

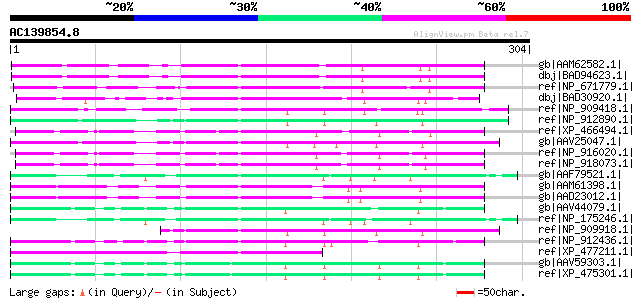
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC139854.8 - phase: 0
(304 letters)
Database: nr
2,540,612 sequences; 863,360,394 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
gb|AAM62582.1| unknown [Arabidopsis thaliana] 86 1e-15
dbj|BAD94623.1| hypothetical protein [Arabidopsis thaliana] gi|1... 86 1e-15
ref|NP_671779.1| expressed protein [Arabidopsis thaliana] 86 1e-15
dbj|BAD30920.1| serine/threonine phosphatase PP7-related-like [O... 79 1e-13
ref|NP_909418.1| P0701D05.29 [Oryza sativa (japonica cultivar-gr... 79 2e-13
ref|NP_912890.1| unnamed protein product [Oryza sativa (japonica... 78 4e-13
ref|XP_466494.1| calcineurin-like phosphoesterase-like protein [... 75 3e-12
gb|AAV25047.1| putative polyprotein [Oryza sativa (japonica cult... 75 3e-12
ref|NP_916020.1| OSJNBb0021A09.19 [Oryza sativa (japonica cultiv... 72 2e-11
ref|NP_918073.1| P0702H08.15 [Oryza sativa (japonica cultivar-gr... 72 2e-11
gb|AAF79521.1| F21D18.16 [Arabidopsis thaliana] gi|25405918|pir|... 72 2e-11
gb|AAM61398.1| unknown [Arabidopsis thaliana] 72 2e-11
gb|AAD23012.1| expressed protein [Arabidopsis thaliana] gi|18400... 72 3e-11
gb|AAV44079.1| putative polyprotein [Oryza sativa (japonica cult... 68 4e-10
ref|NP_175246.1| calcineurin-like phosphoesterase family protein... 65 2e-09
ref|NP_909918.1| putative mutator-like transposase [Oryza sativa... 63 9e-09
ref|NP_912436.1| Putative transposable element [Oryza sativa (ja... 63 9e-09
ref|XP_477211.1| calcineurin-like phosphoesterase-like [Oryza sa... 61 5e-08
gb|AAV59303.1| unknown protein [Oryza sativa (japonica cultivar-... 60 8e-08
ref|XP_475301.1| unknown protein [Oryza sativa (japonica cultiva... 60 8e-08
>gb|AAM62582.1| unknown [Arabidopsis thaliana]
Length = 478
Score = 85.9 bits (211), Expect = 1e-15
Identities = 73/284 (25%), Positives = 128/284 (44%), Gaps = 33/284 (11%)
Query: 2 PMGEMTVTLDDVACLTHLPIEGRMLDHGKKMPKHEGAAQLMTYLGVSQHEAEKICNQKYG 61
P GEMT+TLD+V+ + L ++G+ + K+ K E +Q+ L + E N+
Sbjct: 84 PCGEMTITLDEVSLILGLAVDGKPVVGVKE--KDEDPSQVCLKLLGKLPKGELSGNR--- 138
Query: 62 GYISYPRLRDFYTSYLGRANVLAGTEDPEELEELARVRTYCVRCYLLYLVGCLLFGDRSN 121
++ L++ + A + +E+E Y R YL+Y+VG +F
Sbjct: 139 --VTAKWLKESFAECPKGATM-------KEIE-------YHTRAYLIYIVGSTIFATTDP 182
Query: 122 KRIELIYLTTMADGYAGMRNYSWGGMTLAYLYGELADACRPGHRALGGSVTLLTTWFLAH 181
+I + YL D + Y+WG LA+LY ++ +A + +GG +TLL W H
Sbjct: 183 SKISVDYLILFED-FEKAGEYAWGAAALAFLYRQIGNASQRSQSIIGGCLTLLQCWSYFH 241
Query: 182 FPGFFSVDLNTDYLENYPVAARWK---LPKGHGEGITYWSLLDRIQLDDVCYKPYKEHRE 238
++D +P+A WK + + Y LD + +V + P++ +
Sbjct: 242 ----LNIDRPKRTTRQFPLALLWKGRQQSRSKNDLFKYRKALDDLDPSNVSWCPFEGDLD 297
Query: 239 I--QDFEE--VFWYSGWIMCGVRRVYRHLPKRVLRQYGYVQTIP 278
I Q F++ + S + G + V H P R ++Q+G Q IP
Sbjct: 298 IVPQSFKDNLLLGRSRTKLIGPKVVEWHFPDRCMKQFGLCQVIP 341
>dbj|BAD94623.1| hypothetical protein [Arabidopsis thaliana]
gi|18394553|ref|NP_564042.1| expressed protein
[Arabidopsis thaliana] gi|25371349|pir||E86314 F2H15.15
protein - Arabidopsis thaliana gi|9665070|gb|AAF97272.1|
Contains similarity to a hypothetical protein At2g25010
gi|4559351 from Arabidopsis thaliana chromosome II
gb|AC006585
Length = 478
Score = 85.9 bits (211), Expect = 1e-15
Identities = 73/284 (25%), Positives = 128/284 (44%), Gaps = 33/284 (11%)
Query: 2 PMGEMTVTLDDVACLTHLPIEGRMLDHGKKMPKHEGAAQLMTYLGVSQHEAEKICNQKYG 61
