

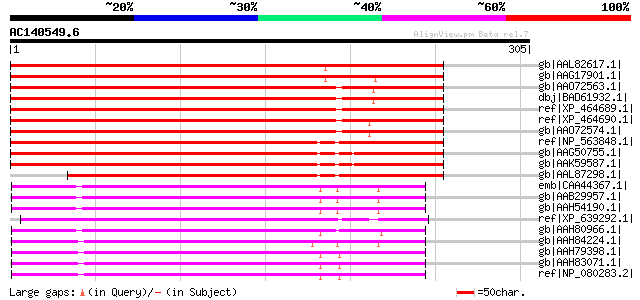
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC140549.6 - phase: 0 /pseudo
(305 letters)
Database: nr
2,540,612 sequences; 863,360,394 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
gb|AAL82617.1| elongation factor 1-gamma [Glycine max] 408 e-112
gb|AAG17901.1| translation elongation factor 1-gamma [Prunus avi... 394 e-108
gb|AAO72563.1| elongation factor 1 gamma-like protein [Oryza sat... 363 4e-99
dbj|BAD61932.1| putative elongation factor 1 gamma [Oryza sativa... 363 4e-99
ref|XP_464689.1| putative elongation factor 1-gamma [Oryza sativ... 360 3e-98
ref|XP_464690.1| Elongation factor 1-gamma [Oryza sativa (japoni... 360 4e-98
gb|AAO72574.1| elongation factor 1 gamma-like protein [Oryza sat... 360 4e-98
ref|NP_563848.1| elongation factor 1B-gamma, putative / eEF-1B g... 360 4e-98
gb|AAG50755.1| elongation factor 1B gamma, putative; tRNA-Undet ... 359 5e-98
gb|AAK59587.1| putative elongation factor 1B gamma [Arabidopsis ... 359 5e-98
gb|AAL87298.1| unknown protein [Arabidopsis thaliana] 301 1e-80
emb|CAA44367.1| elongation factor 1 gamma [Xenopus laevis] gi|10... 172 1e-41
gb|AAB29957.1| elongation factor 1 gamma; EF-1 gamma [Xenopus la... 172 1e-41
gb|AAH54190.1| MGC64329 protein [Xenopus laevis] 172 1e-41
ref|XP_639292.1| hypothetical protein DDB0185297 [Dictyostelium ... 169 7e-41
gb|AAH80966.1| Unknown (protein for IMAGE:6981438) [Xenopus trop... 167 3e-40
gb|AAH84224.1| Unknown (protein for MGC:80886) [Xenopus laevis] 160 3e-38
gb|AAH79398.1| Eef1g protein [Rattus norvegicus] gi|55583762|sp|... 159 7e-38
gb|AAH83071.1| Eukaryotic translation elongation factor 1 gamma ... 159 7e-38
ref|NP_080283.2| eukaryotic translation elongation factor 1 gamm... 159 7e-38
>gb|AAL82617.1| elongation factor 1-gamma [Glycine max]
Length = 420
Score = 408 bits (1048), Expect = e-112
Identities = 204/258 (79%), Positives = 221/258 (85%), Gaps = 3/258 (1%)
Query: 1 MGVSNKTPQFINMNPIGKVPVLETPDGPVFESNAIARYVARL-GHNNLFGSSLIHQAQID 59
MGVSNKTP+F+ +NPIGKVPVLETPDGP+FESNAIARYV RL G N L+GSSLI A I+
Sbjct: 37 MGVSNKTPEFLKINPIGKVPVLETPDGPIFESNAIARYVTRLKGDNTLYGSSLIEYAHIE 96
Query: 60 QWIDFSSLEIDANIMKLYLPRLGFATYFPPVEENAISSLKRALEALNSHLAHNTYLVGHS 119
QWIDFSSLEIDANI+K Y PR+G Y PP EE + SSLKRAL ALN+HLA NTYLVGHS
Sbjct: 97 QWIDFSSLEIDANIIKWYAPRVGRGPYLPPTEEASNSSLKRALGALNTHLASNTYLVGHS 156
Query: 120 VTLADIITTSNLYIGFAKLLVKSFTSEFPHVERYFWTLVNQPNFRKILGQVKQTEAMPPI 179
VTLADII T NLY+GFA +LVKSFTSEFPHVERYFWTLVNQPNFRKI+GQVKQ EA+PP+
Sbjct: 157 VTLADIIMTCNLYLGFANILVKSFTSEFPHVERYFWTLVNQPNFRKIIGQVKQAEAIPPV 216
Query: 180 PSAKQ--QPKESKPKTKDEPKKVAKSEPEKPKVEVEEEAPKPKPKNPLDLLPPSKMILDD 237
SAK+ QPKESK K KDEPKK A PEKPK E EEEAPKPKPKNPLD LPPS MILD+
Sbjct: 217 QSAKKPSQPKESKAKAKDEPKKEANKVPEKPKEEAEEEAPKPKPKNPLDYLPPSPMILDE 276
Query: 238 WKRLYSNTKRNFREVAIK 255
WKRLYSNTK NFREVAIK
Sbjct: 277 WKRLYSNTKTNFREVAIK 294
>gb|AAG17901.1| translation elongation factor 1-gamma [Prunus avium]
gi|13626392|sp|Q9FUM1|EF1G_PRUAV Elongation factor
1-gamma (EF-1-gamma) (eEF-1B gamma)
Length = 422
Score = 394 bits (1011), Expect = e-108
Identities = 194/261 (74%), Positives = 220/261 (83%), Gaps = 6/261 (2%)
Query: 1 MGVSNKTPQFINMNPIGKVPVLETPDGPVFESNAIARYVARLGHNN-LFGSSLIHQAQID 59
MGV+NKTP+++ +NPIGKVP+LETPDGP+FESNAIARYVARL +N L GSSLI A I+
Sbjct: 36 MGVTNKTPEYLKLNPIGKVPLLETPDGPIFESNAIARYVARLKADNPLIGSSLIDYAHIE 95
Query: 60 QWIDFSSLEIDANIMKLYLPRLGFATYFPPVEENAISSLKRALEALNSHLAHNTYLVGHS 119
QWIDF SLEIDANI+ + PR G+A Y PP EE AIS+LKRAL ALN+HLA NTYLVGH
