

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC139708.7 + phase: 0
(159 letters)
Database: nr
2,540,612 sequences; 863,360,394 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
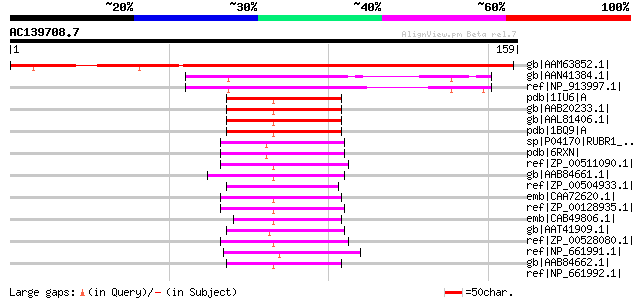
gb|AAM63852.1| unknown [Arabidopsis thaliana] gi|19310697|gb|AAL... 182 3e-45
gb|AAN41384.1| unknown protein [Arabidopsis thaliana] gi|1433489... 61 1e-08
ref|NP_913997.1| unknown protein [Oryza sativa (japonica cultiva... 60 2e-08
pdb|1IU6|A Chain A, Neutron Crystal Structure Of The Rubredoxin ... 50 3e-05
gb|AAB20233.1| rubredoxin, Rd [Pyrococcus furiosus, DSM 3638, P... 49 6e-05
gb|AAL81406.1| rubredoxin [Pyrococcus furiosus DSM 3638] gi|6066... 49 6e-05
pdb|1BQ9|A Chain A, Rubredoxin (Formyl Methionine Mutant) From P... 49 6e-05
sp|P04170|RUBR1_DESDE Rubredoxin 1 (Rd-1) 48 8e-05
pdb|6RXN| Rubredoxin 48 8e-05
ref|ZP_00511090.1| Rubredoxin-type Fe(Cys)4 protein [Chlorobium ... 48 1e-04
gb|AAB84661.1| rubredoxin [Methanothermobacter thermautotrophicu... 47 2e-04
ref|ZP_00504933.1| Flavin reductase-like:Rubredoxin-type Fe(Cys)... 46 3e-04
emb|CAA72620.1| rubredoxin [Clostridium diolis] 46 4e-04
ref|ZP_00128935.1| COG1773: Rubredoxin [Desulfovibrio desulfuric... 46 4e-04
emb|CAB49806.1| rd rubredoxin [Pyrococcus abyssi GE5] gi|1312452... 45 7e-04
gb|AAT41909.1| putative rubrerythrin [Fremyella diplosiphon] 45 7e-04
ref|ZP_00528080.1| Rubredoxin-type Fe(Cys)4 protein [Chlorobium ... 45 9e-04
ref|NP_661991.1| rubredoxin [Chlorobium tepidum TLS] gi|22654089... 45 9e-04
gb|AAB84662.1| rubredoxin [Methanothermobacter thermautotrophicu... 45 9e-04
ref|NP_661992.1| rubredoxin [Chlorobium tepidum TLS] gi|22654090... 44 0.001
>gb|AAM63852.1| unknown [Arabidopsis thaliana] gi|19310697|gb|AAL85079.1| unknown
protein [Arabidopsis thaliana]
gi|15292669|gb|AAK92703.1| unknown protein [Arabidopsis
thaliana] gi|9758248|dbj|BAB08747.1| unnamed protein
product [Arabidopsis thaliana]
gi|18423226|ref|NP_568749.1| rubredoxin family protein
[Arabidopsis thaliana]
Length = 154
Score = 182 bits (461), Expect = 3e-45
Identities = 97/160 (60%), Positives = 115/160 (71%), Gaps = 9/160 (5%)
Query: 1 MALQVQ-ASLYTTRLSPTATPTLLRPLSNPSTLKTSFFSR-SLNLLLHPNQLQLAYGPPR 58
MA+Q+ +LY R S PT P+ ++SF S+ LL + L + PR
Sbjct: 1 MAMQLPPTTLYAGRSSVVLPPT------TPTLQRSSFLPYYSMRLLGNKKSLSKS-SAPR 53
Query: 59 FTMRVASKQAYICRDCGYIYNERTAFDKLPDKYFCPVCGAPKRRFKPYATDVNKKANETD 118
F+MRV+SKQAYICRDCGYIYN+RT FDKLPD YFCPVC APKRRF+ Y DV+K N+ D
Sbjct: 54 FSMRVSSKQAYICRDCGYIYNDRTPFDKLPDNYFCPVCAAPKRRFRAYMPDVSKNVNDKD 113
Query: 119 VRKARKAELQRDEAVGKALPIAVAVGVVVLAGLYFYLNST 158
