

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC140030.7 - phase: 0
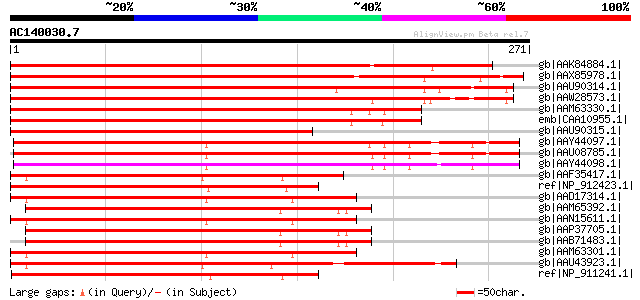
(271 letters)
Database: nr
2,540,612 sequences; 863,360,394 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
gb|AAK84884.1| NAC domain protein NAC2 [Phaseolus vulgaris] 407 e-112
gb|AAX85978.1| NAC1 protein [Glycine max] gi|66394510|gb|AAY4612... 385 e-106
gb|AAU90314.1| putative NAC domain protein NAC2 [Solanum demissum] 317 3e-85
gb|AAW28573.1| putative NAC domain protein NAC2 [Solanum demissum] 313 3e-84
gb|AAM63330.1| NAC domain protein NAC2 [Arabidopsis thaliana] 313 3e-84
emb|CAA10955.1| unnamed protein product [Arabidopsis thaliana] g... 312 7e-84
gb|AAU90315.1| putative NAC domain protein NAC2 [Solanum demissum] 258 1e-67
gb|AAY44097.1| NAC domain transcription factor [Triticum aestivum] 255 1e-66
gb|AAU08785.1| NAC domain transcription factor [Triticum aestivum] 254 1e-66
gb|AAY44098.1| NAC domain transcription factor [Triticum aestivum] 254 2e-66
gb|AAF35417.1| putative jasmonic acid regulatory protein [Arabid... 241 2e-62
ref|NP_912423.1| Putative NAM (no apical meristem) protein [Oryz... 240 3e-62
gb|AAD17314.1| NAC domain protein NAM [Arabidopsis thaliana] gi|... 239 5e-62
gb|AAM65392.1| NAM protein, putative [Arabidopsis thaliana] 239 5e-62
gb|AAN15611.1| NAM-like protein [Arabidopsis thaliana] gi|204666... 239 5e-62
gb|AAP37705.1| At1g61110 [Arabidopsis thaliana] gi|26450820|dbj|... 239 6e-62
gb|AAB71483.1| similar to NAM (gp|X92205|1321924) and CUC2 (gp|A... 239 6e-62
gb|AAM63301.1| NAM-like protein [Arabidopsis thaliana] 238 2e-61
gb|AAU43923.1| NAC domain protein [Lycopersicon esculentum] gi|5... 236 5e-61
ref|NP_911241.1| putative OsNAC5 protein [Oryza sativa (japonica... 236 7e-61
>gb|AAK84884.1| NAC domain protein NAC2 [Phaseolus vulgaris]
Length = 256
Score = 407 bits (1045), Expect = e-112
Identities = 195/255 (76%), Positives = 219/255 (85%), Gaps = 5/255 (1%)
Query: 1 MESSASSELPPGFRFHPTDEELIVHYLCNQATSKPCPASIIPEVDIYKFDPWELPDKSEF 60
M+++ SELPPGFRFHPTDEELIV+YLCNQATSKPCPASIIPEVD+YKFDPWELPDK+EF
Sbjct: 1 MDATTPSELPPGFRFHPTDEELIVYYLCNQATSKPCPASIIPEVDLYKFDPWELPDKTEF 60
Query: 61 EENEWYFFSPRERKYPNGVRPNRATLSGYWKATGTDKAIKSGSKQIGVKKSLVFYKGRPP 120
ENEWYFFSPR+RKYPNGVRPNRAT+SGYWKATGTDKAI SGSK +GVKKSLVFYKGRPP
Sbjct: 61 GENEWYFFSPRDRKYPNGVRPNRATVSGYWKATGTDKAIYSGSKLVGVKKSLVFYKGRPP 120
Query: 121 KGVKTDWIMHEYRLIGSQKQTSKHIGSMRLDDWVLCRIYKKKHMGKTLQQKEDYSTHQFN 180
KG KTDWIMHEYRL S++ ++ IGSMRLDDWVLCRIYKKK+ GKTL+ KE + Q
Sbjct: 121 KGDKTDWIMHEYRLAESKQPVNRKIGSMRLDDWVLCRIYKKKNTGKTLEHKETHPKVQMT 180
Query: 181 DSIITNNDDGELEMMNLTRSCSLTYLLDMNYFGPILSDG---STLDFQINNSNIGIDPYV 237
+ I NND E +MMNL R+ SLTYLLDMNY GPILSDG ST DFQI+N+NIGIDP+V
Sbjct: 181 NLIAANND--EQKMMNLPRTWSLTYLLDMNYLGPILSDGSYCSTFDFQISNANIGIDPFV 238
Query: 238 KPQPVEMTNHYEADS 252
QPVEM N+Y +DS
Sbjct: 239 NSQPVEMANNYVSDS 253
>gb|AAX85978.1| NAC1 protein [Glycine max] gi|66394510|gb|AAY46121.1| NAC domain
protein NAC1 [Glycine max]
Length = 279
Score = 385 bits (988), Expect = e-106