Sbjct: 96 QWIDFGSLEIDANIISWFRPRFGYAVYLPPAEEAAISALKRALGALNTHLASNTYLVGHF 155
Query: 120 VTLADIITTSNLYIGFAKLLVKSFTSEFPHVERYFWTLVNQPNFRKILGQVKQTEAMPPI 179
VTLADII T NL+ GF KL++KSFTSEFPHVERYFWTLVNQP F+K+LG VKQTE++PP+
Sbjct: 156 VTLADIIVTCNLFFGFTKLMIKSFTSEFPHVERYFWTLVNQPKFKKVLGDVKQTESVPPV 215
Query: 180 PSAKQ--QPKESKPKTKDEPKKVAKSEPEKPKVEVE---EEAPKPKPKNPLDLLPPSKMI 234
PSAK+ QPKE+K K K+EPKK AK EP KPK E EEAPKPKPKNPLDLLPPS M+
Sbjct: 216 PSAKKPSQPKETKSKAKEEPKKEAKKEPAKPKAEAAEEVEEAPKPKPKNPLDLLPPSNMV 275
Query: 235 LDDWKRLYSNTKRNFREVAIK 255
LDDWKRLYSNTK NFREVAIK
Sbjct: 276 LDDWKRLYSNTKTNFREVAIK 296
>gb|AAO72563.1| elongation factor 1 gamma-like protein [Oryza sativa (japonica
cultivar-group)]
Length = 413
Score = 363 bits (931), Expect = 4e-99
Identities = 181/258 (70%), Positives = 210/258 (81%), Gaps = 6/258 (2%)
Query: 1 MGVSNKTPQFINMNPIGKVPVLETPDGPVFESNAIARYVARLGHNN-LFGSSLIHQAQID 59
MGVSNKTP+F+ MNP+GK+PVLETP+G VFESNAIARYVARL N+ L GSSLI + I+
Sbjct: 33 MGVSNKTPEFLKMNPLGKIPVLETPEGAVFESNAIARYVARLKDNSSLCGSSLIDYSHIE 92
Query: 60 QWIDFSSLEIDANIMKLYLPRLGFATYFPPVEENAISSLKRALEALNSHLAHNTYLVGHS 119
QW+DFS+ E+DANI + PRLGF Y P +EE AI+SLKR+L ALN+HLA NTYLVGHS
Sbjct: 93 QWMDFSATEVDANIGRWLYPRLGFGPYVPVLEEFAITSLKRSLGALNTHLASNTYLVGHS 152
Query: 120 VTLADIITTSNLYIGFAKLLVKSFTSEFPHVERYFWTLVNQPNFRKILGQVKQTEAMPPI 179
VTLADI+ T NLY GF ++L+KSFTSEFPHVERYFWT+VNQPNF+K++G KQ E++PP+
Sbjct: 153 VTLADIVMTCNLYYGFVRILIKSFTSEFPHVERYFWTMVNQPNFKKVIGDFKQAESVPPV 212
Query: 180 PSAKQQPKESKPKTKDEPKKVAKSEPEKPKVEV--EEEAPKPKPKNPLDLLPPSKMILDD 237
PKESK K E KK A E KPKVE EEEAPKPKPKNPLDLLPPSKMILD+
Sbjct: 213 QKKAAPPKESKAK---EAKKEAPKEAPKPKVEASEEEEAPKPKPKNPLDLLPPSKMILDE 269
Query: 238 WKRLYSNTKRNFREVAIK 255
WKRLYSNTK NFRE+AIK
Sbjct: 270 WKRLYSNTKTNFREIAIK 287
>dbj|BAD61932.1| putative elongation factor 1 gamma [Oryza sativa (japonica
cultivar-group)] gi|54291156|dbj|BAD61828.1| putative
elongation factor 1 gamma [Oryza sativa (japonica
cultivar-group)]
Length = 416
Score = 363 bits (931), Expect = 4e-99
Identities = 181/258 (70%), Positives = 210/258 (81%), Gaps = 6/258 (2%)
Query: 1 MGVSNKTPQFINMNPIGKVPVLETPDGPVFESNAIARYVARLGHNN-LFGSSLIHQAQID 59
MGVSNKTP+F+ MNP+GK+PVLETP+G VFESNAIARYVARL N+ L GSSLI + I+
Sbjct: 36 MGVSNKTPEFLKMNPLGKIPVLETPEGAVFESNAIARYVARLKDNSSLCGSSLIDYSHIE 95
Query: 60 QWIDFSSLEIDANIMKLYLPRLGFATYFPPVEENAISSLKRALEALNSHLAHNTYLVGHS 119
QW+DFS+ E+DANI + PRLGF Y P +EE AI+SLKR+L ALN+HLA NTYLVGHS
Sbjct: 96 QWMDFSATEVDANIGRWLYPRLGFGPYVPVLEEFAITSLKRSLGALNTHLASNTYLVGHS 155
Query: 120 VTLADIITTSNLYIGFAKLLVKSFTSEFPHVERYFWTLVNQPNFRKILGQVKQTEAMPPI 179
VTLADI+ T NLY GF ++L+KSFTSEFPHVERYFWT+VNQPNF+K++G KQ E++PP+
Sbjct: 156 VTLADIVMTCNLYYGFVRILIKSFTSEFPHVERYFWTMVNQPNFKKVIGDFKQAESVPPV 215
Query: 180 PSAKQQPKESKPKTKDEPKKVAKSEPEKPKVEV--EEEAPKPKPKNPLDLLPPSKMILDD 237
PKESK K E KK A E KPKVE EEEAPKPKPKNPLDLLPPSKMILD+
Sbjct: 216 QKKAAPPKESKAK---EAKKEAPKEAPKPKVEASEEEEAPKPKPKNPLDLLPPSKMILDE 272
Query: 238 WKRLYSNTKRNFREVAIK 255
WKRLYSNTK NFRE+AIK
Sbjct: 273 WKRLYSNTKTNFREIAIK 290
>ref|XP_464689.1| putative elongation factor 1-gamma [Oryza sativa (japonica
cultivar-group)] gi|46806490|dbj|BAD17614.1| putative
elongation factor 1-gamma [Oryza sativa (japonica
cultivar-group)]
Length = 414
Score = 360 bits (924), Expect = 3e-98
Identities = 181/256 (70%), Positives = 205/256 (79%), Gaps = 4/256 (1%)
Query: 1 MGVSNKTPQFINMNPIGKVPVLETPDGPVFESNAIARYVARLGHNN-LFGSSLIHQAQID 59
MGVSNKTP+F+ MNPIGK+PVLETPDGPVFESNAIARYV R +N L+GSSLI A I+
Sbjct: 36 MGVSNKTPEFLKMNPIGKIPVLETPDGPVFESNAIARYVTRSKADNPLYGSSLIEYAHIE 95
