

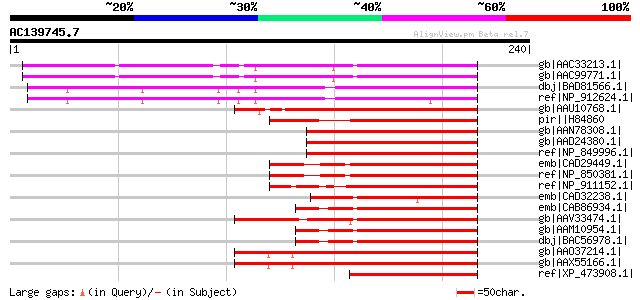
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC139745.7 + phase: 0
(240 letters)
Database: nr
2,540,612 sequences; 863,360,394 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
gb|AAC33213.1| Unknown protein [Arabidopsis thaliana] gi|2644960... 111 2e-23
gb|AAC99771.1| phytochrome-associated protein 3 [Arabidopsis tha... 110 4e-23
dbj|BAD81566.1| putative BP-5 protein [Oryza sativa (japonica cu... 108 2e-22
ref|NP_912624.1| P0498A12.24 [Oryza sativa (japonica cultivar-gr... 101 2e-20
gb|AAU10768.1| hypothetical protein [Oryza sativa (japonica cult... 99 8e-20
pir||H84860 hypothetical protein At2g43010 [imported] - Arabidop... 96 1e-18
gb|AAN78308.1| putative bHLH transcription factor [Arabidopsis t... 93 6e-18
gb|AAD24380.1| unknown protein [Arabidopsis thaliana] gi|2541196... 93 6e-18
ref|NP_849996.1| basic helix-loop-helix (bHLH) family protein [A... 93 6e-18
emb|CAD29449.1| phytochrome interacting factor 4 [Arabidopsis th... 93 7e-18
ref|NP_850381.1| phytochrome-interacting factor 4 (PIF4) / basic... 93 7e-18
ref|NP_911152.1| transcription factor BHLH9-like protein [Oryza ... 92 1e-17
emb|CAD32238.1| BP-5 protein [Oryza sativa] 91 3e-17
emb|CAB86934.1| putative protein [Arabidopsis thaliana] gi|30694... 91 4e-17
gb|AAV33474.1| basic helix-loop-helix protein [Fragaria x ananassa] 91 4e-17
gb|AAM10954.1| putative bHLH transcription factor [Arabidopsis t... 91 4e-17
dbj|BAC56978.1| PIF3 like basic Helix Loop Helix protein [Arabid... 91 4e-17
gb|AAO37214.1| hypothetical protein [Arabidopsis thaliana] 89 8e-17
gb|AAX55166.1| hypothetical protein At2g46970 [Arabidopsis thali... 89 8e-17
ref|XP_473908.1| OSJNBa0058K23.6 [Oryza sativa (japonica cultiva... 88 2e-16
>gb|AAC33213.1| Unknown protein [Arabidopsis thaliana] gi|26449609|dbj|BAC41930.1|
putative transcription factor BHLH8 [Arabidopsis
thaliana] gi|18026964|gb|AAL55715.1| putative
transcription factor BHLH8 [Arabidopsis thaliana]
gi|30681206|ref|NP_849626.1| phytochrome interacting
factor 3 (PIF3) [Arabidopsis thaliana]
gi|15217533|ref|NP_172424.1| phytochrome interacting
factor 3 (PIF3) [Arabidopsis thaliana]
gi|3929586|gb|AAC95156.1| phytochrome interacting factor
3 [Arabidopsis thaliana] gi|25402552|pir||H86228
hypothetical protein [imported] - Arabidopsis thaliana
gi|20532207|sp|O80536|PIF3_ARATH Phytochrome-interacting
factor 3 (Phytochrome-associated protein 3) (Basic
