

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC138464.6 + phase: 0
(250 letters)
Database: nr
2,540,612 sequences; 863,360,394 total letters
Searching..................................................done
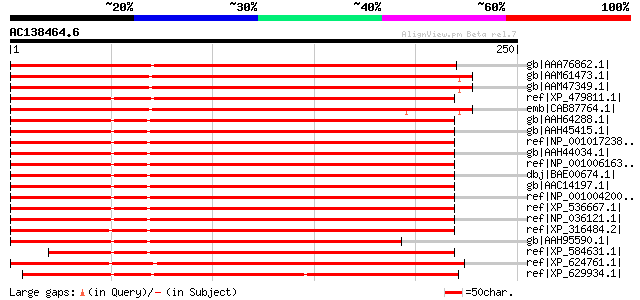
Score E
Sequences producing significant alignments: (bits) Value
gb|AAA76862.1| unknown protein 329 4e-89
gb|AAM61473.1| unknown [Arabidopsis thaliana] gi|18056657|gb|AAL... 292 5e-78
gb|AAM47349.1| AT5g14250/F18O22_40 [Arabidopsis thaliana] gi|169... 292 5e-78
ref|XP_479811.1| putative COP9 complex subunit 3, FUS11 [Oryza s... 288 1e-76
emb|CAB87764.1| putative protein [Arabidopsis thaliana] gi|11290... 276 3e-73
gb|AAH64288.1| COP9 constitutive photomorphogenic homolog subuni... 193 3e-48
gb|AAH45415.1| COP9 constitutive photomorphogenic homolog subuni... 193 4e-48
ref|NP_001017238.1| hypothetical protein LOC549992 [Xenopus trop... 191 2e-47
gb|AAH44034.1| Cops3-prov protein [Xenopus laevis] gi|55976497|s... 189 5e-47
ref|NP_001006163.1| similar to COP9 constitutive photomorphogeni... 188 1e-46
dbj|BAE00674.1| unnamed protein product [Macaca fascicularis] gi... 187 3e-46
gb|AAC14197.1| JAB1-containing signalosome subunit 3 [Homo sapiens] 187 3e-46
ref|NP_001004200.1| COP9 constitutive photomorphogenic homolog s... 187 3e-46
ref|XP_536667.1| PREDICTED: similar to COP9 constitutive photomo... 187 3e-46
ref|NP_036121.1| COP9 (constitutive photomorphogenic), subunit 3... 185 1e-45
ref|XP_316484.2| ENSANGP00000004286 [Anopheles gambiae str. PEST... 180 3e-44
gb|AAH95590.1| Unknown (protein for MGC:111903) [Danio rerio] 180 3e-44
ref|XP_584631.1| PREDICTED: similar to COP9 signalosome complex ... 174 3e-42
ref|XP_624761.1| PREDICTED: similar to COP9 constitutive photomo... 173 3e-42
ref|XP_629934.1| hypothetical protein DDB0219696 [Dictyostelium ... 169 7e-41
>gb|AAA76862.1| unknown protein
Length = 250
Score = 329 bits (844), Expect = 4e-89
Identities = 164/220 (74%), Positives = 190/220 (85%), Gaps = 1/220 (0%)
Query: 1 MICIGQKRFQKALDLLHNVVTAPMTMMNAIVIEAYKKYILVSLIRHGQFSTSLPKYASPL 60
M+CIGQK+F+KAL+LLHNVVTAPM+ +NAI +EAYKKYILVSLI GQFSTS PKY S +
Sbjct: 1 MVCIGQKQFRKALELLHNVVTAPMSTLNAIAVEAYKKYILVSLIHLGQFSTSFPKYTSSV 60
Query: 61 AQRNLKLFCQNPCIELAQTYNNGKVAEVEAFVNANAGRFEADNNLGLAKQVVSSMYKRNI 120
AQRNLK F Q P +EL+ +Y G+++E+E FV N +FE+DNNLGL QVVSSMYKRNI
Sbjct: 61 AQRNLKNFSQ-PYLELSNSYGTGRISELETFVQTNKEKFESDNNLGLVMQVVSSMYKRNI 119
Query: 121 QRLTQTYLTLSLEDIANTVHLNSAKEAEMHVLQMIQDGEIYATINQKDGMVRFLEDPEQY 180
QRLTQTYLTLSL+DIANTV L K+AEMHVLQMI+DGEIYATINQKDGMVRFLEDPEQY
Sbjct: 120 QRLTQTYLTLSLQDIANTVQLRGPKQAEMHVLQMIEDGEIYATINQKDGMVRFLEDPEQY 179
Query: 181 KTCEMIEHIDSSIQRIMALSKKLTATDEQISCDQLYLSKV 220
KTC MIEHIDSSI+R+MA+SKKLT+ DE +SCD +YL KV
Sbjct: 180 KTCGMIEHIDSSIKRLMAVSKKLTSMDELMSCDPMYLGKV 219
>gb|AAM61473.1| unknown [Arabidopsis thaliana] gi|18056657|gb|AAL58102.1| CSN
complex subunit 3 [Arabidopsis thaliana]
gi|18417330|ref|NP_568296.1| COP9 signalosome complex
subunit 3 / CSN complex subunit 3 (CSN3) / FUSCA protein
(FUS11) [Arabidopsis thaliana]
