SYH+M NK+PEYRNGKL+I
Sbjct: 360 HIMEVLSENDLICDDLESISFEDTKVLSNYIEEIVVSALSYHLMNNKDPEYRNGKLVISS 419
Query: 418 NSLSHALGIFQAGKFGDRDSLKLEAQAVTSEKKEEGAAVKPEGKTESPAPAVKTEAEIPT 477
SLSH +F+ GK G R+ LK + + +S K+ + ++KPE KTES V T +
Sbjct: 420 ISLSHGFSLFREGKAGGREKLKQKTKEESS-KEVKAESIKPETKTES----VTTVSSKEE 474
Query: 478 SVRKTDGENSVPASKAEVPDNEFEKRIRPEVIPANEIGVTFSDIGALDETKDSLQELVML 537
++ E P + PDNEFEKRIRPEVIPA EI VTF DIGALDE K+SLQELVML
Sbjct: 475 PEKEAKAEKVTPKAPEVAPDNEFEKRIRPEVIPAEEINVTFKDIGALDEIKESLQELVML 534
Query: 538 PLRRPDLFEGGLLKPCRGILLFGPPGTGKTMLAKAIANEAGASFINVSMSTITSKWFGED 597
PLRRPDLF GGLLKPCRGILLFGPPGTGKTMLAKAIA EAGASFINVSMSTITSKWFGED
Sbjct: 535 PLRRPDLFTGGLLKPCRGILLFGPPGTGKTMLAKAIAKEAGASFINVSMSTITSKWFGED 594
Query: 598 EKNVRALFTLAAKVSPTIIFVDEVDSMLGQRTRVGEHEAMRKIKNEFMSNWDGLTSKSED 657
EKNVRALFTLA+KVSPTIIFVDEVDSMLGQRTRVGEHEAMRKIKNEFMS+WDGL +K +
Sbjct: 595 EKNVRALFTLASKVSPTIIFVDEVDSMLGQRTRVGEHEAMRKIKNEFMSHWDGLMTKPGE 654
Query: 658 RILVLAATNRPFDLDEAIIRRFERRIMVGLPSAENRENILRTLLAKEKVHGGLDFKELAT 717
RILVLAATNRPFDLDEAIIRRFERRIMVGLP+ ENRE ILRTLLAKEKV LD+KELA
Sbjct: 655 RILVLAATNRPFDLDEAIIRRFERRIMVGLPAVENREKILRTLLAKEKVDENLDYKELAM 714
Query: 718 MTEGYSGSDLKNLCTTAAYRPVRELIQQEIQKDSKKKKDAEGQNSQDAQDAKEEVEQERV 777
MTEGY+GSDLKNLCTTAAYRPVRELIQQE KD++KKK E + + + KE ERV
Sbjct: 715 MTEGYTGSDLKNLCTTAAYRPVRELIQQERIKDTEKKKQREPTKAGEEDEGKE----ERV 770
Query: 778 ITLRPLNMQDFKMAKSQVAASFAAEGAGMNELRQWNDLYGEGGSRKKEQLSYFL 831
ITLRPLN QDFK AK+QVAASFAAEGAGM EL+QWN+LYGEGGSRKKEQL+YFL
Sbjct: 771 ITLRPLNRQDFKEAKNQVAASFAAEGAGMGELKQWNELYGEGGSRKKEQLTYFL 824
>dbj|BAD81550.1| unknown protein [Oryza sativa (japonica cultivar-group)]
gi|56784122|dbj|BAD81507.1| unknown protein [Oryza
sativa (japonica cultivar-group)]
Length = 840
Score = 1058 bits (2737), Expect = 0.0
Identities = 561/850 (66%), Positives = 670/850 (78%), Gaps = 29/850 (3%)
Query: 1 MEQKGMLISAA-LSVGVGVGLGLASGQTMFKPNTYSSSSNALTPDKIENEMLRLVVDGRE 59
ME K +++SA + +GVGVGLGLAS + + +T +++E E+ RLVVDG +
Sbjct: 1 MEGKHVVMSAVGIGIGVGVGLGLASAP--WAGGGGQGARVGVTVERVEQELRRLVVDGAD 58
Query: 60 SNVTFDNFPYYLSEQTRVLLTSAAYVHLKHAEVSKYTRNLAPASRTILLSGPAELYQQVL 119
S VTFD FPYYLSEQTRVLLTSAAYVHLK A++S+YTRNLAPASR ILLSGPAELYQQ+L
Sbjct: 59 SRVTFDGFPYYLSEQTRVLLTSAAYVHLKQADISQYTRNLAPASRAILLSGPAELYQQML 118
Query: 120 AKALTHYFEAKLLLFDVTDFSLKIQSRYGS-SNCETSFTRSTSETALARLSDLFGSFALF 178
AKAL HYFEAKLLL D TDF +KI S+YG S+ ++SF RS SET L ++S L GS ++
Sbjct: 119 AKALAHYFEAKLLLLDPTDFLIKIHSKYGGGSSTDSSFKRSISETTLEKVSGLLGSLSIL 178
Query: 179 PQREENQGKIHRQSSGSD--LRQMEAEGSYSKLRRNASASANISSIGLQSNPTNSAPGKH 236
PQ+E+ +G I RQSS +D LR E+ S+ KL+RNAS S+++SS+ Q P N A +
Sbjct: 179 PQKEKPKGTIRRQSSMTDMKLRSSESTSSFPKLKRNASTSSDMSSLASQGPPNNPASLRR 238
Query: 237 ITGWPFDEKILIQTLYKVLLYVSKTYPIVLYMRDADKLLCRSQRIYKLFQTMLTKLSGPI 296
+ W FDEKIL+Q +YKVL VSK PIVLY+RD +K L +S+++Y +F+ +L KL GP+
Sbjct: 239 ASSWTFDEKILVQAVYKVLHSVSKKNPIVLYIRDVEKFLHKSKKMYVMFEKLLNKLEGPV 298
Query: 297 LIIGSRILDSGNECKRVDEMLTSLFPYNIEIKPPEDESRLVSWKSQFEADMKKIQIQDNK 356
L++GSRI+D + + +DE LT+LFPYNIEIKPPE+E+ LVSW SQ E DMK IQ QDN+
Sbjct: 299 LVLGSRIVDMDFD-EELDERLTALFPYNIEIKPPENENHLVSWNSQLEEDMKMIQFQDNR 357
Query: 357 NHIMEVLAANDLDCHDLDSICVADTMVLSNYIEEIIVSAISYHIMKNKEPEYRNGKLIIP 416
NHI EVLA NDL+C DL SIC++DTMVL YIEEI+VSA+SYH+M K+PEYRNGKL++
Sbjct: 358 NHITEVLAENDLECDDLGSICLSDTMVLGRYIEEIVVSAVSYHLMNKKDPEYRNGKLLLS 417
Query: 417 CNSLSHALGIFQAGKFGDRDSLKLEAQAVTSEKKEEGAA-------------VKPEGKTE 463
SLSHAL IFQ K D+DS+KLEA+ S+ + G A + P T
Sbjct: 418 AKSLSHALEIFQENKMYDKDSMKLEAKRDASKVADRGIAPFAAKSETKPATLLPPVPPTA 477
Query: 464 SPAPAVKTEAEIPTSVRKTDGENSVPASKAEV--PDNEFEKRIRPEVIPANEIGVTFSDI 521
+ AP V+++AE P K D N PA+KA PDNEFEKRIRPEVIPANEIGVTF DI
Sbjct: 478 AAAPPVESKAE-PEKFEKKD--NPSPAAKAPEMPPDNEFEKRIRPEVIPANEIGVTFDDI 534
Query: 522 GALDETKDSLQELVMLPLRRPDLFEGGLLKPCRGILLFGPPGTGKTMLAKAIANEAGASF 581
GAL + K+SLQELVMLPLRRPDLF+GGLLKPCRGILLFGPPGTGKTMLAKAIANEA ASF
Sbjct: 535 GALSDIKESLQELVMLPLRRPDLFKGGLLKPCRGILLFGPPGTGKTMLAKAIANEAQASF 594
Query: 582 INVSMSTITSKWFGEDEKNVRALFTLAAKVSPTIIFVDEVDSMLGQRTRVGEHEAMRKIK 641
INVSMSTITSKWFGEDEKNVRALFTLAAKVSPTIIFVDEVDSMLGQR R GEHEAMRKIK
Sbjct: 595 INVSMSTITSKWFGEDEKNVRALFTLAAKVSPTIIFVDEVDSMLGQRNRAGEHEAMRKIK 654
Query: 642 NEFMSNWDGLTSKSEDRILVLAATNRPFDLDEAIIRRFERRIMVGLPSAENRENILRTLL 701
NEFM++WDGL S+ + +ILVLAATNRPFDLDEAIIRRFERRIMVGLPS E+RE ILR+LL
Sbjct: 655 NEFMTHWDGLLSRPDQKILVLAATNRPFDLDEAIIRRFERRIMVGLPSLESRELILRSLL 714
Query: 702 AKEKVHGGLDFKELATMTEGYSGSDLKNLCTTAAYRPVRELIQQEIQKDSKKKKDAEGQN 761
+KEKV GGLD+KELATMTEGYSGSDLKNLCTTAAYRPVRELIQ+E +K+ +KK++ G
Sbjct: 715 SKEKVDGGLDYKELATMTEGYSGSDLKNLCTTAAYRPVRELIQKERKKELEKKREQGG-- 772
Query: 762 SQDAQDAKEEVEQERVITLRPLNMQDFKMAKSQVAASFAAEGAGMNELRQWNDLYGEGGS 821
+A DA + E++ I LRPLNM+D K AK+QVAASFAAEG M EL+QWN+LYGEGGS
Sbjct: 773 --NASDASKMKEKDETIILRPLNMKDLKEAKNQVAASFAAEGTIMGELKQWNELYGEGGS 830
Query: 822 RKKEQLSYFL 831
RKK+QL+YFL
Sbjct: 831 RKKQQLTYFL 840
>pir||G96665 protein F22C12.12 [imported] - Arabidopsis thaliana
gi|6692099|gb|AAF24564.1| F22C12.12 [Arabidopsis
thaliana]
Length = 825
Score = 1030 bits (2662), Expect = 0.0
Identities = 564/865 (65%), Positives = 649/865 (74%), Gaps = 74/865 (8%)
Query: 1 MEQKGMLISA-ALSVGVGVGLGLASGQTMFK-PNTYSSSSNALTPDKIENEMLRLVVDGR 58
M+ K ML+SA + VGVGVGLGLASGQ + K SSS+NA+T DK+E E+LR VVDGR
Sbjct: 1 MDSKQMLLSALGVGVGVGVGLGLASGQAVGKWAGGNSSSNNAVTADKMEKEILRQVVDGR 60
Query: 59 ESNVTFDNFPYYLSEQTRVLLTSAAYVHLKHAEVSKYTRNLAPASRTILLSGPAELYQQV 118
ES +TFD+ EQTRVLLTSAAYVHLKH + SKYTRNL+PASR ILLSGPAELYQQ+
Sbjct: 61 ESKITFDD------EQTRVLLTSAAYVHLKHFDASKYTRNLSPASRAILLSGPAELYQQM 114
Query: 119 LAKALTHYFEAKLLLFDVTDFSLKIQSRYGSSNCETSFTRSTSETALARLSDLFGSFALF 178
LAKAL H+F+AKLLL DV DF+LKIQS+YGS N E+S
Sbjct: 115 LAKALAHFFDAKLLLLDVNDFALKIQSKYGSGNTESSVIAG------------------- 155
Query: 179 PQREENQGKIHRQSSGSDLRQMEAEGSYS--KLRRNASASANISSIGLQSNPTNSAPGKH 236
G + RQSSG D++ EGS + KLRRN+SA+ANIS++ SN AP K
Sbjct: 156 -------GTLRRQSSGVDIKSSSMEGSSNPPKLRRNSSAAANISNLASSSN---QAPLKR 205
Query: 237 ITGWPFDEKILIQTLYKV----------LLYVSKTYPIVLYMRDADKLLCRSQRIYKLFQ 286
+ W FDEK+L+Q+LYKV L YVSK PIVLY+RD + L RSQR Y LFQ
Sbjct: 206 SSSWSFDEKLLVQSLYKVMCKTIKSIKVLAYVSKANPIVLYLRDVENFLFRSQRTYNLFQ 265
Query: 287 TMLTKLSGPILIIGSRILD-SGNECKRVDEMLTSLFPYNIEIKPPEDESRLVSWKSQFEA 345
+L KLSGP+LI+GSRI+D S + + +DE L+++FPYNI+I+PPEDE+ LVSWKSQ E