Identities = 186/279 (66%), Positives = 226/279 (80%), Gaps = 14/279 (5%)
Query: 1 MESSASSELPPGFRFHPTDEELIVHYLCNQATSKPCPASIIPEVDIYKFDPWELPDKSEF 60
ME+ SS LPPGFRFHPTDEELIV+YLCNQA+S+PCPASIIPEVDIYKFDPWELPDK++F
Sbjct: 1 MENRTSSVLPPGFRFHPTDEELIVYYLCNQASSRPCPASIIPEVDIYKFDPWELPDKTDF 60
Query: 61 EENEWYFFSPRERKYPNGVRPNRATLSGYWKATGTDKAIKSGSKQIGVKKSLVFYKGRPP 120
E EWYFFSPRERKYPNGVRPNRAT+SGYWKATGTDKAI SGSK +GVKK+LVFYKG+PP
Sbjct: 61 GEKEWYFFSPRERKYPNGVRPNRATVSGYWKATGTDKAIYSGSKHVGVKKALVFYKGKPP 120
Query: 121 KGVKTDWIMHEYRLIGSQKQTSKHIGSMRLDDWVLCRIYKKKHMGKTLQQKEDYSTHQFN 180
KG+KTDWIMHEYRLIGS++Q ++ +GSMRLDDWVLCRIYKKK++GK+++ KEDY Q N
Sbjct: 121 KGLKTDWIMHEYRLIGSRRQANRQVGSMRLDDWVLCRIYKKKNIGKSMEAKEDYPIAQIN 180
Query: 181 DSIITNNDDGELEMMNLTRSCSLTYLLDMNYFGPI------LSDGSTLDFQINNSNIGID 234
+ N++ E E++ R+ SLT+LL+M+Y GPI S ST DFQIN +N GID
Sbjct: 181 --LTPANNNSEQELVKFPRTSSLTHLLEMDYLGPISHILPDASYNSTFDFQINTANGGID 238
Query: 235 PYVKPQPVEM-----TNHYEADSHSSITNQPIFVKQMHN 268
P+VKPQ VE+ + Y+ +S+I N IFV Q+++
Sbjct: 239 PFVKPQLVEIPYATDSGKYQVKQNSTI-NPTIFVNQVYD 276
>gb|AAU90314.1| putative NAC domain protein NAC2 [Solanum demissum]
Length = 281
Score = 317 bits (811), Expect = 3e-85
Identities = 165/275 (60%), Positives = 204/275 (74%), Gaps = 13/275 (4%)
Query: 1 MESSASSELPPGFRFHPTDEELIVHYLCNQATSKPCPASIIPEVDIYKFDPWELPDKSEF 60
M SS+LPPGFRFHPTDEELI++YL QATS+PCP SIIPE+D+YKFDPWELP+K+EF
Sbjct: 1 MVGKISSDLPPGFRFHPTDEELIMYYLRYQATSRPCPVSIIPEIDVYKFDPWELPEKAEF 60
Query: 61 EENEWYFFSPRERKYPNGVRPNRATLSGYWKATGTDKAIKSGSKQIGVKKSLVFYKGRPP 120
ENEWYFF+PR+RKYPNGVRPNRA +SGYWKATGTDKAI S +K +G+KK+LVFYKG+PP
Sbjct: 61 GENEWYFFTPRDRKYPNGVRPNRAAVSGYWKATGTDKAIYSANKYVGIKKALVFYKGKPP 120
Query: 121 KGVKTDWIMHEYRLIGSQKQT-SKHIGSMRLDDWVLCRIYKKKHMGKTLQ--QKEDYSTH 177
KGVKTDWIMHEYRL S+ QT SK GSMRLDDWVLCRIYKKK++GKT++ + E+
Sbjct: 121 KGVKTDWIMHEYRLSDSKSQTYSKQSGSMRLDDWVLCRIYKKKNLGKTIEMMKVEEEELE 180
Query: 178 QFNDSIITNNDDGELEMMNLTRSCSLTYLLDMNYFGPI---LSDGSTLD---FQINN-SN 230
N SI + G + M L R CSL++LL+++YFG I LSD D + +NN SN
Sbjct: 181 AQNVSINNAIEVGGPQTMKLPRICSLSHLLELDYFGSIPQLLSDNLLYDDQSYTMNNVSN 240
Query: 231 IGIDPYVKPQPVEMTNHYEADSHSSITN--QPIFV 263
V Q + TN+ +++ + N QP+FV
Sbjct: 241 TSNVDQVSSQQ-QNTNNITSNNCNIFFNYQQPLFV 274
>gb|AAW28573.1| putative NAC domain protein NAC2 [Solanum demissum]
Length = 282
Score = 313 bits (802), Expect = 3e-84
Identities = 161/279 (57%), Positives = 203/279 (72%), Gaps = 20/279 (7%)
Query: 1 MESSASSELPPGFRFHPTDEELIVHYLCNQATSKPCPASIIPEVDIYKFDPWELPDKSEF 60
M SS+LPPGFRFHPTDEELI++YL QATS+PCP SIIPE+D+YKFDPWELP+K+EF
Sbjct: 1 MVGKISSDLPPGFRFHPTDEELIMYYLRYQATSRPCPVSIIPEIDVYKFDPWELPEKAEF 60
Query: 61 EENEWYFFSPRERKYPNGVRPNRATLSGYWKATGTDKAIKSGSKQIGVKKSLVFYKGRPP 120
ENEWYFF+PR+RKYPNGVRPNRA +SGYWKATGTDKAI S +K +G+KK+LVFYKG+PP
Sbjct: 61 GENEWYFFTPRDRKYPNGVRPNRAAVSGYWKATGTDKAIYSANKYVGIKKALVFYKGKPP 120
Query: 121 KGVKTDWIMHEYRLIGSQKQT-SKHIGSMRLDDWVLCRIYKKKHMGKTLQQKEDYSTHQF 179
KGVKTDWIMHEYRL S+ QT SK GSMRLDDWVLCRIYKKK++GKT++ +
Sbjct: 121 KGVKTDWIMHEYRLSDSKSQTYSKQSGSMRLDDWVLCRIYKKKNLGKTIEMMKVEEEELE 180
Query: 180 NDSIITNND---DGELEMMNLTRSCSLTYLLDMNYFGPI---LSD-------GSTLDFQI 226
