

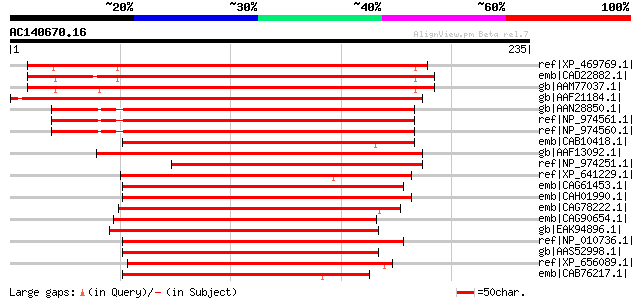
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC140670.16 + phase: 0
(235 letters)
Database: nr
2,540,612 sequences; 863,360,394 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
ref|XP_469769.1| putative transcriptional adaptor [Oryza sativa ... 262 5e-69
emb|CAD22882.1| transcriptional adaptor [Zea mays] 257 2e-67
gb|AAM77037.1| histone acetyltransferase complex component [Zea ... 256 3e-67
gb|AAF21184.1| unknown protein [Arabidopsis thaliana] gi|1453284... 236 5e-61
gb|AAN28850.1| At4g16420/dl4235c [Arabidopsis thaliana] gi|15215... 235 9e-61
ref|NP_974561.1| transcriptional adaptor (ADA2b) [Arabidopsis th... 235 9e-61
ref|NP_974560.1| transcriptional adaptor (ADA2b) [Arabidopsis th... 235 9e-61
emb|CAB10418.1| transcriptional adaptor like protein [Arabidopsi... 217 2e-55
gb|AAF13092.1| unknown protein [Arabidopsis thaliana] 216 5e-55
ref|NP_974251.1| transcriptional adaptor (ADA2a) [Arabidopsis th... 175 8e-43
ref|XP_641229.1| SWIRM domain containing protein [Dictyostelium ... 169 4e-41
emb|CAG61453.1| unnamed protein product [Candida glabrata CBS138... 139 8e-32
emb|CAH01990.1| unnamed protein product [Kluyveromyces lactis NR... 137 3e-31
emb|CAG78222.1| unnamed protein product [Yarrowia lipolytica CLI... 134 2e-30
emb|CAG90654.1| unnamed protein product [Debaryomyces hansenii C... 133 5e-30
gb|EAK94896.1| hypothetical protein CaO19.9867 [Candida albicans... 131 1e-29
ref|NP_010736.1| Transcription coactivator, component of the ADA... 131 2e-29
gb|AAS52998.1| AER318Cp [Ashbya gossypii ATCC 10895] gi|45190920... 131 2e-29
ref|XP_656089.1| transcriptional adaptor ADA2, putative [Entamoe... 130 3e-29
emb|CAB76217.1| SPCC24B10.08c [Schizosaccharomyces pombe] gi|190... 130 4e-29
>ref|XP_469769.1| putative transcriptional adaptor [Oryza sativa (japonica
cultivar-group)] gi|40538991|gb|AAR87248.1| putative
transcriptional adaptor [Oryza sativa (japonica
cultivar-group)]
Length = 570
Score = 262 bits (670), Expect = 5e-69
Identities = 122/186 (65%), Positives = 153/186 (81%), Gaps = 5/186 (2%)
Query: 9 SRSSPIAADD---NRSKRKKVALNADNLETSSAAGMGITTDG-KVSLYHCNYCNKDISGK 64
SR P + DD +RSKR++VA + D ++ SAA G G K +LYHCNYCNKDISGK
Sbjct: 4 SRGVPNSGDDETNHRSKRRRVASSGDAPDSLSAACGGAGEGGGKKALYHCNYCNKDISGK 63
Query: 65 IRIKCAVCQDFDLCIECFSVGVELTPHKSNHPYRVMDNLSFPLICPDWSAEEEMLLLEAL 124
