

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC140721.3 + phase: 0 /pseudo
(1439 letters)
Database: nr
2,540,612 sequences; 863,360,394 total letters
Searching..................................................done
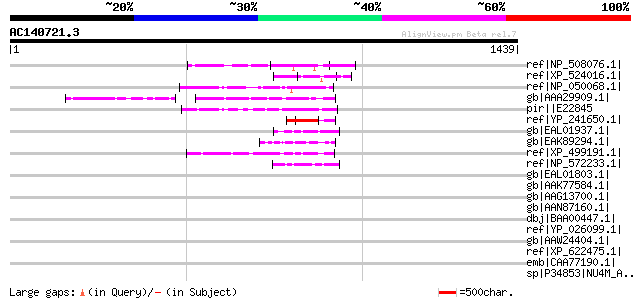
Score E
Sequences producing significant alignments: (bits) Value
ref|NP_508076.1| putative protein, with at least 16 transmembran... 64 3e-08
ref|XP_524016.1| PREDICTED: hypothetical protein XP_524016 [Pan ... 60 5e-07
ref|NP_050068.1| hypothetical protein LetaoMp15 [Leishmania tare... 59 9e-07
gb|AAA29909.1| ORF 3 59 2e-06
pir||E22845 hypothetical protein 4 - Trypanosoma brucei mitochon... 57 5e-06
ref|YP_241650.1| hypothetical protein XC_0549 [Xanthomonas campe... 53 9e-05
gb|EAL01937.1| hypothetical protein CaO19.11807 [Candida albican... 52 1e-04
gb|EAK89294.1| hypothetical protein with signal peptide [Cryptos... 52 1e-04
ref|XP_499191.1| PREDICTED: hypothetical protein XP_499191 [Homo... 51 3e-04
ref|NP_572233.1| CG15780-PA [Drosophila melanogaster] gi|7290595... 51 3e-04
gb|EAL01803.1| hypothetical protein CaO19.4331 [Candida albicans... 49 0.001
gb|AAK77584.1| heme maturase [Tetrahymena thermophila] gi|150276... 49 0.002
gb|AAG13700.1| ABC transporter channel subunit [Malawimonas jako... 48 0.003
gb|AAN87160.1| NADH dehydrogenase subunit 4 [Varroa destructor] 47 0.005
dbj|BAA00447.1| open reading frame (251 AA) [Mus musculus] 47 0.006
ref|YP_026099.1| NADH dehydrogenase subunit 4 [Habronattus orego... 46 0.008
gb|AAW24404.1| NADH dehydrogenase subunit 1 [Vryburgia amaryllidis] 45 0.014
ref|XP_622475.1| PREDICTED: hypothetical protein XP_622475 [Mus ... 45 0.014
emb|CAA77190.1| NADH dehydrogenase subunit 4 [Trichophyton rubru... 45 0.018
sp|P34853|NU4M_APILI NADH-ubiquinone oxidoreductase chain 4 (NAD... 45 0.023
>ref|NP_508076.1| putative protein, with at least 16 transmembrane domains (XA707)
[Caenorhabditis elegans]
Length = 748
Score = 64.3 bits (155), Expect = 3e-08
Identities = 70/271 (25%), Positives = 132/271 (47%), Gaps = 39/271 (14%)
Query: 740 LTSFFIINSLILPFSKFLVVYVMLPLYILIEPNCNLELESLYS*VTSLVIRVLLSMIFNL 799
LT+ + S IL F ++ +++ L + E L ++ ++S + +LL + +
Sbjct: 177 LTTRTLFQSFILLLLLFTILILLIRLGGIFELLIVLIFILIFFSISSNSLLILLIRLITI 236
Query: 800 ERFLC-------QDMLLFMKTISLTLILHPLLLL---IGNIFLIFPLYLILILLS--LPV 847
LC +D+++ + + LIL PL++L +IFLI L+L++ LLS L +
Sbjct: 237 IPILCLIDFSFLKDLIVLRRVGGIHLILRPLIILTIPFIHIFLIIILFLLITLLSIFLRI 296
Query: 848 NLQSLMILLLLLHQL-----------YLLLLLLHLFHLLFPDILPETPLLHLTSKIIFVT 896
L SL I+LL H + ++LLL+L LF LL P HL +++ +
Sbjct: 297 FLGSLSIILLGNHLISLIFRRFLLLIHILLLILLLFFLLIP---------HLRIQLLLIL 347
Query: 897 IFKLRHIIFLIFFPI-----IIFLTIILLLLCLYIHNLSLNPMLKQVNMSVGCMLCKLNF 951
+ + H+IF I+ I II L +++L+ L I + + + + + + + ML + +
Sbjct: 348 LLNVSHLIFNIWLFIRLVLTIILLPLLVLIDILQIFSKVILHINRLLVIILSLMLLRFHR 407
Query: 952 KLLRKL--VLGNLWIYHQMLSLLDVDGFTKS 980
L+ K + L ++ +LS+L F+ S
Sbjct: 408 LLILKTFNLSSLLHLFSDILSILGSSTFSNS 438
Score = 52.0 bits (123), Expect = 1e-04
Identities = 99/408 (24%), Positives = 190/408 (46%), Gaps = 59/408 (14%)
Query: 504 LSFLL-YLKVLVMLFLFPIVIRIVNMYLTMPYGTLDLVICLIKDFI*CLACILILLLIII 562
LSF L +L V+++LFL I+ + ++ + L I+ + L+ L++L+++I
Sbjct: 102 LSFPLHFLFVILLLFL------ILFLLFSLLFRGFSLTAISIRIRL-ALSFSLLILILLI 154
Query: 563 KMLVIFAILPSTNIYLSILVPHMLLLNLNFSI*IFGVLFQLLQFMVIDIFSPF*MTTPDF 622
+L++ + ++I + IL+ + L S + +LF +L ++
Sbjct: 155 FLLLLLSQATISDILIIILIILLTTRTLFQSFILLLLLFTILILLI-------------- 200
Query: 623 SGSFFLKPKLKCLHMLRILLL*FRIIFTPPLNSLELIMALNSCYLLFMLHMALFTRNLV* 682
+L + L I+L+ I F+ NSL LI+ + ++ +L + F+
Sbjct: 201 --------RLGGIFELLIVLIFILIFFSISSNSL-LILLIRLITIIPILCLIDFS----- 246
Query: 683 KLPNKMVELKENINTF*MLVELCFISLNFLLPFGLMLYIMLFSS*IEFLHHYFITNHLTS 742
++ L+ +L L +++ F+ F +++ +L + FL + L+
Sbjct: 247 -FLKDLIVLRRVGGIHLILRPLIILTIPFIHIFLIIILFLLITLLSIFLRIFL--GSLSI 303
Query: 743 FFIINSLI-LPFSKFLVVYVMLPLYILIEPNCNLELESLYS*VTSLVIRVLLSMIFNLER 801
+ N LI L F +FL++ +L L +L+ + + L I++LL ++ N+
Sbjct: 304 ILLGNHLISLIFRRFLLLIHILLLILLL----------FFLLIPHLRIQLLLILLLNVSH 353
Query: 802 FLCQDMLLFMKTISLTLILHPLLLLIGNIFLIFPLYLILI--LLSLPVNLQSLMILLLLL 859
L ++ LF++ + LT+IL PLL+LI +I IF ++ I LL + ++L L LL+
Sbjct: 354 -LIFNIWLFIRLV-LTIILLPLLVLI-DILQIFSKVILHINRLLVIILSLMLLRFHRLLI 410
Query: 860 HQLYLLLLLLHLFHLLFPDILPETPLLHLTSKIIFVTIFKLRHIIFLI 907
+ + L LLH LF DIL ++ F TI L FL+
Sbjct: 411 LKTFNLSSLLH----LFSDILSILGSSTFSNSTSFSTIISLSFSRFLL 454
Score = 48.9 bits (115), Expect = 0.001
Identities = 95/398 (23%), Positives = 170/398 (41%), Gaps = 65/398 (16%)
Query: 556 ILLLIIIKMLVIFAILPSTNIYLSILVPHMLLLNLNFSI*IFGVLFQLLQFMVIDIFSPF 615
+ +L ++ L++ +I + + SI+ L L+F I +LF +L + +F F
Sbjct: 73 LCILHLLSRLILLSISHFSLFHRSIICCE-LSFPLHFLFVIL-LLFLILFLLFSLLFRGF 130
Query: 616 *MTTPDFSGSFFLKPKLKCLHMLRILLL*FRIIFTPPLNSLELIMALNSCYLL--FMLHM 673
+T L L L +L LLL + + LI+ L + L F+L +
Sbjct: 131 SLTAISIRIRLALSFSLLILILLIFLLLLLSQATISDILIIILIILLTTRTLFQSFILLL 190
Query: 674 ALFTRNLV*KLPNKMVELKENINTF*MLVELCFISLNFLLPFGLMLYIMLFSS*I----- 728
LFT ++ ++ L F +L+ L FI + F + +L +++ I
Sbjct: 191 LLFTILIL------LIRLG---GIFELLIVLIFILIFFSISSNSLLILLIRLITIIPILC 241
Query: 729 ----EFLHHYFITNHLTSF-FIINSLILPFSKFLVVYVMLPLYILIEPNCNLELESLYS* 783
FL + + I+ LI+ F+ +++++ L++LI
Sbjct: 242 LIDFSFLKDLIVLRRVGGIHLILRPLIILTIPFIHIFLIIILFLLIT------------- 288
Query: 784 VTSLVIRVLLSMIFNLERFLCQDMLLFMKTISLTLILHPLLLLIGNIFLIFPLYLILILL 843
+ S+ +R+ L +L L + L+ + L++H LLL++ FL+ P I +LL
Sbjct: 289 LLSIFLRIFLG---SLSIILLGNHLISLIFRRFLLLIHILLLILLLFFLLIPHLRIQLLL 345
Query: 844 SLPVNLQSLMILLLLLHQLYLLLLLLHLFHLLFPDILPETPLLHLTSKIIFVTIFKLRHI 903
L +N+ L+ + L +L L ++LL L L+ +L + SK+I HI
Sbjct: 346 ILLLNVSHLIFNIWLFIRLVLTIILLPLLVLI--------DILQIFSKVIL-------HI 390
Query: 904 IFLIFFPIIIFLTIILLLLCLYIHNLSLNPMLKQVNMS 941
L L IIL L+ L H L + LK N+S
Sbjct: 391 NRL--------LVIILSLMLLRFHRLLI---LKTFNLS 417
Score = 40.8 bits (94), Expect = 0.34
Identities = 77/316 (24%), Positives = 149/316 (46%), Gaps = 38/316 (12%)
Query: 154 NYLHIDISLIVLVFILVVVLHLKL-LRFIEMRIRLCNFLQD*MINFLLLELKFFSLILFL 212
++L + + L +++F+L +L L I +RIRL ++F LL L +L L
Sbjct: 107 HFLFVILLLFLILFLLFSLLFRGFSLTAISIRIRLA-------LSFSLLILILLIFLLLL 159
Query: 213 L*TKFIL*LFRKKVIMLLLLL*VYLLIPPFKSMLLIIENLKVVAKASLQISLLDSVLSVI 272
L I + +I+L++LL L F +LL+ L ++ +++ + +L V+
Sbjct: 160 LSQATISDIL---IIILIILLTTRTLFQSFILLLLLFTILILL----IRLGGIFELLIVL 212
Query: 273 VQIIL*IFVT*NMVITMLTNLHQV*MLL-------LKKILKM-----VQCLLLDWVLLQA 320
+ I++ ++ N ++ +L L + +L LK ++ + + +L ++L
Sbjct: 213 IFILIFFSISSNSLLILLIRLITIIPILCLIDFSFLKDLIVLRRVGGIHLILRPLIILTI 272
Query: 321 LLLLVFLKSN*LNLCPCYNNQIWLFQHLLLHKLPPITL--HLPAFPLQ-ILLKVYLLLFL 377
+ +FL + + + +F + L L I L HL + + LL +++LL +
Sbjct: 273 PFIHIFL----IIILFLLITLLSIFLRIFLGSLSIILLGNHLISLIFRRFLLLIHILLLI 328
Query: 378 L*QVNLLFG*LIMVLMNIYVVILLHLVPFIE*NLFMFPYPMVTLC*LILLVMSLSIPSFI 437
L LLF LI L ++ILL V + N+++F ++T+ L LLV+ + F
Sbjct: 329 L----LLFFLLIPHLRIQLLLILLLNVSHLIFNIWLFIRLVLTIILLPLLVLIDILQIFS 384
Query: 438 *VMFFIHLILPSISSL 453
V+ I+ +L I SL
Sbjct: 385 KVILHINRLLVIILSL 400
Score = 38.1 bits (87), Expect = 2.2
Identities = 61/246 (24%), Positives = 106/246 (42%), Gaps = 15/246 (6%)
Query: 799 LERFLCQDMLLFMKTISLTLILHPLLLLIGN-----IFLIFPLYLILILLSLPVNLQSLM 853
+E + M + + + LILH ++ + I + L L+L+ L LP+ L SL
Sbjct: 1 MEHIMRSHMNMRLLDLLSILILHLSTIIFSSSSFQSILELSSLPLLLLSLLLPLRLPSLF 60
Query: 854 ILLLLLHQLYLLLLLLHLFHLLFPDILPETPLLHLTSKIIFVTIFKLRHIIFLIFFPIII 913
L H L +LHL L + L H II + H +F+I +++
Sbjct: 61 NRSLSNHLSIGNLCILHLLSRLILLSISHFSLFH--RSIICCELSFPLHFLFVI---LLL 115
Query: 914 FLTIILLLLCLYIHNLSLNPMLKQVNMSVGCMLCKLNFKLLRKLVLGNLWIYHQMLSLLD 973
FL I+ LL L SL + ++ +++ L L + L+L I ++ +L
Sbjct: 116 FL-ILFLLFSLLFRGFSLTAISIRIRLALSFSLLILILLIFLLLLLSQATISDILIIILI 174