helix-loop-helix protein 8) (bHLH8) (AtbHLH008)
Length = 524
Score = 111 bits (278), Expect = 2e-23
Identities = 79/212 (37%), Positives = 116/212 (54%), Gaps = 9/212 (4%)
Query: 7 KNGKVNFSNFSIPAVFLKSNQQRVQEKESKAVACTKFQQQKHLTLNAATKSNSGYERSRE 66
K VNFS+F PA F K+ + + + K+ Q + L A + S
Sbjct: 194 KPSLVNFSHFLRPATFAKTTNNNLHDTKEKSPQSPPNVFQTRV-LGAKDSEDKVLNESVA 252
Query: 67 LLPTVDEHSEAAASHTNAPRIRGKRKASAINLCNAQKPSSVCSLEA-SNDLNFGVRKSHE 125
D S + + + KA +C++ + SL+ S + +++ H
Sbjct: 253 SATPKDNQKACLISEDSCRKDQESEKAV---VCSSVGSGN--SLDGPSESPSLSLKRKHS 307
Query: 126 DTDDSPYLSDNDEETQENIVKEK-PVREGNRVKRSYRNAKVHNLSERKRRDKINEKIRAL 184
+ D S++ EE + KE P R G KRS R+A+VHNLSER+RRD+INEK+RAL
Sbjct: 308 NIQDIDCHSEDVEEESGDGRKEAGPSRTGLGSKRS-RSAEVHNLSERRRRDRINEKMRAL 366
Query: 185 KELIPNCNKMDKASMLDDAIDYLKTLKLQLQV 216
+ELIPNCNK+DKASMLD+AI+YLK+L+LQ+Q+
Sbjct: 367 QELIPNCNKVDKASMLDEAIEYLKSLQLQVQI 398
>gb|AAC99771.1| phytochrome-associated protein 3 [Arabidopsis thaliana]
Length = 524
Score = 110 bits (274), Expect = 4e-23
Identities = 79/212 (37%), Positives = 115/212 (53%), Gaps = 9/212 (4%)
Query: 7 KNGKVNFSNFSIPAVFLKSNQQRVQEKESKAVACTKFQQQKHLTLNAATKSNSGYERSRE 66
K VNFS+F PA F K+ + + + K+ Q + L A + S
Sbjct: 194 KPSLVNFSHFLRPATFAKTTNNNLHDTKEKSPQSPPNVFQTRV-LGAKDSEDKVLNESVA 252
Query: 67 LLPTVDEHSEAAASHTNAPRIRGKRKASAINLCNAQKPSSVCSLEA-SNDLNFGVRKSHE 125
D S + + + KA +C++ + SL+ S + +++ H
Sbjct: 253 SATPKDNQKACLISEDSCRKDQESEKAV---VCSSVGSGN--SLDGPSESPSLSLKRKHS 307
Query: 126 DTDDSPYLSDNDEETQENIVKEK-PVREGNRVKRSYRNAKVHNLSERKRRDKINEKIRAL 184
+ D S++ EE + KE P R G KRS R A+VHNLSER+RRD+INEK+RAL
Sbjct: 308 NIQDIDCHSEDVEEESGDGRKEAGPSRTGLGSKRS-RLAEVHNLSERRRRDRINEKMRAL 366
Query: 185 KELIPNCNKMDKASMLDDAIDYLKTLKLQLQV 216
+ELIPNCNK+DKASMLD+AI+YLK+L+LQ+Q+
Sbjct: 367 QELIPNCNKVDKASMLDEAIEYLKSLQLQVQI 398
>dbj|BAD81566.1| putative BP-5 protein [Oryza sativa (japonica cultivar-group)]
Length = 565
Score = 108 bits (269), Expect = 2e-22
Identities = 79/233 (33%), Positives = 123/233 (51%), Gaps = 29/233 (12%)
Query: 9 GKVNFSNFSIPAVFLKS---NQQRVQEKESKAVACTKFQQQKHLTLNAATKSNSG----- 60
G +NFS FS PAV ++ + QR Q ++KA T + + + A+ S
Sbjct: 139 GVMNFSLFSRPAVLARATLESAQRTQGTDNKASNVTASNRVESTVVQTASGPRSAPAFAD 198
Query: 61 -----YERSRELLPTVDEHSEAAASHTNAPRIRGKRKASA---INLCNAQKP-------S 105
+ + +P + A N G+ +A ++ A+K S
Sbjct: 199 QRAAAWPPQPKEMPFASTAAAPMAPAVNLHHEMGRDRAGRTMPVHKTEARKAPEATVATS 258
Query: 106 SVCSLEA--SNDLNFGVRKSHEDTDDSPYLSDNDEETQENIVKEKPVREGNRVKRSYRNA 163
