

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC138130.5 + phase: 0 /pseudo
(152 letters)
Database: nr
2,540,612 sequences; 863,360,394 total letters
Searching..................................................done
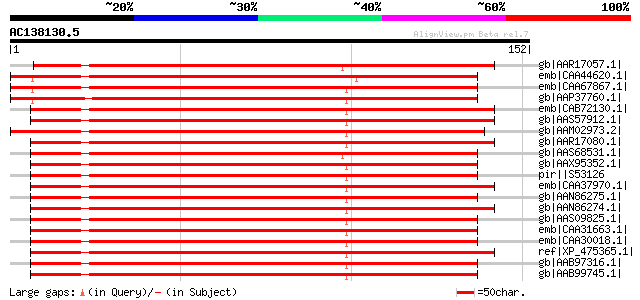
Score E
Sequences producing significant alignments: (bits) Value
gb|AAR17057.1| HSP70 [Vorticella campanula] 191 2e-48
emb|CAA44620.1| Heat Shock 70kD protein [Glycine max] gi|123601|... 191 3e-48
emb|CAA67867.1| heat shock protein hsp70 [Pisum sativum] gi|1076... 190 5e-48
gb|AAP37760.1| At1g16030 [Arabidopsis thaliana] gi|15219109|ref|... 189 9e-48
emb|CAB72130.1| heat shock protein 70 [Cucumis sativus] 189 2e-47
gb|AAS57912.1| 70 kDa heat shock cognate protein 1 [Vigna radiata] 188 2e-47
gb|AAM02973.2| Hsp70 [Crypthecodinium cohnii] 188 3e-47
gb|AAR17080.1| heat shock protein 70-3 [Nicotiana tabacum] 187 6e-47
gb|AAS68531.1| HSP70 [Carchesium polypinum] 187 6e-47
gb|AAX95352.1| dnaK-type molecular chaperone hsp70 - rice (fragm... 186 8e-47
pir||S53126 dnaK-type molecular chaperone hsp70 - rice (fragment) 186 8e-47
emb|CAA37970.1| heat shock protein cognate 70 [Lycopersicon escu... 186 1e-46
gb|AAN86275.1| non-cell-autonomous heat shock cognate protein 70... 186 1e-46
gb|AAN86274.1| non-cell-autonomous heat shock cognate protein 70... 186 1e-46
gb|AAS09825.1| heat shock cognate protein 70 [Thellungiella halo... 186 1e-46
emb|CAA31663.1| hsp70 (AA 6 - 651) [Petunia x hybrida] 186 1e-46
emb|CAA30018.1| heat shock protein 70 [Petunia x hybrida] gi|123... 186 1e-46
ref|XP_475365.1| putative hsp70 [Oryza sativa (japonica cultivar... 186 1e-46
gb|AAB97316.1| cytosolic heat shock 70 protein; HSC70-3 [Spinaci... 186 1e-46
gb|AAB99745.1| HSP70 [Triticum aestivum] 186 1e-46
>gb|AAR17057.1| HSP70 [Vorticella campanula]
Length = 145
Score = 191 bits (486), Expect = 2e-48
Identities = 95/136 (69%), Positives = 111/136 (80%), Gaps = 3/136 (2%)
Query: 8 GIDLGTTYSCIAVWLDENRRVEIIHNDQGNRTTPSFVSFTKDQRLIGDAAKGQATSDPEN 67
GIDLGTTYSC+ +W +N RVEII NDQGNRTTPS+V+FT+ +RLIGDAAK Q +P N
Sbjct: 1 GIDLGTTYSCVGIW--QNERVEIIANDQGNRTTPSYVAFTETERLIGDAAKNQTAKNPIN 58