Query: 60 QWIDFSSLEIDANIMKLYLPRLGFATYFPPVEENAISSLKRALEALNSHLAHNTYLVGHS 119
QW DFS+ E+DANI K PRLG A Y EE AI++LKR+L ALN+HLA NTYLVGHS
Sbjct: 96 QWNDFSATEVDANIGKWLYPRLGIAPYVAVSEEAAIAALKRSLGALNTHLASNTYLVGHS 155
Query: 120 VTLADIITTSNLYIGFAKLLVKSFTSEFPHVERYFWTLVNQPNFRKILGQVKQTEAMPPI 179
VTLADI+ T NLY+GFA+++ KSFTSEFPHVERYFWT+VNQPNF+K+LG VKQ E++PP+
Sbjct: 156 VTLADIVMTCNLYMGFARIMTKSFTSEFPHVERYFWTMVNQPNFKKVLGDVKQAESVPPV 215
Query: 180 PSAKQQPKESKPKTKDEPKKVAKSEPEKPKVEVEEEAPKPKPKNPLDLLPPSKMILDDWK 239
PKE KPK E KK A K E EEEAPKPKPKNPLDLLPPSKMILD+WK
Sbjct: 216 QKKAPPPKEQKPK---EAKKEAPKPKAVEKPEEEEEAPKPKPKNPLDLLPPSKMILDEWK 272
Query: 240 RLYSNTKRNFREVAIK 255
RLYSNTK NFREVAIK
Sbjct: 273 RLYSNTKTNFREVAIK 288
>ref|XP_464690.1| Elongation factor 1-gamma [Oryza sativa (japonica cultivar-group)]
gi|46806491|dbj|BAD17615.1| Elongation factor 1-gamma
[Oryza sativa (japonica cultivar-group)]
gi|3868758|dbj|BAA34206.1| elongation factor 1B gamma
[Oryza sativa] gi|13626515|sp|Q9ZRI7|EF1G_ORYSA
Elongation factor 1-gamma (EF-1-gamma) (eEF-1B gamma)
Length = 418
Score = 360 bits (923), Expect = 4e-98
Identities = 180/260 (69%), Positives = 208/260 (79%), Gaps = 8/260 (3%)
Query: 1 MGVSNKTPQFINMNPIGKVPVLETPDGPVFESNAIARYVARLGHNN-LFGSSLIHQAQID 59
MGVSNKTP+++ MNPIGKVP+LETPDGPVFESNAIARYV R +N L+GSSLI A I+
Sbjct: 36 MGVSNKTPEYLKMNPIGKVPILETPDGPVFESNAIARYVTRSKSDNPLYGSSLIEYAHIE 95
Query: 60 QWIDFSSLEIDANIMKLYLPRLGFATYFPPVEENAISSLKRALEALNSHLAHNTYLVGHS 119
QWIDFS+ E+DAN K PRLGFA Y EE AI++LKR+L ALN+HLA NTYLVGHS
Sbjct: 96 QWIDFSATEVDANTGKWLFPRLGFAPYVAVSEEAAIAALKRSLGALNTHLASNTYLVGHS 155
Query: 120 VTLADIITTSNLYIGFAKLLVKSFTSEFPHVERYFWTLVNQPNFRKILGQVKQTEAMPPI 179
VTLADI+ T NLY+GFA+++ K+FTSEFPHVERYFWT+VNQPNF+K++G VKQ +++P +
Sbjct: 156 VTLADIVMTCNLYMGFARIMTKNFTSEFPHVERYFWTMVNQPNFKKVMGDVKQADSVPQV 215
Query: 180 PSAKQQPKESKPKTKDEPKKVAKSEPEKPKV----EVEEEAPKPKPKNPLDLLPPSKMIL 235
PKE KPK E KK A E KPK E EEEAPKPKPKNPLDLLPPSKMIL
Sbjct: 216 QKKAAAPKEQKPK---EAKKEAPKEAPKPKAAEKPEEEEEAPKPKPKNPLDLLPPSKMIL 272
Query: 236 DDWKRLYSNTKRNFREVAIK 255
D+WKRLYSNTK NFREVAIK
Sbjct: 273 DEWKRLYSNTKTNFREVAIK 292
>gb|AAO72574.1| elongation factor 1 gamma-like protein [Oryza sativa (japonica
cultivar-group)]
Length = 409
Score = 360 bits (923), Expect = 4e-98
Identities = 183/260 (70%), Positives = 207/260 (79%), Gaps = 8/260 (3%)
Query: 1 MGVSNKTPQFINMNPIGKVPVLETPDGPVFESNAIARYVARLGHNN-LFGSSLIHQAQID 59
MGVSNKTP+F+ MNPIGK+PVLETPDGPVFESNAIARYV R +N L+GSSLI A I+
Sbjct: 27 MGVSNKTPEFLKMNPIGKIPVLETPDGPVFESNAIARYVTRSKADNPLYGSSLIEYAHIE 86
Query: 60 QWIDFSSLEIDANIMKLYLPRLGFATYFPPVEENAISSLKRALEALNSHLAHNTYLVGHS 119
QW DFS+ E+DANI K PRLG A Y EE AI++LKR+L ALN+HLA NTYLVGHS
Sbjct: 87 QWNDFSATEVDANIGKWLYPRLGIAPYVAVSEEAAIAALKRSLGALNTHLASNTYLVGHS 146
Query: 120 VTLADIITTSNLYIGFAKLLVKSFTSEFPHVERYFWTLVNQPNFRKILGQVKQTEAMPPI 179
VTLADI+ T NLY+GFA+++ KSFTSEFPHVERYFWT+VNQPNF+K+LG VKQ E++P +
Sbjct: 147 VTLADIVMTCNLYMGFARIMTKSFTSEFPHVERYFWTMVNQPNFKKVLGDVKQAESVPXV 206
Query: 180 PSAKQQPKESKPKTKDEPKKVAKSEPEKPKV----EVEEEAPKPKPKNPLDLLPPSKMIL 235
PKE KPK E KK A E KPK E EEEAPKPKPKNPLDLLPPSKMIL
Sbjct: 207 QKKAPPPKEQKPK---EAKKEAPKEAPKPKAVEKPEEEEEAPKPKPKNPLDLLPPSKMIL 263
Query: 236 DDWKRLYSNTKRNFREVAIK 255
D+WKRLYSNTK NFREVAIK
Sbjct: 264 DEWKRLYSNTKTNFREVAIK 283
>ref|NP_563848.1| elongation factor 1B-gamma, putative / eEF-1B gamma, putative
[Arabidopsis thaliana] gi|2160158|gb|AAB60721.1| Similar
to elongation factor 1-gamma (gb|EF1G_XENLA). ESTs
gb|T20564,gb|T45940,gb|T04527 come from this gene.
