

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC139841.14 + phase: 0 /pseudo
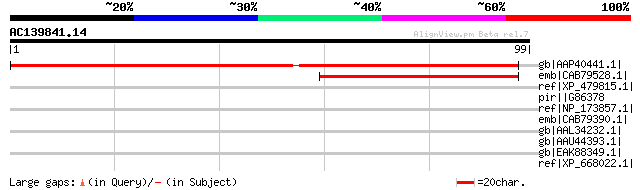
(99 letters)
Database: nr
2,540,612 sequences; 863,360,394 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
gb|AAP40441.1| unknown protein [Arabidopsis thaliana] gi|4678200... 100 7e-21
emb|CAB79528.1| putative protein [Arabidopsis thaliana] gi|44551... 45 6e-04
ref|XP_479815.1| hypothetical protein [Oryza sativa (japonica cu... 40 0.019
pir||G86378 protein F21J9.14 [imported] - Arabidopsis thaliana g... 36 0.21
ref|NP_173857.1| hypothetical protein [Arabidopsis thaliana] 35 0.35
emb|CAB79390.1| putative protein [Arabidopsis thaliana] gi|46782... 35 0.60
gb|AAL34232.1| unknown protein [Arabidopsis thaliana] gi|1433484... 33 1.7
gb|AAU44393.1| hypothetical protein AT1G24480 [Arabidopsis thali... 31 8.6
gb|EAK88349.1| beta transducin ortholog with WD40 repeats [Crypt... 31 8.6
ref|XP_668022.1| WD-repeat membrane protein [Cryptosporidium hom... 31 8.6
>gb|AAP40441.1| unknown protein [Arabidopsis thaliana] gi|4678200|gb|AAD26946.1|
hypothetical protein [Arabidopsis thaliana]
gi|25411695|pir||H84535 hypothetical protein At2g16030
[imported] - Arabidopsis thaliana
gi|15226670|ref|NP_179201.1| expressed protein
[Arabidopsis thaliana]
Length = 231
Score = 100 bits (250), Expect = 7e-21
Identities = 46/97 (47%), Positives = 68/97 (69%), Gaps = 1/97 (1%)
Query: 1 MEKEIETFLRKLSLVAIAIASLTLLILFLQIPNTRVPSEAPSKPHLRFPKSTCDFTSTHL 60
ME+ +E L+++S+V ++I ++ ++I+ LQ P T + EAPSKPH FP+STCD +S
Sbjct: 1 MERNVEKMLKRVSIVFLSIGTVLMVIMILQTPKTCISPEAPSKPHTHFPRSTCD-SSPRQ 59
Query: 61 HLPSHKKNNRLWSSRDWNNKLHSFSRLFLHIRNLGLL 97
HLP KKN R+WSS W ++L+SFS FL R+LG +
Sbjct: 60 HLPLPKKNARIWSSGAWKSRLYSFSTYFLRFRDLGFI 96
>emb|CAB79528.1| putative protein [Arabidopsis thaliana]
gi|4455196|emb|CAB36519.1| putative protein
[Arabidopsis thaliana] gi|15236844|ref|NP_194403.1|
hypothetical protein [Arabidopsis thaliana]
gi|7485347|pir||T04796 hypothetical protein F10M23.70 -
Arabidopsis thaliana
Length = 208
Score = 44.7 bits (104), Expect = 6e-04
Identities = 19/38 (50%), Positives = 25/38 (65%)
Query: 60 LHLPSHKKNNRLWSSRDWNNKLHSFSRLFLHIRNLGLL 97
+HLP KKN R+W S W ++L SFS FL R+LG +
Sbjct: 24 MHLPLPKKNARIWFSGAWKSRLSSFSTYFLRFRDLGFI 61
>ref|XP_479815.1| hypothetical protein [Oryza sativa (japonica cultivar-group)]
gi|42407911|dbj|BAD09051.1| hypothetical protein [Oryza
sativa (japonica cultivar-group)]