Query: 68 TVFDAKRLIGRKFSDSVVQNDIMLWPFRV-IGVDDKPMIIVKYKGQEKRFCPEEISSMIL 126
TVFDAKRLIGRKF+D VVQ DI LWPF+V G DDKP+I+VKYK Q K+F PEEISSM+L
Sbjct: 59 TVFDAKRLIGRKFADPVVQKDIKLWPFKVESGPDDKPVIVVKYKNQTKKFHPEEISSMVL 118
Query: 127 TKMREVAEKFLMSPRK 142
TKM+E AE F+ P K
Sbjct: 119 TKMKETAEAFVGEPIK 134
>emb|CAA44620.1| Heat Shock 70kD protein [Glycine max]
gi|123601|sp|P26413|HSP70_SOYBN Heat shock 70 kDa
protein
Length = 645
Score = 191 bits (485), Expect = 3e-48
Identities = 98/140 (70%), Positives = 113/140 (80%), Gaps = 5/140 (3%)
Query: 1 MMTKS--VVGIDLGTTYSCIAVWLDENRRVEIIHNDQGNRTTPSFVSFTKDQRLIGDAAK 58
M TK +GIDLGTTYSC+ VW +N RVEII NDQGNRTTPS+V+FT +RLIGDAAK
Sbjct: 1 MATKEGKAIGIDLGTTYSCVGVW--QNDRVEIIPNDQGNRTTPSYVAFTDTERLIGDAAK 58
Query: 59 GQATSDPENTVFDAKRLIGRKFSDSVVQNDIMLWPFRVIGVD-DKPMIIVKYKGQEKRFC 117
Q +P+NTVFDAKRLIGR+FSDS VQND+ LWPF+V G DKPMI+V YKG+EK+F
Sbjct: 59 NQVAMNPQNTVFDAKRLIGRRFSDSSVQNDMKLWPFKVGGSPCDKPMIVVNYKGEEKKFS 118
Query: 118 PEEISSMILTKMREVAEKFL 137
EEISSM+L KMREVAE FL
Sbjct: 119 AEEISSMVLVKMREVAEAFL 138
>emb|CAA67867.1| heat shock protein hsp70 [Pisum sativum] gi|1076529|pir||S53498
dnaK-type molecular chaperone HSP71.2 - garden pea
gi|562006|gb|AAA82975.1| PsHSP71.2
Length = 648
Score = 190 bits (483), Expect = 5e-48
Identities = 96/140 (68%), Positives = 113/140 (80%), Gaps = 5/140 (3%)
Query: 1 MMTKS--VVGIDLGTTYSCIAVWLDENRRVEIIHNDQGNRTTPSFVSFTKDQRLIGDAAK 58
M TK +GIDLGTTYSC+ VW +N RVEII NDQGNRTTPS+V+FT +RLIGDAAK
Sbjct: 1 MATKEGKAIGIDLGTTYSCVGVW--QNDRVEIIPNDQGNRTTPSYVAFTDTERLIGDAAK 58
Query: 59 GQATSDPENTVFDAKRLIGRKFSDSVVQNDIMLWPFRVI-GVDDKPMIIVKYKGQEKRFC 117
Q +P+NTVFDAKRLIGR+FSD VQND+ LWPF+V+ G +KPMI+V YKG+EK+F
Sbjct: 59 NQVAMNPQNTVFDAKRLIGRRFSDESVQNDMKLWPFKVVPGPAEKPMIVVNYKGEEKKFA 118
Query: 118 PEEISSMILTKMREVAEKFL 137
EEISSM+L KMREVAE FL
Sbjct: 119 AEEISSMVLIKMREVAEAFL 138
>gb|AAP37760.1| At1g16030 [Arabidopsis thaliana] gi|15219109|ref|NP_173055.1| heat
shock protein 70, putative / HSP70, putative
[Arabidopsis thaliana] gi|6587810|gb|AAF18501.1|
Identical to gb|AJ002551 heat shock protein 70 from
Arabidopsis thaliana and contains a PF|00012 HSP 70
domain. EST gb|F13893 comes from this gene
gi|25082767|gb|AAN71999.1| heat shock protein hsp70,