++ NN G + M L R CSL++LL+++Y+G I LSD T+++
Sbjct: 181 AQNVSINNAIEVGGGPQTMKLPRICSLSHLLELDYYGSIPQLLSDNLLYDDQSYTMNYVS 240
Query: 227 NNSNIGIDPYVKPQPVEMTNHYEADSHSSITN--QPIFV 263
N SN +D Q + TN+ +++ + N QP+FV
Sbjct: 241 NTSN--VDQVSSQQ--QNTNNITSNNCNIFFNYQQPLFV 275
>gb|AAM63330.1| NAC domain protein NAC2 [Arabidopsis thaliana]
Length = 268
Score = 313 bits (802), Expect = 3e-84
Identities = 149/225 (66%), Positives = 177/225 (78%), Gaps = 10/225 (4%)
Query: 1 MESSASSELPPGFRFHPTDEELIVHYLCNQATSKPCPASIIPEVDIYKFDPWELPDKSEF 60
ME ++ S LPPGFRFHPTDEELIV+YL NQ SKPCP SIIPEVDIYKFDPW+LP+K+EF
Sbjct: 1 MEVTSQSTLPPGFRFHPTDEELIVYYLRNQTMSKPCPVSIIPEVDIYKFDPWQLPEKTEF 60
Query: 61 EENEWYFFSPRERKYPNGVRPNRATLSGYWKATGTDKAIKSGSKQIGVKKSLVFYKGRPP 120
ENEWYFFSPRERKYPNGVRPNRA +SGYWKATGTDKAI SGS +GVKK+LVFYKGRPP
Sbjct: 61 GENEWYFFSPRERKYPNGVRPNRAAVSGYWKATGTDKAIHSGSSNVGVKKALVFYKGRPP 120
Query: 121 KGVKTDWIMHEYRLIGSQKQTSKHIGSMRLDDWVLCRIYKKKHMGKTLQQKEDYSTH--- 177
KG+KTDWIMHEYRL S+K ++K GSMRLD+WVLCRIYKK+ K L ++E +
Sbjct: 121 KGIKTDWIMHEYRLHDSRKASTKRSGSMRLDEWVLCRIYKKRGASKLLNEQEGFMDEVLM 180
Query: 178 QFNDSIITN----NDDGELEM---MNLTRSCSLTYLLDMNYFGPI 215
+ ++ N +D E+ M M L R+CSL +LL+M+Y GP+
Sbjct: 181 EDETKVVINEAERRNDEEIMMMTSMKLPRTCSLAHLLEMDYMGPV 225
>emb|CAA10955.1| unnamed protein product [Arabidopsis thaliana]
gi|15293163|gb|AAK93692.1| unknown protein [Arabidopsis
thaliana] gi|13430578|gb|AAK25911.1| unknown protein
[Arabidopsis thaliana] gi|18409291|ref|NP_564966.1| no
apical meristem (NAM) family protein [Arabidopsis
thaliana] gi|50400809|sp|O49255|NAC29_ARATH NAC-domain
containing protein 29 (ANAC029) (NAC2) (NAC-LIKE,
ACTIVATED BY AP3/PI protein) (NAP)
gi|12597796|gb|AAG60108.1| unknown protein [Arabidopsis
thaliana]
Length = 268
Score = 312 bits (799), Expect = 7e-84
Identities = 147/225 (65%), Positives = 174/225 (77%), Gaps = 10/225 (4%)
Query: 1 MESSASSELPPGFRFHPTDEELIVHYLCNQATSKPCPASIIPEVDIYKFDPWELPDKSEF 60
ME ++ S LPPGFRFHPTDEELIV+YL NQ SKPCP SIIPEVDIYKFDPW+LP+K+EF
Sbjct: 1 MEVTSQSTLPPGFRFHPTDEELIVYYLRNQTMSKPCPVSIIPEVDIYKFDPWQLPEKTEF 60
Query: 61 EENEWYFFSPRERKYPNGVRPNRATLSGYWKATGTDKAIKSGSKQIGVKKSLVFYKGRPP 120
ENEWYFFSPRERKYPNGVRPNRA +SGYWKATGTDKAI SGS +GVKK+LVFYKGRPP
Sbjct: 61 GENEWYFFSPRERKYPNGVRPNRAAVSGYWKATGTDKAIHSGSSNVGVKKALVFYKGRPP 120
Query: 121 KGVKTDWIMHEYRLIGSQKQTSKHIGSMRLDDWVLCRIYKKKHMGKTLQQKEDYSTH--- 177
KG+KTDWIMHEYRL S+K ++K GSMRLD+WVLCRIYKK+ K L ++E +
Sbjct: 121 KGIKTDWIMHEYRLHDSRKASTKRNGSMRLDEWVLCRIYKKRGASKLLNEQEGFMDEVLM 180
Query: 178 QFNDSIITNNDDGELE-------MMNLTRSCSLTYLLDMNYFGPI 215
+ ++ N + E M L R+CSL +LL+M+Y GP+
Sbjct: 181 EDETKVVVNEAERRTEEEIMMMTSMKLPRTCSLAHLLEMDYMGPV 225
>gb|AAU90315.1| putative NAC domain protein NAC2 [Solanum demissum]
Length = 163
Score = 258 bits (660), Expect = 1e-67
Identities = 117/159 (73%), Positives = 138/159 (86%), Gaps = 1/159 (0%)
Query: 1 MESSASSELPPGFRFHPTDEELIVHYLCNQATSKPCPASIIPEVDIYKFDPWELPDKSEF 60
M SS+LPPGFRFHPTDEELI++YL QATS+PCP SIIPE+D+YKFDPWELP+K+EF
Sbjct: 1 MVGKISSDLPPGFRFHPTDEELIMYYLRYQATSRPCPVSIIPEIDVYKFDPWELPEKAEF 60
Query: 61 EENEWYFFSPRERKYPNGVRPNRATLSGYWKATGTDKAIKSGSKQIGVKKSLVFYKGRPP 120
