

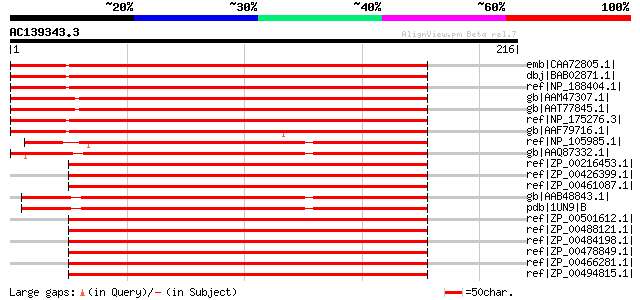
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC139343.3 + phase: 0 /pseudo
(216 letters)
Database: nr
2,540,612 sequences; 863,360,394 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
emb|CAA72805.1| putative 3,4-dihydroxy-2-butanone kinase [Lycope... 294 1e-78
dbj|BAB02871.1| dihydroxyacetone/glycerone kinase-like protein [... 285 8e-76
ref|NP_188404.1| dihydroxyacetone kinase family protein [Arabido... 285 8e-76
gb|AAM47307.1| 3,4-dihydroxy-2-butanone kinase [Oryza sativa (ja... 283 2e-75
gb|AAT77845.1| putative DAK2 domain containing protein [Oryza sa... 283 2e-75
ref|NP_175276.3| dihydroxyacetone kinase family protein [Arabido... 280 2e-74
gb|AAF79716.1| T1N15.4 [Arabidopsis thaliana] 275 6e-73
ref|NP_105985.1| dihydroxyacetone kinase [Mesorhizobium loti MAF... 209 4e-53
gb|AAQ87332.1| Dihydroxyacetone kinase [Rhizobium sp. NGR234] 204 1e-51
ref|ZP_00216453.1| COG2376: Dihydroxyacetone kinase [Burkholderi... 200 3e-50
ref|ZP_00426399.1| Glycerone kinase [Burkholderia vietnamiensis ... 199 3e-50
ref|ZP_00461087.1| Glycerone kinase [Burkholderia cenocepacia HI... 199 6e-50
gb|AAB48843.1| dihydroxyacetone kinase [Citrobacter freundii] gi... 196 3e-49
pdb|1UN9|B Chain B, Crystal Structure Of The Dihydroxyacetone Ki... 196 3e-49
ref|ZP_00501612.1| COG2376: Dihydroxyacetone kinase [Burkholderi... 195 6e-49
ref|ZP_00488121.1| COG2376: Dihydroxyacetone kinase [Burkholderi... 195 6e-49
ref|ZP_00484198.1| COG2376: Dihydroxyacetone kinase [Burkholderi... 195 6e-49
ref|ZP_00478849.1| COG2376: Dihydroxyacetone kinase [Burkholderi... 195 6e-49
ref|ZP_00466281.1| COG2376: Dihydroxyacetone kinase [Burkholderi... 195 6e-49
ref|ZP_00494815.1| COG2376: Dihydroxyacetone kinase [Burkholderi... 194 1e-48
>emb|CAA72805.1| putative 3,4-dihydroxy-2-butanone kinase [Lycopersicon esculentum]
gi|7489046|pir||T06369 probable 3,4-dihydroxy-2-butanone
kinase - tomato gi|7387627|sp|O04059|DHBK_LYCES PUTATIVE
3,4-DIHYDROXY-2-BUTANONE KINASE
Length = 594
Score = 294 bits (753), Expect = 1e-78
Identities = 150/178 (84%), Positives = 158/178 (88%), Gaps = 1/178 (0%)
Query: 1 METYPSLQYLDGYPNVKVVFRKDVYPGPPYDKVAIISGGGSGHEPAHAGFVGEGMLTAAI 60
+E YP LQYLDG+P VKVV R DV G YDKVAIISGGGSGHEPAHAGFVGEGMLTAAI
Sbjct: 24 IENYPGLQYLDGFPEVKVVLRADV-SGAKYDKVAIISGGGSGHEPAHAGFVGEGMLTAAI 82
Query: 61 CGDVFASPTVEAILAGIRAVTGPMGCLLIVKNYTGDRLNFGSAAEQAKSEGYKVESVIVA 120
