

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC140547.14 - phase: 0
(270 letters)
Database: nr
2,540,612 sequences; 863,360,394 total letters
Searching..................................................done
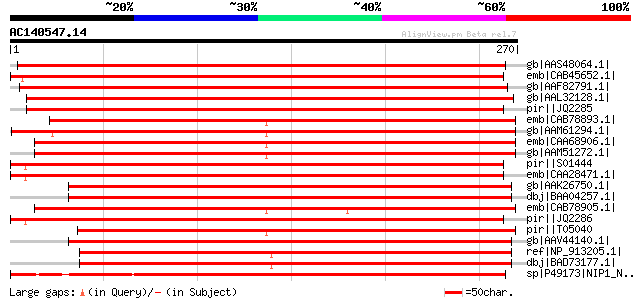
Score E
Sequences producing significant alignments: (bits) Value
gb|AAS48064.1| NIP2 [Medicago truncatula] 404 e-111
emb|CAB45652.1| nodulin26-like intrinsic protein [Pisum sativum] 396 e-109
gb|AAF82791.1| multifunctional transport intrinsic membrane prot... 379 e-104
gb|AAL32128.1| multifunctional aquaporin [Medicago truncatula] 372 e-102
pir||JQ2285 nodulin-26 - soybean 364 2e-99
emb|CAB78893.1| major intrinsic protein (MIP)-like [Arabidopsis ... 361 1e-98
gb|AAM61294.1| major intrinsic protein (MIP)- like [Arabidopsis ... 358 1e-97
emb|CAA68906.1| NLM1 protein (NodLikeMip1) [Arabidopsis thaliana] 348 7e-95
gb|AAM51272.1| putative nodulin-26 protein [Arabidopsis thaliana... 348 7e-95
pir||S01444 nodulin-26 precursor - soybean 344 2e-93
emb|CAA28471.1| nodulin [Glycine max] gi|1352509|sp|P08995|NO26_... 341 1e-92
gb|AAK26750.1| NOD26-like membrane integral protein ZmNIP1-1 [Ze... 341 1e-92
dbj|BAA04257.1| major intrinsic protein [Oryza sativa] gi|502517... 340 2e-92
emb|CAB78905.1| nodulin-26-like protein [Arabidopsis thaliana] g... 340 3e-92
pir||JQ2286 nodulin-26 - soybean 340 3e-92
pir||T05040 nodulin-26-like protein F13C5.200 - Arabidopsis thal... 336 4e-91
gb|AAV44140.1| unknown protein [Oryza sativa (japonica cultivar-... 317 2e-85
ref|NP_913205.1| putative major intrinsic protein [Oryza sativa ... 300 4e-80
dbj|BAD73177.1| putative membrane integral protein ZmNIP1-1 [Ory... 300 4e-80
sp|P49173|NIP1_NICAL Probable aquaporin NIP-type (Pollen-specifi... 291 1e-77
>gb|AAS48064.1| NIP2 [Medicago truncatula]
Length = 269
Score = 404 bits (1037), Expect = e-111
Identities = 197/260 (75%), Positives = 228/260 (86%)
Query: 5 SNGNLDVVMNINDDATKKCDDTTIDDHVPLLQKLVAEVVGTFFLIFAGCAAVVVNLNNDK 64
SN ++V+N+N D + K +D+T LQK VAEV+GT+FLIFAGCA+V+VN NN+
Sbjct: 7 SNATNEIVLNVNKDVSNKSEDSTSHATASFLQKSVAEVIGTYFLIFAGCASVLVNKNNEN 66
Query: 65 VVTLPGISIVWGLAVMVLVYSIGHISGAHFNPAVTIAHTTTGRFPLKQLPAYIIAQVVGS 124
VVTLPGISIVWGLAVMVLVYS+GHISGAHFNPAVTIA +T RFPLKQ+PAY+ AQV GS
Sbjct: 67 VVTLPGISIVWGLAVMVLVYSLGHISGAHFNPAVTIAFASTKRFPLKQVPAYVAAQVFGS 126
Query: 125 TLASGVLKLIFSGKENQFAGTLPAGSDLQAFVVEFIITFFLMFIISGVATDNRAIGELAG 184
TLASG L+LIF+GK NQF GTLPAGSDLQAFV+EFIITF+ MFIISGVATDNRAIGELAG
Sbjct: 127 TLASGTLRLIFTGKHNQFVGTLPAGSDLQAFVIEFIITFYPMFIISGVATDNRAIGELAG 186
Query: 185 LAVGSTVILNVLFAGPITGASMNPARSLGPAIVHHEYRGIWIYMVSPILGALAGTWTYTF 244
+AVGSTV+LNV+FAGPITGASMNPARS+GPA++H EYRGIWIY+VSPILGA+AG W Y
Sbjct: 187 IAVGSTVLLNVMFAGPITGASMNPARSIGPALLHSEYRGIWIYLVSPILGAVAGAWVYNV 246
Query: 245 LRITNKPVRELTKSSSFLKA 264
+R T+KPVRE+TKSSSFLKA
Sbjct: 247 IRYTDKPVREITKSSSFLKA 266
>emb|CAB45652.1| nodulin26-like intrinsic protein [Pisum sativum]
Length = 270
Score = 396 bits (1017), Expect = e-109
Identities = 201/266 (75%), Positives = 229/266 (85%), Gaps = 3/266 (1%)
Query: 1 MGDIS--NGNLDVVMNINDDATKKC-DDTTIDDHVPLLQKLVAEVVGTFFLIFAGCAAVV 57
MGD S N ++V+N+N D + +D+T LLQKLVAEVVGT+FLIFAGCAAV
Sbjct: 1 MGDNSGCNETNEIVVNVNKDVSNITQEDSTAHATASLLQKLVAEVVGTYFLIFAGCAAVA 60
