

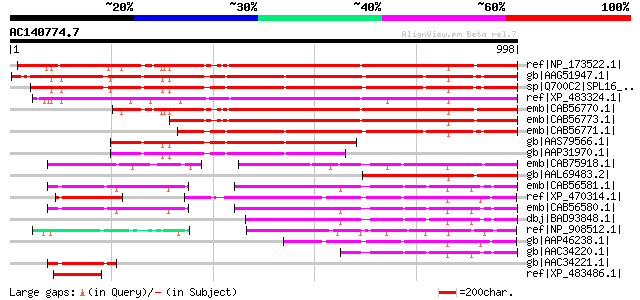
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC140774.7 + phase: 0 /pseudo
(998 letters)
Database: nr
2,540,612 sequences; 863,360,394 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
ref|NP_173522.1| SPL1-Related2 protein (SPL1R2) [Arabidopsis tha... 908 0.0
gb|AAG51947.1| unknown protein; 70902-74753 [Arabidopsis thaliana] 858 0.0
sp|Q700C2|SPL16_ARATH Squamosa-promoter binding-like protein 16 ... 858 0.0
ref|XP_483324.1| putative SPL1-Related2 protein [Oryza sativa (j... 787 0.0
emb|CAB56770.1| SPL1-Related2 protein [Arabidopsis thaliana] 709 0.0
emb|CAB56773.1| Spl1-Related2 protein [Arabidopsis thaliana] 674 0.0
emb|CAB56771.1| SPL1-Related3 protein [Arabidopsis thaliana] 643 0.0
gb|AAS79566.1| At1g76580 [Arabidopsis thaliana] gi|46367500|emb|... 392 e-107
gb|AAP31970.1| At1g76580 [Arabidopsis thaliana] gi|22330667|ref|... 357 1e-96
emb|CAB75918.1| squamosa promoter binding protein-like 12 [Arabi... 289 4e-76
gb|AAL69483.2| putative SPL1-related protein [Arabidopsis thaliana] 287 1e-75
emb|CAB56581.1| squamosa promoter binding protein-like 1 [Arabid... 287 1e-75
ref|XP_470314.1| putative SBP-domain protein [Oryza sativa (japo... 287 1e-75
emb|CAB56580.1| squamosa promoter binding protein-like 1 [Arabid... 287 1e-75
dbj|BAD93848.1| squamosa promoter binding protein-like 1 [Arabid... 274 1e-71
ref|NP_908512.1| unnamed protein product [Oryza sativa (japonica... 272 4e-71
gb|AAP46238.1| unknown protein, 5'-partial [Oryza sativa (japoni... 187 1e-45
gb|AAC34220.1| unknown protein [Arabidopsis thaliana] 175 6e-42
gb|AAC34221.1| putative squamosa-promoter binding protein [Arabi... 150 3e-34
ref|XP_483486.1| SBP-domain protein-like [Oryza sativa (japonica... 131 1e-28
>ref|NP_173522.1| SPL1-Related2 protein (SPL1R2) [Arabidopsis thaliana]
gi|67461574|sp|Q8RY95|SPL14_ARATH Squamosa-promoter
binding-like protein 14 (SPL1-related protein 2)
gi|4836890|gb|AAD30593.1| Unknown protein [Arabidopsis
thaliana]
Length = 1035
Score = 908 bits (2346), Expect = 0.0
Identities = 532/1060 (50%), Positives = 682/1060 (64%), Gaps = 110/1060 (10%)
Query: 16 LSSHQFYDSSNTKKRDL---LSSYDVVHIPN--DNWNPKEWNWDSIRFMTAKSTTVEPQQ 70
+++ F S +KRDL +S+ V P D WN K W+WDS RF AK VE Q+
Sbjct: 8 VAAPMFIHQSLGRKRDLYYPMSNRLVQSQPQRRDEWNSKMWDWDSRRF-EAKPVDVEVQE 66
Query: 71 V----------EESLNLNLGS-----------TGLVRPNKRIRSGSPTSASYPMCQVDNC 109
E L+LNLGS T VRPNK++RSGSP +YPMCQVDNC
Sbjct: 67 FDLTLRNRSGEERGLDLNLGSGLTAVEETTTTTQNVRPNKKVRSGSP-GGNYPMCQVDNC 125
Query: 110 KEDLSKAKDYHRRHKVCEAHSKASKALLGNQMQRFCQQCSRFHPLVEFDEGKRSCRRRLA 169
EDLS AKDYHRRHKVCE HSKA+KAL+G QMQRFCQQCSRFH L EFDEGKRSCRRRLA
Sbjct: 126 TEDLSHAKDYHRRHKVCEVHSKATKALVGKQMQRFCQQCSRFHLLSEFDEGKRSCRRRLA 185
Query: 170 GHNRRRRKT-QPDEVAVGGSPPLNQ------VAANLEIFNLLTAIADGSQGKFEER---- 218
GHNRRRRKT QP+EVA G P N AN+++ LLTA+A +QGK +
Sbjct: 186 GHNRRRRKTTQPEEVASGVVVPGNHDTTNNTANANMDLMALLTALAC-AQGKNAVKPPVG 244
Query: 219 RSQVPDKEQLVQILNRI---PLPADLTAKLLDVGNNLNAKNDNVQMETSPSYHHRDDQLN 275
VPD+EQL+QILN+I PLP DL +KL ++G+ D+ P+ + ++D +N
Sbjct: 245 SPAVPDREQLLQILNKINALPLPMDLVSKLNNIGSLARKNMDH------PTVNPQND-MN 297
Query: 276 NAPPAPLTKDFLAVLSTT--PSTP-----ARNGG---------------NGSTSSADHMR 313
A P+ T D LAVLSTT S+P GG NG T++ +
Sbjct: 298 GASPS--TMDLLAVLSTTLGSSSPDALAILSQGGFGNKDSEKTKLSSYENGVTTNLEKRT 355
Query: 314 --------ERSSGSSQSPNDDSDCQ-EDVRVKLPLQLFGSSPENDSPSKLPSSRKYFSSE 364
ERSS S+QSP+ DSD + +D R L LQLF SSPE++S + SSRKY+SS
Sbjct: 356 FGFSSVGGERSSSSNQSPSQDSDSRGQDTRSSLSLQLFTSSPEDESRPTVASSRKYYSSA 415
Query: 365 SSNPVDERTPSSSPPVVEMNFGLQGGIRGFNSNCISTGFGGNANKETSQSHSCTTIPLDL 424
SSNPV++R+PSSSP + E+ F LQ S +K +S C +PL+L
Sbjct: 416 SSNPVEDRSPSSSPVMQEL-FPLQASPETMRSK---------NHKNSSPRTGC--LPLEL 463
Query: 425 FKGSKSNNMIQQSSSVQSVPFKAGYASSGSDYSPPSLNSDTQDRTGRIMFKLFDKHPSHF 484
F S N + + ++GYASSGSDYSPPSLNSD QDRTG+I+FKL DK PS
Sbjct: 464 FGAS---NRGAADPNFKGFGQQSGYASSGSDYSPPSLNSDAQDRTGKIVFKLLDKDPSQL 520
Query: 485 PGTLRTQIYNWLSTRPSDLESYIRPGCVVLSIYASMSSAAWVQLEENFLQRVDSLIHNSD 544
PGTLR++IYNWLS PS++ESYIRPGCVVLS+Y +MS AAW QLE+ LQR+ L+ NS
Sbjct: 521 PGTLRSEIYNWLSNIPSEMESYIRPGCVVLSVYVAMSPAAWEQLEQKLLQRLGVLLQNSP 580
Query: 545 SDFWRNGRFLVYSGSQLASHKDGRIRMCKPWGTWRSPELISVSPLAIVGGQETSISLKGR 604
SDFWRN RF+V +G QLASHK+G++R K W TW SPELISVSP+A+V G+ETS+ ++GR
Sbjct: 581 SDFWRNARFIVNTGRQLASHKNGKVRCSKSWRTWNSPELISVSPVAVVAGEETSLVVRGR 640
Query: 605 NLSAPGTKIHCTGADCYTSSEVIGSGDPGMVYDEIKLSGFEVQNTSPSVLGRCFIEVENG 664
+L+ G I CT Y + EV + ++DE+ ++ F+VQN P LGRCFIEVENG
Sbjct: 641 SLTNDGISIRCTHMGSYMAMEVTRAVCRQTIFDELNVNSFKVQNVHPGFLGRCFIEVENG 700
Query: 665 FKGNSFPVIIANASICKELRPLESEFDEEEKMCDAISEEHEHHFGRPKSRDEALHFLNEL 724
F+G+SFP+IIANASICKEL L EF K D E+ + P SR+E L FLNEL
Sbjct: 701 FRGDSFPLIIANASICKELNRLGEEF--HPKSQDMTEEQAQSSNRGPTSREEVLCFLNEL 758
Query: 725 GWLFQRERFSNVHEVPDYSLDRFKFVLTFSVERNCCMLVKTLLDMLVDKHFEGEGLSTGS 784
GWLFQ+ + S + E D+SL RFKF+L SVER+ C L++TLLDMLV+++ + L+ +
Sbjct: 759 GWLFQKNQTSELREQSDFSLARFKFLLVCSVERDYCALIRTLLDMLVERNLVNDELNREA 818
Query: 785 VEMLKAIQLLNRAVKRKCTSMVDLLINYSITSKN-DTSKKYVFPPNLEGPGGITPLHLAA 843
++ML IQLLNRAVKRK T MV+LLI+Y + +S+K+VF PN+ GPGGITPLHLAA
Sbjct: 819 LDMLAEIQLLNRAVKRKSTKMVELLIHYLVNPLTLSSSRKFVFLPNITGPGGITPLHLAA 878
Query: 844 STTDSEGVIDSLTNDPQE----CWETLADENGQTPHAYAMMRNNHSYNMLVARKCSDRQR 899
T+ S+ +ID LTNDPQE W TL D GQTP++YA +RNNH+YN LVARK +D++
Sbjct: 879 CTSGSDDMIDLLTNDPQEIGLSSWNTLRDATGQTPYSYAAIRNNHNYNSLVARKLADKRN 938
Query: 900 SEVSVRIDNEIEHPSLGIELMQKRIN-QVKRVGDSCSKCAIAEVRAKRRFSGSRSWLHGP 958
+VS+ I++E+ + + KR++ ++ + SC+ CA ++ +RR SGS+ P
Sbjct: 939 KQVSLNIEHEVVDQT----GLSKRLSLEMNKSSSSCASCATVALKYQRRVSGSQRLFPTP 994
Query: 959 FIHSMLAVAAVCVCVCVLFRGTPYVGSVSPFRWENLNYGT 998
IHSMLAVA VCVCVCV P V S F W L+YG+
Sbjct: 995 IIHSMLAVATVCVCVCVFMHAFPIVRQGSHFSWGGLDYGS 1034
>gb|AAG51947.1| unknown protein; 70902-74753 [Arabidopsis thaliana]
Length = 1020
Score = 858 bits (2217), Expect = 0.0
Identities = 501/1065 (47%), Positives = 662/1065 (62%), Gaps = 121/1065 (11%)
Query: 3 KVAPPLLPLHPPMLSSHQFYDSSNTKKRDLLSSYDVVHIPNDNWNPKEWNWDSIRFMTAK 62
+VA P+ +H PM Y + L+SS +P D+W W WD RF +
Sbjct: 7 QVAAPI-SVHHPMGKKRDLY----VQPHWLVSS----QLPKDDWQMNRWKWDGQRFEAIE 57
Query: 63 STTVEPQ-QVEESLNLNLGS---------TGLVRPNKRIRSGSPTSA-----SYPMCQVD 107
Q ++ L+LNL L RP+K++RSGSP S +YP CQVD
Sbjct: 58 LQGESLQLSNKKGLDLNLPCGFNDVEGTPVDLTRPSKKVRSGSPGSGGGGGGNYPKCQVD 117
Query: 108 NCKEDLSKAKDYHRRHKVCEAHSKASKALLGNQMQRFCQQCSRFHPLVEFDEGKRSCRRR 167
NCKEDLS AKDYHRRHKVCE HSKA+KAL+G QMQRFCQQCSRFH L EFDEGKRSCRRR
Sbjct: 118 NCKEDLSIAKDYHRRHKVCEVHSKATKALVGKQMQRFCQQCSRFHLLSEFDEGKRSCRRR 177
Query: 168 LAGHNRRRRKTQPDEVAVGGSPPLNQVAA-----------NLEIFNLLTAI--ADGSQGK 214
L GHNRRRRKTQPD + +QV A N+++ LLTA+ A G
Sbjct: 178 LDGHNRRRRKTQPDAIT-------SQVVALENRDNTSNNTNMDVMALLTALVCAQGRNEA 230
Query: 215 FEERRSQVPDKEQLVQILNRI---PLPADLTAKLLDVGNNLNAKNDNVQMETSPSYHHRD 271
VP +EQL+QILN+I PLP +LT+KL ++G L KN PS +
Sbjct: 231 TTNGSPGVPQREQLLQILNKIKALPLPMNLTSKLNNIGI-LARKNPE-----QPSPMNPQ 284
Query: 272 DQLNNAPPAPLTKDFLAVLSTTPSTPA-------RNGGNGSTSSADHMR----------- 313