VRKARKAELQRDEAVGKALPI +AVGV+ LA LYFY+NST
Sbjct: 114 VRKARKAELQRDEAVGKALPIGIAVGVLALAALYFYVNST 153
>gb|AAN41384.1| unknown protein [Arabidopsis thaliana] gi|14334892|gb|AAK59624.1|
unknown protein [Arabidopsis thaliana]
gi|18418200|ref|NP_568342.1| rubredoxin family protein
[Arabidopsis thaliana] gi|10177062|dbj|BAB10504.1|
unnamed protein product [Arabidopsis thaliana]
Length = 271
Score = 60.8 bits (146), Expect = 1e-08
Identities = 41/101 (40%), Positives = 51/101 (49%), Gaps = 26/101 (25%)
Query: 56 PPRFTMRVASKQ----AYICRDCGYIYNERTAFDKLPDKYFCPVCGAPKRRFKPYATDVN 111
PPRF ++ Q +IC DCG+IY +FD+ PD Y CP C APK+RF Y DVN
Sbjct: 180 PPRFGRKLTETQKARATHICLDCGFIYTLPKSFDEQPDTYVCPQCIAPKKRFAKY--DVN 237
Query: 112 KKANETDVRKARKAELQRDEAVGKAL-PIAVAVGVVVLAGL 151
+A+G L PI V VG +LAGL
Sbjct: 238 -----------------TGKAIGGGLPPIGVIVG--LLAGL 259
>ref|NP_913997.1| unknown protein [Oryza sativa (japonica cultivar-group)]
gi|51964714|ref|XP_507141.1| PREDICTED P0577B11.138 gene
product [Oryza sativa (japonica cultivar-group)]
gi|28564642|dbj|BAC57824.1| unknown protein [Oryza
sativa (japonica cultivar-group)]
gi|37806461|dbj|BAC99896.1| unknown protein [Oryza
sativa (japonica cultivar-group)]
Length = 260
Score = 60.1 bits (144), Expect = 2e-08
Identities = 36/102 (35%), Positives = 51/102 (49%), Gaps = 25/102 (24%)
Query: 56 PPRFTMRVASKQ----AYICRDCGYIYNERTAFDKLPDKYFCPVCGAPKRRFKPYATDVN 111
PPRF ++ Q +IC DCGYIY F++ PD+Y CP C APK+RF Y +
Sbjct: 169 PPRFGRKLTESQKARATHICLDCGYIYFLPKPFEEQPDEYGCPQCNAPKKRFAKYDAETG 228
Query: 112 KKANETDVRKARKAELQRDEAVGKAL-PIAVAVGVVV-LAGL 151
+ A+G AL PI V V +++ +AG+
Sbjct: 229 R-------------------AIGGALPPITVIVSLIIGIAGV 251
>pdb|1IU6|A Chain A, Neutron Crystal Structure Of The Rubredoxin Mutant From
Pyrococcus Furiosus gi|23200152|pdb|1IU5|A Chain A,
X-Ray Crystal Structure Of The Rubredoxin Mutant From
Pyrococcus Furiosus gi|40889728|pdb|1RWD|A Chain A,
Backbone Nmr Structure Of A Mutant P. Furiosus
Rubredoxin Using Residual Dipolar Couplings
Length = 53
Score = 49.7 bits (117), Expect = 3e-05
Identities = 20/47 (42%), Positives = 29/47 (61%), Gaps = 11/47 (23%)
Query: 69 YICRDCGYIYNER-----------TAFDKLPDKYFCPVCGAPKRRFK 104
Y+C+ CGYIY+E T F+++PD + CP+CGAPK F+
Sbjct: 3 YVCKICGYIYDEDAGDPDNGVSPGTKFEEIPDDWVCPICGAPKSEFE 49
>gb|AAB20233.1| rubredoxin, Rd [Pyrococcus furiosus, DSM 3638, Peptide, 53 aa]
gi|52696107|pdb|1VCX|A Chain A, Neutron Crystal
Structure Of The Wild Type Rubredoxin From Pyrococcus
Furiosus At 1.5a Resolution gi|6729721|pdb|1BRF|A Chain
A, Rubredoxin (Wild Type) From Pyrococcus Furiosus
gi|443341|pdb|1ZRP| Rubredoxin (Zn-Substituted) (Nmr,