P GEMT+TLD+V+ + L ++G+ + K+ K E +Q+ L + E N+
Sbjct: 84 PCGEMTITLDEVSLILGLAVDGKPVVGVKE--KDEDPSQVCLRLLGKLPKGELSGNR--- 138
Query: 62 GYISYPRLRDFYTSYLGRANVLAGTEDPEELEELARVRTYCVRCYLLYLVGCLLFGDRSN 121
++ L++ + A + +E+E Y R YL+Y+VG +F
Sbjct: 139 --VTAKWLKESFAECPKGATM-------KEIE-------YHTRAYLIYIVGSTIFATTDP 182
Query: 122 KRIELIYLTTMADGYAGMRNYSWGGMTLAYLYGELADACRPGHRALGGSVTLLTTWFLAH 181
+I + YL D + Y+WG LA+LY ++ +A + +GG +TLL W H
Sbjct: 183 SKISVDYLILFED-FEKAGEYAWGAAALAFLYRQIGNASQRSQSIIGGCLTLLQCWSYFH 241
Query: 182 FPGFFSVDLNTDYLENYPVAARWK---LPKGHGEGITYWSLLDRIQLDDVCYKPYKEHRE 238
++D +P+A WK + + Y LD + +V + P++ +
Sbjct: 242 ----LNIDRPKRTTRQFPLALLWKGRQQSRSKNDLFKYRKALDDLDPSNVSWCPFEGDLD 297
Query: 239 I--QDFEE--VFWYSGWIMCGVRRVYRHLPKRVLRQYGYVQTIP 278
I Q F++ + S + G + V H P R ++Q+G Q IP
Sbjct: 298 IVPQSFKDNLLLGRSRTKLIGPKVVEWHFPDRCMKQFGLCQVIP 341
>ref|NP_671779.1| expressed protein [Arabidopsis thaliana]
Length = 667
Score = 85.9 bits (211), Expect = 1e-15
Identities = 80/287 (27%), Positives = 125/287 (42%), Gaps = 36/287 (12%)
Query: 3 MGEMTVTLDDVACLTHLPIEGRMLDHGKKMPKHEGAAQLMTYLGVSQHEAEKICNQKYGG 62
+GEMTVTL+D+A L L I+G+ + + +A YLG S N GG
Sbjct: 96 VGEMTVTLEDIALLLGLGIDGKPVIG---LTYTTCSAVCERYLGKSPAS-----NSASGG 147
Query: 63 YISYPRLRDFYTSYLGRANVLAGTEDPEELEELARVRTYCVRCYLLYLVGCLLFGDRSNK 122
+ L+D ++ D EE+ R RT R YLLYLVG +F +
Sbjct: 148 MVKLSWLKDNFSE----------CPDDASFEEVER-RT---RAYLLYLVGSTIFSTTTGN 193
Query: 123 RIELIYLTTMADGYAGMRNYSWGGMTLAYLYGELADACRPGHRALGGSVTLLTTWFLAHF 182
++ ++YL D + ++WG LA+LY L +A + G +TLL W H
Sbjct: 194 KVPVMYLPLFED-FDDAGTFAWGAAALAFLYRALGNASVKSQSTICGCLTLLQCWSYYHL 252
Query: 183 PGFFSVDLNTDYL-ENYPVAARWK----LPKGHGEGITYWSLLDRIQLDDVCYKPYKEHR 237
LN + + + +P +WK P + + + Y LD ++ DV + PY E+
Sbjct: 253 -NIGRPKLNREPIHDQFPFVLKWKGKQNGPTANRDVVFYRKALDVMKPTDVVWLPY-ENM 310
Query: 238 EIQDFEE------VFWYSGWIMCGVRRVYRHLPKRVLRQYGYVQTIP 278
D + + S ++ + RHLP R +Q+ Q IP
Sbjct: 311 NGGDMSDRMRKSLLLGRSKTMLISFDKAERHLPDRCRKQFDLFQDIP 357
>dbj|BAD30920.1| serine/threonine phosphatase PP7-related-like [Oryza sativa
(japonica cultivar-group)]
Length = 619
Score = 79.3 bits (194), Expect = 1e-13
Identities = 83/285 (29%), Positives = 126/285 (44%), Gaps = 41/285 (14%)
Query: 5 EMTVTLDDVACLTHLPIEGRMLDHGKKMPKHEGAAQLMT--YLGVSQHEAEKICNQKYGG 62
EM +L DV+ + +P+ G ++ P + A + M +LG S E++C G
Sbjct: 96 EMAPSLRDVSYILGIPVTGHVVT---AEPIGDEAVRRMCLHFLGESPGNGEQLC-----G 147
Query: 63 YISYPRLRDFYTSYLGRANVLAGTEDPEELEELARVRTYCVRCYLLYLVGCLLFGDRSNK 122
I RL Y + E+P + E+A Y R YLLYLVG LF D
Sbjct: 148 LI---RLTWLYRKFHQLP------ENPT-INEIA----YSTRAYLLYLVGSTLFPDTMRG 193
Query: 123 RIELIYLTTMADGYAGMRNYSWGGMTLAYLYGELADACRPGHRA-LGGSVTLLTTWFLAH 181
+ YL +AD + +R Y+WG LA+LY L+ A P GS TLL W +
Sbjct: 194 FVSPRYLPLLAD-FRKIREYAWGSAALAHLYRGLSVAVTPNATTQFLGSATLLMAWIYEY 252
Query: 182 FPGFFSVDLNTDYLENYPVAARWKL---PKGHGEGITYW-SLLDRIQLDDVCYKPYKEHR 237
P N + L P A RW +G + + W + D +Q DV + PYK+
Sbjct: 253 LPLTQPQQKNQNTL--LPRACRWNFGGATRGQRKKVMEWRKVFDELQFSDVNWNPYKDMN 310
Query: 238 E--IQDF----EEVFWYSGWIMC-GVRRVYRHLPKRVLRQYGYVQ 275
I ++ + + + W++ ++ VY +P R RQ+G Q
Sbjct: 311 PAIIPEYCIAADNICYSRTWLISFNIKEVY--VPDRFSRQFGREQ 353
>ref|NP_909418.1| P0701D05.29 [Oryza sativa (japonica cultivar-group)]
gi|13486671|dbj|BAB39908.1| hypothetical protein~similar
to Oryza sativa chromosome 1, P0499C11.11 [Oryza sativa
(japonica cultivar-group)]
