

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC139601.8 - phase: 0
(268 letters)
Database: nr
2,540,612 sequences; 863,360,394 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
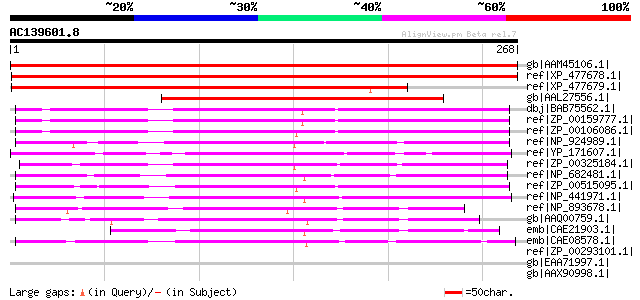
gb|AAM45106.1| unknown protein [Arabidopsis thaliana] gi|1529271... 427 e-118
ref|XP_477678.1| unknown protein [Oryza sativa (japonica cultiva... 414 e-114
ref|XP_477679.1| unknown protein [Oryza sativa (japonica cultiva... 315 1e-84
gb|AAL27556.1| hypothetical protein [Musa acuminata] 244 2e-63
dbj|BAB75562.1| alr3863 [Nostoc sp. PCC 7120] gi|17231355|ref|NP... 161 2e-38
ref|ZP_00159777.1| COG1201: Lhr-like helicases [Anabaena variabi... 159 6e-38
ref|ZP_00106086.1| COG0546: Predicted phosphatases [Nostoc punct... 150 4e-35
ref|NP_924989.1| hypothetical protein glr2043 [Gloeobacter viola... 147 2e-34
ref|YP_171607.1| hypothetical protein syc0897_c [Synechococcus e... 146 5e-34
ref|ZP_00325184.1| COG0546: Predicted phosphatases [Trichodesmiu... 134 3e-30
ref|NP_682481.1| hypothetical protein tlr1691 [Thermosynechococc... 130 4e-29
ref|ZP_00515095.1| conserved hypothetical protein [Crocosphaera ... 128 2e-28
ref|NP_441971.1| hypothetical protein sll0295 [Synechocystis sp.... 127 2e-28
ref|NP_893678.1| hypothetical protein PMM1561 [Prochlorococcus m... 86 1e-15
gb|AAQ00759.1| HAD superfamily hydrolase [Prochlorococcus marinu... 73 9e-12
emb|CAE21903.1| conserved hypothetical protein [Prochlorococcus ... 72 2e-11
emb|CAE08578.1| conserved hypothetical protein [Synechococcus sp... 70 6e-11
ref|ZP_00293101.1| COG0546: Predicted phosphatases [Thermobifida... 40 0.089
gb|EAA71997.1| hypothetical protein FG08797.1 [Gibberella zeae P... 35 1.7
gb|AAX90998.1| ORF010 [Bacteriophage 2638A] gi|66395460|ref|YP_2... 35 2.2
>gb|AAM45106.1| unknown protein [Arabidopsis thaliana] gi|15292713|gb|AAK92725.1|
unknown protein [Arabidopsis thaliana]
gi|3702346|gb|AAC62903.1| expressed protein [Arabidopsis
thaliana] gi|20197209|gb|AAM14973.1| expressed protein
[Arabidopsis thaliana] gi|18406942|ref|NP_566060.1|
expressed protein [Arabidopsis thaliana]
gi|30690184|ref|NP_850443.1| expressed protein
[Arabidopsis thaliana] gi|7486468|pir||T02448
hypothetical protein At2g45990 [imported] - Arabidopsis
thaliana
Length = 268
Score = 427 bits (1097), Expect = e-118
Identities = 203/268 (75%), Positives = 235/268 (86%)
Query: 1 MGHLYALDFDGVLCDTCGETAISALKAAKLRWPDLFGSVDSSTEDWIVEQMIKVRPVVET 60
MG LYALDFDGVLCD+CGE+++SA+KAAK+RWPDLF VDS+ E+WIV+QM VRPVVET
Sbjct: 1 MGDLYALDFDGVLCDSCGESSLSAVKAAKVRWPDLFEGVDSALEEWIVDQMHIVRPVVET 60
Query: 61 GYETLLLVRLLLETRVPSIRKSSVAEGLTVEGILEDWFKLKPIVMEEWNENRDDLIDLFG 120
GYE LLLVRLLLET++PSIRKSSVAEGLTV+GILE W K KP++ME W+E+RD L+DLFG
Sbjct: 61 GYENLLLVRLLLETKIPSIRKSSVAEGLTVDGILESWAKFKPVIMEAWDEDRDALVDLFG 120
Query: 121 KVRDDWLENDFAGWIQGNRFYPGVADALRFASSKVYIVTTKQGRFADALLRELAGITIPP 180
KVRDDW+ D WI NRFYPGV+DAL+FASSK+YIVTTKQGRFA+ALLRE+AG+ IP
Sbjct: 121 KVRDDWINKDLTTWIGANRFYPGVSDALKFASSKIYIVTTKQGRFAEALLREIAGVIIPS 180
Query: 181 ERIYGLGTGPKVETLKKLQKMPEHQGLTLHFVEDRIAALKNVIKEPELDNWNLYLVNWGF 240