[Arabidopsis thaliana] gi|25299533|pir||B86230
hypothetical protein [imported] - Arabidopsis thaliana
gi|13626364|sp|O04487|EF1G_ARATH Probable elongation
factor 1-gamma 1 (EF-1-gamma) (eEF-1B gamma)
Length = 414
Score = 360 bits (923), Expect = 4e-98
Identities = 183/256 (71%), Positives = 210/256 (81%), Gaps = 4/256 (1%)
Query: 1 MGVSNKTPQFINMNPIGKVPVLETPDGPVFESNAIARYVARL-GHNNLFGSSLIHQAQID 59
MGV+NKTP F+ MNPIGKVPVLETP+G VFESNAIARYV+RL G N+L GSSLI AQI+
Sbjct: 36 MGVTNKTPAFLKMNPIGKVPVLETPEGSVFESNAIARYVSRLNGDNSLNGSSLIEYAQIE 95
Query: 60 QWIDFSSLEIDANIMKLYLPRLGFATYFPPVEENAISSLKRALEALNSHLAHNTYLVGHS 119
QWIDFSSLEI A+I++ + PR+GF Y P EE AIS+LKRAL+ALN+HL NTYLVGHS
Sbjct: 96 QWIDFSSLEIYASILRWFGPRMGFMPYSAPAEEGAISTLKRALDALNTHLTSNTYLVGHS 155
Query: 120 VTLADIITTSNLYIGFAKLLVKSFTSEFPHVERYFWTLVNQPNFRKILGQVKQTEAMPPI 179
+TLADIIT NL +GFA ++ K FTSEFPHVERYFWT+VNQPNF K+LG VKQTEA+PPI
Sbjct: 156 ITLADIITVCNLNLGFATVMTKKFTSEFPHVERYFWTVVNQPNFTKVLGDVKQTEAVPPI 215
Query: 180 PSAKQQPKESKPKTKDEPKKVAKSEPEKPKVEVEEEAPKPKPKNPLDLLPPSKMILDDWK 239
S K+ + +KP K+EPKK E PK+ EEEAPKPK KNPLDLLPPS M+LDDWK
Sbjct: 216 AS-KKAAQPAKP--KEEPKKKEAPVAEAPKLAEEEEAPKPKAKNPLDLLPPSPMVLDDWK 272
Query: 240 RLYSNTKRNFREVAIK 255
RLYSNTK NFREVAIK
Sbjct: 273 RLYSNTKSNFREVAIK 288
>gb|AAG50755.1| elongation factor 1B gamma, putative; tRNA-Undet [Arabidopsis
thaliana] gi|24030437|gb|AAN41373.1| putative elongation
factor 1B gamma [Arabidopsis thaliana]
gi|21553395|gb|AAM62488.1| elongation factor 1B gamma,
putative [Arabidopsis thaliana]
gi|21360471|gb|AAM47351.1| At1g57720/T8L23_18
[Arabidopsis thaliana] gi|17978699|gb|AAL47343.1|
unknown protein [Arabidopsis thaliana]
gi|15223649|ref|NP_176084.1| elongation factor 1B-gamma,
putative / eEF-1B gamma, putative [Arabidopsis thaliana]
gi|16226841|gb|AAL16277.1| At1g57720/T8L23_18
[Arabidopsis thaliana] gi|15983511|gb|AAL11623.1|
At1g57720/T8L23_18 [Arabidopsis thaliana]
gi|13877603|gb|AAK43879.1| Unknown protein [Arabidopsis
thaliana] gi|25299534|pir||E96611 probable elongation
factor 1B gamma [imported] - Arabidopsis thaliana
gi|13626393|sp|Q9FVT2|EF1H_ARATH Probable elongation
factor 1-gamma 2 (EF-1-gamma) (eEF-1B gamma)
Length = 413
Score = 359 bits (922), Expect = 5e-98
Identities = 179/256 (69%), Positives = 212/256 (81%), Gaps = 5/256 (1%)
Query: 1 MGVSNKTPQFINMNPIGKVPVLETPDGPVFESNAIARYVARL-GHNNLFGSSLIHQAQID 59
MGV+NK+P+F+ MNPIGKVPVLETP+GP+FESNAIARYV+R G N+L GSSLI A I+
Sbjct: 36 MGVTNKSPEFLKMNPIGKVPVLETPEGPIFESNAIARYVSRKNGDNSLNGSSLIEYAHIE 95
Query: 60 QWIDFSSLEIDANIMKLYLPRLGFATYFPPVEENAISSLKRALEALNSHLAHNTYLVGHS 119
QWIDFSSLEIDAN++K + PR+G+A + P EE AIS+LKR LEALN+HLA NT+LVGHS
Sbjct: 96 QWIDFSSLEIDANMLKWFAPRMGYAPFSAPAEEAAISALKRGLEALNTHLASNTFLVGHS 155
Query: 120 VTLADIITTSNLYIGFAKLLVKSFTSEFPHVERYFWTLVNQPNFRKILGQVKQTEAMPPI 179
VTLADI+T NL +GFA ++ K FTS FPHVERYFWT+VNQP F+K+LG KQTEA+PP+
Sbjct: 156 VTLADIVTICNLNLGFATVMTKKFTSAFPHVERYFWTMVNQPEFKKVLGDAKQTEAVPPV 215
Query: 180 PSAKQQPKESKPKTKDEPKKVAKSEPEKPKVEVEEEAPKPKPKNPLDLLPPSKMILDDWK 239
P+ K+ P+ +KP K+EPKK A E PK EEEAPKPK KNPLDLLPPS M+LDDWK
Sbjct: 216 PT-KKAPQPAKP--KEEPKKAA-PVAEAPKPAEEEEAPKPKAKNPLDLLPPSPMVLDDWK 271
Query: 240 RLYSNTKRNFREVAIK 255
RLYSNTK NFREVAIK
Sbjct: 272 RLYSNTKSNFREVAIK 287
>gb|AAK59587.1| putative elongation factor 1B gamma [Arabidopsis thaliana]
Length = 413
Score = 359 bits (922), Expect = 5e-98
