

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC139841.2 - phase: 0 /pseudo
(285 letters)
Database: nr
2,540,612 sequences; 863,360,394 total letters
Searching..................................................done
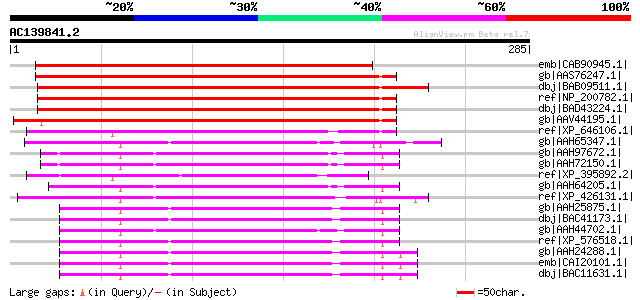
Score E
Sequences producing significant alignments: (bits) Value
emb|CAB90945.1| putative protein [Arabidopsis thaliana] gi|11357... 306 6e-82
gb|AAS76247.1| At3g46180 [Arabidopsis thaliana] gi|42565652|ref|... 305 7e-82
dbj|BAB09511.1| UDP-galactose transporter related protein-like [... 305 1e-81
ref|NP_200782.1| UDP-galactose/UDP-glucose transporter-related [... 303 4e-81
dbj|BAD43224.1| unnamed protein product [Arabidopsis thaliana] g... 303 4e-81
gb|AAV44195.1| unknow protein [Oryza sativa (japonica cultivar-g... 271 2e-71
ref|XP_646106.1| hypothetical protein DDB0190395 [Dictyostelium ... 142 8e-33
gb|AAH65347.1| Hypothetical protein zgc:77349 [Danio rerio] gi|5... 132 9e-30
gb|AAH97672.1| LOC432208 protein [Xenopus laevis] 129 1e-28
gb|AAH72150.1| LOC432208 protein [Xenopus laevis] 129 1e-28
ref|XP_395892.2| PREDICTED: similar to ENSANGP00000012787 [Apis ... 128 2e-28
gb|AAH64205.1| LOC394918 protein [Xenopus tropicalis] 125 1e-27
ref|XP_426131.1| PREDICTED: similar to solute carrier family 35,... 125 2e-27
gb|AAH25875.1| Slc35b2 protein [Mus musculus] gi|22477708|gb|AAH... 124 3e-27
dbj|BAC41173.1| unnamed protein product [Mus musculus] 124 3e-27
gb|AAH44702.1| Slc35b1-prov protein [Xenopus laevis] 123 7e-27
ref|XP_576518.1| PREDICTED: similar to embryonic seven-span tran... 123 7e-27
gb|AAH24288.1| Solute carrier family 35, member B2 [Homo sapiens... 122 1e-26
emb|CAI20101.1| OTTHUMP00000016513 [Homo sapiens] 122 1e-26
dbj|BAC11631.1| unnamed protein product [Homo sapiens] 122 1e-26
>emb|CAB90945.1| putative protein [Arabidopsis thaliana] gi|11357381|pir||T49259
hypothetical protein F12M12.150 - Arabidopsis thaliana
Length = 344
Score = 306 bits (783), Expect = 6e-82
Identities = 142/185 (76%), Positives = 166/185 (88%)
Query: 15 SLDDSNVTRNKLWKVAFAVSGIMLTLVTYGVLQEKIMRVPYGVNKDYFKYSLFLVFCNRI 74
S++++ + KLWK FA+SGIMLTLV YG+LQEKIMRVPYG+ K+YFK+SLFLVFCNR+
Sbjct: 6 SVNEAKEKKKKLWKAVFAISGIMLTLVIYGLLQEKIMRVPYGLKKEYFKHSLFLVFCNRL 65
Query: 75 TTSAVSAAALVARDKALLPVAPIYKYCLVSVSNILTTTCQYEALKYVSFPVQTLAKCAKM 134
TTSAVSAAAL+A K L PVAP+YKYCL+SV+NILTTTCQYEALKYVSFPVQTLAKCAKM
Sbjct: 66 TTSAVSAAALLASKKVLDPVAPVYKYCLISVTNILTTTCQYEALKYVSFPVQTLAKCAKM 125
Query: 135 IPVMIWSALIMQKKYQGTDYLLAFSVTLGCSIFILYPAGTDISPYGRGRENTIWGILLMI 194
IPVM+W LIMQKKY+G DYL+AF VTLGCS+FIL+PAG DISPY +GRENT+WG+ LM+
Sbjct: 126 IPVMVWGTLIMQKKYRGFDYLVAFLVTLGCSVFILFPAGDDISPYNKGRENTVWGVSLMV 185
Query: 195 GYLGY 199
GYLGY
Sbjct: 186 GYLGY 190
>gb|AAS76247.1| At3g46180 [Arabidopsis thaliana] gi|42565652|ref|NP_190204.2|
