

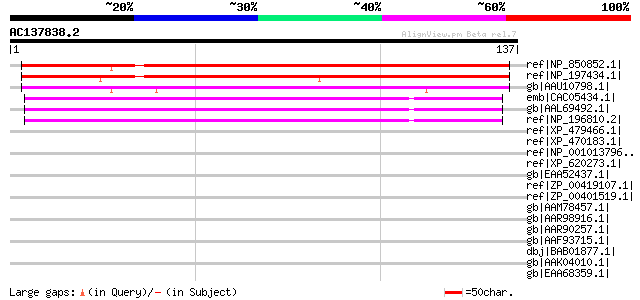
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC137838.2 - phase: 0
(137 letters)
Database: nr
2,540,612 sequences; 863,360,394 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
ref|NP_850852.1| armadillo/beta-catenin repeat family protein / ... 157 5e-38
ref|NP_197434.1| armadillo/beta-catenin repeat family protein / ... 152 1e-36
gb|AAU10798.1| hypothetical protein [Oryza sativa (japonica cult... 130 5e-30
emb|CAC05434.1| putative protein [Arabidopsis thaliana] 79 2e-14
gb|AAL69492.1| unknown protein [Arabidopsis thaliana] 79 2e-14
ref|NP_196810.2| armadillo/beta-catenin repeat family protein / ... 79 2e-14
ref|XP_479466.1| hypothetical protein [Oryza sativa (japonica cu... 39 0.021
ref|XP_470183.1| Putative AMP-binding protein [Oryza sativa (jap... 38 0.060
ref|NP_001013796.1| expressed sequence AW146299 [Mus musculus] g... 35 0.39
ref|XP_620273.1| PREDICTED: expressed sequence AW146299 [Mus mus... 35 0.39
gb|EAA52437.1| hypothetical protein MG05129.4 [Magnaporthe grise... 34 0.67
ref|ZP_00419107.1| type II and III secretion system protein:NolW... 34 0.87
ref|ZP_00401519.1| Glycosyl transferase, family 3:Glycosyl trans... 33 1.1
gb|AAM78457.1| nonribosomal peptide synthetase; NRPS [Hypocrea v... 33 1.1
gb|AAR98916.1| peptide synthetase [Trichoderma asperellum] 33 1.5
gb|AAR90257.1| polyketide synthase [Cochliobolus heterostrophus] 33 1.9
gb|AAF93715.1| aspartokinase, alpha and beta subunits [Vibrio ch... 32 2.5
dbj|BAB01877.1| unnamed protein product [Arabidopsis thaliana] g... 32 2.5
gb|AAK04010.1| unknown [Pasteurella multocida subsp. multocida s... 32 2.5
gb|EAA68359.1| hypothetical protein FG01658.1 [Gibberella zeae P... 32 3.3
>ref|NP_850852.1| armadillo/beta-catenin repeat family protein / BTB/POZ
domain-containing protein [Arabidopsis thaliana]
Length = 710
Score = 157 bits (397), Expect = 5e-38
Identities = 85/134 (63%), Positives = 102/134 (75%), Gaps = 4/134 (2%)
Query: 4 QRRQGHCLSERKGQKRKLDEELP--EDRQISSAPPTADERAALLVEVANQVTVLESTFTW 61
+RR+G ERKGQKRKL+E EDR+IS+ + D ALL EVA QV+VL S F+W
Sbjct: 6 ERREGRSFPERKGQKRKLEEGAAAVEDREISAV--STDGGQALLSEVAAQVSVLNSAFSW 63
Query: 62 NEADRAAAKRATHALADLAKNEEVVNVIVEGGAIPALIKHLQAPPVTDCVQKPLPFEHEV 121
E+DRAAAKRAT LA+LAKNE++VNVIV+GGA+PAL+ HLQAPP D P+EHEV
Sbjct: 64 QESDRAAAKRATQVLAELAKNEDLVNVIVDGGAVPALMTHLQAPPYNDGDLAEKPYEHEV 123
Query: 122 EKGSAFALGLLAVK 135
EKGSAFALGLLA+K
Sbjct: 124 EKGSAFALGLLAIK 137
