

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC139525.8 - phase: 0
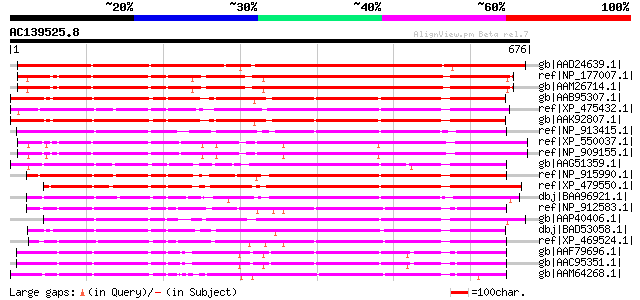
(676 letters)
Database: nr
2,540,612 sequences; 863,360,394 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
gb|AAD24639.1| putative receptor-like protein kinase [Arabidopsi... 866 0.0
ref|NP_177007.1| leucine-rich repeat transmembrane protein kinas... 515 e-144
gb|AAM26714.1| At1g68400/T2E12_5 [Arabidopsis thaliana] gi|14190... 514 e-144
gb|AAB95307.1| putative receptor-like protein kinase [Arabidopsi... 474 e-132
ref|XP_475432.1| putative phytosulfokine receptor kinase [Oryza ... 471 e-131
gb|AAK92807.1| putative receptor protein kinase [Arabidopsis tha... 471 e-131
ref|NP_913415.1| unnamed protein product [Oryza sativa (japonica... 466 e-129
ref|XP_550037.1| putative atypical receptor-like kinase MARK [Or... 465 e-129
ref|NP_909155.1| putative receptor kinase [Oryza sativa (japonic... 465 e-129
gb|AAG51359.1| putative protein kinase; 49514-51513 [Arabidopsis... 461 e-128
ref|NP_915990.1| putative receptor-like protein kinase [Oryza sa... 460 e-128
ref|XP_479550.1| putative receptor-like protein kinase [Oryza sa... 454 e-126
dbj|BAA96921.1| receptor-like protein kinase [Arabidopsis thalia... 449 e-124
ref|NP_912583.1| Putative leucine-rich repeat transmembrane prot... 428 e-118
gb|AAP40406.1| putative leucine-rich repeat transmembrane protei... 427 e-118
dbj|BAD53058.1| receptor-like protein kinase 1-like [Oryza sativ... 426 e-117
ref|XP_469524.1| putative receptor kinase [Oryza sativa] gi|1332... 425 e-117
gb|AAF79696.1| T1N15.9 [Arabidopsis thaliana] gi|18402209|ref|NP... 424 e-117
gb|AAC95351.1| receptor-like protein kinase [Arabidopsis thaliana] 424 e-117
gb|AAM64268.1| receptor kinase, putative [Arabidopsis thaliana] 423 e-117
>gb|AAD24639.1| putative receptor-like protein kinase [Arabidopsis thaliana]
gi|25408487|pir||B84782 probable receptor-like protein
kinase [imported] - Arabidopsis thaliana
gi|15227998|ref|NP_181196.1| leucine-rich repeat
transmembrane protein kinase, putative [Arabidopsis
thaliana]
Length = 672
Score = 866 bits (2238), Expect = 0.0
Identities = 455/675 (67%), Positives = 537/675 (79%), Gaps = 26/675 (3%)
Query: 11 LFLSIYIVPCLTHNDTQALTLFRQQTDTHGQLLTNWTGPEACSASWHGVTCTPNN-RVTT 69
L L +++ L NDT ALTLFR QTDTHG L NWTG +AC++SW GV+C+P++ RVT
Sbjct: 10 LLLLLHLSITLAQNDTNALTLFRLQTDTHGNLAGNWTGSDACTSSWQGVSCSPSSHRVTE 69
Query: 70 LVLPSLNLRGPIDALSSLTHLRLLDLHNNRLNGTVSASLLSNCTNLKLLYLAGNDFSGQI 129
L LPSL+LRGP+ +LSSL LRLLDLH+NRLNGTVS L+NC NL+L+YLAGND SG+I
Sbjct: 70 LSLPSLSLRGPLTSLSSLDQLRLLDLHDNRLNGTVSP--LTNCKNLRLVYLAGNDLSGEI 127
Query: 130 PPEISSLNNLLRLDLSDNNLAGDIPNEISRLTNLLTLRLQNNALSGNIPDLSSIMPNLTE 189
P EIS L ++RLDLSDNN+ G IP EI T +LT+R+QNN L+G IPD S M +L E
Sbjct: 128 PKEISFLKRMIRLDLSDNNIRGVIPREILGFTRVLTIRIQNNELTGRIPDFSQ-MKSLLE 186
Query: 190 LNMTNNEFYGKVPNTMLNKFGDESFSGNEGLCGSKPFQVCSLTENSPPSSEPVQTVPSNP 249
LN++ NE +G V + ++ KFGD SFSGNEGLCGS P VC++T N P SS Q VPSNP
Sbjct: 187 LNVSFNELHGNVSDGVVKKFGDLSFSGNEGLCGSDPLPVCTIT-NDPESSNTDQIVPSNP 245
Query: 250 SSFPATSVIAR-PRSQHHKGLSPGVIVAIVVAICVALLVVTSFVVAHCCAR----GRGVN 304
+S P + V R P H+G+ PG+I A++ CVA++V+ SF A CC R G
Sbjct: 246 TSIPHSPVSVREPEIHSHRGIKPGIIAAVIGG-CVAVIVLVSFGFAFCCGRLDRNGERSK 304
Query: 305 SNSLMGSEAGKRKSYGSEKKVYNSNGGGGDSSDGTSGTDMSKLVFFDRRNGFELEDLLRA 364
S S+ G G E K +S G GG+S D TS TD S+LVFF+RR FEL+DLL+A
Sbjct: 305 SGSVETGFVG-----GGEGKRRSSYGEGGES-DATSATDRSRLVFFERRKQFELDDLLKA 358
Query: 365 SAEMLGKGSLGTVYRAVLDDGST-VAVKRLKDANPCARHEFEQYMDVIGKLKHPNIVKLR 423
SAEMLGKGSLGTVY+AVLDDGST VAVKRLKDANPC R EFEQYM++IG+LKH N+VKLR
Sbjct: 359 SAEMLGKGSLGTVYKAVLDDGSTTVAVKRLKDANPCPRKEFEQYMEIIGRLKHQNVVKLR 418
Query: 424 AYYYAKEEKLLVYDYLSNGSLHALLHGNRGPGRIPLDWTTRISLVLGAARGLARIHTEYS 483
AYYYAKEEKLLVY+YL NGSLH+LLHGNRGPGRIPLDWTTRISL+LGAARGLA+IH EYS
Sbjct: 419 AYYYAKEEKLLVYEYLPNGSLHSLLHGNRGPGRIPLDWTTRISLMLGAARGLAKIHDEYS 478
Query: 484 AAKVPHGNVKSSNVLLDKNGVACISDFGLSLLLNPVHATARLGGYRAPEQTEQKRLSQQA 543
+K+PHGN+KSSNVLLD+NGVA I+DFGLSLLLNPVHA ARLGGYRAPEQ+E KRLSQ+A
Sbjct: 479 ISKIPHGNIKSSNVLLDRNGVALIADFGLSLLLNPVHAIARLGGYRAPEQSEIKRLSQKA 538
Query: 544 DVYSFGVLLLEVLTGKAPSLQYPSPANRPRKV------EEEETVVDLPKWVRSVVREEWT 597
DVYSFGVLLLEVLTGKAPS+ +PSP +RPR EEEE VVDLPKWVRSVV+EEWT
Sbjct: 539 DVYSFGVLLLEVLTGKAPSI-FPSP-SRPRSAASVAVEEEEEAVVDLPKWVRSVVKEEWT 596
Query: 598 GEVFDQELLRYKNIEEELVSMLHVGLACVVQQPEKRPTMVDVVKMIEDIRVEQSPLCEDY 657
EVFD ELLRYKNIEEE+V+MLH+GLACVV QPEKRPTM +VVKM+E+IRVEQSP+ ED+
Sbjct: 597 AEVFDPELLRYKNIEEEMVAMLHIGLACVVPQPEKRPTMAEVVKMVEEIRVEQSPVGEDF 656
Query: 658 DESRNSLSPSIPTTE 672
DESRNS+SPS+ TT+
Sbjct: 657 DESRNSMSPSLATTD 671
>ref|NP_177007.1| leucine-rich repeat transmembrane protein kinase, putative
[Arabidopsis thaliana] gi|25404733|pir||H96707 probable
receptor kinase T2E12.5 [imported] - Arabidopsis
thaliana gi|6714351|gb|AAF26042.1| putative receptor
kinase; 18202-20717 [Arabidopsis thaliana]
Length = 670
Score = 515 bits (1326), Expect = e-144
Identities = 309/670 (46%), Positives = 419/670 (62%), Gaps = 58/670 (8%)
Query: 11 LFLSIYIVPCL----THNDTQALTLFRQQTDTHGQLLTNWTGPEACSASWHGVTCTPNNR 66
L L I + CL + D++ L F+ D+ G+L + T C W GV+C NR
Sbjct: 13 LSLLILLQSCLLSSSSSTDSETLLNFKLTADSTGKLNSWNTTTNPCQ--WTGVSCN-RNR 69
Query: 67 VTTLVLPSLNLRGPIDALSSLTHLRLLDLHNNRLNGTVSASLLSNCTNLKLLYLAGNDFS 126
VT LVL +NL G I +L+SLT LR+L L +N L+G + LSN T LKLL+L+ N FS
Sbjct: 70 VTRLVLEDINLTGSISSLTSLTSLRVLSLKHNNLSGPIPN--LSNLTALKLLFLSNNQFS 127
Query: 127 GQIPPEISSLNNLLRLDLSDNNLAGDIPNEISRLTNLLTLRLQNNALSGNIPDLSSIMPN 186
G P I+SL L RLDLS NN +G IP +++ LT+LLTLRL++N SG IP+++ + +
Sbjct: 128 GNFPTSITSLTRLYRLDLSFNNFSGQIPPDLTDLTHLLTLRLESNRFSGQIPNIN--LSD 185
Query: 187 LTELNMTNNEFYGKVPNTMLNKFGDESFSGNEGLCGSKPFQVCSLTENSP---------- 236
L + N++ N F G++PN+ L++F + F+ N LCG+ P C+ + P
Sbjct: 186 LQDFNVSGNNFNGQIPNS-LSQFPESVFTQNPSLCGA-PLLKCTKLSSDPTKPGRPDEAK 243
Query: 237 --PSSEPVQTVPSNPSSFPATSVIARPRSQHHKGLSPGVIVAIVVAICVALLVVTSFVVA 294
P ++P +TVPS+P TS+ +S + +S ++AI++ + L V S ++
Sbjct: 244 ASPLNKP-ETVPSSP-----TSIHGGDKSNNTSRISTISLIAIILGDFIILSFV-SLLLY 296
Query: 295 HCCARGRGVNSNSLMGSEAGKRKSYGSEKKVYNSN----GGGGDSSDGTSGTDMSKLVFF 350
+C R VN K EK VY+SN +++ D K+VFF
Sbjct: 297 YCFWRQYAVNKKK-------HSKILEGEKIVYSSNPYPTSTQNNNNQNQQVGDKGKMVFF 349
Query: 351 DRRNGFELEDLLRASAEMLGKGSLGTVYRAVLDDGSTVAVKRLKDANPCA-RHEFEQYMD 409
+ FELEDLLRASAEMLGKG GT Y+AVL+DG+ VAVKRLKDA A + EFEQ M+
Sbjct: 350 EGTRRFELEDLLRASAEMLGKGGFGTAYKAVLEDGNEVAVKRLKDAVTVAGKKEFEQQME 409
Query: 410 VIGKLKHPNIVKLRAYYYAKEEKLLVYDYLSNGSLHALLHGNRGPGRIPLDWTTRISLVL 469
V+G+L+H N+V L+AYY+A+EEKLLVYDY+ NGSL LLHGNRGPGR PLDWTTR+ +
Sbjct: 410 VLGRLRHTNLVSLKAYYFAREEKLLVYDYMPNGSLFWLLHGNRGPGRTPLDWTTRLKIAA 469
Query: 470 GAARGLARIHTEYSAAKVPHGNVKSSNVLLDKNGVACISDFGLSLLLNPVHATARLGGYR 529
GAARGLA IH K+ HG++KS+NVLLD++G A +SDFGLS+ P A+ GYR
Sbjct: 470 GAARGLAFIHGSCKTLKLTHGDIKSTNVLLDRSGNARVSDFGLSIFA-PSQTVAKSNGYR 528