40 Structures) gi|442698|pdb|1CAD| Rubredoxin (Reduced)
gi|442697|pdb|1CAA| Rubredoxin (Oxidized)
Length = 53
Score = 48.5 bits (114), Expect = 6e-05
Identities = 20/47 (42%), Positives = 29/47 (61%), Gaps = 11/47 (23%)
Query: 69 YICRDCGYIYNER-----------TAFDKLPDKYFCPVCGAPKRRFK 104
++C+ CGYIY+E T F++LPD + CP+CGAPK F+
Sbjct: 3 WVCKICGYIYDEDAGDPDNGISPGTKFEELPDDWVCPICGAPKSEFE 49
>gb|AAL81406.1| rubredoxin [Pyrococcus furiosus DSM 3638] gi|6066243|gb|AAF03228.1|
rubredoxin [Pyrococcus furiosus]
gi|19857441|sp|P24297|RUBR_PYRFU Rubredoxin (Rd)
gi|18977654|ref|NP_579011.1| rubredoxin [Pyrococcus
furiosus DSM 3638] gi|6729714|pdb|1BQ8|A Chain A,
Rubredoxin (Methionine Mutant) From Pyrococcus Furiosus
Length = 54
Score = 48.5 bits (114), Expect = 6e-05
Identities = 20/47 (42%), Positives = 29/47 (61%), Gaps = 11/47 (23%)
Query: 69 YICRDCGYIYNER-----------TAFDKLPDKYFCPVCGAPKRRFK 104
++C+ CGYIY+E T F++LPD + CP+CGAPK F+
Sbjct: 4 WVCKICGYIYDEDAGDPDNGISPGTKFEELPDDWVCPICGAPKSEFE 50
>pdb|1BQ9|A Chain A, Rubredoxin (Formyl Methionine Mutant) From Pyrococcus
Furiosus
Length = 54
Score = 48.5 bits (114), Expect = 6e-05
Identities = 20/47 (42%), Positives = 29/47 (61%), Gaps = 11/47 (23%)
Query: 69 YICRDCGYIYNER-----------TAFDKLPDKYFCPVCGAPKRRFK 104
++C+ CGYIY+E T F++LPD + CP+CGAPK F+
Sbjct: 4 WVCKICGYIYDEDAGDPDNGISPGTKFEELPDDWVCPICGAPKSEFE 50
>sp|P04170|RUBR1_DESDE Rubredoxin 1 (Rd-1)
Length = 45
Score = 48.1 bits (113), Expect = 8e-05
Identities = 20/43 (46%), Positives = 26/43 (59%), Gaps = 4/43 (9%)
Query: 67 QAYICRDCGYIYN----ERTAFDKLPDKYFCPVCGAPKRRFKP 105
Q Y+C CGY Y+ + FD+LPD + CPVCG K +F P
Sbjct: 2 QKYVCNVCGYEYDPAEHDNVPFDQLPDDWCCPVCGVSKDQFSP 44
>pdb|6RXN| Rubredoxin
Length = 46
Score = 48.1 bits (113), Expect = 8e-05
Identities = 20/43 (46%), Positives = 26/43 (59%), Gaps = 4/43 (9%)
Query: 67 QAYICRDCGYIYN----ERTAFDKLPDKYFCPVCGAPKRRFKP 105
Q Y+C CGY Y+ + FD+LPD + CPVCG K +F P
Sbjct: 3 QKYVCNVCGYEYDPAEHDNVPFDQLPDDWCCPVCGVSKDQFSP 45
>ref|ZP_00511090.1| Rubredoxin-type Fe(Cys)4 protein [Chlorobium limicola DSM 245]
gi|67784787|gb|EAM44160.1| Rubredoxin-type Fe(Cys)4
protein [Chlorobium limicola DSM 245]
Length = 58
Score = 47.8 bits (112), Expect = 1e-04
Identities = 20/51 (39%), Positives = 28/51 (54%), Gaps = 11/51 (21%)
Query: 67 QAYICRDCGYIYNER-----------TAFDKLPDKYFCPVCGAPKRRFKPY 106
Q ++C CGY+Y+ TAF+ +PD + CPVCG K F+PY
Sbjct: 2 QKWVCVPCGYVYDPEVGDLDGGVEPGTAFEDIPDDWVCPVCGVDKTLFEPY 52
>gb|AAB84661.1| rubredoxin [Methanothermobacter thermautotrophicus str. Delta H]
gi|15678183|ref|NP_275298.1| rubredoxin
[Methanothermobacter thermautotrophicus str. Delta H]
