

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC140025.4 + phase: 0
(277 letters)
Database: nr
2,540,612 sequences; 863,360,394 total letters
Searching..................................................done
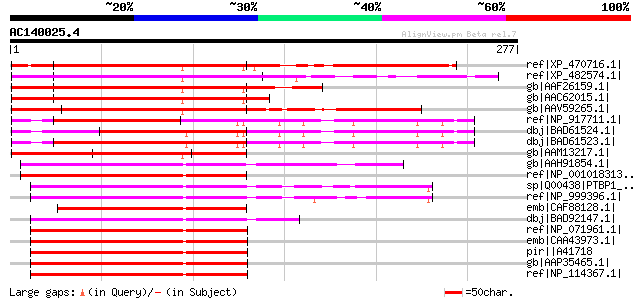
Score E
Sequences producing significant alignments: (bits) Value
ref|XP_470716.1| putative polypyrimidine tract-binding protein [... 244 2e-63
ref|XP_482574.1| putative polypyrimidine tract-binding protein h... 242 1e-62
gb|AAF26159.1| putative polypyrimidine tract-binding protein [Ar... 220 3e-56
gb|AAC62015.1| polypyrimidine tract-binding protein homolog [Ara... 216 4e-55
gb|AAV59265.1| At5g53180 [Arabidopsis thaliana] gi|53749130|gb|A... 203 5e-51
ref|NP_917711.1| putative polypyrimidine tract-binding protein [... 190 3e-47
dbj|BAD61524.1| polypyrimidine tract-binding protein-like [Oryza... 190 3e-47
dbj|BAD61523.1| polypyrimidine tract-binding protein 1-like [Ory... 190 3e-47
gb|AAM13217.1| putative polypyrimidine tract-binding protein [Ar... 159 6e-38
gb|AAH91854.1| Unknown (protein for IMAGE:7152787) [Danio rerio] 102 1e-20
ref|NP_001018313.1| hypothetical protein LOC323490 [Danio rerio]... 102 2e-20
sp|Q00438|PTBP1_RAT Polypyrimidine tract-binding protein 1 (PTB)... 101 3e-20
ref|NP_999396.1| polypyrimidine tract-binding protein [Sus scrof... 100 8e-20
emb|CAF88128.1| unnamed protein product [Tetraodon nigroviridis] 99 2e-19
dbj|BAD92147.1| polypyrimidine tract-binding protein 1 isoform c... 98 2e-19
ref|NP_071961.1| polypyrimidine tract binding protein 1 isoform ... 97 4e-19
emb|CAA43973.1| polypirimidine tract binding protein [Homo sapie... 97 4e-19
pir||A41718 polypyrimidine tract-binding protein PTB-1 - mouse 97 4e-19
gb|AAP35465.1| polypyrimidine tract binding protein 1 [Homo sapi... 97 4e-19
ref|NP_114367.1| polypyrimidine tract-binding protein 1 isoform ... 97 4e-19
>ref|XP_470716.1| putative polypyrimidine tract-binding protein [Oryza sativa]
gi|18921326|gb|AAL82531.1| putative polypyrimidine
tract-binding protein [Oryza sativa]
Length = 464
Score = 244 bits (623), Expect = 2e-63
Identities = 144/258 (55%), Positives = 163/258 (62%), Gaps = 32/258 (12%)
Query: 2 LPVNHSAIEGAAQPAIGPDGKRIETESNVLLASIENMQYAVTVDVINTVFSAFGTVQKIA 61
LPVN SAI+ QPA+G DG+++E E NVLLASIENMQYAVTVDV++TVFSAFGTVQKIA
Sbjct: 220 LPVNSSAID-TLQPAVGADGRKVEAEGNVLLASIENMQYAVTVDVLHTVFSAFGTVQKIA 278
Query: 62 MFEKNGQTQALIQYPDVTTAAAAREALEGHCIYDGGYCKLHLSYSRHTDLNVKAFSDKSR 121
+FEKNG TQALIQYPDVTTA+ A+EALEGHCIYDGGYCKLHLSYSRHTDLNVKA SDKSR
Sbjct: 279 IFEKNGGTQALIQYPDVTTASVAKEALEGHCIYDGGYCKLHLSYSRHTDLNVKAHSDKSR 338
Query: 122 DYTVP-----LVPAPV--------WQ-NPQAAPMYPTNSPAYQTQVPGGSPAYQTQVPGG 167
DYT+P VP P WQ NPQA Y A +P + T G
Sbjct: 339 DYTIPQGAMQAVPQPPGVPTTSAGWQGNPQAGGAYAPPGAA--------APNHGTT---G 387
Query: 168 QVPSWDLTQHAVRPGYVPVPGAYPGQTGAFPTM-PSYGSAAMPTASSPLAQSSHPGAPHN 226
QVP+W + GY P PGAYPGQ + P + G + P A +S P
Sbjct: 388 QVPNW----NPGNSGYAPAPGAYPGQMYSSPMQYGASGGFSAPAAPPQELHTSQQMPPPQ 443
Query: 227 VNLQPSGGSTSGPGSSPH 244
QP G G PH
Sbjct: 444 YGNQPGPAGAPGTG-QPH 460
Score = 101 bits (252), Expect = 2e-20
Identities = 61/112 (54%), Positives = 72/112 (63%), Gaps = 7/112 (6%)
Query: 25 ETESNVLLASIENMQYA-VTVDVINTVFSAFGTVQKIAMFEKNGQTQALIQYPDVTTAAA 83
ET NVLL +IE +Q VT+DVI+ VFSAFG V KIA FEK QALIQY D TA+A