IRIKC+ C DFDLC+ECFSVG E+TPH+SNHPYRVMDNLSFPLICPDW+A+EE+LLLE +
Sbjct: 64 IRIKCSKCPDFDLCVECFSVGAEVTPHRSNHPYRVMDNLSFPLICPDWNADEEILLLEGI 123
Query: 125 DMYGFGNWNDVADNIGTKSKSQCIDHYNTVYVNSPCFPVPDLSHFSGKNKEELLAMAK-G 183
+MYG GNW +VA+++GTK+K+QCIDHY T Y+NSPC+P+PD+SH +GKN++ELLAMAK
Sbjct: 124 EMYGLGNWAEVAEHVGTKTKAQCIDHYTTAYMNSPCYPLPDMSHVNGKNRKELLAMAKVQ 183
Query: 184 NQVKKG 189
+ KKG
Sbjct: 184 GESKKG 189
>emb|CAD22882.1| transcriptional adaptor [Zea mays]
Length = 565
Score = 257 bits (657), Expect = 2e-67
Identities = 122/190 (64%), Positives = 153/190 (80%), Gaps = 7/190 (3%)
Query: 9 SRSSPIAADDN---RSKRKKVALNADNLETSSAAGMGITTDG--KVSLYHCNYCNKDISG 63
SR + DD+ RSKR++VA D ++ SA G+G +G K +LYHCNYCNKDISG
Sbjct: 4 SRGVQNSGDDDTVHRSKRRRVASGGDATDSVSA-GIGGAGEGGGKKALYHCNYCNKDISG 62
Query: 64 KIRIKCAVCQDFDLCIECFSVGVELTPHKSNHPYRVMDNLSFPLICPDWSAEEEMLLLEA 123
KIRIKC+ C DFDLC+ECFSVG E+TPH+SNHPY+VMDNLSFPLICPDW+A+EE+LLLE
Sbjct: 63 KIRIKCSKCPDFDLCVECFSVGAEVTPHRSNHPYKVMDNLSFPLICPDWNADEEILLLEG 122
Query: 124 LDMYGFGNWNDVADNIGTKSKSQCIDHYNTVYVNSPCFPVPDLSHFSGKNKEELLAMAK- 182
++MYG GNW +VA+++GTKSK QCIDHY T Y+NSPC+P+PD+SH +GKN++ELLAMAK
Sbjct: 123 IEMYGLGNWLEVAEHVGTKSKLQCIDHYTTAYMNSPCYPLPDMSHVNGKNRKELLAMAKV 182
Query: 183 GNQVKKGLLL 192
+ KKG L
Sbjct: 183 QGESKKGTSL 192
>gb|AAM77037.1| histone acetyltransferase complex component [Zea mays]
Length = 565
Score = 256 bits (655), Expect = 3e-67
Identities = 120/189 (63%), Positives = 152/189 (79%), Gaps = 5/189 (2%)
Query: 9 SRSSPIAADDN---RSKRKKVALNADNLETSSAA-GMGITTDGKVSLYHCNYCNKDISGK 64
SR + DD+ RSKR++V+ D ++ SA+ G GK +LYHCNYCNKDISGK
Sbjct: 4 SRGVLSSGDDDTGHRSKRRRVSSGGDATDSISASIGGAGEGGGKKALYHCNYCNKDISGK 63
Query: 65 IRIKCAVCQDFDLCIECFSVGVELTPHKSNHPYRVMDNLSFPLICPDWSAEEEMLLLEAL 124
IRIKC+ C DFDLC+ECFSVG E+TPH+SNHPY+VMDNLSFPLICPDW+A+EE+LLLE +
Sbjct: 64 IRIKCSKCPDFDLCVECFSVGAEVTPHRSNHPYKVMDNLSFPLICPDWNADEEILLLEGI 123
Query: 125 DMYGFGNWNDVADNIGTKSKSQCIDHYNTVYVNSPCFPVPDLSHFSGKNKEELLAMAK-G 183
+MYG GNW +VA+++GTKSK QCIDHY + Y+NSPC+P+PD+SH +GKN++ELLAMAK
Sbjct: 124 EMYGLGNWLEVAEHVGTKSKLQCIDHYTSAYMNSPCYPLPDMSHVNGKNRKELLAMAKVQ 183
Query: 184 NQVKKGLLL 192
+ KKG LL
Sbjct: 184 GESKKGTLL 192
>gb|AAF21184.1| unknown protein [Arabidopsis thaliana] gi|14532842|gb|AAK64103.1|
unknown protein [Arabidopsis thaliana]
gi|13430668|gb|AAK25956.1| unknown protein [Arabidopsis
thaliana] gi|13591698|gb|AAK31319.1| transcriptional
