

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC138464.5 + phase: 0
(235 letters)
Database: nr
2,540,612 sequences; 863,360,394 total letters
Searching..................................................done
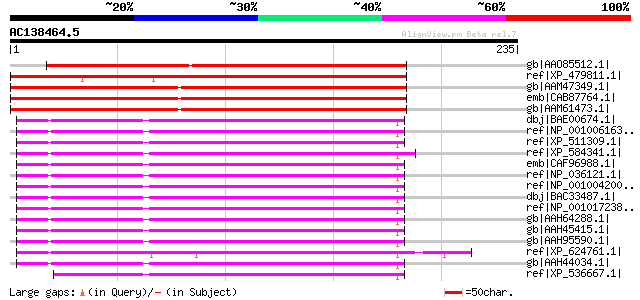
Score E
Sequences producing significant alignments: (bits) Value
gb|AAO85512.1| CSN3 [Nicotiana benthamiana] 209 5e-53
ref|XP_479811.1| putative COP9 complex subunit 3, FUS11 [Oryza s... 205 7e-52
gb|AAM47349.1| AT5g14250/F18O22_40 [Arabidopsis thaliana] gi|169... 186 4e-46
emb|CAB87764.1| putative protein [Arabidopsis thaliana] gi|11290... 182 5e-45
gb|AAM61473.1| unknown [Arabidopsis thaliana] gi|18056657|gb|AAL... 182 5e-45
dbj|BAE00674.1| unnamed protein product [Macaca fascicularis] gi... 93 5e-18
ref|NP_001006163.1| similar to COP9 constitutive photomorphogeni... 93 5e-18
ref|XP_511309.1| PREDICTED: similar to COP9 complex subunit 3 [P... 93 5e-18
ref|XP_584341.1| PREDICTED: similar to COP9 constitutive photomo... 93 5e-18
emb|CAF96988.1| unnamed protein product [Tetraodon nigroviridis] 93 5e-18
ref|NP_036121.1| COP9 (constitutive photomorphogenic), subunit 3... 93 7e-18
ref|NP_001004200.1| COP9 constitutive photomorphogenic homolog s... 93 7e-18
dbj|BAC33487.1| unnamed protein product [Mus musculus] 93 7e-18
ref|NP_001017238.1| hypothetical protein LOC549992 [Xenopus trop... 93 7e-18
gb|AAH64288.1| COP9 constitutive photomorphogenic homolog subuni... 92 2e-17
gb|AAH45415.1| COP9 constitutive photomorphogenic homolog subuni... 92 2e-17
gb|AAH95590.1| Unknown (protein for MGC:111903) [Danio rerio] 92 2e-17
ref|XP_624761.1| PREDICTED: similar to COP9 constitutive photomo... 91 2e-17
gb|AAH44034.1| Cops3-prov protein [Xenopus laevis] gi|55976497|s... 91 3e-17
ref|XP_536667.1| PREDICTED: similar to COP9 constitutive photomo... 91 3e-17
>gb|AAO85512.1| CSN3 [Nicotiana benthamiana]
Length = 175
Score = 209 bits (532), Expect = 5e-53
Identities = 99/167 (59%), Positives = 129/167 (76%), Gaps = 1/167 (0%)
Query: 18 SADIARLHTILKHSDETLRSDSSRLYPILQLIDPSIHSLGFLYLLDAFSAASVSNQQQAE 77
S+DI +LH LK S+E L SD RL L +DP+ HSLGFLY+L+A + + +Q E
Sbjct: 1 SSDITQLHNFLKQSEELLHSDFGRLLSSLAELDPNTHSLGFLYILEACMSFPAAKEQVNE 60
Query: 78 EAVPIIARFINACSVEQIRLAPEKFVSVCKRLKDLVTLLEAPIRGVAPMFTALRKLQVSA 137
V ++ RFIN+C+ EQIRLAP+KF+SVCKR KD + LLEAPIRGVA M TA+RKLQ S+
Sbjct: 61 LLVSVV-RFINSCAAEQIRLAPDKFISVCKRFKDQIILLEAPIRGVASMLTAVRKLQSSS 119
Query: 138 EHLTPLHAEFLMLCLLAKCYKTGLSILDDDVFEVDHPRDLYLYCYYG 184
E LT LH +FL+LC+LAKCYKTG+++L+DD++E+D PRD +LYCYYG
Sbjct: 120 EQLTTLHPDFLLLCVLAKCYKTGITVLEDDIYEIDQPRDFFLYCYYG 166
>ref|XP_479811.1| putative COP9 complex subunit 3, FUS11 [Oryza sativa (japonica
cultivar-group)] gi|42407907|dbj|BAD09047.1| putative