gi|14388969|gb|AAK61872.1| COP9 signalosome subunit 3
[Arabidopsis thaliana] gi|14388971|gb|AAK61873.1| COP9
signalosome subunit 3 [Arabidopsis thaliana]
gi|55976552|sp|Q8W575|CSN3_ARATH COP9 signalosome
complex subunit 3 (Signalosome subunit 3) (FUSCA protein
11) (FUSCA11)
Length = 429
Score = 292 bits (748), Expect = 5e-78
Identities = 156/230 (67%), Positives = 178/230 (76%), Gaps = 3/230 (1%)
Query: 1 MICIGQKRFQKALDLLHNVVTAPMTMMNAIVIEAYKKYILVSLIRHGQFSTSLPKYASPL 60
MICIG KRFQKAL+LL+NVVTAPM +NAI +EAYKKYILVSLI +GQF+ +LPK AS
Sbjct: 189 MICIGLKRFQKALELLYNVVTAPMHQVNAIALEAYKKYILVSLIHNGQFTNTLPKCASTA 248
Query: 61 AQRNLKLFCQNPCIELAQTYNNGKVAEVEAFVNANAGRFEADNNLGLAKQVVSSMYKRNI 120
AQR+ K + P IEL YN+GK+ E+EA V A FE D NLGL KQ VSS+YKRNI
Sbjct: 249 AQRSFKNYT-GPYIELGNCYNDGKIGELEALVVARNAEFEEDKNLGLVKQAVSSLYKRNI 307
Query: 121 QRLTQTYLTLSLEDIANTVHLNSAKEAEMHVLQMIQDGEIYATINQKDGMVRFLEDPEQY 180
RLTQ YLTLSL+DIAN V L +AKEAEMHVLQMIQDG+I+A INQKDGMVRFLEDPEQY
Sbjct: 308 LRLTQKYLTLSLQDIANMVQLGNAKEAEMHVLQMIQDGQIHALINQKDGMVRFLEDPEQY 367
Query: 181 KTCEMIEHIDSSIQRIMALSKKLTATDEQISCDQLYLSKV--SVQSFSFG 228
K+ EMIE +DS IQR + LSK L A DE +SCD LYL KV Q + FG
Sbjct: 368 KSSEMIEIMDSVIQRTIGLSKNLLAMDESLSCDPLYLGKVGRERQRYDFG 417
>gb|AAM47349.1| AT5g14250/F18O22_40 [Arabidopsis thaliana]
gi|16930463|gb|AAL31917.1| AT5g14250/F18O22_40
[Arabidopsis thaliana]
Length = 429
Score = 292 bits (748), Expect = 5e-78
Identities = 156/230 (67%), Positives = 178/230 (76%), Gaps = 3/230 (1%)
Query: 1 MICIGQKRFQKALDLLHNVVTAPMTMMNAIVIEAYKKYILVSLIRHGQFSTSLPKYASPL 60
MICIG KRFQKAL+LL+NVVTAPM +NAI +EAYKKYILVSLI +GQF+ +LPK AS
Sbjct: 189 MICIGLKRFQKALELLYNVVTAPMHQVNAIALEAYKKYILVSLIHNGQFTNTLPKCASTA 248
Query: 61 AQRNLKLFCQNPCIELAQTYNNGKVAEVEAFVNANAGRFEADNNLGLAKQVVSSMYKRNI 120
AQR+ K + P IEL YN+GK+ E+EA V A FE D NLGL KQ VSS+YKRNI
Sbjct: 249 AQRSFKNYT-GPYIELGNCYNDGKIGELEALVVARNAEFEEDKNLGLVKQAVSSLYKRNI 307
Query: 121 QRLTQTYLTLSLEDIANTVHLNSAKEAEMHVLQMIQDGEIYATINQKDGMVRFLEDPEQY 180
RLTQ YLTLSL+DIAN V L +AKEAEMHVLQMIQDG+I+A INQKDGMVRFLEDPEQY
Sbjct: 308 LRLTQKYLTLSLQDIANMVQLGNAKEAEMHVLQMIQDGQIHALINQKDGMVRFLEDPEQY 367
Query: 181 KTCEMIEHIDSSIQRIMALSKKLTATDEQISCDQLYLSKV--SVQSFSFG 228
K+ EMIE +DS IQR + LSK L A DE +SCD LYL KV Q + FG
Sbjct: 368 KSSEMIEIMDSVIQRTIGLSKNLLAMDESLSCDPLYLGKVGRERQRYDFG 417
>ref|XP_479811.1| putative COP9 complex subunit 3, FUS11 [Oryza sativa (japonica
cultivar-group)] gi|42407907|dbj|BAD09047.1| putative
COP9 complex subunit 3, FUS11 [Oryza sativa (japonica
cultivar-group)]
Length = 426
Score = 288 bits (736), Expect = 1e-76
Identities = 148/219 (67%), Positives = 177/219 (80%), Gaps = 2/219 (0%)
Query: 1 MICIGQKRFQKALDLLHNVVTAPMTMMNAIVIEAYKKYILVSLIRHGQFSTSLPKYASPL 60
MI IG K+F ALD LHN VT PM+ +NAI +EAYKKYILVSLI++GQ S PKY S
Sbjct: 188 MIYIGLKKFTIALDFLHNAVTVPMSSLNAIAVEAYKKYILVSLIQNGQVP-SFPKYTSST 246
Query: 61 AQRNLKLFCQNPCIELAQTYNNGKVAEVEAFVNANAGRFEADNNLGLAKQVVSSMYKRNI 120
AQRNLK Q ++L+ Y+ G +E+E ++ NA +F++DNNLGL KQV+SSMYKRNI
Sbjct: 247 AQRNLKNHAQ-VYVDLSTCYSKGNYSELEEYIQLNAEKFQSDNNLGLVKQVLSSMYKRNI 305
