

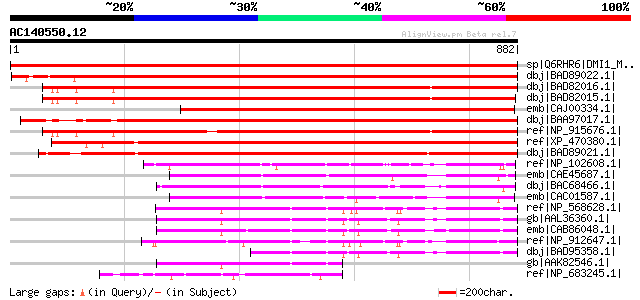
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC140550.12 + phase: 0
(882 letters)
Database: nr
2,540,612 sequences; 863,360,394 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
sp|Q6RHR6|DMI1_MEDTR Putative ion channel DMI-1 (Does not make i... 1736 0.0
dbj|BAD89022.1| POLLUX [Lotus corniculatus var. japonicus] gi|58... 1406 0.0
dbj|BAD82016.1| putative DMI1 protein [Oryza sativa (japonica cu... 1111 0.0
dbj|BAD82015.1| putative DMI1 protein [Oryza sativa (japonica cu... 1108 0.0
emb|CAJ00334.1| DMI1 protein homologue [Pisum sativum] 1107 0.0
dbj|BAA97017.1| unnamed protein product [Arabidopsis thaliana] g... 1095 0.0
ref|NP_915676.1| P0677H08.29 [Oryza sativa (japonica cultivar-gr... 1071 0.0
ref|XP_470380.1| expressed protein [Oryza sativa (japonica culti... 1002 0.0
dbj|BAD89021.1| CASTOR [Lotus corniculatus var. japonicus] gi|58... 998 0.0
ref|NP_102608.1| probable secreted protein [Mesorhizobium loti M... 256 3e-66
emb|CAE45687.1| hypothetical protein [Streptomyces parvulus] 210 2e-52
dbj|BAC68466.1| hypothetical protein [Streptomyces avermitilis M... 203 2e-50
emb|CAC01587.1| putative lipoprotein [Streptomyces coelicolor A3... 187 1e-45
ref|NP_568628.1| phosphotransferase-related [Arabidopsis thalian... 186 3e-45
gb|AAL36360.1| unknown protein [Arabidopsis thaliana] gi|3067983... 182 3e-44
emb|CAB86048.1| putative protein [Arabidopsis thaliana] gi|11281... 176 2e-42
ref|NP_912647.1| Unknown protein [Oryza sativa (japonica cultiva... 150 2e-34
dbj|BAD95358.1| hypothetical protein [Arabidopsis thaliana] 137 2e-30
gb|AAK82546.1| AT5g02940/F9G14_250 [Arabidopsis thaliana] 111 9e-23
ref|NP_683245.1| putative potassium channel protein [Thermosynec... 83 5e-14
>sp|Q6RHR6|DMI1_MEDTR Putative ion channel DMI-1 (Does not make infections protein 1)
gi|44953222|gb|AAS49490.1| DMI1 protein [Medicago
truncatula]
Length = 882
Score = 1736 bits (4495), Expect = 0.0
Identities = 882/882 (100%), Positives = 882/882 (100%)
Query: 1 MAKSNEESSNLNVMNKPPLKKTKTLPSLNLRVSVTPPNPNDNNGIGGTSTTKTDFSEQQW 60
MAKSNEESSNLNVMNKPPLKKTKTLPSLNLRVSVTPPNPNDNNGIGGTSTTKTDFSEQQW
Sbjct: 1 MAKSNEESSNLNVMNKPPLKKTKTLPSLNLRVSVTPPNPNDNNGIGGTSTTKTDFSEQQW 60
Query: 61 NYPSFLGIGSTSRKRRQPPPPPSKPPVNLIPPHPRPLSVNDHNKTTSSLLPQPSSSSITK 120
NYPSFLGIGSTSRKRRQPPPPPSKPPVNLIPPHPRPLSVNDHNKTTSSLLPQPSSSSITK
Sbjct: 61 NYPSFLGIGSTSRKRRQPPPPPSKPPVNLIPPHPRPLSVNDHNKTTSSLLPQPSSSSITK 120
Query: 121 QQQQHSTSSPIFYLLVICCIILVPYSAYLQYKLAKLKDMKLQLCGQIDFCSRNGKTSIQE 180
QQQQHSTSSPIFYLLVICCIILVPYSAYLQYKLAKLKDMKLQLCGQIDFCSRNGKTSIQE
Sbjct: 121 QQQQHSTSSPIFYLLVICCIILVPYSAYLQYKLAKLKDMKLQLCGQIDFCSRNGKTSIQE 180
Query: 181 EVDDDDNADSRTIALYIVLFTLILPFVLYKYLDYLPQIINFLRRTESNKEDVPLKKRVAY 240
EVDDDDNADSRTIALYIVLFTLILPFVLYKYLDYLPQIINFLRRTESNKEDVPLKKRVAY
Sbjct: 181 EVDDDDNADSRTIALYIVLFTLILPFVLYKYLDYLPQIINFLRRTESNKEDVPLKKRVAY 240
Query: 241 MVDVFFSIYPYAKLLALLCATLFLIAFGGLALYAVTGGSMAEALWHSWTYVADAGNHAET 300
MVDVFFSIYPYAKLLALLCATLFLIAFGGLALYAVTGGSMAEALWHSWTYVADAGNHAET
Sbjct: 241 MVDVFFSIYPYAKLLALLCATLFLIAFGGLALYAVTGGSMAEALWHSWTYVADAGNHAET 300
Query: 301 EGTGQRIVSVSISAGGMLIFAMMLGLVSDAISEKVDSLRKGKSEVIERNHVLILGWSDKL 360
EGTGQRIVSVSISAGGMLIFAMMLGLVSDAISEKVDSLRKGKSEVIERNHVLILGWSDKL
Sbjct: 301 EGTGQRIVSVSISAGGMLIFAMMLGLVSDAISEKVDSLRKGKSEVIERNHVLILGWSDKL 360
Query: 361 GSLLKQLAIANKSVGGGVIVVLAEKEKEEMEMDIAKLEFDFMGTSVICRSGSPLILADLK 420
GSLLKQLAIANKSVGGGVIVVLAEKEKEEMEMDIAKLEFDFMGTSVICRSGSPLILADLK
Sbjct: 361 GSLLKQLAIANKSVGGGVIVVLAEKEKEEMEMDIAKLEFDFMGTSVICRSGSPLILADLK 420
Query: 421 KVSVSKARAIIVLAADENADQSDARALRVVLSLAGVKEGLRGHVVVEMSDLDNEPLVKLV 480
KVSVSKARAIIVLAADENADQSDARALRVVLSLAGVKEGLRGHVVVEMSDLDNEPLVKLV
Sbjct: 421 KVSVSKARAIIVLAADENADQSDARALRVVLSLAGVKEGLRGHVVVEMSDLDNEPLVKLV 480
Query: 481 GGELIETVVAHDVIGRLMIQCALQPGLAQIWEDILGFENAEFYIKRWPELDDLLFKDILI 540
GGELIETVVAHDVIGRLMIQCALQPGLAQIWEDILGFENAEFYIKRWPELDDLLFKDILI
Sbjct: 481 GGELIETVVAHDVIGRLMIQCALQPGLAQIWEDILGFENAEFYIKRWPELDDLLFKDILI 540
Query: 541 SFPDAIPCGVKVAADGGKIVINPDDNYVLRDGDEVLVIAEDDDTYAPGPLPEVRKGYFPR 600
SFPDAIPCGVKVAADGGKIVINPDDNYVLRDGDEVLVIAEDDDTYAPGPLPEVRKGYFPR
Sbjct: 541 SFPDAIPCGVKVAADGGKIVINPDDNYVLRDGDEVLVIAEDDDTYAPGPLPEVRKGYFPR 600
Query: 601 IRDPPKYPEKILFCGWRRDIDDMIMVLEAFLAPGSELWMFNEVPEKERERKLAAGELDVF 660
IRDPPKYPEKILFCGWRRDIDDMIMVLEAFLAPGSELWMFNEVPEKERERKLAAGELDVF
Sbjct: 601 IRDPPKYPEKILFCGWRRDIDDMIMVLEAFLAPGSELWMFNEVPEKERERKLAAGELDVF 660
Query: 661 GLENIKLVHREGNAVIRRHLESLPLETFDSILILADESVEDSVAHSDSRSLATLLLIRDI 720
GLENIKLVHREGNAVIRRHLESLPLETFDSILILADESVEDSVAHSDSRSLATLLLIRDI
Sbjct: 661 GLENIKLVHREGNAVIRRHLESLPLETFDSILILADESVEDSVAHSDSRSLATLLLIRDI 720
Query: 721 QSRRLPYRDTKSTSLRLSGFSHNSWIREMQQASDKSIIISEILDSRTRNLVSVSRISDYV 780
QSRRLPYRDTKSTSLRLSGFSHNSWIREMQQASDKSIIISEILDSRTRNLVSVSRISDYV
Sbjct: 721 QSRRLPYRDTKSTSLRLSGFSHNSWIREMQQASDKSIIISEILDSRTRNLVSVSRISDYV 780
Query: 781 LSNELVSMALAMVAEDKQINRVLEELFAEEGNEMCIKPAEFYLFDQEELCFYDIMIRGRT 840
LSNELVSMALAMVAEDKQINRVLEELFAEEGNEMCIKPAEFYLFDQEELCFYDIMIRGRT
Sbjct: 781 LSNELVSMALAMVAEDKQINRVLEELFAEEGNEMCIKPAEFYLFDQEELCFYDIMIRGRT 840
Query: 841 RKEIVIGYRLANQERAIINPSEKSVPRKWSLDDVFVVLASGE 882
RKEIVIGYRLANQERAIINPSEKSVPRKWSLDDVFVVLASGE
Sbjct: 841 RKEIVIGYRLANQERAIINPSEKSVPRKWSLDDVFVVLASGE 882
>dbj|BAD89022.1| POLLUX [Lotus corniculatus var. japonicus]
gi|58430445|dbj|BAD89020.1| ion channel [Lotus
corniculatus var. japonicus]
gi|62287141|sp|Q5H8A5|POLLU_LOTJA Putative ion channel
POLLUX, chloroplast precursor
Length = 917
Score = 1406 bits (3640), Expect = 0.0
Identities = 743/906 (82%), Positives = 787/906 (86%), Gaps = 35/906 (3%)
Query: 4 SNEESSNLNVMNKPPLKKTKTL---PS-------LNLRVSVTPPNPNDNNGIGGTSTTKT 53
++EES NL+ + KPPLKKTKTL PS L+LRVS+ DNN
Sbjct: 20 NDEESPNLSTVIKPPLKKTKTLLPPPSSSSSNRPLHLRVSI------DNNNNNNAPPPPA 73
Query: 54 DFSEQQWNYPSFLGIGSTSRKRRQP---PPPPSKPPVNLIPPHPRPLSVNDHNKTTSSLL 110
DFS+ QWNYPSFLG +T+RKRR PP S + IP N + T ++
Sbjct: 74 DFSDHQWNYPSFLG--TTTRKRRPSSVKPPSTSNLRFDTIPKTKTKTKTNTNTNTNTNTN 131
Query: 111 ----------PQPSSSSITKQQQQHSTSSPIFYLLVICCIILVPYSAYLQYKLAKLKDMK 160
P PSSS + + Q + S PIFYLL+I CII VPYS+YLQYKLAKL+D K
Sbjct: 132 TNTNTDLPPPPVPSSSPVARPQHHNHRSPPIFYLLIITCIIFVPYSSYLQYKLAKLEDHK 191
Query: 161 LQLC--GQIDFCSRNGKTSIQEEVDDDDNAD--SRTIALYIVLFTLILPFVLYKYLDYLP 216
L LC QI F S +G I + D + SR ALYIVLFTLILPF+LYKYLDYLP
Sbjct: 192 LHLCRQSQIHFSSGHGNGKISIPIHDASFSYILSRKAALYIVLFTLILPFLLYKYLDYLP 251
Query: 217 QIINFLRRTESNKEDVPLKKRVAYMVDVFFSIYPYAKLLALLCATLFLIAFGGLALYAVT 276
QIINFLRRT +NKEDVPLKKR+AYM+DVFFSIYPYAKLLALL ATLFLI FGGLALYAVT
Sbjct: 252 QIINFLRRTHNNKEDVPLKKRIAYMLDVFFSIYPYAKLLALLFATLFLIGFGGLALYAVT 311
Query: 277 GGSMAEALWHSWTYVADAGNHAETEGTGQRIVSVSISAGGMLIFAMMLGLVSDAISEKVD 336
GGS+AEALWHSWTYVAD+GNHAET+GTGQR+VSVSIS+GGMLIFAMMLGLVSDAISEKVD
Sbjct: 312 GGSLAEALWHSWTYVADSGNHAETQGTGQRVVSVSISSGGMLIFAMMLGLVSDAISEKVD 371
Query: 337 SLRKGKSEVIERNHVLILGWSDKLGSLLKQLAIANKSVGGGVIVVLAEKEKEEMEMDIAK 396
SLRKGK EVIERNH+LILGWSDKLGSLLKQLAIANKSVGGGVIVVLAEKEKEEMEMDI K
Sbjct: 372 SLRKGKCEVIERNHILILGWSDKLGSLLKQLAIANKSVGGGVIVVLAEKEKEEMEMDITK 431
Query: 397 LEFDFMGTSVICRSGSPLILADLKKVSVSKARAIIVLAADENADQSDARALRVVLSLAGV 456