COP9 complex subunit 3, FUS11 [Oryza sativa (japonica
cultivar-group)]
Length = 426
Score = 205 bits (522), Expect = 7e-52
Identities = 103/186 (55%), Positives = 138/186 (73%), Gaps = 2/186 (1%)
Query: 1 MDPLEPLIAQIQGLSSSSADIARLHTILKHSD-ETLRSDSSRLYPILQLIDPSIHSLGFL 59
M+ +E L+A IQGLS S ++A LH +L+ +D E LR+ S+ L P L + PS HSLGFL
Sbjct: 1 METVETLVAHIQGLSGSGEELAHLHNLLRQADGEPLRAHSAALLPFLAQLHPSAHSLGFL 60
Query: 60 YLLDAF-SAASVSNQQQAEEAVPIIARFINACSVEQIRLAPEKFVSVCKRLKDLVTLLEA 118
YLL+AF S+AS Q + + A F+ +CS EQIRLAP+KF+SVC+ K+ V L A
Sbjct: 61 YLLEAFASSASNLRAQGGGDFLVTTADFLVSCSAEQIRLAPDKFLSVCRVFKNEVMQLNA 120
Query: 119 PIRGVAPMFTALRKLQVSAEHLTPLHAEFLMLCLLAKCYKTGLSILDDDVFEVDHPRDLY 178
PIRG+AP+ A+RK+Q S+E LTP+HA++L+LCLLAK YK GLS+L+DD+ EVD P+DL+
Sbjct: 121 PIRGIAPLRAAIRKIQTSSEELTPIHADYLLLCLLAKQYKAGLSVLEDDILEVDQPKDLF 180
Query: 179 LYCYYG 184
LYCYYG
Sbjct: 181 LYCYYG 186
>gb|AAM47349.1| AT5g14250/F18O22_40 [Arabidopsis thaliana]
gi|16930463|gb|AAL31917.1| AT5g14250/F18O22_40
[Arabidopsis thaliana]
Length = 429
Score = 186 bits (473), Expect = 4e-46
Identities = 94/184 (51%), Positives = 127/184 (68%), Gaps = 1/184 (0%)
Query: 1 MDPLEPLIAQIQGLSSSSADIARLHTILKHSDETLRSDSSRLYPILQLIDPSIHSLGFLY 60
++ +E +I IQGLS S D++ LH +L+ + ++LR++ + L +D S HSLG+LY
Sbjct: 5 VNSVEAVITSIQGLSGSPEDLSALHDLLRGAQDSLRAEPGVNFSTLDQLDASKHSLGYLY 64
Query: 61 LLDAFSAASVSNQQQAEEAVPIIARFINACSVEQIRLAPEKFVSVCKRLKDLVTLLEAPI 120
L+ + VS ++ A E +PIIARFIN+C QIRLA KFVS+CK LKD V L P+
Sbjct: 65 FLEVLTCGPVSKEKAAYE-IPIIARFINSCDAGQIRLASYKFVSLCKILKDHVIALGDPL 123
Query: 121 RGVAPMFTALRKLQVSAEHLTPLHAEFLMLCLLAKCYKTGLSILDDDVFEVDHPRDLYLY 180
RGV P+ A++KLQVS++ LT LH EFL LCL AK YK+G SIL DD+ E+D PRD +LY
Sbjct: 124 RGVGPLLNAVQKLQVSSKRLTALHPEFLQLCLQAKSYKSGFSILSDDIVEIDQPRDFFLY 183
Query: 181 CYYG 184
YYG
Sbjct: 184 SYYG 187
>emb|CAB87764.1| putative protein [Arabidopsis thaliana] gi|11290078|pir||T48598
hypothetical protein F18O22.40 - Arabidopsis thaliana
Length = 459
Score = 182 bits (463), Expect = 5e-45
Identities = 92/184 (50%), Positives = 126/184 (68%), Gaps = 1/184 (0%)
Query: 1 MDPLEPLIAQIQGLSSSSADIARLHTILKHSDETLRSDSSRLYPILQLIDPSIHSLGFLY 60
++ +E +I IQGLS S D++ LH +L+ + ++LR++ + L +D S HSLG+LY
Sbjct: 5 VNSVEAVITSIQGLSGSPEDLSALHDLLRGAQDSLRAEPGVNFSTLDQLDASKHSLGYLY 64
Query: 61 LLDAFSAASVSNQQQAEEAVPIIARFINACSVEQIRLAPEKFVSVCKRLKDLVTLLEAPI 120
L+ + VS ++ A E +PIIARFIN+C QIRLA KFVS+CK LKD V L P+
Sbjct: 65 FLEVLTCGPVSKEKAAYE-IPIIARFINSCDAGQIRLASYKFVSLCKILKDHVIALGDPL 123
Query: 121 RGVAPMFTALRKLQVSAEHLTPLHAEFLMLCLLAKCYKTGLSILDDDVFEVDHPRDLYLY 180
RGV P+ A++KLQVS++ LT LH + L LCL AK YK+G SIL DD+ E+D PRD +LY
Sbjct: 124 RGVGPLLNAVQKLQVSSKRLTALHPDVLQLCLQAKSYKSGFSILSDDIVEIDQPRDFFLY 183
Query: 181 CYYG 184
YYG
Sbjct: 184 SYYG 187