adaptor ADA2a [Arabidopsis thaliana]
gi|18398044|ref|NP_566317.1| transcriptional adaptor
(ADA2a) [Arabidopsis thaliana]
Length = 548
Score = 236 bits (601), Expect = 5e-61
Identities = 107/187 (57%), Positives = 143/187 (76%), Gaps = 1/187 (0%)
Query: 1 MGRCCRAASRSSPIAADDNRSKRKKVALNADNLETSSAAGMGITTDGKVSLYHCNYCNKD 60
MGR + ASR + + +SKRKK++L +N S + G+ + K LY CNYC+KD
Sbjct: 1 MGRS-KLASRPAEEDLNPGKSKRKKISLGPENAAASISTGIEAGNERKPGLYCCNYCDKD 59
Query: 61 ISGKIRIKCAVCQDFDLCIECFSVGVELTPHKSNHPYRVMDNLSFPLICPDWSAEEEMLL 120
+SG +R KCAVC DFDLC+ECFSVGVEL HK++HPYRVMDNLSF L+ DW+A+EE+LL
Sbjct: 60 LSGLVRFKCAVCMDFDLCVECFSVGVELNRHKNSHPYRVMDNLSFSLVTSDWNADEEILL 119
Query: 121 LEALDMYGFGNWNDVADNIGTKSKSQCIDHYNTVYVNSPCFPVPDLSHFSGKNKEELLAM 180
LEA+ YGFGNW +VAD++G+K+ ++CI H+N+ Y+ SPCFP+PDLSH GK+K+ELLAM
Sbjct: 120 LEAIATYGFGNWKEVADHVGSKTTTECIKHFNSAYMQSPCFPLPDLSHTIGKSKDELLAM 179
Query: 181 AKGNQVK 187
+K + VK
Sbjct: 180 SKDSAVK 186
>gb|AAN28850.1| At4g16420/dl4235c [Arabidopsis thaliana] gi|15215640|gb|AAK91365.1|
AT4g16420/dl4235c [Arabidopsis thaliana]
gi|13591700|gb|AAK31320.1| transcriptional adaptor ADA2b
[Arabidopsis thaliana] gi|18414653|ref|NP_567495.1|
transcriptional adaptor (ADA2b) [Arabidopsis thaliana]
Length = 487
Score = 235 bits (599), Expect = 9e-61
Identities = 105/164 (64%), Positives = 134/164 (81%), Gaps = 4/164 (2%)
Query: 20 RSKRKKVALNADNLETSSAAGMGITTDGKVSLYHCNYCNKDISGKIRIKCAVCQDFDLCI 79
R+++KK A N +N E++S G GK Y+C+YC KDI+GKIRIKCAVC DFDLCI
Sbjct: 17 RTRKKKNAANVENFESTSLVP-GAEGGGK---YNCDYCQKDITGKIRIKCAVCPDFDLCI 72
Query: 80 ECFSVGVELTPHKSNHPYRVMDNLSFPLICPDWSAEEEMLLLEALDMYGFGNWNDVADNI 139
EC SVG E+TPHK +HPYRVM NL+FPLICPDWSA++EMLLLE L++YG GNW +VA+++
Sbjct: 73 ECMSVGAEITPHKCDHPYRVMGNLTFPLICPDWSADDEMLLLEGLEIYGLGNWAEVAEHV 132
Query: 140 GTKSKSQCIDHYNTVYVNSPCFPVPDLSHFSGKNKEELLAMAKG 183
GTKSK QC++HY +Y+NSP FP+PD+SH +GKN++EL AMAKG
Sbjct: 133 GTKSKEQCLEHYRNIYLNSPFFPLPDMSHVAGKNRKELQAMAKG 176
>ref|NP_974561.1| transcriptional adaptor (ADA2b) [Arabidopsis thaliana]
Length = 486
Score = 235 bits (599), Expect = 9e-61
Identities = 105/164 (64%), Positives = 134/164 (81%), Gaps = 4/164 (2%)
Query: 20 RSKRKKVALNADNLETSSAAGMGITTDGKVSLYHCNYCNKDISGKIRIKCAVCQDFDLCI 79
R+++KK A N +N E++S G GK Y+C+YC KDI+GKIRIKCAVC DFDLCI
Sbjct: 17 RTRKKKNAANVENFESTSLVP-GAEGGGK---YNCDYCQKDITGKIRIKCAVCPDFDLCI 72