ENEWYFF+PR+RKYPNGVRPNRA +SGYWKATGTDKAI S +K +G+KK+LVFYKG+PP
Sbjct: 61 GENEWYFFTPRDRKYPNGVRPNRAAVSGYWKATGTDKAIYSANKYVGIKKALVFYKGKPP 120
Query: 121 KGVKTDWIMHEYRLIGSQKQT-SKHIGSMRLDDWVLCRI 158
KGVKTDWIMHEYRL S+ QT SK GSMR+ ++CR+
Sbjct: 121 KGVKTDWIMHEYRLSDSKSQTYSKQSGSMRVRTSLICRL 159
>gb|AAY44097.1| NAC domain transcription factor [Triticum aestivum]
Length = 355
Score = 255 bits (651), Expect = 1e-66
Identities = 146/300 (48%), Positives = 183/300 (60%), Gaps = 41/300 (13%)
Query: 3 SSASSELPPGFRFHPTDEELIVHYLCNQATSKPCPASIIPEVDIYKFDPWELPDKSEFEE 62
S+A LPPGFRFHPTDEELIVHYL QA S P P II EV+IYK +PW+LP K+ F E
Sbjct: 7 SAAMPALPPGFRFHPTDEELIVHYLGRQAASMPSPVPIIAEVNIYKCNPWDLPGKALFGE 66
Query: 63 NEWYFFSPRERKYPNGVRPNRATLSGYWKATGTDKAIKS--GSKQIGVKKSLVFYKGRPP 120
NEWYFFSPR+RKYPNG RPNRA SGYWKATGTDKAI S ++ IGVKK+LVFY+G+PP
Sbjct: 67 NEWYFFSPRDRKYPNGARPNRAAGSGYWKATGTDKAILSTPANESIGVKKALVFYRGKPP 126
Query: 121 KGVKTDWIMHEYRLIGSQKQTSKHIG-SMRLDDWVLCRIYKKKHMGKTLQQKEDYSTHQF 179
KGVKTDWIMHEYRL + +T+K G SMRLDDWVLCRI+KK + H+
Sbjct: 127 KGVKTDWIMHEYRLTAADNRTTKRRGSSMRLDDWVLCRIHKKCGNLPNFSSSDQEQEHEQ 186
Query: 180 NDSIITN-------------------NDDGELEM---MNLTRSCSLTYLL---DMNYFGP 214
S + + ND +L+ M + +SCSLT LL D
Sbjct: 187 ESSTVEDSQNNHTVSSPKSEAFDGDGNDHLQLQQFRPMAIAKSCSLTDLLNTVDYAALSH 246
Query: 215 ILSDGSTLDFQINNSNIGIDPYVKPQ--------PVEMTNHYEADSHSSITNQPIFVKQM 266
+L DG+ ++S+ G D + P+ P + T HY +++ + N+ I V Q+
Sbjct: 247 LLLDGA----GASSSDAGADYQLPPENPLIYSQPPWQQTLHYN-NNNGYVNNETIDVPQL 301
>gb|AAU08785.1| NAC domain transcription factor [Triticum aestivum]
Length = 354
Score = 254 bits (650), Expect = 1e-66
Identities = 145/300 (48%), Positives = 183/300 (60%), Gaps = 41/300 (13%)
Query: 3 SSASSELPPGFRFHPTDEELIVHYLCNQATSKPCPASIIPEVDIYKFDPWELPDKSEFEE 62
S+A LPPGFRFHPTDEELIVHYL QA S P P II EV+IYK +PW+LP K+ F E
Sbjct: 6 SAAMPALPPGFRFHPTDEELIVHYLRRQAASMPSPVPIIAEVNIYKCNPWDLPGKALFGE 65
Query: 63 NEWYFFSPRERKYPNGVRPNRATLSGYWKATGTDKAIKS--GSKQIGVKKSLVFYKGRPP 120
NEWYFFSPR+RKYPNG RPNRA SGYWKATGTDKAI S ++ IGVKK+LVFY+G+PP
Sbjct: 66 NEWYFFSPRDRKYPNGARPNRAAGSGYWKATGTDKAILSTPANESIGVKKALVFYRGKPP 125
Query: 121 KGVKTDWIMHEYRLIGSQKQTSKHIG-SMRLDDWVLCRIYKKKHMGKTLQQKEDYSTHQF 179
KGVKTDWIMHEYRL + +T+K G SMRLDDWVLCRI+KK + H+
Sbjct: 126 KGVKTDWIMHEYRLTAADNRTTKRRGSSMRLDDWVLCRIHKKCGNLPNFSSSDQEQEHEQ 185
Query: 180 NDSIITNND-----------------DGELEM-----MNLTRSCSLTYLL---DMNYFGP 214
S + ++ D L++ M + +SCSLT LL D
Sbjct: 186 ESSTVEDSQNNHTVSSPKSEAFDGDGDDHLQLQQFRPMAIAKSCSLTDLLNTVDYAALSH 245
Query: 215 ILSDGSTLDFQINNSNIGIDPYVKPQ--------PVEMTNHYEADSHSSITNQPIFVKQM 266
+L DG+ ++S+ G D + P+ P + T HY +++ + N+ I V Q+
Sbjct: 246 LLLDGA----GASSSDAGADYQLPPENPLIYSQPPWQQTLHYN-NNNGYVNNETIDVPQL 300
>gb|AAY44098.1| NAC domain transcription factor [Triticum aestivum]
Length = 359
Score = 254 bits (648), Expect = 2e-66
Identities = 144/301 (47%), Positives = 181/301 (59%), Gaps = 39/301 (12%)
Query: 3 SSASSELPPGFRFHPTDEELIVHYLCNQATSKPCPASIIPEVDIYKFDPWELPDKSEFEE 62
S+A LPPGFRFHPTDEELIVHYL QA S P P II EV+IYK +PW+LP K+ F E