Query: 530 APEQTEQKRLSQQADVYSFGVLLLEVLTGKAPSLQYPSPANRPRKVEEEETVVDLPKWVR 589
APE + ++ +Q++DVYSFGVLLLE+LTGK P++ + VDLP+WV+
Sbjct: 529 APELIDGRKHTQKSDVYSFGVLLLEILTGKCPNMV---------ETGHSGGAVDLPRWVQ 579
Query: 590 SVVREEWTGEVFDQELLRYKNIEEELVSMLHVGLACVVQQPEKRPTMVDVVKMIEDIR-- 647
SVVREEWT EVFD EL+RYK+IEEE+V +L + +AC + RP M VVK+IEDIR
Sbjct: 580 SVVREEWTAEVFDLELMRYKDIEEEMVGLLQIAMACTAVAADHRPKMGHVVKLIEDIRGG 639
Query: 648 -VEQSPLCED 656
E SP C D
Sbjct: 640 GSEASP-CND 648
>gb|AAM26714.1| At1g68400/T2E12_5 [Arabidopsis thaliana] gi|14190425|gb|AAK55693.1|
At1g68400/T2E12_5 [Arabidopsis thaliana]
Length = 671
Score = 514 bits (1325), Expect = e-144
Identities = 309/671 (46%), Positives = 419/671 (62%), Gaps = 59/671 (8%)
Query: 11 LFLSIYIVPCL----THNDTQALTLFRQQTDTHGQLLTNWTGPEACSASWHGVTCTPNNR 66
L L I + CL + D++ L F+ D+ G+L + T C W GV+C NR
Sbjct: 13 LSLLILLQSCLLSSSSSTDSETLLNFKLTADSTGKLNSWNTTTNPCQ--WTGVSCN-RNR 69
Query: 67 VTTLVLPSLNLRGPIDALSSLTHLRLLDLHNNRLNGTVSASLLSNCTNLKLLYLAGNDFS 126
VT LVL +NL G I +L+SLT LR+L L +N L+G + LSN T LKLL+L+ N FS
Sbjct: 70 VTRLVLEDINLTGSISSLTSLTSLRVLSLKHNNLSGPIPN--LSNLTALKLLFLSNNQFS 127
Query: 127 GQIPPEISSLNNLLRLDLSDNNLAGDIPNEISRLTNLLTLRLQNNALSGNIPDLSSIMPN 186
G P I+SL L RLDLS NN +G IP +++ LT+LLTLRL++N SG IP+++ + +
Sbjct: 128 GNFPTSITSLTRLYRLDLSFNNFSGQIPPDLTDLTHLLTLRLESNRFSGQIPNIN--LSD 185
Query: 187 LTELNMTNNEFYGKVPNTMLNKFGDESFSGNEGLCGSKPFQVCSLTENSP---------- 236
L + N++ N F G++PN+ L++F + F+ N LCG+ P C+ + P
Sbjct: 186 LQDFNVSGNNFNGQIPNS-LSQFPESVFTQNPSLCGA-PLLKCTKLSSDPTKPGRPDEAK 243
Query: 237 --PSSEPVQTVPSNPSSFPATSVIARPRSQHHKGLSPGVIVAIVVAICVALLVVTSFVVA 294
P ++P +TVPS+P TS+ +S + +S ++AI++ + L V S ++
Sbjct: 244 ASPLNKP-ETVPSSP-----TSIHGGDKSNNTSRISTISLIAIILGDFIILSFV-SLLLY 296
Query: 295 HCCARGRGVNSNSLMGSEAGKRKSYGSEKKVYNSN----GGGGDSSDGTSGTDMSKLVFF 350
+C R VN K EK VY+SN +++ D K+VFF
Sbjct: 297 YCFWRQYAVNKKK-------HSKILEGEKIVYSSNPYPTSTQNNNNQNQQVGDKGKMVFF 349
Query: 351 DRRNGFELEDLLRASAEMLGKGSLGTVYRAVLDDGSTVAVKRLKDANPCA--RHEFEQYM 408
+ FELEDLLRASAEMLGKG GT Y+AVL+DG+ VAVKRLKDA A + EFEQ M
Sbjct: 350 EGTRRFELEDLLRASAEMLGKGGFGTAYKAVLEDGNEVAVKRLKDAVTVAGKKKEFEQQM 409
Query: 409 DVIGKLKHPNIVKLRAYYYAKEEKLLVYDYLSNGSLHALLHGNRGPGRIPLDWTTRISLV 468
+V+G+L+H N+V L+AYY+A+EEKLLVYDY+ NGSL LLHGNRGPGR PLDWTTR+ +
Sbjct: 410 EVLGRLRHTNLVSLKAYYFAREEKLLVYDYMPNGSLFWLLHGNRGPGRTPLDWTTRLKIA 469
Query: 469 LGAARGLARIHTEYSAAKVPHGNVKSSNVLLDKNGVACISDFGLSLLLNPVHATARLGGY 528
GAARGLA IH K+ HG++KS+NVLLD++G A +SDFGLS+ P A+ GY
Sbjct: 470 AGAARGLAFIHGSCKTLKLTHGDIKSTNVLLDRSGNARVSDFGLSIFA-PSQTVAKSNGY 528
Query: 529 RAPEQTEQKRLSQQADVYSFGVLLLEVLTGKAPSLQYPSPANRPRKVEEEETVVDLPKWV 588
RAPE + ++ +Q++DVYSFGVLLLE+LTGK P++ + VDLP+WV
Sbjct: 529 RAPELIDGRKHTQKSDVYSFGVLLLEILTGKCPNMV---------ETGHSGGAVDLPRWV 579
Query: 589 RSVVREEWTGEVFDQELLRYKNIEEELVSMLHVGLACVVQQPEKRPTMVDVVKMIEDIR- 647
+SVVREEWT EVFD EL+RYK+IEEE+V +L + +AC + RP M VVK+IEDIR
Sbjct: 580 QSVVREEWTAEVFDLELMRYKDIEEEMVGLLQIAMACTAVAADHRPKMGHVVKLIEDIRG 639
Query: 648 --VEQSPLCED 656
E SP C D
Sbjct: 640 GGSEASP-CND 649
>gb|AAB95307.1| putative receptor-like protein kinase [Arabidopsis thaliana]
gi|60543329|gb|AAX22262.1| At2g26730 [Arabidopsis
thaliana] gi|25407845|pir||B84664 probable receptor-like
protein kinase [imported] - Arabidopsis thaliana
gi|15225780|ref|NP_180241.1| leucine-rich repeat
transmembrane protein kinase, putative [Arabidopsis
thaliana]
Length = 658
Score = 474 bits (1220), Expect = e-132
Identities = 288/660 (43%), Positives = 401/660 (60%), Gaps = 58/660 (8%)
Query: 1 MNYVIMFFFFLFLSIYIVPCLTHNDTQALTLFRQQTDTHGQLLTNWTGPEACSASWHGVT 60
+++V+ F + L V + + QAL F QQ +L N AC+ W GV
Sbjct: 4 ISWVLNSLFSILLLTQRVNSESTAEKQALLTFLQQIPHENRLQWN-ESDSACN--WVGVE 60
Query: 61 CTPN-NRVTTLVLPSLNLRG--PIDALSSLTHLRLLDLHNNRLNGTVSASLLSNCTNLKL 117
C N + + +L LP L G P +L LT LR+L L +NRL+G + + SN T+L+
Sbjct: 61 CNSNQSSIHSLRLPGTGLVGQIPSGSLGRLTELRVLSLRSNRLSGQIPSDF-SNLTHLRS 119
Query: 118 LYLAGNDFSGQIPPEISSLNNLLRLDLSDNNLAGDIPNEISRLTNLLTLRLQNNALSGNI 177
LYL N+FSG+ P + LNNL+RLD+S NN G IP ++ LT+L L L NN SGN+
Sbjct: 120 LYLQHNEFSGEFPTSFTQLNNLIRLDISSNNFTGSIPFSVNNLTHLTGLFLGNNGFSGNL 179
Query: 178 PDLSSIMPNLTELNMTNNEFYGKVPNTMLNKFGDESFSGNEGLCGSKPFQVCSLTENSPP 237
P +S L + N++NN G +P++ L++F ESF+GN LCG P + C SP
Sbjct: 180 PSISL---GLVDFNVSNNNLNGSIPSS-LSRFSAESFTGNVDLCGG-PLKPCKSFFVSP- 233
Query: 238 SSEPVQTVPSNPSSFPATSVIARPRSQHHKGLSPGVIVAIVVAICVALLVVTSFVVAHCC 297
S P PSN R S+ K LS IVAI+VA + L++ + ++ C
Sbjct: 234 SPSPSLINPSN-----------RLSSKKSK-LSKAAIVAIIVASALVALLLLALLLFLCL 281
Query: 298 ARGRGVN-SNSLMGSEAGKRK---------SYGSEKKVYNSNGGGGDSSDGTSGTDMSKL 347
+ RG N + + AG S E+ S+G GG+ T+ +KL
Sbjct: 282 RKRRGSNEARTKQPKPAGVATRNVDLPPGASSSKEEVTGTSSGMGGE-------TERNKL 334
Query: 348 VFFDRR-NGFELEDLLRASAEMLGKGSLGTVYRAVLDDGSTVAVKRLKDANPCARHEFEQ 406
VF + F+LEDLLRASAE+LGKGS+GT Y+AVL++G+TV VKRLKD ++ EFE
Sbjct: 335 VFTEGGVYSFDLEDLLRASAEVLGKGSVGTSYKAVLEEGTTVVVKRLKDVM-ASKKEFET 393
Query: 407 YMDVIGKLKHPNIVKLRAYYYAKEEKLLVYDYLSNGSLHALLHGNRGPGRIPLDWTTRIS 466
M+V+GK+KHPN++ LRAYYY+K+EKLLV+D++ GSL ALLHG+RG GR PLDW R+
Sbjct: 394 QMEVVGKIKHPNVIPLRAYYYSKDEKLLVFDFMPTGSLSALLHGSRGSGRTPLDWDNRMR 453
Query: 467 LVLGAARGLARIHTEYSAAKVPHGNVKSSNVLLDKNGVACISDFGLSLLLNPVHATARLG 526
+ + AARGLA +H +AK+ HGN+K+SN+LL N C+SD+GL+ L + RL
Sbjct: 454 IAITAARGLAHLHV---SAKLVHGNIKASNILLHPNQDTCVSDYGLNQLFSNSSPPNRLA 510
Query: 527 GYRAPEQTEQKRLSQQADVYSFGVLLLEVLTGKAPSLQYPSPANRPRKVEEEETVVDLPK 586
GY APE E ++++ ++DVYSFGVLLLE+LTGK+P+ + E +DLP+
Sbjct: 511 GYHAPEVLETRKVTFKSDVYSFGVLLLELLTGKSPN-----------QASLGEEGIDLPR 559
Query: 587 WVRSVVREEWTGEVFDQELLRYKNIEEELVSMLHVGLACVVQQPEKRPTMVDVVKMIEDI 646
WV SVVREEWT EVFD EL+RY NIEEE+V +L + +ACV P++RP M +V++MIED+
Sbjct: 560 WVLSVVREEWTAEVFDVELMRYHNIEEEMVQLLQIAMACVSTVPDQRPVMQEVLRMIEDV 619
>ref|XP_475432.1| putative phytosulfokine receptor kinase [Oryza sativa (japonica
cultivar-group)] gi|46576015|gb|AAT01376.1| putative
phytosulfokine receptor kinase [Oryza sativa (japonica
cultivar-group)]
Length = 638
Score = 471 bits (1212), Expect = e-131
Identities = 301/661 (45%), Positives = 395/661 (59%), Gaps = 51/661 (7%)
Query: 1 MNYVIMFFF----FLFLSIYIVPCLTHN-DTQALTLFRQQTDTHGQLLTNWTGPEACSAS 55
M ++++ F FLFL I C N D QAL F HG+ L NWT S
Sbjct: 1 MQHLVLIAFLSASFLFLHIPCARCADLNSDRQALLAFAASVP-HGRKL-NWTLTTQVCTS 58
Query: 56 WHGVTCTPNNR-VTTLVLPSLNLRGPI--DALSSLTHLRLLDLHNNRLNGTVSASLLSNC 112
W G+TCTP+ R V L LP++ L GPI D L L L++L L +NRL ++ + S
Sbjct: 59 WVGITCTPDGRRVRELRLPAVGLFGPIPSDTLGKLDALQVLSLRSNRLTISLPPDVAS-I 117
Query: 113 TNLKLLYLAGNDFSGQIPPEISSLNNLLRLDLSDNNLAGDIPNEISRLTNLLTLRLQNNA 172
+L LYL N+ SG IP +SS NL LDLS N+ G+IP ++ +T L L LQNN+
Sbjct: 118 PSLHSLYLQHNNLSGIIPTSLSS--NLTFLDLSYNSFDGEIPLKVQNITQLTALLLQNNS 175
Query: 173 LSGNIPDLSSIMPNLTELNMTNNEFYGKVPNTMLNKFGDESFSGNEGLCGSKPFQVCSLT 232
LSG IPDL +PNL LN++NN G +P + L KF SF GN LCG P + C T
Sbjct: 176 LSGPIPDLH--LPNLRHLNLSNNNLSGPIPPS-LQKFPASSFFGNAFLCGL-PLEPCPGT 231