LEFDFMGTSVICRSGSPLILADLKKVSVSKARAIIVLA+DENADQSDARALRVVLSL GV
Sbjct: 432 LEFDFMGTSVICRSGSPLILADLKKVSVSKARAIIVLASDENADQSDARALRVVLSLTGV 491
Query: 457 KEGLRGHVVVEMSDLDNEPLVKLVGGELIETVVAHDVIGRLMIQCALQPGLAQIWEDILG 516
KEGLRGHVVVEMSDLDNEPLVKLVGGELIETVVAHDVIGRLMIQCALQPGLAQIWEDILG
Sbjct: 492 KEGLRGHVVVEMSDLDNEPLVKLVGGELIETVVAHDVIGRLMIQCALQPGLAQIWEDILG 551
Query: 517 FENAEFYIKRWPELDDLLFKDILISFPDAIPCGVKVAADGGKIVINPDDNYVLRDGDEVL 576
FENAEFYIKRWPELD L FKDILISFPDAIPCGVKVAADGGKIVINPDD+YV+RDGDEVL
Sbjct: 552 FENAEFYIKRWPELDGLSFKDILISFPDAIPCGVKVAADGGKIVINPDDSYVMRDGDEVL 611
Query: 577 VIAEDDDTYAPGPLPEVRKGYFPRIRDPPKYPEKILFCGWRRDIDDMIMVLEAFLAPGSE 636
VIAEDDDTY+PG LPEV KG+FPRI D PKYPEKILFCGWRRDIDDMIMVLEAFLAPGSE
Sbjct: 612 VIAEDDDTYSPGSLPEVLKGFFPRIPDAPKYPEKILFCGWRRDIDDMIMVLEAFLAPGSE 671
Query: 637 LWMFNEVPEKERERKLAAGELDVFGLENIKLVHREGNAVIRRHLESLPLETFDSILILAD 696
LWMFNEVPEKERE+KLAAG LDVFGLENIKLVHREGNAVIRRHLESLPLETFDSILILAD
Sbjct: 672 LWMFNEVPEKEREKKLAAGGLDVFGLENIKLVHREGNAVIRRHLESLPLETFDSILILAD 731
Query: 697 ESVEDSVAHSDSRSLATLLLIRDIQSRRLPYRDTKSTSLRLSGFSHNSWIREMQQASDKS 756
ESVEDSVAHSDSRSLATLLLIRDIQSRRLPY+DTKSTSLRLSGFSHNSWIREMQQASDKS
Sbjct: 732 ESVEDSVAHSDSRSLATLLLIRDIQSRRLPYKDTKSTSLRLSGFSHNSWIREMQQASDKS 791
Query: 757 IIISEILDSRTRNLVSVSRISDYVLSNELVSMALAMVAEDKQINRVLEELFAEEGNEMCI 816
IIISEILDSRTRNLVSVSRISDYVLSNELVSMALAMVAEDKQINRVLEELFAE+GNEMCI
Sbjct: 792 IIISEILDSRTRNLVSVSRISDYVLSNELVSMALAMVAEDKQINRVLEELFAEQGNEMCI 851
Query: 817 KPAEFYLFDQEELCFYDIMIRGRTRKEIVIGYRLANQERAIINPSEKSVPRKWSLDDVFV 876
KPAEFYLFDQEELCFYDIMIRGR R+EI+IGYRLANQERAIINPSEK V RKWSL DVFV
Sbjct: 852 KPAEFYLFDQEELCFYDIMIRGRARQEIIIGYRLANQERAIINPSEKLVARKWSLGDVFV 911
Query: 877 VLASGE 882
V+ASG+
Sbjct: 912 VIASGD 917
>dbj|BAD82016.1| putative DMI1 protein [Oryza sativa (japonica cultivar-group)]
gi|56784216|dbj|BAD81711.1| putative DMI1 protein [Oryza
sativa (japonica cultivar-group)]
Length = 943
Score = 1111 bits (2873), Expect = 0.0
Identities = 587/847 (69%), Positives = 675/847 (79%), Gaps = 22/847 (2%)
Query: 57 EQQWNYPSFLGIGSTS----RKRRQPPPPP------SKPPVNLIPPHPRPLSVNDHNKTT 106
++ W YPSFLG ++ R ++Q P S+ P PP +S + K+
Sbjct: 98 DRDWCYPSFLGPHASRPRPPRSQQQTPTTTAAAAADSRSPTPAAPPQTASVSQREEEKSL 157
Query: 107 SSLLPQPS------SSSITKQQQQHSTSSPIFYLLVICCIILVPYS-AYLQYKLAKLKDM 159
+S++ +P S S QQ+ YL+++ + ++ +S A Q+ A +
Sbjct: 158 ASVVKRPMLLDERRSLSPPPPQQRAPRFDLSPYLVLMLVVTVISFSLAIWQWMKATVLQE 217
Query: 160 KLQLCGQIDFCSRNGKTSI----QEEVDDDDNADSRTIALYIVLFTLILPFVLYKYLDYL 215
K++ C + T + D N+ +A + +P L KY+D L
Sbjct: 218 KIRSCCSVSTVDCKTTTEAFKINGQHGSDFINSADWNLASCSRMLVFAIPVFLVKYIDQL 277
Query: 216 PQIINFLRRTESNKEDVPLKKRVAYMVDVFFSIYPYAKLLALLCATLFLIAFGGLALYAV 275
+ R S +E+VPLKKR+AY VDVFFS +PYAKLLALL AT+ LIA GG+ALY V
Sbjct: 278 RRRNTDSIRLRSTEEEVPLKKRIAYKVDVFFSGHPYAKLLALLLATIILIASGGIALYVV 337
Query: 276 TGGSMAEALWHSWTYVADAGNHAETEGTGQRIVSVSISAGGMLIFAMMLGLVSDAISEKV 335
+G EALW SWT+VAD+GNHA+ G G RIVSVSIS+GGML+FA MLGLVSDAISEKV
Sbjct: 338 SGSGFLEALWLSWTFVADSGNHADQVGLGPRIVSVSISSGGMLVFATMLGLVSDAISEKV 397
Query: 336 DSLRKGKSEVIERNHVLILGWSDKLGSLLKQLAIANKSVGGGVIVVLAEKEKEEMEMDIA 395
DS RKGKSEVIE NH+LILGWSDKLGSLLKQLAIANKS+GGGV+VVLAE++KEEMEMDI
Sbjct: 398 DSWRKGKSEVIEVNHILILGWSDKLGSLLKQLAIANKSIGGGVVVVLAERDKEEMEMDIG 457
Query: 396 KLEFDFMGTSVICRSGSPLILADLKKVSVSKARAIIVLAADENADQSDARALRVVLSLAG 455
KLEFDFMGTSVICRSGSPLILADLKKVSVSKARAIIVLA+DENADQSDARALRVVLSL G
Sbjct: 458 KLEFDFMGTSVICRSGSPLILADLKKVSVSKARAIIVLASDENADQSDARALRVVLSLTG 517
Query: 456 VKEGLRGHVVVEMSDLDNEPLVKLVGGELIETVVAHDVIGRLMIQCALQPGLAQIWEDIL 515
VKEGLRGHVVVEMSDLDNEPLVKLVGGELIETVVAHDVIGRLMIQCALQPGLAQIWEDIL
Sbjct: 518 VKEGLRGHVVVEMSDLDNEPLVKLVGGELIETVVAHDVIGRLMIQCALQPGLAQIWEDIL 577
Query: 516 GFENAEFYIKRWPELDDLLFKDILISFPDAIPCGVKVAADGGKIVINPDDNYVLRDGDEV 575
GFENAEFYIKRWPELD + F D+LISFPDA+PCGVK+A+ GKI++NPD++YVL++GDEV
Sbjct: 578 GFENAEFYIKRWPELDGMRFGDVLISFPDAVPCGVKIASKAGKILMNPDNDYVLQEGDEV 637
Query: 576 LVIAEDDDTYAPGPLPEVRKGYFPRIRDPPKYPEKILFCGWRRDIDDMIMVLEAFLAPGS 635
LVIAEDDDTY P LP+VRKG+ P I PPKYPEKILFCGWRRDI DMIMVLEAFLAPGS
Sbjct: 638 LVIAEDDDTYVPASLPQVRKGFLPNIPTPPKYPEKILFCGWRRDIHDMIMVLEAFLAPGS 697
Query: 636 ELWMFNEVPEKERERKLAAGELDVFGLENIKLVHREGNAVIRRHLESLPLETFDSILILA 695
ELWMFNEVPEKERERKL G +D++GL NIKLVH+EGNAVIRRHLESLPLETFDSILILA
Sbjct: 698 ELWMFNEVPEKERERKLTDGGMDIYGLTNIKLVHKEGNAVIRRHLESLPLETFDSILILA 757
Query: 696 DESVEDSVAHSDSRSLATLLLIRDIQSRRLPYRDTKSTSLRLSGFSHNSWIREMQQASDK 755
DESVEDS+ HSDSRSLATLLLIRDIQS+RLP ++ KS LR +GF H+SWIREMQ ASDK
Sbjct: 758 DESVEDSIVHSDSRSLATLLLIRDIQSKRLPSKELKS-PLRYNGFCHSSWIREMQHASDK 816
Query: 756 SIIISEILDSRTRNLVSVSRISDYVLSNELVSMALAMVAEDKQINRVLEELFAEEGNEMC 815
SIIISEILDSRTRNLVSVS+ISDYVLSNELVSMALAMVAEDKQINRVLEELFAEEGNEMC
Sbjct: 817 SIIISEILDSRTRNLVSVSKISDYVLSNELVSMALAMVAEDKQINRVLEELFAEEGNEMC 876
Query: 816 IKPAEFYLFDQEELCFYDIMIRGRTRKEIVIGYRLANQERAIINPSEKSVPRKWSLDDVF 875
I+ AEFYL++QEEL F+DIM+R R R E+VIGYRLAN ++AIINP +KS RKWSLDDVF
Sbjct: 877 IRSAEFYLYEQEELSFFDIMVRARERDEVVIGYRLANDDQAIINPEQKSEIRKWSLDDVF 936
Query: 876 VVLASGE 882
VV++ G+
Sbjct: 937 VVISKGD 943
>dbj|BAD82015.1| putative DMI1 protein [Oryza sativa (japonica cultivar-group)]
gi|56784215|dbj|BAD81710.1| putative DMI1 protein [Oryza
sativa (japonica cultivar-group)]
gi|62286504|sp|Q5N941|DMI1_ORYSA Putative ion channel
DMI1, chloroplast precursor
Length = 965
Score = 1108 bits (2865), Expect = 0.0
Identities = 586/844 (69%), Positives = 673/844 (79%), Gaps = 22/844 (2%)
Query: 57 EQQWNYPSFLGIGSTS----RKRRQPPPPP------SKPPVNLIPPHPRPLSVNDHNKTT 106
++ W YPSFLG ++ R ++Q P S+ P PP +S + K+
Sbjct: 98 DRDWCYPSFLGPHASRPRPPRSQQQTPTTTAAAAADSRSPTPAAPPQTASVSQREEEKSL 157
Query: 107 SSLLPQPS------SSSITKQQQQHSTSSPIFYLLVICCIILVPYS-AYLQYKLAKLKDM 159
+S++ +P S S QQ+ YL+++ + ++ +S A Q+ A +
Sbjct: 158 ASVVKRPMLLDERRSLSPPPPQQRAPRFDLSPYLVLMLVVTVISFSLAIWQWMKATVLQE 217
Query: 160 KLQLCGQIDFCSRNGKTSI----QEEVDDDDNADSRTIALYIVLFTLILPFVLYKYLDYL 215
K++ C + T + D N+ +A + +P L KY+D L
Sbjct: 218 KIRSCCSVSTVDCKTTTEAFKINGQHGSDFINSADWNLASCSRMLVFAIPVFLVKYIDQL 277
Query: 216 PQIINFLRRTESNKEDVPLKKRVAYMVDVFFSIYPYAKLLALLCATLFLIAFGGLALYAV 275
+ R S +E+VPLKKR+AY VDVFFS +PYAKLLALL AT+ LIA GG+ALY V
Sbjct: 278 RRRNTDSIRLRSTEEEVPLKKRIAYKVDVFFSGHPYAKLLALLLATIILIASGGIALYVV 337
Query: 276 TGGSMAEALWHSWTYVADAGNHAETEGTGQRIVSVSISAGGMLIFAMMLGLVSDAISEKV 335
+G EALW SWT+VAD+GNHA+ G G RIVSVSIS+GGML+FA MLGLVSDAISEKV
Sbjct: 338 SGSGFLEALWLSWTFVADSGNHADQVGLGPRIVSVSISSGGMLVFATMLGLVSDAISEKV 397
Query: 336 DSLRKGKSEVIERNHVLILGWSDKLGSLLKQLAIANKSVGGGVIVVLAEKEKEEMEMDIA 395
DS RKGKSEVIE NH+LILGWSDKLGSLLKQLAIANKS+GGGV+VVLAE++KEEMEMDI
Sbjct: 398 DSWRKGKSEVIEVNHILILGWSDKLGSLLKQLAIANKSIGGGVVVVLAERDKEEMEMDIG 457
Query: 396 KLEFDFMGTSVICRSGSPLILADLKKVSVSKARAIIVLAADENADQSDARALRVVLSLAG 455
KLEFDFMGTSVICRSGSPLILADLKKVSVSKARAIIVLA+DENADQSDARALRVVLSL G
Sbjct: 458 KLEFDFMGTSVICRSGSPLILADLKKVSVSKARAIIVLASDENADQSDARALRVVLSLTG 517
Query: 456 VKEGLRGHVVVEMSDLDNEPLVKLVGGELIETVVAHDVIGRLMIQCALQPGLAQIWEDIL 515
VKEGLRGHVVVEMSDLDNEPLVKLVGGELIETVVAHDVIGRLMIQCALQPGLAQIWEDIL
Sbjct: 518 VKEGLRGHVVVEMSDLDNEPLVKLVGGELIETVVAHDVIGRLMIQCALQPGLAQIWEDIL 577
