

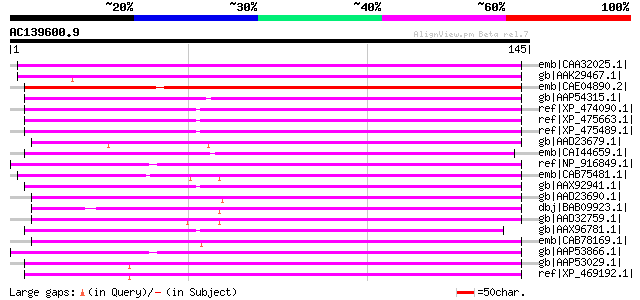
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC139600.9 - phase: 0 /pseudo
(145 letters)
Database: nr
2,540,612 sequences; 863,360,394 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
emb|CAA32025.1| unnamed protein product [Nicotiana tabacum] gi|1... 96 2e-19
gb|AAK29467.1| polyprotein-like [Lycopersicon chilense] 94 9e-19
emb|CAE04890.2| OSJNBa0042I15.12 [Oryza sativa (japonica cultiva... 92 2e-18
gb|AAP54315.1| putative polyprotein [Oryza sativa (japonica cult... 92 3e-18
ref|XP_474090.1| OSJNBa0033G05.13 [Oryza sativa (japonica cultiv... 92 3e-18
ref|XP_475663.1| putative polyprotein [Oryza sativa (japonica cu... 91 6e-18
ref|XP_475489.1| putative polyprotein [Oryza sativa (japonica cu... 91 6e-18
gb|AAD23679.1| putative retroelement pol polyprotein [Arabidopsi... 90 1e-17
emb|CAI44659.1| OSJNBa0061C06.9 [Oryza sativa (japonica cultivar... 89 2e-17
ref|NP_916849.1| retrovirus-related pol polyprotein from transpo... 89 2e-17
emb|CAB75481.1| copia-like polyprotein [Arabidopsis thaliana] gi... 88 4e-17
gb|AAX92941.1| retrotransposon protein, putative, Ty1-copia sub-... 88 4e-17
gb|AAD23690.1| putative retroelement pol polyprotein [Arabidopsi... 87 8e-17
dbj|BAB09923.1| copia-like retrotransposable element [Arabidopsi... 87 1e-16
gb|AAD32759.1| putative retroelement pol polyprotein [Arabidopsi... 87 1e-16
gb|AAX96781.1| retrotransposon protein, putative, Ty1-copia sub-... 86 2e-16
emb|CAB78169.1| putative retrotransposon [Arabidopsis thaliana] ... 84 5e-16
gb|AAP53866.1| putative polyprotein [Oryza sativa (japonica cult... 84 5e-16
gb|AAP53029.1| putative retrotransposon-related protein [Oryza s... 83 1e-15
ref|XP_469192.1| putative polyprotein [Oryza sativa (japonica cu... 81 6e-15
>emb|CAA32025.1| unnamed protein product [Nicotiana tabacum]
gi|130582|sp|P10978|POLX_TOBAC Retrovirus-related Pol
polyprotein from transposon TNT 1-94 [Contains: Protease
; Reverse transcriptase ; Endonuclease]
Length = 1328
Score = 95.9 bits (237), Expect = 2e-19
Identities = 51/141 (36%), Positives = 81/141 (57%)
Query: 3 GSKWDIEKFTGDNDFGLLKVKMEAMLIQQKCEKALKGKGSLPVTMSRAEKTEMVDKARSV 62
G K+++ KF GDN F + +M +LIQQ K L P TM + ++ ++A S
Sbjct: 3 GVKYEVAKFNGDNGFSTWQRRMRDLLIQQGLHKVLDVDSKKPDTMKAEDWADLDERAASA 62