gi|14285763|sp|O26258|RUBR_METTH Probable rubredoxin
(RD)
Length = 63
Score = 46.6 bits (109), Expect = 2e-04
Identities = 22/54 (40%), Positives = 30/54 (54%), Gaps = 11/54 (20%)
Query: 63 VASKQAYICRDCGYIYNER-----------TAFDKLPDKYFCPVCGAPKRRFKP 105
V++ + Y CR CGYIY+ T F+ LP+ + CP CGA K+ FKP
Sbjct: 8 VSAMKRYKCRVCGYIYDPEKGEPRTDTPPGTPFEDLPETWRCPSCGAKKKMFKP 61
>ref|ZP_00504933.1| Flavin reductase-like:Rubredoxin-type Fe(Cys)4 protein [Clostridium
thermocellum ATCC 27405] gi|67850470|gb|EAM46048.1|
Flavin reductase-like:Rubredoxin-type Fe(Cys)4 protein
[Clostridium thermocellum ATCC 27405]
Length = 220
Score = 46.2 bits (108), Expect = 3e-04
Identities = 18/35 (51%), Positives = 21/35 (59%)
Query: 69 YICRDCGYIYNERTAFDKLPDKYFCPVCGAPKRRF 103
Y+C CGY Y F+ LP+ Y CPVCG PK F
Sbjct: 182 YVCSICGYEYTGDIPFEDLPEDYVCPVCGQPKSVF 216
>emb|CAA72620.1| rubredoxin [Clostridium diolis]
Length = 53
Score = 45.8 bits (107), Expect = 4e-04
Identities = 20/49 (40%), Positives = 27/49 (54%), Gaps = 11/49 (22%)
Query: 67 QAYICRDCGYIYNER-----------TAFDKLPDKYFCPVCGAPKRRFK 104
+ YIC CGYIY+E T F+ +PD + CP+CG PK F+
Sbjct: 2 EKYICTVCGYIYDEAAGDPDNGVAPGTKFEDIPDDWVCPLCGVPKSDFE 50
>ref|ZP_00128935.1| COG1773: Rubredoxin [Desulfovibrio desulfuricans G20]
Length = 55
Score = 45.8 bits (107), Expect = 4e-04
Identities = 20/50 (40%), Positives = 27/50 (54%), Gaps = 11/50 (22%)
Query: 67 QAYICRDCGYIYNER-----------TAFDKLPDKYFCPVCGAPKRRFKP 105
Q Y+C CGY Y+ TA++ +P + CPVCGAPK F+P
Sbjct: 5 QKYVCNICGYEYDPAEGDPDNGVAPGTAWEDVPSDWLCPVCGAPKSEFEP 54
>emb|CAB49806.1| rd rubredoxin [Pyrococcus abyssi GE5]
gi|13124529|sp|Q9V099|RUBR_PYRAB Rubredoxin (Rd)
gi|14521100|ref|NP_126575.1| rubredoxin [Pyrococcus
abyssi GE5]
Length = 53
Score = 45.1 bits (105), Expect = 7e-04
Identities = 20/45 (44%), Positives = 26/45 (57%), Gaps = 11/45 (24%)
Query: 71 CRDCGYIYNER-----------TAFDKLPDKYFCPVCGAPKRRFK 104
C+ CGYIY+E T F+ LPD + CP+CGAPK F+
Sbjct: 6 CKICGYIYDEDEGDPDNGISPGTKFEDLPDDWVCPLCGAPKSEFE 50
>gb|AAT41909.1| putative rubrerythrin [Fremyella diplosiphon]
Length = 251
Score = 45.1 bits (105), Expect = 7e-04
Identities = 20/48 (41%), Positives = 27/48 (55%), Gaps = 11/48 (22%)
Query: 69 YICRDCGYIYNE-----------RTAFDKLPDKYFCPVCGAPKRRFKP 105
+ICR C IY+ TAF+ +PD + CP+CGA K+ FKP
Sbjct: 197 WICRQCSMIYDPVAGDPDSGIAPGTAFEDIPDDWRCPICGATKKTFKP 244
>ref|ZP_00528080.1| Rubredoxin-type Fe(Cys)4 protein [Chlorobium phaeobacteroides DSM
266] gi|67775951|gb|EAM35613.1| Rubredoxin-type Fe(Cys)4
protein [Chlorobium phaeobacteroides DSM 266]
Length = 58
Score = 44.7 bits (104), Expect = 9e-04