Query: 233 ENSPPSSEPVQTVPSNPSSFPATSVIARPRSQHHKGLSPGVIVAIVVAICVALLVVTSFV 292
SP P+ P+ SF K LS GVI+AI + LL++ +
Sbjct: 232 APSPSPMSPLP--PNTKKSF-------------WKRLSLGVIIAIAAGGGLLLLILIVVL 276
Query: 293 VAHCCARGRGVNSNSLMGSEAGKRKSYGSEKKVYNSNGGGGDSSDGTSGTDMSKLVFFDR 352
+ R + S GK + G +K SS G + +KL+FF+
Sbjct: 277 LICIFKRKKDGEPGIASFSSKGKAAAGGRAEKSKQEY-----SSSGIQEAERNKLIFFNG 331
Query: 353 RN-GFELEDLLRASAEMLGKGSLGTVYRAVLDDGSTVAVKRLKDANPCARHEFEQYMDVI 411
+ F+LEDLLRASAE+LGKGS GT Y+AVL+DG+TV VKRLK+ + EFEQ M++I
Sbjct: 332 CSYNFDLEDLLRASAEVLGKGSYGTTYKAVLEDGTTVVVKRLKEV-VAGKREFEQQMEII 390
Query: 412 GKL-KHPNIVKLRAYYYAKEEKLLVYDYLSNGSLHALLHGNRGPGRIPLDWTTRISLVLG 470
G++ +H N V+LRAYYY+K+EKLLVYDY++ GSL A LHGNR GR LDW TR+ + L
Sbjct: 391 GRVGQHQNAVQLRAYYYSKDEKLLVYDYMTPGSLCAALHGNRTAGRTTLDWATRVKISLE 450
Query: 471 AARGLARIHTEYSAAKVPHGNVKSSNVLLDKNGVACISDFGLSLLLNPVHATARLGGYRA 530
AARG+A +H E K HGN+KSSN+LL + ACIS+FGL+ L+ H ARL GYRA
Sbjct: 451 AARGIAHLHAE-GGGKFIHGNIKSSNILLSQGLSACISEFGLAQLMAIPHIPARLIGYRA 509
Query: 531 PEQTEQKRLSQQADVYSFGVLLLEVLTGKAPSLQYPSPANRPRKVEEEETVVDLPKWVRS 590
PE E KR +Q++DVYS+GVLLLE+LTGKAP R E+++ LP+WV+S
Sbjct: 510 PEVLETKRQTQKSDVYSYGVLLLEMLTGKAPL----------RSPGREDSIEHLPRWVQS 559
Query: 591 VVREEWTGEVFDQELLRYKNIEEELVSMLHVGLACVVQQPEKRPTMVDVVKMIEDIRVEQ 650
VVREEWT EVFD +LLR+ N E+E+V ML + +ACV P++RP M +VV+ IE+IR
Sbjct: 560 VVREEWTSEVFDADLLRHPNSEDEMVQMLQLAMACVAIVPDQRPRMEEVVRRIEEIRNSS 619
Query: 651 S 651
S
Sbjct: 620 S 620
>gb|AAK92807.1| putative receptor protein kinase [Arabidopsis thaliana]
Length = 658
Score = 471 bits (1212), Expect = e-131
Identities = 287/660 (43%), Positives = 400/660 (60%), Gaps = 58/660 (8%)
Query: 1 MNYVIMFFFFLFLSIYIVPCLTHNDTQALTLFRQQTDTHGQLLTNWTGPEACSASWHGVT 60
+++V+ F + L V + + QAL F QQ +L N AC+ W GV
Sbjct: 4 ISWVLNSLFSILLLTQRVNSESTAEKQALLTFLQQIPHENRLQWN-ESDSACN--WVGVE 60
Query: 61 CTPN-NRVTTLVLPSLNLRG--PIDALSSLTHLRLLDLHNNRLNGTVSASLLSNCTNLKL 117
C N + + +L LP L G P +L LT LR+L L +NRL+G + + SN T+L+
Sbjct: 61 CNSNQSSIHSLRLPGTGLVGQIPSGSLGRLTELRVLSLRSNRLSGQIPSDF-SNLTHLRS 119
Query: 118 LYLAGNDFSGQIPPEISSLNNLLRLDLSDNNLAGDIPNEISRLTNLLTLRLQNNALSGNI 177
LYL N+FSG+ P + LNNL+RLD+S NN G IP ++ LT+L L L NN SGN+
Sbjct: 120 LYLQHNEFSGEFPTSFTQLNNLIRLDISSNNFTGSIPFSVNNLTHLTGLFLGNNGFSGNL 179
Query: 178 PDLSSIMPNLTELNMTNNEFYGKVPNTMLNKFGDESFSGNEGLCGSKPFQVCSLTENSPP 237
P +S L + N++NN G +P++ L++F ESF+GN LCG P + C SP
Sbjct: 180 PSISL---GLVDFNVSNNNLNGSIPSS-LSRFSAESFTGNVDLCGG-PLKPCKSFFVSP- 233
Query: 238 SSEPVQTVPSNPSSFPATSVIARPRSQHHKGLSPGVIVAIVVAICVALLVVTSFVVAHCC 297
S P PSN R S+ K LS IVAI+VA + L++ + ++ C
Sbjct: 234 SPSPSLINPSN-----------RLSSKKSK-LSKAAIVAIIVASALVALLLLALLLFLCL 281
Query: 298 ARGRGVN-SNSLMGSEAGKRK---------SYGSEKKVYNSNGGGGDSSDGTSGTDMSKL 347
+ RG N + + AG S E+ S+G GG+ T+ +KL
Sbjct: 282 RKRRGSNEARTKQPKPAGVATRNVDLPPGASSSKEEVTGTSSGMGGE-------TERNKL 334
Query: 348 VFFDRR-NGFELEDLLRASAEMLGKGSLGTVYRAVLDDGSTVAVKRLKDANPCARHEFEQ 406
VF + F+LEDLLRASAE+LGKGS+GT Y+AVL++G+TV VKRLKD ++ EFE
Sbjct: 335 VFTEGGVYSFDLEDLLRASAEVLGKGSVGTSYKAVLEEGTTVVVKRLKDVM-ASKKEFET 393
Query: 407 YMDVIGKLKHPNIVKLRAYYYAKEEKLLVYDYLSNGSLHALLHGNRGPGRIPLDWTTRIS 466
M+V+GK+K PN++ LRAYYY+K+EKLLV+D++ GSL ALLHG+RG GR PLDW R+
Sbjct: 394 QMEVVGKIKRPNVIPLRAYYYSKDEKLLVFDFMPTGSLSALLHGSRGSGRTPLDWDNRMR 453
Query: 467 LVLGAARGLARIHTEYSAAKVPHGNVKSSNVLLDKNGVACISDFGLSLLLNPVHATARLG 526
+ + AARGLA +H +AK+ HGN+K+SN+LL N C+SD+GL+ L + RL
Sbjct: 454 IAITAARGLAHLHV---SAKLVHGNIKASNILLHPNQDTCVSDYGLNQLFSNSSPPNRLA 510
Query: 527 GYRAPEQTEQKRLSQQADVYSFGVLLLEVLTGKAPSLQYPSPANRPRKVEEEETVVDLPK 586
GY APE E ++++ ++DVYSFGVLLLE+LTGK+P+ + E +DLP+
Sbjct: 511 GYHAPEVLETRKVTFKSDVYSFGVLLLELLTGKSPN-----------QASLGEEGIDLPR 559
Query: 587 WVRSVVREEWTGEVFDQELLRYKNIEEELVSMLHVGLACVVQQPEKRPTMVDVVKMIEDI 646
WV SVVREEWT EVFD EL+RY NIEEE+V +L + +ACV P++RP M +V++MIED+
Sbjct: 560 WVLSVVREEWTAEVFDVELMRYHNIEEEMVQLLQIAMACVSTVPDQRPVMQEVLRMIEDV 619
>ref|NP_913415.1| unnamed protein product [Oryza sativa (japonica cultivar-group)]
gi|9711799|dbj|BAB07903.1| putative receptor-like kinase
[Oryza sativa (japonica cultivar-group)]
gi|7573610|dbj|BAA94519.1| putative receptor-like kinase
[Oryza sativa (japonica cultivar-group)]
Length = 641
Score = 466 bits (1199), Expect = e-129
Identities = 281/646 (43%), Positives = 386/646 (59%), Gaps = 54/646 (8%)
Query: 10 FLFLSIYIVPCLTHNDTQALTLFRQQTDTHG-QLLTNWTGPEACSASWHGVTCTPN-NRV 67
FL L++ +V C D L G + NW +W GVTC+ + +RV
Sbjct: 12 FLALALVVVVCAAEPDADRAALLDFLAGLGGGRGRINWASSPRVCGNWTGVTCSGDGSRV 71
Query: 68 TTLVLPSLNLRGPID--ALSSLTHLRLLDLHNNRLNGTVSASLLSNCTNLKLLYLAGNDF 125
L LP L L GP+ L LT L++L L N L+G LLS +L L+L N F
Sbjct: 72 VALRLPGLGLSGPVPRGTLGRLTALQVLSLRANSLSGEFPEELLS-LASLTGLHLQLNAF 130
Query: 126 SGQIPPEISSLNNLLRLDLSDNNLAGDIPNEISRLTNLLTLRLQNNALSGNIPDLSSIMP 185
SG +PPE++ L L LDLS N G +P +S LT L+ L L NN+LSG +PDL +P
Sbjct: 131 SGALPPELARLRALQVLDLSFNGFNGTLPAALSNLTQLVALNLSNNSLSGRVPDLG--LP 188
Query: 186 NLTELNMTNNEFYGKVPNTMLNKFGDESFSGNEGLCGSKPFQVCSLTENSPPSSEPVQTV 245
L LN++NN G VP ++L +F D +F+GN P S+ P T
Sbjct: 189 ALQFLNLSNNHLDGPVPTSLL-RFNDTAFAGNN--------------VTRPASASPAGTP 233
Query: 246 PSNPSSFPATSVIARPRSQHHKGLSPGVIVAIVVAICVAL-LVVTSFVVAHCCARGRGVN 304
PS + R R LS I+AIVV CVA+ V+ F++A C G G +
Sbjct: 234 PSGSPAAAGAPAKRRVR------LSQAAILAIVVGGCVAVSAVIAVFLIAFCNRSGGGGD 287
Query: 305 S--NSLMGSEAGKRKSYGSEKKVYNSNGGGGDSSDGTSGTDMSKLVFFDRRN-GFELEDL 361
+ ++ ++G++K S + S G + DG +++VFF+ F+LEDL
Sbjct: 288 EEVSRVVSGKSGEKKGRESPE----SKAVIGKAGDG------NRIVFFEGPALAFDLEDL 337
Query: 362 LRASAEMLGKGSLGTVYRAVLDDGSTVAVKRLKDANPCARHEFEQYMDVIGKLKHPNIVK 421
LRASAE+LGKG+ GT YRAVL+D +TV VKRLK+ + R +FEQ M+++G+++H N+ +
Sbjct: 338 LRASAEVLGKGAFGTAYRAVLEDATTVVVKRLKEVS-AGRRDFEQQMELVGRIRHANVAE 396
Query: 422 LRAYYYAKEEKLLVYDYLSNGSLHALLHGNRGPGRIPLDWTTRISLVLGAARGLARIHTE 481
LRAYYY+K+EKLLVYD+ S GS+ +LHG RG R PL+W TR+ + LGAARG+A IHTE
Sbjct: 397 LRAYYYSKDEKLLVYDFYSRGSVSNMLHGKRGEDRTPLNWETRVRIALGAARGIAHIHTE 456
Query: 482 YSAAKVPHGNVKSSNVLLDKNGVACISDFGLSLLLNPVHATARLGGYRAPEQTEQKRLSQ 541
+ K HGN+K+SNV L+ C+SD GL+ L+NP+ A +R GY APE T+ ++ SQ
Sbjct: 457 -NNGKFVHGNIKASNVFLNNQQYGCVSDLGLASLMNPITARSRSLGYCAPEVTDSRKASQ 515
Query: 542 QADVYSFGVLLLEVLTGKAPSLQYPSPANRPRKVEEEETVVDLPKWVRSVVREEWTGEVF 601
+DVYSFGV +LE+LTG++P +Q N VV L +WV+SVVREEWT EVF
Sbjct: 516 CSDVYSFGVFILELLTGRSP-VQITGGGNE---------VVHLVRWVQSVVREEWTAEVF 565
Query: 602 DQELLRYKNIEEELVSMLHVGLACVVQQPEKRPTMVDVVKMIEDIR 647
D EL+RY NIEEE+V ML + +ACV + PE+RP M DVV+M+ED+R
Sbjct: 566 DVELMRYPNIEEEMVEMLQIAMACVSRTPERRPKMSDVVRMLEDVR 611
>ref|XP_550037.1| putative atypical receptor-like kinase MARK [Oryza sativa (japonica
cultivar-group)] gi|53792169|dbj|BAD52802.1| putative
atypical receptor-like kinase MARK [Oryza sativa
(japonica cultivar-group)]
Length = 791
Score = 465 bits (1196), Expect = e-129
Identities = 309/706 (43%), Positives = 404/706 (56%), Gaps = 70/706 (9%)
Query: 11 LFLSIYIVPCLT-----------HNDTQALTLFRQQTDTHGQLLTNW---TGPEACSASW 56