Query: 974 VDGFTKS---NFMLMALLRDIKQDWLPKVTIKLRAWIILIHILQLLNSQQSDL*LLYLPF 1030
+ T++ +F+L+ LL I L ++ I+LI IL + + L +L +
Sbjct: 175 ILLTTRTLFQSFILLLLLFTI-LILLIRLGGIFELLIVLIFILIFFSISSNSLLILLIRL 233
Query: 1031 IICIYI 1036
I I I
Sbjct: 234 ITIIPI 239
>ref|XP_524016.1| PREDICTED: hypothetical protein XP_524016 [Pan troglodytes]
Length = 273
Score = 60.1 bits (144), Expect = 5e-07
Identities = 58/195 (29%), Positives = 99/195 (50%), Gaps = 19/195 (9%)
Query: 749 LILPFSKFLVVYVMLP--LYILIEPNCNLELESLYS*VTSLVIRVLLSMIF-NLERFLCQ 805
++LP L++ +LP L++LI L L + V ++ +L++ + +L L
Sbjct: 1 MVLPHLLHLLITTVLPHLLHLLITTVLPHPLHLLITTVLPHLLHLLITTVLPHLLHLLIT 60
Query: 806 DMLLFMKTISLTLIL-HPLLLLIGNIFLIFPLYLILILLSLPVNLQSLMILLLLLHQLYL 864
+L + + +T +L HPL LLI + P L L++ + ++L L+I +L H L+L
Sbjct: 61 TVLPHLLHLLITTVLPHPLHLLITTVL---PHLLYLLITRVLLHLLHLLITTVLPHLLHL 117
Query: 865 LL--LLLHLFHLLFPDILPE-------TPLLHLTSKIIFVTIFKLRHIIF---LIFFPII 912
L+ +LLHL HLL +LP T LLHL +I + L H++ L+ +
Sbjct: 118 LITKVLLHLLHLLITTVLPHLLHLLITTVLLHLLHLLITTDLLHLLHLLITKVLLHLLHL 177
Query: 913 IFLTIILLLLCLYIH 927
+ T +L LL L I+
Sbjct: 178 LITTGLLHLLHLLIN 192
Score = 52.8 bits (125), Expect = 9e-05
Identities = 49/158 (31%), Positives = 83/158 (52%), Gaps = 15/158 (9%)
Query: 816 LTLILHPLLLLIGNIFLIFPLYLILILLSLPVNLQSLMILLLLLHQLYLLL--LLLHLFH 873
+T +L LL L+ I + P L L++ ++ + L+I +L H LYLL+ +LLHL H
Sbjct: 47 ITTVLPHLLHLL--ITTVLPHLLHLLITTVLPHPLHLLITTVLPHLLYLLITRVLLHLLH 104
Query: 874 LLFPDILPETPLLHLTSKIIFVTIFKLRHIIFLIFFPIIIFLTIILLLLCLYIHNLSLNP 933
LL +LP LLHL +I + L H++ P ++ L I +LL L +H L
Sbjct: 105 LLITTVLPH--LLHL---LITKVLLHLLHLLITTVLPHLLHLLITTVLLHL-LHLLITTD 158
Query: 934 MLKQVNMSVGCMLCKLNFKLLRKLV-LGNLWIYHQMLS 970
+L +++ ++ K+ LL L+ G L + H +++
Sbjct: 159 LLHLLHL----LITKVLLHLLHLLITTGLLHLLHLLIN 192
Score = 42.4 bits (98), Expect = 0.12
Identities = 53/182 (29%), Positives = 91/182 (49%), Gaps = 11/182 (6%)
Query: 700 MLVELCFISLNFLLPFGLMLYIMLFSS*IEFLHHYFITNHLTSFF-IINSLILPFSKFLV 758
+L L + + +LP L L L ++ + L H IT L ++ + +LP L+
Sbjct: 14 VLPHLLHLLITTVLPHPLHL---LITTVLPHLLHLLITTVLPHLLHLLITTVLPHLLHLL 70
Query: 759 VYVMLP--LYILIEPNCNLELESLYS*VTSLVIRVLLSMIF-NLERFLCQDMLLFMKTIS 815
+ +LP L++LI L L + V ++ +L++ + +L L +LL + +
Sbjct: 71 ITTVLPHPLHLLITTVLPHLLYLLITRVLLHLLHLLITTVLPHLLHLLITKVLLHLLHLL 130
Query: 816 LTLILHPLLLLIGNIFLIFPLYLILI--LLSLPVNLQSLMILLLLLHQLYLLLLLLHLFH 873
+T +L LL L+ L+ L+L++ LL L ++L +LL LLH L + LLHL H
Sbjct: 131 ITTVLPHLLHLLITTVLLHLLHLLITTDLLHL-LHLLITKVLLHLLH-LLITTGLLHLLH 188
Query: 874 LL 875
LL
Sbjct: 189 LL 190
>ref|NP_050068.1| hypothetical protein LetaoMp15 [Leishmania tarentolae]
gi|83996|pir||B26696 hypothetical protein 1 (CYb-COII
intergenic region) - Leishmania tarentolae mitochondrion
(fragment) gi|896286|gb|AAA96601.1| NH2 terminus
uncertain
Length = 443
Score = 59.3 bits (142), Expect = 9e-07
Identities = 104/463 (22%), Positives = 200/463 (42%), Gaps = 73/463 (15%)
Query: 483 LVWLNRKMVYTSFIHHANLLIL-SFLLYLKVLVMLFLFPIVIRIVNMYLTMPYGTLDLVI 541
L +L+ +++ I+ L IL S++ ++ +L+ LF F + +N + + +
Sbjct: 28 LFYLSFDLIWLIIINIIILTILDSYICFIFLLLFLFCFFFLFCFLNFDTRFVFMIIIMQY 87
Query: 542 CLIKDFI*CLACILILLLIIIKMLVIFAILPSTNIYLSILVP-HMLLLNL-NFSI*IFGV 599
+I F+ + + I +L + L++F IL S+ IL+ + ++NL NF +
Sbjct: 88 IIIFMFLHVIHILFISILFELFSLLLFLILISSRFGYKILILWYYYMINLINFIL----- 142
Query: 600 LFQLLQFMVIDIFSPF*MTTPDFSGSFFLKPKLKCLHMLRILLL*FRIIFTPPLNSLE-L 658
LF LL +M+++ + DF F + L L + ILL+ F++ + LE L
Sbjct: 143 LFVLLYYMILN----YCFYLCDFCFLIFDEEWLGILCLFYILLILFKLYIAILILFLEQL 198
Query: 659 IMALNSCYLLFMLHMALFTRNLV*KLPNKMVELKENINTF*MLVELCFISLNFLLPFGLM 718
+ L ++ML TF +L CFI + L+ F +
Sbjct: 199 YIRLGVFIFIYML-------------------------TFYVL--FCFILIILLICF--I 229
Query: 719 LYIMLFSS*IEFLHHYFITNHLTSFFIINSLIL----PFSKFLVVYVMLPLYILIEPNCN 774
+ ++F I + L SF I++ L + FS ++++ YI
Sbjct: 230 YFYIIFIKLIIIQSITCVLIGLNSFAIVSLLFVLSVNNFSFLFLIFISTKNYIFY---MY 286
Query: 775 LELESLYS*VTSLVIRVLLSMIF----------NLERFLCQDMLL-FMKTISLTLILHPL 823
L +YS SL+I ++L F N FL + F + +++++ +
Sbjct: 287 LNFHLIYS--LSLIILIILYYFFIVYNMFDIKYNENYFLINFIFFSFFNNLLISIMIACI 344
Query: 824 LLLIGNIFLIFPLYLILILLSLPVNLQSLMILL-----LLLHQLYLLLLLLHLFHLLFPD 878
L IG+I ++F +L + L L ++ + I+ L++ ++ L++++F +
Sbjct: 345 FLCIGSIPIVFGFFLKVFCLLLHLSYLGICIVFFSIIWLIIIYIFYFRLIINIFIFSYQF 404
Query: 879 ILPETPLLHLT--SKIIFVTIFKLRHIIFLIFFPIIIFLTIIL 919
I LH+ S + F+ F IF++FF II +IL
Sbjct: 405 IGFWVVRLHILGFSNLFFILSFS----IFILFFDIINLFDLIL 443
Score = 48.9 bits (115), Expect = 0.001
Identities = 91/405 (22%), Positives = 179/405 (43%), Gaps = 60/405 (14%)
Query: 596 IFGVLFQLLQFMVIDIFSPF*MTTPDFSGSFFLKPKLKCLHMLRILLL*FRIIFTPPLNS 655
+F L ++ F+++ F + D++ F+L L L ++ I++L I +
Sbjct: 3 LFLYLIHIILFLLLYSF----IILCDYTSLFYLSFDLIWLIIINIIIL---TILDSYICF 55
Query: 656 LELIMALNSCYLLFMLHMALFTRNLV*KLPNKMVELKENINTF*ML--VELCFISLNFLL 713
+ L++ L C+ + TR + M+ + + I F L + + FIS+ F L
Sbjct: 56 IFLLLFL-FCFFFLFCFLNFDTRFVF------MIIIMQYIIIFMFLHVIHILFISILFEL 108
Query: 714 PFGLMLYIMLFSS*IEF----LHHYF---ITNHLTSFFIINSLILPFSKFL--------- 757
F L+L+++L SS + L +Y+ + N + F ++ +IL + +L
Sbjct: 109 -FSLLLFLILISSRFGYKILILWYYYMINLINFILLFVLLYYMILNYCFYLCDFCFLIFD 167
Query: 758 --------VVYVMLPLYILIEPNCNLELESLYS*VTSLVIRVLLSMI----FNLERFLCQ 805
+ Y++L L+ L L LE LY + + +L+ F L L
Sbjct: 168 EEWLGILCLFYILLILFKLYIAILILFLEQLYIRLGVFIFIYMLTFYVLFCFILIILLIC 227
Query: 806 DMLLFMKTISLTLILHPLLLLIG-NIFLIFPLYLILILLSLPVNLQSLMILLLLLHQLYL 864
+ ++ I L +I +LIG N F I + +L L VN S + L+ + + Y+
Sbjct: 228 FIYFYIIFIKLIIIQSITCVLIGLNSFAI-----VSLLFVLSVNNFSFLFLIFISTKNYI 282
Query: 865 LLLLLHLFHLLFPDILPETPLLHLTSKIIFVTIFKLRH-----IIFLIFFPIIIFLTIIL 919
+ L+ FHL++ L L+ L I +F +++ +I IFF L I +
Sbjct: 283 FYMYLN-FHLIYS--LSLIILIILYYFFIVYNMFDIKYNENYFLINFIFFSFFNNLLISI 339
Query: 920 LLLCLYIHNLSLNPMLKQVNMSVGCMLCKLNFKLLRKLVLGNLWI 964
++ C+++ S+ P++ + V C+L L++ + + +W+
Sbjct: 340 MIACIFLCIGSI-PIVFGFFLKVFCLLLHLSYLGICIVFFSIIWL 383
>gb|AAA29909.1| ORF 3
Length = 393
Score = 58.5 bits (140), Expect = 2e-06
Identities = 73/402 (18%), Positives = 179/402 (44%), Gaps = 44/402 (10%)
Query: 528 MYLTMPYGTLDLVICLIKDFI*CLACILILLLIIIKMLVIFAILPSTNIYLSILVPHMLL 587
M++ M L +++ ++ + + I+I+L+++ + V F I + +++ L+ +++
Sbjct: 1 MFMMMMMVMLMMMVAVVVMVVMMVMFIMIMLIVVFTITVFF-ISTTIIVFVIFLIVILII 59
Query: 588 LNLNFSI*IFGVLFQLLQFMVIDIFSPF*MTTPDFSGSFFLKPKLKCLHMLRILLL*FRI 647
L + F + IF ++F + ++ P + + F F + L + ++ + ++ I
Sbjct: 60 LAIFFGVTIFLIIFSMTMLVI-----PVLVVSVFFVTIFIIAILLVTIFVVSVFVVTIFI 114
Query: 648 I----FTPPLNSLELIMALNSCYLLFMLHMALFTRNLV*KLPNKMVELKENINTF*MLVE 703
I T + S+ L+ L+ L + +F R ++ + ++ L I +LV
Sbjct: 115 IAVLLVTIFVISILLVTIFVVSILVVTLFITIFVRAIL-VVTILVINLFVTIFVRAILVI 173
Query: 704 LCFISLNFLLPFGLMLYIMLFSS*IEFLHHYFITNHLTSFFIINSLILPFSKFLVVYVML 763
F+++ + L++ +F I + F+T + + +I + F + ++V +
Sbjct: 174 TLFVAIFVRTILVITLFVAIFVRTILVIT-LFVTIFVRTILVITLFVAIFVRTILVITLF 232
Query: 764 PLYILIEPNCNLELESLYS*VTSLVIRVLLSMIFNLERFLCQDMLLFMKTISLTLILHPL 823
+I VT LVI V + I + F+ +++ + IS+ +I +
Sbjct: 233 VTIFIISILL----------VTILVISVFIIAILTISIFVISILIITIFVISVFVITIFV 282
Query: 824 LLLIGNIFLIFPLYLILILLSLPVNLQSLMILLLLLHQLYLLLLLLHLFHLLFPDILPET 883
+ + L+ ++++ + V++ +L+ LL ++++ +L+
Sbjct: 283 VTIFAITVLVISIFIVAFFIVFAVSI----LLVPLLMTIFIIAILV-------------- 324
Query: 884 PLLHLTSKIIFVTIFKLRHIIFLIFFPIIIFLTIILLLLCLY 925
+T +I T+ L II +F I IFLTI++ ++ +Y
Sbjct: 325 ----ITIFVIVFTVSVLAGIIACVFSTITIFLTIVIFIINIY 362
Score = 53.5 bits (127), Expect = 5e-05
Identities = 70/316 (22%), Positives = 153/316 (48%), Gaps = 31/316 (9%)
Query: 158 IDISLIVLVFILVVVLHLKLLRFIEMRIRLCNFLQD*MINFLLLELKFFS---LILFLL* 214
I ++IV V L+V+L + L F + I L F ++ +L+ FF +I LL
Sbjct: 42 ISTTIIVFVIFLIVILII-LAIFFGVTIFLIIFSMTMLVIPVLVVSVFFVTIFIIAILLV 100
