

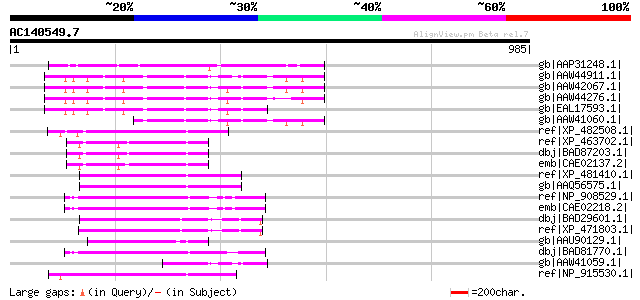
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC140549.7 + phase: 0
(985 letters)
Database: nr
2,540,612 sequences; 863,360,394 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
gb|AAP31248.1| transposase [Fusarium oxysporum f. sp. melonis] 203 2e-50
gb|AAW44911.1| conserved hypothetical protein [Cryptococcus neof... 141 9e-32
gb|AAW42067.1| conserved hypothetical protein [Cryptococcus neof... 139 4e-31
gb|AAW44276.1| conserved hypothetical protein [Cryptococcus neof... 132 7e-29
gb|EAL17593.1| hypothetical protein CNBM0450 [Cryptococcus neofo... 130 3e-28
gb|AAW41060.1| conserved hypothetical protein [Cryptococcus neof... 128 1e-27
ref|XP_482508.1| putative far-red impaired response protein [Ory... 104 2e-20
ref|XP_463702.1| putative far-red impaired response protein [Ory... 103 3e-20
dbj|BAD87203.1| far-red impaired response-like [Oryza sativa (ja... 103 3e-20
emb|CAE02137.2| OSJNBa0074L08.5 [Oryza sativa (japonica cultivar... 102 5e-20
ref|XP_481410.1| far-red impaired response protein -like [Oryza ... 100 2e-19
gb|AAQ56575.1| putative transposase [Oryza sativa (japonica cult... 100 2e-19
ref|NP_908529.1| P0702D12.15 [Oryza sativa (japonica cultivar-gr... 99 5e-19
emb|CAE02218.2| OSJNBb0002N06.8 [Oryza sativa (japonica cultivar... 98 2e-18
dbj|BAD29601.1| putative far-red impaired response protein [Oryz... 97 3e-18
ref|XP_471803.1| OSJNBa0050F15.2 [Oryza sativa (japonica cultiva... 96 8e-18
gb|AAU90129.1| unknown protein [Oryza sativa (japonica cultivar-... 94 3e-17
dbj|BAD81770.1| putative far-red impaired response protein [Oryz... 94 3e-17
gb|AAW41059.1| hypothetical protein CNA05530 [Cryptococcus neofo... 92 8e-17
ref|NP_915530.1| P0529E05.9 [Oryza sativa (japonica cultivar-gro... 91 2e-16
>gb|AAP31248.1| transposase [Fusarium oxysporum f. sp. melonis]
Length = 836
Score = 203 bits (517), Expect = 2e-50
Identities = 154/538 (28%), Positives = 256/538 (46%), Gaps = 31/538 (5%)
Query: 74 NRDELLEWVRRQANKAGFTIVTQRSS-LINPMFRLV--CERSGSHIVPKKKPKHANTGSR 130
+R+EL + A G+ V +RSS N ++ C+R G+ +P + T +R
Sbjct: 23 SREELRSAINAWAAPRGYAFVIKRSSKTANGRTHVIFNCDR-GAGRIPSLSDRRQTT-TR 80
Query: 131 KCGCLFMISGYQSKQTKEWGLNILNGVH--NHPMEPALE--GHILAGRLKEDDKKIVRDL 186
+ GCLF + +S W L G H H EP+ H +L ++ V L
Sbjct: 81 RTGCLFSVLAKESLCKTIWSLRHRPGPHFSQHNHEPSFSEVAHPTLRQLSRQEEITVNQL 140
Query: 187 TKSKMLPRNILIHLKNQRPHCMTNVKQVYNERQQIWKANRGDKKPLQFLISKLEEHNYTY 246
T + + P+ I L+ + + + +YN + + + + L +L E +
Sbjct: 141 TNAGIAPKEIGSFLRITS-NTLATQQDIYNCIAKGRRDLSKGQSNIHALADQLNEEGF-- 197
Query: 247 YSRTQL-ESNTIEDIFWAHPTSIKLFNNFPTVLVMDSTYKTNMYRMPMFEVVGVTSTDLT 305
++R L ES+ + + +AHP S++ +P VL++DSTYKTN ++MP+ ++VGV + T
Sbjct: 198 WNRICLDESSRVTAVLFAHPKSLEYLKTYPEVLILDSTYKTNRFKMPLLDIVGVDACQRT 257
Query: 306 YSVGFGFMTHEKEENFVWVLTMLRKLLSS-KMNMHKVIVTDRDMSLMKAVAHVFPESYAL 364
+ + F F++ E+E +F W L LR + + + VI+TDR ++ M AV+ FP S
Sbjct: 258 FCIAFAFLSGEEEGDFTWALQALRSVYEDHNIGLPSVILTDRCLACMNAVSSCFPGSALF 317
Query: 365 NCFFHVQANVKQRC----VLNCKYPLGFKKDGKEVSNRDVVKKIMNAWKAMVESPNQQLY 420
C +H+ V+ C P G + +E K+ N W +V S + +Y
Sbjct: 318 LCLWHINKAVQSYCRPAFTRGKDNPQGLGGESEE------WKEFFNFWHEIVASTTEDIY 371
Query: 421 ANALVEFKDS-CSDFPIFVDYAMTT-LDEVKDKIVRAWTDHVLHLGCRTTNRVESAHALL 478
L +FK D+ V Y + T LD K V+AW + LH T+RVE H+L+
Sbjct: 372 NERLEKFKKRYIPDYINEVGYILETWLDLYKKSFVKAWVNTHLHFEQYATSRVEGIHSLI 431
Query: 479 KKYLDNSVGDLGTCWEKIHHMLLLQFTAIQTSFGQNVCVLEHRFKDVTLYSGLGGHVSRN 538
K +L++S DL W I +L+ Q + ++ + + R V LYS + G +S
Sbjct: 432 KLHLNHSQVDLFEAWRVIKLVLMNQLSQLEANQARQHISNPIRESRV-LYSNIRGWISHE 490
Query: 539 ALDNIALEEKRCRETLCMDNDICGCVQRTSYGLPCACEIATKLLQEKPILLDEIYHHW 596
AL + + RE L + +C V + GLPCA + L Q +P+LL+ + HW
Sbjct: 491 ALRKVETQ----RERLLKEVPVCTGVFTRTLGLPCAHSLQPLLKQNQPLLLNHFHSHW 544
>gb|AAW44911.1| conserved hypothetical protein [Cryptococcus neoformans var.
neoformans JEC21] gi|58270124|ref|XP_572218.1| conserved
hypothetical protein [Cryptococcus neoformans var.