CGDVFASP V++ILAGIRAVTGPMGCLLIVKNYTGDRLNFG AAEQAKSEGYKVE VIV
Sbjct: 83 CGDVFASPNVDSILAGIRAVTGPMGCLLIVKNYTGDRLNFGLAAEQAKSEGYKVEMVIVG 142
Query: 121 DDCAIPPPLEMVGRRGLAGTILVHKVAGAAAAAGLSLADVAAEARYASENVGTMGVAL 178
DDCA+PPP + GRRGLAGT+LVHKVAGAAAA GL LADVAAEA+ ASE VGTMGVAL
Sbjct: 143 DDCALPPPRGIAGRRGLAGTLLVHKVAGAAAACGLPLADVAAEAKRASEMVGTMGVAL 200
>dbj|BAB02871.1| dihydroxyacetone/glycerone kinase-like protein [Arabidopsis
thaliana]
Length = 618
Score = 285 bits (728), Expect = 8e-76
Identities = 145/178 (81%), Positives = 157/178 (87%), Gaps = 1/178 (0%)
Query: 1 METYPSLQYLDGYPNVKVVFRKDVYPGPPYDKVAIISGGGSGHEPAHAGFVGEGMLTAAI 60
+ETYP LQYLDG+P VKVV R DV YDKVA+ISGGGSGHEPAHAG+VGEGMLTAAI
Sbjct: 24 VETYPGLQYLDGFPEVKVVLRADV-SAAKYDKVAVISGGGSGHEPAHAGYVGEGMLTAAI 82
Query: 61 CGDVFASPTVEAILAGIRAVTGPMGCLLIVKNYTGDRLNFGSAAEQAKSEGYKVESVIVA 120
CGDVFASP V++ILAGIRAVTG GCLLIVKNYTGDRLNFG AAEQAKSEGYKVE+VIV
Sbjct: 83 CGDVFASPPVDSILAGIRAVTGTEGCLLIVKNYTGDRLNFGLAAEQAKSEGYKVETVIVG 142
Query: 121 DDCAIPPPLEMVGRRGLAGTILVHKVAGAAAAAGLSLADVAAEARYASENVGTMGVAL 178
+DCA+PPP + GRRGLAGT+LVHKVAGAAAAAGLSL VAAEA+ ASE VGTMGVAL
Sbjct: 143 EDCALPPPRGIAGRRGLAGTVLVHKVAGAAAAAGLSLEKVAAEAKCASEMVGTMGVAL 200
>ref|NP_188404.1| dihydroxyacetone kinase family protein [Arabidopsis thaliana]
Length = 595
Score = 285 bits (728), Expect = 8e-76
Identities = 145/178 (81%), Positives = 157/178 (87%), Gaps = 1/178 (0%)
Query: 1 METYPSLQYLDGYPNVKVVFRKDVYPGPPYDKVAIISGGGSGHEPAHAGFVGEGMLTAAI 60
+ETYP LQYLDG+P VKVV R DV YDKVA+ISGGGSGHEPAHAG+VGEGMLTAAI
Sbjct: 24 VETYPGLQYLDGFPEVKVVLRADV-SAAKYDKVAVISGGGSGHEPAHAGYVGEGMLTAAI 82
Query: 61 CGDVFASPTVEAILAGIRAVTGPMGCLLIVKNYTGDRLNFGSAAEQAKSEGYKVESVIVA 120
CGDVFASP V++ILAGIRAVTG GCLLIVKNYTGDRLNFG AAEQAKSEGYKVE+VIV
Sbjct: 83 CGDVFASPPVDSILAGIRAVTGTEGCLLIVKNYTGDRLNFGLAAEQAKSEGYKVETVIVG 142
Query: 121 DDCAIPPPLEMVGRRGLAGTILVHKVAGAAAAAGLSLADVAAEARYASENVGTMGVAL 178
+DCA+PPP + GRRGLAGT+LVHKVAGAAAAAGLSL VAAEA+ ASE VGTMGVAL
Sbjct: 143 EDCALPPPRGIAGRRGLAGTVLVHKVAGAAAAAGLSLEKVAAEAKCASEMVGTMGVAL 200
>gb|AAM47307.1| 3,4-dihydroxy-2-butanone kinase [Oryza sativa (japonica
cultivar-group)]
Length = 575
Score = 283 bits (725), Expect = 2e-75
Identities = 143/178 (80%), Positives = 156/178 (87%), Gaps = 1/178 (0%)
Query: 1 METYPSLQYLDGYPNVKVVFRKDVYPGPPYDKVAIISGGGSGHEPAHAGFVGEGMLTAAI 60
+ETYP LQYLDG+P +KVV R DV G YDKVA+ISGGGSGHEP HAGFVG GMLTAA+
Sbjct: 24 VETYPGLQYLDGFPQIKVVLRADVVQGA-YDKVAVISGGGSGHEPTHAGFVGPGMLTAAV 82