LFLS ++ C D AL+ FR D G L +W P C +W
Sbjct: 110 LFLSALLLRCFVCYADGGGGGSLDADVAALSDFRLAADRSGAL-ASWDLAANPAPCG-TW 167
Query: 57 HGVTCTPNNRVTTLVLPSLNLRGP--IDALSSLTHLRLLDLHNNRLNGTVSASLLSNCTN 114
GV+C RVT LVL L G + AL+ L LR+L L N L G + LS
Sbjct: 168 RGVSCA-GGRVTRLVLEGFGLSGDAALPALARLDGLRVLSLKGNGLTGAIPD--LSPLAG 224
Query: 115 LKLLYLAGNDFSGQIPPEISSLNNLLRLDLSDNNLAGDIPNEISRLTNLLTLRLQNNALS 174
LKLL+LAGN SG IPP I +L L RLDLS NNL+G +P E++RL LLTLRL +N LS
Sbjct: 225 LKLLFLAGNSLSGPIPPSIGALYRLYRLDLSFNNLSGVVPPELNRLDRLLTLRLDSNRLS 284
Query: 175 GNIPDLSSIMPNLTELNMTNNEFYGKVPNTMLNKFGDESFSGNEGLCGSKPFQVCSLTEN 234
G I ++ +P L + N++NN G++P M KF +F GN GLC S P C
Sbjct: 285 GGIDGIA--LPVLQDFNVSNNLLTGRIPVAMA-KFPVGAFGGNAGLC-SAPLPPCKDEAQ 340
Query: 235 SPPSSEPVQTVPSNP----SSFPATSVIARPRSQHHKG---LSPGVIVAIVVAICVALLV 287
P +S V + P ++ A+S A+P G +S +VAIV + +
Sbjct: 341 QPNASAAVNASATPPCPPAAAMVASSPSAKPAGAATSGKGKMSCAAVVAIVAGDFAVVGL 400
Query: 288 VTSFVVAHCCARGRGVNSNSLMGSEAGKRKSYGSEKKVYNSNGGGGDSSDGTSGT--DMS 345
V + + R L G + +R G EK VY+S+ G +G +
Sbjct: 401 VAGLLFCYFWPR--------LSGRRSARRLREG-EKIVYSSSPYGATGVVTAAGGTFERG 451
Query: 346 KLVFFDRRNG-----FELEDLLRASAEMLGKGSLGTVYRAVLDDGSTVAVKRLKDANPCA 400
K+VF + + FEL+DLLRASAEMLGKG GT Y+AVL DGS VAVKRL+DA A
Sbjct: 452 KMVFLEDVSSGGGKRFELDDLLRASAEMLGKGGCGTAYKAVLGDGSVVAVKRLRDATAAA 511
Query: 401 --RHEFEQYMDVIGKLKHPNIVKLRAYYYAKEEKLLVYDYLSNGSLHALLHGNRGPGRIP 458
+ +FE +M V+G+L+HPNIV L AYYYA++EKLLVY+++ NGSL +LLHGNRGPGR P
Sbjct: 512 ASKKDFEHHMAVLGRLRHPNIVPLNAYYYARDEKLLVYEFMPNGSLFSLLHGNRGPGRTP 571
Query: 459 LDWTTRISLVLGAARGLARIH----TEYSAAKVPHGNVKSSNVLLDKNGVACISDFGLSL 514
LDW R+ + AARGLA IH ++ HGN+KS+N+LLDK GV ++D GL+
Sbjct: 572 LDWAARMRIASAAARGLAYIHHASRRGSGTPRLAHGNIKSTNILLDKAGVGRLADCGLAQ 631
Query: 515 L-LNPVHATARLGGYRAPEQTEQKR--LSQQADVYSFGVLLLEVLTGKAPSLQYPSPANR 571
L +P A AR GYRAPE R SQ+ DVY+FGV+LLE+LTG+ P + P+
Sbjct: 632 LGSSPAAAAARSAGYRAPEAPPPPRPWASQKGDVYAFGVVLLELLTGRCPGSELPNGG-- 689
Query: 572 PRKVEEEETVVDLPKWVRSVVREEWTGEVFDQELLRYKNIEEELVSMLHVGLACVVQQPE 631
VV+LP+WV+SVVREEWT EVFD EL++ K IEEE+V+ML + L+C P+
Sbjct: 690 --------VVVELPRWVQSVVREEWTSEVFDLELMKDKGIEEEMVAMLQLALSCASAAPD 741
Query: 632 KRPTMVDVVKMIEDIRV--EQSPLCEDYDESRN-SLSPSIPTTEDG 674
+RP + VVKMIE+IR E SP E DES S+S S +E G
Sbjct: 742 QRPKIGYVVKMIEEIRACGEASPSHESMDESSGVSVSDSPAVSEGG 787
>ref|NP_909155.1| putative receptor kinase [Oryza sativa (japonica cultivar-group)]
Length = 697
Score = 465 bits (1196), Expect = e-129
Identities = 309/706 (43%), Positives = 404/706 (56%), Gaps = 70/706 (9%)
Query: 11 LFLSIYIVPCLT-----------HNDTQALTLFRQQTDTHGQLLTNW---TGPEACSASW 56
LFLS ++ C D AL+ FR D G L +W P C +W
Sbjct: 16 LFLSALLLRCFVCYADGGGGGSLDADVAALSDFRLAADRSGAL-ASWDLAANPAPCG-TW 73
Query: 57 HGVTCTPNNRVTTLVLPSLNLRGP--IDALSSLTHLRLLDLHNNRLNGTVSASLLSNCTN 114
GV+C RVT LVL L G + AL+ L LR+L L N L G + LS
Sbjct: 74 RGVSCA-GGRVTRLVLEGFGLSGDAALPALARLDGLRVLSLKGNGLTGAIPD--LSPLAG 130
Query: 115 LKLLYLAGNDFSGQIPPEISSLNNLLRLDLSDNNLAGDIPNEISRLTNLLTLRLQNNALS 174
LKLL+LAGN SG IPP I +L L RLDLS NNL+G +P E++RL LLTLRL +N LS
Sbjct: 131 LKLLFLAGNSLSGPIPPSIGALYRLYRLDLSFNNLSGVVPPELNRLDRLLTLRLDSNRLS 190
Query: 175 GNIPDLSSIMPNLTELNMTNNEFYGKVPNTMLNKFGDESFSGNEGLCGSKPFQVCSLTEN 234
G I ++ +P L + N++NN G++P M KF +F GN GLC S P C
Sbjct: 191 GGIDGIA--LPVLQDFNVSNNLLTGRIPVAMA-KFPVGAFGGNAGLC-SAPLPPCKDEAQ 246
Query: 235 SPPSSEPVQTVPSNP----SSFPATSVIARPRSQHHKG---LSPGVIVAIVVAICVALLV 287
P +S V + P ++ A+S A+P G +S +VAIV + +
Sbjct: 247 QPNASAAVNASATPPCPPAAAMVASSPSAKPAGAATSGKGKMSCAAVVAIVAGDFAVVGL 306
Query: 288 VTSFVVAHCCARGRGVNSNSLMGSEAGKRKSYGSEKKVYNSNGGGGDSSDGTSGT--DMS 345
V + + R L G + +R G EK VY+S+ G +G +
Sbjct: 307 VAGLLFCYFWPR--------LSGRRSARRLREG-EKIVYSSSPYGATGVVTAAGGTFERG 357
Query: 346 KLVFFDRRNG-----FELEDLLRASAEMLGKGSLGTVYRAVLDDGSTVAVKRLKDANPCA 400
K+VF + + FEL+DLLRASAEMLGKG GT Y+AVL DGS VAVKRL+DA A
Sbjct: 358 KMVFLEDVSSGGGKRFELDDLLRASAEMLGKGGCGTAYKAVLGDGSVVAVKRLRDATAAA 417
Query: 401 --RHEFEQYMDVIGKLKHPNIVKLRAYYYAKEEKLLVYDYLSNGSLHALLHGNRGPGRIP 458
+ +FE +M V+G+L+HPNIV L AYYYA++EKLLVY+++ NGSL +LLHGNRGPGR P
Sbjct: 418 ASKKDFEHHMAVLGRLRHPNIVPLNAYYYARDEKLLVYEFMPNGSLFSLLHGNRGPGRTP 477
Query: 459 LDWTTRISLVLGAARGLARIH----TEYSAAKVPHGNVKSSNVLLDKNGVACISDFGLSL 514
LDW R+ + AARGLA IH ++ HGN+KS+N+LLDK GV ++D GL+
Sbjct: 478 LDWAARMRIASAAARGLAYIHHASRRGSGTPRLAHGNIKSTNILLDKAGVGRLADCGLAQ 537
Query: 515 L-LNPVHATARLGGYRAPEQTEQKR--LSQQADVYSFGVLLLEVLTGKAPSLQYPSPANR 571
L +P A AR GYRAPE R SQ+ DVY+FGV+LLE+LTG+ P + P+
Sbjct: 538 LGSSPAAAAARSAGYRAPEAPPPPRPWASQKGDVYAFGVVLLELLTGRCPGSELPNGG-- 595
Query: 572 PRKVEEEETVVDLPKWVRSVVREEWTGEVFDQELLRYKNIEEELVSMLHVGLACVVQQPE 631
VV+LP+WV+SVVREEWT EVFD EL++ K IEEE+V+ML + L+C P+
Sbjct: 596 --------VVVELPRWVQSVVREEWTSEVFDLELMKDKGIEEEMVAMLQLALSCASAAPD 647
Query: 632 KRPTMVDVVKMIEDIRV--EQSPLCEDYDESRN-SLSPSIPTTEDG 674
+RP + VVKMIE+IR E SP E DES S+S S +E G
Sbjct: 648 QRPKIGYVVKMIEEIRACGEASPSHESMDESSGVSVSDSPAVSEGG 693
>gb|AAG51359.1| putative protein kinase; 49514-51513 [Arabidopsis thaliana]
gi|42572323|ref|NP_974257.1| leucine-rich repeat
transmembrane protein kinase, putative [Arabidopsis
thaliana] gi|15231955|ref|NP_187480.1| leucine-rich
repeat transmembrane protein kinase, putative
[Arabidopsis thaliana]
Length = 640
Score = 461 bits (1187), Expect = e-128
Identities = 285/659 (43%), Positives = 394/659 (59%), Gaps = 56/659 (8%)
Query: 1 MNYVIMFFFFLFLSIYIVPCLT---HNDTQALTLFRQQTDTHGQLLTNWTGPEACSASWH 57
M +I F FL ++ ++ CL+ +D QAL F +L NW ASW
Sbjct: 1 MMKIIAAFLFLLVTTFVSRCLSADIESDKQALLEFASLVPHSRKL--NWNSTIPICASWT 58
Query: 58 GVTCTPNN-RVTTLVLPSLNLRGPID--ALSSLTHLRLLDLHNNRLNGTVSASLLSNCTN 114
G+TC+ NN RVT L LP L GP+ L LR++ L +N L G + + +LS
Sbjct: 59 GITCSKNNARVTALRLPGSGLYGPLPEKTFEKLDALRIISLRSNHLQGNIPSVILS-LPF 117
Query: 115 LKLLYLAGNDFSGQIPPEISSLNNLLRLDLSDNNLAGDIPNEISRLTNLLTLRLQNNALS 174
++ LY N+FSG IPP +S + L+ LDLS N+L+G+IP + LT L L LQNN+LS
Sbjct: 118 IRSLYFHENNFSGTIPPVLS--HRLVNLDLSANSLSGNIPTSLQNLTQLTDLSLQNNSLS 175
Query: 175 GNIPDLSSIMPNLTELNMTNNEFYGKVPNTMLNKFGDESFSGNEGLCGSKPFQVCSLTEN 234
G IP+L P L LN++ N G VP+++ F SF GN LCG+ P C
Sbjct: 176 GPIPNLP---PRLKYLNLSFNNLNGSVPSSV-KSFPASSFQGNSLLCGA-PLTPCPENTT 230
Query: 235 SPPSSEPVQTVPSNPSSFPATSVIARPRSQHHKGLSPGVIVAIVVAICVALLVVTSFVVA 294
+P S P+ P+ P T+ I R ++ K LS G IV I V V L ++ + ++
Sbjct: 231 APSPS------PTTPTEGPGTTNIGRGTAK--KVLSTGAIVGIAVGGSVLLFIILA-IIT 281
Query: 295 HCCARGRGVNSNSLMGSEAGKRKSYGSEKKVYNSNGGGGDSSDGTSGTDMSKLVFFDRRN 354
CCA+ R + G+ + + K S+ + G + +KLVFF+ +
Sbjct: 282 LCCAKKR----------DGGQDSTAVPKAKPGRSDNKAEEFGSGVQEAEKNKLVFFEGSS 331
Query: 355 -GFELEDLLRASAEMLGKGSLGTVYRAVLDDGSTVAVKRLKDANPCARHEFEQYMDVIGK 413
F+LEDLLRASAE+LGKGS GT Y+A+L++G+TV VKRLK+ + EFEQ M+ +G+
Sbjct: 332 YNFDLEDLLRASAEVLGKGSYGTTYKAILEEGTTVVVKRLKEV-AAGKREFEQQMEAVGR 390
Query: 414 LK-HPNIVKLRAYYYAKEEKLLVYDYLSNGSLHALLHGNRGPGRIPLDWTTRISLVLGAA 472
+ H N+ LRAYY++K+EKLLVYDY G+ LLHGN GR LDW TR+ + L AA