neoformans JEC21]
Length = 920
Score = 141 bits (356), Expect = 9e-32
Identities = 143/579 (24%), Positives = 242/579 (41%), Gaps = 89/579 (15%)
Query: 66 FSDDVKSKNRDELLEWVRRQANKAGFTIVTQRSSLINP---------MFRLVCERSGSHI 116
F + +++DE V+ A GF V +RS P + + C +
Sbjct: 91 FDEQAYYQSKDECKGAVQAYAASQGFAAVIRRSKRKKPTKTHKEGRELIQYSCIFGDEYR 150
Query: 117 VPK----KKPKHANTGSRKCGCLFMISGYQSKQT--------KEWGLNILNGVHNHPMEP 164
+ K + + K GC F ++ + K W ++N HNHP
Sbjct: 151 CTRGDSFKDVRIRTIQTVKNGCPFAAVAWEMAEAEVKEHGGDKCWTWKVVNASHNHPPVA 210
Query: 165 ALEGHIL--AGRLKEDDKKIVRDLTKSKMLPRNILIHLKNQRPHCMTNVKQV----YNER 218
+ +L A R E K+++ L S+ +I+ + + P M + + + R
Sbjct: 211 NIGIALLRRASRTPEA-KELINTLIDSRTPFPSIISTMSQRFPTVMLTSEDLHRLAFRRR 269
Query: 219 QQIWKANRGDKKPLQFLISKLEEHNYTYYSRTQLESNTIEDIFWAHPTSIKLFNNFPTVL 278
+ + ++ L+S E H +R +L + + T+ L + FP VL
Sbjct: 270 ADLRAGSSASAACIRQLVSNDELHRPFVNTRDELIG-----LVYTKRTARALIHRFPEVL 324
Query: 279 VMDSTYKTNMYRMPMFEVVGVTSTDLTYSVGFGFMTHEKEENFVWVLTMLRKLLSSKMNM 338
D TYKTN+YRMPM +VG TST +TY+ G M E + L +L+ K ++
Sbjct: 325 FFDCTYKTNLYRMPMLHIVGSTSTGMTYTAGVILMLRETTNWYTQALNSFFELV-GKPDV 383
Query: 339 HKVIVTDRDMSLMKAVAHVFPESYALNCFFHVQANVKQRCVLNCKYPLGFKKDGKEVSNR 398
KV++TDRD +L+ A+ V P++Y +CF+H+Q NVK + C V
Sbjct: 384 -KVVITDRDPALINALMSVLPKAYRFSCFWHLQENVKSN-IRPC-----------FVDEE 430
Query: 399 DVVKKIMNAWKAMVES---PNQQLYANALVEFKDSCSDFPIFVDYAMTTLDEVKDKIVRA 455
++V + N KA +E ++LY + + Y + LDE+K++ V A
Sbjct: 431 NIVMAVSNFCKAWLEEGYRKMEELYPG---------QKYARAISY-IRGLDEIKERFVHA 480
Query: 456 WTDHVLHLGCRTTNRVESAHALLKKYLDNSVGDLGTCWEKIHHMLLLQFTAIQTSFGQNV 515
+ + LH G +R+E HA LKK +D GD LLL ++ T F Q
Sbjct: 481 YINKQLHFGQTGNSRLEGQHATLKKSIDTKYGD-----------LLLVIGSLSTYFDQQW 529
Query: 516 CVLEHRFK---------DVTLYSGLGGHVSRNALDNIALEEKRCRETL---------CMD 557
+ R + + + L G +SR A+ +A + + L +
Sbjct: 530 LKILKRIELERTRCATHIPSTFVRLKGSISRAAMKLLAEQLTLAKRYLGDYDEGVDDFEE 589
Query: 558 NDICGCVQRTSYGLPCACEIATKLLQEKPILLDEIYHHW 596
+ C S+GLPCA + + + + + + D I+ HW
Sbjct: 590 SHPCSGSFTKSHGLPCAHRLISFVRERRHLEKDHIHPHW 628
>gb|AAW42067.1| conserved hypothetical protein [Cryptococcus neoformans var.
neoformans JEC21] gi|50260537|gb|EAL23192.1|
hypothetical protein CNBA5360 [Cryptococcus neoformans
var. neoformans B-3501A] gi|50259285|gb|EAL21958.1|
hypothetical protein CNBC0980 [Cryptococcus neoformans
var. neoformans B-3501A] gi|50256296|gb|EAL19021.1|
hypothetical protein CNBH1230 [Cryptococcus neoformans
var. neoformans B-3501A] gi|57229113|gb|AAW45547.1|
conserved hypothetical protein [Cryptococcus neoformans
var. neoformans JEC21] gi|58271396|ref|XP_572854.1|
conserved hypothetical protein [Cryptococcus neoformans
var. neoformans JEC21] gi|58264436|ref|XP_569374.1|
conserved hypothetical protein [Cryptococcus neoformans
var. neoformans JEC21]
Length = 932
Score = 139 bits (351), Expect = 4e-31
Identities = 144/582 (24%), Positives = 242/582 (40%), Gaps = 83/582 (14%)
Query: 66 FSDDVKSKNRDELLEWVRRQANKAGFTIVTQRSSLINP---------MFRLVCERSGSHI 116
F + +++DE V+ A GF V +RS P + + C +
Sbjct: 91 FDEQAYYQSKDECKGAVQAYAASQGFAAVIRRSKRKKPTKTHKEGRELIQYSCIFGDEYR 150
Query: 117 VPK----KKPKHANTGSRKCGCLFMISGYQSKQT--------KEWGLNILNGVHNHPMEP 164
+ K + + K GC F ++ + K W ++N HNHP
Sbjct: 151 CTRGDSFKDVRIRTIQTVKNGCPFAAVAWEMAEAEVKEHGGDKCWTWKVVNASHNHPPVA 210
Query: 165 ALEGHIL--AGRLKEDDKKIVRDLTKSKMLPRNILIHLKNQRPHCMTNVKQV----YNER 218
+ +L A R E K+++ L S+ +I+ + + P M + + + R
Sbjct: 211 NIGIALLRRASRTPEA-KELINTLIDSRTPFPSIISTMSQRFPTVMLTSEDLHRLAFRRR 269
Query: 219 QQIWKANRGDKKPLQFLISKLEEHNYTYYSRTQLESNTIEDIFWAHPTSIKLFNNFPTVL 278
+ + ++ L+S E H +R +L + + T+ L + FP VL
Sbjct: 270 ADLRAGSSASAACIRQLVSNDELHRPFVNTRDELIG-----LVYTKRTARALIHRFPEVL 324
Query: 279 VMDSTYKTNMYRMPMFEVVGVTSTDLTYSVGFGFMTHEKEENFVWVLTMLRKLLSSKMNM 338
D TYKTN+YRMPM +VG TST +TY+ G M E + L +L+ K ++
Sbjct: 325 FFDCTYKTNLYRMPMLHIVGSTSTGMTYTAGVILMLRETTNWYTQALNSFFELV-GKPDV 383
Query: 339 HKVIVTDRDMSLMKAVAHVFPESYALNCFFHVQANVKQRCVLNCKYPLGFKKDGKEVSNR 398
KV++TDRD +L+ A+ V P++Y +CF+H+Q NVK + C V
Sbjct: 384 -KVVITDRDPALINALMSVLPKAYRFSCFWHLQENVKSN-IRPC-----------FVDEE 430
Query: 399 DVVKKIMNAWKAMV------ESPNQQLYANALVEFKDSCSDFPIFVDYAMTTLDEVKDKI 452
++V + N KA V +S Q +E + + Y + LDE+K++
Sbjct: 431 NIVMAVSNFCKAWVRYCVHAKSDEQLEEGYRKMEELYPGQKYARAISY-IRGLDEIKERF 489
Query: 453 VRAWTDHVLHLGCRTTNRVESAHALLKKYLDNSVGDLGTCWEKIHHMLLLQFTAIQTSFG 512
V A+ + LH G +R+E HA LKK +D GD LLL ++ T F
Sbjct: 490 VHAYINKQLHFGQTGNSRLEGQHATLKKSIDTKYGD-----------LLLVIGSLSTYFD 538
Query: 513 QNVCVLEHRFK---------DVTLYSGLGGHVSRNALDNIALEEKRCRETL--------- 554
Q + R + + + L G +SR A+ +A + + L
Sbjct: 539 QQWLKILKRIELERTRCATHIPSTFVRLKGSISRAAMKLLAEQLTLAKRYLGDYDEGVDD 598
Query: 555 CMDNDICGCVQRTSYGLPCACEIATKLLQEKPILLDEIYHHW 596
++ C S+GLPCA + + + + + + D I+ HW
Sbjct: 599 FEESHPCSGSFTKSHGLPCAHRLISFVRERRHLEKDHIHPHW 640
>gb|AAW44276.1| conserved hypothetical protein [Cryptococcus neoformans var.