ERIYGLG+GPKVE LK LQ PEHQGLTLHFVEDR+A LKNVIKEPELD W+LYL WG+
Sbjct: 181 ERIYGLGSGPKVEVLKLLQDKPEHQGLTLHFVEDRLATLKNVIKEPELDKWSLYLGTWGY 240
Query: 241 NTQKERDEAAANPRIQLIDLSDFSSKLK 268
NT+KER EAA PRIQ+I+LS FS+KLK
Sbjct: 241 NTEKERAEAAGIPRIQVIELSTFSNKLK 268
>ref|XP_477678.1| unknown protein [Oryza sativa (japonica cultivar-group)]
gi|22324441|dbj|BAC10357.1| unknown protein [Oryza
sativa (japonica cultivar-group)]
Length = 269
Score = 414 bits (1064), Expect = e-114
Identities = 192/267 (71%), Positives = 232/267 (85%)
Query: 2 GHLYALDFDGVLCDTCGETAISALKAAKLRWPDLFGSVDSSTEDWIVEQMIKVRPVVETG 61
G LYALDFDGVLCD+CGE+++SA+KAAK+RWP +F VD++ E+WIVEQM +RPVVETG
Sbjct: 3 GDLYALDFDGVLCDSCGESSLSAVKAAKVRWPWVFEQVDAAMEEWIVEQMYTLRPVVETG 62
Query: 62 YETLLLVRLLLETRVPSIRKSSVAEGLTVEGILEDWFKLKPIVMEEWNENRDDLIDLFGK 121
YE LLLVRLL+E R+PS R+SSVA+GL+++ ILE+W KLKP +M EWNE+RD L+DLFG
Sbjct: 63 YENLLLVRLLIEIRIPSARRSSVADGLSIQEILENWLKLKPTIMSEWNEDRDSLVDLFGS 122
Query: 122 VRDDWLENDFAGWIQGNRFYPGVADALRFASSKVYIVTTKQGRFADALLRELAGITIPPE 181
+RDDW+END +GWI NRFYPG ADAL+F+SS+VYIVTTKQGRFA+ALL+ELAGI P E
Sbjct: 123 IRDDWIENDLSGWIGANRFYPGTADALKFSSSEVYIVTTKQGRFAEALLKELAGIEFPSE 182
Query: 182 RIYGLGTGPKVETLKKLQKMPEHQGLTLHFVEDRIAALKNVIKEPELDNWNLYLVNWGFN 241
RIYGLGTGPKV+ L++LQ+MP+HQGLTLHFVEDR+A LKNVIKEP LD WNLYLVNWG+N
Sbjct: 183 RIYGLGTGPKVKVLQQLQQMPQHQGLTLHFVEDRLATLKNVIKEPALDQWNLYLVNWGYN 242
Query: 242 TQKERDEAAANPRIQLIDLSDFSSKLK 268
T KER++A RIQ+IDL FS KLK
Sbjct: 243 TPKEREDAEGISRIQVIDLPGFSQKLK 269
>ref|XP_477679.1| unknown protein [Oryza sativa (japonica cultivar-group)]
gi|51963388|ref|XP_506290.1| PREDICTED P0025D09.105-2
gene product [Oryza sativa (japonica cultivar-group)]
gi|38194220|dbj|BAC83355.2| unknown protein [Oryza
sativa (japonica cultivar-group)]
Length = 294
Score = 315 bits (806), Expect = 1e-84
Identities = 150/234 (64%), Positives = 185/234 (78%), Gaps = 25/234 (10%)
Query: 2 GHLYALDFDGVLCDTCGETAISALKAAKLRWPDLFGSVDSSTEDWIVEQMIKVRPVVETG 61
G LYALDFDGVLCD+CGE+++SA+KAAK+RWP +F VD++ E+WIVEQM +RPVVETG
Sbjct: 3 GDLYALDFDGVLCDSCGESSLSAVKAAKVRWPWVFEQVDAAMEEWIVEQMYTLRPVVETG 62
Query: 62 YETLLLVRLLLETRVPSIRKSSVAEGLTVEGILEDWFKLKPIVMEEWNENRDDLIDLFGK 121
YE LLLVRLL+E R+PS R+SSVA+GL+++ ILE+W KLKP +M EWNE+RD L+DLFG
Sbjct: 63 YENLLLVRLLIEIRIPSARRSSVADGLSIQEILENWLKLKPTIMSEWNEDRDSLVDLFGS 122
Query: 122 VRDDWLENDFAGWIQGNRFYPGVADALRFASSKVYIVTTKQGRFADALLRELAGITIPPE 181
+RDDW+END +GWI NRFYPG ADAL+F+SS+VYIVTTKQGRFA+ALL+ELAGI P E
Sbjct: 123 IRDDWIENDLSGWIGANRFYPGTADALKFSSSEVYIVTTKQGRFAEALLKELAGIEFPSE 182
Query: 182 RIYGLGTG-------------------------PKVETLKKLQKMPEHQGLTLH 210
RIYGLGTG PKV+ L++LQ+MP+HQGLTLH
Sbjct: 183 RIYGLGTGLVQYFFYFLFSPVNHFIESVSLCSSPKVKVLQQLQQMPQHQGLTLH 236
>gb|AAL27556.1| hypothetical protein [Musa acuminata]
Length = 151
Score = 244 bits (623), Expect = 2e-63
Identities = 114/149 (76%), Positives = 134/149 (89%)
Query: 81 KSSVAEGLTVEGILEDWFKLKPIVMEEWNENRDDLIDLFGKVRDDWLENDFAGWIQGNRF 140
K VA+GLTVE ILE+W +LKPI+M+EW+E RD LIDLFG+VRD+W++ND +GWI NRF
Sbjct: 1 KLGVADGLTVEAILENWSQLKPIIMKEWDEERDALIDLFGRVRDEWIDNDLSGWIGANRF 60