Length = 792
Score = 79.0 bits (193), Expect = 2e-13
Identities = 90/318 (28%), Positives = 130/318 (40%), Gaps = 55/318 (17%)
Query: 1 MPMGEMTVTLDDVACLTHLPIEGRMLDHGKKMPKHEGAAQLMTYLGVSQHEAEKICNQKY 60
MP GE+T+TL DVA + LPI G + P++E + YLG +A +
Sbjct: 278 MPCGEITITLQDVAMILGLPIAGHAVTVNPTEPQNELVER---YLG----KAPPPDRPRP 330
Query: 61 GGYISYPRLRDFYTSYLGRANVLAGTEDPEELEELARVRTYCVRCYLLYLVGCLLFGDRS 120
G +S+ R N D E +++ AR Y+L L+ LLF D S
Sbjct: 331 GLRVSWVR---------AEFNNCPEDADEETIKQHARA-------YILSLISGLLFPDAS 374
Query: 121 NKRIELIYLTTMADGYAGMRNYSWGGMTLAYLYGELADACR--PGHRALGGSVTLLTTWF 178
+AD + +YSWG TLAYLY + DACR L G + LL W
Sbjct: 375 GDLYTFYPFPLIAD-LENIGSYSWGSATLAYLYRAMCDACRRQSDGSNLTGCLLLLQFWS 433
Query: 179 LAHFP---------GFFSVDLNTDYLENYPVAARWKL--------PKGHGEGITYWSLLD 221
HFP + +V+ D + V RW + P E T + D
Sbjct: 434 WEHFPIGRPDLVKLKYPNVEELEDERDRPTVGLRWVVGVCARRAAPARCYEHFT--NEFD 491
Query: 222 RIQLDDVCYKPYKEHR--EIQ-----DFEEVFWYSGWIMCGVRRVYRHLPKRVLRQYGYV 274
+ D V + PY+E R E+Q + W + + V ++P+RV+RQ+G
Sbjct: 492 LLTDDQVVWCPYREERMKELQLAPICTQDSHLWLTRAPLLYFFMVEIYMPERVMRQFGLH 551
Query: 275 QTIPRHPTDVRDLPTAFH 292
Q P +RD H
Sbjct: 552 QVC---PPPLRDTSAELH 566
>ref|NP_912890.1| unnamed protein product [Oryza sativa (japonica cultivar-group)]
Length = 1230
Score = 77.8 bits (190), Expect = 4e-13
Identities = 84/313 (26%), Positives = 125/313 (39%), Gaps = 38/313 (12%)
Query: 1 MPMGEMTVTLDDVACLTHLPIEGRMLDHGKKMPKHEGAAQLMTYLGVSQHEAEKICNQKY 60
+P GE+TVTL+DVA + LPI G+ + + YLG+ A Q
Sbjct: 592 LPCGELTVTLEDVAMILGLPIRGQAVTGDTASGNWRERVE--EYLGLEPPVAPDGQRQTK 649
Query: 61 GGYISYPRLRDFYTSYLGRANVLAGTEDPEELEELARVRTYCVRCYLLYLVGCLLFGDRS 120
+ LR + + P E +E A V+ YC R Y+LY+ G +LF D
Sbjct: 650 TSGVPLSWLRANFG------------QCPAEADE-ATVQRYC-RAYVLYIFGSILFPDSG 695
Query: 121 NKRIELIYLTTMADGYAGMRNYSWGGMTLAYLYGELADACR--PGHRALGGSVTLLTTWF 178
++L +AD + +SWG LA+LY +L DACR G L G V LL W
Sbjct: 696 GDMASWMWLPLLAD-WDEAGTFSWGSAALAWLYRQLCDACRRQGGDANLAGCVWLLQVWM 754
Query: 179 LAHFP------GFFSVDLNTDYLENYPVAARWKLPKGHGEG------ITYWSLLDRIQLD 226
P + D VA W+ G Y + +D +Q +
Sbjct: 755 WMRLPVGRPMWRTHQAWPHQDADRRPTVAHLWESVPSPVVGRRNLAYYHYTNEMDYLQPE 814
Query: 227 DVCYKPYKEHREIQ-------DFEEVFWYSGWIMCGVRRVYRHLPKRVLRQYGYVQTIPR 279
V + PY+ ++ E+ + V + RV+RQ+G +QTIP
Sbjct: 815 HVVWMPYQAQEVLELELNPMCHIEDALKTLRCPLICFYAVEFQMCHRVMRQFGRLQTIPH 874
Query: 280 HPTDVRDLPTAFH 292
+ DL F+
Sbjct: 875 RFSTSIDLHNHFN 887
>ref|XP_466494.1| calcineurin-like phosphoesterase-like protein [Oryza sativa
(japonica cultivar-group)] gi|50726478|dbj|BAD34087.1|
calcineurin-like phosphoesterase-like protein [Oryza
sativa (japonica cultivar-group)]
Length = 766
Score = 74.7 bits (182), Expect = 3e-12
Identities = 88/296 (29%), Positives = 129/296 (42%), Gaps = 48/296 (16%)
Query: 4 GEMTVTLDDVACLTHLPIEGRMLDHGKKMPKHEGAAQLMTYLGVSQHEAEKICNQKYGGY 63
GEMTVTL DV+ L LPI+GR + H+ A ++ LG H+ + + +
Sbjct: 153 GEMTVTLQDVSMLLALPIDGRPV---CSTTDHDYAQMVIDCLG---HD-PRGPSMPGKSF 205
Query: 64 ISYPRLRDFYTSYLGRANVLAGTEDPEELEELARVRTYCVRCYLLYLVGCLLFGDRSNKR 123
+ Y L+ + E PE ++ R VR Y+L L+ +LF D + R
Sbjct: 206 LHYKWLKKHF------------YELPEGTDDQTVERH--VRAYILSLLCGVLFPDGTG-R 250
Query: 124 IELIYLTTMADGYAGMRNYSWGGMTLAYLYGELAD-ACRPGHRALGGSVTLLTTWF---- 178
+ LIYL +AD + + YSWG LA+LY L A + +GGS+ LL W
Sbjct: 251 MSLIYLPLIAD-LSLVGTYSWGSAVLAFLYRSLCSVASSHNIKNIGGSLLLLQLWSWERS 309