neoformans JEC21] gi|58268854|ref|XP_571583.1| conserved
hypothetical protein [Cryptococcus neoformans var.
neoformans JEC21]
Length = 895
Score = 132 bits (331), Expect = 7e-29
Identities = 140/573 (24%), Positives = 233/573 (40%), Gaps = 102/573 (17%)
Query: 66 FSDDVKSKNRDELLEWVRRQANKAGFTIVTQRSSLINP---------MFRLVCERSGSHI 116
F + +++DE V+ A GF V +RS P + + C +
Sbjct: 91 FDEQAYYQSKDECKGAVQAYAASQGFAAVIRRSKRKKPTKTHKEGRELIQYSCIFGDEYR 150
Query: 117 VPK----KKPKHANTGSRKCGCLFMISGYQSKQT--------KEWGLNILNGVHNHPMEP 164
+ K + + K GC F ++ + K W ++N HNHP
Sbjct: 151 CTRGDSFKDVRIRTIQTVKNGCPFAAVAWEMAEAEVKEHGGDKCWTWKVVNASHNHPPVA 210
Query: 165 ALEGHIL--AGRLKEDDKKIVRDLTKSKMLPRNILIHLKNQRPHCMTNVKQV----YNER 218
+ +L A R E K+++ L S+ +I+ + + P M + + + R
Sbjct: 211 NIGIALLRRASRTPEA-KELINTLIDSRTPFPSIISTMSQRFPTVMLTSEDLHRLAFRRR 269
Query: 219 QQIWKANRGDKKPLQFLISKLEEHNYTYYSRTQLESNTIEDIFWAHPTSIKLFNNFPTVL 278
+ + ++ L+S E H +R +L + + T+ L + FP VL
Sbjct: 270 ADLRAGSSASAACIRQLVSNDELHRPFVNTRDELIG-----LVYTKRTARALIHRFPEVL 324
Query: 279 VMDSTYKTNMYRMPMFEVVGVTSTDLTYSVGFGFMTHEKEENFVWVLTMLRKLLSSKMNM 338
D TYKTN+YRMPM +VG TST +TY+ G M E + L +L+ K ++
Sbjct: 325 FFDCTYKTNLYRMPMLHIVGSTSTGMTYTAGVILMLRETTNWYTQALNSFFELV-GKPDV 383
Query: 339 HKVIVTDRDMSLMKAVAHVFPESYALNCFFHVQANVKQRCVLNCKYPLGFKKDGKEVSNR 398
KV++TDRD +L+ A+ V P++Y +CF+H+Q NVK + C V
Sbjct: 384 -KVVITDRDPALINALMSVLPKAYRFSCFWHLQENVKSN-IRPC-----------FVDEE 430
Query: 399 DVVKKIMNAWKAMV------ESPNQQLYANALVEFKDSCSDFPIFVDYAMTTLDEVKDKI 452
++V + N KA V +S Q +E + + Y + LDE+K++
Sbjct: 431 NIVMAVSNFCKAWVRYCVHAKSDEQLEEGYRKMEELYPGQKYARAISY-IRGLDEIKERF 489
Query: 453 VRAWTDHVLHLGCRTTNRVESAHALLKKYLDNSVGDLGTCWEKIHHMLLLQFTAIQTSFG 512
V A+ + LH G +R+E HA LKK +D GDL LLL+
Sbjct: 490 VHAYINKQLHFGQTGNSRLEGQHATLKKSIDTKYGDL----------LLLK--------- 530
Query: 513 QNVCVLEHRFKDVTLYSGLGGHVSRNALDNIALEEKRCRETL---------CMDNDICGC 563
G +SR A+ +A + + L ++ C
Sbjct: 531 --------------------GSISRAAMKLLAEQLTLAKRYLGDYDEGVDDFEESHPCSG 570
Query: 564 VQRTSYGLPCACEIATKLLQEKPILLDEIYHHW 596
S+GLPCA + + + + + + D I+ HW
Sbjct: 571 SFTKSHGLPCAHRLISFVRERRHLEKDHIHPHW 603
>gb|EAL17593.1| hypothetical protein CNBM0450 [Cryptococcus neoformans var.
neoformans B-3501A] gi|57226908|gb|AAW43368.1| conserved
hypothetical protein [Cryptococcus neoformans var.
neoformans JEC21] gi|58267038|ref|XP_570675.1| conserved
hypothetical protein [Cryptococcus neoformans var.
neoformans JEC21]
Length = 580
Score = 130 bits (326), Expect = 3e-28
Identities = 120/457 (26%), Positives = 197/457 (42%), Gaps = 54/457 (11%)
Query: 66 FSDDVKSKNRDELLEWVRRQANKAGFTIVTQRSSLINP---------MFRLVCERSGSHI 116
F + +++DE V+ A GF V +RS P + + C +
Sbjct: 91 FDEQAYYQSKDECKGAVQAYAASQGFAAVIRRSKRKKPTKTHKEGRELIQYSCIFGDEYR 150
Query: 117 VPK----KKPKHANTGSRKCGCLFMISGYQSKQT--------KEWGLNILNGVHNHPMEP 164
+ K + + K GC F ++ + K W ++N HNHP
Sbjct: 151 CTRGDSFKDVRIRTIQTVKNGCPFAAVAWEMAEAEVKEHGGDKCWTWKVVNASHNHPPVA 210
Query: 165 ALEGHIL--AGRLKEDDKKIVRDLTKSKMLPRNILIHLKNQRPHCMTNVKQV----YNER 218
+ +L A R E K+++ L S+ +I+ + + P M + + + R
Sbjct: 211 NIGIALLRRASRTPEA-KELINTLIDSRTPFPSIISTMSQRFPTVMLTSEDLHRLAFRRR 269
Query: 219 QQIWKANRGDKKPLQFLISKLEEHNYTYYSRTQLESNTIEDIFWAHPTSIKLFNNFPTVL 278
+ + ++ L+S E H +R +L + + T+ L + FP VL
Sbjct: 270 ADLRAGSSASAACIRQLVSNDELHRPFVNTRDELIG-----LVYTKRTARALIHRFPEVL 324
Query: 279 VMDSTYKTNMYRMPMFEVVGVTSTDLTYSVGFGFMTHEKEENFVWVLTMLRKLLSSKMNM 338
D TYKTN+YRMPM +VG TST +TY+ G M E + L +L+ K ++
Sbjct: 325 FFDCTYKTNLYRMPMLHIVGSTSTGMTYTAGVILMLRETTNWYTQALNSFFELV-GKPDV 383
Query: 339 HKVIVTDRDMSLMKAVAHVFPESYALNCFFHVQANVKQRCVLNCKYPLGFKKDGKEVSNR 398
KV++TDRD +L+ A+ V P++Y +CF+H+Q NVK + C V
Sbjct: 384 -KVVITDRDPALINALMSVLPKAYRFSCFWHLQENVKSN-IRPC-----------FVDEE 430
Query: 399 DVVKKIMNAWKAMV------ESPNQQLYANALVEFKDSCSDFPIFVDYAMTTLDEVKDKI 452
++V + N KA V +S Q +E + + Y + LDE+K++
Sbjct: 431 NIVMAVSNFCKAWVRYCVHAKSDEQLEEGYRKMEELYPGQKYARAISY-IRGLDEIKERF 489
Query: 453 VRAWTDHVLHLGCRTTNRVESAHALLKKYLDNSVGDL 489
V A+ + LH G +R+E HA LKK +D GDL
Sbjct: 490 VHAYINKQLHFGQTGNSRLEGQHATLKKSIDTKYGDL 526
>gb|AAW41060.1| conserved hypothetical protein [Cryptococcus neoformans var.
neoformans JEC21] gi|58258933|ref|XP_566879.1| conserved
hypothetical protein [Cryptococcus neoformans var.