UDP-galactose/UDP-glucose transporter-related
[Arabidopsis thaliana] gi|40823122|gb|AAR92260.1|
At3g46180 [Arabidopsis thaliana]
Length = 347
Score = 305 bits (782), Expect = 7e-82
Identities = 146/198 (73%), Positives = 172/198 (86%), Gaps = 1/198 (0%)
Query: 15 SLDDSNVTRNKLWKVAFAVSGIMLTLVTYGVLQEKIMRVPYGVNKDYFKYSLFLVFCNRI 74
S++++ + KLWK FA+SGIMLTLV YG+LQEKIMRVPYG+ K+YFK+SLFLVFCNR+
Sbjct: 6 SVNEAKEKKKKLWKAVFAISGIMLTLVIYGLLQEKIMRVPYGLKKEYFKHSLFLVFCNRL 65
Query: 75 TTSAVSAAALVARDKALLPVAPIYKYCLVSVSNILTTTCQYEALKYVSFPVQTLAKCAKM 134
TTSAVSAAAL+A K L PVAP+YKYCL+SV+NILTTTCQYEALKYVSFPVQTLAKCAKM
Sbjct: 66 TTSAVSAAALLASKKVLDPVAPVYKYCLISVTNILTTTCQYEALKYVSFPVQTLAKCAKM 125
Query: 135 IPVMIWSALIMQKKYQGTDYLLAFSVTLGCSIFILYPAGTDISPYGRGRENTIWGILLMI 194
IPVM+W LIMQKKY+G DYL+AF VTLGCS+FIL+PAG DISPY +GRENT+WG+ LM+
Sbjct: 126 IPVMVWGTLIMQKKYRGFDYLVAFLVTLGCSVFILFPAGDDISPYNKGRENTVWGVSLMV 185
Query: 195 GYLGYHYKTQSLFAFRLF 212
GYLG+ T S F +LF
Sbjct: 186 GYLGFDGFT-STFQDKLF 202
>dbj|BAB09511.1| UDP-galactose transporter related protein-like [Arabidopsis
thaliana]
Length = 345
Score = 305 bits (780), Expect = 1e-81
Identities = 152/215 (70%), Positives = 176/215 (81%), Gaps = 1/215 (0%)
Query: 16 LDDSNVTRNKLWKVAFAVSGIMLTLVTYGVLQEKIMRVPYGVNKDYFKYSLFLVFCNRIT 75
L + V NKLWK FAVSGIM TLV YGVLQEKIMRVPYGVNK++FK+SLFLVFCNR+T
Sbjct: 6 LVNGGVKENKLWKGVFAVSGIMSTLVIYGVLQEKIMRVPYGVNKEFFKHSLFLVFCNRLT 65
Query: 76 TSAVSAAALVARDKALLPVAPIYKYCLVSVSNILTTTCQYEALKYVSFPVQTLAKCAKMI 135
TSAVSA AL+A K L PVAP+YKYCL+SV+NILTTTCQYEALKYVSFPVQTLAKCAKMI
Sbjct: 66 TSAVSAGALLASKKVLDPVAPVYKYCLISVTNILTTTCQYEALKYVSFPVQTLAKCAKMI 125
Query: 136 PVMIWSALIMQKKYQGTDYLLAFSVTLGCSIFILYPAGTDISPYGRGRENTIWGILLMIG 195
PVM+W LIMQKKY+G DYL+AF VTLGCS+FIL+PAG D+SPY +GRENT+WG+ LM G
Sbjct: 126 PVMVWGTLIMQKKYKGFDYLVAFLVTLGCSVFILFPAGDDVSPYNKGRENTVWGVSLMAG 185
Query: 196 YLGYHYKTQSLFAFRLFSLAYFVSFLSSLLESHSF 230
YLG+ T S F +LF + S S +L S+ +
Sbjct: 186 YLGFDGFT-STFQDKLFKPSKRRSMWSIILCSYHY 219
>ref|NP_200782.1| UDP-galactose/UDP-glucose transporter-related [Arabidopsis
thaliana] gi|44917565|gb|AAS49107.1| At5g59740
[Arabidopsis thaliana]
Length = 344
Score = 303 bits (776), Expect = 4e-81
Identities = 148/197 (75%), Positives = 168/197 (85%), Gaps = 1/197 (0%)
Query: 16 LDDSNVTRNKLWKVAFAVSGIMLTLVTYGVLQEKIMRVPYGVNKDYFKYSLFLVFCNRIT 75
L + V NKLWK FAVSGIM TLV YGVLQEKIMRVPYGVNK++FK+SLFLVFCNR+T
Sbjct: 6 LVNGGVKENKLWKGVFAVSGIMSTLVIYGVLQEKIMRVPYGVNKEFFKHSLFLVFCNRLT 65
Query: 76 TSAVSAAALVARDKALLPVAPIYKYCLVSVSNILTTTCQYEALKYVSFPVQTLAKCAKMI 135
TSAVSA AL+A K L PVAP+YKYCL+SV+NILTTTCQYEALKYVSFPVQTLAKCAKMI
Sbjct: 66 TSAVSAGALLASKKVLDPVAPVYKYCLISVTNILTTTCQYEALKYVSFPVQTLAKCAKMI 125
Query: 136 PVMIWSALIMQKKYQGTDYLLAFSVTLGCSIFILYPAGTDISPYGRGRENTIWGILLMIG 195
PVM+W LIMQKKY+G DYL+AF VTLGCS+FIL+PAG D+SPY +GRENT+WG+ LM G