Sbjct: 110 ETAGNVLLVTIEGVQANDVTIDVIHLVFSAFGFVHKIATFEKAAGFQALIQYTDAATASA 169
Query: 84 AREALEGHCI------YDGGYCKLHLSYSRHTDLNVKAFSDKSRDYTVPLVP 129
AREAL+G I C L +S+S H DLN+K S +SRDYT P +P
Sbjct: 170 AREALDGRSIPRYLLPEHVTSCCLRISFSAHKDLNIKFQSHRSRDYTNPYLP 221
>ref|XP_482574.1| putative polypyrimidine tract-binding protein homolog [Oryza sativa
(japonica cultivar-group)] gi|42407521|dbj|BAD10638.1|
putative polypyrimidine tract-binding protein homolog
[Oryza sativa (japonica cultivar-group)]
Length = 461
Score = 242 bits (617), Expect = 1e-62
Identities = 143/270 (52%), Positives = 164/270 (59%), Gaps = 35/270 (12%)
Query: 2 LPVNHSAIEGAAQPAIGPDGKRIETESNVLLASIENMQYAVTVDVINTVFSAFGTVQKIA 61
LPVN +AIEG AQP +GPDGK E ESNVLLASIENMQYAVTVDV++TVFSAFGTVQKIA
Sbjct: 216 LPVNPTAIEGIAQPTLGPDGKIKEPESNVLLASIENMQYAVTVDVLHTVFSAFGTVQKIA 275
Query: 62 MFEKNGQTQALIQYPDVTTAAAAREALEGHCIYDGGYCKLHLSYSRHTDLNVKAFSDKSR 121
MFEKNG QALIQYPD+TTAA A++ALEGHCIYDGGYCKLHLSYSRHTDLNVKA ++SR
Sbjct: 276 MFEKNGGMQALIQYPDITTAAVAKQALEGHCIYDGGYCKLHLSYSRHTDLNVKAHDERSR 335
Query: 122 DYTVPLVPAPVWQNPQAAPMYPTNSPAYQTQVPG----GSPAYQTQVPGGQVPSWDLTQH 177
DYTV P+ Q AP T A+Q P GS A T P GQVP+W+
Sbjct: 336 DYTVSSDPSAQMQAAAQAPGPSTPGVAWQNTAPSASFYGSTAAAT--PVGQVPAWNPNMQ 393
Query: 178 AVRPGYVPVPGAYPGQTGAFPTMPSYGSAAMPTASSPLAQSSHPGAPHNVNLQPSGGSTS 237
A GA+ + A+PT P MP + PH + S G+
Sbjct: 394 A---------GAFGSASSAYPTQP-----MMPGS-----------VPHYPGIGSSSGALP 428
Query: 238 GPGSSPHMQQNLGAQGMVRPGAPPNVRPGG 267
+ H G V P AP + P G
Sbjct: 429 VSFQASHQMPQYG----VPPAAPHHAPPAG 454
Score = 88.6 bits (218), Expect = 2e-16
Identities = 54/121 (44%), Positives = 71/121 (58%), Gaps = 11/121 (9%)
Query: 25 ETESNVLLASIENMQ-YAVTVDVINTVFSAFGTVQKIAMFEKNGQTQALIQYPDVTTAAA 83
++ NVLL + E +Q +++DVI+ VFSAFG V KIA FEK QALIQY D TA
Sbjct: 106 DSSGNVLLVTFEGVQPNDISIDVIHLVFSAFGFVHKIATFEKAAGFQALIQYTDAPTALE 165
Query: 84 AREALEGHCI------YDGGYCKLHLSYSRHTDLNVKAFSDKSRDYTVPLVPAPVWQNPQ 137
A+ +L+G I C L +++S H DLN+K S +SRDYT P +P NP
Sbjct: 166 AKNSLDGRSIPRYLLPEHVPTCHLRITFSAHKDLNIKFQSHRSRDYTNPYLPV----NPT 221
Query: 138 A 138
A
Sbjct: 222 A 222
>gb|AAF26159.1| putative polypyrimidine tract-binding protein [Arabidopsis
thaliana] gi|15232047|ref|NP_186764.1| polypyrimidine
tract-binding protein, putative / heterogeneous nuclear
ribonucleoprotein, putative [Arabidopsis thaliana]
Length = 399
Score = 220 bits (561), Expect = 3e-56
Identities = 116/186 (62%), Positives = 133/186 (71%), Gaps = 25/186 (13%)
Query: 2 LPVNHSAIEGAAQPAIGPDGKRIETESNVLLASIENMQYAVTVDVINTVFSAFGTVQKIA 61
LPVN +A++G+ QPA+G DGK++E++SNVLL IENMQYAVTVDV++TVFSA+GTVQKIA
Sbjct: 216 LPVNQTAMDGSMQPALGADGKKVESQSNVLLGLIENMQYAVTVDVLHTVFSAYGTVQKIA 275
Query: 62 MFEKNGQTQALIQYPDVTTAAAAREALEGHCIYDGGYCKLHLSYSRHTDLNVKAFSDKSR 121
+FEKNG TQALIQY D+ TAA A+EALEGHCIYDGGYCKL LSYSRHTDLNVKAFSDKSR
Sbjct: 276 IFEKNGSTQALIQYSDIPTAAMAKEALEGHCIYDGGYCKLRLSYSRHTDLNVKAFSDKSR 335
Query: 122 DYTVP---------------LVPAPVWQNPQAAPMYPTNSPAYQTQVPGGSP-AYQTQVP 165
DYT+P P WQNPQA Y GGSP Y + P
Sbjct: 336 DYTLPDLSLLVAQKGPAVSGSAPPAGWQNPQAQSQYSGY---------GGSPYMYPSSDP 386
Query: 166 GGQVPS 171
G PS
Sbjct: 387 NGASPS 392
Score = 101 bits (252), Expect = 2e-20
Identities = 57/112 (50%), Positives = 73/112 (64%), Gaps = 7/112 (6%)
Query: 25 ETESNVLLASIENMQ-YAVTVDVINTVFSAFGTVQKIAMFEKNGQTQALIQYPDVTTAAA 83