neoformans JEC21]
Length = 678
Score = 128 bits (321), Expect = 1e-27
Identities = 109/386 (28%), Positives = 173/386 (44%), Gaps = 55/386 (14%)
Query: 235 LISKLEEHNYTYYSRTQLESNTIEDIFWAHPTSIKLFNNFPTVLVMDSTYKTNMYRMPMF 294
L+S E H +R +L + + T+ L + FP VL D TYKTN+YRMPM
Sbjct: 32 LVSNDELHRPFVNTRDELIG-----LVYTKRTARALIHRFPEVLFFDCTYKTNLYRMPML 86
Query: 295 EVVGVTSTDLTYSVGFGFMTHEKEENFVWVLTMLRKLLSSKMNMHKVIVTDRDMSLMKAV 354
+VG TST +TY+ G M E + L +L+ K ++ KV++TDRD +L+ A+
Sbjct: 87 HIVGSTSTGMTYTAGVILMLRETTNWYTQALNSFFELV-GKPDV-KVVITDRDPALINAL 144
Query: 355 AHVFPESYALNCFFHVQANVKQRCVLNCKYPLGFKKDGKEVSNRDVVKKIMNAWKAMV-- 412
V P++Y +CF+H+Q NVK + C V ++V + N KA V
Sbjct: 145 MSVLPKAYRFSCFWHLQENVKSN-IRPC-----------FVDEENIVMAVSNFCKAWVRY 192
Query: 413 ----ESPNQQLYANALVEFKDSCSDFPIFVDYAMTTLDEVKDKIVRAWTDHVLHLGCRTT 468
+S Q +E + + Y + LDE+K++ V A+ + LH G
Sbjct: 193 CVHAKSDEQLEEGYRKMEELYPGQKYARAISY-IRGLDEIKERFVHAYINKQLHFGQTGN 251
Query: 469 NRVESAHALLKKYLDNSVGDLGTCWEKIHHMLLLQFTAIQTSFGQNVCVLEHRFK----- 523
+R+E HA LKK +D GD LLL ++ T F Q + R +
Sbjct: 252 SRLEGQHATLKKSIDTKYGD-----------LLLVIGSLSTYFDQQWLKILKRIELERTR 300
Query: 524 ----DVTLYSGLGGHVSRNALDNIALEEKRCRETL---------CMDNDICGCVQRTSYG 570
+ + L G +SR A+ +A + + L ++ C S+G
Sbjct: 301 CATHIPSTFVRLKGSISRAAMKLLAEQLTLAKRYLGDYDEGVDDFEESHPCSGSFTKSHG 360
Query: 571 LPCACEIATKLLQEKPILLDEIYHHW 596
LPCA + + + + + + D I+ HW
Sbjct: 361 LPCAHRLISFVRERRHLEKDHIHPHW 386
>ref|XP_482508.1| putative far-red impaired response protein [Oryza sativa (japonica
cultivar-group)] gi|25553698|dbj|BAC24942.1| putative
far-red impaired response protein [Oryza sativa
(japonica cultivar-group)]
Length = 1148
Score = 104 bits (259), Expect = 2e-20
Identities = 91/370 (24%), Positives = 152/370 (40%), Gaps = 36/370 (9%)
Query: 73 KNRDELLEWVRRQANKAGFTIVT----------QRSSLINPMFRLVCERSGSHIVPKKKP 122
KN E E+ A AGF+IV + + FR C R G K
Sbjct: 369 KNITEAHEFFNFYALLAGFSIVRAHNYHTTSKKRNGEVTRVTFR--CNRQGKPTSQSKSV 426
Query: 123 KHANT----------GSRKCGCLFMISGYQSKQTKEWGLNILNGVHNHPMEPALEGHILA 172
T + C C +IS ++ + W ++ + HNH + P E L
Sbjct: 427 SAEETVVSERNTNENDATDCKCALVIS----ERNEIWNISRVQLDHNHQLSPRDEVRFLK 482
Query: 173 GR--LKEDDKKIVRDLTKSKMLPRNILIHLKNQRPHCMT---NVKQVYNERQQIWKANRG 227
+ ++K ++R L + + R++++ L R + K + N R I K
Sbjct: 483 SHKHMTTEEKMLIRTLKECNIPTRHMIVILSVLRGGLTSLPYTKKDISNVRTTINKETSS 542
Query: 228 DK--KPLQFLISKLEEHNYTYYSRTQLESNTIEDIFWAHPTSIKLFNNFPTVLVMDSTYK 285
+ K L+F K E+ + +Y ES ++++FW S + + + V+ D+TY
Sbjct: 543 NDIMKTLEFFRRKKEKDPHFFYEFDLDESKKVKNLFWTDGRSREWYEKYGDVVSFDTTYF 602
Query: 286 TNMYRMPMFEVVGVTSTDLTYSVGFGFMTHEKEENFVWVLTMLRKLLSSKMNMHKVIVTD 345
TN Y +P VG+ T G F+ E E F W+ K +S K K I+TD
Sbjct: 603 TNKYNLPFAPFVGIYGHGNTIVFGCAFLHDETSETFKWLFRTFLKAMSQK--EPKTIITD 660
Query: 346 RDMSLMKAVAHVFPESYALNCFFHVQANVKQRCVLNCKYPLGFKKDGKEVSNRDVVKKIM 405
+D ++ A+A VF + NCFFH+ K G + +++ N V ++
Sbjct: 661 QDGAMRSAIAQVFQNAKHRNCFFHIVKKAFNLSGNLLKAKEGLYDEYEDIINNSVTEEEF 720
Query: 406 N-AWKAMVES 414
W+ M++S
Sbjct: 721 EYLWQEMIDS 730
>ref|XP_463702.1| putative far-red impaired response protein [Oryza sativa (japonica
cultivar-group)]
Length = 1114
Score = 103 bits (257), Expect = 3e-20
Identities = 74/285 (25%), Positives = 122/285 (41%), Gaps = 23/285 (8%)
Query: 109 CERSGSHIVPKKKPKHANTGSRK--------CGCLFMISGYQSKQTKEWGLNILNGVHNH 160
C R G + +KK K R C C+ + + W + +N HNH
Sbjct: 401 CYRYGKNDDGQKKKKKKTQQQRNTQVIDRTDCKCMLTVR----LEGDVWRILSMNLNHNH 456
Query: 161 PMEPALEGHILAGR--LKEDDKKIVRDLTKSKMLPRNILIHLKNQR------PHCMTNVK 212
+ P E L + +++KK++R L + + RN++ L R P+ +V
Sbjct: 457 TLTPNREAKFLRSHKHMTDEEKKLIRTLKQCNIPTRNMISILSFLRGGIAALPYTRKDVS 516
Query: 213 QVYNERQQIWKANRGDKKPLQFLISKLEEHNYTYYSRTQLESNTIEDIFWAHPTSIKLFN 272
V + N + +++L K + YY T E+N ++ +FW S +L+
Sbjct: 517 NVGTAINSETR-NTDMNQVMEYLRQKEAKDPGYYYKLTLDENNKVKSMFWTDGRSTQLYE 575
Query: 273 NFPTVLVMDSTYKTNMYRMPMFEVVGVTSTDLTYSVGFGFMTHEKEENFVWVLTMLRKLL 332
+ + D+TYKTN Y MP +VGVT F+ E E F WV +
Sbjct: 576 QYGEFVSFDTTYKTNRYNMPFAPIVGVTGHGNICIFACAFLGDETTETFKWVFETFLTAM 635
Query: 333 SSKMNMHKVIVTDRDMSLMKAVAHVFPESYALNCFFHVQANVKQR 377
K + I+TD+D+++ A+ VFP S NC FH+ ++R
Sbjct: 636 GGK--HPETIITDQDLAMRAAIRQVFPNSKHRNCLFHILKKCRER 678
>dbj|BAD87203.1| far-red impaired response-like [Oryza sativa (japonica
cultivar-group)]
Length = 1130