Sbjct: 126 PVMVWGTLIMQKKYKGFDYLVAFLVTLGCSVFILFPAGDDVSPYNKGRENTVWGVSLMAG 185
Query: 196 YLGYHYKTQSLFAFRLF 212
YLG+ T S F +LF
Sbjct: 186 YLGFDGFT-STFQDKLF 201
>dbj|BAD43224.1| unnamed protein product [Arabidopsis thaliana]
gi|51969060|dbj|BAD43222.1| unnamed protein product
[Arabidopsis thaliana]
Length = 344
Score = 303 bits (776), Expect = 4e-81
Identities = 148/197 (75%), Positives = 168/197 (85%), Gaps = 1/197 (0%)
Query: 16 LDDSNVTRNKLWKVAFAVSGIMLTLVTYGVLQEKIMRVPYGVNKDYFKYSLFLVFCNRIT 75
L + V NKLWK FAVSGIM TLV YGVLQEKIMRVPYGVNK++FK+SLFLVFCNR+T
Sbjct: 6 LVNGGVKENKLWKGVFAVSGIMSTLVIYGVLQEKIMRVPYGVNKEFFKHSLFLVFCNRLT 65
Query: 76 TSAVSAAALVARDKALLPVAPIYKYCLVSVSNILTTTCQYEALKYVSFPVQTLAKCAKMI 135
TSAVSA AL+A K L PVAP+YKYCL+SV+NILTTTCQYEALKYVSFPVQTLAKCAKMI
Sbjct: 66 TSAVSAGALLASKKVLDPVAPVYKYCLISVTNILTTTCQYEALKYVSFPVQTLAKCAKMI 125
Query: 136 PVMIWSALIMQKKYQGTDYLLAFSVTLGCSIFILYPAGTDISPYGRGRENTIWGILLMIG 195
PVM+W LIMQKKY+G DYL+AF VTLGCS+FIL+PAG D+SPY +GRENT+WG+ LM G
Sbjct: 126 PVMVWGTLIMQKKYKGFDYLVAFLVTLGCSVFILFPAGDDVSPYNKGRENTVWGVSLMAG 185
Query: 196 YLGYHYKTQSLFAFRLF 212
YLG+ T S F +LF
Sbjct: 186 YLGFDGFT-STFQDKLF 201
>gb|AAV44195.1| unknow protein [Oryza sativa (japonica cultivar-group)]
gi|52353517|gb|AAU44083.1| unknown protein [Oryza sativa
(japonica cultivar-group)]
Length = 360
Score = 271 bits (692), Expect = 2e-71
Identities = 140/223 (62%), Positives = 164/223 (72%), Gaps = 14/223 (6%)
Query: 3 MAETTTSSSSSSSL-------------DDSNVTRNKLWKVAFAVSGIMLTLVTYGVLQEK 49
MA+ +SS+S+S+ D R + + FAV GIM TL+ YG+LQEK
Sbjct: 1 MADAASSSASASAAVAGTLPVAAAATGKDKEDRRRLVGRCGFAVVGIMSTLLIYGLLQEK 60
Query: 50 IMRVPYGVNKDYFKYSLFLVFCNRITTSAVSAAALVARDKALLPVAPIYKYCLVSVSNIL 109
IMRVPYG K++F+YSLFLVFCNRITTS VSA L A K+L PVAP+ KYC+VSVSNIL
Sbjct: 61 IMRVPYGAEKEFFRYSLFLVFCNRITTSTVSALVLTASKKSLDPVAPLQKYCVVSVSNIL 120
Query: 110 TTTCQYEALKYVSFPVQTLAKCAKMIPVMIWSALIMQKKYQGTDYLLAFSVTLGCSIFIL 169
TTTCQYEALKYVSFPVQTLAKCAKMIPVMIW +IM+KKY G DY A VT+GCS+FIL
Sbjct: 121 TTTCQYEALKYVSFPVQTLAKCAKMIPVMIWGTIIMRKKYGGKDYFFAVVVTVGCSLFIL 180
Query: 170 YPAGTDISPYGRGRENTIWGILLMIGYLGYHYKTQSLFAFRLF 212
YPA D SP+ RGRENTIWG+ LM+GYLG+ T S F +LF
Sbjct: 181 YPASMDASPFNRGRENTIWGVSLMLGYLGFDGFT-STFQDKLF 222
>ref|XP_646106.1| hypothetical protein DDB0190395 [Dictyostelium discoideum]
gi|60474214|gb|EAL72151.1| hypothetical protein
DDB0190395 [Dictyostelium discoideum]
Length = 359
Score = 142 bits (359), Expect = 8e-33
Identities = 83/206 (40%), Positives = 124/206 (59%), Gaps = 9/206 (4%)
Query: 10 SSSSSSLDDSNVTRNKLWKVAFAVSGIMLTLVTYGVLQEKIMRVPY---GVNKDYFKYSL 66
SS+++S ++ V + K+A A GIM + + YG+LQE++M VPY +++YF S
Sbjct: 10 SSTTNSNNNEKVQLSYNMKLALATGGIMGSFLLYGILQERLMVVPYKNADGSEEYFTDST 69
Query: 67 FLVFCNRITTSAVSAAALVARDKALLPVAPIYKYCLVSVSNILTTTCQYEALKYVSFPVQ 126
FLV NR+ + ++ ++ R ++L VAP++KY V++SN T CQYEALKYV+FP Q