Length = 260
Score = 39.7 bits (91), Expect = 0.019
Identities = 25/88 (28%), Positives = 43/88 (48%), Gaps = 12/88 (13%)
Query: 13 SLVAIAIASLTLLILFLQIPNTRVP--------SEAPSKPHLRFPK----STCDFTSTHL 60
SL+ + I L+LL+L+ + P S P PH+R + S D+ L
Sbjct: 17 SLLLLLIPFLSLLLLYSYSSSPPPPPVAFAVPLSPTPPSPHIRMRRAGFRSYEDYLRHQL 76
Query: 61 HLPSHKKNNRLWSSRDWNNKLHSFSRLF 88
+ + R+W++RDW+ K+ +F+R F
Sbjct: 77 NKTLDPRLRRVWATRDWHRKVDAFARAF 104
>pir||G86378 protein F21J9.14 [imported] - Arabidopsis thaliana
gi|9743354|gb|AAF97978.1| F21J9.14 [Arabidopsis
thaliana]
Length = 239
Score = 36.2 bits (82), Expect = 0.21
Identities = 24/87 (27%), Positives = 38/87 (43%), Gaps = 6/87 (6%)
Query: 19 IASLTLLILFLQIPNTRVPSEAPSKPHLRFPKSTCDFTS------THLHLPSHKKNNRLW 72
I L+ +FL +P S KP + +TS L+ + + +W
Sbjct: 7 ILKYVLVSIFLTLPLILFFSIQLRKPEKELIRIRPGYTSYDYYIQRQLNKTLNPRLRTIW 66
Query: 73 SSRDWNNKLHSFSRLFLHIRNLGLLPK 99
+RDW+ K+ FSR F ++ GLL K
Sbjct: 67 MTRDWDRKIKVFSRFFQDLKRQGLLSK 93
>ref|NP_173857.1| hypothetical protein [Arabidopsis thaliana]
Length = 239
Score = 35.4 bits (80), Expect = 0.35
Identities = 25/91 (27%), Positives = 38/91 (41%), Gaps = 8/91 (8%)
Query: 12 LSLVAIAIASLTLLILFLQIPNTRVPSEAPSKPHLRFPKSTCDFT---STHLHLPSHKKN 68
L V ++I LILF I P K +R + L+ + +
Sbjct: 8 LKYVLVSIFLTLPLILFFSIQ-----LRTPEKELIRIRPGYTSYDYYIQRQLNKTLNPRF 62
Query: 69 NRLWSSRDWNNKLHSFSRLFLHIRNLGLLPK 99
+W +RDW+ K+ FSR F ++ GLL K
Sbjct: 63 RTIWMTRDWDRKIKVFSRFFQDLKRQGLLSK 93
>emb|CAB79390.1| putative protein [Arabidopsis thaliana] gi|4678259|emb|CAB41120.1|
putative protein [Arabidopsis thaliana]
gi|7486528|pir||T06664 hypothetical protein F6I7.10 -
Arabidopsis thaliana
Length = 942
Score = 34.7 bits (78), Expect = 0.60
Identities = 29/110 (26%), Positives = 47/110 (42%), Gaps = 13/110 (11%)
Query: 1 MEKEIETFLRKLSLVAIAIASLTLLILFL-QIPNT----------RVPSEAPSKPHLRFP 49
+E ++ + KL SL+ LI+FL P T V S A +R
Sbjct: 692 LEANMKFVIPKLPFTLSLFLSLSFLIIFLFAFPTTFLRRPLSSSSSVTSVADEGIRIRHH 751
Query: 50 --KSTCDFTSTHLHLPSHKKNNRLWSSRDWNNKLHSFSRLFLHIRNLGLL 97
S + L+ + K ++W++RDW+ K+ FS F + + GLL
Sbjct: 752 GYSSYEAYIKHQLNKTQNPKLRKVWTTRDWDRKVRVFSTFFRRLSDRGLL 801
>gb|AAL34232.1| unknown protein [Arabidopsis thaliana] gi|14334844|gb|AAK59600.1|
unknown protein [Arabidopsis thaliana]
gi|22328908|ref|NP_680738.1| expressed protein
[Arabidopsis thaliana]
Length = 247
Score = 33.1 bits (74), Expect = 1.7
Identities = 13/38 (34%), Positives = 23/38 (60%)