putative [Arabidopsis thaliana] gi|25295976|pir||B86295
hypothetical protein T24D18.14 [imported] - Arabidopsis
thaliana
Length = 646
Score = 189 bits (481), Expect = 9e-48
Identities = 96/140 (68%), Positives = 114/140 (80%), Gaps = 5/140 (3%)
Query: 1 MMTKS--VVGIDLGTTYSCIAVWLDENRRVEIIHNDQGNRTTPSFVSFTKDQRLIGDAAK 58
M TKS +GIDLGTTYSC+ VW+ N RVEII NDQGNRTTPS+V+FT +RLIGDAAK
Sbjct: 1 MATKSEKAIGIDLGTTYSCVGVWM--NDRVEIIPNDQGNRTTPSYVAFTDTERLIGDAAK 58
Query: 59 GQATSDPENTVFDAKRLIGRKFSDSVVQNDIMLWPFRVI-GVDDKPMIIVKYKGQEKRFC 117
Q +P+NTVFDAKRLIGRKFSD VQ+DI+ WPF+V+ G +KPMI+V YK +EK+F
Sbjct: 59 NQVALNPQNTVFDAKRLIGRKFSDPSVQSDILHWPFKVVSGPGEKPMIVVSYKNEEKQFS 118
Query: 118 PEEISSMILTKMREVAEKFL 137
PEEISSM+L KM+EVAE FL
Sbjct: 119 PEEISSMVLVKMKEVAEAFL 138
>emb|CAB72130.1| heat shock protein 70 [Cucumis sativus]
Length = 647
Score = 189 bits (479), Expect = 2e-47
Identities = 97/137 (70%), Positives = 112/137 (80%), Gaps = 3/137 (2%)
Query: 7 VGIDLGTTYSCIAVWLDENRRVEIIHNDQGNRTTPSFVSFTKDQRLIGDAAKGQATSDPE 66
+GIDLGTTYSC+ VW ++ RVEII NDQGNRTTPS+V+FT +RLIGDAAK Q T +P
Sbjct: 10 IGIDLGTTYSCVGVW--QHDRVEIIANDQGNRTTPSYVAFTDTERLIGDAAKNQVTMNPI 67
Query: 67 NTVFDAKRLIGRKFSDSVVQNDIMLWPFRVI-GVDDKPMIIVKYKGQEKRFCPEEISSMI 125
NTVFDAKRLIGR+FSDS VQ+DI LWPF+VI G DKPMIIV YKG EK+F EEISSM+
Sbjct: 68 NTVFDAKRLIGRRFSDSSVQSDIKLWPFKVIPGPGDKPMIIVSYKGGEKQFAAEEISSMV 127
Query: 126 LTKMREVAEKFLMSPRK 142
L KMRE+AE +L S K
Sbjct: 128 LIKMREIAEAYLGSTVK 144
>gb|AAS57912.1| 70 kDa heat shock cognate protein 1 [Vigna radiata]
Length = 649
Score = 188 bits (478), Expect = 2e-47
Identities = 94/137 (68%), Positives = 112/137 (81%), Gaps = 3/137 (2%)
Query: 7 VGIDLGTTYSCIAVWLDENRRVEIIHNDQGNRTTPSFVSFTKDQRLIGDAAKGQATSDPE 66
+GIDLGTTYSC+ VW ++ RVEII NDQGNRTTPS+V+FT +RLIGDAAK Q +P
Sbjct: 10 IGIDLGTTYSCVGVW--QHDRVEIIANDQGNRTTPSYVAFTDSERLIGDAAKNQVAMNPT 67
Query: 67 NTVFDAKRLIGRKFSDSVVQNDIMLWPFRVI-GVDDKPMIIVKYKGQEKRFCPEEISSMI 125
NTVFDAKRLIGR+FSD+ VQ+DI LWPF+VI G DKPMI+V YKG+EK+F EEISSM+
Sbjct: 68 NTVFDAKRLIGRRFSDASVQSDIKLWPFKVISGAGDKPMIVVNYKGEEKQFSAEEISSMV 127
Query: 126 LTKMREVAEKFLMSPRK 142
L KM+E+AE FL S K
Sbjct: 128 LIKMKEIAEAFLGSTVK 144
>gb|AAM02973.2| Hsp70 [Crypthecodinium cohnii]
Length = 647
