

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC139355.5 - phase: 0
(672 letters)
Database: nr
2,540,612 sequences; 863,360,394 total letters
Searching..................................................done
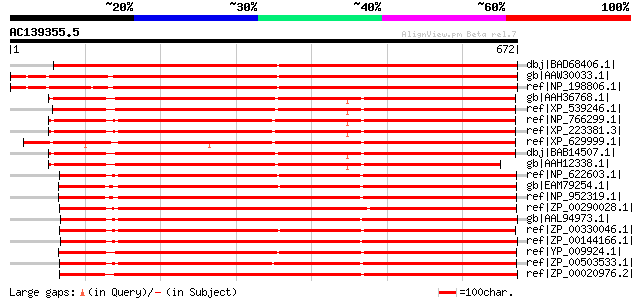
Score E
Sequences producing significant alignments: (bits) Value
dbj|BAD68406.1| putative GTP-binding membrane protein LepA [Oryz... 1033 0.0
gb|AAW30033.1| At5g39900 [Arabidopsis thaliana] gi|56461750|gb|A... 1033 0.0
ref|NP_198806.1| GTP-binding protein LepA, putative [Arabidopsis... 1024 0.0
gb|AAH36768.1| Hypothetical protein FLJ13220 [Homo sapiens] 703 0.0
ref|XP_539246.1| PREDICTED: similar to Hypothetical protein FLJ1... 702 0.0
ref|NP_766299.1| hypothetical protein LOC231279 isoform a [Mus m... 700 0.0
ref|XP_223381.3| PREDICTED: similar to expressed sequence AA4075... 697 0.0
ref|XP_629999.1| hypothetical protein DDB0184025 [Dictyostelium ... 697 0.0
dbj|BAB14507.1| unnamed protein product [Homo sapiens] gi|113454... 694 0.0
gb|AAH12338.1| FLJ13220 protein [Homo sapiens] 680 0.0
ref|NP_622603.1| Membrane GTPase LepA [Thermoanaerobacter tengco... 668 0.0
gb|EAM79254.1| Small GTP-binding protein domain:GTP-binding prot... 660 0.0
ref|NP_952319.1| GTP-binding protein LepA [Geobacter sulfurreduc... 660 0.0
ref|ZP_00290028.1| COG0481: Membrane GTPase LepA [Magnetococcus ... 649 0.0
gb|AAL94973.1| GTP-binding protein lepA [Fusobacterium nucleatum... 648 0.0
ref|ZP_00330046.1| COG0481: Membrane GTPase LepA [Moorella therm... 647 0.0
ref|ZP_00144166.1| GTP-binding protein lepA [Fusobacterium nucle... 643 0.0
ref|YP_009924.1| GTP-binding protein LepA [Desulfovibrio vulgari... 642 0.0
ref|ZP_00503533.1| Small GTP-binding protein domain:GTP-binding ... 640 0.0
ref|ZP_00020976.2| COG0481: Membrane GTPase LepA [Chloroflexus a... 638 0.0
>dbj|BAD68406.1| putative GTP-binding membrane protein LepA [Oryza sativa (japonica
cultivar-group)]
Length = 663
Score = 1033 bits (2672), Expect = 0.0
Identities = 510/615 (82%), Positives = 567/615 (91%), Gaps = 2/615 (0%)
Query: 58 DLSQYPPELVRNFSIIAHVDHGKSTLADRLLELTGTIKKGLGQPQYLDKLQVERERGITV 117
+L YPPE VRNFSIIAHVDHGKSTLADRLLELTGTIKKG GQPQYLDKLQVERERGITV
Sbjct: 48 ELGLYPPERVRNFSIIAHVDHGKSTLADRLLELTGTIKKGHGQPQYLDKLQVERERGITV 107
Query: 118 KAQTATMFYKNIINGDDFKDGKESSNYLLNLIDTPGHVDFSYEVSRSLAACQGVLLVVDA 177
KAQTATMFY++ N D ++ +YLLNLIDTPGHVDFSYEVSRSLAACQG LLVVDA
Sbjct: 108 KAQTATMFYRHANNQLPASDQPDAPSYLLNLIDTPGHVDFSYEVSRSLAACQGALLVVDA 167
Query: 178 AQGVQAQTVANFYLAFESNLAIIPVINKIDQPTADPDRVKGQLKSMFDLDPSDALLTSAK 237
AQGVQAQT+ANFYLAFESNL+IIPVINKIDQPTADPD VK QLK +FD+DPS+ALLTSAK
Sbjct: 168 AQGVQAQTIANFYLAFESNLSIIPVINKIDQPTADPDNVKAQLKRLFDIDPSEALLTSAK 227
Query: 238 TGVGLEHVLPAVIERIPPPPGKSESSLRMLLLDSYFDEYRGVICHVAVVDGALRKGDKIS 297
TG GL VLPAVIERIP PPGK +S +RMLLLDSY+DEY+GVICHVAVVDGAL KGDKI+
Sbjct: 228 TGQGLSQVLPAVIERIPSPPGKCDSPVRMLLLDSYYDEYKGVICHVAVVDGALHKGDKIA 287
Query: 298 SAATGKSYEAMDIGIMHPELTPTGILFTGQVGYVITGMRTTKEARIGDTIYHTKSTVDVE 357
SAATG++YE +D+GIMHPELTPTG+L+TGQVGYVI+GMR+TKEARIGDT++ KS V E
Sbjct: 288 SAATGRTYEVLDVGIMHPELTPTGVLYTGQVGYVISGMRSTKEARIGDTLHQAKSIV--E 345
Query: 358 PLPGFKAAKHMVFSGLFPADGSDFEALSHAIEKLTCNDASVSVTKETSTALGLGFRCGFL 417
PLPGFK A+HMVFSGL+PADGSDF+ALSHAIEKLTCNDASVSVTKETSTALG+GFRCGFL
Sbjct: 346 PLPGFKPARHMVFSGLYPADGSDFDALSHAIEKLTCNDASVSVTKETSTALGMGFRCGFL 405
Query: 418 GLLHMDVFHQRLEQEYGAHIISTVPTVPYIYEYSDGSKLEVQNPAALPSNPKQRVVACWE 477
GLLHMDVFHQRLEQE+GA +IST+PTVPYI+EY DGSK++V+NPAAL SNP +R+ ACWE
Sbjct: 406 GLLHMDVFHQRLEQEHGAQVISTIPTVPYIFEYGDGSKVQVENPAALASNPGKRIAACWE 465
Query: 478 PTVIATIVMPSEYVGPVITLLSERRGEQLEYSFIDSQRVFMKYRLPLREIVIDFYNELKS 537
PTVIATI++PSEYVGPVI L SERRGEQ EY+FID+QR +KYRLPLREI++DFYNELKS
Sbjct: 466 PTVIATIIIPSEYVGPVIMLCSERRGEQQEYTFIDAQRALLKYRLPLREIIVDFYNELKS 525
Query: 538 ITSGYASFDYEDSDYQPSDLVKLDILLNGQAVDAMATIVHNSKSYRVGRELVEKLKKVID 597
ITSGYA+FDYEDS+YQ SDLVK+DILLNGQ VDAMATIVHN K+ RVGRELV+KLKK I+
Sbjct: 526 ITSGYATFDYEDSEYQQSDLVKMDILLNGQPVDAMATIVHNQKAQRVGRELVDKLKKFIE 585
Query: 598 RQMFEIIIQAAIGSKIIARETVTAMRKNVLAKCYGGDITRKKKLLEKQKEGKKRMKRVGS 657
RQMFEI IQAA+GSK+IARET++AMRKNVLAKCYGGDITRKKKLLEKQKEGKKRMKRVGS
Sbjct: 586 RQMFEITIQAAVGSKVIARETLSAMRKNVLAKCYGGDITRKKKLLEKQKEGKKRMKRVGS 645
Query: 658 VDVPQEAFHELLKVS 672
VD+PQEAFHELLKVS
Sbjct: 646 VDIPQEAFHELLKVS 660
>gb|AAW30033.1| At5g39900 [Arabidopsis thaliana] gi|56461750|gb|AAV91331.1|
At5g39900 [Arabidopsis thaliana]
gi|10176983|dbj|BAB10215.1| GTP-binding membrane protein
LepA homolog [Arabidopsis thaliana]
Length = 663
Score = 1033 bits (2670), Expect = 0.0
Identities = 522/674 (77%), Positives = 597/674 (88%), Gaps = 13/674 (1%)
Query: 1 MGFLRKASKTLKQSNY-VSLLFNFNPLSSRITHERFSITRAL-FCTQSRQNYTKEKAIID 58
MG + +ASKTLK S +S+LFN + S+R + +A F + SRQ+ ++ ID
Sbjct: 1 MGSMYRASKTLKSSRQALSILFN-SLNSNRQNPTCIGLYQAYGFSSDSRQS--SKEPTID 57
Query: 59 LSQYPPELVRNFSIIAHVDHGKSTLADRLLELTGTIKKGLGQPQYLDKLQVERERGITVK 118
L+++P E +RNFSIIAH+DHGKSTLADRL+ELTGTIKKG GQPQYLDKLQVERERGITVK
Sbjct: 58 LTKFPSEKIRNFSIIAHIDHGKSTLADRLMELTGTIKKGHGQPQYLDKLQVERERGITVK 117
Query: 119 AQTATMFYKNIINGDDFKDGKESSNYLLNLIDTPGHVDFSYEVSRSLAACQGVLLVVDAA 178
AQTATMFY+N + + +E+S YLLNLIDTPGHVDFSYEVSRSL+ACQG LLVVDAA
Sbjct: 118 AQTATMFYENKV------EDQEASGYLLNLIDTPGHVDFSYEVSRSLSACQGALLVVDAA 171
Query: 179 QGVQAQTVANFYLAFESNLAIIPVINKIDQPTADPDRVKGQLKSMFDLDPSDALLTSAKT 238
QGVQAQTVANFYLAFE+NL I+PVINKIDQPTADP+RVK QLKSMFDLD D LL SAKT
Sbjct: 172 QGVQAQTVANFYLAFEANLTIVPVINKIDQPTADPERVKAQLKSMFDLDTEDVLLVSAKT 231
Query: 239 GVGLEHVLPAVIERIPPPPGKSESSLRMLLLDSYFDEYRGVICHVAVVDGALRKGDKISS 298
G+GLEHVLPAVIERIPPPPG SES LRMLL DS+F+EY+GVIC+V+VVDG L KGDK+S
Sbjct: 232 GLGLEHVLPAVIERIPPPPGISESPLRMLLFDSFFNEYKGVICYVSVVDGMLSKGDKVSF 291
Query: 299 AATGKSYEAMDIGIMHPELTPTGILFTGQVGYVITGMRTTKEARIGDTIYHTKSTVDVEP 358
AA+G+SYE +D+GIMHPELT TG+L TGQVGY++TGMRTTKEARIGDTIY TK+TV EP
Sbjct: 292 AASGQSYEVLDVGIMHPELTSTGMLLTGQVGYIVTGMRTTKEARIGDTIYRTKTTV--EP 349
Query: 359 LPGFKAAKHMVFSGLFPADGSDFEALSHAIEKLTCNDASVSVTKETSTALGLGFRCGFLG 418
LPGFK +HMVFSG++PADGSDFEAL HA+EKLTCNDASVSV KETSTALG+GFRCGFLG
Sbjct: 350 LPGFKPVRHMVFSGVYPADGSDFEALGHAMEKLTCNDASVSVAKETSTALGMGFRCGFLG 409
Query: 419 LLHMDVFHQRLEQEYGAHIISTVPTVPYIYEYSDGSKLEVQNPAALPSNPKQRVVACWEP 478
LLHMDVFHQRLEQEYG +IST+PTVPY +EYSDGSKL+VQNPAALPSNPK RV A WEP
Sbjct: 410 LLHMDVFHQRLEQEYGTQVISTIPTVPYTFEYSDGSKLQVQNPAALPSNPKYRVTASWEP 469