Query: 215 TKFIL*LFRKKVIMLLLLL*VYLLIPPFKSMLLIIENLKVVAKASLQISLLDSVLSVIVQ 274
T F++ +F + ++ +LL +I + ++ L V ++ + + V +++
Sbjct: 101 TIFVVSVFVVTIFIIAVLLVTIFVISILLVTIFVVSILVVTLFITIFVRAILVVTILVIN 160
Query: 275 IIL*IFVT*NMVITMLTNLHQV*MLLLKKILKMVQCLLLDWVLLQALLLLVFLKSN*LNL 334
+ + IFV +VIT+ + +L ++ + + + +L+ L + +F+++ +
Sbjct: 161 LFVTIFVRAILVITLFVAIFVRTIL----VITLFVAIFVRTILVITLFVTIFVRTILV-- 214
Query: 335 CPCYNNQIWLFQHLLLHKLPPITLHLPAFPLQILLKVYLLLFLL*QVNLLFG*LIMVL-M 393
I LF + + + ITL + F + ILL L++ + +I +L +
Sbjct: 215 -------ITLFVAIFVRTILVITLFVTIFIISILLVTILVISVF---------IIAILTI 258
Query: 394 NIYVVILLHLVPFIE*NLFMFPYPMVTLC*LILLVMSLSIPSFI*VMFFIHLILPSISSL 453
+I+V+ +L + F+ ++F+ +VT+ + +LV+S+ I +F V L++P + ++
Sbjct: 259 SIFVISILIITIFVI-SVFVITIFVVTIFAITVLVISIFIVAFFIVFAVSILLVPLLMTI 317
Query: 454 *QNVVIH*LVSFIFLL 469
+I LV IF++
Sbjct: 318 ---FIIAILVITIFVI 330
Score = 47.0 bits (110), Expect = 0.005
Identities = 66/323 (20%), Positives = 151/323 (46%), Gaps = 30/323 (9%)
Query: 363 FPLQILLKVYLLLFLL*QVNLLFG*LIMVLMNIYVVILLHLVPFIE*NLFMFPYPMVTLC 422
F + I L ++ + L+ V ++ + + + +++ + +V +F+ +VT+
Sbjct: 64 FGVTIFLIIFSMTMLVIPVLVVSVFFVTIFIIAILLVTIFVVSVFVVTIFIIAVLLVTIF 123
Query: 423 *LILLVMSLSIPSFI*VMFFIHLILPSISSL*QNVVIH*LVSFIFLLKNVLYRTNNL*R* 482
+ +L++++ + S + V FI + + +I +V+ LV +F+
Sbjct: 124 VISILLVTIFVVSILVVTLFITIFVRAI------LVVTILVINLFVT------------- 164
Query: 483 LVWLNRKMVYTSFIH---HANLLILSFL-LYLKVLVMLFLFPIVIRIVNMYLTMPYGTLD 538
+++ +V T F+ L+I F+ ++++ ++++ LF + + +T+
Sbjct: 165 -IFVRAILVITLFVAIFVRTILVITLFVAIFVRTILVITLFVTIFVRTILVITLFVAIFV 223
Query: 539 LVICLIKDFI*CLACILILLLIIIKMLVIFAILPSTNIYLSILVPHMLLLNLNFSI*IFG 598
I +I F+ ++L+ I++ + I AIL + +SIL+ + ++++ F I IF
Sbjct: 224 RTILVITLFVTIFIISILLVTILVISVFIIAILTISIFVISILIITIFVISV-FVITIFV 282
Query: 599 VLFQLLQFMVIDIFSPF*MTTPDFSGSFFLKPKLKCLHMLRILLL-*FRIIFTPPLNSLE 657
V + +VI IF F+ S L P L + ++ IL++ F I+FT ++ L
Sbjct: 283 VTIFAITVLVISIFIVAFFIV--FAVSILLVPLLMTIFIIAILVITIFVIVFT--VSVLA 338
Query: 658 LIMALNSCYLLFMLHMALFTRNL 680
I+A + L + +F N+
Sbjct: 339 GIIACVFSTITIFLTIVIFIINI 361
Score = 42.0 bits (97), Expect = 0.15
Identities = 69/377 (18%), Positives = 176/377 (46%), Gaps = 27/377 (7%)
Query: 165 LVFILVVVLHLKLLRFIEMRIRLCNFLQD*MINFLLLELKFFS-----LILFLL*TKFIL 219
+ ++++V+ + ++ + M + + F+ +I + + F S ++FL+ IL
Sbjct: 1 MFMMMMMVMLMMMVAVVVMVVMMVMFIMIMLIVVFTITVFFISTTIIVFVIFLIVILIIL 60
Query: 220 *LFRKKVIMLLLLL*VYLLIPPFKSMLLIIENLKVVAKASLQISLLDSVLSVIVQIIL*I 279
+F I L++ L+IP + + + ++A + I ++ SV V + II +
Sbjct: 61 AIFFGVTIFLIIFSMTMLVIPVLVVSVFFV-TIFIIAILLVTIFVV-SVFVVTIFIIAVL 118
Query: 280 FVT*NMVITMLTNLHQV*MLLLKKILKM-VQCLLLDWVLLQALLLLVFLKSN*LNLCPCY 338
VT ++ +L + V +L++ + + V+ +L+ +L+ L + +F+++ +
Sbjct: 119 LVTIFVISILLVTIFVVSILVVTLFITIFVRAILVVTILVINLFVTIFVRAILV------ 172
Query: 339 NNQIWLFQHLLLHKLPPITLHLPAFPLQILLKVYLLLFLL*QVNLLFG*LIMVLMNIYVV 398
I LF + + + ITL + F ++ +L + L + + + L+ + + + +V
Sbjct: 173 ---ITLFVAIFVRTILVITLFVAIF-VRTILVITLFVTIFVRTILVITLFVAIFVRTILV 228
Query: 399 ILLHLVPFIE*NLFMFPYPMVTLC*LILLVMSLSIPS------FI*VMFFIHLILPSISS 452
I L + FI ++ + ++++ + +L +S+ + S F+ +F I + + +I +
Sbjct: 229 ITLFVTIFII-SILLVTILVISVFIIAILTISIFVISILIITIFVISVFVITIFVVTIFA 287
Query: 453 L*QNVVIH*LVSFIFLLKNVLYRTNNL*R*LVWLNRKMVYTSFIHHANLLILSFLLYLKV 512
+ V+ +V+F + + L +++ +V T F+ + +L+ ++
Sbjct: 288 ITVLVISIFIVAFFIVFAVSILLVPLL--MTIFIIAILVITIFVIVFTVSVLAGIIACVF 345
Query: 513 LVMLFLFPIVIRIVNMY 529
+ IVI I+N+Y
Sbjct: 346 STITIFLTIVIFIINIY 362
Score = 38.1 bits (87), Expect = 2.2
Identities = 50/235 (21%), Positives = 111/235 (46%), Gaps = 27/235 (11%)
Query: 700 MLVELCFISLNFLLPFGL-MLYIMLFSS*IEFLHHYFITNHLTSFFIINSLILPFSKFLV 758
+++ + +S+ F+ F + +L + +F + + + I L + F+I+ L++ F+V
Sbjct: 78 LVIPVLVVSVFFVTIFIIAILLVTIFVVSVFVVTIFIIAVLLVTIFVISILLVTI--FVV 135
Query: 759 VYVMLPLYILIEPNCNLELESLYS*VTSLVIRVLLSMIFNLERFLCQDMLLFMKTISLTL 818
+++ L+I I L VT LVI + +++ + + +F++TI
Sbjct: 136 SILVVTLFITIFVRAILV-------VTILVINLFVTIFVRAILVITLFVAIFVRTI---- 184
Query: 819 ILHPLLLLIGNIFLIFPLYLILILLSLPVNLQSLMILLLLLHQLYLLLLLLHLFHLLFPD 878
L+I IF +++I L + + +++++++ L + +L++ LF +F
Sbjct: 185 ------LVITLFVAIFVRTILVITLFVTIFVRTILVITLFVAIFVRTILVITLFVTIFII 238
Query: 879 ILPETPLLHLTSKIIFV---TIFKLRHIIFLIF----FPIIIFLTIILLLLCLYI 926
+ +L ++ II + +IF + +I IF F I IF+ I + L I
Sbjct: 239 SILLVTILVISVFIIAILTISIFVISILIITIFVISVFVITIFVVTIFAITVLVI 293
>pir||E22845 hypothetical protein 4 - Trypanosoma brucei mitochondrion
Length = 445
Score = 57.0 bits (136), Expect = 5e-06
Identities = 99/449 (22%), Positives = 183/449 (40%), Gaps = 59/449 (13%)
Query: 488 RKMVYTSFIH-HANLLILSFLLYLKVLVMLFL-FPIVIRIVNMY-LTMPYGTLDLVICLI 544
+K ++ IH L+I SF++ + L F ++ ++N++ +T+ LD IC I
Sbjct: 3 KKSLFLYIIHIFIFLIIYSFIILCDYTTLTLLSFDLLWLLINLFWITL----LDSYICFI 58
Query: 545 KD--FI*CLACILILLLIIIKMLVIFAILPSTNIYLSILVPHMLLLNLNFSI*IFGVLFQ 602
F+ C L + L I I+ I+L I + H++++++ F I F
Sbjct: 59 FILLFLFCFTLFFCFLSFDTRFLFIIIIIQYIIIFLFIFINHIIIISILFEI------FS 112
Query: 603 LLQFMVIDIFSPF*MTTPDFSGSFFLKPKLKCLHMLRILLL*FRIIFTPPLNSLELIMAL 662
LL F++ + + F + L+++ +LL + F M L
Sbjct: 113 LLLFLL--------LMSSRFGYKILVLWYYYMLNLINFILLFILLYF----------MIL 154
Query: 663 NSCYLLFMLHMALFTRNLV*KLPNKMVELKENINTF*MLVELCFISLNFLLPFGLMLYIM 722
N C+ L +F E + F L+ L + + FL+ F LYI
Sbjct: 155 NYCFFLCDFCFLVFDE-----------EWLGILCLFYTLLILFKLYIAFLILFMEQLYIR 203
Query: 723 LFSS*IEFLHHYFITNHLTSFFIINSLILPFSKFLVVYVMLPLYILIEPNCNLELESL-- 780
L F+ Y +T ++ FI+ L++ F F ++++ L L+ +C L L
Sbjct: 204 LG----VFIFIYMLTFYILFCFILIILLISFIYFYILFIKLLLF----QSCTCVLIGLNS 255
Query: 781 YS*VTSLVIRVLLSMIFNLERFLCQDMLLFMKTISLTLILHPLLLLIGNIFLIFPLYLIL 840
++ V+ L + + + F F+ +F ++ LI L+L+ I+ F +Y I
Sbjct: 256 FAIVSLLFVLSVNNFCFLFLIFISTKNYIFYLYLNFHLIYSISLVLLIIIYYFFIIYNIF 315
Query: 841 ILLSLPVNLQSLMILLLLLHQLYLLLLLLHLFHLLFPDILPETPLLHLTSKIIFVTIFKL 900
N + + +L+ LLL L I P++ +F + +L
Sbjct: 316 D-FKYNENYFLINFIFFSFFNNFLISLLLACLFLCIGAI----PIVFGFFIKVFCLLLQL 370
Query: 901 RHIIFLIFFPIIIFLTIILLLLCLYIHNL 929
++ I F II+L II + I N+
Sbjct: 371 SYLCICIGFFFIIWLIIIYIFYFRLIVNI 399
Score = 37.0 bits (84), Expect = 4.9
Identities = 92/445 (20%), Positives = 183/445 (40%), Gaps = 83/445 (18%)
Query: 168 ILVVVLHLKLLRFIEMRIRLCNFLQD*MINFLLLELKFFSLILFLL*TKFIL*LFRKKVI 227
+ + ++H+ + I I LC++ +++F LL L +L L +I +F ++
Sbjct: 6 LFLYIIHIFIFLIIYSFIILCDYTTLTLLSFDLLWL-LINLFWITLLDSYICFIF---IL 61
Query: 228 MLLLLL*VYLLIPPFKSMLLIIENLKVVAKASLQISLLDSVLSVIVQIIL*IFVT*NMVI 287
+ L ++ F + L I ++ + I L IF+ ++I
Sbjct: 62 LFLFCFTLFFCFLSFDTRFLFI-----------------IIIIQYIIIFLFIFINHIIII 104
Query: 288 TMLTNLHQV*MLLLKKILKMVQCLLLDWV-----LLQALLLLVFLKSN*LNLCP-----C 337
++L + + + LL + +L+ W L+ +LL + L LN C C
Sbjct: 105 SILFEIFSLLLFLLLMSSRFGYKILVLWYYYMLNLINFILLFILLYFMILNYCFFLCDFC 164
Query: 338 YN--NQIWL------FQHLLLHKLPPITLHLPAFPLQILLKVYLLLFLL*QVNLLFG*LI 389
+ ++ WL + L+L KL L L L I L V++ +++L +LF ++
Sbjct: 165 FLVFDEEWLGILCLFYTLLILFKLYIAFLILFMEQLYIRLGVFIFIYML-TFYILFCFIL 223
Query: 390 MVLMNIYV---VILLHLVPF-------IE*NLF-----MFPYPMVTLC*LILLVMSLSIP 434
++L+ ++ ++ + L+ F I N F +F + C L L+ +S
Sbjct: 224 IILLISFIYFYILFIKLLLFQSCTCVLIGLNSFAIVSLLFVLSVNNFCFLFLIFISTKN- 282
Query: 435 SFI*VMFFIHLILPSISSL*QNVVIH*LVSFIFLLKNVL-YRTNNL*R*LVWLNRKMVYT 493
+F+++L I S+ ++V+ ++ + F++ N+ ++ N N ++
Sbjct: 283 ----YIFYLYLNFHLIYSI--SLVLLIIIYYFFIIYNIFDFKYNE--------NYFLINF 328
Query: 494 SFIHHANLLILSFLLYLKVLVMLFLFPIVIRIVNMYLTMPYGTLDLVICLIKDFI*CLAC 553
F N ++S LL LFL I IV +G V CL+ C
Sbjct: 329 IFFSFFNNFLISLLL-----ACLFLCIGAIPIV-------FGFFIKVFCLLLQLSYLCIC 376
Query: 554 ILILLLIIIKMLVIFAILPSTNIYL 578
I +I + ++ IF NI++
Sbjct: 377 IGFFFIIWLIIIYIFYFRLIVNIFI 401
>ref|YP_241650.1| hypothetical protein XC_0549 [Xanthomonas campestris pv. campestris
str. 8004] gi|66572220|gb|AAY47630.1| conserved
hypothetical protein [Xanthomonas campestris pv.