Query: 516 GFENAEFYIKRWPELDDLLFKDILISFPDAIPCGVKVAADGGKIVINPDDNYVLRDGDEV 575
GFENAEFYIKRWPELD + F D+LISFPDA+PCGVK+A+ GKI++NPD++YVL++GDEV
Sbjct: 578 GFENAEFYIKRWPELDGMRFGDVLISFPDAVPCGVKIASKAGKILMNPDNDYVLQEGDEV 637
Query: 576 LVIAEDDDTYAPGPLPEVRKGYFPRIRDPPKYPEKILFCGWRRDIDDMIMVLEAFLAPGS 635
LVIAEDDDTY P LP+VRKG+ P I PPKYPEKILFCGWRRDI DMIMVLEAFLAPGS
Sbjct: 638 LVIAEDDDTYVPASLPQVRKGFLPNIPTPPKYPEKILFCGWRRDIHDMIMVLEAFLAPGS 697
Query: 636 ELWMFNEVPEKERERKLAAGELDVFGLENIKLVHREGNAVIRRHLESLPLETFDSILILA 695
ELWMFNEVPEKERERKL G +D++GL NIKLVH+EGNAVIRRHLESLPLETFDSILILA
Sbjct: 698 ELWMFNEVPEKERERKLTDGGMDIYGLTNIKLVHKEGNAVIRRHLESLPLETFDSILILA 757
Query: 696 DESVEDSVAHSDSRSLATLLLIRDIQSRRLPYRDTKSTSLRLSGFSHNSWIREMQQASDK 755
DESVEDS+ HSDSRSLATLLLIRDIQS+RLP ++ KS LR +GF H+SWIREMQ ASDK
Sbjct: 758 DESVEDSIVHSDSRSLATLLLIRDIQSKRLPSKELKS-PLRYNGFCHSSWIREMQHASDK 816
Query: 756 SIIISEILDSRTRNLVSVSRISDYVLSNELVSMALAMVAEDKQINRVLEELFAEEGNEMC 815
SIIISEILDSRTRNLVSVS+ISDYVLSNELVSMALAMVAEDKQINRVLEELFAEEGNEMC
Sbjct: 817 SIIISEILDSRTRNLVSVSKISDYVLSNELVSMALAMVAEDKQINRVLEELFAEEGNEMC 876
Query: 816 IKPAEFYLFDQEELCFYDIMIRGRTRKEIVIGYRLANQERAIINPSEKSVPRKWSLDDVF 875
I+ AEFYL++QEEL F+DIM+R R R E+VIGYRLAN ++AIINP +KS RKWSLDDVF
Sbjct: 877 IRSAEFYLYEQEELSFFDIMVRARERDEVVIGYRLANDDQAIINPEQKSEIRKWSLDDVF 936
Query: 876 VVLA 879
VV++
Sbjct: 937 VVIS 940
>emb|CAJ00334.1| DMI1 protein homologue [Pisum sativum]
Length = 580
Score = 1107 bits (2863), Expect = 0.0
Identities = 569/580 (98%), Positives = 575/580 (99%)
Query: 298 AETEGTGQRIVSVSISAGGMLIFAMMLGLVSDAISEKVDSLRKGKSEVIERNHVLILGWS 357
AETEG GQRIVSVSISAGGMLIFAMMLGLVSDAISEKVDSLRKGKSEVIERNHVLILGWS
Sbjct: 1 AETEGMGQRIVSVSISAGGMLIFAMMLGLVSDAISEKVDSLRKGKSEVIERNHVLILGWS 60
Query: 358 DKLGSLLKQLAIANKSVGGGVIVVLAEKEKEEMEMDIAKLEFDFMGTSVICRSGSPLILA 417
DKLGSLLKQLAIANKSVGGGVIVVLAEKEKEEMEMDIAKLEFDFMGTSVICRSGSPLILA
Sbjct: 61 DKLGSLLKQLAIANKSVGGGVIVVLAEKEKEEMEMDIAKLEFDFMGTSVICRSGSPLILA 120
Query: 418 DLKKVSVSKARAIIVLAADENADQSDARALRVVLSLAGVKEGLRGHVVVEMSDLDNEPLV 477
DLKKVSVSKARAIIVLA+DENADQSDARALRVVLSL GVKEGLRGHVVVEMSDLDNEPLV
Sbjct: 121 DLKKVSVSKARAIIVLASDENADQSDARALRVVLSLTGVKEGLRGHVVVEMSDLDNEPLV 180
Query: 478 KLVGGELIETVVAHDVIGRLMIQCALQPGLAQIWEDILGFENAEFYIKRWPELDDLLFKD 537
KLVGGELIETVVAHDVIGRLMIQCALQPGLAQIWEDILGFENAEFYIKRWP LD LLFKD
Sbjct: 181 KLVGGELIETVVAHDVIGRLMIQCALQPGLAQIWEDILGFENAEFYIKRWPXLDGLLFKD 240
Query: 538 ILISFPDAIPCGVKVAADGGKIVINPDDNYVLRDGDEVLVIAEDDDTYAPGPLPEVRKGY 597
ILISFPDAIPCGVKV+ADGGKIVINPDDNYVLRDGDEVLVIAEDDDTYAPGPLPEVRKGY
Sbjct: 241 ILISFPDAIPCGVKVSADGGKIVINPDDNYVLRDGDEVLVIAEDDDTYAPGPLPEVRKGY 300
Query: 598 FPRIRDPPKYPEKILFCGWRRDIDDMIMVLEAFLAPGSELWMFNEVPEKERERKLAAGEL 657
FPRIRDPPKYPEKILFCGWRRDIDDMIMVLEAFLAPGSELWMFNEVPEK+RERKLAAGEL
Sbjct: 301 FPRIRDPPKYPEKILFCGWRRDIDDMIMVLEAFLAPGSELWMFNEVPEKQRERKLAAGEL 360
Query: 658 DVFGLENIKLVHREGNAVIRRHLESLPLETFDSILILADESVEDSVAHSDSRSLATLLLI 717
DVFGLENIKLVHREGNAVIRRHLESLPLETFDSILILADESVEDSVAHSDSRSLATLLLI
Sbjct: 361 DVFGLENIKLVHREGNAVIRRHLESLPLETFDSILILADESVEDSVAHSDSRSLATLLLI 420
Query: 718 RDIQSRRLPYRDTKSTSLRLSGFSHNSWIREMQQASDKSIIISEILDSRTRNLVSVSRIS 777
RDIQSRRLPYRDTKSTSLRLSGFSHNSWIREMQQASDKSIIISEILDSRTRNLVSVSRIS
Sbjct: 421 RDIQSRRLPYRDTKSTSLRLSGFSHNSWIREMQQASDKSIIISEILDSRTRNLVSVSRIS 480
Query: 778 DYVLSNELVSMALAMVAEDKQINRVLEELFAEEGNEMCIKPAEFYLFDQEELCFYDIMIR 837
DYVLSNELVSMALAMVAEDKQINRVLEELFAEEGNEMCIKPAEFYLFDQEELCFYDIMIR
Sbjct: 481 DYVLSNELVSMALAMVAEDKQINRVLEELFAEEGNEMCIKPAEFYLFDQEELCFYDIMIR 540
Query: 838 GRTRKEIVIGYRLANQERAIINPSEKSVPRKWSLDDVFVV 877
GRTRKEIVIGYRLA+QERA+INPSEKS+ RKWSLDDVFVV
Sbjct: 541 GRTRKEIVIGYRLASQERALINPSEKSMTRKWSLDDVFVV 580
>dbj|BAA97017.1| unnamed protein product [Arabidopsis thaliana]
gi|15240588|ref|NP_199807.1| expressed protein
[Arabidopsis thaliana] gi|62286581|sp|Q9LTX4|DMI1_ARATH
Putative ion channel DMI1
Length = 824
Score = 1095 bits (2832), Expect = 0.0
Identities = 575/863 (66%), Positives = 677/863 (77%), Gaps = 51/863 (5%)
Query: 20 KKTKTLPSLNLRVSVTPPNPNDNNGIGGTSTTKTDFSEQQWNYPSFLGIGSTSRKRRQPP 79
+++KTLP+ + R +P P G T + D S P+ +P
Sbjct: 13 QRSKTLPTNSYRRFTSPVIPTFRYDDGDTVSYDGDDSSNLPTVPN---------PEEKPV 63
Query: 80 PPPSKPPVNLIPPHPRPLSVNDHNKTTSSLLPQPSSSSITKQQQQHSTSSPIFYLLVICC 139
P PS+ P S IT+ Q S + + L C
Sbjct: 64 PVPSQSP----------------------------SQRITRLWTQFSLT----HCLKFIC 91
Query: 140 IILVPYSAYLQYKLAKLKDMKLQLCGQIDFCSRNGKTSIQEEVDDDDNADSRTIALYIVL 199
Y +L+ K+++L+ + L + + S N + + +SR + + V+
Sbjct: 92 SCSFTYVMFLRSKVSRLEAENIILLTRCNSSSDNNEM---------EETNSRAVVFFSVI 142
Query: 200 FTLILPFVLYKYLDYLPQIINFLRRTESNKEDVPLKKRVAYMVDVFFSIYPYAKLLALLC 259
T +LPF+LY YLD L + N LRRT KEDVPLKKR+AY +DV FS+YPYAKLLALL
Sbjct: 143 ITFVLPFLLYMYLDDLSHVKNLLRRTNQKKEDVPLKKRLAYSLDVCFSVYPYAKLLALLL 202
Query: 260 ATLFLIAFGGLALYAVTGGSMAEALWHSWTYVADAGNHAETEGTGQRIVSVSISAGGMLI 319
AT+ LI +GGLALYAV+ + EALW SWT+VAD+G+HA+ G G RIVSV+ISAGGMLI
Sbjct: 203 ATVVLIVYGGLALYAVSDCGVDEALWLSWTFVADSGSHADRVGVGARIVSVAISAGGMLI 262
Query: 320 FAMMLGLVSDAISEKVDSLRKGKSEVIERNHVLILGWSDKLGSLLKQLAIANKSVGGGVI 379
FA MLGL+SDAIS+ VDSLRKGKSEV+E NH+LILGWSDKLGSLLKQLAIANKS+GGGV+
Sbjct: 263 FATMLGLISDAISKMVDSLRKGKSEVLESNHILILGWSDKLGSLLKQLAIANKSIGGGVV 322
Query: 380 VVLAEKEKEEMEMDIAKLEFDFMGTSVICRSGSPLILADLKKVSVSKARAIIVLAADENA 439
VVLAE++KEEME DIAK EFD MGTSVICRSGSPLILADLKKVSVS ARAIIVL +DENA
Sbjct: 323 VVLAERDKEEMETDIAKFEFDLMGTSVICRSGSPLILADLKKVSVSNARAIIVLGSDENA 382
Query: 440 DQSDARALRVVLSLAGVKEGLRGHVVVEMSDLDNEPLVKLVGGELIETVVAHDVIGRLMI 499
DQSDARALRVVLSL GVKEG +GHVVVEM DLDNEPLVKLVGGE IETVVAHDVIGRLMI
Sbjct: 383 DQSDARALRVVLSLTGVKEGWKGHVVVEMCDLDNEPLVKLVGGERIETVVAHDVIGRLMI 442
Query: 500 QCALQPGLAQIWEDILGFENAEFYIKRWPELDDLLFKDILISFPDAIPCGVKVAADGGKI 559
QCALQPGLAQIWEDILGFENAEFYIK+WP+LD F+D+LISFP+AIPCGVKVAAD GKI
Sbjct: 443 QCALQPGLAQIWEDILGFENAEFYIKKWPQLDGYCFEDVLISFPNAIPCGVKVAAD-GKI 501
Query: 560 VINPDDNYVLRDGDEVLVIAEDDDTYAPGPLPEVRKGYFPRIRDPPKYPEKILFCGWRRD 619
V+NP D+YVL++GDE+LVIAEDDDTYAPG LPEVR +FP+++DPPKYPEKILFCGWRRD
Sbjct: 502 VLNPSDDYVLKEGDEILVIAEDDDTYAPGSLPEVRMCHFPKMQDPPKYPEKILFCGWRRD 561
Query: 620 IDDMIMVLEAFLAPGSELWMFNEVPEKERERKLAAGELDVFGLENIKLVHREGNAVIRRH 679
IDDMI VLEA LAPGSELWMFNEVP++ERE+KL L++ L NIKLVHR+GNAVIRRH
Sbjct: 562 IDDMIKVLEALLAPGSELWMFNEVPDQEREKKLTDAGLNISKLVNIKLVHRQGNAVIRRH 621
Query: 680 LESLPLETFDSILILADESVEDSVAHSDSRSLATLLLIRDIQSRRLPYRDTKSTSLRLSG 739
LESLPLETFDSILILA++S+E+S+ HSDSRSLATLLLIRDIQS+RLPY+D KS++LR+SG
Sbjct: 622 LESLPLETFDSILILAEQSLENSIVHSDSRSLATLLLIRDIQSKRLPYKDAKSSALRISG 681
Query: 740 FSHNSWIREMQQASDKSIIISEILDSRTRNLVSVSRISDYVLSNELVSMALAMVAEDKQI 799
F + WIR+MQQASDKSI+ISEILDSRT+NLVSVSRISDYVLSNELVSMALAMVAEDKQI
Sbjct: 682 FPNCCWIRKMQQASDKSIVISEILDSRTKNLVSVSRISDYVLSNELVSMALAMVAEDKQI 741
Query: 800 NRVLEELFAEEGNEMCIKPAEFYLFDQEELCFYDIMIRGRTRKEIVIGYRLANQERAIIN 859
NRVL+ELFAE+GNE+CI+PAEFY++DQEE+CFYDIM R R R+EI+IGYRLA E+A+IN
Sbjct: 742 NRVLKELFAEKGNELCIRPAEFYIYDQEEVCFYDIMRRARQRQEIIIGYRLAGMEQAVIN 801
Query: 860 PSEKSVPRKWSLDDVFVVLASGE 882
P++KS KWSLDDVFVV+AS +
Sbjct: 802 PTDKSKLTKWSLDDVFVVIASSQ 824
>ref|NP_915676.1| P0677H08.29 [Oryza sativa (japonica cultivar-group)]
Length = 927
Score = 1072 bits (2771), Expect = 0.0