Query: 58 VNLNNDKVVTLPGISIVWGLAVMVLVYSIGHISGAHFNPAVTIAHTTTGRFPLKQLPAYI 117
VN NND VVTLPGISIVWGLAVMVLVYS+GHISGAHFNPAVTIA TT RFPLKQ+PAYI
Sbjct: 61 VNKNNDNVVTLPGISIVWGLAVMVLVYSLGHISGAHFNPAVTIAFATTRRFPLKQVPAYI 120
Query: 118 IAQVVGSTLASGVLKLIFSGKENQFAGTLPAGSDLQAFVVEFIITFFLMFIISGVATDNR 177
AQV GSTLASG L+L+FSGK +QF GTL AGS+LQAFV+EFIITF+LMFIISGVATDNR
Sbjct: 121 AAQVFGSTLASGTLRLLFSGKHDQFVGTLAAGSNLQAFVMEFIITFYLMFIISGVATDNR 180
Query: 178 AIGELAGLAVGSTVILNVLFAGPITGASMNPARSLGPAIVHHEYRGIWIYMVSPILGALA 237
AIGELAG+AVGSTV+LNV+FAGPITGASMNPARS+GPA VH+EYRGIWIYM+SPI+GA++
Sbjct: 181 AIGELAGIAVGSTVLLNVMFAGPITGASMNPARSIGPAFVHNEYRGIWIYMISPIVGAVS 240
Query: 238 GTWTYTFLRITNKPVRELTKSSSFLK 263
G W Y +R T+KPVRE+TKS SFLK
Sbjct: 241 GAWVYNVIRYTDKPVREITKSGSFLK 266
>gb|AAF82791.1| multifunctional transport intrinsic membrane protein 2; LIMP2
[Lotus japonicus]
Length = 270
Score = 379 bits (974), Expect = e-104
Identities = 183/260 (70%), Positives = 223/260 (85%)
Query: 6 NGNLDVVMNINDDATKKCDDTTIDDHVPLLQKLVAEVVGTFFLIFAGCAAVVVNLNNDKV 65
NG +VV+N+N DA+K + + + V LQK++AE+VGT+F IFAGCA++VVN NND V
Sbjct: 11 NGAHEVVVNVNKDASKTIEVSDTNFTVSFLQKVIAELVGTYFFIFAGCASIVVNKNNDNV 70
Query: 66 VTLPGISIVWGLAVMVLVYSIGHISGAHFNPAVTIAHTTTGRFPLKQLPAYIIAQVVGST 125
VTLPGI++VWGLAVMVLVYS+GHISGAHFNPA TIA +T RFP KQ+PAY+ AQV+GST
Sbjct: 71 VTLPGIALVWGLAVMVLVYSLGHISGAHFNPAATIAFASTKRFPWKQVPAYVSAQVLGST 130
Query: 126 LASGVLKLIFSGKENQFAGTLPAGSDLQAFVVEFIITFFLMFIISGVATDNRAIGELAGL 185
LASG L+LIFSGK NQFAG LP GS+LQAFV+EFIITFFL+FI+ GVATD+RAIGE+AG+
Sbjct: 131 LASGTLRLIFSGKHNQFAGALPTGSNLQAFVIEFIITFFLIFILFGVATDDRAIGEVAGI 190
Query: 186 AVGSTVILNVLFAGPITGASMNPARSLGPAIVHHEYRGIWIYMVSPILGALAGTWTYTFL 245
VGSTV+LNVLFAGPITGASMNPARS+G A VH+EYRGIWIY++SP LGA+AG W Y +
Sbjct: 191 VVGSTVLLNVLFAGPITGASMNPARSIGSAFVHNEYRGIWIYLLSPTLGAVAGAWVYNIV 250
Query: 246 RITNKPVRELTKSSSFLKAV 265
R T+KP+RE+TK+ SFLK +
Sbjct: 251 RYTDKPLREITKNVSFLKGI 270
>gb|AAL32128.1| multifunctional aquaporin [Medicago truncatula]
Length = 276
Score = 372 bits (955), Expect = e-102
Identities = 177/259 (68%), Positives = 221/259 (84%)
Query: 10 DVVMNINDDATKKCDDTTIDDHVPLLQKLVAEVVGTFFLIFAGCAAVVVNLNNDKVVTLP 69
+VV++ N D++ C + VP LQKL+AE+VGT+FLIFAGCA++VVN +ND VVTLP
Sbjct: 13 EVVLDTNKDSSDTCKGSGSFVSVPFLQKLIAEMVGTYFLIFAGCASIVVNKDNDNVVTLP 72
Query: 70 GISIVWGLAVMVLVYSIGHISGAHFNPAVTIAHTTTGRFPLKQLPAYIIAQVVGSTLASG 129
GI+IVWGL ++VL+YS+GHISGAHFNPAVTIA TT RFPL Q+PAYI AQ++G+TLASG
Sbjct: 73 GIAIVWGLTLLVLIYSLGHISGAHFNPAVTIAFATTRRFPLLQVPAYISAQLLGATLASG 132
Query: 130 VLKLIFSGKENQFAGTLPAGSDLQAFVVEFIITFFLMFIISGVATDNRAIGELAGLAVGS 189
LKLIFSG + F+GTLP+GS+LQAFV+EFI TF+LMF ISGVATD RAIGELAG+A+GS
Sbjct: 133 TLKLIFSGAHDHFSGTLPSGSNLQAFVLEFITTFYLMFTISGVATDTRAIGELAGIAIGS 192
Query: 190 TVILNVLFAGPITGASMNPARSLGPAIVHHEYRGIWIYMVSPILGALAGTWTYTFLRITN 249
T++LNV+ AGP+TGASMNP R+LGPA VH+EYRGIWIY++SPILGA+AG W Y +R TN
Sbjct: 193 TLLLNVMIAGPVTGASMNPVRTLGPAFVHNEYRGIWIYLLSPILGAIAGAWVYNTVRYTN 252
Query: 250 KPVRELTKSSSFLKAVSKG 268
KP+RE+T+S+SFLK +G
Sbjct: 253 KPLREITQSASFLKEAGRG 271
>pir||JQ2285 nodulin-26 - soybean
Length = 271
Score = 364 bits (934), Expect = 2e-99
Identities = 175/254 (68%), Positives = 214/254 (83%)
Query: 10 DVVMNINDDATKKCDDTTIDDHVPLLQKLVAEVVGTFFLIFAGCAAVVVNLNNDKVVTLP 69
+VV+N+ D +K + + VP LQKLVAEVVGT+FLIFAGCA+VVVN NND VVTLP