Query: 121 QRLTQTYLTLSLEDIANTVHLNSAKEAEMHVLQMIQDGEIYATINQKDGMVRFLEDPEQY 180
QRLTQTYLTLSLEDIA++V L + KEAEMHVL+MI+DGEI+ATINQKDGMV F EDPEQY
Sbjct: 306 QRLTQTYLTLSLEDIASSVQLKTPKEAEMHVLRMIEDGEIHATINQKDGMVSFHEDPEQY 365
Query: 181 KTCEMIEHIDSSIQRIMALSKKLTATDEQISCDQLYLSK 219
K+CEM+EHIDSSIQR+MALSKKL++ DE ISCD YL K
Sbjct: 366 KSCEMVEHIDSSIQRLMALSKKLSSIDENISCDPAYLMK 404
>emb|CAB87764.1| putative protein [Arabidopsis thaliana] gi|11290078|pir||T48598
hypothetical protein F18O22.40 - Arabidopsis thaliana
Length = 459
Score = 276 bits (707), Expect = 3e-73
Identities = 156/260 (60%), Positives = 178/260 (68%), Gaps = 33/260 (12%)
Query: 1 MICIGQKRFQKALDLLHNVVTAPMTMMNAIVIEAYKKYILVSLIRHGQFSTSLPKYASPL 60
MICIG KRFQKAL+LL+NVVTAPM +NAI +EAYKKYILVSLI +GQF+ +LPK AS
Sbjct: 189 MICIGLKRFQKALELLYNVVTAPMHQVNAIALEAYKKYILVSLIHNGQFTNTLPKCASTA 248
Query: 61 AQRNLKLFCQNPCIELAQTYNNGKVAEVEAFVNANAGRFEADNNLGLAKQVVSSMYKRNI 120
AQR+ K + P IEL YN+GK+ E+EA V A FE D NLGL KQ VSS+YKRNI
Sbjct: 249 AQRSFKNYT-GPYIELGNCYNDGKIGELEALVVARNAEFEEDKNLGLVKQAVSSLYKRNI 307
Query: 121 QRLTQTYLTLSLEDIANTVHLNSAKEAEMHVLQMIQDGEIYATINQKDGMVRFLEDPEQY 180
RLTQ YLTLSL+DIAN V L +AKEAEMHVLQMIQDG+I+A INQKDGMVRFLEDPEQY
Sbjct: 308 LRLTQKYLTLSLQDIANMVQLGNAKEAEMHVLQMIQDGQIHALINQKDGMVRFLEDPEQY 367
Query: 181 KTCEMIEHIDSSIQ------------------------------RIMALSKKLTATDEQI 210
K+ EMIE +DS IQ R + LSK L A DE +
Sbjct: 368 KSSEMIEIMDSVIQSTWNQVDMEMTVLPKESSSIKQVMCFDKNFRTIGLSKNLLAMDESL 427
Query: 211 SCDQLYLSKV--SVQSFSFG 228
SCD LYL KV Q + FG
Sbjct: 428 SCDPLYLGKVGRERQRYDFG 447
>gb|AAH64288.1| COP9 constitutive photomorphogenic homolog subunit 3 [Danio rerio]
gi|55976433|sp|Q6P2U9|CSN3_BRARE COP9 signalosome
complex subunit 3 (Signalosome subunit 3)
Length = 423
Score = 193 bits (491), Expect = 3e-48
Identities = 104/219 (47%), Positives = 145/219 (65%), Gaps = 2/219 (0%)
Query: 1 MICIGQKRFQKALDLLHNVVTAPMTMMNAIVIEAYKKYILVSLIRHGQFSTSLPKYASPL 60
MI G K F++AL +T P ++ I++EAYKKYILVSLI HG+ LPKY S +
Sbjct: 190 MIYTGLKNFERALYFFEQAITTPAMAVSHIMLEAYKKYILVSLILHGKVQ-QLPKYTSQI 248
Query: 61 AQRNLKLFCQNPCIELAQTYNNGKVAEVEAFVNANAGRFEADNNLGLAKQVVSSMYKRNI 120
R +K N ELAQ Y AE+ A VN ++ F DNN GL KQ +SS+YK+NI
Sbjct: 249 VGRFIKPL-SNAYHELAQIYATNNPAELRALVNKHSETFTRDNNTGLVKQCLSSLYKKNI 307
Query: 121 QRLTQTYLTLSLEDIANTVHLNSAKEAEMHVLQMIQDGEIYATINQKDGMVRFLEDPEQY 180
QRLT+T+LTLSL+D+A+ V L+ +EAE +VL MI+DGEIYA+INQKDGMV F ++PE+Y
Sbjct: 308 QRLTKTFLTLSLQDMASRVQLSGPQEAEKYVLHMIEDGEIYASINQKDGMVCFHDNPEKY 367
Query: 181 KTCEMIEHIDSSIQRIMALSKKLTATDEQISCDQLYLSK 219
M+ ID + + + L +KL + D++I+ + ++ K
Sbjct: 368 NNPAMLHKIDQEMLKCIELDEKLKSMDQEITVNPQFVQK 406
>gb|AAH45415.1| COP9 constitutive photomorphogenic homolog subunit 3 [Danio rerio]
gi|53292634|ref|NP_955979.1| COP9 constitutive
photomorphogenic homolog subunit 3 [Danio rerio]
Length = 423
Score = 193 bits (490), Expect = 4e-48
Identities = 104/219 (47%), Positives = 145/219 (65%), Gaps = 2/219 (0%)
Query: 1 MICIGQKRFQKALDLLHNVVTAPMTMMNAIVIEAYKKYILVSLIRHGQFSTSLPKYASPL 60
MI G K F++AL +T P ++ I++EAYKKYILVSLI HG+ LPKY S +
Sbjct: 190 MIYTGLKNFERALYFFEQAITTPAMAVSHIMLEAYKKYILVSLILHGKVQ-QLPKYTSQI 248