Query: 80 ECFSVGVELTPHKSNHPYRVMDNLSFPLICPDWSAEEEMLLLEALDMYGFGNWNDVADNI 139
EC SVG E+TPHK +HPYRVM NL+FPLICPDWSA++EMLLLE L++YG GNW +VA+++
Sbjct: 73 ECMSVGAEITPHKCDHPYRVMGNLTFPLICPDWSADDEMLLLEGLEIYGLGNWAEVAEHV 132
Query: 140 GTKSKSQCIDHYNTVYVNSPCFPVPDLSHFSGKNKEELLAMAKG 183
GTKSK QC++HY +Y+NSP FP+PD+SH +GKN++EL AMAKG
Sbjct: 133 GTKSKEQCLEHYRNIYLNSPFFPLPDMSHVAGKNRKELQAMAKG 176
>ref|NP_974560.1| transcriptional adaptor (ADA2b) [Arabidopsis thaliana]
Length = 483
Score = 235 bits (599), Expect = 9e-61
Identities = 105/164 (64%), Positives = 134/164 (81%), Gaps = 4/164 (2%)
Query: 20 RSKRKKVALNADNLETSSAAGMGITTDGKVSLYHCNYCNKDISGKIRIKCAVCQDFDLCI 79
R+++KK A N +N E++S G GK Y+C+YC KDI+GKIRIKCAVC DFDLCI
Sbjct: 17 RTRKKKNAANVENFESTSLVP-GAEGGGK---YNCDYCQKDITGKIRIKCAVCPDFDLCI 72
Query: 80 ECFSVGVELTPHKSNHPYRVMDNLSFPLICPDWSAEEEMLLLEALDMYGFGNWNDVADNI 139
EC SVG E+TPHK +HPYRVM NL+FPLICPDWSA++EMLLLE L++YG GNW +VA+++
Sbjct: 73 ECMSVGAEITPHKCDHPYRVMGNLTFPLICPDWSADDEMLLLEGLEIYGLGNWAEVAEHV 132
Query: 140 GTKSKSQCIDHYNTVYVNSPCFPVPDLSHFSGKNKEELLAMAKG 183
GTKSK QC++HY +Y+NSP FP+PD+SH +GKN++EL AMAKG
Sbjct: 133 GTKSKEQCLEHYRNIYLNSPFFPLPDMSHVAGKNRKELQAMAKG 176
>emb|CAB10418.1| transcriptional adaptor like protein [Arabidopsis thaliana]
gi|7268392|emb|CAB78684.1| transcriptional adaptor like
protein [Arabidopsis thaliana] gi|7485207|pir||H71430
hypothetical protein - Arabidopsis thaliana
Length = 480
Score = 217 bits (553), Expect = 2e-55
Identities = 94/139 (67%), Positives = 117/139 (83%), Gaps = 7/139 (5%)
Query: 52 YHCNYCNKDISGKIRIKCAVCQDFDLCIECFSVGVELTPHKSNHPYRVMDNLSFPLICPD 111
Y+C+YC KDI+GKIRIKCAVC DFDLCIEC SVG E+TPHK +HPYRVM NL+FPLICPD
Sbjct: 31 YNCDYCQKDITGKIRIKCAVCPDFDLCIECMSVGAEITPHKCDHPYRVMGNLTFPLICPD 90
Query: 112 WSAEEEMLLLEALDMYGFGNWNDVADNIGTKSKSQCIDHYNTVYVNSPCFPVP------- 164
WSA++EMLLLE L++YG GNW +VA+++GTKSK QC++HY +Y+NSP FP+P
Sbjct: 91 WSADDEMLLLEGLEIYGLGNWAEVAEHVGTKSKEQCLEHYRNIYLNSPFFPLPEKRLFSQ 150
Query: 165 DLSHFSGKNKEELLAMAKG 183
D+SH +GKN++EL AMAKG
Sbjct: 151 DMSHVAGKNRKELQAMAKG 169
>gb|AAF13092.1| unknown protein [Arabidopsis thaliana]
Length = 527
Score = 216 bits (549), Expect = 5e-55
Identities = 93/148 (62%), Positives = 121/148 (80%)
Query: 40 GMGITTDGKVSLYHCNYCNKDISGKIRIKCAVCQDFDLCIECFSVGVELTPHKSNHPYRV 99
G+ + K LY CNYC+KD+SG +R KCAVC DFDLC+ECFSVGVEL HK++HPYRV
Sbjct: 18 GIEAGNERKPGLYCCNYCDKDLSGLVRFKCAVCMDFDLCVECFSVGVELNRHKNSHPYRV 77