SVCS S++L ++ + + D+D + + +++ + G R + R A
Sbjct: 259 SVCSGNGAGSDELWRQQKRKCQAQAECSASQDDDLDDEPGVLR----KSGTRSTKRSRTA 314
Query: 164 KVHNLSERKRRDKINEKIRALKELIPNCNKMDKASMLDDAIDYLKTLKLQLQV 216
+VHNLSER+RRD+INEK+RAL+ELIPNCNK+DKASMLD+AI+YLKTL+LQ+Q+
Sbjct: 315 EVHNLSERRRRDRINEKMRALQELIPNCNKIDKASMLDEAIEYLKTLQLQVQM 367
>ref|NP_912624.1| P0498A12.24 [Oryza sativa (japonica cultivar-group)]
Length = 895
Score = 101 bits (252), Expect = 2e-20
Identities = 79/239 (33%), Positives = 123/239 (51%), Gaps = 35/239 (14%)
Query: 9 GKVNFSNFSIPAVFLKS---NQQRVQEKESKAVACTKFQQQKHLTLNAATKSNSG----- 60
G +NFS FS PAV ++ + QR Q ++KA T + + + A+ S
Sbjct: 475 GVMNFSLFSRPAVLARATLESAQRTQGTDNKASNVTASNRVESTVVQTASGPRSAPAFAD 534
Query: 61 -----YERSRELLPTVDEHSEAAASHTNAPRIRGKRKASA---INLCNAQKP-------S 105
+ + +P + A N G+ +A ++ A+K S
Sbjct: 535 QRAAAWPPQPKEMPFASTAAAPMAPAVNLHHEMGRDRAGRTMPVHKTEARKAPEATVATS 594
Query: 106 SVCSLEA--SNDLNFGVRKSHEDTDDSPYLSDNDEETQENIVKEKPVREGNRVKRSYRNA 163
SVCS S++L ++ + + D+D + + +++ + G R + R A
Sbjct: 595 SVCSGNGAGSDELWRQQKRKCQAQAECSASQDDDLDDEPGVLR----KSGTRSTKRSRTA 650
Query: 164 KVHNLSERKRRDKINEKIRALKELIPNCNK------MDKASMLDDAIDYLKTLKLQLQV 216
+VHNLSER+RRD+INEK+RAL+ELIPNCNK +DKASMLD+AI+YLKTL+LQ+Q+
Sbjct: 651 EVHNLSERRRRDRINEKMRALQELIPNCNKACYVLQIDKASMLDEAIEYLKTLQLQVQM 709
>gb|AAU10768.1| hypothetical protein [Oryza sativa (japonica cultivar-group)]
gi|50511445|gb|AAT77368.1| hypothetical protein [Oryza
sativa (japonica cultivar-group)]
Length = 505
Score = 99.4 bits (246), Expect = 8e-20
Identities = 57/113 (50%), Positives = 78/113 (68%), Gaps = 4/113 (3%)
Query: 105 SSVCSLEASN-DLNFGVRKSHEDTDDSPYLSDNDEETQENIVKEKPVREGNRVKRSYRNA 163
SSVCS LN+ R SH + + + + DE ++ + R R + R A
Sbjct: 281 SSVCSGNGDRRQLNW--RDSHNN-QSAEWSASQDELDLDDELAGVHRRSAARSSKRSRTA 337
Query: 164 KVHNLSERKRRDKINEKIRALKELIPNCNKMDKASMLDDAIDYLKTLKLQLQV 216
+VHNLSER+RRD+INEK+RAL+ELIPNCNK+DKASML++AI+YLKTL+LQ+Q+
Sbjct: 338 EVHNLSERRRRDRINEKMRALQELIPNCNKIDKASMLEEAIEYLKTLQLQVQM 390
>pir||H84860 hypothetical protein At2g43010 [imported] - Arabidopsis thaliana
Length = 423
Score = 95.5 bits (236), Expect = 1e-18
Identities = 51/96 (53%), Positives = 69/96 (71%), Gaps = 14/96 (14%)
Query: 121 RKSHEDTDDSPYLSDNDEETQENIVKEKPVREGNRVKRSYRNAKVHNLSERKRRDKINEK 180
RK TD+S LSD+++ + N R R A+VHNLSER+RRD+INE+
Sbjct: 226 RKRINHTDESVSLSDSNQRSGSN--------------RRSRAAEVHNLSERRRRDRINER 271
Query: 181 IRALKELIPNCNKMDKASMLDDAIDYLKTLKLQLQV 216