>ref|NP_197434.1| armadillo/beta-catenin repeat family protein / BTB/POZ
domain-containing protein [Arabidopsis thaliana]
Length = 636
Score = 152 bits (385), Expect = 1e-36
Identities = 85/135 (62%), Positives = 102/135 (74%), Gaps = 5/135 (3%)
Query: 4 QRRQGHCLSERKGQKRKLDE--ELPEDRQISSAPPTADERAALLVEVANQVTVLESTFTW 61
+RR+G ERKGQKRKL+E EDR+IS+ + D ALL EVA QV+VL S F+W
Sbjct: 6 ERREGRSFPERKGQKRKLEEGAAAVEDREISAV--STDGGQALLSEVAAQVSVLNSAFSW 63
Query: 62 NEADRAAAKRATHALADLAKN-EEVVNVIVEGGAIPALIKHLQAPPVTDCVQKPLPFEHE 120
E+DRAAAKRAT LA+LAKN E++VNVIV+GGA+PAL+ HLQAPP D P+EHE
Sbjct: 64 QESDRAAAKRATQVLAELAKNAEDLVNVIVDGGAVPALMTHLQAPPYNDGDLAEKPYEHE 123
Query: 121 VEKGSAFALGLLAVK 135
VEKGSAFALGLLA+K
Sbjct: 124 VEKGSAFALGLLAIK 138
>gb|AAU10798.1| hypothetical protein [Oryza sativa (japonica cultivar-group)]
Length = 752
Score = 130 bits (328), Expect = 5e-30
Identities = 82/171 (47%), Positives = 97/171 (55%), Gaps = 39/171 (22%)
Query: 4 QRRQGHCLSERKGQKRKLDEELP---------------------------------EDRQ 30
Q++Q RKGQKRKL++E E+ +
Sbjct: 5 QQQQPPHRPRRKGQKRKLEDEAAASASAAAAAAAAAAAAATATPSSLGSAGADDDNEEEE 64
Query: 31 ISSAPPTA---DERAALLVEVANQVTVLESTFTWNEADRAAAKRATHALADLAKNEEVVN 87
SA P +AAL EV QV L F+W ADRAAAKRATH LA+LAKNEEVVN
Sbjct: 65 DGSAGPEICCRHSQAALAREVRTQVDALHRCFSWRHADRAAAKRATHVLAELAKNEEVVN 124
Query: 88 VIVEGGAIPALIKHLQAPPVTDCV---QKPLPFEHEVEKGSAFALGLLAVK 135
VIVEGGA+PAL+ HL+ PP + Q+P PFEHEVEKG+AFALGLLAVK
Sbjct: 125 VIVEGGAVPALVCHLKEPPAVAVLQEEQQPRPFEHEVEKGAAFALGLLAVK 175
>emb|CAC05434.1| putative protein [Arabidopsis thaliana]
Length = 706
Score = 79.3 bits (194), Expect = 2e-14
Identities = 50/129 (38%), Positives = 70/129 (53%), Gaps = 1/129 (0%)
Query: 5 RRQGHCLSERKGQKRKLDEELPEDRQISSAPPTADERAALLVEVANQVTVLESTFTWNEA 64
+RQ + KRKL ++ DE L+V + V VL S+F+ +
Sbjct: 6 KRQRTTRLAARNLKRKLSHNTDGAPIVTQLIDIDDEPIDLVVAIRRHVEVLNSSFSDPDF 65
Query: 65 DRAAAKRATHALADLAKNEEVVNVIVEGGAIPALIKHLQAPPVTDCVQKPLPFEHEVEKG 124
D A K A +ADLAK +E V +IVE GAIPAL+++L++P V C P EH++EK
Sbjct: 66 DHEAVKEAAADIADLAKIDENVEIIVENGAIPALVRYLESPLVV-CGNVPKSCEHKLEKD 124
Query: 125 SAFALGLLA 133
A ALGL+A
Sbjct: 125 CALALGLIA 133
>gb|AAL69492.1| unknown protein [Arabidopsis thaliana]
Length = 736
Score = 79.3 bits (194), Expect = 2e-14
Identities = 50/129 (38%), Positives = 70/129 (53%), Gaps = 1/129 (0%)
Query: 5 RRQGHCLSERKGQKRKLDEELPEDRQISSAPPTADERAALLVEVANQVTVLESTFTWNEA 64
+RQ + KRKL ++ DE L+V + V VL S+F+ +
Sbjct: 33 KRQRTTRLAARNLKRKLSHNTDGAPIVTQLIDIDDEPIDLVVAIRRHVEVLNSSFSDPDF 92
