

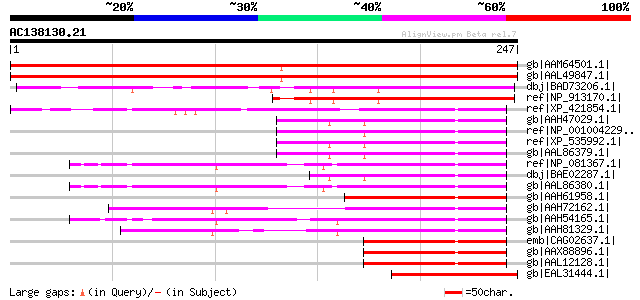
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC138130.21 - phase: 0 /pseudo
(247 letters)
Database: nr
2,540,612 sequences; 863,360,394 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
gb|AAM64501.1| unknown [Arabidopsis thaliana] gi|9279713|dbj|BAB... 374 e-102
gb|AAL49847.1| unknown protein [Arabidopsis thaliana] 374 e-102
dbj|BAD73206.1| unknown protein [Oryza sativa (japonica cultivar... 167 3e-40
ref|NP_913170.1| B1015E06.6 [Oryza sativa (japonica cultivar-gro... 153 4e-36
ref|XP_421854.1| PREDICTED: similar to mitotic phosphoprotein 44... 66 8e-10
gb|AAH47029.1| NUP35 protein [Homo sapiens] gi|21717634|gb|AAM76... 62 2e-08
ref|NP_001004229.1| mitotic phosphoprotein 44 [Rattus norvegicus... 62 2e-08
ref|XP_535992.1| PREDICTED: hypothetical protein XP_535992 [Cani... 62 2e-08
gb|AAL86379.1| mitotic phosphoprotein 44 [Homo sapiens] 62 2e-08
ref|NP_081367.1| nucleoporin 35 [Mus musculus] gi|29179550|gb|AA... 61 3e-08
dbj|BAE02287.1| unnamed protein product [Macaca fascicularis] 61 3e-08
gb|AAL86380.1| mitotic phosphoprotein 44 [Mus musculus] 61 3e-08
gb|AAH61958.1| Unknown (protein for MGC:65979) [Danio rerio] gi|... 60 4e-08
gb|AAH72162.1| MGC80197 protein [Xenopus laevis] 60 5e-08
gb|AAH54165.1| MGC64281 protein [Xenopus laevis] 60 5e-08
gb|AAH81329.1| MGC89479 protein [Xenopus tropicalis] gi|56118408... 60 7e-08
emb|CAG02637.1| unnamed protein product [Tetraodon nigroviridis] 60 7e-08
gb|AAX88896.1| unknown [Homo sapiens] 59 1e-07
gb|AAL12128.1| mitotic phosphoprotein 44 [Xenopus laevis] 58 2e-07
gb|EAL31444.1| GA19671-PA [Drosophila pseudoobscura] 58 2e-07
>gb|AAM64501.1| unknown [Arabidopsis thaliana] gi|9279713|dbj|BAB01270.1| unnamed
protein product [Arabidopsis thaliana]
gi|26983840|gb|AAN86172.1| unknown protein [Arabidopsis
thaliana] gi|18401087|ref|NP_566542.1| mitotic
phosphoprotein N' end (MPPN) family protein [Arabidopsis
thaliana] gi|2062172|gb|AAB63646.1| unknown protein
[Arabidopsis thaliana]
Length = 329
Score = 374 bits (959), Expect = e-102
Identities = 181/248 (72%), Positives = 204/248 (81%), Gaps = 1/248 (0%)
Query: 1 MSTTAQRTPRSGRQSLFFQDLASPVNARRGKFSSPGQSGASSSLWRENFGNSDLPPPPFF 60
MS A RTP+SGRQSL FQDLASPV+ARRGKFSSPGQ+ A S+LWRENFG SDLPPPP +