Identities = 573/847 (67%), Positives = 660/847 (77%), Gaps = 38/847 (4%)
Query: 57 EQQWNYPSFLGIGSTS----RKRRQPPPPP------SKPPVNLIPPHPRPLSVNDHNKTT 106
++ W YPSFLG ++ R ++Q P S+ P PP +S + K+
Sbjct: 98 DRDWCYPSFLGPHASRPRPPRSQQQTPTTTAAAAADSRSPTPAAPPQTASVSQREEEKSL 157
Query: 107 SSLLPQPS------SSSITKQQQQHSTSSPIFYLLVICCIILVPYS-AYLQYKLAKLKDM 159
+S++ +P S S QQ+ YL+++ + ++ +S A Q+ A +
Sbjct: 158 ASVVKRPMLLDERRSLSPPPPQQRAPRFDLSPYLVLMLVVTVISFSLAIWQWMKATVLQE 217
Query: 160 KLQLCGQIDFCSRNGKTSI----QEEVDDDDNADSRTIALYIVLFTLILPFVLYKYLDYL 215
K++ C + T + D N+ +A + +P L KY+D L
Sbjct: 218 KIRSCCSVSTVDCKTTTEAFKINGQHGSDFINSADWNLASCSRMLVFAIPVFLVKYIDQL 277
Query: 216 PQIINFLRRTESNKEDVPLKKRVAYMVDVFFSIYPYAKLLALLCATLFLIAFGGLALYAV 275
+ R S +E+VPLKKR+AY VDVFFS +PYAKLLALL AT+ LIA GG+ALY V
Sbjct: 278 RRRNTDSIRLRSTEEEVPLKKRIAYKVDVFFSGHPYAKLLALLLATIILIASGGIALYVV 337
Query: 276 TGGSMAEALWHSWTYVADAGNHAETEGTGQRIVSVSISAGGMLIFAMMLGLVSDAISEKV 335
+G EALW SWT+VAD+GNHA+ G G RIVSVSIS+GGML+FA MLGLVSDAISEKV
Sbjct: 338 SGSGFLEALWLSWTFVADSGNHADQVGLGPRIVSVSISSGGMLVFATMLGLVSDAISEKV 397
Query: 336 DSLRKGKSEVIERNHVLILGWSDKLGSLLKQLAIANKSVGGGVIVVLAEKEKEEMEMDIA 395
DS RKGKSE GSLLKQLAIANKS+GGGV+VVLAE++KEEMEMDI
Sbjct: 398 DSWRKGKSE----------------GSLLKQLAIANKSIGGGVVVVLAERDKEEMEMDIG 441
Query: 396 KLEFDFMGTSVICRSGSPLILADLKKVSVSKARAIIVLAADENADQSDARALRVVLSLAG 455
KLEFDFMGTSVICRSGSPLILADLKKVSVSKARAIIVLA+DENADQSDARALRVVLSL G
Sbjct: 442 KLEFDFMGTSVICRSGSPLILADLKKVSVSKARAIIVLASDENADQSDARALRVVLSLTG 501
Query: 456 VKEGLRGHVVVEMSDLDNEPLVKLVGGELIETVVAHDVIGRLMIQCALQPGLAQIWEDIL 515
VKEGLRGHVVVEMSDLDNEPLVKLVGGELIETVVAHDVIGRLMIQCALQPGLAQIWEDIL
Sbjct: 502 VKEGLRGHVVVEMSDLDNEPLVKLVGGELIETVVAHDVIGRLMIQCALQPGLAQIWEDIL 561
Query: 516 GFENAEFYIKRWPELDDLLFKDILISFPDAIPCGVKVAADGGKIVINPDDNYVLRDGDEV 575
GFENAEFYIKRWPELD + F D+LISFPDA+PCGVK+A+ GKI++NPD++YVL++GDEV
Sbjct: 562 GFENAEFYIKRWPELDGMRFGDVLISFPDAVPCGVKIASKAGKILMNPDNDYVLQEGDEV 621
Query: 576 LVIAEDDDTYAPGPLPEVRKGYFPRIRDPPKYPEKILFCGWRRDIDDMIMVLEAFLAPGS 635
LVIAEDDDTY P LP+VRKG+ P I PPKYPEKILFCGWRRDI DMIMVLEAFLAPGS
Sbjct: 622 LVIAEDDDTYVPASLPQVRKGFLPNIPTPPKYPEKILFCGWRRDIHDMIMVLEAFLAPGS 681
Query: 636 ELWMFNEVPEKERERKLAAGELDVFGLENIKLVHREGNAVIRRHLESLPLETFDSILILA 695
ELWMFNEVPEKERERKL G +D++GL NIKLVH+EGNAVIRRHLESLPLETFDSILILA
Sbjct: 682 ELWMFNEVPEKERERKLTDGGMDIYGLTNIKLVHKEGNAVIRRHLESLPLETFDSILILA 741
Query: 696 DESVEDSVAHSDSRSLATLLLIRDIQSRRLPYRDTKSTSLRLSGFSHNSWIREMQQASDK 755
DESVEDS+ HSDSRSLATLLLIRDIQS+RLP ++ KS LR +GF H+SWIREMQ ASDK
Sbjct: 742 DESVEDSIVHSDSRSLATLLLIRDIQSKRLPSKELKS-PLRYNGFCHSSWIREMQHASDK 800
Query: 756 SIIISEILDSRTRNLVSVSRISDYVLSNELVSMALAMVAEDKQINRVLEELFAEEGNEMC 815
SIIISEILDSRTRNLVSVS+ISDYVLSNELVSMALAMVAEDKQINRVLEELFAEEGNEMC
Sbjct: 801 SIIISEILDSRTRNLVSVSKISDYVLSNELVSMALAMVAEDKQINRVLEELFAEEGNEMC 860
Query: 816 IKPAEFYLFDQEELCFYDIMIRGRTRKEIVIGYRLANQERAIINPSEKSVPRKWSLDDVF 875
I+ AEFYL++QEEL F+DIM+R R R E+VIGYRLAN ++AIINP +KS RKWSLDDVF
Sbjct: 861 IRSAEFYLYEQEELSFFDIMVRARERDEVVIGYRLANDDQAIINPEQKSEIRKWSLDDVF 920
Query: 876 VVLASGE 882
VV++ G+
Sbjct: 921 VVISKGD 927
>ref|XP_470380.1| expressed protein [Oryza sativa (japonica cultivar-group)]
gi|62286620|sp|Q75LD5|DMI1L_ORYSA Putative ion channel
DMI1-like, chloroplast precursor
gi|41469646|gb|AAS07369.1| expressed protein [Oryza
sativa (japonica cultivar-group)]
Length = 893
Score = 1002 bits (2591), Expect = 0.0
Identities = 524/827 (63%), Positives = 630/827 (75%), Gaps = 22/827 (2%)
Query: 74 KRRQPPPPPSKPPVNLIPPHPRPLSVNDHNKTTSSLLPQPSSSSITKQQQQHSTSSPI-- 131
+++Q PPP+ PP PR V + T + +++++ Q H + S
Sbjct: 71 QQKQQQPPPTTPPPAPRRRDPRYAGVRRGDVRTLTAEKAAAAAAVPTAAQVHGSKSAASA 130
Query: 132 ----------FYLLVICCIILVPYSAYLQYKLAKLKDM------KLQLCGQIDFCSRNGK 175
+V+C LV ++ L ++ LK KLQ C +
Sbjct: 131 TTLRWSGMVSVAAIVLCFSSLVRSNSSLHDQVHHLKAQLAEATTKLQSCITESSMDMSSI 190
Query: 176 TSIQEEVDDDDNADSRTIALYIVLFTLILPFVLYKYLDYLPQIINFLRRTESNKEDVPLK 235
S Q N + +L + L TL P ++ KY+D + LR ++ ++E+VP+
Sbjct: 191 LSYQSNNSTSQNRGLKNFSLLLSLSTLYAPLLILKYMDLFLK----LRSSQDSEEEVPIN 246
Query: 236 KRVAYMVDVFFSIYPYAKLLALLCATLFLIAFGGLALYAVTGGSMAEALWHSWTYVADAG 295
KR+AY VD+F S+ PYAK L LL ATL LI GGLALY V S+ + LW SWT+VAD+G
Sbjct: 247 KRLAYRVDIFLSLQPYAKPLVLLVATLLLIGLGGLALYGVNDDSLLDCLWLSWTFVADSG 306
Query: 296 NHAETEGTGQRIVSVSISAGGMLIFAMMLGLVSDAISEKVDSLRKGKSEVIERNHVLILG 355
NHA EG G ++VSVSIS GGML+FAMMLGLV+D+ISEK DSLRKG+SEVIE++H L+LG
Sbjct: 307 NHANAEGFGPKLVSVSISIGGMLVFAMMLGLVTDSISEKFDSLRKGRSEVIEQSHTLVLG 366
Query: 356 WSDKLGSLLKQLAIANKSVGGGVIVVLAEKEKEEMEMDIAKLEFDFMGTSVICRSGSPLI 415
WSDKLGSLL Q+AIAN+S+GGG IVV+AEK+KEEME DIAK+EFD GT++ICRSGSPLI
Sbjct: 367 WSDKLGSLLNQIAIANESLGGGTIVVMAEKDKEEMEADIAKMEFDLKGTAIICRSGSPLI 426
Query: 416 LADLKKVSVSKARAIIVLAADENADQSDARALRVVLSLAGVKEGLRGHVVVEMSDLDNEP 475
LADLKKVSVSKARAI+VLA + NADQSDARALR VLSL GVKEGLRGH+VVE+SDLDNE
Sbjct: 427 LADLKKVSVSKARAIVVLAEEGNADQSDARALRTVLSLTGVKEGLRGHIVVELSDLDNEV 486
Query: 476 LVKLVGGELIETVVAHDVIGRLMIQCALQPGLAQIWEDILGFENAEFYIKRWPELDDLLF 535
LVKLVGG+L+ETVVAHDVIGRLMIQCA QPGLAQIWEDILGFEN EFYIKRWP+LD + F
Sbjct: 487 LVKLVGGDLVETVVAHDVIGRLMIQCARQPGLAQIWEDILGFENCEFYIKRWPQLDGMQF 546
Query: 536 KDILISFPDAIPCGVKVAADGGKIVINPDDNYVLRDGDEVLVIAEDDDTYAPGPLPEVRK 595
+D+LISFPDAIPCG+KVA+ GGKI++NPDD YVL++GDEVLVIAEDDDTYAP PLP+V +
Sbjct: 547 EDVLISFPDAIPCGIKVASYGGKIILNPDDFYVLQEGDEVLVIAEDDDTYAPAPLPKVMR 606
Query: 596 GYFPRIRDPPKYPEKILFCGWRRDIDDMIMVLEAFLAPGSELWMFNEVPEKERERKLAAG 655
GY P+ PK PE+ILFCGWRRD++DMIMVL+AFLAPGSELWMFN+VPE +RERKL G
Sbjct: 607 GYLPKDFVVPKSPERILFCGWRRDMEDMIMVLDAFLAPGSELWMFNDVPEMDRERKLIDG 666
Query: 656 ELDVFGLENIKLVHREGNAVIRRHLESLPLETFDSILILADESVEDSVAHSDSRSLATLL 715
LD LENI LVHREGNAVIRRHLESLPLE+FDSILILADESVEDS +DSRSLATLL
Sbjct: 667 GLDFSRLENITLVHREGNAVIRRHLESLPLESFDSILILADESVEDSAIQADSRSLATLL 726
Query: 716 LIRDIQSRRLPYRDTKSTSLRLSGFSHNSWIREMQQASDKSIIISEILDSRTRNLVSVSR 775
LIRDIQ++RLP+R+ + + F SWI EMQQASDKS+IISEILD RT+NL+SVS+
Sbjct: 727 LIRDIQAKRLPFREAMVSHVTRGSFCEGSWIGEMQQASDKSVIISEILDPRTKNLLSVSK 786
Query: 776 ISDYVLSNELVSMALAMVAEDKQINRVLEELFAEEGNEMCIKPAEFYLFDQEELCFYDIM 835
ISDYVLSNELVSMALAMVAED+QIN VLEELFAE+GNEM I+PA+ YL + EEL F+++M
Sbjct: 787 ISDYVLSNELVSMALAMVAEDRQINDVLEELFAEQGNEMQIRPADLYLREDEELNFFEVM 846
Query: 836 IRGRTRKEIVIGYRLANQERAIINPSEKSVPRKWSLDDVFVVLASGE 882
+RGR RKEIVIGYRL + ERAIINP +K R+WS DVFVV+ E
Sbjct: 847 LRGRQRKEIVIGYRLVDAERAIINPPDKVSRRRWSAKDVFVVITEKE 893
>dbj|BAD89021.1| CASTOR [Lotus corniculatus var. japonicus]
gi|58430443|dbj|BAD89019.1| ion channel [Lotus
corniculatus var. japonicus]
gi|62286596|sp|Q5H8A6|CASTO_LOTJA Putative ion channel
CASTOR, chloroplast precursor
Length = 853
Score = 998 bits (2580), Expect = 0.0
Identities = 529/840 (62%), Positives = 641/840 (75%), Gaps = 31/840 (3%)
Query: 50 TTKTDFSEQQWNYPSFLGIGSTSRKRRQPPPPPS---KPPVNLIPPHPRPLSVNDHNKTT 106
T S YPS GI R + P S KP ++++ P +S N+ N +
Sbjct: 38 TNSNTHSAPSSTYPS--GIRHRRRVKFSRTPTTSSNEKPQISIVSDKPSAISKNNLNWLS 95
Query: 107 SSLLPQPSSSSITKQQQQHSTSSPIFYLLVICCIILVPYSAYLQYKLAKLKD--MKLQLC 164
Q + L ++ ++L+ + +L+ ++ KL+ ++L C
Sbjct: 96 -----------------QFGLQFALVTLTIVFLLLLLLRNTHLESQVNKLQGEILRLHAC 138