campestris str. 8004]
Length = 103
Score = 52.8 bits (125), Expect = 9e-05
Identities = 41/114 (35%), Positives = 63/114 (54%), Gaps = 15/114 (13%)
Query: 811 MKTISLTLILHPLLLLIGNIFLIFPLYLILILLSLPVNLQSLMILLLLLHQLYLLLLLLH 870
M+T L L+L LLLL+ + L+ L L+L+LL L + L L++LLLLL L LLLLLL
Sbjct: 3 MRTAMLLLLLLLLLLLLLLLLLLLLLLLLLLLLLLLLLLLLLLLLLLLLLLLLLLLLLLL 62
Query: 871 LFHLLFPDILPETPLLHLTSKIIFVTIFKLRHIIFLIFFPIIIFLTIILLLLCL 924
L LL ++ + + L ++ L+ +++ L ++LLLL L
Sbjct: 63 LLLLLL---------------LLLLLLLLLLLLLLLLLLLLLLLLLLLLLLLLL 101
Score = 51.2 bits (121), Expect = 3e-04
Identities = 38/91 (41%), Positives = 57/91 (61%)
Query: 785 TSLVIRVLLSMIFNLERFLCQDMLLFMKTISLTLILHPLLLLIGNIFLIFPLYLILILLS 844
T++++ +LL ++ L L +LL + + L L+L LLLL+ + L+ L L+L+LL
Sbjct: 5 TAMLLLLLLLLLLLLLLLLLLLLLLLLLLLLLLLLLLLLLLLLLLLLLLLLLLLLLLLLL 64
Query: 845 LPVNLQSLMILLLLLHQLYLLLLLLHLFHLL 875
L + L L++LLLLL L LLLLLL L LL
Sbjct: 65 LLLLLLLLLLLLLLLLLLLLLLLLLLLLLLL 95
Score = 49.7 bits (117), Expect = 7e-04
Identities = 38/89 (42%), Positives = 55/89 (61%)
Query: 787 LVIRVLLSMIFNLERFLCQDMLLFMKTISLTLILHPLLLLIGNIFLIFPLYLILILLSLP 846
L++ +LL ++ L L +LL + + L L+L LLLL+ + L+ L L+L+LL L
Sbjct: 8 LLLLLLLLLLLLLLLLLLLLLLLLLLLLLLLLLLLLLLLLLLLLLLLLLLLLLLLLLLLL 67
Query: 847 VNLQSLMILLLLLHQLYLLLLLLHLFHLL 875
+ L L++LLLLL L LLLLLL L LL
Sbjct: 68 LLLLLLLLLLLLLLLLLLLLLLLLLLLLL 96
Score = 49.7 bits (117), Expect = 7e-04
Identities = 38/89 (42%), Positives = 55/89 (61%)
Query: 787 LVIRVLLSMIFNLERFLCQDMLLFMKTISLTLILHPLLLLIGNIFLIFPLYLILILLSLP 846
L++ +LL ++ L L +LL + + L L+L LLLL+ + L+ L L+L+LL L
Sbjct: 10 LLLLLLLLLLLLLLLLLLLLLLLLLLLLLLLLLLLLLLLLLLLLLLLLLLLLLLLLLLLL 69
Query: 847 VNLQSLMILLLLLHQLYLLLLLLHLFHLL 875
+ L L++LLLLL L LLLLLL L LL
Sbjct: 70 LLLLLLLLLLLLLLLLLLLLLLLLLLLLL 98
Score = 49.7 bits (117), Expect = 7e-04
Identities = 38/89 (42%), Positives = 55/89 (61%)
Query: 787 LVIRVLLSMIFNLERFLCQDMLLFMKTISLTLILHPLLLLIGNIFLIFPLYLILILLSLP 846
L++ +LL ++ L L +LL + + L L+L LLLL+ + L+ L L+L+LL L
Sbjct: 14 LLLLLLLLLLLLLLLLLLLLLLLLLLLLLLLLLLLLLLLLLLLLLLLLLLLLLLLLLLLL 73
Query: 847 VNLQSLMILLLLLHQLYLLLLLLHLFHLL 875
+ L L++LLLLL L LLLLLL L LL
Sbjct: 74 LLLLLLLLLLLLLLLLLLLLLLLLLLLLL 102
Score = 49.7 bits (117), Expect = 7e-04
Identities = 38/89 (42%), Positives = 55/89 (61%)
Query: 787 LVIRVLLSMIFNLERFLCQDMLLFMKTISLTLILHPLLLLIGNIFLIFPLYLILILLSLP 846
L++ +LL ++ L L +LL + + L L+L LLLL+ + L+ L L+L+LL L
Sbjct: 12 LLLLLLLLLLLLLLLLLLLLLLLLLLLLLLLLLLLLLLLLLLLLLLLLLLLLLLLLLLLL 71
Query: 847 VNLQSLMILLLLLHQLYLLLLLLHLFHLL 875
+ L L++LLLLL L LLLLLL L LL
Sbjct: 72 LLLLLLLLLLLLLLLLLLLLLLLLLLLLL 100
Score = 49.7 bits (117), Expect = 7e-04
Identities = 38/89 (42%), Positives = 55/89 (61%)
Query: 787 LVIRVLLSMIFNLERFLCQDMLLFMKTISLTLILHPLLLLIGNIFLIFPLYLILILLSLP 846
L++ +LL ++ L L +LL + + L L+L LLLL+ + L+ L L+L+LL L
Sbjct: 9 LLLLLLLLLLLLLLLLLLLLLLLLLLLLLLLLLLLLLLLLLLLLLLLLLLLLLLLLLLLL 68
Query: 847 VNLQSLMILLLLLHQLYLLLLLLHLFHLL 875
+ L L++LLLLL L LLLLLL L LL
Sbjct: 69 LLLLLLLLLLLLLLLLLLLLLLLLLLLLL 97
Score = 49.7 bits (117), Expect = 7e-04
Identities = 38/89 (42%), Positives = 55/89 (61%)
Query: 787 LVIRVLLSMIFNLERFLCQDMLLFMKTISLTLILHPLLLLIGNIFLIFPLYLILILLSLP 846
L++ +LL ++ L L +LL + + L L+L LLLL+ + L+ L L+L+LL L
Sbjct: 13 LLLLLLLLLLLLLLLLLLLLLLLLLLLLLLLLLLLLLLLLLLLLLLLLLLLLLLLLLLLL 72
Query: 847 VNLQSLMILLLLLHQLYLLLLLLHLFHLL 875
+ L L++LLLLL L LLLLLL L LL
Sbjct: 73 LLLLLLLLLLLLLLLLLLLLLLLLLLLLL 101
Score = 49.7 bits (117), Expect = 7e-04
Identities = 38/89 (42%), Positives = 55/89 (61%)
Query: 787 LVIRVLLSMIFNLERFLCQDMLLFMKTISLTLILHPLLLLIGNIFLIFPLYLILILLSLP 846
L++ +LL ++ L L +LL + + L L+L LLLL+ + L+ L L+L+LL L
Sbjct: 11 LLLLLLLLLLLLLLLLLLLLLLLLLLLLLLLLLLLLLLLLLLLLLLLLLLLLLLLLLLLL 70
Query: 847 VNLQSLMILLLLLHQLYLLLLLLHLFHLL 875
+ L L++LLLLL L LLLLLL L LL
Sbjct: 71 LLLLLLLLLLLLLLLLLLLLLLLLLLLLL 99
Score = 49.3 bits (116), Expect = 0.001
Identities = 42/118 (35%), Positives = 62/118 (51%), Gaps = 22/118 (18%)
Query: 807 MLLFMKTISLTLILHPLLLLIGNIFLIFPLYLILILLSLPVNLQSLMILLLLLHQLYLLL 866
MLL + + L L+L LLLL+ + L+ L L+L+LL L + L L++LLLLL L LLL
Sbjct: 7 MLLLLLLLLLLLLLLLLLLLLLLLLLLLLLLLLLLLLLLLLLLLLLLLLLLLLLLLLLLL 66
Query: 867 LLLHLFHLLFPDILPETPLLHLTSKIIFVTIFKLRHIIFLIFFPIIIFLTIILLLLCL 924
LLL L LL +L ++ L+ +++ L ++LLLL L
Sbjct: 67 LLLLLLLLLLLLLL----------------------LLLLLLLLLLLLLLLLLLLLLL 102
Score = 48.9 bits (115), Expect = 0.001
Identities = 36/95 (37%), Positives = 56/95 (58%)
Query: 832 LIFPLYLILILLSLPVNLQSLMILLLLLHQLYLLLLLLHLFHLLFPDILPETPLLHLTSK 891
L+ L L+L+LL L + L L++LLLLL L LLLLLL L LL +L LL L
Sbjct: 8 LLLLLLLLLLLLLLLLLLLLLLLLLLLLLLLLLLLLLLLLLLLLLLLLLLLLLLLLLLLL 67
Query: 892 IIFVTIFKLRHIIFLIFFPIIIFLTIILLLLCLYI 926
++ + + L ++ L+ +++ L ++LLLL L +
Sbjct: 68 LLLLLLLLLLLLLLLLLLLLLLLLLLLLLLLLLLL 102
Score = 48.5 bits (114), Expect = 0.002
Identities = 39/95 (41%), Positives = 56/95 (58%)
Query: 807 MLLFMKTISLTLILHPLLLLIGNIFLIFPLYLILILLSLPVNLQSLMILLLLLHQLYLLL 866
+LL + + L L+L LLLL+ + L+ L L+L+LL L + L L++LLLLL L LLL
Sbjct: 9 LLLLLLLLLLLLLLLLLLLLLLLLLLLLLLLLLLLLLLLLLLLLLLLLLLLLLLLLLLLL 68
Query: 867 LLLHLFHLLFPDILPETPLLHLTSKIIFVTIFKLR 901
LLL L LL +L LL L ++ + + LR
Sbjct: 69 LLLLLLLLLLLLLLLLLLLLLLLLLLLLLLLLLLR 103
Score = 48.1 bits (113), Expect = 0.002
Identities = 35/95 (36%), Positives = 56/95 (58%)
Query: 832 LIFPLYLILILLSLPVNLQSLMILLLLLHQLYLLLLLLHLFHLLFPDILPETPLLHLTSK 891
++ L L+L+LL L + L L++LLLLL L LLLLLL L LL +L LL L
Sbjct: 7 MLLLLLLLLLLLLLLLLLLLLLLLLLLLLLLLLLLLLLLLLLLLLLLLLLLLLLLLLLLL 66
Query: 892 IIFVTIFKLRHIIFLIFFPIIIFLTIILLLLCLYI 926
++ + + L ++ L+ +++ L ++LLLL L +
Sbjct: 67 LLLLLLLLLLLLLLLLLLLLLLLLLLLLLLLLLLL 101
Score = 47.8 bits (112), Expect = 0.003
Identities = 36/89 (40%), Positives = 53/89 (59%)
Query: 787 LVIRVLLSMIFNLERFLCQDMLLFMKTISLTLILHPLLLLIGNIFLIFPLYLILILLSLP 846
+ +R + ++ L L +LL + + L L+L LLLL+ + L+ L L+L+LL L
Sbjct: 1 MAMRTAMLLLLLLLLLLLLLLLLLLLLLLLLLLLLLLLLLLLLLLLLLLLLLLLLLLLLL 60
Query: 847 VNLQSLMILLLLLHQLYLLLLLLHLFHLL 875
+ L L++LLLLL L LLLLLL L LL
Sbjct: 61 LLLLLLLLLLLLLLLLLLLLLLLLLLLLL 89
Score = 44.3 bits (103), Expect = 0.031
Identities = 35/97 (36%), Positives = 56/97 (57%)
Query: 840 LILLSLPVNLQSLMILLLLLHQLYLLLLLLHLFHLLFPDILPETPLLHLTSKIIFVTIFK 899
++LL L + L L++LLLLL L LLLLLL L LL +L LL L ++ + +
Sbjct: 7 MLLLLLLLLLLLLLLLLLLLLLLLLLLLLLLLLLLLLLLLLLLLLLLLLLLLLLLLLLLL 66
Query: 900 LRHIIFLIFFPIIIFLTIILLLLCLYIHNLSLNPMLK 936
L ++ L+ +++ L ++LLLL L + L L +L+
Sbjct: 67 LLLLLLLLLLLLLLLLLLLLLLLLLLLLLLLLLLLLR 103
Score = 38.9 bits (89), Expect = 1.3
Identities = 27/92 (29%), Positives = 55/92 (59%)
Query: 499 ANLLILSFLLYLKVLVMLFLFPIVIRIVNMYLTMPYGTLDLVICLIKDFI*CLACILILL 558
A LL+L LL L +L++L L +++ ++ + L + L L++ L+ + L +L+LL
Sbjct: 6 AMLLLLLLLLLLLLLLLLLLLLLLLLLLLLLLLLLLLLLLLLLLLLLLLLLLLLLLLLLL 65
Query: 559 LIIIKMLVIFAILPSTNIYLSILVPHMLLLNL 590
L+++ +L++ +L + L +L+ +LLL L
Sbjct: 66 LLLLLLLLLLLLLLLLLLLLLLLLLLLLLLLL 97
Score = 38.1 bits (87), Expect = 2.2
Identities = 26/90 (28%), Positives = 54/90 (59%)
Query: 501 LLILSFLLYLKVLVMLFLFPIVIRIVNMYLTMPYGTLDLVICLIKDFI*CLACILILLLI 560
LL+L LL L +L++L L +++ ++ + L + L L++ L+ + L +L+LLL+
Sbjct: 11 LLLLLLLLLLLLLLLLLLLLLLLLLLLLLLLLLLLLLLLLLLLLLLLLLLLLLLLLLLLL 70
Query: 561 IIKMLVIFAILPSTNIYLSILVPHMLLLNL 590
++ +L++ +L + L +L+ +LLL L
Sbjct: 71 LLLLLLLLLLLLLLLLLLLLLLLLLLLLLL 100
Score = 38.1 bits (87), Expect = 2.2
Identities = 26/90 (28%), Positives = 54/90 (59%)
Query: 501 LLILSFLLYLKVLVMLFLFPIVIRIVNMYLTMPYGTLDLVICLIKDFI*CLACILILLLI 560
LL+L LL L +L++L L +++ ++ + L + L L++ L+ + L +L+LLL+
Sbjct: 10 LLLLLLLLLLLLLLLLLLLLLLLLLLLLLLLLLLLLLLLLLLLLLLLLLLLLLLLLLLLL 69
Query: 561 IIKMLVIFAILPSTNIYLSILVPHMLLLNL 590
++ +L++ +L + L +L+ +LLL L
Sbjct: 70 LLLLLLLLLLLLLLLLLLLLLLLLLLLLLL 99
Score = 38.1 bits (87), Expect = 2.2
Identities = 26/90 (28%), Positives = 54/90 (59%)
Query: 501 LLILSFLLYLKVLVMLFLFPIVIRIVNMYLTMPYGTLDLVICLIKDFI*CLACILILLLI 560
LL+L LL L +L++L L +++ ++ + L + L L++ L+ + L +L+LLL+
Sbjct: 9 LLLLLLLLLLLLLLLLLLLLLLLLLLLLLLLLLLLLLLLLLLLLLLLLLLLLLLLLLLLL 68
Query: 561 IIKMLVIFAILPSTNIYLSILVPHMLLLNL 590
++ +L++ +L + L +L+ +LLL L
Sbjct: 69 LLLLLLLLLLLLLLLLLLLLLLLLLLLLLL 98
Score = 38.1 bits (87), Expect = 2.2
Identities = 26/90 (28%), Positives = 54/90 (59%)
Query: 501 LLILSFLLYLKVLVMLFLFPIVIRIVNMYLTMPYGTLDLVICLIKDFI*CLACILILLLI 560
LL+L LL L +L++L L +++ ++ + L + L L++ L+ + L +L+LLL+
Sbjct: 13 LLLLLLLLLLLLLLLLLLLLLLLLLLLLLLLLLLLLLLLLLLLLLLLLLLLLLLLLLLLL 72
Query: 561 IIKMLVIFAILPSTNIYLSILVPHMLLLNL 590
++ +L++ +L + L +L+ +LLL L
Sbjct: 73 LLLLLLLLLLLLLLLLLLLLLLLLLLLLLL 102
Score = 38.1 bits (87), Expect = 2.2
Identities = 26/90 (28%), Positives = 54/90 (59%)