++AL+ELIP+C+K DKAS+LD+AIDYLK+L+LQLQV
Sbjct: 272 MKALQELIPHCSKTDKASILDEAIDYLKSLQLQLQV 307
>gb|AAN78308.1| putative bHLH transcription factor [Arabidopsis thaliana]
gi|28372351|dbj|BAC56979.1| PIF3 like basic Helix Loop
Helix protein [Arabidopsis thaliana]
gi|30680909|ref|NP_179608.2| basic helix-loop-helix
(bHLH) family protein [Arabidopsis thaliana]
Length = 478
Score = 93.2 bits (230), Expect = 6e-18
Identities = 44/79 (55%), Positives = 61/79 (76%)
Query: 138 EETQENIVKEKPVREGNRVKRSYRNAKVHNLSERKRRDKINEKIRALKELIPNCNKMDKA 197
+ET+ + K R + R A+VHNLSERKRRD+INE+++AL+ELIP CNK DKA
Sbjct: 261 DETESRSEETKQARVSTTSTKRSRAAEVHNLSERKRRDRINERMKALQELIPRCNKSDKA 320
Query: 198 SMLDDAIDYLKTLKLQLQV 216
SMLD+AI+Y+K+L+LQ+Q+
Sbjct: 321 SMLDEAIEYMKSLQLQIQM 339
>gb|AAD24380.1| unknown protein [Arabidopsis thaliana] gi|25411969|pir||A84586
hypothetical protein At2g20180 [imported] - Arabidopsis
thaliana
Length = 490
Score = 93.2 bits (230), Expect = 6e-18
Identities = 44/79 (55%), Positives = 61/79 (76%)
Query: 138 EETQENIVKEKPVREGNRVKRSYRNAKVHNLSERKRRDKINEKIRALKELIPNCNKMDKA 197
+ET+ + K R + R A+VHNLSERKRRD+INE+++AL+ELIP CNK DKA
Sbjct: 261 DETESRSEETKQARVSTTSTKRSRAAEVHNLSERKRRDRINERMKALQELIPRCNKSDKA 320
Query: 198 SMLDDAIDYLKTLKLQLQV 216
SMLD+AI+Y+K+L+LQ+Q+
Sbjct: 321 SMLDEAIEYMKSLQLQIQM 339
>ref|NP_849996.1| basic helix-loop-helix (bHLH) family protein [Arabidopsis thaliana]
Length = 407
Score = 93.2 bits (230), Expect = 6e-18
Identities = 44/79 (55%), Positives = 61/79 (76%)
Query: 138 EETQENIVKEKPVREGNRVKRSYRNAKVHNLSERKRRDKINEKIRALKELIPNCNKMDKA 197
+ET+ + K R + R A+VHNLSERKRRD+INE+++AL+ELIP CNK DKA
Sbjct: 190 DETESRSEETKQARVSTTSTKRSRAAEVHNLSERKRRDRINERMKALQELIPRCNKSDKA 249
Query: 198 SMLDDAIDYLKTLKLQLQV 216
SMLD+AI+Y+K+L+LQ+Q+
Sbjct: 250 SMLDEAIEYMKSLQLQIQM 268
>emb|CAD29449.1| phytochrome interacting factor 4 [Arabidopsis thaliana]
gi|18026966|gb|AAL55716.1| putative transcription factor
BHLH9 [Arabidopsis thaliana]
gi|28201855|sp|Q8W2F3|PIF4_ARATH Phytochrome-interacting
factor 4 (Basic helix-loop-helix protein 9) (bHLH9)
(AtbHLH009) (Short under red-light 2)
gi|30689218|ref|NP_565991.2| phytochrome-interacting
factor 4 (PIF4) / basic helix-loop-helix protein 9
(bHLH9) / short under red-light 2 (SRL2) [Arabidopsis
thaliana]
Length = 430
Score = 92.8 bits (229), Expect = 7e-18
Identities = 53/96 (55%), Positives = 69/96 (71%), Gaps = 9/96 (9%)
Query: 121 RKSHEDTDDSPYLSDNDEETQENIVKEKPVREGNRVKRSYRNAKVHNLSERKRRDKINEK 180