Query: 63 IVLCLGDKVLRKVAKESTAASMWPKLEALYMTKSLTHQQFLKQ*LYSFKMVKSKAIMELL 122
I L L D V+ + E TA +W +LE+LYM+K+LT++ +LK+ LY+ M + + L
Sbjct: 63 IRLHLSDDVVNNIIDEDTARGIWTRLESLYMSKTLTNKLYLKKQLYALHMSEGTNFLSHL 122
Query: 123 VEFNKIIGDLENIEVRLEDVD 143
FN +I L N+ V++E+ D
Sbjct: 123 NVFNGLITQLANLGVKIEEED 143
>gb|AAK29467.1| polyprotein-like [Lycopersicon chilense]
Length = 1328
Score = 93.6 bits (231), Expect = 9e-19
Identities = 53/142 (37%), Positives = 83/142 (58%), Gaps = 1/142 (0%)
Query: 3 GSKWDIEKFTGDND-FGLLKVKMEAMLIQQKCEKALKGKGSLPVTMSRAEKTEMVDKARS 61
G K+++ KF GD F + + +M+ +LIQQ KAL GK P +M + E+ +KA S
Sbjct: 3 GVKYEVAKFNGDKPVFSMWQRRMKDLLIQQGLHKALGGKSKKPESMKLEDWEELDEKAAS 62
Query: 62 VIVLCLGDKVLRKVAKESTAASMWPKLEALYMTKSLTHQQFLKQ*LYSFKMVKSKAIMEL 121
I L L D V+ + E +A +W KLE LYM+K+LT++ +LK+ LY+ M + +
Sbjct: 63 AIRLHLTDDVVNNIVDEESACGIWTKLENLYMSKTLTNKLYLKKQLYTLHMDEGTNFLSH 122
Query: 122 LVEFNKIIGDLENIEVRLEDVD 143
L N +I L N+ V++E+ D
Sbjct: 123 LNVLNGLITQLANLGVKIEEED 144
>emb|CAE04890.2| OSJNBa0042I15.12 [Oryza sativa (japonica cultivar-group)]
Length = 432
Score = 92.4 bits (228), Expect = 2e-18
Identities = 50/139 (35%), Positives = 86/139 (60%), Gaps = 2/139 (1%)
Query: 5 KWDIEKFTGDNDFGLLKVKMEAMLIQQKCEKALKGKGSLPVTMSRAEKTEMVDKARSVIV 64
K+D+ KF G +FGL + +++ +L QQ+ KALKG P M + EM +A + I
Sbjct: 43 KFDLVKFDGFGNFGLWQTRVKDLLAQQEVSKALKGVK--PAKMEDDDCEEMQLQAAATIR 100
Query: 65 LCLGDKVLRKVAKESTAASMWPKLEALYMTKSLTHQQFLKQ*LYSFKMVKSKAIMELLVE 124
CL D+++ V +E++ +W KLEA +M+K+LT++++LKQ LY KM + + +
Sbjct: 101 RCLSDQIMYHVMEETSPKIIWEKLEAQFMSKTLTNKRYLKQKLYGLKMQEGADLTAHVNV 160
Query: 125 FNKIIGDLENIEVRLEDVD 143
FN+++ DL ++V ++D D
Sbjct: 161 FNQLVTDLVKMDVEVDDED 179
>gb|AAP54315.1| putative polyprotein [Oryza sativa (japonica cultivar-group)]
gi|37535452|ref|NP_922028.1| putative polyprotein [Oryza
sativa (japonica cultivar-group)]
gi|22094359|gb|AAM91886.1| putative polyprotein [Oryza
sativa (japonica cultivar-group)]
Length = 1280
Score = 92.0 bits (227), Expect = 3e-18
Identities = 56/139 (40%), Positives = 80/139 (57%), Gaps = 1/139 (0%)
Query: 5 KWDIEKFTGDNDFGLLKVKMEAMLIQQKCEKALKGKGSLPVTMSRAEKTEMVDKARSVIV 64
K+D+ D F L +VKM A+L QQ + AL G S EK + KA S I
Sbjct: 40 KYDLPLLDRDTRFSLWQVKMRAVLAQQDLDDALSGFDKRTQDWSNDEKKKD-RKAMSYIH 98
Query: 65 LCLGDKVLRKVAKESTAASMWPKLEALYMTKSLTHQQFLKQ*LYSFKMVKSKAIMELLVE 124