Query: 479 TVIATIVMPSEYVGPVITLLSERRGEQLEYSFIDSQRVFMKYRLPLREIVIDFYNELKSI 538
TVIATI++PSEYVG VI L S+RRG+QLEY+FID+QRVF+KY+LPLREIV+DFY+ELKSI
Sbjct: 470 TVIATIILPSEYVGAVINLCSDRRGQQLEYTFIDAQRVFLKYQLPLREIVVDFYDELKSI 529
Query: 539 TSGYASFDYEDSDYQPSDLVKLDILLNGQAVDAMATIVHNSKSYRVGRELVEKLKKVIDR 598
TSGYASFDYED++YQ SDLVKLDILLNGQAVDA+ATIVH K+YRVG+ELVEKLK I+R
Sbjct: 530 TSGYASFDYEDAEYQASDLVKLDILLNGQAVDALATIVHKQKAYRVGKELVEKLKNYIER 589
Query: 599 QMFEIIIQAAIGSKIIARETVTAMRKNVLAKCYGGDITRKKKLLEKQKEGKKRMKRVGSV 658
QMFE++IQAAIGSKIIAR+T++AMRKNVLAKCYGGDITRKKKLLEKQKEGKKRMKRVGSV
Sbjct: 590 QMFEVMIQAAIGSKIIARDTISAMRKNVLAKCYGGDITRKKKLLEKQKEGKKRMKRVGSV 649
Query: 659 DVPQEAFHELLKVS 672
D+P EAF ++LKVS
Sbjct: 650 DIPHEAFQQILKVS 663
>ref|NP_198806.1| GTP-binding protein LepA, putative [Arabidopsis thaliana]
Length = 661
Score = 1024 bits (2648), Expect = 0.0
Identities = 520/674 (77%), Positives = 595/674 (88%), Gaps = 15/674 (2%)
Query: 1 MGFLRKASKTLKQSNY-VSLLFNFNPLSSRITHERFSITRAL-FCTQSRQNYTKEKAIID 58
MG + +ASKTLK S +S+LFN + S+R + +A F + SRQ+ ++ ID
Sbjct: 1 MGSMYRASKTLKSSRQALSILFN-SLNSNRQNPTCIGLYQAYGFSSDSRQS--SKEPTID 57
Query: 59 LSQYPPELVRNFSIIAHVDHGKSTLADRLLELTGTIKKGLGQPQYLDKLQVERERGITVK 118
L+++P E +RNFSIIAH+DHGKSTLADRL+ELTGTIKKG GQPQYLDKLQ RERGITVK
Sbjct: 58 LTKFPSEKIRNFSIIAHIDHGKSTLADRLMELTGTIKKGHGQPQYLDKLQ--RERGITVK 115
Query: 119 AQTATMFYKNIINGDDFKDGKESSNYLLNLIDTPGHVDFSYEVSRSLAACQGVLLVVDAA 178
AQTATMFY+N + + +E+S YLLNLIDTPGHVDFSYEVSRSL+ACQG LLVVDAA
Sbjct: 116 AQTATMFYENKV------EDQEASGYLLNLIDTPGHVDFSYEVSRSLSACQGALLVVDAA 169
Query: 179 QGVQAQTVANFYLAFESNLAIIPVINKIDQPTADPDRVKGQLKSMFDLDPSDALLTSAKT 238
QGVQAQTVANFYLAFE+NL I+PVINKIDQPTADP+RVK QLKSMFDLD D LL SAKT
Sbjct: 170 QGVQAQTVANFYLAFEANLTIVPVINKIDQPTADPERVKAQLKSMFDLDTEDVLLVSAKT 229
Query: 239 GVGLEHVLPAVIERIPPPPGKSESSLRMLLLDSYFDEYRGVICHVAVVDGALRKGDKISS 298
G+GLEHVLPAVIERIPPPPG SES LRMLL DS+F+EY+GVIC+V+VVDG L KGDK+S
Sbjct: 230 GLGLEHVLPAVIERIPPPPGISESPLRMLLFDSFFNEYKGVICYVSVVDGMLSKGDKVSF 289
Query: 299 AATGKSYEAMDIGIMHPELTPTGILFTGQVGYVITGMRTTKEARIGDTIYHTKSTVDVEP 358
AA+G+SYE +D+GIMHPELT TG+L TGQVGY++TGMRTTKEARIGDTIY TK+TV EP
Sbjct: 290 AASGQSYEVLDVGIMHPELTSTGMLLTGQVGYIVTGMRTTKEARIGDTIYRTKTTV--EP 347
Query: 359 LPGFKAAKHMVFSGLFPADGSDFEALSHAIEKLTCNDASVSVTKETSTALGLGFRCGFLG 418
LPGFK +HMVFSG++PADGSDFEAL HA+EKLTCNDASVSV KETSTALG+GFRCGFLG
Sbjct: 348 LPGFKPVRHMVFSGVYPADGSDFEALGHAMEKLTCNDASVSVAKETSTALGMGFRCGFLG 407
Query: 419 LLHMDVFHQRLEQEYGAHIISTVPTVPYIYEYSDGSKLEVQNPAALPSNPKQRVVACWEP 478
LLHMDVFHQRLEQEYG +IST+PTVPY +EYSDGSKL+VQNPAALPSNPK RV A WEP
Sbjct: 408 LLHMDVFHQRLEQEYGTQVISTIPTVPYTFEYSDGSKLQVQNPAALPSNPKYRVTASWEP 467
Query: 479 TVIATIVMPSEYVGPVITLLSERRGEQLEYSFIDSQRVFMKYRLPLREIVIDFYNELKSI 538
TVIATI++PSEYVG VI L S+RRG+QLEY+FID+QRVF+KY+LPLREIV+DFY+ELKSI
Sbjct: 468 TVIATIILPSEYVGAVINLCSDRRGQQLEYTFIDAQRVFLKYQLPLREIVVDFYDELKSI 527
Query: 539 TSGYASFDYEDSDYQPSDLVKLDILLNGQAVDAMATIVHNSKSYRVGRELVEKLKKVIDR 598
TSGYASFDYED++YQ SDLVKLDILLNGQAVDA+ATIVH K+YRVG+ELVEKLK I+R
Sbjct: 528 TSGYASFDYEDAEYQASDLVKLDILLNGQAVDALATIVHKQKAYRVGKELVEKLKNYIER 587
Query: 599 QMFEIIIQAAIGSKIIARETVTAMRKNVLAKCYGGDITRKKKLLEKQKEGKKRMKRVGSV 658
QMFE++IQAAIGSKIIAR+T++AMRKNVLAKCYGGDITRKKKLLEKQKEGKKRMKRVGSV
Sbjct: 588 QMFEVMIQAAIGSKIIARDTISAMRKNVLAKCYGGDITRKKKLLEKQKEGKKRMKRVGSV 647
Query: 659 DVPQEAFHELLKVS 672
D+P EAF ++LKVS
Sbjct: 648 DIPHEAFQQILKVS 661
>gb|AAH36768.1| Hypothetical protein FLJ13220 [Homo sapiens]
Length = 669
Score = 703 bits (1814), Expect = 0.0
Identities = 364/628 (57%), Positives = 465/628 (73%), Gaps = 27/628 (4%)
Query: 52 KEKAIIDLSQYPPELVRNFSIIAHVDHGKSTLADRLLELTGTIKKGLGQPQYLDKLQVER 111
KEK +D+S++P E +RNFSI+AHVDHGKSTLADRLLELTGTI K Q LDKLQVER
Sbjct: 55 KEK--LDMSRFPVENIRNFSIVAHVDHGKSTLADRLLELTGTIDKTKNNKQVLDKLQVER 112
Query: 112 ERGITVKAQTATMFYKNIINGDDFKDGKESSNYLLNLIDTPGHVDFSYEVSRSLAACQGV 171
ERGITVKAQTA++FY E YLLNLIDTPGHVDFSYEVSRSL+ACQGV
Sbjct: 113 ERGITVKAQTASLFYNC-----------EGKQYLLNLIDTPGHVDFSYEVSRSLSACQGV 161
Query: 172 LLVVDAAQGVQAQTVANFYLAFESNLAIIPVINKIDQPTADPDRVKGQLKSMFDLDPSDA 231
LLVVDA +G+QAQTVANF+LAFE+ L++IPVINKID ADP+RV+ Q++ +FD+ +
Sbjct: 162 LLVVDANEGIQAQTVANFFLAFEAQLSVIPVINKIDLKNADPERVENQIEKVFDIPSDEC 221
Query: 232 LLTSAKTGVGLEHVLPAVIERIPPPPGKSESSLRMLLLDSYFDEYRGVICHVAVVDGALR 291
+ SAK G +E VL A+IERIPPP ++ LR L+ DS FD+YRGVI +VA+ DG +
Sbjct: 222 IKISAKLGTNVESVLQAIIERIPPPKVHRKNPLRALVFDSTFDQYRGVIANVALFDGVVS 281
Query: 292 KGDKISSAATGKSYEAMDIGIMHPELTPTGILFTGQVGYVITGMRTTKEARIGDTIYHTK 351
KGDKI SA T K+YE ++G+++P PT L+ GQVGY+I GM+ EA+IGDT+ K
Sbjct: 282 KGDKIVSAHTQKTYEVNEVGVLNPNEQPTHKLYAGQVGYLIAGMKDVTEAQIGDTLCLHK 341
Query: 352 STVDVEPLPGFKAAKHMVFSGLFPADGSDFEALSHAIEKLTCNDASVSVTKETSTALGLG 411
VEPLPGFK+AK MVF+G++P D S++ L AIEKLT ND+SV+V +++S ALG G
Sbjct: 342 Q--PVEPLPGFKSAKPMVFAGMYPLDQSEYNNLKSAIEKLTLNDSSVTVHRDSSLALGAG 399
Query: 412 FRCGFLGLLHMDVFHQRLEQEYGAHIISTVPTVPY---------IYEYSDGSKLEVQNPA 462
+R GFLGLLHM+VF+QRLEQEY A +I T PTVPY I E+ + ++ + NPA
Sbjct: 400 WRLGFLGLLHMEVFNQRLEQEYNASVILTTPTVPYKAVLSSSKLIKEHRE-KEITIINPA 458
Query: 463 ALPSNPKQRVVACWEPTVIATIVMPSEYVGPVITLLSERRGEQLEYSFIDSQRVFMKYRL 522
P K +V EP V+ TI+ P EY G ++ L RR Q FID RV +KY
Sbjct: 459 QFPD--KSKVTEYLEPVVLGTIITPDEYTGKIMMLCEARRAVQKNMIFIDQNRVMLKYLF 516
Query: 523 PLREIVIDFYNELKSITSGYASFDYEDSDYQPSDLVKLDILLNGQAVDAMATIVHNSKSY 582
PL EIV+DFY+ LKS++SGYASFDYED+ YQ ++LVK+DILLNG V+ + T+VH K++
Sbjct: 517 PLNEIVVDFYDSLKSLSSGYASFDYEDAGYQTAELVKMDILLNGNTVEELVTVVHKDKAH 576
Query: 583 RVGRELVEKLKKVIDRQMFEIIIQAAIGSKIIARETVTAMRKNVLAKCYGGDITRKKKLL 642
+G+ + E+LK + RQ+FEI IQAAIGSKIIARETV A RKNVLAKCYGGDITRK KLL
Sbjct: 577 SIGKAICERLKDSLPRQLFEIAIQAAIGSKIIARETVKAYRKNVLAKCYGGDITRKMKLL 636
Query: 643 EKQKEGKKRMKRVGSVDVPQEAFHELLK 670
++Q EGKK+++++G+V+VP++AF ++LK
Sbjct: 637 KRQAEGKKKLRKIGNVEVPKDAFIKVLK 664
>ref|XP_539246.1| PREDICTED: similar to Hypothetical protein FLJ13220 [Canis
familiaris]
Length = 887
Score = 702 bits (1811), Expect = 0.0
Identities = 363/623 (58%), Positives = 463/623 (74%), Gaps = 25/623 (4%)
Query: 57 IDLSQYPPELVRNFSIIAHVDHGKSTLADRLLELTGTIKKGLGQPQYLDKLQVERERGIT 116
ID+S++ E RNFSIIAHVDHGKSTLADRLLELTG I K Q LDKLQVERERGIT
Sbjct: 276 IDMSRFSVENTRNFSIIAHVDHGKSTLADRLLELTGAIDKTKNNKQVLDKLQVERERGIT 335
Query: 117 VKAQTATMFYKNIINGDDFKDGKESSNYLLNLIDTPGHVDFSYEVSRSLAACQGVLLVVD 176