>gb|AAM61473.1| unknown [Arabidopsis thaliana] gi|18056657|gb|AAL58102.1| CSN
complex subunit 3 [Arabidopsis thaliana]
gi|18417330|ref|NP_568296.1| COP9 signalosome complex
subunit 3 / CSN complex subunit 3 (CSN3) / FUSCA protein
(FUS11) [Arabidopsis thaliana]
gi|14388969|gb|AAK61872.1| COP9 signalosome subunit 3
[Arabidopsis thaliana] gi|14388971|gb|AAK61873.1| COP9
signalosome subunit 3 [Arabidopsis thaliana]
gi|55976552|sp|Q8W575|CSN3_ARATH COP9 signalosome
complex subunit 3 (Signalosome subunit 3) (FUSCA protein
11) (FUSCA11)
Length = 429
Score = 182 bits (463), Expect = 5e-45
Identities = 92/184 (50%), Positives = 126/184 (68%), Gaps = 1/184 (0%)
Query: 1 MDPLEPLIAQIQGLSSSSADIARLHTILKHSDETLRSDSSRLYPILQLIDPSIHSLGFLY 60
++ +E +I IQGLS S D++ LH +L+ + ++LR++ + L +D S HSLG+LY
Sbjct: 5 VNSVEAVITSIQGLSGSPEDLSALHDLLRGAQDSLRAEPGVNFSTLDQLDASKHSLGYLY 64
Query: 61 LLDAFSAASVSNQQQAEEAVPIIARFINACSVEQIRLAPEKFVSVCKRLKDLVTLLEAPI 120
L+ + VS ++ A E +PIIARFIN+C QIRLA KFVS+CK LKD V L P+
Sbjct: 65 FLEVLTCGPVSKEKAAYE-IPIIARFINSCDAGQIRLASYKFVSLCKILKDHVIALGDPL 123
Query: 121 RGVAPMFTALRKLQVSAEHLTPLHAEFLMLCLLAKCYKTGLSILDDDVFEVDHPRDLYLY 180
RGV P+ A++KLQVS++ LT LH + L LCL AK YK+G SIL DD+ E+D PRD +LY
Sbjct: 124 RGVGPLLNAVQKLQVSSKRLTALHPDVLQLCLQAKSYKSGFSILSDDIVEIDQPRDFFLY 183
Query: 181 CYYG 184
YYG
Sbjct: 184 SYYG 187
>dbj|BAE00674.1| unnamed protein product [Macaca fascicularis]
gi|55725035|emb|CAH89385.1| hypothetical protein [Pongo
pygmaeus] gi|12804885|gb|AAH01891.1| COP9 constitutive
photomorphogenic homolog subunit 3 [Homo sapiens]
gi|23238222|ref|NP_003644.2| COP9 constitutive
photomorphogenic homolog subunit 3 [Homo sapiens]
gi|55976621|sp|Q9UNS2|CSN3_HUMAN COP9 signalosome
complex subunit 3 (Signalosome subunit 3) (SGN3)
(JAB1-containing signalosome subunit 3)
gi|5257136|gb|AAD41247.1| COP9 complex subunit 3 [Homo
sapiens]
Length = 423
Score = 93.2 bits (230), Expect = 5e-18
Identities = 57/184 (30%), Positives = 93/184 (49%), Gaps = 7/184 (3%)
Query: 4 LEPLIAQIQGLSSSSADIARLHTILKHSDETLRSDSSRLYPILQLIDPSIHSLGFLYLLD 63
LE + ++ LS+ + +L ++ S E L + S L +L +D HSLG L +L
Sbjct: 5 LEQFVNSVRQLSAQG-QMTQLCELINKSGELLAKNLSHLDTVLGALDVQEHSLGVLAVL- 62
Query: 64 AFSAASVSNQQQAEEAVPIIARFINACSVEQIRLAPEKFVSVCKRLKDLVTLLEAPIRGV 123
F S+ + E + FI+ C+ E IR A + F +C +L + + + P+RG+
Sbjct: 63 -FVKFSMPSVPDFETLFSQVQLFISTCNGEHIRYATDTFAGLCHQLTNALVERKQPLRGI 121
Query: 124 APMFTALRKLQVSAEHLTPLHAEFLMLCLLAKCYKTGLSILDDDVFEVDHPRDLY----L 179
+ A+ K+Q++ LT +HA+ LCLLAKC+K L LD D+ ++ Y
Sbjct: 122 GILKQAIDKMQMNTNQLTSIHADLCQLCLLAKCFKPALPYLDVDMMDICKENGAYDAKHF 181
Query: 180 YCYY 183
CYY
Sbjct: 182 LCYY 185
>ref|NP_001006163.1| similar to COP9 constitutive photomorphogenic homolog subunit 3;