Sbjct: 266 KLLQKLSGPVLILGSRIVDLSSEDAQEIDEKLSAVFPYNIDIRPPEDETHLVSWKSQLER 325
Query: 346 DMKKIQIQDNKNHIMEVLAANDLDCHDLDSICVADTMVLSNYIEEIIVSAISYHIMKNKE 405
DM IQ QDN+NHIMEVL+ NDL C DL+SI DT VLSNYIEEI+VSA+SYH+M NK+
Sbjct: 326 DMNMIQTQDNRNHIMEVLSENDLICDDLESISFEDTKVLSNYIEEIVVSALSYHLMNNKD 385
Query: 406 PEYRNGKLIIPCNSLSHALGIFQAGKFGDRDSLKLEAQAVTSEKKEEGAAVKPEGKTESP 465
PEYRNGKL+I SLSH +F+ GK G R+ LK + + +S K+ + ++KPE KTES
Sbjct: 386 PEYRNGKLVISSISLSHGFSLFREGKAGGREKLKQKTKEESS-KEVKAESIKPETKTES- 443
Query: 466 APAVKTEAEIPTSVRKTDGENSVPASKAEVPDNEFEKRIRPEVIPANEIGVTFSDIGALD 525
V T + ++ E P + PDNEFEKRIRPEVIPA EI VTF DIGALD
Sbjct: 444 ---VTTVSSKEEPEKEAKAEKVTPKAPEVAPDNEFEKRIRPEVIPAEEINVTFKDIGALD 500
Query: 526 ETKDSLQELVMLPLRRPDLFEGGLLKPCRGILLFGPPGTGKTMLAKAIANEAGASFINVS 585
E K+SLQELVMLPLRRPDLF GGLLKPCRGILLFGPPGTGKTMLAKAIA EAGASFINVS
Sbjct: 501 EIKESLQELVMLPLRRPDLFTGGLLKPCRGILLFGPPGTGKTMLAKAIAKEAGASFINVS 560
Query: 586 MSTITSKWFGEDEKNVRALFTLAAKVSPTIIFVDEVDSMLGQRTRVGEHEAMRKIKNEFM 645
MSTITSKWFGEDEKNVRALFTLA+KVSPTIIFVDEVDSMLGQRTRVGEHEAMRKIKNEFM
Sbjct: 561 MSTITSKWFGEDEKNVRALFTLASKVSPTIIFVDEVDSMLGQRTRVGEHEAMRKIKNEFM 620
Query: 646 SNWDGLTSKSEDRILVLAATNRPFDLDEAIIRRFERRIMVGLPSAENRENILRTLLAKEK 705
S+WDGL +K +RILVLAATNRPFDLDEAIIRRFERRIMVGLP+ ENRE ILRTLLAKEK
Sbjct: 621 SHWDGLMTKPGERILVLAATNRPFDLDEAIIRRFERRIMVGLPAVENREKILRTLLAKEK 680
Query: 706 VHGGLDFKELATMTEGYSGSDLKNLCTTAAYRPVRELIQQEIQKDSKKKK---------- 755
V LD+KELA MTEGY+GSDLKNLCTTAAYRPVRELIQQE KD+ +
Sbjct: 681 VDENLDYKELAMMTEGYTGSDLKNLCTTAAYRPVRELIQQERIKDTVRNNISLRLFLYTS 740
Query: 756 -------DAEGQNSQDAQDAKEEVE--QERVITLRPLNMQDFKMAKSQVAASFAAEGAGM 806
D E + ++ A EE E +ERVITLRPLN QDFK AK+QVAASFAAEGAGM
Sbjct: 741 IFILVLTDCEKKKQREPTKAGEEDEGKEERVITLRPLNRQDFKEAKNQVAASFAAEGAGM 800
Query: 807 NELRQWNDLYGEGGSRKKEQLSYFL 831
EL+QWN+LYGEGGSRKKEQL+YFL
Sbjct: 801 GELKQWNELYGEGGSRKKEQLTYFL 825
>ref|NP_913449.1| P0492F05.26 [Oryza sativa (japonica cultivar-group)]
Length = 810
Score = 1008 bits (2607), Expect = 0.0
Identities = 536/840 (63%), Positives = 640/840 (75%), Gaps = 49/840 (5%)
Query: 10 AALSVGVGVGLGLASGQTMFKPNTYSSSSNALTPDKIENEMLRLVVDGRESNVTFDNFPY 69
+A+ +G+GVG+GL + + +T +++E E+ RLVVDG +S VTFD FPY
Sbjct: 2 SAVGIGIGVGVGLGLASAPWAGGGGQGARVGVTVERVEQELRRLVVDGADSRVTFDGFPY 61
Query: 70 YLSEQTRVLLTSAAYVHLKHAEVSKYTRNLAPASRTILLSGPAELYQQVLAKALTHYFEA 129
YLSEQTRVLLTSAAYVHLK A++S+YTRNLAPASR ILLSGPAELYQQ+LAKAL HYFEA
Sbjct: 62 YLSEQTRVLLTSAAYVHLKQADISQYTRNLAPASRAILLSGPAELYQQMLAKALAHYFEA 121
Query: 130 KLLLFDVTDFSLKIQSRYGS-SNCETSFTRSTSETALARLSDLFGSFALFPQREENQGKI 188
KLLL D TDF +KI S+YG S+ ++SF RS SET L ++S L GS ++ PQ+E+ +G I
Sbjct: 122 KLLLLDPTDFLIKIHSKYGGGSSTDSSFKRSISETTLEKVSGLLGSLSILPQKEKPKGTI 181
Query: 189 HRQSSGSD--LRQMEAEGSYSKLRRNASASANISSIGLQSNPTNSAPGKHITGWPFDEKI 246
RQSS +D LR E+ S+ KL+RNAS S+++SS+ Q P N A + + W FDEKI
Sbjct: 182 RRQSSMTDMKLRSSESTSSFPKLKRNASTSSDMSSLASQGPPNNPASLRRASSWTFDEKI 241
Query: 247 LIQTLYKVLLYVSKTYPIVLYMRDADKLLCRSQRIYKLFQTMLTKLSGPILIIGSRILDS 306
L+Q +YKVL VSK PIVLY+RD +K L +S+++Y +F+ +L KL GP+L++GSRI+D
Sbjct: 242 LVQAVYKVLHSVSKKNPIVLYIRDVEKFLHKSKKMYVMFEKLLNKLEGPVLVLGSRIVDM 301
Query: 307 GNECKRVDEMLTSLFPYNIEIKPPEDESRLVSWKSQFEADMKKIQIQDNKNHIMEVLAAN 366
+ + +DE LT+LFPYNIEIKPPE+E+ LVSW SQ E DMK IQ QDN+NHI EVLA N
Sbjct: 302 DFD-EELDERLTALFPYNIEIKPPENENHLVSWNSQLEEDMKMIQFQDNRNHITEVLAEN 360
Query: 367 DLDCHDLDSICVADTMVLSNYIEEIIVSAISYHIMKNKEPEYRNGKLIIPCNSLSHALGI 426
DL+C DL SIC++DTMVL YIEEI+VSA+SYH+M K+PEYRNGKL++ SLSHAL I
Sbjct: 361 DLECDDLGSICLSDTMVLGRYIEEIVVSAVSYHLMNKKDPEYRNGKLLLSAKSLSHALEI 420
Query: 427 FQAGKFGDRDSLKLEAQAVTSEKKEEGAA-------------VKPEGKTESPAPAVKTEA 473
FQ K D+DS+KLEA+ S+ + G A + P T + AP V+++A
Sbjct: 421 FQENKMYDKDSMKLEAKRDASKVADRGIAPFAAKSETKPATLLPPVPPTAAAAPPVESKA 480
Query: 474 EIPTSVRKTDGENSVPASKAEV--PDNEFEKRIRPEVIPANEIGVTFSDIGALDETKDSL 531
E P K D N PA+KA PDNEFEKRIRPEVIPANEIGVTF DIGAL + K+SL
Sbjct: 481 E-PEKFEKKD--NPSPAAKAPEMPPDNEFEKRIRPEVIPANEIGVTFDDIGALSDIKESL 537
Query: 532 QELVMLPLRRPDLFEGGLLKPCRGILLFGPPGTGKTMLAKAIANEAGASFINVSMSTITS 591
QELVMLPLRRPDLF+GGLLKPCRGILLFGPPGTGKTMLAKAIANEA ASFINVSMSTITS
Sbjct: 538 QELVMLPLRRPDLFKGGLLKPCRGILLFGPPGTGKTMLAKAIANEAQASFINVSMSTITS 597
Query: 592 KWFGEDEKNVRALFTLAAKVSPTIIFVDEVDSMLGQRTRVGEHEAMRKIKNEFMSNWDGL 651
KWFGEDEKNVRALFTLAAKVSPTIIFVDEVDSMLGQR R GEHEAMRKIKNEFM++WDGL
Sbjct: 598 KWFGEDEKNVRALFTLAAKVSPTIIFVDEVDSMLGQRNRAGEHEAMRKIKNEFMTHWDGL 657
Query: 652 TSKSEDRILVLAATNRPFDLDEAIIRRFERRIMVGLPSAENRENILRTLLAKEKVHGGLD 711
S+ + +ILVLAATNRPFDLDEAIIRRFERRIMVGLPS E+RE ILR+LL+KEKV GGLD
Sbjct: 658 LSRPDQKILVLAATNRPFDLDEAIIRRFERRIMVGLPSLESRELILRSLLSKEKVDGGLD 717
Query: 712 FKELATMTEGYSGSDLKNLCTTAAYRPVRELIQQEIQKDSKKKKDAEGQNSQDAQDAKEE 771
+KELATMTEGYSGSDL K+KK +G N+ DA K
Sbjct: 718 YKELATMTEGYSGSDL------------------------KEKKREQGGNASDASKMK-- 751
Query: 772 VEQERVITLRPLNMQDFKMAKSQVAASFAAEGAGMNELRQWNDLYGEGGSRKKEQLSYFL 831
E++ I LRPLNM+D K AK+QVAASFAAEG M EL+QWN+LYGEGGSRKK+QL+YFL
Sbjct: 752 -EKDETIILRPLNMKDLKEAKNQVAASFAAEGTIMGELKQWNELYGEGGSRKKQQLTYFL 810
>gb|AAL32670.1| similar to homeobox protein [Arabidopsis thaliana]
Length = 752
Score = 1005 bits (2598), Expect = 0.0
Identities = 533/758 (70%), Positives = 612/758 (80%), Gaps = 14/758 (1%)
Query: 1 MEQKGMLISA-ALSVGVGVGLGLASGQTMFK-PNTYSSSSNALTPDKIENEMLRLVVDGR 58
M+ K ML+SA + VGVGVGLGLASGQ + K SSS+NA+T DK+E E+LR VVDGR
Sbjct: 1 MDSKQMLLSALGVGVGVGVGLGLASGQAVGKWAGGNSSSNNAVTADKMEKEILRQVVDGR 60
Query: 59 ESNVTFDNFPYYLSEQTRVLLTSAAYVHLKHAEVSKYTRNLAPASRTILLSGPAELYQQV 118
ES +TFD FPYYLSEQTRVLLTSAAYVHLKH + SKYTRNL+PASR ILLSGPAELYQQ+
Sbjct: 61 ESKITFDEFPYYLSEQTRVLLTSAAYVHLKHFDASKYTRNLSPASRAILLSGPAELYQQM 120
Query: 119 LAKALTHYFEAKLLLFDVTDFSLKIQSRYGSSNCETS-FTRSTSETALARLSDLFGSFAL 177
LAKAL H+F+AKLLL DV DF LKIQS+YGS N E+S F RS SE+AL +LS LF SF++
Sbjct: 121 LAKALAHFFDAKLLLLDVNDFGLKIQSKYGSGNTESSSFKRSPSESALEQLSGLFSSFSI 180
Query: 178 FPQREENQ--GKIHRQSSGSDLRQMEAEGSYS--KLRRNASASANISSIGLQSNPTNSAP 233
PQREE++ G + RQSSG D++ EGS + KLRRN+SA+ANIS++ SN SAP
Sbjct: 181 LPQREESKAGGTLRRQSSGVDIKSSSMEGSSNPPKLRRNSSAAANISNLASSSNQV-SAP 239
Query: 234 GKHITGWPFDEKILIQTLYKVLLYVSKTYPIVLYMRDADKLLCRSQRIYKLFQTMLTKLS 293
K + W FDEK+L+Q+LYKVL YVSK PIVLY+RD + L RSQR Y LFQ +L KLS
Sbjct: 240 LKRSSSWSFDEKLLVQSLYKVLAYVSKANPIVLYLRDVENFLFRSQRTYNLFQKLLQKLS 299