Query: 61 CGDVFASPTVEAILAGIRAVTGPMGCLLIVKNYTGDRLNFGSAAEQAKSEGYKVESVIVA 120
GDVF SP V++ILA IRAVTGPMGCLLIVKNYTGDRLNFG AAEQAKSEGYK+E VIV
Sbjct: 83 SGDVFTSPPVDSILAAIRAVTGPMGCLLIVKNYTGDRLNFGLAAEQAKSEGYKMEMVIVG 142
Query: 121 DDCAIPPPLEMVGRRGLAGTILVHKVAGAAAAAGLSLADVAAEARYASENVGTMGVAL 178
DDCA+PPP + GRRGLAGTILVHKVAGAAA AGLSLA+VAAEA++ASE VGTMGVAL
Sbjct: 143 DDCALPPPRGIAGRRGLAGTILVHKVAGAAADAGLSLAEVAAEAKHASEVVGTMGVAL 200
>gb|AAT77845.1| putative DAK2 domain containing protein [Oryza sativa (japonica
cultivar-group)]
Length = 594
Score = 283 bits (725), Expect = 2e-75
Identities = 143/178 (80%), Positives = 156/178 (87%), Gaps = 1/178 (0%)
Query: 1 METYPSLQYLDGYPNVKVVFRKDVYPGPPYDKVAIISGGGSGHEPAHAGFVGEGMLTAAI 60
+ETYP LQYLDG+P +KVV R DV G YDKVA+ISGGGSGHEP HAGFVG GMLTAA+
Sbjct: 24 VETYPGLQYLDGFPQIKVVLRADVVQGA-YDKVAVISGGGSGHEPTHAGFVGPGMLTAAV 82
Query: 61 CGDVFASPTVEAILAGIRAVTGPMGCLLIVKNYTGDRLNFGSAAEQAKSEGYKVESVIVA 120
GDVF SP V++ILA IRAVTGPMGCLLIVKNYTGDRLNFG AAEQAKSEGYK+E VIV
Sbjct: 83 SGDVFTSPPVDSILAAIRAVTGPMGCLLIVKNYTGDRLNFGLAAEQAKSEGYKMEMVIVG 142
Query: 121 DDCAIPPPLEMVGRRGLAGTILVHKVAGAAAAAGLSLADVAAEARYASENVGTMGVAL 178
DDCA+PPP + GRRGLAGTILVHKVAGAAA AGLSLA+VAAEA++ASE VGTMGVAL
Sbjct: 143 DDCALPPPRGIAGRRGLAGTILVHKVAGAAADAGLSLAEVAAEAKHASEVVGTMGVAL 200
>ref|NP_175276.3| dihydroxyacetone kinase family protein [Arabidopsis thaliana]
Length = 593
Score = 280 bits (716), Expect = 2e-74
Identities = 140/178 (78%), Positives = 157/178 (87%), Gaps = 1/178 (0%)
Query: 1 METYPSLQYLDGYPNVKVVFRKDVYPGPPYDKVAIISGGGSGHEPAHAGFVGEGMLTAAI 60
+ETYP LQYLDG P +KVV R DV YDKVA+ISGGGSGHEPA AG+VGEGMLTAAI
Sbjct: 24 VETYPGLQYLDGLPEIKVVLRADV-SAANYDKVAVISGGGSGHEPAQAGYVGEGMLTAAI 82
Query: 61 CGDVFASPTVEAILAGIRAVTGPMGCLLIVKNYTGDRLNFGSAAEQAKSEGYKVESVIVA 120
CGDVFASPTV++ILA IRAVTGP GCLL+V NYTGDRLNFG AAEQAK+EG+ +E+VIV
Sbjct: 83 CGDVFASPTVDSILARIRAVTGPKGCLLVVTNYTGDRLNFGLAAEQAKTEGFDIETVIVG 142
Query: 121 DDCAIPPPLEMVGRRGLAGTILVHKVAGAAAAAGLSLADVAAEARYASENVGTMGVAL 178
DDCA+PPP + GRRGLAGT+LVHKVAGAAAAAGLSLA+VAAEA++ASE VGTMGVAL
Sbjct: 143 DDCALPPPRGVAGRRGLAGTVLVHKVAGAAAAAGLSLAEVAAEAKHASEMVGTMGVAL 200
>gb|AAF79716.1| T1N15.4 [Arabidopsis thaliana]
Length = 625
Score = 275 bits (703), Expect = 6e-73
Identities = 140/180 (77%), Positives = 157/180 (86%), Gaps = 3/180 (1%)
Query: 1 METYPSLQYLDGYPNVKVVFRKDVYPGPPYDKVAIISGGGSGHEPAHAGFVGEGMLTAAI 60
+ETYP LQYLDG P +KVV R DV YDKVA+ISGGGSGHEPA AG+VGEGMLTAAI
