

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
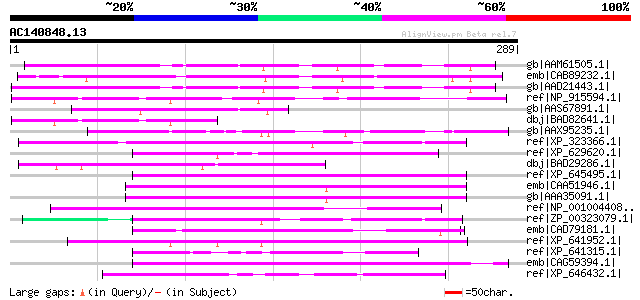
Query= AC140848.13 + phase: 0
(289 letters)
Database: nr
2,540,612 sequences; 863,360,394 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
gb|AAM61505.1| unknown [Arabidopsis thaliana] 170 5e-41
emb|CAB89232.1| putative protein [Arabidopsis thaliana] gi|20465... 169 6e-41
gb|AAD21443.1| expressed protein [Arabidopsis thaliana] gi|18491... 168 2e-40
ref|NP_915594.1| P0489B03.23 [Oryza sativa (japonica cultivar-gr... 86 2e-15
gb|AAS67891.1| calcium/calmodulin protein kinase [Nicotiana taba... 67 6e-10
dbj|BAD82641.1| unknown protein [Oryza sativa (japonica cultivar... 62 1e-08
gb|AAX95235.1| conserved hypothetical protein [Oryza sativa (jap... 60 9e-08
ref|XP_323366.1| predicted protein [Neurospora crassa] gi|289187... 54 4e-06
ref|XP_629620.1| NAD+ kinase family protein [Dictyostelium disco... 53 9e-06
dbj|BAD29286.1| unknown protein [Oryza sativa (japonica cultivar... 53 9e-06
ref|XP_645495.1| hypothetical protein DDB0168484 [Dictyostelium ... 53 1e-05
emb|CAA51946.1| unnamed protein product [Saccharomyces cerevisia... 51 4e-05
gb|AAA35091.1| selected as a weak suppressor of a mutant of the ... 51 4e-05
ref|NP_001004408.1| hypothetical protein LOC424547 [Gallus gallu... 50 7e-05
ref|ZP_00323079.1| hypothetical protein PpenA01001008 [Pediococc... 50 7e-05
emb|CAD79181.1| hypothetical protein-signal peptide prediction [... 49 2e-04
ref|XP_641952.1| hypothetical protein DDB0206180 [Dictyostelium ... 49 2e-04
ref|XP_641315.1| hypothetical protein DDB0206452 [Dictyostelium ... 49 2e-04
emb|CAG59394.1| unnamed protein product [Candida glabrata CBS138... 49 2e-04
ref|XP_646432.1| hypothetical protein DDB0190700 [Dictyostelium ... 48 4e-04
>gb|AAM61505.1| unknown [Arabidopsis thaliana]
Length = 263
Score = 170 bits (430), Expect = 5e-41
Identities = 118/277 (42%), Positives = 153/277 (54%), Gaps = 40/277 (14%)
Query: 9 SACAGGSTDRKIACETQADDLKTDPPPESSHHHP-DSPPESFWLSRDEENEWLDRNFIYE 67
S C GGS RKI CET ADD P +S P D PPES++LS D + EWL N ++
Sbjct: 12 SVCIGGSDHRKIVCETLADDSTIPPYYNNSAVSPSDFPPESYFLSNDAQLEWLSDNAFFD 71
Query: 68 RKESTKGNSSSTNLNPNSNSNSNSTSNSQRFANLKSKTSMIGLPKPQKPSFDAKNLRNHK 127
RK+S KGNS N NPNSN +SQRF LKSK S+IGLPKPQK F+ R H
Sbjct: 72 RKDSQKGNSGILNSNPNSN------PSSQRFL-LKSKASIIGLPKPQKTCFNEAKQRRHA 124
Query: 128 PSNIKLFPKRSASVGK---SFVEPSSPKVSCMGKVRSKRGKTSTVTAAAKAAVKEKTART 184
N ++ KR S K S +EPSSPKVSC+G+VRS+R + ++ ++K++R
Sbjct: 125 GKN-RVILKRVGSRIKTDISLLEPSSPKVSCIGRVRSRRER-------SRRMHRQKSSRV 176
Query: 185 E--RKQSKHSFFKSFRAIFRSGGRNKHDRMNDSSALDSSSITKNTNSVSDSKARDSTASL 242
E + K F SFRAIFR G K D SA ++ + T+NT+ +
Sbjct: 177 EPVNRVKKPGFMASFRAIFRIKGGCK-----DVSARETHTSTRNTHDIRSRLP------- 224
Query: 243 NDVSFVESVTRNSVSEGEP--PGLGVMMRFTSGRRSE 277
E+V ++ GEP PGLG M RF SGRR++
Sbjct: 225 -----AEAVEKSIFDGGEPVVPGLGGMTRFASGRRAD 256
>emb|CAB89232.1| putative protein [Arabidopsis thaliana] gi|20465291|gb|AAM20049.1|
unknown protein [Arabidopsis thaliana]
gi|18377716|gb|AAL67008.1| unknown protein [Arabidopsis
thaliana] gi|15231297|ref|NP_190839.1| expressed protein
[Arabidopsis thaliana] gi|11357760|pir||T49024
hypothetical protein F3C22.110 - Arabidopsis thaliana
Length = 289
Score = 169 bits (429), Expect = 6e-41
Identities = 125/302 (41%), Positives = 164/302 (53%), Gaps = 41/302 (13%)
Query: 5 ETLVSACAGGSTDRKIACETQADDLKTDPPPESSHHHP------DSPPESFWLSRDEENE 58
+ + SAC GGS DRKI+CET ADD + P +S P D PPES+ LS++ + E
Sbjct: 3 QVVASACTGGS-DRKISCETLADD--NEDSPHNSKIRPVSISAVDFPPESYSLSKEAQLE 59
Query: 59 WLDRNFIYERKESTKGNSSSTNLNPNSNSNSNSTSNSQRFANLKSKTSMIGLPKPQKPSF 118
WL+ N +ERKES KGNSS+ NPN+N NS+S S LKSK S+I LPKPQK F
Sbjct: 60 WLNDNAFFERKESQKGNSSAPISNPNTNPNSSSHRIS-----LKSKASIIRLPKPQKTCF 114
Query: 119 -DAKNLRNHKPSNIKLFPKRSASVGKS---FVEPSSPKVSCMGKVRSKRGKTSTVTAAAK 174
+AK RN + + + PKR S KS EP SPKVSC+G+VRSKR + ++
Sbjct: 115 NEAKKRRNCRIARTLMIPKRIGSRLKSDPTLSEPCSPKVSCIGRVRSKRDR-------SR 167
Query: 175 AAVKEKTARTERKQ------SKHSFFKSFRAIFRSGGRNKHDRMNDSSALDSSSITKNTN 228
++K+ RT + K FF SFRAIFR+GG K D + + +
Sbjct: 168 RMQRQKSGRTNSFKDKPVPVKKPGFFASFRAIFRTGGGCK-DLSASGAHAPRRDVVVSPP 226
Query: 229 SVSDSKARDSTASLNDVSFVESV-------TRNSVSEGEP--PGLGVMMRFTSGRRSESW 279
VS ++ D L +S +R S+ GEP PGLG M RFTSGRR +
Sbjct: 227 RVSVRRSTDIRGRLPPGDVGKSSPQRNSTGSRRSIDGGEPVLPGLGGMTRFTSGRRPDLL 286
Query: 280 VD 281
V+
Sbjct: 287 VE 288
>gb|AAD21443.1| expressed protein [Arabidopsis thaliana] gi|18491123|gb|AAL69530.1|
At2g36220/F2H17.17 [Arabidopsis thaliana]
gi|15450733|gb|AAK96638.1| At2g36220/F2H17.17
[Arabidopsis thaliana] gi|25408452|pir||B84778
hypothetical protein At2g36220 [imported] - Arabidopsis
thaliana gi|18404079|ref|NP_565838.1| expressed protein
[Arabidopsis thaliana]
Length = 263
Score = 168 bits (425), Expect = 2e-40
Identities = 119/285 (41%), Positives = 157/285 (54%), Gaps = 41/285 (14%)
Query: 2 LQVETLVSA-CAGGSTDRKIACETQADDLKTDPPPESSHHHP-DSPPESFWLSRDEENEW 59
+ +E VS+ C GGS RKI CET ADD P +S P D PPES++LS D + EW
Sbjct: 4 VDIEAFVSSVCIGGSDHRKIVCETLADDSTIPPYYNNSAVSPSDFPPESYFLSNDAQLEW 63
Query: 60 LDRNFIYERKESTKGNSSSTNLNPNSNSNSNSTSNSQRFANLKSKTSMIGLPKPQKPSFD 119
L N ++RK+S KGNS N NPNSN +SQRF LKSK S+IGLPKPQK F+
Sbjct: 64 LSDNAFFDRKDSQKGNSGILNSNPNSN------PSSQRFL-LKSKASIIGLPKPQKTCFN 116
Query: 120 AKNLRNHKPSNIKLFPKRSASVGK---SFVEPSSPKVSCMGKVRSKRGKTSTVTAAAKAA 176
R H N ++ KR S K S +EPSSPKVSC+G+VRS+R + ++
Sbjct: 117 EAKQRRHAGKN-RVILKRVGSRIKTDISLLEPSSPKVSCIGRVRSRRER-------SRRM 168
Query: 177 VKEKTARTE--RKQSKHSFFKSFRAIFRSGGRNKHDRMNDSSALDSSSITKNTNSVSDSK 234
++K++R E + K F SFRAIFR G K D SA ++ + T+NT+ +
Sbjct: 169 HRQKSSRVEPVNRVKKPGFMASFRAIFRIKGGCK-----DVSARETHTSTRNTHDIRSRL 223
Query: 235 ARDSTASLNDVSFVESVTRNSVSEGEP--PGLGVMMRFTSGRRSE 277
E+ ++ GEP PGLG M RF SGRR++
Sbjct: 224 P------------AEADEKSVFDGGEPVVPGLGGMTRFASGRRAD 256
>ref|NP_915594.1| P0489B03.23 [Oryza sativa (japonica cultivar-group)]
Length = 300
Score = 85.5 bits (210), Expect = 2e-15
Identities = 88/308 (28%), Positives = 131/308 (41%), Gaps = 81/308 (26%)
Query: 2 LQVETLVSACAG-GSTDRKIACET--------QADDLKTDPPPESSHHHPDSPPESFWLS 52
+ +E+LV AG G+ DRK++CET A + PPP PD PPES +
Sbjct: 4 VDLESLVCGVAGAGAGDRKVSCETVIAAGESGDASPPRMPPPPPPPD--PDFPPESITIP 61
Query: 53 RDEENEWLDRNFIYERKESTKGNSSSTNLNPNSNSNSN-----STSNSQRFANL-KSKTS 106
+E + + N IY+R +STKG STN + ++SN S SNS R A + T+
Sbjct: 62 IGDEVAFSELNPIYDRDDSTKG---STNPKSAAGASSNPIPAKSRSNSTRIAGAPAAATT 118
Query: 107 MIGLPKPQKPSFDAKNLRNHKPSNIKLFP-KRSAS----------VGKSFVEPSSPKVSC 155
GLP +P+F + +PS ++ P KRS S G EP SPKVSC
Sbjct: 119 FFGLPASIRPAFTRR-----RPSQGRILPDKRSGSRGGGGGGSSRRGDGEEEPRSPKVSC 173
Query: 156 MGKVRSKRGKTSTVTAAAKAAVKEKTARTERKQSKHSFFKSFRAIFRSGGRNKHDRMNDS 215
+GKV S R E+ R+ ++ +++ A+ GG
Sbjct: 174 IGKVLSDR---------------ERYGRSRGRR----WWRGLVAVLLCGGGCSCQGGGRR 214
Query: 216 SALDSSSITKNTNSVSDSKARDSTASLNDVSFVESVTRNSVSEGEPPGLGVMMRFTSGRR 275
A ++ ++ + D K G+ M RF SGRR
Sbjct: 215 HARKKVALDEDHHDGDDDK--------------------------QAGIAAMRRFKSGRR 248
Query: 276 SESWVDDS 283
+ SWV+++
Sbjct: 249 TASWVEEA 256