L L + +L++V KE TAA +W KLE + MTK LT + LKQ L+ K+ ++M+ L
Sbjct: 99 LHLSNNILQEVLKEETAAGLWLKLEQICMTKDLTSKMHLKQKLFLHKLQDDGSVMDHLST 158
Query: 125 FNKIIGDLENIEVRLEDVD 143
F +I+ DLE+IEV+ ++ D
Sbjct: 159 FKEIVADLESIEVKYDEED 177
>ref|XP_474090.1| OSJNBa0033G05.13 [Oryza sativa (japonica cultivar-group)]
gi|38344889|emb|CAD41912.2| OSJNBa0033G05.13 [Oryza
sativa (japonica cultivar-group)]
Length = 1181
Score = 91.7 bits (226), Expect = 3e-18
Identities = 55/139 (39%), Positives = 81/139 (57%), Gaps = 1/139 (0%)
Query: 5 KWDIEKFTGDNDFGLLKVKMEAMLIQQKCEKALKGKGSLPVTMSRAEKTEMVDKARSVIV 64
K+D+ D F L +VKM A+L QQ+ + AL G S EK + KA S I
Sbjct: 5 KYDLPLLDRDTRFSLWQVKMRAVLAQQELDDALSGFDKRTQDWSNDEK-KRDRKAMSYIH 63
Query: 65 LCLGDKVLRKVAKESTAASMWPKLEALYMTKSLTHQQFLKQ*LYSFKMVKSKAIMELLVE 124
L L + +L++V KE TAA +W KLE + MTK LT + LKQ L+ K+ ++M+ L
Sbjct: 64 LHLSNNILQEVLKEETAAGLWLKLEQICMTKDLTSKMHLKQKLFLHKLQDDGSVMDHLSA 123
Query: 125 FNKIIGDLENIEVRLEDVD 143
F +I+ DLE++EV+ ++ D
Sbjct: 124 FKEIVADLESMEVKYDEKD 142
>ref|XP_475663.1| putative polyprotein [Oryza sativa (japonica cultivar-group)]
gi|48475188|gb|AAT44257.1| putative polyprotein [Oryza
sativa (japonica cultivar-group)]
Length = 1211
Score = 90.9 bits (224), Expect = 6e-18
Identities = 55/139 (39%), Positives = 79/139 (56%), Gaps = 1/139 (0%)
Query: 5 KWDIEKFTGDNDFGLLKVKMEAMLIQQKCEKALKGKGSLPVTMSRAEKTEMVDKARSVIV 64
K+D+ D F L +VKM A+L QQ + AL G S EK + K S I
Sbjct: 5 KYDLPLLDRDTRFSLWQVKMRAVLAQQDLDDALSGFDKRTQDWSNDEK-KRDRKTMSYIH 63
Query: 65 LCLGDKVLRKVAKESTAASMWPKLEALYMTKSLTHQQFLKQ*LYSFKMVKSKAIMELLVE 124
L L + +L++V KE TAA +W KLE + MTK LT + LKQ L+ K+ ++M+ L
Sbjct: 64 LHLSNNILQEVLKEETAAGLWLKLEQICMTKDLTSKMHLKQKLFLHKLQDDGSVMDHLSA 123
Query: 125 FNKIIGDLENIEVRLEDVD 143
F KI+ DLE++EV+ ++ D
Sbjct: 124 FKKIVADLESMEVKYDEED 142
>ref|XP_475489.1| putative polyprotein [Oryza sativa (japonica cultivar-group)]
gi|48475213|gb|AAT44282.1| putative polyprotein [Oryza
sativa (japonica cultivar-group)]
Length = 1243
Score = 90.9 bits (224), Expect = 6e-18
Identities = 55/139 (39%), Positives = 81/139 (57%), Gaps = 1/139 (0%)
Query: 5 KWDIEKFTGDNDFGLLKVKMEAMLIQQKCEKALKGKGSLPVTMSRAEKTEMVDKARSVIV 64
K+D+ D F L +VKM A+L QQ + AL G S EK + KA S I
Sbjct: 5 KYDLPLLDRDTRFSLWQVKMRAVLAQQDLDDALSGFDKRTQDWSNDEK-KRDRKAISYIH 63
Query: 65 LCLGDKVLRKVAKESTAASMWPKLEALYMTKSLTHQQFLKQ*LYSFKMVKSKAIMELLVE 124