COP9 complex subunit 3; JAB1-containing signalosome
subunit 3; COP9 (constitutive photomorphogenic,
Arabidopsis, homolog) subunit 3 [Gallus gallus]
gi|53133626|emb|CAG32142.1| hypothetical protein [Gallus
gallus]
Length = 423
Score = 93.2 bits (230), Expect = 5e-18
Identities = 57/184 (30%), Positives = 94/184 (50%), Gaps = 7/184 (3%)
Query: 4 LEPLIAQIQGLSSSSADIARLHTILKHSDETLRSDSSRLYPILQLIDPSIHSLGFLYLLD 63
LE + ++ LS+ + +L ++ S E L + S L +L +D HSLG L +L
Sbjct: 5 LEQFVNSVRQLSAQG-QMTQLCELINKSGELLAKNLSHLDTVLGALDVQEHSLGVLAVL- 62
Query: 64 AFSAASVSNQQQAEEAVPIIARFINACSVEQIRLAPEKFVSVCKRLKDLVTLLEAPIRGV 123
F S+ + E + FI+ C+ E IR A + F +C +L + + + P+RG+
Sbjct: 63 -FVKFSMPSIPDFETLFSQVQLFISTCNGEHIRYATDTFAGLCHQLTNALVERKQPLRGI 121
Query: 124 APMFTALRKLQVSAEHLTPLHAEFLMLCLLAKCYKTGLSILDDDVFEVDHPRDLY----L 179
+ + A+ K+Q++ LT +HA+ LCLLAKC+K L LD D+ ++ Y
Sbjct: 122 SILRQAIDKMQMNTNQLTSIHADLCQLCLLAKCFKPALPYLDVDMMDICKENGAYDAKHF 181
Query: 180 YCYY 183
CYY
Sbjct: 182 LCYY 185
>ref|XP_511309.1| PREDICTED: similar to COP9 complex subunit 3 [Pan troglodytes]
Length = 458
Score = 93.2 bits (230), Expect = 5e-18
Identities = 57/184 (30%), Positives = 93/184 (49%), Gaps = 7/184 (3%)
Query: 4 LEPLIAQIQGLSSSSADIARLHTILKHSDETLRSDSSRLYPILQLIDPSIHSLGFLYLLD 63
LE + ++ LS+ + +L ++ S E L + S L +L +D HSLG L +L
Sbjct: 5 LEQFVNSVRQLSAQG-QMTQLCELINKSGELLAKNLSHLDTVLGALDVQEHSLGVLAVL- 62
Query: 64 AFSAASVSNQQQAEEAVPIIARFINACSVEQIRLAPEKFVSVCKRLKDLVTLLEAPIRGV 123
F S+ + E + FI+ C+ E IR A + F +C +L + + + P+RG+
Sbjct: 63 -FVKFSMPSVPDFETLFSQVQLFISTCNGEHIRYATDTFAGLCHQLTNALVERKQPLRGI 121
Query: 124 APMFTALRKLQVSAEHLTPLHAEFLMLCLLAKCYKTGLSILDDDVFEVDHPRDLY----L 179
+ A+ K+Q++ LT +HA+ LCLLAKC+K L LD D+ ++ Y
Sbjct: 122 GILKQAIDKMQMNTNQLTSIHADLCQLCLLAKCFKPALPYLDVDMMDICKENGAYDAKHF 181
Query: 180 YCYY 183
CYY
Sbjct: 182 LCYY 185
>ref|XP_584341.1| PREDICTED: similar to COP9 constitutive photomorphogenic homolog
subunit 3, partial [Bos taurus]
Length = 263
Score = 93.2 bits (230), Expect = 5e-18
Identities = 58/191 (30%), Positives = 96/191 (49%), Gaps = 9/191 (4%)
Query: 4 LEPLIAQIQGLSSSSADIARLHTILKHSDETLRSDSSRLYPILQLIDPSIHSLGFLYLLD 63
LE + ++ LS+ + +L ++ S E L + S L +L +D HSLG L +L
Sbjct: 5 LEQFVNSVRQLSAQG-QMTQLCELINKSGELLAKNLSHLDTVLGALDVQEHSLGVLAVL- 62
Query: 64 AFSAASVSNQQQAEEAVPIIARFINACSVEQIRLAPEKFVSVCKRLKDLVTLLEAPIRGV 123
F S+ + E + FI+ C+ E IR A + F +C +L + + + P+RG+
Sbjct: 63 -FVKFSMPSVPDFETLFSQVQLFISTCNGEHIRYATDTFAGLCHQLTNALVERKQPLRGI 121
Query: 124 APMFTALRKLQVSAEHLTPLHAEFLMLCLLAKCYKTGLSILDDDVFEVDHPRDLY----- 178
+ A+ K+Q++ LT +HA+ LCLLAKC+K L LD D+ ++ Y
Sbjct: 122 GVLRQAIDKMQMNTNQLTSVHADLCQLCLLAKCFKPALPYLDVDMVDICKENGAYDAKHF 181
Query: 179 -LYCYYGCVAF 188
Y YYG + +
Sbjct: 182 LCYYYYGGMVY 192