Query: 165 GQIDFCSRNGKTSIQEEVDDDDNADS--RTIALYIVLFTLILPFVLYKYLDYLPQIINFL 222
Q+D + + T+ + + + ++ R +AL++ L++P +++KY+DY+ +
Sbjct: 139 HQLDTLNVSSSTAHKSQDTHPCSCENFKRNLALFLSFMLLLIPLIIFKYIDYVSRS---- 194
Query: 223 RRTESNKEDVPLKKRVAYMVDVFFSIYPYAKLLALLCATLFLIAFGGLALYAVTGGSMAE 282
R +E+ E V L K++AY VDVF S+YPYAK L LL ATL LI GGL L+ VT +
Sbjct: 195 RLSENISEQVSLNKQIAYRVDVFLSVYPYAKPLVLLVATLLLIFLGGLTLFGVTTEDLGH 254
Query: 283 ALWHSWTYVADAGNHAETEGTGQRIVSVSISAGGMLIFAMMLGLVSDAISEKVDSLRKGK 342
LW SWTYVAD+GNHA +EG G R+V+VSIS GGMLIFAMMLGLVSDAISEK DSLRKGK
Sbjct: 255 CLWLSWTYVADSGNHASSEGIGPRLVAVSISFGGMLIFAMMLGLVSDAISEKFDSLRKGK 314
Query: 343 SEVIERNHVLILGWSDKLGSLLKQLAIANKSVGGGVIVVLAEKEKEEMEMDIAKLEFDFM 402
SEV+E+NH LILGWSDKLGSLL QLAIAN+S+GGG I V+AE++KE+ME+DI K+EFDF
Sbjct: 315 SEVVEQNHTLILGWSDKLGSLLNQLAIANESLGGGTIAVMAERDKEDMELDIGKMEFDFK 374
Query: 403 GTSVICRSGSPLILADLKKVSVSKARAIIVLAADENADQSDARALRVVLSLAGVKEGLRG 462
GTSVICRSGSPLILADLKKVSVSKAR IIVLA D NADQSDARALR VLSL GVKEGLRG
Sbjct: 375 GTSVICRSGSPLILADLKKVSVSKARTIIVLAEDGNADQSDARALRTVLSLTGVKEGLRG 434
Query: 463 HVVVEMSDLDNEPLVKLVGGELIETVVAHDVIGRLMIQCALQPGLAQIWEDILGFENAEF 522
H+VVEMSDLDNE LVKLVGG+L+ETVVAHDVIGRLMIQCA QPGLAQIWEDILGFEN EF
Sbjct: 435 HIVVEMSDLDNEVLVKLVGGDLVETVVAHDVIGRLMIQCARQPGLAQIWEDILGFENCEF 494
Query: 523 YIKRWPELDDLLFKDILISFPDAIPCGVKVAADGGKIVINPDDNYVLRDGDEVLVIAEDD 582
YIKRWP+LD +LF+D+LISFP AIPCG+KVA+ GGKI++NPDD+YVL++GDEVLVIAEDD
Sbjct: 495 YIKRWPQLDGMLFEDVLISFPAAIPCGIKVASYGGKIILNPDDSYVLQEGDEVLVIAEDD 554
Query: 583 DTYAPGPLPEVRKGYFPRIRDPPKYPEKILFCGWRRDIDDMIMVLEAFLAPGSELWMFNE 642
DTYAP PLP VR+G P+ PK PE+ILFCGWRRD++DMI VL+A LAP SELWMFN+
Sbjct: 555 DTYAPAPLPMVRRGSLPKDFVYPKSPERILFCGWRRDMEDMITVLDASLAPDSELWMFND 614
Query: 643 VPEKERERKLAAGELDVFGLENIKLVHREGNAVIRRHLESLPLETFDSILILADESVEDS 702
VPEKERE+KL G LD+ LENI LV+REGNAVIRRHLESLPLE+FDSILILADESVEDS
Sbjct: 615 VPEKEREKKLIDGGLDISRLENISLVNREGNAVIRRHLESLPLESFDSILILADESVEDS 674
Query: 703 VAHSDSRSLATLLLIRDIQSRRLPYRDTKSTSLRLSGFSHNSWIREMQQASDKSIIISEI 762
+DSRSLATLLLIRDIQ+RRLPY ++ + FS SWI EM+QASDK++IISEI
Sbjct: 675 AIQADSRSLATLLLIRDIQARRLPY-VAMASQTQGGNFSKGSWIGEMKQASDKTVIISEI 733
Query: 763 LDSRTRNLVSVSRISDYVLSNELVSMALAMVAEDKQINRVLEELFAEEGNEMCIKPAEFY 822
LD RT+NL+S+S+ISDYVLSNELVSMALAMVAED+QIN VLEELFAEEGNEM I+ A+ Y
Sbjct: 734 LDPRTKNLLSMSKISDYVLSNELVSMALAMVAEDRQINDVLEELFAEEGNEMHIRQADIY 793
Query: 823 LFDQEELCFYDIMIRGRTRKEIVIGYRLANQERAIINPSEKSVPRKWSLDDVFVVLASGE 882
L + EE+ FY+IM+R R R+EI+IGYRLAN ERA+INP K+ RKWSL DVFVV+ E
Sbjct: 794 LREGEEMSFYEIMLRARQRREILIGYRLANAERAVINPPAKTGRRKWSLKDVFVVITEKE 853
>ref|NP_102608.1| probable secreted protein [Mesorhizobium loti MAFF303099]
gi|14021783|dbj|BAB48394.1| probable secreted protein
[Mesorhizobium loti MAFF303099]
Length = 633
Score = 256 bits (653), Expect = 3e-66
Identities = 202/669 (30%), Positives = 327/669 (48%), Gaps = 67/669 (10%)
Query: 234 LKKRVAYMVDVFFSIYPYAKLLALLCATLFLIAFGGLALYAVT-----GG---SMAEALW 285
L R+ Y D + P A L+ L LI A AVT GG + EA W
Sbjct: 7 LGTRLRYGFDKSMAAGPIA-LIGWLAVVSLLIIIAAAAFLAVTRIAPEGGEPLNFFEAFW 65
Query: 286 HSWTYVADAGNHAETEGTGQRIVSVSISAGGMLIFAMMLGLVSDAISEKVDSLRKGKSEV 345
S D+G G R+V + ++ G+ + + ++G++S + K+D LRKG+S V
Sbjct: 66 ESLMRTLDSGTMGGDTGWAFRLVMLVVTLAGIFVVSALIGVLSAGVDGKLDELRKGRSRV 125
Query: 346 IERNHVLILGWSDKLGSLLKQLAIANKSVGGGVIVVLAEKEKEEMEMDIAKLEFDFMGTS 405
+E +H +IL WS + ++ +L IAN S IVV+A +K ME +IA T
Sbjct: 126 LESDHTIILNWSPSIFDVISELVIANASRRRPRIVVMANMDKVAMEDEIAAKVGKLGNTR 185
Query: 406 VICRSGSPLILADLKKVSVSKARAIIVLAADENADQSDARALRVVLSLAGVKEGLRG--- 462
+ICRSG P L DL V+ +R++IVL+ D D D++ ++ VL+L V + R
Sbjct: 186 IICRSGDPTDLYDLAIVNPQTSRSVIVLSPD--GDDPDSQVIKTVLAL--VNDPSRRTDP 241
Query: 463 -HVVVEMSDLDNEPLVKLVGGELIETVVAHDVIGRLMIQCALQPGLAQIWEDILGFENAE 521
++ E+ D N + ++VGG ++ V+A +I R+++ + Q GL+ ++ ++L F+ E
Sbjct: 242 YNIAAEIRDGKNAEVARVVGGAEVQLVLADQLISRIVVHSSRQSGLSGVYSELLDFDGCE 301
Query: 522 FYIKRWPELDDLLFKDILISFPDAIPCGVKVAADGGKIVINPDDNYVLRDGDEVLVIAED 581
Y PEL F + ++++ G + GG++ +NP V+ ++IAED
Sbjct: 302 IYTTTQPELAGKTFGEAVMAYEHCALIG--LCDQGGRVNLNPPSELVIGKDMRAIIIAED 359
Query: 582 DDTYAPGPLPEVRKGYFPRIRDP---PKYPEKILFCGWRRDIDDMIMVLEAFLAPGSELW 638
D PG K IRDP PE+ L GW R + L ++APGS L
Sbjct: 360 DAAIRPGSAG--IKIDTAAIRDPRPVEAKPERTLILGWNRRGPIITYELSRYVAPGSILT 417
Query: 639 MFNEVPEKERERKLAAGELDVFGLENIKLVHREGNAVIRRHLESLPLETFDSILILADES 698
+ + P E+E AG L V G +N+ + R + L SL + ++D +L+L S
Sbjct: 418 IAADTPGLEQE---VAG-LPVAG-DNLSVSCRITDTSSSTALSSLDVPSYDHVLVLG-YS 471
Query: 699 VEDSVAHSDSRSLATLLLIRDIQSRRLPYRDTKSTSLRLSGFSHNSWIREMQQASDKSII 758
+ +D+R+L TLL +R I L +S I
Sbjct: 472 ETMAAQPADTRTLVTLLHLRKI---------ADDAGLHIS-------------------I 503
Query: 759 ISEILDSRTRNLVSVSRISDYVLSNELVSMALAMVAEDKQINRVLEELFAEEGNEMCIKP 818
+SE++D R R L +V++ D+V+SN LVS+ LA +E++ + + ++L E+G+E+ ++P
Sbjct: 504 VSEMIDVRNRELAAVTKADDFVVSNRLVSLMLAQASENQYLAAIFDDLLDEQGSEIYMRP 563
Query: 819 AEFYLFDQEELCFYDIMIRGRTRKEIVIGYRLAN----QERAI----INPSEKSVPRKWS 870
Y+ + L F+ I R R EI IGYR +RA+ +NP KS +
Sbjct: 564 VADYVAIDQPLTFWTIAESARLRGEIAIGYRRIRAGDADQRALGGVTVNPL-KSEALTYL 622
Query: 871 LDDVFVVLA 879
+D +VLA
Sbjct: 623 PEDRIIVLA 631
>emb|CAE45687.1| hypothetical protein [Streptomyces parvulus]
Length = 638
Score = 210 bits (534), Expect = 2e-52
Identities = 167/620 (26%), Positives = 290/620 (45%), Gaps = 56/620 (9%)
Query: 279 SMAEALWHSWTYVADAGNHAETEGTGQR-IVSVSISAGGMLIFAMMLGLVSDAISEKVDS 337
S+AE L W + GT R ++SV ++ +L + ++GL++ A++E++ S
Sbjct: 56 SLAERLAEVWRLTGETLRLGGATGTPLRAMLSVLLALVTLLYVSTLVGLITTALTERLTS 115
Query: 338 LRKGKSEVIERNHVLILGWSDKLGSLLKQLAIANKSVGGGVIVVLAEKEKEEMEMDIAKL 397
LR+G+S V+E+ H ++LGWS+++ +++ +L AN + G +VVLA+++K ME +
Sbjct: 116 LRRGRSTVLEQGHAVVLGWSEQVFTVVSELVAANVNQRGAAVVVLADRDKTVMEESLGTK 175
Query: 398 EFDFMGTSVICRSGSPLILADLKKVSVSKARAIIVLAADENADQSDARALRVVLSLAGVK 457
GT +ICRSG A L S + A ++VL DE +DA ++ +L+L
Sbjct: 176 VGSCGGTRLICRSGPTTDPAVLPLTSPATAGVVLVLPPDE--PHADAEVVKTLLALRAAL 233
Query: 458 EGL--RGHVVVEMSDLDNEPLVKLVGGELIETVVAHDVIGRLMIQCALQPGLAQIWEDIL 515
G R VV + D L G + + V RL++Q A +PG++ + ++L
Sbjct: 234 AGAKPRPPVVAAVRDDRYRLAACLAAGPDGVVLESDTVTARLIVQAARRPGISLVHRELL 293
Query: 516 GFENAEFYIKRWPELDDLLFKDILISFPDAIPCGVKVAADGGKIVINPDDNYVLRDGDEV 575
F EFY+ P L F ++L+S+ G+ G ++NP + D +
Sbjct: 294 DFAGDEFYLISEPALTGRPFGEVLLSYSTTSVVGLM---RGCTPLLNPPPTTPVAPDDLL 350
Query: 576 LVIAEDDDTYAPGPLPEVRKGYFPRIRDPPKYP-EKILFCGWRRDIDDMIMVLEAFLAPG 634
+VI DDDT E + R P P E+IL GW R ++ L PG
Sbjct: 351 VVITGDDDTARLDDCAESVEKAAVASRPPTPAPAERILLLGWNRRAPLVVDQLHRRARPG 410
Query: 635 SELWMFNEVPEKERERKLAAGELDVFGLEN----IKLVHREGNAVIRRHLESLPLETFDS 690
S + + E P + R+++ E D EN + L G+ L L + ++DS
Sbjct: 411 SAVDVVAE-PGEATIREISEAEADSGNGENGGNGLSLALHHGDITRPETLRRLDVHSYDS 469
Query: 691 ILILA-DESVEDSVAHSDSRSLATLLLIRDIQS---RRLPYRDTKSTSLRLSGFSHNSWI 746
+++L D + D+R+L TLLL+R ++ R LP
Sbjct: 470 VIVLGRDPAPGQPPDDPDNRTLVTLLLLRQLEEATGRELP-------------------- 509
Query: 747 REMQQASDKSIIISEILDSRTRNLVSVSRISDYVLSNELVSMALAMVAEDKQINRVLEEL 806
+++E++D R R L + +D ++S +L+ + ++ +++++ + V EEL
Sbjct: 510 -----------VVTELIDDRNRALAPIGPGADVIISGKLIGLLMSQISQNRHLAAVFEEL 558