Query: 294 GPILIIGSRILD-SGNECKRVDEMLTSLFPYNIEIKPPEDESRLVSWKSQFEADMKKIQI 352
GP+LI+GSRI+D S + + +DE L+++FPYNI+I+PPEDE+ LVSWKSQ E DM IQ
Sbjct: 300 GPVLILGSRIVDLSSEDAQEIDEKLSAVFPYNIDIRPPEDETHLVSWKSQLERDMNMIQT 359
Query: 353 QDNKNHIMEVLAANDLDCHDLDSICVADTMVLSNYIEEIIVSAISYHIMKNKEPEYRNGK 412
QDN+NHIMEVL+ NDL C DL+SI DT VLSNYIEEI+VSA+SYH+M NK+PEYRNGK
Sbjct: 360 QDNRNHIMEVLSENDLICDDLESISFEDTKVLSNYIEEIVVSALSYHLMNNKDPEYRNGK 419
Query: 413 LIIPCNSLSHALGIFQAGKFGDRDSLKLEAQAVTSEKKEEGAAVKPEGKTESPAPAVKTE 472
L+I SLSH +F+ GK G R+ LK + + +S K+ + ++KPE KTES V T
Sbjct: 420 LVISSISLSHGFSLFREGKAGGREKLKQKTKEESS-KEVKAESIKPETKTES----VTTV 474
Query: 473 AEIPTSVRKTDGENSVPASKAEVPDNEFEKRIRPEVIPANEIGVTFSDIGALDETKDSLQ 532
+ ++ E P + PDNEFEKRIRPEVIPA EI VTF DIGALDE K+SLQ
Sbjct: 475 SSKEEPEKEAKAEKVTPKAPEVAPDNEFEKRIRPEVIPAEEINVTFKDIGALDEIKESLQ 534
Query: 533 ELVMLPLRRPDLFEGGLLKPCRGILLFGPPGTGKTMLAKAIANEAGASFINVSMSTITSK 592
ELVMLPLRRPDLF GGLLKPCRGILLFGPPGTGKTMLAKAIA EAGASFINVSMSTITSK
Sbjct: 535 ELVMLPLRRPDLFTGGLLKPCRGILLFGPPGTGKTMLAKAIAKEAGASFINVSMSTITSK 594
Query: 593 WFGEDEKNVRALFTLAAKVSPTIIFVDEVDSMLGQRTRVGEHEAMRKIKNEFMSNWDGLT 652
WFGEDEKNVRALFTLA+KVSPTIIFVDEVDSMLGQRTRVGEHEAMRKIKNEFMS+WDGL
Sbjct: 595 WFGEDEKNVRALFTLASKVSPTIIFVDEVDSMLGQRTRVGEHEAMRKIKNEFMSHWDGLM 654
Query: 653 SKSEDRILVLAATNRPFDLDEAIIRRFERRIMVGLPSAENRENILRTLLAKEKVHGGLDF 712
+K +RILVLAATNRPFDLDEAIIRRFERRIMVGLP+ ENRE ILRTLLAKEKV LD+
Sbjct: 655 TKPGERILVLAATNRPFDLDEAIIRRFERRIMVGLPAVENREKILRTLLAKEKVDENLDY 714
Query: 713 KELATMTEGYSGSDLKNLCTTAAYRPVRELIQQEIQKD 750
KELA MTEGY+GSDLKNLCTTAAYRPVRELIQQE KD
Sbjct: 715 KELAMMTEGYTGSDLKNLCTTAAYRPVRELIQQERIKD 752
>ref|XP_475967.1| unknown protein [Oryza sativa (japonica cultivar-group)]
gi|48843801|gb|AAT47060.1| unknown protein [Oryza sativa
(japonica cultivar-group)]
Length = 855
Score = 1003 bits (2593), Expect = 0.0
Identities = 534/875 (61%), Positives = 658/875 (75%), Gaps = 64/875 (7%)
Query: 1 MEQKGMLISAALSVGVGVGLGLASGQTMFKPNTYSSSSNALTPDKIENEMLRLVVDGRES 60
MEQ+ + +SA LSVGVGVGLGLAS + + + ++E E+ RLVVDGRE
Sbjct: 1 MEQRNLFVSA-LSVGVGVGLGLASARWAAPGSGEGGGGAGIGVAELEAELRRLVVDGREG 59
Query: 61 NVTFDNFPYYLSEQTRVLLTSAAYVHLKHAEVSKYTRNLAPASRTILLSGPAELYQQVLA 120
+VTFD F YYLSE+T+ +L SAA+VHLK A++SK+ RNL ASR ILLSGP E Y Q LA
Sbjct: 60 DVTFDEFRYYLSERTKEVLISAAFVHLKQADLSKHIRNLCAASRAILLSGPTEPYLQSLA 119
Query: 121 KALTHYFEAKLLLFDVTDFSLKIQSRYGSSNCETSFTRSTSETALARLSDLFGSFALFPQ 180
+AL+HY++A+LL+ DVTDFSL+IQS+YGSS+ + ++S SET R+SDL GSF +FP+
Sbjct: 120 RALSHYYKAQLLILDVTDFSLRIQSKYGSSSKGLAQSQSISETTFGRMSDLIGSFTIFPK 179
Query: 181 REENQGKIHRQSSGSDLRQMEAEGSYSK--LRRNASASANISSIGLQSNPTNSAPGKHIT 238
E + + RQ+S +D+R +E S + LR+NAS S++IS + Q + +S + +
Sbjct: 180 SAEPRESLQRQTSSADVRSRGSEASSNAPPLRKNASMSSDISDVSSQCS-AHSVSARRTS 238
Query: 239 GWPFDEKILIQTLYKVLLYVSKTYPIVLYMRDADKLLCRSQRIYKLFQTMLTKLSGPILI 298
W FDEK+LIQ+LYKV++ V++ P++LY+RD D+LL RSQR Y LFQ ML KL+G +LI
Sbjct: 239 SWCFDEKVLIQSLYKVMVSVAENNPVILYIRDVDQLLHRSQRTYSLFQKMLAKLTGQVLI 298
Query: 299 IGSRILDSGNECKRVDEMLTSLFPYNIEIKPPEDESRLVSWKSQFEADMKKIQIQDNKNH 358
+GSR+LDS ++ VDE ++SLFP++++IKPPE+E+ L SWK+Q E D KKIQIQDN+NH
Sbjct: 299 LGSRLLDSDSDHTDVDERVSSLFPFHVDIKPPEEETHLDSWKTQMEEDTKKIQIQDNRNH 358
Query: 359 IMEVLAANDLDCHDLDSICVADTMVLSNYIEEIIVSAISYHIMKNKEPEYRNGKLIIPCN 418
I+EVL+ANDLDC DL SIC ADTMVLSNYIEEIIVSA+SYH++ NK+PEY+NGKL++
Sbjct: 359 IIEVLSANDLDCDDLSSICQADTMVLSNYIEEIIVSAVSYHMIHNKDPEYKNGKLVLSSK 418
Query: 419 SLSHALGIFQAGKFGDRDSLKLEAQAVTSEKKEEGAAVKPEGKTESPAPAVKTEAEIPTS 478
SLSH L IFQ FG +++LKLE +GA + +TE A +
Sbjct: 419 SLSHGLSIFQESGFGGKETLKLEDDL-------KGATGPKKSETEKSA-----------T 460
Query: 479 VRKTDGENSVPASKAEVPDNEFEKRIRPEVIPANEIGVTFSDIGALDETKDSLQELVMLP 538
V DG+ +P K E+PDNEFEKRIRPEVIPA+EIGVTF DIGAL + K+SLQELVMLP
Sbjct: 461 VPLKDGDGPLPPPKPEIPDNEFEKRIRPEVIPASEIGVTFDDIGALADIKESLQELVMLP 520
Query: 539 LRRPDLFEGGLLKPCRGILLFGPPGTGKTMLAKAIANEAGASFINVSMSTITSKWFGEDE 598
LRRPDLF+GGLLKPCRGILLFGPPGTGKTMLAKAIAN+AGASFINVSMSTITSKWFGEDE
Sbjct: 521 LRRPDLFKGGLLKPCRGILLFGPPGTGKTMLAKAIANDAGASFINVSMSTITSKWFGEDE 580
Query: 599 KNVRALFTLAAKVSPTIIFVDEVDSMLGQRTRVGEHEAMRKIKNEFMSNWDGLTSKSEDR 658
KNVRALF+LAAKV+PTIIFVDEVDSMLGQR R GEHEAMRKIKNEFMS+WDGL SKS +R
Sbjct: 581 KNVRALFSLAAKVAPTIIFVDEVDSMLGQRARCGEHEAMRKIKNEFMSHWDGLLSKSGER 640
Query: 659 ILVLAATNRPFDLDEAIIRRFERRIMVGLPSAENRENILRTLLAKEKVHGGLDFKELATM 718
ILVLAATNRPFDLDEAIIRRFERRIMVGLP+ ++RE ILRTLL+KEKV +D+KELATM
Sbjct: 641 ILVLAATNRPFDLDEAIIRRFERRIMVGLPTLDSRELILRTLLSKEKVAEDIDYKELATM 700
Query: 719 TEGYSGSDLKNLCTTAAYRPVRELIQQEIQKDSK------KKKDAEGQNSQDAQDAKE-- 770
TEGYSGSDLKNLC TAAYRPVREL+++E +K+ + K+K A +NS+ + KE
Sbjct: 701 TEGYSGSDLKNLCVTAAYRPVRELLKREREKEMERRANEAKEKAATAENSESPESKKEKE 760
Query: 771 --------EVEQER--------------------------VITLRPLNMQDFKMAKSQVA 796
E E+ER I LRPL M+D + AK+QVA
Sbjct: 761 NSENPESKEKEKERKENSENKEEKTENKQDNSKAEGGTEGTIDLRPLTMEDLRQAKNQVA 820
Query: 797 ASFAAEGAGMNELRQWNDLYGEGGSRKKEQLSYFL 831
ASFA EGA MNEL+QWNDLYGEGGSRKK+QL+YFL
Sbjct: 821 ASFATEGAVMNELKQWNDLYGEGGSRKKQQLTYFL 855
>emb|CAB79602.1| putative protein [Arabidopsis thaliana] gi|4455359|emb|CAB36769.1|
putative protein [Arabidopsis thaliana]
gi|15234347|ref|NP_194529.1| AAA-type ATPase family
protein [Arabidopsis thaliana] gi|7487959|pir||T02901
MSP1 protein homolog T13J8.110 - Arabidopsis thaliana
Length = 726
Score = 933 bits (2412), Expect = 0.0
Identities = 511/833 (61%), Positives = 612/833 (73%), Gaps = 109/833 (13%)
Query: 1 MEQKGMLISAALSVGVGVGLGLASGQTMFK-PNTYSSSSNALTPDKIENEMLRLVVDGRE 59
MEQK +L SA L VGVG+G+GLASGQ++ K N S+ + LT +KIE E++R +VDGRE
Sbjct: 1 MEQKSVLFSA-LGVGVGLGIGLASGQSLGKWANGSISAEDGLTGEKIEQELVRQIVDGRE 59
Query: 60 SNVTFDNFPYYLSEQTRVLLTSAAYVHLKHAEVSKYTRNLAPASRTILLSGPAELYQQVL 119
S+VTFD FPYYLSE+TR+LLTSAAYVHLK +++SK+TRNLAP S+ ILLSGPA+ Q +
Sbjct: 60 SSVTFDEFPYYLSEKTRLLLTSAAYVHLKQSDISKHTRNLAPGSKAILLSGPADTEQVWM 119
Query: 120 AKALTHYFEAKLLLFDVTDFSLKIQSRYGSSNCETSFTRSTSETALARLSDLFGSFALFP 179
+ T + R TS L S F + P
Sbjct: 120 RQERT-----------------------------WTLRRHTSGNDLH--SRGFDVTSQPP 148
Query: 180 QREENQGKIHRQSSGSDLRQMEAEGSYSKLRRNASASANISSIGLQSNPTNSAPGKHITG 239
+ + N S+ SD+ + + S + + ++ SAN+
Sbjct: 149 RLKRN------ASAASDMSSISSR-SATSVSASSKRSANLC------------------- 182
Query: 240 WPFDEKILIQTLYKVLLYVSKTYPIVLYMRDADKLLCRSQRIYKLFQTMLTKLSGPILII 299
FDE++ +Q+LYKVL+ +S+T PI++Y+RD +K LC+S+R YKLFQ +LTKLSGP+L++