+ +N A +P T D LA LS + + A GG G+ S D +
Sbjct: 285 NSMNGAS-SPSTMDLLAALSASLGSSAPEAIAFLSQGGFGNKESNDRTKLTSSDHSATTS 343
Query: 314 --------------ERSSGSSQSPNDDSDCQ-EDVRVKLPLQLFGSSPENDSPSKLPSSR 358
ER+S ++ SP+ SD + +D R L LQLF SSPE +S K+ SS
Sbjct: 344 LEKKTLEFPSFGGGERTSSTNHSPSQYSDSRGQDTRSSLSLQLFTSSPEEESRPKVASST 403
Query: 359 KYFSSESSNPVDERTPSSSPPVVEMNFGLQGGIRGFNSNCISTGFGGNANKETSQSHSCT 418
KY+SS SSNPV++R+PSSSP + E+ F + N K+TS S +
Sbjct: 404 KYYSSASSNPVEDRSPSSSPVMQEL----------FPLHTSPETRRYNNYKDTSTSPRTS 453
Query: 419 TIPLDLFKGSKSNNMIQQSSSVQSVPFKAGYASSGSDYSPPSLNSDTQDRTGRIMFKLFD 478
+PL+LF SN + + + ++GYASSGSDYSPPSLNS+ Q+RTG+I FKLF+
Sbjct: 454 CLPLELF--GASNRGATANPNYNVLRHQSGYASSGSDYSPPSLNSNAQERTGKISFKLFE 511
Query: 479 KHPSHFPGTLRTQIYNWLSTRPSDLESYIRPGCVVLSIYASMSSAAWVQLEENFLQRVDS 538
K PS P TLRT+I+ WLS+ PSD+ES+IRPGCV+LS+Y +MS++AW QLEEN LQRV S
Sbjct: 512 KDPSQLPNTLRTEIFRWLSSFPSDMESFIRPGCVILSVYVAMSASAWEQLEENLLQRVRS 571
Query: 539 LIHNSDSDFWRNGRFLVYSGSQLASHKDGRIRMCKPWGTWRSPELISVSPLAIVGGQETS 598
L+ DS+FW N RFLV +G QLASHK G IR+ K W T PELI+VSPLA+V G+ET+
Sbjct: 572 LVQ--DSEFWSNSRFLVNAGRQLASHKHGEIRLSKSWRTLNLPELITVSPLAVVAGEETA 629
Query: 599 ISLKGRNLSAPGTKIHCTGADCYTSSEVIGSGDPGMVYDEIKLSGFEVQNTSPSVLGRCF 658
+ ++GRNL+ G ++ C Y S EV G DE+ +S F+VQ+ S LGRCF
Sbjct: 630 LIVRGRNLTNDGMRLRCAHMGNYASMEVTGREHRLTKVDELNVSSFQVQSASSVSLGRCF 689
Query: 659 IEVENGFKGNSFPVIIANASICKELRPLESEFDEEEKMCDAISEEHEHHFGRPKSRDEAL 718
IE+ENG +G++FP+IIANA+ICKEL LE EF ++ + EE + RP+SR+E L
Sbjct: 690 IELENGLRGDNFPLIIANATICKELNRLEEEFHPKD-----VIEEQIQNLDRPRSREEVL 744
Query: 719 HFLNELGWLFQRERFSNVHEVPDYSLDRFKFVLTFSVERNCCMLVKTLLDMLVDKHFEGE 778
FLNELGWLFQR+ S++H PD+SL RFKF+L SVER+ C L++T+LDM+V+++ +
Sbjct: 745 CFLNELGWLFQRKWTSDIHGEPDFSLPRFKFLLVCSVERDYCSLIRTVLDMMVERNLGKD 804
Query: 779 G-LSTGSVEMLKAIQLLNRAVKRKCTSMVDLLINYSITSKNDTSKKYVFPPNLEGPGGIT 837
G L+ S++ML IQLLNRA+KR+ T M + LI+YS+ N +++ ++F P++ GPG IT
Sbjct: 805 GLLNKESLDMLADIQLLNRAIKRRNTKMAETLIHYSV---NPSTRNFIFLPSIAGPGDIT 861
Query: 838 PLHLAASTTDSEGVIDSLTNDPQE----CWETLADENGQTPHAYAMMRNNHSYNMLVARK 893
PLHLAAST+ S+ +ID+LTNDPQE CW TL D GQTP +YA MR+NHSYN LVARK
Sbjct: 862 PLHLAASTSSSDDMIDALTNDPQEIGLSCWNTLVDATGQTPFSYAAMRDNHSYNTLVARK 921
Query: 894 CSDRQRSEVSVRIDNEIEHPSLGIELMQKRINQVKRVGDSCSKCAIAEVRAKRRFSGSRS 953
+D++ ++S+ I+N I+ I L ++ +++KR SC+ CA ++ +R+ SGSR
Sbjct: 922 LADKRNGQISLNIENGIDQ----IGLSKRLSSELKR---SCNTCASVALKYQRKVSGSRR 974
Query: 954 WLHGPFIHSMLAVAAVCVCVCVLFRGTPYVGSVSPFRWENLNYGT 998
P IHSMLAVA VCVCVCV P V S F W L+YG+
Sbjct: 975 LFPTPIIHSMLAVATVCVCVCVFMHAFPMVRQGSHFSWGGLDYGS 1019
>sp|Q700C2|SPL16_ARATH Squamosa-promoter binding-like protein 16 (SPL1-related protein 3)
Length = 988
Score = 858 bits (2217), Expect = 0.0
Identities = 492/1027 (47%), Positives = 648/1027 (62%), Gaps = 112/1027 (10%)
Query: 41 IPNDNWNPKEWNWDSIRFMTAKSTTVEPQ-QVEESLNLNLGS---------TGLVRPNKR 90
+P D+W W WD RF + Q ++ L+LNL L RP+K+
Sbjct: 4 LPKDDWQMNRWKWDGQRFEAIELQGESLQLSNKKGLDLNLPCGFNDVEGTPVDLTRPSKK 63
Query: 91 IRSGSPTSA-----SYPMCQVDNCKEDLSKAKDYHRRHKVCEAHSKASKALLGNQMQRFC 145
+RSGSP S +YP CQVDNCKEDLS AKDYHRRHKVCE HSKA+KAL+G QMQRFC
Sbjct: 64 VRSGSPGSGGGGGGNYPKCQVDNCKEDLSIAKDYHRRHKVCEVHSKATKALVGKQMQRFC 123
Query: 146 QQCSRFHPLVEFDEGKRSCRRRLAGHNRRRRKTQPDEVAVGGSPPLNQVAA--------- 196
QQCSRFH L EFDEGKRSCRRRL GHNRRRRKTQPD + +QV A
Sbjct: 124 QQCSRFHLLSEFDEGKRSCRRRLDGHNRRRRKTQPDAIT-------SQVVALENRDNTSN 176
Query: 197 --NLEIFNLLTAI--ADGSQGKFEERRSQVPDKEQLVQILNRI---PLPADLTAKLLDVG 249
N+++ LLTA+ A G VP +EQL+QILN+I PLP +LT+KL ++G
Sbjct: 177 NTNMDVMALLTALVCAQGRNEATTNGSPGVPQREQLLQILNKIKALPLPMNLTSKLNNIG 236
Query: 250 NNLNAKNDNVQMETSPSYHHRDDQLNNAPPAPLTKDFLAVLSTTPSTPA-------RNGG 302
L KN PS + + +N A +P T D LA LS + + A GG
Sbjct: 237 I-LARKNPE-----QPSPMNPQNSMNGAS-SPSTMDLLAALSASLGSSAPEAIAFLSQGG 289
Query: 303 NGSTSSADHMR-------------------------ERSSGSSQSPNDDSDCQ-EDVRVK 336
G+ S D + ER+S ++ SP+ SD + +D R
Sbjct: 290 FGNKESNDRTKLTSSDHSATTSLEKKTLEFPSFGGGERTSSTNHSPSQYSDSRGQDTRSS 349
Query: 337 LPLQLFGSSPENDSPSKLPSSRKYFSSESSNPVDERTPSSSPPVVEMNFGLQGGIRGFNS 396
L LQLF SSPE +S K+ SS KY+SS SSNPV++R+PSSSP + E+ F
Sbjct: 350 LSLQLFTSSPEEESRPKVASSTKYYSSASSNPVEDRSPSSSPVMQEL----------FPL 399
Query: 397 NCISTGFGGNANKETSQSHSCTTIPLDLFKGSKSNNMIQQSSSVQSVPFKAGYASSGSDY 456
+ N K+TS S + +PL+LF SN + + + ++GYASSGSDY
Sbjct: 400 HTSPETRRYNNYKDTSTSPRTSCLPLELF--GASNRGATANPNYNVLRHQSGYASSGSDY 457
Query: 457 SPPSLNSDTQDRTGRIMFKLFDKHPSHFPGTLRTQIYNWLSTRPSDLESYIRPGCVVLSI 516
SPPSLNS+ Q+RTG+I FKLF+K PS P TLRT+I+ WLS+ PSD+ES+IRPGCV+LS+
Sbjct: 458 SPPSLNSNAQERTGKISFKLFEKDPSQLPNTLRTEIFRWLSSFPSDMESFIRPGCVILSV 517
Query: 517 YASMSSAAWVQLEENFLQRVDSLIHNSDSDFWRNGRFLVYSGSQLASHKDGRIRMCKPWG 576
Y +MS++AW QLEEN LQRV SL+ DS+FW N RFLV +G QLASHK GRIR+ K W
Sbjct: 518 YVAMSASAWEQLEENLLQRVRSLVQ--DSEFWSNSRFLVNAGRQLASHKHGRIRLSKSWR 575
Query: 577 TWRSPELISVSPLAIVGGQETSISLKGRNLSAPGTKIHCTGADCYTSSEVIGSGDPGMVY 636
T PELI+VSPLA+V G+ET++ ++GRNL+ G ++ C Y S EV G
Sbjct: 576 TLNLPELITVSPLAVVAGEETALIVRGRNLTNDGMRLRCAHMGNYASMEVTGREHRLTKV 635
Query: 637 DEIKLSGFEVQNTSPSVLGRCFIEVENGFKGNSFPVIIANASICKELRPLESEFDEEEKM 696
DE+ +S F+VQ+ S LGRCFIE+ENG +G++FP+IIANA+ICKEL LE EF ++
Sbjct: 636 DELNVSSFQVQSASSVSLGRCFIELENGLRGDNFPLIIANATICKELNRLEEEFHPKD-- 693
Query: 697 CDAISEEHEHHFGRPKSRDEALHFLNELGWLFQRERFSNVHEVPDYSLDRFKFVLTFSVE 756
+ EE + RP+SR+E L FLNELGWLFQR+ S++H PD+SL RFKF+L SVE
Sbjct: 694 ---VIEEQIQNLDRPRSREEVLCFLNELGWLFQRKWTSDIHGEPDFSLPRFKFLLVCSVE 750
Query: 757 RNCCMLVKTLLDMLVDKHFEGEG-LSTGSVEMLKAIQLLNRAVKRKCTSMVDLLINYSIT 815
R+ C L++T+LDM+V+++ +G L+ S++ML IQLLNRA+KR+ T M + LI+YS+
Sbjct: 751 RDYCSLIRTVLDMMVERNLGKDGLLNKESLDMLADIQLLNRAIKRRNTKMAETLIHYSV- 809
Query: 816 SKNDTSKKYVFPPNLEGPGGITPLHLAASTTDSEGVIDSLTNDPQE----CWETLADENG 871
N +++ ++F P++ GPG ITPLHLAAST+ S+ +ID+LTNDPQE CW TL D G
Sbjct: 810 --NPSTRNFIFLPSIAGPGDITPLHLAASTSSSDDMIDALTNDPQEIGLSCWNTLVDATG 867
Query: 872 QTPHAYAMMRNNHSYNMLVARKCSDRQRSEVSVRIDNEIEHPSLGIELMQKRINQVKRVG 931
QTP +YA MR+NHSYN LVARK +D++ ++S+ I+N I+ I L ++ +++KR
Sbjct: 868 QTPFSYAAMRDNHSYNTLVARKLADKRNGQISLNIENGIDQ----IGLSKRLSSELKR-- 921
Query: 932 DSCSKCAIAEVRAKRRFSGSRSWLHGPFIHSMLAVAAVCVCVCVLFRGTPYVGSVSPFRW 991
SC+ CA ++ +R+ SGSR P IHSMLAVA VCVCVCV P V S F W
Sbjct: 922 -SCNTCASVALKYQRKVSGSRRLFPTPIIHSMLAVATVCVCVCVFMHAFPMVRQGSHFSW 980
Query: 992 ENLNYGT 998
L+YG+
Sbjct: 981 GGLDYGS 987
>ref|XP_483324.1| putative SPL1-Related2 protein [Oryza sativa (japonica
cultivar-group)] gi|42408812|dbj|BAD10073.1| putative
SPL1-Related2 protein [Oryza sativa (japonica
cultivar-group)]
Length = 1140
Score = 787 bits (2033), Expect = 0.0
Identities = 478/1101 (43%), Positives = 634/1101 (57%), Gaps = 158/1101 (14%)
Query: 45 NWNPKEWNWDSIRFMTAKSTTV----------EPQQVEES-------------------- 74
NWNP+ W+WDS R +TAK ++ QQ ++S
Sbjct: 50 NWNPRMWDWDS-RALTAKPSSDALRVNAGLSHHQQQQQQSPPAAAKAAEALRQGGGGSGG 108
Query: 75 LNLNLG-------------------------------STGLVRPNKRIRSGSPTSAS--- 100
LNL LG +VRP+KR+RSGSP SAS
Sbjct: 109 LNLQLGLREDAATPMDVSPAATTVSSSPSPPASSAPAQEPVVRPSKRVRSGSPGSASGGG 168