>gb|AAS67891.1| calcium/calmodulin protein kinase [Nicotiana tabacum]
gi|25053813|gb|AAN71903.1| calcium/calmodulin protein
kinase 1 [Nicotiana tabacum]
Length = 1415
Score = 67.0 bits (162), Expect = 6e-10
Identities = 45/129 (34%), Positives = 60/129 (45%), Gaps = 6/129 (4%)
Query: 36 ESSHHHPDSPPESFWLSRDEENEWLDRNFIYERKESTK--GNSSSTNLNPNSNSNSNSTS 93
+S + D PPESFW+ +D E +W D N +R S G N + S S+ + TS
Sbjct: 30 KSPENFLDLPPESFWIPKDSEQDWFDENATIQRMTSMMKLGFFGKANHHSKSFSHRSFTS 89
Query: 94 NSQRFANLKSKTSMIGLPKPQKPSFDAKNLRNHKPSNIKLFPKRSASVGKSF---VEPSS 150
TS+ LP+ +K S NL+ K LF RS K EP S
Sbjct: 90 TLFNHHQKPKSTSLFALPQSKKTSSTEGNLKQKKVPK-SLFRSRSEPGRKGIRHVREPGS 148
Query: 151 PKVSCMGKV 159
PKVSC+G+V
Sbjct: 149 PKVSCIGRV 157
>dbj|BAD82641.1| unknown protein [Oryza sativa (japonica cultivar-group)]
gi|56784813|dbj|BAD82034.1| unknown protein [Oryza
sativa (japonica cultivar-group)]
Length = 249
Score = 62.4 bits (150), Expect = 1e-08
Identities = 48/132 (36%), Positives = 68/132 (51%), Gaps = 20/132 (15%)
Query: 2 LQVETLVSACAG-GSTDRKIACET--------QADDLKTDPPPESSHHHPDSPPESFWLS 52
+ +E+LV AG G+ DRK++CET A + PPP PD PPES +
Sbjct: 4 VDLESLVCGVAGAGAGDRKVSCETVIAAGESGDASPPRMPPPPPPPD--PDFPPESITIP 61
Query: 53 RDEENEWLDRNFIYERKESTKGNSSSTNLNPNSNSNSN-----STSNSQRFANL-KSKTS 106
+E + + N IY+R +STKG STN + ++SN S SNS R A + T+
Sbjct: 62 IGDEVAFSELNPIYDRDDSTKG---STNPKSAAGASSNPIPAKSRSNSTRIAGAPAAATT 118
Query: 107 MIGLPKPQKPSF 118
GLP +P+F
Sbjct: 119 FFGLPASIRPAF 130
>gb|AAX95235.1| conserved hypothetical protein [Oryza sativa (japonica
cultivar-group)] gi|62732731|gb|AAX94850.1| expressed
protein [Oryza sativa (japonica cultivar-group)]
Length = 241
Score = 59.7 bits (143), Expect = 9e-08
Identities = 73/255 (28%), Positives = 110/255 (42%), Gaps = 51/255 (20%)
Query: 45 PPESFWLSRDEENEWLDRNFIYERKESTKGNSSSTNLNPNSNSNSNSTSNSQRFA-NLKS 103
PPES L ++ W D +Y+R +S K N++ + N +++ SQRF+ NLK
Sbjct: 12 PPESTRLRIGDDIAWSDVGGVYDRDDSLKENTNPKCILKNHLPGAHN-GGSQRFSGNLKP 70
Query: 104 KTS-MIGLPKPQKPSFDAKNLRNHKPSNIKLFPKRSASVG------KSFV---EPSSPKV 153
+ +IG+ K KN R+H P+ +FPK+ A G K+ V EP+SPKV
Sbjct: 71 TAAPIIGISG--KLGQGGKN-RHHPPA---MFPKKVAVTGGGGRNPKAAVPEHEPTSPKV 124
Query: 154 SCMGKVRSKRGKTSTVTAAAKAAVKEKTARTERKQSK----HSFFKSFRAIFRSGGRNKH 209
SC+GKV S R E+ R R + +FR R+
Sbjct: 125 SCIGKVLSDR---------------ERARRGRRPAGRMVPAGGCCPGLGGLFR---RSHS 166
Query: 210 DRMNDSSALDSSSITKNTNSVSDSKARDSTASLNDVSFVESVTRNSVSEGEPPGLGVMMR 269
+ N +D S + + ++ V+ T + + PGLG MMR
Sbjct: 167 RKKNAVECVDQSPPPLPPWASRRGEPKE----------VKEAT-PAAAAAMAPGLGGMMR 215
Query: 270 FTSGRRSESWVDDSE 284
F SGRR+ W + E
Sbjct: 216 FASGRRAADWATEME 230
>ref|XP_323366.1| predicted protein [Neurospora crassa] gi|28918756|gb|EAA28426.1|
predicted protein [Neurospora crassa]
Length = 471
Score = 54.3 bits (129), Expect = 4e-06
Identities = 64/261 (24%), Positives = 114/261 (43%), Gaps = 16/261 (6%)
Query: 6 TLVSACAGGSTDRKIACETQADDLKTDPPPESSHHHPDSPPESFWLSRDEENEWLDRNFI 65
+LVS K A T DD +D SS S +S S DE+ +
Sbjct: 62 SLVSIFQTWEAKPKQAGSTSGDDSDSDDESSSSSESESSDSDSDSDSEDEKKKEAPA--- 118
Query: 66 YERKESTKGNSSSTNLNPNSNSNSNSTSNSQRFANLKSKTSMIGLPKPQKPSFDAKNLRN 125
++ ES+ +SSS++ + +S+S+S+S+ + + + + K + S + + +
Sbjct: 119 -KKAESSSSSSSSSDSSSDSSSDSSSSEDEAKKTKAAAPATNPLKRKAESSSESSSSSSD 177
Query: 126 HKPSNIKLFPKRSASVGKSFVEPSSPKVSCMGKVRSKRGKTSTVTA------AAKAAVKE 179
S+ + K V SS S S +S+ + AAKAA +
Sbjct: 178 SSSSDSSSSEDEAPKAKKQKVAESSSSSSESSSDSSSDSSSSSESEDDKKKPAAKAASPK 237
Query: 180 KTARTERKQSKHSFFKSFRAIFRSGGRNKHDRMNDSSALDSSSITKNTNSVSDSKARDST 239
K A ++++S S S + + +DSS+ DSSS + +++S SDS + DS+
Sbjct: 238 KAAAAKKEESSDSSSSD-----SSSSDSSSESSSDSSSSDSSSDSDSSSSDSDSSS-DSS 291
Query: 240 ASLNDVSFVESVTRNSVSEGE 260
+ + S +S + +S SE E
Sbjct: 292 SDSDSSSDSDSDSDSSESEAE 312
Score = 37.0 bits (84), Expect = 0.65
Identities = 50/254 (19%), Positives = 95/254 (36%), Gaps = 17/254 (6%)
Query: 30 KTDPPPESSHHHPDSPPESFWLSRDEENEWLDRNFIYERKESTKGNSSSTNLNPNSNSNS 89
K + ESS DS S DE + K+ SSS++ +S+S+S
Sbjct: 164 KAESSSESSSSSSDSSSSDSSSSEDEAPK---------AKKQKVAESSSSSSESSSDSSS 214
Query: 90 NSTSNSQRFANLKSKTSMIGLPKPQKPSFDAKNLRNHKPSNIKLFPKRSASVGKSFVEPS 149
+S+S+S+ + K + PK + ++ + + +S S + S
Sbjct: 215 DSSSSSESEDDKKKPAAKAASPKKAAAAKKEESSDSSSSDSSSSDSSSESSSDSSSSDSS 274
Query: 150 SPKVSCMGKVRSKRGKTSTVTAAAKAAVKEKTARTE--RKQSKHSFFKSFRAIFRSGGRN 207
S S S +S +++ + ++ +E +K+ K K+ ++ S +
Sbjct: 275 SDSDSSSSDSDSSSDSSSDSDSSSDSDSDSDSSESEAEKKEVKKEAKKATKSDSSSSDSS 334
Query: 208 KHDRMNDSSALDSSSITKNTNSVSDSKARDSTASL------NDVSFVESVTRNSVSEGEP 261
D +DSS+ + + + DS+A+L V E T N P
Sbjct: 335 SSDSSSDSSSDSEDEKKTDKKKAASTNGSDSSATLKASTESTPVPTTEEKTNNRGKANGP 394
Query: 262 PGLGVMMRFTSGRR 275
P + F+ R+
Sbjct: 395 PPKKEIQPFSRVRK 408
Score = 35.0 bits (79), Expect = 2.5
Identities = 27/122 (22%), Positives = 50/122 (40%)
Query: 136 KRSASVGKSFVEPSSPKVSCMGKVRSKRGKTSTVTAAAKAAVKEKTARTERKQSKHSFFK 195
K+S K+ +PS+ ++ + K ++ T K K + S S F+
Sbjct: 9 KKSGDKKKAVAQPSAQLLAAVEKFLTENNFGDAATEFTKQITKNGWEKKSSDASLVSIFQ 68
Query: 196 SFRAIFRSGGRNKHDRMNDSSALDSSSITKNTNSVSDSKARDSTASLNDVSFVESVTRNS 255
++ A + G D + SSS +++++S SDS + D ES + +S
Sbjct: 69 TWEAKPKQAGSTSGDDSDSDDESSSSSESESSDSDSDSDSEDEKKKEAPAKKAESSSSSS 128
Query: 256 VS 257
S
Sbjct: 129 SS 130
>ref|XP_629620.1| NAD+ kinase family protein [Dictyostelium discoideum]
gi|60463004|gb|EAL61200.1| NAD+ kinase family protein
[Dictyostelium discoideum]
Length = 857
Score = 53.1 bits (126), Expect = 9e-06
Identities = 43/177 (24%), Positives = 84/177 (47%), Gaps = 7/177 (3%)
Query: 71 STKGNSSSTNLNPNSNSNSNSTSNSQRFANLKSKTSMIGLPKPQKPS--FDAKNLRNHKP 128
+ N+++ N N N+N+N+N+ +N+ +L SK +I LP Q+ S + N+ N+KP
Sbjct: 142 NNNNNNNNNNNNNNNNNNNNNNNNNSSPLSLYSKKELIPLPAIQEESNNYQNNNITNNKP 201
Query: 129 SNIKLFPKRSASVGKSFVEPSSPKVSCMGKVRSKRGKTSTVTAAAKAAVKEKTARTERKQ 188
N+K+ + + EP+ VS S + ++ + + + T +K+
Sbjct: 202 -NLKVTLNNN---NNTTTEPTKRIVSTSSSSSSDDEEPIPISPVSNSFLSPLNFTTGKKK 257
Query: 189 SKHSFFKSFRAIFRSGG-RNKHDRMNDSSALDSSSITKNTNSVSDSKARDSTASLND 244
SK + F + S NK N+ D+++I N N S++ + + +S +D
Sbjct: 258 SKKNQFYYVSTYYSSCDLPNKVMDENEIEEEDNNNIISNNNKASNNNKKSNFSSHDD 314
>dbj|BAD29286.1| unknown protein [Oryza sativa (japonica cultivar-group)]
gi|50251404|dbj|BAD28431.1| unknown protein [Oryza
sativa (japonica cultivar-group)]
Length = 214
Score = 53.1 bits (126), Expect = 9e-06
Identities = 49/188 (26%), Positives = 80/188 (42%), Gaps = 14/188 (7%)
Query: 6 TLVSACAGGSTDRKIACETQ---ADDLKTDPPPESSH--------HHPDSPPESFWLSRD 54
TL S GG + + CE + +DD D S + P SP ES WL
Sbjct: 5 TLDSLLHGGGGEPEDECEDEFSGSDDDGEDGGGGSEEWGGDVDGEYDPYSPAESLWLRIG 64
Query: 55 EENEWLDRNFIYERKESTKGNSSSTNLNPNSNSNSNSTSNSQRFANLKSKTSMI--GLPK 112
E+ +W + + ER++STKG S+ + S + + + + +++ GLP
Sbjct: 65 EDIDWSEVGAVLEREDSTKGASNPKSAAACSCAGAPAARMPTCAGGGGTAKAVVIAGLPA 124
Query: 113 PQKPSFDAKNLRNHKPSNIKLFPKRSASVGKSFVEPSSPKVSCMGKVRSKRGKTSTVTAA 172
+ + ++ R + + + A EP SPKVSC+G VRS+ A
Sbjct: 125 AARKA-SREHERRRRLGRARARARVFAGDAVEVAEPGSPKVSCLGGVRSRARAQQPCCPA 183
Query: 173 AKAAVKEK 180
A AA + +
Sbjct: 184 AAAAGRRR 191
>ref|XP_645495.1| hypothetical protein DDB0168484 [Dictyostelium discoideum]
gi|42734036|gb|AAS38911.1| similar to Leishmania major.