Sbjct: 391 ISPHVNVAPLRAYYFSKDEKLLVYDYYQGGNFSMLLHGNNEGGRAALDWETRLRICLEAA 450
Query: 473 RGLARIHTEYSAAKVPHGNVKSSNVLLDKNGVACISDFGLSLLLNPVHAT---ARLGGYR 529
RG++ IH+ S AK+ HGN+KS NVLL + C+SDFG++ L++ H T +R GYR
Sbjct: 451 RGISHIHSA-SGAKLLHGNIKSPNVLLTQELHVCVSDFGIAPLMS--HHTLIPSRSLGYR 507
Query: 530 APEQTEQKRLSQQADVYSFGVLLLEVLTGKAPSLQYPSPANRPRKVEEEETVVDLPKWVR 589
APE E ++ +Q++DVYSFGVLLLE+LTGKA K E VVDLPKWV+
Sbjct: 508 APEAIETRKHTQKSDVYSFGVLLLEMLTGKAAG-----------KTTGHEEVVDLPKWVQ 556
Query: 590 SVVREEWTGEVFDQELLRYK-NIEEELVSMLHVGLACVVQQPEKRPTMVDVVKMIEDIR 647
SVVREEWTGEVFD EL++ + N+EEE+V ML + +ACV + P+ RP+M +VV M+E+IR
Sbjct: 557 SVVREEWTGEVFDVELIKQQHNVEEEMVQMLQIAMACVSKHPDSRPSMEEVVNMMEEIR 615
>ref|NP_915990.1| putative receptor-like protein kinase [Oryza sativa (japonica
cultivar-group)] gi|21104781|dbj|BAB93368.1| putative
receptor-like protein kinase [Oryza sativa (japonica
cultivar-group)] gi|15128407|dbj|BAB62593.1| putative
receptor-like protein kinase [Oryza sativa (japonica
cultivar-group)]
Length = 637
Score = 460 bits (1183), Expect = e-128
Identities = 282/633 (44%), Positives = 386/633 (60%), Gaps = 52/633 (8%)
Query: 23 HNDTQALTLFRQQTDTHGQLLTNWTGPEACSASWHGVTCTPNN-RVTTLVLPSLNLRGPI 81
++D QAL F HG+ L NW+ SW GVTCTP+N RV TL LP++ L GP+
Sbjct: 28 NSDKQALLAFAASLP-HGRKL-NWSSAAPVCTSWVGVTCTPDNSRVQTLRLPAVGLFGPL 85
Query: 82 --DALSSLTHLRLLDLHNNRLNGTVSASLLSNCTNLKLLYLAGNDFSGQIPPEISSLNNL 139
D L L L +L L +NR+ + + + +L LYL N+ SG IP ++S L
Sbjct: 86 PSDTLGKLDALEVLSLRSNRITVDLPPEV-GSIPSLHSLYLQHNNLSGIIPTSLTS--TL 142
Query: 140 LRLDLSDNNLAGDIPNEISRLTNLLTLRLQNNALSGNIPDLSSIMPNLTELNMTNNEFYG 199
LDLS N G+IP + LT L L LQNN+LSG IPDL +P L LN++NN G
Sbjct: 143 TFLDLSYNTFDGEIPLRVQNLTQLTALLLQNNSLSGPIPDLQ--LPKLRHLNLSNNNLSG 200
Query: 200 KVPNTMLNKFGDESFSGNEGLCGSKPFQVCSLTENSPPSSEPVQTVPSNPSSFPATSVIA 259
+P + L +F SF GN LCG P Q C T SP S P++PS
Sbjct: 201 PIPPS-LQRFPANSFLGNAFLCGF-PLQPCPGTAPSPSPS------PTSPSP-------G 245
Query: 260 RPRSQHHKGLSPGVIVAIVVAICVALLVVTSFVVAHCCARGRGVNSNSLMGSEAGKRKSY 319
+ + K + GVI+A+ A V LL++ ++ C + + + S GK +
Sbjct: 246 KAKKGFWKRIRTGVIIALAAAGGVLLLILIVLLLI-CIFKRKKSTEPTTASSSKGKTVAG 304
Query: 320 G---SEKKVYNSNGGGGDSSDGTSGTDMSKLVFFDRRN-GFELEDLLRASAEMLGKGSLG 375
G + K+ Y+S G + +KLVFF+ + F+LEDLLRASAE+LGKGS G
Sbjct: 305 GRGENPKEEYSS---------GVQEAERNKLVFFEGCSYNFDLEDLLRASAEVLGKGSYG 355
Query: 376 TVYRAVLDDGSTVAVKRLKDANPCARHEFEQYMDVIGKL-KHPNIVKLRAYYYAKEEKLL 434
T Y+AVL+DG+TV VKRLK+ + +FEQ M+++G++ +H N+V LRAYYY+K+EKLL
Sbjct: 356 TTYKAVLEDGTTVVVKRLKEV-VVGKKDFEQQMEIVGRVGQHQNVVPLRAYYYSKDEKLL 414
Query: 435 VYDYLSNGSLHALLHGNRGPGRIPLDWTTRISLVLGAARGLARIHTEYSAAKVPHGNVKS 494
VYDY+ +GSL +LHGN+ G+ PLDW TR+ + LG ARG+A +H E K HGN+KS
Sbjct: 415 VYDYIPSGSLAVVLHGNKATGKAPLDWETRVKISLGVARGIAHLHAE-GGGKFIHGNLKS 473
Query: 495 SNVLLDKNGVACISDFGLSLLLNPVHATARLGGYRAPEQTEQKRLSQQADVYSFGVLLLE 554
SN+LL +N C+S+FGL+ L+ A ARL GYRAPE E K+ +Q++DVYSFGVL+LE
Sbjct: 474 SNILLSQNLDGCVSEFGLAQLMTIPPAPARLVGYRAPEVLETKKPTQKSDVYSFGVLVLE 533
Query: 555 VLTGKAPSLQYPSPANRPRKVEEEETVVDLPKWVRSVVREEWTGEVFDQELLRYKNIEEE 614
+LTGKAP R E+++ LP+WV+SVVREEWT EVFD +LLR+ NIE+E
Sbjct: 534 MLTGKAPL----------RSPGREDSIEHLPRWVQSVVREEWTAEVFDVDLLRHPNIEDE 583
Query: 615 LVSMLHVGLACVVQQPEKRPTMVDVVKMIEDIR 647
+V ML V +ACV P++RP M +V++ I +IR
Sbjct: 584 MVQMLQVAMACVAAPPDQRPKMDEVIRRIVEIR 616
>ref|XP_479550.1| putative receptor-like protein kinase [Oryza sativa (japonica
cultivar-group)] gi|51979072|ref|XP_507413.1| PREDICTED
OSJNBa0008J01.18 gene product [Oryza sativa (japonica
cultivar-group)] gi|51963652|ref|XP_506571.1| PREDICTED
OSJNBa0008J01.18 gene product [Oryza sativa (japonica
cultivar-group)] gi|33146664|dbj|BAC80010.1| putative
receptor-like protein kinase [Oryza sativa (japonica
cultivar-group)]
Length = 640
Score = 454 bits (1167), Expect = e-126
Identities = 276/632 (43%), Positives = 391/632 (61%), Gaps = 56/632 (8%)
Query: 45 NWTGPEACSASWHGVTCTPN-NRVTTLVLPSLNLRG--PIDALSSLTHLRLLDLHNNRLN 101
NW+ + SWHGV C+ + + + L +P L G P + L L L++L L +NRL
Sbjct: 52 NWSQSTSL-CSWHGVKCSGDQSHIFELRVPGAGLIGAIPPNTLGKLDSLQVLSLRSNRLA 110
Query: 102 GTVSASLLSNCTNLKLLYLAGNDFSGQIPPEISSLN-NLLRLDLSDNNLAGDIPNEISRL 160
G++ + + + +L+ +YL N+FSG +P S LN NL +DLS N+ G+IP + L
Sbjct: 111 GSLPSDV-TTLPSLRSIYLQHNNFSGDLP---SFLNPNLSVVDLSYNSFTGEIPISLQNL 166
Query: 161 TNLLTLRLQNNALSGNIPDLSSIMPNLTELNMTNNEFYGKVPNTMLNKFGDESFSGNEGL 220
+ L L LQ N+LSG+IPDL +P+L LN++NN+ G++P + L F + SF GN GL
Sbjct: 167 SQLSVLNLQENSLSGSIPDLK--LPSLRLLNLSNNDLKGQIPQS-LQTFPNGSFLGNPGL 223
Query: 221 CGSKPFQVCSLTENSPPSSEPVQTVPSNPSSFPATSVIARPRSQHH-KGLSPGVIVAIVV 279
CG P C L ++ PS P++PSS P P S HH K G I+A+ V
Sbjct: 224 CGP-PLAKCLLPDSPTPS-------PASPSSAPT------PMSAHHEKKFGAGFIIAVAV 269
Query: 280 A-ICVALLVVTSFVVAHCCARGRGVNSNSLMGSEAGKRKSYGSEKKVYNSNGGGGDSSDG 338
V + VV VV + +G+ + G G R SEK + S G
Sbjct: 270 GGFAVLMFVVVVLVVCNSKRKGKKESGVDYKGKGTGVR----SEKPKQ-------EFSSG 318
Query: 339 TSGTDMSKLVFFDRRN-GFELEDLLRASAEMLGKGSLGTVYRAVLDDGSTVAVKRLKDAN 397
+ +KLVF + + F+LEDLLRASAE+LGKGS GT Y+A+L+DG+ V VKRLKD
Sbjct: 319 VQIAEKNKLVFLEGCSYTFDLEDLLRASAEVLGKGSYGTAYKAILEDGTVVVVKRLKDV- 377
Query: 398 PCARHEFEQYMDVIGKL-KHPNIVKLRAYYYAKEEKLLVYDYLSNGSLHALLHGNRG-PG 455
+ EFEQ M++IG+L KH N+V LRAYYY+K+EKL+VYDYL+NGS LHG RG
Sbjct: 378 VAGKKEFEQQMELIGRLGKHANLVPLRAYYYSKDEKLIVYDYLTNGSFSTKLHGIRGVTE 437
Query: 456 RIPLDWTTRISLVLGAARGLARIHTEYSAAKVPHGNVKSSNVLLDKNGVACISDFGLSLL 515
+ PLDW+TR+ ++LG A G+A +H E AK+ HGN+KS+N+LLD++ + +SD+GL+ L
Sbjct: 438 KTPLDWSTRVKIILGTAYGIAHVHAE-GGAKLTHGNIKSTNILLDQDYSSYVSDYGLTAL 496
Query: 516 LN-PVHATARLGGYRAPEQTEQKRLSQQADVYSFGVLLLEVLTGKAPSLQYPSPANRPRK 574
++ P +A+ + GYRAPE E ++++Q++DVYSFGVLL+E+LTGKAP +
Sbjct: 497 MSVPANASRVVVGYRAPETIENRKITQKSDVYSFGVLLMEMLTGKAPL-----------Q 545
Query: 575 VEEEETVVDLPKWVRSVVREEWTGEVFDQELLRYKNIEEELVSMLHVGLACVVQQPEKRP 634
+ + VVDLP+WV SVVREEWT EVFD EL++ +NIEEELV ML + +AC + P++RP
Sbjct: 546 SQGNDDVVDLPRWVHSVVREEWTAEVFDVELIKQQNIEEELVQMLQIAMACTSRSPDRRP 605
Query: 635 TMVDVVKMIEDIRVEQSPLCEDYDESRNSLSP 666
+M DV++MIE +R S DE +P
Sbjct: 606 SMEDVIRMIEGLRHSASESRASSDEKMKDSNP 637
>dbj|BAA96921.1| receptor-like protein kinase [Arabidopsis thaliana]
gi|18086391|gb|AAL57654.1| unknown protein [Arabidopsis
thaliana] gi|15237162|ref|NP_200638.1| leucine-rich
repeat transmembrane protein kinase, putative
[Arabidopsis thaliana] gi|24797034|gb|AAN64529.1|
At5g58299/At5g58299 [Arabidopsis thaliana]
Length = 654
Score = 449 bits (1154), Expect = e-124
Identities = 279/653 (42%), Positives = 390/653 (58%), Gaps = 60/653 (9%)
Query: 23 HNDTQALTLFRQQTDTHGQLLTNWTGPEACSASWHGVTCTPNNR-VTTLVLPSLNLRGPI 81
++D QAL F +L NW SW GVTCT + V L LP + L GPI
Sbjct: 46 NSDRQALLAFAASVPHLRRL--NWNSTNHICKSWVGVTCTSDGTSVHALRLPGIGLLGPI 103
Query: 82 --DALSSLTHLRLLDLHNNRLNGTVSASLLSNCTNLKLLYLAGNDFSGQIPPEISSLNNL 139
+ L L LR+L L +N L+G + + S +L +YL N+FSG++P +S N+
Sbjct: 104 PPNTLGKLESLRILSLRSNLLSGNLPPDIHS-LPSLDYIYLQHNNFSGEVPSFVSRQLNI 162
