

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC140913.1 - phase: 0 /pseudo
(695 letters)
Database: nr
2,540,612 sequences; 863,360,394 total letters
Searching..................................................done
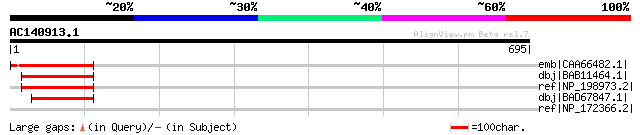
Score E
Sequences producing significant alignments: (bits) Value
emb|CAA66482.1| transcription factor [Vicia faba] gi|7488864|pir... 169 3e-40
dbj|BAB11464.1| transcription factor-like protein [Arabidopsis t... 81 1e-13
ref|NP_198973.2| zinc finger (MIZ type) family protein [Arabidop... 81 1e-13
dbj|BAD67847.1| putative transcription factor [Oryza sativa (jap... 62 8e-08
ref|NP_172366.2| zinc finger (MIZ type) family protein [Arabidop... 40 0.34
>emb|CAA66482.1| transcription factor [Vicia faba] gi|7488864|pir||T12184 probable
transcription factor - fava bean
Length = 828
Score = 169 bits (428), Expect = 3e-40
Identities = 89/112 (79%), Positives = 95/112 (84%), Gaps = 2/112 (1%)
Query: 1 METNTSSPLSTLPESGAMTATANPVSPSLVNLYRITKVLERLATHFVPGNRSDAFEFFNL 60
ME+NTSS + P+ A T NPVSPSLVNLYRITKVL+RLA HF PGNRSD+FEFFNL
Sbjct: 1 MESNTSSSMP--PDLAATNGTTNPVSPSLVNLYRITKVLDRLAFHFQPGNRSDSFEFFNL 58
Query: 61 CLSLSRGIDYALANGEVPLKANELPILMKQMYQRKTDDHSQAAVMVLMISVK 112
CLSLSRGIDYALANGE P KANELP LMKQMYQRKTD+ S AAVMVLMISVK
Sbjct: 59 CLSLSRGIDYALANGEPPPKANELPTLMKQMYQRKTDELSLAAVMVLMISVK 110
>dbj|BAB11464.1| transcription factor-like protein [Arabidopsis thaliana]
Length = 719
Score = 81.3 bits (199), Expect = 1e-13
Identities = 41/96 (42%), Positives = 60/96 (61%)
Query: 17 AMTATANPVSPSLVNLYRITKVLERLATHFVPGNRSDAFEFFNLCLSLSRGIDYALANGE 76
A T + SLVN +R+ V +RL H G + D EF C+S ++GID+A+AN +
Sbjct: 11 AGTGLREKTAASLVNSFRLASVTQRLRYHIQDGAKVDPKEFQICCISFAKGIDFAIANND 70
Query: 77 VPLKANELPILMKQMYQRKTDDHSQAAVMVLMISVK 112
+P K E P L+KQ+ + TD +++ A+MVLMISVK
Sbjct: 71 IPKKVEEFPWLLKQLCRHGTDVYTKTALMVLMISVK 106
>ref|NP_198973.2| zinc finger (MIZ type) family protein [Arabidopsis thaliana]
Length = 703
Score = 81.3 bits (199), Expect = 1e-13
Identities = 41/96 (42%), Positives = 60/96 (61%)
Query: 17 AMTATANPVSPSLVNLYRITKVLERLATHFVPGNRSDAFEFFNLCLSLSRGIDYALANGE 76
A T + SLVN +R+ V +RL H G + D EF C+S ++GID+A+AN +
Sbjct: 11 AGTGLREKTAASLVNSFRLASVTQRLRYHIQDGAKVDPKEFQICCISFAKGIDFAIANND 70
Query: 77 VPLKANELPILMKQMYQRKTDDHSQAAVMVLMISVK 112
+P K E P L+KQ+ + TD +++ A+MVLMISVK
Sbjct: 71 IPKKVEEFPWLLKQLCRHGTDVYTKTALMVLMISVK 106
>dbj|BAD67847.1| putative transcription factor [Oryza sativa (japonica
cultivar-group)]
Length = 872
Score = 61.6 bits (148), Expect = 8e-08
Identities = 33/85 (38%), Positives = 55/85 (63%), Gaps = 2/85 (2%)
Query: 30 VNLYRITKVLERLATHFVPGNRS--DAFEFFNLCLSLSRGIDYALANGEVPLKANELPIL 87
+N R+ + +RL THF G + + + +L + +RGID+AL++G+VP A+E+P +
Sbjct: 34 MNARRLVMIGDRLRTHFRGGGGTVLEPPDLAHLVYAFARGIDFALSSGDVPTVASEIPSI 93
Query: 88 MKQMYQRKTDDHSQAAVMVLMISVK 112
+K++Y D Q++VMVLMIS K
Sbjct: 94 LKKVYLVGKDQFLQSSVMVLMISCK 118
>ref|NP_172366.2| zinc finger (MIZ type) family protein [Arabidopsis thaliana]
Length = 842
Score = 39.7 bits (91), Expect = 0.34
Identities = 21/46 (45%), Positives = 30/46 (64%), Gaps = 1/46 (2%)
Query: 68 IDYALANGEVPLKANELPILMKQMYQRKTDDH-SQAAVMVLMISVK 112
ID A+ EVP EL +++ + +RK DD+ ++A VM LMISVK
Sbjct: 44 IDAAIGRNEVPGNIQELALILNNVCRRKCDDYQTRAVVMALMISVK 89
Database: nr
Posted date: Jul 5, 2005 12:34 AM
Number of letters in database: 863,360,394
Number of sequences in database: 2,540,612
Lambda K H
0.357 0.160 0.557
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 970,347,483
Number of Sequences: 2540612
Number of extensions: 34130797
Number of successful extensions: 143936
Number of sequences better than 10.0: 5
Number of HSP's better than 10.0 without gapping: 4
Number of HSP's successfully gapped in prelim test: 1
Number of HSP's that attempted gapping in prelim test: 143929
Number of HSP's gapped (non-prelim): 6
length of query: 695
length of database: 863,360,394
effective HSP length: 135
effective length of query: 560
effective length of database: 520,377,774
effective search space: 291411553440
effective search space used: 291411553440
T: 11
A: 40
X1: 14 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 37 (21.7 bits)
S2: 79 (35.0 bits)
Medicago: description of AC140913.1