Query: 179 -----LAHFPGFFSVDLNTDYLENYPVAARWKLPKGHGEGIT-----YWSLLDRIQLDDV 228
L P D+ D L PV RW + E T Y L+ + D V
Sbjct: 310 HVGRPLVRSPVCPETDIPQDVL---PVGFRWVGARTQSENATRCLKQYRDELNLQRADQV 366
Query: 229 CYKPYKEHREIQDFEEV------FWYSGWIMCGVRRVYRHLPKRVLRQYGYVQTIP 278
++PY H E + W + + V +LP+RV+RQ+G Q+IP
Sbjct: 367 KWEPYL-HIESASLPLLCTKNADLWLTQAPLINFPIVEMYLPERVMRQFGLRQSIP 421
>gb|AAV25047.1| putative polyprotein [Oryza sativa (japonica cultivar-group)]
Length = 1754
Score = 74.7 bits (182), Expect = 3e-12
Identities = 84/308 (27%), Positives = 126/308 (40%), Gaps = 38/308 (12%)
Query: 1 MPMGEMTVTLDDVACLTHLPIEGRMLDHGKKMPKHEGAAQLMTYLGVSQHEAEKICNQKY 60
+P GE+TVTL+DVA + LPI G+ + + YLG+ A Q
Sbjct: 974 LPCGELTVTLEDVAMILGLPIRGQAVTGDTASGNWRERVE--EYLGLEPPVAPDGQRQTK 1031
Query: 61 GGYISYPRLRDFYTSYLGRANVLAGTEDPEELEELARVRTYCVRCYLLYLVGCLLFGDRS 120
+ LR + + P E +E A V+ YC R Y+LY+ G +LF D
Sbjct: 1032 TSGVPLSWLRANFG------------QCPAEADE-ATVQRYC-RAYVLYIFGSILFPDSG 1077
Query: 121 NKRIELIYLTTMADGYAGMRNYSWGGMTLAYLYGELADACR--PGHRALGGSVTLLTTW- 177
++L +AD + +SWG LA+LY +L DACR G L G V LL W
Sbjct: 1078 GDMASWMWLPLLAD-WDEAGTFSWGSAALAWLYRQLCDACRRQGGDANLAGCVWLLQVWM 1136
Query: 178 ---FLAHFPGFFSVDL--NTDYLENYPVAARWKLPKGHGEG------ITYWSLLDRIQLD 226
L P + + + D VA W+ G Y + +D +Q +
Sbjct: 1137 WMRLLVGRPMWRTHQAWPHQDADRRPTVAHLWESVPSPVVGRRNLAYCHYTNEMDYLQPE 1196
Query: 227 DVCYKPYKEHREIQ-------DFEEVFWYSGWIMCGVRRVYRHLPKRVLRQYGYVQTIPR 279
V + PY+ ++ E+ + V + RV+RQ+G +QTIP
Sbjct: 1197 HVVWMPYQAQEVLELELNPMCHIEDALKTLRCPLICFYAVEFQMCHRVMRQFGRLQTIPH 1256
Query: 280 HPTDVRDL 287
+ DL
Sbjct: 1257 RFSTSIDL 1264
>ref|NP_916020.1| OSJNBb0021A09.19 [Oryza sativa (japonica cultivar-group)]
Length = 761
Score = 72.4 bits (176), Expect = 2e-11
Identities = 87/296 (29%), Positives = 127/296 (42%), Gaps = 48/296 (16%)
Query: 4 GEMTVTLDDVACLTHLPIEGRMLDHGKKMPKHEGAAQLMTYLGVSQHEAEKICNQKYGGY 63
GEM VTL DVA L LPI+GR + H+ A ++ LG H+ + + +
Sbjct: 170 GEMAVTLQDVAMLLALPIDGRPV---CSTTDHDYAQMVIDCLG---HD-PRGPSMPGKSF 222
Query: 64 ISYPRLRDFYTSYLGRANVLAGTEDPEELEELARVRTYCVRCYLLYLVGCLLFGDRSNKR 123
+ Y L+ + E PE ++ R VR Y+L L+ +LF D + R
Sbjct: 223 LHYKWLKKHF------------YELPEGADDQTVERH--VRAYILSLLCGVLFPDGTG-R 267
Query: 124 IELIYLTTMADGYAGMRNYSWGGMTLAYLYGELAD-ACRPGHRALGGSVTLLTTWF---- 178
+ LIYL +AD + + YSWG LA+LY L A + +GGS+ LL W
Sbjct: 268 MSLIYLPLIAD-LSRVGTYSWGSAVLAFLYRSLCSVASSHNIKNIGGSLLLLQLWSWERS 326
Query: 179 -----LAHFPGFFSVDLNTDYLENYPVAARWKLPKGHGEGIT-----YWSLLDRIQLDDV 228
L P D+ D PV RW + E T Y L+ + D V
Sbjct: 327 HVGRPLVRSPLCPETDIPQDL---PPVGFRWVGARTQSENATRCLKQYRDELNLQRADQV 383
Query: 229 CYKPYKEHREIQDF------EEVFWYSGWIMCGVRRVYRHLPKRVLRQYGYVQTIP 278
++PY H E + W + + V +LP+RV+RQ+G Q+IP
Sbjct: 384 KWEPYL-HIESSSLPLLCTKDADLWLTQAPLINFPIVEMYLPERVMRQFGLRQSIP 438
>ref|NP_918073.1| P0702H08.15 [Oryza sativa (japonica cultivar-group)]
Length = 718
Score = 72.4 bits (176), Expect = 2e-11
Identities = 87/296 (29%), Positives = 127/296 (42%), Gaps = 48/296 (16%)
Query: 4 GEMTVTLDDVACLTHLPIEGRMLDHGKKMPKHEGAAQLMTYLGVSQHEAEKICNQKYGGY 63
GEM VTL DVA L LPI+GR + H+ A ++ LG H+ + + +
Sbjct: 184 GEMAVTLQDVAMLLALPIDGRPV---CSTTDHDYAQMVIDCLG---HD-PRGPSMPGKSF 236
Query: 64 ISYPRLRDFYTSYLGRANVLAGTEDPEELEELARVRTYCVRCYLLYLVGCLLFGDRSNKR 123
+ Y L+ + E PE ++ R VR Y+L L+ +LF D + R
Sbjct: 237 LHYKWLKKHF------------YELPEGADDQTVERH--VRAYILSLLCGVLFPDGTG-R 281
Query: 124 IELIYLTTMADGYAGMRNYSWGGMTLAYLYGELAD-ACRPGHRALGGSVTLLTTWF---- 178