L L + +L++V KE TAA +W KLE + MTK LT + LKQ L+ K+ +++M+ L
Sbjct: 64 LHLSNNILQEVLKEETAAGLWLKLEQICMTKDLTSKMHLKQKLFLHKLQDDESVMDHLSA 123
Query: 125 FNKIIGDLENIEVRLEDVD 143
F +I+ DLE++EV+ ++ D
Sbjct: 124 FKEIVADLESMEVKYDEDD 142
>gb|AAD23679.1| putative retroelement pol polyprotein [Arabidopsis thaliana]
gi|25412027|pir||G84599 probable retroelement pol
polyprotein [imported] - Arabidopsis thaliana
Length = 838
Score = 90.1 bits (222), Expect = 1e-17
Identities = 60/149 (40%), Positives = 87/149 (58%), Gaps = 12/149 (8%)
Query: 7 DIEKFTGDNDFGLLKVKMEA------MLIQQKCEKALKGKGSLPVTMSRAEKTE------ 54
++EK G+ D+ L K K+ A +L + ++A++ + S T S KTE
Sbjct: 7 EVEKLDGEGDYVLWKEKLLAHIELLGLLEGLEEDEAIEEEESTAETDSLLTKTEDKVLKE 66
Query: 55 MVDKARSVIVLCLGDKVLRKVAKESTAASMWPKLEALYMTKSLTHQQFLKQ*LYSFKMVK 114
KARS ++L LG+ VLRKV KE TAA M L+ L+M KSL ++ +LKQ LY +KM
Sbjct: 67 KRGKARSTVILSLGNHVLRKVIKEKTAAGMIRVLDKLFMAKSLPNRIYLKQRLYGYKMSD 126
Query: 115 SKAIMELLVEFNKIIGDLENIEVRLEDVD 143
S I E + +F K+I DLEN++V + D D
Sbjct: 127 SMTIEENVNDFFKLISDLENVKVSVPDED 155
>emb|CAI44659.1| OSJNBa0061C06.9 [Oryza sativa (japonica cultivar-group)]
Length = 1007
Score = 89.4 bits (220), Expect = 2e-17
Identities = 53/137 (38%), Positives = 79/137 (56%), Gaps = 1/137 (0%)
Query: 5 KWDIEKFTGDNDFGLLKVKMEAMLIQQKCEKALKGKGSLPVTMSRAEKTEMVDKARSVIV 64
K+D+ D F L +VKM A+L QQ + AL G S+ EK KA S I
Sbjct: 5 KYDLPLLDRDTRFSLWQVKMRAVLAQQDLDDALSGFDKRTQDWSKDEKKRDC-KAMSYIH 63
Query: 65 LCLGDKVLRKVAKESTAASMWPKLEALYMTKSLTHQQFLKQ*LYSFKMVKSKAIMELLVE 124
+ L + +L++V KE TAA +W KLE + MTK LT + LKQ L+ K+ ++M+ L
Sbjct: 64 VHLSNNILQEVLKEETAAGLWLKLEQICMTKDLTSKMHLKQKLFLHKLQDDGSVMDHLFA 123
Query: 125 FNKIIGDLENIEVRLED 141
F +I+ DLE++EV+ ++
Sbjct: 124 FKEIVADLESMEVKYDE 140
>ref|NP_916849.1| retrovirus-related pol polyprotein from transposon TNT 1-94-like
[Oryza sativa (japonica cultivar-group)]
Length = 425
Score = 89.0 bits (219), Expect = 2e-17
Identities = 51/143 (35%), Positives = 85/143 (58%), Gaps = 2/143 (1%)
Query: 1 TMGSKWDIEKFTGDNDFGLLKVKMEAMLIQQKCEKALKGKGSLPVTMSRAEKTEMVDKAR 60
T+ SK+++ KF G +F L +++++ +L QQ KAL+ ++P M + EM +A
Sbjct: 4 TVVSKFEMVKFDGTGNFVLWQMRLKDLLAQQGISKALEE--TMPEKMDAGKWEEMKAQAA 61
Query: 61 SVIVLCLGDKVLRKVAKESTAASMWPKLEALYMTKSLTHQQFLKQ*LYSFKMVKSKAIME 120
+ I L L D V+ +V E T +W KL +LYM+KSLT + +LKQ LY +M + + +