VKAQTA++FY E YLLNLIDTPGHVDFSYEVSRSL+ACQGVLLVVD
Sbjct: 336 VKAQTASLFYNC-----------EGKQYLLNLIDTPGHVDFSYEVSRSLSACQGVLLVVD 384
Query: 177 AAQGVQAQTVANFYLAFESNLAIIPVINKIDQPTADPDRVKGQLKSMFDLDPSDALLTSA 236
A +G+QAQTVANF+LAFE+ L++IPVINKID ADP+RV+ Q++ +FD+ ++ + SA
Sbjct: 385 ANEGIQAQTVANFFLAFEAQLSVIPVINKIDLKNADPERVEKQIEKVFDIPRNECIKISA 444
Query: 237 KTGVGLEHVLPAVIERIPPPPGKSESSLRMLLLDSYFDEYRGVICHVAVVDGALRKGDKI 296
K G +E VL AVIERIPPP ++SL+ L+ DS FD+YRGVI +VA+ DG + KGDKI
Sbjct: 445 KLGTNVERVLQAVIERIPPPKVHRKNSLKALVFDSTFDQYRGVIANVALFDGMVSKGDKI 504
Query: 297 SSAATGKSYEAMDIGIMHPELTPTGILFTGQVGYVITGMRTTKEARIGDTIYHTKSTVDV 356
SA T K+YE ++G+++P PT L+ GQVGY+I GM+ EA+IGDT+Y K V
Sbjct: 505 VSAHTKKTYEVNEVGVLNPNEQPTHKLYAGQVGYLIAGMKDVTEAQIGDTLYLHKQ--PV 562
Query: 357 EPLPGFKAAKHMVFSGLFPADGSDFEALSHAIEKLTCNDASVSVTKETSTALGLGFRCGF 416
EPLPGFK+AK MVF+G++P D S++ L A+EKLT ND+SV+V +++S ALG G+R GF
Sbjct: 563 EPLPGFKSAKPMVFAGMYPIDQSEYNNLKSAVEKLTINDSSVTVHRDSSLALGAGWRLGF 622
Query: 417 LGLLHMDVFHQRLEQEYGAHIISTVPTVPY---------IYEYSDGSKLEVQNPAALPSN 467
LGLLHM+VF+QRLEQEY A +I T PTVPY I EY + ++ + NPA P
Sbjct: 623 LGLLHMEVFNQRLEQEYNASVILTTPTVPYKAVLSSAKLIKEYRE-KEITIINPAQFPD- 680
Query: 468 PKQRVVACWEPTVIATIVMPSEYVGPVITLLSERRGEQLEYSFIDSQRVFMKYRLPLREI 527
K +V EP V+ TI+ P EY+G V+ L RR Q +ID RV +KY PL EI
Sbjct: 681 -KSKVTEYLEPVVLGTIITPDEYIGKVMMLCQARRAVQKNMMYIDQNRVMLKYLFPLNEI 739
Query: 528 VIDFYNELKSITSGYASFDYEDSDYQPSDLVKLDILLNGQAVDAMATIVHNSKSYRVGRE 587
V+DFY+ LKS++SGYASFDYED+ YQ ++LVK+DILLNG V+ + TIVH K++ VG+
Sbjct: 740 VVDFYDSLKSLSSGYASFDYEDAGYQTAELVKMDILLNGNIVEELVTIVHKDKAHSVGKV 799
Query: 588 LVEKLKKVIDRQMFEIIIQAAIGSKIIARETVTAMRKNVLAKCYGGDITRKKKLLEKQKE 647
+ E+LK + RQ+FEI IQAAIGSKIIARETV A RKNVLAKCYGGDITRK KLL++Q E
Sbjct: 800 ICERLKDSLPRQLFEIAIQAAIGSKIIARETVKAYRKNVLAKCYGGDITRKMKLLKRQAE 859
Query: 648 GKKRMKRVGSVDVPQEAFHELLK 670
GKK+++++G+++VP++AF ++L+
Sbjct: 860 GKKKLRKIGNIEVPKDAFIKVLR 882
>ref|NP_766299.1| hypothetical protein LOC231279 isoform a [Mus musculus]
gi|26351195|dbj|BAC39234.1| unnamed protein product [Mus
musculus]
Length = 651
Score = 700 bits (1807), Expect = 0.0
Identities = 362/628 (57%), Positives = 468/628 (73%), Gaps = 27/628 (4%)
Query: 52 KEKAIIDLSQYPPELVRNFSIIAHVDHGKSTLADRLLELTGTIKKGLGQPQYLDKLQVER 111
KEK D+S++P E +RNFSIIAHVDHGKSTLADRLLELTGTI K Q LDKLQVER
Sbjct: 37 KEKP--DMSRFPVEDIRNFSIIAHVDHGKSTLADRLLELTGTIDKTKKNKQVLDKLQVER 94
Query: 112 ERGITVKAQTATMFYKNIINGDDFKDGKESSNYLLNLIDTPGHVDFSYEVSRSLAACQGV 171
ERGITVKAQTA++FY GK+ YLLNLIDTPGHVDFSYEVSRSL+ACQGV
Sbjct: 95 ERGITVKAQTASLFYSF--------GGKQ---YLLNLIDTPGHVDFSYEVSRSLSACQGV 143
Query: 172 LLVVDAAQGVQAQTVANFYLAFESNLAIIPVINKIDQPTADPDRVKGQLKSMFDLDPSDA 231
LLVVDA +G+QAQTVANF+LAFE+ L++IPVINKID ADP+RV Q++ +FD+ +
Sbjct: 144 LLVVDANEGIQAQTVANFFLAFEAQLSVIPVINKIDLKNADPERVGKQIEKVFDIPSEEC 203
Query: 232 LLTSAKTGVGLEHVLPAVIERIPPPPGKSESSLRMLLLDSYFDEYRGVICHVAVVDGALR 291
+ SAK G ++ VL AVIERIPPP E+ L+ L+ DS FD+YRGVI ++A+ DG +
Sbjct: 204 IKISAKLGTNVDSVLQAVIERIPPPKVHRENPLKALVFDSTFDQYRGVIANIALFDGVVS 263
Query: 292 KGDKISSAATGKSYEAMDIGIMHPELTPTGILFTGQVGYVITGMRTTKEARIGDTIYHTK 351
KGDKI SA T K+YE ++GI++P PT L+ GQVG++I GM+ EA+IGDT+Y
Sbjct: 264 KGDKIVSAHTKKAYEVNEVGILNPNEQPTHKLYAGQVGFLIAGMKDVTEAQIGDTLY--L 321
Query: 352 STVDVEPLPGFKAAKHMVFSGLFPADGSDFEALSHAIEKLTCNDASVSVTKETSTALGLG 411
VEPLPGFK+AK MVF+G++P D S++ L AIEKLT ND+SV+V +++S ALG G
Sbjct: 322 HNHPVEPLPGFKSAKPMVFAGVYPIDQSEYNNLKSAIEKLTLNDSSVTVHRDSSLALGAG 381
Query: 412 FRCGFLGLLHMDVFHQRLEQEYGAHIISTVPTVPY---------IYEYSDGSKLEVQNPA 462
+R GFLGLLHM+VF+QRLEQEY A +I T PTVPY I EY + ++ + NPA
Sbjct: 382 WRLGFLGLLHMEVFNQRLEQEYNASVILTTPTVPYKAVLSSAKLIKEYKE-KEITIINPA 440
Query: 463 ALPSNPKQRVVACWEPTVIATIVMPSEYVGPVITLLSERRGEQLEYSFIDSQRVFMKYRL 522
P K +V EP V+ T++ P+EY G ++ L RR Q +FID RV +KY
Sbjct: 441 QFPE--KSQVTEYLEPVVLGTVITPTEYTGKIMALCQARRAIQKNMTFIDENRVMLKYLF 498
Query: 523 PLREIVIDFYNELKSITSGYASFDYEDSDYQPSDLVKLDILLNGQAVDAMATIVHNSKSY 582
PL EIV+DFY+ LKS++SGYASFDYED+ YQ ++LVK+DILLNG V+ + T+VH K+Y
Sbjct: 499 PLNEIVVDFYDSLKSLSSGYASFDYEDAGYQTAELVKMDILLNGNMVEELVTVVHREKAY 558
Query: 583 RVGRELVEKLKKVIDRQMFEIIIQAAIGSKIIARETVTAMRKNVLAKCYGGDITRKKKLL 642
VG+ + E+LK+ + RQ++EI IQAA+GSK+IARETV A RKNVLAKCYGGDITRK KLL
Sbjct: 559 TVGKSICERLKESLPRQLYEIAIQAAVGSKVIARETVKAYRKNVLAKCYGGDITRKMKLL 618
Query: 643 EKQKEGKKRMKRVGSVDVPQEAFHELLK 670
++Q EGKK+++++G++++P++AF ++LK
Sbjct: 619 KRQSEGKKKLRKIGNIEIPKDAFIKVLK 646
>ref|XP_223381.3| PREDICTED: similar to expressed sequence AA407526 isoform a [Rattus
norvegicus]
Length = 725
Score = 697 bits (1799), Expect = 0.0
Identities = 362/628 (57%), Positives = 463/628 (73%), Gaps = 27/628 (4%)
Query: 52 KEKAIIDLSQYPPELVRNFSIIAHVDHGKSTLADRLLELTGTIKKGLGQPQYLDKLQVER 111
KEK D+S++P E +RNFSIIAHVDHGKSTLADRLLELTGTI K Q LDKLQVER
Sbjct: 111 KEKP--DMSRFPVEDIRNFSIIAHVDHGKSTLADRLLELTGTIDKTQKNKQVLDKLQVER 168
Query: 112 ERGITVKAQTATMFYKNIINGDDFKDGKESSNYLLNLIDTPGHVDFSYEVSRSLAACQGV 171
ERGITVKAQTA++FY GK+ YLLNLIDTPGHVDFSYEVSRSL+ACQGV
Sbjct: 169 ERGITVKAQTASLFYSF--------GGKQ---YLLNLIDTPGHVDFSYEVSRSLSACQGV 217
Query: 172 LLVVDAAQGVQAQTVANFYLAFESNLAIIPVINKIDQPTADPDRVKGQLKSMFDLDPSDA 231
LLVVDA +G+QAQTVANF+LAFE+ L++IPVINKID ADP+RV Q++ +FD+ +
Sbjct: 218 LLVVDANEGIQAQTVANFFLAFEAQLSVIPVINKIDLKNADPERVGKQIEKVFDIPSEEC 277
Query: 232 LLTSAKTGVGLEHVLPAVIERIPPPPGKSESSLRMLLLDSYFDEYRGVICHVAVVDGALR 291
+ SAK G ++ VL AVIERIPPP + L+ L+ DS FD+YRGV+ +A+ DG L
Sbjct: 278 IKISAKLGTNVDSVLQAVIERIPPPKVHRANPLKALVFDSTFDQYRGVVASIALFDGMLS 337
Query: 292 KGDKISSAATGKSYEAMDIGIMHPELTPTGILFTGQVGYVITGMRTTKEARIGDTIYHTK 351
KGDKI SA T K+YE +GI++P PT L+ GQVG++I GM+ EA+IGDT+Y
Sbjct: 338 KGDKIVSAHTKKTYEVNQVGILNPNEQPTHKLYAGQVGFLIAGMKDVTEAQIGDTLY--L 395
Query: 352 STVDVEPLPGFKAAKHMVFSGLFPADGSDFEALSHAIEKLTCNDASVSVTKETSTALGLG 411
VEPLPGFK+AK MVF+G++P D S++ L AIEKLT ND+SV+V +++S ALG G
Sbjct: 396 HNHPVEPLPGFKSAKPMVFAGVYPIDQSEYNNLKSAIEKLTLNDSSVTVHRDSSLALGAG 455
Query: 412 FRCGFLGLLHMDVFHQRLEQEYGAHIISTVPTVPY---------IYEYSDGSKLEVQNPA 462
+R GFLGLLHM+VF+QRLEQEY A +I T PTVPY I EY + ++ + NPA
Sbjct: 456 WRLGFLGLLHMEVFNQRLEQEYNASVILTTPTVPYKAVLSSAKLIKEYKE-KEITIINPA 514
Query: 463 ALPSNPKQRVVACWEPTVIATIVMPSEYVGPVITLLSERRGEQLEYSFIDSQRVFMKYRL 522
P K +V EP V+ T++ P+EY G ++ L RR Q +FID RV +KY
Sbjct: 515 QFPD--KSKVTEYLEPVVLGTVITPTEYTGKIMELCQARRAIQKNMTFIDENRVMLKYLF 572
Query: 523 PLREIVIDFYNELKSITSGYASFDYEDSDYQPSDLVKLDILLNGQAVDAMATIVHNSKSY 582