Sbjct: 183 --FDERLFLQSLYKVLVSISETNPIIIYLRDVEK-LCQSERFYKLFQRLLTKLSGPVLVL 239
Query: 300 GSRILDSGNECKRVDEMLTSLFPYNIEIKPPEDESRLVSWKSQFEADMKKIQIQDNKNHI 359
GSR+L+ ++C+ V E +++LFPYNIEI+PPEDE++L+SWK++FE DMK IQ QDNKNHI
Sbjct: 240 GSRLLEPEDDCQEVGEGISALFPYNIEIRPPEDENQLMSWKTRFEDDMKVIQFQDNKNHI 299
Query: 360 MEVLAANDLDCHDLDSICVADTMVLSNYIEEIIVSAISYHIMKNKEPEYRNGKLIIPCNS 419
EVLAANDL+C DL SIC ADTM LS++IEEI+VSAISYH+M NKEPEY+NG+L+I NS
Sbjct: 300 AEVLAANDLECDDLGSICHADTMFLSSHIEEIVVSAISYHLMNNKEPEYKNGRLVISSNS 359
Query: 420 LSHALGIFQAGKFGDRDSLKLEAQAVTSEKKEEGAAVKPEGKTESPAPAVKTEAEIPTSV 479
LSH L I Q G+ DSLKL+
Sbjct: 360 LSHGLNILQEGQGCFEDSLKLDTNI----------------------------------- 384
Query: 480 RKTDGENSVPASKAEVPDNEFEKRIRPEVIPANEIGVTFSDIGALDETKDSLQELVMLPL 539
SK PDNEFEKRIRPEVIPANEIGVTF+DIG+LDETK+SLQELVMLPL
Sbjct: 385 ----------DSKEVAPDNEFEKRIRPEVIPANEIGVTFADIGSLDETKESLQELVMLPL 434
Query: 540 RRPDLFEGGLLKPCRGILLFGPPGTGKTMLAKAIANEAGASFINVSMSTITSKWFGEDEK 599
RRPDLF+GGLLKPCRGILLFGPPGTGKTM+AKAIANEAGASFINVSMSTITSKWFGEDEK
Sbjct: 435 RRPDLFKGGLLKPCRGILLFGPPGTGKTMMAKAIANEAGASFINVSMSTITSKWFGEDEK 494
Query: 600 NVRALFTLAAKVSPTIIFVDEVDSMLGQRTRVGEHEAMRKIKNEFMSNWDGLTSKSEDRI 659
NVRALFTLAAKVSPTIIFVDEVDSMLGQRTRVGEHEAMRKIKNEFM++WDGL S + DRI
Sbjct: 495 NVRALFTLAAKVSPTIIFVDEVDSMLGQRTRVGEHEAMRKIKNEFMTHWDGLMSNAGDRI 554
Query: 660 LVLAATNRPFDLDEAIIRRFERRIMVGLPSAENRENILRTLLAKEKVHGGLDFKELATMT 719
LVLAATNRPFDLDEAIIRRFERRIMVGLPS E+RE ILRTLL+KEK LDF+ELA MT
Sbjct: 555 LVLAATNRPFDLDEAIIRRFERRIMVGLPSVESREKILRTLLSKEKTE-NLDFQELAQMT 613
Query: 720 EGYSGSDLKNLCTTAAYRPVRELIQQEIQKDSKKKKDAEGQ-NSQDAQDAKEEVEQERVI 778
+GYSGSDLKN CTTAAYRPVRELI+QE KD +++K E + NS++ +AKEEV +ER I
Sbjct: 614 DGYSGSDLKNFCTTAAYRPVRELIKQECLKDQERRKREEAEKNSEEGSEAKEEVSEERGI 673
Query: 779 TLRPLNMQDFKMAKSQVAASFAAEGAGMNELRQWNDLYGEGGSRKKEQLSYFL 831
TLRPL+M+D K+AKSQVAASFAAEGAGMNEL+QWNDLYGEGGSRKKEQLSYFL
Sbjct: 674 TLRPLSMEDMKVAKSQVAASFAAEGAGMNELKQWNDLYGEGGSRKKEQLSYFL 726
>ref|NP_917758.1| P0501G01.20 [Oryza sativa (japonica cultivar-group)]
gi|12313686|dbj|BAB21091.1| cell division cycle gene
CDC48-like [Oryza sativa (japonica cultivar-group)]
Length = 812
Score = 743 bits (1918), Expect = 0.0
Identities = 412/868 (47%), Positives = 558/868 (63%), Gaps = 96/868 (11%)
Query: 3 QKGMLISAALSVGVGVGLGLASGQTMFKPNTYSSSSNALTPDKIENEMLRLVVDGRESNV 62
+ G +I++A+ VGVGVG+G+ + T ++ T ++E E+ LVVDGR+ V
Sbjct: 2 EHGSIIASAVGVGVGVGVGIGLVSSRL---TGLATGGGATAAEVEAELRCLVVDGRDVGV 58
Query: 63 TFDNFPYYLSEQTRVLLTSAAYVHLKHAEVSKYTRNLAPASRTILLSGPAELYQQVLAKA 122
+FD+FPYYLSEQ+++ LTS A+VHL + + R L+ +SRTILL GP+E Y Q LAKA
Sbjct: 59 SFDDFPYYLSEQSKLALTSTAFVHLSPTILPNHIRVLSASSRTILLCGPSEAYLQSLAKA 118
Query: 123 LTHYFEAKLLLFDVTDFSLKIQSRYGSSNCETSFTRSTSETALARLSDLFGSFALFPQRE 182
L + F A+LLL DV DF+ K+ +YG + + RS +E A R+S L G+F LF ++E
Sbjct: 119 LANQFSARLLLLDVIDFACKLHHKYGGPSNTQTRERSMTEAAFDRVSSLVGAFNLFRKKE 178
Query: 183 E--NQGKIHRQSSGSDLRQMEAEGSYSKLRRNASASANISSIGLQSNPTNSAPGKHITG- 239
E G + R++ DLR S S +S + + + + K++
Sbjct: 179 EPTGTGPLSRETGILDLRT-----STCCPHNTPSVRVQLSLVPPEKDHDPES-SKYLASV 232
Query: 240 ---WPFDEKILIQTLYKVLLYVSKTYPIVLYMRDADKLLCRSQRIYKLFQTMLTKLSGPI 296
W +EK+LIQ+LYK+++ S+ P++LY+RD D LL S++ Y +FQ ML KLSG +
Sbjct: 233 KPCWSLNEKVLIQSLYKIIVSASEISPVILYIRDVDDLLGSSEKAYCMFQKMLKKLSGRV 292
Query: 297 LIIGSRILDSGNECKRVDEMLTSLFPYNIEIKPPEDESRLVSWKSQFEADMKKIQIQDNK 356
++IGS+ LD + + ++E + +LFP +E KPP+D+ L WK+Q E D Q +
Sbjct: 293 IVIGSQFLDDDEDREDIEESVCALFPCILETKPPKDKVLLEKWKTQMEEDSNNNNNQVVQ 352
Query: 357 NHIMEVLAANDLDCHDLDSICVADTM-VLSNYIEEIIVSAISYHIMKNKEPEYRNGKLII 415
N+I EVLA N+L+C DL SI D ++ Y+EEII ++SYH+M NK P+YRNG L+I
Sbjct: 353 NYIAEVLAENNLECEDLSSINADDDCKIIVAYLEEIITPSVSYHLMNNKNPKYRNGNLVI 412
Query: 416 PCNSLSHALGIFQAGKFGDRDSLKLEAQAVTSEKKEEGAAVKPEGKTESPAPAVKTEAEI 475
SLSH L IFQ +D T E K+E V
Sbjct: 413 SSESLSHGLRIFQESNDLGKD---------TVEAKDETEMV------------------- 444
Query: 476 PTSVRKTDGENSVPASKAEVPDNEFEKRIRPEVIPANEIGVTFSDIGALDETKDSLQELV 535
VPDNE+EK+IRP VIPANEIGVTF DIGAL + K+ L ELV
Sbjct: 445 -------------------VPDNEYEKKIRPTVIPANEIGVTFDDIGALADIKECLHELV 485
Query: 536 MLPLRRPDLFEGGLLKPCRGILLFGPPGTGKTMLAKAIANEAGASFINVSMSTITSKWFG 595
MLPL+RPD F+GGLLKPC+G+LLFGPPGTGKTMLAKA+AN AGASF+N+SM+++TSKW+G
Sbjct: 486 MLPLQRPDFFKGGLLKPCKGVLLFGPPGTGKTMLAKALANAAGASFLNISMASMTSKWYG 545
Query: 596 EDEKNVRALFTLAAKVSPTIIFVDEVDSMLGQRTRVGEHEAMRKIKNEFMSNWDGLTSKS 655
E EK ++ALF+LAAK++P IIF+DEVDSMLG+R E+EA R++KNEFM++WDGL SKS
Sbjct: 546 ESEKCIQALFSLAAKLAPAIIFIDEVDSMLGKRDNHSENEASRRVKNEFMAHWDGLLSKS 605
Query: 656 EDRILVLAATNRPFDLDEAIIRRFERRIMVGLPSAENRENILRTLLAKEKVHGGLDFKEL 715
+RILVLAATNRPFDLD+A+IRRFE RIMVGLP+ E+RE IL+TLL+KE V +DFKEL
Sbjct: 606 NERILVLAATNRPFDLDDAVIRRFEHRIMVGLPTLESRELILKTLLSKETVE-NIDFKEL 664
Query: 716 ATMTEGYSGSDLKNLCTTAAYRPVRELIQQEIQK-------------DSKKKKDAEGQNS 762
A MTEGY+ SDLKN+C TAAY PVREL+Q+E K K K G S
Sbjct: 665 AKMTEGYTSSDLKNICVTAAYHPVRELLQKEKNKVKKETAPETKQEPKEKTKIQENGTKS 724
Query: 763 QDAQDAKEEVEQER-------------------VITLRPLNMQDFKMAKSQVAASFAAEG 803
D++ K++++ + TLRPLNM+D + AK +VAASFA+EG
Sbjct: 725 SDSKTEKDKLDNKEGKKDKPADKKDKSDKGDAGETTLRPLNMEDLRKAKDEVAASFASEG 784
Query: 804 AGMNELRQWNDLYGEGGSRKKEQLSYFL 831
MN++++WN+LYG+GGSRK+EQL+YFL
Sbjct: 785 VVMNQIKEWNELYGKGGSRKREQLTYFL 812
>gb|AAN46222.1| unknown protein [Arabidopsis lyrata] gi|28188591|gb|AAN46221.1|
unknown protein [Arabidopsis lyrata]
Length = 316
Score = 521 bits (1341), Expect = e-146
Identities = 267/317 (84%), Positives = 290/317 (91%), Gaps = 2/317 (0%)
Query: 491 SKAEVPDNEFEKRIRPEVIPANEIGVTFSDIGALDETKDSLQELVMLPLRRPDLFEGGLL 550
SK PDNEFEKRIRPEVIPANEIGVTF+DIG+LDETK+SLQELVMLPLRRPDLF+GGLL
Sbjct: 1 SKEVAPDNEFEKRIRPEVIPANEIGVTFADIGSLDETKESLQELVMLPLRRPDLFKGGLL 60
Query: 551 KPCRGILLFGPPGTGKTMLAKAIANEAGASFINVSMSTITSKWFGEDEKNVRALFTLAAK 610
KPCRGILLFGPPGTGKTM+AKAIANEAGASFINVSMSTITSKWFGEDEKNVRALFTLAAK
Sbjct: 61 KPCRGILLFGPPGTGKTMMAKAIANEAGASFINVSMSTITSKWFGEDEKNVRALFTLAAK 120
Query: 611 VSPTIIFVDEVDSMLGQRTRVGEHEAMRKIKNEFMSNWDGLTSKSEDRILVLAATNRPFD 670
VSPTIIFVDEVDSMLGQRTRVGEHEAMRKIKNEFM++WDGL S + DRILVLAATNRPFD
Sbjct: 121 VSPTIIFVDEVDSMLGQRTRVGEHEAMRKIKNEFMTHWDGLMSNAGDRILVLAATNRPFD 180
Query: 671 LDEAIIRRFERRIMVGLPSAENRENILRTLLAKEKVHGGLDFKELATMTEGYSGSDLKNL 730
LDEAIIRRFERRIMVGLPS E+RE ILRTLL+KEK LDF ELA MT+GYSGSDLKN
Sbjct: 181 LDEAIIRRFERRIMVGLPSVESREKILRTLLSKEKTE-NLDFHELAQMTDGYSGSDLKNF 239