Sbjct: 1 MSAAAHRTPKSGRQSLLFQDLASPVSARRGKFSSPGQAAAVSALWRENFGGSDLPPPPMY 60
Query: 61 TLEDRSDFSPESGILDYQMSPETKSDHKTPIQTPNREFSTPAKGKSEASTSYALRGVQQN 120
TL+DRSDFSPESGI DY SP+ KSD +TP Q+ + TP KGK EAS S++L QQ+
Sbjct: 61 TLDDRSDFSPESGIADYSASPDAKSDRRTPFQSSGKNIVTPGKGKLEASPSFSLLNAQQS 120
Query: 121 QQGSLSLNWWS-SPAKSGGEQEEKGRGSPVEGVVQPGALITLPPPPEVARPEVQRNSLPA 179
QQ S S +WWS S A S EQ++KG+GSPVEGVVQPGAL+TLPPP EVARPEVQR +P
Sbjct: 121 QQVSGSPSWWSQSKAGSSTEQDDKGKGSPVEGVVQPGALVTLPPPREVARPEVQRQIIPT 180
Query: 180 GNLNEEEWVTVYGFSPNDTNLVLREFEKCGEILKHVPGPRDANWMHILYQNRSAAQNALN 239
GNL+EEEWVTVYGFSP DTNLVLREFEKCG +LKHVPGPR+ANWMHILYQNRS A ALN
Sbjct: 181 GNLDEEEWVTVYGFSPGDTNLVLREFEKCGMVLKHVPGPRNANWMHILYQNRSDAHKALN 240
Query: 240 KNGIQING 247
K G+ ING
Sbjct: 241 KAGMMING 248
>gb|AAL49847.1| unknown protein [Arabidopsis thaliana]
Length = 329
Score = 374 bits (959), Expect = e-102
Identities = 181/248 (72%), Positives = 204/248 (81%), Gaps = 1/248 (0%)
Query: 1 MSTTAQRTPRSGRQSLFFQDLASPVNARRGKFSSPGQSGASSSLWRENFGNSDLPPPPFF 60
MS A RTP+SGRQSL FQDLASPV+ARRGKFSSPGQ+ A S+LWRENFG SDLPPPP +
Sbjct: 1 MSAAAHRTPKSGRQSLLFQDLASPVSARRGKFSSPGQAAAVSALWRENFGGSDLPPPPMY 60
Query: 61 TLEDRSDFSPESGILDYQMSPETKSDHKTPIQTPNREFSTPAKGKSEASTSYALRGVQQN 120
TL+DRSDFSPESGI DY SP+ KSD +TP Q+ + TP KGK EAS S++L QQ+
Sbjct: 61 TLDDRSDFSPESGIADYSASPDAKSDRRTPFQSSGKNIVTPGKGKLEASPSFSLLNAQQS 120
Query: 121 QQGSLSLNWWS-SPAKSGGEQEEKGRGSPVEGVVQPGALITLPPPPEVARPEVQRNSLPA 179
QQ S S +WWS S A S EQ++KG+GSPVEGVVQPGAL+TLPPP EVARPEVQR +P
Sbjct: 121 QQVSGSPSWWSQSKAGSSTEQDDKGKGSPVEGVVQPGALVTLPPPREVARPEVQRQIIPT 180
Query: 180 GNLNEEEWVTVYGFSPNDTNLVLREFEKCGEILKHVPGPRDANWMHILYQNRSAAQNALN 239
GNL+EEEWVTVYGFSP DTNLVLREFEKCG +LKHVPGPR+ANWMHILYQNRS A ALN
Sbjct: 181 GNLDEEEWVTVYGFSPGDTNLVLREFEKCGMVLKHVPGPRNANWMHILYQNRSDAHKALN 240
Query: 240 KNGIQING 247
K G+ ING
Sbjct: 241 KAGMMING 248
>dbj|BAD73206.1| unknown protein [Oryza sativa (japonica cultivar-group)]
Length = 327
Score = 167 bits (422), Expect = 3e-40
Identities = 117/266 (43%), Positives = 140/266 (51%), Gaps = 55/266 (20%)
Query: 4 TAQRTPRSGRQSLFFQDLASPVNARRGKFSSPGQSGASSSLWRENFGNSDLPPPP--FFT 61
+A R F +DL+SP+ S +SL RE G++ PPPP
Sbjct: 2 SASRPDHRRHHPPFLRDLSSPI--------SSAVRLPPASLRRETQGSTTPPPPPPPLLF 53