Query: 61 AQRNLKLFCQNPCIELAQTYNNGKVAEVEAFVNANAGRFEADNNLGLAKQVVSSMYKRNI 120
R +K N ELAQ Y AE+ A VN ++ F DNN GL KQ +SS+YK+NI
Sbjct: 249 VGRFIKPL-SNAYHELAQIYATNNPAELRALVNKHSETFTRDNNSGLVKQCLSSLYKKNI 307
Query: 121 QRLTQTYLTLSLEDIANTVHLNSAKEAEMHVLQMIQDGEIYATINQKDGMVRFLEDPEQY 180
QRLT+T+LTLSL+D+A+ V L+ +EAE +VL MI+DGEIYA+INQKDGMV F ++PE+Y
Sbjct: 308 QRLTKTFLTLSLQDMASRVQLSGPQEAEKYVLHMIEDGEIYASINQKDGMVCFHDNPEKY 367
Query: 181 KTCEMIEHIDSSIQRIMALSKKLTATDEQISCDQLYLSK 219
M+ ID + + + L +KL + D++I+ + ++ K
Sbjct: 368 NNPAMLHKIDQEMLKCIELDEKLKSMDQEITVNPQFVQK 406
>ref|NP_001017238.1| hypothetical protein LOC549992 [Xenopus tropicalis]
Length = 423
Score = 191 bits (485), Expect = 2e-47
Identities = 103/219 (47%), Positives = 146/219 (66%), Gaps = 2/219 (0%)
Query: 1 MICIGQKRFQKALDLLHNVVTAPMTMMNAIVIEAYKKYILVSLIRHGQFSTSLPKYASPL 60
MI G K F++AL +T P ++ I++EAYKKYILVSLI HG+ LPKY S +
Sbjct: 190 MIYTGLKNFERALYFYEQAITTPAMAVSHIMLEAYKKYILVSLILHGKVQ-QLPKYTSQI 248
Query: 61 AQRNLKLFCQNPCIELAQTYNNGKVAEVEAFVNANAGRFEADNNLGLAKQVVSSMYKRNI 120
R +K N ELAQ Y+ AE+ V+ + F DNN+GL KQ +SS+YK+NI
Sbjct: 249 VGRFIKPL-SNAYHELAQVYSTNNPAELRNLVSKHNETFTRDNNMGLVKQCLSSLYKKNI 307
Query: 121 QRLTQTYLTLSLEDIANTVHLNSAKEAEMHVLQMIQDGEIYATINQKDGMVRFLEDPEQY 180
QRLT+T+LTLSL+D+A+ V L+ A+EAE +VL MI+DGEI+A+INQKDGMV F ++PE+Y
Sbjct: 308 QRLTKTFLTLSLQDMASRVQLSGAQEAEKYVLYMIEDGEIFASINQKDGMVCFHDNPEKY 367
Query: 181 KTCEMIEHIDSSIQRIMALSKKLTATDEQISCDQLYLSK 219
M+ +ID + R + L +L A D++I+ + ++ K
Sbjct: 368 NNPAMLHNIDQEMLRCIDLDDRLKAMDQEITVNPQFVQK 406
>gb|AAH44034.1| Cops3-prov protein [Xenopus laevis]
gi|55976497|sp|Q7ZTN8|CSN3_XENLA COP9 signalosome
complex subunit 3 (Signalosome subunit 3)
Length = 423
Score = 189 bits (481), Expect = 5e-47
Identities = 102/219 (46%), Positives = 145/219 (65%), Gaps = 2/219 (0%)
Query: 1 MICIGQKRFQKALDLLHNVVTAPMTMMNAIVIEAYKKYILVSLIRHGQFSTSLPKYASPL 60
MI G K F++A+ +T P ++ I++EAYKKYILVSLI HG+ LPKY S +
Sbjct: 190 MIYTGLKNFERAMYFYEQAITTPAMAVSHIMLEAYKKYILVSLILHGKVQ-QLPKYTSQV 248
Query: 61 AQRNLKLFCQNPCIELAQTYNNGKVAEVEAFVNANAGRFEADNNLGLAKQVVSSMYKRNI 120
R +K N ELAQ Y+ AE+ V+ + F DNN+GL KQ +SS+YK+NI
Sbjct: 249 VGRFIKPL-SNAYHELAQVYSTNNPAELRNLVSKHNETFTRDNNMGLVKQCLSSLYKKNI 307
Query: 121 QRLTQTYLTLSLEDIANTVHLNSAKEAEMHVLQMIQDGEIYATINQKDGMVRFLEDPEQY 180
QRLT+T+LTLSL+D+A+ V L+ A+EAE +VL MI+DGEI+A+INQKDGMV F + PE+Y
Sbjct: 308 QRLTKTFLTLSLQDMASRVQLSGAQEAEKYVLYMIEDGEIFASINQKDGMVCFHDSPEKY 367
Query: 181 KTCEMIEHIDSSIQRIMALSKKLTATDEQISCDQLYLSK 219
M+ +ID + R + L +L A D++I+ + ++ K
Sbjct: 368 NNPAMLHNIDQEMLRCIDLDDRLKAMDQEITVNPQFVQK 406
>ref|NP_001006163.1| similar to COP9 constitutive photomorphogenic homolog subunit 3;
COP9 complex subunit 3; JAB1-containing signalosome
subunit 3; COP9 (constitutive photomorphogenic,
Arabidopsis, homolog) subunit 3 [Gallus gallus]
gi|53133626|emb|CAG32142.1| hypothetical protein [Gallus
gallus]
Length = 423
Score = 188 bits (477), Expect = 1e-46
Identities = 100/219 (45%), Positives = 147/219 (66%), Gaps = 2/219 (0%)
Query: 1 MICIGQKRFQKALDLLHNVVTAPMTMMNAIVIEAYKKYILVSLIRHGQFSTSLPKYASPL 60