Sbjct: 62 ATIRLSLSDSVMYQVMDEKTPKEIWDKLASLYMSKSLTSKLYLKQQLYGLQMQEESDLRK 121
Query: 121 LLVEFNKIIGDLENIEVRLEDVD 143
+ FN+++ DL ++V L D D
Sbjct: 122 HVDVFNQLVVDLSKLDVNLYDED 144
>emb|CAB75481.1| copia-like polyprotein [Arabidopsis thaliana]
gi|11278366|pir||T47492 copia-like polyprotein -
Arabidopsis thaliana
Length = 1363
Score = 88.2 bits (217), Expect = 4e-17
Identities = 58/157 (36%), Positives = 89/157 (55%), Gaps = 17/157 (10%)
Query: 3 GSKWDIEKFTGDNDFGLLKVKMEAMLIQQKCEKALKGKGSLPVTMSR------------A 50
G++ ++EKF G D+ + K K+ A + L+ + P+ R
Sbjct: 3 GARIEVEKFDGRGDYTMWKEKLLAHIDMLGLSAVLR-ESETPMGKERDSEKSDEDEKEER 61
Query: 51 EKTEMVD----KARSVIVLCLGDKVLRKVAKESTAASMWPKLEALYMTKSLTHQQFLKQ* 106
EK E + KARS IVL + D+VLRK+ KE++AA+M L+ LYM+K+L ++ +LKQ
Sbjct: 62 EKMEAFEEKKRKARSTIVLSVSDRVLRKIKKETSAAAMLEALDRLYMSKALPNRIYLKQK 121
Query: 107 LYSFKMVKSKAIMELLVEFNKIIGDLENIEVRLEDVD 143
LYSFKM ++ +I + EF I+ DLEN+ V + D D
Sbjct: 122 LYSFKMSENLSIEGNIDEFLHIVADLENLNVLVSDED 158
>gb|AAX92941.1| retrotransposon protein, putative, Ty1-copia sub-class [Oryza
sativa (japonica cultivar-group)]
Length = 2340
Score = 88.2 bits (217), Expect = 4e-17
Identities = 55/139 (39%), Positives = 79/139 (56%), Gaps = 1/139 (0%)
Query: 5 KWDIEKFTGDNDFGLLKVKMEAMLIQQKCEKALKGKGSLPVTMSRAEKTEMVDKARSVIV 64
K+D+ D F L +VKM A+L QQ + AL G S EK + KA S I
Sbjct: 212 KYDLPLLYRDTRFSLWQVKMRAVLAQQDLDDALSGFDKRTHDWSNDEK-KRDRKAMSYIH 270
Query: 65 LCLGDKVLRKVAKESTAASMWPKLEALYMTKSLTHQQFLKQ*LYSFKMVKSKAIMELLVE 124
L L + +L++V KE AA +W KLE + MTK LT + LKQ L+ K+ ++M+ L
Sbjct: 271 LHLSNNILQEVLKEEIAAGLWLKLEQICMTKDLTSKMHLKQTLFLHKLQDDGSVMDHLSA 330
Query: 125 FNKIIGDLENIEVRLEDVD 143
F +II DLE++EV+ ++ D
Sbjct: 331 FKEIIADLESMEVKYDEED 349
>gb|AAD23690.1| putative retroelement pol polyprotein [Arabidopsis thaliana]
gi|25301702|pir||E84601 probable retroelement pol
polyprotein [imported] - Arabidopsis thaliana
Length = 1333
Score = 87.0 bits (214), Expect = 8e-17
Identities = 58/152 (38%), Positives = 85/152 (55%), Gaps = 15/152 (9%)
Query: 7 DIEKFTGDNDFGLLKVKMEAMLIQQKCEKALKGKGSLPVTMSRAEKTEMVDK-------- 58
++EKF G D+ + K K+ A L ALK + L ++ + TE +K
Sbjct: 7 EVEKFDGRGDYTMWKEKLMAHLDILGLSVALKEEDDLVEKVAEMQLTEEEEKEEVLRREL 66
Query: 59 -------ARSVIVLCLGDKVLRKVAKESTAASMWPKLEALYMTKSLTHQQFLKQ*LYSFK 111
ARS IVL + D+VLRK+ KE +AA+M L+ LYM+K+L ++ + KQ LYSFK