Sbjct: 24 VETYPGLQYLDGLPEIKVVLRADV-SAANYDKVAVISGGGSGHEPAQAGYVGEGMLTAAI 82
Query: 61 CGDVFASPTVEAILAGIRAVTGPMGCLLIVKNYTGDRLNFGSAAEQAKSEGYKVE--SVI 118
CGDVFASPTV++ILA IRAVTGP GCLL+V NYTGDRLNFG AAEQAK+EG+ +E +VI
Sbjct: 83 CGDVFASPTVDSILARIRAVTGPKGCLLVVTNYTGDRLNFGLAAEQAKTEGFDIEVTTVI 142
Query: 119 VADDCAIPPPLEMVGRRGLAGTILVHKVAGAAAAAGLSLADVAAEARYASENVGTMGVAL 178
V DDCA+PPP + GRRGLAGT+LVHKVAGAAAAAGLSLA+VAAEA++ASE VGTMGVAL
Sbjct: 143 VGDDCALPPPRGVAGRRGLAGTVLVHKVAGAAAAAGLSLAEVAAEAKHASEMVGTMGVAL 202
>ref|NP_105985.1| dihydroxyacetone kinase [Mesorhizobium loti MAFF303099]
gi|14025170|dbj|BAB51771.1| dihydroxyacetone kinase
[Mesorhizobium loti MAFF303099]
Length = 547
Score = 209 bits (532), Expect = 4e-53
Identities = 105/174 (60%), Positives = 141/174 (80%), Gaps = 11/174 (6%)
Query: 7 LQYLDGYPNVKVVFRKDVYPGPPYDK--VAIISGGGSGHEPAHAGFVGEGMLTAAICGDV 64
L LDGYP +KVV R D +DK V+++SGGG+GHEP+HAGFVG+GMLTAA+ G++
Sbjct: 28 LARLDGYPEIKVVLRAD------WDKSRVSVVSGGGAGHEPSHAGFVGKGMLTAAVSGEI 81
Query: 65 FASPTVEAILAGIRAVTGPMGCLLIVKNYTGDRLNFGSAAEQAKSEGYKVESVIVADDCA 124
FASP+VEA+LA IRAVTGP GCLLIVKNYTGDRLNFG AAE+A++EG++VE VIVADD A
Sbjct: 82 FASPSVEAVLAAIRAVTGPAGCLLIVKNYTGDRLNFGLAAEKARAEGFRVEMVIVADDIA 141
Query: 125 IPPPLEMVGRRGLAGTILVHKVAGAAAAAGLSLADVAAEARYASENVGTMGVAL 178
+P ++ RG+AGT+ VHK++G + AG +LAD+AA AR A++++ ++G++L
Sbjct: 142 LP---DIAQPRGVAGTLFVHKISGHLSEAGHNLADIAAAARAAAKDIVSLGMSL 192
>gb|AAQ87332.1| Dihydroxyacetone kinase [Rhizobium sp. NGR234]
Length = 547
Score = 204 bits (519), Expect = 1e-51
Identities = 108/180 (60%), Positives = 136/180 (75%), Gaps = 9/180 (5%)
Query: 1 METYP--SLQYLDGYPNVKVVFRKDVYPGPPYDKVAIISGGGSGHEPAHAGFVGEGMLTA 58
++T P SL LD YP++KVV R D KVA+ISGGG+GHEP+HAGFVG GMLTA
Sbjct: 23 LQTAPAGSLARLDTYPDIKVVLRGDWDKA----KVAVISGGGAGHEPSHAGFVGRGMLTA 78
Query: 59 AICGDVFASPTVEAILAGIRAVTGPMGCLLIVKNYTGDRLNFGSAAEQAKSEGYKVESVI 118
A+ G++FASP+VEA+L IRA TGP GCLLIVKNYTGDRLNFG AAE+A+ EG++VE VI
Sbjct: 79 AVSGEIFASPSVEAVLTAIRAATGPKGCLLIVKNYTGDRLNFGLAAEKARVEGFRVEMVI 138
Query: 119 VADDCAIPPPLEMVGRRGLAGTILVHKVAGAAAAAGLSLADVAAEARYASENVGTMGVAL 178
VADD A+P ++V RG+AGT+ VHK+AG A G L VAA AR A+ ++ ++GV+L
Sbjct: 139 VADDIALP---DIVQPRGVAGTLFVHKIAGHLAECGAGLETVAAAARSAARDIVSLGVSL 195
>ref|ZP_00216453.1| COG2376: Dihydroxyacetone kinase [Burkholderia cepacia R18194]
Length = 552
Score = 200 bits (508), Expect = 3e-50
Identities = 102/153 (66%), Positives = 122/153 (79%)
Query: 26 PGPPYDKVAIISGGGSGHEPAHAGFVGEGMLTAAICGDVFASPTVEAILAGIRAVTGPMG 85