Query: 62 LEDRSDFSPESGILDYQMSPETKSDHKTPIQTPNREFSTPAKGKSEASTSYALRGVQQNQ 121
L+D S SP SP TP Q S P + +S LR
Sbjct: 54 LDDLSHHSP---------SPRPA----TPPQAAAMSPSPPPPHRGGLFSSTPLRS----- 95
Query: 122 QGSLS-LNWWSSPAKSGGEQEEKGR-GSPVEGVVQPG------------------ALITL 161
GS S WWSS +EE R GSPV+GVVQP LITL
Sbjct: 96 NGSPSPAAWWSS------SREEMAREGSPVDGVVQPQQQPSPTTASGQQSQQQKVTLITL 149
Query: 162 PPPPEVARPEVQRNSLP-AGNLNEEEWVTVYGFSPNDTNLVLREFEKCGEILKHVPGPRD 220
PPP EVARPE+ ++S P AG ++EEEWVTV+GF P DTNLVLREFEKCG +L+HVPGPRD
Sbjct: 150 PPPREVARPEMPKDSTPSAGRVDEEEWVTVFGFLPGDTNLVLREFEKCGIVLRHVPGPRD 209
Query: 221 ANWMHILYQNRSAAQNALNKNGIQIN 246
ANWMHILYQ+R AQ AL K+G Q+N
Sbjct: 210 ANWMHILYQSRHDAQKALTKHGQQLN 235
>ref|NP_913170.1| B1015E06.6 [Oryza sativa (japonica cultivar-group)]
Length = 439
Score = 153 bits (387), Expect = 4e-36
Identities = 82/138 (59%), Positives = 95/138 (68%), Gaps = 26/138 (18%)
Query: 129 WWSSPAKSGGEQEEKGR-GSPVEGVVQPG------------------ALITLPPPPEVAR 169
WWSS +EE R GSPV+GVVQP LITLPPP EVAR
Sbjct: 30 WWSS------SREEMAREGSPVDGVVQPQQQPSPTTASGQQSQQQKVTLITLPPPREVAR 83
Query: 170 PEVQRNSLP-AGNLNEEEWVTVYGFSPNDTNLVLREFEKCGEILKHVPGPRDANWMHILY 228
PE+ ++S P AG ++EEEWVTV+GF P DTNLVLREFEKCG +L+HVPGPRDANWMHILY
Sbjct: 84 PEMPKDSTPSAGRVDEEEWVTVFGFLPGDTNLVLREFEKCGIVLRHVPGPRDANWMHILY 143
Query: 229 QNRSAAQNALNKNGIQIN 246
Q+R AQ AL K+G Q+N
Sbjct: 144 QSRHDAQKALTKHGQQLN 161
>ref|XP_421854.1| PREDICTED: similar to mitotic phosphoprotein 44 [Gallus gallus]
Length = 325
Score = 66.2 bits (160), Expect = 8e-10
Identities = 69/255 (27%), Positives = 107/255 (41%), Gaps = 40/255 (15%)
Query: 1 MSTTAQRTPRSGRQSLFFQDLASPVNARRGKFSSPGQSGASSSLWRENFGNSDLPPPPFF 60
M+ A P +G + + L SP + + G +S+ + F DLP P
Sbjct: 1 MACFAMDPPPAGAEPM---TLGSPTSPKPG----------ASAQFLPGFLMGDLPAP--V 45
Query: 61 TLEDRSDFSPESGILDYQM------SPETK--SDHKT-----PIQTPNREFSTPAKGKSE 107
T + R+ P G +D + SP HK P+++ E S+P G S
Sbjct: 46 TPQPRALSGPAVGAMDVRSPLLAGGSPPQPVVPTHKDKSGAPPVRSIYDELSSPGLGSSP 105
Query: 108 ASTSYALRGVQQNQQGSLSLNWWSSPAKSGGEQEEKGRGSPVEGVVQPGALITLPPPPEV 167
++ A G Q L N S+P + G P + + P L
Sbjct: 106 LTSRRA--GSFSLTQSPLVGNMPSTPGTASSMFSPASIGQPRKTTLSPAQLDPF------ 157
Query: 168 ARPEVQRNSLPAGNLNEEEWVTVYGFSPNDTNLVLREFEKCGEILKHVPGPRDANWMHIL 227
Q +SL + + ++ WVTV+GF + +L +F + G ILKHV NWMHI