Ppg3 [Dictyostelium discoideum]
gi|60473584|gb|EAL71525.1| hypothetical protein
DDB0168484 [Dictyostelium discoideum]
Length = 374
Score = 52.8 bits (125), Expect = 1e-05
Identities = 42/190 (22%), Positives = 86/190 (45%)
Query: 71 STKGNSSSTNLNPNSNSNSNSTSNSQRFANLKSKTSMIGLPKPQKPSFDAKNLRNHKPSN 130
S+ +SSS++ + +S+S+S+S+S+S ++ S +S S + + + S+
Sbjct: 184 SSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSS 243
Query: 131 IKLFPKRSASVGKSFVEPSSPKVSCMGKVRSKRGKTSTVTAAAKAAVKEKTARTERKQSK 190
S+S S SS S S +S+ ++++ ++ ++ + S
Sbjct: 244 SSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSS 303
Query: 191 HSFFKSFRAIFRSGGRNKHDRMNDSSALDSSSITKNTNSVSDSKARDSTASLNDVSFVES 250
S S + S + + SS+ SSS + +++S S S + S++S + S S
Sbjct: 304 SSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSS 363
Query: 251 VTRNSVSEGE 260
+ +S S GE
Sbjct: 364 SSSSSSSSGE 373
>emb|CAA51946.1| unnamed protein product [Saccharomyces cerevisiae]
gi|6322945|ref|NP_013018.1| Nucleolar, serine-rich
protein with a role in preribosome assembly or
transport; may function as a chaperone of small
nucleolar ribonucleoprotein particles (snoRNPs);
immunologically and structurally to rat Nopp140; Srp40p
[Saccharomyces cerevisiae] gi|486581|emb|CAA82171.1|
SRP40 [Saccharomyces cerevisiae]
gi|548976|sp|P32583|SRP40_YEAST Suppressor protein SRP40
Length = 406
Score = 50.8 bits (120), Expect = 4e-05
Identities = 48/206 (23%), Positives = 92/206 (44%), Gaps = 12/206 (5%)
Query: 67 ERKESTKGNSSSTNLNPNSNSNSNSTSNSQRFANLKSKTSMIGLPKPQKPSFDAKNLRNH 126
+ KE + +SSS++ + +S+S+S+S+S+S + S +S S D+ + +
Sbjct: 17 KEKEIEEKSSSSSSSSSSSSSSSSSSSSSSSSSGESSSSSSSSSSSSSSDSSDSSDSESS 76
Query: 127 KPSNIKLFPKRSASVGKSFVEPSSPKVSCMGKVRSKRGKTSTVTAAAKAAVKE------- 179
S+ S+S +S E S S ++S+ + + K
Sbjct: 77 SSSSSSSSSSSSSSDSESSSESDSSSSGSSSSSSSSSDESSSESESEDETKKRARESDNE 136
Query: 180 -----KTARTERKQSKHSFFKSFRAIFRSGGRNKHDRMNDSSALDSSSITKNTNSVSDSK 234
K A+TE + S S S + S + + +DSS+ SSS ++S SDS+
Sbjct: 137 DAKETKKAKTEPESSSSSESSSSGSSSSSESESGSESDSDSSSSSSSSSDSESDSESDSQ 196
Query: 235 ARDSTASLNDVSFVESVTRNSVSEGE 260
+ S++S + S +S + +S S+ +
Sbjct: 197 SSSSSSSSDSSSDSDSSSSDSSSDSD 222
Score = 40.4 bits (93), Expect = 0.059
Identities = 43/192 (22%), Positives = 81/192 (41%), Gaps = 4/192 (2%)
Query: 67 ERKESTKGNSSSTNLNPNSNSNSNSTSNSQRFANLKSKTSMIGLPKPQKPSFDAKNLRNH 126
E S+ +SSS++ + +S S+S S S+S ++ S +S + + K R
Sbjct: 74 ESSSSSSSSSSSSSSSSDSESSSESDSSSSGSSSSSSSSSDESSSESESEDETKKRARES 133
Query: 127 KPSNIKLFPKRSASVGKSFVEPSSPKVSCMGKVRSKRGKTSTVTAAAKAAVKEKTARTER 186
+ K K S SS + S G S ++ + + + ++ ++ +E
Sbjct: 134 DNEDAKETKKAKTEPESS----SSSESSSSGSSSSSESESGSESDSDSSSSSSSSSDSES 189
Query: 187 KQSKHSFFKSFRAIFRSGGRNKHDRMNDSSALDSSSITKNTNSVSDSKARDSTASLNDVS 246
S S + S + + SS DSSS + +++S SDS + S+ S + S
Sbjct: 190 DSESDSQSSSSSSSSDSSSDSDSSSSDSSSDSDSSSSSSSSSSDSDSDSDSSSDSDSSGS 249
Query: 247 FVESVTRNSVSE 258
S + +S S+
Sbjct: 250 SDSSSSSDSSSD 261
Score = 40.0 bits (92), Expect = 0.077
Identities = 51/245 (20%), Positives = 95/245 (37%), Gaps = 20/245 (8%)
Query: 17 DRKIACETQADDLKTDPPPESSHHHPDSPPESFWLSRDEENEWLDRNFIYERKESTKGNS 76
+ K + + + + SS S S S + D + + + S+ +S
Sbjct: 22 EEKSSSSSSSSSSSSSSSSSSSSSSSSSGESSSSSSSSSSSSSSDSSDSSDSESSSSSSS 81
Query: 77 SSTNLNPNSNSNSNSTSNSQRFANLKSKTSMIGLPKPQKPSFDAKNLRNHKPSN------ 130
SS++ + +S+S S+S S+S + S +S + S D R + N
Sbjct: 82 SSSSSSSSSDSESSSESDSSSSGSSSSSSSSSDESSSESESEDETKKRARESDNEDAKET 141
Query: 131 --IKLFPKRSASVGKSFVEPSSPKVSCMGKVRSKRGKTSTVTAAAKAAVKEKTARTERKQ 188
K P+ S+S S SS S G +S+ +++ + E +++
Sbjct: 142 KKAKTEPESSSSSESSSSGSSSSSESESGSESDSDSSSSSSSSSDSESDSESDSQSSSSS 201
Query: 189 SKH------------SFFKSFRAIFRSGGRNKHDRMNDSSALDSSSITKNTNSVSDSKAR 236
S S S + S + D +DSS+ SS + +++S SDS +
Sbjct: 202 SSSDSSSDSDSSSSDSSSDSDSSSSSSSSSSDSDSDSDSSSDSDSSGSSDSSSSSDSSSD 261
Query: 237 DSTAS 241
+ST+S
Sbjct: 262 ESTSS 266
Score = 39.7 bits (91), Expect = 0.10
Identities = 57/278 (20%), Positives = 116/278 (41%), Gaps = 16/278 (5%)
Query: 9 SACAGGSTDRKIACETQADDLKTDPPPESSHHHPDSPPESFWLSRDEENEWLDRNFIYER 68
S+ + S+D + + E+ + + SS S ES ++ E + + +
Sbjct: 83 SSSSSSSSDSESSSESDSSSSGSSSSSSSSSDESSSESESEDETKKRARESDNEDAKETK 142
Query: 69 KESTKGNSSSTNLNPNSNSNSNSTSNSQRFANLKSKTSMIGLPKPQKPSFDAKNLRNHKP 128
K T+ SSS++ + +S S+S+S S S ++ S +S S + + +
Sbjct: 143 KAKTEPESSSSSESSSSGSSSSSESESGSESDSDSSSS----------SSSSSDSESDSE 192
Query: 129 SNIKLFPKRSASVGKSFVEPSSPKVSCMGKVRSKRGKTSTVTAAAKAAVKEKTARTERKQ 188
S+ + S+S S + SS S S +S+ +++ + + ++ ++
Sbjct: 193 SDSQSSSSSSSSDSSSDSDSSSSDSSS----DSDSSSSSSSSSSDSDSDSDSSSDSDSSG 248
Query: 189 SKHSFFKSFRAIFRSGGRNKHDRMNDSSALDSSSITKNTNSVSDSKARDSTASLNDVSFV 248
S S S + S + D +DS + SS + + +SKA ++ AS N+ +
Sbjct: 249 SSDSSSSSDSSSDESTSSDSSDSDSDSDSGSSSELETKEATADESKAEETPASSNESTPS 308
Query: 249 ESVTRNSVSEGEPPGLGVMMRFTSGRRSESWVDDSEIH 286
S + ++ P G + R+ S VD S+I+
Sbjct: 309 ASSSSSANKLNIPAGTDEIKE--GQRKHFSRVDRSKIN 344
Score = 39.3 bits (90), Expect = 0.13
Identities = 47/247 (19%), Positives = 93/247 (37%), Gaps = 10/247 (4%)
Query: 9 SACAGGSTDRKIACETQADDLKTDPPPESSHHHPDSPPESFWLSRDEENEWLDRNFIYER 68
S+ + S+ + + + + + SS DS S D E+ +
Sbjct: 32 SSSSSSSSSSSSSSSSSSGESSSSSSSSSSSSSSDSSDSS-----DSESSSSSSSSSSSS 86
Query: 69 KESTKGNSSSTNLNPNSNSNSNSTSNSQRFANLKSKTSMIGLPKPQKPSFDAKNLRNHKP 128
S+ SSS + + +S S+S+S+S+S ++ + + DAK + K
Sbjct: 87 SSSSDSESSSESDSSSSGSSSSSSSSSDESSSESESEDETKKRARESDNEDAKETKKAKT 146
Query: 129 SNIKLFPKRSASVGKSFVEPSSPKVSCMGKVRSKRGKTSTVTAAAKAAVKEKTARTERKQ 188
P+ S+S S SS S G +S+ +++ + E +++
Sbjct: 147 E-----PESSSSSESSSSGSSSSSESESGSESDSDSSSSSSSSSDSESDSESDSQSSSSS 201
Query: 189 SKHSFFKSFRAIFRSGGRNKHDRMNDSSALDSSSITKNTNSVSDSKARDSTASLNDVSFV 248
S + + + SS+ S +++S SDS ++S +D S
Sbjct: 202 SSSDSSSDSDSSSSDSSSDSDSSSSSSSSSSDSDSDSDSSSDSDSSGSSDSSSSSDSSSD 261
Query: 249 ESVTRNS 255
ES + +S
Sbjct: 262 ESTSSDS 268
Score = 35.4 bits (80), Expect = 1.9
Identities = 32/120 (26%), Positives = 50/120 (41%), Gaps = 2/120 (1%)
Query: 165 KTSTVTAAAKAAVKEKTARTERKQSKHSFFKSFRAIFRSGGRNKHDRMNDSSALDSSSIT 224
K V K +VKEK E K S S S + S + +SS+ SSS +
Sbjct: 4 KKIKVDEVPKLSVKEK--EIEEKSSSSSSSSSSSSSSSSSSSSSSSSSGESSSSSSSSSS 61
Query: 225 KNTNSVSDSKARDSTASLNDVSFVESVTRNSVSEGEPPGLGVMMRFTSGRRSESWVDDSE 284
+++ SDS +S++S + S S + +S S E +S S+ +SE
Sbjct: 62 SSSSDSSDSSDSESSSSSSSSSSSSSSSSDSESSSESDSSSSGSSSSSSSSSDESSSESE 121
Score = 34.7 bits (78), Expect = 3.2
Identities = 31/147 (21%), Positives = 59/147 (40%)
Query: 132 KLFPKRSASVGKSFVEPSSPKVSCMGKVRSKRGKTSTVTAAAKAAVKEKTARTERKQSKH 191
K ++S+S S SS S S +S+ ++++ ++ + + ++ + S
Sbjct: 19 KEIEEKSSSSSSSSSSSSSSSSSSSSSSSSSGESSSSSSSSSSSSSSDSSDSSDSESSSS 78
Query: 192 SFFKSFRAIFRSGGRNKHDRMNDSSALDSSSITKNTNSVSDSKARDSTASLNDVSFVESV 251
S S + S + + + SS SSS + + S S+S++ D T S E
Sbjct: 79 SSSSSSSSSSSSDSESSSESDSSSSGSSSSSSSSSDESSSESESEDETKKRARESDNEDA 138
Query: 252 TRNSVSEGEPPGLGVMMRFTSGRRSES 278
++ EP +SG S S
Sbjct: 139 KETKKAKTEPESSSSSESSSSGSSSSS 165
>gb|AAA35091.1| selected as a weak suppressor of a mutant of the subunit AC40 of
DNA dependant RNA polymerase I and III
Length = 406
Score = 50.8 bits (120), Expect = 4e-05
Identities = 48/206 (23%), Positives = 92/206 (44%), Gaps = 12/206 (5%)
Query: 67 ERKESTKGNSSSTNLNPNSNSNSNSTSNSQRFANLKSKTSMIGLPKPQKPSFDAKNLRNH 126
+ KE + +SSS++ + +S+S+S+S+S+S + S +S S D+ + +
Sbjct: 17 KEKEIEEKSSSSSSSSSSSSSSSSSSSSSSSSSGESSSSSSSSSSSSSSDSSDSSDSESS 76
Query: 127 KPSNIKLFPKRSASVGKSFVEPSSPKVSCMGKVRSKRGKTSTVTAAAKAAVKE------- 179
S+ S+S +S E S S ++S+ + + K
Sbjct: 77 SSSSSSSSSSSSSSDSESSSESDSSSSGSSSSSSSSSDESSSESESEDETKKRARESDNE 136
Query: 180 -----KTARTERKQSKHSFFKSFRAIFRSGGRNKHDRMNDSSALDSSSITKNTNSVSDSK 234
K A+TE + S S S + S + + +DSS+ SSS ++S SDS+
Sbjct: 137 DAKETKKAKTEPESSSSSESSSSGSSSSSESESGSESDSDSSSSSSSSSDSESDSESDSQ 196
Query: 235 ARDSTASLNDVSFVESVTRNSVSEGE 260