Sbjct: 70 FLVLSNRVFAALMAIVIVLKRGESLKNVAPLHKYVGVALSNFCATWCQYEALKYVNFPTQ 129
Query: 127 TLAKCAKMIPVMIWSALIMQKKYQGTDYLLAFSVTLGCSIFILYPAGTDISPYGRGRENT 186
TL KC KM+PVM+ I KKY DY +A ++T GC IF L ++ NT
Sbjct: 130 TLGKCGKMLPVMLVGTFISGKKYGLKDYSIALTITTGCMIFFLTGKISN-----NESSNT 184
Query: 187 IWGILLMIGYLGYHYKTQSLFAFRLF 212
+GI+LM Y+ + T S F ++F
Sbjct: 185 SYGIILMALYMFFDSFT-STFQEKMF 209
>gb|AAH65347.1| Hypothetical protein zgc:77349 [Danio rerio]
gi|58475142|gb|AAH90054.1| Zgc:64182 protein [Danio
rerio] gi|45387691|ref|NP_991198.1| hypothetical protein
zgc:77349 [Danio rerio]
Length = 435
Score = 132 bits (333), Expect = 9e-30
Identities = 89/249 (35%), Positives = 138/249 (54%), Gaps = 28/249 (11%)
Query: 9 SSSSSSSLDDSNVTRNKLWKVAFAVSGIMLTLVTYGVLQEKIMRVPYGVNK-----DYFK 63
+S + + +S+ + + +K+ F +G+ ++ +T+GVLQE++M YG ++ + F+
Sbjct: 96 ASPAGRNESESSSSARQAFKLIFCAAGLQVSYLTWGVLQERVMTRSYGSSEAEGSGERFR 155
Query: 64 YSLFLVFCNRITTSAVSAAALVARDKALLPVAPIYKYCLVSVSNILTTTCQYEALKYVSF 123
S FLVF NRI VS V K AP+YKY S+SNIL++ CQYEALK++SF
Sbjct: 156 DSQFLVFMNRILALTVSGLWCVLF-KQPRHGAPMYKYSFASLSNILSSWCQYEALKFISF 214
Query: 124 PVQTLAKCAKMIPVMIWSALIMQKKYQGTDYLLAFSVTLGCSIFILYPAGTDISPYGRGR 183
P Q LAK +K+IPVM+ ++ +K Y+ +YL A ++LG S+F+L + TD P
Sbjct: 215 PTQVLAKASKVIPVMLMGKIVSRKSYEYWEYLTAVLISLGVSMFLL-SSSTDKHP---ST 270
Query: 184 ENTIWGILLMIGYLG------------YHYK---TQSLFAFRLFSLAYFVSFLSSLLESH 228
T G+L++ GY+ + YK Q +F LFS + V SLLE
Sbjct: 271 VTTFSGVLILAGYIVFDSFTSNWQDNLFKYKMSSVQMMFGVNLFSCLFTV---GSLLEQG 327
Query: 229 SFHI*LLFI 237
+F L F+
Sbjct: 328 AFFNSLAFM 336
>gb|AAH97672.1| LOC432208 protein [Xenopus laevis]
Length = 439
Score = 129 bits (324), Expect = 1e-28
Identities = 79/205 (38%), Positives = 122/205 (58%), Gaps = 14/205 (6%)
Query: 18 DSNVTRNKLWKVAFAVSGIMLTLVTYGVLQEKIMRVPYGVNK----DYFKYSLFLVFCNR 73
D + T+ L K+ F +G+ ++ +T+GVLQE++M YG + + F+ S FLVF NR
Sbjct: 110 DPSTTQQAL-KLLFCAAGLQVSYLTWGVLQERVMTRTYGSEEGGPGERFRDSQFLVFMNR 168
Query: 74 ITTSAVSAAALVARDKALLPVAPIYKYCLVSVSNILTTTCQYEALKYVSFPVQTLAKCAK 133
I V A + K AP+YKY S+SNIL++ CQYEALK++SFP Q LAK +K
Sbjct: 169 ILALTV-AGLYCSVTKQPRHGAPMYKYSFASLSNILSSWCQYEALKFISFPTQVLAKASK 227
Query: 134 MIPVMIWSALIMQKKYQGTDYLLAFSVTLGCSIFILYPAGTDISPYGRGRENTIWGILLM 193
+IPVM+ L+ K Y+ +YL A +++G S+F+L G D P+G T G++++
Sbjct: 228 VIPVMLMGKLVSHKSYEYWEYLTAVLISVGVSMFLLSNGGGD-RPWG---VTTFSGVVIL 283
Query: 194 IGYLGYHYKT----QSLFAFRLFSL 214
GY+ + T SLF +++ S+
Sbjct: 284 AGYIVFDSFTSNWQDSLFKYKMSSV 308
>gb|AAH72150.1| LOC432208 protein [Xenopus laevis]
Length = 458
Score = 129 bits (324), Expect = 1e-28
Identities = 79/205 (38%), Positives = 122/205 (58%), Gaps = 14/205 (6%)
Query: 18 DSNVTRNKLWKVAFAVSGIMLTLVTYGVLQEKIMRVPYGVNK----DYFKYSLFLVFCNR 73
D + T+ L K+ F +G+ ++ +T+GVLQE++M YG + + F+ S FLVF NR
Sbjct: 129 DPSTTQQAL-KLLFCAAGLQVSYLTWGVLQERVMTRTYGSEEGGPGERFRDSQFLVFMNR 187