+ LIYL +AD + + YSWG LA+LY L A + +GGS+ LL W
Sbjct: 282 MSLIYLPLIAD-LSRVGTYSWGSAVLAFLYRSLCSVASSHNIKNIGGSLLLLQLWSWERS 340
Query: 179 -----LAHFPGFFSVDLNTDYLENYPVAARWKLPKGHGEGIT-----YWSLLDRIQLDDV 228
L P D+ D PV RW + E T Y L+ + D V
Sbjct: 341 HVGRPLVRSPLCPETDIPQDV---PPVGFRWVGARTQSENATRCLKQYRDELNLQRADQV 397
Query: 229 CYKPYKEHREIQDF------EEVFWYSGWIMCGVRRVYRHLPKRVLRQYGYVQTIP 278
++PY H E + W + + V +LP+RV+RQ+G Q+IP
Sbjct: 398 KWEPYL-HIESSSLPLLCTKDADLWLTQAPLINFPIVEMYLPERVMRQFGLRQSIP 452
>gb|AAF79521.1| F21D18.16 [Arabidopsis thaliana] gi|25405918|pir||D96521 protein
F21D18.16 [imported] - Arabidopsis thaliana
Length = 1340
Score = 72.4 bits (176), Expect = 2e-11
Identities = 86/322 (26%), Positives = 130/322 (39%), Gaps = 58/322 (18%)
Query: 1 MPMGEMTVTLDDVACLTHLPIEGRMLDHGKKMPKHEGAAQLMTYLGVSQHEAEKICNQKY 60
+P GE+TVTL DV L L ++G + G +++ +C
Sbjct: 101 LPAGEITVTLQDVNILLGLRVDGPAVT------------------GSTKYNWADLCEDLL 142
Query: 61 GGYISYPRLRDFYTSYLG----RANVLAGTEDPEELEELARVRTYC-VRCYLLYLVGCLL 115
G P +D + S++ R N DP+E V C R ++L L+ L
Sbjct: 143 G---HRPGPKDLHGSHVSLAWLRENFRNLPADPDE------VTLKCHTRAFVLALMSGFL 193
Query: 116 FGDRSNKRIELIYLTTMADGYAGMRNYSWGGMTLAYLYGELADACRPGHRALGGSVTLLT 175
+GD+S + L +L + D + + SWG TLA LY EL A + + G + LL
Sbjct: 194 YGDKSKHDVALTFLPLLRD-FDEVAKLSWGSATLALLYRELCRASKRTVSTICGPLVLLQ 252
Query: 176 TWFLAHF----PGFFSVDLNTDYLENY------PVAARWKLPKGHGE----GITYW-SLL 220
W PG D+ Y++ P+ RW+ H E G+ ++
Sbjct: 253 LWAWERLHVGRPGRLK-DVGASYMDGIDGPLPDPLGCRWRASLSHKENPRGGLDFYRDQF 311
Query: 221 DRIQLDDVCYKPY-----KEHREIQDFEEVFWYSGWIMCGVRRVYRHLPKRVLRQYGYVQ 275
D+ + + V ++PY + I E W + + V H P RVLRQ+G Q
Sbjct: 312 DQQKDEQVIWQPYTPDLLAKIPLICVSGENIWRTVAPLICFDVVEWHRPDRVLRQFGLHQ 371
Query: 276 TIPRHPTDVRDLPTAFHCADVR 297
TIP P D A H D R
Sbjct: 372 TIPA-PCDNE---KALHAIDKR 389
>gb|AAM61398.1| unknown [Arabidopsis thaliana]
Length = 509
Score = 72.0 bits (175), Expect = 2e-11
Identities = 73/287 (25%), Positives = 117/287 (40%), Gaps = 35/287 (12%)
Query: 1 MPMGEMTVTLDDVACLTHLPIEGRMLDHGKKMPKHEGAAQLMTYLGVSQHEAEKICNQKY 60
+P+GEMT+TLD+VA + L I+G + G K+ LG A K N
Sbjct: 92 LPLGEMTITLDEVALVLGLEIDGDPIV-GSKVDDEVAMDMCGRLLGKLPSAANKEVN--- 147
Query: 61 GGYISYPRLRDFYTSYLGRANVLAGTEDPEELE-ELARVRTYCVRCYLLYLVGCLLFGDR 119
S +L ++ +E PE+ ++ + T R YLLYL+G +F
Sbjct: 148 ---CSRVKLNWLKRTF---------SECPEDASFDVVKCHT---RAYLLYLIGSTIFATT 192
Query: 120 SNKRIELIYLTTMADGYAGMRNYSWGGMTLAYLYGELADACRPGHRALGGSVTLLTTWFL 179
++ + YL D + Y+WG LA LY L +A + G +TLL W
Sbjct: 193 DGDKVSVKYLPLFED-FDQAGRYAWGAAALACLYRALGNASLKSQSNICGCLTLLQCW-- 249
Query: 180 AHFPGFFSVDLNTDYLEN--YPVAARW--KLPKGHGEGITYWSLLDRIQLDDVCYKPYKE 235
+F +D+ +P+A W K + + Y LD + + + PY+
Sbjct: 250 ----SYFHLDIGRPEKSEACFPLALLWKGKGSRSKTDLSEYRRELDDLDPSKITWCPYER 305
Query: 236 HREI----QDFEEVFWYSGWIMCGVRRVYRHLPKRVLRQYGYVQTIP 278
+ + + S + ++ H P R LRQ+G Q IP
Sbjct: 306 FENLIPPHIKAKLILGRSKTTLVCFEKIELHFPDRCLRQFGKRQPIP 352
>gb|AAD23012.1| expressed protein [Arabidopsis thaliana]
gi|18400697|ref|NP_565582.1| expressed protein
[Arabidopsis thaliana] gi|25371347|pir||B84643
hypothetical protein At2g25010 [imported] - Arabidopsis
thaliana
Length = 509
Score = 71.6 bits (174), Expect = 3e-11
Identities = 73/287 (25%), Positives = 117/287 (40%), Gaps = 35/287 (12%)
Query: 1 MPMGEMTVTLDDVACLTHLPIEGRMLDHGKKMPKHEGAAQLMTYLGVSQHEAEKICNQKY 60
+P+GEMT+TLD+VA + L I+G + G K+ LG A K N