Score = 103 bits (257), Expect = 3e-20
Identities = 74/285 (25%), Positives = 122/285 (41%), Gaps = 23/285 (8%)
Query: 109 CERSGSHIVPKKKPKHANTGSRK--------CGCLFMISGYQSKQTKEWGLNILNGVHNH 160
C R G + +KK K R C C+ + + W + +N HNH
Sbjct: 417 CYRYGKNDDGQKKKKKKTQQQRNTQVIDRTDCKCMLTVR----LEGDVWRILSMNLNHNH 472
Query: 161 PMEPALEGHILAGR--LKEDDKKIVRDLTKSKMLPRNILIHLKNQR------PHCMTNVK 212
+ P E L + +++KK++R L + + RN++ L R P+ +V
Sbjct: 473 TLTPNREAKFLRSHKHMTDEEKKLIRTLKQCNIPTRNMISILSFLRGGIAALPYTRKDVS 532
Query: 213 QVYNERQQIWKANRGDKKPLQFLISKLEEHNYTYYSRTQLESNTIEDIFWAHPTSIKLFN 272
V + N + +++L K + YY T E+N ++ +FW S +L+
Sbjct: 533 NVGTAINSETR-NTDMNQVMEYLRQKEAKDPGYYYKLTLDENNKVKSMFWTDGRSTQLYE 591
Query: 273 NFPTVLVMDSTYKTNMYRMPMFEVVGVTSTDLTYSVGFGFMTHEKEENFVWVLTMLRKLL 332
+ + D+TYKTN Y MP +VGVT F+ E E F WV +
Sbjct: 592 QYGEFVSFDTTYKTNRYNMPFAPIVGVTGHGNICIFACAFLGDETTETFKWVFETFLTAM 651
Query: 333 SSKMNMHKVIVTDRDMSLMKAVAHVFPESYALNCFFHVQANVKQR 377
K + I+TD+D+++ A+ VFP S NC FH+ ++R
Sbjct: 652 GGK--HPETIITDQDLAMRAAIRQVFPNSKHRNCLFHILKKCRER 694
>emb|CAE02137.2| OSJNBa0074L08.5 [Oryza sativa (japonica cultivar-group)]
gi|50926620|ref|XP_473257.1| OSJNBa0074L08.5 [Oryza
sativa (japonica cultivar-group)]
Length = 1061
Score = 102 bits (255), Expect = 5e-20
Identities = 75/288 (26%), Positives = 123/288 (42%), Gaps = 29/288 (10%)
Query: 109 CERSGSHIVPKKKPKHANTGSRK--------CGCLFMISGYQSKQTKEWGLNILNGVHNH 160
C R G + +KK K R C C+ + W + +N HNH
Sbjct: 348 CYRYGKNDDGQKKKKKKTQQQRNTQVIDRTDCKCMLTVR----LDGDVWRILSMNLNHNH 403
Query: 161 PMEPALEGHILAGR--LKEDDKKIVRDLTKSKMLPRNILIHLKNQR------PHC---MT 209
+ P E L + +++KK++R L + + RN++ L R P+ ++
Sbjct: 404 TLTPNREAKFLRSHKHMTDEEKKLIRTLKQCNIPTRNMISILSFLRGGIAALPYTGKDVS 463
Query: 210 NVKQVYNERQQIWKANRGDKKPLQFLISKLEEHNYTYYSRTQLESNTIEDIFWAHPTSIK 269
NV N + N + +++L K + YY T E+N ++ +FW S +
Sbjct: 464 NVGTAINSETR----NTDMNQVMEYLRQKEAKDPGYYYKLTLDENNKVKSMFWTDGRSTQ 519
Query: 270 LFNNFPTVLVMDSTYKTNMYRMPMFEVVGVTSTDLTYSVGFGFMTHEKEENFVWVLTMLR 329
L+ + + D+TYKTN Y MP +VGVT F+ E E F WV
Sbjct: 520 LYEQYGEFVSFDTTYKTNRYNMPFAPIVGVTGHGNICIFACAFLGDETTETFKWVFETFL 579
Query: 330 KLLSSKMNMHKVIVTDRDMSLMKAVAHVFPESYALNCFFHVQANVKQR 377
+ K + I+TD+D+++ A+ VFP S NC FH+ ++R
Sbjct: 580 TAMGGK--HPETIITDQDLAMRAAIRQVFPNSKHRNCLFHILKKCRER 625
>ref|XP_481410.1| far-red impaired response protein -like [Oryza sativa (japonica
cultivar-group)] gi|35215248|dbj|BAC92598.1| putative
far-red impaired response protein [Oryza sativa
(japonica cultivar-group)] gi|35215057|dbj|BAC92415.1|
putative far-red impaired response protein [Oryza sativa
(japonica cultivar-group)]
Length = 1132
Score = 100 bits (249), Expect = 2e-19
Identities = 80/319 (25%), Positives = 135/319 (42%), Gaps = 13/319 (4%)
Query: 132 CGCLFMISGYQSKQTKEWGLNILNGVHNHPMEPALEGHILAGR--LKEDDKKIVRDLTKS 189
C C+ M+ + W + L+ HNH + P E L + +K+++R L +
Sbjct: 479 CKCMMMVEKMFALGDV-WQIATLDLKHNHALCPRREAKFLKSHKNMTIKEKRLIRTLKEC 537
Query: 190 KMLPRNILIHLKNQR---PHCMTNVKQVYNERQQIWKANRGD--KKPLQFLISKLEEHNY 244
+ R++++ L R P K V N I R + K+ L +L K E
Sbjct: 538 NIPTRSMIVILSFLRGGLPALPYTKKDVSNVGTAINSETRNNDMKQVLAYLRKKEIEDPG 597
Query: 245 TYYSRTQLESNTIEDIFWAHPTSIKLFNNFPTVLVMDSTYKTNMYRMPMFEVVGVTSTDL 304
Y E+N + +FW S +L+ + + D+TY+TN Y MP VGV
Sbjct: 598 ISYKFKLDENNKVTSMFWTDGRSTQLYEEYGDCISFDTTYRTNRYNMPFAPFVGVIGHGT 657
Query: 305 TYSVGFGFMTHEKEENFVWVLTMLRKLLSSKMNMHKVIVTDRDMSLMKAVAHVFPESYAL 364
T G F+ E E F WV + K K I+TD+D ++ A+A VF +
Sbjct: 658 TCLFGCAFLGDETAETFKWVFETFATAMGGK--HPKTIITDQDNAMRSAIAQVFQNTKHR 715
Query: 365 NCFFHVQANVKQR--CVLNCKYPLGFKKDGKEV-SNRDVVKKIMNAWKAMVESPNQQLYA 421
NC FH++ N +++ + + K + ++ SN + + W M+E N Q
Sbjct: 716 NCLFHIKKNCREKTGSMFSQKSNKNLYDEYDDILSNCLTEAEFESLWPQMIEKFNLQNVN 775
Query: 422 NALVEFKDSCSDFPIFVDY 440
+ +K+ P++ Y
Sbjct: 776 YLKIMWKNRAQFVPVYFKY 794
>gb|AAQ56575.1| putative transposase [Oryza sativa (japonica cultivar-group)]
Length = 1037
Score = 100 bits (249), Expect = 2e-19
Identities = 80/319 (25%), Positives = 135/319 (42%), Gaps = 13/319 (4%)
Query: 132 CGCLFMISGYQSKQTKEWGLNILNGVHNHPMEPALEGHILAGR--LKEDDKKIVRDLTKS 189
C C+ M+ + W + L+ HNH + P E L + +K+++R L +
Sbjct: 384 CKCMMMVEKMFALGDV-WQIATLDLKHNHALCPRREAKFLKSHKNMTIKEKRLIRTLKEC 442
Query: 190 KMLPRNILIHLKNQR---PHCMTNVKQVYNERQQIWKANRGD--KKPLQFLISKLEEHNY 244