Query: 74 ITTSAVSAAALVARDKALLPVAPIYKYCLVSVSNILTTTCQYEALKYVSFPVQTLAKCAK 133
I V A + K AP+YKY S+SNIL++ CQYEALK++SFP Q LAK +K
Sbjct: 188 ILALTV-AGLYCSVTKQPRHGAPMYKYSFASLSNILSSWCQYEALKFISFPTQVLAKASK 246
Query: 134 MIPVMIWSALIMQKKYQGTDYLLAFSVTLGCSIFILYPAGTDISPYGRGRENTIWGILLM 193
+IPVM+ L+ K Y+ +YL A +++G S+F+L G D P+G T G++++
Sbjct: 247 VIPVMLMGKLVSHKSYEYWEYLTAVLISVGVSMFLLSNGGGD-RPWG---VTTFSGVVIL 302
Query: 194 IGYLGYHYKT----QSLFAFRLFSL 214
GY+ + T SLF +++ S+
Sbjct: 303 AGYIVFDSFTSNWQDSLFKYKMSSV 327
>ref|XP_395892.2| PREDICTED: similar to ENSANGP00000012787 [Apis mellifera]
Length = 442
Score = 128 bits (321), Expect = 2e-28
Identities = 77/191 (40%), Positives = 113/191 (58%), Gaps = 10/191 (5%)
Query: 10 SSSSSSLDDSNVTRNKLWKVAFAVSGIMLTLVTYGVLQEKIMRVPY---GVNKDYFKYSL 66
SS+ S+ T++ L + + G+ ++ +T+G LQEKIM Y NKD F+ S
Sbjct: 107 SSTPSNHSQRTFTQDALL-LLYCFLGLQISYLTWGYLQEKIMTQEYEDVAGNKDRFQDSQ 165
Query: 67 FLVFCNRITTSAVSAAALVARDKALLPVAPIYKYCLVSVSNILTTTCQYEALKYVSFPVQ 126
FLVF NRI +S L+ R + AP+YKY S+SNI+++ CQYEALKYVSFP Q
Sbjct: 166 FLVFVNRILAFLMSGLYLIIRRQPQHK-APLYKYAFCSLSNIMSSWCQYEALKYVSFPTQ 224
Query: 127 TLAKCAKMIPVMIWSALIMQKKYQGTDYLLAFSVTLGCSIFILYPAGTDISPYGRGRENT 186
LAK +K+IPVMI ++ Y+ +Y+ A +++G ++F+L D S Y T
Sbjct: 225 VLAKASKIIPVMIMGKIVSHTTYEYYEYVTAILISIGMTLFML-----DSSDYKNDGATT 279
Query: 187 IWGILLMIGYL 197
+ GI+L+ GYL
Sbjct: 280 VSGIILLGGYL 290
>gb|AAH64205.1| LOC394918 protein [Xenopus tropicalis]
Length = 453
Score = 125 bits (314), Expect = 1e-27
Identities = 76/201 (37%), Positives = 118/201 (57%), Gaps = 13/201 (6%)
Query: 22 TRNKLWKVAFAVSGIMLTLVTYGVLQEKIMRVPYGVNK----DYFKYSLFLVFCNRITTS 77
T + K+ F +G+ ++ +T+GVLQE++M YG + + F+ S FLVF NRI
Sbjct: 127 TAQQALKLLFCAAGLQVSYLTWGVLQERVMTRTYGSEEGSPGERFRDSQFLVFMNRILAL 186
Query: 78 AVSAAALVARDKALLPVAPIYKYCLVSVSNILTTTCQYEALKYVSFPVQTLAKCAKMIPV 137
V A + K AP+YKY S+SNIL++ CQYEALK++SFP Q LAK +K+IPV
Sbjct: 187 TV-AGVYCSISKQPRHGAPMYKYSFASLSNILSSWCQYEALKFISFPTQVLAKASKVIPV 245
Query: 138 MIWSALIMQKKYQGTDYLLAFSVTLGCSIFILYPAGTDISPYGRGRENTIWGILLMIGYL 197
M+ L+ K Y+ +YL A +++G S+F+L G + P G T G++++ GY+
Sbjct: 246 MLMGKLVSHKTYEYWEYLTAVLISVGVSMFLLSNGGGN-RPSG---VTTFSGVVILSGYI 301
Query: 198 GYHYKT----QSLFAFRLFSL 214
+ T SLF +++ S+
Sbjct: 302 VFDSFTSNWQDSLFKYKMSSV 322
>ref|XP_426131.1| PREDICTED: similar to solute carrier family 35, member B2;
3-phosphoadenosine 5-phosphosulfate transporter [Gallus
gallus]
Length = 395
Score = 125 bits (313), Expect = 2e-27
Identities = 88/249 (35%), Positives = 130/249 (51%), Gaps = 29/249 (11%)
Query: 5 ETTTSSSSSSSLDDSNVTRNKLWKVAFAVSGIMLTLVTYGVLQEKIMRVPYGVNK----D 60
ET S + + T ++ K+ F +G+ ++ +T+GV+QE++M YG + +
Sbjct: 51 ETEDGSLPPRAEPTESSTARQVLKLLFCAAGLQVSYLTWGVVQERVMTRTYGATETDPGE 110
Query: 61 YFKYSLFLVFCNRITTSAVSAAALVARDKALLPVAPIYKYCLVSVSNILTTTCQYEALKY 120
FK S FLVF NRI V A A K A +YKY S+SNIL++ CQYEALKY