Query: 100 MDNLSFPLICPDWSAEEEMLLLEALDMYGFGNWNDVADNIGTKSKSQCIDHYNTVYVNSP 159
MDNLSF L+ DW+A+EE+LLLEA+ YGFGNW +VAD++G+K+ ++CI H+N+ Y+ SP
Sbjct: 78 MDNLSFSLVTSDWNADEEILLLEAIATYGFGNWKEVADHVGSKTTTECIKHFNSAYMQSP 137
Query: 160 CFPVPDLSHFSGKNKEELLAMAKGNQVK 187
CFP+PDLSH GK+K+ELLAM+K + VK
Sbjct: 138 CFPLPDLSHTIGKSKDELLAMSKDSAVK 165
>ref|NP_974251.1| transcriptional adaptor (ADA2a) [Arabidopsis thaliana]
Length = 477
Score = 175 bits (444), Expect = 8e-43
Identities = 75/114 (65%), Positives = 98/114 (85%)
Query: 74 DFDLCIECFSVGVELTPHKSNHPYRVMDNLSFPLICPDWSAEEEMLLLEALDMYGFGNWN 133
DFDLC+ECFSVGVEL HK++HPYRVMDNLSF L+ DW+A+EE+LLLEA+ YGFGNW
Sbjct: 2 DFDLCVECFSVGVELNRHKNSHPYRVMDNLSFSLVTSDWNADEEILLLEAIATYGFGNWK 61
Query: 134 DVADNIGTKSKSQCIDHYNTVYVNSPCFPVPDLSHFSGKNKEELLAMAKGNQVK 187
+VAD++G+K+ ++CI H+N+ Y+ SPCFP+PDLSH GK+K+ELLAM+K + VK
Sbjct: 62 EVADHVGSKTTTECIKHFNSAYMQSPCFPLPDLSHTIGKSKDELLAMSKDSAVK 115
>ref|XP_641229.1| SWIRM domain containing protein [Dictyostelium discoideum]
gi|60469272|gb|EAL67266.1| myb domain-containing protein
[Dictyostelium discoideum]
Length = 914
Score = 169 bits (429), Expect = 4e-41
Identities = 74/134 (55%), Positives = 98/134 (72%), Gaps = 2/134 (1%)
Query: 51 LYHCNYCNKDISGKIRIKCAVCQDFDLCIECFSVGVELTPHKSNHPYRVMDNLSFPLICP 110
LYHC+YC KDISG +RI+C+VC DFDLC+ECFSVGVE+TPH++ H Y V+DN+ FP+
Sbjct: 383 LYHCDYCQKDISGVVRIRCSVCTDFDLCLECFSVGVEITPHRNFHDYHVVDNMHFPMFTD 442
Query: 111 DWSAEEEMLLLEALDMYGFGNWNDVADNIGTKSKS--QCIDHYNTVYVNSPCFPVPDLSH 168
DW A+EE+LLLEA+++YG GNWN+V++N+G SKS +C HY Y+NS P+PD S
Sbjct: 443 DWGADEELLLLEAIELYGLGNWNEVSENVGAHSKSPLECKAHYFAHYLNSSTSPLPDTSK 502
Query: 169 FSGKNKEELLAMAK 182
N+ AK
Sbjct: 503 VLTTNENVHFKRAK 516
>emb|CAG61453.1| unnamed protein product [Candida glabrata CBS138]
gi|50292119|ref|XP_448492.1| unnamed protein product
[Candida glabrata]
Length = 445
Score = 139 bits (349), Expect = 8e-32
Identities = 53/127 (41%), Positives = 87/127 (67%)
Query: 52 YHCNYCNKDISGKIRIKCAVCQDFDLCIECFSVGVELTPHKSNHPYRVMDNLSFPLICPD 111
+HC+ C+ D + ++RI CAVC ++DLC+ CFS G+ H+ H YR+++ S+P++CPD
Sbjct: 5 FHCDVCSSDCTHRVRISCAVCPEYDLCVPCFSQGLYNGNHRPFHDYRIIETNSYPILCPD 64
Query: 112 WSAEEEMLLLEALDMYGFGNWNDVADNIGTKSKSQCIDHYNTVYVNSPCFPVPDLSHFSG 171
W A+EE+ L++ G GNW+D+A++IG++ K + DHY Y+NSP +P+PD++
Sbjct: 65 WGADEELALIKGAQSLGLGNWHDIAEHIGSREKDEVRDHYIEYYINSPFYPIPDITKDIQ 124
Query: 172 KNKEELL 178
+EE L
Sbjct: 125 VPQEEFL 131