Sbjct: 67 LEEKRRKARSAIVLSVTDRVLRKIKKEQSAAAMLGVLDKLYMSKALPNRIYQKQKLYSFK 126
Query: 112 MVKSKAIMELLVEFNKIIGDLENIEVRLEDVD 143
M ++ +I + EF +II DLEN V + D D
Sbjct: 127 MSENLSIEGNIDEFLRIIADLENTNVLVSDED 158
>dbj|BAB09923.1| copia-like retrotransposable element [Arabidopsis thaliana]
Length = 1342
Score = 86.7 bits (213), Expect = 1e-16
Identities = 58/160 (36%), Positives = 84/160 (52%), Gaps = 26/160 (16%)
Query: 7 DIEKFTGDNDFGLLKVKMEAMLIQQKCEKALKGKGSLPVTMSRAEKTEMVD--------- 57
++EKF GD D+ L K E +L + L+G G + TE+ D
Sbjct: 7 EVEKFDGDGDYILWK---EKLLAHMEMLGLLEGLGEEEEAVVEDSTTEISDGGNQDPETA 63
Query: 58 --------------KARSVIVLCLGDKVLRKVAKESTAASMWPKLEALYMTKSLTHQQFL 103
KARS I+L LG+ VLRKV K+ TAA M L+ L+M KSL ++ +L
Sbjct: 64 TSKLEDKILKEKRGKARSTIILSLGNNVLRKVIKQKTAAGMIKVLDQLFMAKSLPNRIYL 123
Query: 104 KQ*LYSFKMVKSKAIMELLVEFNKIIGDLENIEVRLEDVD 143
KQ LY +KM ++ + E + +F K+I DLEN++V + D D
Sbjct: 124 KQRLYGYKMSENMTMEENVNDFFKLISDLENVKVVVPDED 163
>gb|AAD32759.1| putative retroelement pol polyprotein [Arabidopsis thaliana]
gi|25301696|pir||F84486 probable retroelement pol
polyprotein [imported] - Arabidopsis thaliana
Length = 1356
Score = 86.7 bits (213), Expect = 1e-16
Identities = 60/152 (39%), Positives = 87/152 (56%), Gaps = 15/152 (9%)
Query: 7 DIEKFTGDNDFGLLKVKMEAMLIQQKCEKALKGKGSLPVTMS-----------RAEKTEM 55
++EKF G D+ + K K+ A + ALK S S + EK E
Sbjct: 7 EVEKFDGRGDYTMWKEKLLAHMDILGLNTALKESESTGEKKSVLDESDEDYEEKLEKFEA 66
Query: 56 VD----KARSVIVLCLGDKVLRKVAKESTAASMWPKLEALYMTKSLTHQQFLKQ*LYSFK 111
++ KARS IVL + D+VLRK+ KESTAA+M L+ LYM+K+L ++ + KQ LYSFK
Sbjct: 67 LEEKKKKARSAIVLSVTDRVLRKIKKESTAAAMLLALDKLYMSKALPNRIYPKQKLYSFK 126
Query: 112 MVKSKAIMELLVEFNKIIGDLENIEVRLEDVD 143
M ++ ++ + EF +II DLEN+ V + D D
Sbjct: 127 MSENLSVEGNIDEFLQIITDLENMNVIISDED 158
>gb|AAX96781.1| retrotransposon protein, putative, Ty1-copia sub-class [Oryza
sativa (japonica cultivar-group)]
Length = 1170
Score = 85.5 bits (210), Expect = 2e-16
Identities = 52/134 (38%), Positives = 78/134 (57%), Gaps = 1/134 (0%)
Query: 5 KWDIEKFTGDNDFGLLKVKMEAMLIQQKCEKALKGKGSLPVTMSRAEKTEMVDKARSVIV 64
K+D+ D F L +VKM+A+L QQ + AL G S EK + KA S I
Sbjct: 5 KYDLSLLDRDIRFSLWQVKMQAVLAQQDLDDALSGFDKTTQDWSGDEK-KRGRKALSYIH 63
Query: 65 LCLGDKVLRKVAKESTAASMWPKLEALYMTKSLTHQQFLKQ*LYSFKMVKSKAIMELLVE 124
L L + +L+ V K+ TAA +W KLE + MTK LT +LKQ L+ K+ +++M+ L