RK TD+S LSD I + R G+ R R A+VHNLSER+RRD+INE+
Sbjct: 226 RKRINHTDESVSLSDA-------IGNKSNQRSGSN--RRSRAAEVHNLSERRRRDRINER 276
Query: 181 IRALKELIPNCNKMDKASMLDDAIDYLKTLKLQLQV 216
++AL+ELIP+C+K DKAS+LD+AIDYLK+L+LQLQV
Sbjct: 277 MKALQELIPHCSKTDKASILDEAIDYLKSLQLQLQV 312
>ref|NP_850381.1| phytochrome-interacting factor 4 (PIF4) / basic helix-loop-helix
protein 9 (bHLH9) / short under red-light 2 (SRL2)
[Arabidopsis thaliana]
Length = 428
Score = 92.8 bits (229), Expect = 7e-18
Identities = 53/96 (55%), Positives = 69/96 (71%), Gaps = 9/96 (9%)
Query: 121 RKSHEDTDDSPYLSDNDEETQENIVKEKPVREGNRVKRSYRNAKVHNLSERKRRDKINEK 180
RK TD+S LSD I + R G+ R R A+VHNLSER+RRD+INE+
Sbjct: 226 RKRINHTDESVSLSDA-------IGNKSNQRSGSN--RRSRAAEVHNLSERRRRDRINER 276
Query: 181 IRALKELIPNCNKMDKASMLDDAIDYLKTLKLQLQV 216
++AL+ELIP+C+K DKAS+LD+AIDYLK+L+LQLQV
Sbjct: 277 MKALQELIPHCSKTDKASILDEAIDYLKSLQLQLQV 312
>ref|NP_911152.1| transcription factor BHLH9-like protein [Oryza sativa (japonica
cultivar-group)] gi|24059945|dbj|BAC21408.1|
transcription factor BHLH9-like protein [Oryza sativa
(japonica cultivar-group)] gi|23495742|dbj|BAC19953.1|
transcription factor BHLH9-like protein [Oryza sativa
(japonica cultivar-group)]
Length = 417
Score = 92.4 bits (228), Expect = 1e-17
Identities = 49/96 (51%), Positives = 73/96 (76%), Gaps = 10/96 (10%)
Query: 121 RKSHEDTDDSPYLSDNDEETQENIVKEKPVREGNRVKRSYRNAKVHNLSERKRRDKINEK 180
++ E+ DS LS+ +ET+ + ++P KR R A+VHNLSER+RRD+INEK
Sbjct: 198 KRGREELVDS--LSEVADETRPS---KRPA-----AKRRTRAAEVHNLSERRRRDRINEK 247
Query: 181 IRALKELIPNCNKMDKASMLDDAIDYLKTLKLQLQV 216
+RAL+EL+P+CNK DKAS+LD+AI+YLK+L++Q+Q+
Sbjct: 248 LRALQELVPHCNKTDKASILDEAIEYLKSLQMQVQI 283
>emb|CAD32238.1| BP-5 protein [Oryza sativa]
Length = 335
Score = 90.9 bits (224), Expect = 3e-17
Identities = 48/79 (60%), Positives = 64/79 (80%), Gaps = 3/79 (3%)
Query: 140 TQENIVKEKPVREGNRVKRSYRNAKVHNLSERKRRDKINEKIRALKEL--IPNCNKMDKA 197
++ +V KP + +RS R A+VHNLSER+RRD+INEK+RAL+EL IP+CNK DKA
Sbjct: 146 SRSRLVARKPPAKMTTARRS-RAAEVHNLSERRRRDRINEKMRALQELELIPHCNKTDKA 204
Query: 198 SMLDDAIDYLKTLKLQLQV 216
SMLD+AI+YLK+L+LQL+V
Sbjct: 205 SMLDEAIEYLKSLQLQLRV 223
>emb|CAB86934.1| putative protein [Arabidopsis thaliana]
gi|30694919|ref|NP_851021.1| basic helix-loop-helix
(bHLH) family protein [Arabidopsis thaliana]
gi|11357514|pir||T47788 hypothetical protein F17J16.110
- Arabidopsis thaliana
Length = 442
Score = 90.5 bits (223), Expect = 4e-17