>emb|CAH01990.1| unnamed protein product [Kluyveromyces lactis NRRL Y-1140]
gi|50303311|ref|XP_451597.1| unnamed protein product
[Kluyveromyces lactis]
Length = 435
Score = 137 bits (344), Expect = 3e-31
Identities = 52/131 (39%), Positives = 88/131 (66%)
Query: 52 YHCNYCNKDISGKIRIKCAVCQDFDLCIECFSVGVELTPHKSNHPYRVMDNLSFPLICPD 111
+HC+ C+ D + ++RI CAVC ++DLC+ CF+ G+ H+ +H Y+V++ S+P++C D
Sbjct: 5 FHCDICSADCTNRVRITCAVCPEYDLCVPCFAKGLYNGKHRPSHDYKVIETNSYPILCED 64
Query: 112 WSAEEEMLLLEALDMYGFGNWNDVADNIGTKSKSQCIDHYNTVYVNSPCFPVPDLSHFSG 171
W A+EE+LL++ G GNW D+AD+IG++ K + DHY Y+NS +P+PD++
Sbjct: 65 WGADEELLLIKGAQTLGLGNWQDIADHIGSRDKEEVYDHYLKFYLNSEHYPIPDITKDIN 124
Query: 172 KNKEELLAMAK 182
+ +E+ L K
Sbjct: 125 EPQEQFLEKRK 135
>emb|CAG78222.1| unnamed protein product [Yarrowia lipolytica CLIB99]
gi|50556010|ref|XP_505413.1| hypothetical protein
[Yarrowia lipolytica]
Length = 463
Score = 134 bits (337), Expect = 2e-30
Identities = 56/136 (41%), Positives = 87/136 (63%), Gaps = 8/136 (5%)
Query: 50 SLYHCNYCNKDISGKIRIKCAVCQDFDLCIECFSVGVELTPHKSNHPYRVMDNLSFPLIC 109
S +HC+ C+ D + +RI+CA CQD+DLC++CFS G HK H Y +++ ++P+
Sbjct: 5 SRFHCDVCSCDCTNLVRIRCAECQDYDLCVQCFSQGASSGVHKPYHNYMIIEQHAYPIFT 64
Query: 110 PDWSAEEEMLLLEALDMYGFGNWNDVADNIGTKSKSQCIDHYNTVYVNSPCFPVPDL--- 166
DW A+EE+LL+E +M G GNW D+AD+IG +SK + +HY VY++SP +P+P +
Sbjct: 65 EDWGADEELLLIEGAEMQGLGNWQDIADHIGGRSKEEVGEHYKEVYLDSPDYPLPPMKRK 124
Query: 167 -----SHFSGKNKEEL 177
+ F+ KE L
Sbjct: 125 FEITAAEFAANKKERL 140
>emb|CAG90654.1| unnamed protein product [Debaryomyces hansenii CBS767]
gi|50427117|ref|XP_462166.1| unnamed protein product
[Debaryomyces hansenii]
Length = 447
Score = 133 bits (334), Expect = 5e-30
Identities = 51/119 (42%), Positives = 83/119 (68%)
Query: 48 KVSLYHCNYCNKDISGKIRIKCAVCQDFDLCIECFSVGVELTPHKSNHPYRVMDNLSFPL 107
K LYHC+ C+ D + +IRI+CA+C D+DLC+ CF+ G HK H Y+V++ ++P+
Sbjct: 4 KTRLYHCDVCSSDCTNRIRIQCAICADYDLCVPCFASGSATGDHKPWHDYQVIEQNTYPI 63
Query: 108 ICPDWSAEEEMLLLEALDMYGFGNWNDVADNIGTKSKSQCIDHYNTVYVNSPCFPVPDL 166
DW A+EE+LL++ + +G GNW D+AD+IG +SK + +HY +Y+NS +P+P++
Sbjct: 64 FDKDWGADEELLLVQGCETFGLGNWQDIADHIGNRSKDEVKNHYFDIYLNSKEYPLPEM 122
>gb|EAK94896.1| hypothetical protein CaO19.9867 [Candida albicans SC5314]
gi|46435456|gb|EAK94837.1| hypothetical protein
CaO19.2331 [Candida albicans SC5314]
Length = 445
Score = 131 bits (330), Expect = 1e-29