Score = 188 bits (477), Expect = 3e-47
Identities = 95/140 (67%), Positives = 112/140 (79%), Gaps = 3/140 (2%)
Query: 1 MMTKSVVGIDLGTTYSCIAVWLDENRRVEIIHNDQGNRTTPSFVSFTKDQRLIGDAAKGQ 60
M K+ +GIDLGTTYSC+ VW +N VEII NDQGNRTTPS+V+FT +RLIGDAAK Q
Sbjct: 1 MAKKTAIGIDLGTTYSCVGVW--KNDGVEIIANDQGNRTTPSYVAFTDTERLIGDAAKNQ 58
Query: 61 ATSDPENTVFDAKRLIGRKFSDSVVQNDIMLWPFRVI-GVDDKPMIIVKYKGQEKRFCPE 119
+PENTVFDAKRLIGRKF+D +VQ DI LWPF+VI G DKPMI+V +G+EK+F PE
Sbjct: 59 VARNPENTVFDAKRLIGRKFADPIVQADIKLWPFKVIAGAGDKPMIVVNAQGEEKKFHPE 118
Query: 120 EISSMILTKMREVAEKFLMS 139
EISSMIL KM+E AE +L S
Sbjct: 119 EISSMILLKMKETAEAYLGS 138
>gb|AAR17080.1| heat shock protein 70-3 [Nicotiana tabacum]
Length = 648
Score = 187 bits (474), Expect = 6e-47
Identities = 94/137 (68%), Positives = 111/137 (80%), Gaps = 3/137 (2%)
Query: 7 VGIDLGTTYSCIAVWLDENRRVEIIHNDQGNRTTPSFVSFTKDQRLIGDAAKGQATSDPE 66
+GIDLGTTYSC+ VW ++ RVEII NDQGNRTTPS+V FT +RLIGDAAK Q +P
Sbjct: 10 IGIDLGTTYSCVGVW--QHDRVEIIANDQGNRTTPSYVGFTDSERLIGDAAKNQVAMNPI 67
Query: 67 NTVFDAKRLIGRKFSDSVVQNDIMLWPFRVI-GVDDKPMIIVKYKGQEKRFCPEEISSMI 125
NTVFDAKRLIGR+FSD+ VQ+DI LWPF+VI G DKPMI+V YKG+EK+F EEISSM+
Sbjct: 68 NTVFDAKRLIGRRFSDASVQSDIKLWPFKVISGPGDKPMIVVNYKGEEKQFAAEEISSMV 127
Query: 126 LTKMREVAEKFLMSPRK 142
L KM+E+AE FL S K
Sbjct: 128 LIKMKEIAEAFLGSTVK 144
>gb|AAS68531.1| HSP70 [Carchesium polypinum]
Length = 146
Score = 187 bits (474), Expect = 6e-47
Identities = 89/132 (67%), Positives = 110/132 (82%), Gaps = 3/132 (2%)
Query: 7 VGIDLGTTYSCIAVWLDENRRVEIIHNDQGNRTTPSFVSFTKDQRLIGDAAKGQATSDPE 66
+GIDLGTTYSC+ +W +N RV+II NDQGNRTTPS+V+FT +RLIGDAAK Q +P
Sbjct: 1 IGIDLGTTYSCVGIW--QNERVQIIANDQGNRTTPSYVAFTDTERLIGDAAKNQTAKNPI 58
Query: 67 NTVFDAKRLIGRKFSDSVVQNDIMLWPFRV-IGVDDKPMIIVKYKGQEKRFCPEEISSMI 125
NTVFDAKRLIGRKF+D +VQ DI LWPF+V G DDKP+I+VK+KG+ K+F PEEISSM+
Sbjct: 59 NTVFDAKRLIGRKFNDPIVQKDIKLWPFKVESGPDDKPVIVVKFKGETKKFHPEEISSMV 118
Query: 126 LTKMREVAEKFL 137
LTKM++ A+ FL
Sbjct: 119 LTKMKQTAQSFL 130
>gb|AAX95352.1| dnaK-type molecular chaperone hsp70 - rice (fragment) [Oryza sativa
(japonica cultivar-group)]
Length = 649
Score = 186 bits (473), Expect = 8e-47
Identities = 92/132 (69%), Positives = 110/132 (82%), Gaps = 3/132 (2%)