Identities = 179/256 (69%), Positives = 212/256 (81%), Gaps = 5/256 (1%)
Query: 1 MGVSNKTPQFINMNPIGKVPVLETPDGPVFESNAIARYVARL-GHNNLFGSSLIHQAQID 59
MGV+NK+P+F+ MNPIGKVPVLETP+GP+FESNAIARYV+R G N+L GSSLI A I+
Sbjct: 36 MGVTNKSPEFLKMNPIGKVPVLETPEGPIFESNAIARYVSRKNGDNSLNGSSLIEYAHIE 95
Query: 60 QWIDFSSLEIDANIMKLYLPRLGFATYFPPVEENAISSLKRALEALNSHLAHNTYLVGHS 119
QWIDFSSLEIDAN++K + PR+G+A + P EE AIS+LKR LEALN+HLA NT+LVGHS
Sbjct: 96 QWIDFSSLEIDANMLKWFAPRMGYAPFSAPAEEAAISALKRGLEALNTHLASNTFLVGHS 155
Query: 120 VTLADIITTSNLYIGFAKLLVKSFTSEFPHVERYFWTLVNQPNFRKILGQVKQTEAMPPI 179
VTLADI+T NL +GFA ++ K FTS FPHVERYFWT+VNQP F+K+LG KQTEA+PP+
Sbjct: 156 VTLADIVTICNLNLGFATVMTKEFTSAFPHVERYFWTMVNQPEFKKVLGDAKQTEAVPPV 215
Query: 180 PSAKQQPKESKPKTKDEPKKVAKSEPEKPKVEVEEEAPKPKPKNPLDLLPPSKMILDDWK 239
P+ K+ P+ +KP K+EPKK A E PK EEEAPKPK KNPLDLLPPS M+LDDWK
Sbjct: 216 PT-KKAPQPAKP--KEEPKKAA-PVAEAPKPAEEEEAPKPKAKNPLDLLPPSPMVLDDWK 271
Query: 240 RLYSNTKRNFREVAIK 255
RLYSNTK NFREVAIK
Sbjct: 272 RLYSNTKSNFREVAIK 287
>gb|AAL87298.1| unknown protein [Arabidopsis thaliana]
Length = 345
Score = 301 bits (772), Expect = 1e-80
Identities = 155/222 (69%), Positives = 179/222 (79%), Gaps = 4/222 (1%)
Query: 35 IARYVARL-GHNNLFGSSLIHQAQIDQWIDFSSLEIDANIMKLYLPRLGFATYFPPVEEN 93
IARYV+RL G N+L GSSLI AQI+QWIDFSSLEI A+I++ + PR+GF Y P EE
Sbjct: 1 IARYVSRLNGDNSLNGSSLIEYAQIEQWIDFSSLEIYASILRWFGPRMGFMPYSAPAEEG 60
Query: 94 AISSLKRALEALNSHLAHNTYLVGHSVTLADIITTSNLYIGFAKLLVKSFTSEFPHVERY 153
AIS+LKRAL+ALN+HL NTYLVGHS+TLADIIT NL +GFA ++ K FTSEFPHVERY
Sbjct: 61 AISTLKRALDALNTHLTSNTYLVGHSITLADIITVCNLNLGFATVMTKKFTSEFPHVERY 120
Query: 154 FWTLVNQPNFRKILGQVKQTEAMPPIPSAKQQPKESKPKTKDEPKKVAKSEPEKPKVEVE 213
FWT+VNQPNF K+LG VKQTEA+PPI S K+ + +KP K+EPKK E PK+ E
Sbjct: 121 FWTVVNQPNFTKVLGDVKQTEAVPPIAS-KKAAQPAKP--KEEPKKKEAPVAEAPKLAEE 177
Query: 214 EEAPKPKPKNPLDLLPPSKMILDDWKRLYSNTKRNFREVAIK 255
EEAPKPK KNPLDLLPPS M+LDDWKRLYSNTK NFREVAIK
Sbjct: 178 EEAPKPKAKNPLDLLPPSPMVLDDWKRLYSNTKSNFREVAIK 219
>emb|CAA44367.1| elongation factor 1 gamma [Xenopus laevis] gi|104038|pir||S20060
translation elongation factor eEF-1 gamma chain -
African clawed frog gi|119166|sp|P26642|EF1G_XENLA
Elongation factor 1-gamma type 1 (EF-1-gamma) (eEF-1B
gamma) (p47)
Length = 436
Score = 172 bits (436), Expect = 1e-41
Identities = 105/261 (40%), Positives = 145/261 (55%), Gaps = 21/261 (8%)
Query: 2 GVSNKTPQFINMNPIGKVPVLETPDG-PVFESNAIARYVARLGHNNLFGSSLIHQAQIDQ 60
GV+NKTP+F+ P+GKVP E DG +FES+AIA YV G++ L G++ +HQAQ+ Q
Sbjct: 41 GVTNKTPEFLKKFPLGKVPAFEGKDGFCLFESSAIAHYV---GNDELRGTTRLHQAQVIQ 97
Query: 61 WIDFSSLEIDANIMKLYLPRLGFATYFPPVEENAISSLKRALEALNSHLAHNTYLVGHSV 120
W+ FS I P LG Y E A +K L L+SHL T+LVG +
Sbjct: 98 WVSFSDSHIVPPASAWVFPTLGIMQYNKQATEQAKEGIKTVLGVLDSHLQTRTFLVGERI 157
Query: 121 TLADIITTSNLYIGFAKLLVKSFTSEFPHVERYFWTLVNQPNFRKILGQVKQTEAMPPIP 180
TLADI T +L + ++L SF F +V R+F T VNQP FR +LG+VK + M
Sbjct: 158 TLADITVTCSLLWLYKQVLEPSFRQPFGNVTRWFVTCVNQPEFRAVLGEVKLCDKMAQFD 217
Query: 181 S--------AKQQPKESKP-----KTKDEPKKVAKSEPEKPKVEVEEE----APKPKPKN 223
+ K+ PK+ KP K K+E KK A + P+ +++E A +PK K+
Sbjct: 218 AKKFAEMQPKKETPKKEKPAKEPKKEKEEKKKAAPTPAPAPEDDLDESEKALAAEPKSKD 277
Query: 224 PLDLLPPSKMILDDWKRLYSN 244
P LP S I+D++KR YSN
Sbjct: 278 PYAHLPKSSFIMDEFKRKYSN 298
>gb|AAB29957.1| elongation factor 1 gamma; EF-1 gamma [Xenopus laevis]
gi|2134057|pir||I51237 translation elongation factor