Sbjct: 12 EVVVNVTKDTSKTMEPSDSFVSVPFLQKLVAEVVGTYFLIFAGCASVVVNKNNDNVVTLP 71
Query: 70 GISIVWGLAVMVLVYSIGHISGAHFNPAVTIAHTTTGRFPLKQLPAYIIAQVVGSTLASG 129
GI+I WGL V VLVY++GHISGAHFNPAVTIA +T RFPL Q+PAY+ AQ++GSTLASG
Sbjct: 72 GIAIAWGLVVTVLVYTVGHISGAHFNPAVTIAFASTRRFPLMQVPAYVAAQLLGSTLASG 131
Query: 130 VLKLIFSGKENQFAGTLPAGSDLQAFVVEFIITFFLMFIISGVATDNRAIGELAGLAVGS 189
LKL+F GK +QF+GTLP G++LQAFV EFIITF LMF+ISGVATDNRA+GELAG+A+GS
Sbjct: 132 TLKLLFMGKHDQFSGTLPNGTNLQAFVFEFIITFLLMFVISGVATDNRAVGELAGIAIGS 191
Query: 190 TVILNVLFAGPITGASMNPARSLGPAIVHHEYRGIWIYMVSPILGALAGTWTYTFLRITN 249
T++LNV+ GP+TGASMNP RSLGPAIVH EYRGIWIY+++P++GA+AG Y +R T+
Sbjct: 192 TILLNVIIGGPVTGASMNPVRSLGPAIVHGEYRGIWIYLLAPVVGAIAGALVYNTIRYTD 251
Query: 250 KPVRELTKSSSFLK 263
KP+ E TKS+SFLK
Sbjct: 252 KPLSETTKSASFLK 265
>emb|CAB78893.1| major intrinsic protein (MIP)-like [Arabidopsis thaliana]
gi|2832619|emb|CAA16748.1| major intrinsic protein
(MIP)-like [Arabidopsis thaliana]
gi|11071656|emb|CAC14597.1| aquaglyceroporin
[Arabidopsis thaliana] gi|23197776|gb|AAN15415.1| major
intrinsic protein (MIP)- like [Arabidopsis thaliana]
gi|18252891|gb|AAL62372.1| major intrinsic protein
(MIP)- like [Arabidopsis thaliana]
gi|32363340|sp|Q8LFP7|NIP12_ARATH Aquaporin NIP1.2
(NOD26-like intrinsic protein 1.2) (Nodulin-26-like
major intrinsic protein 2) (AtNLM2) (NLM2 protein)
(NodLikeMip2) gi|15234059|ref|NP_193626.1|
aquaglyceroporin / NOD26-like major intrinsic protein 2
(NLM2) [Arabidopsis thaliana] gi|7449724|pir||T05028
nodulin-26-like protein F13C5.80 - Arabidopsis thaliana
Length = 294
Score = 361 bits (927), Expect = 1e-98
Identities = 176/255 (69%), Positives = 215/255 (84%), Gaps = 7/255 (2%)
Query: 22 KCDDTTIDDHVPLLQKLVAEVVGTFFLIFAGCAAVVVNLNNDKVVTLPGISIVWGLAVMV 81
K D+ + VP LQKL+AEV+GT+FLIFAGCAAV VN +DK VTLPGI+IVWGL VMV
Sbjct: 38 KKQDSLLSISVPFLQKLMAEVLGTYFLIFAGCAAVAVNTQHDKAVTLPGIAIVWGLTVMV 97
Query: 82 LVYSIGHISGAHFNPAVTIAHTTTGRFPLKQLPAYIIAQVVGSTLASGVLKLIF------ 135
LVYS+GHISGAHFNPAVTIA + GRFPLKQ+PAY+I+QV+GSTLA+ L+L+F
Sbjct: 98 LVYSLGHISGAHFNPAVTIAFASCGRFPLKQVPAYVISQVIGSTLAAATLRLLFGLDQDV 157
Query: 136 -SGKENQFAGTLPAGSDLQAFVVEFIITFFLMFIISGVATDNRAIGELAGLAVGSTVILN 194
SGK + F GTLP+GS+LQ+FV+EFIITF+LMF+ISGVATDNRAIGELAGLAVGSTV+LN
Sbjct: 158 CSGKHDVFVGTLPSGSNLQSFVIEFIITFYLMFVISGVATDNRAIGELAGLAVGSTVLLN 217
Query: 195 VLFAGPITGASMNPARSLGPAIVHHEYRGIWIYMVSPILGALAGTWTYTFLRITNKPVRE 254
V+ AGP++GASMNP RSLGPA+V+ YRG+WIY+VSPI+GA++G W Y +R T+KP+RE
Sbjct: 218 VIIAGPVSGASMNPGRSLGPAMVYSCYRGLWIYIVSPIVGAVSGAWVYNMVRYTDKPLRE 277
Query: 255 LTKSSSFLKAVSKGA 269
+TKS SFLK V G+
Sbjct: 278 ITKSGSFLKTVRNGS 292
>gb|AAM61294.1| major intrinsic protein (MIP)- like [Arabidopsis thaliana]
Length = 293
Score = 358 bits (918), Expect = 1e-97
Identities = 180/283 (63%), Positives = 224/283 (78%), Gaps = 15/283 (5%)
Query: 2 GDISNGNLDVVMNINDDATK--------KCDDTTIDDHVPLLQKLVAEVVGTFFLIFAGC 53
GD +G + V + D+ + K D+ + VP LQKL+AEV+GT+FLIFAGC
Sbjct: 9 GDARDGAVVVNLKEEDEQQQQQAIHKPLKKQDSLLSISVPFLQKLMAEVLGTYFLIFAGC 68
Query: 54 AAVVVNLNNDKVVTLPGISIVWGLAVMVLVYSIGHISGAHFNPAVTIAHTTTGRFPLKQL 113
AAV VN +DK VTL GI+IVWGL VMVLVYS+GHISGAHFNPAVTIA + GRFPLKQ+
Sbjct: 69 AAVAVNTQHDKAVTLLGIAIVWGLTVMVLVYSLGHISGAHFNPAVTIAFASCGRFPLKQV 128
Query: 114 PAYIIAQVVGSTLASGVLKLIF-------SGKENQFAGTLPAGSDLQAFVVEFIITFFLM 166