+ S++S + S +S + +S S+ +
Sbjct: 197 SSSSSSSSDSSSDSDSSSSDSSSDSD 222
Score = 40.4 bits (93), Expect = 0.059
Identities = 43/192 (22%), Positives = 81/192 (41%), Gaps = 4/192 (2%)
Query: 67 ERKESTKGNSSSTNLNPNSNSNSNSTSNSQRFANLKSKTSMIGLPKPQKPSFDAKNLRNH 126
E S+ +SSS++ + +S S+S S S+S ++ S +S + + K R
Sbjct: 74 ESSSSSSSSSSSSSSSSDSESSSESDSSSSGSSSSSSSSSDESSSESESEDETKKRARES 133
Query: 127 KPSNIKLFPKRSASVGKSFVEPSSPKVSCMGKVRSKRGKTSTVTAAAKAAVKEKTARTER 186
+ K K S SS + S G S ++ + + + ++ ++ +E
Sbjct: 134 DNEDAKETKKAKTEPESS----SSSESSSSGSSSSSESESGSESDSDSSSSSSSSSDSES 189
Query: 187 KQSKHSFFKSFRAIFRSGGRNKHDRMNDSSALDSSSITKNTNSVSDSKARDSTASLNDVS 246
S S + S + + SS DSSS + +++S SDS + S+ S + S
Sbjct: 190 DSESDSQSSSSSSSSDSSSDSDSSSSDSSSDSDSSSSSSSSSSDSDSDSDSSSDSDSSGS 249
Query: 247 FVESVTRNSVSE 258
S + +S S+
Sbjct: 250 SDSSSSSDSSSD 261
Score = 40.0 bits (92), Expect = 0.077
Identities = 51/245 (20%), Positives = 95/245 (37%), Gaps = 20/245 (8%)
Query: 17 DRKIACETQADDLKTDPPPESSHHHPDSPPESFWLSRDEENEWLDRNFIYERKESTKGNS 76
+ K + + + + SS S S S + D + + + S+ +S
Sbjct: 22 EEKSSSSSSSSSSSSSSSSSSSSSSSSSGESSSSSSSSSSSSSSDSSDSSDSESSSSSSS 81
Query: 77 SSTNLNPNSNSNSNSTSNSQRFANLKSKTSMIGLPKPQKPSFDAKNLRNHKPSN------ 130
SS++ + +S+S S+S S+S + S +S + S D R + N
Sbjct: 82 SSSSSSSSSDSESSSESDSSSSGSSSSSSSSSDESSSESESEDETKKRARESDNEDAKET 141
Query: 131 --IKLFPKRSASVGKSFVEPSSPKVSCMGKVRSKRGKTSTVTAAAKAAVKEKTARTERKQ 188
K P+ S+S S SS S G +S+ +++ + E +++
Sbjct: 142 KKAKTEPESSSSSESSSSGSSSSSESESGSESDSDSSSSSSSSSDSESDSESDSQSSSSS 201
Query: 189 SKH------------SFFKSFRAIFRSGGRNKHDRMNDSSALDSSSITKNTNSVSDSKAR 236
S S S + S + D +DSS+ SS + +++S SDS +
Sbjct: 202 SSSDSSSDSDSSSSDSSSDSDSSSSSSSSSSDSDSDSDSSSDSDSSGSSDSSSSSDSSSD 261
Query: 237 DSTAS 241
+ST+S
Sbjct: 262 ESTSS 266
Score = 39.7 bits (91), Expect = 0.10
Identities = 57/278 (20%), Positives = 116/278 (41%), Gaps = 16/278 (5%)
Query: 9 SACAGGSTDRKIACETQADDLKTDPPPESSHHHPDSPPESFWLSRDEENEWLDRNFIYER 68
S+ + S+D + + E+ + + SS S ES ++ E + + +
Sbjct: 83 SSSSSSSSDSESSSESDSSSSGSSSSSSSSSDESSSESESEDETKKRARESDNEDAKETK 142
Query: 69 KESTKGNSSSTNLNPNSNSNSNSTSNSQRFANLKSKTSMIGLPKPQKPSFDAKNLRNHKP 128
K T+ SSS++ + +S S+S+S S S ++ S +S S + + +
Sbjct: 143 KAKTEPESSSSSESSSSGSSSSSESESGSESDSDSSSS----------SSSSSDSESDSE 192
Query: 129 SNIKLFPKRSASVGKSFVEPSSPKVSCMGKVRSKRGKTSTVTAAAKAAVKEKTARTERKQ 188
S+ + S+S S + SS S S +S+ +++ + + ++ ++
Sbjct: 193 SDSQSSSSSSSSDSSSDSDSSSSDSSS----DSDSSSSSSSSSSDSDSDSDSSSDSDSSG 248
Query: 189 SKHSFFKSFRAIFRSGGRNKHDRMNDSSALDSSSITKNTNSVSDSKARDSTASLNDVSFV 248
S S S + S + D +DS + SS + + +SKA ++ AS N+ +
Sbjct: 249 SSDSSSSSDSSSDESTSSDSSDSDSDSDSGSSSELETKEATADESKAEETPASSNESTPS 308
Query: 249 ESVTRNSVSEGEPPGLGVMMRFTSGRRSESWVDDSEIH 286
S + ++ P G + R+ S VD S+I+
Sbjct: 309 ASSSSSANKLNIPAGTDEIKE--GQRKHFSRVDRSKIN 344
Score = 39.3 bits (90), Expect = 0.13
Identities = 47/247 (19%), Positives = 93/247 (37%), Gaps = 10/247 (4%)
Query: 9 SACAGGSTDRKIACETQADDLKTDPPPESSHHHPDSPPESFWLSRDEENEWLDRNFIYER 68
S+ + S+ + + + + + SS DS S D E+ +
Sbjct: 32 SSSSSSSSSSSSSSSSSSGESSSSSSSSSSSSSSDSSDSS-----DSESSSSSSSSSSSS 86
Query: 69 KESTKGNSSSTNLNPNSNSNSNSTSNSQRFANLKSKTSMIGLPKPQKPSFDAKNLRNHKP 128
S+ SSS + + +S S+S+S+S+S ++ + + DAK + K
Sbjct: 87 SSSSDSESSSESDSSSSGSSSSSSSSSDESSSESESEDETKKRARESDNEDAKETKKAKT 146
Query: 129 SNIKLFPKRSASVGKSFVEPSSPKVSCMGKVRSKRGKTSTVTAAAKAAVKEKTARTERKQ 188
P+ S+S S SS S G +S+ +++ + E +++
Sbjct: 147 E-----PESSSSSESSSSGSSSSSESESGSESDSDSSSSSSSSSDSESDSESDSQSSSSS 201
Query: 189 SKHSFFKSFRAIFRSGGRNKHDRMNDSSALDSSSITKNTNSVSDSKARDSTASLNDVSFV 248
S + + + SS+ S +++S SDS ++S +D S
Sbjct: 202 SSSDSSSDSDSSSSDSSSDSDSSSSSSSSSSDSDSDSDSSSDSDSSGSSDSSSSSDSSSD 261
Query: 249 ESVTRNS 255
ES + +S
Sbjct: 262 ESTSSDS 268
Score = 35.4 bits (80), Expect = 1.9
Identities = 32/120 (26%), Positives = 50/120 (41%), Gaps = 2/120 (1%)
Query: 165 KTSTVTAAAKAAVKEKTARTERKQSKHSFFKSFRAIFRSGGRNKHDRMNDSSALDSSSIT 224
K V K +VKEK E K S S S + S + +SS+ SSS +
Sbjct: 4 KKIKVDEVPKLSVKEK--EIEEKSSSSSSSSSSSSSSSSSSSSSSSSSGESSSSSSSSSS 61
Query: 225 KNTNSVSDSKARDSTASLNDVSFVESVTRNSVSEGEPPGLGVMMRFTSGRRSESWVDDSE 284
+++ SDS +S++S + S S + +S S E +S S+ +SE
Sbjct: 62 SSSSDSSDSSDSESSSSSSSSSSSSSSSSDSESSSESDSSSSGSSSSSSSSSDESSSESE 121
Score = 34.7 bits (78), Expect = 3.2
Identities = 31/147 (21%), Positives = 59/147 (40%)
Query: 132 KLFPKRSASVGKSFVEPSSPKVSCMGKVRSKRGKTSTVTAAAKAAVKEKTARTERKQSKH 191
K ++S+S S SS S S +S+ ++++ ++ + + ++ + S
Sbjct: 19 KEIEEKSSSSSSSSSSSSSSSSSSSSSSSSSGESSSSSSSSSSSSSSDSSDSSDSESSSS 78
Query: 192 SFFKSFRAIFRSGGRNKHDRMNDSSALDSSSITKNTNSVSDSKARDSTASLNDVSFVESV 251
S S + S + + + SS SSS + + S S+S++ D T S E
Sbjct: 79 SSSSSSSSSSSSDSESSSESDSSSSGSSSSSSSSSDESSSESESEDETKKRARESDNEDA 138
Query: 252 TRNSVSEGEPPGLGVMMRFTSGRRSES 278
++ EP +SG S S
Sbjct: 139 KETKKAKTEPESSSSSESSSSGSSSSS 165
>ref|NP_001004408.1| hypothetical protein LOC424547 [Gallus gallus]
gi|3123014|sp|P87498|VIT1_CHICK Vitellogenin I precursor
(Minor vitellogenin) [Contains: Lipovitellin I (LVI);
Phosvitin (PV); Lipovitellin II (LVII); YGP42]
gi|7442003|pir||T29088 vitellogenin I precursor
[validated] - chicken gi|1871444|dbj|BAA13973.1|
vitellogenin I [Gallus gallus]
Length = 1912
Score = 50.1 bits (118), Expect = 7e-05
Identities = 47/223 (21%), Positives = 96/223 (42%), Gaps = 24/223 (10%)
Query: 24 TQADDLKTDPPPESSHHHPDSPPESFWLSRDEENEWLDRNFIYERKESTKGNSSSTNLNP 83
+++ + T P S + S + + +++ ++ F S+ G+SSS++ +
Sbjct: 1104 SESSESTTSTPSSSDSDNRASQGDPQINLKSRQSKANEKKFYPFGDSSSSGSSSSSSSSS 1163
Query: 84 NSNSNSNSTSNSQRFANLKSKTSMIGLPKPQKPSFDAKNLRNHKPSNIKLFPKRSASVGK 143
+S+S+S+S+S S ++ S +S K +++ ++++ S+ S+S K
Sbjct: 1164 SSSSDSSSSSRSSSSSDSSSSSSSSSSSSSSKSKSSSRSSKSNRSSSSSNSKDSSSSSSK 1223
Query: 144 SFVEPSSPKVSCMGKVRSKRGKTSTVTAAAKAAVKEKTARTERKQSKHSFFKSFRAIFRS 203
S + SS S R K K S T+ A+ E
Sbjct: 1224 SNSKGSSSSSSKASGTRQKAKKQSKTTSFPHASAAE------------------------ 1259
Query: 204 GGRNKHDRMNDSSALDSSSITKNTNSVSDSKARDSTASLNDVS 246
G R+ H++ ++ + SSS ++NS S S + S++ + VS
Sbjct: 1260 GERSVHEQKQETQSSSSSSSRASSNSRSTSSSTSSSSESSGVS 1302
Score = 46.2 bits (108), Expect = 0.001
Identities = 50/227 (22%), Positives = 87/227 (38%), Gaps = 20/227 (8%)
Query: 71 STKGNSSSTNLNPNSNSNSNSTSNSQRFANLKSKTSMIGLPK------PQKPSFDAKNLR 124
S +S ST P+S+ + N S NLKS+ S K + +
Sbjct: 1102 SISESSESTTSTPSSSDSDNRASQGDPQINLKSRQSKANEKKFYPFGDSSSSGSSSSSSS 1161
Query: 125 NHKPSNIKLFPKRSASVGKSFVEPSSPKVSCMGKVRSKRGKTSTVTAAAKAAVKEKTART 184
+ S+ RS+S S SS S K +S + + +++ + K+ ++ +
Sbjct: 1162 SSSSSSDSSSSSRSSSSSDSSSSSSSSSSSSSSKSKSSSRSSKSNRSSSSSNSKDSSSSS 1221
Query: 185 ERKQSKHSFFKSFRAIFRSGGRNKHDRMNDSSALDSSSITKNTNSVSDSK--ARDSTASL 242
+ SK S S +A SG R K + + +++ +S + SV + K + S++S
Sbjct: 1222 SKSNSKGSSSSSSKA---SGTRQKAKKQSKTTSFPHASAAEGERSVHEQKQETQSSSSSS 1278
Query: 243 NDVSFVESVTRNSVSEGEPPGLGVMMRFTSGRRSESWVDDSEIHVSR 289
+ S T +S S +SG W D E R
Sbjct: 1279 SRASSNSRSTSSSTSSSSE---------SSGVSHRQWKQDREAETKR 1316
Score = 35.8 bits (81), Expect = 1.5
Identities = 57/264 (21%), Positives = 109/264 (40%), Gaps = 57/264 (21%)
Query: 9 SACAGGSTDRKIACETQADDLKTDPPPESSHHHPDSPPESFWLSRDEENEWLDRNFIYER 68
S+ + S+ + + +D SS S +S SR ++ +R+
Sbjct: 1157 SSSSSSSSSSSDSSSSSRSSSSSDSSSSSSSSSSSSSSKSKSSSRSSKS---NRSSSSSN 1213
Query: 69 KESTKGNSSSTNLNPNSNSNSNSTSNSQRFANLKSKTSMIGLPKPQKPSFDAKNLRNHKP 128
+ + +SS +N + S+S+S+ S +++ A +SKT+
Sbjct: 1214 SKDSSSSSSKSN-SKGSSSSSSKASGTRQKAKKQSKTTS--------------------- 1251
Query: 129 SNIKLFPKRSASVGKSFVE------PSSPKVSCMGKVRSKRGKTSTVTAAAKAAV----- 177
FP SA+ G+ V SS S S+ +ST +++ + V
Sbjct: 1252 -----FPHASAAEGERSVHEQKQETQSSSSSSSRASSNSRSTSSSTSSSSESSGVSHRQW 1306
Query: 178 -KEKTARTERKQSK---HSFF-----------KSFRAIFRSGGRNKHDRMNDSSALDSSS 222
+++ A T+R +S+ HS + K +R FRS + H SS+ SSS