Identities = 48/122 (39%), Positives = 84/122 (68%)
Query: 46 DGKVSLYHCNYCNKDISGKIRIKCAVCQDFDLCIECFSVGVELTPHKSNHPYRVMDNLSF 105
D + L+HC+ C+ D + +IRI+CA+C D+DLC+ CF+ G+ HK H Y++++ ++
Sbjct: 2 DSRTKLFHCDVCSSDCTNRIRIQCAICTDYDLCVPCFAAGLTTGDHKPWHDYQIIEQNTY 61
Query: 106 PLICPDWSAEEEMLLLEALDMYGFGNWNDVADNIGTKSKSQCIDHYNTVYVNSPCFPVPD 165
P+ DW A+EE+LL++ + G GNW D+AD+IG +SK + +HY +Y+ S +P+P+
Sbjct: 62 PIFDRDWGADEELLLIQGCETSGLGNWADIADHIGNRSKEEVAEHYFKIYLESKDYPLPE 121
Query: 166 LS 167
++
Sbjct: 122 MN 123
>ref|NP_010736.1| Transcription coactivator, component of the ADA and SAGA
transcriptional adaptor/HAT (histone acetyltransferase)
complexes; Ada2p [Saccharomyces cerevisiae]
gi|399006|sp|Q02336|ADA2_YEAST Transcriptional adapter 2
gi|927705|gb|AAB64871.1| Ada2p: probable transcriptional
adaptor; YDR448W; CAI: 0.12 [Saccharomyces cerevisiae]
gi|170991|gb|AAA34393.1| ADA2
Length = 434
Score = 131 bits (329), Expect = 2e-29
Identities = 48/127 (37%), Positives = 83/127 (64%)
Query: 52 YHCNYCNKDISGKIRIKCAVCQDFDLCIECFSVGVELTPHKSNHPYRVMDNLSFPLICPD 111
+HC+ C+ D + ++R+ CA+C ++DLC+ CFS G H+ H YR+++ S+P++CPD
Sbjct: 5 FHCDVCSADCTNRVRVSCAICPEYDLCVPCFSQGSYTGKHRPYHDYRIIETNSYPILCPD 64
Query: 112 WSAEEEMLLLEALDMYGFGNWNDVADNIGTKSKSQCIDHYNTVYVNSPCFPVPDLSHFSG 171
W A+EE+ L++ G GNW D+AD+IG++ K + +HY Y+ S +P+PD++
Sbjct: 65 WGADEELQLIKGAQTLGLGNWQDIADHIGSRGKEEVKEHYLKYYLESKYYPIPDITQNIH 124
Query: 172 KNKEELL 178
++E L
Sbjct: 125 VPQDEFL 131
>gb|AAS52998.1| AER318Cp [Ashbya gossypii ATCC 10895] gi|45190920|ref|NP_985174.1|
AER318Cp [Eremothecium gossypii]
Length = 435
Score = 131 bits (329), Expect = 2e-29
Identities = 48/116 (41%), Positives = 78/116 (66%)
Query: 52 YHCNYCNKDISGKIRIKCAVCQDFDLCIECFSVGVELTPHKSNHPYRVMDNLSFPLICPD 111
+HC+ C+ D + ++RI CAVC ++DLC+ CFS G+ H+ H YR+++ S+P++C D
Sbjct: 5 FHCDICSSDCTNRVRISCAVCPEYDLCVPCFSQGLYNGNHRPYHDYRIIETNSYPILCDD 64
Query: 112 WSAEEEMLLLEALDMYGFGNWNDVADNIGTKSKSQCIDHYNTVYVNSPCFPVPDLS 167
W A+EE L++ G GNW D+ADNIG++ K + +HY Y+ S +P+PD++
Sbjct: 65 WGADEEQALIKGAQTLGLGNWQDIADNIGSREKEEVYEHYVKYYLKSDFYPIPDIT 120
>ref|XP_656089.1| transcriptional adaptor ADA2, putative [Entamoeba histolytica
HM-1:IMSS] gi|67470426|ref|XP_651177.1| transcriptional
adaptor ADA2, putative [Entamoeba histolytica HM-1:IMSS]
gi|56473270|gb|EAL50705.1| transcriptional adaptor ADA2,
putative [Entamoeba histolytica HM-1:IMSS]