MI G K F++AL +T P ++ I++E+YKKYILVSLI G+ LPKY S +
Sbjct: 190 MIYTGLKNFERALYFYEQAITTPAMAVSHIMLESYKKYILVSLILLGKVQ-QLPKYTSQI 248
Query: 61 AQRNLKLFCQNPCIELAQTYNNGKVAEVEAFVNANAGRFEADNNLGLAKQVVSSMYKRNI 120
R +K N ELAQ Y+ K +E+ VN ++ F DNN+GL KQ +SS+YK+NI
Sbjct: 249 VGRFIKPL-SNAYHELAQVYSTNKPSELRNLVNKHSETFTRDNNMGLVKQCLSSLYKKNI 307
Query: 121 QRLTQTYLTLSLEDIANTVHLNSAKEAEMHVLQMIQDGEIYATINQKDGMVRFLEDPEQY 180
QRLT+T+LTLSL+D+A+ V L+ +EAE +VL MI+DGEI+A+INQKDGMV F ++PE+Y
Sbjct: 308 QRLTKTFLTLSLQDMASRVQLSGPQEAEKYVLHMIEDGEIFASINQKDGMVCFHDNPEKY 367
Query: 181 KTCEMIEHIDSSIQRIMALSKKLTATDEQISCDQLYLSK 219
M+ +ID + + + L ++L A D++I+ + ++ K
Sbjct: 368 NNPAMLHNIDQEMLKCIELDERLKAMDQEITVNPQFVQK 406
>dbj|BAE00674.1| unnamed protein product [Macaca fascicularis]
gi|55725035|emb|CAH89385.1| hypothetical protein [Pongo
pygmaeus] gi|12804885|gb|AAH01891.1| COP9 constitutive
photomorphogenic homolog subunit 3 [Homo sapiens]
gi|23238222|ref|NP_003644.2| COP9 constitutive
photomorphogenic homolog subunit 3 [Homo sapiens]
gi|55976621|sp|Q9UNS2|CSN3_HUMAN COP9 signalosome
complex subunit 3 (Signalosome subunit 3) (SGN3)
(JAB1-containing signalosome subunit 3)
gi|5257136|gb|AAD41247.1| COP9 complex subunit 3 [Homo
sapiens]
Length = 423
Score = 187 bits (474), Expect = 3e-46
Identities = 99/219 (45%), Positives = 146/219 (66%), Gaps = 2/219 (0%)
Query: 1 MICIGQKRFQKALDLLHNVVTAPMTMMNAIVIEAYKKYILVSLIRHGQFSTSLPKYASPL 60
MI G K F++AL +T P ++ I++E+YKKYILVSLI G+ LPKY S +
Sbjct: 190 MIYTGLKNFERALYFYEQAITTPAMAVSHIMLESYKKYILVSLILLGKVQ-QLPKYTSQI 248
Query: 61 AQRNLKLFCQNPCIELAQTYNNGKVAEVEAFVNANAGRFEADNNLGLAKQVVSSMYKRNI 120
R +K N ELAQ Y+ +E+ VN ++ F DNN+GL KQ +SS+YK+NI
Sbjct: 249 VGRFIKPL-SNAYHELAQVYSTNNPSELRNLVNKHSETFTRDNNMGLVKQCLSSLYKKNI 307
Query: 121 QRLTQTYLTLSLEDIANTVHLNSAKEAEMHVLQMIQDGEIYATINQKDGMVRFLEDPEQY 180
QRLT+T+LTLSL+D+A+ V L+ +EAE +VL MI+DGEI+A+INQKDGMV F ++PE+Y
Sbjct: 308 QRLTKTFLTLSLQDMASRVQLSGPQEAEKYVLHMIEDGEIFASINQKDGMVSFHDNPEKY 367
Query: 181 KTCEMIEHIDSSIQRIMALSKKLTATDEQISCDQLYLSK 219
M+ +ID + + + L ++L A D++I+ + ++ K
Sbjct: 368 NNPAMLHNIDQEMLKCIELDERLKAMDQEITVNPQFVQK 406
>gb|AAC14197.1| JAB1-containing signalosome subunit 3 [Homo sapiens]
Length = 403
Score = 187 bits (474), Expect = 3e-46
Identities = 99/219 (45%), Positives = 146/219 (66%), Gaps = 2/219 (0%)
Query: 1 MICIGQKRFQKALDLLHNVVTAPMTMMNAIVIEAYKKYILVSLIRHGQFSTSLPKYASPL 60
MI G K F++AL +T P ++ I++E+YKKYILVSLI G+ LPKY S +
Sbjct: 170 MIYTGLKNFERALYFYEQAITTPAMAVSHIMLESYKKYILVSLILLGKVQ-QLPKYTSQI 228
Query: 61 AQRNLKLFCQNPCIELAQTYNNGKVAEVEAFVNANAGRFEADNNLGLAKQVVSSMYKRNI 120
R +K N ELAQ Y+ +E+ VN ++ F DNN+GL KQ +SS+YK+NI
Sbjct: 229 VGRFIKPL-SNAYHELAQVYSTNNPSELRNLVNKHSETFTRDNNMGLVKQCLSSLYKKNI 287
Query: 121 QRLTQTYLTLSLEDIANTVHLNSAKEAEMHVLQMIQDGEIYATINQKDGMVRFLEDPEQY 180
QRLT+T+LTLSL+D+A+ V L+ +EAE +VL MI+DGEI+A+INQKDGMV F ++PE+Y
Sbjct: 288 QRLTKTFLTLSLQDMASRVQLSGPQEAEKYVLHMIEDGEIFASINQKDGMVSFHDNPEKY 347
Query: 181 KTCEMIEHIDSSIQRIMALSKKLTATDEQISCDQLYLSK 219
M+ +ID + + + L ++L A D++I+ + ++ K
Sbjct: 348 NNPAMLHNIDQEMLKCIELDERLKAMDQEITVNPQFVQK 386