Query: 7 VGIDLGTTYSCIAVWLDENRRVEIIHNDQGNRTTPSFVSFTKDQRLIGDAAKGQATSDPE 66
+GIDLGTTYSC+ VW ++ RVEII NDQGNRTTPS+V FT +RLIGDAAK Q +P
Sbjct: 10 IGIDLGTTYSCVGVW--QHDRVEIIANDQGNRTTPSYVGFTDSERLIGDAAKNQVAMNPI 67
Query: 67 NTVFDAKRLIGRKFSDSVVQNDIMLWPFRVI-GVDDKPMIIVKYKGQEKRFCPEEISSMI 125
NTVFDAKRLIGR+FSD+ VQ+DI LWPF+VI G DKPMI+V+YKG+EK+F EEISSM+
Sbjct: 68 NTVFDAKRLIGRRFSDASVQSDIKLWPFKVIAGPGDKPMIVVQYKGEEKQFAAEEISSMV 127
Query: 126 LTKMREVAEKFL 137
L KMRE+AE +L
Sbjct: 128 LIKMREIAEAYL 139
>pir||S53126 dnaK-type molecular chaperone hsp70 - rice (fragment)
Length = 649
Score = 186 bits (473), Expect = 8e-47
Identities = 92/132 (69%), Positives = 110/132 (82%), Gaps = 3/132 (2%)
Query: 7 VGIDLGTTYSCIAVWLDENRRVEIIHNDQGNRTTPSFVSFTKDQRLIGDAAKGQATSDPE 66
+GIDLGTTYSC+ VW ++ RVEII NDQGNRTTPS+V FT +RLIGDAAK Q +P
Sbjct: 10 IGIDLGTTYSCVGVW--QHDRVEIIANDQGNRTTPSYVGFTDSERLIGDAAKNQVAMNPI 67
Query: 67 NTVFDAKRLIGRKFSDSVVQNDIMLWPFRVI-GVDDKPMIIVKYKGQEKRFCPEEISSMI 125
NTVFDAKRLIGR+FSD+ VQ+DI LWPF+VI G DKPMI+V+YKG+EK+F EEISSM+
Sbjct: 68 NTVFDAKRLIGRRFSDASVQSDIKLWPFKVIAGPGDKPMIVVQYKGEEKQFAAEEISSMV 127
Query: 126 LTKMREVAEKFL 137
L KMRE+AE +L
Sbjct: 128 LIKMREIAEAYL 139
>emb|CAA37970.1| heat shock protein cognate 70 [Lycopersicon esculentum]
gi|123613|sp|P24629|HSP71_LYCES Heat shock cognate 70
kDa protein 1
Length = 650
Score = 186 bits (472), Expect = 1e-46
Identities = 94/137 (68%), Positives = 110/137 (79%), Gaps = 3/137 (2%)
Query: 7 VGIDLGTTYSCIAVWLDENRRVEIIHNDQGNRTTPSFVSFTKDQRLIGDAAKGQATSDPE 66
+GIDLGTTYSC+ VW ++ RVEII NDQGNRTTPS+V FT +RLIGDAAK Q +P
Sbjct: 10 IGIDLGTTYSCVGVW--QHDRVEIIANDQGNRTTPSYVGFTDTERLIGDAAKNQVALNPI 67
Query: 67 NTVFDAKRLIGRKFSDSVVQNDIMLWPFRVI-GVDDKPMIIVKYKGQEKRFCPEEISSMI 125
NTVFDAKRLIGR+FSD+ VQ D+ LWPF+VI G DKPMI+V YKG+EK F EEISSM+
Sbjct: 68 NTVFDAKRLIGRRFSDASVQEDMKLWPFKVIPGPGDKPMIVVTYKGEEKEFAAEEISSMV 127
Query: 126 LTKMREVAEKFLMSPRK 142
LTKM+E+AE FL S K
Sbjct: 128 LTKMKEIAEAFLGSTVK 144
>gb|AAN86275.1| non-cell-autonomous heat shock cognate protein 70 [Cucurbita
maxima]
Length = 652
Score = 186 bits (472), Expect = 1e-46
Identities = 91/132 (68%), Positives = 111/132 (83%), Gaps = 3/132 (2%)