Sbjct: 1307 KQDREAETKRVKSQFNSHSSYDIPNEWETYLPKVYRLRFRSAHTHWHSGHRTSSS-SSSS 1365
Query: 223 ITKNTNSVSDSKARDSTASLNDVS 246
+++ +S S+S + DS++ + +S
Sbjct: 1366 SSESGSSHSNSSSSDSSSRRSHMS 1389
>ref|ZP_00323079.1| hypothetical protein PpenA01001008 [Pediococcus pentosaceus ATCC
25745]
Length = 2334
Score = 50.1 bits (118), Expect = 7e-05
Identities = 59/254 (23%), Positives = 101/254 (39%), Gaps = 29/254 (11%)
Query: 8 VSACAGGSTDRKIACETQADDLKTDPPPESSHHHPDSPPESFWLSRDEENEWLDRNFIYE 67
VS S I ++ +D + T +S H S S S +
Sbjct: 772 VSQSISTSKSDSITSDSISDSISTSISDSTSTSHSTSDSVSTSNSNSDS----------- 820
Query: 68 RKESTKGNSSSTNLNPN-SNSNSNSTSNSQRFANLKSKTSMIGLPKPQKPSFDAKNLRNH 126
K ++ S+ST+++ + S+SNS STS S+ + S + K S N
Sbjct: 821 -KSKSESRSTSTSISDSISDSNSKSTSESRSTSTSSSDSKSDSASKSDSISKSDSITSNS 879
Query: 127 KPSNIKLFPKRSASVG--KSFVEPSSPKVSCMGKVRSKRGKTSTVTAAAKAAVKEKTART 184
+I S+S KS E S S + K+++ + + +V + T+
Sbjct: 880 ISESISTSNSDSSSKSDSKSTSESRSTSTSVSDSISDSNSKSTSESRSTSTSVSDSTS-- 937
Query: 185 ERKQSKHSFFKSFRAIFRSGGRNKHDRMNDSSALDSSSITKNTNSVSDSKARDSTASLND 244
+ + HS S +DS++ +S + S+SDSK+ DS + +
Sbjct: 938 DSTSTSHSTSDSVST-----------SNSDSNSKSTSESRSTSTSISDSKS-DSASKSDS 985
Query: 245 VSFVESVTRNSVSE 258
VS +S+T NS+SE
Sbjct: 986 VSKSDSITSNSISE 999
Score = 47.4 bits (111), Expect = 5e-04
Identities = 49/188 (26%), Positives = 77/188 (40%), Gaps = 16/188 (8%)
Query: 71 STKGNSSSTNLNPNSNSNSNSTSNSQRFANLKSKTSMIGLPKPQKPSFDAKNLRNHKPSN 130
ST ++S + NS+SNS STS S+ + S + K S N +
Sbjct: 941 STSHSTSDSVSTSNSDSNSKSTSESRSTSTSISDSKSDSASKSDSVSKSDSITSNSISES 1000
Query: 131 IKLFPKRSASVGKSFVEPSSPKVSCMGKVRSKRGKTSTVTAAAKAAVKEKTARTERKQSK 190
I S+S S +S VS ST +A+ + K + T S
Sbjct: 1001 ISTSKSDSSSKSMSDSRSASTSVS-----------DSTSDSASTSHSKSDSVSTSNSDSS 1049
Query: 191 HSFFKSFRAIFRSGGRNKHDRMNDSSALDSSSITKNTNSVSDSKARDSTASLNDVSFVES 250
++ S R+ ++DS + S + SVSDSK+ DS + + +S +S
Sbjct: 1050 ----SKSDSVSTSDSRSTSTSVSDSISKSMSDSRSTSTSVSDSKS-DSESKSDSISKSDS 1104
Query: 251 VTRNSVSE 258
+T NS+SE
Sbjct: 1105 ITSNSISE 1112
Score = 45.8 bits (107), Expect = 0.001
Identities = 51/219 (23%), Positives = 93/219 (42%), Gaps = 21/219 (9%)
Query: 43 DSPPESFWLSRDEENEWLDRNFIYERKESTKGNSSSTNLNPN---SNSNSNSTSNSQRFA 99
DS +S S + N + K ++ S+ST+++ + S S S+STS S
Sbjct: 1482 DSTSDSASTSHSTSDSVSTSNSDSDSKSTSDSRSASTSVSDSKSDSESKSDSTSKSDSIT 1541
Query: 100 NLKSKTSMIGLPKPQKPSFDAKNLRNHKPSNIKLFPKRSASVGKSFVEPSSPKVSCMGKV 159
+ S+ D+K++ + + + S SV S + +S S V
Sbjct: 1542 SNSISESISTSNSDSSSKSDSKSISDSRST--------STSVSDSTSDSASTSHSTSDSV 1593
Query: 160 RSKRGKTSTVTAAAKAAVKEKTARTERKQSKHSFFKSFRAIFRSGGRNKHDRMNDSSALD 219
TS + +K+ + ++ T S + + S +K +DSS+
Sbjct: 1594 -----STSNSDSDSKSMSESRSTSTSVSDSTSDSASTSHSTSDSVSTSK----SDSSSKS 1644
Query: 220 SSSITKNTNSVSDSKARDSTASLNDVSFVESVTRNSVSE 258
+S + S+SDSK+ DS + + +S +S+T NS+SE
Sbjct: 1645 TSDSRSTSTSISDSKS-DSASKSDSISKSDSITSNSISE 1682
Score = 45.1 bits (105), Expect = 0.002
Identities = 58/254 (22%), Positives = 103/254 (39%), Gaps = 21/254 (8%)
Query: 4 VETLVSACAGGSTDRKIACETQADDLKTDPPPESSHHHPDSPPESFWLSRDEENEWLDRN 63
V T S ST + T D K+D +S DS +S ++ + +E + +
Sbjct: 1257 VSTSNSDSDSKSTSDSRSASTSVSDSKSDSASKS-----DSTSKSDSITSNSISESISTS 1311
Query: 64 FIYERKESTKGNSSSTNLNPNSNSNSNSTSNSQRFANLKSKTSMIGLPKPQKPSFDAKNL 123
S+K +S ST+ +S S S S SNS +N KS + S
Sbjct: 1312 ---NSDSSSKSDSKSTS---DSRSTSTSVSNSISDSNSKSTSDSRSTSTSVSDSTSDSVS 1365
Query: 124 RNHKPSNIKLFPKRSASVGKSFVEPSSPKVSCMGKVRSKRGKTSTVTAAAKAAVKEKTAR 183
+H S+ + S S KS + S S + K+++ + + +V + T+
Sbjct: 1366 TSHSTSD-SVSTSNSDSDSKSASDSRSTSTSVSNSISDSNSKSTSDSRSTSTSVSDSTS- 1423
Query: 184 TERKQSKHSFFKSFRAIFRSGGRNKHDRMNDSSALDSSSITKNTNSVSDSKARDSTASLN 243
+ + HS S + D + S++ S+ T +NS+SDS ++ ++ S +
Sbjct: 1424 -DSVSTSHSTSDSVST-------SNSDSDSKSASDSRSTSTSVSNSISDSNSKSTSDSRS 1475
Query: 244 DVSFVESVTRNSVS 257
+ V T +S S
Sbjct: 1476 TSTSVSDSTSDSAS 1489
Score = 45.1 bits (105), Expect = 0.002
Identities = 46/221 (20%), Positives = 93/221 (41%), Gaps = 23/221 (10%)
Query: 60 LDRNFIYERKESTKGNSSSTNLNPNSNSNSNSTSNSQRFANLKSKTSMIGLPKPQKPSFD 119
L ++ + ST G++S +N S+S S S S S+ + S + + K + S
Sbjct: 663 LSQSISASKSTSTSGSTSVSNSKSTSDSVSQSISTSKSDSTSASGSVSDSVSKSKSDSIT 722
Query: 120 AKNLRNHKPSNIKLFPKRSASVGKSFVEPSSPKVSCMGKVRSKRGKTSTVTAAAKAAVKE 179
+ ++ N +++ S SV +S S S V + + + +V+ + + +
Sbjct: 723 SDSISNSISTSVSGSKSTSDSVSQSISTSKSTSTSGSTSVSNSKSTSDSVSQSISTSKSD 782
Query: 180 K-----------TARTERKQSKHSFFKSFRAIFRSGGRNKHDRMNDSSALDSSSITKNTN 228
T+ ++ + HS S + S +K + S++ S ++N
Sbjct: 783 SITSDSISDSISTSISDSTSTSHSTSDSV-STSNSNSDSKSKSESRSTSTSISDSISDSN 841
Query: 229 SVSDSKARDSTASLND-----------VSFVESVTRNSVSE 258
S S S++R ++ S +D +S +S+T NS+SE
Sbjct: 842 SKSTSESRSTSTSSSDSKSDSASKSDSISKSDSITSNSISE 882
Score = 44.7 bits (104), Expect = 0.003
Identities = 60/264 (22%), Positives = 112/264 (41%), Gaps = 26/264 (9%)
Query: 15 STDRKIACETQADDLKTDPPPESSHHHPDSPPESFWLSR---DEENEWLDRNFIYERKES 71
ST + T D +D +SH DS S S D ++ R+ +S
Sbjct: 1889 STSDSRSASTSVSDSTSDST-STSHSTSDSVSTSNSDSSSKSDSKSASDSRSTSTSVSDS 1947
Query: 72 TKGNSSSTNLNPNSNSNSNSTSNSQRFANLKSKTSMIGLPKPQKPSF-----DAKNLRNH 126
T ++S+++ +S S SNS S+S+ ++ +S ++ I S D+ + N
Sbjct: 1948 TSNSTSASHSTSDSVSTSNSDSDSKSMSDSRSTSTSISDSTSDSTSTSHSTSDSVSTSNS 2007
Query: 127 ----KPSNIKLFPKRSAS--VGKSFVEPSSPKVSCMGKVRSKRGKTSTVTAAAKAAVKEK 180
K ++ RSAS V S + +S S V + +S+ + + A+
Sbjct: 2008 DSSSKSDSVSTSDSRSASTSVSDSTSDSASTSHSTSDSVSTSNSDSSSKSDSKSASDSRS 2067
Query: 181 TARTERKQSKHSFFKSFR-----AIFRSGGRNKHDRMNDSSALDSSSITKNTNSVSDSKA 235
T+ + + +S S + S +K D ++ S + +S+ ++ SVS S +
Sbjct: 2068 TSTSVSDSTSNSTSASHSTSDSVSTSNSDSSSKSDSISTSESRSASTSVSDSTSVSTSDS 2127
Query: 236 RDSTASLNDVSFVESVTRNSVSEG 259
R ++ S++D T NS+S+G
Sbjct: 2128 RSTSTSISDS------TSNSISDG 2145
Score = 42.4 bits (98), Expect = 0.016
Identities = 64/261 (24%), Positives = 106/261 (40%), Gaps = 30/261 (11%)
Query: 24 TQADDLKTDPPPESSHHHPDSPPESFWLSRDEENEWLDRNFIYERKESTKGNSSSTNLNP 83
+++D + T +S DS +S SR D E K + S S +
Sbjct: 1050 SKSDSVSTSDSRSTSTSVSDSISKSMSDSRSTSTSVSDSKSDSESKSDSISKSDS--ITS 1107
Query: 84 NSNSNSNSTSNSQRFANLKSKTSMIGLPKPQKPSFDAKNLRNHKPSNIKLFPKRSASVGK 143
NS S S STSNS ++ SK++ S + + S K S S+ +
Sbjct: 1108 NSISESISTSNSDSISDSNSKSTSDSRSTSTSISDSKSDSASKSDSVSKSDSITSDSISE 1167
Query: 144 SFVEPSSPKVS-CMGKVRSKRGKTSTVTAAAK---AAVKEKTARTERKQSKHSFFKSFRA 199
S +S +S K S TST + +K A+ T+ + + S KS +
Sbjct: 1168 SISTSNSDSISDSNSKSTSDSRSTSTSVSDSKSDSASTSHSTSDSVSTSNSDSSSKS-DS 1226
Query: 200 IFRSGGRNKHDRMNDSSALDSSSITKNTN-----SVSDSKAR-----------------D 237
+ S R+ ++DS++ DS+S + +T+ S SDS ++ D
Sbjct: 1227 VSTSDSRSTSTSISDSTS-DSASTSHSTSDSVSTSNSDSDSKSTSDSRSASTSVSDSKSD 1285
Query: 238 STASLNDVSFVESVTRNSVSE 258
S + + S +S+T NS+SE
Sbjct: 1286 SASKSDSTSKSDSITSNSISE 1306
Score = 42.0 bits (97), Expect = 0.020
Identities = 55/240 (22%), Positives = 98/240 (39%), Gaps = 4/240 (1%)
Query: 24 TQADDLKTDPPPES-SHHHPDSPPESFWLSRDEENEWLDRNFIYERKESTKGNSSST-NL 81
+++D + ++ ES S DS +S SR D +K +S ST N
Sbjct: 987 SKSDSITSNSISESISTSKSDSSSKSMSDSRSASTSVSDSTSDSASTSHSKSDSVSTSNS 1046
Query: 82 NPNSNSNSNSTSNSQRFANLKSKTSMIGLPKPQKPSFDAKNLRNHKPSNIKLFPKRSASV 141
+ +S S+S STS+S+ + S + + + S + ++ S K +
Sbjct: 1047 DSSSKSDSVSTSDSRSTSTSVSDSISKSMSDSRSTSTSVSDSKSDSESKSDSISKSDSIT 1106
Query: 142 GKSFVEPSSPKVS-CMGKVRSKRGKTSTVTAAAKAAVKEKTARTERKQSKHSFFKSFRAI 200
S E S S + SK S T+ + + K +A SK S +I
Sbjct: 1107 SNSISESISTSNSDSISDSNSKSTSDSRSTSTSISDSKSDSASKSDSVSKSDSITS-DSI 1165
Query: 201 FRSGGRNKHDRMNDSSALDSSSITKNTNSVSDSKARDSTASLNDVSFVESVTRNSVSEGE 260
S + D ++DS++ +S + SVSDSK+ ++ S + V + +S S+ +
Sbjct: 1166 SESISTSNSDSISDSNSKSTSDSRSTSTSVSDSKSDSASTSHSTSDSVSTSNSDSSSKSD 1225
Score = 41.6 bits (96), Expect = 0.027
Identities = 53/254 (20%), Positives = 105/254 (40%), Gaps = 21/254 (8%)
Query: 15 STDRKIACETQADDLKTDPPPESSHHHPDSPPESFWLSRDEENEWL---DRNFIYERKES 71
ST + T D K+D +SH DS S S + + R+ +S