PL EIV+DFY+ LKS++SGYASFDYED+ YQ +DL+K+DILLNG V+ + T+VH K+Y
Sbjct: 573 PLNEIVVDFYDSLKSLSSGYASFDYEDAGYQTADLIKMDILLNGNIVEELVTVVHREKAY 632
Query: 583 RVGRELVEKLKKVIDRQMFEIIIQAAIGSKIIARETVTAMRKNVLAKCYGGDITRKKKLL 642
VG+ + E+LK + RQ+FEI +QAAIGSK+IARETV A RKNVLAKCYGGDITRK KLL
Sbjct: 633 TVGKSICERLKDSLPRQLFEIAVQAAIGSKVIARETVKAYRKNVLAKCYGGDITRKMKLL 692
Query: 643 EKQKEGKKRMKRVGSVDVPQEAFHELLK 670
++Q EGKK+++++G++++P+ AF ++LK
Sbjct: 693 KRQAEGKKKLRKIGNIEIPKNAFIKVLK 720
>ref|XP_629999.1| hypothetical protein DDB0184025 [Dictyostelium discoideum]
gi|60463403|gb|EAL61590.1| hypothetical protein
DDB0184025 [Dictyostelium discoideum]
Length = 2122
Score = 697 bits (1798), Expect = 0.0
Identities = 366/673 (54%), Positives = 472/673 (69%), Gaps = 36/673 (5%)
Query: 19 LLFNFNPLSSRITHERFSITRAL--FCTQSRQNYTKEKAIIDLSQYPPELVRNFSIIAHV 76
LL ++R+++ S T ++ FC++S + EK +DLS Y + +RNFSIIAH+
Sbjct: 1459 LLRRTTTTTTRLSYINNSPTLSIRSFCSKSTTITSDEK--LDLSGYTTDRIRNFSIIAHI 1516
Query: 77 DHGKSTLADRLLELTGTIKKGL----------------GQPQYLDKLQVERERGITVKAQ 120
DHGK+TL+ +LL LTGT+ K + + QYLDKLQVE+ERGITVKAQ
Sbjct: 1517 DHGKTTLSTKLLSLTGTLPKSIYGSGEDSLVQKEKSEERREQYLDKLQVEKERGITVKAQ 1576
Query: 121 TATMFYKNIINGDDFKDGKESSNYLLNLIDTPGHVDFSYEVSRSLAACQGVLLVVDAAQG 180
T TM Y+N +G D YLLNLIDTPGHVDFSYEVSRSL ACQG LLVVDA QG
Sbjct: 1577 TCTMKYRNKEDGKD---------YLLNLIDTPGHVDFSYEVSRSLMACQGALLVVDAVQG 1627
Query: 181 VQAQTVANFYLAFESNLAIIPVINKIDQPTADPDRVKGQLKSMFDLDPSDALLTSAKTGV 240
VQAQT+AN+YLA +S L +IPVINKID PTAD +RVK +LK F P DA+L SAKTGV
Sbjct: 1628 VQAQTMANYYLALDSGLEVIPVINKIDLPTADVERVKQELKDAFGFHPDDAVLVSAKTGV 1687
Query: 241 GLEHVLPAVIERIPPPPGKSESS---LRMLLLDSYFDEYRGVICHVAVVDGALRKGDKIS 297
G+ +LPAVI+RIPPP ++S + LL DS+FD +RGVIC + VVDG ++KGD I
Sbjct: 1688 GITDILPAVIDRIPPPQEPTKSPKVPFKALLFDSWFDRFRGVICLIKVVDGKVKKGDSIV 1747
Query: 298 SAATGKSYEAMDIGIMHPELTPTGILFTGQVGYVITGMRTTKEARIGDTIYHTKSTVDVE 357
SA ++YE D+GIMHPE P LFTGQVGY+ GM+T+KEAR+GDT Y VE
Sbjct: 1748 SAGNKQTYEVFDVGIMHPEQRPAPSLFTGQVGYITPGMKTSKEARVGDTFY--MKDFPVE 1805
Query: 358 PLPGFKAAKHMVFSGLFPADGSDFEALSHAIEKLTCNDASVSVTKETSTALGLGFRCGFL 417
PLPGF+ AK MVF+G++P D D+ L + EKL D+S+S+T ETS ALG+GFRCGFL
Sbjct: 1806 PLPGFQPAKQMVFAGIYPVDSLDYTQLRESFEKLMLTDSSISMTNETSVALGMGFRCGFL 1865
Query: 418 GLLHMDVFHQRLEQEYGAHIISTVPTVPYIYEYSDGSKLEVQNPAALPSNPKQRVVACWE 477
GLLHMDV QRLEQEYG +I+T PTVPY +DG ++ + NPA PS + + +E
Sbjct: 1866 GLLHMDVVLQRLEQEYGQVVIATPPTVPYRCLLTDGKEILISNPAGYPS--AEHLSKTFE 1923
Query: 478 PTVIATIVMPSEYVGPVITLLSERRGEQLEYSFIDSQRVFMKYRLPLREIVIDFYNELKS 537
P V + +P+EY G V+ L + RG ++ R + + PL EIV DFY+ LK
Sbjct: 1924 PMVTGHVTLPTEYFGAVVKLCMDSRGILTNQETLEGNRTRITFNFPLGEIVTDFYDNLKR 1983
Query: 538 ITSGYASFDYEDSDYQPSDLVKLDILLNGQAVDAMATIVHNSKSYRVGRELVEKLKKVID 597
I+SGYAS DYED+ YQ SD+ K+ +LLNG+ VD+++ IVH S + R R LV++L+KVID
Sbjct: 1984 ISSGYASLDYEDNGYQESDVAKVRVLLNGEEVDSLSAIVHKSNAQRYSRSLVKRLRKVID 2043
Query: 598 RQMFEIIIQAAIGSKIIARETVTAMRKNVLAKCYGGDITRKKKLLEKQKEGKKRMKRVGS 657
RQMF++ IQA +GS + ARET++AMRK+V AKCYGGDITR++KLL+KQKEGKKRMK++G
Sbjct: 2044 RQMFQVNIQAMVGSDVQARETISAMRKDVTAKCYGGDITRRRKLLDKQKEGKKRMKQMGC 2103
Query: 658 VDVPQEAFHELLK 670
V++ QE F +L+K
Sbjct: 2104 VELSQEGFLKLMK 2116
>dbj|BAB14507.1| unnamed protein product [Homo sapiens] gi|11345460|ref|NP_068746.1|
hypothetical protein LOC60558 [Homo sapiens]
Length = 669
Score = 694 bits (1791), Expect = 0.0
Identities = 361/628 (57%), Positives = 461/628 (72%), Gaps = 27/628 (4%)
Query: 52 KEKAIIDLSQYPPELVRNFSIIAHVDHGKSTLADRLLELTGTIKKGLGQPQYLDKLQVER 111
KEK D+S++P E +RNFSI+AHVDHGKSTL DRLLELTGTI K Q LDKLQVER
Sbjct: 55 KEKP--DMSRFPVENIRNFSIVAHVDHGKSTLTDRLLELTGTIDKTKNNKQVLDKLQVER 112
Query: 112 ERGITVKAQTATMFYKNIINGDDFKDGKESSNYLLNLIDTPGHVDFSYEVSRSLAACQGV 171
ERGITVKAQTA++FY E YLLNLIDTPGHVDFSYEVSRSL+ACQGV
Sbjct: 113 ERGITVKAQTASLFYNC-----------EGKQYLLNLIDTPGHVDFSYEVSRSLSACQGV 161
Query: 172 LLVVDAAQGVQAQTVANFYLAFESNLAIIPVINKIDQPTADPDRVKGQLKSMFDLDPSDA 231
LLVVDA +G+QAQTVANF+LAFE+ L++IPVINKID ADP+RV+ Q++ +FD+ +
Sbjct: 162 LLVVDANEGIQAQTVANFFLAFEAQLSVIPVINKIDLKNADPERVENQIEKVFDIPSDEC 221
Query: 232 LLTSAKTGVGLEHVLPAVIERIPPPPGKSESSLRMLLLDSYFDEYRGVICHVAVVDGALR 291
+ SAK G +E VL A+IERIPPP ++ LR L+ DS FD+YRGVI +VA+ DG +
Sbjct: 222 IKISAKLGTNVESVLQAIIERIPPPKVHRKNPLRALVFDSTFDQYRGVIANVALFDGVVS 281
Query: 292 KGDKISSAATGKSYEAMDIGIMHPELTPTGILFTGQVGYVITGMRTTKEARIGDTIYHTK 351
KGDKI SA T K+YE ++G+++P PT L+ GQVGY+I GM+ EA+IGDT+ K
Sbjct: 282 KGDKIVSAHTQKTYEVNEVGVLNPNEQPTHKLYAGQVGYLIAGMKDVTEAQIGDTLCLHK 341
Query: 352 STVDVEPLPGFKAAKHMVFSGLFPADGSDFEALSHAIEKLTCNDASVSVTKETSTALGLG 411
VEPLPGFK+AK MVF+G++P D S++ L AIEKLT ND+SV+V +++S ALG G
Sbjct: 342 Q--PVEPLPGFKSAKPMVFAGMYPLDQSEYNNLKSAIEKLTLNDSSVTVHRDSSLALGAG 399
Query: 412 FRCGFLGLLHMDVFHQRLEQEYGAHIISTVPTVPY---------IYEYSDGSKLEVQNPA 462
+R GFLGLLHM+VF+QR EQEY A +I T PTVPY I E+ + ++ + NPA
Sbjct: 400 WRLGFLGLLHMEVFNQRPEQEYNASVILTTPTVPYKAVLSSSKLIKEHRE-KEITIINPA 458
Query: 463 ALPSNPKQRVVACWEPTVIATIVMPSEYVGPVITLLSERRGEQLEYSFIDSQRVFMKYRL 522
P K +V EP V+ TI+ P EY G ++ L RR Q FID RV +KY
Sbjct: 459 QFPD--KSKVTEYLEPVVLGTIITPDEYTGKIMMLCEARRAVQKNMIFIDQNRVMLKYLF 516
Query: 523 PLREIVIDFYNELKSITSGYASFDYEDSDYQPSDLVKLDILLNGQAVDAMATIVHNSKSY 582
PL EIV+DFY+ LKS++SGYASFDYED+ YQ ++LVK+DILLNG V+ + T+VH K++
Sbjct: 517 PLNEIVVDFYDSLKSLSSGYASFDYEDAGYQTAELVKMDILLNGNTVEELVTVVHKDKAH 576
Query: 583 RVGRELVEKLKKVIDRQMFEIIIQAAIGSKIIARETVTAMRKNVLAKCYGGDITRKKKLL 642
+G+ + E+LK + RQ+FEI IQAAIGSKIIARETV A RKNVLAKC GGDITRK KLL
Sbjct: 577 SIGKAICERLKDSLPRQLFEIAIQAAIGSKIIARETVKAYRKNVLAKCCGGDITRKMKLL 636
Query: 643 EKQKEGKKRMKRVGSVDVPQEAFHELLK 670
++Q EGKK+++++G+V+VP++AF ++LK
Sbjct: 637 KRQAEGKKKLRKIGNVEVPKDAFIKVLK 664
>gb|AAH12338.1| FLJ13220 protein [Homo sapiens]
Length = 645
Score = 680 bits (1755), Expect = 0.0
Identities = 356/608 (58%), Positives = 445/608 (72%), Gaps = 27/608 (4%)
Query: 52 KEKAIIDLSQYPPELVRNFSIIAHVDHGKSTLADRLLELTGTIKKGLGQPQYLDKLQVER 111
KEK D+S++P E +RNFSI+AHVDHGKSTLADRLLELTGTI K Q LDKLQVER
Sbjct: 55 KEKP--DMSRFPVENIRNFSIVAHVDHGKSTLADRLLELTGTIDKTKNNKQVLDKLQVER 112
Query: 112 ERGITVKAQTATMFYKNIINGDDFKDGKESSNYLLNLIDTPGHVDFSYEVSRSLAACQGV 171
ERGITVKAQTA++FY E YLLNLIDTPGHVDFSYEVSRSL+ACQGV
Sbjct: 113 ERGITVKAQTASLFYNC-----------EGKQYLLNLIDTPGHVDFSYEVSRSLSACQGV 161
Query: 172 LLVVDAAQGVQAQTVANFYLAFESNLAIIPVINKIDQPTADPDRVKGQLKSMFDLDPSDA 231