Sbjct: 92 LPLGEMTITLDEVALVLGLEIDGDPIV-GSKVGDEVAMDMCGRLLGKLPSAANKEVN--- 147
Query: 61 GGYISYPRLRDFYTSYLGRANVLAGTEDPEELE-ELARVRTYCVRCYLLYLVGCLLFGDR 119
S +L ++ +E PE+ ++ + T R YLLYL+G +F
Sbjct: 148 ---CSRVKLNWLKRTF---------SECPEDASFDVVKCHT---RAYLLYLIGSTIFATT 192
Query: 120 SNKRIELIYLTTMADGYAGMRNYSWGGMTLAYLYGELADACRPGHRALGGSVTLLTTWFL 179
++ + YL D + Y+WG LA LY L +A + G +TLL W
Sbjct: 193 DGDKVSVKYLPLFED-FDQAGRYAWGAAALACLYRALGNASLKSQSNICGCLTLLQCW-- 249
Query: 180 AHFPGFFSVDLNTDYLEN--YPVAARW--KLPKGHGEGITYWSLLDRIQLDDVCYKPYKE 235
+F +D+ +P+A W K + + Y LD + + + PY+
Sbjct: 250 ----SYFHLDIGRPEKSEACFPLALLWKGKGSRSKTDLSEYRRELDDLDPSKITWCPYER 305
Query: 236 HREI----QDFEEVFWYSGWIMCGVRRVYRHLPKRVLRQYGYVQTIP 278
+ + + S + ++ H P R LRQ+G Q IP
Sbjct: 306 FENLIPPHIKAKLILGRSKTTLVCFEKIELHFPDRCLRQFGKRQPIP 352
>gb|AAV44079.1| putative polyprotein [Oryza sativa (japonica cultivar-group)]
Length = 1567
Score = 67.8 bits (164), Expect = 4e-10
Identities = 73/288 (25%), Positives = 116/288 (39%), Gaps = 31/288 (10%)
Query: 1 MPMGEMTVTLDDVACLTHLPIEGRMLDHGKKMPKHEGAAQLMTYLGVSQHEAEKICNQKY 60
+P GEMT+TL DVA + LP+ G + + P AQ+ G+ + Q
Sbjct: 1042 LPSGEMTITLQDVAMILALPLRGHAVTGRTETPGWH--AQVQQLFGIPLN-----IEQGQ 1094
Query: 61 GGYISYPRLRDFYTSYLGRANVLAGTEDPEELEELARVRTYCVRCYLLYLVGCLLFGDRS 120
GG + S+L + N +D E R Y R Y+L+L+G +LF D
Sbjct: 1095 GG---KKKQNGIPLSWLSQ-NFSHLDDDAEPW----RAECY-ARAYILHLLGGVLFPDAG 1145
Query: 121 NKRIELIYLTTMADGYAGMRNYSWGGMTLAYLYGELADAC--RPGHRALGGSVTLLTTWF 178
I++ +A+ + +SWG LA+ Y +L +AC + + G V L+ W
Sbjct: 1146 GDIASAIWIPLVAN-LGDLGRFSWGSAVLAWTYRQLCEACCRQAPSSNMSGCVLLIQMWM 1204
Query: 179 LAHFPGFFSVDLNTDYLENYPVAARWKLPKGHGEGITYWSLLDRIQLDDVCYKPYKEHRE 238
P ++ YL A + Y + +D +Q + + PY +
Sbjct: 1205 WLRLPNEPDMEKTVAYLFESTATAHAHRDVAYKH---YVNEMDYLQPQHIEWLPYHTNEA 1261
Query: 239 --------IQDFEEVFWYSGWIMCGVRRVYRHLPKRVLRQYGYVQTIP 278
+ F Y ++C V HLP RV+RQ+G Q P
Sbjct: 1262 SSLTLNSMCNRDSDYFMYQCPLIC-FWAVEYHLPHRVMRQFGKKQDWP 1308
>ref|NP_175246.1| calcineurin-like phosphoesterase family protein [Arabidopsis
thaliana]
Length = 1338
Score = 65.1 bits (157), Expect = 2e-09
Identities = 86/323 (26%), Positives = 130/323 (39%), Gaps = 62/323 (19%)
Query: 1 MPMGEMTVTLDDVACLTHLPIEGRMLDHGKKMPKHEGAAQLMTYLGVSQHEAEKICNQKY 60
+P GE+TVTL DV L L ++G + G +++ +C
Sbjct: 101 LPAGEITVTLQDVNILLGLRVDGPAVT------------------GSTKYNWADLCEDLL 142
Query: 61 GGYISYPRLRDFYTSYLG----RANVLAGTEDPEELEELARVRTYC-VRCYLLYLVGCLL 115
G P +D + S++ R N DP+E V C R ++L L+ L
Sbjct: 143 G---HRPGPKDLHGSHVSLAWLRENFRNLPADPDE------VTLKCHTRAFVLALMSGFL 193
Query: 116 FGDRSNKRIELIYLTTMADGYAGMRNYSWGGMTLAYLYGELADACRPGHRALGGSVTLLT 175
+GD+S + L +L + D + + SWG TLA LY EL A + + G + LL
Sbjct: 194 YGDKSKHDVALTFLPLLRD-FDEVAKLSWGSATLALLYRELCRASKRTVSTICGPLVLLQ 252
Query: 176 TWFLAHF----PGFFSVDLNTDYLENY------PVAARWKL-----PKGHGEGITYW-SL 219
W PG D+ Y++ P+ R L P+G G+ ++
Sbjct: 253 LWAWERLHVGRPGRLK-DVGASYMDGIDGPLPDPLGCRASLSHKENPRG---GLDFYRDQ 308
Query: 220 LDRIQLDDVCYKPY-----KEHREIQDFEEVFWYSGWIMCGVRRVYRHLPKRVLRQYGYV 274
D+ + + V ++PY + I E W + + V H P RVLRQ+G
Sbjct: 309 FDQQKDEQVIWQPYTPDLLAKIPLICVSGENIWRTVAPLICFDVVEWHRPDRVLRQFGLH 368
Query: 275 QTIPRHPTDVRDLPTAFHCADVR 297
QTIP P D A H D R
Sbjct: 369 QTIPA-PCDNE---KALHAIDKR 387
>ref|NP_909918.1| putative mutator-like transposase [Oryza sativa (japonica
cultivar-group)] gi|29150382|gb|AAO72391.1| mutator-like