Sbjct: 1184 STSDSRSTSTSVSDSKSDSA-STSHSTSDSVSTSNSDSSSKSDSVSTSDSRSTSTSISDS 1242
Query: 72 TKGNSSSTNLNPNSNSNSNSTSNSQRFANLKSKTSMIGLPKPQKPSFDAKNLRNHKPSNI 131
T ++S+++ +S S SNS S+S+ ++ +S ++ + K S ++ ++
Sbjct: 1243 TSDSASTSHSTSDSVSTSNSDSDSKSTSDSRSASTSVSDSKSDSASKSDSTSKSDSITSN 1302
Query: 132 KLFPK--------RSASVGKSFVEPSSPKVSCMGKVRSKRGKTSTVTAAAKAAVKEKTAR 183
+ S S KS + S S + K+++ + + +V + T+
Sbjct: 1303 SISESISTSNSDSSSKSDSKSTSDSRSTSTSVSNSISDSNSKSTSDSRSTSTSVSDSTS- 1361
Query: 184 TERKQSKHSFFKSFRAIFRSGGRNKHDRMNDSSALDSSSITKNTNSVSDSKARDSTASLN 243
+ + HS S + D + S++ S+ T +NS+SDS ++ ++ S +
Sbjct: 1362 -DSVSTSHSTSDSVST-------SNSDSDSKSASDSRSTSTSVSNSISDSNSKSTSDSRS 1413
Query: 244 DVSFVESVTRNSVS 257
+ V T +SVS
Sbjct: 1414 TSTSVSDSTSDSVS 1427
Score = 41.6 bits (96), Expect = 0.027
Identities = 41/200 (20%), Positives = 80/200 (39%), Gaps = 4/200 (2%)
Query: 62 RNFIYERKESTKGNSSSTNLNPNSNSNSNSTSNSQRFANLKSKTSMIGLPKPQKPSFDAK 121
R+ + ST G++S +N S+S S S S S+ + S + + K + S +
Sbjct: 528 RSVSTSKSTSTSGSTSVSNSKSTSDSVSQSISTSKSNSASASGSVSDSVSKSKSDSITSD 587
Query: 122 NLRNHKPSNIKLFPKRSASVGKSFVEPSSPKVSCMGKVRSKRGKTSTVT----AAAKAAV 177
++ N +++ S S +S S S V + + + +V+ + +
Sbjct: 588 SISNSISTSVSGSKSTSDSASQSISTSKSTSTSGSTSVSNSKSMSDSVSQSISTSKSTST 647
Query: 178 KEKTARTERKQSKHSFFKSFRAIFRSGGRNKHDRMNDSSALDSSSITKNTNSVSDSKARD 237
T+ + K + S +S A + N S DS S + +T+ + A
Sbjct: 648 SGSTSVSNSKSTSDSLSQSISASKSTSTSGSTSVSNSKSTSDSVSQSISTSKSDSTSASG 707
Query: 238 STASLNDVSFVESVTRNSVS 257
S + S +S+T +S+S
Sbjct: 708 SVSDSVSKSKSDSITSDSIS 727
Score = 40.8 bits (94), Expect = 0.045
Identities = 55/251 (21%), Positives = 94/251 (36%), Gaps = 11/251 (4%)
Query: 15 STDRKIACETQADDLKTDPPPESSHHHPDSPPESFWLSRDEENEWLDRNFIYERKESTKG 74
S I + ++ + T SS S ES S + D ST
Sbjct: 1670 SKSDSITSNSISESISTSKSDSSSKSDSKSTSESRSASTSVSDSTSDS---ISTSHSTSD 1726
Query: 75 NSSSTNLNPNSNSNSNSTSNSQRFANLKSKTSMIGLPKPQKPSFDAKNLRNHKPSNIKLF 134
+ S++N + +S S+S STS S R A+ S + D+ + N S+
Sbjct: 1727 SVSTSNSDSSSKSDSKSTSES-RSASTSVSDSTSDSTSTSHSTSDSVSTSNSDSSSKSAS 1785
Query: 135 PKRSASVGKS-FVEPSSPKVSCMGKVRSKRGKTSTVTAAAKAAVKEKTARTERKQSKHSF 193
RS S S + S+ K + + S ST + + + + T S
Sbjct: 1786 DSRSTSTSVSDSISDSNSKSTSDSRSASTSVSDSTSDSTSTSHSTSDSVSTSNSDSDSKS 1845
Query: 194 FKSFRAIFRSGGRNKHDRMNDSSALDSSSITKNTNS------VSDSKARDSTASLNDVSF 247
R+ S + D + S + S T N++S VS S +R ++ S++D +
Sbjct: 1846 MSDSRSTSTSISDSTSDSTSTSHSTSDSVSTSNSDSSSKSDSVSTSDSRSASTSVSDSTS 1905
Query: 248 VESVTRNSVSE 258
+ T +S S+
Sbjct: 1906 DSTSTSHSTSD 1916
Score = 39.7 bits (91), Expect = 0.10
Identities = 49/243 (20%), Positives = 95/243 (38%), Gaps = 8/243 (3%)
Query: 24 TQADDLKTDPPPESSHHHPDSPPESFWLSRDEENEWLDRNFIYERKESTKGNSSSTNLNP 83
+++D + T +S DS +S S + N + K ++ S+ST+++
Sbjct: 1222 SKSDSVSTSDSRSTSTSISDSTSDSASTSHSTSDSVSTSNSDSDSKSTSDSRSASTSVSD 1281
Query: 84 N---SNSNSNSTSNSQRFANLKSKTSMIGLPKPQKPSFDAKNLRNHKPSNIKLFPKRSAS 140
+ S S S+STS S + S+ D+K+ + + ++ + S S
Sbjct: 1282 SKSDSASKSDSTSKSDSITSNSISESISTSNSDSSSKSDSKSTSDSRSTSTSVSNSISDS 1341
Query: 141 VGKSFVEPSSPKVSCMGKVR---SKRGKTSTVTAAAKAAVKEKTARTERKQSKH--SFFK 195
KS + S S S TS + + + K+A R S +
Sbjct: 1342 NSKSTSDSRSTSTSVSDSTSDSVSTSHSTSDSVSTSNSDSDSKSASDSRSTSTSVSNSIS 1401
Query: 196 SFRAIFRSGGRNKHDRMNDSSALDSSSITKNTNSVSDSKARDSTASLNDVSFVESVTRNS 255
+ S R+ ++DS++ S+ ++SVS S + + S +D + NS
Sbjct: 1402 DSNSKSTSDSRSTSTSVSDSTSDSVSTSHSTSDSVSTSNSDSDSKSASDSRSTSTSVSNS 1461
Query: 256 VSE 258
+S+
Sbjct: 1462 ISD 1464
Score = 36.2 bits (82), Expect = 1.1
Identities = 46/204 (22%), Positives = 85/204 (41%), Gaps = 25/204 (12%)
Query: 63 NFIYERKESTKGNSSSTNLNPNSNSNSNSTSNSQRFANL------KSKTSMIGLPKPQKP 116
N + K T+ NSS+ + + + NS S TS S + KS +S + + +
Sbjct: 76 NATTDAKSETQSNSSAKSTSADKNSASQKTSESTSLSTSASTSQSKSLSSSLKEAQTKST 135
Query: 117 SFDAKNLRNHKPSNIKLFPKRSASVGKSFVEPSSPKVSCMGKVRSKRGKTSTVTAAAKAA 176
S DA + +++ S K ++ + S K + KTS TA++ A
Sbjct: 136 SKDATKVADNQQSKQKDGKEQKNA-------KSDAKQDVKQTAKDDNVKTSQSTASSSTA 188
Query: 177 VKEKTARTERKQSKHSFFKSFRAIFRSGGRNKHDRMNDSSALDSSSITKNTNSVSDSKAR 236
+ + + + S S SG + +S++ S N+ VSDS+ R
Sbjct: 189 SNQGSEHAQINDNISSLSNS-----TSGS------ITESASGSLSVSLSNSLFVSDSRKR 237
Query: 237 DSTASLNDVSFVESVTRNSVSEGE 260
D ++ N S +S++ N+ ++ E
Sbjct: 238 DKDSASNSQSLAKSLS-NAKAQAE 260
Score = 35.0 bits (79), Expect = 2.5
Identities = 47/201 (23%), Positives = 83/201 (40%), Gaps = 14/201 (6%)
Query: 68 RKESTKGNSSSTNLNPNSNS------NSNSTSNSQRFANLKSKTSMIGLPKPQKPSFDAK 121
+ ST G++S +N S+S S STS S + SK++ L + S
Sbjct: 615 KSTSTSGSTSVSNSKSMSDSVSQSISTSKSTSTSGSTSVSNSKSTSDSLSQSISASKSTS 674
Query: 122 NLRNHKPSNIKLFPKRSASVGKSFVEPSSPKVSCMGKVRSKRGKTSTVTAAAKAAVKE-K 180
+ SN K S SV +S S S G V K+ + + + +
Sbjct: 675 TSGSTSVSNSK---STSDSVSQSISTSKSDSTSASGSVSDSVSKSKSDSITSDSISNSIS 731
Query: 181 TARTERKQSKHSFFKSF---RAIFRSGGRNKHDRMNDSSALDSS-SITKNTNSVSDSKAR 236
T+ + K + S +S ++ SG + + + S ++ S S +K+ + SDS +
Sbjct: 732 TSVSGSKSTSDSVSQSISTSKSTSTSGSTSVSNSKSTSDSVSQSISTSKSDSITSDSISD 791
Query: 237 DSTASLNDVSFVESVTRNSVS 257
+ S++D + T +SVS
Sbjct: 792 SISTSISDSTSTSHSTSDSVS 812
Score = 34.7 bits (78), Expect = 3.2
Identities = 53/259 (20%), Positives = 101/259 (38%), Gaps = 26/259 (10%)
Query: 15 STDRKIACETQADDLKTDPPPESSHHHPDSPPESFWLSRDEENEWLDRNFIYERKESTKG 74
ST + T D +D +SH DS S + D ++ ++ R ST
Sbjct: 1699 STSESRSASTSVSDSTSDSI-STSHSTSDSVSTS---NSDSSSKSDSKSTSESRSASTSV 1754
Query: 75 NSSSTNLNPNSNSNSNSTSNSQRFANLKSKTSMIGLPKPQKPSFDAKNLRNHKPSNIKLF 134
+ S+++ S+S S+S S S ++ KS + S N ++ S
Sbjct: 1755 SDSTSDSTSTSHSTSDSVSTSNSDSSSKSASDSRSTSTSVSDSISDSNSKSTSDSR---- 1810
Query: 135 PKRSASVGKSFVEPSSPKVSCMGKVRSKRG----KTSTVTAAAKAAVKEKTARTERKQSK 190
S SV S + +S S V + K+ + + + ++ + T+ + +
Sbjct: 1811 -SASTSVSDSTSDSTSTSHSTSDSVSTSNSDSDSKSMSDSRSTSTSISDSTS--DSTSTS 1867
Query: 191 HSFFKSFR-----------AIFRSGGRNKHDRMNDSSALDSSSITKNTNSVSDSKARDST 239
HS S ++ S R+ ++DS++ +S+ ++SVS S + S+
Sbjct: 1868 HSTSDSVSTSNSDSSSKSDSVSTSDSRSASTSVSDSTSDSTSTSHSTSDSVSTSNSDSSS 1927
Query: 240 ASLNDVSFVESVTRNSVSE 258
S + + T SVS+
Sbjct: 1928 KSDSKSASDSRSTSTSVSD 1946
Score = 33.1 bits (74), Expect = 9.4
Identities = 36/187 (19%), Positives = 76/187 (40%), Gaps = 12/187 (6%)
Query: 70 ESTKGNSSSTNLNPNSNSNSNSTSNSQRFANLKSKTSMIGLPKPQKPSFDAKNLRNHKPS 129
+S ++S ++ +S S S+STS+S +N S + K + D+++
Sbjct: 2020 DSRSASTSVSDSTSDSASTSHSTSDSVSTSNSDSSSK-----SDSKSASDSRSTSTSVSD 2074
Query: 130 NIKLFPKRSASVGKSFVEPSSPKVSCMGKVRSKRGKTSTVTAAAKAAVKEKTARTERKQS 189
+ S S S +S S + + ++++ + + +V +R+
Sbjct: 2075 STSNSTSASHSTSDSVSTSNSDSSSKSDSISTSESRSASTSVSDSTSVSTSDSRSTSTSI 2134
Query: 190 KHSFFKSFRAIFRSGGRNKHDRMN----DSSALDSSSITKNTNSVSDSKARDSTASLNDV 245
S S I G HD + +S++ S T ++ S+S S + ++ S N
Sbjct: 2135 SDSTSNS---ISDGNGSTSHDNHHGSGSESNSESGSQSTSDSQSISTSISDSTSDSKNSG 2191
Query: 246 SFVESVT 252
S +S++
Sbjct: 2192 SASQSIS 2198
>emb|CAD79181.1| hypothetical protein-signal peptide prediction [Rhodopirellula
baltica SH 1] gi|32477034|ref|NP_870028.1| hypothetical
protein-signal peptide prediction [Rhodopirellula
baltica SH 1]
Length = 669
Score = 48.9 bits (115), Expect = 2e-04
Identities = 48/191 (25%), Positives = 87/191 (45%), Gaps = 23/191 (12%)
Query: 71 STKGNSSSTNLNPNSNSNSNSTSNSQRFANLKSKTSMIGLPKPQKPSFDAKNLRNHKPSN 130
S+ +SSS++ +P+S+S+S S+S+S + S +SM G S + +L + S+
Sbjct: 20 SSDSSSSSSSSSPSSSSSSGSSSSS---SGSSSSSSMSGSSSSSMSSSSSSSLSSSSSSS 76
Query: 131 IKLFPKRSASVGKSFVEPSSPKVSCMGKVRSKRGKTSTVTAAAKAAVKEKTARTERKQSK 190
S S S PSS S S +S+ +++ + ++ ++ S
Sbjct: 77 SGSSSSSSGSSSSSSGSPSSSSSSGSSSSSSSGSSSSSSKSSSSSDSSSNSSSSDSSDSS 136
Query: 191 HSFFKSFRAIFRSGGRNKHDRMNDSSALDSSSITKNTNSVSDSKARDSTASLND--VSFV 248
HS S SS+ S S T +++SVS S++ DS+ S + + +
Sbjct: 137 HSSSAS------------------SSSDSSDSSTSSSSSVSSSQSSDSSDSSDSSTSTSL 178
Query: 249 ESVTRNSVSEG 259
SV+ +S S+G
Sbjct: 179 PSVSSSSSSDG 189