>ref|NP_001004200.1| COP9 constitutive photomorphogenic homolog subunit 3 [Rattus
norvegicus] gi|51260717|gb|AAH79143.1| COP9 constitutive
photomorphogenic homolog subunit 3 [Rattus norvegicus]
Length = 423
Score = 187 bits (474), Expect = 3e-46
Identities = 99/219 (45%), Positives = 146/219 (66%), Gaps = 2/219 (0%)
Query: 1 MICIGQKRFQKALDLLHNVVTAPMTMMNAIVIEAYKKYILVSLIRHGQFSTSLPKYASPL 60
MI G K F++AL +T P ++ I++E+YKKYILVSLI G+ LPKY S +
Sbjct: 190 MIYTGLKNFERALYFYEQAITTPAMAVSHIMLESYKKYILVSLILLGKVQ-QLPKYTSQI 248
Query: 61 AQRNLKLFCQNPCIELAQTYNNGKVAEVEAFVNANAGRFEADNNLGLAKQVVSSMYKRNI 120
R +K N ELAQ Y+ +E+ VN ++ F DNN+GL KQ +SS+YK+NI
Sbjct: 249 VGRFIKPL-SNAYHELAQVYSTNNPSELRNLVNKHSETFTRDNNMGLVKQCLSSLYKKNI 307
Query: 121 QRLTQTYLTLSLEDIANTVHLNSAKEAEMHVLQMIQDGEIYATINQKDGMVRFLEDPEQY 180
QRLT+T+LTLSL+D+A+ V L+ +EAE +VL MI+DGEI+A+INQKDGMV F ++PE+Y
Sbjct: 308 QRLTKTFLTLSLQDMASRVQLSGPQEAEKYVLHMIEDGEIFASINQKDGMVSFHDNPEKY 367
Query: 181 KTCEMIEHIDSSIQRIMALSKKLTATDEQISCDQLYLSK 219
M+ +ID + + + L ++L A D++I+ + ++ K
Sbjct: 368 NNPAMLHNIDQEMLKCIELDERLKAMDQEITVNPQFVQK 406
>ref|XP_536667.1| PREDICTED: similar to COP9 constitutive photomorphogenic homolog
subunit 3 [Canis familiaris]
Length = 414
Score = 187 bits (474), Expect = 3e-46
Identities = 99/219 (45%), Positives = 146/219 (66%), Gaps = 2/219 (0%)
Query: 1 MICIGQKRFQKALDLLHNVVTAPMTMMNAIVIEAYKKYILVSLIRHGQFSTSLPKYASPL 60
MI G K F++AL +T P ++ I++E+YKKYILVSLI G+ LPKY S +
Sbjct: 181 MIYTGLKNFERALYFYEQAITTPAMAVSHIMLESYKKYILVSLILLGKVQ-QLPKYTSQI 239
Query: 61 AQRNLKLFCQNPCIELAQTYNNGKVAEVEAFVNANAGRFEADNNLGLAKQVVSSMYKRNI 120
R +K N ELAQ Y+ +E+ VN ++ F DNN+GL KQ +SS+YK+NI
Sbjct: 240 VGRFIKPL-SNAYHELAQVYSTNNPSELRNLVNKHSETFTRDNNMGLVKQCLSSLYKKNI 298
Query: 121 QRLTQTYLTLSLEDIANTVHLNSAKEAEMHVLQMIQDGEIYATINQKDGMVRFLEDPEQY 180
QRLT+T+LTLSL+D+A+ V L+ +EAE +VL MI+DGEI+A+INQKDGMV F ++PE+Y
Sbjct: 299 QRLTKTFLTLSLQDMASRVQLSGPQEAEKYVLHMIEDGEIFASINQKDGMVSFHDNPEKY 358
Query: 181 KTCEMIEHIDSSIQRIMALSKKLTATDEQISCDQLYLSK 219
M+ +ID + + + L ++L A D++I+ + ++ K
Sbjct: 359 NNPAMLHNIDQEMLKCIELDERLKAMDQEITVNPQFVQK 397
>ref|NP_036121.1| COP9 (constitutive photomorphogenic), subunit 3 [Mus musculus]
gi|45829710|gb|AAH68179.1| COP9 (constitutive
photomorphogenic), subunit 3 [Mus musculus]
gi|55976220|sp|O88543|CSN3_MOUSE COP9 signalosome
complex subunit 3 (Signalosome subunit 3) (SGN3)
(JAB1-containing signalosome subunit 3)
gi|3309168|gb|AAC33900.1| COP9 complex subunit 3 [Mus
musculus]
Length = 423
Score = 185 bits (469), Expect = 1e-45
Identities = 98/219 (44%), Positives = 146/219 (65%), Gaps = 2/219 (0%)
Query: 1 MICIGQKRFQKALDLLHNVVTAPMTMMNAIVIEAYKKYILVSLIRHGQFSTSLPKYASPL 60
MI G K F++AL +T P ++ I++E+YKKYILVSLI G+ LPKY S +
Sbjct: 190 MIYTGLKNFERALYFYEQAITTPAMAVSHIMLESYKKYILVSLILLGKVQ-QLPKYTSQI 248
Query: 61 AQRNLKLFCQNPCIELAQTYNNGKVAEVEAFVNANAGRFEADNNLGLAKQVVSSMYKRNI 120
R +K N ELAQ Y+ +E+ V+ ++ F DNN+GL KQ +SS+YK+NI
Sbjct: 249 VGRFIKPL-SNAYHELAQVYSTNNPSELRNLVSKHSETFTRDNNMGLVKQCLSSLYKKNI 307
Query: 121 QRLTQTYLTLSLEDIANTVHLNSAKEAEMHVLQMIQDGEIYATINQKDGMVRFLEDPEQY 180