Query: 101 ---------------YPMCQVDNCKEDLSKAKDYHRRHKVCEAHSKASKALLGNQMQRFC 145
YPMCQVD+C+ DL+ AKDYHRRHKVCE H K +KAL+GNQMQRFC
Sbjct: 169 GGGGGGGNSGGGGGSYPMCQVDDCRADLTNAKDYHRRHKVCEIHGKTTKALVGNQMQRFC 228
Query: 146 QQCSRFHPLVEFDEGKRSCRRRLAGHNRRRRKTQPDEVAVGGSPPLNQV-AANL--EIFN 202
QQCSRFHPL EFDEGKRSCRRRLAGHNRRRRKTQP +VA P NQ AAN +I N
Sbjct: 229 QQCSRFHPLSEFDEGKRSCRRRLAGHNRRRRKTQPTDVASQLLLPGNQENAANRTQDIVN 288
Query: 203 LLTAIA--DGSQGKFEERRSQVPDKEQLVQILNRI-------PLPADLTAKLLDVGNNLN 253
L+T IA GS +PDK+ LVQI+++I ++ +D+ + +
Sbjct: 289 LITVIARLQGSNVGKLPSIPPIPDKDNLVQIISKINSINNGNSASKSPPSEAVDLNASHS 348
Query: 254 AKNDNVQMETS---PSYHHRDDQLN---------NAPPAPLTKDFLAVLSTTPST--PAR 299
+ D+VQ T+ + D Q N + P T D LAVLST +T P
Sbjct: 349 QQQDSVQRTTNGFEKQTNGLDKQTNGFDKQADGFDKQAVPSTMDLLAVLSTALATSNPDS 408
Query: 300 NGGNGSTSSADHMRERSSGSSQSP-NDDSDCQEDVRV----------------------- 335
N SS +S S P N + ++ +RV
Sbjct: 409 NTSQSQGSSDSSGNNKSKSQSTEPANVVNSHEKSIRVFSATRKNDALERSPEMYKQPDQE 468
Query: 336 ---KLPLQLFGSSPENDSPSKLPSSRKYFSSESSNPVDERTPSSSPPVVEMNFGLQGGIR 392
L L+LFGS+ E D P K+ ++ KY SSESSNP+DER+PSSSPPV F IR
Sbjct: 469 TPPYLSLRLFGST-EEDVPCKMDTANKYLSSESSNPLDERSPSSSPPVTHKFFP----IR 523
Query: 393 GFNSNCISTGFGGN-ANKETSQSHSCTTIPLDLFKGSKSNNMIQQSSSVQSVPFKAGYAS 451
+ + +G + A E S S + PL+LFK S+ ++ S + +++ Y S
Sbjct: 524 SVDEDARIADYGEDIATVEVSTSRAWRAPPLELFKDSERPI---ENGSPPNPAYQSCYTS 580
Query: 452 SG-SDYSPPSLNSDTQDRTGRIMFKLFDKHPSHFPGTLRTQIYNWLSTRPSDLESYIRPG 510
+ SD+SP + NSD QDRTGRI+FKLF K PS PG LR +I NWL P+++E YIRPG
Sbjct: 581 TSCSDHSPSTSNSDGQDRTGRIIFKLFGKEPSTIPGNLRGEIVNWLKHSPNEMEGYIRPG 640
Query: 511 CVVLSIYASMSSAAWVQLEENFLQRVDSLIHNSDSDFWRNGRFLVYSGSQLASHKDGRIR 570
C+VLS+Y SM + AW +LEEN LQRV++L+ SD DFWR GRFLV + +QL S+KDG R
Sbjct: 641 CLVLSMYLSMPAIAWDELEENLLQRVNTLVQGSDLDFWRKGRFLVRTDAQLVSYKDGATR 700
Query: 571 MCKPWGTWRSPELISVSPLAIVGGQETSISLKGRNLSAPGTKIHCTGADCYTSSEVIGSG 630
+ K W TW +PEL VSP+A+VGG++TS+ LKGRNL+ PGT+IHCT Y S EV+ S
Sbjct: 701 LSKSWRTWNTPELTFVSPIAVVGGRKTSLILKGRNLTIPGTQIHCTSTGKYISKEVLCSA 760
Query: 631 DPGMVYDEIKLSGFEVQNTSPSVLGRCFIEVENGFKGNSFPVIIANASICKELRPLESEF 690
PG +YD+ + F++ +LGR FIEVEN F+GNSFPVIIAN+S+C+ELR LE+E
Sbjct: 761 YPGTIYDDSGVETFDLPGEPHLILGRYFIEVENRFRGNSFPVIIANSSVCQELRSLEAEL 820
Query: 691 DEEEKMCDAISEEHEHHFGRPKSRDEALHFLNELGWLFQRERFSNVHEVPD--------Y 742
E + D S++ H R K +DE LHFLNELGWLFQ+ S E D +
Sbjct: 821 -EGSQFVDGSSDDQAHDARRLKPKDEVLHFLNELGWLFQKAAASTSAEKSDSSGLDLMYF 879
Query: 743 SLDRFKFVLTFSVERNCCMLVKTLLDMLVDKHFEGEGLSTGSVEMLKAIQLLNRAVKRKC 802
S RF+++L FS ER+ C L KTLL++L + + LS ++EML I LLNRAVKRK
Sbjct: 880 STARFRYLLLFSSERDWCSLTKTLLEILAKRSLASDELSQETLEMLSEIHLLNRAVKRKS 939
Query: 803 TSMVDLLINYSITSKNDTSKKYVFPPNLEGPGGITPLHLAASTTDSEGVIDSLTNDPQE- 861
+ M LL+ + + +D SK Y F PN+ GPGG+TPLHLAAS D+ ++D+LT+DPQ+
Sbjct: 940 SHMARLLVQFVVVCPDD-SKLYPFLPNVAGPGGLTPLHLAASIEDAVDIVDALTDDPQQI 998
Query: 862 ---CWETLADENGQTPHAYAMMRNNHSYNMLVARKCSDRQRSEVSVRIDNEIEHPSLGIE 918
CW + D++GQ+P YA +RNN++YN LVA+K DR+ ++V++ + E H
Sbjct: 999 GLSCWHSALDDDGQSPETYAKLRNNNAYNELVAQKLVDRKNNQVTIMVGKEEIHMDQSGN 1058
Query: 919 LMQKRINQVKRVG-DSCSKCAIAEVRAKRRFSGSRSWLHGPFIHSMLAVAAVCVCVCVLF 977
+ +K + ++ + SC++CAI + RR SR L P+IHSMLA+AAVCVCVCV
Sbjct: 1059 VGEKNKSAIQALQIRSCNQCAILDAGLLRRPMHSRGLLARPYIHSMLAIAAVCVCVCVFM 1118
Query: 978 RGTPYVGSVSPFRWENLNYGT 998
R S F+WE L++GT
Sbjct: 1119 RALLRFNSGRSFKWERLDFGT 1139
>emb|CAB56770.1| SPL1-Related2 protein [Arabidopsis thaliana]
Length = 812
Score = 709 bits (1829), Expect = 0.0
Identities = 411/840 (48%), Positives = 543/840 (63%), Gaps = 75/840 (8%)
Query: 203 LLTAIADGSQGKFEER----RSQVPDKEQLVQILNRI---PLPADLTAKLLDVGNNLNAK 255
LLTA+A +QGK + VPD+EQL+QILN+I PLP DL +KL ++G+
Sbjct: 3 LLTALAC-AQGKNAVKPPVGSPAVPDREQLLQILNKINALPLPMDLVSKLNNIGSLARKN 61
Query: 256 NDNVQMETSPSYHHRDDQLNNAPPAPLTKDFLAVLSTT--PSTP-----ARNGG------ 302
D+ P+ + ++D +N A P+ T D LAVLSTT S+P GG
Sbjct: 62 MDH------PTVNPQND-MNGASPS--TMDLLAVLSTTLGSSSPDALAILSQGGFGNKDS 112
Query: 303 ---------NGSTSSADHMR--------ERSSGSSQSPNDDSDCQ-EDVRVKLPLQLFGS 344
NG T++ + ERSS S+QSP+ DSD + +D R L LQLF S
Sbjct: 113 EKTKLSSYENGVTTNLEKRTFGFSSVGGERSSSSNQSPSQDSDSRGQDTRSSLSLQLFTS 172
Query: 345 SPENDSPSKLPSSRKYFSSESSNPVDERTPSSSPPVVEMNFGLQGGIRGFNSNCISTGFG 404
SPE++S + SSRKY+SS SSNPV++R+PSSSP + E+ F LQ S
Sbjct: 173 SPEDESRPTVASSRKYYSSASSNPVEDRSPSSSPVMQEL-FPLQASPETMRSK------- 224
Query: 405 GNANKETSQSHSCTTIPLDLFKGSKSNNMIQQSSSVQSVPFKAGYASSGSDYSPPSLNSD 464
+K +S C +PL+LF S N + + ++GYASSGSDYSPPSLNSD
Sbjct: 225 --NHKNSSPRTGC--LPLELFGAS---NRGAADPNFKGFGQQSGYASSGSDYSPPSLNSD 277
Query: 465 TQDRTGRIMFKLFDKHPSHFPGTLRTQIYNWLSTRPSDLESYIRPGCVVLSIYASMSSAA 524
QDRTG+I+FKL DK PS PGTLR++IYNWLS PS++ESYIRPGCVVLS+Y +MS AA
Sbjct: 278 AQDRTGKIVFKLLDKDPSQLPGTLRSEIYNWLSNIPSEMESYIRPGCVVLSVYVAMSPAA 337
Query: 525 WVQLEENFLQRVDSLIHNSDSDFWRNGRFLVYSGSQLASHKDGRIRMCKPWGTWRSPELI 584
W QLE+ LQR+ L+ NS SDFWRN RF+V +G QLASHK+G++R K W TW SPELI
Sbjct: 338 WEQLEQKLLQRLGVLLQNSPSDFWRNARFIVNTGRQLASHKNGKVRCSKSWRTWNSPELI 397
Query: 585 SVSPLAIVGGQETSISLKGRNLSAPGTKIHCTGADCYTSSEVIGSGDPGMVYDEIKLSGF 644
SVSP+A+V G+ETS+ ++GR+L+ G I CT Y + EV + ++DE+ ++ F
Sbjct: 398 SVSPVAVVAGEETSLVVRGRSLTNDGISIRCTHMGSYMAMEVTRAVCRQTIFDELNVNSF 457
Query: 645 EVQNTSPSVLGRCFIEVENGFKGNSFPVIIANASICKELRPLESEFDEEEKMCDAISEEH 704
+VQN P LGRCFIEVENGF+G+SFP+IIANASICKEL L EF K D E+
Sbjct: 458 KVQNVHPGFLGRCFIEVENGFRGDSFPLIIANASICKELNRLGEEF--HPKSQDMTEEQA 515
Query: 705 EHHFGRPKSRDEALHFLNELGWLFQRERFSNVHEVPDYSLDRFKFVLTFSVERNCCMLVK 764
+ P SR+E L FLNELGWLFQ+ + S + E D+SL RFKF+L SVER+ C L++
Sbjct: 516 QSSNRGPTSREEVLCFLNELGWLFQKNQTSELREQSDFSLARFKFLLVCSVERDYCALIR 575
Query: 765 TLLDMLVDKHFEGEGLSTGSVEMLKAIQLLNRAVKRKCTSMVDLLINYSIT-SKNDTSKK 823
TLLDMLV+++ + L+ +++ML IQLLNRAVKRK T MV+LLI+Y + S +S+K
Sbjct: 576 TLLDMLVERNLVNDELNREALDMLAEIQLLNRAVKRKSTKMVELLIHYLVNPSALSSSRK 635
Query: 824 YVFPPNLEGPGGITPLHLAASTTDSEGVIDSLTNDPQE----CWETLADENGQTPHAYAM 879
+VF PN+ GPGGITPLHLAA T+ S+ +ID LTNDPQE W TL D GQTP++YA
Sbjct: 636 FVFLPNITGPGGITPLHLAACTSGSDDMIDLLTNDPQEIGLSSWNTLRDATGQTPYSYAA 695
Query: 880 MRNNHSYNMLVARKCSDRQRSEVSVRIDNEIEHPSLGIELMQKRIN-QVKRVGDSCSKCA 938
+RNNH+YN LVARK +D++ +VS+ I++E+ + + KR++ ++ + SC+ CA
Sbjct: 696 IRNNHNYNSLVARKLADKRNKQVSLNIEHEVVDQT----GLSKRLSLEMNKSSSSCASCA 751
Query: 939 IAEVRAKRRFSGSRSWLHGPFIHSMLAVAAVCVCVCVLFRGTPYVGSVSPFRWENLNYGT 998
++ +RR SGS+ P IHSMLAVA VCVCVCV P V S F W L+YG+
Sbjct: 752 TVALKYQRRVSGSQRLFPTPIIHSMLAVATVCVCVCVFMHAFPIVRQGSHFSWGGLDYGS 811
>emb|CAB56773.1| Spl1-Related2 protein [Arabidopsis thaliana]
Length = 712
Score = 674 bits (1740), Expect = 0.0
Identities = 361/692 (52%), Positives = 472/692 (68%), Gaps = 28/692 (4%)
Query: 314 ERSSGSSQSPNDDSDCQ-EDVRVKLPLQLFGSSPENDSPSKLPSSRKYFSSESSNPVDER 372
ERSS S+QSP+ DSD + +D R L LQLF SSPE++S + SSRKY+SS SSNPV++R
Sbjct: 41 ERSSSSNQSPSQDSDSRGQDTRSSLSLQLFTSSPEDESRPTVASSRKYYSSASSNPVEDR 100
Query: 373 TPSSSPPVVEMNFGLQGGIRGFNSNCISTGFGGNANKETSQSHSCTTIPLDLFKGSKSNN 432
+PSSSP + E+ F LQ S +K +S C +PL+LF S N
Sbjct: 101 SPSSSPVMQEL-FPLQASPETMRSK---------NHKNSSPRTGC--LPLELFGAS---N 145