Query: 7 VGIDLGTTYSCIAVWLDENRRVEIIHNDQGNRTTPSFVSFTKDQRLIGDAAKGQATSDPE 66
+GIDLGTTYSC+ VW ++ RVEII NDQGNRTTPS+V+FT ++RLIGDAAK Q +P
Sbjct: 10 IGIDLGTTYSCVGVW--QHDRVEIIANDQGNRTTPSYVAFTDNERLIGDAAKNQVAMNPI 67
Query: 67 NTVFDAKRLIGRKFSDSVVQNDIMLWPFRVI-GVDDKPMIIVKYKGQEKRFCPEEISSMI 125
NTVFDAKRLIGR+FSDS VQ+DI LWPF+VI G DKPMI+V YKG+EK+F EE+SSM+
Sbjct: 68 NTVFDAKRLIGRRFSDSSVQSDIKLWPFKVIAGPGDKPMIVVNYKGEEKQFSAEEVSSMV 127
Query: 126 LTKMREVAEKFL 137
L KM+E+AE +L
Sbjct: 128 LIKMKEIAEAYL 139
>gb|AAN86274.1| non-cell-autonomous heat shock cognate protein 70 [Cucurbita
maxima]
Length = 647
Score = 186 bits (472), Expect = 1e-46
Identities = 93/137 (67%), Positives = 112/137 (80%), Gaps = 3/137 (2%)
Query: 7 VGIDLGTTYSCIAVWLDENRRVEIIHNDQGNRTTPSFVSFTKDQRLIGDAAKGQATSDPE 66
+GIDLGTTYSC+ VW ++ RVEII NDQGNRTTPS+V+FT +RLIGDAAK Q +P
Sbjct: 10 IGIDLGTTYSCVGVW--QHDRVEIIANDQGNRTTPSYVAFTDSERLIGDAAKNQVAMNPI 67
Query: 67 NTVFDAKRLIGRKFSDSVVQNDIMLWPFRVI-GVDDKPMIIVKYKGQEKRFCPEEISSMI 125
NTVFDAKRLIGR+FSD+ VQ+DI LWPF+VI G DKPMI+V YKG++K+F EEISSM+
Sbjct: 68 NTVFDAKRLIGRRFSDAPVQSDIKLWPFKVIAGPGDKPMIVVSYKGEDKQFAAEEISSMV 127
Query: 126 LTKMREVAEKFLMSPRK 142
L KMRE+AE +L S K
Sbjct: 128 LIKMREIAEAYLGSTVK 144
>gb|AAS09825.1| heat shock cognate protein 70 [Thellungiella halophila]
Length = 651
Score = 186 bits (472), Expect = 1e-46
Identities = 92/132 (69%), Positives = 110/132 (82%), Gaps = 3/132 (2%)
Query: 7 VGIDLGTTYSCIAVWLDENRRVEIIHNDQGNRTTPSFVSFTKDQRLIGDAAKGQATSDPE 66
+GIDLGTTYSC+ VW ++ RVEII NDQGNRTTPS+V+FT +RLIGDAAK Q +P
Sbjct: 10 IGIDLGTTYSCVGVW--QHDRVEIIANDQGNRTTPSYVAFTDSERLIGDAAKNQVAMNPI 67
Query: 67 NTVFDAKRLIGRKFSDSVVQNDIMLWPFRVI-GVDDKPMIIVKYKGQEKRFCPEEISSMI 125
NTVFDAKRLIGR+FSDS VQ+D+ LWPF++I G DKPMIIV YKG+EK+F EEISSM+
Sbjct: 68 NTVFDAKRLIGRRFSDSSVQSDMKLWPFKIIPGPADKPMIIVNYKGEEKQFAAEEISSMV 127
Query: 126 LTKMREVAEKFL 137
L KMRE+AE +L
Sbjct: 128 LIKMREIAEAYL 139
>emb|CAA31663.1| hsp70 (AA 6 - 651) [Petunia x hybrida]
Length = 646
Score = 186 bits (471), Expect = 1e-46
Identities = 92/132 (69%), Positives = 109/132 (81%), Gaps = 3/132 (2%)
Query: 7 VGIDLGTTYSCIAVWLDENRRVEIIHNDQGNRTTPSFVSFTKDQRLIGDAAKGQATSDPE 66
+GIDLGTTYSC+ VW ++ RVEII NDQGNRTTPS+V FT +RLIGDAAK Q +P