Sbjct: 111 KFKDSQFLVFMNRILAFTV-AGLYCALTKQPRHGAAMYKYSFASLSNILSSWCQYEALKY 169
Query: 121 VSFPVQTLAKCAKMIPVMIWSALIMQKKYQGTDYLLAFSVTLGCSIFILYPA-GTDISPY 179
+SFP Q LAK +K+IPVM+ L+ K Y+ +YL A +++G S+F+L A +S
Sbjct: 170 ISFPTQVLAKASKVIPVMMMGKLVSHKSYEYWEYLTAALISVGVSMFLLSSAPNRHVSTV 229
Query: 180 GRGRENTIWGILLMIGYLGYH------------YK---TQSLFAFRLFSLAYFVSFL--- 221
T G++L+ GY+ + YK Q +F +FS + V L
Sbjct: 230 -----TTFSGVVLLAGYIVFDSFTSNWQDALFTYKMSPVQMMFGVNVFSCLFTVGSLLEQ 284
Query: 222 SSLLESHSF 230
+LLES F
Sbjct: 285 GALLESVHF 293
>gb|AAH25875.1| Slc35b2 protein [Mus musculus] gi|22477708|gb|AAH36992.1| Slc35b2
protein [Mus musculus] gi|15778614|gb|AAL07487.1|
embryonic seven-span transmembrane protein-like protein
[Mus musculus] gi|67461578|sp|Q91ZN5|S35B2_MOUSE
Adenosine 3'-phospho 5'-phosphosulfate transporter 1
(PAPS transporter 1) (Solute carrier family 35, member
B2) gi|20900129|ref|XP_128634.1| PREDICTED: solute
carrier family 35, member B2 [Mus musculus]
Length = 431
Score = 124 bits (311), Expect = 3e-27
Identities = 71/195 (36%), Positives = 115/195 (58%), Gaps = 13/195 (6%)
Query: 28 KVAFAVSGIMLTLVTYGVLQEKIMRVPYGVNK----DYFKYSLFLVFCNRITTSAVSAAA 83
K+ F SG+ ++ +T+G+LQE++M YG ++F S FLV NR+ V+
Sbjct: 111 KLVFCASGLQVSYLTWGILQERVMTGSYGATATSPGEHFTDSQFLVLMNRVLALVVAGLY 170
Query: 84 LVARDKALLPVAPIYKYCLVSVSNILTTTCQYEALKYVSFPVQTLAKCAKMIPVMIWSAL 143
V R K AP+Y+Y S+SN+L++ CQYEALK+VSFP Q LAK +K+IPVM+ L
Sbjct: 171 CVLR-KQPRHGAPMYRYSFASLSNVLSSWCQYEALKFVSFPTQVLAKASKVIPVMMMGKL 229
Query: 144 IMQKKYQGTDYLLAFSVTLGCSIFILYPAGTDISPYGRGRENTIWGILLMIGYLGYHYKT 203
+ ++ Y+ +YL A +++G S+F+L S T+ G++L+ GY+ + T
Sbjct: 230 VSRRSYEHWEYLTAGLISIGVSMFLLSSGPEPRS----SPATTLSGLVLLAGYIAFDSFT 285
Query: 204 ----QSLFAFRLFSL 214
+LFA+++ S+
Sbjct: 286 SNWQDALFAYKMSSV 300
>dbj|BAC41173.1| unnamed protein product [Mus musculus]
Length = 349
Score = 124 bits (311), Expect = 3e-27
Identities = 71/195 (36%), Positives = 115/195 (58%), Gaps = 13/195 (6%)
Query: 28 KVAFAVSGIMLTLVTYGVLQEKIMRVPYGVNK----DYFKYSLFLVFCNRITTSAVSAAA 83
K+ F SG+ ++ +T+G+LQE++M YG ++F S FLV NR+ V+
Sbjct: 62 KLVFCASGLQVSYLTWGILQERVMTGSYGATATSPGEHFTDSQFLVLMNRVLALVVAGLY 121
Query: 84 LVARDKALLPVAPIYKYCLVSVSNILTTTCQYEALKYVSFPVQTLAKCAKMIPVMIWSAL 143
V R K AP+Y+Y S+SN+L++ CQYEALK+VSFP Q LAK +K+IPVM+ L
Sbjct: 122 CVLR-KQPRHGAPMYRYSFASLSNVLSSWCQYEALKFVSFPTQVLAKASKVIPVMMMGKL 180
Query: 144 IMQKKYQGTDYLLAFSVTLGCSIFILYPAGTDISPYGRGRENTIWGILLMIGYLGYHYKT 203
+ ++ Y+ +YL A +++G S+F+L S T+ G++L+ GY+ + T
Sbjct: 181 VSRRSYEHWEYLTAGLISIGVSMFLLSSGPEPRS----SPATTLSGLVLLAGYIAFDSFT 236
Query: 204 ----QSLFAFRLFSL 214
+LFA+++ S+
Sbjct: 237 SNWQDALFAYKMSSV 251
>gb|AAH44702.1| Slc35b1-prov protein [Xenopus laevis]
Length = 439
Score = 123 bits (308), Expect = 7e-27
Identities = 75/195 (38%), Positives = 115/195 (58%), Gaps = 13/195 (6%)