Identities = 46/84 (54%), Positives = 66/84 (77%), Gaps = 5/84 (5%)
Query: 133 LSDNDEETQENIVKEKPVREGNRVKRSYRNAKVHNLSERKRRDKINEKIRALKELIPNCN 192
L+ D++T N K + +RS R A+VHNLSER+RRD+INE+++AL+ELIP+C+
Sbjct: 233 LTSTDDQTMGN----KSSQRSGSTRRS-RAAEVHNLSERRRRDRINERMKALQELIPHCS 287
Query: 193 KMDKASMLDDAIDYLKTLKLQLQV 216
+ DKAS+LD+AIDYLK+L++QLQV
Sbjct: 288 RTDKASILDEAIDYLKSLQMQLQV 311
>gb|AAV33474.1| basic helix-loop-helix protein [Fragaria x ananassa]
Length = 298
Score = 90.5 bits (223), Expect = 4e-17
Identities = 55/114 (48%), Positives = 76/114 (66%), Gaps = 6/114 (5%)
Query: 105 SSVCSLEASNDLNFGVRKSHEDTDDSPYLSDNDEETQENIVKEKPVREGNRV--KRSYRN 162
++V + N + V S + D+ S EE E +V+E V+ G R KRS R
Sbjct: 90 TAVVTASGPNVSSSSVGASENEADEYDCES---EEGLEALVEEAAVKSGGRSSSKRS-RA 145
Query: 163 AKVHNLSERKRRDKINEKIRALKELIPNCNKMDKASMLDDAIDYLKTLKLQLQV 216
A+VHNLSE++RR +INEK++AL+ LIPN NK DKASMLD+AI+YLK L+LQ+Q+
Sbjct: 146 AEVHNLSEKRRRSRINEKMKALQNLIPNSNKTDKASMLDEAIEYLKQLQLQVQM 199
>gb|AAM10954.1| putative bHLH transcription factor [Arabidopsis thaliana]
Length = 442
Score = 90.5 bits (223), Expect = 4e-17
Identities = 46/84 (54%), Positives = 66/84 (77%), Gaps = 5/84 (5%)
Query: 133 LSDNDEETQENIVKEKPVREGNRVKRSYRNAKVHNLSERKRRDKINEKIRALKELIPNCN 192
L+ D++T N K + +RS R A+VHNLSER+RRD+INE+++AL+ELIP+C+
Sbjct: 233 LTSTDDQTMGN----KSSQRSGSTRRS-RAAEVHNLSERRRRDRINERMKALQELIPHCS 287
Query: 193 KMDKASMLDDAIDYLKTLKLQLQV 216
+ DKAS+LD+AIDYLK+L++QLQV
Sbjct: 288 RTDKASILDEAIDYLKSLQMQLQV 311
>dbj|BAC56978.1| PIF3 like basic Helix Loop Helix protein [Arabidopsis thaliana]
gi|30694924|ref|NP_191465.3| basic helix-loop-helix
(bHLH) family protein [Arabidopsis thaliana]
Length = 444
Score = 90.5 bits (223), Expect = 4e-17
Identities = 46/84 (54%), Positives = 66/84 (77%), Gaps = 5/84 (5%)
Query: 133 LSDNDEETQENIVKEKPVREGNRVKRSYRNAKVHNLSERKRRDKINEKIRALKELIPNCN 192
L+ D++T N K + +RS R A+VHNLSER+RRD+INE+++AL+ELIP+C+
Sbjct: 233 LTSTDDQTMGN----KSSQRSGSTRRS-RAAEVHNLSERRRRDRINERMKALQELIPHCS 287
Query: 193 KMDKASMLDDAIDYLKTLKLQLQV 216
+ DKAS+LD+AIDYLK+L++QLQV
Sbjct: 288 RTDKASILDEAIDYLKSLQMQLQV 311
>gb|AAO37214.1| hypothetical protein [Arabidopsis thaliana]
Length = 416
Score = 89.4 bits (220), Expect = 8e-17
Identities = 48/115 (41%), Positives = 77/115 (66%), Gaps = 3/115 (2%)
Query: 105 SSVCSLEASNDLNF-GVRKSHEDTDD--SPYLSDNDEETQENIVKEKPVREGNRVKRSYR 161
SS S S DL+ +++ + D ++ S YLS+N ++ ++ + R V + R
Sbjct: 170 SSKFSRGTSRDLSCCSLKRKYGDIEEEESTYLSNNSDDESDDAKTQVHARTRKPVTKRKR 229