PAY+I+QV+GSTLA+ L+L+F SGK + F GTLP+GS+LQ+FV+EFIITF+LM
Sbjct: 129 PAYVISQVIGSTLAAATLRLLFGLDQDVCSGKHDVFVGTLPSGSNLQSFVIEFIITFYLM 188
Query: 167 FIISGVATDNRAIGELAGLAVGSTVILNVLFAGPITGASMNPARSLGPAIVHHEYRGIWI 226
F+ISGVATDNRAIGELAGLAVGSTV+LNV+ AGP++GASMNP RSLGPA+V+ YRG+WI
Sbjct: 189 FVISGVATDNRAIGELAGLAVGSTVLLNVIIAGPVSGASMNPGRSLGPAMVYSCYRGLWI 248
Query: 227 YMVSPILGALAGTWTYTFLRITNKPVRELTKSSSFLKAVSKGA 269
Y+VSPI+GA++G W Y +R T+KP+RE+TKS SFLK V G+
Sbjct: 249 YIVSPIVGAVSGAWVYNMVRYTDKPLREITKSGSFLKTVRNGS 291
>emb|CAA68906.1| NLM1 protein (NodLikeMip1) [Arabidopsis thaliana]
Length = 279
Score = 348 bits (894), Expect = 7e-95
Identities = 169/263 (64%), Positives = 212/263 (80%), Gaps = 7/263 (2%)
Query: 14 NINDDATKKCDDTTIDDHVPLLQKLVAEVVGTFFLIFAGCAAVVVNLNNDKVVTLPGISI 73
+I++ K D+ + VP LQKL+AE +GT+FL+F GCA+VVVN+ ND VVTLPGI+I
Sbjct: 16 DIHNPRPLKKQDSLLSVSVPFLQKLIAEFLGTYFLVFTGCASVVVNMQNDNVVTLPGIAI 75
Query: 74 VWGLAVMVLVYSIGHISGAHFNPAVTIAHTTTGRFPLKQLPAYIIAQVVGSTLASGVLKL 133
VWGL +MVL+YS+GHISGAH NPAVTIA + GRFPLKQ+PAY+I+QV+GSTLA+ L+L
Sbjct: 76 VWGLTIMVLIYSLGHISGAHINPAVTIAFASCGRFPLKQVPAYVISQVIGSTLAAATLRL 135
Query: 134 IF-------SGKENQFAGTLPAGSDLQAFVVEFIITFFLMFIISGVATDNRAIGELAGLA 186
+F SGK + F G+ P GSDLQAF +EFI+TF+LMFIISGVATDNRAIGELAGLA
Sbjct: 136 LFGLDHDVCSGKHDVFIGSSPVGSDLQAFTMEFIVTFYLMFIISGVATDNRAIGELAGLA 195
Query: 187 VGSTVILNVLFAGPITGASMNPARSLGPAIVHHEYRGIWIYMVSPILGALAGTWTYTFLR 246
+GSTV+LNVL A P++ ASMNP RSLGPA+V+ Y+GIWIY+V+P LGA+AG W Y +R
Sbjct: 196 IGSTVLLNVLIAAPVSSASMNPGRSLGPALVYGCYKGIWIYLVAPTLGAIAGAWVYNTVR 255
Query: 247 ITNKPVRELTKSSSFLKAVSKGA 269
T+KP+RE+TKS SFLK V G+
Sbjct: 256 YTDKPLREITKSGSFLKTVRIGS 278
>gb|AAM51272.1| putative nodulin-26 protein [Arabidopsis thaliana]
gi|17380644|gb|AAL36152.1| putative nodulin-26 protein
[Arabidopsis thaliana] gi|21536734|gb|AAM61066.1|
nodulin-26-like protein [Arabidopsis thaliana]
gi|18415224|ref|NP_567572.1| major intrinsic family
protein / MIP family protein [Arabidopsis thaliana]
gi|32363362|sp|Q8VZW1|NI11_ARATH Aquaporin NIP1.1
(NOD26-like intrinsic protein 1.1) (Nodulin-26-like
major intrinsic protein 1) (AtNLM1) (NLM1 protein)
(NodLikeMip1)
Length = 296
Score = 348 bits (894), Expect = 7e-95
Identities = 169/263 (64%), Positives = 212/263 (80%), Gaps = 7/263 (2%)
Query: 14 NINDDATKKCDDTTIDDHVPLLQKLVAEVVGTFFLIFAGCAAVVVNLNNDKVVTLPGISI 73
+I++ K D+ + VP LQKL+AE +GT+FL+F GCA+VVVN+ ND VVTLPGI+I
Sbjct: 33 DIHNPRPLKKQDSLLSVSVPFLQKLIAEFLGTYFLVFTGCASVVVNMQNDNVVTLPGIAI 92
Query: 74 VWGLAVMVLVYSIGHISGAHFNPAVTIAHTTTGRFPLKQLPAYIIAQVVGSTLASGVLKL 133
VWGL +MVL+YS+GHISGAH NPAVTIA + GRFPLKQ+PAY+I+QV+GSTLA+ L+L
Sbjct: 93 VWGLTIMVLIYSLGHISGAHINPAVTIAFASCGRFPLKQVPAYVISQVIGSTLAAATLRL 152
Query: 134 IF-------SGKENQFAGTLPAGSDLQAFVVEFIITFFLMFIISGVATDNRAIGELAGLA 186
+F SGK + F G+ P GSDLQAF +EFI+TF+LMFIISGVATDNRAIGELAGLA
Sbjct: 153 LFGLDHDVCSGKHDVFIGSSPVGSDLQAFTMEFIVTFYLMFIISGVATDNRAIGELAGLA 212
Query: 187 VGSTVILNVLFAGPITGASMNPARSLGPAIVHHEYRGIWIYMVSPILGALAGTWTYTFLR 246
+GSTV+LNVL A P++ ASMNP RSLGPA+V+ Y+GIWIY+V+P LGA+AG W Y +R
Sbjct: 213 IGSTVLLNVLIAAPVSSASMNPGRSLGPALVYGCYKGIWIYLVAPTLGAIAGAWVYNTVR 272
Query: 247 ITNKPVRELTKSSSFLKAVSKGA 269
T+KP+RE+TKS SFLK V G+
Sbjct: 273 YTDKPLREITKSGSFLKTVRIGS 295
>pir||S01444 nodulin-26 precursor - soybean
Length = 271
Score = 344 bits (882), Expect = 2e-93