Query: 28 KVAFAVSGIMLTLVTYGVLQEKIMRVPYGVNK----DYFKYSLFLVFCNRITTSAVSAAA 83
K+ F +G+ ++ +T+GVLQE++M YG + + F+ S FLVF NRI V A
Sbjct: 119 KLLFCTAGLQVSYLTWGVLQERVMTRTYGSEEGGPGERFRDSQFLVFMNRILALTV-AGL 177
Query: 84 LVARDKALLPVAPIYKYCLVSVSNILTTTCQYEALKYVSFPVQTLAKCAKMIPVMIWSAL 143
+ K AP+YKY S+SNIL++ CQYEALK++SFP Q LAK +K+IPVM+ L
Sbjct: 178 YCSVTKQPRHGAPMYKYSFASLSNILSSWCQYEALKFISFPTQVLAKASKVIPVMLMGKL 237
Query: 144 IMQKKYQGTDYLLAFSVTLGCSIFILYPAGTDISPYGRGRENTIWGILLMIGYLGYHYKT 203
+ K Y+ +YL A +++G S+F+L G P G T G++++ GY+ + T
Sbjct: 238 VSHKSYEYWEYLTAVLISVGVSMFLL-SNGEGNRPSG---VTTFSGLVILAGYIVFDSFT 293
Query: 204 ----QSLFAFRLFSL 214
SLF +++ S+
Sbjct: 294 SNWQDSLFKYKMSSV 308
>ref|XP_576518.1| PREDICTED: similar to embryonic seven-span transmembrane
protein-like protein [Rattus norvegicus]
Length = 492
Score = 123 bits (308), Expect = 7e-27
Identities = 70/195 (35%), Positives = 115/195 (58%), Gaps = 13/195 (6%)
Query: 28 KVAFAVSGIMLTLVTYGVLQEKIMRVPYGVNK----DYFKYSLFLVFCNRITTSAVSAAA 83
K+ F +G+ ++ +T+GVLQE++M YG ++F S FLV NR+ V+
Sbjct: 172 KLVFCAAGLQVSYLTWGVLQERVMTGSYGATATSPGEHFTDSQFLVLMNRVLALVVAGLY 231
Query: 84 LVARDKALLPVAPIYKYCLVSVSNILTTTCQYEALKYVSFPVQTLAKCAKMIPVMIWSAL 143
+ R K AP+Y+Y S+SN+L++ CQYEALK+VSFP Q LAK +K+IPVM+ L
Sbjct: 232 CILR-KQPRHGAPMYRYSFASLSNVLSSWCQYEALKFVSFPTQVLAKASKVIPVMMMGKL 290
Query: 144 IMQKKYQGTDYLLAFSVTLGCSIFILYPAGTDISPYGRGRENTIWGILLMIGYLGYHYKT 203
+ ++ Y+ +YL A +++G S+F+L S T+ G++L+ GY+ + T
Sbjct: 291 VSRRSYEHWEYLTAGLISIGVSMFLLSSGPEPRS----SPATTLSGLVLLAGYIAFDSFT 346
Query: 204 ----QSLFAFRLFSL 214
+LFA+++ S+
Sbjct: 347 SNWQDALFAYKMSSV 361
>gb|AAH24288.1| Solute carrier family 35, member B2 [Homo sapiens]
gi|32480471|dbj|BAC79117.1| adenosine 3'-phospho
5'-phosphosulfate (PAPS) transporter [Homo sapiens]
gi|67461576|sp|Q8TB61|S35B2_HUMAN Adenosine 3'-phospho
5'-phosphosulfate transporter 1 (PAPS transporter 1)
(Solute carrier family 35, member B2) (Putative
NF-kappa-B activating protein PM15)
gi|30026034|ref|NP_835361.1| solute carrier family 35,
member B2 [Homo sapiens]
Length = 432
Score = 122 bits (306), Expect = 1e-26
Identities = 74/207 (35%), Positives = 120/207 (57%), Gaps = 15/207 (7%)
Query: 28 KVAFAVSGIMLTLVTYGVLQEKIMRVPYGVNK----DYFKYSLFLVFCNRITTSAVSAAA 83
K+ F +G+ ++ +T+GVLQE++M YG + F S FLV NR+ V+ +
Sbjct: 111 KLLFCATGLQVSYLTWGVLQERVMTRSYGATATSPGERFTDSQFLVLMNRVLALIVAGLS 170
Query: 84 LVARDKALLPVAPIYKYCLVSVSNILTTTCQYEALKYVSFPVQTLAKCAKMIPVMIWSAL 143
V K AP+Y+Y S+SN+L++ CQYEALK+VSFP Q LAK +K+IPVM+ L
Sbjct: 171 CVLC-KQPRHGAPMYRYSFASLSNVLSSWCQYEALKFVSFPTQVLAKASKVIPVMLMGKL 229
Query: 144 IMQKKYQGTDYLLAFSVTLGCSIFILYPAGTDISPYGRGRENTIWGILLMIGYLGYHYKT 203
+ ++ Y+ +YL A +++G S+F+L S T+ G++L+ GY+ + T
Sbjct: 230 VSRRSYEHWEYLTATLISIGVSMFLLSSGPEPRS----SPATTLSGLILLAGYIAFDSFT 285
Query: 204 ----QSLFAFRLFS--LAYFVSFLSSL 224