+ NVLL + E ++ + V++DVI+ VFSAFG V KIA FEK QAL+Q+ DV TA+A
Sbjct: 106 DVPGNVLLVTFEGVESHEVSIDVIHLVFSAFGFVHKIATFEKAAGFQALVQFTDVETASA 165
Query: 84 AREALEGHCI------YDGGYCKLHLSYSRHTDLNVKAFSDKSRDYTVPLVP 129
AR AL+G I G C L +SYS HTDLN+K S +SRDYT P +P
Sbjct: 166 ARSALDGRSIPRYLLSAHVGSCSLRMSYSAHTDLNIKFQSHRSRDYTNPYLP 217
>gb|AAC62015.1| polypyrimidine tract-binding protein homolog [Arabidopsis thaliana]
gi|11358627|pir||T51814 polypyrimidine tract-binding
protein homolog [imported] - Arabidopsis thaliana
Length = 418
Score = 216 bits (551), Expect = 4e-55
Identities = 107/156 (68%), Positives = 123/156 (78%), Gaps = 15/156 (9%)
Query: 2 LPVNHSAIEGAAQPAIGPDGKRIETESNVLLASIENMQYAVTVDVINTVFSAFGTVQKIA 61
LPVN +A++G+ QPA+G DGK++E++SNVLL IENMQYAVTVDV++TVFSA+GTVQKIA
Sbjct: 216 LPVNQTAMDGSMQPALGADGKKVESQSNVLLGLIENMQYAVTVDVLHTVFSAYGTVQKIA 275
Query: 62 MFEKNGQTQALIQYPDVTTAAAAREALEGHCIYDGGYCKLHLSYSRHTDLNVKAFSDKSR 121
+FEKNG TQALIQY D+ TAA A+EALEGHCIYDGGYCKL LSYSRHTDLNVKAFSDKSR
Sbjct: 276 IFEKNGSTQALIQYSDIPTAAMAKEALEGHCIYDGGYCKLRLSYSRHTDLNVKAFSDKSR 335
Query: 122 DYTVP---------------LVPAPVWQNPQAAPMY 142
DYT+P P WQNPQA Y
Sbjct: 336 DYTLPDLSLLVAQKGPAVSGSAPPAGWQNPQAQSQY 371
Score = 101 bits (252), Expect = 2e-20
Identities = 57/112 (50%), Positives = 73/112 (64%), Gaps = 7/112 (6%)
Query: 25 ETESNVLLASIENMQ-YAVTVDVINTVFSAFGTVQKIAMFEKNGQTQALIQYPDVTTAAA 83
+ NVLL + E ++ + V++DVI+ VFSAFG V KIA FEK QAL+Q+ DV TA+A
Sbjct: 106 DVPGNVLLVTFEGVESHEVSIDVIHLVFSAFGFVHKIATFEKAAGFQALVQFTDVETASA 165
Query: 84 AREALEGHCI------YDGGYCKLHLSYSRHTDLNVKAFSDKSRDYTVPLVP 129
AR AL+G I G C L +SYS HTDLN+K S +SRDYT P +P
Sbjct: 166 ARSALDGRSIPRYLLSAHVGSCSLRMSYSAHTDLNIKFQSHRSRDYTNPYLP 217
>gb|AAV59265.1| At5g53180 [Arabidopsis thaliana] gi|53749130|gb|AAU90050.1|
At5g53180 [Arabidopsis thaliana]
gi|9757999|dbj|BAB08421.1| polypyrimidine tract-binding
RNA transport protein-like [Arabidopsis thaliana]
gi|15238677|ref|NP_200130.1| polypyrimidine
tract-binding protein, putative / heterogeneous nuclear
ribonucleoprotein, putative [Arabidopsis thaliana]
Length = 429
Score = 203 bits (516), Expect = 5e-51
Identities = 118/225 (52%), Positives = 139/225 (61%), Gaps = 13/225 (5%)
Query: 2 LPVNHSAIEGAAQPAIGPDGKRIETESNVLLASIENMQYAVTVDVINTVFSAFGTVQKIA 61
LPV SAI+ Q A+G DGK++E ESNVLLASIENMQYAVT+DV++ VF+AFG VQKIA
Sbjct: 217 LPVAPSAIDSTGQVAVGVDGKKMEPESNVLLASIENMQYAVTLDVLHMVFAAFGEVQKIA 276
Query: 62 MFEKNGQTQALIQYPDVTTAAAAREALEGHCIYDGGYCKLHLSYSRHTDLNVKAFSDKSR 121
MF+KNG QALIQY DV TA A+EALEGHCIYDGG+CKLH++YSRHTDL++K +D+SR
Sbjct: 277 MFDKNGGVQALIQYSDVQTAVVAKEALEGHCIYDGGFCKLHITYSRHTDLSIKVNNDRSR 336
Query: 122 DYTVPLVPAPVWQNPQAAPMYPTNSPAYQTQVPGGSPAYQTQVPGGQVPSWDLTQHAVRP 181
DYT+P P P+ Q P P Y N Y GGS Q Q P G W V+P
Sbjct: 337 DYTMPNPPVPMPQQPVQNP-YAGNPQQY--HAAGGSHHQQQQQPQG---GW------VQP 384
Query: 182 GYVPVPGAYPGQTGAFPTMPSYGSAAM-PTASSPLAQSSHPGAPH 225
G G G + PS S P P PG H
Sbjct: 385 GGQGSMGMGGGGHNHYMAPPSSSSMHQGPGGHMPPQHYGGPGPMH 429
Score = 90.1 bits (222), Expect = 6e-17
Identities = 53/108 (49%), Positives = 66/108 (61%), Gaps = 7/108 (6%)
Query: 29 NVLLASIENMQYA-VTVDVINTVFSAFGTVQKIAMFEKNGQTQALIQYPDVTTAAAAREA 87
NVLL +IE V++DV++ VFSAFG V KI FEK QAL+Q+ D TA AA+ A
Sbjct: 111 NVLLVTIEGDDARMVSIDVLHLVFSAFGFVHKITTFEKTAGYQALVQFTDAETATAAKLA 170
Query: 88 LEGHCI------YDGGYCKLHLSYSRHTDLNVKAFSDKSRDYTVPLVP 129