Sbjct: 7 SAAMPALPPGFRFHPTDEELIVHYLGRQAASMPSPVPIIAEVNIYKCNPWDLPGKALFGE 66
Query: 63 NEWYFFSPRERKYPNGVRPNRATLSGYWKATGTDKAIKS--GSKQIGVKKSLVFYKGRPP 120
NEWYFFSPR+RKYPNG RPNRA SGYWKATGTDKAI S ++ IGVKK+LVFY+G+PP
Sbjct: 67 NEWYFFSPRDRKYPNGARPNRAAGSGYWKATGTDKAILSTPANESIGVKKALVFYRGKPP 126
Query: 121 KGVKTDWIMHEYRLIGSQKQTSKHIG-SMRLDDWVLCRIYKKKHMGKTLQQKEDYSTHQF 179
KGVKTDWIMHEYRL + +T+K G SMRLDDWVLCRI+KK + + H+
Sbjct: 127 KGVKTDWIMHEYRLTAADNRTTKRRGSSMRLDDWVLCRIHKKCNNLHNFSSSDQEQEHEQ 186
Query: 180 NDSIITNND------------------DGELEM-----MNLTRSCSLTYLL---DMNYFG 213
S + D +L++ M + +SCSLT LL D
Sbjct: 187 ESSTTVEDSHNNHTVSSPKSEAFDGDGDDQLQLQQFRPMAIAKSCSLTDLLNTVDYAALS 246
Query: 214 PILSDGSTLDFQINNSNIGIDPYVKPQ--------PVEMTNHYEADSHSSITNQPIFVKQ 265
+L DG+ ++S+ G D + P+ P + T HY +++ + I V Q
Sbjct: 247 HLLLDGAGAG--ASSSDAGADYQLPPENPLIYSQPPWQQTLHYNNNNNGYVNIDTIDVPQ 304
Query: 266 M 266
+
Sbjct: 305 I 305
>gb|AAF35417.1| putative jasmonic acid regulatory protein [Arabidopsis thaliana]
gi|15795116|dbj|BAB02380.1| jasmonic acid regulatory
protein-like [Arabidopsis thaliana]
gi|28827466|gb|AAO50577.1| putative jasmonic acid
regulatory protein [Arabidopsis thaliana]
gi|28393364|gb|AAO42106.1| putative jasmonic acid
regulatory protein [Arabidopsis thaliana]
gi|15232606|ref|NP_188170.1| no apical meristem (NAM)
family protein (NAC2) [Arabidopsis thaliana]
gi|12060426|dbj|BAB20600.1| AtNAC2 [Arabidopsis
thaliana]
Length = 364
Score = 241 bits (614), Expect = 2e-62
Identities = 115/191 (60%), Positives = 138/191 (72%), Gaps = 17/191 (8%)
Query: 1 MESSASS--------ELPPGFRFHPTDEELIVHYLCNQATSKPCPASIIPEVDIYKFDPW 52
MES+ SS LPPGFRFHPTDEEL+VHYL +A S P P +II EVD+YKFDPW
Sbjct: 1 MESTDSSGGPPPPQPNLPPGFRFHPTDEELVVHYLKRKAASAPLPVAIIAEVDLYKFDPW 60
Query: 53 ELPDKSEFEENEWYFFSPRERKYPNGVRPNRATLSGYWKATGTDKAI--KSGSKQIGVKK 110
ELP K+ F E EWYFFSPR+RKYPNG RPNRA SGYWKATGTDK + G++++GVKK
Sbjct: 61 ELPAKASFGEQEWYFFSPRDRKYPNGARPNRAATSGYWKATGTDKPVLASDGNQKVGVKK 120
Query: 111 SLVFYKGRPPKGVKTDWIMHEYRLIGSQKQT-------SKHIGSMRLDDWVLCRIYKKKH 163
+LVFY G+PPKGVK+DWIMHEYRLI ++ S+RLDDWVLCRIYKK +
Sbjct: 121 ALVFYSGKPPKGVKSDWIMHEYRLIENKPNNRPPGCDFGNKKNSLRLDDWVLCRIYKKNN 180
Query: 164 MGKTLQQKEDY 174
+ + +D+
Sbjct: 181 ASRHVDNDKDH 191
>ref|NP_912423.1| Putative NAM (no apical meristem) protein [Oryza sativa (japonica
cultivar-group)] gi|24899399|gb|AAN64999.1| Putative NAM
(no apical meristem) protein [Oryza sativa (japonica
cultivar-group)]
Length = 392
Score = 240 bits (612), Expect = 3e-62
Identities = 114/173 (65%), Positives = 129/173 (73%), Gaps = 12/173 (6%)
Query: 1 MESSASSELPPGFRFHPTDEELIVHYLCNQATSKPCPASIIPEVDIYKFDPWELPDKSEF 60
M S + LPPGFRFHPTDEELIVHYL N+A S PCP SII +VDIYKFDPW+LP K +
Sbjct: 1 MVLSNPAMLPPGFRFHPTDEELIVHYLRNRAASSPCPVSIIADVDIYKFDPWDLPSKENY 60
Query: 61 EENEWYFFSPRERKYPNGVRPNRATLSGYWKATGTDKAIKSG-----SKQIGVKKSLVFY 115
+ EWYFFSPR+RKYPNG+RPNRA SGYWKATGTDK I S ++ +GVKK+LVFY
Sbjct: 61 GDREWYFFSPRDRKYPNGIRPNRAAGSGYWKATGTDKPIHSSGGAATNESVGVKKALVFY 120
Query: 116 KGRPPKGVKTDWIMHEYRLIGSQKQTSK-------HIGSMRLDDWVLCRIYKK 161
KGRPPKG KT+WIMHEYRL + + SMRLDDWVLCRIYKK