Query: 141 YPGVADALRFASSKVYIVTTKQGRFADALLRELAGITIPPERIYGLGTGPKVETLKKLQK 200
YPGVADALRFASS++YIVTTKQ RFADALLRELAG+TIP ERIYGLGTGPKV+ LK+LQ+
Sbjct: 61 YPGVADALRFASSQLYIVTTKQARFADALLRELAGVTIPAERIYGLGTGPKVKVLKQLQE 120
Query: 201 MPEHQGLTLHFVEDRIAALKNVIKEPELD 229
MPEHQGL+LHFVEDR+A LKNVIKEP +
Sbjct: 121 MPEHQGLSLHFVEDRLATLKNVIKEPSFE 149
>dbj|BAB75562.1| alr3863 [Nostoc sp. PCC 7120] gi|17231355|ref|NP_487903.1|
hypothetical protein alr3863 [Nostoc sp. PCC 7120]
gi|25354705|pir||AH2288 hypothetical protein alr3863
[imported] - Nostoc sp. (strain PCC 7120)
Length = 261
Score = 161 bits (408), Expect = 2e-38
Identities = 89/265 (33%), Positives = 156/265 (58%), Gaps = 22/265 (8%)
Query: 4 LYALDFDGVLCDTCGETAISALKAAKLRWPDLFGSVDSSTEDWIVEQMIKVRPVVETGYE 63
+ ALDFDGV+CD I + A + ++ +++ D + + ++RPV+ETG+E
Sbjct: 8 ILALDFDGVICDGL----IEYFEVAWRTYCQIWSPAENTPPDDLALRFYRLRPVIETGWE 63
Query: 64 TLLLVRLLLETRVPSIRKSSVAEGLTVEGILEDWFKLKPIVMEEWNENRDDLIDLFGKVR 123
+L++ L++ G + + IL++W + P ++ + ++ +R
Sbjct: 64 MPVLIKALVD-------------GNSDDQILQEWTSITPKILLDDKLQAKEIATKLDGLR 110
Query: 124 DDWLENDFAGWIQGNRFYPGVADALRFASS---KVYIVTTKQGRFADALLRELAGITIPP 180
D+W+ ND GW+ +RFY GV + L+ A + K+YIVTTK+GRF + LL + G+ +P
Sbjct: 111 DEWIANDLDGWLSLHRFYQGVIEKLKIAVASEVKLYIVTTKEGRFVEQLLHQ-EGVDLPR 169
Query: 181 ERIYGLGTG-PKVETLKKLQKMPEHQGLTLHFVEDRIAALKNVIKEPELDNWNLYLVNWG 239
+ I+G PK E +++L + +H+ ++L FVEDRI L+ V ++ +L++ L+L +WG
Sbjct: 170 DAIFGKEVKRPKYEIIRELIQAADHEPVSLWFVEDRIKTLQLVQQQSDLEDVKLFLADWG 229
Query: 240 FNTQKERDEAAANPRIQLIDLSDFS 264
+NTQ ER A ++PRIQL+ LS F+
Sbjct: 230 YNTQSERKAAQSDPRIQLLSLSQFA 254
>ref|ZP_00159777.1| COG1201: Lhr-like helicases [Anabaena variabilis ATCC 29413]
Length = 261
Score = 159 bits (403), Expect = 6e-38
Identities = 91/265 (34%), Positives = 151/265 (56%), Gaps = 22/265 (8%)
Query: 4 LYALDFDGVLCDTCGETAISALKAAKLRWPDLFGSVDSSTEDWIVEQMIKVRPVVETGYE 63
+ ALDFDGV+CD I + A + L+ D D + + ++RPV+ETG+E
Sbjct: 8 ILALDFDGVICDGL----IEYFEVAWRTYCQLWSPADDIPPDDLALRFYRLRPVIETGWE 63
Query: 64 TLLLVRLLLETRVPSIRKSSVAEGLTVEGILEDWFKLKPIVMEEWNENRDDLIDLFGKVR 123
+L++ L++ G + + IL++W + P ++ + ++ +R
Sbjct: 64 MPVLIKALVD-------------GNSDDQILQEWTSITPKILLDDKLQAKEIATKLDALR 110
Query: 124 DDWLENDFAGWIQGNRFYPGVADALRFASS---KVYIVTTKQGRFADALLRELAGITIPP 180
D W+ ND GW+ +RFY GV + L+ + K+YIVTTK+GRF + LL + G+ +P
Sbjct: 111 DQWIANDLDGWLSLHRFYQGVIEKLKITVASEVKLYIVTTKEGRFVEQLLHQ-EGVDLPR 169
Query: 181 ERIYGLGTG-PKVETLKKLQKMPEHQGLTLHFVEDRIAALKNVIKEPELDNWNLYLVNWG 239
+ I+G PK E L++L + +H+ ++L FVEDRI L+ V ++ +L++ L+L +WG
Sbjct: 170 DSIFGKEVKRPKYEILRELIQAADHKPVSLWFVEDRIKTLQLVQQQTDLEDVKLFLADWG 229
Query: 240 FNTQKERDEAAANPRIQLIDLSDFS 264
+NTQ ER A +PRIQL+ LS F+
Sbjct: 230 YNTQSERKAAQNDPRIQLLSLSQFA 254
>ref|ZP_00106086.1| COG0546: Predicted phosphatases [Nostoc punctiforme PCC 73102]
Length = 261
Score = 150 bits (379), Expect = 4e-35
Identities = 84/265 (31%), Positives = 150/265 (55%), Gaps = 22/265 (8%)
Query: 4 LYALDFDGVLCDTCGETAISALKAAKLRWPDLFGSVDSSTEDWIVEQMIKVRPVVETGYE 63
+ ALDFDGV+CD I + A + +++ + + D + + ++RPV+ETG+E
Sbjct: 8 ILALDFDGVICDGL----IEYFEVAWRTYCEIWSPANDTPGDDLALRFYRLRPVIETGWE 63