L+G I G C L ++YS HTDL VK S +SRDYT P +P
Sbjct: 171 LDGRSIPRYLLAETVGQCSLKITYSAHTDLTVKFQSHRSRDYTNPYLP 218
>ref|NP_917711.1| putative polypyrimidine tract-binding protein [Oryza sativa
(japonica cultivar-group)]
Length = 522
Score = 190 bits (483), Expect = 3e-47
Identities = 132/282 (46%), Positives = 157/282 (54%), Gaps = 42/282 (14%)
Query: 2 LPVNHSAIEGAAQPAIGPDGKRIETESNVLLASIENMQYAVTVDVINTVFSAFGTVQKIA 61
LPV SAI+G+ GPDGK+ E ESNVLLAS+ENMQY VT+DV++ VFSAFG VQKIA
Sbjct: 183 LPVAPSAIDGS-----GPDGKKQEAESNVLLASVENMQYVVTIDVLHEVFSAFGFVQKIA 237
Query: 62 MFEKNGQTQALIQYPDVTTAAAAREALEGHCIYDGGYCKLHLSYSRHTDLNVKAFSDKSR 121
+FEKN QALIQYPD+ TA AA+EALEGH IY+GGYCKLHL++SRHTDLNVK +++ R
Sbjct: 238 IFEKNSGFQALIQYPDIQTAVAAKEALEGHSIYEGGYCKLHLTFSRHTDLNVKVNNERGR 297
Query: 122 DY----TVP-------LVPAPVWQNP-QAAPMYPTNS--PAYQTQVPGGSPA--YQTQVP 165
DY T P L P PV+ AP T + P T P G+P+ Y + P
Sbjct: 298 DYTGGNTAPTSNQPSILGPQPVYSGAYNNAPSSATGAVVPPGTTLTPPGAPSHPYTSSEP 357
Query: 166 GGQVPSWDLTQHAVRPGYVPV---PGAYPGQTGAFPT-MPSYGSAAMPTASSPL-AQSSH 220
Q P AV G P+ G G G P P YGS P S+ H
Sbjct: 358 LPQTP-------AVPSGGAPLYTSQGILQGPPGVPPAQFPGYGSPQFPPGSAQAQMHQQH 410
Query: 221 P-----GAPHNVNLQPSGGS---TSGPGSSPHMQQNLGAQGM 254
P P +N QP GS G G P +Q G Q M
Sbjct: 411 PVQGSQQMPGQMNHQPPPGSFMQYPGDGGRP-VQDAPGQQAM 451
Score = 71.2 bits (173), Expect = 3e-11
Identities = 39/70 (55%), Positives = 50/70 (70%), Gaps = 1/70 (1%)
Query: 25 ETESNVLLASIEN-MQYAVTVDVINTVFSAFGTVQKIAMFEKNGQTQALIQYPDVTTAAA 83
E NVLL S+E + AV++DV++ VFSAFG VQKIA FEK QALIQ+ D TA++
Sbjct: 105 EAAGNVLLVSMEGVLPDAVSIDVLHLVFSAFGFVQKIATFEKASGYQALIQFCDTETASS 164
Query: 84 AREALEGHCI 93
A+ AL+G CI
Sbjct: 165 AKAALDGRCI 174
>dbj|BAD61524.1| polypyrimidine tract-binding protein-like [Oryza sativa (japonica
cultivar-group)]
Length = 448
Score = 190 bits (483), Expect = 3e-47
Identities = 132/282 (46%), Positives = 157/282 (54%), Gaps = 42/282 (14%)
Query: 2 LPVNHSAIEGAAQPAIGPDGKRIETESNVLLASIENMQYAVTVDVINTVFSAFGTVQKIA 61
LPV SAI+G+ GPDGK+ E ESNVLLAS+ENMQY VT+DV++ VFSAFG VQKIA
Sbjct: 109 LPVAPSAIDGS-----GPDGKKQEAESNVLLASVENMQYVVTIDVLHEVFSAFGFVQKIA 163
Query: 62 MFEKNGQTQALIQYPDVTTAAAAREALEGHCIYDGGYCKLHLSYSRHTDLNVKAFSDKSR 121
+FEKN QALIQYPD+ TA AA+EALEGH IY+GGYCKLHL++SRHTDLNVK +++ R
Sbjct: 164 IFEKNSGFQALIQYPDIQTAVAAKEALEGHSIYEGGYCKLHLTFSRHTDLNVKVNNERGR 223
Query: 122 DY----TVP-------LVPAPVWQNP-QAAPMYPTNS--PAYQTQVPGGSPA--YQTQVP 165
DY T P L P PV+ AP T + P T P G+P+ Y + P
Sbjct: 224 DYTGGNTAPTSNQPSILGPQPVYSGAYNNAPSSATGAVVPPGTTLTPPGAPSHPYTSSEP 283
Query: 166 GGQVPSWDLTQHAVRPGYVPV---PGAYPGQTGAFPT-MPSYGSAAMPTASSPL-AQSSH 220
Q P AV G P+ G G G P P YGS P S+ H
Sbjct: 284 LPQTP-------AVPSGGAPLYTSQGILQGPPGVPPAQFPGYGSPQFPPGSAQAQMHQQH 336
Query: 221 P-----GAPHNVNLQPSGGS---TSGPGSSPHMQQNLGAQGM 254
P P +N QP GS G G P +Q G Q M
Sbjct: 337 PVQGSQQMPGQMNHQPPPGSFMQYPGDGGRP-VQDAPGQQAM 377
Score = 80.1 bits (196), Expect = 6e-14
Identities = 46/86 (53%), Positives = 55/86 (63%), Gaps = 6/86 (6%)
Query: 50 VFSAFGTVQKIAMFEKNGQTQALIQYPDVTTAAAAREALEGHCIYD------GGYCKLHL 103
VFSAFG VQKIA FEK QALIQ+ D TA++A+ AL+G CI C L +
Sbjct: 25 VFSAFGFVQKIATFEKASGYQALIQFCDTETASSAKAALDGRCIPSYLLPELDVPCTLRI 84
Query: 104 SYSRHTDLNVKAFSDKSRDYTVPLVP 129
+YS HT LNVK S +SRDYT P +P
Sbjct: 85 NYSAHTVLNVKFQSHRSRDYTNPYLP 110
>dbj|BAD61523.1| polypyrimidine tract-binding protein 1-like [Oryza sativa (japonica