LLVVDA +G+QAQTVANF+LAFE+ L++IPVINKID ADP+RV+ Q++ +FD+ +
Sbjct: 162 LLVVDANEGIQAQTVANFFLAFEAQLSVIPVINKIDLKNADPERVENQIEKVFDIPSDEC 221
Query: 232 LLTSAKTGVGLEHVLPAVIERIPPPPGKSESSLRMLLLDSYFDEYRGVICHVAVVDGALR 291
+ SAK G +E VL A+IERIPPP ++ LR L+ DS FD+YRGVI +VA+ DG +
Sbjct: 222 IKISAKLGTNVESVLQAIIERIPPPKVHRKNPLRALVFDSTFDQYRGVIANVALFDGVVS 281
Query: 292 KGDKISSAATGKSYEAMDIGIMHPELTPTGILFTGQVGYVITGMRTTKEARIGDTIYHTK 351
KGDKI SA T K+YE ++G+++P PT L+ GQVGY+I GM+ EA+IGDT+ K
Sbjct: 282 KGDKIVSAHTQKTYEVNEVGVLNPNEQPTHKLYAGQVGYLIAGMKDVTEAQIGDTLCLHK 341
Query: 352 STVDVEPLPGFKAAKHMVFSGLFPADGSDFEALSHAIEKLTCNDASVSVTKETSTALGLG 411
VEPLPGFK+AK MVF+G++P D S++ L AIEKLT ND+SV+V +++S ALG G
Sbjct: 342 Q--PVEPLPGFKSAKPMVFAGMYPLDQSEYNNLKSAIEKLTLNDSSVTVHRDSSLALGAG 399
Query: 412 FRCGFLGLLHMDVFHQRLEQEYGAHIISTVPTVPY---------IYEYSDGSKLEVQNPA 462
+R GFLGLLHM+VF+QRLEQEY A +I T PTVPY I E+ + ++ + NPA
Sbjct: 400 WRLGFLGLLHMEVFNQRLEQEYNASVILTTPTVPYKAVLSSSKLIKEHRE-KEITIINPA 458
Query: 463 ALPSNPKQRVVACWEPTVIATIVMPSEYVGPVITLLSERRGEQLEYSFIDSQRVFMKYRL 522
P K +V EP V+ TI+ P EY G ++ L RR Q FID RV +KY
Sbjct: 459 QFPD--KSKVTEYLEPVVLGTIITPDEYTGKIMMLCEARRAVQKNMIFIDQNRVMLKYLF 516
Query: 523 PLREIVIDFYNELKSITSGYASFDYEDSDYQPSDLVKLDILLNGQAVDAMATIVHNSKSY 582
PL EIV+DFY+ LKS++SGYASFDYED+ YQ ++LVK+DILLNG V+ + T+VH K++
Sbjct: 517 PLNEIVVDFYDSLKSLSSGYASFDYEDAGYQTAELVKMDILLNGNTVEELVTVVHKDKAH 576
Query: 583 RVGRELVEKLKKVIDRQMFEIIIQAAIGSKIIARETVTAMRKNVLAKCYGGDITRKKKLL 642
+G+ + E+LK + RQ+FEI IQAAIGSKIIARETV A RKNVLAKCYGGDITRK KLL
Sbjct: 577 SIGKAICERLKDSLPRQLFEIAIQAAIGSKIIARETVKAYRKNVLAKCYGGDITRKMKLL 636
Query: 643 EKQKEGKK 650
++Q EGKK
Sbjct: 637 KRQAEGKK 644
>ref|NP_622603.1| Membrane GTPase LepA [Thermoanaerobacter tengcongensis MB4]
gi|20515955|gb|AAM24207.1| Membrane GTPase LepA
[Thermoanaerobacter tengcongensis MB4]
gi|24211904|sp|Q8RB72|LEPA_THETN GTP-binding protein
lepA
Length = 603
Score = 668 bits (1723), Expect = 0.0
Identities = 334/605 (55%), Positives = 442/605 (72%), Gaps = 13/605 (2%)
Query: 67 VRNFSIIAHVDHGKSTLADRLLELTGTIKKGLGQPQYLDKLQVERERGITVKAQTATMFY 126
+RNF IIAH+DHGKSTLADRL+E TG + + Q LD L++ERERGIT+K + M Y
Sbjct: 8 IRNFCIIAHIDHGKSTLADRLIEKTGFLSEREMDKQILDNLELERERGITIKLKPVRMIY 67
Query: 127 KNIINGDDFKDGKESSNYLLNLIDTPGHVDFSYEVSRSLAACQGVLLVVDAAQGVQAQTV 186
K KDG E Y LNLIDTPGHVDF+YEVSRS+AAC+G LLVVDA+QG++AQT+
Sbjct: 68 KA-------KDGNE---YELNLIDTPGHVDFTYEVSRSIAACEGALLVVDASQGIEAQTL 117
Query: 187 ANFYLAFESNLAIIPVINKIDQPTADPDRVKGQLKSMFDLDPSDALLTSAKTGVGLEHVL 246
AN YLA E +L IIPVINKID P+ADP+RVK +++ + +D S+ALLTSAK G+G+E VL
Sbjct: 118 ANVYLALEHDLEIIPVINKIDLPSADPERVKREIEDIIGIDASEALLTSAKEGIGIEEVL 177
Query: 247 PAVIERIPPPPGKSESSLRMLLLDSYFDEYRGVICHVAVVDGALRKGDKISSAATGKSYE 306
A+++RIPPP G + L+ L+ DS++D Y+G I V +VDG +++G +I +TGK +E
Sbjct: 178 EAIVKRIPPPQGDEDKPLKALIFDSFYDNYKGAISFVRIVDGKVKRGMRIKMFSTGKVFE 237
Query: 307 AMDIGIMHPELTPTGILFTGQVGYVITGMRTTKEARIGDTIYHTKSTVDVEPLPGFKAAK 366
++G P+L P L G+VGY+ ++ K+ R+GDTI + D PLPG+K
Sbjct: 238 VTEVGSFRPDLYPVEELKAGEVGYIAANIKNVKDTRVGDTITDADNPAD-SPLPGYKEVV 296
Query: 367 HMVFSGLFPADGSDFEALSHAIEKLTCNDASVSVTKETSTALGLGFRCGFLGLLHMDVFH 426
MVF G++PADG D+E L A+EKL NDAS+ +TS ALG GFRCGFLGLLHM++
Sbjct: 297 PMVFCGIYPADGEDYENLKEALEKLQLNDASLVFEPDTSAALGFGFRCGFLGLLHMEIVQ 356
Query: 427 QRLEQEYGAHIISTVPTVPYIYEYSDGSKLEVQNPAALPSNPKQRVVACWEPTVIATIVM 486
+RLE+EY ++++T P+V Y ++G LE+ NP +P P ++ EP V ATI++
Sbjct: 357 ERLEREYNLNLVTTAPSVIYKVYKTNGEVLELDNPTKMP--PPTQIDHIEEPIVEATIMV 414
Query: 487 PSEYVGPVITLLSERRGEQLEYSFIDSQRVFMKYRLPLREIVIDFYNELKSITSGYASFD 546
P++YVGPV+ L ERRG L +I+ RV +KY +PL EI+ DF++ LKS T GYAS D
Sbjct: 415 PTDYVGPVMELCQERRGVYLGMEYIEKTRVLLKYEMPLNEIIYDFFDALKSRTRGYASLD 474
Query: 547 YEDSDYQPSDLVKLDILLNGQAVDAMATIVHNSKSYRVGRELVEKLKKVIDRQMFEIIIQ 606
YE Y+ SDLVKLDIL+NG+ VDA++ IVH K+Y GR++VEKLK+ I R +FEI IQ
Sbjct: 475 YEFKGYKKSDLVKLDILVNGEVVDALSMIVHKDKAYEKGRKIVEKLKENIPRHLFEIPIQ 534
Query: 607 AAIGSKIIARETVTAMRKNVLAKCYGGDITRKKKLLEKQKEGKKRMKRVGSVDVPQEAFH 666
AAIGS+IIARETV A+RKNVLAKCYGGD+TRKKKLLEKQKEGKKRM+++G+V++PQEAF
Sbjct: 535 AAIGSRIIARETVKALRKNVLAKCYGGDVTRKKKLLEKQKEGKKRMRQIGTVEIPQEAFM 594
Query: 667 ELLKV 671
+LK+
Sbjct: 595 SILKL 599
>gb|EAM79254.1| Small GTP-binding protein domain:GTP-binding protein LepA
[Geobacter metallireducens GS-15]
Length = 629
Score = 660 bits (1703), Expect = 0.0
Identities = 331/607 (54%), Positives = 442/607 (72%), Gaps = 13/607 (2%)
Query: 65 ELVRNFSIIAHVDHGKSTLADRLLELTGTIKKGLGQPQYLDKLQVERERGITVKAQTATM 124
E +RNFSIIAH+DHGKSTLADRLLE TG + + Q Q+LDK+ +ERERGIT+KAQT +
Sbjct: 34 ENIRNFSIIAHIDHGKSTLADRLLEYTGALSERERQDQFLDKMDLERERGITIKAQTVRL 93
Query: 125 FYKNIINGDDFKDGKESSNYLLNLIDTPGHVDFSYEVSRSLAACQGVLLVVDAAQGVQAQ 184
Y+ DD KD Y+LNLIDTPGHVDF+YEVSRSLAAC+G LLVVDA+QGV+AQ
Sbjct: 94 NYR----ADDGKD------YVLNLIDTPGHVDFTYEVSRSLAACEGGLLVVDASQGVEAQ 143
Query: 185 TVANFYLAFESNLAIIPVINKIDQPTADPDRVKGQLKSMFDLDPSDALLTSAKTGVGLEH 244
T+AN YLA ++NL + PV+NKID P A+P+RVK +++ + LD DA+L SAK G+G
Sbjct: 144 TLANVYLAIDNNLEVFPVLNKIDLPAAEPERVKHEIEEIIGLDAHDAVLASAKEGIGTRE 203
Query: 245 VLPAVIERIPPPPGKSESSLRMLLLDSYFDEYRGVICHVAVVDGALRKGDKISSAATGKS 304
+L ++++IPPP G + L+ LL DS++D+Y+GVI ++DG L+KGDKI +TG+S
Sbjct: 204 ILEEIVKKIPPPEGDPAAPLKALLFDSWYDQYQGVIILARLIDGILKKGDKIQLVSTGRS 263
Query: 305 YEAMDIGIMHPELTPTGILFTGQVGYVITGMRTTKEARIGDTIYHTKSTVDVEPLPGFKA 364
YEA+ +G+ P + L G+VG+VI G++ +A+IGDT+ HT PL GFK
Sbjct: 264 YEALKVGVFAPVMREVPQLSAGEVGFVIAGIKDVADAKIGDTVTHTLKPCTT-PLGGFKE 322
Query: 365 AKHMVFSGLFPADGSDFEALSHAIEKLTCNDASVSVTKETSTALGLGFRCGFLGLLHMDV 424
K MVFSGL+P D S +E L A+ KL ND+S S ETS ALG GFRCGFLGLLHM++
Sbjct: 323 VKPMVFSGLYPIDTSQYEQLRDALAKLKLNDSSFSYEPETSLALGFGFRCGFLGLLHMEI 382
Query: 425 FHQRLEQEYGAHIISTVPTVPYIYEYSDGSKLEVQNPAALPSNPKQRVVACWEPTVIATI 484
+RLE+E+ +I+T PTV Y G + +++ LP P Q + EP ++A+I
Sbjct: 383 IQERLEREFNLDLITTAPTVVYRVHRIKGDMISIESANQLP--PTQEIDYVEEPFILASI 440
Query: 485 VMPSEYVGPVITLLSERRGEQLEYSFIDSQRVFMKYRLPLREIVIDFYNELKSITSGYAS 544
P+E+VG ++ L E+RG Q E ++ RV + Y LPL E+V+DFY+ LKSIT GYAS
Sbjct: 441 HTPNEFVGGILALCEEKRGVQREIKYLTPTRVMIIYELPLNEVVLDFYDRLKSITKGYAS 500
Query: 545 FDYEDSDYQPSDLVKLDILLNGQAVDAMATIVHNSKSYRVGRELVEKLKKVIDRQMFEII 604
DYE DY+ S+LV+++I++NG+ VDA++ I+H K+Y GR+LV K+K++I RQMFE+
Sbjct: 501 LDYEHLDYRRSELVRMNIMINGEVVDALSLIIHRDKAYYRGRDLVSKMKELIPRQMFEVA 560
Query: 605 IQAAIGSKIIARETVTAMRKNVLAKCYGGDITRKKKLLEKQKEGKKRMKRVGSVDVPQEA 664