Query: 731 CTTAAYRPVRELIQQEIQKDSKKKKDAEGQ-NSQDAQDAKEEVEQERVITLRPLNMQDFK 789
CTTAAYRPVRELI+QE KD ++KK E + +S++ + KEEV +ERVITLRPL+M+D K
Sbjct: 240 CTTAAYRPVRELIKQECLKDQERKKKEEAEKSSEEGSETKEEVSEERVITLRPLSMEDMK 299
Query: 790 MAKSQVAASFAAEGAGM 806
+AKSQVAASFAAEGAGM
Sbjct: 300 VAKSQVAASFAAEGAGM 316
>gb|AAN46220.1| unknown protein [Arabidopsis thaliana] gi|28188587|gb|AAN46219.1|
unknown protein [Arabidopsis thaliana]
gi|28188585|gb|AAN46218.1| unknown protein [Arabidopsis
thaliana] gi|28188583|gb|AAN46217.1| unknown protein
[Arabidopsis thaliana] gi|28188581|gb|AAN46216.1|
unknown protein [Arabidopsis thaliana]
gi|28188579|gb|AAN46215.1| unknown protein [Arabidopsis
thaliana] gi|28188577|gb|AAN46214.1| unknown protein
[Arabidopsis thaliana] gi|28188575|gb|AAN46213.1|
unknown protein [Arabidopsis thaliana]
gi|28188573|gb|AAN46212.1| unknown protein [Arabidopsis
thaliana]
Length = 316
Score = 521 bits (1341), Expect = e-146
Identities = 267/317 (84%), Positives = 291/317 (91%), Gaps = 2/317 (0%)
Query: 491 SKAEVPDNEFEKRIRPEVIPANEIGVTFSDIGALDETKDSLQELVMLPLRRPDLFEGGLL 550
SK PDNEFEKRIRPEVIPANEIGVTF+DIG+LDETK+SLQELVMLPLRRPDLF+GGLL
Sbjct: 1 SKEVAPDNEFEKRIRPEVIPANEIGVTFADIGSLDETKESLQELVMLPLRRPDLFKGGLL 60
Query: 551 KPCRGILLFGPPGTGKTMLAKAIANEAGASFINVSMSTITSKWFGEDEKNVRALFTLAAK 610
KPCRGILLFGPPGTGKTM+AKAIANEAGASFINVSMSTITSKWFGEDEKNVRALFTLAAK
Sbjct: 61 KPCRGILLFGPPGTGKTMMAKAIANEAGASFINVSMSTITSKWFGEDEKNVRALFTLAAK 120
Query: 611 VSPTIIFVDEVDSMLGQRTRVGEHEAMRKIKNEFMSNWDGLTSKSEDRILVLAATNRPFD 670
VSPTIIFVDEVDSMLGQRTRVGEHEAMRKIKNEFM++WDGL S + DRILVLAATNRPFD
Sbjct: 121 VSPTIIFVDEVDSMLGQRTRVGEHEAMRKIKNEFMTHWDGLMSNAGDRILVLAATNRPFD 180
Query: 671 LDEAIIRRFERRIMVGLPSAENRENILRTLLAKEKVHGGLDFKELATMTEGYSGSDLKNL 730
LDEAIIRRFERRIMVGLPS E+RE ILRTLL+KEK LDF+ELA MT+GYSGSDLKN
Sbjct: 181 LDEAIIRRFERRIMVGLPSVESREKILRTLLSKEKTE-NLDFQELAQMTDGYSGSDLKNF 239
Query: 731 CTTAAYRPVRELIQQEIQKDSKKKKDAEGQ-NSQDAQDAKEEVEQERVITLRPLNMQDFK 789
CTTAAYRPVRELI+QE KD +++K E + NS++ +AKEEV +ER ITLRPL+M+D K
Sbjct: 240 CTTAAYRPVRELIKQECLKDQERRKREEAEKNSEEGSEAKEEVSEERGITLRPLSMEDMK 299
Query: 790 MAKSQVAASFAAEGAGM 806
+AKSQVAASFAAEGAGM
Sbjct: 300 VAKSQVAASFAAEGAGM 316
>gb|AAN46211.1| unknown protein [Arabidopsis thaliana]
Length = 316
Score = 518 bits (1335), Expect = e-145
Identities = 266/317 (83%), Positives = 290/317 (90%), Gaps = 2/317 (0%)
Query: 491 SKAEVPDNEFEKRIRPEVIPANEIGVTFSDIGALDETKDSLQELVMLPLRRPDLFEGGLL 550
SK PDNEFEKRIRPEVIPANEIGVTF+DIG+LDETK+SLQELVMLPLRRPDLF+GGLL
Sbjct: 1 SKEVAPDNEFEKRIRPEVIPANEIGVTFADIGSLDETKESLQELVMLPLRRPDLFKGGLL 60
Query: 551 KPCRGILLFGPPGTGKTMLAKAIANEAGASFINVSMSTITSKWFGEDEKNVRALFTLAAK 610
KPCRGILLFGPPGTGKTM+AKAIANEAGASFINVSMSTITSKWFGEDEKNVRALFTLAAK
Sbjct: 61 KPCRGILLFGPPGTGKTMMAKAIANEAGASFINVSMSTITSKWFGEDEKNVRALFTLAAK 120
Query: 611 VSPTIIFVDEVDSMLGQRTRVGEHEAMRKIKNEFMSNWDGLTSKSEDRILVLAATNRPFD 670
VSPTIIFVDEVDSMLGQRTRVGEHEAMRKIKNEFM++WDGL S + DRILVLAATNRPFD
Sbjct: 121 VSPTIIFVDEVDSMLGQRTRVGEHEAMRKIKNEFMTHWDGLMSNAGDRILVLAATNRPFD 180
Query: 671 LDEAIIRRFERRIMVGLPSAENRENILRTLLAKEKVHGGLDFKELATMTEGYSGSDLKNL 730
LDEAIIRRFERRIMVGLPS E+RE ILRTLL+KEK LDF+ELA MT+GYSGSDLKN
Sbjct: 181 LDEAIIRRFERRIMVGLPSVESREKILRTLLSKEKTE-NLDFQELAQMTDGYSGSDLKNF 239
Query: 731 CTTAAYRPVRELIQQEIQKDSKKKKDAEGQ-NSQDAQDAKEEVEQERVITLRPLNMQDFK 789
CTTAAYRPVRELI+QE KD +++K E + NS++ +AKEEV +ER ITLRPL+M+D K
Sbjct: 240 CTTAAYRPVRELIKQECLKDQERRKREEAEKNSEEGSEAKEEVSEERGITLRPLSMEDMK 299
Query: 790 MAKSQVAASFAAEGAGM 806
+AK QVAASFAAEGAGM
Sbjct: 300 VAKIQVAASFAAEGAGM 316
>ref|XP_479469.1| putative MSP1(mitochondrial sorting of proteins) protein [Oryza
sativa (japonica cultivar-group)]
gi|33146850|dbj|BAC79845.1| putative MSP1(mitochondrial
sorting of proteins) protein [Oryza sativa (japonica
cultivar-group)]
Length = 1081
Score = 513 bits (1321), Expect = e-143
Identities = 316/835 (37%), Positives = 463/835 (54%), Gaps = 120/835 (14%)
Query: 46 IENEMLRLVVDGRESNVTFDNFPYYLSEQTRVLLTSAAYVHLKHAEVSKYTRNLAPASRT 105
++ ++ ++VV+ + + +FD+FPYYLSE T+ L S+AYV+L E K+T++++ +
Sbjct: 318 LKEDLKKVVVNASDISDSFDSFPYYLSENTKNALLSSAYVNLCCKESIKWTKHISSLCQR 377
Query: 106 ILLSGPA--ELYQQVLAKALTHYFEAKLLLFDVTDFSLKIQSRYGSSNCETSFTRSTSET 163
+LLSGPA E+YQ+ L KALT +F AKLL+ D SL + S S+ +
Sbjct: 378 VLLSGPAGSEIYQESLVKALTKHFGAKLLIIDP---SLLASGQSSKSKESESYKKGDRVR 434
Query: 164 ALARLSDLFGSFALFPQREENQGKIHRQSSGSDLRQMEAEGSYSKL--RRNASASANISS 221
+ + L QR + G S ++R E SK+ R + I
Sbjct: 435 YIGSVQST--GIILEGQRAPDYG------SQGEVRLPFEENESSKVGVRFDKKIPGGIDL 486
Query: 222 IG-LQSNPTNSAPGKHIT----GWPFDEKILIQTLYKVLLYVSKTYPIVLYMRDADKLLC 276
G + + P + GW K +Y+ S+ P++L+++D +K+ C
Sbjct: 487 GGNCEVDRGFFCPVDSLCLDGPGWEDRAKHPFDVIYEFASEESQHGPLILFLKDVEKM-C 545
Query: 277 RSQRIYKLFQTMLTKLSGPILIIGS----------------------------------- 301
+ Y + + + I+GS
Sbjct: 546 GNSYSYHGLKNKIESFPAGVFIVGSQIHTDSRKDKSNSGSPFLSKFPYSQAILDLTFQDS 605
Query: 302 --RILDSGNECKRVDEMLTSLFPYNIEIKPPEDESRLVSWKSQFEADMKKIQIQDNKNHI 359
R+ D E ++ + LT LFP + I+ P+DE L WK + D++ ++ + N + I
Sbjct: 606 FGRVNDKNKEALKIAKHLTKLFPNKVTIQTPQDELELSQWKQLLDRDVEILKAKANTSKI 665
Query: 360 MEVLAANDLDCHDLD-SICVADTMVLSNYIEEIIVSAISYHIMKNKEPEYRN-GKLIIPC 417
L N L+C D++ S CV D ++ + +++++ A+S+ + P N G L +
Sbjct: 666 QSFLTRNGLECADIETSACVKDRILTNECVDKVVGYALSHQFKHSTIPTRENDGLLALSG 725
Query: 418 NSLSHALGIFQAGKFGDRDSLKLEAQAVTSEKKEEGAAVKPEGKTESPAPAVKTEAEIPT 477
SL H + + DS++ + + +++K
Sbjct: 726 ESLKHGVELL--------DSMQSDPKKKSTKK---------------------------- 749
Query: 478 SVRKTDGENSVPASKAEVPDNEFEKRIRPEVIPANEIGVTFSDIGALDETKDSLQELVML 537
+ K +NEFEKR+ +VIP +EIGVTF DIGAL+ K++L+ELVML
Sbjct: 750 ------------SLKDVTTENEFEKRLLGDVIPPDEIGVTFEDIGALENVKETLKELVML 797
Query: 538 PLRRPDLF-EGGLLKPCRGILLFGPPGTGKTMLAKAIANEAGASFINVSMSTITSKWFGE 596
PL+RP+LF +G L+KPC+GILLFGPPGTGKTMLAKA+A EAGA+FIN+SMS+I SKWFGE
Sbjct: 798 PLQRPELFSKGQLMKPCKGILLFGPPGTGKTMLAKAVATEAGANFINISMSSIASKWFGE 857
Query: 597 DEKNVRALFTLAAKVSPTIIFVDEVDSMLGQRTRVGEHEAMRKIKNEFMSNWDGLTSKSE 656
EK V+A+F+LA+K++P++IFVDEVD MLG+R GEHEAMRK+KNEFM NWDGL +K +
Sbjct: 858 GEKYVKAVFSLASKIAPSVIFVDEVDGMLGRRENPGEHEAMRKMKNEFMVNWDGLRTKDK 917
Query: 657 DRILVLAATNRPFDLDEAIIRRFERRIMVGLPSAENRENILRTLLAKEKVHGGLDFKELA 716
+R+LVLAATNRPFDLDEA++RR RR+MV LP A NR+ IL +LAKE + +D + LA
Sbjct: 918 ERVLVLAATNRPFDLDEAVVRRLPRRLMVNLPDASNRKKILSVILAKEDLADDVDLEALA 977