cultivar-group)]
Length = 554
Score = 190 bits (483), Expect = 3e-47
Identities = 132/282 (46%), Positives = 157/282 (54%), Gaps = 42/282 (14%)
Query: 2 LPVNHSAIEGAAQPAIGPDGKRIETESNVLLASIENMQYAVTVDVINTVFSAFGTVQKIA 61
LPV SAI+G+ GPDGK+ E ESNVLLAS+ENMQY VT+DV++ VFSAFG VQKIA
Sbjct: 215 LPVAPSAIDGS-----GPDGKKQEAESNVLLASVENMQYVVTIDVLHEVFSAFGFVQKIA 269
Query: 62 MFEKNGQTQALIQYPDVTTAAAAREALEGHCIYDGGYCKLHLSYSRHTDLNVKAFSDKSR 121
+FEKN QALIQYPD+ TA AA+EALEGH IY+GGYCKLHL++SRHTDLNVK +++ R
Sbjct: 270 IFEKNSGFQALIQYPDIQTAVAAKEALEGHSIYEGGYCKLHLTFSRHTDLNVKVNNERGR 329
Query: 122 DY----TVP-------LVPAPVWQNP-QAAPMYPTNS--PAYQTQVPGGSPA--YQTQVP 165
DY T P L P PV+ AP T + P T P G+P+ Y + P
Sbjct: 330 DYTGGNTAPTSNQPSILGPQPVYSGAYNNAPSSATGAVVPPGTTLTPPGAPSHPYTSSEP 389
Query: 166 GGQVPSWDLTQHAVRPGYVPV---PGAYPGQTGAFPT-MPSYGSAAMPTASSPL-AQSSH 220
Q P AV G P+ G G G P P YGS P S+ H
Sbjct: 390 LPQTP-------AVPSGGAPLYTSQGILQGPPGVPPAQFPGYGSPQFPPGSAQAQMHQQH 442
Query: 221 P-----GAPHNVNLQPSGGS---TSGPGSSPHMQQNLGAQGM 254
P P +N QP GS G G P +Q G Q M
Sbjct: 443 PVQGSQQMPGQMNHQPPPGSFMQYPGDGGRP-VQDAPGQQAM 483
Score = 96.3 bits (238), Expect = 8e-19
Identities = 57/112 (50%), Positives = 72/112 (63%), Gaps = 7/112 (6%)
Query: 25 ETESNVLLASIEN-MQYAVTVDVINTVFSAFGTVQKIAMFEKNGQTQALIQYPDVTTAAA 83
E NVLL S+E + AV++DV++ VFSAFG VQKIA FEK QALIQ+ D TA++
Sbjct: 105 EAAGNVLLVSMEGVLPDAVSIDVLHLVFSAFGFVQKIATFEKASGYQALIQFCDTETASS 164
Query: 84 AREALEGHCIYD------GGYCKLHLSYSRHTDLNVKAFSDKSRDYTVPLVP 129
A+ AL+G CI C L ++YS HT LNVK S +SRDYT P +P
Sbjct: 165 AKAALDGRCIPSYLLPELDVPCTLRINYSAHTVLNVKFQSHRSRDYTNPYLP 216
>gb|AAM13217.1| putative polypyrimidine tract-binding protein [Arabidopsis
thaliana] gi|24899823|gb|AAN65126.1| putative
polypyrimidine tract-binding protein [Arabidopsis
thaliana]
Length = 189
Score = 159 bits (403), Expect = 6e-38
Identities = 74/98 (75%), Positives = 89/98 (90%)
Query: 2 LPVNHSAIEGAAQPAIGPDGKRIETESNVLLASIENMQYAVTVDVINTVFSAFGTVQKIA 61
LPVN +A++G+ QPA+G DGK++E++SNVLL IENMQYAVTVDV++TVFSA+GTVQKIA
Sbjct: 91 LPVNQTAMDGSMQPALGADGKKVESQSNVLLGLIENMQYAVTVDVLHTVFSAYGTVQKIA 150
Query: 62 MFEKNGQTQALIQYPDVTTAAAAREALEGHCIYDGGYC 99
+FEKNG TQALIQY D+ TAA A+EALEGHCIYDGGYC
Sbjct: 151 IFEKNGSTQALIQYSDIPTAAMAKEALEGHCIYDGGYC 188
Score = 89.0 bits (219), Expect = 1e-16
Identities = 49/90 (54%), Positives = 58/90 (64%), Gaps = 6/90 (6%)
Query: 46 VINTVFSAFGTVQKIAMFEKNGQTQALIQYPDVTTAAAAREALEGHCI------YDGGYC 99
+I VFSAFG V KIA FEK QAL+Q+ DV TA+AAR AL+G I G C
Sbjct: 3 IIGQVFSAFGFVHKIATFEKAAGFQALVQFTDVETASAARSALDGRSIPRYLLSAHVGSC 62
Query: 100 KLHLSYSRHTDLNVKAFSDKSRDYTVPLVP 129
L +SYS HTDLN+K S +SRDYT P +P
Sbjct: 63 SLRMSYSAHTDLNIKFQSHRSRDYTNPYLP 92
>gb|AAH91854.1| Unknown (protein for IMAGE:7152787) [Danio rerio]
Length = 586
Score = 102 bits (254), Expect = 1e-20
Identities = 73/209 (34%), Positives = 106/209 (49%), Gaps = 15/209 (7%)
Query: 7 SAIEGAAQPAIGPDGKRIETESNVLLASIENMQYAVTVDVINTVFSAFGTVQKIAMFEKN 66
+A++ + G D + S VL +EN+ Y VT+DV++ +FS FGTV K+ F KN
Sbjct: 189 NAVQTGSLTLGGVDPSGMTGPSPVLRVIVENLFYPVTLDVLHQIFSKFGTVLKVITFTKN 248
Query: 67 GQTQALIQYPDVTTAAAAREALEGHCIYDGGYCKLHLSYSRHTDLNVKAFSDKSRDYTVP 126
Q QAL+Q+ D TA A+ AL+G IY+ G C L +S+S+ T LNVK +DKSRDYT P
Sbjct: 249 NQFQALLQFTDGLTAQHAKLALDGQNIYN-GCCTLRISFSKLTSLNVKYNNDKSRDYTRP 307