Query: 64 TLLLVRLLLETRVPSIRKSSVAEGLTVEGILEDWFKLKPIVMEEWNENRDDLIDLFGKVR 123
+L++ L++ G+ E I +W + P ++ ++ R
Sbjct: 64 MPVLIKALVD-------------GIPDEKIFHEWLSIAPQLLLNDKLQAREIAAKLDNQR 110
Query: 124 DDWLENDFAGWIQGNRFYPGVADALRF---ASSKVYIVTTKQGRFADALLRELAGITIPP 180
D+W+ D GW+ +RFYPGV + ++ + K+YIVTTK+GRF LL++ G+ +P
Sbjct: 111 DEWITTDLDGWLSLHRFYPGVVEKIKLTLDSGVKLYIVTTKEGRFVQQLLQQ-EGVNLPT 169
Query: 181 ERIYGLGTG-PKVETLKKLQKMPEHQGLTLHFVEDRIAALKNVIKEPELDNWNLYLVNWG 239
I+G PK E L++L++ E++ ++L FVEDR+ L+ V ++ +L++ L+L +WG
Sbjct: 170 AAIFGKEVKRPKYEILRELKQQAENKPVSLWFVEDRLKTLQLVQQQTDLEDVKLFLADWG 229
Query: 240 FNTQKERDEAAANPRIQLIDLSDFS 264
+NTQ ER+ A + +IQ++ LS F+
Sbjct: 230 YNTQAEREAAQNDLQIQVLSLSQFA 254
>ref|NP_924989.1| hypothetical protein glr2043 [Gloeobacter violaceus PCC 7421]
gi|35212610|dbj|BAC89984.1| glr2043 [Gloeobacter
violaceus PCC 7421]
Length = 261
Score = 147 bits (372), Expect = 2e-34
Identities = 100/268 (37%), Positives = 140/268 (51%), Gaps = 29/268 (10%)
Query: 4 LYALDFDGVLCDTCGETAISALKAAKLRW--PDLFGSVDSSTEDWIVEQMIKVRPVVETG 61
L ALDFDGVLCD E +A + + W PD + + E ++RPVVETG
Sbjct: 8 LLALDFDGVLCDGLLEYFQTAWQVYRRLWTPPDNLAPPAA-----VAELFYRLRPVVETG 62
Query: 62 YETLLLVRLLLETRVPSIRKSSVAEGLTVEGILEDWFKLKPIVMEEWNENRDDLIDLFGK 121
+E LLV S++ G+ E IL DW + ++ + + L +
Sbjct: 63 WEMPLLV-------------SAIVGGVEPEAILADWGGISQQLLAQSGVSAPQLAGEVDR 109
Query: 122 VRDDWLENDFAGWIQGNRFYPGVADALR----FASSKVYIVTTKQGRFADALLRELAGIT 177
RD W+ D GW+Q +R YPGVA LR V+++TTK+ RF LL E AG+
Sbjct: 110 TRDAWIARDLEGWLQLHRLYPGVAGRLRALCEHPQPAVFMITTKESRFV-LLLLEQAGVD 168
Query: 178 IPPERIYGLGTG-PKVETLKKLQKMPEHQGLTLHFVEDRIAALKNVIKEPELDNWNLYLV 236
P ERI+G T PK ETL KL + + FVEDR+A L+ V + EL + LYL
Sbjct: 169 WPGERIFGKDTQQPKTETLAKLLGAGYER---IWFVEDRLATLEKVARLAELASVQLYLA 225
Query: 237 NWGFNTQKERDEAAANPRIQLIDLSDFS 264
+WG+NT ER+ A+ RI+L++L F+
Sbjct: 226 DWGYNTPTERERVRADSRIRLLNLEQFA 253
>ref|YP_171607.1| hypothetical protein syc0897_c [Synechococcus elongatus PCC 6301]
gi|56685865|dbj|BAD79087.1| hypothetical protein
[Synechococcus elongatus PCC 6301]
gi|53762877|ref|ZP_00163313.2| COG0546: Predicted
phosphatases [Synechococcus elongatus PCC 7942]
Length = 254
Score = 146 bits (369), Expect = 5e-34
Identities = 99/266 (37%), Positives = 148/266 (55%), Gaps = 26/266 (9%)
Query: 1 MGHLYALDFDGVLCDTCGETAISALKAAKLRWPDLFGSVDSSTEDWIVEQMIKVRPVVET 60
M L ALDFDGVLCD E ++A +A R P + S ED + +RPV+E
Sbjct: 1 MASLLALDFDGVLCDGLREYFLAAWQAGCDRDPSWPEVPEPSLED----RFRLLRPVIEQ 56
Query: 61 GYETLLLVRLLLETRVPSIRKSSVAEGLTVEGILEDWFKLKPIVMEEWNENRDDLIDLFG 120
G+E +L++ L R+ + AE +L DW +L V+++W +L
Sbjct: 57 GWEMPVLLQALR-------REVADAE------VLADWPQLCDRVLKDWGLTTTELSQAMD 103
Query: 121 KVRDDWLENDFAGWIQGNRFYPGVADALRFASSKVYIVTTKQGRFADALLRELAGITIPP 180
+VRD W++ D W+Q + FYPGVA+ L ++ I++TK GRF LL ++ + PP
Sbjct: 104 RVRDRWIKRDRKEWLQLHHFYPGVAERLAQQTAPWVIISTKDGRFIAELLEQIPNLQ-PP 162
Query: 181 ERIYGLGTG-PKVETLKKLQKMPEHQGLTLHFVEDRIAALKNVIKEPELDNWNLYLVNWG 239
IYG G PK +TL +LQ E + FVEDR+ AL+ +L++ +LYL +WG
Sbjct: 163 LAIYGKEVGVPKTQTLIQLQVEFEQ----IAFVEDRLPALE---AAAQLESVDLYLADWG 215