QRLT+T+LTLSL+D+A+ V L+ +EAE +VL MI+DGEI+A+INQKDGMV F ++PE+Y
Sbjct: 308 QRLTKTFLTLSLQDMASRVQLSGPQEAEKYVLHMIEDGEIFASINQKDGMVSFHDNPEKY 367
Query: 181 KTCEMIEHIDSSIQRIMALSKKLTATDEQISCDQLYLSK 219
M+ +ID + + + L ++L A D++I+ + ++ K
Sbjct: 368 NNPAMLHNIDQEMLKCIELDERLKAMDQEITVNPQFVQK 406
>ref|XP_316484.2| ENSANGP00000004286 [Anopheles gambiae str. PEST]
gi|55239217|gb|EAA11895.2| ENSANGP00000004286 [Anopheles
gambiae str. PEST]
Length = 432
Score = 180 bits (457), Expect = 3e-44
Identities = 96/219 (43%), Positives = 140/219 (63%), Gaps = 2/219 (0%)
Query: 1 MICIGQKRFQKALDLLHNVVTAPMTMMNAIVIEAYKKYILVSLIRHGQFSTSLPKYASPL 60
MI K +++AL V+ P M+ I++E+YKKYILVSLI HG+ +PKY+S +
Sbjct: 193 MIYSAVKNYERALYFFEVAVSTPALAMSHIMLESYKKYILVSLILHGKV-LPIPKYSSQV 251
Query: 61 AQRNLKLFCQNPCIELAQTYNNGKVAEVEAFVNANAGRFEADNNLGLAKQVVSSMYKRNI 120
R +K + +L+ YN EV VN F+ D N+GL KQVV+S+YK+NI
Sbjct: 252 ITRFMKPL-SHAYHDLSSAYNTSSADEVRNVVNKYRDTFQRDTNMGLVKQVVASLYKKNI 310
Query: 121 QRLTQTYLTLSLEDIANTVHLNSAKEAEMHVLQMIQDGEIYATINQKDGMVRFLEDPEQY 180
QRLT+T+LTLSL D+A+ V L+ EAE ++L MI+ GEI+ATINQKDGMV F +DP++Y
Sbjct: 311 QRLTKTFLTLSLADVASRVQLSGPAEAEKYILNMIKSGEIFATINQKDGMVVFKDDPQEY 370
Query: 181 KTCEMIEHIDSSIQRIMALSKKLTATDEQISCDQLYLSK 219
E+ E + I +M L+K++ DE+I + +++ K
Sbjct: 371 NDQEVFEMVQHQISLVMGLNKQILKMDEEIMLNPVFVKK 409
>gb|AAH95590.1| Unknown (protein for MGC:111903) [Danio rerio]
Length = 381
Score = 180 bits (457), Expect = 3e-44
Identities = 98/193 (50%), Positives = 129/193 (66%), Gaps = 2/193 (1%)
Query: 1 MICIGQKRFQKALDLLHNVVTAPMTMMNAIVIEAYKKYILVSLIRHGQFSTSLPKYASPL 60
MI G K F++AL +T P ++ I++EAYKKYILVSLI HG+ LPKY S +
Sbjct: 190 MIYTGLKNFERALYFFEQAITTPAMAVSHIMLEAYKKYILVSLILHGKVQ-QLPKYTSQI 248
Query: 61 AQRNLKLFCQNPCIELAQTYNNGKVAEVEAFVNANAGRFEADNNLGLAKQVVSSMYKRNI 120
R +K N ELAQ Y AE+ A VN ++ F DNN GL KQ +SS+YK+NI
Sbjct: 249 VGRFIKPL-SNAYHELAQIYATNNPAELRALVNKHSETFTRDNNSGLVKQCLSSLYKKNI 307
Query: 121 QRLTQTYLTLSLEDIANTVHLNSAKEAEMHVLQMIQDGEIYATINQKDGMVRFLEDPEQY 180
QRLT+T+LTLSL+D+A+ V L+ +EAE +VL MI+DGEIYA+INQKDGMV F ++PE+Y
Sbjct: 308 QRLTKTFLTLSLQDMASRVQLSGPQEAEKYVLHMIEDGEIYASINQKDGMVCFHDNPEKY 367
Query: 181 KTCEMIEHIDSSI 193
M+ ID +
Sbjct: 368 NNPAMLHKIDQEM 380
>ref|XP_584631.1| PREDICTED: similar to COP9 signalosome complex subunit 3
(Signalosome subunit 3) (SGN3) (JAB1-containing
signalosome subunit 3), partial [Bos taurus]
Length = 216
Score = 174 bits (440), Expect = 3e-42
Identities = 91/200 (45%), Positives = 137/200 (68%), Gaps = 2/200 (1%)
Query: 20 VTAPMTMMNAIVIEAYKKYILVSLIRHGQFSTSLPKYASPLAQRNLKLFCQNPCIELAQT 79
+T P ++ I++E+YKKYILVSLI G+ LPKY S + R +K N ELAQ
Sbjct: 2 ITTPAMAVSHIMLESYKKYILVSLILLGKVQ-QLPKYTSQIVGRFIKPL-SNAYHELAQV 59
Query: 80 YNNGKVAEVEAFVNANAGRFEADNNLGLAKQVVSSMYKRNIQRLTQTYLTLSLEDIANTV 139
Y+ +E+ + V ++ F DNN+GL KQ +SS+YK+NIQRLT+T+LTLSL+D+A+ V
Sbjct: 60 YSTNNPSELRSLVTKHSEIFTRDNNMGLVKQCLSSLYKKNIQRLTKTFLTLSLQDMASRV 119
Query: 140 HLNSAKEAEMHVLQMIQDGEIYATINQKDGMVRFLEDPEQYKTCEMIEHIDSSIQRIMAL 199
L+ +EAE +VL MI+DGEI+A+INQKDGMV F ++PE+Y M+ +ID + + + L
Sbjct: 120 QLSGPQEAEKYVLHMIEDGEIFASINQKDGMVSFHDNPEKYNNPAMLHNIDQEMLKCIEL 179