Query: 433 MIQQSSSVQSVPFKAGYASSGSDYSPPSLNSDTQDRTGRIMFKLFDKHPSHFPGTLRTQI 492
+ + ++GYASSGSDYSPPSLNSD QDRTG+I+FKL DK PS PGTLR++I
Sbjct: 146 RGAADPNFKGFGQQSGYASSGSDYSPPSLNSDAQDRTGKIVFKLLDKDPSQLPGTLRSEI 205
Query: 493 YNWLSTRPSDLESYIRPGCVVLSIYASMSSAAWVQLEENFLQRVDSLIHNSDSDFWRNGR 552
YNWLS PS++ESYIRPGCVVLS+Y +MS AAW QLE+ LQR+ L+ NS SDFWRN R
Sbjct: 206 YNWLSNIPSEMESYIRPGCVVLSVYVAMSPAAWEQLEQKLLQRLGVLLQNSPSDFWRNAR 265
Query: 553 FLVYSGSQLASHKDGRIRMCKPWGTWRSPELISVSPLAIVGGQETSISLKGRNLSAPGTK 612
F+V +G QLASHK+G++R K W TW SPELISVSP+A+V G+ETS+ ++GR+L+ G
Sbjct: 266 FIVNTGRQLASHKNGKVRCSKSWRTWNSPELISVSPVAVVAGEETSLVVRGRSLTNDGIS 325
Query: 613 IHCTGADCYTSSEVIGSGDPGMVYDEIKLSGFEVQNTSPSVLGRCFIEVENGFKGNSFPV 672
I CT Y + EV + ++DE+ ++ F+VQN P LGRCFIEVENGF+G+SFP+
Sbjct: 326 IRCTHMGSYMAMEVTRAVCRQTIFDELNVNSFKVQNVHPGFLGRCFIEVENGFRGDSFPL 385
Query: 673 IIANASICKELRPLESEFDEEEKMCDAISEEHEHHFGRPKSRDEALHFLNELGWLFQRER 732
IIANASICKEL L EF K D E+ + P SR+E L FLNELGWLFQ+ +
Sbjct: 386 IIANASICKELNRLGEEF--HPKSQDMTEEQAQSSNRGPTSREEVLCFLNELGWLFQKNQ 443
Query: 733 FSNVHEVPDYSLDRFKFVLTFSVERNCCMLVKTLLDMLVDKHFEGEGLSTGSVEMLKAIQ 792
S + E D+SL RFKF+L SVER+ C L++TLLDMLV+++ + L+ +++ML IQ
Sbjct: 444 TSELREQSDFSLARFKFLLVCSVERDYCALIRTLLDMLVERNLVNDELNREALDMLAEIQ 503
Query: 793 LLNRAVKRKCTSMVDLLINYSITSKN-DTSKKYVFPPNLEGPGGITPLHLAASTTDSEGV 851
LLNRAVKRK T MV+LLI+Y + +S+K+VF PN+ GPGGITPLHLAA T+ S+ +
Sbjct: 504 LLNRAVKRKSTKMVELLIHYLVNPLTLSSSRKFVFLPNITGPGGITPLHLAACTSGSDDM 563
Query: 852 IDSLTNDPQE----CWETLADENGQTPHAYAMMRNNHSYNMLVARKCSDRQRSEVSVRID 907
ID LTNDPQE W TL D GQTP++YA +RNNH+YN LVARK +D++ +VS+ I+
Sbjct: 564 IDLLTNDPQEIGLSSWNTLRDATGQTPYSYAAIRNNHNYNSLVARKLADKRNKQVSLNIE 623
Query: 908 NEIEHPSLGIELMQKRIN-QVKRVGDSCSKCAIAEVRAKRRFSGSRSWLHGPFIHSMLAV 966
+E+ + + KR++ ++ + SC+ CA ++ +RR SGS+ P IHSMLAV
Sbjct: 624 HEVVDQT----GLSKRLSLEMNKSSSSCASCATVALKYQRRVSGSQRLFPTPIIHSMLAV 679
Query: 967 AAVCVCVCVLFRGTPYVGSVSPFRWENLNYGT 998
A VCVCVCV P V S F W L+YG+
Sbjct: 680 ATVCVCVCVFMHAFPIVRQGSHFSWGGLDYGS 711
>emb|CAB56771.1| SPL1-Related3 protein [Arabidopsis thaliana]
Length = 647
Score = 643 bits (1658), Expect = 0.0
Identities = 342/673 (50%), Positives = 460/673 (67%), Gaps = 34/673 (5%)
Query: 331 EDVRVKLPLQLFGSSPENDSPSKLPSSRKYFSSESSNPVDERTPSSSPPVVEMNFGLQGG 390
+D R L LQLF SSPE +S K+ SS KY+SS SSNPV++R+PSSSP + E+
Sbjct: 3 QDTRSSLSLQLFTSSPEEESRPKVASSTKYYSSASSNPVEDRSPSSSPVMQEL------- 55
Query: 391 IRGFNSNCISTGFGGNANKETSQSHSCTTIPLDLFKGSKSNNMIQQSSSVQSVPFKAGYA 450
F + N K+TS S + +PL+LF SN + + + ++GYA
Sbjct: 56 ---FPLHTSPETRRYNNYKDTSTSPRTSCLPLELF--GASNRGATANPNYNVLRHQSGYA 110
Query: 451 SSGSDYSPPSLNSDTQDRTGRIMFKLFDKHPSHFPGTLRTQIYNWLSTRPSDLESYIRPG 510
SSGSDYSPPSLNS+ Q+RTG+I FKLF+K PS P TLRT+I+ WLS+ PSD+ES+IRPG
Sbjct: 111 SSGSDYSPPSLNSNAQERTGKISFKLFEKDPSQLPNTLRTEIFRWLSSFPSDMESFIRPG 170
Query: 511 CVVLSIYASMSSAAWVQLEENFLQRVDSLIHNSDSDFWRNGRFLVYSGSQLASHKDGRIR 570
CV+LS+Y +MS++AW QLEEN LQRV SL+ DS+FW N RFLV +G QLASHK GRIR
Sbjct: 171 CVILSVYVAMSASAWEQLEENLLQRVRSLVQ--DSEFWSNSRFLVNAGRQLASHKHGRIR 228
Query: 571 MCKPWGTWRSPELISVSPLAIVGGQETSISLKGRNLSAPGTKIHCTGADCYTSSEVIGSG 630
+ K W T PELI+VSPLA+V G+ET++ ++GRNL+ G ++ C Y S EV G
Sbjct: 229 LSKSWRTLNLPELITVSPLAVVAGEETALIVRGRNLTNDGMRLRCAHMGNYASMEVTGRE 288
Query: 631 DPGMVYDEIKLSGFEVQNTSPSVLGRCFIEVENGFKGNSFPVIIANASICKELRPLESEF 690
DE+ +S F+VQ+ S LGRCFIE+ENG +G++FP+IIANA+ICKEL LE EF
Sbjct: 289 HRLTKVDELNVSSFQVQSASSVSLGRCFIELENGLRGDNFPLIIANATICKELNRLEEEF 348
Query: 691 DEEEKMCDAISEEHEHHFGRPKSRDEALHFLNELGWLFQRERFSNVHEVPDYSLDRFKFV 750
++ + EE + RP+SR+E L FLNELGWLFQR+ S++H PD+SL RFKF+
Sbjct: 349 HPKD-----VIEEQIQNLDRPRSREEVLCFLNELGWLFQRKWTSDIHGEPDFSLPRFKFL 403
Query: 751 LTFSVERNCCMLVKTLLDMLVDKHFEGEG-LSTGSVEMLKAIQLLNRAVKRKCTSMVDLL 809
L SVER+ C L++T+LDM+V+++ +G L+ S++ML IQLLNRA+KR+ T M + L
Sbjct: 404 LVCSVERDYCSLIRTVLDMMVERNLGKDGLLNKESLDMLADIQLLNRAIKRRNTKMAETL 463
Query: 810 INYSITSKNDTSKKYVFPPNLEGPGGITPLHLAASTTDSEGVIDSLTNDPQE----CWET 865
I+YS+ N +++ ++F P++ GPG ITPLHLAAST+ S+ +ID+LTNDPQE CW T
Sbjct: 464 IHYSV---NPSTRNFIFLPSIAGPGDITPLHLAASTSSSDDMIDALTNDPQEIGLSCWNT 520
Query: 866 LADENGQTPHAYAMMRNNHSYNMLVARKCSDRQRSEVSVRIDNEIEHPSLGIELMQKRIN 925
L D GQTP +YA MR+NHSYN LVARK +D++ ++S+ I+N I+ I L ++ +
Sbjct: 521 LVDATGQTPFSYAAMRDNHSYNTLVARKLADKRNGQISLNIENGIDQ----IGLSKRLSS 576
Query: 926 QVKRVGDSCSKCAIAEVRAKRRFSGSRSWLHGPFIHSMLAVAAVCVCVCVLFRGTPYVGS 985
++KR SC+ CA ++ +R+ SGSR P IHSMLAVA VCVCVCV P V
Sbjct: 577 ELKR---SCNTCASVALKYQRKVSGSRRLFPTPIIHSMLAVATVCVCVCVFMHAFPMVRQ 633
Query: 986 VSPFRWENLNYGT 998
S F W L+YG+
Sbjct: 634 GSHFSWGGLDYGS 646
>gb|AAS79566.1| At1g76580 [Arabidopsis thaliana] gi|46367500|emb|CAG25876.1|
hypothetical protein [Arabidopsis thaliana]
Length = 503
Score = 392 bits (1007), Expect = e-107
Identities = 237/524 (45%), Positives = 318/524 (60%), Gaps = 59/524 (11%)
Query: 198 LEIFNLLTAI--ADGSQGKFEERRSQVPDKEQLVQILNRI---PLPADLTAKLLDVGNNL 252
+++ LLTA+ A G VP +EQL+QILN+I PLP +LT+KL ++G L
Sbjct: 1 MDVMALLTALVCAQGRNEATTNGSPGVPQREQLLQILNKIKALPLPMNLTSKLNNIGI-L 59
Query: 253 NAKNDNVQMETSPSYHHRDDQLNNAPPAPLTKDFLAVLSTTPSTPA-------RNGGNGS 305
KN PS + + +N A +P T D LA LS + + A GG G+
Sbjct: 60 ARKNPE-----QPSPMNPQNSMNGAS-SPSTMDLLAALSASLGSSAPEAIAFLSQGGFGN 113
Query: 306 TSSADHMR-------------------------ERSSGSSQSPNDDSDCQ-EDVRVKLPL 339
S D + ER+S ++ SP+ SD + +D R L L
Sbjct: 114 KESNDRTKLTSSDHSATTSLEKKTLEFPSFGGGERTSSTNHSPSQYSDSRGQDTRSSLSL 173
Query: 340 QLFGSSPENDSPSKLPSSRKYFSSESSNPVDERTPSSSPPVVEMNFGLQGGIRGFNSNCI 399
QLF SSPE +S K+ SS KY+SS SSNPV++R+PSSSP + E+ F +
Sbjct: 174 QLFTSSPEEESRPKVASSTKYYSSASSNPVEDRSPSSSPVMQEL----------FPLHTS 223
Query: 400 STGFGGNANKETSQSHSCTTIPLDLFKGSKSNNMIQQSSSVQSVPFKAGYASSGSDYSPP 459
N K+TS S + +PL+LF SN + + + ++GYASSGSDYSPP
Sbjct: 224 PETRRYNNYKDTSTSPRTSCLPLELF--GASNRGATANPNYNVLRHQSGYASSGSDYSPP 281
Query: 460 SLNSDTQDRTGRIMFKLFDKHPSHFPGTLRTQIYNWLSTRPSDLESYIRPGCVVLSIYAS 519
SLNS+ Q+RTG+I FKLF+K PS P TLRT+I+ WLS+ PSD+ES+IRPGCV+LS+Y +
Sbjct: 282 SLNSNAQERTGKISFKLFEKDPSQLPNTLRTEIFRWLSSFPSDMESFIRPGCVILSVYVA 341
Query: 520 MSSAAWVQLEENFLQRVDSLIHNSDSDFWRNGRFLVYSGSQLASHKDGRIRMCKPWGTWR 579
MS++AW QLEEN LQRV SL+ DS+FW N RFLV +G QLASHK GRIR+ K W T
Sbjct: 342 MSASAWEQLEENLLQRVRSLVQ--DSEFWSNSRFLVNAGRQLASHKHGRIRLSKSWRTLN 399
Query: 580 SPELISVSPLAIVGGQETSISLKGRNLSAPGTKIHCTGADCYTSSEVIGSGDPGMVYDEI 639
PELI+VSPLA+V G+ET++ ++GRNL+ G ++ C Y S EV G DE+
Sbjct: 400 LPELITVSPLAVVAGEETALIVRGRNLTNDGMRLRCAHMGNYASMEVTGREHRLTKVDEL 459
Query: 640 KLSGFEVQNTSPSVLGRCFIEVENGFKGNSFPVIIANASICKEL 683
+S F+VQ+ S LGRCFIE+ENG +G++FP+IIANA+ICKEL
Sbjct: 460 NVSSFQVQSASSVSLGRCFIELENGLRGDNFPLIIANATICKEL 503
>gb|AAP31970.1| At1g76580 [Arabidopsis thaliana] gi|22330667|ref|NP_177784.2|
SPL1-Related3 protein (SPL1R3) [Arabidopsis thaliana]
gi|17065188|gb|AAL32748.1| Unknown protein [Arabidopsis
thaliana]
Length = 488
Score = 357 bits (915), Expect = 1e-96
Identities = 221/501 (44%), Positives = 296/501 (58%), Gaps = 59/501 (11%)
Query: 198 LEIFNLLTAI--ADGSQGKFEERRSQVPDKEQLVQILNRI---PLPADLTAKLLDVGNNL 252