Identities = 164/265 (61%), Positives = 214/265 (79%), Gaps = 2/265 (0%)
Query: 1 MGDISNG--NLDVVMNINDDATKKCDDTTIDDHVPLLQKLVAEVVGTFFLIFAGCAAVVV 58
M D S G + +VV+N+ + ++ + VP LQKLVAE VGT+FLIFAGCA++VV
Sbjct: 1 MADYSAGTESQEVVVNVTKNTSETIQRSDSLVSVPFLQKLVAEAVGTYFLIFAGCASLVV 60
Query: 59 NLNNDKVVTLPGISIVWGLAVMVLVYSIGHISGAHFNPAVTIAHTTTGRFPLKQLPAYII 118
N N ++T PGI+IVWGL + VLVY++GHISG HFNPAVTIA +T RFPL Q+PAY++
Sbjct: 61 NENYYNMITFPGIAIVWGLVLTVLVYTVGHISGGHFNPAVTIAFASTRRFPLIQVPAYVV 120
Query: 119 AQVVGSTLASGVLKLIFSGKENQFAGTLPAGSDLQAFVVEFIITFFLMFIISGVATDNRA 178
AQ++GS LASG L+L+F G +QF+GT+P G++LQAFV EFI+TFFLMF+I GVATDNRA
Sbjct: 121 AQLLGSILASGTLRLLFMGNHDQFSGTVPNGTNLQAFVFEFIMTFFLMFVICGVATDNRA 180
Query: 179 IGELAGLAVGSTVILNVLFAGPITGASMNPARSLGPAIVHHEYRGIWIYMVSPILGALAG 238
+GELAG+A+GST++LNV+ GP+TGASMNPARSLGPA VH EY GIWIY+++P++GA+AG
Sbjct: 181 VGELAGIAIGSTLLLNVIIGGPVTGASMNPARSLGPAFVHGEYEGIWIYLLAPVVGAIAG 240
Query: 239 TWTYTFLRITNKPVRELTKSSSFLK 263
W Y +R T+KP+ E+TKS+SFLK
Sbjct: 241 AWVYNIVRYTDKPLSEITKSASFLK 265
>emb|CAA28471.1| nodulin [Glycine max] gi|1352509|sp|P08995|NO26_SOYBN Nodulin-26
(N-26)
Length = 271
Score = 341 bits (875), Expect = 1e-92
Identities = 163/265 (61%), Positives = 212/265 (79%), Gaps = 2/265 (0%)
Query: 1 MGDISNG--NLDVVMNINDDATKKCDDTTIDDHVPLLQKLVAEVVGTFFLIFAGCAAVVV 58
M D S G + +VV+N+ + ++ + VP LQKLVAE VGT+FLIFAGCA++VV
Sbjct: 1 MADYSAGTESQEVVVNVTKNTSETIQRSDSLVSVPFLQKLVAEAVGTYFLIFAGCASLVV 60
Query: 59 NLNNDKVVTLPGISIVWGLAVMVLVYSIGHISGAHFNPAVTIAHTTTGRFPLKQLPAYII 118
N N ++T PGI+IVWGL + VLVY++GHISG HFNPAVTIA +T RFPL Q+PAY++
Sbjct: 61 NENYYNMITFPGIAIVWGLVLTVLVYTVGHISGGHFNPAVTIAFASTRRFPLIQVPAYVV 120
Query: 119 AQVVGSTLASGVLKLIFSGKENQFAGTLPAGSDLQAFVVEFIITFFLMFIISGVATDNRA 178
AQ++GS LASG L+L+F G +QF+GT+P G++LQAFV EFI+TFFLMF+I GVATDNRA
Sbjct: 121 AQLLGSILASGTLRLLFMGNHDQFSGTVPNGTNLQAFVFEFIMTFFLMFVICGVATDNRA 180
Query: 179 IGELAGLAVGSTVILNVLFAGPITGASMNPARSLGPAIVHHEYRGIWIYMVSPILGALAG 238
+GE AG+A+GST++LNV+ GP+TGASMNPARSLGPA VH EY GIWIY+++P++GA+AG
Sbjct: 181 VGEFAGIAIGSTLLLNVIIGGPVTGASMNPARSLGPAFVHGEYEGIWIYLLAPVVGAIAG 240
Query: 239 TWTYTFLRITNKPVRELTKSSSFLK 263
W Y +R T+KP+ E TKS+SFLK
Sbjct: 241 AWVYNIVRYTDKPLSETTKSASFLK 265
>gb|AAK26750.1| NOD26-like membrane integral protein ZmNIP1-1 [Zea mays]
Length = 282
Score = 341 bits (875), Expect = 1e-92
Identities = 152/236 (64%), Positives = 202/236 (85%)
Query: 32 VPLLQKLVAEVVGTFFLIFAGCAAVVVNLNNDKVVTLPGISIVWGLAVMVLVYSIGHISG 91
VP +QK++AE+ GT+FL+FAGC AV +N + + +T PG++IVWGLAVMV+VY++GHISG
Sbjct: 40 VPFIQKIIAEIFGTYFLMFAGCGAVTINASKNGQITFPGVAIVWGLAVMVMVYAVGHISG 99
Query: 92 AHFNPAVTIAHTTTGRFPLKQLPAYIIAQVVGSTLASGVLKLIFSGKENQFAGTLPAGSD 151
AHFNPAVT+A T+GRFP +QLPAY++AQ++G+TLASG L+L+F G+ F GTLP GS+
Sbjct: 100 AHFNPAVTLAFATSGRFPWRQLPAYVLAQMLGATLASGTLRLMFGGRHEHFPGTLPTGSE 159
Query: 152 LQAFVVEFIITFFLMFIISGVATDNRAIGELAGLAVGSTVILNVLFAGPITGASMNPARS 211
+Q+ V+E I TF+LMF+ISGVATDNRAIGELAGLAVG+T++LNVL AGP++GASMNPARS
Sbjct: 160 VQSLVIEIITTFYLMFVISGVATDNRAIGELAGLAVGATILLNVLIAGPVSGASMNPARS 219
Query: 212 LGPAIVHHEYRGIWIYMVSPILGALAGTWTYTFLRITNKPVRELTKSSSFLKAVSK 267
+GPA+V EY IW+Y+V P++GA+AG W Y +R TNKP+RE+TKS+SFLK+ S+
Sbjct: 220 VGPALVSGEYTSIWVYVVGPVVGAVAGAWAYNLIRFTNKPLREITKSTSFLKSTSR 275
>dbj|BAA04257.1| major intrinsic protein [Oryza sativa] gi|50251783|dbj|BAD27715.1|