Query: 240 FNTQKERDEAAANPRIQLIDLSDFSS 265
+NT ++R +A + RIQL+ L+DFSS
Sbjct: 216 YNTDRDRQQAMTSDRIQLLRLTDFSS 241
>ref|ZP_00325184.1| COG0546: Predicted phosphatases [Trichodesmium erythraeum IMS101]
Length = 268
Score = 134 bits (337), Expect = 3e-30
Identities = 87/263 (33%), Positives = 139/263 (52%), Gaps = 25/263 (9%)
Query: 6 ALDFDGVLCDTCGETAISALKAAKLRWPDLFGSVDSSTEDWIVEQMIKVRPVVETGYETL 65
ALDFDGVLC+ E +A + W D + ++E+ ++RPV+E G+E
Sbjct: 12 ALDFDGVLCNGLSEYFQTAWRTYSQFWQ----ISDEIPLNDLMEKFYRLRPVIEIGWEMP 67
Query: 66 LLVRLLLETRVPSIRKSSVAEGLTVEGILEDWFKLKPIVMEEWNENRDDLIDLFGKVRDD 125
LL+R L+ G+ + I ++W + ++ + N + + RD+
Sbjct: 68 LLIRALIL-------------GIEEDTIFQEWQAIAEKIVIQENLDPWKIGACLDNTRDE 114
Query: 126 WLENDFAGWIQGNRFYPGVADALR---FASSKVYIVTTKQGRFADALLRELAGITIPPER 182
W+ D GW+ ++FYPGV + L+ + K I+TTK+GRFA +LL ++ G+ +P
Sbjct: 115 WIVKDLEGWLSLHQFYPGVVEKLKELMVSEVKPIIITTKEGRFARSLLHKV-GVNLPEAD 173
Query: 183 IYGLGTG-PKVETLK-KLQKMPEHQGLTLHFVEDRIAALKNVIKEPELDNWNLYLVNWGF 240
I G + PK ETLK L K+ T+ F+EDR+ L ++ K P+L L+L +WG+
Sbjct: 174 IIGKESKRPKYETLKILLAKLGART--TIWFIEDRLKTLLSIQKHPDLQEVELFLADWGY 231
Query: 241 NTQKERDEAAANPRIQLIDLSDF 263
NTQKER+ A P I L+ + F
Sbjct: 232 NTQKERNSVAQYPSIHLLSSAQF 254
>ref|NP_682481.1| hypothetical protein tlr1691 [Thermosynechococcus elongatus BP-1]
gi|22295416|dbj|BAC09243.1| tlr1691 [Thermosynechococcus
elongatus BP-1]
Length = 260
Score = 130 bits (327), Expect = 4e-29
Identities = 90/264 (34%), Positives = 135/264 (51%), Gaps = 24/264 (9%)
Query: 4 LYALDFDGVLCDTCGETAISALKAAKLRWPDLFGSVDSSTEDWIVEQMIKVRPVVETGYE 63
L ALDFDGVLC+ E ++ + + WP+ + + + + ++RPV+ G+E
Sbjct: 8 LLALDFDGVLCNGLREYFQTSWRVYQQVWPEPLLGISL---EQLEREFGQLRPVITVGWE 64
Query: 64 TLLLVRLLLETRVPSIRKSSVAEGLTVEGILEDWFKLKPIVMEEWNENRDDLIDLFGKVR 123
LL+R ++ G + IL+DW K++ ++ ++ DL +R
Sbjct: 65 MPLLLR-------------AIVAGTPAQQILQDWPKVRDRLLATYHLTAADLGARVDGLR 111
Query: 124 DDWLENDFAGWIQGNRFYPGVADALRFASSK---VYIVTTKQGRFADALLRELAGITIPP 180
D W+E D+ W+ + FY GV AL+ ++ + IVTTK+ RF LL E AG++ P
Sbjct: 112 DRWIETDWQSWLALHDFYDGVIAALQHWQAQGQALAIVTTKEQRFVTYLL-EQAGLSFPS 170
Query: 181 ERIYGLGTG-PKVETLKKLQKMPEHQGLTLHFVEDRIAALKNVIKEPELDNWNLYLVNWG 239
E IYG PK L+ LQ G L FVEDR+ AL V PEL L+L WG
Sbjct: 171 EAIYGKEQQQPKPVILQALQST---YGAPLWFVEDRLGALLQVAVTPELGQTELFLAAWG 227
Query: 240 FNTQKERDEAAANPRIQLIDLSDF 263
+ T +R +A A+PRI L+ L F
Sbjct: 228 YTTAGDRAQAEAHPRIHLLSLEQF 251
>ref|ZP_00515095.1| conserved hypothetical protein [Crocosphaera watsonii WH 8501]
gi|67856689|gb|EAM51930.1| conserved hypothetical
protein [Crocosphaera watsonii WH 8501]
Length = 263
Score = 128 bits (321), Expect = 2e-28
Identities = 81/265 (30%), Positives = 133/265 (49%), Gaps = 22/265 (8%)
Query: 4 LYALDFDGVLCDTCGETAISALKAAKLRWPDLFGSVDSSTEDWIVEQMIKVRPVVETGYE 63
+ ALDFDGV+C+ E + L+ + W D + E W K+RPV+ETG+E
Sbjct: 9 ILALDFDGVICNGLKEYFQTTLRTYQKLWKD---DSQNDLEIW-ANSFYKLRPVIETGWE 64
Query: 64 TLLLVRLLLETRVPSIRKSSVAEGLTVEGILEDWFKLKPIVMEEWNENRDDLIDLFGKVR 123
+L+R L+ + I +W + ++ + N N+ ++ VR
Sbjct: 65 MPILLRALVLQ-------------YEQDNIESNWHNVCSEIVTKENLNKQQVMSALDGVR 111