+++ LLTA+ A G VP +EQL+QILN+I PLP +LT+KL ++G L
Sbjct: 1 MDVMALLTALVCAQGRNEATTNGSPGVPQREQLLQILNKIKALPLPMNLTSKLNNIGI-L 59
Query: 253 NAKNDNVQMETSPSYHHRDDQLNNAPPAPLTKDFLAVLSTTPSTPA-------RNGGNGS 305
KN PS + + +N A +P T D LA LS + + A GG G+
Sbjct: 60 ARKNPE-----QPSPMNPQNSMNGAS-SPSTMDLLAALSASLGSSAPEAIAFLSQGGFGN 113
Query: 306 TSSADHMR-------------------------ERSSGSSQSPNDDSDCQ-EDVRVKLPL 339
S D + ER+S ++ SP+ SD + +D R L L
Sbjct: 114 KESNDRTKLTSSDHSATTSLEKKTLEFPSFGGGERTSSTNHSPSQYSDSRGQDTRSSLSL 173
Query: 340 QLFGSSPENDSPSKLPSSRKYFSSESSNPVDERTPSSSPPVVEMNFGLQGGIRGFNSNCI 399
QLF SSPE +S K+ SS KY+SS SSNPV++R+PSSSP + E+ F +
Sbjct: 174 QLFTSSPEEESRPKVASSTKYYSSASSNPVEDRSPSSSPVMQEL----------FPLHTS 223
Query: 400 STGFGGNANKETSQSHSCTTIPLDLFKGSKSNNMIQQSSSVQSVPFKAGYASSGSDYSPP 459
N K+TS S + +PL+LF SN + + + ++GYASSGSDYSPP
Sbjct: 224 PETRRYNNYKDTSTSPRTSCLPLELF--GASNRGATANPNYNVLRHQSGYASSGSDYSPP 281
Query: 460 SLNSDTQDRTGRIMFKLFDKHPSHFPGTLRTQIYNWLSTRPSDLESYIRPGCVVLSIYAS 519
SLNS+ Q+RTG+I FKLF+K PS P TLRT+I+ WLS+ PSD+ES+IRPGCV+LS+Y +
Sbjct: 282 SLNSNAQERTGKISFKLFEKDPSQLPNTLRTEIFRWLSSFPSDMESFIRPGCVILSVYVA 341
Query: 520 MSSAAWVQLEENFLQRVDSLIHNSDSDFWRNGRFLVYSGSQLASHKDGRIRMCKPWGTWR 579
MS++AW QLEEN LQRV SL+ DS+FW N RFLV +G QLASHK GRIR+ K W T
Sbjct: 342 MSASAWEQLEENLLQRVRSLV--QDSEFWSNSRFLVNAGRQLASHKHGRIRLSKSWRTLN 399
Query: 580 SPELISVSPLAIVGGQETSISLKGRNLSAPGTKIHCTGADCYTSSEVIGSGDPGMVYDEI 639
PELI+VSPLA+V G+ET++ ++GRNL+ G ++ C Y S EV G DE+
Sbjct: 400 LPELITVSPLAVVAGEETALIVRGRNLTNDGMRLRCAHMGNYASMEVTGREHRLTKVDEL 459
Query: 640 KLSGFEVQNTSPSVLGRCFIE 660
+S F+VQ+ S LGRCFIE
Sbjct: 460 NVSSFQVQSASSVSLGRCFIE 480
>emb|CAB75918.1| squamosa promoter binding protein-like 12 [Arabidopsis thaliana]
gi|6006403|emb|CAB56769.1| squamosa promoter binding
protein-like 12 [Arabidopsis thaliana]
gi|6006395|emb|CAB56768.1| squamosa promoter binding
protein-like 12 [Arabidopsis thaliana]
gi|28392866|gb|AAO41870.1| putative squamosa promoter
binding protein 12 [Arabidopsis thaliana]
gi|67461584|sp|Q9S7P5|SPL12_ARATH Squamosa-promoter
binding-like protein 12 gi|15232241|ref|NP_191562.1|
squamosa promoter-binding protein-like 12 (SPL12)
[Arabidopsis thaliana]
Length = 927
Score = 289 bits (739), Expect = 4e-76
Identities = 199/573 (34%), Positives = 296/573 (50%), Gaps = 53/573 (9%)
Query: 450 ASSGSDYSPPSLNSDTQDRTGRIMFKLFDKHPSHFPGTLRTQIYNWLSTRPSDLESYIRP 509
+ S SD SP S + D Q RT RI+FKLF K P+ FP LR QI NWL+ P+D+ESYIRP
Sbjct: 383 SDSASDQSPSSSSGDAQSRTDRIVFKLFGKEPNDFPVALRGQILNWLAHTPTDMESYIRP 442
Query: 510 GCVVLSIYASMSSAAWVQLEENFLQRVDSLIHNSDSDFWRNGRFLVYSGSQLASHKDGRI 569
GC+VL+IY A+W +L + + L+ SD W +G + +QLA +G++
Sbjct: 443 GCIVLTIYLRQDEASWEELCCDLSFSLRRLLDLSDDPLWTDGWLYLRVQNQLAFAFNGQV 502
Query: 570 RM--CKPWGTWRSPELISVSPLAIVGGQETSISLKGRNLSAPGTKIHCTGADCYTSSEVI 627
+ P + ++I+V PLA+ ++ ++KG NL PGT++ CT + E
Sbjct: 503 VLDTSLPLRSHDYSQIITVRPLAVT--KKAQFTVKGINLRRPGTRLLCTVEGTHLVQEAT 560
Query: 628 GSG----DPGMVYDEIKLSGFEVQNTSPSVLGRCFIEVEN--GFKGNSFPVIIA-NASIC 680
G D +EI F + P GR F+E+E+ G + FP I++ + IC
Sbjct: 561 QGGMEERDDLKENNEIDFVNFSCE--MPIASGRGFMEIEDQGGLSSSFFPFIVSEDEDIC 618
Query: 681 KELRPLESEFDEEEKMCDAISEEHEHHFGRPKSRDEALHFLNELGWLFQRERFSNVHEVP 740
E+R LES + F S +A+ F++E+GWL R +
Sbjct: 619 SEIRRLESTLE----------------FTGTDSAMQAMDFIHEIGWLLHRSELKSRLAAS 662
Query: 741 D------YSLDRFKFVLTFSVERNCCMLVKTLLDMLVDKHFEGEGLSTGSVEMLKAIQLL 794
D +SL RFKF++ FS++R C ++K LL++L FE + L + LL
Sbjct: 663 DHNPEDLFSLIRFKFLIEFSMDREWCCVMKKLLNIL----FEEGTVDPSPDAALSELCLL 718
Query: 795 NRAVKRKCTSMVDLLINYSITSKNDTSKKYVFPPNLEGPGGITPLHLAASTTDSEGVIDS 854
+RAV++ MV++L+ +S KN T +F P+ GPGG+TPLH+AA SE V+D+
Sbjct: 719 HRAVRKNSKPMVEMLLRFSPKKKNQTLAG-LFRPDAAGPGGLTPLHIAAGKDGSEDVLDA 777
Query: 855 LTNDPQ----ECWETLADENGQTPHAYAMMRNNHSYNMLVARKCSDR--QRSEVSVRIDN 908
LT DP + W+ D G TP YA +R + SY LV RK S + + V V I
Sbjct: 778 LTEDPGMTGIQAWKNSRDNTGFTPEDYARLRGHFSYIHLVQRKLSRKPIAKEHVVVNIPE 837
Query: 909 --EIEHPSLGIELMQKRINQVKRVGDSCSKCAIAEVRAKRRFSGSRSWLHGPFIHSMLAV 966
IEH M ++ ++ ++C + + + + +S + P + SM+A+
Sbjct: 838 SFNIEHKQEKRSPMDSSSLEITQI----NQCKLCDHKRVFVTTHHKSVAYRPAMLSMVAI 893
Query: 967 AAVCVCVCVLFRGTPYVGSV-SPFRWENLNYGT 998
AAVCVCV +LF+ P V V PFRWE L YGT
Sbjct: 894 AAVCVCVALLFKSCPEVLYVFQPFRWELLEYGT 926
Score = 149 bits (376), Expect = 5e-34
Identities = 114/321 (35%), Positives = 151/321 (46%), Gaps = 41/321 (12%)
Query: 75 LNLNLGSTGLVRPN-KRIRSGSPTSASYPMCQVDNCKEDLSKAKDYHRRHKVCEAHSKAS 133
L LNLG + K+ + G + CQVDNC DLSK KDYHRRHKVCE HSKA+
Sbjct: 97 LTLNLGGNNIEGNGVKKTKLGGGIPSRAICCQVDNCGADLSKVKDYHRRHKVCEIHSKAT 156
Query: 134 KALLGNQMQRFCQQCSRFHPLVEFDEGKRSCRRRLAGHNRRRRKTQPDEVAVGGSPPLNQ 193
AL+G MQRFCQQCSRFH L EFDEGKRSCRRRLAGHN+RRRK PD + G S +Q
Sbjct: 157 TALVGGIMQRFCQQCSRFHVLEEFDEGKRSCRRRLAGHNKRRRKANPDTIGNGTSMSDDQ 216
Query: 194 VAANLEIFNLLTAIADGSQGKFEERRSQVPDKEQLVQILNRIPLPAD-----LTAKLLDV 248
+N + LL +++ + Q D++ L +L + A LL
Sbjct: 217 -TSNYMLITLLKILSN----IHSNQSDQTGDQDLLSHLLKSLVSQAGEHIGRNLVGLLQG 271
Query: 249 GNNLNAKND--NVQMETSPSYHHRDDQLNNAPPAPLTKDFLAVLSTTPSTPARNGGNGST 306
G L A + N+ S R+D +++ +S TP
Sbjct: 272 GGGLQASQNIGNLSALLSLEQAPREDIKHHS------------VSETPWQEVYANSAQER 319
Query: 307 SSADHMRERSSGSSQSPND---DSDCQEDVRVKLPLQLFGSSPENDSPSKL-------PS 356
+ D ++ + ND DSD D+ P P N + S L S
Sbjct: 320 VAPDRSEKQVKVNDFDLNDIYIDSDDTTDIERSSP------PPTNPATSSLDYHQDSRQS 373
Query: 357 SRKYFSSESSNPVDERTPSSS 377
S S +S+ +++PSSS
Sbjct: 374 SPPQTSRRNSDSASDQSPSSS 394
>gb|AAL69483.2| putative SPL1-related protein [Arabidopsis thaliana]
Length = 314
Score = 287 bits (735), Expect = 1e-75
Identities = 152/310 (49%), Positives = 207/310 (66%), Gaps = 10/310 (3%)
Query: 695 KMCDAISEEHEHHFGRPKSRDEALHFLNELGWLFQRERFSNVHEVPDYSLDRFKFVLTFS 754
K D E+ + P SR+E L FLNELGWLFQ+ + S + E D+SL RFKF+L S
Sbjct: 8 KSQDMTEEQAQSSNRGPTSREEVLCFLNELGWLFQKNQTSELREQSDFSLARFKFLLVCS 67
Query: 755 VERNCCMLVKTLLDMLVDKHFEGEGLSTGSVEMLKAIQLLNRAVKRKCTSMVDLLINYSI 814
VER+ C L++TLLDMLV+++ + L+ +++ML IQLLNRAVKRK T MV+LLI+Y +
Sbjct: 68 VERDYCALIRTLLDMLVERNLVNDELNREALDMLAEIQLLNRAVKRKSTKMVELLIHYLV 127
Query: 815 TSKN-DTSKKYVFPPNLEGPGGITPLHLAASTTDSEGVIDSLTNDPQE----CWETLADE 869
+S+K+VF PN+ GPGGITPLHLAA T+ S+ +ID LTNDPQE W TL D
Sbjct: 128 NPLTLSSSRKFVFLPNITGPGGITPLHLAACTSGSDDMIDLLTNDPQEIGLSSWNTLRDA 187
Query: 870 NGQTPHAYAMMRNNHSYNMLVARKCSDRQRSEVSVRIDNEIEHPSLGIELMQKRIN-QVK 928
GQTP++YA +RNNH+YN LVARK +D++ +VS+ I++E+ + + KR++ ++
Sbjct: 188 TGQTPYSYAAIRNNHNYNSLVARKLADKRNKQVSLNIEHEVVDQT----GLSKRLSLEMN 243
Query: 929 RVGDSCSKCAIAEVRAKRRFSGSRSWLHGPFIHSMLAVAAVCVCVCVLFRGTPYVGSVSP 988
+ SC+ CA ++ +RR SGS+ P IHSMLAVA VCVCVCV P V S
Sbjct: 244 KSSSSCASCATVALKYQRRVSGSQRLFPTPIIHSMLAVATVCVCVCVFMHAFPIVRQGSH 303
Query: 989 FRWENLNYGT 998
F W L+YG+
Sbjct: 304 FSWGGLDYGS 313
>emb|CAB56581.1| squamosa promoter binding protein-like 1 [Arabidopsis thaliana]
gi|3650274|emb|CAA09698.1| squamosa-promoter binding
protein-like 1 [Arabidopsis thaliana]
gi|67461551|sp|Q9SMX9|SPL1_ARATH Squamosa-promoter
binding-like protein 1 gi|30690591|ref|NP_850468.1|
squamosa promoter-binding protein-like 1 (SPL1)