Query: 807 FAEEGNEMCIKPAEFYLFDQEELCFYDIMIRGRTRKEIVIGYR------LANQERAIINP 860
F+ EG + ++PA YL F ++ R R E IGYR INP
Sbjct: 559 FSAEGAGVRLRPATDYLLPGSTTSFATVVAAARRRGECAIGYRDHADASTRPHYGVRINP 618
Query: 861 SEKSVPRKWSLDDVFVVLAS 880
++ R+W+ +D VV+ +
Sbjct: 619 PKRE-RRRWTAEDEVVVIGT 637
>dbj|BAC68466.1| hypothetical protein [Streptomyces avermitilis MA-4680]
gi|29827297|ref|NP_821931.1| hypothetical protein SAV756
[Streptomyces avermitilis MA-4680]
Length = 656
Score = 203 bits (517), Expect = 2e-50
Identities = 180/642 (28%), Positives = 298/642 (46%), Gaps = 61/642 (9%)
Query: 255 LALLCATLFLIAFGGLALYAVTGG--SMAEALWHSWTYVADAGNHAETEGTGQR-IVSVS 311
L ++C + ++ L ++ G S++ L W A+ GT R ++S
Sbjct: 35 LVIICLAV-VVPVSALLVWTDPGSPRSLSGRLAAVWRSSAETLRLGTVTGTPLRMLLSAL 93
Query: 312 ISAGGMLIFAMMLGLVSDAISEKVDSLRKGKSEVIERNHVLILGWSDKLGSLLKQLAIAN 371
+ +L + ++G+++ ++E++ L +G+S V+E+ H ++LGWSD++ +++ +L A
Sbjct: 94 LGLVALLCVSTLVGVITTGLAERLAELSRGRSTVLEQGHAVVLGWSDQVSTVVGELVAAQ 153
Query: 372 KSVGGGVIVVLAEKEKEEMEMDIAKLEFDFMGTSVICRSGSPLILADLKKVSVSKARAII 431
S +VVLAE++K EME +A T ++CRSG L VS A ++
Sbjct: 154 SSYRPRAVVVLAERDKTEMERALAAHVGPAGRTRLVCRSGPASDPGVLALVSPQTASTVL 213
Query: 432 VLAADENADQSDARALRVVLSLAGV-KEGLRGHVVVE--MSDLDNEPLVKLVGGELIETV 488
VL + E +DA LRV+L+L V EG G V+ + D P +L G +
Sbjct: 214 VLPSGE--PTADAEVLRVLLALRAVLGEGTGGPPVLAAVLDDRYRAP-ARLAAGPRGTVL 270
Query: 489 VAHDVIGRLMIQCALQPGLAQIWEDILGFENAEFYIKRWPELDDLLFKDILISFPDAIPC 548
V RL+ QC +PGL+ + D+L F EF++ F L+ A C
Sbjct: 271 ETDTVTARLIAQCVGRPGLSLVLRDLLDFAGDEFHLAEATAFHGGPFGAALLG--HATSC 328
Query: 549 GVKVAADGGKIVINPDDNYVLRDGDEVLVIAEDDDTYAPGPLPE-VRKGYFPRIRDPPKY 607
V + G+ ++NP ++ G ++V+ DD + P V + PP+
Sbjct: 329 VVGLLTAEGRTLLNPPAATLVAPGSRLVVLTRDDGSARPEDCRHLVEPSAIAMAQPPPED 388
Query: 608 PEKILFCGWRRDIDDMIMVLEAFLAPGSELWMFNE--VPEKERERKLAAGELDVFGLENI 665
+L GW R ++ L PGS L + + VP RE + A L V ++
Sbjct: 389 AAHLLLLGWNRRAPLVVNQLRRTARPGSVLDVVTDRAVPGPTREPESGAARLAV-RFQSA 447
Query: 666 KLVHREGNAVIRRHLESLPLETFDSILILADESVEDSVAHSDSRSLATLLLIRDIQSRRL 725
L E L L L +D +++L E D D R+L TLL +R ++ R
Sbjct: 448 DLSRPE-------TLLGLDLAPYDGLIVLGPEP-GDGPDRPDDRTLVTLLALRLLEER-- 497
Query: 726 PYRDTKSTSLRLSGFSHNSWIREMQQASDKSIIISEILDSRTRNLVSVSRISDYVLSNEL 785
+G REM+ +I+E+ D R R L V+ SD ++S EL
Sbjct: 498 ------------TG-------REMR-------VITELTDDRNRPLAPVNPGSDVIVSGEL 531
Query: 786 VSMALAMVAEDKQINRVLEELFAEEGNEMCIKPAEFYLFDQEELCFYDIMIRGRTRKEIV 845
+ LA +A+++ + V +ELF+ +G+ +C++ A Y+ E F ++ R R E
Sbjct: 532 TGLLLAQIAQNRHLAPVFDELFSADGSTICLRQAGHYVRTGCETTFATVVAAARDRGECA 591
Query: 846 IGYRLANQERAI-------INPSEKSVPRKWSLDDVFVVLAS 880
IGYR + RAI +NP +KS R+WS DD VV+A+
Sbjct: 592 IGYR-RHDRRAIPPDHGIRLNP-DKSERREWSADDQVVVVAT 631
>emb|CAC01587.1| putative lipoprotein [Streptomyces coelicolor A3(2)]
gi|21225466|ref|NP_631245.1| putative lipoprotein
[Streptomyces coelicolor A3(2)]
Length = 672
Score = 187 bits (475), Expect = 1e-45
Identities = 156/614 (25%), Positives = 270/614 (43%), Gaps = 57/614 (9%)
Query: 279 SMAEALWHSWTYVADAGNHAETEGTGQR-IVSVSISAGGMLIFAMMLGLVSDAISEKVDS 337
S+ E L W A+ G R ++SV + +L + ++G+++ + ++++
Sbjct: 57 SLTERLVAVWRTSAETLRLGGVTGAPLRMLLSVFLGLIALLCVSTLVGVITTGLGDRLEE 116
Query: 338 LRKGKSEVIERNHVLILGWSDKLGSLLKQLAIANKSVGGGVIVVLAEKEKEEMEMDIAKL 397
LR+G+S V+E+ H ++LGWSD++ +++ ++ I+ G + VLA+++ M D+
Sbjct: 117 LRRGRSRVLEKGHAVVLGWSDQVFTVVGEMVISQVGRVRGAVAVLADRDSAVMASDLNAA 176
Query: 398 EFDFMGTSVICRSGSPLILADLKKVSVSKARAIIVLAADENADQSDARALRVVLSL-AGV 456
G V+CR+G+P+ A L ++ + A ++VL D++AD DA +RV+L+L A +
Sbjct: 177 LGVTRGVRVVCRTGAPIDPAALALLTPAAAHCVLVLPGDDDAD--DAEVVRVLLALRALL 234
Query: 457 KEGLRGHVVVEMSDLDNEPLVKLVGGELIETVVAHDVIGRLMIQCALQPGLAQIWEDILG 516
G VV + D +L G + RL++Q A PGL + D+L
Sbjct: 235 GAGAGPPVVAAVRDERFLTAARLAAGPRGFVLDVESTAARLLVQAARHPGLVRALRDLLD 294
Query: 517 FENAEFYIKRWPELDDLLFKDILISFPDAIPCGVKVAADGGKIVINPDDNYVLRDGDEVL 576
AEF++ P+ L F +I + +A C V A G+ ++ P GD ++
Sbjct: 295 LTGAEFHVVHAPDALGLTFAEISSRYEEA--CAVGYLAADGRALLTPASGARCGPGDRLI 352
Query: 577 VIAEDDDTYAPGPLPEVRKGYFPRIR----DPPKYPEKILFCGWRRDIDDMIMVLEAFLA 632
V+A DD P P + D + K L GW R ++ L
Sbjct: 353 VVARDD---RPPVAKREGTAVDPTVMADRPDRQRSFSKTLLLGWNRRAPLVMESLSRTAQ 409
Query: 633 PGSELWMFNEVPEKERERKLAAGELDVFGLENIKLVHREGNAVIRRHLESLPLETFDSIL 692
PGS L V + + AG + E + + + G L +L L +DS++
Sbjct: 410 PGSHL----HVVSGADDGPVTAGAVGPTDDERVAVTYHVGEPTRPDTLRALDLFGYDSVI 465
Query: 693 ILADESVEDSVAHSDSRSLATLLLIRDIQ---SRRLPYRDTKSTSLRLSGFSHNSWIREM 749
LA ++ A D + L TLL +R ++ R LP
Sbjct: 466 ALAPDAGPHR-ARPDDQLLLTLLNLRAVEEETGRALP----------------------- 501
Query: 750 QQASDKSIIISEILDSRTRNLVSVSRISDYVLSNELVSMALAMVAEDKQINRVLEELFAE 809
+++E+ D R+ L + D V+ EL S+ + +A++ + V EELFA
Sbjct: 502 --------VVAELTDHRSLALAPLGPEGDAVVRGELTSLVMTRIAQNTGMAAVFEELFAA 553
Query: 810 EGNEMCIKPAEFYLFDQEELCFYDIMIRGRTRKEIVIGY-----RLANQERAIINPSEKS 864
G + ++PA Y+ F ++ TR E VIGY R +E + K+
Sbjct: 554 RGGALALRPASHYVLPGRVASFATVVASALTRGECVIGYRAHDARTPRREADVRLAPGKA 613
Query: 865 VPRKWSLDDVFVVL 878
R WS D +V+
Sbjct: 614 ERRVWSSSDEILVV 627
>ref|NP_568628.1| phosphotransferase-related [Arabidopsis thaliana]
gi|16974323|gb|AAL31146.1| AT5g02940/F9G14_250
[Arabidopsis thaliana] gi|15450498|gb|AAK96542.1|
AT5g02940/F9G14_250 [Arabidopsis thaliana]
Length = 817
Score = 186 bits (472), Expect = 3e-45
Identities = 158/685 (23%), Positives = 317/685 (46%), Gaps = 96/685 (14%)
Query: 254 LLALLCATLFLIAFGGLALYAVTGG-SMAEALWHSWTYVADAGNHAETEGTGQRIVSVSI 312
L+AL+ A + + GGL + + + LW +W + + H + + +R++ +
Sbjct: 166 LVALMIACVSFVIIGGLLFFKFRKDLPLEDCLWEAWACLISSSTHLKQKTRIERVIGFVL 225
Query: 313 SAGGMLIFAMMLGLVSDAISEKVDSLRKG-KSEVIERNHVLILGWSDKLGSLLKQL---- 367
+ G+L ++ +L +++ + LR+G + +V+E +H++I G + L +LKQL
Sbjct: 226 AIWGILFYSRLLSTMTEQFRYNMTKLREGAQMQVLEADHIIICGINSHLPFILKQLNSYH 285
Query: 368 ----AIANKSVGGGVIVVLAEKEKEEMEMDIAKLEFDFMGTSVICRSGSPLILADLKKVS 423
+ + ++++++ +++M+ DF ++ +S S + ++ +
Sbjct: 286 EHAVRLGTATARKQRLLLMSDTPRKQMDKLAEAYSKDFNHIDILTKSCSLNLTKSFERAA 345
Query: 424 VSKARAIIVLAADENADQSDARALRVVLSLAGVKEGLRGHVVVEMSDLDNEPLVKLVGGE 483
S ARAII+L + + D A VL+L +++ +VE+S + L+K + G
Sbjct: 346 ASMARAIIILPTKGDRYEVDTDAFLSVLALQPIQKMESIPTIVEVSSPNTYDLLKSISGL 405
Query: 484 LIETVVAHDVIGRLMIQCALQPGLAQIWEDILGFENAEFYIKRWPELDDLLFKDILISFP 543
+E V +V +L +QC+ Q L +I+ +L + F + +P L ++ + + F
Sbjct: 406 KVEPV--ENVTSKLFVQCSRQKDLIKIYRHLLNYSKNVFNLCSFPNLVGTKYRQLRLGFQ 463
Query: 544 DAIPCGVKVAADGGKIVINPDDNYVLRDGDEVLVIA-----EDDDTYAPGPLPEV----- 593
+ + CG+ GK+ +P+DN L + D++L IA + Y L +
Sbjct: 464 EVVVCGL---LRDGKVNFHPNDNEELMETDKLLFIAPLNWKKKQLLYTDMKLENITVPTD 520
Query: 594 -RKGYFPRIR------------------DPPKYP-EKILFCGWRRDIDDMIMVLEAFLAP 633
RK F + R D K P E IL GWR D+ MI + +L P
Sbjct: 521 TRKQVFEKKRSRLSKIIMRPRKSLSKGSDSVKGPTESILLLGWRGDVVQMIEEFDNYLGP 580
Query: 634 GSELWMFNEVPEKERERKLAAGELDVFGLENIKLVHREGN---------AVIR-----RH 679
GS + + ++V ++R R + ++NI++ H+ GN ++R R
Sbjct: 581 GSSMEILSDVSLEDRRR--VGDSIGSVKIKNIQVSHKVGNPLNYDTLKQTIMRMKSKYRK 638
Query: 680 LESLPLETFDSILILADES-VEDSVAHSDSRSLATLLLIRDIQSRRLPYRDTKSTSLRLS 738
+++PL +IL+++D + + +D +S +LLL I + +L
Sbjct: 639 GKNIPL----TILVISDRDWLLGDPSRADKQSAYSLLLAESICN-------------KLG 681