+ R++++ L R P K V N I R + K+ L +L K E
Sbjct: 443 NIPTRSMIVILSFLRGGLPALPYTKKDVSNVGTAINSETRNNDMKQVLAYLRKKEIEDPG 502
Query: 245 TYYSRTQLESNTIEDIFWAHPTSIKLFNNFPTVLVMDSTYKTNMYRMPMFEVVGVTSTDL 304
Y E+N + +FW S +L+ + + D+TY+TN Y MP VGV
Sbjct: 503 ISYKFKLDENNKVTSMFWTDGRSTQLYEEYGDCISFDTTYRTNRYNMPFAPFVGVIGHGT 562
Query: 305 TYSVGFGFMTHEKEENFVWVLTMLRKLLSSKMNMHKVIVTDRDMSLMKAVAHVFPESYAL 364
T G F+ E E F WV + K K I+TD+D ++ A+A VF +
Sbjct: 563 TCLFGCAFLGDETAETFKWVFETFATAMGGK--HPKTIITDQDNAMRSAIAQVFQNTKHR 620
Query: 365 NCFFHVQANVKQR--CVLNCKYPLGFKKDGKEV-SNRDVVKKIMNAWKAMVESPNQQLYA 421
NC FH++ N +++ + + K + ++ SN + + W M+E N Q
Sbjct: 621 NCLFHIKKNCREKTGSMFSQKSNKNLYDEYDDILSNCLTEAEFESLWPQMIEKFNLQNVN 680
Query: 422 NALVEFKDSCSDFPIFVDY 440
+ +K+ P++ Y
Sbjct: 681 YLKIMWKNRAQFVPVYFKY 699
>ref|NP_908529.1| P0702D12.15 [Oryza sativa (japonica cultivar-group)]
Length = 832
Score = 99.4 bits (246), Expect = 5e-19
Identities = 90/391 (23%), Positives = 165/391 (42%), Gaps = 37/391 (9%)
Query: 104 MFRLVCERSGSHIVPK-KKPKHANTGSRKCGCLFMISGYQSKQTKEWGLNILNGVHNHPM 162
M ++ C G + PK KKP S + GC M+ +S+ W + + HNHPM
Sbjct: 113 MRQICCSHQGRN--PKTKKP------SVRIGCPAMMKINRSRAGSGWSVTKVVSAHNHPM 164
Query: 163 EPAL---EGHILAGRLKEDDKKIVRDLTKSKMLPRNILIHLKNQR--PHCMTNVKQVYNE 217
+ ++ + + ++ E + I+ ++ S M N+ L P + ++ +
Sbjct: 165 KKSVGVTKNYQSHNQIDEGTRGIIEEMVDSSMSLTNMYGMLSGMHGGPSMVPFTRKAMDR 224
Query: 218 RQQIWKANRGD---KKPLQFLISKLEEHNYTYYSRTQLESNTIEDIFWAHPTSIKLFNNF 274
+ + +K L L + +YS E+ +++IFW+H S F +F
Sbjct: 225 LAYAIRRDESSDDMQKTLDVLKDLQKRSKNFFYSIQVDEACRVKNIFWSHAVSRLNFEHF 284
Query: 275 PTVLVMDSTYKTNMYRMPMFEVVGVTSTDLTYSVGFGFMTHEKEENFVWVLTMLRKLLSS 334
V+ D+TYKTN Y MP VGV + + G + E EE+F W+ ++ ++
Sbjct: 285 GDVITFDTTYKTNKYNMPFAPFVGVNNHFQSTFFGCALLREETEESFTWLFNTFKECMNG 344
Query: 335 KMNMHKVIVTDRDMSLMKAVAHVFPESYALNCFFHVQANVKQRCVLNCKYPLGFKKDGKE 394
K+ + I+TD S+ A+ VFP + C +HV K+ G
Sbjct: 345 KVPIG--ILTDNCPSMAAAIRTVFPNTIHRVCKWHVLKKAKEFM-------------GNI 389
Query: 395 VSNRDVVKKIMNAWKAMVESPNQQLYANALVEFKDSCSDFPIFVDYAMTTLDEVKDKIVR 454
S R KK + K + ++ ++ + A + D+ + + + +++ K
Sbjct: 390 YSKRHTFKKAFH--KVLTQTLTEEEFVAA---WHKLIRDYNLEKSVYLRHIWDIRRKWAF 444
Query: 455 AWTDHVLHLGCRTTNRVESAHALLKKYLDNS 485
+ H G TT R ESA+ + K ++ S
Sbjct: 445 VYFSHRFFAGMTTTQRSESANHVFKMFVSPS 475
>emb|CAE02218.2| OSJNBb0002N06.8 [Oryza sativa (japonica cultivar-group)]
gi|50923349|ref|XP_472035.1| OSJNBb0002N06.8 [Oryza
sativa (japonica cultivar-group)]
Length = 885
Score = 97.8 bits (242), Expect = 2e-18
Identities = 90/391 (23%), Positives = 164/391 (41%), Gaps = 37/391 (9%)
Query: 104 MFRLVCERSGSHIVPK-KKPKHANTGSRKCGCLFMISGYQSKQTKEWGLNILNGVHNHPM 162
M ++ C G + PK KKP S + GC M+ +S W + + HNHPM
Sbjct: 166 MRQICCSHQGRN--PKTKKP------SVRIGCPAMMKINRSGAGSGWSVTKVVSTHNHPM 217
Query: 163 EPAL---EGHILAGRLKEDDKKIVRDLTKSKMLPRNILIHLKNQR--PHCMTNVKQVYNE 217
+ ++ + + ++ E + I+ ++ S M N+ L P + ++ +
Sbjct: 218 KKSVGVTKNYQSHNQIDEGTRGIIEEMVDSSMSLTNMYGMLSGMHGGPSMVPFTRKAMDR 277
Query: 218 RQQIWKANRGD---KKPLQFLISKLEEHNYTYYSRTQLESNTIEDIFWAHPTSIKLFNNF 274
+ + +K L L + +YS E+ +++IFW+H S F +F
Sbjct: 278 LAYAIRRDESSDDMQKTLDVLKDLQKRSKNFFYSIQVDEACRVKNIFWSHAVSRLNFEHF 337
Query: 275 PTVLVMDSTYKTNMYRMPMFEVVGVTSTDLTYSVGFGFMTHEKEENFVWVLTMLRKLLSS 334
V+ D+TYKTN Y MP VGV + + G + E EE+F W+ ++ ++
Sbjct: 338 GDVITFDTTYKTNKYNMPFAPFVGVNNHFQSTFFGCALLREETEESFTWLFNTFKECMNG 397
Query: 335 KMNMHKVIVTDRDMSLMKAVAHVFPESYALNCFFHVQANVKQRCVLNCKYPLGFKKDGKE 394
K+ + I+TD S+ A+ VFP + C +HV K+ G
Sbjct: 398 KVPIG--ILTDNCPSMAAAIRTVFPNTIHRVCKWHVLKKAKEFM-------------GNI 442
Query: 395 VSNRDVVKKIMNAWKAMVESPNQQLYANALVEFKDSCSDFPIFVDYAMTTLDEVKDKIVR 454
S R KK + K + ++ ++ + A + D+ + + + +++ K
Sbjct: 443 YSKRHTFKKAFH--KVLTQTLTEEEFVAA---WHKLIRDYNLEKSVYLRHIWDIRRKWAF 497
Query: 455 AWTDHVLHLGCRTTNRVESAHALLKKYLDNS 485
+ H G TT R ESA+ + K ++ S
Sbjct: 498 VYFSHRFFAGMTTTQRSESANHVFKMFVSPS 528
>dbj|BAD29601.1| putative far-red impaired response protein [Oryza sativa (japonica
cultivar-group)] gi|50251625|dbj|BAD29488.1| putative
far-red impaired response protein [Oryza sativa
(japonica cultivar-group)]
Length = 688
Score = 96.7 bits (239), Expect = 3e-18
Identities = 84/362 (23%), Positives = 159/362 (43%), Gaps = 40/362 (11%)
Query: 132 CGCLFMISGYQSKQTKEWGLNILNGVHNHPM-EPALEGHILAGRLKEDDKKIVR-DLTKS 189