[Arabidopsis thaliana]
Length = 881
Score = 287 bits (734), Expect = 1e-75
Identities = 198/579 (34%), Positives = 296/579 (50%), Gaps = 54/579 (9%)
Query: 442 SVPFKAGYASSGSDYSPPSLNSDTQDRTGRIMFKLFDKHPSHFPGTLRTQIYNWLSTRPS 501
S P + + S SD SP S + D Q RTGRI+FKLF K P+ FP LR QI +WLS P+
Sbjct: 334 SPPQTSRNSDSASDQSPSSSSEDAQMRTGRIVFKLFGKEPNEFPIVLRGQILDWLSHSPT 393
Query: 502 DLESYIRPGCVVLSIYASMSSAAWVQLEENFLQRVDSLIHNSDSDFWRNGRFLVYSGSQL 561
D+ESYIRPGC+VL+IY + AW +L ++ + L+ SD W G V +QL
Sbjct: 394 DMESYIRPGCIVLTIYLRQAETAWEELSDDLGFSLGKLLDLSDDPLWTTGWIYVRVQNQL 453
Query: 562 ASHKDGRIRMCKPWG--TWRSPELISVSPLAIVGGQETSISLKGRNLSAPGTKIHCTGAD 619
A +G++ + + +ISV PLAI ++ ++KG NL GT++ C+
Sbjct: 454 AFVYNGQVVVDTSLSLKSRDYSHIISVKPLAIAATEKAQFTVKGMNLRQRGTRLLCSVEG 513
Query: 620 CYTSSEVIGSGDPGMVYDEIKLSGFEVQNTS-----PSVLGRCFIEVEN-GFKGNSFP-V 672
Y E D+ K + V+ + P + GR F+E+E+ G + FP +
Sbjct: 514 KYLIQETTHDSTT-REDDDFKDNSEIVECVNFSCDMPILSGRGFMEIEDQGLSSSFFPFL 572
Query: 673 IIANASICKELRPLESEFDEEEKMCDAISEEHEHHFGRPKSRDEALHFLNELGWLFQRER 732
++ + +C E+R LE+ + F S +A+ F++E+GWL R +
Sbjct: 573 VVEDDDVCSEIRILETTLE----------------FTGTDSAKQAMDFIHEIGWLLHRSK 616
Query: 733 FSNVHEVPD-YSLDRFKFVLTFSVERNCCMLVKTLLDMLVDKHFEGEGLSTGSVEMLKAI 791
P + L RF++++ FS++R C +++ LL+M D GE S+ S L +
Sbjct: 617 LGESDPNPGVFPLIRFQWLIEFSMDREWCAVIRKLLNMFFDGAV-GE-FSSSSNATLSEL 674
Query: 792 QLLNRAVKRKCTSMVDLLINYSITSKNDTSKKYVFPPNLEGPGGITPLHLAASTTDSEGV 851
LL+RAV++ MV++L+ Y + ++ +F P+ GP G+TPLH+AA SE V
Sbjct: 675 CLLHRAVRKNSKPMVEMLLRYIPKQQRNS----LFRPDAAGPAGLTPLHIAAGKDGSEDV 730
Query: 852 IDSLTNDPQ----ECWETLADENGQTPHAYAMMRNNHSYNMLVARKCSDRQRSEVSVRI- 906
+D+LT DP E W+T D G TP YA +R + SY L+ RK + + +E V +
Sbjct: 731 LDALTEDPAMVGIEAWKTCRDSTGFTPEDYARLRGHFSYIHLIQRKINKKSTTEDHVVVN 790
Query: 907 ------DNEIEHPSLGIELMQKRINQVKRVGDSCSKCAIAEVRAKRRFSGSRSWLHGPFI 960
D E + P G I Q+ C C V R RS + P +
Sbjct: 791 IPVSFSDREQKEPKSGPMASALEITQI-----PCKLCDHKLVYGTTR----RSVAYRPAM 841
Query: 961 HSMLAVAAVCVCVCVLFRGTPYVGSV-SPFRWENLNYGT 998
SM+A+AAVCVCV +LF+ P V V PFRWE L+YGT
Sbjct: 842 LSMVAIAAVCVCVALLFKSCPEVLYVFQPFRWELLDYGT 880
Score = 155 bits (392), Expect = 6e-36
Identities = 113/305 (37%), Positives = 153/305 (50%), Gaps = 38/305 (12%)
Query: 74 SLNLNLGSTGLVRPNKRIRSGSPTSASYPMCQVDNCKEDLSKAKDYHRRHKVCEAHSKAS 133
+LNLN S GL P K+ +SG+ +CQV+NC+ DLSK KDYHRRHKVCE HSKA+
Sbjct: 84 TLNLNGESDGLF-PAKKTKSGA-------VCQVENCEADLSKVKDYHRRHKVCEMHSKAT 135
Query: 134 KALLGNQMQRFCQQCSRFHPLVEFDEGKRSCRRRLAGHNRRRRKTQPDEVAVGGSPPLNQ 193
A +G +QRFCQQCSRFH L EFDEGKRSCRRRLAGHN+RRRKT P+ A G P +
Sbjct: 136 SATVGGILQRFCQQCSRFHLLQEFDEGKRSCRRRLAGHNKRRRKTNPEPGANGN--PSDD 193
Query: 194 VAANLEIFNLLTAIAD-----GSQGKFEERRSQVPDK--EQLVQILNRIPLPADLTAKLL 246
++N + LL +++ G Q + EQL + L + L + L
Sbjct: 194 HSSNYLLITLLKILSNMHNHTGDQDLMSHLLKSLVSHAGEQLGKNLVELLLQGGGSQGSL 253
Query: 247 DVGNNLNAKNDNVQMETSPSYHHRDDQL---NNAPPAPLTKDF--LAVLSTTPSTPARNG 301
++GN+ + E + R D N + DF + + T
Sbjct: 254 NIGNSALLGIEQAPQEELKQFSARQDGTATENRSEKQVKMNDFDLNDIYIDSDDTDVERS 313
Query: 302 ---GNGSTSSADH------------MRERSSGSSQSPNDDSDCQEDVRVKLPLQLFGSSP 346
N +TSS D+ R S S QSP+ S+ + ++ +LFG P
Sbjct: 314 PPPTNPATSSLDYPSWIHQSSPPQTSRNSDSASDQSPSSSSEDAQMRTGRIVFKLFGKEP 373
Query: 347 ENDSP 351
N+ P
Sbjct: 374 -NEFP 377
>ref|XP_470314.1| putative SBP-domain protein [Oryza sativa (japonica
cultivar-group)] gi|40714694|gb|AAR88600.1| putative
SBP-domain protein [Oryza sativa (japonica
cultivar-group)]
Length = 969
Score = 287 bits (734), Expect = 1e-75
Identities = 215/685 (31%), Positives = 336/685 (48%), Gaps = 72/685 (10%)
Query: 344 SSPENDSPSKLPSSRKYFSSESSNPVDERTPSSS---PPVVEM-NFGLQ---GGIRGFNS 396
+ P N + + +S S PV ++P+ + PP + +F L GG+ GF
Sbjct: 324 AGPSNSKMPFVNGDQCGLASSSVVPVQSKSPTVATPDPPACKFKDFDLNDTYGGMEGFED 383
Query: 397 NCISTGFGGNANKETSQSHSCTTIPLDLFKGSKSNNMIQ--QSSSVQSVPFKAGYASSGS 454
G+ G+ P FK + S N S QS P +G + S S
Sbjct: 384 -----GYEGS--------------PTPAFKTTDSPNCPSWMHQDSTQSPPQTSGNSDSTS 424
Query: 455 DYSPPSLNSDTQDRTGRIMFKLFDKHPSHFPGTLRTQIYNWLSTRPSDLESYIRPGCVVL 514
S S N D Q RT +I+FKLF+K PS P LR+QI WLS+ P+D+ESYIRPGC++L
Sbjct: 425 AQSLSSSNGDAQCRTDKIVFKLFEKVPSDLPPVLRSQILGWLSSSPTDIESYIRPGCIIL 484
Query: 515 SIYASMSSAAWVQLEENFLQRVDSLIHNSDSDFWRNGRFLVYSGSQLASHKDGRIRMCKP 574
++Y + +AW +L +N +D L+++S +FW +G V Q+A +G++ + +P
Sbjct: 485 TVYLRLVESAWKELSDNMSSYLDKLLNSSTGNFWASGLVFVMVRHQIAFMHNGQLMLDRP 544
Query: 575 W--GTWRSPELISVSPLAIVGGQETSISLKGRNLSAPGTKIHCTGADCYTSSEVIGSGDP 632
+++ V P+A + + ++G NL + +++ C+ I D
Sbjct: 545 LANSAHHYCKILCVRPIAAPFSTKVNFRVEGLNLVSDSSRLICS-----FEGSCIFQEDT 599
Query: 633 GMVYDEIKLSGFEVQN---TSPSVLGRCFIEVEN-GFKGNSFPVIIANASICKELRPLES 688
+ D+++ E N PS GR F+EVE+ GF FP IIA IC E+ LES
Sbjct: 600 DNIVDDVEHDDIEYLNFCCPLPSSRGRGFVEVEDGGFSNGFFPFIIAEQDICSEVCELES 659
Query: 689 EFDEEEKMCDAISEEHEHHFGRPKSRDEALHFLNELGWLFQRER-FSNVHEVP--DYSLD 745
F+ S HE +R++AL FLNELGWL R S +VP +++
Sbjct: 660 IFE---------SSSHE-QADDDNARNQALEFLNELGWLLHRANIISKQDKVPLASFNIW 709
Query: 746 RFKFVLTFSVERNCCMLVKTLLDMLVDKHFEG--EGLSTGSVEMLKAIQLLNRAVKRKCT 803
RF+ + F++ER C + K LLD L F G + S E++ + LL+ AV+ K
Sbjct: 710 RFRNLGIFAMEREWCAVTKLLLDFL----FTGLVDIGSQSPEEVVLSENLLHAAVRMKSA 765
Query: 804 SMVDLLINYSIT-SKNDTSKKYVFPPNLEGPGGITPLHLAASTTDSEGVIDSLTNDPQ-- 860
MV L+ Y S T++ ++F P+ +GP TPLH+AA+T D+E V+D+LTNDP
Sbjct: 766 QMVRFLLGYKPNESLKRTAETFLFRPDAQGPSKFTPLHIAAATDDAEDVLDALTNDPGLV 825
Query: 861 --ECWETLADENGQTPHAYAMMRNNHSYNMLVARKCSDR-QRSEVSVRIDNEIEHPSLGI 917
W D G TP YA R N +Y +V +K + + V + + + I HP +
Sbjct: 826 GINTWRNARDGAGFTPEDYARQRGNDAYLNMVEKKINKHLGKGHVVLGVPSSI-HPVITD 884
Query: 918 ELMQKRIN-----QVKRVGDSCSKCAIAEVRAKRRFSGSRSWLHGPFIHSMLAVAAVCVC 972
+ ++ V SC+ C+ + S +R++L+ P + +++ +A +CVC
Sbjct: 885 GVKPGEVSLEIGMTVPPPAPSCNACSRQALMYPN--STARTFLYRPAMLTVMGIAVICVC 942
Query: 973 VCVLFRGTPYVGSVSPFRWENLNYG 997
V +L P V + FRWE L G
Sbjct: 943 VGLLLHTCPKVYAAPTFRWELLERG 967
Score = 154 bits (390), Expect = 1e-35
Identities = 77/132 (58%), Positives = 94/132 (70%), Gaps = 4/132 (3%)
Query: 90 RIRSGSPTSASYPMCQVDNCKEDLSKAKDYHRRHKVCEAHSKASKALLGNQMQRFCQQCS 149
R++ GSP+ P CQV+ C DL+ +DYHRRHKVCE H+KA+ A++GN +QRFCQQCS
Sbjct: 141 RVQGGSPSG---PACQVEGCTADLTGVRDYHRRHKVCEMHAKATTAVVGNTVQRFCQQCS 197
Query: 150 RFHPLVEFDEGKRSCRRRLAGHNRRRRKTQPDEVAVGGSPPLNQVAANLEIFNLLTAIAD 209
RFHPL EFDEGKRSCRRRLAGHNRRRRKT+P EVAVGGS ++ + LL A+
Sbjct: 198 RFHPLQEFDEGKRSCRRRLAGHNRRRRKTRP-EVAVGGSAFTEDKISSYLLLGLLGVCAN 256
Query: 210 GSQGKFEERRSQ 221
+ E R Q
Sbjct: 257 LNADNAEHLRGQ 268
>emb|CAB56580.1| squamosa promoter binding protein-like 1 [Arabidopsis thaliana]
Length = 881
Score = 287 bits (734), Expect = 1e-75
Identities = 198/579 (34%), Positives = 296/579 (50%), Gaps = 54/579 (9%)
Query: 442 SVPFKAGYASSGSDYSPPSLNSDTQDRTGRIMFKLFDKHPSHFPGTLRTQIYNWLSTRPS 501
S P + + S SD SP S + D Q RTGRI+FKLF K P+ FP LR QI +WLS P+
Sbjct: 334 SPPQTSRNSDSASDQSPSSSSEDAQMRTGRIVFKLFGKEPNEFPIVLRGQILDWLSHSPT 393
Query: 502 DLESYIRPGCVVLSIYASMSSAAWVQLEENFLQRVDSLIHNSDSDFWRNGRFLVYSGSQL 561
D+ESYIRPGC+VL+IY + AW +L ++ + L+ SD W G V +QL