Query: 501 LLILSFLLYLKVLVMLFLFPIVIRIVNMYLTMPYGTLDLVICLIKDFI*CLACILILLLI 560
LL+L LL L +L++L L +++ ++ + L + L L++ L+ + L +L+LLL+
Sbjct: 12 LLLLLLLLLLLLLLLLLLLLLLLLLLLLLLLLLLLLLLLLLLLLLLLLLLLLLLLLLLLL 71
Query: 561 IIKMLVIFAILPSTNIYLSILVPHMLLLNL 590
++ +L++ +L + L +L+ +LLL L
Sbjct: 72 LLLLLLLLLLLLLLLLLLLLLLLLLLLLLL 101
>gb|EAL01937.1| hypothetical protein CaO19.11807 [Candida albicans SC5314]
Length = 191
Score = 52.0 bits (123), Expect = 1e-04
Identities = 44/188 (23%), Positives = 89/188 (46%), Gaps = 23/188 (12%)
Query: 748 SLILPFSKFLVVYVMLPLYILIEPNCNLELESLYS*VTSLVIRVLLSMIFNLERFLCQDM 807
+ ++ F KF+ +++ E++ + ++V R L MI L F+ +
Sbjct: 3 TFLMVFKKFITIFIF---------------ETIMFFLFTMVFRFLFFMILFLFLFM---I 44
Query: 808 LLFMKTISLTLILHPLLLLIGNIFLIFPLYLILILLSLPVNLQSLMILLLLLHQLYLLLL 867
L F+ + +IL L++L FL+ +L+++ LS+ ++L +L + + L +
Sbjct: 45 LFFLMILFFLMILFFLMILF---FLMILFFLMVLFLSMVLSLSVFFLLFMFFFLFFFLSM 101
Query: 868 LLHLFHLLFPDILPETPLLHLTSKIIFVTIFKLRHIIFLIFFPIIIFLTIILLLLCLYIH 927
+ LF +F +L L + F+ + +IFL F I +FL+ + LLL +++
Sbjct: 102 VFFLFLFMF--LLFFMFFLFFMFLLFFMFLLFFMFMIFLFTFMIFLFLSFMFLLLFMFMI 159
Query: 928 NLSLNPML 935
L L M+
Sbjct: 160 FLLLLIMI 167
Score = 47.8 bits (112), Expect = 0.003
Identities = 45/171 (26%), Positives = 81/171 (47%), Gaps = 10/171 (5%)
Query: 711 FLLPFGLMLYIMLFSS*IEFLHH------YFITNHLTSFFIINSLILPFSKFLVVYVMLP 764
FL+ F + I +F + + FL +F+ L F I+ L++ F ++ ++M+
Sbjct: 4 FLMVFKKFITIFIFETIMFFLFTMVFRFLFFMILFLFLFMILFFLMILFFLMILFFLMIL 63
Query: 765 LYILIEPNCNLELESLYS*VTSLVIRVLLSMIFNLERFLCQDMLLFMKTISLTLILHPLL 824
+++I + S V SL + LL M F L FL LF+ L +
Sbjct: 64 FFLMI---LFFLMVLFLSMVLSLSVFFLLFMFFFLFFFLSMVFFLFLFMFLLFFMFFLFF 120
Query: 825 L-LIGNIFLIFPLYLILILLSLPVNLQSLMILLLLLHQLYLLLLLLHLFHL 874
+ L+ +FL+F +++I + + S M LLL + ++LLLL++ L L
Sbjct: 121 MFLLFFMFLLFFMFMIFLFTFMIFLFLSFMFLLLFMFMIFLLLLIMILLLL 171
Score = 40.4 bits (93), Expect = 0.44
Identities = 44/175 (25%), Positives = 81/175 (46%), Gaps = 12/175 (6%)
Query: 698 F*MLVELCFISLNFLLPFGLMLYIML--FSS*IEFLHHYFITNHLTSFFIIN-SLILPFS 754
F M+ F + FL F ++ ++M+ F + FL F L ++ S++L S
Sbjct: 25 FTMVFRFLFFMILFLFLFMILFFLMILFFLMILFFLMILFFLMILFFLMVLFLSMVLSLS 84
Query: 755 KFLVVYVMLPLYILIEPNCNLELESLYS*VTSLVIRVLLSMIFNLERFLCQDMLLFMKTI 814
F ++++ L+ + L L + + L L F+ ++F+ T
Sbjct: 85 VFFLLFMFFFLFFFLSMVFFLFLFMFLLFFMFFLFFMFLLFFMFLLFFM---FMIFLFTF 141
Query: 815 SLTLILHPLLLLIGNIFLIFPLYLILILLSLPVNLQSLMILLLLLHQLYLLLLLL 869
+ L L + LL+ +F+IF L LI+ILL L SLM+L L ++ L+ + ++
Sbjct: 142 MIFLFLSFMFLLLF-MFMIFLLLLIMILLLL-----SLMVLFLFINYLWFFMFIV 190
Score = 38.5 bits (88), Expect = 1.7
Identities = 45/202 (22%), Positives = 83/202 (40%), Gaps = 42/202 (20%)
Query: 367 ILLKVYLLLFLL*QVNLLFG*LIMVLMNIYVVILLHLVPFIE*NLFMFPYPMVTLC*LIL 426
I+ ++ ++F +LF L M+L + ++ L ++ F+ LF + +
Sbjct: 20 IMFFLFTMVFRFLFFMILFLFLFMILFFLMILFFLMILFFLM-ILFFLMILFFLMVLFLS 78
Query: 427 LVMSLSIPSFI*VMFFIHLILPSISSL*QNVVIH*LVSFIFLLKNVLYRTNNL*R*LVWL 486
+V+SLS+ + + FF+ L +V F+FL +L+
Sbjct: 79 MVLSLSVFFLLFMFFFLFFFLS-------------MVFFLFLFMFLLF------------ 113
Query: 487 NRKMVYTSFIHHANLLILSFLLYLKVLVMLFLFPIVIRIVNMYLTMPYGTLDLVICLIKD 546
+ F+ LL FLL+ ++ LF F I + + M+L L
Sbjct: 114 -----FMFFLFFMFLLFFMFLLFFMFMIFLFTFMIFLFLSFMFL-----------LLFMF 157
Query: 547 FI*CLACILILLLIIIKMLVIF 568
I L I+ILLL+ + +L +F
Sbjct: 158 MIFLLLLIMILLLLSLMVLFLF 179
>gb|EAK89294.1| hypothetical protein with signal peptide [Cryptosporidium parvum]
gi|66358938|ref|XP_626647.1| hypothetical protein
cgd3_620 [Cryptosporidium parvum]
Length = 242
Score = 52.0 bits (123), Expect = 1e-04
Identities = 65/219 (29%), Positives = 97/219 (43%), Gaps = 42/219 (19%)
Query: 708 SLNFLLPFGLMLYIMLFSS*IEFLHHYFITNHLTSFFIINSLILPFSKFLVVYVMLPLYI 767
S F+L FGL+L+ LF +H +FI L F+ L+ +++++ L+I
Sbjct: 58 SFLFILFFGLLLFFYLF------VHLFFILFFLRFLFV---LLFILCHLFILHLLFVLFI 108
Query: 768 LIEPNCNLELESLYS*VTSLVIRVLLSMIFNLERF-LCQDMLLFMKTISLTLILHPLLLL 826
L C L + L+ IR L + F F LC LF+ + L + LL
Sbjct: 109 L----CLLFI-FLFILCLLFFIRFLFFLRFLFVLFILCH---LFILHLLFVLFIICLLFF 160
Query: 827 IGNIFLIFPLYLILILLSLPVNLQSLMILLLLLHQLYLLLLLLHLFHLLFPDILPETPLL 886
I +F++F L + IL +L ++ +L LL LYLL LL H LF
Sbjct: 161 IRFLFVLFILCHLFIL-----HLLFVLFILCLLSFLYLLFSLLFFLHFLF---------- 205
Query: 887 HLTSKIIFVTIFKLRHIIFLIFFPIIIFLTIILLLLCLY 925
+ +F L I+ L+F II L +L LL LY
Sbjct: 206 -------VLLLFVLLFILCLLF--IICLLFALLFLLPLY 235
Score = 45.8 bits (107), Expect = 0.011
Identities = 58/246 (23%), Positives = 106/246 (42%), Gaps = 42/246 (17%)
Query: 809 LFMKTISLTLILHPLLLLIGNIFLIFPLYLILILLSLPVNLQSLMILLLLLHQLYLLLLL 868
+F +SL P FL + +L+ L V+L ++ L L L +L
Sbjct: 37 IFSSCLSLFSFFSPSFSFSPFSFLFILFFGLLLFFYLFVHLFFILFFLRFLFVLLFILCH 96
Query: 869 LHLFHLLFPDILPETPLLHLTSKIIFVTIFKLRHIIFLIFFPIIIFLTIILLLLCLYIHN 928
L + HLLF +L + ++F+ +F L + F+ F + FL ++ +L L+I +
Sbjct: 97 LFILHLLF--------VLFILC-LLFIFLFILCLLFFIRFLFFLRFLFVLFILCHLFILH 147
Query: 929 LSLNPMLKQVNMSVGCMLCKLNFKLLRKLVLGNLWIYHQM--LSLLDVDGFTKSNFMLMA 986
L + + C+L + F L +L +L+I H + L +L + F F L+
Sbjct: 148 LLFV-------LFIICLLFFIRF-LFVLFILCHLFILHLLFVLFILCLLSFLYLLFSLLF 199
Query: 987 LLRDIKQDWLPKVTIKLRAWIILIHILQLLNSQQSDL*LLYLPFIICIYINLMSIMPFFM 1046
L + +++L+ +L L +L L FIIC+ L+ ++P +
Sbjct: 200 FLHFL--------------FVLLLFVL---------LFILCLLFIICLLFALLFLLPLYW 236
Query: 1047 VNFRKM 1052
V + M
Sbjct: 237 VVYLTM 242
Score = 39.7 bits (91), Expect = 0.75
Identities = 66/268 (24%), Positives = 116/268 (42%), Gaps = 29/268 (10%)
Query: 576 IYLSILVPHMLLLNLNFSI*IFGVLFQLLQF-MVIDIFSPF*MTTPDFSGSFFLKPKLKC 634
++LS +P ++ + + +F V + + +I IFS FS SF P
Sbjct: 1 VFLSKGIPPLIKFPIFAGMVVFAVKLPVEELESLISIFSSCLSLFSFFSPSFSFSP-FSF 59
Query: 635 LHMLRILLL*FRIIFTPPLNSLELIMALNSCYLLFMLHMALFTRNLV*KLPNKMVELKEN 694
L +L LL F +F ++ ++ L ++L + LF +L+ L
Sbjct: 60 LFILFFGLLLFFYLF---VHLFFILFFLRFLFVLLFILCHLFILHLLFVL---------- 106
Query: 695 INTF*MLVELCFISLNFLLPFGLMLYIMLFSS*IEFLHHYFITNHLTSFFIINSLILPFS 754
F + + F+ + LL F L+ + F + L H FI + L FII +L F
Sbjct: 107 ---FILCLLFIFLFILCLLFFIRFLFFLRFLFVLFILCHLFILHLLFVLFII--CLLFFI 161
Query: 755 KFLVVYVMLPLYILIEPNCNLELESLYS*VTSLVIRVLLSMIFNLERFLCQDMLLFMKTI 814
+FL V L+IL C+L + L + L + L ++F+L FL +L + +
Sbjct: 162 RFLFV-----LFIL----CHLFILHLLFVLFILCLLSFLYLLFSLLFFLHFLFVLLLFVL 212
Query: 815 SLTLILHPLLLLIGNIFLIFPLYLILIL 842
L L ++ L+ + + PLY ++ L
Sbjct: 213 LFILCLLFIICLLFALLFLLPLYWVVYL 240
>ref|XP_499191.1| PREDICTED: hypothetical protein XP_499191 [Homo sapiens]
Length = 397
Score = 51.2 bits (121), Expect = 3e-04
Identities = 76/425 (17%), Positives = 184/425 (42%), Gaps = 47/425 (11%)
Query: 502 LILSFLLYLKVLVMLFLFPIVIRIVNMYLTMPYGTLDLVICLIKDFI*CLACILILLLII 561
++ L ++ ++ M +F ++ + +++++ + TL + + F+ + L+ + ++
Sbjct: 1 MVFRTLAFVTLVFMSLVFVTLVFVTLVFMSLVFMTLVFMSLV---FVTLVFMSLVFMSLV 57
Query: 562 IKMLVIFAILPSTNIYLSILVPHMLLLNLNFSI*IF-GVLFQLLQFMVIDIFSPF*MTTP 620
LV +++ T +++S++ ++ + L F +F +LF + F + F T
Sbjct: 58 FMTLVFMSLVFMTLVFMSLVFMSLVFMTLVFMTLVFVSLLFMTMVFRTL-AFVTLVFVTL 116
Query: 621 DFSGSFFLKPKLKCLHMLRILLL*FRIIFTPPLNSLELIMALNSCYLLFM--LHMALFTR 678
F F+ L + ++ + L+ ++F + + M+L L+FM + M L
Sbjct: 117 VFMSLVFMT--LVFMSLVFVTLVFVTLVFMSLVFMTLVFMSLVFMTLVFMSLVFMTLVFM 174
Query: 679 NLV*KLPNKMVELKENINTF*MLV--ELCFISLNFL-LPFGLMLYIMLFSS*IEFLHHYF 735
+LV + L F LV L F+SL F+ L F ++++ L + F+ F
Sbjct: 175 SLV------FMSLVFMTLVFMSLVFMSLVFMSLVFMTLVFMCLVFMTLVFMFLVFMSLVF 228
Query: 736 ITNHLTSFFIINSLILPFSKFLVVYVMLPLYILIEPNCNLELESLYS*VTSLVIRVLLSM 795
+T S + + + +V++ L L+ SL + V +S+
Sbjct: 229 MTLVFMSLVFMTLVFMSLVFMTLVFMSLVFMSLV-------FMSLVFMSLVFMSLVFMSL 281
Query: 796 IFNLERFLCQDMLLFMKTISLTLILHPLLLLIGNIFLIFPLYLILILLSLPVNLQSLMIL 855
+F F+ L+FM + ++L+ L+ + L+F + + L+ + + SL+ +
Sbjct: 282 VFMTLVFMS---LVFMSLVFMSLVFMTLVFMC----LVFMTLVFMFLVFMSLVFMSLVFM 334
Query: 856 LLLLHQLYLLLLLLHLFHLLFPDILPETPLLHLTSKIIFVTIFKLRHIIFLIFFPIIIFL 915
L+ L + L+ +F ++F+++ + + + F ++F+
Sbjct: 335 TLVFMSLVFMSLVFMTLVFMF---------------LVFMSLVFMTLVFMFLVFTSLVFM 379
Query: 916 TIILL 920
T++ +
Sbjct: 380 TLVFI 384
Score = 41.6 bits (96), Expect = 0.20
Identities = 65/371 (17%), Positives = 158/371 (42%), Gaps = 58/371 (15%)
Query: 555 LILLLIIIKMLVIFAILPSTNIYLSILVPHMLLLNLNFSI*IF-GVLFQLLQFMVIDIFS 613
L + ++ LV ++ T +++S++ ++ ++L F +F ++F L FM + S
Sbjct: 6 LAFVTLVFMSLVFVTLVFVTLVFMSLVFMTLVFMSLVFVTLVFMSLVFMSLVFMTLVFMS 65
Query: 614 PF*MTTPDFSGSFFLKPKLKCLHMLRILLL*FRIIFTPPLNSLELIMALNSCYLLFMLHM 673
MT S F + ++ + L+ ++F ++ L + M + + ++ +
Sbjct: 66 LVFMTLVFMSLVF--------MSLVFMTLVFMTLVF---VSLLFMTMVFRTLAFVTLVFV 114
Query: 674 ALFTRNLV*KLPNKMVELKENINTF*MLVELCFISLNFL-LPFGLMLYIMLFSS*IEFLH 732
L +LV + L F+SL F+ L F ++++ L + F+
Sbjct: 115 TLVFMSLV-------------------FMTLVFMSLVFVTLVFVTLVFMSLVFMTLVFMS 155
Query: 733 HYFITNHLTSFFIINSLILPFSKFLVVYVMLPLYILIEPNCNLELESLYS*VTSLVIRVL 792