CGCL + ++++ W + + H H + P + R+ D KK +L S
Sbjct: 88 CGCLAKLEIERNEEKGVWFVKEFDNQHTHELANPDHVAFLGVHRVMSDSKKAQAVELRMS 147
Query: 190 KMLPRNILIHLKNQRPHCMTN---VKQVYN--ERQQIWKANRGDKKP-LQFLISKLEEHN 243
+ P ++ ++N +K +YN R ++ D + L++L K EE
Sbjct: 148 GLRPFQVMEVMENNHDELDEVGFVMKDLYNFFTRYEMKNIKGHDAEDVLKYLTRKQEEDA 207
Query: 244 YTYYSRTQLESNTIEDIFWAHPTSIKLFNNFPTVLVMDSTYKTNMYRMPMFEVVGVTSTD 303
++ T E + ++FWA S + F V++ DSTY+ N Y +P +GV
Sbjct: 208 EFFFKYTTDEEGRLRNVFWADAESRLDYAAFGGVVIFDSTYRVNKYNLPFIPFIGVNHHR 267
Query: 304 LTYSVGFGFMTHEKEENFVWVLTMLRKLLSSKMNMH-KVIVTDRDMSLMKAVAHVFPESY 362
T G G +++E ++ W +L L + +H K ++TD D+++ KA++ V P +Y
Sbjct: 268 STTIFGCGILSNESVNSYCW---LLETFLEAMRQVHPKSLITDGDLAMAKAISKVMPGAY 324
Query: 363 ALNCFFHVQANVKQRCVLNCKYPLGFKKDGKEVSNRDVVKKIMNAWKAMVESPNQQLYAN 422
C +H++ N+ + + + P K+ K + ES +++ +
Sbjct: 325 HRLCTWHIEENMSR----HLRKP-----------------KLDELRKLIYESMDEEEFER 363
Query: 423 ALVEFKDSCSDFPIFVDYAMTTLDEVKDKIVRAWTDHVLHLGCRTTNRVES----AHALL 478
+FK++ + + + +++K A+TD LG R+ R ES H LL
Sbjct: 364 RWADFKENGGTGN---EQWIALMYRLREKWAAAYTDGKYLLGMRSNQRSESLNSKLHTLL 420
Query: 479 KK 480
K+
Sbjct: 421 KR 422
>ref|XP_471803.1| OSJNBa0050F15.2 [Oryza sativa (japonica cultivar-group)]
gi|38345091|emb|CAD40514.2| OSJNBa0050F15.2 [Oryza
sativa (japonica cultivar-group)]
Length = 688
Score = 95.5 bits (236), Expect = 8e-18
Identities = 84/363 (23%), Positives = 158/363 (43%), Gaps = 40/363 (11%)
Query: 131 KCGCLFMISGYQSKQTKEWGLNILNGVHNHPM-EPALEGHILAGRLKEDDKKIVR-DLTK 188
+CGCL + ++++ W + + H H + P + R+ D KK +L
Sbjct: 87 RCGCLAKLEIERNEEKGVWFVKEFDNQHTHELANPDHVAFLGVHRVMSDSKKAQAVELRM 146
Query: 189 SKMLPRNILIHLKNQRPHCMTN---VKQVYN--ERQQIWKANRGDKKP-LQFLISKLEEH 242
S + P ++ ++N +K +YN R + D + L++L K EE
Sbjct: 147 SGLRPFQVMEVMENNHDELDEVGFVMKDLYNFFTRYDMKNIKGHDAEDVLKYLTRKQEED 206
Query: 243 NYTYYSRTQLESNTIEDIFWAHPTSIKLFNNFPTVLVMDSTYKTNMYRMPMFEVVGVTST 302
++ T E + ++FWA S + F V++ DSTY+ N Y +P +GV
Sbjct: 207 AEFFFKYTTDEEGRLRNVFWADAESRLDYAAFGGVVIFDSTYRVNKYNLPFIPFIGVNHH 266
Query: 303 DLTYSVGFGFMTHEKEENFVWVLTMLRKLLSSKMNMH-KVIVTDRDMSLMKAVAHVFPES 361
T G G +++E ++ W +L L + +H K ++TD D+++ KA++ V P +
Sbjct: 267 RSTTIFGCGILSNESVNSYCW---LLETFLEAMRQVHPKSLITDGDLAMAKAISKVMPGA 323
Query: 362 YALNCFFHVQANVKQRCVLNCKYPLGFKKDGKEVSNRDVVKKIMNAWKAMVESPNQQLYA 421
Y C +H++ N+ + + + P K+ K + ES +++ +
Sbjct: 324 YHRLCTWHIEENMSR----HLRKP-----------------KLDELRKLIYESMDEEEFE 362
Query: 422 NALVEFKDSCSDFPIFVDYAMTTLDEVKDKIVRAWTDHVLHLGCRTTNRVES----AHAL 477
+FK++ + + +++K A+TD LG R+ R ES H L
Sbjct: 363 RRWADFKENGGTGN---GQWIALMYRLREKWAAAYTDGKYLLGMRSNQRSESLNSKLHTL 419
Query: 478 LKK 480
LK+
Sbjct: 420 LKR 422
>gb|AAU90129.1| unknown protein [Oryza sativa (japonica cultivar-group)]
Length = 904
Score = 93.6 bits (231), Expect = 3e-17
Identities = 62/236 (26%), Positives = 108/236 (45%), Gaps = 14/236 (5%)
Query: 149 WGLNILNGVHNHPMEPALEGHILAGR--LKEDDKKIVRDLTKSKMLPRNILIHLKNQRP- 205
W + LN HNHP+ P E L + + +K ++R L K + R I+ L R
Sbjct: 431 WYITRLNLEHNHPLSPPEERKFLWSHKHMIDQEKLLIRTLNKINVPTRMIMSVLSYVRGG 490
Query: 206 -HCMTNVKQVYNERQQIWKANRGDKKPLQ---FLISKLEEHNYTYYSRTQLESNTIEDIF 261
+ K+ + + + G +Q F K+ E Y+ E+N ++ +F
Sbjct: 491 LFAVPYTKKAMSNYRDFVRRESGKNDMMQCLDFFEKKISEDPLFYFRFRTDENNVVKSLF 550
Query: 262 WAHPTSIKLFNNFPTVLVMDSTYKTNMYRMPMFEVVGVTSTDLTYSVGFGFMTHEKEENF 321
W+ S K + F ++ D+TYKTN Y +P VG+TS G+ F+ E
Sbjct: 551 WSDRNSRKFYEMFGDIVSFDTTYKTNRYDLPFAPFVGITSHGDNCLFGYAFLQDE----- 605
Query: 322 VWVLTMLRKLLSSKMNMHKVIVTDRDMSLMKAVAHVFPESYALNCFFHVQANVKQR 377
V+ + + K + I+TD+D+++ A+ VFP++ NC FH+ +N + +
Sbjct: 606 TMVVQYVLDCMGGK--VPATIITDQDLAMKAAIVIVFPDTVHRNCMFHMLSNARDK 659
>dbj|BAD81770.1| putative far-red impaired response protein [Oryza sativa (japonica
cultivar-group)]
Length = 883
Score = 93.6 bits (231), Expect = 3e-17
Identities = 89/392 (22%), Positives = 161/392 (40%), Gaps = 39/392 (9%)
Query: 104 MFRLVCERSGSHIVPK-KKPKHANTGSRKCGCLFMISGYQSKQTKEWGLNILNGVHNHPM 162
M ++ C G + PK KKP S + GC M+ S W + + HNHPM
Sbjct: 164 MRQICCSHQGRN--PKTKKP------SVRIGCPAMMKINMSGAGSGWSVTKVVSAHNHPM 215
Query: 163 EPAL---EGHILAGRLKEDDKKIVRDLTKSKMLPRNILIHLKNQR--PHCMTNVKQVYNE 217
+ ++ + + ++ E + I+ ++ S M N+ L P + ++ +
Sbjct: 216 KKSVGVTKNYQSHNQIDEGTRGIIEEMVDSSMSLTNMYGMLSGMHGGPSMVPFTRKAMDR 275