major intrinsic protein [Oryza sativa (japonica
cultivar-group)] gi|1076748|pir||S52003 major intrinsic
protein - rice
Length = 284
Score = 340 bits (873), Expect = 2e-92
Identities = 155/236 (65%), Positives = 201/236 (84%)
Query: 32 VPLLQKLVAEVVGTFFLIFAGCAAVVVNLNNDKVVTLPGISIVWGLAVMVLVYSIGHISG 91
VP +QK++AE+ GT+FLIFAGC AV +N + + +T PG++IVWGLAVMV+VY++GHISG
Sbjct: 44 VPFIQKIIAEIFGTYFLIFAGCGAVTINQSKNGQITFPGVAIVWGLAVMVMVYAVGHISG 103
Query: 92 AHFNPAVTIAHTTTGRFPLKQLPAYIIAQVVGSTLASGVLKLIFSGKENQFAGTLPAGSD 151
AHFNPAVT+A T RFP +Q+PAY AQ++G+TLA+G L+L+F G+ F GTLPAGSD
Sbjct: 104 AHFNPAVTLAFATCRRFPWRQVPAYAAAQMLGATLAAGTLRLMFGGRHEHFPGTLPAGSD 163
Query: 152 LQAFVVEFIITFFLMFIISGVATDNRAIGELAGLAVGSTVILNVLFAGPITGASMNPARS 211
+Q+ V+EFIITF+LMF+ISGVATDNRAIGELAGLAVG+T++LNVL AGPI+GASMNPARS
Sbjct: 164 VQSLVLEFIITFYLMFVISGVATDNRAIGELAGLAVGATILLNVLIAGPISGASMNPARS 223
Query: 212 LGPAIVHHEYRGIWIYMVSPILGALAGTWTYTFLRITNKPVRELTKSSSFLKAVSK 267
LGPA++ EYR IW+Y+V P+ GA+AG W Y +R TNKP+RE+TKS SFLK++++
Sbjct: 224 LGPAMIGGEYRSIWVYIVGPVAGAVAGAWAYNIIRFTNKPLREITKSGSFLKSMNR 279
>emb|CAB78905.1| nodulin-26-like protein [Arabidopsis thaliana]
gi|7228236|emb|CAA16760.2| nodulin-26-like protein
[Arabidopsis thaliana] gi|25324917|pir||H85214
nodulin-26-like protein [imported] - Arabidopsis
thaliana
Length = 308
Score = 340 bits (871), Expect = 3e-92
Identities = 169/275 (61%), Positives = 212/275 (76%), Gaps = 19/275 (6%)
Query: 14 NINDDATKKCDDTTIDDHVPLLQKLVAEVVGTFFLIFAGCAAVVVNLNNDKVVTLPGISI 73
+I++ K D+ + VP LQKL+AE +GT+FL+F GCA+VVVN+ ND VVTLPGI+I
Sbjct: 33 DIHNPRPLKKQDSLLSVSVPFLQKLIAEFLGTYFLVFTGCASVVVNMQNDNVVTLPGIAI 92
Query: 74 VWGLAVMVLVYSIGHISGAHFNPAVTIAHTTTGRFPLKQLPAYIIAQVVGSTLASGVLKL 133
VWGL +MVL+YS+GHISGAH NPAVTIA + GRFPLKQ+PAY+I+QV+GSTLA+ L+L
Sbjct: 93 VWGLTIMVLIYSLGHISGAHINPAVTIAFASCGRFPLKQVPAYVISQVIGSTLAAATLRL 152
Query: 134 IF-------SGKENQFAGTLPAGSDLQAFVVEFIITFFLMFIISGVATDNRA-------- 178
+F SGK + F G+ P GSDLQAF +EFI+TF+LMFIISGVATDNRA
Sbjct: 153 LFGLDHDVCSGKHDVFIGSSPVGSDLQAFTMEFIVTFYLMFIISGVATDNRAKLNIGTKC 212
Query: 179 ----IGELAGLAVGSTVILNVLFAGPITGASMNPARSLGPAIVHHEYRGIWIYMVSPILG 234
IGELAGLA+GSTV+LNVL A P++ ASMNP RSLGPA+V+ Y+GIWIY+V+P LG
Sbjct: 213 CNIQIGELAGLAIGSTVLLNVLIAAPVSSASMNPGRSLGPALVYGCYKGIWIYLVAPTLG 272
Query: 235 ALAGTWTYTFLRITNKPVRELTKSSSFLKAVSKGA 269
A+AG W Y +R T+KP+RE+TKS SFLK V G+
Sbjct: 273 AIAGAWVYNTVRYTDKPLREITKSGSFLKTVRIGS 307
>pir||JQ2286 nodulin-26 - soybean
Length = 271
Score = 340 bits (871), Expect = 3e-92
Identities = 162/265 (61%), Positives = 211/265 (79%), Gaps = 2/265 (0%)
Query: 1 MGDISNG--NLDVVMNINDDATKKCDDTTIDDHVPLLQKLVAEVVGTFFLIFAGCAAVVV 58
M D S G + +VV+N+ + ++ + VP LQKLVAE VGT+FLIFAGCA++VV
Sbjct: 1 MADYSAGTESQEVVVNVTKNTSETIQRSDSLVSVPFLQKLVAEAVGTYFLIFAGCASLVV 60
Query: 59 NLNNDKVVTLPGISIVWGLAVMVLVYSIGHISGAHFNPAVTIAHTTTGRFPLKQLPAYII 118
N N ++T PGI+IVWGL + VLVY++GHISG HFNPAVTIA +T RFPL Q+PAY++
Sbjct: 61 NENYYNMITFPGIAIVWGLVLTVLVYTVGHISGGHFNPAVTIAFASTRRFPLIQVPAYVV 120
Query: 119 AQVVGSTLASGVLKLIFSGKENQFAGTLPAGSDLQAFVVEFIITFFLMFIISGVATDNRA 178
AQ++GS LASG L+L+F G +QF+GT+P G++LQAFV EFI+TFFLMF+I GVATDNRA
Sbjct: 121 AQLLGSILASGTLRLLFMGNHDQFSGTVPNGTNLQAFVFEFIMTFFLMFVICGVATDNRA 180
Query: 179 IGELAGLAVGSTVILNVLFAGPITGASMNPARSLGPAIVHHEYRGIWIYMVSPILGALAG 238