F+T S + + + +V++ L L+ + V
Sbjct: 156 LVFMTLVFMSLVFMTLVFMSLVFMSLVFMTLVFMSLV-----------------FMSLVF 198
Query: 793 LSMIFNLERFLCQDMLLFMKTISLTLILHPLLLL-IGNIFLIFPLYLILILLSLPVNLQS 851
+S++F F+C L+FM + + L+ L+ + + + L+F + + L+ + + S
Sbjct: 199 MSLVFMTLVFMC---LVFMTLVFMFLVFMSLVFMTLVFMSLVFMTLVFMSLVFMTLVFMS 255
Query: 852 LMILLLLLHQLYLLLLLLHLFHLLFPDILPETPLLHLTSKIIFVTIFKLRHIIFLIFFPI 911
L+ + L+ L + L+ L+F ++ T + ++F+++ + + + F
Sbjct: 256 LVFMSLVFMSLVFMSLVF--MSLVFMSLVFMTLVF---MSLVFMSLVFMSLVFMTLVFMC 310
Query: 912 IIFLTIILLLL 922
++F+T++ + L
Sbjct: 311 LVFMTLVFMFL 321
Score = 41.2 bits (95), Expect = 0.26
Identities = 76/398 (19%), Positives = 171/398 (42%), Gaps = 54/398 (13%)
Query: 247 LIIENLKVVAKASLQISLLDSVLSVIVQIIL*IFVT*NMVITMLTNLHQV*MLLLKKILK 306
L+ +L V + + + V +V + L +FVT +V L + V M L+ L
Sbjct: 11 LVFMSLVFVTLVFVTLVFMSLVFMTLVFMSL-VFVT--LVFMSLVFMSLVFMTLVFMSLV 67
Query: 307 MVQCLLLDWVLLQALLL-LVFLKSN*LNLCPCYNNQIWLFQHLLLHKLPPITLHLPAFPL 365
+ + + V + + + LVF+ ++L LF ++ L +TL
Sbjct: 68 FMTLVFMSLVFMSLVFMTLVFMTLVFVSL---------LFMTMVFRTLAFVTLVFVTLVF 118
Query: 366 QILLKVYLLLFLL*QVNLLFG*LIMVLMNIYVVILLHLV------------PFIE*NLFM 413
L+ + L+ L V L+F L+ + + ++ + LV + +L
Sbjct: 119 MSLVFMTLVFMSLVFVTLVFVTLVFMSLVFMTLVFMSLVFMTLVFMSLVFMTLVFMSLVF 178
Query: 414 FPYPMVTLC*LILLVMSLSIPSFI*V---------MFFIHLILPSISSL*QNVVIH*LVS 464
+TL + L+ MSL S + + M + + L +S + +V LV
Sbjct: 179 MSLVFMTLVFMSLVFMSLVFMSLVFMTLVFMCLVFMTLVFMFLVFMSLVFMTLVFMSLVF 238
Query: 465 FIFLLKNVLYRTNNL*R*LVWLN---RKMVYTSFIHHANLLILSFLLYLKVLVMLFLFPI 521
+ ++++ T LV+++ +V+ S + + L+ L+++ ++ M +F
Sbjct: 239 MTLVFMSLVFMT------LVFMSLVFMSLVFMSLVFMS--LVFMSLVFMSLVFMTLVFMS 290
Query: 522 VIRIVNMYLTMPYGTLDLVICLIKDFI*CLACILILLLIIIKMLVIFAILPSTNIYLSIL 581
++ + +++++ + TL + +CL+ L+ + ++ LV +++ T +++S++
Sbjct: 291 LVFMSLVFMSLVFMTL-VFMCLV-------FMTLVFMFLVFMSLVFMSLVFMTLVFMSLV 342
Query: 582 VPHMLLLNLNFSI*IF-GVLFQLLQFMVIDIFSPF*MT 618
++ + L F +F ++F L FM + S MT
Sbjct: 343 FMSLVFMTLVFMFLVFMSLVFMTLVFMFLVFTSLVFMT 380
>ref|NP_572233.1| CG15780-PA [Drosophila melanogaster] gi|7290595|gb|AAF46045.1|
CG15780-PA [Drosophila melanogaster]
Length = 640
Score = 50.8 bits (120), Expect = 3e-04
Identities = 47/194 (24%), Positives = 101/194 (51%), Gaps = 21/194 (10%)
Query: 745 IINSLILPFSKFLVVYVMLPLYILIEPNCNLELESLYS*VTSLVIRVLLSMIFNLERFLC 804
+++ +L K + +L L +L+E LEL L+ L++ ++L M+F L+
Sbjct: 78 VLHLQVLMLVKIWLQLQVLVLVLLLE-EMMLELHVLFL----LLMEMVLQMVFQLQELF- 131
Query: 805 QDMLLFMKTISLTLILHPLLLLIGNIFLIFP---LYLILILLSLPVNLQSLMILLLLLHQ 861
+LL + L ++L ++L + +FL+ L L ++L+ + + LQ L +LL+ +
Sbjct: 132 --LLLMEMVLQLQVLLMEMVLQLQELFLLLMEMVLQLQVLLMEMVLQLQELFLLLMEM-- 187
Query: 862 LYLLLLLLHLFHLLFPDILPETPLLHLTSKIIFVTIFKLRHIIFLIFFPIIIFLTIILLL 921
+L L + LL +++ + +L L ++ + +L+ ++FLI +++ L +++LL
Sbjct: 188 ----VLQLQVLFLLLMEMVLQLQVLFL---LLMEMVLQLQ-VLFLILLEMVLQLLLLVLL 239
Query: 922 LCLYIHNLSLNPML 935
L + + L P L
Sbjct: 240 LMVVVAVKVLAPQL 253
Score = 46.6 bits (109), Expect = 0.006
Identities = 71/299 (23%), Positives = 136/299 (44%), Gaps = 63/299 (21%)
Query: 298 MLLLKKILKMVQCLLLDWVL----LQALLLLVFLKSN*LNLCPCYNNQIWLFQHLLLHKL 353
MLLL +++ L+L V+ LQ L+L++ L+ L + ++ L LL+ +L
Sbjct: 15 MLLLVEMVLQFHALVLQLVVIGMQLQVLVLVLVLEEIGLQVLKLLLEEMVLQLLLLVLQL 74
Query: 354 PPITLHLPAFPLQ---ILLKVYLLLFLL*QVNLLFG*LIMVLMNIYVVILLHLVPFIE*N 410
+ LHL L + L+V +L+ LL ++ L L ++LM + + ++ L
Sbjct: 75 MEMVLHLQVLMLVKIWLQLQVLVLVLLLEEMMLELHVLFLLLMEMVLQMVFQLQ-----E 129
Query: 411 LFMFPYPMVTLC*LILLVMSLSIPSFI*VMFFIHLILPSISSL*QNVVIH*LVSFIFLLK 470
LF+ MV ++L+ M L + F+ L++
Sbjct: 130 LFLLLMEMVLQLQVLLMEMVLQLQEL----------------------------FLLLME 161
Query: 471 NVLYRTNNL*R*LVWLNRKMVYTSFIHHANLLILSFLLYLKVLVMLFLFPIVIRIVNMYL 530
VL L L +MV + LL++ +L L+VL +L L +V+++ ++L
Sbjct: 162 MVLQ--------LQVLLMEMVLQ--LQELFLLLMEMVLQLQVLFLL-LMEMVLQLQVLFL 210
Query: 531 TMPYGTLDLVICLIKDFI*CLACILILLLIIIKMLVIFAILPSTNIYLSILVPHMLLLN 589
+ +++V+ L F+ L +L LLL+++ ++V+ A+ +L P +LLL+
Sbjct: 211 LL----MEMVLQLQVLFLILLEMVLQLLLLVLLLMVVVAV--------KVLAPQLLLLD 257
Score = 46.2 bits (108), Expect = 0.008
Identities = 45/177 (25%), Positives = 103/177 (57%), Gaps = 15/177 (8%)
Query: 162 LIVLVFILVVVLHLKLLRFIEMRIRLCNFLQD*MINFLLLELKFFSLILFLL*TKFIL*L 221
L++++ ++ +VLHL++L +++ ++L + ++ ++LEL +LFLL + +L +
Sbjct: 68 LLLVLQLMEMVLHLQVLMLVKIWLQLQVLVLVLLLEEMMLELH----VLFLLLMEMVLQM 123
Query: 222 -FRKKVIMLLLLL*VYLLIPPFKSMLLIIENLKVVAK---ASLQISLLDSVLS------V 271
F+ + + LLL+ V L M+L ++ L ++ LQ+ L++ VL +
Sbjct: 124 VFQLQELFLLLMEMVLQLQVLLMEMVLQLQELFLLLMEMVLQLQVLLMEMVLQLQELFLL 183
Query: 272 IVQIIL*IFVT*NMVITMLTNLHQV*MLLLKKILKM-VQCLLLDWVLLQALLLLVFL 327
+++++L + V +++ M+ L + +LL++ +L++ V L+L ++LQ LLL++ L
Sbjct: 184 LMEMVLQLQVLFLLLMEMVLQLQVLFLLLMEMVLQLQVLFLILLEMVLQLLLLVLLL 240
Score = 45.4 bits (106), Expect = 0.014
Identities = 49/216 (22%), Positives = 111/216 (50%), Gaps = 28/216 (12%)
Query: 755 KFLVVYVMLPLYILIEPNCNLELESLYS*VTSLVIRVLL------SMIFNLERFLCQDML 808
+ +V+ + L + +L+ + L+ L + +V+++LL M+ +L+ + +
Sbjct: 31 QLVVIGMQLQVLVLVLVLEEIGLQVLKLLLEEMVLQLLLLVLQLMEMVLHLQVLMLVKIW 90
Query: 809 LFMKTISLTLILHPLLLLIGNIFL---------IFPLY-LILILLSLPVNLQSLMI-LLL 857
L ++ + L L+L ++L + +FL +F L L L+L+ + + LQ L++ ++L
Sbjct: 91 LQLQVLVLVLLLEEMMLELHVLFLLLMEMVLQMVFQLQELFLLLMEMVLQLQVLLMEMVL 150
Query: 858 LLHQLYLLL--LLLHLFHLLFPDILPETPL------LHLTSKIIFVTIFKL---RHIIFL 906
L +L+LLL ++L L LL +L L + L +++F+ + ++ ++FL
Sbjct: 151 QLQELFLLLMEMVLQLQVLLMEMVLQLQELFLLLMEMVLQLQVLFLLLMEMVLQLQVLFL 210
Query: 907 IFFPIIIFLTIILLLLCLYIHNLSLNPMLKQVNMSV 942
+ +++ L ++ L+L + L L +L V ++V
Sbjct: 211 LLMEMVLQLQVLFLILLEMVLQLLLLVLLLMVVVAV 246
Score = 44.3 bits (103), Expect = 0.031
Identities = 66/297 (22%), Positives = 131/297 (43%), Gaps = 53/297 (17%)
Query: 243 KSMLLIIENLKVVAKASLQISLLDSVLSVIVQIIL*IFVT*NMVITMLTNLHQV*MLLLK 302
K MLL++E + LQ+ ++ L V+V + L+L+
Sbjct: 13 KMMLLLVEMVLQFHALVLQLVVIGMQLQVLVLV-----------------------LVLE 49
Query: 303 KILKMVQCLLLDWVLLQALLLLVFLKSN*LNLCPCYNNQIWLFQHLLLHKLPPITLHLPA 362
+I V LLL+ ++LQ LLL++ L L+L +IWL +L+ + L L
Sbjct: 50 EIGLQVLKLLLEEMVLQLLLLVLQLMEMVLHLQVLMLVKIWLQLQVLV-----LVLLLEE 104
Query: 363 FPLQILLKVYLLLFLL*QVNLLFG*LIMVLMNIYV---VILLHLVPFIE*NLFMFPYPMV 419
L++ + LL+ ++ Q+ L ++LM + + V+L+ +V ++ LF+ MV
Sbjct: 105 MMLELHVLFLLLMEMVLQMVFQLQELFLLLMEMVLQLQVLLMEMVLQLQ-ELFLLLMEMV 163
Query: 420 TLC*LILLVMSLSIPSFI*VMFFIHLILPSISSL*QNVVIH*LVSFIFLLKNVLYRTNNL 479
++L+ M L + ++ + L L + L +V+ V F+ L++ VL
Sbjct: 164 LQLQVLLMEMVLQLQELFLLLMEMVLQLQVLFLLLMEMVLQLQVLFLLLMEMVLQ----- 218
Query: 480 *R*LVWLNRKMVYTSFIHHANLLILSFLLYLKVLVMLFLFPIVIRIVNMYLTMPYGT 536
+ L++L +L L +LV+L + + ++++ L + G+
Sbjct: 219 ----------------LQVLFLILLEMVLQLLLLVLLLMVVVAVKVLAPQLLLLDGS 259
Score = 36.2 bits (82), Expect = 8.3
Identities = 50/235 (21%), Positives = 111/235 (46%), Gaps = 41/235 (17%)
Query: 809 LFMKTISLTLILHPLLLLIGNIFLIFPLYLILILLSLPVNLQSLMILLLLLHQ--LYLLL 866
+ + + + L H L+L + ++ + L +++L L + L +L LLL + L LLL
Sbjct: 14 MMLLLVEMVLQFHALVLQL----VVIGMQLQVLVLVLVLEEIGLQVLKLLLEEMVLQLLL 69
Query: 867 LLLHLFH-----------------------LLFPDILPETPLLH-LTSKIIFVTIFKLRH 902
L+L L LL +++ E +L L +++ +F+L+
Sbjct: 70 LVLQLMEMVLHLQVLMLVKIWLQLQVLVLVLLLEEMMLELHVLFLLLMEMVLQMVFQLQE 129
Query: 903 IIFLIFFPIIIFLTIILLLLCLYIHNLSLNPMLKQVNMSVGCMLCKLNFKLLRK-LVLGN 961
+FL+ +++ L ++L+ + L + L L +L ++ + + +L ++ +L L+L
Sbjct: 130 -LFLLLMEMVLQLQVLLMEMVLQLQELFL--LLMEMVLQLQVLLMEMVLQLQELFLLLME 186
Query: 962 LWIYHQMLSLLDVDGFTKSNFMLMALLRDIKQDWLPKVTIKLRAWIILIHILQLL 1016
+ + Q+L LL ++ + + + L+ + Q +++ I+L +LQLL
Sbjct: 187 MVLQLQVLFLLLMEMVLQLQVLFLLLMEMVLQ-------LQVLFLILLEMVLQLL 234
>gb|EAL01803.1| hypothetical protein CaO19.4331 [Candida albicans SC5314]
Length = 185
Score = 48.9 bits (115), Expect = 0.001
Identities = 36/144 (25%), Positives = 70/144 (48%), Gaps = 6/144 (4%)
Query: 792 LLSMIFNLERFLCQDMLLFMKTISLTLILHPLLLLIGNIFLIFPLYLILILLSLPVNLQS 851
L +M+F F+ + LFM L ++ ++L FL+ +L+++ LS+ ++L
Sbjct: 24 LFTMVFRFLFFMILFLFLFMILFFLMILFFLMILF----FLMILFFLMVLFLSMVLSLSV 79
Query: 852 LMILLLLLHQLYLLLLLLHLFHLLFPDILPETPLLHLTSKIIFVTIFKLRHIIFLIFFPI 911
+L + + L ++ LF +F +L L + F+ + +IFL F I
Sbjct: 80 FFLLFMFFFLFFFLSMVFFLFLFMF--LLFFMFFLFFMFLLFFMFLLFFMFMIFLFTFMI 137
Query: 912 IIFLTIILLLLCLYIHNLSLNPML 935
+FL+ + LLL +++ L L M+
Sbjct: 138 FLFLSFMFLLLFMFMIFLLLLIMI 161
Score = 47.8 bits (112), Expect = 0.003
Identities = 44/171 (25%), Positives = 80/171 (46%), Gaps = 16/171 (9%)
Query: 711 FLLPFGLMLYIMLFSS*IEFLHH------YFITNHLTSFFIINSLILPFSKFLVVYVMLP 764
FL+ F + I +F + + FL +F+ L F I+ L++ F ++ ++M+
Sbjct: 4 FLMVFKKFITIFIFETIMFFLFTMVFRFLFFMILFLFLFMILFFLMILFFLMILFFLMIL 63
Query: 765 LYILIEPNCNLELESLYS*VTSLVIRVLLSMIFNLERFLCQDMLLFMKTISLTLILHPLL 824
++++ S V SL + LL M F L FL LF+ L +