Sbjct: 121 KGRPPKGTKTNWIMHEYRLAAADAHAANTYRPMKFRNTSMRLDDWVLCRIYKK 173
>gb|AAD17314.1| NAC domain protein NAM [Arabidopsis thaliana]
gi|15219110|ref|NP_175696.1| no apical meristem (NAM)
family protein [Arabidopsis thaliana]
gi|4325282|gb|AAD17313.1| NAC domain protein NAM
[Arabidopsis thaliana] gi|12324644|gb|AAG52280.1|
NAM-like protein; 59502-58357 [Arabidopsis thaliana]
gi|25405609|pir||A96570 NAM-like protein, 59502-58357
[imported] - Arabidopsis thaliana
gi|50401213|sp|Q9ZNU2|NAC18_ARATH NAC-domain containing
protein 18 (ANAC018) (NO APICAL MERISTEM protein)
(AtNAM)
Length = 320
Score = 239 bits (611), Expect = 5e-62
Identities = 118/197 (59%), Positives = 139/197 (69%), Gaps = 16/197 (8%)
Query: 1 MESSASS--------ELPPGFRFHPTDEELIVHYLCNQATSKPCPASIIPEVDIYKFDPW 52
MES+ SS LPPGFRFHPTDEEL++HYL +A S P P +II +VD+YKFDPW
Sbjct: 1 MESTDSSGGPPPPQPNLPPGFRFHPTDEELVIHYLKRKADSVPLPVAIIADVDLYKFDPW 60
Query: 53 ELPDKSEFEENEWYFFSPRERKYPNGVRPNRATLSGYWKATGTDKAIKS----GSKQIGV 108
ELP K+ F E EWYFFSPR+RKYPNG RPNRA SGYWKATGTDK + S GSK++GV
Sbjct: 61 ELPAKASFGEQEWYFFSPRDRKYPNGARPNRAATSGYWKATGTDKPVISTGGGGSKKVGV 120
Query: 109 KKSLVFYKGRPPKGVKTDWIMHEYRLIGSQKQTSKHIG----SMRLDDWVLCRIYKKKHM 164
KK+LVFY G+PPKGVK+DWIMHEYRL ++ G S+RLDDWVLCRIYKK +
Sbjct: 121 KKALVFYSGKPPKGVKSDWIMHEYRLTDNKPTHICDFGNKKNSLRLDDWVLCRIYKKNNS 180
Query: 165 GKTLQQKEDYSTHQFND 181
+ + H ND
Sbjct: 181 TASRHHHHLHHIHLDND 197
>gb|AAM65392.1| NAM protein, putative [Arabidopsis thaliana]
Length = 323
Score = 239 bits (611), Expect = 5e-62
Identities = 118/202 (58%), Positives = 140/202 (68%), Gaps = 21/202 (10%)
Query: 9 LPPGFRFHPTDEELIVHYLCNQATSKPCPASIIPEVDIYKFDPWELPDKSEFEENEWYFF 68
LPPGFRFHPTDEEL+VHYL +A S P P SII E+D+YKFDPWELP K+ F E+EWYFF
Sbjct: 16 LPPGFRFHPTDEELVVHYLKKKAASVPLPVSIIAEIDLYKFDPWELPSKASFGEHEWYFF 75
Query: 69 SPRERKYPNGVRPNRATLSGYWKATGTDKAI-KSGSKQIGVKKSLVFYKGRPPKGVKTDW 127
SPR+RKYPNGVRPNRA SGYWKATGTDK I S ++GVKK+LVFY G+PPKG+KTDW
Sbjct: 76 SPRDRKYPNGVRPNRAATSGYWKATGTDKPIFTCNSHKVGVKKALVFYGGKPPKGIKTDW 135
Query: 128 IMHEYRLIGSQKQ--------TSKHIGSMRLDDWVLCRIYKKKHMGKTLQQ----KEDY- 174
IMHEYRL T+ S+RLDDWVLCRIYKK + + +ED
Sbjct: 136 IMHEYRLTDGNLSTAAKPPDLTTTRKNSLRLDDWVLCRIYKKNSSQRPTMERVLLREDLM 195
Query: 175 -------STHQFNDSIITNNDD 189
S + + S++ NND+
Sbjct: 196 EGMLSKSSANSSSTSVLDNNDN 217
>gb|AAN15611.1| NAM-like protein [Arabidopsis thaliana] gi|20466640|gb|AAM20637.1|
NAM-like protein [Arabidopsis thaliana]
Length = 320
Score = 239 bits (611), Expect = 5e-62
Identities = 118/197 (59%), Positives = 139/197 (69%), Gaps = 16/197 (8%)
Query: 1 MESSASS--------ELPPGFRFHPTDEELIVHYLCNQATSKPCPASIIPEVDIYKFDPW 52
MES+ SS LPPGFRFHPTDEEL++HYL +A S P P +II +VD+YKFDPW
Sbjct: 1 MESTDSSGGPPPPQPNLPPGFRFHPTDEELVIHYLKRKADSVPLPVAIIADVDLYKFDPW 60
Query: 53 ELPDKSEFEENEWYFFSPRERKYPNGVRPNRATLSGYWKATGTDKAIKS----GSKQIGV 108
ELP K+ F E EWYFFSPR+RKYPNG RPNRA SGYWKATGTDK + S GSK++GV
Sbjct: 61 ELPAKASFGEQEWYFFSPRDRKYPNGARPNRAATSGYWKATGTDKPVISTGGGGSKKVGV 120
Query: 109 KKSLVFYKGRPPKGVKTDWIMHEYRLIGSQKQTSKHIG----SMRLDDWVLCRIYKKKHM 164
KK+LVFY G+PPKGVK+DWIMHEYRL ++ G S+RLDDWVLCRIYKK +