Query: 127 LVPAPVWQNPQAAPMYPTNSPAYQTQVPGGSPAYQTQVPGGQVPSWDLTQHAVRPGYVPV 186
+P + P ++ A PG A P++ + Q +
Sbjct: 308 DLP-----TGDSQPALEHHAVAAAFAAPGIISAAPYASAHAFPPAFAIQQAGL------- 355
Query: 187 PGAYPGQTGAFPTMPSYGSAAMPTASSPL 215
PG GA ++ G+AA A+S L
Sbjct: 356 --TIPGVPGALASLTIPGAAAAAAAASRL 382
>ref|NP_001018313.1| hypothetical protein LOC323490 [Danio rerio]
gi|63100731|gb|AAH95372.1| Hypothetical protein
LOC323490 [Danio rerio]
Length = 574
Score = 102 bits (253), Expect = 2e-20
Identities = 56/123 (45%), Positives = 78/123 (62%), Gaps = 1/123 (0%)
Query: 7 SAIEGAAQPAIGPDGKRIETESNVLLASIENMQYAVTVDVINTVFSAFGTVQKIAMFEKN 66
+A++ DG + ++S VL +EN+ Y VT+DV++ +FS FGTV KI F KN
Sbjct: 170 NAVQTGGMSLAAIDGAGMGSQSPVLRVIVENLFYPVTLDVLHQIFSKFGTVLKIITFTKN 229
Query: 67 GQTQALIQYPDVTTAAAAREALEGHCIYDGGYCKLHLSYSRHTDLNVKAFSDKSRDYTVP 126
Q QAL+QY D TA A+ +L+G IY+ C L +S+S+ T LNVK +DKSRDYT P
Sbjct: 230 NQFQALVQYSDGMTAQHAKLSLDGQNIYN-ACCTLRISFSKLTSLNVKYNNDKSRDYTRP 288
Query: 127 LVP 129
+P
Sbjct: 289 DLP 291
>sp|Q00438|PTBP1_RAT Polypyrimidine tract-binding protein 1 (PTB) (Heterogeneous nuclear
ribonucleoprotein I) (hnRNP I) (Pyrimidine-binding
protein) (PYBP)
Length = 555
Score = 101 bits (251), Expect = 3e-20
Identities = 79/225 (35%), Positives = 112/225 (49%), Gaps = 23/225 (10%)
Query: 12 AAQPAIGPDGKRIETESNVLLASIENMQYAVTVDVINTVFSAFGTVQKIAMFEKNGQTQA 71
AA A G + +S VL +EN+ Y VT+DV++ +FS FGTV KI F KN Q QA
Sbjct: 164 AASAAAVDAGMAMAGQSPVLRIIVENLFYPVTLDVLHQIFSKFGTVLKIITFTKNNQFQA 223
Query: 72 LIQYPDVTTAAAAREALEGHCIYDGGYCKLHLSYSRHTDLNVKAFSDKSRDYTVPLVPAP 131
L+QY D +A A+ +L+G IY+ C L + +S+ T LNVK +DKSRDYT P +P+
Sbjct: 224 LLQYADPVSAQHAKLSLDGQNIYN-ACCTLRIDFSKLTSLNVKYNNDKSRDYTRPDLPS- 281
Query: 132 VWQNPQAAPMYPTNSPAYQTQVPGGSPAYQTQVP-GGQVPSWDLTQHAVRPGYVPVPGAY 190
+ T + A+ G+P + P G VPS H P + +
Sbjct: 282 ---GDSQPSLDQTMAAAF------GAPGIMSASPYAGAVPS-----HLCHPSRAGL--SV 325
Query: 191 PGQTGAFPTMPSYGSAAMPTASSPLAQSSHPGAPHNV----NLQP 231
P GA + +AA A+ +A GA ++V NL P
Sbjct: 326 PNVHGALAPLAIPSAAAAAAAAGRIAIPGLAGAGNSVLLVSNLNP 370
>ref|NP_999396.1| polypyrimidine tract-binding protein [Sus scrofa]
gi|1122433|emb|CAA63597.1| polypyrimidine tract-binding
protein [Sus scrofa] gi|2500586|sp|Q29099|PTBP1_PIG
Polypyrimidine tract-binding protein 1 (PTB)
(Heterogeneous nuclear ribonucleoprotein I) (hnRNP I)
Length = 557
Score = 99.8 bits (247), Expect = 8e-20
Identities = 79/227 (34%), Positives = 114/227 (49%), Gaps = 26/227 (11%)
Query: 12 AAQPAIGPDGKRIETESNVLLASIENMQYAVTVDVINTVFSAFGTVQKIAMFEKNGQTQA 71
AA A G + +S VL +EN+ Y VT+DV++ +FS FGTV KI F KN Q QA
Sbjct: 165 AASAAAVDAGMAMAGQSPVLRIIVENLFYPVTLDVLHQIFSKFGTVLKIITFTKNNQFQA 224
Query: 72 LIQYPDVTTAAAAREALEGHCIYDGGYCKLHLSYSRHTDLNVKAFSDKSRDYTVPLVPAP 131
L+QY D +A A+ +L+G IY+ C L + +S+ T LNVK +DKSRDYT P +P+
Sbjct: 225 LLQYADPVSAQHAKLSLDGQNIYN-ACCTLRIDFSKLTSLNVKYNNDKSRDYTRPDLPS- 282
Query: 132 VWQNPQAAPMYPTNSPAYQTQVPGGSPAYQTQVP---GGQVPSWDLTQHAVRPGYVPVPG 188
+ T + A+ G+P + P G P++ + Q A V VP
Sbjct: 283 ---GDNQPSLDQTMAAAF------GAPGIMSASPYAGAGFPPTFAIPQAAT----VSVPN 329
Query: 189 AYPGQTGAFPTMPSYGSAAMPTASSPLAQSSHPGAPHNV----NLQP 231
+ GA + +AA A+ +A GA ++V NL P
Sbjct: 330 VH----GALAPLAIPSAAARAAAAGRIAIPGLAGAGNSVLLVSNLNP 372
>emb|CAF88128.1| unnamed protein product [Tetraodon nigroviridis]
Length = 217