Query: 739 GFSHNSWIREMQQASDKSIIISEILDSRT-RNLVSVSRISDYVLSNELVSMALAMVAEDK 797
HN + SEI+DS+ + + + ++ + E++S+ A VAE+
Sbjct: 682 VKVHN--------------LASEIVDSKLGKQITGLKPSLTFIAAEEVMSLVTAQVAENS 727
Query: 798 QINRVLEELFAEEGNEMCIKPAEFYLFDQEELCFYDIMIRGRTRKEIVIGYRLANQERAI 857
++N V +++ +G+E+ +K E Y+ + E F ++ R R+E+ IGY ++ +
Sbjct: 728 ELNEVWKDILDADGDEIYVKDVELYMKEGENPSFTELSERAWLRREVAIGYIKGGKK--M 785
Query: 858 INPSEKSVPRKWSLDDVFVVLASGE 882
INP K+ P +DD +V++ E
Sbjct: 786 INPVPKNEPLSLEMDDSLIVISELE 810
>gb|AAL36360.1| unknown protein [Arabidopsis thaliana] gi|30679833|ref|NP_195914.2|
expressed protein [Arabidopsis thaliana]
Length = 813
Score = 182 bits (463), Expect = 3e-44
Identities = 156/685 (22%), Positives = 317/685 (45%), Gaps = 98/685 (14%)
Query: 255 LALLCATLFLIAFGGLALYAVTGG-SMAEALWHSWTYVADAGNHAETEGTGQRIVSVSIS 313
+ LL + GGL + S+ + LW +W + +A H E + +R++ ++
Sbjct: 163 VVLLITCFSFVIIGGLFFFKFRKDTSLEDCLWEAWACLVNADTHLEQKTRFERLIGFVLA 222
Query: 314 AGGMLIFAMMLGLVSDAISEKVDSLRKGKS-EVIERNHVLILGWSDKLGSLLKQL----- 367
G++ ++ +L +++ + +R+G +V+E +H++I G + L +LKQL
Sbjct: 223 IWGIVFYSRLLSTMTEQFRYHMKKVREGAHMQVLESDHIIICGINSHLPFILKQLNSYQQ 282
Query: 368 ---AIANKSVGGGVIVVLAEKEKEEMEMDIAKLEFDFMGTSVICRSGSPLILADLKKVSV 424
+ + ++++++ ++EM+ DF ++ +S S + ++ +
Sbjct: 283 HAVRLGTTTARKQTLLLMSDTPRKEMDKLAEAYAKDFDQLDILTKSCSLNMTKSFERAAA 342
Query: 425 SKARAIIVLAADENADQSDARALRVVLSLAGVKEGLRGHVVVEMSDLDNEPLVKLVGGEL 484
ARAII+L + + D A VL+L +++ +VE+S + L+K + G
Sbjct: 343 CMARAIIILPTKGDRYEVDTDAFLSVLALEPIQKMESIPTIVEVSSSNMYDLLKSISGLK 402
Query: 485 IETVVAHDVIGRLMIQCALQPGLAQIWEDILGFENAEFYIKRWPELDDLLFKDILISFPD 544
+E V + +L +QC+ Q L +I+ +L + F + +P L + ++ + + F +
Sbjct: 403 VEPV--ENSTSKLFVQCSRQKDLIKIYRHLLNYSKNVFNLCSFPNLTGMKYRQLRLGFQE 460
Query: 545 AIPCGVKVAADGGKIVINPDDNYVLRDGDEVLVIA-------------------EDDDTY 585
+ CG+ GK+ +P+D+ L + D++L IA E DDT
Sbjct: 461 VVVCGI---LRDGKVNFHPNDDEELMETDKLLFIAPLKKDFLYTDMKTENMTVDETDDTR 517
Query: 586 APGPLPEVRKGYFPRIRDPP-----------KYP-EKILFCGWRRDIDDMIMVLEAFLAP 633
+ E +K +I P K P E IL GWR D+ +MI +++L P
Sbjct: 518 KQ--VYEEKKSRLEKIITRPSKSLSKGSDSFKGPKESILLLGWRGDVVNMIKEFDSYLGP 575
Query: 634 GSELWMFNEVPEKER---ERKLAAGELDVFGLENIKLVHREGNAV-----------IRRH 679
GS L + ++VP ++R ++ +A G+ ++NI++ H GN + ++
Sbjct: 576 GSSLEILSDVPLEDRRGVDQSIATGK-----IKNIQVSHSVGNHMDYDTLKESIMHMQNK 630
Query: 680 LESLPLETFDSILILAD-ESVEDSVAHSDSRSLATLLLIRDIQSRRLPYRDTKSTSLRLS 738
E + +I++++D + + + +D +S TLLL I + +L
Sbjct: 631 YEKGEEDIRLTIVVISDRDLLLGDPSRADKQSAYTLLLAETICN-------------KLG 677
Query: 739 GFSHNSWIREMQQASDKSIIISEILDSRT-RNLVSVSRISDYVLSNELVSMALAMVAEDK 797
HN + SEI+D++ + + + ++ + E++S+ A VAE+
Sbjct: 678 VKVHN--------------LASEIVDTKLGKQITRLKPSLTFIAAEEVMSLVTAQVAENS 723
Query: 798 QINRVLEELFAEEGNEMCIKPAEFYLFDQEELCFYDIMIRGRTRKEIVIGYRLANQERAI 857
++N V +++ EG+E+ +K E Y+ + E F ++ R R+E+ IGY ++ I
Sbjct: 724 ELNEVWKDILDAEGDEIYVKDIELYMKEGENPSFTELSERAWLRREVAIGYIKGGKK--I 781
Query: 858 INPSEKSVPRKWSLDDVFVVLASGE 882
INP K+ P ++D +V++ E
Sbjct: 782 INPVPKTEPVSLEMEDSLIVISELE 806
>emb|CAB86048.1| putative protein [Arabidopsis thaliana] gi|11281135|pir||T48315
hypothetical protein F9G14.250 - Arabidopsis thaliana
Length = 776
Score = 176 bits (447), Expect = 2e-42
Identities = 153/684 (22%), Positives = 314/684 (45%), Gaps = 88/684 (12%)
Query: 255 LALLCATLFLIAFGGLALYAVTGG-SMAEALWHSWTYVADAGNHAETEGTGQRIVSVSIS 313
+ LL + GGL + S+ + LW +W + +A H E + +R++ ++
Sbjct: 118 VVLLITCFSFVIIGGLFFFKFRKDTSLEDCLWEAWACLVNADTHLEQKTRFERLIGFVLA 177
Query: 314 AGGMLIFAMMLGLVSDAISEKVDSLRKGKS-EVIERNHVLILGWSDKLGSLLKQL----- 367
G++ ++ +L +++ + +R+G +V+E +H++I G + L +LKQL
Sbjct: 178 IWGIVFYSRLLSTMTEQFRYHMKKVREGAHMQVLESDHIIICGINSHLPFILKQLNSYQQ 237
Query: 368 ---AIANKSVGGGVIVVLAEKEKEEMEMDIAKLEFDFMGTSVICRSGSPLILADLKKVSV 424
+ + ++++++ ++EM+ DF ++ +S + + ++ +
Sbjct: 238 HAVRLGTTTARKQTLLLMSDTPRKEMDKLAEAYAKDFDQLDILTKSLN--MTKSFERAAA 295
Query: 425 SKARAIIVLAADENADQSDARALRVVLSLAGVKEGLRGHVVVEMSDLDNEPLVKLVGGEL 484
ARAII+L + + D A VL+L +++ +VE+S + L+K + G
Sbjct: 296 CMARAIIILPTKGDRYEVDTDAFLSVLALEPIQKMESIPTIVEVSSSNMYDLLKSISGLK 355
Query: 485 IETVVAHDVIGRLMIQCALQPGLAQIWEDILGFENAEFYIKRWPELDDLLFKDILISFPD 544
+E V + +L +QC+ Q L +I+ +L + F + +P L + ++ + + F +
Sbjct: 356 VEPV--ENSTSKLFVQCSRQKDLIKIYRHLLNYSKNVFNLCSFPNLTGMKYRQLRLGFQE 413
Query: 545 AIPCGVKVAADGGKIVINPDDNYVLRDGDEVLVIA-------------------EDDDTY 585
+ CG+ GK+ +P+D+ L + D++L IA E DDT
Sbjct: 414 VVVCGI---LRDGKVNFHPNDDEELMETDKLLFIAPLKKDFLYTDMKTENMTVDETDDTR 470
Query: 586 APGPLPEVRKGYFPRIRDPP-----------KYP-EKILFCGWRRDIDDMIMVLEAFLAP 633
+ E +K +I P K P E IL GWR D+ +MI +++L P
Sbjct: 471 KQ--VYEEKKSRLEKIITRPSKSLSKGSDSFKGPKESILLLGWRGDVVNMIKEFDSYLGP 528
Query: 634 GSELWMFNEVPEKER---ERKLAAGELDVFGLENIKLVHREGNAV-----------IRRH 679
GS L + ++VP ++R ++ +A G++ +NI++ H GN + ++
Sbjct: 529 GSSLEILSDVPLEDRRGVDQSIATGKI-----KNIQVSHSVGNHMDYDTLKESIMHMQNK 583
Query: 680 LESLPLETFDSILILADESVE-DSVAHSDSRSLATLLLIRDIQSRRLPYRDTKSTSLRLS 738
E + +I++++D + + +D +S TLLL I ++ L
Sbjct: 584 YEKGEEDIRLTIVVISDRDLLLGDPSRADKQSAYTLLLAETICNK-------------LG 630
Query: 739 GFSHNSWIREMQQASDKSIIISEILDSRTRNLVSVSRISDYVLSNELVSMALAMVAEDKQ 798
HN + K ++ D++ + + ++ + E++S+ A VAE+ +
Sbjct: 631 VKVHNLASEIVDTKLGKQLLKH---DTKPYQITRLKPSLTFIAAEEVMSLVTAQVAENSE 687
Query: 799 INRVLEELFAEEGNEMCIKPAEFYLFDQEELCFYDIMIRGRTRKEIVIGYRLANQERAII 858
+N V +++ EG+E+ +K E Y+ + E F ++ R R+E+ IGY ++ II
Sbjct: 688 LNEVWKDILDAEGDEIYVKDIELYMKEGENPSFTELSERAWLRREVAIGYIKGGKK--II 745
Query: 859 NPSEKSVPRKWSLDDVFVVLASGE 882
NP K+ P ++D +V++ E
Sbjct: 746 NPVPKTEPVSLEMEDSLIVISELE 769
>ref|NP_912647.1| Unknown protein [Oryza sativa (japonica cultivar-group)]
gi|22773250|gb|AAN06856.1| Unknown protein [Oryza sativa
(japonica cultivar-group)]
Length = 1293
Score = 150 bits (379), Expect = 2e-34
Identities = 168/737 (22%), Positives = 317/737 (42%), Gaps = 138/737 (18%)
Query: 229 KEDVPLKKRVAY--MVDVFFSI----YPY---------AKLLALLCATLFLIAFGGLALY 273
K+ VPLK V++ + D+ +SI Y + ++ L+ L+ GG +
Sbjct: 151 KKPVPLKLDVSFPQLPDIRWSISRLYYLFNSQLERNIALSIITLMITCFSLVVVGGFLFH 210
Query: 274 AVTGG--SMAEALWHSWTYVADAGNHAETEGTGQRIVSVSISAGGMLIFAMMLGLVSDAI 331
S+ E W +W + + H + +R++ ++ G+L ++ +L ++
Sbjct: 211 KFRKNQQSLEECFWEAWACLISSSTHLRQKTRIERVLGFFLAIWGILFYSRLLSATTEQF 270
Query: 332 SEKVDSLRKG-KSEVIERNHVLILGWSDKLGSLLKQL-AIANKSVGGGVIVVLAEKEKEE 389
++ +R+G + +VIE +H++I G + L S+L QL S+ G A K++
Sbjct: 271 RIQMHKVREGAQQQVIEDDHIIICGVNSHLPSILNQLNKFHESSIRLG--TATARKQRIL 328
Query: 390 MEMDIAKLEFDFMGTS---------VICRSGSPLILADLKKVSVSKARAIIVLAADENAD 440
+ D+ + + + +G S V +S S + ++ + +KA++II+L A
Sbjct: 329 LLSDLPRKQIEKLGDSFAKDLNHIDVFTKSCSLSLTKSFERAAANKAKSIIILPAKNERY 388
Query: 441 QSDARALRVVLSLAGVKEGLRGHVVVEMSDLDNEPLVKLVGGELIETVVAHDVIGRLMIQ 500
+ D A +L+L + + +VE S+ L+K + G ++ V +L +Q
Sbjct: 389 EVDTDAFLSLLALQSLPQIASIPTIVEASNSTTCDLLKSITGLNVQPV--EMAASKLFVQ 446
Query: 501 CALQPGLAQIWEDILGFENAEFYIKRWPELDDLLFKDILISFPDAIPCGVKVAADGGKIV 560
C+ Q EN F + + E+ + + D+ PDA+ CG+ G +
Sbjct: 447 CSRQK------------ENV-FNLFSFREVVGMKYVDVRRRIPDAVVCGI---FRSGMMH 490
Query: 561 INPDDNYVLRDGDEVLVIA------EDDDTYAPGP---------------------LPEV 593