Query: 717 TMTEGYSGSDLKNLCTTAAYRPVRELIQQEIQKDSKKKKDAEGQNSQDAQDAKEEVEQER 776
+T+GYSGSD+KNLC TAA+ P+RE++++E K++ AE +N K
Sbjct: 978 NLTDGYSGSDMKNLCVTAAHCPIREILERE----KKERASAEAEN-------KPLPPPRS 1026
Query: 777 VITLRPLNMQDFKMAKSQVAASFAAEGAGMNELRQWNDLYGEGGSRKKEQLSYFL 831
+R L M DFK A QV AS ++ M EL QWNDLYGEGGSRKK LSYF+
Sbjct: 1027 SSDVRSLRMNDFKHAHEQVCASITSDSRNMTELIQWNDLYGEGGSRKKTSLSYFM 1081
>ref|NP_171788.2| AAA-type ATPase family protein [Arabidopsis thaliana]
Length = 1252
Score = 486 bits (1250), Expect = e-135
Identities = 271/652 (41%), Positives = 389/652 (59%), Gaps = 112/652 (17%)
Query: 220 SSIGLQSNPTNSAPGKHITGWPFDEKILIQTLYKVLLYVSKTYPIVLYMRDADKLLCRSQ 279
SS+ L+S+ ++ A +K+ I +++V S+ ++L+++D +K + +
Sbjct: 673 SSLRLESSSSDDA-----------DKLAINEIFEVAFNESERGSLILFLKDIEKSVSGNT 721
Query: 280 RIYKLFQTMLTKLSGPILII---------------------------------------G 300
+Y ++ L L I++I G
Sbjct: 722 DVYITLKSKLENLPENIVVIASQTQLDNRKEKSHPGGFLFTKFGSNQTALLDLAFPDTFG 781
Query: 301 SRILDSGNECKRVDEMLTSLFPYNIEIKPPEDESRLVSWKSQFEADMKKIQIQDNKNHIM 360
R+ D E + + +T LFP + I+ PEDE+ LV WK + E D + ++ Q N I
Sbjct: 782 GRLQDRNTEMPKAVKQITRLFPNKVTIQLPEDEASLVDWKDKLERDTEILKAQANITSIR 841
Query: 361 EVLAANDLDCHDLDSICVADTMVLSNYIEEIIVSAISYHIMKNKEPEYRNGKLIIPCNSL 420
VL+ N L C D++ +C+ D + S+ +E+++ A ++H+M EP ++ KLII S+
Sbjct: 842 AVLSKNQLVCPDIEILCIKDQTLPSDSVEKVVGFAFNHHLMNCSEPTVKDNKLIISAESI 901
Query: 421 SHALGIFQAGKFGDRDSLKLEAQAVTSEKKEEGAAVKPEGKTESPAPAVKTEAEIPTSVR 480
++ L + + +E K ++K
Sbjct: 902 TYGLQLLHE---------------IQNENKSTKKSLKDV--------------------- 925
Query: 481 KTDGENSVPASKAEVPDNEFEKRIRPEVIPANEIGVTFSDIGALDETKDSLQELVMLPLR 540
V +NEFEK++ +VIP ++IGV+FSDIGAL+ KD+L+ELVMLPL+
Sbjct: 926 --------------VTENEFEKKLLSDVIPPSDIGVSFSDIGALENVKDTLKELVMLPLQ 971
Query: 541 RPDLF-EGGLLKPCRGILLFGPPGTGKTMLAKAIANEAGASFINVSMSTITSKWFGEDEK 599
RP+LF +G L KP +GILLFGPPGTGKTMLAKA+A EAGA+FIN+SMS+ITSKWFGE EK
Sbjct: 972 RPELFGKGQLTKPTKGILLFGPPGTGKTMLAKAVATEAGANFINISMSSITSKWFGEGEK 1031
Query: 600 NVRALFTLAAKVSPTIIFVDEVDSMLGQRTRVGEHEAMRKIKNEFMSNWDGLTSKSEDRI 659
V+A+F+LA+K++P++IFVDEVDSMLG+R GEHEAMRK+KNEFM NWDGL +K ++R+
Sbjct: 1032 YVKAVFSLASKIAPSVIFVDEVDSMLGRRENPGEHEAMRKMKNEFMINWDGLRTKDKERV 1091
Query: 660 LVLAATNRPFDLDEAIIRRFERRIMVGLPSAENRENILRTLLAKEKVHGGLDFKELATMT 719
LVLAATNRPFDLDEA+IRR RR+MV LP + NR IL +LAKE++ +D + +A MT
Sbjct: 1092 LVLAATNRPFDLDEAVIRRLPRRLMVNLPDSANRSKILSVILAKEEMAEDVDLEAIANMT 1151
Query: 720 EGYSGSDLKNLCTTAAYRPVRELIQQEIQKDSKKKKDAEGQNSQDAQDAKEEVEQERVIT 779
+GYSGSDLKNLC TAA+ P+RE+ ++K+ K++ A+ +N Q
Sbjct: 1152 DGYSGSDLKNLCVTAAHLPIREI----LEKEKKERSVAQAENRAMPQLYSS-------TD 1200
Query: 780 LRPLNMQDFKMAKSQVAASFAAEGAGMNELRQWNDLYGEGGSRKKEQLSYFL 831
+RPLNM DFK A QV AS A++ + MNEL+QWN+LYGEGGSRKK LSYF+
Sbjct: 1201 VRPLNMNDFKTAHDQVCASVASDSSNMNELQQWNELYGEGGSRKKTSLSYFM 1252
Score = 68.6 bits (166), Expect = 8e-10
Identities = 40/103 (38%), Positives = 66/103 (63%), Gaps = 4/103 (3%)
Query: 37 SSNALTPDKIENEMLRL-VVDGRESNVTFDNFPYYLSEQTRVLLTSAAYVHLKHA-EVSK 94
S++ LT + + LR +++ ++ V+F+NFPY+LS T+ +L + Y H+K+ E ++
Sbjct: 421 SASVLTRRQAHKDSLRGGILNPQDIEVSFENFPYFLSGTTKDVLMISTYAHIKYGKEYAE 480
Query: 95 YTRNLAPASRTILLSGP--AELYQQVLAKALTHYFEAKLLLFD 135
Y +L A ILLSGP +E+YQ++LAKAL AKL++ D
Sbjct: 481 YASDLPTACPRILLSGPSGSEIYQEMLAKALAKQCGAKLMIVD 523
>gb|AAM13995.1| unknown protein [Arabidopsis thaliana]
Length = 1265
Score = 476 bits (1225), Expect = e-132
Identities = 268/627 (42%), Positives = 382/627 (60%), Gaps = 100/627 (15%)
Query: 244 EKILIQTLYKVLLYVSKTYPIVLYMRDADKLLCRSQRIYKLFQTMLTKLSGPILIIGSRI 303
+K+ + +++V L S+ ++L+++D +K L + +Y ++ L L I++I S+
Sbjct: 700 DKLAVNEIFEVALSESEGGSLILFLKDIEKSLVGNSDVYATLKSKLETLPENIVVIASQT 759
Query: 304 -LDS----------------GNECKRVD---------------------EMLTSLFPYNI 325
LDS GN+ +D + +T LFP I
Sbjct: 760 QLDSRKEKSHPGGFLFTKFGGNQTALLDLAFPDNFGKLHDRSKETPKSMKQITRLFPNKI 819
Query: 326 EIKPPEDESRLVSWKSQFEADMKKIQIQDNKNHIMEVLAANDLDCHDLDSICVADTMVLS 385
I+ P++E+ L WK + + D + +++Q N I+ VLA N LDC DL ++C+ D + S
Sbjct: 820 AIQLPQEEALLSDWKEKLDRDTEILKVQANITSILAVLAKNKLDCPDLGTLCIKDQTLPS 879
Query: 386 NYIEEIIVSAISYHIMKNKEPEYRNGKLIIPCNSLSHALGIFQAGKFGDRDSLKLEAQAV 445
+E+++ A +H+M EP ++ KL+I S+S+ L +
Sbjct: 880 ESVEKVVGWAFGHHLMICTEPIVKDNKLVISAESISYGLQTLHD---------------I 924
Query: 446 TSEKKEEGAAVKPEGKTESPAPAVKTEAEIPTSVRKTDGENSVPASKAEVPDNEFEKRIR 505
+E K S++K+ K V +NEFEK++
Sbjct: 925 QNENK---------------------------SLKKS--------LKDVVTENEFEKKLL 949
Query: 506 PEVIPANEIGVTFSDIGALDETKDSLQELVMLPLRRPDLFEGG-LLKPCRGILLFGPPGT 564
+VIP ++IGV+F DIGAL+ K++L+ELVMLPL+RP+LF+ G L KP +GILLFGPPGT
Sbjct: 950 SDVIPPSDIGVSFDDIGALENVKETLKELVMLPLQRPELFDKGQLTKPTKGILLFGPPGT 1009
Query: 565 GKTMLAKAIANEAGASFINVSMSTITSKWFGEDEKNVRALFTLAAKVSPTIIFVDEVDSM 624
GKTMLAKA+A EAGA+FIN+SMS+ITSKWFGE EK V+A+F+LA+K++P++IFVDEVDSM
Sbjct: 1010 GKTMLAKAVATEAGANFINISMSSITSKWFGEGEKYVKAVFSLASKIAPSVIFVDEVDSM 1069
Query: 625 LGQRTRVGEHEAMRKIKNEFMSNWDGLTSKSEDRILVLAATNRPFDLDEAIIRRFERRIM 684
LG+R GEHEAMRK+KNEFM NWDGL +K +R+LVLAATNRPFDLDEA+IRR RR+M
Sbjct: 1070 LGRRENPGEHEAMRKMKNEFMVNWDGLRTKDRERVLVLAATNRPFDLDEAVIRRLPRRLM 1129
Query: 685 VGLPSAENRENILRTLLAKEKVHGGLDFKELATMTEGYSGSDLKNLCTTAAYRPVRELIQ 744
V LP A NR IL +LAKE++ +D + +A MT+GYSGSDLKNLC TAA+ P+RE+
Sbjct: 1130 VNLPDATNRSKILSVILAKEEIAPDVDLEAIANMTDGYSGSDLKNLCVTAAHFPIREI-- 1187
Query: 745 QEIQKDSKKKKDAEGQNSQDAQDAKEEVEQERVITLRPLNMQDFKMAKSQVAASFAAEGA 804
++K+ K+K A+ +N + +R L M DFK A QV AS +++ +
Sbjct: 1188 --LEKEKKEKTAAQAEN-------RPTPPLYSCTDVRSLTMNDFKAAHDQVCASVSSDSS 1238
Query: 805 GMNELRQWNDLYGEGGSRKKEQLSYFL 831
MNEL+QWN+LYGEGGSRKK LSYF+
Sbjct: 1239 NMNELQQWNELYGEGGSRKKTSLSYFM 1265
Score = 67.0 bits (162), Expect = 2e-09
Identities = 35/85 (41%), Positives = 57/85 (66%), Gaps = 3/85 (3%)
Query: 54 VVDGRESNVTFDNFPYYLSEQTRVLLTSAAYVHLK-HAEVSKYTRNLAPASRTILLSGP- 111
V++ + +++F+NFPYYLS T+ +L + YVH+ ++ + + +L A ILLSGP
Sbjct: 450 VLNAQNIDISFENFPYYLSATTKGVLMISMYVHMNGGSKYANFATDLTTACPRILLSGPS 509
Query: 112 -AELYQQVLAKALTHYFEAKLLLFD 135
+E+YQ++LAKAL F AKL++ D
Sbjct: 510 SSEIYQEMLAKALAKQFGAKLMIVD 534
>ref|NP_567238.2| AAA-type ATPase family protein [Arabidopsis thaliana]
Length = 1265
Score = 476 bits (1225), Expect = e-132
Identities = 268/627 (42%), Positives = 382/627 (60%), Gaps = 100/627 (15%)