Score = 46.6 bits (109), Expect = 8e-04
Identities = 42/187 (22%), Positives = 78/187 (41%), Gaps = 10/187 (5%)
Query: 71 STKGNSSSTNLNPNSNSNSNSTSNSQRFANLKSKTSMIGLPKPQKPSFDAKNLRNHKPSN 130
S+ + SS++ + +S S+S+S+ +S + S +SM G S + +L + S+
Sbjct: 17 SSSSSDSSSSSSSSSPSSSSSSGSSSSSSGSSSSSSMSGSSSSSMSSSSSSSLSSSSSSS 76
Query: 131 IKLFPKRSASVGKSFVEPSSPKVSCMGKVRSKRGKTSTVTAAAKAAVKEKTARTERKQSK 190
S S S PSS S S +S+ +++ + ++ ++ S
Sbjct: 77 SGSSSSSSGSSSSSSGSPSSSSSSGSSSSSSSGSSSSSSKSSSSSDSSSNSSSSDSSDSS 136
Query: 191 HSFFKSFRAIFRSGGRNKHDRMNDSSALDSSSITKNTNSVSDSKARDSTASLNDVSFVES 250
HS S + D + S++ SS + ++ SDS ++ SL VS S
Sbjct: 137 HS----------SSASSSSDSSDSSTSSSSSVSSSQSSDSSDSSDSSTSTSLPSVSSSSS 186
Query: 251 VTRNSVS 257
+S S
Sbjct: 187 SDGSSSS 193
Score = 45.1 bits (105), Expect = 0.002
Identities = 52/253 (20%), Positives = 98/253 (38%), Gaps = 39/253 (15%)
Query: 6 TLVSACAGGSTDRKIACETQADDLKTDPPPESSHHHPDSPPESFWLSRDEENEWLDRNFI 65
+L S S+D A +D +SS+ +S L
Sbjct: 222 SLPSISGSSSSDGSSASSDSSDSSSNSQSSDSSNSESSDSSDSISLPS------------ 269
Query: 66 YERKESTKGNSSSTNLNPNSNSNSNSTSNSQRFANLKSKTSMIGLPKPQKPSFDAKNLRN 125
S+ G+SS+++ +SNS+SNS S+ + ++ + LP S A + +
Sbjct: 270 ISGSSSSAGSSSASS--DSSNSSSNSQSSDSSTSESNDSSASVSLPSVSGSSSSASDSSS 327
Query: 126 HKPSNIKLFPKRSASVGKSFVEPSSPKVSCMGKVRSKRGKTSTVTAAAKAAVKEKTARTE 185
+ + +S +S S+P VS S+ +++ ++ ++++ E +
Sbjct: 328 SESQSSDSGSSSDSSTSQSSESASTPSVSSSQSSTSESASSTSESSNSESSQSESSTSES 387
Query: 186 RKQSKHSFFKSFRAIFRSGGRNKHDRMNDSSALDSSSITKNTNSVSDSKARDSTASLNDV 245
R +S+ +DS +L S S +T+S SDS++ DS +
Sbjct: 388 RSRSE----------------------SDSESLPSYSWNSSTSSDSDSESSDSESESQSQ 425
Query: 246 SFVESVTRNSVSE 258
S T SVS+
Sbjct: 426 S---DSTSQSVSD 435
Score = 39.7 bits (91), Expect = 0.10
Identities = 42/192 (21%), Positives = 81/192 (41%), Gaps = 10/192 (5%)
Query: 71 STKGNSSSTNLNPNSNSNSNSTSNSQRFA--NLKSKTSMIGLPKPQKPSFDAKNLRNHKP 128
S+ + SS+N + +SNS S+ +S + ++ +S G S A + ++
Sbjct: 238 SSDSSDSSSNSQSSDSSNSESSDSSDSISLPSISGSSSSAG-------SSSASSDSSNSS 290
Query: 129 SNIKLFPKRSASVGKSFVEPSSPKVSCMGKVRSKRGKTSTVTAAAKAAVKEKTARTERKQ 188
SN + ++ S S P VS S + + ++ + ++ T+++
Sbjct: 291 SNSQSSDSSTSESNDSSASVSLPSVSGSSSSASDSSSSESQSSDSGSSSDSSTSQSSESA 350
Query: 189 SKHSFFKSFRAIFRSGGRNKHDRMNDSSALDSSSITKNTNSVSDSKARDSTASLNDVSFV 248
S S S + S ++SS +SS+ + S SDS++ S S N +
Sbjct: 351 STPSVSSSQSSTSESASSTSESSNSESSQSESSTSESRSRSESDSESLPS-YSWNSSTSS 409
Query: 249 ESVTRNSVSEGE 260
+S + +S SE E
Sbjct: 410 DSDSESSDSESE 421
Score = 37.0 bits (84), Expect = 0.65
Identities = 48/220 (21%), Positives = 86/220 (38%), Gaps = 9/220 (4%)
Query: 71 STKGNSSSTNLNPNSNSNSNSTSNSQRFANLKSKTS----MIGLPK-PQKPSFDAKNLRN 125
S +SSS + +S+ +S+S+SNSQ + S +S LP S D + +
Sbjct: 180 SVSSSSSSDGSSSSSSGSSDSSSNSQSSDSSNSDSSDSSDSTSLPSISGSSSSDGSSASS 239
Query: 126 HKPSNIKLFPKRSASVGKSFVEPSSPKVSCMGKVRSKRGKTSTVTAAAKAAVKEKTARTE 185
+ +S +S S + + S G +S + ++ ++ +++ +
Sbjct: 240 DSSDSSSNSQSSDSSNSESSDSSDSISLPSISGSSSSAGSSSASSDSSNSSSNSQSSDSS 299
Query: 186 RKQSKHSFFK-SFRAIFRSGGRNKHDRMNDSSALDSSSITKNTNSVSDSKARDSTASLND 244
+S S S ++ S ++S + DS S + ++ S S A + S +
Sbjct: 300 TSESNDSSASVSLPSVSGSSSSASDSSSSESQSSDSGSSSDSSTSQSSESASTPSVSSSQ 359
Query: 245 VSFVESVTRNSVSEGEPPGLGVMMRFTSGRRSESWVDDSE 284
S ES + S S S RSES DSE
Sbjct: 360 SSTSESASSTSESSNSESSQSESSTSESRSRSES---DSE 396
Score = 35.8 bits (81), Expect = 1.5
Identities = 41/191 (21%), Positives = 80/191 (41%), Gaps = 5/191 (2%)
Query: 52 SRDEENEWLDRNFIYERKESTKGNSSSTNLNPNSNSNSNSTSNSQRFANLKSKTSMIGLP 111
S + +E + + ST ++SST+ + NS S+ + +S S+ + +S++ LP
Sbjct: 342 STSQSSESASTPSVSSSQSSTSESASSTSESSNSESSQSESSTSE--SRSRSESDSESLP 399
Query: 112 KPQKPSFDAKNLRNHKPSNIKLFPKRSASVGKSFVEPSSPKVSCMGKVRSKRGKTSTVTA 171
S + + + + +S S +S + S S RS + + +A
Sbjct: 400 SYSWNSSTSSDSDSESSDSESESQSQSDSTSQSVSDSGSDSQSESSIDRSS--DSQSYSA 457
Query: 172 AAKAAVKEKTARTERKQSKHSFFKSFRAIFRSGGRNKHDRMNDSSALDSSS-ITKNTNSV 230
+A A+ + + + + S S G + S DS S T ++S
Sbjct: 458 SASASYSDSASASGSQSGSASGSGSNSGSDSGSGSVSASESDSGSGSDSDSRSTSTSDSE 517
Query: 231 SDSKARDSTAS 241
SDS++ DST++
Sbjct: 518 SDSRSDDSTSA 528
Score = 35.0 bits (79), Expect = 2.5
Identities = 56/288 (19%), Positives = 107/288 (36%), Gaps = 16/288 (5%)
Query: 9 SACAGGSTDRKIACETQADDLKTDPPPESSHHHPDSPPESFWLSRDEENEWLDRNFIYER 68
S+ A S+D + + + + + +SS DS S S + +
Sbjct: 138 SSSASSSSDSSDSSTSSSSSVSSSQSSDSS----DSSDSSTSTSLPSVSSSSSSDGSSSS 193
Query: 69 KESTKGNSSSTNLNPNSNSNSNSTSNSQRFANLKSKTSMIGLPKPQKPSFDAKNLRNHKP 128
+ +SS++ + +SNS+S+ +S+S ++ +S G S + N ++
Sbjct: 194 SSGSSDSSSNSQSSDSSNSDSSDSSDSTSLPSISGSSSSDGSSASSDSSDSSSNSQSSDS 253
Query: 129 SN-----------IKLFPKRSASVGKSFVEPSSPKVSCMGKVRSKRGKTSTVTAAAKAAV 177
SN + S+S G S S S + S ++A+ +
Sbjct: 254 SNSESSDSSDSISLPSISGSSSSAGSSSASSDSSNSSSNSQSSDSSTSESNDSSASVSLP 313
Query: 178 KEKTARTERKQSKHSFFKSFRAIFRSGGRNKHDRMNDSSALDSSSITKNTNSVSD-SKAR 236
+ + S S +S + S + S+ SSS + + S S S++
Sbjct: 314 SVSGSSSSASDSSSSESQSSDSGSSSDSSTSQSSESASTPSVSSSQSSTSESASSTSESS 373
Query: 237 DSTASLNDVSFVESVTRNSVSEGEPPGLGVMMRFTSGRRSESWVDDSE 284
+S +S ++ S ES +R+ P +S SES +SE
Sbjct: 374 NSESSQSESSTSESRSRSESDSESLPSYSWNSSTSSDSDSESSDSESE 421
Score = 34.3 bits (77), Expect = 4.2
Identities = 58/291 (19%), Positives = 109/291 (36%), Gaps = 16/291 (5%)
Query: 9 SACAGGSTDRKIACETQADDLKTDPPPESSHHHPDSPPESFWLSRDEENEWLDRNFIYER 68
S+ + GS+ + +D +SS S S S D +
Sbjct: 104 SSSSSGSSSSSSKSSSSSDSSSNSSSSDSSDSSHSSSASSSSDSSDSSTSSSSSVSSSQS 163
Query: 69 KESTKGNSSSTNLNPNSNSNSNSTSNSQRFANLKSKTSMIGLPKPQKPSFDAKNLRNHKP 128
+S+ + SST+ + S S+S+S+ S ++ S +S S D+ N +
Sbjct: 164 SDSSDSSDSSTSTSLPSVSSSSSSDGSSSSSSGSSDSSS------NSQSSDSSNSDSSDS 217
Query: 129 SNIKLFPK---RSASVGKSFVEPSSPKVSCMGKVRSKRGKTSTVTAAAKAAVKEKTARTE 185
S+ P S+S G S SS S S ++S + + ++ +
Sbjct: 218 SDSTSLPSISGSSSSDGSSASSDSSDSSSNSQSSDSSNSESSDSSDSISLPSISGSSSSA 277
Query: 186 RKQSKHSFFKSFRAIFRSGGRNKHDRMNDSSALDSSSITKNTNSVSDSKARDSTASLNDV 245
S S + + +S + + + S+++ S++ +++S SDS + +S +S +
Sbjct: 278 GSSSASSDSSNSSSNSQSSDSSTSESNDSSASVSLPSVSGSSSSASDSSSSESQSSDSGS 337
Query: 246 SF-------VESVTRNSVSEGEPPGLGVMMRFTSGRRSESWVDDSEIHVSR 289
S ES + SVS + + SES +S SR
Sbjct: 338 SSDSSTSQSSESASTPSVSSSQSSTSESASSTSESSNSESSQSESSTSESR 388
>ref|XP_641952.1| hypothetical protein DDB0206180 [Dictyostelium discoideum]
gi|60470006|gb|EAL67987.1| hypothetical protein
DDB0206180 [Dictyostelium discoideum]
Length = 1350
Score = 48.9 bits (115), Expect = 2e-04
Identities = 48/251 (19%), Positives = 103/251 (40%), Gaps = 23/251 (9%)
Query: 34 PPESSHHHPDSPPESFWLSRDEENEWLDRNFIYERKESTKGNSSSTNLNPNSNSNSN--- 90
P + H +SPP + ++ N + N + N+++ N N N+N+N+N
Sbjct: 336 PSQQIPHISNSPPSPYQITSYNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNN 395
Query: 91 --------STSNSQRFANLKSKTSMIGLPKPQKPS---FDAKNLRNHKPSNIKLFPKRSA 139
+++N+ N+ + ++I K P+ NL N+ +N +S
Sbjct: 396 NNNNNNNSNSNNNSSMGNISEELNLIYANKSSTPTNMPVVINNLGNNNNNNNNNIGTQSI 455
Query: 140 SVG---------KSFVEPSSPKVSCMGKVRSKRGKTSTVTAAAKAAVKEKTARTERKQSK 190
S +S EP S + +++ TS+VT + + E + +
Sbjct: 456 SSSSTPLIQSRKRSICEPPSNDLLINVSMQNTSSTTSSVTIPSPLSSFEIPFEKQIESQV 515
Query: 191 HSFFKSFRAIFRSGGRNKHDRMNDSSALDSSSITKNTNSVSDSKARDSTASLNDVSFVES 250
S F+SF + N ++ N+++ ++++ + NTN+ +++ + L E
Sbjct: 516 LSSFESFNNSNNNNNNNNNNNNNNNNNNNNNNNSNNTNTNNNNNSLRKDILLQISDTQEV 575
Query: 251 VTRNSVSEGEP 261
+ R+ + E P
Sbjct: 576 LYRSLLKENIP 586
Score = 40.0 bits (92), Expect = 0.077
Identities = 19/73 (26%), Positives = 34/73 (46%)
Query: 34 PPESSHHHPDSPPESFWLSRDEENEWLDRNFIYERKESTKGNSSSTNLNPNSNSNSNSTS 93
P S HH + P ++ S N + N + N+++ N N N+N+N+N+ +
Sbjct: 192 PMSPSQHHTNMPTVMYYNSNSNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNN 251
Query: 94 NSQRFANLKSKTS 106
N+ +N S S
Sbjct: 252 NNNSNSNSNSNNS 264
>ref|XP_641315.1| hypothetical protein DDB0206452 [Dictyostelium discoideum]
gi|60469343|gb|EAL67337.1| hypothetical protein