>emb|CAF96988.1| unnamed protein product [Tetraodon nigroviridis]
Length = 302
Score = 93.2 bits (230), Expect = 5e-18
Identities = 58/184 (31%), Positives = 93/184 (50%), Gaps = 7/184 (3%)
Query: 4 LEPLIAQIQGLSSSSADIARLHTILKHSDETLRSDSSRLYPILQLIDPSIHSLGFLYLLD 63
LE + ++ LS+ + +L ++ S E L + S L +L +D HSLG L +L
Sbjct: 5 LEQFVNNVRQLSAQG-QMTQLCELINKSGELLAKNLSHLDTVLGALDVQEHSLGVLAVL- 62
Query: 64 AFSAASVSNQQQAEEAVPIIARFINACSVEQIRLAPEKFVSVCKRLKDLVTLLEAPIRGV 123
F S+ N E + FI+ C+ E IR A + F +C +L + + + P+RGV
Sbjct: 63 -FVKFSMPNIPDFETLFSQVQLFISTCNGEHIRYATDTFAGLCHQLTNALVERKQPLRGV 121
Query: 124 APMFTALRKLQVSAEHLTPLHAEFLMLCLLAKCYKTGLSILDDDVFEVDHPRDLY----L 179
+ A+ K+Q++ LT +HA+ LCLLAKC+K L L+ D+ ++ Y
Sbjct: 122 VILKQAIDKMQMNTNQLTSVHADLCQLCLLAKCFKPALPFLELDMMDICKENGAYDAKHF 181
Query: 180 YCYY 183
CYY
Sbjct: 182 LCYY 185
>ref|NP_036121.1| COP9 (constitutive photomorphogenic), subunit 3 [Mus musculus]
gi|45829710|gb|AAH68179.1| COP9 (constitutive
photomorphogenic), subunit 3 [Mus musculus]
gi|55976220|sp|O88543|CSN3_MOUSE COP9 signalosome
complex subunit 3 (Signalosome subunit 3) (SGN3)
(JAB1-containing signalosome subunit 3)
gi|3309168|gb|AAC33900.1| COP9 complex subunit 3 [Mus
musculus]
Length = 423
Score = 92.8 bits (229), Expect = 7e-18
Identities = 57/184 (30%), Positives = 93/184 (49%), Gaps = 7/184 (3%)
Query: 4 LEPLIAQIQGLSSSSADIARLHTILKHSDETLRSDSSRLYPILQLIDPSIHSLGFLYLLD 63
LE + ++ LS+ + +L ++ S E L + S L +L +D HSLG L +L
Sbjct: 5 LEQFVNSVRQLSAQG-QMTQLCELINKSGELLAKNLSHLDTVLGALDVQEHSLGVLAVL- 62
Query: 64 AFSAASVSNQQQAEEAVPIIARFINACSVEQIRLAPEKFVSVCKRLKDLVTLLEAPIRGV 123
F S+ + E + FI+ C+ E IR A + F +C +L + + + P+RG+
Sbjct: 63 -FVKFSMPSVPDFETLFSQVQLFISTCNGEHIRYATDTFAGLCHQLTNALVERKQPLRGI 121
Query: 124 APMFTALRKLQVSAEHLTPLHAEFLMLCLLAKCYKTGLSILDDDVFEVDHPRDLY----L 179
+ A+ K+Q++ LT +HA+ LCLLAKC+K L LD D+ ++ Y
Sbjct: 122 GILKQAIDKMQMNTNQLTSVHADLCQLCLLAKCFKPALPYLDVDMMDICKENGAYDAKHF 181
Query: 180 YCYY 183
CYY
Sbjct: 182 LCYY 185
>ref|NP_001004200.1| COP9 constitutive photomorphogenic homolog subunit 3 [Rattus
norvegicus] gi|51260717|gb|AAH79143.1| COP9 constitutive
photomorphogenic homolog subunit 3 [Rattus norvegicus]
Length = 423
Score = 92.8 bits (229), Expect = 7e-18
Identities = 57/184 (30%), Positives = 93/184 (49%), Gaps = 7/184 (3%)
Query: 4 LEPLIAQIQGLSSSSADIARLHTILKHSDETLRSDSSRLYPILQLIDPSIHSLGFLYLLD 63
LE + ++ LS+ + +L ++ S E L + S L +L +D HSLG L +L
Sbjct: 5 LEQFVNSVRQLSAQG-QMTQLCELINKSGELLAKNLSHLDTVLGALDVQEHSLGVLAVL- 62
Query: 64 AFSAASVSNQQQAEEAVPIIARFINACSVEQIRLAPEKFVSVCKRLKDLVTLLEAPIRGV 123
F S+ + E + FI+ C+ E IR A + F +C +L + + + P+RG+
Sbjct: 63 -FVKFSMPSVPDFETLFSQVQLFISTCNGEHIRYATDTFAGLCHQLTNALVERKQPLRGI 121
Query: 124 APMFTALRKLQVSAEHLTPLHAEFLMLCLLAKCYKTGLSILDDDVFEVDHPRDLY----L 179