Sbjct: 64 LHLSNNILQAVLKDETAAGLWLKLEQICMTKDLTSNMYLKQKLFLHKLQDDESVMDHLST 123
Query: 125 FNKIIGDLENIEVR 138
F +II +LE++E++
Sbjct: 124 FKEIIAELESMEMK 137
>emb|CAB78169.1| putative retrotransposon [Arabidopsis thaliana]
gi|4539406|emb|CAB40039.1| putative retrotransposon
[Arabidopsis thaliana] gi|7444416|pir||T04181
hypothetical protein F7L13.40 - Arabidopsis thaliana
Length = 1230
Score = 84.3 bits (207), Expect = 5e-16
Identities = 56/150 (37%), Positives = 82/150 (54%), Gaps = 13/150 (8%)
Query: 7 DIEKFTGDNDFGLLKVKMEAMLIQQKCEKALKGKGSLPVTMSRAEK-------------T 53
++EKF G D+ L K K+ A + AL+ S+ + E+
Sbjct: 7 EMEKFDGHGDYTLWKEKLMAHMDLLGLTVALRETQSVSDPLESEEEGKESEKGDKEALME 66
Query: 54 EMVDKARSVIVLCLGDKVLRKVAKESTAASMWPKLEALYMTKSLTHQQFLKQ*LYSFKMV 113
E KARS IVL + D+VLRK KE TA SM L+ LYM+K+L ++ +LKQ LYS+KM
Sbjct: 67 EKRQKARSTIVLSVSDQVLRKSKKEKTAPSMLEALDKLYMSKALPNRIYLKQKLYSYKMQ 126
Query: 114 KSKAIMELLVEFNKIIGDLENIEVRLEDVD 143
++ ++ + EF ++I DLEN V + D D
Sbjct: 127 ENLSVEGNIDEFLRLIADLENTNVLVSDED 156
>gb|AAP53866.1| putative polyprotein [Oryza sativa (japonica cultivar-group)]
gi|37534554|ref|NP_921579.1| putative polyprotein [Oryza
sativa (japonica cultivar-group)]
Length = 382
Score = 84.3 bits (207), Expect = 5e-16
Identities = 47/143 (32%), Positives = 86/143 (59%), Gaps = 2/143 (1%)
Query: 1 TMGSKWDIEKFTGDNDFGLLKVKMEAMLIQQKCEKALKGKGSLPVTMSRAEKTEMVDKAR 60
T SK+++ KF G +F L +++++ +L QQ KAL+ ++P + + EM +A
Sbjct: 4 TAVSKFEVVKFDGTGNFVLWQMRLKDLLAQQGISKALQE--TMPEKIDADKWNEMKAQAA 61
Query: 61 SVIVLCLGDKVLRKVAKESTAASMWPKLEALYMTKSLTHQQFLKQ*LYSFKMVKSKAIME 120
+ I L L D V+ +V E + +W KL +L+M+KSLT + +LKQ LY ++ + + +
Sbjct: 62 ATIRLSLSDSVMYQVMDEKSPKEIWDKLASLHMSKSLTSKLYLKQQLYGLQVQEESDLRK 121
Query: 121 LLVEFNKIIGDLENIEVRLEDVD 143
+ FN+++ DL ++V+L+D D
Sbjct: 122 HVDVFNQLVVDLSKLDVKLDDED 144
>gb|AAP53029.1| putative retrotransposon-related protein [Oryza sativa (japonica
cultivar-group)] gi|37532880|ref|NP_920742.1| putative
retrotransposon-related protein [Oryza sativa (japonica
cultivar-group)] gi|22655747|gb|AAN04164.1| Putative
retrotransposon protein [Oryza sativa (japonica
cultivar-group)] gi|16905223|gb|AAL31093.1| putative
retrotransposon-related protein [Oryza sativa]
Length = 1229
Score = 83.2 bits (204), Expect = 1e-15
Identities = 51/140 (36%), Positives = 82/140 (58%), Gaps = 1/140 (0%)