EF-1 gamma - African clawed frog
Length = 436
Score = 172 bits (436), Expect = 1e-41
Identities = 105/261 (40%), Positives = 145/261 (55%), Gaps = 21/261 (8%)
Query: 2 GVSNKTPQFINMNPIGKVPVLETPDG-PVFESNAIARYVARLGHNNLFGSSLIHQAQIDQ 60
GV+NKTP+F+ P+GKVP E DG +FES+AIA YV G++ L G++ +HQAQ+ Q
Sbjct: 41 GVTNKTPEFLKKFPLGKVPAFEGKDGFCLFESSAIAHYV---GNDELRGTTRLHQAQVIQ 97
Query: 61 WIDFSSLEIDANIMKLYLPRLGFATYFPPVEENAISSLKRALEALNSHLAHNTYLVGHSV 120
W+ FS I P LG Y E A +K L L+SHL T+LVG +
Sbjct: 98 WVSFSDSHIVPPASAWVFPTLGIMQYNKQATEQAKEEIKTVLGVLDSHLQTRTFLVGERI 157
Query: 121 TLADIITTSNLYIGFAKLLVKSFTSEFPHVERYFWTLVNQPNFRKILGQVKQTEAMPPIP 180
TLADI T +L + ++L SF F +V R+F T VNQP FR +LG+VK + M
Sbjct: 158 TLADITVTCSLLWLYKQVLEPSFRQPFGNVTRWFVTCVNQPEFRAVLGEVKLCDKMAQFD 217
Query: 181 S--------AKQQPKESKP-----KTKDEPKKVAKSEPEKPKVEVEEE----APKPKPKN 223
+ K+ PK+ KP K K+E KK A + P+ +++E A +PK K+
Sbjct: 218 AKKFAEMQPKKETPKKEKPAKEPKKEKEEKKKAAPTPAPAPEDDLDESEKALAAEPKSKD 277
Query: 224 PLDLLPPSKMILDDWKRLYSN 244
P LP S I+D++KR YSN
Sbjct: 278 PYAHLPKSSFIMDEFKRKYSN 298
>gb|AAH54190.1| MGC64329 protein [Xenopus laevis]
Length = 436
Score = 172 bits (436), Expect = 1e-41
Identities = 105/261 (40%), Positives = 145/261 (55%), Gaps = 21/261 (8%)
Query: 2 GVSNKTPQFINMNPIGKVPVLETPDG-PVFESNAIARYVARLGHNNLFGSSLIHQAQIDQ 60
GV+NKTP+F+ P+GKVP E DG +FES+AIA YV G++ L G++ +HQAQ+ Q
Sbjct: 41 GVTNKTPEFLKKFPLGKVPAFEGKDGFCLFESSAIAHYV---GNDELRGTTRLHQAQVIQ 97
Query: 61 WIDFSSLEIDANIMKLYLPRLGFATYFPPVEENAISSLKRALEALNSHLAHNTYLVGHSV 120
W+ FS I P LG Y E A +K L L+SHL T+LVG +
Sbjct: 98 WVSFSDSHIVPPASAWVFPTLGIMQYNKQATEQAKEEIKTVLGVLDSHLQTRTFLVGERI 157
Query: 121 TLADIITTSNLYIGFAKLLVKSFTSEFPHVERYFWTLVNQPNFRKILGQVKQTEAMPPIP 180
TLADI T +L + ++L SF F +V R+F T VNQP FR +LG+VK + M
Sbjct: 158 TLADITVTCSLLWLYKQVLEPSFRQPFGNVTRWFVTCVNQPEFRAVLGEVKLCDKMAQFD 217
Query: 181 S--------AKQQPKESKP-----KTKDEPKKVAKSEPEKPKVEVEEE----APKPKPKN 223
+ K+ PK+ KP K K+E KK A + P+ +++E A +PK K+
Sbjct: 218 AKKFAEMQPKKETPKKEKPAKEPKKEKEEKKKAAPTPAPAPEDDLDESEKALAAEPKSKD 277
Query: 224 PLDLLPPSKMILDDWKRLYSN 244
P LP S I+D++KR YSN
Sbjct: 278 PYAHLPKSSFIMDEFKRKYSN 298
>ref|XP_639292.1| hypothetical protein DDB0185297 [Dictyostelium discoideum]
gi|60467920|gb|EAL65933.1| hypothetical protein
DDB0185297 [Dictyostelium discoideum]
Length = 1020
Score = 169 bits (429), Expect = 7e-41
Identities = 94/240 (39%), Positives = 137/240 (56%), Gaps = 7/240 (2%)
Query: 7 TPQFINMNPIGKVPVLETPDGPVFESNAIARYVARLGHNNLFGSSLIHQAQIDQWIDFSS 66
T +F P+GKVP L T GP+FESN +ARYVARL ++ ++GS A DQWID+++
Sbjct: 651 TEEFKTNFPLGKVPALLTEQGPLFESNTMARYVARLNNSTIYGSDAYTAALNDQWIDYAA 710
Query: 67 LEIDANIMKLYLPRLGFATYFPPVEENAISSLKRALEALNSHLAHNTYLVGHSVTLADII 126
EID N + LG+ Y A ++K+ L L++ L + +L G V LADII
Sbjct: 711 NEIDPNAIPWLYAILGYYDYNAKDSNKAKENMKKVLAFLDAQLLNKPFLTGFRVALADII 770
Query: 127 TTSNLYIGFAKLLVKSFTSEFPHVERYFWTLVNQPNFRKILGQVKQTEAMPPIPSAKQQP 186
+L+ + + +F S + +V R+F T +NQPNF+ ++G+ E M + K++
Sbjct: 771 VVCSLFNFYKMVFEPTFRSPYVNVNRWFTTCINQPNFKAVIGEFALCEKMMVYVAPKKEV 830
Query: 187 KESKPKTKDEPKKVAKSEPEKPKVEVEEEAPKPKPKNPLDLLPPSKMILDDWKRLYSNTK 246
KE KPK PKK K + +V+ KPK KNPLD L PS +LD++KR YSN +
Sbjct: 831 KEEKPKA--APKKEVKEDAGDDEVD-----EKPKKKNPLDELAPSTFVLDEFKRTYSNNE 883
>gb|AAH80966.1| Unknown (protein for IMAGE:6981438) [Xenopus tropicalis]
Length = 435
Score = 167 bits (424), Expect = 3e-40