Sbjct: 394 DMESYIRPGCIVLTIYLRQAETAWEELSDDLGFSLGKLLDLSDDPLWTTGWIYVRVQNQL 453
Query: 562 ASHKDGRIRMCKPWG--TWRSPELISVSPLAIVGGQETSISLKGRNLSAPGTKIHCTGAD 619
A +G++ + + +ISV PLAI ++ ++KG NL GT++ C+
Sbjct: 454 AFVYNGQVVVDTSLSLKSRDYSHIISVKPLAIAATEKAQFTVKGMNLRRRGTRLLCSVEG 513
Query: 620 CYTSSEVIGSGDPGMVYDEIKLSGFEVQNTS-----PSVLGRCFIEVEN-GFKGNSFP-V 672
Y E D+ K + V+ + P + GR F+E+E+ G + FP +
Sbjct: 514 KYLIQETTHDSTT-REDDDFKDNSEIVECVNFSCDMPILSGRGFMEIEDQGLSSSFFPFL 572
Query: 673 IIANASICKELRPLESEFDEEEKMCDAISEEHEHHFGRPKSRDEALHFLNELGWLFQRER 732
++ + +C E+R LE+ + F S +A+ F++E+GWL R +
Sbjct: 573 VVEDDDVCSEIRILETTLE----------------FTGTDSAKQAMDFIHEIGWLLHRSK 616
Query: 733 FSNVHEVPD-YSLDRFKFVLTFSVERNCCMLVKTLLDMLVDKHFEGEGLSTGSVEMLKAI 791
P + L RF++++ FS++R C +++ LL+M D GE S+ S L +
Sbjct: 617 LGESDPNPGVFPLIRFQWLIEFSMDREWCAVIRKLLNMFFDGAV-GE-FSSSSNATLSEL 674
Query: 792 QLLNRAVKRKCTSMVDLLINYSITSKNDTSKKYVFPPNLEGPGGITPLHLAASTTDSEGV 851
LL+RAV++ MV++L+ Y + ++ +F P+ GP G+TPLH+AA SE V
Sbjct: 675 CLLHRAVRKNSKPMVEMLLRYIPKQQRNS----LFRPDAAGPAGLTPLHIAAGKDGSEDV 730
Query: 852 IDSLTNDPQ----ECWETLADENGQTPHAYAMMRNNHSYNMLVARKCSDRQRSEVSVRI- 906
+D+LT DP E W+T D G TP YA +R + SY L+ RK + + +E V +
Sbjct: 731 LDALTEDPAMVGIEAWKTCRDSTGFTPEDYARLRGHFSYIHLIQRKINKKSTTEDHVVVN 790
Query: 907 ------DNEIEHPSLGIELMQKRINQVKRVGDSCSKCAIAEVRAKRRFSGSRSWLHGPFI 960
D E + P G I Q+ C C V R RS + P +
Sbjct: 791 IPVSFSDREQKEPKSGPMASALEITQI-----PCKLCDHKLVYGTTR----RSVAYRPAM 841
Query: 961 HSMLAVAAVCVCVCVLFRGTPYVGSV-SPFRWENLNYGT 998
SM+A+AAVCVCV +LF+ P V V PFRWE L+YGT
Sbjct: 842 LSMVAIAAVCVCVALLFKSCPEVLYVFQPFRWELLDYGT 880
Score = 152 bits (384), Expect = 5e-35
Identities = 112/305 (36%), Positives = 152/305 (49%), Gaps = 38/305 (12%)
Query: 74 SLNLNLGSTGLVRPNKRIRSGSPTSASYPMCQVDNCKEDLSKAKDYHRRHKVCEAHSKAS 133
+LNLN S GL P K+ +SG+ +CQV+NC+ DLSK KDYHRRHKVCE HSKA+
Sbjct: 84 TLNLNGESDGLF-PAKKTKSGA-------VCQVENCEADLSKVKDYHRRHKVCEMHSKAT 135
Query: 134 KALLGNQMQRFCQQCSRFHPLVEFDEGKRSCRRRLAGHNRRRRKTQPDEVAVGGSPPLNQ 193
A + +QRFCQQCSRFH L EFDEGKRSCRRRLAGHN+RRRKT P+ A G P +
Sbjct: 136 SATVEGILQRFCQQCSRFHLLQEFDEGKRSCRRRLAGHNKRRRKTNPEPGANGN--PSDD 193
Query: 194 VAANLEIFNLLTAIAD-----GSQGKFEERRSQVPDK--EQLVQILNRIPLPADLTAKLL 246
++N + LL +++ G Q + EQL + L + L + L
Sbjct: 194 HSSNYLLITLLKILSNMHNHTGDQDLMSHLLKSLVSHAGEQLGKNLVELLLQGGGSQGSL 253
Query: 247 DVGNNLNAKNDNVQMETSPSYHHRDDQL---NNAPPAPLTKDF--LAVLSTTPSTPARNG 301
++GN+ + E + R D N + DF + + T
Sbjct: 254 NIGNSALLGIEQAPQEELKQFSARQDGTATENRSEKQVKMNDFDLNDIYIDSDDTDVERS 313
Query: 302 ---GNGSTSSADH------------MRERSSGSSQSPNDDSDCQEDVRVKLPLQLFGSSP 346
N +TSS D+ R S S QSP+ S+ + ++ +LFG P
Sbjct: 314 PPPTNPATSSLDYPSWIHQSSPPQTSRNSDSASDQSPSSSSEDAQMRTGRIVFKLFGKEP 373
Query: 347 ENDSP 351
N+ P
Sbjct: 374 -NEFP 377
>dbj|BAD93848.1| squamosa promoter binding protein-like 1 [Arabidopsis thaliana]
Length = 528
Score = 274 bits (700), Expect = 1e-71
Identities = 189/556 (33%), Positives = 285/556 (50%), Gaps = 54/556 (9%)
Query: 465 TQDRTGRIMFKLFDKHPSHFPGTLRTQIYNWLSTRPSDLESYIRPGCVVLSIYASMSSAA 524
+Q RTGRI+FKLF K P+ FP LR QI +WLS P+D+ESYIRPGC+VL+IY + A
Sbjct: 4 SQMRTGRIVFKLFGKEPNEFPIVLRGQILDWLSHSPTDMESYIRPGCIVLTIYLRQAETA 63
Query: 525 WVQLEENFLQRVDSLIHNSDSDFWRNGRFLVYSGSQLASHKDGRIRMCKPWG--TWRSPE 582
W +L ++ + L+ SD W G V +QLA +G++ + +
Sbjct: 64 WEELSDDLGFSLGKLLDLSDDPLWTTGWIYVRVQNQLAFVYNGQVVVDTSLSLKSRDYSH 123
Query: 583 LISVSPLAIVGGQETSISLKGRNLSAPGTKIHCTGADCYTSSEVIGSGDPGMVYDEIKLS 642
+ISV PLAI ++ ++KG NL GT++ C+ Y E D+ K +
Sbjct: 124 IISVKPLAIAATEKAQFTVKGMNLRQRGTRLLCSVEGKYLIQETTHDSTT-REDDDFKDN 182
Query: 643 GFEVQNTS-----PSVLGRCFIEVEN-GFKGNSFP-VIIANASICKELRPLESEFDEEEK 695
V+ + P + GR F+E+E+ G + FP +++ + +C E+R LE+ +
Sbjct: 183 SEIVECVNFSCDMPILSGRGFMEIEDQGLSSSFFPFLVVEDDDVCSEIRILETTLE---- 238
Query: 696 MCDAISEEHEHHFGRPKSRDEALHFLNELGWLFQRERFSNVHEVPD-YSLDRFKFVLTFS 754
F S +A+ F++E+GWL R + P + L RF++++ FS
Sbjct: 239 ------------FTGTDSAKQAMDFIHEIGWLLHRSKLGESDPNPGVFPLIRFQWLIEFS 286
Query: 755 VERNCCMLVKTLLDMLVDKHFEGEGLSTGSVEMLKAIQLLNRAVKRKCTSMVDLLINYSI 814
++R C +++ LL+M D GE S+ S L + LL+RAV++ MV++L+ Y
Sbjct: 287 MDREWCAVIRKLLNMFFDGAV-GE-FSSSSNATLSELCLLHRAVRKNSKPMVEMLLRYIP 344
Query: 815 TSKNDTSKKYVFPPNLEGPGGITPLHLAASTTDSEGVIDSLTNDPQ----ECWETLADEN 870
+ ++ +F P+ GP G+TPLH+AA SE V+D+LT DP E W+T D
Sbjct: 345 KQQRNS----LFRPDAAGPAGLTPLHIAAGKDGSEDVLDALTEDPAMVGIEAWKTCRDST 400
Query: 871 GQTPHAYAMMRNNHSYNMLVARKCSDRQRSEVSVRI-------DNEIEHPSLGIELMQKR 923
G TP YA +R + SY L+ RK + + +E V + D E + P G
Sbjct: 401 GFTPEDYARLRGHFSYIHLIQRKINKKSTTEDHVVVNIPVSFSDREQKEPKSGPMASALE 460
Query: 924 INQVKRVGDSCSKCAIAEVRAKRRFSGSRSWLHGPFIHSMLAVAAVCVCVCVLFRGTPYV 983
I Q+ C C V R RS + P + SM+A+AAVCVCV +LF+ P V
Sbjct: 461 ITQI-----PCKLCDHKLVYGTTR----RSVAYRPAMLSMVAIAAVCVCVALLFKSCPEV 511
Query: 984 GSV-SPFRWENLNYGT 998
V PFRWE L+YGT
Sbjct: 512 LYVFQPFRWELLDYGT 527
>ref|NP_908512.1| unnamed protein product [Oryza sativa (japonica cultivar-group)]
gi|8468036|dbj|BAA96636.1| putative squamosa promoter
binding protein-like 1 [Oryza sativa (japonica
cultivar-group)]
Length = 862
Score = 272 bits (696), Expect = 4e-71
Identities = 183/580 (31%), Positives = 289/580 (49%), Gaps = 74/580 (12%)
Query: 466 QDRTGRIMFKLFDKHPSHFPGTLRTQIYNWLSTRPSDLESYIRPGCVVLSIYASMSSAAW 525
++RT +I+FKLF K P+ FP LR QI +WLS PSD+ESYIRPGC++L+IY + + W
Sbjct: 307 ENRTDKIVFKLFGKEPNDFPSDLRAQILSWLSNCPSDIESYIRPGCIILTIYMRLPNWMW 366
Query: 526 VQLEENFLQRVDSLIHNSDSDFWRNGRFLVYSGSQLASHKDGRIRMCKPW--GTWRSPEL 583
+L + + LI S WR G L +G + + PW ++
Sbjct: 367 DKLAADPAHWIQKLISLSTDTLWRTGWMYARVQDYLTLSCNGNLMLASPWQPAIGNKHQI 426
Query: 584 ISVSPLAIVGGQETSISLKGRNLSAPGTKIHCTGADCYTSSEVIGSGDPGMVYDEIKLSG 643
+ ++P+A+ + S+KG N++ P TK+ C Y E + D+ K+
Sbjct: 427 LFITPIAVACSSTANFSVKGLNIAQPTTKLLCIFGGKYLIQEATEK-----LLDDTKMQR 481
Query: 644 ----FEVQNTSPSVLGRCFIEVENGFKGN-SFPVIIANASICKELRPLES-----EFDEE 693
+ PS GR FIEVE+ + + SFP ++A +C E+R LE FD
Sbjct: 482 GPQCLTFSCSFPSTSGRGFIEVEDLDQSSLSFPFVVAEEDVCSEIRTLEHLLNLVSFD-- 539
Query: 694 EKMCDAISEEHEHHFGRPKSRDEALHFLNELGWLFQRERFSNVHEVP-----DYSLDRFK 748
D + E+++ SRD AL+FL+E GW QR E P + RF+
Sbjct: 540 ----DTLVEKND----LLASRDRALNFLHEFGWFLQRSHIRATSETPKDCTEGFPAARFR 591
Query: 749 FVLTFSVERNCCMLVKTLLDMLVDKHFEGEGLSTGSVEMLKAIQLLNRAVKRKCTSMVDL 808
++L+F+V+R C ++K LLD L + + ST VE + L+ AV ++ ++D
Sbjct: 592 WLLSFAVDREFCAVIKKLLDTLFQGGVDLDVQST--VEFVLKQDLVFVAVNKRSKPLIDF 649
Query: 809 LINYSITS------KNDTSKKYVFPPNLEGPGGITPLHLAASTTDSEGVIDSLTNDPQE- 861
L+ Y+ +S ++ +++F P++ GP ITPLH+AA+ +D+ GV+D+LT+DPQ+
Sbjct: 650 LLTYTTSSAPMDGTESAAPAQFLFTPDIAGPSDITPLHIAATYSDTAGVLDALTDDPQQL 709
Query: 862 ---CWETLADENGQTPHAYAMMRNNHSYNMLVARKCSDR-QRSEVSVRIDNEIEHPSLGI 917
W+ D G TP YA R + SY +V K R ++ VSV I + +
Sbjct: 710 GIKAWKNARDATGLTPEDYARKRGHESYIEMVQNKIDSRLPKAHVSVTISS----TTSTT 765
Query: 918 ELMQKRINQVKRVGD-----------------SCSKC--AIAEVRAKRRFSGSRSWLHGP 958
+ +K +Q K SC +C +A RF +R P
Sbjct: 766 DFTEKHASQSKTTDQTAFDVEKGQQISTKPPLSCRQCLPELAYRHHLNRFLSTR-----P 820
Query: 959 FIHSMLAVAAVCVCVCVLFRGTPYVGSV-SPFRWENLNYG 997