P P VAI+SGGGSGHEPAH G+VGEGML+AA+CG+VF SP+ +A+LA IRA GP G
Sbjct: 27 PEPAQRPVAILSGGGSGHEPAHGGYVGEGMLSAAVCGEVFTSPSTDAVLAAIRASAGPNG 86
Query: 86 CLLIVKNYTGDRLNFGSAAEQAKSEGYKVESVIVADDCAIPPPLEMVGRRGLAGTILVHK 145
LLIVKNYTGDRLNFG AAE A++EG VE+VIVADD ++ +E RRG+AGT+L+HK
Sbjct: 87 ALLIVKNYTGDRLNFGLAAELARAEGIPVETVIVADDVSLRGRVERGQRRGIAGTVLIHK 146
Query: 146 VAGAAAAAGLSLADVAAEARYASENVGTMGVAL 178
+AGAAAA GL LA VAA AR A+ +GTMGVAL
Sbjct: 147 LAGAAAARGLPLARVAAIARDAAAELGTMGVAL 179
>ref|ZP_00426399.1| Glycerone kinase [Burkholderia vietnamiensis G4]
gi|67530134|gb|EAM26980.1| Glycerone kinase
[Burkholderia vietnamiensis G4]
Length = 569
Score = 199 bits (507), Expect = 3e-50
Identities = 100/153 (65%), Positives = 121/153 (78%)
Query: 26 PGPPYDKVAIISGGGSGHEPAHAGFVGEGMLTAAICGDVFASPTVEAILAGIRAVTGPMG 85
P P VA++SGGGSGHEPAH G+VGEGML+AA+CG+VF SP+ +A+LA IRA GP G
Sbjct: 41 PEPSRRPVAVLSGGGSGHEPAHGGYVGEGMLSAAVCGEVFTSPSTDAVLAAIRASAGPNG 100
Query: 86 CLLIVKNYTGDRLNFGSAAEQAKSEGYKVESVIVADDCAIPPPLEMVGRRGLAGTILVHK 145
LL+VKNYTGDRLNFG AAE A++EG VE VIVADD ++ +E RRG+AGT+L+HK
Sbjct: 101 ALLVVKNYTGDRLNFGLAAELARAEGIPVEMVIVADDVSLRGVVERGQRRGIAGTVLIHK 160
Query: 146 VAGAAAAAGLSLADVAAEARYASENVGTMGVAL 178
+AGAAAA GL LA VAA AR A+ +GTMGVAL
Sbjct: 161 LAGAAAARGLPLARVAAVAREAAAEIGTMGVAL 193
>ref|ZP_00461087.1| Glycerone kinase [Burkholderia cenocepacia HI2424]
gi|67656658|ref|ZP_00454040.1| Glycerone kinase
[Burkholderia cenocepacia AU 1054]
gi|67102577|gb|EAM19708.1| Glycerone kinase
[Burkholderia cenocepacia HI2424]
gi|67095843|gb|EAM13370.1| Glycerone kinase
[Burkholderia cenocepacia AU 1054]
Length = 626
Score = 199 bits (505), Expect = 6e-50
Identities = 101/153 (66%), Positives = 122/153 (79%)
Query: 26 PGPPYDKVAIISGGGSGHEPAHAGFVGEGMLTAAICGDVFASPTVEAILAGIRAVTGPMG 85
P P VAI+SGGGSGHEPAH G+VGEGML+AA+CG+VF SP+ +A+LA IRA GP G
Sbjct: 98 PEPSQRPVAILSGGGSGHEPAHGGYVGEGMLSAAVCGEVFTSPSTDAVLAAIRASAGPNG 157
Query: 86 CLLIVKNYTGDRLNFGSAAEQAKSEGYKVESVIVADDCAIPPPLEMVGRRGLAGTILVHK 145
LLIVKNYTGDRLNFG AAE A++EG VE+VIVADD ++ +E RRG+AGT+L+HK
Sbjct: 158 ALLIVKNYTGDRLNFGLAAELARAEGIPVETVIVADDVSLRGRVERGQRRGIAGTVLIHK 217
Query: 146 VAGAAAAAGLSLADVAAEARYASENVGTMGVAL 178
+AGAAAA GL LA VA+ AR A+ +GTMGVAL
Sbjct: 218 LAGAAAARGLPLARVASIARDAAAELGTMGVAL 250
>gb|AAB48843.1| dihydroxyacetone kinase [Citrobacter freundii]
gi|38493068|pdb|1UN8|B Chain B, Crystal Structure Of The
Dihydroxyacetone Kinase Of C. Freundii (Native Form)