Query: 244 EKILIQTLYKVLLYVSKTYPIVLYMRDADKLLCRSQRIYKLFQTMLTKLSGPILIIGSRI 303
+K+ + +++V L S+ ++L+++D +K L + +Y ++ L L I++I S+
Sbjct: 700 DKLAVNEIFEVALSESEGGSLILFLKDIEKSLVGNSDVYATLKSKLETLPENIVVIASQT 759
Query: 304 -LDS----------------GNECKRVD---------------------EMLTSLFPYNI 325
LDS GN+ +D + +T LFP I
Sbjct: 760 QLDSRKEKSHPGGFLFTKFGGNQTALLDLAFPDNFGKLHDRSKETPKSMKQITRLFPNKI 819
Query: 326 EIKPPEDESRLVSWKSQFEADMKKIQIQDNKNHIMEVLAANDLDCHDLDSICVADTMVLS 385
I+ P++E+ L WK + + D + +++Q N I+ VLA N LDC DL ++C+ D + S
Sbjct: 820 AIQLPQEEALLSDWKEKLDRDTEILKVQANITSILAVLAKNKLDCPDLGTLCIKDQTLPS 879
Query: 386 NYIEEIIVSAISYHIMKNKEPEYRNGKLIIPCNSLSHALGIFQAGKFGDRDSLKLEAQAV 445
+E+++ A +H+M EP ++ KL+I S+S+ L +
Sbjct: 880 ESVEKVVGWAFGHHLMICTEPIVKDNKLVISAESISYGLQTLHD---------------I 924
Query: 446 TSEKKEEGAAVKPEGKTESPAPAVKTEAEIPTSVRKTDGENSVPASKAEVPDNEFEKRIR 505
+E K S++K+ K V +NEFEK++
Sbjct: 925 QNENK---------------------------SLKKS--------LKDVVTENEFEKKLL 949
Query: 506 PEVIPANEIGVTFSDIGALDETKDSLQELVMLPLRRPDLFEGG-LLKPCRGILLFGPPGT 564
+VIP ++IGV+F DIGAL+ K++L+ELVMLPL+RP+LF+ G L KP +GILLFGPPGT
Sbjct: 950 SDVIPPSDIGVSFDDIGALENVKETLKELVMLPLQRPELFDKGQLTKPTKGILLFGPPGT 1009
Query: 565 GKTMLAKAIANEAGASFINVSMSTITSKWFGEDEKNVRALFTLAAKVSPTIIFVDEVDSM 624
GKTMLAKA+A EAGA+FIN+SMS+ITSKWFGE EK V+A+F+LA+K++P++IFVDEVDSM
Sbjct: 1010 GKTMLAKAVATEAGANFINISMSSITSKWFGEGEKYVKAVFSLASKIAPSVIFVDEVDSM 1069
Query: 625 LGQRTRVGEHEAMRKIKNEFMSNWDGLTSKSEDRILVLAATNRPFDLDEAIIRRFERRIM 684
LG+R GEHEAMRK+KNEFM NWDGL +K +R+LVLAATNRPFDLDEA+IRR RR+M
Sbjct: 1070 LGRRENPGEHEAMRKMKNEFMVNWDGLRTKDRERVLVLAATNRPFDLDEAVIRRLPRRLM 1129
Query: 685 VGLPSAENRENILRTLLAKEKVHGGLDFKELATMTEGYSGSDLKNLCTTAAYRPVRELIQ 744
V LP A NR IL +LAKE++ +D + +A MT+GYSGSDLKNLC TAA+ P+RE+
Sbjct: 1130 VNLPDATNRSKILSVILAKEEIAPDVDLEAIANMTDGYSGSDLKNLCVTAAHFPIREI-- 1187
Query: 745 QEIQKDSKKKKDAEGQNSQDAQDAKEEVEQERVITLRPLNMQDFKMAKSQVAASFAAEGA 804
++K+ K+K A+ +N + +R L M DFK A QV AS +++ +
Sbjct: 1188 --LEKEKKEKTAAQAEN-------RPTPPLYSCTDVRSLTMNDFKAAHDQVCASVSSDSS 1238
Query: 805 GMNELRQWNDLYGEGGSRKKEQLSYFL 831
MNEL+QWN+LYGEGGSRKK LSYF+
Sbjct: 1239 NMNELQQWNELYGEGGSRKKTSLSYFM 1265
Score = 67.0 bits (162), Expect = 2e-09
Identities = 35/85 (41%), Positives = 57/85 (66%), Gaps = 3/85 (3%)
Query: 54 VVDGRESNVTFDNFPYYLSEQTRVLLTSAAYVHLK-HAEVSKYTRNLAPASRTILLSGP- 111
V++ + +++F+NFPYYLS T+ +L + YVH+ ++ + + +L A ILLSGP
Sbjct: 450 VLNAQNIDISFENFPYYLSATTKGVLMISMYVHMNGGSKYANFATDLTTACPRILLSGPS 509
Query: 112 -AELYQQVLAKALTHYFEAKLLLFD 135
+E+YQ++LAKAL F AKL++ D
Sbjct: 510 GSEIYQEMLAKALAKQFGAKLMIVD 534
>ref|NP_194217.2| AAA-type ATPase family protein [Arabidopsis thaliana]
Length = 1122
Score = 449 bits (1154), Expect = e-124
Identities = 258/604 (42%), Positives = 360/604 (58%), Gaps = 84/604 (13%)
Query: 245 KILIQTLYKVLLYVSKTYPIVLYMRDADKLLCRSQRIYKLFQTMLTKLSGPILIIGS--- 301
++L+ TL++V+ S+T P +L+++DA+K + + +Y FQ L L +++I S
Sbjct: 584 RLLVNTLFEVVHSESRTCPFILFLKDAEKSVAGNFDLYSAFQIRLEYLPENVIVICSQTH 643
Query: 302 ----------RILDSGNECKRVDEMLTSLFPYNIEIKPPEDESRLVSWKSQFEADMKKIQ 351
R G E E+L LF I I+ P+DE RL WK Q + D + +
Sbjct: 644 SDHLKVKDIGRQKKQGKEVPHATELLAELFENKITIQMPQDEKRLTLWKHQMDRDAETSK 703
Query: 352 IQDNKNHIMEVLAANDLDCHDLDS----ICVADTMVLSNYIEEIIVSAISYHIMKNKEPE 407
++ N NH+ VL L C L++ +C+ D + + +E+II A HI +K P+
Sbjct: 704 VKSNFNHLRMVLRRRGLGCEGLETTWSRMCLKDLTLQRDSVEKIIGWAFGNHI--SKNPD 761
Query: 408 YRNGKLIIPCNSLSHALGIFQAGKFGDRDSLKLEAQAVTSEKKEEGAAVKPEGKTESPAP 467
K+ + S+
Sbjct: 762 TDPAKVTLSRESI----------------------------------------------- 774
Query: 468 AVKTEAEIPTSVRKTDGENSVPASKAEVPDNEFEKRIRPEVIPANEIGVTFSDIGALDET 527
E + + D + S + K V +N FEKR+ +VI ++I VTF DIGAL++
Sbjct: 775 ------EFGIGLLQNDLKGSTSSKKDIVVENVFEKRLLSDVILPSDIDVTFDDIGALEKV 828
Query: 528 KDSLQELVMLPLRRPDLF-EGGLLKPCRGILLFGPPGTGKTMLAKAIANEAGASFINVSM 586
KD L+ELVMLPL+RP+LF +G L KPC+GILLFGPPGTGKTMLAKA+A EA A+FIN+SM
Sbjct: 829 KDILKELVMLPLQRPELFCKGELTKPCKGILLFGPPGTGKTMLAKAVAKEADANFINISM 888
Query: 587 STITSKWFGEDEKNVRALFTLAAKVSPTIIFVDEVDSMLGQRTRVGEHEAMRKIKNEFMS 646
S+ITSKWFGE EK V+A+F+LA+K+SP++IFVDEVDSMLG+R EHEA RKIKNEFM
Sbjct: 889 SSITSKWFGEGEKYVKAVFSLASKMSPSVIFVDEVDSMLGRREHPREHEASRKIKNEFMM 948
Query: 647 NWDGLTSKSEDRILVLAATNRPFDLDEAIIRRFERRIMVGLPSAENRENILRTLLAKEKV 706
+WDGLT++ +R+LVLAATNRPFDLDEA+IRR RR+MVGLP NR IL+ +LAKE +
Sbjct: 949 HWDGLTTQERERVLVLAATNRPFDLDEAVIRRLPRRLMVGLPDTSNRAFILKVILAKEDL 1008
Query: 707 HGGLDFKELATMTEGYSGSDLKNLCTTAAYRPVRELIQQEIQKDSKKKKDAEGQNSQDAQ 766
LD E+A+MT GYSGSDLKNLC TAA+RP++E++++E K+++DA
Sbjct: 1009 SPDLDIGEIASMTNGYSGSDLKNLCVTAAHRPIKEILEKE-----KRERDAA------LA 1057
Query: 767 DAKEEVEQERVITLRPLNMQDFKMAKSQVAASFAAEGAGMNELRQWNDLYGEGGSRKKEQ 826
K LR LN++DF+ A V+AS ++E A M L+QWN L+GEGGS K++
Sbjct: 1058 QGKVPPPLSGSSDLRALNVEDFRDAHKWVSASVSSESATMTALQQWNKLHGEGGSGKQQS 1117
Query: 827 LSYF 830
S++
Sbjct: 1118 FSFY 1121
Score = 85.5 bits (210), Expect = 7e-15
Identities = 47/103 (45%), Positives = 61/103 (58%), Gaps = 2/103 (1%)
Query: 35 SSSSNALTPDKIENEMLRLVVDGRESNVTFDNFPYYLSEQTRVLLTSAAYVHLKHAEVSK 94
S+S N L + V G V+F NFPYYLSE T+ L A+Y+HLK E +
Sbjct: 277 STSGNGLQSAIFREAIQAGFVRGENMEVSFKNFPYYLSEYTKAALLYASYIHLKKKEYVQ 336
Query: 95 YTRNLAPASRTILLSGPA--ELYQQVLAKALTHYFEAKLLLFD 135
+ ++ P + ILLSGPA E+YQ+ LAKAL EAKLL+FD
Sbjct: 337 FVSDMTPMNPRILLSGPAGSEIYQETLAKALARDLEAKLLIFD 379
>gb|AAF02877.1| Unknown protein [Arabidopsis thaliana] gi|25518408|pir||C86159
hypothetical protein F22D16.11 - Arabidopsis thaliana
Length = 1217
Score = 417 bits (1073), Expect = e-115
Identities = 209/341 (61%), Positives = 268/341 (78%), Gaps = 12/341 (3%)
Query: 492 KAEVPDNEFEKRIRPEVIPANEIGVTFSDIGALDETKDSLQELVMLPLRRPDLF-EGGLL 550
K V +NEFEK++ +VIP ++IGV+FSDIGAL+ KD+L+ELVMLPL+RP+LF +G L
Sbjct: 888 KDVVTENEFEKKLLSDVIPPSDIGVSFSDIGALENVKDTLKELVMLPLQRPELFGKGQLT 947
Query: 551 KPCRGILLFGPPGTGKTMLAKAIANEAGASFINVSMSTITSKWFGEDEKNVRALFTLAAK 610
KP +GILLFGPPGTGKTMLAKA+A EAGA+FIN+SMS+ITSKWFGE EK V+A+F+LA+K
Sbjct: 948 KPTKGILLFGPPGTGKTMLAKAVATEAGANFINISMSSITSKWFGEGEKYVKAVFSLASK 1007
Query: 611 VSPTIIFVDEVDSMLGQRTRVGEHEAMRKIKNEFMSNWDGLTSKSEDRILVLAATNRPFD 670
++P++IFVDEVDSMLG+R GEHEAMRK+KNEFM NWDGL +K ++R+LVLAATNRPFD
Sbjct: 1008 IAPSVIFVDEVDSMLGRRENPGEHEAMRKMKNEFMINWDGLRTKDKERVLVLAATNRPFD 1067
Query: 671 LDEAIIRRFERRIMVGLPSAENRENILRTLLAKEKVHGGLDFKELATMTEGYSGSDLKNL 730