Query: 218 RQQIWKANRGD---KKPLQFLISKLEEHNYTYYSRTQLESNTIEDIFWAHPTSIKLFNNF 274
+ + +K L L + +YS E+ +++IFW+H S F +F
Sbjct: 276 VAYAIRRDESSDDMQKTLDVLKDLQKRSKNFFYSIQVDEACRVKNIFWSHAVSRLNFEHF 335
Query: 275 PTVLVMDSTYKTNMYRMPMFEVVGVTSTDLTYSVGFGFMTHEKEENFVWVLTMLRKLLSS 334
V+ D+TYKTN Y MP VGV + + G + E EE+F W+ ++ ++
Sbjct: 336 GDVITFDTTYKTNKYNMPFAPFVGVNNHFQSTFFGCALLREETEESFTWLFNTFKECMNG 395
Query: 335 KMNMHKVIVTDRDMSLMKAVAHVFPESYALNCFFHVQANVKQRCVLNCKYPLGFKKDGKE 394
K+ + I+TD S+ A+ VFP + C HV K+ FKK +
Sbjct: 396 KVPIG--ILTDNCPSMAAAIRTVFPNTIHRVCKSHVLKKAKEFMGNIYSKCHTFKKAFHK 453
Query: 395 VSNRDVV-KKIMNAWKAMVESPNQQLYANALVEFKDSCSDFPIFVDYAMTTLDEVKDKIV 453
V + + ++ + AW ++ D+ + + + +++ K
Sbjct: 454 VLTQTLTEEEFVAAWHKLIR-------------------DYNLEKSVYLRHIWDIRRKWA 494
Query: 454 RAWTDHVLHLGCRTTNRVESAHALLKKYLDNS 485
+ H G TT R ESA+ + K ++ S
Sbjct: 495 FVYFSHRFFAGMTTTQRSESANHVFKMFVSPS 526
>gb|AAW41059.1| hypothetical protein CNA05530 [Cryptococcus neoformans var.
neoformans JEC21] gi|58258931|ref|XP_566878.1|
hypothetical protein CNA05530 [Cryptococcus neoformans
var. neoformans JEC21]
Length = 206
Score = 92.0 bits (227), Expect = 8e-17
Identities = 64/202 (31%), Positives = 100/202 (48%), Gaps = 27/202 (13%)
Query: 291 MPMFEVVGVTSTDLTYSVGFGFMTHEKEENFVWVLTMLRKLLSSKMNMHKVIVTDRDMSL 350
MPM +VG TST +TY+ G M E + L +L+ K ++ KV++TDRD +L
Sbjct: 1 MPMLHIVGSTSTGMTYTAGVILMLRETTNWYTQALNSFFELVG-KPDV-KVVITDRDPAL 58
Query: 351 MKAVAHVFPESYALNCFFHVQANVKQRCVLNCKYPLGFKKDGKEVSNRDVVKKIMNAWKA 410
+ A+ V P++Y +CF+H+Q NVK + C V ++V + N KA
Sbjct: 59 INALMSVLPKAYRFSCFWHLQENVKSN-IRPC-----------FVDEENIVMAVSNFCKA 106
Query: 411 MVES---PNQQLYANALVEFKDSCSDFPIFVDYAMTTLDEVKDKIVRAWTDHVLHLGCRT 467
+E ++LY + + Y + LDE+K++ V A+ + LH G
Sbjct: 107 WLEEGYRKMEELYPG---------QKYARAISY-IRGLDEIKERFVHAYINKQLHFGQTG 156
Query: 468 TNRVESAHALLKKYLDNSVGDL 489
+R+E HA LKK +D GDL
Sbjct: 157 NSRLEGQHATLKKSIDTKYGDL 178
>ref|NP_915530.1| P0529E05.9 [Oryza sativa (japonica cultivar-group)]
Length = 773
Score = 90.5 bits (223), Expect = 2e-16
Identities = 88/371 (23%), Positives = 153/371 (40%), Gaps = 18/371 (4%)
Query: 74 NRDELLEWVRRQANKAGFTIVT----QRSSLINPMFRLVCERSGSHIVPKKKPKHANTGS 129
+ D E+ R A GF+I + S+ + + VC R G +
Sbjct: 159 DEDIAYEFYNRYAGDVGFSIRKFWHDKSSTNVIRTKKFVCSREGFNKRNTSDACQRKRAD 218
Query: 130 RKCGCLFMISGYQSKQTKEWGLNILNGVHNHPMEPALEGHILAG--RLKEDDKKIVRDLT 187
+ GC+ ++ + T ++ + + HNH + + H+L R+ E K + L
Sbjct: 219 TRVGCMAEMT-IKITPTGKYAIASFSNTHNHELITPSKAHLLRSQRRMTEAQKAQIDILN 277
Query: 188 KSKMLPRN---ILIHLKNQRPHCMTNVKQVYN--ERQQIWKANRGDKKP-LQFLISKLEE 241
S + + ++ R K N +++ GD LQ+L K E
Sbjct: 278 DSGVRSKEGHEVMSRQAGGRQSLTFTRKDYKNYLRSKRMKSIQEGDTGAILQYLQDKQME 337
Query: 242 HNYTYYSRTQLESNTIEDIFWAHPTSIKLFNNFPTVLVMDSTYKTNMYRMPMFEVVGVTS 301
+ +Y+ E + +IFWA S+ F+ F V+ D+TY+TN Y P VGV
Sbjct: 338 NPSFFYAIQVDEDEMMTNIFWADARSVLDFDYFGDVICFDTTYRTNNYGRPFALFVGVNH 397
Query: 302 TDLTYSVGFGFMTHEKEENFVWVLTMLRKLLSSKMNMHKVIVTDRDMSLMKAVAHVFPES 361
T G + E F W+ ++ +S K + I+TD+ +++ A+ VFP S
Sbjct: 398 HKQTVVFGAALLYDETTSTFEWLFETFKRAMSGK--EPRTILTDQCAAIINAIGTVFPNS 455
Query: 362 YALNCFFHVQANVKQRCVLNCKYPLGFKKD-GKEVSNRDVVKKIMNAWKAMVE--SPNQQ 418
C +H+ N + FK D GK V + + V + ++AW M+E + N
Sbjct: 456 THRLCVWHMYQNAAVHLSHIFQGSKTFKNDFGKCVFDFEEVDEFISAWNKMIEEYNLNDN 515
Query: 419 LYANALVEFKD 429
+ + L E K+
Sbjct: 516 QWLHHLFEIKE 526
Database: nr
Posted date: Jul 5, 2005 12:34 AM
Number of letters in database: 863,360,394
Number of sequences in database: 2,540,612
Lambda K H
0.319 0.135 0.411
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 1,762,522,266
Number of Sequences: 2540612
Number of extensions: 78176630
Number of successful extensions: 227068
Number of sequences better than 10.0: 584
Number of HSP's better than 10.0 without gapping: 264
Number of HSP's successfully gapped in prelim test: 324
Number of HSP's that attempted gapping in prelim test: 225602
Number of HSP's gapped (non-prelim): 1130
length of query: 985
length of database: 863,360,394
effective HSP length: 138
effective length of query: 847
effective length of database: 512,755,938
effective search space: 434304279486
effective search space used: 434304279486
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 80 (35.4 bits)
Medicago: description of AC140549.7