gi|38493067|pdb|1UN8|A Chain A, Crystal Structure Of The
Dihydroxyacetone Kinase Of C. Freundii (Native Form)
gi|1169288|sp|P45510|DAK_CITFR Dihydroxyacetone kinase
(Glycerone kinase) (DHA kinase)
Length = 552
Score = 196 bits (499), Expect = 3e-49
Identities = 101/173 (58%), Positives = 130/173 (74%), Gaps = 7/173 (4%)
Query: 6 SLQYLDGYPNVKVVFRKDVYPGPPYDKVAIISGGGSGHEPAHAGFVGEGMLTAAICGDVF 65
+L L+ P +++V R+D+ + VA+ISGGGSGHEPAH GF+G+GMLTAA+CGDVF
Sbjct: 28 NLARLESDPAIRIVVRRDLNK----NNVAVISGGGSGHEPAHVGFIGKGMLTAAVCGDVF 83
Query: 66 ASPTVEAILAGIRAVTGPMGCLLIVKNYTGDRLNFGSAAEQAKSEGYKVESVIVADDCAI 125
ASP+V+A+L I+AVTG GCLLIVKNYTGDRLNFG AAE+A+ GY VE +IV DD ++
Sbjct: 84 ASPSVDAVLTAIQAVTGEAGCLLIVKNYTGDRLNFGLAAEKARRLGYNVEMLIVGDDISL 143
Query: 126 PPPLEMVGRRGLAGTILVHKVAGAAAAAGLSLADVAAEARYASENVGTMGVAL 178
P + RG+AGTILVHK+AG A G +LA V EA+YA+ N ++GVAL
Sbjct: 144 P---DNKHPRGIAGTILVHKIAGYFAERGYNLATVLREAQYAASNTFSLGVAL 193
>pdb|1UN9|B Chain B, Crystal Structure Of The Dihydroxyacetone Kinase From C.
Freundii In Complex With Amp-Pnp And Mg2+
gi|38493069|pdb|1UN9|A Chain A, Crystal Structure Of The
Dihydroxyacetone Kinase From C. Freundii In Complex With
Amp-Pnp And Mg2+
Length = 552
Score = 196 bits (499), Expect = 3e-49
Identities = 101/173 (58%), Positives = 130/173 (74%), Gaps = 7/173 (4%)
Query: 6 SLQYLDGYPNVKVVFRKDVYPGPPYDKVAIISGGGSGHEPAHAGFVGEGMLTAAICGDVF 65
+L L+ P +++V R+D+ + VA+ISGGGSGHEPAH GF+G+GMLTAA+CGDVF
Sbjct: 28 NLARLESDPAIRIVVRRDLNK----NNVAVISGGGSGHEPAHVGFIGKGMLTAAVCGDVF 83
Query: 66 ASPTVEAILAGIRAVTGPMGCLLIVKNYTGDRLNFGSAAEQAKSEGYKVESVIVADDCAI 125
ASP+V+A+L I+AVTG GCLLIVKNYTGDRLNFG AAE+A+ GY VE +IV DD ++
Sbjct: 84 ASPSVDAVLTAIQAVTGEAGCLLIVKNYTGDRLNFGLAAEKARRLGYNVEMLIVGDDISL 143
Query: 126 PPPLEMVGRRGLAGTILVHKVAGAAAAAGLSLADVAAEARYASENVGTMGVAL 178
P + RG+AGTILVHK+AG A G +LA V EA+YA+ N ++GVAL
Sbjct: 144 P---DNKHPRGIAGTILVHKIAGYFAERGYNLATVLREAQYAASNTFSLGVAL 193
>ref|ZP_00501612.1| COG2376: Dihydroxyacetone kinase [Burkholderia pseudomallei S13]
Length = 570
Score = 195 bits (496), Expect = 6e-49
Identities = 99/153 (64%), Positives = 118/153 (76%)
Query: 26 PGPPYDKVAIISGGGSGHEPAHAGFVGEGMLTAAICGDVFASPTVEAILAGIRAVTGPMG 85
P P VA+ISGGGSGHEPAH G+VG GML+AA+CG+VF SP+ +A+LA IRA GP G
Sbjct: 41 PEPAKRAVAVISGGGSGHEPAHGGYVGAGMLSAAVCGEVFTSPSADAVLAAIRATAGPHG 100
Query: 86 CLLIVKNYTGDRLNFGSAAEQAKSEGYKVESVIVADDCAIPPPLEMVGRRGLAGTILVHK 145
LLIVKNYTGDRLNFG AAE A++EG VE V+VADD ++ E RRG+AGT+LVHK