Query: 140 LRLDLSDNNLAGDIPNEISRLTNLLTLRLQNNALSGNIPDLSSIMPNLTELNMTNNEFYG 199
L DLS N+ G IP L L L LQNN LSG +P+L ++ +L LN++NN G
Sbjct: 163 L--DLSFNSFTGKIPATFQNLKQLTGLSLQNNKLSGPVPNLDTV--SLRRLNLSNNHLNG 218
Query: 200 KVPNTMLNKFGDESFSGNEGLCGSKPFQVCSLTENSPPSSEPVQTVPSNPSSFPATSVIA 259
+P+ L F SFSGN LCG P Q C+ T + PPS P + P P FP
Sbjct: 219 SIPSA-LGGFPSSSFSGNTLLCGL-PLQPCA-TSSPPPSLTPHISTPPLPP-FP------ 268
Query: 260 RPRSQHHKGLSPGVIVAIVVAICV---ALLVVTSFVVAHCCARGRGVNSNSLMGSEAGKR 316
H +G + V+ ++ I ALL++ + ++ CC + + +S++ +
Sbjct: 269 -----HKEGSKRKLHVSTIIPIAAGGAALLLLITVIILCCCIKKKDKREDSIVKVKTLTE 323
Query: 317 KSYGSEKKVYNSNGGGGDSSDGTSGTDMSKLVFFDRRN-GFELEDLLRASAEMLGKGSLG 375
K+ K+ + S G + +KLVFF+ + F+LEDLLRASAE+LGKGS G
Sbjct: 324 KA----KQEFGS---------GVQEPEKNKLVFFNGCSYNFDLEDLLRASAEVLGKGSYG 370
Query: 376 TVYRAVLDDGSTVAVKRLKDANPCARHEFEQYMDVIGKL-KHPNIVKLRAYYYAKEEKLL 434
T Y+AVL++ +TV VKRLK+ + EFEQ M++I ++ HP++V LRAYYY+K+EKL+
Sbjct: 371 TAYKAVLEESTTVVVKRLKEV-AAGKREFEQQMEIISRVGNHPSVVPLRAYYYSKDEKLM 429
Query: 435 VYDYLSNGSLHALLHGNRGPGRIPLDWTTRISLVLGAARGLARIHTEYSAAKVPHGNVKS 494
V DY G+L +LLHGNRG + PLDW +R+ + L AA+G+A +H K HGN+KS
Sbjct: 430 VCDYYPAGNLSSLLHGNRGSEKTPLDWDSRVKITLSAAKGIAHLHAA-GGPKFSHGNIKS 488
Query: 495 SNVLLDKNGVACISDFGLSLLLNPVHATARLGGYRAPEQTEQKRLSQQADVYSFGVLLLE 554
SNV++ + ACISDFGL+ L+ A R GYRAPE E ++ + ++DVYSFGVL+LE
Sbjct: 489 SNVIMKQESDACISDFGLTPLMAVPIAPMRGAGYRAPEVMETRKHTHKSDVYSFGVLILE 548
Query: 555 VLTGKAPSLQYPSPANRPRKVEEEETVVDLPKWVRSVVREEWTGEVFDQELLRYKNIEEE 614
+LTGK+P +Q PS + +VDLP+WV+SVVREEWT EVFD EL+R++NIEEE
Sbjct: 549 MLTGKSP-VQSPS----------RDDMVDLPRWVQSVVREEWTSEVFDIELMRFQNIEEE 597
Query: 615 LVSMLHVGLACVVQQPEKRPTMVDVVKMIEDIRVEQS----PLCEDYDESRNS 663
+V ML + +ACV Q PE RPTM DVV+MIE+IRV S P +D + ++S
Sbjct: 598 MVQMLQIAMACVAQVPEVRPTMDDVVRMIEEIRVSDSETTRPSSDDNSKPKDS 650
>ref|NP_912583.1| Putative leucine-rich repeat transmembrane protein kinase [Oryza
sativa (japonica cultivar-group)]
gi|22748334|gb|AAN05336.1| Putative leucine-rich repeat
transmembrane protein kinase [Oryza sativa (japonica
cultivar-group)]
Length = 675
Score = 428 bits (1100), Expect = e-118
Identities = 272/658 (41%), Positives = 367/658 (55%), Gaps = 65/658 (9%)
Query: 23 HNDTQALTLFRQQTDTHGQLLTNWTGPEACSASWHGVTCTPNNRVTTLVLPSLNLRG--P 80
+ D QAL R L +W + +W GVTC + RVT L LP L G P
Sbjct: 29 NTDAQALQALRSAVGKSA--LPSWNSSTP-TCNWQGVTCE-SGRVTELRLPGAGLMGTLP 84
Query: 81 IDALSSLTHLRLLDLHNNRLNGTVSASLLSNCTNLKLLYLAGNDFSGQIPPEISSLNNLL 140
+ L +L+ LR L L N L G + L S L+ +Y N FSG++P + +L NL+
Sbjct: 85 SNVLGNLSALRTLSLRYNALTGPIPDDL-SRLPELRAIYFQHNSFSGEVPASVFTLKNLV 143
Query: 141 RLDLSDNNLAGDIPNEISRLTNLLTLRLQNNALSGNIPDLSSIMPNLTELNMTNNEFYGK 200
RLDL+ N +G+I + ++L L TL L N+ +G IP L +P L++ N++ N+ G
Sbjct: 144 RLDLAGNKFSGEISPDFNKLNRLGTLFLDGNSFTGEIPKLD--LPTLSQFNVSYNKLNGS 201
Query: 201 VPNTMLNKFGDESFSGNEGLCGSKPFQVC-SLTENSPPSSEPVQTVPSNPSSFPATSVIA 259
+P + L K +SF G GLCG P +C T +P S VQ + S
Sbjct: 202 IPRS-LRKMPKDSFLGT-GLCGG-PLGLCPGETALTPAGSPEVQPAGGGAADAGGAS--- 255
Query: 260 RPRSQHHKGLSPGVIVAIVVAICVALLVVTSFVVAHCCARGRGVNSNSLMGSEAGKRKSY 319
S K LS G I I + +L++ + + C R + +S E G+
Sbjct: 256 ---SGTKKKLSGGAIAGIAIGCVFGVLLLLALIFLLC--RKKSSSSTPATAVEKGRDLQM 310
Query: 320 GS---EKKVYNSNGGGGDSSDGTSGT------------------------DMSKLVFFDR 352
E K N + G + G + KL+FF
Sbjct: 311 APMDMEPKGQNGSAAGNGAHVGAAAAAPAAATSAAVAAAAAAAKTGGATGGSKKLIFFGP 370
Query: 353 RNG---FELEDLLRASAEMLGKGSLGTVYRAVLDDGSTVAVKRLKDANPCARHEFEQYMD 409
F+LEDLLRASAE+LGKG+ GT Y+AV++ GS VAVKRLKD + EF + +
Sbjct: 371 MAAAPPFDLEDLLRASAEVLGKGAFGTAYKAVMESGSAVAVKRLKDVD-LPEPEFRERIA 429
Query: 410 VIGKLKHPNIVKLRAYYYAKEEKLLVYDYLSNGSLHALLHGNRGPGRIPLDWTTRISLVL 469
IG ++H +V LRAYY++K+EKLLVYDY+S GSL ALLHGNR GR PLDW TR ++ L
Sbjct: 430 AIGAVQHELVVPLRAYYFSKDEKLLVYDYMSMGSLSALLHGNRASGRTPLDWETRSAIAL 489
Query: 470 GAARGLARIHTEYSAAKVPHGNVKSSNVLLDKNGVACISDFGLSLLLNPVHATARLGGYR 529
AARG+A IH+ A HGN+KSSNVLL KN A +SD GL L+ P + R+ GYR
Sbjct: 490 AAARGVAHIHSTGPTAS--HGNIKSSNVLLTKNYEARVSDHGLPTLVGPSFSPTRVSGYR 547
Query: 530 APEQTEQKRLSQQADVYSFGVLLLEVLTGKAPSLQYPSPANRPRKVEEEETVVDLPKWVR 589
APE T+ +R+SQ+ADVYSFGVLLLE+LTGKAP+ V EE +DLP+WV+
Sbjct: 548 APEVTDIRRVSQKADVYSFGVLLLELLTGKAPT----------HAVVNEEG-LDLPRWVQ 596
Query: 590 SVVREEWTGEVFDQELLRYKNIEEELVSMLHVGLACVVQQPEKRPTMVDVVKMIEDIR 647
SVVREEWT EVFDQELLRY+N+EEE+V +L + + C Q P++RP+M +V I++IR
Sbjct: 597 SVVREEWTAEVFDQELLRYQNVEEEMVQLLQLAIDCSAQHPDRRPSMSEVAARIDEIR 654
>gb|AAP40406.1| putative leucine-rich repeat transmembrane protein kinase
[Arabidopsis thaliana] gi|7269223|emb|CAB81292.1|
putative receptor kinase [Arabidopsis thaliana]
gi|4454043|emb|CAA23040.1| putative receptor kinase
[Arabidopsis thaliana] gi|26451766|dbj|BAC42978.1|
putative receptor kinase [Arabidopsis thaliana]
gi|15236593|ref|NP_194105.1| leucine-rich repeat
transmembrane protein kinase, putative [Arabidopsis
thaliana] gi|7446453|pir||T05606 protein kinase homolog
F9D16.210 - Arabidopsis thaliana
Length = 638
Score = 427 bits (1098), Expect = e-118
Identities = 251/637 (39%), Positives = 373/637 (58%), Gaps = 55/637 (8%)
Query: 45 NWTGPEACSASWHGVTCTPN-NRVTTLVLPSLNLRG--PIDALSSLTHLRLLDLHNNRLN 101
NW W GVTC + +R+ + LP + L G P + +S L+ LR+L L +N ++
Sbjct: 47 NWNETSQVCNIWTGVTCNQDGSRIIAVRLPGVGLNGQIPPNTISRLSALRVLSLRSNLIS 106
Query: 102 GTVSASLLSNCTNLKLLYLAGNDFSGQIPPEISSLNNLLRLDLSDNNLAGDIPNEISRLT 161
G + +L LYL N+ SG +P + S NL ++LS+N G IP+ +SRL
Sbjct: 107 GEFPKDFVE-LKDLAFLYLQDNNLSGPLPLDFSVWKNLTSVNLSNNGFNGTIPSSLSRLK 165
Query: 162 NLLTLRLQNNALSGNIPDLSSIMPNLTELNMTNN-EFYGKVPNTMLNKFGDESFSGNEGL 220
+ +L L NN LSG+IPDLS ++ +L ++++NN + G +P+ L +F S++G + +
Sbjct: 166 RIQSLNLANNTLSGDIPDLS-VLSSLQHIDLSNNYDLAGPIPD-WLRRFPFSSYTGIDII 223
Query: 221 CGSKPFQVCSLTENSPPSSEPVQTVPSNPSSFPATSVIARPRSQHHKGLSPGVIVAIVVA 280
PP P PS +P GLS V + IV+A
Sbjct: 224 ---------------PPGGNYTLVTPPPPSE----QTHQKPSKARFLGLSETVFLLIVIA 264
Query: 281 ICVALLVVTSFVVAHCCARGRGVNSNSLMGSEAGKRKSYGSEKKVYNSNGGGGDSSDGTS 340
+ + ++ +FV+ C R + + ++ S+ K+ G +
Sbjct: 265 VSIVVITALAFVLTVCYVRRKLRRGDGVI-----------SDNKLQKKGGMSPEKFVSRM 313
Query: 341 GTDMSKLVFFDRRN-GFELEDLLRASAEMLGKGSLGTVYRAVLDDGSTVAVKRLKDANPC 399
++L FF+ N F+LEDLLRASAE+LGKG+ GT Y+AVL+D ++VAVKRLKD
Sbjct: 314 EDVNNRLSFFEGCNYSFDLEDLLRASAEVLGKGTFGTTYKAVLEDATSVAVKRLKDV-AA 372
Query: 400 ARHEFEQYMDVIGKLKHPNIVKLRAYYYAKEEKLLVYDYLSNGSLHALLHGNRGPGRIPL 459
+ +FEQ M++IG +KH N+V+L+AYYY+K+EKL+VYDY S GS+ +LLHGNRG RIPL
Sbjct: 373 GKRDFEQQMEIIGGIKHENVVELKAYYYSKDEKLMVYDYFSRGSVASLLHGNRGENRIPL 432
Query: 460 DWTTRISLVLGAARGLARIHTEYSAAKVPHGNVKSSNVLLDKNGVACISDFGLSLLLNPV 519
DW TR+ + +GAA+G+ARIH E + K+ HGN+KSSN+ L+ C+SD GL+ +++P+
Sbjct: 433 DWETRMKIAIGAAKGIARIHKE-NNGKLVHGNIKSSNIFLNSESNGCVSDLGLTAVMSPL 491
Query: 520 -HATARLGGYRAPEQTEQKRLSQQADVYSFGVLLLEVLTGKAPSLQYPSPANRPRKVEEE 578
+R GYRAPE T+ ++ SQ +DVYSFGV+LLE+LTGK+P
Sbjct: 492 APPISRQAGYRAPEVTDTRKSSQLSDVYSFGVVLLELLTGKSPI-----------HTTAG 540