+ S++A+AAVCVCV ++ +G P++G + PFRW +L G
Sbjct: 821 AVLSLVAIAAVCVCVGLIMQGPPHIGGMRGPFRWNSLRSG 860
Score = 118 bits (295), Expect = 1e-24
Identities = 106/360 (29%), Positives = 143/360 (39%), Gaps = 93/360 (25%)
Query: 46 WNPKEWNWDSIRFMTAKST-----------------------------TVEPQQVEESLN 76
W+ +W WDS F+ S + E EE +N
Sbjct: 12 WDLNDWRWDSNLFLATPSNASPSKCSRRELGRAEGEIDFGVVDKRRRVSPEDDDGEECIN 71
Query: 77 LNL---------GSTGLVRPNKRIRSGSPTSASYPMCQVDNCKEDLSKAKDYHRRHKVCE 127
G G ++ R G+ S+S P CQVD C +LS A+DY++RHKVCE
Sbjct: 72 AATTNGDDGQISGQRGRSSEDEMPRQGT-CSSSGPCCQVDGCTVNLSSARDYNKRHKVCE 130
Query: 128 AHSKASKALLGNQMQRFCQQCSRFHPLVEFDEGKRSCRRRLAGHNRRRRKTQPDEVAVGG 187
H+K+ + N RFCQQCSRFH L EFDEGK+SCR RLA HNRRRRK Q V
Sbjct: 131 VHTKSGVVRIKNVEHRFCQQCSRFHFLQEFDEGKKSCRSRLAQHNRRRRKVQ-----VQA 185
Query: 188 SPPLNQVAANLEIFNLLTAIADGSQGKFEERRS-QVPDKEQLVQILNRIPLPADLTAKLL 246
+N + N + N L + G S Q+ L ++ + A L
Sbjct: 186 GVDVNSLHENHSLSNTLLLLLKQLSGLDSSGPSEQINGPNYLTNLVKNL-------AALA 238
Query: 247 DVGNNLNAKNDNVQMETSPSYHHRDDQLNNAPPAPLTKDFLAVLSTTPSTPARNGGNGST 306
N D L NA A A+ S T + A+ GN
Sbjct: 239 GTQRN-------------------QDMLKNANSA-------AIASHTGNYVAK--GNSLH 270
Query: 307 SSADHMRERSSGSSQSPNDDSDCQ------------EDVRVKLPLQLFGSSPENDSPSKL 354
S H+ + +++ P + Q E+ K+ +LFG P ND PS L
Sbjct: 271 DSRPHIPVGTESTAEEPTVERRVQNFDLNDAYVEGDENRTDKIVFKLFGKEP-NDFPSDL 329
>gb|AAP46238.1| unknown protein, 5'-partial [Oryza sativa (japonica
cultivar-group)] gi|50919619|ref|XP_470170.1| unknown
protein, 5'-partial [Oryza sativa (japonica
cultivar-group)]
Length = 462
Score = 187 bits (476), Expect = 1e-45
Identities = 146/481 (30%), Positives = 235/481 (48%), Gaps = 44/481 (9%)
Query: 539 LIHNSDSDFWRNGRFLVYSGSQLASHKDGRIRMCKPWGTWRSP--ELISVSPLAIVGGQE 596
L+++S +FW +G V Q+A +G++ + +P +++ V P+A +
Sbjct: 2 LLNSSTGNFWASGLVFVMVRHQIAFMHNGQLMLDRPLANSAHHYCKILCVRPIAAPFSTK 61
Query: 597 TSISLKGRNLSAPGTKIHCTGADCYTSSEVIGSGDPGMVYDEIKLSGFEVQNTS---PSV 653
+ ++G NL + +++ C+ I D + D+++ E N PS
Sbjct: 62 VNFRVEGLNLVSDSSRLICS-----FEGSCIFQEDTDNIVDDVEHDDIEYLNFCCPLPSS 116
Query: 654 LGRCFIEVENG-FKGNSFPVIIANASICKELRPLESEFDEEEKMCDAISEEHEHHFGRPK 712
GR F+EVE+G F FP IIA IC E+ LES F+ S HE
Sbjct: 117 RGRGFVEVEDGGFSNGFFPFIIAEQDICSEVCELESIFE---------SSSHEQA-DDDN 166
Query: 713 SRDEALHFLNELGWLFQRERF-SNVHEVP--DYSLDRFKFVLTFSVERNCCMLVKTLLDM 769
+R++AL FLNELGWL R S +VP +++ RF+ + F++ER C + K LLD
Sbjct: 167 ARNQALEFLNELGWLLHRANIISKQDKVPLASFNIWRFRNLGIFAMEREWCAVTKLLLDF 226
Query: 770 LVDKHFEG--EGLSTGSVEMLKAIQLLNRAVKRKCTSMVDLLINYSIT-SKNDTSKKYVF 826
L F G + S E++ + LL+ AV+ K MV L+ Y S T++ ++F
Sbjct: 227 L----FTGLVDIGSQSPEEVVLSENLLHAAVRMKSAQMVRFLLGYKPNESLKRTAETFLF 282
Query: 827 PPNLEGPGGITPLHLAASTTDSEGVIDSLTNDPQ----ECWETLADENGQTPHAYAMMRN 882
P+ +GP TPLH+AA+T D+E V+D+LTNDP W D G TP YA R
Sbjct: 283 RPDAQGPSKFTPLHIAAATDDAEDVLDALTNDPGLVGINTWRNARDGAGFTPEDYARQRG 342
Query: 883 NHSYNMLVARKCSDR-QRSEVSVRIDNEIEHPSLGIELMQKRIN-----QVKRVGDSCSK 936
N +Y +V +K + + V + + + I HP + + ++ V SC+
Sbjct: 343 NDAYLNMVEKKINKHLGKGHVVLGVPSSI-HPVITDGVKPGEVSLEIGMTVPPPAPSCNA 401
Query: 937 CAIAEVRAKRRFSGSRSWLHGPFIHSMLAVAAVCVCVCVLFRGTPYVGSVSPFRWENLNY 996
C+ + S +R++L+ P + +++ +A +CVCV +L P V + FRWE L
Sbjct: 402 CSRQALMYPN--STARTFLYRPAMLTVMGIAVICVCVGLLLHTCPKVYAAPTFRWELLER 459
Query: 997 G 997
G
Sbjct: 460 G 460
>gb|AAC34220.1| unknown protein [Arabidopsis thaliana]
Length = 383
Score = 175 bits (444), Expect = 6e-42
Identities = 122/363 (33%), Positives = 186/363 (50%), Gaps = 46/363 (12%)
Query: 651 PSVLGRCFIEVEN-GFKGNSFP-VIIANASICKELRPLESEFDEEEKMCDAISEEHEHHF 708
P + GR F+E+E+ G + FP +++ + +C E+R LE+ + F
Sbjct: 51 PILSGRGFMEIEDQGLSSSFFPFLVVEDDDVCSEIRILETTLE----------------F 94
Query: 709 GRPKSRDEALHFLNELGWLFQRERFSNVHEVPD-YSLDRFKFVLTFSVERNCCMLVKTLL 767
S +A+ F++E+GWL R + P + L RF++++ FS++R C +++ LL
Sbjct: 95 TGTDSAKQAMDFIHEIGWLLHRSKLGESDPNPGVFPLIRFQWLIEFSMDREWCAVIRKLL 154
Query: 768 DMLVDKHFEGEGLSTGSVEMLKAIQLLNRAVKRKCTSMVDLLINYSITSKNDTSKKYVFP 827
+M D GE S+ S L + LL+RAV++ MV++L+ Y + ++ +F
Sbjct: 155 NMFFDGAV-GE-FSSSSNATLSELCLLHRAVRKNSKPMVEMLLRYIPKQQRNS----LFR 208
Query: 828 PNLEGPGGITPLHLAASTTDSEGVIDSLTNDPQ----ECWETLADENGQTPHAYAMMRNN 883
P+ GP G+TPLH+AA SE V+D+LT DP E W+T D G TP YA +R +
Sbjct: 209 PDAAGPAGLTPLHIAAGKDGSEDVLDALTEDPAMVGIEAWKTCRDSTGFTPEDYARLRGH 268
Query: 884 HSYNMLVARKCSDRQRSEVSVRI-------DNEIEHPSLGIELMQKRINQVKRVGDSCSK 936
SY L+ RK + + +E V + D E + P G I Q+ C
Sbjct: 269 FSYIHLIQRKINKKSTTEDHVVVNIPVSFSDREQKEPKSGPMASALEITQI-----PCKL 323
Query: 937 CAIAEVRAKRRFSGSRSWLHGPFIHSMLAVAAVCVCVCVLFRGTPYVGSV-SPFRWENLN 995
C V R RS + P + SM+A+AAVCVCV +LF+ P V V PFRWE L+
Sbjct: 324 CDHKLVYGTTR----RSVAYRPAMLSMVAIAAVCVCVALLFKSCPEVLYVFQPFRWELLD 379
Query: 996 YGT 998
YGT
Sbjct: 380 YGT 382
>gb|AAC34221.1| putative squamosa-promoter binding protein [Arabidopsis thaliana]
Length = 240
Score = 150 bits (378), Expect = 3e-34
Identities = 78/136 (57%), Positives = 97/136 (70%), Gaps = 10/136 (7%)
Query: 74 SLNLNLGSTGLVRPNKRIRSGSPTSASYPMCQVDNCKEDLSKAKDYHRRHKVCEAHSKAS 133
+LNLN S GL P K+ +SG+ +CQV+NC+ DLSK KDYHRRHKVCE HSKA+
Sbjct: 84 TLNLNGESDGLF-PAKKTKSGA-------VCQVENCEADLSKVKDYHRRHKVCEMHSKAT 135
Query: 134 KALLGNQMQRFCQQCSRFHPLVEFDEGKRSCRRRLAGHNRRRRKTQPDEVAVGGSPPLNQ 193
A +G +QRFCQQCSRFH L EFDEGKRSCRRRLAGHN+RRRKT P+ A G P +
Sbjct: 136 SATVGGILQRFCQQCSRFHLLQEFDEGKRSCRRRLAGHNKRRRKTNPEPGANGN--PSDD 193
Query: 194 VAANLEIFNLLTAIAD 209
++N + LL +++
Sbjct: 194 HSSNYLLITLLKILSN 209
>ref|XP_483486.1| SBP-domain protein-like [Oryza sativa (japonica cultivar-group)]
gi|42761373|dbj|BAD11641.1| SBP-domain protein-like
[Oryza sativa (japonica cultivar-group)]
Length = 455
Score = 131 bits (330), Expect = 1e-28
Identities = 62/96 (64%), Positives = 68/96 (70%), Gaps = 1/96 (1%)
Query: 87 PNKRIRSGSPTSASY-PMCQVDNCKEDLSKAKDYHRRHKVCEAHSKASKALLGNQMQRFC 145
P KR R + P C VD CKEDLSK +DYHRRHKVCEAHSK ++ + RFC
Sbjct: 100 PAKRPRGAAAAGQQQCPSCAVDGCKEDLSKCRDYHRRHKVCEAHSKTPLVVVSGREMRFC 159
Query: 146 QQCSRFHPLVEFDEGKRSCRRRLAGHNRRRRKTQPD 181
QQCSRFH L EFDE KRSCR+RL GHNRRRRK QPD
Sbjct: 160 QQCSRFHLLQEFDEAKRSCRKRLDGHNRRRRKPQPD 195
Database: nr
Posted date: Jul 5, 2005 12:34 AM
Number of letters in database: 863,360,394
Number of sequences in database: 2,540,612
Lambda K H
0.316 0.132 0.394
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 1,785,064,170
Number of Sequences: 2540612
Number of extensions: 79083341
Number of successful extensions: 225451
Number of sequences better than 10.0: 481
Number of HSP's better than 10.0 without gapping: 95
Number of HSP's successfully gapped in prelim test: 397
Number of HSP's that attempted gapping in prelim test: 223235
Number of HSP's gapped (non-prelim): 1455
length of query: 998
length of database: 863,360,394
effective HSP length: 138
effective length of query: 860
effective length of database: 512,755,938
effective search space: 440970106680
effective search space used: 440970106680
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.6 bits)
S2: 80 (35.4 bits)
Medicago: description of AC140774.7