IQAAIG+K+IARETV A+RK+VLAKCYGGDITRK+KLLEKQKEGKKRMK VG+V++PQEA
Sbjct: 561 IQAAIGAKVIARETVKALRKDVLAKCYGGDITRKRKLLEKQKEGKKRMKNVGNVELPQEA 620
Query: 665 FHELLKV 671
F +LKV
Sbjct: 621 FLAILKV 627
>ref|NP_952319.1| GTP-binding protein LepA [Geobacter sulfurreducens PCA]
gi|39983248|gb|AAR34642.1| GTP-binding protein LepA
[Geobacter sulfurreducens PCA]
gi|46396178|sp|P60789|LEPA_GEOSL GTP-binding protein
lepA
Length = 600
Score = 660 bits (1703), Expect = 0.0
Identities = 329/607 (54%), Positives = 445/607 (73%), Gaps = 13/607 (2%)
Query: 65 ELVRNFSIIAHVDHGKSTLADRLLELTGTIKKGLGQPQYLDKLQVERERGITVKAQTATM 124
+ +RNFSIIAH+DHGKSTLADRLLE TG + + Q Q+LDK+ +ERERGIT+KAQT +
Sbjct: 4 DTIRNFSIIAHIDHGKSTLADRLLEYTGALTEREMQDQFLDKMDLERERGITIKAQTVRL 63
Query: 125 FYKNIINGDDFKDGKESSNYLLNLIDTPGHVDFSYEVSRSLAACQGVLLVVDAAQGVQAQ 184
Y+ DD D Y+LNLIDTPGHVDF+YEVSRSLAAC+G LLVVDA+QGV+AQ
Sbjct: 64 TYR----ADDGND------YILNLIDTPGHVDFTYEVSRSLAACEGALLVVDASQGVEAQ 113
Query: 185 TVANFYLAFESNLAIIPVINKIDQPTADPDRVKGQLKSMFDLDPSDALLTSAKTGVGLEH 244
T+AN YLA ++NL + PV+NKID P A+P+RVK +++ + LD DA++ SAK G+G
Sbjct: 114 TLANVYLAIDNNLEVFPVLNKIDLPAAEPERVKHEIEEIIGLDAHDAVMASAKEGIGTRE 173
Query: 245 VLPAVIERIPPPPGKSESSLRMLLLDSYFDEYRGVICHVAVVDGALRKGDKISSAATGKS 304
+L ++++IPPP G + L+ LL DS++D+Y+GVI V V+DG ++KGDKI +TG+S
Sbjct: 174 ILEEIVKKIPPPEGDPAAPLKALLFDSWYDQYQGVIILVRVIDGTVKKGDKIQLVSTGRS 233
Query: 305 YEAMDIGIMHPELTPTGILFTGQVGYVITGMRTTKEARIGDTIYHTKSTVDVEPLPGFKA 364
YEA+ +G+ P + L G+VG++I G++ +A+IGDT+ H V PL GFK
Sbjct: 234 YEALKVGVFAPVMREVPQLAAGEVGFLIAGIKDVADAKIGDTVTHALKPC-VTPLGGFKE 292
Query: 365 AKHMVFSGLFPADGSDFEALSHAIEKLTCNDASVSVTKETSTALGLGFRCGFLGLLHMDV 424
K MVFSGL+P D + +E L A+ KL ND+S S ETS ALG GFRCGFLGLLHM++
Sbjct: 293 VKPMVFSGLYPIDTAQYEQLRDALAKLKLNDSSFSFEPETSLALGFGFRCGFLGLLHMEI 352
Query: 425 FHQRLEQEYGAHIISTVPTVPYIYEYSDGSKLEVQNPAALPSNPKQRVVACWEPTVIATI 484
+RLE+E+ +I+T PTV Y G + +++ LP P Q + EP ++A+I
Sbjct: 353 IQERLEREFNLDLITTAPTVVYKVHRIKGDVITIESANQLP--PLQEIDYIEEPFILASI 410
Query: 485 VMPSEYVGPVITLLSERRGEQLEYSFIDSQRVFMKYRLPLREIVIDFYNELKSITSGYAS 544
P+E+VG +++L E+RG Q E ++ RV + Y LPL E+V+DFY+ LKSIT GYAS
Sbjct: 411 HTPNEFVGGILSLCEEKRGVQREIKYLTPTRVMIIYELPLNEVVLDFYDRLKSITKGYAS 470
Query: 545 FDYEDSDYQPSDLVKLDILLNGQAVDAMATIVHNSKSYRVGRELVEKLKKVIDRQMFEII 604
DYE +Y+ SDLV+++IL+NG+AVDA++ I+H K+Y GR+LV K+K++I RQMFE++
Sbjct: 471 LDYEHLNYRRSDLVRMNILINGEAVDALSLIIHRDKAYYRGRDLVSKMKELIPRQMFEVV 530
Query: 605 IQAAIGSKIIARETVTAMRKNVLAKCYGGDITRKKKLLEKQKEGKKRMKRVGSVDVPQEA 664
IQAAIG+K+IARETV A+RK+VLAKCYGGDITRK+KLLEKQKEGKKRMK VG+V++PQEA
Sbjct: 531 IQAAIGAKVIARETVKALRKDVLAKCYGGDITRKRKLLEKQKEGKKRMKNVGNVELPQEA 590
Query: 665 FHELLKV 671
F +LKV
Sbjct: 591 FLAILKV 597
>ref|ZP_00290028.1| COG0481: Membrane GTPase LepA [Magnetococcus sp. MC-1]
Length = 598
Score = 649 bits (1673), Expect = 0.0
Identities = 320/605 (52%), Positives = 442/605 (72%), Gaps = 13/605 (2%)
Query: 67 VRNFSIIAHVDHGKSTLADRLLELTGTIKKGLGQPQYLDKLQVERERGITVKAQTATMFY 126
+RNFSIIAH+DHGKSTLADRL++ TG + + + Q LD + +ERERGIT+KAQ+ + Y
Sbjct: 6 IRNFSIIAHIDHGKSTLADRLIQFTGALSERDMKEQVLDSMDIERERGITIKAQSVRLNY 65
Query: 127 KNIINGDDFKDGKESSNYLLNLIDTPGHVDFSYEVSRSLAACQGVLLVVDAAQGVQAQTV 186
K KDGK Y+LNLIDTPGHVDF+YEVSRSLAAC+G LL+VDAAQGV+AQT+
Sbjct: 66 KA-------KDGK---TYVLNLIDTPGHVDFTYEVSRSLAACEGALLIVDAAQGVEAQTM 115
Query: 187 ANFYLAFESNLAIIPVINKIDQPTADPDRVKGQLKSMFDLDPSDALLTSAKTGVGLEHVL 246
AN YLA E +L IIPV+NKID P+A+P+RV+ Q++ + LD S+A+L SAK+G+G+E +L
Sbjct: 116 ANVYLALEHDLEIIPVLNKIDLPSAEPERVREQIEEVIGLDASEAILASAKSGIGIEEIL 175
Query: 247 PAVIERIPPPPGKSESSLRMLLLDSYFDEYRGVICHVAVVDGALRKGDKISSAATGKSYE 306
A+++R+PPP G ++ + L++DS++D Y GV+ V DG L+ G+KI G Y
Sbjct: 176 EAIVKRVPPPKGDLDAPAKALVVDSWYDNYLGVVSLARVYDGVLKAGEKIRFMGVGMDYP 235
Query: 307 AMDIGIMHPELTPTGILFTGQVGYVITGMRTTKEARIGDTIYHTKSTVDVEPLPGFKAAK 366
++G + P LT L G+VG ++ G++ +AR+GDTI + LPGF+ AK
Sbjct: 236 LDNVGFLTPVLTKVAELKAGEVGCIMAGIKKLSDARVGDTITTVRRPC-ASQLPGFQPAK 294
Query: 367 HMVFSGLFPADGSDFEALSHAIEKLTCNDASVSVTKETSTALGLGFRCGFLGLLHMDVFH 426
MVF+GL+P D +D+E L A+EKL NDAS++ ETS ALG GFRCGFLG+LHM++
Sbjct: 295 SMVFAGLYPVDSADYEDLKDALEKLAINDASLNYEVETSPALGFGFRCGFLGMLHMEIIQ 354
Query: 427 QRLEQEYGAHIISTVPTVPYIYEYSDGSKLEVQNPAALPSNPKQRVVACWEPTVIATIVM 486
+RLE+E+ +++T PTV Y + +DG +++++P LP K+ + EP ++A I++
Sbjct: 355 ERLEREFDLDLVTTAPTVVYRVKQTDGQIMDIRSPGDLPPTTKREYIE--EPYILANIMV 412
Query: 487 PSEYVGPVITLLSERRGEQLEYSFIDSQRVFMKYRLPLREIVIDFYNELKSITSGYASFD 546
P+EY+GPV+ L ++RRG Q + S+I RV ++Y +P+ E+V+DF++ LKS+T GYAS D
Sbjct: 413 PAEYMGPVMQLCTDRRGTQKDMSYISDTRVMVQYEMPMSEVVMDFFDRLKSMTKGYASLD 472
Query: 547 YEDSDYQPSDLVKLDILLNGQAVDAMATIVHNSKSYRVGRELVEKLKKVIDRQMFEIIIQ 606
Y DY+ SDLVKLDIL+NG VDA++ IVH + S GREL +K+K++I RQMF++ +Q
Sbjct: 473 YHLLDYRMSDLVKLDILINGDPVDALSVIVHRNISQYRGRELAKKMKELIHRQMFDVAVQ 532
Query: 607 AAIGSKIIARETVTAMRKNVLAKCYGGDITRKKKLLEKQKEGKKRMKRVGSVDVPQEAFH 666
A IG KIIARETV A+RKNV AKCYGGDITRK+KLLEKQK GKKRMK+VG V++PQEAF
Sbjct: 533 ACIGGKIIARETVKALRKNVTAKCYGGDITRKRKLLEKQKAGKKRMKQVGKVEIPQEAFL 592
Query: 667 ELLKV 671
+LKV
Sbjct: 593 AVLKV 597
>gb|AAL94973.1| GTP-binding protein lepA [Fusobacterium nucleatum subsp. nucleatum
ATCC 25586] gi|19704112|ref|NP_603674.1| GTP-binding
protein lepA [Fusobacterium nucleatum subsp. nucleatum
ATCC 25586] gi|24211906|sp|Q8RFD1|LEPA_FUSNN GTP-binding
protein lepA
Length = 604
Score = 648 bits (1672), Expect = 0.0
Identities = 329/605 (54%), Positives = 437/605 (71%), Gaps = 12/605 (1%)
Query: 68 RNFSIIAHVDHGKSTLADRLLELTGTIKKGLGQPQYLDKLQVERERGITVKAQTATMFYK 127
RNFSIIAH+DHGKST+ADRLLE TGT+ + + Q LD + +ERE+GIT+KAQ T+FYK
Sbjct: 11 RNFSIIAHIDHGKSTIADRLLEYTGTVSERDMKEQILDSMDLEREKGITIKAQAVTLFYK 70
Query: 128 NIINGDDFKDGKESSNYLLNLIDTPGHVDFSYEVSRSLAACQGVLLVVDAAQGVQAQTVA 187
K+G+E Y LNLIDTPGHVDF YEVSRSLAAC+G LLVVDAAQGV+AQT+A
Sbjct: 71 A-------KNGEE---YELNLIDTPGHVDFIYEVSRSLAACEGALLVVDAAQGVEAQTLA 120
Query: 188 NFYLAFESNLAIIPVINKIDQPTADPDRVKGQLKSMFDLDPSDALLTSAKTGVGLEHVLP 247
N YLA E+NL I+P+INKID P A+P++VK +++ + L DA+L SAK G+G+E +L
Sbjct: 121 NVYLAIENNLEILPIINKIDLPAAEPEKVKREIEDIIGLPADDAVLASAKNGIGIEDILE 180
Query: 248 AVIERIPPPPGKSESSLRMLLLDSYFDEYRGVICHVAVVDGALRKGDKISSAATGKSYEA 307
A++ RIP P ++ L+ L+ DSYFD+YRGVI +V V+DG ++KGDKI +T K E
Sbjct: 181 AIVHRIPAPNYDEDAPLKALIFDSYFDDYRGVITYVKVLDGNIKKGDKIKIWSTEKELEV 240