Query: 124 DDWLENDFAGWIQGNRFYPGVADALRF---ASSKVYIVTTKQGRFADALLRELAGITIPP 180
D W++ D W+ + FYPGV + L +S+ +YIVTTK+GRF LL++ ++ P
Sbjct: 112 DHWIQTDLDNWLALHEFYPGVLEKLGKLLDSSTLLYIVTTKEGRFVKQLLKK-QNLSFPE 170
Query: 181 ERIYGLGTG-PKVETLKKLQKMPEHQGLTLHFVEDRIAALKNVIKEPELDNWNLYLVNWG 239
+ I+G PK +TL+++ K+ + L F+ED + L V + L NL+L +WG
Sbjct: 171 DHIFGKEVKQPKFDTLRQILKINQETPNNLWFIEDLLKTLNKVKSQEYLTEVNLFLADWG 230
Query: 240 FNTQKERDEAAANPRIQLIDLSDFS 264
+NT K + + I L+ L FS
Sbjct: 231 YNTIKSHELVKQDSTINLLSLYTFS 255
>ref|NP_441971.1| hypothetical protein sll0295 [Synechocystis sp. PCC 6803]
gi|1001418|dbj|BAA10041.1| sll0295 [Synechocystis sp.
PCC 6803] gi|7469406|pir||S76063 hypothetical protein -
Synechocystis sp. (strain PCC 6803)
Length = 268
Score = 127 bits (320), Expect = 2e-28
Identities = 86/269 (31%), Positives = 129/269 (46%), Gaps = 24/269 (8%)
Query: 3 HLYALDFDGVLCDTCGETAISALKAAKLRWPDLFGSVDSSTEDWIVEQMIKVRPVVETGY 62
HL LDFDGVLCD E ++ + + WPDL D + +RPV+ETG+
Sbjct: 12 HLLVLDFDGVLCDGLQEYFQTSCQVCRQIWPDL----PREKLDRQRDNFYFLRPVIETGW 67
Query: 63 ETLLLVRLLLETRVPSIRKSSVAEGLTVEGILEDWFKLKPIVMEEWNENRDDLIDLFGKV 122
E LL++ L A G+ I W + + + + L + +V
Sbjct: 68 EMPLLLKAL-------------ATGVEPAAIEAAWPAVAQTLQRQEQIGKSQLAPVLDQV 114
Query: 123 RDDWLENDFAGWIQGNRFYPGVADALRFASSK-----VYIVTTKQGRFADALLRELAGIT 177
RD+++ ND A W+ + FYPGV L +Y+VTTK+GRF LL+ +
Sbjct: 115 RDNYIHNDLAYWLGLHHFYPGVIGQLNHWLQSPYPQWLYVVTTKEGRFVQQLLKNQK-VD 173
Query: 178 IPPERIYGLGTG-PKVETLKKLQKMPEHQGLTLHFVEDRIAALKNVIKEPELDNWNLYLV 236
P +I G PK +TL++L+ + G L FVED + L+ V +P L+ +L+L
Sbjct: 174 FPLGQIIGKEIKQPKFKTLEQLRVKHQCDGDRLWFVEDMLTTLETVANQPALEQTSLFLA 233
Query: 237 NWGFNTQKERDEAAANPRIQLIDLSDFSS 265
+WG+NT R A R L+ L FS+
Sbjct: 234 DWGYNTPDSRGLAKQKKRFHLLSLQQFSA 262
>ref|NP_893678.1| hypothetical protein PMM1561 [Prochlorococcus marinus subsp.
pastoris str. CCMP1986] gi|33634335|emb|CAE20020.1|
conserved hypothetical protein [Prochlorococcus marinus
subsp. pastoris str. CCMP1986]
Length = 258
Score = 85.9 bits (211), Expect = 1e-15
Identities = 71/242 (29%), Positives = 118/242 (48%), Gaps = 21/242 (8%)
Query: 4 LYALDFDGVLCDTCGETAISALKAAK--LRWPDLFGSVDSSTEDWIVEQMIKVRPVVETG 61
L+ DFDGV+ D E S+L A + + P + +D + + I++RP V+ G
Sbjct: 6 LFLFDFDGVIVDGMNEYWHSSLLAFEKFINSPKIL--IDQNLYKQVSNTFIEMRPWVKYG 63
Query: 62 YETLLLVRLLLETRVPSIRKSSVAEGLTVEGILEDWFKLKPIVMEEWNENRDDLIDLFGK 121
+E L++V ++++ P ++ + L + + V+ E + +DL K
Sbjct: 64 WEMLIIVHQIIKSEDPLNNQNKI-------NFLNKYHQNCQKVLLENSWVAEDLQKCLDK 116
Query: 122 VRDDWLENDFAGWIQGNR-FYPGVA--DALRFASSKVYIVTTKQGRFADALLRELAGITI 178
R +ENDF WI+ +R FY + + L+ K I+TTK FA +L +L I
Sbjct: 117 ARKYQIENDFDNWIRLHRPFYEVIVFIEKLKKEKIKTGIITTKGKIFAGKILEKL---NI 173
Query: 179 PPERIYGLGTGPKVETLKKLQKMPEHQGLTLHFVEDRIAALKNVIKEPELDNWNLYLVNW 238
PE I+G +G KVE + +L + E G F+EDR L ++ + P N YL +W
Sbjct: 174 YPELIFGYESGTKVEIISELWREYEIMG----FIEDRRNTLLDIKQNPVTSNIPCYLADW 229
Query: 239 GF 240
G+
Sbjct: 230 GY 231
>gb|AAQ00759.1| HAD superfamily hydrolase [Prochlorococcus marinus subsp. marinus
str. CCMP1375] gi|33241164|ref|NP_876106.1| HAD
superfamily hydrolase [Prochlorococcus marinus subsp.