transposase [Oryza sativa (japonica cultivar-group)]
gi|28209524|gb|AAO37542.1| putative mutator-like
transposase [Oryza sativa (japonica cultivar-group)]
Length = 1381
Score = 63.2 bits (152), Expect = 9e-09
Identities = 62/220 (28%), Positives = 91/220 (41%), Gaps = 24/220 (10%)
Query: 89 PEELEELARVRTYCVRCYLLYLVGCLLFGDRSNKRIELIYLTTMADGYAGMRNYSWGGMT 148
P E +E A V+ YC R Y+LY+ G +LF D ++L +AD + +SWG
Sbjct: 804 PAEADE-ATVQRYC-RAYVLYIFGSILFPDSGGDMASWMWLPLLAD-WDEAGTFSWGSAA 860
Query: 149 LAYLYGELADACR--PGHRALGGSVTLLTTWFLAHFP------GFFSVDLNTDYLENYPV 200
LA+LY +L DACR G L G V LL W P + D V
Sbjct: 861 LAWLYRQLCDACRRQGGDANLAGCVWLLQVWMWMRLPVGRPMWRTHQAWPHQDADRCPTV 920
Query: 201 AARWKLPKGHGEG------ITYWSLLDRIQLDDVCYKPYKEHREIQ-------DFEEVFW 247
A W+ G Y + +D +Q + V + PY+ ++ E+
Sbjct: 921 AHLWESVPSPVVGRRNLAYYHYTNEMDYLQPEHVVWMPYQAQEVLELELNPMCHIEDALK 980
Query: 248 YSGWIMCGVRRVYRHLPKRVLRQYGYVQTIPRHPTDVRDL 287
+ V + RV+RQ+G +QTIP + DL
Sbjct: 981 TLRCPLICFYAVEFQMCHRVMRQFGRLQTIPHRFSTSIDL 1020
>ref|NP_912436.1| Putative transposable element [Oryza sativa (japonica
cultivar-group)] gi|27476096|gb|AAO17027.1| Putative
transposable element [Oryza sativa (japonica
cultivar-group)]
Length = 663
Score = 63.2 bits (152), Expect = 9e-09
Identities = 78/305 (25%), Positives = 125/305 (40%), Gaps = 50/305 (16%)
Query: 1 MPMGEMTVTLDDVACLTHLPIEGRMLDHGKKMPKHEGAAQLMTYLGVSQHEAEKICNQKY 60
+P GEMT+TL DVA + LP+ G + + P AQ+ G+ + Q
Sbjct: 92 LPSGEMTITLQDVAMILALPLRGHAVTGRTETPGWH--AQVQQLFGIPLN-----IEQGQ 144
Query: 61 GGYISYPRLRDFYTSYLGRANVLAGTEDPEELEELARVRTYCVRCYLLYLVGCLLFGDRS 120
GG + S+L + N +D E RV Y R Y+L+L+G +LF D
Sbjct: 145 GGK---KKQNGIPLSWLSQ-NFSHLDDDAEPW----RVECYA-RAYILHLLGGVLFPDAG 195
Query: 121 NKRIELIYLTTMADGYAGMRNYSWGGMTLAYLYGELADAC--RPGHRALGGSVTLLTTWF 178
I++ +A+ + +SWG LA+ Y +L +AC + + G V L+ W
Sbjct: 196 GDIASAIWIPLVAN-LGDLGRFSWGSAVLAWTYRQLCEACCRQAPSSNMSGCVLLIQIWM 254
Query: 179 LAHFP------------GFFS---VDLNTDYL-ENYPVA-ARWKLPKGHGEGITYWSLLD 221
P +++ ++ YL E+ A A W + H Y + +D
Sbjct: 255 WLRLPVGRPKWRQSFTPWYYNEPDMEKTVAYLFESTATAHAHWDVAYKH-----YVNEMD 309
Query: 222 RIQLDDVCYKPYKEHRE--------IQDFEEVFWYSGWIMCGVRRVYRHLPKRVLRQYGY 273
+Q + + PY + + F Y ++C Y HLP RV+RQ+G
Sbjct: 310 CLQPQHIEWLPYHTNEASSLTLNLMCNRDSDYFMYQCPLICFWAVEY-HLPHRVMRQFGK 368
Query: 274 VQTIP 278
Q P
Sbjct: 369 KQDWP 373
>ref|XP_477211.1| calcineurin-like phosphoesterase-like [Oryza sativa (japonica
cultivar-group)] gi|50510004|dbj|BAD30617.1|
calcineurin-like phosphoesterase-like [Oryza sativa
(japonica cultivar-group)] gi|33146683|dbj|BAC80078.1|
calcineurin-like phosphoesterase-like [Oryza sativa
(japonica cultivar-group)]
Length = 327
Score = 60.8 bits (146), Expect = 5e-08
Identities = 47/184 (25%), Positives = 79/184 (42%), Gaps = 9/184 (4%)
Query: 1 MPMGEMTVTLDDVACLTHLPIEGRMLDHGKKMPKHEGAAQLMTYLGVSQHEAEKICNQKY 60
+P+GEMT+TL DV+CL LPI GR + + +L+ QH +K ++
Sbjct: 87 LPVGEMTITLQDVSCLWGLPIHGRPITGQADGSWVDMIERLLGIPMEEQHMKQKKRKKED 146
Query: 61 G-GYISYPRLRDFYTSYLGRANVLAGTEDPEELEELARVRTYCVRCYLLYLVGCLLFGDR 119
+SY R + R V+ E+ + R +L ++G ++F D
Sbjct: 147 DMTMVSYSRYSISLSKLRDRFRVMPKNATEREI-------NWYTRALVLDIIGSMVFTDT 199
Query: 120 SNKRIELIYLTTMADGYAGMRNYSWGGMTLAYLYGELADACRPGHRALGGSVTLLTTWFL 179
S + +YL M + + Y+WG L+ LY +L+ A + + LL W
Sbjct: 200 SGDGVPAMYLQFMMN-LSEQTEYNWGAAALSMLYRQLSIASEKERAEISRPLLLLQLWSW 258
Query: 180 AHFP 183
+ P
Sbjct: 259 SRLP 262