Sbjct: 158 ---YTQGDSLTSEDQLDDTWVTVFGFPQASASYILLQFAQYGNILKHVMS-NTGNWMHIR 213
Query: 228 YQNRSAAQNALNKNG 242
YQ++ A+ AL+K+G
Sbjct: 214 YQSKLQARKALSKDG 228
>gb|AAH47029.1| NUP35 protein [Homo sapiens] gi|21717634|gb|AAM76704.1| nucleoporin
Nup35 [Homo sapiens] gi|38197538|gb|AAH61698.1|
Nucleoporin 35kDa [Homo sapiens]
gi|31982904|ref|NP_612142.2| nucleoporin 35kDa isoform a
[Homo sapiens]
Length = 326
Score = 61.6 bits (148), Expect = 2e-08
Identities = 40/126 (31%), Positives = 62/126 (48%), Gaps = 15/126 (11%)
Query: 131 SSPAKSGGEQEEKGRGSPVEGVVQ-PGALITLPPPPEVARPE-------------VQRNS 176
S+P S + SP+ GV PG ++ P + +P Q +S
Sbjct: 105 STPLTSRRQPNISVMQSPLVGVTSTPGTGQSMFSPASIGQPRKTTLSPAQLDPFYTQGDS 164
Query: 177 LPAGNLNEEEWVTVYGFSPNDTNLVLREFEKCGEILKHVPGPRDANWMHILYQNRSAAQN 236
L + + ++ WVTV+GF + +L +F + G ILKHV NWMHI YQ++ A+
Sbjct: 165 LTSEDHLDDSWVTVFGFPQASASYILLQFAQYGNILKHVMS-NTGNWMHIRYQSKLQARK 223
Query: 237 ALNKNG 242
AL+K+G
Sbjct: 224 ALSKDG 229
>ref|NP_001004229.1| mitotic phosphoprotein 44 [Rattus norvegicus]
gi|51260703|gb|AAH78914.1| Mitotic phosphoprotein 44
[Rattus norvegicus]
Length = 325
Score = 61.6 bits (148), Expect = 2e-08
Identities = 39/125 (31%), Positives = 61/125 (48%), Gaps = 14/125 (11%)
Query: 131 SSPAKSGGEQEEKGRGSPVEGVVQPGALITLPPPPEVARPE-------------VQRNSL 177
S+P S + SP+ GV P ++ P + +P Q +SL
Sbjct: 105 STPLPSRRQANISVLQSPLVGVTTPVTGQSMFSPANIGQPRKTTLSPAQLDPFYTQGDSL 164
Query: 178 PAGNLNEEEWVTVYGFSPNDTNLVLREFEKCGEILKHVPGPRDANWMHILYQNRSAAQNA 237
+ + ++ WVTV+GF + +L +F + G ILKHV NWMHI YQ++ A+ A
Sbjct: 165 TSEDHLDDTWVTVFGFPQASASYILLQFAQYGNILKHVMS-NTGNWMHIRYQSKLQARKA 223
Query: 238 LNKNG 242
L+K+G
Sbjct: 224 LSKDG 228
>ref|XP_535992.1| PREDICTED: hypothetical protein XP_535992 [Canis familiaris]
Length = 401
Score = 61.6 bits (148), Expect = 2e-08
Identities = 41/129 (31%), Positives = 64/129 (48%), Gaps = 18/129 (13%)
Query: 131 SSPAKSGGEQEEKGRGSPVEGVVQ----PGALITLPPPPEVARPE-------------VQ 173
S+P S + SP+ GV+ PGA ++ P + +P Q
Sbjct: 177 STPLTSRRQPNISVMQSPLVGVMTTTPTPGAGQSMFSPANIGQPRKTTLSPAQLDPFYTQ 236
Query: 174 RNSLPAGNLNEEEWVTVYGFSPNDTNLVLREFEKCGEILKHVPGPRDANWMHILYQNRSA 233
+SL + + ++ WVTV+GF + +L +F + G ILKHV NWMHI YQ++
Sbjct: 237 GDSLTSEDHLDDTWVTVFGFPQASASYILLQFAQYGNILKHVMS-NTGNWMHIRYQSKLQ 295
Query: 234 AQNALNKNG 242