Sbjct: 64 FFLMV---------LFLSMVLSLSVFFLLFMFFFLFFFLSMVFFLFLFMFLLFFMFFLFF 114
Query: 825 L-LIGNIFLIFPLYLILILLSLPVNLQSLMILLLLLHQLYLLLLLLHLFHL 874
+ L+ +FL+F +++I + + S M LLL + ++LLLL++ L L
Sbjct: 115 MFLLFFMFLLFFMFMIFLFTFMIFLFLSFMFLLLFMFMIFLLLLIMILLLL 165
Score = 44.7 bits (104), Expect = 0.023
Identities = 37/179 (20%), Positives = 83/179 (45%), Gaps = 19/179 (10%)
Query: 748 SLILPFSKFLVVYVMLPLYILIEPNCNLELESLYS*VTSLVIRVLLSMIFNLERFLCQDM 807
+ ++ F KF+ +++ + + ++ + +++ + L MI L M
Sbjct: 3 TFLMVFKKFITIFIFETIMFFL-------FTMVFRFLFFMILFLFLFMILFFLMILFFLM 55
Query: 808 LLFMKTIS--LTLILHPLLLLIGNIFLIFPLYLILILLSLPVNLQSLMILLLLLHQLYLL 865
+LF I L ++ ++L + FL+F + + LS+ + L L + L+ +
Sbjct: 56 ILFFLMILFFLMVLFLSMVLSLSVFFLLFMFFFLFFFLSM------VFFLFLFMFLLFFM 109
Query: 866 LLLLHLFHLLFPDILPETPLLHLTSKIIFVTIFKLRHIIFLIFFPIIIFLTIILLLLCL 924
L +F L F +L ++ L + +IF+ + +FL+ F +IFL +++++L L
Sbjct: 110 FFLFFMFLLFFMFLLFFMFMIFLFTFMIFLFL----SFMFLLLFMFMIFLLLLIMILLL 164
Score = 40.8 bits (94), Expect = 0.34
Identities = 43/172 (25%), Positives = 79/172 (45%), Gaps = 12/172 (6%)
Query: 698 F*MLVELCFISLNFLLPFGLMLYIMLFSS*IEFLHHYFITNHLTSFFIINSLILPFSKFL 757
F M+ F + FL F ++ ++M+ + L I L F+ S++L S F
Sbjct: 25 FTMVFRFLFFMILFLFLFMILFFLMILFF-LMILFFLMILFFLMVLFL--SMVLSLSVFF 81
Query: 758 VVYVMLPLYILIEPNCNLELESLYS*VTSLVIRVLLSMIFNLERFLCQDMLLFMKTISLT 817
++++ L+ + L L + + L L F+ ++F+ T +
Sbjct: 82 LLFMFFFLFFFLSMVFFLFLFMFLLFFMFFLFFMFLLFFMFLLFFM---FMIFLFTFMIF 138
Query: 818 LILHPLLLLIGNIFLIFPLYLILILLSLPVNLQSLMILLLLLHQLYLLLLLL 869
L L + LL+ +F+IF L LI+ILL L SLM+L L ++ L+ + ++
Sbjct: 139 LFLSFMFLLLF-MFMIFLLLLIMILLLL-----SLMVLFLFINYLWFFMFIV 184
Score = 39.7 bits (91), Expect = 0.75
Identities = 42/181 (23%), Positives = 80/181 (43%), Gaps = 13/181 (7%)
Query: 389 IMVLMNIYVVILLHLVPFIE*NL-FMFPYPMVTLC*LILLVMSLSIPSFI*VMFFIHLIL 447
+MV + + + F + F F + M+ L +++ L I F+ ++FF+ +IL
Sbjct: 5 LMVFKKFITIFIFETIMFFLFTMVFRFLFFMILFLFLFMILFFLMILFFLMILFFL-MIL 63
Query: 448 PSISSL*QNVVIH*LVSFIFLLKNVLYRTNNL*R*LVWLNRKMVYTSFIHHANLLILSFL 507
+ L ++V+ V F+ + L+ ++ L + + F+ LL FL
Sbjct: 64 FFLMVLFLSMVLSLSVFFLLFMFFFLFFFLSMVFFLFLFMFLLFFMFFLFFMFLLFFMFL 123
Query: 508 LYLKVLVMLFLFPIVIRIVNMYLTMPYGTLDLVICLIKDFI*CLACILILLLIIIKMLVI 567
L+ ++ LF F I + + M+L L I L I+ILLL+ + +L +
Sbjct: 124 LFFMFMIFLFTFMIFLFLSFMFL-----------LLFMFMIFLLLLIMILLLLSLMVLFL 172
Query: 568 F 568
F
Sbjct: 173 F 173
>gb|AAK77584.1| heme maturase [Tetrahymena thermophila]
gi|15027653|ref|NP_149387.1| heme maturase [Tetrahymena
thermophila]
Length = 518
Score = 48.5 bits (114), Expect = 0.002
Identities = 100/469 (21%), Positives = 202/469 (42%), Gaps = 69/469 (14%)
Query: 528 MYLTMPYGTLDLVICLIKDFI*CLACILILLLIIIKMLV--------IFAILPSTNIYLS 579
MYL + + +I + ++ + + I+ L++IK V I+ L + I L+
Sbjct: 1 MYLDLIFIYTLTLITIFNNYFKKIIFVFIISLLLIKSYVFYYYYFESIYIALLNNLINLN 60
Query: 580 ---ILVPHMLLLNLNFSI*IFGVLFQLLQFMVIDIFS----PF*MTTPDFSGSFFLKPKL 632
IL+ ++N+NF F +LF +L +I+ ++ + + +G F + P L
Sbjct: 61 YNIILIFLFYIINVNFKFLNFFILFFILNCELINNYTNNIESYNLNINLLNGLFLIHPIL 120
Query: 633 KCLHMLRIL-LL*FRIIFTPPLNSLELIMALNSCYLLFMLHMALFTRNLV*KLPNKMVEL 691
R L +L F +++ I+ Y +F L + +NL+ K+ ++ +
Sbjct: 121 IYYFYSRFLNILNFTLLYNN-------ILISKKKYNIFNLININYIKNLI-KVNIHIILV 172
Query: 692 KENINTF*MLVELCFIS-LNFLLPFGLMLYIMLFSS*IEFLHHYFITNHLTSFFIINSLI 750
++ + EL + + N+ L + L+I+L S I +H+ +I L S + SL+
Sbjct: 173 AISLGCWWAFQELNWGTWWNWDLVELINLFILL--SFIFLIHNNYIYIKLNSINQLYSLL 230
Query: 751 LPFSKFLVVYVMLPLYILIE--------PNCNLELESLYS*VTSLVIRVLLSMIFNLERF 802
+ + ++ ++L Y LI+ N N ++ +Y + + I N ++F
Sbjct: 231 IKY----LICILLVRYNLIQSIHNFLSIDNLNQYIQYVYYCLCFFLYFYFFLKIINTKKF 286
Query: 803 LCQDMLLFMKTISLTLILHPLLLLIGNIFLIFPLYLILILLSLPVNLQSLMILLLLLHQL 862
F+K L NI Y+I+I L + +L I +
Sbjct: 287 YKFKYYNFIK-------------LTSNIV----FYIIIINLFYNFKVNTLQIFIYSYINT 329
Query: 863 YLLLLLLHLFHLLFPDILPETPLLHLTSKIIF-----VTIFKLRHIIFLIFFPIIIFLTI 917
++LL+L +F+++ P L++ K+IF V I L+ F F+ +
Sbjct: 330 FILLVLCSIFYIIL--YQPSIIKLYMNFKLIFSWISNVFIISLKTFFFYRSIKKYKFIHL 387
Query: 918 ILLLLCLYIHNLSLNPMLKQVNMSVGCMLCKLNFKLLRKLVLGNLWIYH 966
+++ L+I SLN + ++ + +LNF L++ N++IY+
Sbjct: 388 FIVIFILFI---SLNLLSFNISFLNNHQIQELNF---FDLIIKNMYIYN 430
>gb|AAG13700.1| ABC transporter channel subunit [Malawimonas jakobiformis]
gi|11466650|ref|NP_066333.1| ABC transporter channel
subunit [Malawimonas jakobiformis]
Length = 218
Score = 47.8 bits (112), Expect = 0.003
Identities = 61/220 (27%), Positives = 102/220 (45%), Gaps = 39/220 (17%)
Query: 704 LCFISLNFLLPFGLMLYIMLFSS*IEFLHHYFITNHLTSFFIINSLILP-FSKFLVVYVM 762
L ++ L FL F +L I + ++ +F+ NH FFII+ LI K Y
Sbjct: 21 LSYVILLFLFLFISVLIIGNNVDILNYISIFFLNNH---FFIISILISSNIFKRNGDYKT 77
Query: 763 LPLYILIEPNCNLELESLYS*VTSLVIRVLLSMIFNLERFLCQDMLLFMKTISLTLILHP 822
L LY+ N N++LE + VI+++L +F F I + ++
Sbjct: 78 LRLYL----NSNIKLEHYF------VIKLILYWLF------------FFLPIITIIYINI 115
Query: 823 LLLLIGN---IFLIFPLYLILILLSLPVNLQSLMILLLLLHQLYLLLLLLHLFHLLFPDI 879
+++ I N I LI +YLI L+++ +N S ++ + + +L LLF I
Sbjct: 116 IIMNINNNYDILLIIIIYLIYGLITIFINTVSYLVSFSIKN---------NLISLLFNQI 166
Query: 880 LPETPLLHLTSKIIFVTIFKLRHIIFLIFFPIIIFLTIIL 919
L PL+ L + II ++ + + I+L II+ LTI L
Sbjct: 167 L-NIPLIILCTNIITLSKNEFNYGIYLTGLLIILILTIFL 205
>gb|AAN87160.1| NADH dehydrogenase subunit 4 [Varroa destructor]
Length = 433
Score = 47.0 bits (110), Expect = 0.005
Identities = 95/460 (20%), Positives = 187/460 (40%), Gaps = 78/460 (16%)
Query: 496 IHHANLLILSFLLYLKVLVMLFLFPIVIRIVNMYLTMPYGTLDLVICLIKDFI*CLACIL 555
+++ NL +++ L Y+ + M+ + + I+ M+L+M Y +
Sbjct: 24 MNNINLFMINNLFYIDEMSMMLIILSIWIILLMFLSMKYNKM---------------LFT 68
Query: 556 ILLLIIIKMLVIFAILPSTNIYLSILVPHMLLLNLNFSI*IFGVLFQLLQFMV-IDIFSP 614
+ +++I +++ F+I +N+ L ++ M ++ + F I +G + + + + +++
Sbjct: 69 LFNILLITLILSFSI---SNLILFYIMFEMTIIPMLFFIMFWGYQMERFEASIYLLLYTI 125
Query: 615 F*MTTPDFSGSFFLKPKLKCLHMLRILLL*FRIIFTPPLNSLELIMALNSCYLLFMLHMA 674
F GS L + L+ L + F L + E++ L L F++ M
Sbjct: 126 F--------GSLPLLLMILMLNNHYSLSMFFWTFLNIKLPNFEMMFLL----LAFLIKMP 173
Query: 675 LFTRNLV*KLPNKMVELKENINTF*MLVELCFISLNFLLPFGLMLYIMLFSS*IEFLHHY 734
++ +L LP VE + + LL M+YI F S I +++
Sbjct: 174 IYGFHLW--LPKAHVEAP---------IAGSMLLAGILLNLVDMVYIEFFMS-INWMNIK 221
Query: 735 FITNHLTSFFIINSLILPFSKFLVVYVMLPLYILIEPNCNLELESLYS*VTSLVIRVLLS 794
+ + II ++I+ S + + + + I C++ S+V+ L+S
Sbjct: 222 LLEIFIWPLTIIGAIII--SIMCITQIDIKMLIAYSSICHM----------SIVVGGLMS 269
Query: 795 ---------MIFNLERFLCQDMLLFMKTISLTLILHPLLLLIGNIFLIFPLYLILILLSL 845
++ + LC L FM I LLL+ + IFP+ + L+
Sbjct: 270 GNFWGQSGALLMMISHGLCSSSLFFMANIYYERFFTRNLLLLKGLMNIFPVLGFFMFLNC 329
Query: 846 PVNLQS--LMILL--LLLHQLYLLLLLLHLFHLLFPDILPETPLLHLTS----------K 891
+NL S M LL +LL+ + +L++ L+ L ++L S K
Sbjct: 330 VINLGSPPTMNLLSEILLYGSIMKWNILNMPLLMILSFLSSFYSIYLFSITQHGKFIFHK 389
Query: 892 IIFVTIFKLRHIIFLIFFPIIIFLTIILLLLCLYIHNLSL 931
IF K +II FP+ I+L + L + Y L L
Sbjct: 390 AIFSPSIKEYNIIIFHLFPLFIYLFKMELFMFFYFKKLLL 429
>dbj|BAA00447.1| open reading frame (251 AA) [Mus musculus]
Length = 250
Score = 46.6 bits (109), Expect = 0.006
Identities = 56/241 (23%), Positives = 115/241 (47%), Gaps = 32/241 (13%)
Query: 700 MLVELCFISLNFLLPFGLMLYIMLFSS*IEF-LHHYFITNHLTSFFIINSLILPFSKF-L 757
M E ++S+ L F L L + ++ S I ++H I+ +L+ + L + S L
Sbjct: 1 MFTEHIYLSIYLSLSFSLSLSLSIYLSIICLSIYHLSISIYLSIYLSYVCLSIYLSIICL 60
Query: 758 VVYVMLPLYILIEPNCNLE---LESLYS*V--TSLVIRVLLSMIFNLERFLCQDMLLFMK 812
VY+ L +Y+ I + L L S+Y + S+ + + LS+ ++ + D L M
Sbjct: 61 SVYIYLSIYLSIYLSIYLSYVCLSSIYLSIYHLSIYLSIYLSIYLSIYLSIMSDCLSIM- 119
Query: 813 TISLTLILHPLLLLIGNIFLIFPLYLILILLSLPVNLQSLMILLLLLHQLYLLLLLLHLF 872
++ L++ L ++ +YL +I LS+ +++ + L + L + L + L ++
Sbjct: 120 SVCLSICLS---------YVCLSIYLSIICLSVYLSIYLCIYLSIYLSYVCLSSICLSIY 170
Query: 873 HLLFPDILPETPLLHLTSKIIFVT----IFKLRHIIFLIFFPII---IFLTIILLLLCLY 925
++ L ++ I+++ I L ++ LI+ PII ++L+ I L +C+Y
Sbjct: 171 LSIY--------LSYVCLSSIYLSYAVYIIYLSYVCLLIYLPIIYLSMYLSSISLCVCMY 222
Query: 926 I 926
I
Sbjct: 223 I 223
>ref|YP_026099.1| NADH dehydrogenase subunit 4 [Habronattus oregonensis]
gi|46850140|gb|AAT02494.1| NADH dehydrogenase subunit 4
[Habronattus oregonensis]
Length = 429
Score = 46.2 bits (108), Expect = 0.008
Identities = 49/218 (22%), Positives = 107/218 (48%), Gaps = 31/218 (14%)
Query: 807 MLLFMKT--ISLTLILHPLLLLIGNIFLIFPLYLILILLSLPVNLQSLMILLLLLHQLYL 864
+++F+KT IS+++I+ ++ L+ F +++ +++ P+ S++++LL L + L
Sbjct: 12 IMMFIKTNLISVSIIIMSIMSLMMLTFNSKSIFMNSLMMMDPL---SILMVLLSLISVIL 68
Query: 865 LLLLLHLFHLLFPDILPETPLLHLTSKIIFVTIFKLRHII-FLIFFPIIIFLTIILLLLC 923
++L ++ L+ P I+P +I +T F ++I+ F IFF +++ T++L+ +
Sbjct: 69 IILSSNMLTLITPTIIP--------IMLILITTFSSKNILMFYIFFELVLIPTMLLITM- 119
Query: 924 LYIHNLSLNPMLKQVNMSVGCMLCKLNFKLLRKLVLGNLWIYHQMLSLLDVDGFTKSNFM 983