Query: 200 SKKLTATDEQISCDQLYLSK 219
++L A D++I+ + ++ K
Sbjct: 180 DERLKAMDQEITVNPQFVQK 199
>ref|XP_624761.1| PREDICTED: similar to COP9 constitutive photomorphogenic homolog
subunit 3 [Apis mellifera]
Length = 445
Score = 173 bits (439), Expect = 3e-42
Identities = 95/224 (42%), Positives = 138/224 (61%), Gaps = 2/224 (0%)
Query: 1 MICIGQKRFQKALDLLHNVVTAPMTMMNAIVIEAYKKYILVSLIRHGQFSTSLPKYASPL 60
MI K + +AL VT P ++ I++EAYKKYILVSLI HG+ +LP+Y S +
Sbjct: 196 MIYTALKNYDRALYFFEVCVTTPAMAVSYIMLEAYKKYILVSLILHGKV-LNLPRYTSQV 254
Query: 61 AQRNLKLFCQNPCIELAQTYNNGKVAEVEAFVNANAGRFEADNNLGLAKQVVSSMYKRNI 120
R +K Q ELA Y EV+ + F D+N+GL KQV+S +YK+NI
Sbjct: 255 VNRYMKPLGQQ-YQELATAYQMNSCEEVQNIITKYQQLFTRDHNMGLVKQVLSYLYKKNI 313
Query: 121 QRLTQTYLTLSLEDIANTVHLNSAKEAEMHVLQMIQDGEIYATINQKDGMVRFLEDPEQY 180
QRLT+T+LTLSL D+A+ V L+ +AE ++L MI+DGEI+ATINQKDGMV F +DPE+Y
Sbjct: 314 QRLTKTFLTLSLSDVASRVQLSGPADAEKYILNMIEDGEIFATINQKDGMVVFHDDPEKY 373
Query: 181 KTCEMIEHIDSSIQRIMALSKKLTATDEQISCDQLYLSKVSVQS 224
+ +M+ ++ + M L K++ +E++ Y+ K Q+
Sbjct: 374 NSPQMLAKLEKEMAACMELDKRVLEMEEEVVLTPQYVRKACGQN 417
>ref|XP_629934.1| hypothetical protein DDB0219696 [Dictyostelium discoideum]
gi|60463354|gb|EAL61545.1| hypothetical protein
DDB0219696 [Dictyostelium discoideum]
Length = 418
Score = 169 bits (428), Expect = 7e-41
Identities = 91/215 (42%), Positives = 138/215 (63%), Gaps = 3/215 (1%)
Query: 7 KRFQKALDLLHNVVTAPMTMMNAIVIEAYKKYILVSLIRHGQFSTSLPKYASPLAQRNLK 66
K+++KA++ V+TAP + ++AI +EAYKKY++V LI+ G PK + QRN+K
Sbjct: 188 KKYKKAIEAFKFVITAPASALSAITVEAYKKYLIVYLIQFGSVQ-HFPKCTPAVVQRNIK 246
Query: 67 LFCQNPCIELAQTYNNGKVAEVEAFVNANAGRFEADNNLGLAKQVVSSMYKRNIQRLTQT 126
C+ P E Q+++NG + ++ ++ A F+ D+N GL K + S Y+RNI++LTQT
Sbjct: 247 SHCK-PYTEFVQSFSNGNINDIMNKASSGAEFFQKDSNWGLVKLSIKSTYRRNIKKLTQT 305
Query: 127 YLTLSLEDIANTVHLNSAKEAEMHVLQMIQDGEIYATINQKDGMVRFLEDPEQYKTCEMI 186
++TLS+ DIA V L AK AE VL+MI++GEI+ATINQKDGMV F E E + ++
Sbjct: 306 FMTLSINDIAEKVSLPKAK-AEQFVLKMIEEGEIFATINQKDGMVTFDECFEDFSGPTIL 364
Query: 187 EHIDSSIQRIMALSKKLTATDEQISCDQLYLSKVS 221
+D +I I+ L K+ DE+IS YL K++
Sbjct: 365 NDLDRNINNIVGLESKIKEMDEKISLSNNYLKKLA 399
Database: nr
Posted date: Jul 5, 2005 12:34 AM
Number of letters in database: 863,360,394
Number of sequences in database: 2,540,612
Lambda K H
0.320 0.133 0.370
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 354,299,092
Number of Sequences: 2540612
Number of extensions: 12729604
Number of successful extensions: 33052
Number of sequences better than 10.0: 175
Number of HSP's better than 10.0 without gapping: 94
Number of HSP's successfully gapped in prelim test: 82
Number of HSP's that attempted gapping in prelim test: 32899
Number of HSP's gapped (non-prelim): 188
length of query: 250
length of database: 863,360,394
effective HSP length: 125
effective length of query: 125
effective length of database: 545,783,894
effective search space: 68222986750
effective search space used: 68222986750
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.9 bits)
S2: 73 (32.7 bits)
Medicago: description of AC138464.6