+GE AG+A+GST++LNV+ GP+TGASMNP RSLGPA VH EY GIWIY+++P++GA+AG
Sbjct: 181 VGEFAGIAIGSTLLLNVIIGGPVTGASMNPVRSLGPAFVHGEYEGIWIYLLAPVVGAIAG 240
Query: 239 TWTYTFLRITNKPVRELTKSSSFLK 263
W Y +R T+KP+ E TKS+SFLK
Sbjct: 241 AWVYNIVRYTDKPLSETTKSASFLK 265
>pir||T05040 nodulin-26-like protein F13C5.200 - Arabidopsis thaliana
(fragment)
Length = 241
Score = 336 bits (862), Expect = 4e-91
Identities = 161/240 (67%), Positives = 200/240 (83%), Gaps = 7/240 (2%)
Query: 37 KLVAEVVGTFFLIFAGCAAVVVNLNNDKVVTLPGISIVWGLAVMVLVYSIGHISGAHFNP 96
+L+AE +GT+FL+F GCA+VVVN+ ND VVTLPGI+IVWGL +MVL+YS+GHISGAH NP
Sbjct: 1 QLIAEFLGTYFLVFTGCASVVVNMQNDNVVTLPGIAIVWGLTIMVLIYSLGHISGAHINP 60
Query: 97 AVTIAHTTTGRFPLKQLPAYIIAQVVGSTLASGVLKLIF-------SGKENQFAGTLPAG 149
AVTIA + GRFPLKQ+PAY+I+QV+GSTLA+ L+L+F SGK + F G+ P G
Sbjct: 61 AVTIAFASCGRFPLKQVPAYVISQVIGSTLAAATLRLLFGLDHDVCSGKHDVFIGSSPVG 120
Query: 150 SDLQAFVVEFIITFFLMFIISGVATDNRAIGELAGLAVGSTVILNVLFAGPITGASMNPA 209
SDLQAF +EFI+TF+LMFIISGVATDNRAIGELAGLA+GSTV+LNVL A P++ ASMNP
Sbjct: 121 SDLQAFTMEFIVTFYLMFIISGVATDNRAIGELAGLAIGSTVLLNVLIAAPVSSASMNPG 180
Query: 210 RSLGPAIVHHEYRGIWIYMVSPILGALAGTWTYTFLRITNKPVRELTKSSSFLKAVSKGA 269
RSLGPA+V+ Y+GIWIY+V+P LGA+AG W Y +R T+KP+RE+TKS SFLK V G+
Sbjct: 181 RSLGPALVYGCYKGIWIYLVAPTLGAIAGAWVYNTVRYTDKPLREITKSGSFLKTVRIGS 240
>gb|AAV44140.1| unknown protein [Oryza sativa (japonica cultivar-group)]
Length = 286
Score = 317 bits (812), Expect = 2e-85
Identities = 147/236 (62%), Positives = 186/236 (78%)
Query: 32 VPLLQKLVAEVVGTFFLIFAGCAAVVVNLNNDKVVTLPGISIVWGLAVMVLVYSIGHISG 91
V QK++AE++GTFFLIFAGCAAV VN VT PGI I WGLAVMV+VYS+GHISG
Sbjct: 50 VQFAQKVIAEILGTFFLIFAGCAAVAVNKRTGGTVTFPGICITWGLAVMVMVYSVGHISG 109
Query: 92 AHFNPAVTIAHTTTGRFPLKQLPAYIIAQVVGSTLASGVLKLIFSGKENQFAGTLPAGSD 151
AH NPAVT+A T GRFP +++PAY AQV GS AS L+ +F G F GT PAGSD
Sbjct: 110 AHLNPAVTLAFATCGRFPWRRVPAYAAAQVAGSAAASAALRALFGGAPEHFFGTAPAGSD 169
Query: 152 LQAFVVEFIITFFLMFIISGVATDNRAIGELAGLAVGSTVILNVLFAGPITGASMNPARS 211
+Q+ +EFIITF+LMF++SGVATDNRAIGELAGLAVG+TV++NVLFAGPI+GASMNPAR+
Sbjct: 170 VQSLAMEFIITFYLMFVVSGVATDNRAIGELAGLAVGATVLVNVLFAGPISGASMNPART 229
Query: 212 LGPAIVHHEYRGIWIYMVSPILGALAGTWTYTFLRITNKPVRELTKSSSFLKAVSK 267
+GPAI+ Y GIW+Y+ P+ GA+AG W Y +R T+KP+RE+T ++SF+++ +
Sbjct: 230 IGPAIILGRYTGIWVYIAGPVFGAVAGAWAYNLIRFTDKPLREITMTASFIRSTRR 285
>ref|NP_913205.1| putative major intrinsic protein [Oryza sativa (japonica
cultivar-group)]
Length = 246
Score = 300 bits (767), Expect = 4e-80
Identities = 142/236 (60%), Positives = 185/236 (78%), Gaps = 6/236 (2%)
Query: 38 LVAEVVGTFFLIFAGCAAVVVNLNNDKVVTLPGISIVWGLAVMVLVYSIGHISGAHFNPA 97
++AE++GT+F+IFAGC AVVVN + VT PGI VWGL VMVLVY++ HISGAHFNPA
Sbjct: 9 ILAEILGTYFMIFAGCGAVVVNQSTGGAVTFPGICAVWGLVVMVLVYTVSHISGAHFNPA 68
Query: 98 VTIAHTTTGRFPLKQLPAYIIAQVVGSTLASGVLKLIFSGK------ENQFAGTLPAGSD 151
VT+A T GRF KQ+P+Y++AQV+GST+AS L+++F G E+ F GT PAGS
Sbjct: 69 VTVAFATCGRFRWKQVPSYVVAQVLGSTMASLTLRVVFGGGGGGARGEHLFFGTTPAGSM 128
Query: 152 LQAFVVEFIITFFLMFIISGVATDNRAIGELAGLAVGSTVILNVLFAGPITGASMNPARS 211
QA +EF+I+FFLMF++SGVATDNRAIGELAGLAVG+TV +NVLFAGP+TGASMNPARS
Sbjct: 129 AQAAALEFVISFFLMFVVSGVATDNRAIGELAGLAVGATVAVNVLFAGPVTGASMNPARS 188
Query: 212 LGPAIVHHEYRGIWIYMVSPILGALAGTWTYTFLRITNKPVRELTKSSSFLKAVSK 267
LGPA+V Y G+W+Y+ +P+ G + G W Y LR T+KP+R++ ++SFL+ S+