Sbjct: 101 ALLIVKNYTGDRLNFGLAAELARAEGIPVEIVVVADDVSLRALTERGRRRGIAGTVLVHK 160
Query: 146 VAGAAAAAGLSLADVAAEARYASENVGTMGVAL 178
+AGAAA GL+LA VAA A A+ +GTMGVAL
Sbjct: 161 LAGAAAERGLTLAQVAAAANAAAARLGTMGVAL 193
>ref|ZP_00488121.1| COG2376: Dihydroxyacetone kinase [Burkholderia pseudomallei 668]
Length = 570
Score = 195 bits (496), Expect = 6e-49
Identities = 99/153 (64%), Positives = 118/153 (76%)
Query: 26 PGPPYDKVAIISGGGSGHEPAHAGFVGEGMLTAAICGDVFASPTVEAILAGIRAVTGPMG 85
P P VA+ISGGGSGHEPAH G+VG GML+AA+CG+VF SP+ +A+LA IRA GP G
Sbjct: 41 PEPAKRAVAVISGGGSGHEPAHGGYVGAGMLSAAVCGEVFTSPSADAVLAAIRATAGPHG 100
Query: 86 CLLIVKNYTGDRLNFGSAAEQAKSEGYKVESVIVADDCAIPPPLEMVGRRGLAGTILVHK 145
LLIVKNYTGDRLNFG AAE A++EG VE V+VADD ++ E RRG+AGT+LVHK
Sbjct: 101 ALLIVKNYTGDRLNFGLAAELARAEGIPVEIVVVADDVSLRALTERGRRRGIAGTVLVHK 160
Query: 146 VAGAAAAAGLSLADVAAEARYASENVGTMGVAL 178
+AGAAA GL+LA VAA A A+ +GTMGVAL
Sbjct: 161 LAGAAAERGLTLAQVAAAANAAAARLGTMGVAL 193
>ref|ZP_00484198.1| COG2376: Dihydroxyacetone kinase [Burkholderia pseudomallei 1710b]
Length = 570
Score = 195 bits (496), Expect = 6e-49
Identities = 99/153 (64%), Positives = 118/153 (76%)
Query: 26 PGPPYDKVAIISGGGSGHEPAHAGFVGEGMLTAAICGDVFASPTVEAILAGIRAVTGPMG 85
P P VA+ISGGGSGHEPAH G+VG GML+AA+CG+VF SP+ +A+LA IRA GP G
Sbjct: 41 PEPAKRAVAVISGGGSGHEPAHGGYVGAGMLSAAVCGEVFTSPSADAVLAAIRATAGPHG 100
Query: 86 CLLIVKNYTGDRLNFGSAAEQAKSEGYKVESVIVADDCAIPPPLEMVGRRGLAGTILVHK 145
LLIVKNYTGDRLNFG AAE A++EG VE V+VADD ++ E RRG+AGT+LVHK
Sbjct: 101 ALLIVKNYTGDRLNFGLAAELARAEGIPVEIVVVADDVSLRALTERGRRRGIAGTVLVHK 160
Query: 146 VAGAAAAAGLSLADVAAEARYASENVGTMGVAL 178
+AGAAA GL+LA VAA A A+ +GTMGVAL
Sbjct: 161 LAGAAAERGLTLAQVAAAANTAAARLGTMGVAL 193
>ref|ZP_00478849.1| COG2376: Dihydroxyacetone kinase [Burkholderia pseudomallei 1710a]
Length = 498
Score = 195 bits (496), Expect = 6e-49
Identities = 99/153 (64%), Positives = 118/153 (76%)
Query: 26 PGPPYDKVAIISGGGSGHEPAHAGFVGEGMLTAAICGDVFASPTVEAILAGIRAVTGPMG 85
P P VA+ISGGGSGHEPAH G+VG GML+AA+CG+VF SP+ +A+LA IRA GP G
Sbjct: 41 PEPAKRAVAVISGGGSGHEPAHGGYVGAGMLSAAVCGEVFTSPSADAVLAAIRATAGPHG 100
Query: 86 CLLIVKNYTGDRLNFGSAAEQAKSEGYKVESVIVADDCAIPPPLEMVGRRGLAGTILVHK 145
LLIVKNYTGDRLNFG AAE A++EG VE V+VADD ++ E RRG+AGT+LVHK
Sbjct: 101 ALLIVKNYTGDRLNFGLAAELARAEGIPVEIVVVADDVSLRALTERGRRRGIAGTVLVHK 160
Query: 146 VAGAAAAAGLSLADVAAEARYASENVGTMGVAL 178
+AGAAA GL+LA VAA A A+ +GTMGVAL
Sbjct: 161 LAGAAAERGLTLAQVAAAANTAAARLGTMGVAL 193