DDB0206452 [Dictyostelium discoideum]
Length = 2176
Score = 48.9 bits (115), Expect = 2e-04
Identities = 39/163 (23%), Positives = 80/163 (48%), Gaps = 28/163 (17%)
Query: 71 STKGNSSSTNLNPNSNSNSNSTSNSQRFANLKSKTSMIGLPKPQKPSFDAKNLRNHKPSN 130
S ++S++N N NSNSNSNS SNS N+ S ++ G+ P K ++KP
Sbjct: 315 SNSNSNSNSNSNSNSNSNSNSNSNSNNSNNVNS-NNINGVQAPPK---------DYKPEP 364
Query: 131 IKLFPKRSASVGKSFVEPSSPKVSCMGKVRSKRGKTSTVTAAAKAAVKEKTARTERKQSK 190
+ PK + +GK S + K+ S+ KT++ ++++ ++ + +T+ + +
Sbjct: 365 VN--PKEMSFIGKG----GSQILKLKKKITSRIDKTTSSSSSSSSSSSKNQNQTQNQNNV 418
Query: 191 HSFFKSFRAIFRSGGRNKHDRMNDSSALDSSSITKNTNSVSDS 233
+G N ++ N+++ ++++ N N+ S+S
Sbjct: 419 ------------NGTNNNNNNNNNNNNNNNNNNNNNNNNNSNS 449
Score = 39.3 bits (90), Expect = 0.13
Identities = 54/258 (20%), Positives = 100/258 (37%), Gaps = 32/258 (12%)
Query: 6 TLVSACAGGSTDRKIACETQADDLKTDPPPESSHHHPDSPPESFWLSRDEENEWLDRNFI 65
T S A T + + + + T PPP SS S E S +
Sbjct: 141 TPTSTIASTPTPTTVISDVTSPSVPTSPPPPSSTSTSTSTNEQTLTSAQISSS------- 193
Query: 66 YERKESTKGNSSSTNLNPNSNSNSNSTSNSQRFANLKSKTSMIGLPKPQKPSFDAKNLRN 125
S N ++ N N+N+N+N NS + + S+ G+ S ++ ++ +
Sbjct: 194 ----PSISNNMNNITNNNNNNNNNNGNRNS----IMVNSNSIDGI-----ASMNSNSITS 240
Query: 126 HKPSNIKLFPKRSASVGKSFVEPSSPKVSCMGKVRSKRGKTSTVTAAAKAAVKEKTARTE 185
S ++F + ++PSS + ++R K + + + T
Sbjct: 241 ESTSVFEIFTIDCWTPVDRDIKPSSNSPQILSELRVK------IMVKIRDIYFDTDTNTN 294
Query: 186 RKQSKHSFFKSFRAIFRSGGRNKHDRMNDSSALDSSSITKNTNSVSDSKARDS-TASLND 244
QS S I GG N + N +S +S+S N+NS S+S + +S + N+
Sbjct: 295 SAQSIDD--SSSSGIIGGGGTNSNSNSNSNSNSNSNS---NSNSNSNSNSNNSNNVNSNN 349
Query: 245 VSFVESVTRNSVSEGEPP 262
++ V++ ++ E P
Sbjct: 350 INGVQAPPKDYKPEPVNP 367
Score = 37.0 bits (84), Expect = 0.65
Identities = 41/207 (19%), Positives = 83/207 (39%), Gaps = 11/207 (5%)
Query: 34 PPESSHHHPDSPPESFWLSRDEENEWLDRNFIYERKESTKGNSSSTNLNPNSNSNSNSTS 93
PP+ P +P E ++ + + I R + T +SSS++ + + N N
Sbjct: 356 PPKDYKPEPVNPKEMSFIGKGGSQILKLKKKITSRIDKTTSSSSSSSSSSSKNQNQTQNQ 415
Query: 94 NSQRFANLKSKTSMIGLPKPQKPSFDAKNLRNHKPSNIKLFPKRS--ASVGKSFVEPSSP 151
N+ N + + + + N N+ ++ + P S ++ + EP++P
Sbjct: 416 NNVNGTNNNNNNNN---NNNNNNNNNNNNNNNNNSNSCNITPSNSDHNNIAGNQSEPTTP 472
Query: 152 KVSCMGKVRSKRGKTSTVTAAAKAAVKEKTARTERKQSKHSFFKSFRAIFRSGGRNKHDR 211
+ +S ++STVT A + + + T + + + +I S +
Sbjct: 473 PLHS----QSISSQSSTVTTAPPSPITQNNIATTTTTTTTTTTTTTASIAGSPNVQTTNT 528
Query: 212 MNDSSALDSSS--ITKNTNSVSDSKAR 236
N + SS IT N +S++ KAR
Sbjct: 529 TNSTGNNSGSSFLITNNKDSIAIEKAR 555
>emb|CAG59394.1| unnamed protein product [Candida glabrata CBS138]
gi|50288077|ref|XP_446467.1| unnamed protein product
[Candida glabrata]
Length = 371
Score = 48.5 bits (114), Expect = 2e-04
Identities = 48/214 (22%), Positives = 92/214 (42%), Gaps = 12/214 (5%)
Query: 71 STKGNSSSTNLNPNSNSNSNSTSNSQRFANLKSKTSMIGLPKPQKPSFDAKNLRNHKPSN 130
S+ +SSS++ + +S+S+S S+S S ++ S +S D+++ + S+
Sbjct: 38 SSSSSSSSSSSSSSSSSDSESSSTSSSSSSDSSSSSSSESSSSSDSGSDSESSSSSSSSS 97
Query: 131 IKLFPKRSASVGKSFVEPSSPKVSCMGKVRSKRGKTSTVTAAAKAAVKEKTARTERKQSK 190
S+ SS S S S+ ++++ ++ ++ + S
Sbjct: 98 SSSSSSSSSDSDSDSDSSSSSSSSSSSSSSSDSDSDSSSSSSSSSSDSSSSSSSSDSDSD 157
Query: 191 HSFFKSFRAIFRSGGRNKHDRMNDSSALDSSSITKNTNSVSDSKARDSTASLNDVSFVES 250
S S S + D ++S + SSS + + +S SDS + DS++S +D S +S
Sbjct: 158 SSSSSSSSDSSSSSSSSDSDSDSESDSSSSSSSSSSDSSSSDSSSSDSSSSDSDSSSSDS 217
Query: 251 VTRNSVSEGEPPGLGVMMRFTSGRRSESWVDDSE 284
+ +S S+ G SES +DSE
Sbjct: 218 DSSSSDSDSSDSG------------SESDTEDSE 239
Score = 40.8 bits (94), Expect = 0.045
Identities = 47/216 (21%), Positives = 91/216 (41%), Gaps = 11/216 (5%)
Query: 69 KESTKGNSSSTNLNPNSNSNSNSTSNSQRFANLKSKTSMIGLPKPQKPSFDAKNLRNHKP 128
KE+ +SSS++ S+S+S+S+S+S ++ S +S S + +
Sbjct: 14 KEAEASSSSSSS----SSSSSSSSSSSSSSSSSSSSSSSSSSSSSDSESSSTSSSSSSDS 69
Query: 129 SNIKLFPKRSASVGKSFVEPSSPKVSCMGKVRSKRGKTSTVTAAAKAAVKEKTARTERKQ 188
S+ S+S S E SS S +S+ +++ + + ++ +
Sbjct: 70 SSSSSSESSSSSDSGSDSESSSSS-------SSSSSSSSSSSSSDSDSDSDSSSSSSSSS 122
Query: 189 SKHSFFKSFRAIFRSGGRNKHDRMNDSSALDSSSITKNTNSVSDSKARDSTASLNDVSFV 248
S S S S + D + SS+ DS S + +++S SDS + S++ + S
Sbjct: 123 SSSSSSDSDSDSSSSSSSSSSDSSSSSSSSDSDSDSSSSSSSSDSSSSSSSSDSDSDSES 182
Query: 249 ESVTRNSVSEGEPPGLGVMMRFTSGRRSESWVDDSE 284
+S + +S S + +S S+S DS+
Sbjct: 183 DSSSSSSSSSSDSSSSDSSSSDSSSSDSDSSSSDSD 218
Score = 37.7 bits (86), Expect = 0.38
Identities = 52/228 (22%), Positives = 93/228 (39%), Gaps = 26/228 (11%)
Query: 37 SSHHHPDSPPESFWLSRDEENEWLDRNFIYERKESTKGNSSSTNLNPNSNSNSNSTSNSQ 96
SS S S S D E+ + + S+ SSS++ + + + +S+S+S+S
Sbjct: 38 SSSSSSSSSSSSSSSSSDSESSSTSSSSSSDSSSSSSSESSSSSDSGSDSESSSSSSSSS 97
Query: 97 RFANLKSKTSMIGLPKPQKPSFDAKNLRNHKPSNIKLFPKRSASVGKSFVEPSSPKVSCM 156
++ S + S + + + S+ S+S S SS S
Sbjct: 98 SSSSSSSSSDSDSDSDSSSSSSSSSSSSSSSDSD-----SDSSSSSSSSSSDSSSSSSSS 152
Query: 157 GKVRSKRGKTSTVTAAAKAAVKEKTARTERKQSKHSFFKSFRAIFRSGGRNKHDRMNDSS 216
+S+ +++ ++ + + +E S S S +DSS
Sbjct: 153 DSDSDSSSSSSSSDSSSSSSSSDSDSDSESDSSSSSSSSS----------------SDSS 196
Query: 217 ALDSSSITKNTNSVSDSKARDSTASLNDV----SFVESVTRNSVSEGE 260
+ DSSS + +++S SDS + DS +S +D S ES T +S SE E
Sbjct: 197 SSDSSS-SDSSSSDSDSSSSDSDSSSSDSDSSDSGSESDTEDSESESE 243
Score = 34.3 bits (77), Expect = 4.2
Identities = 32/150 (21%), Positives = 62/150 (41%), Gaps = 1/150 (0%)
Query: 135 PKRSASVGKSFVEPSSPKVSCMGKVRSKRGKTSTVTAAAKAAVKEKTARTERKQSKHSFF 194
PK SA ++ SS S S +S+ ++++ ++ ++ +E + S
Sbjct: 7 PKLSAKAKEAEASSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSDSESSSTSSSSS 66
Query: 195 KSFRAIFRSGGRNKHDRMNDSSALDSSSITKNTNSVSDSKARDSTASLNDVSFVESVTRN 254
+ S + D +DS + SSS + +++S S S DS + + S S + +
Sbjct: 67 SDSSSSSSSESSSSSDSGSDSESSSSSSSSSSSSSSSSSSDSDSDSD-SSSSSSSSSSSS 125
Query: 255 SVSEGEPPGLGVMMRFTSGRRSESWVDDSE 284
S S+ + +S S S DS+
Sbjct: 126 SSSDSDSDSSSSSSSSSSDSSSSSSSSDSD 155
>ref|XP_646432.1| hypothetical protein DDB0190700 [Dictyostelium discoideum]
gi|60474394|gb|EAL72331.1| hypothetical protein
DDB0190700 [Dictyostelium discoideum]
Length = 788
Score = 47.8 bits (112), Expect = 4e-04
Identities = 44/195 (22%), Positives = 85/195 (43%), Gaps = 21/195 (10%)
Query: 54 DEENEWLDRNFIYERKESTKGNSSSTNLNPNSNSNSNSTSNSQRFANLKSKTSMIGLPKP 113
DE++E+ D + N+++ N N N+N+N+N+ +N+ N +K + +
Sbjct: 105 DEDDEYYDSGNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNYNKNNNEEIDDY 164
Query: 114 QKPSFDAKNLRNHKPSNIKLFPKRSASVGKSFVEPSSPKVSCMGKVRSKRGKTSTVTAAA 173
+ D +N +N N+++ K + +E K GK + K K
Sbjct: 165 GEDEDDEENQQN----NLQIDEKENE------IENEKGKEKENGKEKEKEKK-------- 206
Query: 174 KAAVKEKTARTERKQSKHSFFKSFRAIFRSGGRNKHDRMNDSSALDSSSITKNTNSVSDS 233
K KEK E++Q +H + + +S N ++ ND+ + + K TN +S
Sbjct: 207 KEKEKEKEKEKEKEQQQHQQQQQQQ---QSINNNNNNNKNDNKSNNFKDALKITNQSDES 263
Query: 234 KARDSTASLNDVSFV 248
+ +T + N+ S V
Sbjct: 264 SSIITTHTPNESSTV 278
Database: nr
Posted date: Jul 5, 2005 12:34 AM
Number of letters in database: 863,360,394
Number of sequences in database: 2,540,612
Lambda K H
0.307 0.121 0.337
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 489,886,575
Number of Sequences: 2540612
Number of extensions: 20188906
Number of successful extensions: 282043
Number of sequences better than 10.0: 2062
Number of HSP's better than 10.0 without gapping: 584
Number of HSP's successfully gapped in prelim test: 1584
Number of HSP's that attempted gapping in prelim test: 213814
Number of HSP's gapped (non-prelim): 21280
length of query: 289
length of database: 863,360,394
effective HSP length: 127
effective length of query: 162
effective length of database: 540,702,670
effective search space: 87593832540
effective search space used: 87593832540
T: 11
A: 40
X1: 16 ( 7.1 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (21.6 bits)
S2: 74 (33.1 bits)
Medicago: description of AC140848.13