LDEA+IRR RR+MV LP + NR IL +LAKE++ +D + +A MT+GYSGSDLKNL
Sbjct: 1068 LDEAVIRRLPRRLMVNLPDSANRSKILSVILAKEEMAEDVDLEAIANMTDGYSGSDLKNL 1127
Query: 731 CTTAAYRPVRELIQQEIQKDSKKKKDAEGQNSQDAQDAKEEVEQERVITLRPLNMQDFKM 790
C TAA+ P+RE+ ++K+ K++ A+ +N Q +RPLNM DFK
Sbjct: 1128 CVTAAHLPIREI----LEKEKKERSVAQAENRAMPQLYSS-------TDVRPLNMNDFKT 1176
Query: 791 AKSQVAASFAAEGAGMNELRQWNDLYGEGGSRKKEQLSYFL 831
A QV AS A++ + MNEL+QWN+LYGEGGSRKK LSYF+
Sbjct: 1177 AHDQVCASVASDSSNMNELQQWNELYGEGGSRKKTSLSYFM 1217
Score = 57.8 bits (138), Expect = 1e-06
Identities = 40/131 (30%), Positives = 66/131 (49%), Gaps = 32/131 (24%)
Query: 37 SSNALTPDKIENEMLRL-VVDGRESNVTFDNFPYYLSEQTRVLLTSAAYVHLKHA-EVSK 94
S++ LT + + LR +++ ++ V+F+NFPY+LS T+ +L + Y H+K+ E ++
Sbjct: 430 SASVLTRRQAHKDSLRGGILNPQDIEVSFENFPYFLSGTTKDVLMISTYAHIKYGKEYAE 489
Query: 95 YTRNLAPASRTILLSGP------------------------------AELYQQVLAKALT 124
Y +L A ILLSGP +E+YQ++LAKAL
Sbjct: 490 YASDLPTACPRILLSGPSGKLWTSIVYESFVSHFHFPNKFSYGIFEGSEIYQEMLAKALA 549
Query: 125 HYFEAKLLLFD 135
AKL++ D
Sbjct: 550 KQCGAKLMIVD 560
Score = 44.7 bits (104), Expect = 0.013
Identities = 38/158 (24%), Positives = 77/158 (48%), Gaps = 21/158 (13%)
Query: 220 SSIGLQSNPTNSAPGKHITGWPFDEKILIQTLYKVLLYVSKTYPIVLYMRDADKLLCRSQ 279
SS+ L+S+ ++ A +K+ I +++V S+ ++L+++D +K + +
Sbjct: 708 SSLRLESSSSDDA-----------DKLAINEIFEVAFNESERGSLILFLKDIEKSVSGNT 756
Query: 280 RIYKLFQTMLTKLSGPILIIGSRI-LDSGNECKRVDEMLTSLFPYN----IEIKPPEDES 334
+Y ++ L L I++I S+ LD+ E L + F N +++ P DE+
Sbjct: 757 DVYITLKSKLENLPENIVVIASQTQLDNRKEKSHPGGFLFTKFGSNQTALLDLAFP-DEA 815
Query: 335 RLVSWKSQFEADMKKIQIQDN----KNHIMEVLAANDL 368
LV WK + E D + ++ Q N + H++ L N +
Sbjct: 816 SLVDWKDKLERDTEILKAQANITSIRAHLVICLIENHM 853
>emb|CAB79395.1| putative protein [Arabidopsis thaliana] gi|4678264|emb|CAB41125.1|
putative protein [Arabidopsis thaliana]
gi|23296350|gb|AAN13049.1| unknown protein [Arabidopsis
thaliana] gi|7486530|pir||T06669 hypothetical protein
F6I7.60 - Arabidopsis thaliana
Length = 442
Score = 410 bits (1055), Expect = e-113
Identities = 231/506 (45%), Positives = 313/506 (61%), Gaps = 71/506 (14%)
Query: 330 PEDESRLVSWKSQFEADMKKIQIQDNKNHIMEVLAANDLDCHDLDS----ICVADTMVLS 385
P+DE RL WK Q + D + +++ N NH+ VL L C L++ +C+ D +
Sbjct: 2 PQDEKRLTLWKHQMDRDAETSKVKSNFNHLRMVLRRRGLGCEGLETTWSRMCLKDLTLQR 61
Query: 386 NYIEEIIVSAISYHIMKNKEPEYRNGKLIIPCNSLSHALGIFQAGKFGDRDSLKLEAQAV 445
+ +E+II A HI +K P+ K+ + S+
Sbjct: 62 DSVEKIIGWAFGNHI--SKNPDTDPAKVTLSRESI------------------------- 94
Query: 446 TSEKKEEGAAVKPEGKTESPAPAVKTEAEIPTSVRKTDGENSVPASKAEVPDNEFEKRIR 505
E + + D + S + K V +N FEKR+
Sbjct: 95 ----------------------------EFGIGLLQNDLKGSTSSKKDIVVENVFEKRLL 126
Query: 506 PEVIPANEIGVTFSDIGALDETKDSLQELVMLPLRRPDLF-EGGLLKPCRGILLFGPPGT 564
+VI ++I VTF DIGAL++ KD L+ELVMLPL+RP+LF +G L KPC+GILLFGPPGT
Sbjct: 127 SDVILPSDIDVTFDDIGALEKVKDILKELVMLPLQRPELFCKGELTKPCKGILLFGPPGT 186
Query: 565 GKTMLAKAIANEAGASFINVSMSTITSKWFGEDEKNVRALFTLAAKVSPTIIFVDEVDSM 624
GKTMLAKA+A EA A+FIN+SMS+ITSKWFGE EK V+A+F+LA+K+SP++IFVDEVDSM
Sbjct: 187 GKTMLAKAVAKEADANFINISMSSITSKWFGEGEKYVKAVFSLASKMSPSVIFVDEVDSM 246
Query: 625 LGQRTRVGEHEAMRKIKNEFMSNWDGLTSKSEDRILVLAATNRPFDLDEAIIRRFERRIM 684
LG+R EHEA RKIKNEFM +WDGLT++ +R+LVLAATNRPFDLDEA+IRR RR+M
Sbjct: 247 LGRREHPREHEASRKIKNEFMMHWDGLTTQERERVLVLAATNRPFDLDEAVIRRLPRRLM 306
Query: 685 VGLPSAENRENILRTLLAKEKVHGGLDFKELATMTEGYSGSDLKNLCTTAAYRPVRELIQ 744
VGLP NR IL+ +LAKE + LD E+A+MT GYSGSDLKNLC TAA+RP++E+++
Sbjct: 307 VGLPDTSNRAFILKVILAKEDLSPDLDIGEIASMTNGYSGSDLKNLCVTAAHRPIKEILE 366
Query: 745 QEIQKDSKKKKDAEGQNSQDAQDAKEEVEQERVITLRPLNMQDFKMAKSQVAASFAAEGA 804
+E K+++DA K LR LN++DF+ A V+AS ++E A
Sbjct: 367 KE-----KRERDAA------LAQGKVPPPLSGSSDLRALNVEDFRDAHKWVSASVSSESA 415
Query: 805 GMNELRQWNDLYGEGGSRKKEQLSYF 830
M L+QWN L+GEGGS K++ S++
Sbjct: 416 TMTALQQWNKLHGEGGSGKQQSFSFY 441
>gb|AAC19276.1| T14P8.7 [Arabidopsis thaliana] gi|7269007|emb|CAB80740.1| AT4g02470
[Arabidopsis thaliana] gi|7487016|pir||T01303
hypothetical protein T14P8.7 - Arabidopsis thaliana
Length = 371
Score = 410 bits (1053), Expect = e-112
Identities = 205/341 (60%), Positives = 264/341 (77%), Gaps = 12/341 (3%)
Query: 492 KAEVPDNEFEKRIRPEVIPANEIGVTFSDIGALDETKDSLQELVMLPLRRPDLFEGG-LL 550
K V +NEFEK++ +VIP ++IGV+F DIGAL+ K++L+ELVMLPL+RP+LF+ G L
Sbjct: 42 KDVVTENEFEKKLLSDVIPPSDIGVSFDDIGALENVKETLKELVMLPLQRPELFDKGQLT 101
Query: 551 KPCRGILLFGPPGTGKTMLAKAIANEAGASFINVSMSTITSKWFGEDEKNVRALFTLAAK 610
KP +GILLFGPPGTGKTMLAKA+A EAGA+FIN+SMS+ITSKWFGE EK V+A+F+LA+K
Sbjct: 102 KPTKGILLFGPPGTGKTMLAKAVATEAGANFINISMSSITSKWFGEGEKYVKAVFSLASK 161
Query: 611 VSPTIIFVDEVDSMLGQRTRVGEHEAMRKIKNEFMSNWDGLTSKSEDRILVLAATNRPFD 670
++P++IFVDEVDSMLG+R GEHEAMRK+KNEFM NWDGL +K +R+LVLAATNRPFD
Sbjct: 162 IAPSVIFVDEVDSMLGRRENPGEHEAMRKMKNEFMVNWDGLRTKDRERVLVLAATNRPFD 221
Query: 671 LDEAIIRRFERRIMVGLPSAENRENILRTLLAKEKVHGGLDFKELATMTEGYSGSDLKNL 730
LDEA+IRR RR+MV LP A NR IL +LAKE++ +D + +A MT+GYSGSDLKNL
Sbjct: 222 LDEAVIRRLPRRLMVNLPDATNRSKILSVILAKEEIAPDVDLEAIANMTDGYSGSDLKNL 281
Query: 731 CTTAAYRPVRELIQQEIQKDSKKKKDAEGQNSQDAQDAKEEVEQERVITLRPLNMQDFKM 790
C TAA+ P+RE+ ++K+ K+K A+ +N + +R L M DFK
Sbjct: 282 CVTAAHFPIREI----LEKEKKEKTAAQAEN-------RPTPPLYSCTDVRSLTMNDFKA 330
Query: 791 AKSQVAASFAAEGAGMNELRQWNDLYGEGGSRKKEQLSYFL 831
A QV AS +++ + MNEL+QWN+LYGEGGSRKK LSYF+
Sbjct: 331 AHDQVCASVSSDSSNMNELQQWNELYGEGGSRKKTSLSYFM 371
Database: nr
Posted date: Jul 5, 2005 12:34 AM
Number of letters in database: 863,360,394
Number of sequences in database: 2,540,612
Lambda K H
0.315 0.132 0.366
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 1,326,160,830
Number of Sequences: 2540612
Number of extensions: 55802261
Number of successful extensions: 192315
Number of sequences better than 10.0: 5067
Number of HSP's better than 10.0 without gapping: 2891
Number of HSP's successfully gapped in prelim test: 2181
Number of HSP's that attempted gapping in prelim test: 180641
Number of HSP's gapped (non-prelim): 8048
length of query: 831
length of database: 863,360,394
effective HSP length: 137
effective length of query: 694
effective length of database: 515,296,550
effective search space: 357615805700
effective search space used: 357615805700
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (22.0 bits)
S2: 80 (35.4 bits)
Medicago: description of AC140913.6