Identities = 105/264 (39%), Positives = 142/264 (53%), Gaps = 25/264 (9%)
Query: 2 GVSNKTPQFINMNPIGKVPVLETPDG-PVFESNAIARYVARLGHNNLFGSSLIHQAQIDQ 60
GV+NKT +F+ P+GKVP E DG +FES+AIA YV G++ L G++ +HQAQ+ Q
Sbjct: 38 GVTNKTSEFLKKFPLGKVPAFEGNDGFCLFESSAIAHYV---GNDELRGTNRLHQAQVIQ 94
Query: 61 WIDFSSLEIDANIMKLYLPRLGFATYFPPVEENAISSLKRALEALNSHLAHNTYLVGHSV 120
W+ FS I P LG Y E A +K L L+SHL T+LVG V
Sbjct: 95 WVSFSDSHIVPPASAWVFPTLGIMQYNKQATEQAKEEIKAVLGVLDSHLKTRTFLVGERV 154
Query: 121 TLADIITTSNLYIGFAKLLVKSFTSEFPHVERYFWTLVNQPNFRKILGQVKQTEAMPPIP 180
TLADI T +L + ++L SF F +V R+F T VNQP FR +LG+VK + M
Sbjct: 155 TLADIAVTCSLLWLYKQVLEPSFRQPFGNVTRWFVTCVNQPEFRAVLGEVKLCDKMAQFD 214
Query: 181 S---AKQQPKESKPKTKDEPKKVAKSEPEKPKVEVEEEAP-----------------KPK 220
+ A+ QPK+ PK K++P K K E E+ K AP +PK
Sbjct: 215 AKKFAEMQPKKETPK-KEKPAKEPKKEKEEKKKATPAPAPAPAPEDDMDESEKALAAEPK 273
Query: 221 PKNPLDLLPPSKMILDDWKRLYSN 244
K+P LP S ++D++KR YSN
Sbjct: 274 SKDPYAHLPKSSFVMDEFKRKYSN 297
>gb|AAH84224.1| Unknown (protein for MGC:80886) [Xenopus laevis]
Length = 447
Score = 160 bits (406), Expect = 3e-38
Identities = 101/262 (38%), Positives = 143/262 (54%), Gaps = 22/262 (8%)
Query: 2 GVSNKTPQFINMNPIGKVPVLETPDG-PVFESNAIARYVARLGHNNLFGSS-LIHQAQID 59
G++NKTP+F+ P+GKVP E DG +FES+AIA YVA ++ L GS+ +HQAQ+
Sbjct: 51 GLTNKTPEFLKKFPLGKVPAFEGNDGFCLFESSAIAHYVA---NDELRGSNNRLHQAQVI 107
Query: 60 QWIDFSSLEIDANIMKLYLPRLGFATYFPPVEENAISSLKRALEALNSHLAHNTYLVGHS 119
QW+ FS + P LG + E A +K L L+SHL T+LVG
Sbjct: 108 QWVGFSDSHVVPPASAWVFPTLGIMQFNKQATEQAKEEIKTVLGVLDSHLQTRTFLVGER 167
Query: 120 VTLADIITTSNLYIGFAKLLVKSFTSEFPHVERYFWTLVNQPNFRKILGQVKQTEAM--- 176
+TLADI +L + ++L SF + +V R+F T VNQP FR +LG+VK + M
Sbjct: 168 ITLADITLACSLLWLYKQVLEPSFRQPYGNVTRWFVTCVNQPEFRAVLGEVKLCDKMAQF 227
Query: 177 -----PPIPSAKQQPKESKP-----KTKDEPKKVAKSEPEKPKVEVEEE----APKPKPK 222
+ K+ PK+ KP K K+E KK + P+ E++E A +PK K
Sbjct: 228 DAKKFAEVQPKKETPKKEKPAKEPKKEKEEKKKATPAPAPAPEDELDESEKALAAEPKSK 287
Query: 223 NPLDLLPPSKMILDDWKRLYSN 244
+P LP S I+D++KR YSN
Sbjct: 288 DPYAHLPKSSFIMDEFKRKYSN 309
>gb|AAH79398.1| Eef1g protein [Rattus norvegicus] gi|55583762|sp|Q68FR6|EF1G_RAT
Elongation factor 1-gamma (EF-1-gamma) (eEF-1B gamma)
Length = 437
Score = 159 bits (403), Expect = 7e-38
Identities = 103/262 (39%), Positives = 138/262 (52%), Gaps = 22/262 (8%)
Query: 2 GVSNKTPQFINMNPIGKVPVLETPDG-PVFESNAIARYVARLGHNNLFGSSLIHQAQIDQ 60
G +N+TP+F+ P GKVP E DG VFESNAIA YV+ + L GS+ AQ+ Q
Sbjct: 41 GQTNRTPEFLRKFPAGKVPAFEGDDGFCVFESNAIAYYVS---NEELRGSTPEAAAQVVQ 97
Query: 61 WIDFSSLEIDANIMKLYLPRLGFATYFPPVEENAISSLKRALEALNSHLAHNTYLVGHSV 120
W+ F+ +I P LG + ENA +KR L L++HL T+LVG V
Sbjct: 98 WVSFADSDIVPPASTWVFPTLGIMHHNKQATENAKEEVKRILGLLDTHLKTRTFLVGERV 157
Query: 121 TLADIITTSNLYIGFAKLLVKSFTSEFPHVERYFWTLVNQPNFRKILGQVKQTEAMPPIP 180
TLADI L + ++L SF FP+ R+F T +NQP FR ILG+VK E M
Sbjct: 158 TLADITVVCTLLWLYKQVLEPSFRQAFPNTNRWFLTCINQPQFRAILGEVKLCEKMAQFD 217
Query: 181 S---AKQQPKESKPK--------------TKDEPKKVAKSEPEKPKVEVEEE-APKPKPK 222
+ A+ QPK+ P+ + E KK A PE+ E E+ A +PK K
Sbjct: 218 AKKFAESQPKKDTPRKEKGSREEKQKPQTERKEEKKAAAPAPEEEMDECEQALAAEPKAK 277
Query: 223 NPLDLLPPSKMILDDWKRLYSN 244
+P LP S +LD++KR YSN
Sbjct: 278 DPFAHLPKSTFVLDEFKRKYSN 299
>gb|AAH83071.1| Eukaryotic translation elongation factor 1 gamma [Mus musculus]
gi|13626388|sp|Q9D8N0|EF1G_MOUSE Elongation factor