>ref|ZP_00466281.1| COG2376: Dihydroxyacetone kinase [Burkholderia pseudomallei 1655]
Length = 525
Score = 195 bits (496), Expect = 6e-49
Identities = 99/153 (64%), Positives = 118/153 (76%)
Query: 26 PGPPYDKVAIISGGGSGHEPAHAGFVGEGMLTAAICGDVFASPTVEAILAGIRAVTGPMG 85
P P VA+ISGGGSGHEPAH G+VG GML+AA+CG+VF SP+ +A+LA IRA GP G
Sbjct: 41 PEPAKRAVAVISGGGSGHEPAHGGYVGAGMLSAAVCGEVFTSPSADAVLAAIRATAGPHG 100
Query: 86 CLLIVKNYTGDRLNFGSAAEQAKSEGYKVESVIVADDCAIPPPLEMVGRRGLAGTILVHK 145
LLIVKNYTGDRLNFG AAE A++EG VE V+VADD ++ E RRG+AGT+LVHK
Sbjct: 101 ALLIVKNYTGDRLNFGLAAELARAEGIPVEIVVVADDVSLRALTERGRRRGIAGTVLVHK 160
Query: 146 VAGAAAAAGLSLADVAAEARYASENVGTMGVAL 178
+AGAAA GL+LA VAA A A+ +GTMGVAL
Sbjct: 161 LAGAAAERGLTLAQVAAAANAAAARLGTMGVAL 193
>ref|ZP_00494815.1| COG2376: Dihydroxyacetone kinase [Burkholderia pseudomallei
Pasteur]
Length = 570
Score = 194 bits (493), Expect = 1e-48
Identities = 98/153 (64%), Positives = 118/153 (77%)
Query: 26 PGPPYDKVAIISGGGSGHEPAHAGFVGEGMLTAAICGDVFASPTVEAILAGIRAVTGPMG 85
P P VA+ISGGGSGHEPAH G+VG GML+AA+CG+VF SP+ +A+LA IRA GP G
Sbjct: 41 PEPAKRAVAVISGGGSGHEPAHGGYVGAGMLSAAVCGEVFTSPSADAVLAAIRATAGPHG 100
Query: 86 CLLIVKNYTGDRLNFGSAAEQAKSEGYKVESVIVADDCAIPPPLEMVGRRGLAGTILVHK 145
LLIVKNYTGDRLNFG AAE A+++G VE V+VADD ++ E RRG+AGT+LVHK
Sbjct: 101 ALLIVKNYTGDRLNFGLAAELARADGIPVEIVVVADDVSLRALTERGRRRGIAGTVLVHK 160
Query: 146 VAGAAAAAGLSLADVAAEARYASENVGTMGVAL 178
+AGAAA GL+LA VAA A A+ +GTMGVAL
Sbjct: 161 LAGAAAERGLTLAQVAAAANAAAARLGTMGVAL 193
Database: nr
Posted date: Jul 5, 2005 12:34 AM
Number of letters in database: 863,360,394
Number of sequences in database: 2,540,612
Lambda K H
0.317 0.137 0.398
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 319,021,281
Number of Sequences: 2540612
Number of extensions: 12962801
Number of successful extensions: 37583
Number of sequences better than 10.0: 221
Number of HSP's better than 10.0 without gapping: 210
Number of HSP's successfully gapped in prelim test: 12
Number of HSP's that attempted gapping in prelim test: 37247
Number of HSP's gapped (non-prelim): 225
length of query: 216
length of database: 863,360,394
effective HSP length: 123
effective length of query: 93
effective length of database: 550,865,118
effective search space: 51230455974
effective search space used: 51230455974
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.6 bits)
S2: 72 (32.3 bits)
Medicago: description of AC139343.3