Query: 308 MDIGIMHPELTPTGILFTGQVGYVITGMRTTKEARIGDTIYHTKSTVDVEPLPGFKAAKH 367
++ GI P + T IL TG VGY+ITG++T + R+GDTI K+ + PL GFK A+
Sbjct: 241 LEAGIFSPTMKSTDILSTGSVGYIITGVKTIHDTRVGDTITSVKNPA-LFPLAGFKPAQS 299
Query: 368 MVFSGLFPADGSDFEALSHAIEKLTCNDASVSVTKETSTALGLGFRCGFLGLLHMDVFHQ 427
MVF+G++P D+E L A+EKL NDAS++ ETS ALG GFRCGFLGLLHM++ +
Sbjct: 300 MVFAGVYPLFTDDYEELREALEKLQLNDASLTFVPETSIALGFGFRCGFLGLLHMEIIVE 359
Query: 428 RLEQEYGAHIISTVPTVPYIYEYSDGSKLEVQNPAALPSNPKQRVVACWEPTVIATIVMP 487
RL +EY +IST P+V Y + + + NP P +P + + EP + +++P
Sbjct: 360 RLRREYNIDLISTTPSVEYKVSIDNQEEKVIDNPCEFP-DPGRGKITIQEPYIRGKVIVP 418
Query: 488 SEYVGPVITLLSERRGEQLEYSFIDSQRVFMKYRLPLREIVIDFYNELKSITSGYASFDY 547
EYVG V+ L E+RG + ++D R + Y LPL EIVIDFY++LKS T GYASF+Y
Sbjct: 419 KEYVGNVMELCQEKRGIFISMDYLDETRSMLSYELPLAEIVIDFYDKLKSRTKGYASFEY 478
Query: 548 EDSDYQPSDLVKLDILLNGQAVDAMATIVHNSKSYRVGRELVEKLKKVIDRQMFEIIIQA 607
E S+Y+ S+LVK+DIL++G+ VDA + I HN ++ G+ + +KL +VI RQ FEI IQA
Sbjct: 479 ELSEYKISNLVKVDILVSGKPVDAFSFIAHNDNAFHRGKAICQKLSEVIPRQQFEIPIQA 538
Query: 608 AIGSKIIARETVTAMRKNVLAKCYGGDITRKKKLLEKQKEGKKRMKRVGSVDVPQEAFHE 667
A+GSKIIARET+ A RKNV+AKCYGGDITRKKKLLEKQKEGKKRMK +G+V++PQEAF
Sbjct: 539 ALGSKIIARETIKAYRKNVIAKCYGGDITRKKKLLEKQKEGKKRMKSIGNVEIPQEAFVS 598
Query: 668 LLKVS 672
+LK++
Sbjct: 599 VLKLN 603
>ref|ZP_00330046.1| COG0481: Membrane GTPase LepA [Moorella thermoacetica ATCC 39073]
Length = 602
Score = 647 bits (1670), Expect = 0.0
Identities = 327/604 (54%), Positives = 434/604 (71%), Gaps = 13/604 (2%)
Query: 67 VRNFSIIAHVDHGKSTLADRLLELTGTIKKGLGQPQYLDKLQVERERGITVKAQTATMFY 126
+RNF IIAH+DHGKSTLADRLLE TG + K Q LD + +ERERGIT+K Q + Y
Sbjct: 8 IRNFCIIAHIDHGKSTLADRLLEYTGALSKREMVDQVLDTMDLERERGITIKLQAVRLHY 67
Query: 127 KNIINGDDFKDGKESSNYLLNLIDTPGHVDFSYEVSRSLAACQGVLLVVDAAQGVQAQTV 186
K +DG+E Y+LNLIDTPGHVDF+YEVSRSLAAC+G LLVVDAAQG++AQT+
Sbjct: 68 KA-------RDGQE---YVLNLIDTPGHVDFTYEVSRSLAACEGALLVVDAAQGIEAQTL 117
Query: 187 ANFYLAFESNLAIIPVINKIDQPTADPDRVKGQLKSMFDLDPSDALLTSAKTGVGLEHVL 246
AN YLA E NL IIPVINKID P+A+P+RV+ +++ + LD S+A+L SAKTGVG E +L
Sbjct: 118 ANVYLALEHNLEIIPVINKIDLPSAEPERVRREIEDVIGLDASEAILASAKTGVGTEEIL 177
Query: 247 PAVIERIPPPPGKSESSLRMLLLDSYFDEYRGVICHVAVVDGALRKGDKISSAATGKSYE 306
A+I R+PPP G E+ L+ L+ DS FD YRG I + VV G +RKGD+I ATG +E
Sbjct: 178 EAIIRRVPPPRGDGEAPLQALIFDSIFDSYRGAIPYFRVVQGRVRKGDRIRFMATGAEFE 237
Query: 307 AMDIGIMHPELTPTGILFTGQVGYVITGMRTTKEARIGDTIYHTKSTVDVEPLPGFKAAK 366
++G+ P P L G+VG++ ++ K+ R+GDTI + PLPG++
Sbjct: 238 VNEVGVFTPAPRPVESLAAGEVGFLSASIKNVKDTRVGDTITSAERPAPA-PLPGYRKVM 296
Query: 367 HMVFSGLFPADGSDFEALSHAIEKLTCNDASVSVTKETSTALGLGFRCGFLGLLHMDVFH 426
MV+ GLFP + ++ L A+EKL NDAS++ ETS ALG GFRCGFLGLLHM++
Sbjct: 297 PMVYCGLFPVESERYDDLRDALEKLQLNDASLTFETETSVALGFGFRCGFLGLLHMEIIQ 356
Query: 427 QRLEQEYGAHIISTVPTVPYIYEYSDGSKLEVQNPAALPSNPKQRVVACWEPTVIATIVM 486
+RLE+EYG +I+T P+V Y ++GS + V NP ALP+ + EP V ATI+
Sbjct: 357 ERLEREYGLELITTAPSVVYRVVGTNGSVIMVDNPTALPA--PNLIDHIEEPFVEATIMT 414
Query: 487 PSEYVGPVITLLSERRGEQLEYSFIDSQRVFMKYRLPLREIVIDFYNELKSITSGYASFD 546
P ++VGPV+ L E+RG L ++ +RV +KY LPL EI+ DF+++LKS T GYAS D
Sbjct: 415 PKDFVGPVMELCQEKRGSFLNMDYLSEKRVALKYDLPLAEIIYDFFDQLKSRTRGYASLD 474
Query: 547 YEDSDYQPSDLVKLDILLNGQAVDAMATIVHNSKSYRVGRELVEKLKKVIDRQMFEIIIQ 606
Y Y+PS+LVK+DIL+N + VDA++ I H ++Y+ GR LVE+L+++I RQ+F++ IQ
Sbjct: 475 YSLKGYRPSELVKMDILVNNEVVDALSLITHRDQAYQRGRALVERLRQLIPRQLFDVPIQ 534
Query: 607 AAIGSKIIARETVTAMRKNVLAKCYGGDITRKKKLLEKQKEGKKRMKRVGSVDVPQEAFH 666
AAIGS++IARET+ A+RKNVLAKCYGGD+TRK+KLLEKQKEGKKRMK+VG+VD+PQEAF
Sbjct: 535 AAIGSRVIARETIPALRKNVLAKCYGGDVTRKRKLLEKQKEGKKRMKQVGTVDIPQEAFM 594
Query: 667 ELLK 670
+LK
Sbjct: 595 AVLK 598
>ref|ZP_00144166.1| GTP-binding protein lepA [Fusobacterium nucleatum subsp. vincentii
ATCC 49256] gi|27887139|gb|EAA24245.1| GTP-binding
protein lepA [Fusobacterium nucleatum subsp. vincentii
ATCC 49256]
Length = 604
Score = 643 bits (1658), Expect = 0.0
Identities = 325/605 (53%), Positives = 434/605 (71%), Gaps = 12/605 (1%)
Query: 68 RNFSIIAHVDHGKSTLADRLLELTGTIKKGLGQPQYLDKLQVERERGITVKAQTATMFYK 127
RNFSIIAH+DHGKST+ADRLLE TGT+ + + Q LD + +ERE+GIT+KAQ T+FYK
Sbjct: 11 RNFSIIAHIDHGKSTIADRLLEYTGTVSERDMKEQILDSMDLEREKGITIKAQAVTLFYK 70
Query: 128 NIINGDDFKDGKESSNYLLNLIDTPGHVDFSYEVSRSLAACQGVLLVVDAAQGVQAQTVA 187
K+G+E Y LNLIDTPGHVDF YEVSRSLAAC+G LLVVDAAQGV+AQT+A
Sbjct: 71 A-------KNGQE---YELNLIDTPGHVDFIYEVSRSLAACEGALLVVDAAQGVEAQTLA 120
Query: 188 NFYLAFESNLAIIPVINKIDQPTADPDRVKGQLKSMFDLDPSDALLTSAKTGVGLEHVLP 247
N YLA E+NL I+P+INKID P A+P++VK +++ + L DA+L SAK G+G+E +L
Sbjct: 121 NVYLAIENNLEILPIINKIDLPAAEPEKVKREIEDIIGLPADDAVLASAKNGIGIEDILE 180
Query: 248 AVIERIPPPPGKSESSLRMLLLDSYFDEYRGVICHVAVVDGALRKGDKISSAATGKSYEA 307
A++ +IP P + L+ L+ DS+FD+YRGVI +V V+DG + KGDKI +T K E
Sbjct: 181 AIVHKIPAPNYDENAPLKALIFDSFFDDYRGVITYVKVLDGKIEKGDKIKVWSTEKELEV 240
Query: 308 MDIGIMHPELTPTGILFTGQVGYVITGMRTTKEARIGDTIYHTKSTVDVEPLPGFKAAKH 367
++ GI P + T +L +G VGY+ITG++T + R+GDTI K+ + PL GFK A+
Sbjct: 241 LEAGIFSPTMKSTDVLSSGSVGYIITGVKTIHDTRVGDTITSAKNPA-LFPLAGFKPAQS 299
Query: 368 MVFSGLFPADGSDFEALSHAIEKLTCNDASVSVTKETSTALGLGFRCGFLGLLHMDVFHQ 427
MVF+G++P D+E L A+EKL NDAS++ ETS ALG GFRCGFLGLLHM++ +
Sbjct: 300 MVFAGVYPLFTDDYEELREALEKLQLNDASLTFVPETSIALGFGFRCGFLGLLHMEIIVE 359
Query: 428 RLEQEYGAHIISTVPTVPYIYEYSDGSKLEVQNPAALPSNPKQRVVACWEPTVIATIVMP 487
RL +EY +IST P+V Y + + + NP P +P + + EP + +++P
Sbjct: 360 RLRREYNIDLISTTPSVKYKVSIDNQEEKVIDNPCEFP-DPGRGKITIQEPYIRGKVIVP 418
Query: 488 SEYVGPVITLLSERRGEQLEYSFIDSQRVFMKYRLPLREIVIDFYNELKSITSGYASFDY 547
EYVG V+ L E+RG + ++D R + Y LPL EIVIDFY++LKS T GYASF+Y
Sbjct: 419 KEYVGNVMELCQEKRGIFISMDYLDETRSMLSYELPLAEIVIDFYDKLKSRTKGYASFEY 478
Query: 548 EDSDYQPSDLVKLDILLNGQAVDAMATIVHNSKSYRVGRELVEKLKKVIDRQMFEIIIQA 607
E SDY+ S+LVK+DIL++G+ VDA + I HN ++ G+ + +KL +VI RQ FEI IQA
Sbjct: 479 ELSDYRVSNLVKVDILVSGKPVDAFSFIAHNDNAFHRGKAICQKLSEVIPRQQFEIPIQA 538
Query: 608 AIGSKIIARETVTAMRKNVLAKCYGGDITRKKKLLEKQKEGKKRMKRVGSVDVPQEAFHE 667
+GSKIIARET+ A RKNV+AKCYGGDITRKKKLLEKQKEGKKRMK +G+V++PQEAF
Sbjct: 539 TLGSKIIARETIKAYRKNVIAKCYGGDITRKKKLLEKQKEGKKRMKSIGNVEIPQEAFVS 598
Query: 668 LLKVS 672
+LK++
Sbjct: 599 VLKLN 603
>ref|YP_009924.1| GTP-binding protein LepA [Desulfovibrio vulgaris subsp. vulgaris
str. Hildenborough] gi|46448529|gb|AAS95183.1|
GTP-binding protein LepA [Desulfovibrio vulgaris subsp.