Query: 579 ETVVDLPKWVRSVVREEWTGEVFDQELLRYKNIEEELVSMLHVGLACVVQQPEKRPTMVD 638
+ ++ L +WV SVVREEWT EVFD ELLRY NIEEE+V ML + ++CVV+ ++RP M D
Sbjct: 541 DEIIHLVRWVHSVVREEWTAEVFDIELLRYTNIEEEMVEMLQIAMSCVVKAADQRPKMSD 600
Query: 639 VVKMIEDI---RVEQSPLCEDYDESRNSLSPSIPTTE 672
+V++IE++ R P E +S N S + +E
Sbjct: 601 LVRLIENVGNRRTSIEPEPELKPKSENGASETSTPSE 637
>dbj|BAD53058.1| receptor-like protein kinase 1-like [Oryza sativa (japonica
cultivar-group)] gi|53792194|dbj|BAD52827.1|
receptor-like protein kinase 1-like [Oryza sativa
(japonica cultivar-group)]
Length = 684
Score = 426 bits (1095), Expect = e-117
Identities = 279/649 (42%), Positives = 375/649 (56%), Gaps = 58/649 (8%)
Query: 24 NDTQALTLFRQQTDTHGQLLTNWTGPEACSASWHGVTCTPNNRVTTLV-LPSLNLRG--P 80
++ AL F T +L N + AC W GVTC N V LP + L G P
Sbjct: 33 SERSALLAFLAATPHERRLGWN-SSTSACG--WVGVTCDAGNATVVQVRLPGVGLIGAIP 89
Query: 81 IDALSSLTHLRLLDLHNNRLNGTVSASLLSNCTNLKLLYLAGNDFSGQIPPEISSLNNLL 140
L LT+L++L L +NR+ G + +L L+LL+L N SG IPP +S L L
Sbjct: 90 PGTLGRLTNLQVLSLRSNRILGGIPDDVLQ-LPQLRLLFLQNNLLSGAIPPAVSKLAALE 148
Query: 141 RLDLSDNNLAGDIPNEISRLTNLLTLRLQNNALSGNIPDLSSIMPNLTELNMTNNEFYGK 200
RL LS NNL+G IP ++ LT+L LRL N LSGNIP +S + +L N+++N G
Sbjct: 149 RLVLSSNNLSGPIPFTLNNLTSLRALRLDGNKLSGNIPSIS--IQSLVVFNVSDNNLNGS 206
Query: 201 VPNTMLNKFGDESFSGNEGLCGSKPFQVCSLTENSPPSSEPVQTVPSNPSSFPATSVIAR 260
+P + L +F E F+GN LCGS P C SP S V +P+ P + ++
Sbjct: 207 IPAS-LARFPAEDFAGNLQLCGS-PLPPCKSFFPSPSPSPGV-----SPADVPGAASSSK 259
Query: 261 PRSQHHKGLSPGVIVAIVV-AICVALLVVTSFVVAHCCARGRGVNSNSLMGSEAGKRKSY 319
R LS I IVV A+ +ALL++ + V+ R RG + + A
Sbjct: 260 KRR-----LSGAAIAGIVVGAVVLALLLLVAAVLCAVSKRRRGASEGPKSTTAAAAGAGA 314
Query: 320 GSEKKVYNSNGGGGDSSDGTSGTDM------------------SKLVFFDRRNG--FELE 359
+ + V G G +S DM S+LVF + G F+LE
Sbjct: 315 AAARGVPPPGSGEGTGMTSSSKEDMGGASGSAAAAVAAVAAEPSRLVFVGKGAGYSFDLE 374
Query: 360 DLLRASAEMLGKGSLGTVYRAVLDDGSTVAVKRLKDANPCARHEFEQYMDVIGKLKHPNI 419
DLLRASAE+LGKGS+GT Y+AVL++G+TV VKRLKD AR EF+ +MD +GK++H N+
Sbjct: 375 DLLRASAEVLGKGSVGTSYKAVLEEGTTVVVKRLKDV-AVARREFDAHMDALGKVEHRNV 433
Query: 420 VKLRAYYYAKEEKLLVYDYLSNGSLHALLHGNRGPGRIPLDWTTRISLVLGAARGLARIH 479
+ +RAYY++K+EKLLV+DYL NGSL A+LHG+RG G+ PLDW R+ L AARGLA +H
Sbjct: 434 LPVRAYYFSKDEKLLVFDYLPNGSLSAMLHGSRGSGKTPLDWDARMRSALSAARGLAHLH 493
Query: 480 TEYSAAKVPHGNVKSSNVLLDKNG-VACISDFGLSLLLNPVHATARLGGYRAPEQTEQKR 538
T +S + HGNVKSSNVLL + A +SDF L + P A GGYRAPE + +R
Sbjct: 494 TVHS---LVHGNVKSSNVLLRPDADAAALSDFCLHPIFAPSSARPGAGGYRAPEVVDTRR 550
Query: 539 LSQQADVYSFGVLLLEVLTGKAPSLQYPSPANRPRKVEEEETVVDLPKWVRSVVREEWTG 598
+ +ADVYS GVLLLE+LTGK+P+ E + +DLP+WV+SVVREEWT
Sbjct: 551 PTYKADVYSLGVLLLELLTGKSPT----------HASLEGDGTLDLPRWVQSVVREEWTA 600
Query: 599 EVFDQELLRY-KNIEEELVSMLHVGLACVVQQPEKRPTMVDVVKMIEDI 646
EVFD EL+R + EEE+V++L V +ACV P+ RP DVV+MIE+I
Sbjct: 601 EVFDVELVRLGASAEEEMVALLQVAMACVATVPDARPDAPDVVRMIEEI 649
>ref|XP_469524.1| putative receptor kinase [Oryza sativa] gi|13324792|gb|AAK18840.1|
putative receptor kinase [Oryza sativa]
Length = 686
Score = 425 bits (1093), Expect = e-117
Identities = 275/641 (42%), Positives = 358/641 (54%), Gaps = 47/641 (7%)
Query: 25 DTQALTLFRQQTDTHGQLLTNWTGPEACSA-SWHGVTCTPNNRVTTLVLPSLNLRG--PI 81
D +AL FR H W G + A SW GVTC RV L LP L G P
Sbjct: 36 DARALLAFRDAVGRH----VAWNGSDPGGACSWTGVTCE-GGRVAVLRLPGAALAGRVPE 90
Query: 82 DALSSLTHLRLLDLHNNRLNGTVSASLLSNCTNLKLLYLAGNDFSGQIPPEISSLNNLLR 141
L +LT L L L N L G + L S L+ ++L GN SG+ P +L L+R
Sbjct: 91 GTLGNLTALHTLSLRLNALAGALPGDLTSAAA-LRNVFLNGNRLSGEFPRAFLALQGLVR 149
Query: 142 LDLSDNNLAGDIPNEISRLTNLLTLRLQNNALSGNIPDLSSIMPNLTELNMTNNEFYGKV 201
L + N+L+G IP + LT L L L+NN SG IPDL L + N++ N+ G +
Sbjct: 150 LAIGGNDLSGSIPPALGNLTRLKVLLLENNRFSGEIPDLKQ---PLQQFNVSFNQLNGSI 206
Query: 202 PNTMLNKFGDESFSGNEGLCGSKPFQVC--SLTENSPPSSEPVQTVPSNPSSFPATSVIA 259
P T L +F G GLCG P C ++ + P +PV P+N +
Sbjct: 207 PAT-LRTMPRSAFLGT-GLCGG-PLGPCPGEVSPSPAPGEQPVSPTPANNGDKGGNGGES 263
Query: 260 RPRSQHHKGLSPGVIVAIVVAICVALLVVTSFVVAHCCARGRGVNSNSLMG---SEAGKR 316
+S K LS G I I + V ++ ++ CC GR + M S A
Sbjct: 264 GKKS---KKLSGGAIAGIAIGSAVGAALLLFLLICLCCRSGRTKTRSMEMPPPPSSAPAV 320
Query: 317 KSYGSEKKVYNSNGG-------GGDSSDGTSGTDMSKLVFFDRRNG---FELEDLLRASA 366
+ G + S G + T KL+FF F+LEDLLRASA
Sbjct: 321 VAAGRKPPEMTSAAAVAPMATVGNPHAPLGQSTSGKKLIFFGSAAAVAPFDLEDLLRASA 380
Query: 367 EMLGKGSLGTVYRAVLDDGSTVAVKRLKDANPCARHEFEQYMDVIGKLKHPNIVKLRAYY 426
E+LGKG+ GT Y+AVL+ G+TVAVKRLKD EF + IG+L+H IV LRAYY
Sbjct: 381 EVLGKGAFGTTYKAVLESGATVAVKRLKDVT-LTEPEFRDRIADIGELQHEFIVPLRAYY 439
Query: 427 YAKEEKLLVYDYLSNGSLHALLHGNRGPGRIPLDWTTRISLVLGAARGLARIHTEYSAAK 486
Y+K+EKLLVYD++ GSL A+LHGNRG GR PL+W TR S+ L AARG+ IH+ S+A
Sbjct: 440 YSKDEKLLVYDFMPMGSLSAVLHGNRGSGRTPLNWETRSSIALAAARGVEYIHSTSSSAS 499
Query: 487 VPHGNVKSSNVLLDKNGVACISDFGLSLLLNPVHATARLGGYRAPEQTEQKRLSQQADVY 546
HGN+KSSNVLL+K+ A +SD GLS L+ P A +R GYRAPE T+ +R+SQ+ADVY
Sbjct: 500 --HGNIKSSNVLLNKSYQARLSDNGLSALVGPSSAPSRASGYRAPEVTDPRRVSQKADVY 557
Query: 547 SFGVLLLEVLTGKAPSLQYPSPANRPRKVEEEETVVDLPKWVRSVVREEWTGEVFDQELL 606
SFGVLLLE+LTGKAPS + + VDLP+WV+SVVR EWT EVFD ELL
Sbjct: 558 SFGVLLLELLTGKAPS-----------QAALNDEGVDLPRWVQSVVRSEWTAEVFDMELL 606
Query: 607 RYKNIEEELVSMLHVGLACVVQQPEKRPTMVDVVKMIEDIR 647
RY+N+EE++V +L + + CV Q P+ RP+M VV IE+I+
Sbjct: 607 RYQNVEEQMVQLLQLAIDCVAQVPDARPSMPHVVLRIEEIK 647
>gb|AAF79696.1| T1N15.9 [Arabidopsis thaliana] gi|18402209|ref|NP_564528.1|
leucine-rich repeat transmembrane protein kinase,
putative [Arabidopsis thaliana] gi|25405930|pir||G96524
protein T1N15.9 [imported] - Arabidopsis thaliana
Length = 655
Score = 424 bits (1089), Expect = e-117
Identities = 277/664 (41%), Positives = 384/664 (57%), Gaps = 63/664 (9%)
Query: 10 FLFLSIYIVPCLTHNDTQA-LTLFRQQTDTHGQLLTNWTGPEACSASWHGVTCTPNNRVT 68
FL L + +P + D A T G W + +W GV C +NRVT
Sbjct: 17 FLSLLLLSLPLPSTQDLNADRTALLSLRSAVGGRTFRWNIKQTSPCNWAGVKCE-SNRVT 75
Query: 69 TLVLPSLNLRG--PIDALSSLTHLRLLDLHNNRLNGTVSASLLSNCTNLKLLYLAGNDFS 126
L LP + L G P +LT LR L L N L+G++ L S +NL+ LYL GN FS
Sbjct: 76 ALRLPGVALSGDIPEGIFGNLTQLRTLSLRLNALSGSLPKDL-STSSNLRHLYLQGNRFS 134
Query: 127 GQIPPEISSLNNLLRLDLSDNNLAGDIPNEISRLTNLLTLRLQNNALSGNIPDLSSIMPN 186
G+IP + SL++L+RL+L+ N+ G+I + + LT L TL L+NN LSG+IPDL +P
Sbjct: 135 GEIPEVLFSLSHLVRLNLASNSFTGEISSGFTNLTKLKTLFLENNQLSGSIPDLD--LP- 191
Query: 187 LTELNMTNNEFYGKVPNTMLNKFGDESFSGNEGLCGSKPFQVCSLTENSPPSSEPVQTVP 246
L + N++NN G +P L +F +SF LCG KP ++C P E V + P
Sbjct: 192 LVQFNVSNNSLNGSIPKN-LQRFESDSFL-QTSLCG-KPLKLC-------PDEETVPSQP 241
Query: 247 SNPSSFPATSVIARPRSQHHKGLSPGVIVAIVVAICVALLVVTSFVVAHCCA----RGRG 302
++ + SV + LS G I IV+ V ++ ++ C R R
Sbjct: 242 TSGGNRTPPSVEGSEEKKKKNKLSGGAIAGIVIGCVVGFALIVLILMVLCRKKSNKRSRA 301
Query: 303 VNSNSLMGSEAGKRKSYGSEKKVYNSN------------GGGGDSSDGTSGTDMSKLVFF 350
V+ +++ E + G ++ V N N G G +S+G +G KLVFF
Sbjct: 302 VDISTIKQQEP---EIPGDKEAVDNGNVYSVSAAAAAAMTGNGKASEG-NGPATKKLVFF 357
Query: 351 DRRNG-FELEDLLRASAEMLGKGSLGTVYRAVLDDGSTVAVKRLKDANPCARHEFEQYMD 409