Score = 98.6 bits (244), Expect = 2e-19
Identities = 54/103 (52%), Positives = 70/103 (67%), Gaps = 1/103 (0%)
Query: 27 ESNVLLASIENMQYAVTVDVINTVFSAFGTVQKIAMFEKNGQTQALIQYPDVTTAAAARE 86
+S VL +EN+ Y VT+DV++ +FS FGTV KI F KN Q QALIQY D TA A+
Sbjct: 19 QSPVLRVIVENLFYPVTLDVLHQIFSKFGTVLKIITFTKNNQFQALIQYADSMTAQHAKL 78
Query: 87 ALEGHCIYDGGYCKLHLSYSRHTDLNVKAFSDKSRDYTVPLVP 129
+L+G IY+ C L +S+S+ T LNVK +DKSRDYT P +P
Sbjct: 79 SLDGQNIYN-ACCTLRISFSKLTSLNVKYNNDKSRDYTRPDLP 120
>dbj|BAD92147.1| polypyrimidine tract-binding protein 1 isoform c variant [Homo
sapiens]
Length = 329
Score = 98.2 bits (243), Expect = 2e-19
Identities = 59/147 (40%), Positives = 85/147 (57%), Gaps = 5/147 (3%)
Query: 12 AAQPAIGPDGKRIETESNVLLASIENMQYAVTVDVINTVFSAFGTVQKIAMFEKNGQTQA 71
AA A G + +S VL +EN+ Y VT+DV++ +FS FGTV KI F KN Q QA
Sbjct: 182 AASAAAVDAGMAMAGQSPVLRIIVENLFYPVTLDVLHQIFSKFGTVLKIITFTKNNQFQA 241
Query: 72 LIQYPDVTTAAAAREALEGHCIYDGGYCKLHLSYSRHTDLNVKAFSDKSRDYTVPLVPAP 131
L+QY D +A A+ +L+G IY+ C L + +S+ T LNVK +DKSRDYT P +P+
Sbjct: 242 LLQYADPVSAQHAKLSLDGQNIYN-ACCTLRIDFSKLTSLNVKYNNDKSRDYTRPDLPS- 299
Query: 132 VWQNPQAAPMYPTNSPAYQTQVPGGSP 158
+ T + A+ ++P +P
Sbjct: 300 ---GDSQPSLDQTMAAAFGKRLPDAAP 323
>ref|NP_071961.1| polypyrimidine tract binding protein 1 isoform b [Rattus
norvegicus] gi|57002|emb|CAA43202.1| pyrimidine binding
protein 1 [Rattus norvegicus]
Length = 530
Score = 97.4 bits (241), Expect = 4e-19
Identities = 55/119 (46%), Positives = 75/119 (62%), Gaps = 1/119 (0%)
Query: 12 AAQPAIGPDGKRIETESNVLLASIENMQYAVTVDVINTVFSAFGTVQKIAMFEKNGQTQA 71
AA A G + +S VL +EN+ Y VT+DV++ +FS FGTV KI F KN Q QA
Sbjct: 164 AASAAAVDAGMAMAGQSPVLRIIVENLFYPVTLDVLHQIFSKFGTVLKIITFTKNNQFQA 223
Query: 72 LIQYPDVTTAAAAREALEGHCIYDGGYCKLHLSYSRHTDLNVKAFSDKSRDYTVPLVPA 130
L+QY D +A A+ +L+G IY+ C L + +S+ T LNVK +DKSRDYT P +P+
Sbjct: 224 LLQYADPVSAQHAKLSLDGQNIYN-ACCTLRIDFSKLTSLNVKYNNDKSRDYTRPDLPS 281
>emb|CAA43973.1| polypirimidine tract binding protein [Homo sapiens]
gi|35774|emb|CAA43056.1| polypyrimidine tract-binding
protein (pPTB) [Homo sapiens] gi|13325140|gb|AAH04383.1|
Polypyrimidine tract-binding protein 1, isoform c [Homo
sapiens] gi|131528|sp|P26599|PTBP1_HUMAN Polypyrimidine
tract-binding protein 1 (PTB) (Heterogeneous nuclear
ribonucleoprotein I) (hnRNP I) (57 kDa RNA-binding
protein PPTB-1) gi|4096061|gb|AAC99798.1| PTB_HUMAN;
PTB; HETEROGENEOUS NUCLEA; HNRNP I; 57 KD RNA-BINDING
PROTEIN PPTB-1 [Homo sapiens]
gi|14165466|ref|NP_114368.1| polypyrimidine
tract-binding protein 1 isoform c [Homo sapiens]
Length = 531
Score = 97.4 bits (241), Expect = 4e-19
Identities = 55/119 (46%), Positives = 75/119 (62%), Gaps = 1/119 (0%)
Query: 12 AAQPAIGPDGKRIETESNVLLASIENMQYAVTVDVINTVFSAFGTVQKIAMFEKNGQTQA 71
AA A G + +S VL +EN+ Y VT+DV++ +FS FGTV KI F KN Q QA
Sbjct: 165 AASAAAVDAGMAMAGQSPVLRIIVENLFYPVTLDVLHQIFSKFGTVLKIITFTKNNQFQA 224
Query: 72 LIQYPDVTTAAAAREALEGHCIYDGGYCKLHLSYSRHTDLNVKAFSDKSRDYTVPLVPA 130
L+QY D +A A+ +L+G IY+ C L + +S+ T LNVK +DKSRDYT P +P+
Sbjct: 225 LLQYADPVSAQHAKLSLDGQNIYN-ACCTLRIDFSKLTSLNVKYNNDKSRDYTRPDLPS 282
>pir||A41718 polypyrimidine tract-binding protein PTB-1 - mouse
Length = 528
Score = 97.4 bits (241), Expect = 4e-19
Identities = 55/119 (46%), Positives = 75/119 (62%), Gaps = 1/119 (0%)
Query: 12 AAQPAIGPDGKRIETESNVLLASIENMQYAVTVDVINTVFSAFGTVQKIAMFEKNGQTQA 71