vulgaris str. Hildenborough]
gi|60390041|sp|Q72E76|LEPA_DESVH GTP-binding protein
lepA
Length = 601
Score = 642 bits (1656), Expect = 0.0
Identities = 323/607 (53%), Positives = 432/607 (70%), Gaps = 13/607 (2%)
Query: 65 ELVRNFSIIAHVDHGKSTLADRLLELTGTIKKGLGQPQYLDKLQVERERGITVKAQTATM 124
E +RNFSIIAH+DHGKSTLADR+LE+TG + + QYLD++ +ERERGIT+KAQT +
Sbjct: 5 EHIRNFSIIAHIDHGKSTLADRILEVTGLVSDREKRDQYLDRMDLERERGITIKAQTVRI 64
Query: 125 FYKNIINGDDFKDGKESSNYLLNLIDTPGHVDFSYEVSRSLAACQGVLLVVDAAQGVQAQ 184
+ + K Y+LNLIDTPGHVDF+YEVSRSLAAC G LLVVDA+QGV+AQ
Sbjct: 65 PFTS----------KTGRKYILNLIDTPGHVDFNYEVSRSLAACDGALLVVDASQGVEAQ 114
Query: 185 TVANFYLAFESNLAIIPVINKIDQPTADPDRVKGQLKSMFDLDPSDALLTSAKTGVGLEH 244
T+AN YLA + + IIPV+NKID P++D DRVK +++ LD +DA+ SAKTG+ ++
Sbjct: 115 TLANVYLALDHDHDIIPVLNKIDLPSSDVDRVKAEIEESIGLDCTDAIAVSAKTGMNVDK 174
Query: 245 VLPAVIERIPPPPGKSESSLRMLLLDSYFDEYRGVICHVAVVDGALRKGDKISSAATGKS 304
VL A++ER+P P G + L+ L+ DS++D Y+GV+ V+DG LRKGD++ AT KS
Sbjct: 175 VLEAIVERLPAPEGNLNAPLKALIFDSWYDSYQGVVVLFRVMDGVLRKGDRVRMFATEKS 234
Query: 305 YEAMDIGIMHPELTPTGILFTGQVGYVITGMRTTKEARIGDTIYHTKSTVDVEPLPGFKA 364
YE + +G+ P++ L G+VG++ ++ +A++GDTI HT EP+PGFK
Sbjct: 235 YEVIRLGVFSPDIVDVAELGAGEVGFLCANIKELGDAKVGDTITHTDRPAS-EPVPGFKE 293
Query: 365 AKHMVFSGLFPADGSDFEALSHAIEKLTCNDASVSVTKETSTALGLGFRCGFLGLLHMDV 424
+ MVF GL+P D +++E L ++EKL NDA+ S ETS ALG GFRCGFLGLLHM++
Sbjct: 294 VQPMVFCGLYPTDAAEYEPLKASLEKLQLNDAAFSYEPETSQALGFGFRCGFLGLLHMEI 353
Query: 425 FHQRLEQEYGAHIISTVPTVPYIYEYSDGSKLEVQNPAALPSNPKQRVVACWEPTVIATI 484
+RLE+E+ +I+T P+V Y E DG ++ NP+ LP R+ + +EP V I
Sbjct: 354 IQERLEREFQVDLIATAPSVIYKVETVDGKTQDIDNPSKLPD--PTRITSLYEPYVRMDI 411
Query: 485 VMPSEYVGPVITLLSERRGEQLEYSFIDSQRVFMKYRLPLREIVIDFYNELKSITSGYAS 544
+P+E+VG V+ L E+RG Q +I + RV + Y LP EIV DF++ LKS T GYAS
Sbjct: 412 HVPNEFVGNVLKLCEEKRGIQKNMGYIAANRVVITYELPFAEIVFDFFDRLKSGTKGYAS 471
Query: 545 FDYEDSDYQPSDLVKLDILLNGQAVDAMATIVHNSKSYRVGRELVEKLKKVIDRQMFEII 604
DYE DY+ S LV+LDIL+NG+AVDA+A IVH K+Y GR L KLK+ I RQ+FE+
Sbjct: 472 MDYEPVDYRESSLVRLDILINGEAVDALAVIVHRDKAYHYGRALALKLKRTIPRQLFEVA 531
Query: 605 IQAAIGSKIIARETVTAMRKNVLAKCYGGDITRKKKLLEKQKEGKKRMKRVGSVDVPQEA 664
IQAAIG K+IARET++AMRKNV AKCYGGDITRK+KLLEKQKEGK+RMKR+G+V++PQEA
Sbjct: 532 IQAAIGQKVIARETISAMRKNVTAKCYGGDITRKRKLLEKQKEGKRRMKRMGNVELPQEA 591
Query: 665 FHELLKV 671
F L+V
Sbjct: 592 FLAALQV 598
>ref|ZP_00503533.1| Small GTP-binding protein domain:GTP-binding protein LepA
[Clostridium thermocellum ATCC 27405]
gi|67851650|gb|EAM47213.1| Small GTP-binding protein
domain:GTP-binding protein LepA [Clostridium
thermocellum ATCC 27405]
Length = 603
Score = 640 bits (1651), Expect = 0.0
Identities = 329/606 (54%), Positives = 428/606 (70%), Gaps = 14/606 (2%)
Query: 67 VRNFSIIAHVDHGKSTLADRLLELTGTIKKGLGQPQYLDKLQVERERGITVKAQTATMFY 126
+RNF IIAH+DHGKSTLADRLLE+TG + + + Q LD +++ERERGIT+KAQ M Y
Sbjct: 9 IRNFCIIAHIDHGKSTLADRLLEMTGVLTEREMEDQVLDTMEIERERGITIKAQAVRMVY 68
Query: 127 KNIINGDDFKDGKESSNYLLNLIDTPGHVDFSYEVSRSLAACQGVLLVVDAAQGVQAQTV 186
K KDG+E Y+LNLIDTPGHVDF+YEVSRSLAAC+G +LVVDAAQG++AQT+
Sbjct: 69 KA-------KDGEE---YILNLIDTPGHVDFNYEVSRSLAACEGAILVVDAAQGIEAQTL 118
Query: 187 ANFYLAFESNLAIIPVINKIDQPTADPDRVKGQLKSMFDLDPSDALLTSAKTGVGLEHVL 246
AN YLA + +L I+PVINKID P+A PD VK +++ + LD S+A L SAK G+ +E VL
Sbjct: 119 ANVYLALDHDLEILPVINKIDLPSAQPDVVKKEIEDVIGLDASEAPLISAKNGINIEAVL 178
Query: 247 PAVIERIPPPPGKSESSLRMLLLDSYFDEYRGVICHVAVVDGALRKGDKISSAATGKSYE 306
+V++ +PPP G + LR L+ DSY+D Y+GVI ++ V +G L+ GDK+ T K +
Sbjct: 179 ESVVKNVPPPEGDENAPLRALIFDSYYDSYKGVIVYIRVKEGTLKLGDKVRMMYTNKEFT 238
Query: 307 AMDIGIMHPE-LTPTGILFTGQVGYVITGMRTTKEARIGDTIYHTKSTVDVEPLPGFKAA 365
+IG M P L P L G+VGY ++ K+ R+GDT+ T EPLPG+K
Sbjct: 239 VTEIGYMKPGGLVPGTQLSAGEVGYFAASIKNVKDTRVGDTVT-TADNPAKEPLPGYKKV 297
Query: 366 KHMVFSGLFPADGSDFEALSHAIEKLTCNDASVSVTKETSTALGLGFRCGFLGLLHMDVF 425
MVF G++PADGS + L A+EKL NDAS++ E+S ALG GFRCGFLGLLHM++
Sbjct: 298 NPMVFCGIYPADGSKYGDLRDALEKLQLNDASLTFEPESSVALGFGFRCGFLGLLHMEII 357
Query: 426 HQRLEQEYGAHIISTVPTVPYIYEYSDGSKLEVQNPAALPSNPKQRVVACWEPTVIATIV 485
+RLE+E+ +++T P+V Y ++G L + NP LP P + EP V ATI+
Sbjct: 358 QERLEREFDFDLVTTAPSVIYKVTKTNGETLYIDNPTNLP--PPAEIKTMEEPIVKATIM 415
Query: 486 MPSEYVGPVITLLSERRGEQLEYSFIDSQRVFMKYRLPLREIVIDFYNELKSITSGYASF 545
P+EYVG ++ L ERRG + ++ID RV + Y LPL EI+ DF++ LKS T GYAS
Sbjct: 416 TPTEYVGNIMELAQERRGIYKDMTYIDEGRVMLTYELPLNEIIYDFFDALKSRTKGYASL 475
Query: 546 DYEDSDYQPSDLVKLDILLNGQAVDAMATIVHNSKSYRVGRELVEKLKKVIDRQMFEIII 605
DYE Y+ SDLVKLDI+LNG+ VDA++ IVH K+Y GR + EKLK+ I RQ FE+ I
Sbjct: 476 DYEMLGYRESDLVKLDIMLNGEIVDALSFIVHREKAYARGRRIAEKLKEAIPRQQFEVPI 535
Query: 606 QAAIGSKIIARETVTAMRKNVLAKCYGGDITRKKKLLEKQKEGKKRMKRVGSVDVPQEAF 665
QA IG KIIARETV A RK+VLAKCYGGDITRKKKLLEKQKEGKKRM+++G+V+VPQEAF
Sbjct: 536 QACIGGKIIARETVKAYRKDVLAKCYGGDITRKKKLLEKQKEGKKRMRQIGTVEVPQEAF 595
Query: 666 HELLKV 671
+LK+
Sbjct: 596 MSVLKL 601
>ref|ZP_00020976.2| COG0481: Membrane GTPase LepA [Chloroflexus aurantiacus]
Length = 607
Score = 638 bits (1645), Expect = 0.0
Identities = 317/606 (52%), Positives = 434/606 (71%), Gaps = 13/606 (2%)
Query: 67 VRNFSIIAHVDHGKSTLADRLLELTGTIKKGLGQPQYLDKLQVERERGITVKAQTATMFY 126
+RNFSIIAHVDHGK+TL+DRLLE T TI + + Q LD L +ERE+GIT+K + M Y
Sbjct: 7 IRNFSIIAHVDHGKTTLSDRLLEATRTISEREQREQLLDSLDLEREKGITIKLKPVRMEY 66
Query: 127 KNIINGDDFKDGKESSNYLLNLIDTPGHVDFSYEVSRSLAACQGVLLVVDAAQGVQAQTV 186
+ + Y LNLIDTPGHVDFSYEV+RSLAAC+G LLVVDA+QG++AQT+
Sbjct: 67 R----------AADGQLYQLNLIDTPGHVDFSYEVNRSLAACEGALLVVDASQGIEAQTL 116
Query: 187 ANFYLAFESNLAIIPVINKIDQPTADPDRVKGQLKSMFDLDPSDALLTSAKTGVGLEHVL 246
AN YLA E++L IIPV+NKID P A P+ V +++++ + DA++ SAK G+G+ +L
Sbjct: 117 ANTYLALENDLVIIPVVNKIDLPGAHPEEVAQEIETVIGIPAEDAIMASAKEGIGIAEIL 176
Query: 247 PAVIERIPPPPGKSESSLRMLLLDSYFDEYRGVICHVAVVDGALRKGDKISSAATGKSYE 306
A++ RIPPP G+ + R L+ DS++D Y+GVI +V VVDGA D++ TG E
Sbjct: 177 EAIVRRIPPPRGQRQQPARALIFDSHYDPYKGVIAYVRVVDGAFTSRDRVRFMGTGAQAE 236
Query: 307 AMDIGIMHPELTPTGILFTGQVGYVITGMRTTKEARIGDTIYHTKSTVDVEPLPGFKAAK 366
+++GI P +TP L G+VGY+ TG+++ + ++GDTI + EPLPG++ AK
Sbjct: 237 TLELGIFRPAMTPLNELVAGEVGYIATGLKSVADCQVGDTITLADAPA-AEPLPGYRPAK 295
Query: 367 HMVFSGLFPADGSDFEALSHAIEKLTCNDASVSVTKETSTALGLGFRCGFLGLLHMDVFH 426
MVF+GL+P D + + L A+E+L NDA++S E+S ALG GFRCGFLGLLHM++
Sbjct: 296 PMVFAGLYPIDSNAYPELREALERLKLNDAALSFQPESSAALGFGFRCGFLGLLHMEIVR 355
Query: 427 QRLEQEYGAHIISTVPTVPYIYEYSDGSKLEVQNPAALPSNPKQRVVACWEPTVIATIVM 486
+RLE+EY ++ T P+V Y SDG + V NP+ +P + RV A EP V +++
Sbjct: 356 ERLEREYKLDLLITAPSVEYQVLLSDGEIVTVANPSEMPD--QSRVEAIEEPVVEISVIA 413
Query: 487 PSEYVGPVITLLSERRGEQLEYSFIDSQRVFMKYRLPLREIVIDFYNELKSITSGYASFD 546
P Y+G ++ L++ RRG + ++D RV +KYR+PL EIVIDFY++LKS T GYAS D
Sbjct: 414 PVRYIGTIMELVTTRRGTFISMDYLDPTRVLLKYRMPLAEIVIDFYDQLKSRTQGYASLD 473
Query: 547 YEDSDYQPSDLVKLDILLNGQAVDAMATIVHNSKSYRVGRELVEKLKKVIDRQMFEIIIQ 606
Y + YQ +DLVKLDIL+NGQ VDA++ IVH +Y+ GR+LVE+L+++I RQMFE+ IQ
Sbjct: 474 YHLAGYQEADLVKLDILVNGQPVDALSLIVHRDFAYQRGRDLVERLRQLIPRQMFEVPIQ 533
Query: 607 AAIGSKIIARETVTAMRKNVLAKCYGGDITRKKKLLEKQKEGKKRMKRVGSVDVPQEAFH 666
AAIGSK+IARET+ A+RK+VLAKCYGGD+TRK+KLLEKQKEGKKRMK VGSV++PQEAF
Sbjct: 534 AAIGSKVIARETIRALRKDVLAKCYGGDVTRKRKLLEKQKEGKKRMKMVGSVEIPQEAFM 593
Query: 667 ELLKVS 672
+L ++
Sbjct: 594 AILSLN 599
Database: nr
Posted date: Jul 5, 2005 12:34 AM
Number of letters in database: 863,360,394
Number of sequences in database: 2,540,612
Lambda K H
0.319 0.136 0.383
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 1,094,341,194
Number of Sequences: 2540612
Number of extensions: 46278272
Number of successful extensions: 147478
Number of sequences better than 10.0: 5811
Number of HSP's better than 10.0 without gapping: 2637
Number of HSP's successfully gapped in prelim test: 3178
Number of HSP's that attempted gapping in prelim test: 131864
Number of HSP's gapped (non-prelim): 9493
length of query: 672
length of database: 863,360,394
effective HSP length: 135
effective length of query: 537
effective length of database: 520,377,774
effective search space: 279442864638
effective search space used: 279442864638
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 79 (35.0 bits)
Medicago: description of AC139355.5