1-gamma (EF-1-gamma) (eEF-1B gamma)
gi|26349423|dbj|BAC38351.1| unnamed protein product [Mus
musculus] gi|26341388|dbj|BAC34356.1| unnamed protein
product [Mus musculus] gi|12841703|dbj|BAB25320.1|
unnamed protein product [Mus musculus]
Length = 437
Score = 159 bits (403), Expect = 7e-38
Identities = 103/262 (39%), Positives = 138/262 (52%), Gaps = 22/262 (8%)
Query: 2 GVSNKTPQFINMNPIGKVPVLETPDG-PVFESNAIARYVARLGHNNLFGSSLIHQAQIDQ 60
G +N+TP+F+ P GKVP E DG VFESNAIA YV+ + L GS+ AQ+ Q
Sbjct: 41 GQTNRTPEFLRKFPAGKVPAFEGDDGFCVFESNAIAYYVS---NEELRGSTPEAAAQVVQ 97
Query: 61 WIDFSSLEIDANIMKLYLPRLGFATYFPPVEENAISSLKRALEALNSHLAHNTYLVGHSV 120
W+ F+ +I P LG + ENA +KR L L++HL T+LVG V
Sbjct: 98 WVSFADSDIVPPASTWVFPTLGIMHHNKQATENAKEEVKRILGLLDTHLKTRTFLVGERV 157
Query: 121 TLADIITTSNLYIGFAKLLVKSFTSEFPHVERYFWTLVNQPNFRKILGQVKQTEAMPPIP 180
TLADI L + ++L SF FP+ R+F T +NQP FR ILG+VK E M
Sbjct: 158 TLADITVVCTLLWLYKQVLEPSFRQAFPNTNRWFLTCINQPQFRAILGEVKLCEKMAQFD 217
Query: 181 S---AKQQPKESKPK--------------TKDEPKKVAKSEPEKPKVEVEEE-APKPKPK 222
+ A+ QPK+ P+ + E KK A PE+ E E+ A +PK K
Sbjct: 218 AKKFAESQPKKDTPRKEKGSREEKQKPQAERKEEKKAAAPAPEEEMDECEQALAAEPKAK 277
Query: 223 NPLDLLPPSKMILDDWKRLYSN 244
+P LP S +LD++KR YSN
Sbjct: 278 DPFAHLPKSTFVLDEFKRKYSN 299
>ref|NP_080283.2| eukaryotic translation elongation factor 1 gamma [Mus musculus]
gi|16506251|gb|AAL23895.1| elongation factor-like
protein [Mus musculus]
Length = 437
Score = 159 bits (403), Expect = 7e-38
Identities = 103/262 (39%), Positives = 138/262 (52%), Gaps = 22/262 (8%)
Query: 2 GVSNKTPQFINMNPIGKVPVLETPDG-PVFESNAIARYVARLGHNNLFGSSLIHQAQIDQ 60
G +N+TP+F+ P GKVP E DG VFESNAIA YV+ + L GS+ AQ+ Q
Sbjct: 41 GQTNRTPEFLRKFPAGKVPAFEGDDGFCVFESNAIAYYVS---NEELRGSTPEAAAQVVQ 97
Query: 61 WIDFSSLEIDANIMKLYLPRLGFATYFPPVEENAISSLKRALEALNSHLAHNTYLVGHSV 120
W+ F+ +I P LG + ENA +KR L L++HL T+LVG V
Sbjct: 98 WVSFADSDIVPPASTWVFPTLGIMHHNKQATENAKEEVKRILGLLDTHLKTRTFLVGERV 157
Query: 121 TLADIITTSNLYIGFAKLLVKSFTSEFPHVERYFWTLVNQPNFRKILGQVKQTEAMPPIP 180
TLADI L + ++L SF FP+ R+F T +NQP FR ILG+VK E M
Sbjct: 158 TLADITVVCTLLWLYRQVLEPSFRQAFPNTNRWFLTCINQPQFRAILGEVKLCEKMAQFD 217
Query: 181 S---AKQQPKESKPK--------------TKDEPKKVAKSEPEKPKVEVEEE-APKPKPK 222
+ A+ QPK+ P+ + E KK A PE+ E E+ A +PK K
Sbjct: 218 AKKFAESQPKKDTPRKEKGSREEKQKPQAERKEEKKAAAPAPEEEMDECEQALAAEPKAK 277
Query: 223 NPLDLLPPSKMILDDWKRLYSN 244
+P LP S +LD++KR YSN
Sbjct: 278 DPFAHLPKSTFVLDEFKRKYSN 299
Database: nr
Posted date: Jul 5, 2005 12:34 AM
Number of letters in database: 863,360,394
Number of sequences in database: 2,540,612
Lambda K H
0.317 0.134 0.393
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 524,329,434
Number of Sequences: 2540612
Number of extensions: 22649005
Number of successful extensions: 158383
Number of sequences better than 10.0: 2472
Number of HSP's better than 10.0 without gapping: 653
Number of HSP's successfully gapped in prelim test: 1874
Number of HSP's that attempted gapping in prelim test: 140890
Number of HSP's gapped (non-prelim): 11105
length of query: 305
length of database: 863,360,394
effective HSP length: 127
effective length of query: 178
effective length of database: 540,702,670
effective search space: 96245075260
effective search space used: 96245075260
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 75 (33.5 bits)
Medicago: description of AC140549.6