Query: 65 DRAAAKRATHALADLAKNEEVVNVIVEGGAIPALIKHLQAPPVTDCVQKPLPFEHEVEKG 124
D A K A +ADLAK +E V +IVE GAIPAL+++L++P V C P EH++EK
Sbjct: 93 DHEAVKEAAADIADLAKIDENVEIIVENGAIPALVRYLESPLVV-CGNVPKSCEHKLEKD 151
Query: 125 SAFALGLLA 133
A ALGL+A
Sbjct: 152 CALALGLIA 160
>ref|NP_196810.2| armadillo/beta-catenin repeat family protein / BTB/POZ
domain-containing protein [Arabidopsis thaliana]
Length = 709
Score = 79.3 bits (194), Expect = 2e-14
Identities = 50/129 (38%), Positives = 70/129 (53%), Gaps = 1/129 (0%)
Query: 5 RRQGHCLSERKGQKRKLDEELPEDRQISSAPPTADERAALLVEVANQVTVLESTFTWNEA 64
+RQ + KRKL ++ DE L+V + V VL S+F+ +
Sbjct: 6 KRQRTTRLAARNLKRKLSHNTDGAPIVTQLIDIDDEPIDLVVAIRRHVEVLNSSFSDPDF 65
Query: 65 DRAAAKRATHALADLAKNEEVVNVIVEGGAIPALIKHLQAPPVTDCVQKPLPFEHEVEKG 124
D A K A +ADLAK +E V +IVE GAIPAL+++L++P V C P EH++EK
Sbjct: 66 DHEAVKEAAADIADLAKIDENVEIIVENGAIPALVRYLESPLVV-CGNVPKSCEHKLEKD 124
Query: 125 SAFALGLLA 133
A ALGL+A
Sbjct: 125 CALALGLIA 133
>ref|XP_479466.1| hypothetical protein [Oryza sativa (japonica cultivar-group)]
gi|22831132|dbj|BAC15993.1| hypothetical protein [Oryza
sativa (japonica cultivar-group)]
Length = 54
Score = 39.3 bits (90), Expect = 0.021
Identities = 22/49 (44%), Positives = 29/49 (58%), Gaps = 9/49 (18%)
Query: 60 TWNEADRAAA--KRATHALADLAKNEEVVNVIVEGGAIPALIKHLQAPP 106
TW AA+ K+ HA EEV NVI++G A+PAL+ HL+ PP
Sbjct: 8 TWASQRSAASTPKQGPHA-------EEVANVIIKGSAMPALVFHLKGPP 49
>ref|XP_470183.1| Putative AMP-binding protein [Oryza sativa (japonica
cultivar-group)] gi|20502991|gb|AAM22700.1| Putative
AMP-binding protein [Oryza sativa (japonica
cultivar-group)]
Length = 565
Score = 37.7 bits (86), Expect = 0.060
Identities = 23/79 (29%), Positives = 36/79 (45%), Gaps = 2/79 (2%)
Query: 1 MEFQRRQGHCLSERKGQKRKLDEELPEDRQISSAPPTADERAALLVEVANQVTVLESTFT 60
ME R G+C + R + R+ D LP D ++ A R + +V + + T TFT
Sbjct: 7 MEVDARSGYCAATRTFRSRRADVPLPADPEVDVVSFLASRRHSGVVALVDAATGRRITFT 66
Query: 61 --WNEADRAAAKRATHALA 77
W AA+ A H ++
Sbjct: 67 ELWRAVAGAASALAAHPVS 85
>ref|NP_001013796.1| expressed sequence AW146299 [Mus musculus]
gi|61660291|gb|AAX50192.1| importin alpha 2 [Mus
musculus]
Length = 499
Score = 35.0 bits (79), Expect = 0.39
Identities = 30/103 (29%), Positives = 50/103 (48%), Gaps = 5/103 (4%)
Query: 6 RQGHCLSERKGQKRKLDEELPEDRQISSAPPTADERAALLVEVANQVTVLESTFTWNEAD 65
RQ S + +K + DE++ + R I ++D + LV+ N T+ + N +D
Sbjct: 25 RQQRIASSLQLRKTRKDEQVLKRRNIDLF--SSDMVSQALVKEVN-FTLDDIIQAVNSSD 81
Query: 66 RAAAKRATHALADLAKNEEV--VNVIVEGGAIPALIKHLQAPP 106