Sbjct: 5 IGIDLGTTYSCVGVW--QHDRVEIIANDQGNRTTPSYVGFTDTERLIGDAAKNQVAMNPI 62
Query: 67 NTVFDAKRLIGRKFSDSVVQNDIMLWPFRVI-GVDDKPMIIVKYKGQEKRFCPEEISSMI 125
NTVFDAKRLIGR+FSD VQ+DI LWPF+VI G DKPMI+V YKG+EK+F EEISSM+
Sbjct: 63 NTVFDAKRLIGRRFSDPSVQSDIKLWPFKVIPGPGDKPMIVVTYKGEEKQFAAEEISSMV 122
Query: 126 LTKMREVAEKFL 137
LTKM+E+AE +L
Sbjct: 123 LTKMKEIAEAYL 134
>emb|CAA30018.1| heat shock protein 70 [Petunia x hybrida]
gi|123650|sp|P09189|HSP7C_PETHY Heat shock cognate 70
kDa protein
Length = 651
Score = 186 bits (471), Expect = 1e-46
Identities = 92/132 (69%), Positives = 109/132 (81%), Gaps = 3/132 (2%)
Query: 7 VGIDLGTTYSCIAVWLDENRRVEIIHNDQGNRTTPSFVSFTKDQRLIGDAAKGQATSDPE 66
+GIDLGTTYSC+ VW ++ RVEII NDQGNRTTPS+V FT +RLIGDAAK Q +P
Sbjct: 10 IGIDLGTTYSCVGVW--QHDRVEIIANDQGNRTTPSYVGFTDTERLIGDAAKNQVAMNPI 67
Query: 67 NTVFDAKRLIGRKFSDSVVQNDIMLWPFRVI-GVDDKPMIIVKYKGQEKRFCPEEISSMI 125
NTVFDAKRLIGR+FSD VQ+DI LWPF+VI G DKPMI+V YKG+EK+F EEISSM+
Sbjct: 68 NTVFDAKRLIGRRFSDPSVQSDIKLWPFKVIPGPGDKPMIVVTYKGEEKQFAAEEISSMV 127
Query: 126 LTKMREVAEKFL 137
LTKM+E+AE +L
Sbjct: 128 LTKMKEIAEAYL 139
>ref|XP_475365.1| putative hsp70 [Oryza sativa (japonica cultivar-group)]
gi|47900318|gb|AAT39165.1| putative hsp70 [Oryza sativa
(japonica cultivar-group)]
Length = 646
Score = 186 bits (471), Expect = 1e-46
Identities = 92/137 (67%), Positives = 112/137 (81%), Gaps = 3/137 (2%)
Query: 7 VGIDLGTTYSCIAVWLDENRRVEIIHNDQGNRTTPSFVSFTKDQRLIGDAAKGQATSDPE 66
+GIDLGTTYSC+ VW ++ RVEII NDQGNRTTPS+V+FT +RLIGDAAK Q +P
Sbjct: 9 IGIDLGTTYSCVGVW--QHDRVEIIANDQGNRTTPSYVAFTDTERLIGDAAKNQVAMNPT 66
Query: 67 NTVFDAKRLIGRKFSDSVVQNDIMLWPFRVI-GVDDKPMIIVKYKGQEKRFCPEEISSMI 125
NTVFDAKRLIGR+FSD VQ+D+ LWPF+V+ G DKPMI+V+YKG+EK+F EEISSM+
Sbjct: 67 NTVFDAKRLIGRRFSDPSVQSDMKLWPFKVVPGPGDKPMIVVQYKGEEKQFAAEEISSMV 126
Query: 126 LTKMREVAEKFLMSPRK 142
L KMRE+AE +L S K
Sbjct: 127 LIKMREIAEAYLGSSIK 143
>gb|AAB97316.1| cytosolic heat shock 70 protein; HSC70-3 [Spinacia oleracea]
gi|2660770|gb|AAB88133.1| cytosolic heat shock 70
protein [Spinacia oleracea] gi|2660768|gb|AAB88132.1|
cytosolic heat shock 70 protein [Spinacia oleracea]
gi|11277116|pir||T45517 heat shock protein 70, cytosolic
[imported] - spinach
Length = 651