>gb|AAV59303.1| unknown protein [Oryza sativa (japonica cultivar-group)]
Length = 725
Score = 60.1 bits (144), Expect = 8e-08
Identities = 74/300 (24%), Positives = 114/300 (37%), Gaps = 40/300 (13%)
Query: 1 MPMGEMTVTLDDVACLTHLPIEGRMLDHGKKMPKHEGAAQLMTYLGVSQHEAEKICNQKY 60
+P GEMT+TL DVA + LP+ G + + P AQ+ G+ + Q
Sbjct: 140 LPSGEMTITLQDVAMILALPLRGHAVTGRTETPGWH--AQVQQLFGIPLN-----IEQGQ 192
Query: 61 GGYISYPRLRDFYTSYLGRANVLAGTEDPEELEELARVRTYCVRCYLLYLVGCLLFGDRS 120
GG + S+L + N +D E RV Y R Y+L+L+G +LF D
Sbjct: 193 GG---KKKQNGIPLSWLSQ-NFSHLDDDAEPW----RVECY-ARAYILHLLGGVLFPDAG 243
Query: 121 NKRIELIYLTTMADGYAGMRNYSWGGMTLAYLYGELADAC--RPGHRALGGSVTLLTTWF 178
I++ + + +SWG LA+ Y +L +AC + + G V L+ W
Sbjct: 244 GDIASAIWI-PLVTNLGDLGRFSWGSAVLAWTYRQLCEACCRQAPSSNMSGCVLLIQMWM 302
Query: 179 LAHFP--------GFFSVDLNTDYLENYPVAARWKLPKGHGEGIT----YWSLLDRIQLD 226
P F N +E H Y + +D +Q
Sbjct: 303 WLRLPIGRPKWRQSFTPWPYNEPDMEKTVAYLFESTATAHAHRDVAYKHYVNEMDCLQPQ 362
Query: 227 DVCYKPYKEHRE--------IQDFEEVFWYSGWIMCGVRRVYRHLPKRVLRQYGYVQTIP 278
+ + PY + + F Y ++C V HLP RV+RQ+G Q P
Sbjct: 363 HIEWLPYHTNEASSLTLNSMCNRDSDYFMYQCPLIC-FWAVEYHLPHRVMRQFGKKQDWP 421
>ref|XP_475301.1| unknown protein [Oryza sativa (japonica cultivar-group)]
gi|49328188|gb|AAT58884.1| unknown protein [Oryza sativa
(japonica cultivar-group)]
Length = 1564
Score = 60.1 bits (144), Expect = 8e-08
Identities = 74/300 (24%), Positives = 114/300 (37%), Gaps = 40/300 (13%)
Query: 1 MPMGEMTVTLDDVACLTHLPIEGRMLDHGKKMPKHEGAAQLMTYLGVSQHEAEKICNQKY 60
+P GEMT+TL DVA + LP+ G + + P AQ+ G+ + Q
Sbjct: 979 LPSGEMTITLQDVAMILALPLRGHAVTGRTETPGWH--AQVQQLFGIPLN-----IEQGQ 1031
Query: 61 GGYISYPRLRDFYTSYLGRANVLAGTEDPEELEELARVRTYCVRCYLLYLVGCLLFGDRS 120
GG + S+L + N +D E RV Y R Y+L+L+G +LF D
Sbjct: 1032 GG---KKKQNGIPLSWLSQ-NFSHLDDDAEPW----RVECY-ARAYILHLLGGVLFPDAG 1082
Query: 121 NKRIELIYLTTMADGYAGMRNYSWGGMTLAYLYGELADAC--RPGHRALGGSVTLLTTWF 178
I++ + + +SWG LA+ Y +L +AC + + G V L+ W
Sbjct: 1083 GDIASAIWI-PLVTNLGDLGRFSWGSAVLAWTYRQLCEACCRQAPSSNMSGCVLLIQMWM 1141
Query: 179 LAHFP--------GFFSVDLNTDYLENYPVAARWKLPKGHGEGIT----YWSLLDRIQLD 226
P F N +E H Y + +D +Q
Sbjct: 1142 WLRLPIGRPKWRQSFTPWPYNEPDMEKTVAYLFESTATAHAHRDVAYKHYVNEMDCLQPQ 1201
Query: 227 DVCYKPYKEHRE--------IQDFEEVFWYSGWIMCGVRRVYRHLPKRVLRQYGYVQTIP 278
+ + PY + + F Y ++C V HLP RV+RQ+G Q P
Sbjct: 1202 HIEWLPYHTNEASSLTLNSMCNRDSDYFMYQCPLIC-FWAVEYHLPHRVMRQFGKKQDWP 1260
Database: nr
Posted date: Jul 5, 2005 12:34 AM
Number of letters in database: 863,360,394
Number of sequences in database: 2,540,612
Lambda K H
0.325 0.142 0.463
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 582,125,096
Number of Sequences: 2540612
Number of extensions: 25831727
Number of successful extensions: 55768
Number of sequences better than 10.0: 133
Number of HSP's better than 10.0 without gapping: 19
Number of HSP's successfully gapped in prelim test: 114
Number of HSP's that attempted gapping in prelim test: 55508
Number of HSP's gapped (non-prelim): 238
length of query: 304
length of database: 863,360,394
effective HSP length: 127
effective length of query: 177
effective length of database: 540,702,670
effective search space: 95704372590
effective search space used: 95704372590
T: 11
A: 40
X1: 15 ( 7.0 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 40 (21.5 bits)
S2: 75 (33.5 bits)
Medicago: description of AC139854.8