Query: 162 NAKVHNLSERKRRDKINEKIRALKELIPNCNKMDKASMLDDAIDYLKTLKLQLQV 216
+ +VH L ERKRRD+ N+K+RAL++L+PNC K DKAS+LD+AI Y++TL+LQ+Q+
Sbjct: 230 STEVHKLYERKRRDEFNKKMRALQDLLPNCYKDDKASLLDEAIKYMRTLQLQVQM 284
>gb|AAX55166.1| hypothetical protein At2g46970 [Arabidopsis thaliana]
gi|22535492|dbj|BAC10689.1| PIF3 like basic Helix Loop
Helix protein [Arabidopsis thaliana]
gi|42569994|ref|NP_182220.2| basic helix-loop-helix
(bHLH) protein, putative [Arabidopsis thaliana]
Length = 416
Score = 89.4 bits (220), Expect = 8e-17
Identities = 48/115 (41%), Positives = 77/115 (66%), Gaps = 3/115 (2%)
Query: 105 SSVCSLEASNDLNF-GVRKSHEDTDD--SPYLSDNDEETQENIVKEKPVREGNRVKRSYR 161
SS S S DL+ +++ + D ++ S YLS+N ++ ++ + R V + R
Sbjct: 170 SSKFSRGTSRDLSCCSLKRKYGDIEEEESTYLSNNSDDESDDAKTQVHARTRKPVTKRKR 229
Query: 162 NAKVHNLSERKRRDKINEKIRALKELIPNCNKMDKASMLDDAIDYLKTLKLQLQV 216
+ +VH L ERKRRD+ N+K+RAL++L+PNC K DKAS+LD+AI Y++TL+LQ+Q+
Sbjct: 230 STEVHKLYERKRRDEFNKKMRALQDLLPNCYKDDKASLLDEAIKYMRTLQLQVQM 284
>ref|XP_473908.1| OSJNBa0058K23.6 [Oryza sativa (japonica cultivar-group)]
gi|38344324|emb|CAE02150.2| OSJNBa0058K23.6 [Oryza
sativa (japonica cultivar-group)]
Length = 181
Score = 88.2 bits (217), Expect = 2e-16
Identities = 40/59 (67%), Positives = 52/59 (87%)
Query: 158 RSYRNAKVHNLSERKRRDKINEKIRALKELIPNCNKMDKASMLDDAIDYLKTLKLQLQV 216
R R+A+ HN SER+RRD+INEK++AL+EL+PNC K DK SMLD+AIDYLK+L+LQLQ+
Sbjct: 10 RRSRSAEFHNFSERRRRDRINEKLKALQELLPNCTKTDKVSMLDEAIDYLKSLQLQLQM 68
Database: nr
Posted date: Jul 5, 2005 12:34 AM
Number of letters in database: 863,360,394
Number of sequences in database: 2,540,612
Lambda K H
0.315 0.130 0.358
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 365,891,058
Number of Sequences: 2540612
Number of extensions: 14263858
Number of successful extensions: 66800
Number of sequences better than 10.0: 1686
Number of HSP's better than 10.0 without gapping: 769
Number of HSP's successfully gapped in prelim test: 921
Number of HSP's that attempted gapping in prelim test: 65322
Number of HSP's gapped (non-prelim): 1936
length of query: 240
length of database: 863,360,394
effective HSP length: 124
effective length of query: 116
effective length of database: 548,324,506
effective search space: 63605642696
effective search space used: 63605642696
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.6 bits)
S2: 73 (32.7 bits)
Medicago: description of AC139745.7