Score = 186 bits (471), Expect = 1e-46
Identities = 91/132 (68%), Positives = 109/132 (81%), Gaps = 3/132 (2%)
Query: 7 VGIDLGTTYSCIAVWLDENRRVEIIHNDQGNRTTPSFVSFTKDQRLIGDAAKGQATSDPE 66
+GIDLGTTYSC+ VW ++ RVEII NDQGNRTTPS+V+FT +RLIGDAAK Q +P
Sbjct: 10 IGIDLGTTYSCVGVW--QHDRVEIIANDQGNRTTPSYVAFTDSERLIGDAAKNQVAMNPI 67
Query: 67 NTVFDAKRLIGRKFSDSVVQNDIMLWPFRVI-GVDDKPMIIVKYKGQEKRFCPEEISSMI 125
NTVFDAKRLIGR+FSD+ VQ+D+ LWPF+VI G DKPMI+V YKG++K F EEISSM+
Sbjct: 68 NTVFDAKRLIGRRFSDASVQSDMKLWPFKVIPGAGDKPMIVVSYKGEDKEFAAEEISSMV 127
Query: 126 LTKMREVAEKFL 137
L KMRE+AE FL
Sbjct: 128 LIKMREIAEAFL 139
>gb|AAB99745.1| HSP70 [Triticum aestivum]
Length = 648
Score = 186 bits (471), Expect = 1e-46
Identities = 92/132 (69%), Positives = 109/132 (81%), Gaps = 3/132 (2%)
Query: 7 VGIDLGTTYSCIAVWLDENRRVEIIHNDQGNRTTPSFVSFTKDQRLIGDAAKGQATSDPE 66
+GIDLGTTYSC+ VW ++ RVEII NDQGNRTTPS+V+FT +RLIGDAAK Q +P
Sbjct: 9 IGIDLGTTYSCVGVW--QHDRVEIIANDQGNRTTPSYVAFTDTERLIGDAAKNQVAMNPT 66
Query: 67 NTVFDAKRLIGRKFSDSVVQNDIMLWPFRVI-GVDDKPMIIVKYKGQEKRFCPEEISSMI 125
NTVFDAKRLIGR+FSD VQ+D+ LWPF+VI G DKPMI+V YKG+EK+F EEISSM+
Sbjct: 67 NTVFDAKRLIGRRFSDPSVQSDMKLWPFKVIPGPADKPMIVVNYKGEEKQFAAEEISSMV 126
Query: 126 LTKMREVAEKFL 137
L KMRE+AE FL
Sbjct: 127 LIKMREIAEAFL 138
Database: nr
Posted date: Jul 5, 2005 12:34 AM
Number of letters in database: 863,360,394
Number of sequences in database: 2,540,612
Lambda K H
0.322 0.137 0.408
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 248,498,101
Number of Sequences: 2540612
Number of extensions: 9338952
Number of successful extensions: 21804
Number of sequences better than 10.0: 2051
Number of HSP's better than 10.0 without gapping: 1913
Number of HSP's successfully gapped in prelim test: 138
Number of HSP's that attempted gapping in prelim test: 17149
Number of HSP's gapped (non-prelim): 2096
length of query: 152
length of database: 863,360,394
effective HSP length: 128
effective length of query: 24
effective length of database: 538,162,058
effective search space: 12915889392
effective search space used: 12915889392
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.9 bits)
S2: 67 (30.4 bits)
Medicago: description of AC138130.5