marinus str. CCMP1375]
Length = 258
Score = 72.8 bits (177), Expect = 9e-12
Identities = 62/252 (24%), Positives = 112/252 (43%), Gaps = 27/252 (10%)
Query: 4 LYALDFDGVLCDTCGETAISALKAAKLRWPDLFGSVDSSTEDWIVEQMI----KVRPVVE 59
+ DFDGV+ D E S+ KA + + G++D T+D + +M ++RP V+
Sbjct: 6 ILVFDFDGVIVDGLLEYWDSSRKA----FLKIQGALD--TDDQLPLEMPHEFRQLRPWVK 59
Query: 60 TGYETLLLVRLLLETRVPSIRKSSVAEGLTVEGILEDWFKLKPIVMEEWNENRDDLIDLF 119
G+E +LL L IRK S ++ K ++ W L +
Sbjct: 60 NGWEMVLLTAEL-------IRKDSPLSMHGAFHFANEYHKNCHTALKTWGWEPKQLQNAL 112
Query: 120 GKVRDDWLENDFAGWIQGNRFYPGVADALRFASSKVY---IVTTKQGRFADALLRELAGI 176
+R + ++ D W+ ++ +P +A+ + ++ ++TTK F LL
Sbjct: 113 DNIRKETIKTDKKKWLASHKLFPNIAERIHQLENESVDFGVLTTKSAEFTSELLNHF--- 169
Query: 177 TIPPERIYGLGTGPKVETLKKLQKMPEHQGLTLHFVEDRIAALKNVIKEPELDNWNLYLV 236
+ P +YG +G K L ++ K +G F+EDR A L+ V+ P + + YL
Sbjct: 170 NLHPNFLYGHESGQKTTVLLQISKDHSVRG----FIEDRRATLETVLNTPGISSIPCYLA 225
Query: 237 NWGFNTQKERDE 248
+WG+ +R +
Sbjct: 226 DWGYLKPDDRKD 237
>emb|CAE21903.1| conserved hypothetical protein [Prochlorococcus marinus str. MIT
9313] gi|33863995|ref|NP_895555.1| hypothetical protein
PMT1728 [Prochlorococcus marinus str. MIT 9313]
Length = 240
Score = 72.0 bits (175), Expect = 2e-11
Identities = 59/209 (28%), Positives = 94/209 (44%), Gaps = 19/209 (9%)
Query: 54 VRPVVETGYETLLLVRLLLETRVPSIRKSSVAEGLTVEGILEDWFKLKPIVMEEWNENRD 113
+RP + G+E +LL LL + P +R + A + D+ ++ W
Sbjct: 36 LRPWIHQGWEMVLLAAELLRSDGPLLRHGAKAFSV-------DYHLRCQQALDAWGWQPG 88
Query: 114 DLIDLFGKVRDDWLENDFAGWIQGNRFYPGVADALRFASSK---VYIVTTKQGRFADALL 170
L + +VR LE D W+ +R +PGV + LR + + ++TTK F LL
Sbjct: 89 QLQEALEQVRRSALEADRLNWLARHRPFPGVIERLRGLHDEGFDLVVLTTKGAEFTAELL 148
Query: 171 RELAGITIPPERIYGLGTGPKVETLKKLQKMPEHQGLTLHFVEDRIAALKNVIKEPELDN 230
+ + P +YG +G K E L +L +G FVEDR L+ V+ P L +
Sbjct: 149 K---CFQLAPHGLYGHESGSKTEVLLRLAAERPLRG----FVEDRRVTLETVLATPGLSS 201
Query: 231 WNLYLVNWGFNTQKERDEAAANPRIQLID 259
YL +WG+ K D A I+L++
Sbjct: 202 LPCYLASWGY--LKPEDSRALPMGIRLLE 228
>emb|CAE08578.1| conserved hypothetical protein [Synechococcus sp. WH 8102]
gi|33866595|ref|NP_898154.1| hypothetical protein
SYNW2063 [Synechococcus sp. WH 8102]
Length = 249
Score = 70.1 bits (170), Expect = 6e-11
Identities = 73/268 (27%), Positives = 118/268 (43%), Gaps = 32/268 (11%)
Query: 4 LYALDFDGVLCDTCGETAISALKAAKLRWPDLFGSVDSSTEDWIVEQMIKVRPVVETGYE 63
L DFDGV+ D E SA AA+ L D + + ++RP V G+E
Sbjct: 6 LLVFDFDGVIVDGMAEYWWSAWMAAQR----LNAEPQGLGSDAVPQGFRRLRPWVHHGWE 61
Query: 64 TLLLVRLLLETRVPSIRKSSVAEGLTVEGILEDWFKLKPIVMEEWNENRDDLIDLFGKVR 123
+LL + + L E + D+ + + ++ + L + + R
Sbjct: 62 MVLLAAEMPQ--------------LDPERWVVDYATEQDMALQRRGWSASLLQEALDQTR 107
Query: 124 DDWLENDFAGWIQGNRFYPGVADALR-FASSKV--YIVTTKQGRFADALLRELAGITIPP 180
+ +D A W+ ++ +PG+ D L+ F V ++TTK F LL L + P
Sbjct: 108 QQAVSSDRAAWLGLHQPFPGLVDRLQAFQEEGVDWAVLTTKTAAFTAELLESLG---LRP 164
Query: 181 ERIYGLGTGPKVETLKKLQKMPEHQGLTLHFVEDRIAALKNVIKEPELDNWNLYLVNWGF 240
R+ G GPK E L +LQ+ + + FVEDR A L+ V L + +L +WG+
Sbjct: 165 WRLDGREAGPKPEVLLRLQR----ERVLAGFVEDRRATLETVRDTDGLQSLPCWLASWGY 220