Query: 5 KWDIEKFTGDNDFGLLKVKMEAMLIQQK-CEKALKGKGSLPVTMSRAEKTEMVDKARSVI 63
K+D+ F L +VKM A+L Q ++AL+ G T AE+ KA S+I
Sbjct: 2 KYDLPLQDYKTRFSLWQVKMRAVLAQTSDLDEALESFGKKKTTEWTAEEKRKDRKALSLI 61
Query: 64 VLCLGDKVLRKVAKESTAASMWPKLEALYMTKSLTHQQFLKQ*LYSFKMVKSKAIMELLV 123
L L + +L+KV +E TAA +W KLE++ M+K LT + +K L+S K+ +S +++ +
Sbjct: 62 QLHLSNDILQKVLQEKTAAELWFKLESICMSKDLTSKMHIKMKLFSHKLQESGSVLNHIS 121
Query: 124 EFNKIIGDLENIEVRLEDVD 143
F +II DL ++EV+ +D D
Sbjct: 122 VFKEIIADLVSMEVQFDDED 141
>ref|XP_469192.1| putative polyprotein [Oryza sativa (japonica cultivar-group)]
gi|53370655|gb|AAU89150.1| integrase core domain
containing protein [Oryza sativa (japonica
cultivar-group)] gi|40538906|gb|AAR87163.1| putative
polyprotein [Oryza sativa (japonica cultivar-group)]
Length = 1322
Score = 80.9 bits (198), Expect = 6e-15
Identities = 48/140 (34%), Positives = 82/140 (58%), Gaps = 1/140 (0%)
Query: 5 KWDIEKFTGDNDFGLLKVKMEAMLIQQK-CEKALKGKGSLPVTMSRAEKTEMVDKARSVI 63
K+D+ F L +VKM A+L Q ++AL+ G T AE+ KA S+I
Sbjct: 5 KYDLPLLDYKTRFSLWQVKMRAVLAQTSDLDEALESFGKKKTTEWTAEEKRKDRKALSLI 64
Query: 64 VLCLGDKVLRKVAKESTAASMWPKLEALYMTKSLTHQQFLKQ*LYSFKMVKSKAIMELLV 123
L L + +L++V ++ TAA +W KLE++ M+K LT + +K L+S K+ +S +++ +
Sbjct: 65 QLHLSNDILQEVLQKKTAAELWLKLESICMSKDLTSKMHIKMKLFSHKLHESGSVLNHIS 124
Query: 124 EFNKIIGDLENIEVRLEDVD 143
F +I+ DL ++EV+ +D D
Sbjct: 125 VFKEIVADLVSMEVQFDDED 144
Database: nr
Posted date: Jul 5, 2005 12:34 AM
Number of letters in database: 863,360,394
Number of sequences in database: 2,540,612
Lambda K H
0.322 0.135 0.377
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 198,073,174
Number of Sequences: 2540612
Number of extensions: 6136301
Number of successful extensions: 14500
Number of sequences better than 10.0: 254
Number of HSP's better than 10.0 without gapping: 173
Number of HSP's successfully gapped in prelim test: 81
Number of HSP's that attempted gapping in prelim test: 14288
Number of HSP's gapped (non-prelim): 263
length of query: 145
length of database: 863,360,394
effective HSP length: 121
effective length of query: 24
effective length of database: 555,946,342
effective search space: 13342712208
effective search space used: 13342712208
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.9 bits)
S2: 67 (30.4 bits)
Medicago: description of AC139600.9