+ A+ K+Q++ LT +HA+ LCLLAKC+K L LD D+ ++ Y
Sbjct: 122 GILKQAIDKMQMNTNQLTSVHADLCQLCLLAKCFKPALPYLDVDMMDICKENGAYDAKHF 181
Query: 180 YCYY 183
CYY
Sbjct: 182 LCYY 185
>dbj|BAC33487.1| unnamed protein product [Mus musculus]
Length = 313
Score = 92.8 bits (229), Expect = 7e-18
Identities = 57/184 (30%), Positives = 93/184 (49%), Gaps = 7/184 (3%)
Query: 4 LEPLIAQIQGLSSSSADIARLHTILKHSDETLRSDSSRLYPILQLIDPSIHSLGFLYLLD 63
LE + ++ LS+ + +L ++ S E L + S L +L +D HSLG L +L
Sbjct: 5 LEQFVNSVRQLSAQG-QMTQLCELINKSGELLAKNLSHLDTVLGALDVQEHSLGVLAVL- 62
Query: 64 AFSAASVSNQQQAEEAVPIIARFINACSVEQIRLAPEKFVSVCKRLKDLVTLLEAPIRGV 123
F S+ + E + FI+ C+ E IR A + F +C +L + + + P+RG+
Sbjct: 63 -FVKFSMPSVPDFETLFSQVQLFISTCNGEHIRYATDTFAGLCHQLTNALVERKQPLRGI 121
Query: 124 APMFTALRKLQVSAEHLTPLHAEFLMLCLLAKCYKTGLSILDDDVFEVDHPRDLY----L 179
+ A+ K+Q++ LT +HA+ LCLLAKC+K L LD D+ ++ Y
Sbjct: 122 GILKQAIDKMQMNTNQLTSVHADLCQLCLLAKCFKPALPYLDVDMMDICKENGAYDAKHF 181
Query: 180 YCYY 183
CYY
Sbjct: 182 LCYY 185
>ref|NP_001017238.1| hypothetical protein LOC549992 [Xenopus tropicalis]
Length = 423
Score = 92.8 bits (229), Expect = 7e-18
Identities = 58/184 (31%), Positives = 94/184 (50%), Gaps = 7/184 (3%)
Query: 4 LEPLIAQIQGLSSSSADIARLHTILKHSDETLRSDSSRLYPILQLIDPSIHSLGFLYLLD 63
LE + ++ LSS + +L ++ S E L + S L +L +D HSLG L +L
Sbjct: 5 LEQFVNSVRQLSSQG-QMTQLCELINKSGELLAKNLSHLDTVLGALDVQEHSLGVLAVL- 62
Query: 64 AFSAASVSNQQQAEEAVPIIARFINACSVEQIRLAPEKFVSVCKRLKDLVTLLEAPIRGV 123
F S+ + E + FI+ C+ E IR A + F +C +L + + + P+RG+
Sbjct: 63 -FVKFSMPSIPDFETLFSQVQLFISTCNGEHIRYATDTFAGLCHQLTNALVERKQPLRGI 121
Query: 124 APMFTALRKLQVSAEHLTPLHAEFLMLCLLAKCYKTGLSILDDDVFEVDHPRDLY----L 179
+ A+ K+Q++A LT +HA+ L LLAKC+K L+ LD D+ ++ Y
Sbjct: 122 CVLRQAIDKMQMNANQLTSIHADLCQLSLLAKCFKPALAYLDVDMMDICKENGAYDAKPF 181
Query: 180 YCYY 183
CYY
Sbjct: 182 LCYY 185
>gb|AAH64288.1| COP9 constitutive photomorphogenic homolog subunit 3 [Danio rerio]
gi|55976433|sp|Q6P2U9|CSN3_BRARE COP9 signalosome
complex subunit 3 (Signalosome subunit 3)
Length = 423
Score = 91.7 bits (226), Expect = 2e-17
Identities = 56/184 (30%), Positives = 94/184 (50%), Gaps = 7/184 (3%)
Query: 4 LEPLIAQIQGLSSSSADIARLHTILKHSDETLRSDSSRLYPILQLIDPSIHSLGFLYLLD 63
LE + ++ LS+ + +L ++ S E L + S L +L +D HSLG L +L
Sbjct: 5 LEQFVNNVRQLSAQG-QMTQLCELINKSGELLAKNLSHLDTVLGALDIQEHSLGVLAVL- 62
Query: 64 AFSAASVSNQQQAEEAVPIIARFINACSVEQIRLAPEKFVSVCKRLKDLVTLLEAPIRGV 123
F S+ N E + FI+ C+ E IR A + F +C +L + + + P+RG+
Sbjct: 63 -FVKFSMPNIPDFETLFSQVQLFISTCNGEHIRYATDTFAGLCHQLTNALVERKQPLRGI 121
Query: 124 APMFTALRKLQVSAEHLTPLHAEFLMLCLLAKCYKTGLSILDDDVFEVDHPRDLY----L 179
+ + A+ K+Q++ LT +HA+ LCLLAKC+K + L+ D+ ++ Y
Sbjct: 122 SILKQAIDKMQMNTNQLTSVHADLCQLCLLAKCFKPAVPFLELDMMDICKENGAYDAKHF 181
Query: 180 YCYY 183