Sbjct: 121 KKALVFYSGKPPKGVKSDWIMHEYRLTDNKPTHICDFGNKKNSLRLDDWVLCRIYKKNNS 180
Query: 165 GKTLQQKEDYSTHQFND 181
+ + H ND
Sbjct: 181 TASRHHHHLHHIHLDND 197
>gb|AAP37705.1| At1g61110 [Arabidopsis thaliana] gi|26450820|dbj|BAC42518.1|
unknown protein [Arabidopsis thaliana]
gi|18407026|ref|NP_564771.1| no apical meristem (NAM)
family protein [Arabidopsis thaliana]
Length = 323
Score = 239 bits (610), Expect = 6e-62
Identities = 118/202 (58%), Positives = 140/202 (68%), Gaps = 21/202 (10%)
Query: 9 LPPGFRFHPTDEELIVHYLCNQATSKPCPASIIPEVDIYKFDPWELPDKSEFEENEWYFF 68
LPPGFRFHPTDEEL+VHYL +A S P P SII E+D+YKFDPWELP K+ F E+EWYFF
Sbjct: 16 LPPGFRFHPTDEELVVHYLKKKADSVPLPVSIIAEIDLYKFDPWELPSKASFGEHEWYFF 75
Query: 69 SPRERKYPNGVRPNRATLSGYWKATGTDKAI-KSGSKQIGVKKSLVFYKGRPPKGVKTDW 127
SPR+RKYPNGVRPNRA SGYWKATGTDK I S ++GVKK+LVFY G+PPKG+KTDW
Sbjct: 76 SPRDRKYPNGVRPNRAATSGYWKATGTDKPIFTCNSHKVGVKKALVFYGGKPPKGIKTDW 135
Query: 128 IMHEYRLIGSQKQ--------TSKHIGSMRLDDWVLCRIYKKKHMGKTLQQ----KEDY- 174
IMHEYRL T+ S+RLDDWVLCRIYKK + + +ED
Sbjct: 136 IMHEYRLTDGNLSTAAKPPDLTTTRKNSLRLDDWVLCRIYKKNSSQRPTMERVLLREDLM 195
Query: 175 -------STHQFNDSIITNNDD 189
S + + S++ NND+
Sbjct: 196 EGMLSKSSANSSSTSVLDNNDN 217
>gb|AAB71483.1| similar to NAM (gp|X92205|1321924) and CUC2 (gp|AB002560|1944132)
proteins [Arabidopsis thaliana] gi|25352286|pir||H96636
hypothetical protein F11P17.16 [imported] - Arabidopsis
thaliana
Length = 300
Score = 239 bits (610), Expect = 6e-62
Identities = 118/202 (58%), Positives = 140/202 (68%), Gaps = 21/202 (10%)
Query: 9 LPPGFRFHPTDEELIVHYLCNQATSKPCPASIIPEVDIYKFDPWELPDKSEFEENEWYFF 68
LPPGFRFHPTDEEL+VHYL +A S P P SII E+D+YKFDPWELP K+ F E+EWYFF
Sbjct: 13 LPPGFRFHPTDEELVVHYLKKKADSVPLPVSIIAEIDLYKFDPWELPSKASFGEHEWYFF 72
Query: 69 SPRERKYPNGVRPNRATLSGYWKATGTDKAI-KSGSKQIGVKKSLVFYKGRPPKGVKTDW 127
SPR+RKYPNGVRPNRA SGYWKATGTDK I S ++GVKK+LVFY G+PPKG+KTDW
Sbjct: 73 SPRDRKYPNGVRPNRAATSGYWKATGTDKPIFTCNSHKVGVKKALVFYGGKPPKGIKTDW 132
Query: 128 IMHEYRLIGSQKQ--------TSKHIGSMRLDDWVLCRIYKKKHMGKTLQQ----KEDY- 174
IMHEYRL T+ S+RLDDWVLCRIYKK + + +ED
Sbjct: 133 IMHEYRLTDGNLSTAAKPPDLTTTRKNSLRLDDWVLCRIYKKNSSQRPTMERVLLREDLM 192
Query: 175 -------STHQFNDSIITNNDD 189
S + + S++ NND+
Sbjct: 193 EGMLSKSSANSSSTSVLDNNDN 214
>gb|AAM63301.1| NAM-like protein [Arabidopsis thaliana]
Length = 320
Score = 238 bits (606), Expect = 2e-61
Identities = 117/197 (59%), Positives = 138/197 (69%), Gaps = 16/197 (8%)
Query: 1 MESSASS--------ELPPGFRFHPTDEELIVHYLCNQATSKPCPASIIPEVDIYKFDPW 52
MES+ SS LPPGFRFHPTDEEL++HYL +A S P P +II +VD+YKFDPW
Sbjct: 1 MESTDSSGGPPPPQPNLPPGFRFHPTDEELVIHYLKRKADSVPLPVAIIADVDLYKFDPW 60
Query: 53 ELPDKSEFEENEWYFFSPRERKYPNGVRPNRATLSGYWKATGTDKAIKS----GSKQIGV 108
ELP K+ F E EWYFFSPR+RKYPNG RPNRA SGYWKATGTDK + S GSK++GV
Sbjct: 61 ELPAKASFGEQEWYFFSPRDRKYPNGARPNRAATSGYWKATGTDKPVISTGGGGSKKVGV 120
Query: 109 KKSLVFYKGRPPKGVKTDWIMHEYRLIGSQKQTSKHIG----SMRLDDWVLCRIYKKKHM 164
KK+LVFY G+PPKGVK+DWIMHEYRL ++ G S+RLDDWV CRIYKK +
Sbjct: 121 KKALVFYSGKPPKGVKSDWIMHEYRLTDNKPTHICDFGNKKNSLRLDDWVXCRIYKKNNS 180
Query: 165 GKTLQQKEDYSTHQFND 181
+ + H ND
Sbjct: 181 TASRHHHHLHHIHLDND 197