RAT A ++ E +N+I+E G IP L+ L+A P
Sbjct: 82 PILHFRATRAAREMISQENTPPLNLIIEAGLIPKLVDFLKATP 124
>ref|XP_620273.1| PREDICTED: expressed sequence AW146299 [Mus musculus]
Length = 520
Score = 35.0 bits (79), Expect = 0.39
Identities = 30/103 (29%), Positives = 50/103 (48%), Gaps = 5/103 (4%)
Query: 6 RQGHCLSERKGQKRKLDEELPEDRQISSAPPTADERAALLVEVANQVTVLESTFTWNEAD 65
RQ S + +K + DE++ + R I ++D + LV+ N T+ + N +D
Sbjct: 25 RQQRIASSLQLRKTRKDEQVLKRRNIDLF--SSDMVSQALVKEVN-FTLDDIIQAVNSSD 81
Query: 66 RAAAKRATHALADLAKNEEV--VNVIVEGGAIPALIKHLQAPP 106
RAT A ++ E +N+I+E G IP L+ L+A P
Sbjct: 82 PILHFRATRAAREMISQENTPPLNLIIEAGLIPKLVDFLKATP 124
>gb|EAA52437.1| hypothetical protein MG05129.4 [Magnaporthe grisea 70-15]
gi|39940222|ref|XP_359648.1| hypothetical protein
MG05129.4 [Magnaporthe grisea 70-15]
Length = 2011
Score = 34.3 bits (77), Expect = 0.67
Identities = 28/99 (28%), Positives = 46/99 (46%), Gaps = 3/99 (3%)
Query: 2 EFQRRQGHCLSERKGQKRKLDEELPEDR--QISSAPPTADERAALLVEVANQVTVLESTF 59
+ +++ L + K ++RK E+ ++R Q + A+E AA E A E+
Sbjct: 931 KLSKKEQEKLDKEKEKERKKAEKAEKERAKQEAEEAARAEEEAARAAEEAAAREAEEAAQ 990
Query: 60 TWNEADRAAAKRATHALADLAKNEEVVNVIVEGGAIPAL 98
EA+ AAA+ A A A AK E ++ E + AL
Sbjct: 991 AAKEAEEAAAREAEEA-AQAAKEAEEAEILKEEAELKAL 1028
>ref|ZP_00419107.1| type II and III secretion system protein:NolW-like [Azotobacter
vinelandii AvOP] gi|67085000|gb|EAM04477.1| type II and
III secretion system protein:NolW-like [Azotobacter
vinelandii AvOP]
Length = 717
Score = 33.9 bits (76), Expect = 0.87
Identities = 30/105 (28%), Positives = 53/105 (49%), Gaps = 12/105 (11%)
Query: 12 SERKGQKRKLDEELP-EDRQISSAPPTADERAALLVEVANQVT-----VLESTFTWNEAD 65
S+++ K EE+P ++ S A T+ + AAL +EV Q+T ++E T ++AD
Sbjct: 570 SDKETAKILTGEEIPYQEASSSGATSTSFKEAALSLEVTPQITPDNRIIMEVKVTKDQAD 629
Query: 66 RAAAKRATHALADLAKNEEVVNVIVEGG---AIPALIKHLQAPPV 107
A +A + + + KNE V++ G I + ++QA V
Sbjct: 630 FA---KALNGVPPINKNEVNAKVLIGDGETIVIGGVFSNIQAKSV 671
>ref|ZP_00401519.1| Glycosyl transferase, family 3:Glycosyl transferase, family 3
[Anaeromyxobacter dehalogenans 2CP-C]
gi|66776731|gb|EAL77839.1| Glycosyl transferase, family
3:Glycosyl transferase, family 3 [Anaeromyxobacter
dehalogenans 2CP-C]
Length = 435
Score = 33.5 bits (75), Expect = 1.1
Identities = 33/111 (29%), Positives = 50/111 (44%), Gaps = 3/111 (2%)
Query: 24 ELPEDRQISSAPPTADERAALLVEVANQVTVLESTFTWNEADRAAAKRATHALADLAKNE 83
E+ E ++ AD RA + VE+ ++ + A RAA +RA + LAK E