CYY
Sbjct: 182 LCYY 185
>gb|AAH45415.1| COP9 constitutive photomorphogenic homolog subunit 3 [Danio rerio]
gi|53292634|ref|NP_955979.1| COP9 constitutive
photomorphogenic homolog subunit 3 [Danio rerio]
Length = 423
Score = 91.7 bits (226), Expect = 2e-17
Identities = 56/184 (30%), Positives = 94/184 (50%), Gaps = 7/184 (3%)
Query: 4 LEPLIAQIQGLSSSSADIARLHTILKHSDETLRSDSSRLYPILQLIDPSIHSLGFLYLLD 63
LE + ++ LS+ + +L ++ S E L + S L +L +D HSLG L +L
Sbjct: 5 LEQFVNNVRQLSAQG-QMTQLCELINKSGELLSKNLSHLDTVLGALDIQEHSLGVLAVL- 62
Query: 64 AFSAASVSNQQQAEEAVPIIARFINACSVEQIRLAPEKFVSVCKRLKDLVTLLEAPIRGV 123
F S+ N E + FI+ C+ E IR A + F +C +L + + + P+RG+
Sbjct: 63 -FVKFSMPNIPDFETLFSQVQLFISTCNGEHIRYATDTFAGLCHQLTNALVERKQPLRGI 121
Query: 124 APMFTALRKLQVSAEHLTPLHAEFLMLCLLAKCYKTGLSILDDDVFEVDHPRDLY----L 179
+ + A+ K+Q++ LT +HA+ LCLLAKC+K + L+ D+ ++ Y
Sbjct: 122 SILKQAIDKMQMNTNQLTSVHADLCQLCLLAKCFKPAVPFLELDMMDICKENGAYDAKHF 181
Query: 180 YCYY 183
CYY
Sbjct: 182 LCYY 185
>gb|AAH95590.1| Unknown (protein for MGC:111903) [Danio rerio]
Length = 381
Score = 91.7 bits (226), Expect = 2e-17
Identities = 56/184 (30%), Positives = 94/184 (50%), Gaps = 7/184 (3%)
Query: 4 LEPLIAQIQGLSSSSADIARLHTILKHSDETLRSDSSRLYPILQLIDPSIHSLGFLYLLD 63
LE + ++ LS+ + +L ++ S E L + S L +L +D HSLG L +L
Sbjct: 5 LEQFVNNVRQLSAQG-QMTQLCELINKSGELLAKNLSHLDTVLGALDIQEHSLGVLAVL- 62
Query: 64 AFSAASVSNQQQAEEAVPIIARFINACSVEQIRLAPEKFVSVCKRLKDLVTLLEAPIRGV 123
F S+ N E + FI+ C+ E IR A + F +C +L + + + P+RG+
Sbjct: 63 -FVKFSMPNIPDFETLFSQVQLFISTCNGEHIRYATDTFAGLCHQLTNALVERKQPLRGI 121
Query: 124 APMFTALRKLQVSAEHLTPLHAEFLMLCLLAKCYKTGLSILDDDVFEVDHPRDLY----L 179
+ + A+ K+Q++ LT +HA+ LCLLAKC+K + L+ D+ ++ Y
Sbjct: 122 SILKQAIDKMQMNTNQLTSVHADLCQLCLLAKCFKPAVPFLELDMMDICKENGAYDAKHF 181
Query: 180 YCYY 183
CYY
Sbjct: 182 LCYY 185
>ref|XP_624761.1| PREDICTED: similar to COP9 constitutive photomorphogenic homolog
subunit 3 [Apis mellifera]
Length = 445
Score = 91.3 bits (225), Expect = 2e-17
Identities = 64/225 (28%), Positives = 107/225 (47%), Gaps = 18/225 (8%)
Query: 4 LEPLIAQIQGLSSSSADIARLHTILKHSDETLRSDSSRLYPILQLIDPSIHSLGFLYLLD 63
LE + ++ LS + L I+ S + L + L +L+ ++ HSLG L +L
Sbjct: 5 LELFVNNVRTLSKQG-NFRELSEIISKSTDVLIKNGQHLDNVLETLNLQEHSLGILAVLC 63
Query: 64 A-FSAASVSNQQQAEEAVPIIAR---FINACSVEQIRLAPEKFVSVCKRLKDLVTLLEAP 119
FS + + A+ P+ ++ FI C+ EQ+R A + + +C + L+ P
Sbjct: 64 VKFSLPNPNGANNADAYKPLFSQVQEFIIGCNGEQVRFASDIYAELCHLFTQTLVELQIP 123
Query: 120 IRGVAPMFTALRKLQVSAEHLTPLHAEFLMLCLLAKCYKTGLSILDDDVFEVDHPRDLY- 178
+RG+ + A+RK+Q+ LT +HA+ LCLL+KC+K L LD DV ++ +
Sbjct: 124 LRGIELLCRAIRKIQLFDSQLTSIHADLCQLCLLSKCFKPALEFLDIDVTGINQEEGQFD 183
Query: 179 -----LYCYYGCVAFFFSFELEKRDRC----RICSYISVLHFSYI 214
LY YYG + + L+ DR +C + SYI