Sbjct: 189 LGPAMVAGRYGGVWVYVAAPVSGTVCGAWAYNLLRFTDKPLRDIANTASFLRRSSR 244
>dbj|BAD73177.1| putative membrane integral protein ZmNIP1-1 [Oryza sativa (japonica
cultivar-group)]
Length = 303
Score = 300 bits (767), Expect = 4e-80
Identities = 142/236 (60%), Positives = 185/236 (78%), Gaps = 6/236 (2%)
Query: 38 LVAEVVGTFFLIFAGCAAVVVNLNNDKVVTLPGISIVWGLAVMVLVYSIGHISGAHFNPA 97
++AE++GT+F+IFAGC AVVVN + VT PGI VWGL VMVLVY++ HISGAHFNPA
Sbjct: 66 ILAEILGTYFMIFAGCGAVVVNQSTGGAVTFPGICAVWGLVVMVLVYTVSHISGAHFNPA 125
Query: 98 VTIAHTTTGRFPLKQLPAYIIAQVVGSTLASGVLKLIFSGK------ENQFAGTLPAGSD 151
VT+A T GRF KQ+P+Y++AQV+GST+AS L+++F G E+ F GT PAGS
Sbjct: 126 VTVAFATCGRFRWKQVPSYVVAQVLGSTMASLTLRVVFGGGGGGARGEHLFFGTTPAGSM 185
Query: 152 LQAFVVEFIITFFLMFIISGVATDNRAIGELAGLAVGSTVILNVLFAGPITGASMNPARS 211
QA +EF+I+FFLMF++SGVATDNRAIGELAGLAVG+TV +NVLFAGP+TGASMNPARS
Sbjct: 186 AQAAALEFVISFFLMFVVSGVATDNRAIGELAGLAVGATVAVNVLFAGPVTGASMNPARS 245
Query: 212 LGPAIVHHEYRGIWIYMVSPILGALAGTWTYTFLRITNKPVRELTKSSSFLKAVSK 267
LGPA+V Y G+W+Y+ +P+ G + G W Y LR T+KP+R++ ++SFL+ S+
Sbjct: 246 LGPAMVAGRYGGVWVYVAAPVSGTVCGAWAYNLLRFTDKPLRDIANTASFLRRSSR 301
>sp|P49173|NIP1_NICAL Probable aquaporin NIP-type (Pollen-specific membrane integral
protein) gi|665948|gb|AAA62235.1| putative membrane
integral protein
Length = 270
Score = 291 bits (745), Expect = 1e-77
Identities = 145/264 (54%), Positives = 192/264 (71%), Gaps = 5/264 (1%)
Query: 1 MGDISNGNLDVVMNINDDATKKCDDTTIDDHVPLLQKLVAEVVGTFFLIFAGCAAVVVNL 60
+ + GN+ N +D C ++ V +LQKL+AE +GT+F+IFAGC +V VN
Sbjct: 12 ISQMEEGNIHSASN-SDSNVGFCSSVSV---VVILQKLIAEAIGTYFVIFAGCGSVAVNK 67
Query: 61 NNDKVVTLPGISIVWGLAVMVLVYSIGHISGAHFNPAVTIAHTTTGRFPLKQLPAYIIAQ 120
V T PGI + WGL VMV+VY++G+ISGAHFNPAVTI + GRFP KQ+P YIIAQ
Sbjct: 68 IYGSV-TFPGICVTWGLIVMVMVYTVGYISGAHFNPAVTITFSIFGRFPWKQVPLYIIAQ 126
Query: 121 VVGSTLASGVLKLIFSGKENQFAGTLPAGSDLQAFVVEFIITFFLMFIISGVATDNRAIG 180
++GS LASG L L+F + GT+P GS+ Q+ +E II+F LMF+ISGVATD+RAIG
Sbjct: 127 LMGSILASGTLALLFDVTPQAYFGTVPVGSNGQSLAIEIIISFLLMFVISGVATDDRAIG 186
Query: 181 ELAGLAVGSTVILNVLFAGPITGASMNPARSLGPAIVHHEYRGIWIYMVSPILGALAGTW 240
++AG+AVG T+ LNV AGPI+GASMNPARS+GPAIV H Y G+W+Y+V PI+G LAG +
Sbjct: 187 QVAGIAVGMTITLNVFVAGPISGASMNPARSIGPAIVKHVYTGLWVYVVGPIIGTLAGAF 246
Query: 241 TYTFLRITNKPVRELTKSSSFLKA 264
Y +R T+KP+REL KS+S L++
Sbjct: 247 VYNLIRSTDKPLRELAKSASSLRS 270
Database: nr
Posted date: Jul 5, 2005 12:34 AM
Number of letters in database: 863,360,394
Number of sequences in database: 2,540,612
Lambda K H
0.323 0.139 0.407
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 434,539,765
Number of Sequences: 2540612
Number of extensions: 17829002
Number of successful extensions: 59563
Number of sequences better than 10.0: 1289
Number of HSP's better than 10.0 without gapping: 1162
Number of HSP's successfully gapped in prelim test: 127
Number of HSP's that attempted gapping in prelim test: 55563
Number of HSP's gapped (non-prelim): 1668
length of query: 270
length of database: 863,360,394
effective HSP length: 126
effective length of query: 144
effective length of database: 543,243,282
effective search space: 78227032608
effective search space used: 78227032608
T: 11
A: 40
X1: 16 ( 7.5 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.9 bits)
S2: 74 (33.1 bits)
Medicago: description of AC140547.14