gi|56467881|gb|EAL45791.1| transcriptional adaptor ADA2,
putative [Entamoeba histolytica HM-1:IMSS]
Length = 293
Score = 130 bits (327), Expect = 3e-29
Identities = 59/121 (48%), Positives = 79/121 (64%), Gaps = 1/121 (0%)
Query: 54 CNYCNKDISGKIRIKCAVCQDFDLCIECFSVGVELTPHKSNHPYRVMDNLSFPLICPDWS 113
CN CNK I+ RI C C +FDLC+ECFS G E+ HK+NH YRV+ +L FPL+ DW
Sbjct: 12 CNSCNKVITTMTRITCVECDNFDLCLECFSQGKEIGKHKNNHSYRVIPSLHFPLLSSDWG 71
Query: 114 AEEEMLLLEALDMYGFGNWNDVADNIGTKSKSQCIDHYNTVYVNSPCFPVPDLSH-FSGK 172
A+EE++LLEA++ G NW +V + + TK+ +C HY Y+NS P+PDL F K
Sbjct: 72 ADEELMLLEAIEQKGLDNWPEVENFVKTKTAKECRSHYYDYYLNSKTHPLPDLEESFLKK 131
Query: 173 N 173
N
Sbjct: 132 N 132
>emb|CAB76217.1| SPCC24B10.08c [Schizosaccharomyces pombe]
gi|19075511|ref|NP_588011.1| hypothetical protein
SPCC24B10.08c [Schizosaccharomyces pombe 972h-]
gi|11288471|pir||T50415 ada2-like protein [imported] -
fission yeast (Schizosaccharomyces pombe)
Length = 437
Score = 130 bits (326), Expect = 4e-29
Identities = 52/113 (46%), Positives = 76/113 (67%), Gaps = 1/113 (0%)
Query: 52 YHCNYCNKDISGKIRIKCAVCQDFDLCIECFSVGVELTPHKSNHPYRVMDNLSFPLICPD 111
YHCN C +DI+ I I+C C DFDLCI CF+ G L H +HPYR+++ S+P+ +
Sbjct: 6 YHCNVCAQDITRSIHIRCVECVDFDLCIPCFTSGASLGTHHPSHPYRIIETNSYPIFDEN 65
Query: 112 WSAEEEMLLLEALDMYGFGNWNDVADNIG-TKSKSQCIDHYNTVYVNSPCFPV 163
W A+EE+LL++A + G GNW D+AD +G ++K +C DHY Y+ S C+P+
Sbjct: 66 WGADEELLLIDACETLGLGNWADIADYVGNARTKEECRDHYLKTYIESDCYPL 118
Database: nr
Posted date: Jul 5, 2005 12:34 AM
Number of letters in database: 863,360,394
Number of sequences in database: 2,540,612
Lambda K H
0.321 0.135 0.425
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 406,265,825
Number of Sequences: 2540612
Number of extensions: 16634831
Number of successful extensions: 41087
Number of sequences better than 10.0: 600
Number of HSP's better than 10.0 without gapping: 338
Number of HSP's successfully gapped in prelim test: 262
Number of HSP's that attempted gapping in prelim test: 40429
Number of HSP's gapped (non-prelim): 734
length of query: 235
length of database: 863,360,394
effective HSP length: 124
effective length of query: 111
effective length of database: 548,324,506
effective search space: 60864020166
effective search space used: 60864020166
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.9 bits)
S2: 73 (32.7 bits)
Medicago: description of AC140670.16