Sbjct: 252 EVAEVLELLRGGGPADLRA-VTVELTAEMLLAGGVAAELAAARAAVERAIADGSGLAKLE 310
Query: 84 EVVNVIVEGGAIPALIKHLQAPPVTDCVQKPLPFEHEVEKGSAFALGLLAV 134
E+ V +GG + A+ + P D P P V+ A+GL AV
Sbjct: 311 EI--VAAQGGDVAAIRDPSRLPRAADPYPVPAPAAGFVQAVDTEAVGLAAV 359
>gb|AAM78457.1| nonribosomal peptide synthetase; NRPS [Hypocrea virens]
Length = 20925
Score = 33.5 bits (75), Expect = 1.1
Identities = 22/71 (30%), Positives = 38/71 (52%), Gaps = 3/71 (4%)
Query: 22 DEELPEDRQISSAPPTADERAALLVEVANQVTVLESTFTWNEADRAAAKRATHALADL-A 80
+ E+P R I S T E+ ++ + V + FT+ + D+AA + A H +AD
Sbjct: 7887 NNEIP--RLIESCFHTVIEQQTIIRPESPAVHGWDGNFTYGQLDQAANRLANHLIADYEI 7944
Query: 81 KNEEVVNVIVE 91
KN+E+++V E
Sbjct: 7945 KNDELIHVCFE 7955
>gb|AAR98916.1| peptide synthetase [Trichoderma asperellum]
Length = 489
Score = 33.1 bits (74), Expect = 1.5
Identities = 23/73 (31%), Positives = 36/73 (48%), Gaps = 3/73 (4%)
Query: 17 QKRKLDEELPEDRQISSAPPTADERAALLVEVANQVTVLESTFTWNEADRAAAKRATHAL 76
Q + E+PE I S T E A A + + +FT+ E DRAA + A H +
Sbjct: 71 QSEIFNSEMPE--AIESCFHTLVEMQAARTPDATAICAWDGSFTYRELDRAANRLAHHLM 128
Query: 77 AD-LAKNEEVVNV 88
A+ K +E+++V
Sbjct: 129 AEHSVKLDEIIHV 141
>gb|AAR90257.1| polyketide synthase [Cochliobolus heterostrophus]
Length = 2141
Score = 32.7 bits (73), Expect = 1.9
Identities = 27/100 (27%), Positives = 49/100 (49%), Gaps = 4/100 (4%)
Query: 22 DEELPEDRQISSAPPTADERAALLVEVANQVTVLESTFTWNEADRAAAKRATHALADLAK 81
+E + D S+ PTA+ +A+ VTV + ++ W + + R T A L +
Sbjct: 747 EEAICTDDTFSNLQPTAEYFSAMAGRKVEPVTVQKPSY-WASSAKFRV-RFTSAAKALLR 804
Query: 82 NEEVVNVIVEGGAIPALIKHLQAPPVTDCVQKPLPFEHEV 121
++ NV+VE G P L+ L++ + ++K +P EV
Sbjct: 805 SKPSPNVVVEIGPNPTLVGSLKS--ILSEIKKEIPHPIEV 842
>gb|AAF93715.1| aspartokinase, alpha and beta subunits [Vibrio cholerae O1 biovar
eltor str. N16961] gi|15640569|ref|NP_230198.1|
aspartokinase, alpha and beta subunits [Vibrio cholerae
O1 biovar eltor str. N16961] gi|11260672|pir||A82311
aspartokinase, alpha and beta chains VC0547 [imported] -
Vibrio cholerae (strain N16961 serogroup O1)
Length = 395
Score = 32.3 bits (72), Expect = 2.5
Identities = 16/54 (29%), Positives = 26/54 (47%)
Query: 57 STFTWNEADRAAAKRATHALADLAKNEEVVNVIVEGGAIPALIKHLQAPPVTDC 110
STF N+ ATHA+ +A ++ + VE + P++IK Q + C
Sbjct: 232 STFEVNQGSLIKGSAATHAVCGIALQRDMALIRVESESFPSIIKQCQMLGIEVC 285
>dbj|BAB01877.1| unnamed protein product [Arabidopsis thaliana]
gi|15231056|ref|NP_188652.1| armadillo/beta-catenin
repeat family protein [Arabidopsis thaliana]
Length = 475
Score = 32.3 bits (72), Expect = 2.5