AA A G + +S VL +EN+ Y VT+DV++ +FS FGTV KI F KN Q QA
Sbjct: 164 AASAAAVDAGMAMAGQSPVLRIIVENLFYPVTLDVLHQIFSKFGTVLKIITFTKNNQFQA 223
Query: 72 LIQYPDVTTAAAAREALEGHCIYDGGYCKLHLSYSRHTDLNVKAFSDKSRDYTVPLVPA 130
L+QY D +A A+ +L+G IY+ C L + +S+ T LNVK +DKSRDYT P +P+
Sbjct: 224 LLQYADPVSAQHAKLSLDGQNIYN-ACCTLRIDFSKLTSLNVKYNNDKSRDYTRPDLPS 281
>gb|AAP35465.1| polypyrimidine tract binding protein 1 [Homo sapiens]
gi|32354|emb|CAA47386.1| nuclear ribonucleoprotein [Homo
sapiens] gi|35772|emb|CAA46444.1| polypirimidine tract
binding protein [Homo sapiens]
gi|61359442|gb|AAX41719.1| polypyrimidine tract binding
protein 1 [synthetic construct]
gi|61359435|gb|AAX41718.1| polypyrimidine tract binding
protein 1 [synthetic construct]
gi|12803183|gb|AAH02397.1| Polypyrimidine tract-binding
protein 1, isoform a [Homo sapiens]
gi|15489171|gb|AAH13694.1| Polypyrimidine tract-binding
protein 1, isoform a [Homo sapiens]
gi|4506243|ref|NP_002810.1| polypyrimidine tract-binding
protein 1 isoform a [Homo sapiens] gi|107220|pir||S26294
polypyrimidine tract-binding protein PTB-1 [validated] -
human
Length = 557
Score = 97.4 bits (241), Expect = 4e-19
Identities = 55/119 (46%), Positives = 75/119 (62%), Gaps = 1/119 (0%)
Query: 12 AAQPAIGPDGKRIETESNVLLASIENMQYAVTVDVINTVFSAFGTVQKIAMFEKNGQTQA 71
AA A G + +S VL +EN+ Y VT+DV++ +FS FGTV KI F KN Q QA
Sbjct: 165 AASAAAVDAGMAMAGQSPVLRIIVENLFYPVTLDVLHQIFSKFGTVLKIITFTKNNQFQA 224
Query: 72 LIQYPDVTTAAAAREALEGHCIYDGGYCKLHLSYSRHTDLNVKAFSDKSRDYTVPLVPA 130
L+QY D +A A+ +L+G IY+ C L + +S+ T LNVK +DKSRDYT P +P+
Sbjct: 225 LLQYADPVSAQHAKLSLDGQNIYN-ACCTLRIDFSKLTSLNVKYNNDKSRDYTRPDLPS 282
>ref|NP_114367.1| polypyrimidine tract-binding protein 1 isoform b [Homo sapiens]
gi|35770|emb|CAA46443.1| polypirimidine tract binding
protein [Homo sapiens]
Length = 550
Score = 97.4 bits (241), Expect = 4e-19
Identities = 55/119 (46%), Positives = 75/119 (62%), Gaps = 1/119 (0%)
Query: 12 AAQPAIGPDGKRIETESNVLLASIENMQYAVTVDVINTVFSAFGTVQKIAMFEKNGQTQA 71
AA A G + +S VL +EN+ Y VT+DV++ +FS FGTV KI F KN Q QA
Sbjct: 165 AASAAAVDAGMAMAGQSPVLRIIVENLFYPVTLDVLHQIFSKFGTVLKIITFTKNNQFQA 224
Query: 72 LIQYPDVTTAAAAREALEGHCIYDGGYCKLHLSYSRHTDLNVKAFSDKSRDYTVPLVPA 130
L+QY D +A A+ +L+G IY+ C L + +S+ T LNVK +DKSRDYT P +P+
Sbjct: 225 LLQYADPVSAQHAKLSLDGQNIYN-ACCTLRIDFSKLTSLNVKYNNDKSRDYTRPDLPS 282
Database: nr
Posted date: Jul 5, 2005 12:34 AM
Number of letters in database: 863,360,394
Number of sequences in database: 2,540,612
Lambda K H
0.312 0.130 0.396
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 567,293,718
Number of Sequences: 2540612
Number of extensions: 28103171
Number of successful extensions: 100678
Number of sequences better than 10.0: 3344
Number of HSP's better than 10.0 without gapping: 228
Number of HSP's successfully gapped in prelim test: 3236
Number of HSP's that attempted gapping in prelim test: 91160
Number of HSP's gapped (non-prelim): 9291
length of query: 277
length of database: 863,360,394
effective HSP length: 126
effective length of query: 151
effective length of database: 543,243,282
effective search space: 82029735582
effective search space used: 82029735582
T: 11
A: 40
X1: 16 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (21.9 bits)
S2: 74 (33.1 bits)
Medicago: description of AC140025.4