A+ AL+K+G
Sbjct: 296 ARKALSKDG 304
>gb|AAL86379.1| mitotic phosphoprotein 44 [Homo sapiens]
Length = 326
Score = 61.6 bits (148), Expect = 2e-08
Identities = 40/126 (31%), Positives = 62/126 (48%), Gaps = 15/126 (11%)
Query: 131 SSPAKSGGEQEEKGRGSPVEGVVQ-PGALITLPPPPEVARPE-------------VQRNS 176
S+P S + SP+ GV PG ++ P + +P Q +S
Sbjct: 105 STPLTSRRQPNISVMQSPLVGVTSTPGTGQSMFSPASIGQPRKTTLSPAQLDPFYTQGDS 164
Query: 177 LPAGNLNEEEWVTVYGFSPNDTNLVLREFEKCGEILKHVPGPRDANWMHILYQNRSAAQN 236
L + + ++ WVTV+GF + +L +F + G ILKHV NWMHI YQ++ A+
Sbjct: 165 LTSEDHLDDSWVTVFGFPQASASYILLQFAQYGNILKHVMS-NTGNWMHIRYQSKLQARK 223
Query: 237 ALNKNG 242
AL+K+G
Sbjct: 224 ALSKDG 229
>ref|NP_081367.1| nucleoporin 35 [Mus musculus] gi|29179550|gb|AAH48814.1|
Nucleoporin 35 [Mus musculus]
gi|26354815|dbj|BAC41034.1| unnamed protein product [Mus
musculus] gi|12843826|dbj|BAB26128.1| unnamed protein
product [Mus musculus]
Length = 325
Score = 60.8 bits (146), Expect = 3e-08
Identities = 59/223 (26%), Positives = 100/223 (44%), Gaps = 25/223 (11%)
Query: 30 GKFSSPGQSGASSSLWRENFGNSDLPPPPFFTLEDRSDFSPESGILDYQMSPETKSDHKT 89
G +SP ++GA++ + F DLP P T + RS P G+++ +
Sbjct: 21 GSPTSP-KTGANAQ-FLPGFLMGDLPAP--VTPQPRSISGPSVGVMEMRSPLLAGGSPPQ 76
Query: 90 PIQTPNREFS--TPAKGKSEASTSYALRGVQQNQQGSLSLNWWSSPAKSGGEQEEKGRGS 147
P+ +++ S P + + +S L + +++ SP G +
Sbjct: 77 PVVPAHKDKSGAPPVRSIYDDISSPGLGSTPLTSRRQANISLLQSPLV--------GATT 128
Query: 148 PVEG--------VVQPGALITLPPPPEVARPEVQRNSLPAGNLNEEEWVTVYGFSPNDTN 199
PV G + QP T P ++ Q +SL + + ++ WVTV+GF +
Sbjct: 129 PVPGQSMFSPANIGQPRK--TTLSPAQLDPFYTQGDSLTSEDHLDDTWVTVFGFPQASAS 186
Query: 200 LVLREFEKCGEILKHVPGPRDANWMHILYQNRSAAQNALNKNG 242
+L +F + G ILKHV NWMHI YQ++ A+ AL+K+G
Sbjct: 187 YILLQFAQYGNILKHVMS-NTGNWMHIRYQSKLQARKALSKDG 228
>dbj|BAE02287.1| unnamed protein product [Macaca fascicularis]
Length = 208
Score = 60.8 bits (146), Expect = 3e-08
Identities = 37/110 (33%), Positives = 57/110 (51%), Gaps = 15/110 (13%)
Query: 147 SPVEGVVQ-PGALITLPPPPEVARPE-------------VQRNSLPAGNLNEEEWVTVYG 192
SP+ GV PG ++ P + +P Q +SL + + ++ WVTV+G
Sbjct: 3 SPLVGVTSTPGTGQSMFSPASIGQPRKTTLSPAQLDPFYTQGDSLTSEDHLDDSWVTVFG 62
Query: 193 FSPNDTNLVLREFEKCGEILKHVPGPRDANWMHILYQNRSAAQNALNKNG 242
F + +L +F + G ILKHV NWMHI YQ++ A+ AL+K+G