>gb|AAU43923.1| NAC domain protein [Lycopersicon esculentum]
gi|52353036|gb|AAU43922.1| NAC domain protein
[Lycopersicon esculentum]
Length = 355
Score = 236 bits (602), Expect = 5e-61
Identities = 123/251 (49%), Positives = 161/251 (64%), Gaps = 26/251 (10%)
Query: 1 MESSASS-------ELPPGFRFHPTDEELIVHYLCNQATSKPCPASIIPEVDIYKFDPWE 53
MES+ SS +LPPGFRFHPTDEELIVHYL + P P II E+D+YKFDPWE
Sbjct: 1 MESTDSSTGTRHQPQLPPGFRFHPTDEELIVHYLKKRVAGAPIPVDIIGEIDLYKFDPWE 60
Query: 54 LPDKSEFEENEWYFFSPRERKYPNGVRPNRATLSGYWKATGTDKAI--KSGSKQIGVKKS 111
LP K+ F E EW+FFSPR+RKYPNG RPNRA SGYWKATGTDK + G++++GVKK+
Sbjct: 61 LPAKAIFGEQEWFFFSPRDRKYPNGARPNRAATSGYWKATGTDKPVFTSGGTQKVGVKKA 120
Query: 112 LVFYKGRPPKGVKTDWIMHEYRLI---------GSQKQTSKHIGSMRLDDWVLCRIYKKK 162
LVFY G+PPKGVKT+WIMHEYR++ G + GS+RLDDWVLCRIYKK
Sbjct: 121 LVFYGGKPPKGVKTNWIMHEYRVVENKTNNKPLGCDNIVANKKGSLRLDDWVLCRIYKKN 180
Query: 163 HMGKTLQQKEDYSTHQFNDSIITNNDDGELEMMNLTRSCSLTYLLDMNYFGPILSDGSTL 222
+ +++ H SI N + L+ + + ++ + N + I+++ + +
Sbjct: 181 NTQRSID-----DLHDMLGSIPQNVPNSILQGIKPSNYGTILLENESNMYDGIMNNTNDI 235
Query: 223 DFQINNSNIGI 233
INN+N I
Sbjct: 236 ---INNNNRSI 243
>ref|NP_911241.1| putative OsNAC5 protein [Oryza sativa (japonica cultivar-group)]
gi|24417196|dbj|BAC22555.1| putative OsNAC5 protein
[Oryza sativa (japonica cultivar-group)]
gi|27817885|dbj|BAC55651.1| putative OsNAC5 protein
[Oryza sativa (japonica cultivar-group)]
Length = 425
Score = 236 bits (601), Expect = 7e-61
Identities = 111/179 (62%), Positives = 131/179 (73%), Gaps = 19/179 (10%)
Query: 2 ESSASSELPPGFRFHPTDEELIVHYLCNQATSKPCPASIIPEVDIYKFDPWELPDKSEFE 61
+ ++ ELPPGFRFHPTDEEL+VHYL +A S P P +II EVD+YKFDPW+LP+K+ F
Sbjct: 22 QPGSAPELPPGFRFHPTDEELVVHYLKKKAASVPLPVTIIAEVDLYKFDPWDLPEKANFG 81
Query: 62 ENEWYFFSPRERKYPNGVRPNRATLSGYWKATGTDKAIKSGS---KQIGVKKSLVFYKGR 118
E EWYFFSPR+RKYPNG RPNRA SGYWKATGTDK I S +++GVKK+LVFY+G+
Sbjct: 82 EQEWYFFSPRDRKYPNGARPNRAATSGYWKATGTDKPIMSSGSTREKVGVKKALVFYRGK 141
Query: 119 PPKGVKTDWIMHEYRLIGSQKQT----------------SKHIGSMRLDDWVLCRIYKK 161
PPKGVKT+WIMHEYRL + SK S+RLDDWVLCRIYKK
Sbjct: 142 PPKGVKTNWIMHEYRLTDTSSSAAAVATTRRPPPPITGGSKGAVSLRLDDWVLCRIYKK 200
Database: nr
Posted date: Jul 5, 2005 12:34 AM
Number of letters in database: 863,360,394
Number of sequences in database: 2,540,612
Lambda K H
0.316 0.134 0.411
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 517,538,078
Number of Sequences: 2540612
Number of extensions: 23234630
Number of successful extensions: 45876
Number of sequences better than 10.0: 385
Number of HSP's better than 10.0 without gapping: 321
Number of HSP's successfully gapped in prelim test: 64
Number of HSP's that attempted gapping in prelim test: 44909
Number of HSP's gapped (non-prelim): 404
length of query: 271
length of database: 863,360,394
effective HSP length: 126
effective length of query: 145
effective length of database: 543,243,282
effective search space: 78770275890
effective search space used: 78770275890
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.6 bits)
S2: 74 (33.1 bits)
Medicago: description of AC140030.7