Identities = 22/70 (31%), Positives = 33/70 (46%), Gaps = 3/70 (4%)
Query: 30 QISSAPPTADERAALLVEVANQVTVLESTFTWNEADRAAAKRATHALADLAKNEEVVNVI 89
+I SA T E +LV ++E+ N A R +RA HA+ + ++
Sbjct: 174 EILSALTTIRESRRVLVHSGGLKFLVEAAKVGNLASR---ERACHAIGLIGVTRRARRIL 230
Query: 90 VEGGAIPALI 99
VE G IPAL+
Sbjct: 231 VEAGVIPALV 240
>gb|AAK04010.1| unknown [Pasteurella multocida subsp. multocida str. Pm70]
gi|15603791|ref|NP_246865.1| hypothetical protein PM1926
[Pasteurella multocida subsp. multocida str. Pm70]
gi|20139707|sp|Q9CJR6|RLPA_PASMU RlpA-like protein
precursor
Length = 294
Score = 32.3 bits (72), Expect = 2.5
Identities = 16/43 (37%), Positives = 24/43 (55%)
Query: 72 ATHALADLAKNEEVVNVIVEGGAIPALIKHLQAPPVTDCVQKP 114
A+ LA LAKN+E VN++++G L +H + V KP
Sbjct: 173 ASATLAKLAKNQEAVNMLLQGEDTVELTQHTEEKTVKAATTKP 215
>gb|EAA68359.1| hypothetical protein FG01658.1 [Gibberella zeae PH-1]
gi|46109552|ref|XP_381834.1| hypothetical protein
FG01658.1 [Gibberella zeae PH-1]
Length = 669
Score = 32.0 bits (71), Expect = 3.3
Identities = 31/126 (24%), Positives = 56/126 (43%), Gaps = 11/126 (8%)
Query: 15 KGQKRKLDEELPEDRQISSAPPTADERAALLVEVANQVTVLESTFT----WNEADRAAAK 70
+G RK DE E+R++S PP+ E +L +F+ ++E+D+ +
Sbjct: 490 RGAARKKDENETEEREVSFKPPSFFAPQESHDEANGLADLLSQSFSLGQEYSESDQGSVT 549
Query: 71 RATHALADLAKNEEVVNVIVEGGAIPALIKHLQAPPVTDCVQKPLPFEHEVEKGSAFALG 130
R + + A + + V A+ ++ L A PLPF EV+ A G
Sbjct: 550 RPSSSHAPGSPQFGRLGVEFITIAVLLVLWGLTA-------AFPLPFGREVQLALLSAAG 602
Query: 131 LLAVKV 136
++A++V
Sbjct: 603 IIALRV 608
Database: nr
Posted date: Jul 5, 2005 12:34 AM
Number of letters in database: 863,360,394
Number of sequences in database: 2,540,612
Lambda K H
0.315 0.131 0.366
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 218,622,120
Number of Sequences: 2540612
Number of extensions: 7777198
Number of successful extensions: 24550
Number of sequences better than 10.0: 65
Number of HSP's better than 10.0 without gapping: 19
Number of HSP's successfully gapped in prelim test: 46
Number of HSP's that attempted gapping in prelim test: 24501
Number of HSP's gapped (non-prelim): 88
length of query: 137
length of database: 863,360,394
effective HSP length: 113
effective length of query: 24
effective length of database: 576,271,238
effective search space: 13830509712
effective search space used: 13830509712
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.6 bits)
S2: 67 (30.4 bits)
Medicago: description of AC137838.2