P Q ++ + + LL ++ N + LL++ FT S+ M
Sbjct: 120 -----NGKQPERLQASIYLIMYTIVASLPLLMMIIFFNPF---NSFPLLNITLFTYSSIM 171
Query: 984 LMALLRDIKQD------WLPKVTIK--LRAWIILIHIL 1013
+ + +K WLPK ++ L +IL +L
Sbjct: 172 FIMMAFLVKMPMFLTHLWLPKAHVEAPLEGSMILAAVL 209
>gb|AAW24404.1| NADH dehydrogenase subunit 1 [Vryburgia amaryllidis]
Length = 303
Score = 45.4 bits (106), Expect = 0.014
Identities = 67/274 (24%), Positives = 130/274 (46%), Gaps = 54/274 (19%)
Query: 702 VELCFISLNFLLPFG-----LMLYIMLFSS*IEFLHHYFITNHLT-SFFIINSLILPFSK 755
++L F +LL F L +Y+ S + ++ I N + F II+ +IL +K
Sbjct: 52 IKLLFKDNIYLLMFNYLNYYLFIYLFFSLSFLLWIFIPMIINLINFKFMIISLMILLVNK 111
Query: 756 FLVVYVMLPLYILIEPNCNLELESLYS*VTSLVIRVLLSMIFNLERFLCQDMLLFMKTIS 815
LV +M+ L S+YS + SL+ ++ ++ F +LFM
Sbjct: 112 ILVFILMI-----------WSLNSIYS-ILSLIRLLIQNLSFE---------ILFM---- 146
Query: 816 LTLILHPLLLLIGNIFLIFPLYLILILLSLPVNLQSLMILLLLLHQLYLLLLLLHLFHLL 875
+++ +++LI N LI ++ +L N+ + + L+ +++L++L+ L +
Sbjct: 147 --MMIFYIVILIKNFNLI-------MIFNLNNNINMIFFMSFLIFYIWMLIILVELNRIP 197
Query: 876 FPDILPETPL---LHLT-SKIIFVTIFKLRH--IIFLIFFPIIIFLTIILLLLCLYIHNL 929
F + E+ L L+L S I F+ +F + + IIF++ +IIF + L+ Y+ N
Sbjct: 198 FDFLESESELISGLNLEFSSINFMILFLIEYLDIIFMMILTLIIFFNLNLM---NYLFNF 254
Query: 930 SLNPMLKQVNMSVGCMLCKLNFKLLRKLVLGNLW 963
L ML V + G + + +++ + L+L LW
Sbjct: 255 ILIIMLIMVLVFRGFL---VRYRIDKLLIL--LW 283
Score = 41.2 bits (95), Expect = 0.26
Identities = 60/254 (23%), Positives = 119/254 (46%), Gaps = 57/254 (22%)
Query: 706 FISLNFLLPFGLMLYIMLFSS*IEFLHHYFITNHLTSFFIINSLILPFSKFLVVYVMLPL 765
FI L F L F L ++I + + I F F II+ +IL +K LV +M+
Sbjct: 73 FIYLFFSLSFLLWIFIPMIINLINF-----------KFMIISLMILLVNKILVFILMI-- 119
Query: 766 YILIEPNCNLELESLYS*VTSLVIRVLLSMIFN-LERFLCQDMLLFMKTISLTLILH--- 821
L S+YS + SL+ ++ ++ F L + +++ +K +L +I +
Sbjct: 120 ---------WSLNSIYS-ILSLIRLLIQNLSFEILFMMMIFYIVILIKNFNLIMIFNLNN 169
Query: 822 PLLLLIGNIFLIFPLYLILILLSLP----------------VNLQ----SLMILLLL--L 859
+ ++ FLIF +++++IL+ L +NL+ + MIL L+ L
Sbjct: 170 NINMIFFMSFLIFYIWMLIILVELNRIPFDFLESESELISGLNLEFSSINFMILFLIEYL 229
Query: 860 HQLYLLLLLLHLF------HLLFPDILPETPLLHLTSK--IIFVTIFKLRHIIFLIFFPI 911
+++++L L +F + LF IL ++ L + ++ I KL +++ FFP+
Sbjct: 230 DIIFMMILTLIIFFNLNLMNYLFNFILIIMLIMVLVFRGFLVRYRIDKLLILLWKFFFPL 289
Query: 912 IIFLTIILLLLCLY 925
++ I ++ L ++
Sbjct: 290 LMNYLIFMMFLKMF 303
>ref|XP_622475.1| PREDICTED: hypothetical protein XP_622475 [Mus musculus]
Length = 195
Score = 45.4 bits (106), Expect = 0.014
Identities = 39/164 (23%), Positives = 81/164 (48%), Gaps = 14/164 (8%)
Query: 793 LSMIFNLERFLCQDMLLFMKTISLTLILHPLLLLIGNIFLIFPLYLILILLSLPVNLQSL 852
+ ++ L + L +LL ++ + L P +LL + L+ L +LLS+ + L
Sbjct: 1 MGLLLMLSQLLLPSVLLTVRILQSLSQLIPPVLLTAGLLLMLSQILPPVLLSMGL----L 56
Query: 853 MILLLLLHQLYLLLLLLHLFHLLFPDILPETPLLHLTSKIIFVTIFKLRHIIFL--IFFP 910
++L LLH L+L +LL + L P +L +L S++I + ++ L I P
Sbjct: 57 LMLSQLLHNLFLTAVLLLMLSQLLPSVLLTVWILQSLSQLIPPVLLTAGLLLMLSKILPP 116
Query: 911 IIIFLTIILLLLCLYIHNL--------SLNPMLKQVNMSVGCML 946
+++ + ++L+L L + ++ SL+ ++ V ++ G +L
Sbjct: 117 VLLSMGLLLMLSQLLLPSVLLTVWILQSLSQLIPPVLLTAGLLL 160
Score = 37.0 bits (84), Expect = 4.9
Identities = 62/196 (31%), Positives = 93/196 (46%), Gaps = 26/196 (13%)
Query: 745 IINSLILPFSKFLVVYVMLPLYILIEPNCNLELESLYS*VTSLVIRVLLSM--------- 795
+++ L+LP S L V ++ L LI P L L ++ ++ VLLSM
Sbjct: 6 MLSQLLLP-SVLLTVRILQSLSQLIPPV--LLTAGLLLMLSQILPPVLLSMGLLLMLSQL 62
Query: 796 IFNLERFLCQDMLLFMK----TISLTL-ILHPLLLLIGNIFLIFPLYLILILLSLPVNLQ 850
+ NL FL +LL + ++ LT+ IL L LI + L L L+L + PV L
Sbjct: 63 LHNL--FLTAVLLLMLSQLLPSVLLTVWILQSLSQLIPPVLLTAGLLLMLSKILPPVLLS 120
Query: 851 SLMILLLLLHQLYLLLLLLHLFHL-----LFPDILPETPLLHLTSKIIFVTIFKLRHIIF 905
M LLL+L QL L +LL ++ L L P +L LL + SKI+ + + ++
Sbjct: 121 --MGLLLMLSQLLLPSVLLTVWILQSLSQLIPPVLLTAGLLLMLSKILPPVLLSMGLLLM 178
Query: 906 LIFFPIIIFLTIILLL 921
L + LT LLL
Sbjct: 179 LSQLIPPVQLTAGLLL 194
>emb|CAA77190.1| NADH dehydrogenase subunit 4 [Trichophyton rubrum]
gi|30179829|sp|Q36834|NU4M_TRIRU NADH-ubiquinone
oxidoreductase chain 4 (NADH dehydrogenase subunit 4)
gi|7432364|pir||T14246 NADH2 dehydrogenase (ubiquinone)
(EC 1.6.5.3) chain 4 - dermatophytic fungus
(Trichophyton rubrum) mitochondrion
Length = 494
Score = 45.1 bits (105), Expect = 0.018
Identities = 55/194 (28%), Positives = 99/194 (50%), Gaps = 23/194 (11%)
Query: 744 FIINSLILPFSKFLVVYVMLPLYILIEPNCNLELESLYS*VTSLVIRVLLSMI-FNLERF 802
++ + L++P L+VY +L +YI + + N + +T+L+I ++LSMI F L F
Sbjct: 9 YLFSLLLIP----LIVYFLL-IYIRLRYSNNSNNQIKTIGLTTLIINLILSMIIFILFDF 63
Query: 803 LCQDMLLFMKTISLTLILHPLLLLIGNIFLIFPLYLILIL-LSLPVNLQSL--------M 853
+ L ++ I + L L I I + F L +I+ +SL N S+ +
Sbjct: 64 SSKQFQLQLEEI-YKISYFDLYLGIDGISIYFLLLTTMIMPISLVANWNSIDSKNVLSFV 122
Query: 854 ILLLLLHQLYL---LLLLLHLFHLLFPDILPETPLLHLTSKIIFVTIFKLRHIIFLIFFP 910
I++LLL L L L+L + LF++ F ILP PL L +F + K+R +L +
Sbjct: 123 IIILLLETLLLAVFLVLDILLFYIFFESILP--PLFLLIG--LFGSSDKVRASFYLFLYT 178
Query: 911 IIIFLTIILLLLCL 924
++ L ++L ++ +
Sbjct: 179 LLGSLFMLLSIITM 192
>sp|P34853|NU4M_APILI NADH-ubiquinone oxidoreductase chain 4 (NADH dehydrogenase subunit 4)
gi|552446|gb|AAB96806.1| NADH dehydrogenase subunit 4
[Apis mellifera ligustica]
gi|5834934|ref|NP_008090.1|ND4_10414 NADH dehydrogenase
subunit 4 [Apis mellifera ligustica]
Length = 447
Score = 44.7 bits (104), Expect = 0.023
Identities = 73/327 (22%), Positives = 153/327 (46%), Gaps = 47/327 (14%)
Query: 743 FFIINSLILPFSKFLVVYVMLPLYILIEPNCNLELESLYS*VTSLVIRVLLSMIFNLERF 802
+ ++ ++L ++Y+ + ++ N NL + +L ++I +LL++ FNL
Sbjct: 2 YLLLLLIMLNMLMMSMIYLFMLFMNKMKNNLNLIIGNL------IIINLLLNL-FNLNWI 54
Query: 803 LCQDMLLFMKTISLTLILHPLLLLIGNIFLIFPLYLILILLSLPVNLQSLMILLLLLHQL 862
D + +S + + L++L IF LI +SL N + + + LLL +
Sbjct: 55 ---DWIYIFCNLSFNMYSYGLIMLTLWIFG-------LIFISLNNNSLNCLFMNLLL-MI 103
Query: 863 YLLLLLLHLFHLLFPDILPETPLLHLTSKIIFVTIFKLRHI-------IFLIFFPIIIFL 915
LLL+ L + LLF + E LL +IF + K + +L+F+ +I L
Sbjct: 104 SLLLVFLSMNLLLFY-LFYEFGLL-----LIFYLVVKWGYSENRWLSGFYLMFYTMIFSL 157
Query: 916 TIILLLLCLYIHNLSLNPML-KQVNMSVGCMLCKLNFKLLRKLVLGNLWIYHQMLSLLDV 974
++ ++ +Y+ + SLN ML + +N+++ ML + L+ LV ++++H L V
Sbjct: 158 PMLYIIYYIYLIDYSLNFMLMEMLNLNLNMML--FIYLLMSFLVKIPIYLFHGWLLKAHV 215
Query: 975 DGFTKSNFMLMALLRDIKQDWLPKVTIKLRAWIILIH------------ILQLLNSQQSD 1022
+ + +L +++ + + ++ I + ILI IL L+ Q D
Sbjct: 216 EAPYYGSMILASIMLKLGGYGMLRLMIIYKNEFILIQKILVMINSFGVLILSLMCLSQFD 275
Query: 1023 L-*LLYLPFIICIYINLMSIMPFFMVN 1048
+ ++ + I+ + + +MS+M F ++
Sbjct: 276 MKSIIAISSIVHMGLMIMSMMTFLKIS 302
Score = 37.4 bits (85), Expect = 3.7
Identities = 75/325 (23%), Positives = 142/325 (43%), Gaps = 41/325 (12%)
Query: 555 LILLLIIIKMLVIFAILPSTNIYLSILVPHMLLLNLNF---SI*IFGVLFQLLQFMVIDI 611
L+LLLI++ ML++ + IYL +L + + NLN ++ I +L L ID
Sbjct: 3 LLLLLIMLNMLMM------SMIYLFMLFMNKMKNNLNLIIGNLIIINLLLNLFNLNWIDW 56
Query: 612 FSPF*MTTPDFSGSFFLKPKLKCLHMLRILLL*FRIIFTPPLNSLELIMALNSCYLLFML 671
F + SF + + L I L F + LN L + + L LL L
Sbjct: 57 IYIF------CNLSFNMYSYGLIMLTLWIFGLIFISLNNNSLNCLFMNLLLMISLLLVFL 110
Query: 672 HMALFTRNLV*KLPNKMVELKENINTF*MLVELCFISLNFLLPFGLMLYIMLFSS*IEFL 731
M L L + ++ F ++V+ + +L F LM Y M+FS + ++
Sbjct: 111 SMNLLLFYLFYEFGLLLI--------FYLVVKWGYSENRWLSGFYLMFYTMIFSLPMLYI 162
Query: 732 HHY-FITNHLTSFFIINSLILPFSKFLVVY------VMLPLYILIEPNCNLELESLYS*V 784
+Y ++ ++ +F ++ L L + L +Y V +P+Y+ +E+ Y
Sbjct: 163 IYYIYLIDYSLNFMLMEMLNLNLNMMLFIYLLMSFLVKIPIYLFHGWLLKAHVEAPY--- 219
Query: 785 TSLVIRVLLSMIFNLERFLCQDMLLFMKTISLTLILHPLLLLIGNIFLIFPLYLILILLS 844
+L S++ L + ML M IL +L++ N F + L L L+ LS
Sbjct: 220 --YGSMILASIMLKLGGY---GMLRLMIIYKNEFILIQKILVMINSFGV--LILSLMCLS 272
Query: 845 LPVNLQSLMILLLLLHQLYLLLLLL 869
+++S++ + ++H +++ ++
Sbjct: 273 -QFDMKSIIAISSIVHMGLMIMSMM 296
Database: nr
Posted date: Jul 5, 2005 12:34 AM
Number of letters in database: 863,360,394
Number of sequences in database: 2,540,612
Lambda K H
0.356 0.163 0.531
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 1,945,224,440
Number of Sequences: 2540612
Number of extensions: 72342325
Number of successful extensions: 496815
Number of sequences better than 10.0: 253
Number of HSP's better than 10.0 without gapping: 8
Number of HSP's successfully gapped in prelim test: 255
Number of HSP's that attempted gapping in prelim test: 494339
Number of HSP's gapped (non-prelim): 1469
length of query: 1439
length of database: 863,360,394
effective HSP length: 141
effective length of query: 1298
effective length of database: 505,134,102
effective search space: 655664064396
effective search space used: 655664064396
T: 11
A: 40
X1: 14 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 37 (21.6 bits)
S2: 82 (36.2 bits)
Medicago: description of AC140721.3