+P ++ VL + D++L+IA T++ P EV
Sbjct: 491 FHPCEDEVLTEKDKLLLIAPVSWRRRAQSTFSNSPNGAQNSSHYSESTEGQRSSSMALEV 550
Query: 594 RKGYFPRIRDPPKYP------------EKILFCGWRRDIDDMIMVLEAFLAPGSELWMFN 641
+ IR P E +L GWR + DMI + +L PGS L + +
Sbjct: 551 NETRLNSIRKRPSKTLSKSNDYTLGPREHVLIVGWRPKVTDMIREYDNYLGPGSVLEILS 610
Query: 642 EVPEKERERKLAAGELDVFGLENIKLVHREG---------NAVI-----RRHLESLPLET 687
E P KER L L+NIK+ H+ G A+I R+H +++P
Sbjct: 611 ETPIKERSS--IVNPLMQKQLKNIKVNHQVGCPMNYDTLKEAIIKFKKSRKHDQNVPF-- 666
Query: 688 FDSILILADES-VEDSVAHSDSRSLATLLLIRDIQSRRLPYRDTKSTSLRLSGFSHNSWI 746
S+++++D+ + A D + TLLL +I + D K L
Sbjct: 667 --SVVVISDKDWLGGDTAQVDKQLAYTLLLAENICQK----HDIKVEHL----------- 709
Query: 747 REMQQASDKSIIISEILDSRTRNLVSVSRIS-DYVLSNELVSMALAMVAEDKQINRVLEE 805
+SEI+D+ +S + S ++ + E++S+ A VA ++N V ++
Sbjct: 710 ------------VSEIVDTGLGKQMSRIKPSLSFIGAEEVMSLVTAQVAGSSELNEVWKD 757
Query: 806 LFAEEGNEMCIKPAEFYLFDQEELCFYDIMIRGRTRKEIVIGYRLANQERAIINPSEKSV 865
+ EG+E+ IK FY+ + E++ F ++ R R+E+ +GY ++ INP+ K
Sbjct: 758 ILNAEGDEIYIKEIGFYMKEGEKISFSELTERAILRREVAVGY--VKGKKQYINPTNKLE 815
Query: 866 PRKWSLDDVFVVLASGE 882
+ + D +V++ E
Sbjct: 816 LLSFEMTDQLIVISEFE 832
>dbj|BAD95358.1| hypothetical protein [Arabidopsis thaliana]
Length = 481
Score = 137 bits (344), Expect = 2e-30
Identities = 123/510 (24%), Positives = 237/510 (46%), Gaps = 88/510 (17%)
Query: 420 KKVSVSKARAIIVLAADENADQSDARALRVVLSLAGVKEGLRGHVVVEMSDLDNEPLVKL 479
++ + ARAII+L + + D A VL+L +++ +VE+S + L+K
Sbjct: 6 ERAAACMARAIIILPTKGDRYEVDTDAFLSVLALEPIQKMESIPTIVEVSSSNMYDLLKS 65
Query: 480 VGGELIETVVAHDVIGRLMIQCALQPGLAQIWEDILGFENAEFYIKRWPELDDLLFKDIL 539
+ G +E V + +L +QC+ Q L +I+ +L + F + +P L + ++ +
Sbjct: 66 ISGLKVEPV--ENSTSKLFVQCSRQKDLIKIYRHLLNYSKNVFNLCSFPNLTGMKYRQLR 123
Query: 540 ISFPDAIPCGVKVAADGGKIVINPDDNYVLRDGDEVLVIA-------------------E 580
+ F + + CG+ GK+ +P+D+ L + D++L IA E
Sbjct: 124 LGFQEVVVCGI---LRDGKVNFHPNDDEELMETDKLLFIAPLKKDFLYTDMKTENMTVDE 180
Query: 581 DDDTYAPGPLPEVRKGYFPRIRDPP-----------KYP-EKILFCGWRRDIDDMIMVLE 628
DDT + E +K +I P K P E IL GWR D+ +MI +
Sbjct: 181 TDDTRKQ--VYEEKKSRLEKIITRPSKSLSKGSDSFKGPKESILLLGWRGDVVNMIKEFD 238
Query: 629 AFLAPGSELWMFNEVPEKER---ERKLAAGELDVFGLENIKLVHREGNAV---------- 675
++L PGS L + ++VP ++R ++ +A G+ ++NI++ H GN +
Sbjct: 239 SYLGPGSSLEILSDVPLEDRRGVDQSIATGK-----IKNIQVSHSVGNHMDYDTLKESIM 293
Query: 676 -IRRHLESLPLETFDSILILAD-ESVEDSVAHSDSRSLATLLLIRDIQSRRLPYRDTKST 733
++ E + +I++++D + + + +D +S TLLL I +
Sbjct: 294 HMQNKYEKGEEDIRLTIVVISDRDLLLGDPSRADKQSAYTLLLAETICN----------- 342
Query: 734 SLRLSGFSHNSWIREMQQASDKSIIISEILDSRT-RNLVSVSRISDYVLSNELVSMALAM 792
+L HN + SEI+D++ + + + ++ + E++S+ A
Sbjct: 343 --KLGVKVHN--------------LASEIVDTKLGKQITRLKPSLTFIAAEEVMSLVTAQ 386
Query: 793 VAEDKQINRVLEELFAEEGNEMCIKPAEFYLFDQEELCFYDIMIRGRTRKEIVIGYRLAN 852
VAE+ ++N V +++ EG+E+ +K E Y+ + E F ++ R R+E+ IGY
Sbjct: 387 VAENSELNEVWKDILDAEGDEIYVKDIELYMKEGENPSFTELSERAWLRREVAIGYIKGG 446
Query: 853 QERAIINPSEKSVPRKWSLDDVFVVLASGE 882
++ IINP K+ P ++D +V++ E
Sbjct: 447 KK--IINPVPKTEPVSLEMEDSLIVISELE 474
>gb|AAK82546.1| AT5g02940/F9G14_250 [Arabidopsis thaliana]
Length = 522
Score = 111 bits (278), Expect = 9e-23
Identities = 74/335 (22%), Positives = 162/335 (48%), Gaps = 15/335 (4%)
Query: 255 LALLCATLFLIAFGGLALYAVTGG-SMAEALWHSWTYVADAGNHAETEGTGQRIVSVSIS 313
+ LL + GGL + S+ + LW +W + +A H E + +R++ ++
Sbjct: 163 VVLLITCFSFVIIGGLFFFKFRKDTSLEDCLWEAWACLVNADTHLEQKTRFERLIGFVLA 222
Query: 314 AGGMLIFAMMLGLVSDAISEKVDSLRKGKS-EVIERNHVLILGWSDKLGSLLKQL----- 367
G++ ++ +L +++ + +R+G +V+E +H++I G + L +LKQL
Sbjct: 223 IWGIVFYSRLLSTMTEQFRYHMKKVREGAHMQVLESDHIIICGINSHLPFILKQLNSYQQ 282
Query: 368 ---AIANKSVGGGVIVVLAEKEKEEMEMDIAKLEFDFMGTSVICRSGSPLILADLKKVSV 424
+ + ++++++ ++EM+ DF ++ +S S + ++ +
Sbjct: 283 HAVRLGTTTARKQTLLLMSDTPRKEMDKLAEAYAKDFDQLDILTKSCSLNMTKSFERAAA 342
Query: 425 SKARAIIVLAADENADQSDARALRVVLSLAGVKEGLRGHVVVEMSDLDNEPLVKLVGGEL 484
ARAII+L + + D A VL+L +++ +VE+S + L+K + G
Sbjct: 343 CMARAIIILPTKGDRYEVDTDAFLSVLALEPIQKMESIPTIVEVSSSNMYDLLKSISGLK 402
Query: 485 IETVVAHDVIGRLMIQCALQPGLAQIWEDILGFENAEFYIKRWPELDDLLFKDILISFPD 544
+E V + +L +QC+ Q L +I+ +L + F + +P L + ++ + + F +
Sbjct: 403 VEPV--ENSTSKLFVQCSRQKDLIKIYRHLLNYSKNVFNLCSFPNLTGMKYRQLRLGFQE 460
Query: 545 AIPCGVKVAADGGKIVINPDDNYVLRDGDEVLVIA 579
+ CG+ GK+ +P+D+ L + D++L IA
Sbjct: 461 VVVCGI---LRDGKVNFHPNDDEELMETDKLLFIA 492
>ref|NP_683245.1| putative potassium channel protein [Thermosynechococcus elongatus
BP-1] gi|22296183|dbj|BAC10007.1| tll2456
[Thermosynechococcus elongatus BP-1]
Length = 452
Score = 82.8 bits (203), Expect = 5e-14
Identities = 112/447 (25%), Positives = 194/447 (43%), Gaps = 56/447 (12%)
Query: 156 LKDMKLQLCGQIDFCSRNGKTSIQEEVDDDDNADSRTIALYIVLFTLILPFVLYKYLDYL 215
L D+ L+LC ++ C R ++ R Y + ILP L L
Sbjct: 32 LLDLPLRLCFVVELCLRFAIARKKQ----------RFFRHYWLDLVAILPMPPQWALFRL 81
Query: 216 PQIINFLRRTESNKEDVPLKKRVAYMVDVFFSIYPYAKLLALLCATLFLIAFGGLALYAV 275
++ R + + + + + Y+ +Y A++ ALL L ++ FGGLA Y +
Sbjct: 82 LPLLRLPRAS------ILMNRNLHYIAPQTSGLYG-AQISALLIIVLIML-FGGLAFYII 133
Query: 276 TGGS------MAEALWHSWTYVADA---GNHAETEGTGQRIVSVSISAGGMLIFAMMLGL 326
G S + +ALW+S+ + A G + +T RIV++ + G+ +FA+ G+
Sbjct: 134 EGTSNPDIKTLGDALWYSFFSLVSAEPIGAYPQTHAG--RIVTLVVVLSGLTLFAVFTGV 191
Query: 327 VSDAISEKVDSLRKGKSEVIE--RNHVLILGWSDKLGSLLKQLAIANKSVGGGVIVVLAE 384
VS + +++ S+ K+ ++ RNH+++ GW+ +L++L + IV++AE
Sbjct: 192 VSAFMVQRLQSVMSIKNLDLDELRNHIILCGWNRSAPLVLEELQ-TDPQTRYAPIVMIAE 250
Query: 385 KEK----EEMEMDIAKLEFDFMGTSVICRSGSPLILADLKKVSVSKARAIIVLAADENAD 440
E+ E +D +L F SG + L+KV + A I+LA
Sbjct: 251 LEQLPLNELQTVDQNRLYF---------YSGDYTRIDVLEKVQIYHASRAILLADTSRPR 301
Query: 441 QSDARALRVVLSLAGVKEGLRGHVVVEMSDLDNEPLVKLVGGELIETVVAHDVIGRLMIQ 500
R R VL+ + E L + LD V+L + + VVA ++ G L+
Sbjct: 302 SDQDRDARTVLAALTI-EKLNPAIYTCAQLLDRNNNVQLQAAGVEDVVVADEMAGHLIGN 360
Query: 501 CALQPGLAQIWEDILGFE-NAEFYIKRWPELDDLLFKDI------LISFPDAIPCGVKVA 553
G ++ ++L +FY R P L K L + DA+ V+
Sbjct: 361 AVRNQGAMDVFAELLTVRIGNQFY--RLPLPSTLAGKTFWYAQQQLKAQDDALLIAVERR 418
Query: 554 ADG-GKIVINPDDNYVLRDGDEVLVIA 579
+G + +NP NY L+ GD V+VIA
Sbjct: 419 IEGRRRTYMNPPMNYELQVGDYVVVIA 445
Database: nr
Posted date: Jul 5, 2005 12:34 AM
Number of letters in database: 863,360,394
Number of sequences in database: 2,540,612
Lambda K H
0.319 0.137 0.394
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 1,507,308,903
Number of Sequences: 2540612
Number of extensions: 67401106
Number of successful extensions: 300411
Number of sequences better than 10.0: 319
Number of HSP's better than 10.0 without gapping: 84
Number of HSP's successfully gapped in prelim test: 279
Number of HSP's that attempted gapping in prelim test: 297992
Number of HSP's gapped (non-prelim): 1342
length of query: 882
length of database: 863,360,394
effective HSP length: 137
effective length of query: 745
effective length of database: 515,296,550
effective search space: 383895929750
effective search space used: 383895929750
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 80 (35.4 bits)
Medicago: description of AC140550.12