Sbjct: 63 FPQASASYILLQFAQYGNILKHVMS-NTGNWMHIRYQSKLQARKALSKDG 111
>gb|AAL86380.1| mitotic phosphoprotein 44 [Mus musculus]
Length = 325
Score = 60.8 bits (146), Expect = 3e-08
Identities = 59/223 (26%), Positives = 100/223 (44%), Gaps = 25/223 (11%)
Query: 30 GKFSSPGQSGASSSLWRENFGNSDLPPPPFFTLEDRSDFSPESGILDYQMSPETKSDHKT 89
G +SP ++GA++ + F DLP P T + RS P G+++ +
Sbjct: 21 GSPTSP-KTGANAQ-FLPGFLMGDLPAP--VTPQPRSISGPSVGVMEMRSPLLAGGSPPQ 76
Query: 90 PIQTPNREFS--TPAKGKSEASTSYALRGVQQNQQGSLSLNWWSSPAKSGGEQEEKGRGS 147
P+ +++ S P + + +S L + +++ SP G +
Sbjct: 77 PVVPAHKDKSGAPPVRSIYDDISSPGLGSTPLTSRRQANISLLQSPLV--------GATT 128
Query: 148 PVEG--------VVQPGALITLPPPPEVARPEVQRNSLPAGNLNEEEWVTVYGFSPNDTN 199
PV G + QP T P ++ Q +SL + + ++ WVTV+GF +
Sbjct: 129 PVPGQSMFSPANIGQPRK--TTLSPAQLDPFYTQGDSLTSEDHLDDTWVTVFGFPQASAS 186
Query: 200 LVLREFEKCGEILKHVPGPRDANWMHILYQNRSAAQNALNKNG 242
+L +F + G ILKHV NWMHI YQ++ A+ AL+K+G
Sbjct: 187 YILLQFAQYGNILKHVMS-NTGNWMHIRYQSKLQARKALSKDG 228
>gb|AAH61958.1| Unknown (protein for MGC:65979) [Danio rerio]
gi|41056043|ref|NP_956360.1| Unknown (protein for
MGC:65979) [Danio rerio]
Length = 308
Score = 60.5 bits (145), Expect = 4e-08
Identities = 29/79 (36%), Positives = 48/79 (60%), Gaps = 1/79 (1%)
Query: 164 PPEVARPEVQRNSLPAGNLNEEEWVTVYGFSPNDTNLVLREFEKCGEILKHVPGPRDANW 223
P +V Q ++L + + ++ W+TV+GF P + +L +F + G ILKHV NW
Sbjct: 137 PAQVDPFFTQGDALSSEDQLDDTWITVFGFPPASASYILLQFAQYGNILKHVMS-NTGNW 195
Query: 224 MHILYQNRSAAQNALNKNG 242
MH+ YQ++ A+ AL+K+G
Sbjct: 196 MHVQYQSKLQARKALSKDG 214
>gb|AAH72162.1| MGC80197 protein [Xenopus laevis]
Length = 318
Score = 60.1 bits (144), Expect = 5e-08
Identities = 52/198 (26%), Positives = 81/198 (40%), Gaps = 42/198 (21%)
Query: 49 FGNSDLPPPPFFTLEDRSDFSPESGILDYQMSPETKSDHKTPIQTPNRE-FSTPAKG--- 104
F P P T +D+S P I D SP S +T N TP G
Sbjct: 61 FSGGSPPQPVVPTHKDKSGAPPVRSIYDDIASPGLGSTPLNARKTANFSVLHTPLSGVMP 120
Query: 105 KSEASTSYALRGVQQNQQGSLSLNWWSSPAKSGGEQEEKGRGSPVEGVVQPGALITLPPP 164
S AS+ ++ + Q ++ +LS P
Sbjct: 121 SSPASSVFSPATIGQKRKNTLS-------------------------------------P 143
Query: 165 PEVARPEVQRNSLPAGNLNEEEWVTVYGFSPNDTNLVLREFEKCGEILKHVPGPRDANWM 224
++ Q ++L + + ++ WVTVYGF + +L +F + G I+KHV + NWM
Sbjct: 144 AQMDPFYTQGDALTSDDQLDDTWVTVYGFPQASASYILLQFAQYGNIIKHVMS-NNGNWM 202
Query: 225 HILYQNRSAAQNALNKNG 242