Query: 241 NTQKERDEAAANPR-IQLIDLSDFSSKL 267
+R++ PR IQLID ++ L
Sbjct: 221 LKPSDREDL---PRGIQLIDQDRLATPL 245
>ref|ZP_00293101.1| COG0546: Predicted phosphatases [Thermobifida fusca]
Length = 265
Score = 39.7 bits (91), Expect = 0.089
Identities = 36/141 (25%), Positives = 59/141 (41%), Gaps = 8/141 (5%)
Query: 123 RDDWLENDFAGWIQGNRFYPGVADALRFASSKVYIVTTKQGRFADALLRELAGITIPPER 182
RD E D W+ + YPG+A+ L + + IVT K A+L + G+
Sbjct: 127 RDLLRERDAQFWLGMHTLYPGIAELLVRHAGRTAIVTAKDTLSVRAIL-DFHGLGHTVAA 185
Query: 183 IYGLGTGPKVETLKKLQKMPEHQGLTLHFVEDRIAALKNVIKEPELDNWNLYLVNWGFNT 242
+ G K +++L + F++D + ++ V W WG+ T
Sbjct: 186 VVG-ECSDKAGAVRELCEQAGIPPSAAVFIDDNLTNVRRVAATGARSLW----ARWGYGT 240
Query: 243 QKERDEAAA--NPRIQLIDLS 261
+ EAAA P I+L DL+
Sbjct: 241 PEHAAEAAALRIPEIRLADLA 261
>gb|EAA71997.1| hypothetical protein FG08797.1 [Gibberella zeae PH-1]
gi|46128991|ref|XP_388973.1| hypothetical protein
FG08797.1 [Gibberella zeae PH-1]
Length = 663
Score = 35.4 bits (80), Expect = 1.7
Identities = 27/115 (23%), Positives = 53/115 (45%), Gaps = 25/115 (21%)
Query: 5 YALDFDGVLCDTCGETA--ISALKAAKLRWPDLFGSVDSSTEDWIVEQMIKVRPVVETGY 62
Y D + V + G A + A K+AK+ S+EDW ++Q+ ++ P +
Sbjct: 11 YKYDAEDVWIEPWGPNAFRVRATKSAKM-----------SSEDWALQQLKEITPKISISE 59
Query: 63 ETLLLVRLLLETRVPSIRKSSVAEGLTVEGILEDWFKLKPIVMEEWNENRDDLID 117
++ + ++ R+ + K ++ T +G +++EE+ NR DLID
Sbjct: 60 DSATITNGNIKARISRLGKLTIE---TADG---------KLLLEEYCRNRKDLID 102
>gb|AAX90998.1| ORF010 [Bacteriophage 2638A] gi|66395460|ref|YP_239829.1| ORF010
[Bacteriophage 2638A]
Length = 388
Score = 35.0 bits (79), Expect = 2.2
Identities = 37/160 (23%), Positives = 63/160 (39%), Gaps = 35/160 (21%)
Query: 105 MEEWNENRDDLIDLF---GKVRDDWLENDFAGWIQGNRFYP---GVADALRFASSKVYIV 158
+ E+ E+ D+++ K RD + F + + P G D + +A + I+
Sbjct: 85 LREYVEHYVDMVEELVNDAKARDKDVITMFETRLDLGTYVPESFGTGDVIVYAGGVLEII 144
Query: 159 TTKQGRFADALLRELAGITIPPERIYGLGTGPKVETLKKLQKMPEHQGLTLHFVEDRIAA 218
K G+ E++ I P R+YGLG + L + I
Sbjct: 145 DLKYGKGV-----EVSAINNPQLRLYGLGAYELLSVL------------------EDIHT 181
Query: 219 LKNVIKEPELDNWNL------YLVNWGFNTQKERDEAAAN 252
+K I +P LDN++ L+NWG K + + A N
Sbjct: 182 IKMTIIQPRLDNYSTEELEAKSLINWGLEYVKPKAKLAYN 221
Database: nr
Posted date: Jul 5, 2005 12:34 AM
Number of letters in database: 863,360,394
Number of sequences in database: 2,540,612
Lambda K H
0.319 0.139 0.419
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 462,628,316
Number of Sequences: 2540612
Number of extensions: 19178537
Number of successful extensions: 51626
Number of sequences better than 10.0: 34
Number of HSP's better than 10.0 without gapping: 15
Number of HSP's successfully gapped in prelim test: 19
Number of HSP's that attempted gapping in prelim test: 51550
Number of HSP's gapped (non-prelim): 35
length of query: 268
length of database: 863,360,394
effective HSP length: 126
effective length of query: 142
effective length of database: 543,243,282
effective search space: 77140546044
effective search space used: 77140546044
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 74 (33.1 bits)
Medicago: description of AC139601.8