Sbjct: 184 SKYFLLYYYYGGMIY---TALKNYDRALYFFEVCVTTPAMAVSYI 225
>gb|AAH44034.1| Cops3-prov protein [Xenopus laevis]
gi|55976497|sp|Q7ZTN8|CSN3_XENLA COP9 signalosome
complex subunit 3 (Signalosome subunit 3)
Length = 423
Score = 90.9 bits (224), Expect = 3e-17
Identities = 57/184 (30%), Positives = 93/184 (49%), Gaps = 7/184 (3%)
Query: 4 LEPLIAQIQGLSSSSADIARLHTILKHSDETLRSDSSRLYPILQLIDPSIHSLGFLYLLD 63
LE + ++ LSS + +L ++ S E L + S L +L +D HSLG L +L
Sbjct: 5 LEQFVNSVRQLSSQG-QMTQLCELINKSGELLAKNLSHLDTVLGALDVQEHSLGVLAVL- 62
Query: 64 AFSAASVSNQQQAEEAVPIIARFINACSVEQIRLAPEKFVSVCKRLKDLVTLLEAPIRGV 123
F S+ + E + FI+ C+ E IR A + F +C +L + + + P+RG+
Sbjct: 63 -FVKFSMPSIPDFETLFSQVQLFISTCNGEHIRYATDTFAGLCHQLTNALVERKQPLRGI 121
Query: 124 APMFTALRKLQVSAEHLTPLHAEFLMLCLLAKCYKTGLSILDDDVFEVDHPRDLY----L 179
+ A+ K+Q++A LT +H + L LLAKC+K L+ LD D+ ++ Y
Sbjct: 122 CVIRQAIDKMQMNANQLTSIHGDLCQLSLLAKCFKPALAYLDVDMMDICKENGAYDAKPF 181
Query: 180 YCYY 183
CYY
Sbjct: 182 LCYY 185
>ref|XP_536667.1| PREDICTED: similar to COP9 constitutive photomorphogenic homolog
subunit 3 [Canis familiaris]
Length = 414
Score = 90.5 bits (223), Expect = 3e-17
Identities = 53/167 (31%), Positives = 86/167 (50%), Gaps = 6/167 (3%)
Query: 21 IARLHTILKHSDETLRSDSSRLYPILQLIDPSIHSLGFLYLLDAFSAASVSNQQQAEEAV 80
+ +L ++ S E L + S L +L +D HSLG L +L F S+ + E
Sbjct: 12 MTQLCELINKSGELLAKNLSHLDTVLGALDVQEHSLGVLAVL--FVKFSMPSVPDFETLF 69
Query: 81 PIIARFINACSVEQIRLAPEKFVSVCKRLKDLVTLLEAPIRGVAPMFTALRKLQVSAEHL 140
+ FI+ C+ E IR A + F +C +L + + + P+RG++ + A+ K+Q++ L
Sbjct: 70 SQVQLFISTCNGEHIRYATDTFAGLCHQLTNALVERKQPLRGISILKQAIDKMQMNTNQL 129
Query: 141 TPLHAEFLMLCLLAKCYKTGLSILDDDVFEVDHPRDLY----LYCYY 183
T +HA+ LCLLAKC+K L LD D+ ++ Y CYY
Sbjct: 130 TSIHADLCQLCLLAKCFKPALPYLDVDMMDICKENGAYDAKHFLCYY 176
Database: nr
Posted date: Jul 5, 2005 12:34 AM
Number of letters in database: 863,360,394
Number of sequences in database: 2,540,612
Lambda K H
0.329 0.142 0.425
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 368,873,133
Number of Sequences: 2540612
Number of extensions: 14301714
Number of successful extensions: 41823
Number of sequences better than 10.0: 45
Number of HSP's better than 10.0 without gapping: 30
Number of HSP's successfully gapped in prelim test: 15
Number of HSP's that attempted gapping in prelim test: 41779
Number of HSP's gapped (non-prelim): 45
length of query: 235
length of database: 863,360,394
effective HSP length: 124
effective length of query: 111
effective length of database: 548,324,506
effective search space: 60864020166
effective search space used: 60864020166
T: 11
A: 40
X1: 15 ( 7.1 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 40 (21.8 bits)
S2: 73 (32.7 bits)
Medicago: description of AC138464.5