Query: 60 LHLPSHKKNNRLWSSRDWNNKLHSFSRLFLHIRNLGLL 97
L+ + K ++W++RDW+ K+ FS F + + GLL
Sbjct: 69 LNKTQNPKLRKVWTTRDWDRKVRVFSTFFRRLSDRGLL 106
>gb|AAU44393.1| hypothetical protein AT1G24480 [Arabidopsis thaliana]
Length = 173
Score = 30.8 bits (68), Expect = 8.6
Identities = 12/26 (46%), Positives = 17/26 (65%)
Query: 74 SRDWNNKLHSFSRLFLHIRNLGLLPK 99
+RDW+ K+ FSR F ++ GLL K
Sbjct: 2 TRDWDRKIKXFSRFFQDLKRQGLLSK 27
>gb|EAK88349.1| beta transducin ortholog with WD40 repeats [Cryptosporidium parvum]
gi|66362288|ref|XP_628108.1| beta transducin ortholog
with WD40 repeats [Cryptosporidium parvum]
Length = 981
Score = 30.8 bits (68), Expect = 8.6
Identities = 20/67 (29%), Positives = 33/67 (48%), Gaps = 3/67 (4%)
Query: 12 LSLVAIAIASLTLLILFLQIPNTRVPSEAPSKPHLR--FPKSTCDFTSTHLHLPSHKKNN 69
+S I ++L ++ +I P E P KP F +T +F+ST +H PS + +N
Sbjct: 770 MSFSGIPFSTLQAILFIDEIKERNKPIEPPKKPESAPFFLPNTTEFSST-IHKPSKESDN 828
Query: 70 RLWSSRD 76
S+D
Sbjct: 829 EEIISKD 835
>ref|XP_668022.1| WD-repeat membrane protein [Cryptosporidium hominis]
gi|54659199|gb|EAL37788.1| WD-repeat membrane protein
[Cryptosporidium hominis]
Length = 981
Score = 30.8 bits (68), Expect = 8.6
Identities = 20/67 (29%), Positives = 33/67 (48%), Gaps = 3/67 (4%)
Query: 12 LSLVAIAIASLTLLILFLQIPNTRVPSEAPSKPHLR--FPKSTCDFTSTHLHLPSHKKNN 69
+S I ++L ++ +I P E P KP F +T +F+ST +H PS + +N
Sbjct: 770 MSFSGIPFSTLQAILFIDEIKERNKPIEPPKKPESAPFFLPNTSEFSST-IHKPSKESDN 828
Query: 70 RLWSSRD 76
S+D
Sbjct: 829 EEVISKD 835
Database: nr
Posted date: Jul 5, 2005 12:34 AM
Number of letters in database: 863,360,394
Number of sequences in database: 2,540,612
Lambda K H
0.323 0.136 0.416
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 173,870,738
Number of Sequences: 2540612
Number of extensions: 6857459
Number of successful extensions: 14192
Number of sequences better than 10.0: 10
Number of HSP's better than 10.0 without gapping: 7
Number of HSP's successfully gapped in prelim test: 3
Number of HSP's that attempted gapping in prelim test: 14184
Number of HSP's gapped (non-prelim): 10
length of query: 99
length of database: 863,360,394
effective HSP length: 75
effective length of query: 24
effective length of database: 672,814,494
effective search space: 16147547856
effective search space used: 16147547856
T: 11
A: 40
X1: 16 ( 7.5 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (22.0 bits)
S2: 68 (30.8 bits)
Medicago: description of AC139841.14