HI YQ++ A+ AL+K+G
Sbjct: 203 HIQYQSKLQARKALSKDG 220
>gb|AAH54165.1| MGC64281 protein [Xenopus laevis]
Length = 285
Score = 60.1 bits (144), Expect = 5e-08
Identities = 55/219 (25%), Positives = 94/219 (42%), Gaps = 21/219 (9%)
Query: 30 GKFSSPGQSGASSSLWRENFGNSDLPPPPFFTLEDRSDFSPESGILDYQMSPETKSDHKT 89
G +SP S + L F D+P P T + R P GI++ + +
Sbjct: 19 GSPTSPKPSAGAQFL--PGFLLGDIPTP--VTPQPR----PSLGIMEVRSPLHSGGSPPQ 70
Query: 90 PIQTPNREFS--TPAKGKSEASTSYALRGVQQNQQGSLSLNWWSSPAKSGGEQEEKGRGS 147
P+ +++ S P + + S L +N + S + +P S
Sbjct: 71 PVLPTHKDKSGAPPVRSIYDDVASPGLGSTPRNTRKMASFSVLHTPLSGAIPS------S 124
Query: 148 PVEGVVQPGAL----ITLPPPPEVARPEVQRNSLPAGNLNEEEWVTVYGFSPNDTNLVLR 203
P V P + T P ++ Q ++L + + ++ WVTV+GF + +L
Sbjct: 125 PASNVFSPATIGQSRKTTLSPAQMDPFYTQGDALTSDDQLDDTWVTVFGFPQASASYILL 184
Query: 204 EFEKCGEILKHVPGPRDANWMHILYQNRSAAQNALNKNG 242
+F + G I+KHV + NWMHI YQ++ A+ AL+K+G
Sbjct: 185 QFAQYGNIIKHVMS-NNGNWMHIQYQSKLQARKALSKDG 222
>gb|AAH81329.1| MGC89479 protein [Xenopus tropicalis]
gi|56118408|ref|NP_001008124.1| MGC89479 protein
[Xenopus tropicalis]
Length = 330
Score = 59.7 bits (143), Expect = 7e-08
Identities = 53/193 (27%), Positives = 80/193 (40%), Gaps = 32/193 (16%)
Query: 55 PPPPFFTLEDRSDFSPESGILDYQMSPETKSDHKTPIQTPNRE-FSTPAKGKSEASTSYA 113
P P T +D+S P I D SP S P +T + TP G +S
Sbjct: 67 PQPVVPTHKDKSGAPPVRSIYDDIASPGLGSTPLNPRKTASFSVLHTPLSGVIPSSPEC- 125
Query: 114 LRGVQQNQQGSLSLNWWSSPAKSGGEQEEKGRGSPVEGVVQPGAL----ITLPPPPEVAR 169
+N GS R P V P + T P ++
Sbjct: 126 -----KNISGS--------------------RKRPASSVFSPATIGHSRKTTLSPAQMDP 160
Query: 170 PEVQRNSLPAGNLNEEEWVTVYGFSPNDTNLVLREFEKCGEILKHVPGPRDANWMHILYQ 229
Q ++L + + ++ WVTV+GF + +L +F + G I+KHV + NWMHI YQ
Sbjct: 161 FYTQGDALTSDDQLDDTWVTVFGFPQASASYILLQFAQYGNIIKHVMS-NNGNWMHIQYQ 219
Query: 230 NRSAAQNALNKNG 242
++ A+ AL+K+G
Sbjct: 220 SKLQARKALSKDG 232
>emb|CAG02637.1| unnamed protein product [Tetraodon nigroviridis]
Length = 293
Score = 59.7 bits (143), Expect = 7e-08
Identities = 28/70 (40%), Positives = 43/70 (61%), Gaps = 1/70 (1%)
Query: 173 QRNSLPAGNLNEEEWVTVYGFSPNDTNLVLREFEKCGEILKHVPGPRDANWMHILYQNRS 232
Q SL + + ++ WVTV+GF P + +L +F + G ILKH NWMH+ YQ++
Sbjct: 140 QGESLSSDDRLDQSWVTVFGFPPASASYILLQFAQYGNILKHTMA-SPGNWMHLQYQSKL 198
Query: 233 AAQNALNKNG 242