+LFA+++ S + + V+F S L
Sbjct: 286 SNWQDALFAYKMSSVQMMFGVNFFSCL 312
>emb|CAI20101.1| OTTHUMP00000016513 [Homo sapiens]
Length = 480
Score = 122 bits (306), Expect = 1e-26
Identities = 74/207 (35%), Positives = 120/207 (57%), Gaps = 15/207 (7%)
Query: 28 KVAFAVSGIMLTLVTYGVLQEKIMRVPYGVNK----DYFKYSLFLVFCNRITTSAVSAAA 83
K+ F +G+ ++ +T+GVLQE++M YG + F S FLV NR+ V+ +
Sbjct: 159 KLLFCATGLQVSYLTWGVLQERVMTRSYGATATSPGERFTDSQFLVLMNRVLALIVAGLS 218
Query: 84 LVARDKALLPVAPIYKYCLVSVSNILTTTCQYEALKYVSFPVQTLAKCAKMIPVMIWSAL 143
V K AP+Y+Y S+SN+L++ CQYEALK+VSFP Q LAK +K+IPVM+ L
Sbjct: 219 CVLC-KQPRHGAPMYRYSFASLSNVLSSWCQYEALKFVSFPTQVLAKASKVIPVMLMGKL 277
Query: 144 IMQKKYQGTDYLLAFSVTLGCSIFILYPAGTDISPYGRGRENTIWGILLMIGYLGYHYKT 203
+ ++ Y+ +YL A +++G S+F+L S T+ G++L+ GY+ + T
Sbjct: 278 VSRRSYEHWEYLTATLISIGVSMFLLSSGPEPRS----SPATTLSGLILLAGYIAFDSFT 333
Query: 204 ----QSLFAFRLFS--LAYFVSFLSSL 224
+LFA+++ S + + V+F S L
Sbjct: 334 SNWQDALFAYKMSSVQMMFGVNFFSCL 360
>dbj|BAC11631.1| unnamed protein product [Homo sapiens]
Length = 432
Score = 122 bits (306), Expect = 1e-26
Identities = 74/207 (35%), Positives = 120/207 (57%), Gaps = 15/207 (7%)
Query: 28 KVAFAVSGIMLTLVTYGVLQEKIMRVPYGVNK----DYFKYSLFLVFCNRITTSAVSAAA 83
K+ F +G+ ++ +T+GVLQE++M YG + F S FLV NR+ V+ +
Sbjct: 111 KLLFCATGLQVSYLTWGVLQERVMTRSYGATATSPGERFTDSQFLVLMNRVLALIVAGLS 170
Query: 84 LVARDKALLPVAPIYKYCLVSVSNILTTTCQYEALKYVSFPVQTLAKCAKMIPVMIWSAL 143
V K AP+Y+Y S+SN+L++ CQYEALK+VSFP Q LAK +K+IPVM+ L
Sbjct: 171 CVLC-KQPRHGAPMYRYSFASLSNVLSSWCQYEALKFVSFPTQVLAKASKVIPVMLMGKL 229
Query: 144 IMQKKYQGTDYLLAFSVTLGCSIFILYPAGTDISPYGRGRENTIWGILLMIGYLGYHYKT 203
+ ++ Y+ +YL A +++G S+F+L S T+ G++L+ GY+ + T
Sbjct: 230 VSRRSYEHWEYLTATLISIGVSMFLLSSGPEPRS----SPATTLSGLILLAGYIAFDSFT 285
Query: 204 ----QSLFAFRLFS--LAYFVSFLSSL 224
+LFA+++ S + + V+F S L
Sbjct: 286 SNWQDALFAYKMSSVQMMFGVNFFSCL 312
Database: nr
Posted date: Jul 5, 2005 12:34 AM
Number of letters in database: 863,360,394
Number of sequences in database: 2,540,612
Lambda K H
0.334 0.143 0.434
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 409,610,630
Number of Sequences: 2540612
Number of extensions: 15343011
Number of successful extensions: 63089
Number of sequences better than 10.0: 120
Number of HSP's better than 10.0 without gapping: 109
Number of HSP's successfully gapped in prelim test: 11
Number of HSP's that attempted gapping in prelim test: 62851
Number of HSP's gapped (non-prelim): 133
length of query: 285
length of database: 863,360,394
effective HSP length: 126
effective length of query: 159
effective length of database: 543,243,282
effective search space: 86375681838
effective search space used: 86375681838
T: 11
A: 40
X1: 15 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 39 (21.6 bits)
S2: 74 (33.1 bits)
Medicago: description of AC139841.2