Identities = 18/51 (35%), Positives = 27/51 (52%), Gaps = 11/51 (21%)
Query: 67 QAYICRDCGYIYNER-----------TAFDKLPDKYFCPVCGAPKRRFKPY 106
+ ++C CGY+Y+ T F+ +PD + CPVCG K F+PY
Sbjct: 2 EKWVCVPCGYVYDPEIGDPDSGVEPGTPFENIPDDWCCPVCGVDKSLFEPY 52
>ref|NP_661991.1| rubredoxin [Chlorobium tepidum TLS]
gi|22654089|sp|P58992|RUBR1_CHLTE Rubredoxin 1 (Rd 1)
gi|21647067|gb|AAM72333.1| rubredoxin [Chlorobium
tepidum TLS]
Length = 69
Score = 44.7 bits (104), Expect = 9e-04
Identities = 19/54 (35%), Positives = 29/54 (53%), Gaps = 11/54 (20%)
Query: 68 AYICRDCGYIYNERTA-----------FDKLPDKYFCPVCGAPKRRFKPYATDV 110
+++C +CGYIY+ FDKLPD + CPVC PK +F + + +
Sbjct: 16 SWMCAECGYIYDPAEGNLETNIRPGMPFDKLPDDWSCPVCNHPKNQFTKFISQL 69
>gb|AAB84662.1| rubredoxin [Methanothermobacter thermautotrophicus str. Delta H]
gi|15678184|ref|NP_275299.1| rubredoxin
[Methanothermobacter thermautotrophicus str. Delta H]
gi|7430812|pir||B69075 rubredoxin - Methanobacterium
thermoautotrophicum (strain Delta H)
Length = 53
Score = 44.7 bits (104), Expect = 9e-04
Identities = 20/47 (42%), Positives = 26/47 (54%), Gaps = 11/47 (23%)
Query: 69 YICRDCGYIYNER-----------TAFDKLPDKYFCPVCGAPKRRFK 104
Y+C+ CGYIY+ T F+ LPD + CPVCG K +FK
Sbjct: 4 YVCQMCGYIYDPEEGDPVSGIEAGTPFEDLPDDWVCPVCGVGKDQFK 50
>ref|NP_661992.1| rubredoxin [Chlorobium tepidum TLS]
gi|22654090|sp|P58993|RUBR2_CHLTE Rubredoxin 2 (Rd 2)
gi|21647068|gb|AAM72334.1| rubredoxin [Chlorobium
tepidum TLS]
Length = 52
Score = 44.3 bits (103), Expect = 0.001
Identities = 20/44 (45%), Positives = 23/44 (51%), Gaps = 11/44 (25%)
Query: 71 CRDCGYIYNERT-----------AFDKLPDKYFCPVCGAPKRRF 103
C CGYIYN T +F+ LPD + CPVCGA K F
Sbjct: 6 CNICGYIYNPETGDPEGDIPAGTSFESLPDSWMCPVCGAGKEEF 49
Database: nr
Posted date: Jul 5, 2005 12:34 AM
Number of letters in database: 863,360,394
Number of sequences in database: 2,540,612
Lambda K H
0.322 0.136 0.402
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 260,293,909
Number of Sequences: 2540612
Number of extensions: 9781622
Number of successful extensions: 34906
Number of sequences better than 10.0: 251
Number of HSP's better than 10.0 without gapping: 87
Number of HSP's successfully gapped in prelim test: 164
Number of HSP's that attempted gapping in prelim test: 34666
Number of HSP's gapped (non-prelim): 263
length of query: 159
length of database: 863,360,394
effective HSP length: 117
effective length of query: 42
effective length of database: 566,108,790
effective search space: 23776569180
effective search space used: 23776569180
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.9 bits)
S2: 69 (31.2 bits)
Medicago: description of AC139708.7