F+LEDLLRASAE+LGKG+ GT Y+AVLD + VAVKRLKD A EF++ ++
Sbjct: 358 GNATKVFDLEDLLRASAEVLGKGTFGTAYKAVLDAVTVVAVKRLKDVM-MADKEFKEKIE 416
Query: 410 VIGKLKHPNIVKLRAYYYAKEEKLLVYDYLSNGSLHALLHGNRGPGRIPLDWTTRISLVL 469
++G + H N+V LRAYY++++EKLLVYD++ GSL ALLHGNRG GR PL+W R + +
Sbjct: 417 LVGAMDHENLVPLRAYYFSRDEKLLVYDFMPMGSLSALLHGNRGAGRSPLNWDVRSRIAI 476
Query: 470 GAARGLARIHTEYSAAKVPHGNVKSSNVLLDKNGVACISDFGLSLLL-----NPVHATAR 524
GAARGL +H++ HGN+KSSN+LL K+ A +SDFGL+ L+ NP AT
Sbjct: 477 GAARGLDYLHSQ--GTSTSHGNIKSSNILLTKSHDAKVSDFGLAQLVGSSATNPNRAT-- 532
Query: 525 LGGYRAPEQTEQKRLSQQADVYSFGVLLLEVLTGKAPSLQYPSPANRPRKVEEEETVVDL 584
GYRAPE T+ KR+SQ+ DVYSFGV+LLE++TGKAPS V EE VDL
Sbjct: 533 --GYRAPEVTDPKRVSQKGDVYSFGVVLLELITGKAPS----------NSVMNEEG-VDL 579
Query: 585 PKWVRSVVREEWTGEVFDQELLRYKNIEEELVS-MLHVGLACVVQQPEKRPTMVDVVKMI 643
P+WV+SV R+EW EVFD ELL EEE+++ M+ +GL C Q P++RP M +VV+ +
Sbjct: 580 PRWVKSVARDEWRREVFDSELLSLATDEEEMMAEMVQLGLECTSQHPDQRPEMSEVVRKM 639
Query: 644 EDIR 647
E++R
Sbjct: 640 ENLR 643
>gb|AAC95351.1| receptor-like protein kinase [Arabidopsis thaliana]
Length = 645
Score = 424 bits (1089), Expect = e-117
Identities = 277/664 (41%), Positives = 384/664 (57%), Gaps = 63/664 (9%)
Query: 10 FLFLSIYIVPCLTHNDTQA-LTLFRQQTDTHGQLLTNWTGPEACSASWHGVTCTPNNRVT 68
FL L + +P + D A T G W + +W GV C +NRVT
Sbjct: 7 FLSLLLLSLPLPSTQDLNADRTALLSLRSAVGGRTFRWNIKQTSPCNWAGVKCE-SNRVT 65
Query: 69 TLVLPSLNLRG--PIDALSSLTHLRLLDLHNNRLNGTVSASLLSNCTNLKLLYLAGNDFS 126
L LP + L G P +LT LR L L N L+G++ L S +NL+ LYL GN FS
Sbjct: 66 ALRLPGVALSGDIPEGIFGNLTQLRTLSLRLNALSGSLPKDL-STSSNLRHLYLQGNRFS 124
Query: 127 GQIPPEISSLNNLLRLDLSDNNLAGDIPNEISRLTNLLTLRLQNNALSGNIPDLSSIMPN 186
G+IP + SL++L+RL+L+ N+ G+I + + LT L TL L+NN LSG+IPDL +P
Sbjct: 125 GEIPEVLFSLSHLVRLNLASNSFTGEISSGFTNLTKLKTLFLENNQLSGSIPDLD--LP- 181
Query: 187 LTELNMTNNEFYGKVPNTMLNKFGDESFSGNEGLCGSKPFQVCSLTENSPPSSEPVQTVP 246
L + N++NN G +P L +F +SF LCG KP ++C P E V + P
Sbjct: 182 LVQFNVSNNSLNGSIPKN-LQRFESDSFL-QTSLCG-KPLKLC-------PDEETVPSQP 231
Query: 247 SNPSSFPATSVIARPRSQHHKGLSPGVIVAIVVAICVALLVVTSFVVAHCCA----RGRG 302
++ + SV + LS G I IV+ V ++ ++ C R R
Sbjct: 232 TSGGNRTPPSVEGSEEKKKKNKLSGGAIAGIVIGCVVGFALIVLILMVLCRKKSNKRSRA 291
Query: 303 VNSNSLMGSEAGKRKSYGSEKKVYNSN------------GGGGDSSDGTSGTDMSKLVFF 350
V+ +++ E + G ++ V N N G G +S+G +G KLVFF
Sbjct: 292 VDISTIKQQEP---EIPGDKEAVDNGNVYSVSAAAAAAMTGNGKASEG-NGPATKKLVFF 347
Query: 351 DRRNG-FELEDLLRASAEMLGKGSLGTVYRAVLDDGSTVAVKRLKDANPCARHEFEQYMD 409
F+LEDLLRASAE+LGKG+ GT Y+AVLD + VAVKRLKD A EF++ ++
Sbjct: 348 GNATKVFDLEDLLRASAEVLGKGTFGTAYKAVLDAVTVVAVKRLKDVM-MADKEFKEKIE 406
Query: 410 VIGKLKHPNIVKLRAYYYAKEEKLLVYDYLSNGSLHALLHGNRGPGRIPLDWTTRISLVL 469
++G + H N+V LRAYY++++EKLLVYD++ GSL ALLHGNRG GR PL+W R + +
Sbjct: 407 LVGAMDHENLVPLRAYYFSRDEKLLVYDFMPMGSLSALLHGNRGAGRSPLNWDVRSRIAI 466
Query: 470 GAARGLARIHTEYSAAKVPHGNVKSSNVLLDKNGVACISDFGLSLLL-----NPVHATAR 524
GAARGL +H++ HGN+KSSN+LL K+ A +SDFGL+ L+ NP AT
Sbjct: 467 GAARGLNYLHSQ--GTSTSHGNIKSSNILLTKSHDAKVSDFGLAQLVGSSATNPNRAT-- 522
Query: 525 LGGYRAPEQTEQKRLSQQADVYSFGVLLLEVLTGKAPSLQYPSPANRPRKVEEEETVVDL 584
GYRAPE T+ KR+SQ+ DVYSFGV+LLE++TGKAPS V EE VDL
Sbjct: 523 --GYRAPEVTDPKRVSQKGDVYSFGVVLLELITGKAPS----------NSVMNEEG-VDL 569
Query: 585 PKWVRSVVREEWTGEVFDQELLRYKNIEEELVS-MLHVGLACVVQQPEKRPTMVDVVKMI 643
P+WV+SV R+EW EVFD ELL EEE+++ M+ +GL C Q P++RP M +VV+ +
Sbjct: 570 PRWVKSVARDEWRREVFDSELLSLATDEEEMMAEMVQLGLECTSQHPDQRPEMSEVVRKM 629
Query: 644 EDIR 647
E++R
Sbjct: 630 ENLR 633
>gb|AAM64268.1| receptor kinase, putative [Arabidopsis thaliana]
Length = 639
Score = 423 bits (1088), Expect = e-117
Identities = 281/665 (42%), Positives = 383/665 (57%), Gaps = 52/665 (7%)
Query: 1 MNYVIMFFFFLFLSIYIVPCLTH--NDTQALTLFRQQTDTHGQLLTNWTGPEACSASWHG 58
M+ + +FF L LS+ + P + D AL FR L W + +W G
Sbjct: 1 MSNLSIFFSILLLSLPL-PSIGDLAADKSALLSFRSAVGGRTLL---WDVKQTSPCNWTG 56
Query: 59 VTCTPNNRVTTLVLPSLNLRG--PIDALSSLTHLRLLDLHNNRLNGTVSASLLSNCTNLK 116
V C RVT L LP L G P +LT LR L L N L G++ L C++L+
Sbjct: 57 VLCD-GGRVTALRLPGETLSGHIPEGIFGNLTQLRTLSLRLNGLTGSLPLDL-GRCSDLR 114
Query: 117 LLYLAGNDFSGQIPPEISSLNNLLRLDLSDNNLAGDIPNEISRLTNLLTLRLQNNALSGN 176
LYL GN FSG+IP + SL+NL+RL+L++N +G+I + LT L TL L+NN LSG+
Sbjct: 115 RLYLQGNRFSGEIPEVLFSLSNLVRLNLAENEFSGEISSGFKNLTRLKTLYLENNKLSGS 174
Query: 177 IPDLSSIMPNLTELNMTNNEFYGKVPNTMLNKFGDESFSGNEGLCGSKPFQVCSLTENSP 236
+ DL +L + N++NN G +P + L KF +SF G LCG KP VCS E +
Sbjct: 175 LLDLDL---SLDQFNVSNNLLNGSIPKS-LQKFDSDSFVGTS-LCG-KPLVVCS-NEGTV 227
Query: 237 PSSEPVQTVPSNPSSFPATSVIARPRSQHHKGLSPGVIVAIVVAICVALLVVTSFVVAHC 296
PS P + + P T V R + K LS G I IV+ V L ++ ++
Sbjct: 228 PSQ------PISVGNIPGT-VEGREEKKKRKKLSGGAIAGIVIGCVVGLSLIVMILMVLF 280
Query: 297 CARG----RGVNSNSLMGSEAG----KRKSYGSEKKVY-NSNGGGGDSSDGTSGTDMSKL 347
+G R ++ ++ E K E + Y N + + + M KL
Sbjct: 281 RKKGNERTRAIDLATIKHHEVEIPGEKAAVEAPENRSYVNEYSPSAVKAVEVNSSGMKKL 340
Query: 348 VFFDRRNG-FELEDLLRASAEMLGKGSLGTVYRAVLDDGSTVAVKRLKDANPCARHEFEQ 406
VFF F+LEDLLRASAE+LGKG+ GT Y+AVLD + VAVKRLKD R EF++
Sbjct: 341 VFFGNATKVFDLEDLLRASAEVLGKGTFGTAYKAVLDAVTLVAVKRLKDVTMADR-EFKE 399
Query: 407 YMDVIGKLKHPNIVKLRAYYYAKEEKLLVYDYLSNGSLHALLHGNRGPGRIPLDWTTRIS 466
++V+G + H N+V LRAYYY+ +EKLLVYD++ GSL ALLHGN+G GR PL+W R
Sbjct: 400 KIEVVGAMDHENLVPLRAYYYSGDEKLLVYDFMPMGSLSALLHGNKGAGRPPLNWEVRSG 459
Query: 467 LVLGAARGLARIHTEYSAAKVPHGNVKSSNVLLDKNGVACISDFGLSLLLNPVHATA-RL 525
+ LGAARGL +H++ + HGNVKSSN+LL + A +SDFGL+ L++ T R
Sbjct: 460 IALGAARGLDYLHSQDPLSS--HGNVKSSNILLTNSHDARVSDFGLAQLVSASSTTPNRA 517
Query: 526 GGYRAPEQTEQKRLSQQADVYSFGVLLLEVLTGKAPSLQYPSPANRPRKVEEEETVVDLP 585
GYRAPE T+ +R+SQ+ADVYSFGV+LLE+LTGKAPS V EE +DL
Sbjct: 518 TGYRAPEVTDPRRVSQKADVYSFGVVLLELLTGKAPS----------NSVMNEEG-MDLA 566
Query: 586 KWVRSVVREEWTGEVFDQELLRYK---NIEEELVSMLHVGLACVVQQPEKRPTMVDVVKM 642
+WV SV REEW EVFD EL+ + ++EEE+ ML +G+ C Q P+KRP MV+VV+
Sbjct: 567 RWVHSVAREEWRNEVFDSELMSIETVVSVEEEMAEMLQLGIDCTEQHPDKRPVMVEVVRR 626
Query: 643 IEDIR 647
I+++R
Sbjct: 627 IQELR 631
Database: nr
Posted date: Jul 5, 2005 12:34 AM
Number of letters in database: 863,360,394
Number of sequences in database: 2,540,612
Lambda K H
0.316 0.134 0.392
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 1,200,807,111
Number of Sequences: 2540612
Number of extensions: 53880283
Number of successful extensions: 255059
Number of sequences better than 10.0: 20774
Number of HSP's better than 10.0 without gapping: 5028
Number of HSP's successfully gapped in prelim test: 15829
Number of HSP's that attempted gapping in prelim test: 182863
Number of HSP's gapped (non-prelim): 40138
length of query: 676
length of database: 863,360,394
effective HSP length: 135
effective length of query: 541
effective length of database: 520,377,774
effective search space: 281524375734
effective search space used: 281524375734
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.6 bits)
S2: 79 (35.0 bits)
Medicago: description of AC139525.8