A+ AL+K+G
Sbjct: 199 QARKALSKDG 208
>gb|AAX88896.1| unknown [Homo sapiens]
Length = 193
Score = 58.9 bits (141), Expect = 1e-07
Identities = 29/70 (41%), Positives = 44/70 (62%), Gaps = 1/70 (1%)
Query: 173 QRNSLPAGNLNEEEWVTVYGFSPNDTNLVLREFEKCGEILKHVPGPRDANWMHILYQNRS 232
Q +SL + + ++ WVTV+GF + +L +F + G ILKHV NWMHI YQ++
Sbjct: 28 QGDSLTSEDHLDDSWVTVFGFPQASASYILLQFAQYGNILKHVMS-NTGNWMHIRYQSKL 86
Query: 233 AAQNALNKNG 242
A+ AL+K+G
Sbjct: 87 QARKALSKDG 96
>gb|AAL12128.1| mitotic phosphoprotein 44 [Xenopus laevis]
Length = 318
Score = 58.2 bits (139), Expect = 2e-07
Identities = 27/70 (38%), Positives = 45/70 (63%), Gaps = 1/70 (1%)
Query: 173 QRNSLPAGNLNEEEWVTVYGFSPNDTNLVLREFEKCGEILKHVPGPRDANWMHILYQNRS 232
Q ++L + + ++ WVTV+GF + +L +F + G I+KHV + NWMHI YQ++
Sbjct: 152 QGDALTSDDQLDDTWVTVFGFPQASASYILLQFAQYGNIIKHVMS-NNGNWMHIQYQSKL 210
Query: 233 AAQNALNKNG 242
A+ AL+K+G
Sbjct: 211 QARKALSKDG 220
>gb|EAL31444.1| GA19671-PA [Drosophila pseudoobscura]
Length = 330
Score = 58.2 bits (139), Expect = 2e-07
Identities = 23/61 (37%), Positives = 39/61 (63%)
Query: 187 WVTVYGFSPNDTNLVLREFEKCGEILKHVPGPRDANWMHILYQNRSAAQNALNKNGIQIN 246
WVT++G+ P ++++VL+ F CG I++ P++ NWMH+ + +R + ALN N I
Sbjct: 187 WVTIFGYVPGNSSMVLQHFTLCGTIMEVSHAPKNGNWMHVRFSSRIESDKALNFNEKVIG 246
Query: 247 G 247
G
Sbjct: 247 G 247
Database: nr
Posted date: Jul 5, 2005 12:34 AM
Number of letters in database: 863,360,394
Number of sequences in database: 2,540,612
Lambda K H
0.310 0.129 0.385
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 510,905,482
Number of Sequences: 2540612
Number of extensions: 24856996
Number of successful extensions: 53954
Number of sequences better than 10.0: 143
Number of HSP's better than 10.0 without gapping: 32
Number of HSP's successfully gapped in prelim test: 113
Number of HSP's that attempted gapping in prelim test: 53827
Number of HSP's gapped (non-prelim): 180
length of query: 247
length of database: 863,360,394
effective HSP length: 125
effective length of query: 122
effective length of database: 545,783,894
effective search space: 66585635068
effective search space used: 66585635068
T: 11
A: 40
X1: 16 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (21.7 bits)
S2: 73 (32.7 bits)
Medicago: description of AC138130.21


