

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC137838.10 - phase: 0
(500 letters)
Database: nr
2,540,612 sequences; 863,360,394 total letters
Searching..................................................done
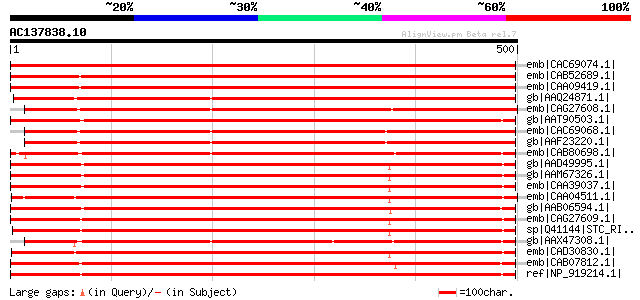
Score E
Sequences producing significant alignments: (bits) Value
emb|CAC69074.1| STP13 protein [Arabidopsis thaliana] gi|15240313... 736 0.0
emb|CAB52689.1| hexose transporter [Lycopersicon esculentum] 722 0.0
emb|CAA09419.1| hexose transporter protein [Lycopersicon esculen... 711 0.0
gb|AAQ24871.1| monosaccharide transporter 4 [Oryza sativa (japon... 700 0.0
emb|CAG27608.1| monosaccharide transporter [Populus tremula x Po... 602 e-170
gb|AAT90503.1| monosaccharide transport protein 1 [Zea mays] 590 e-167
emb|CAC69068.1| STP8 protein [Arabidopsis thaliana] gi|15240279|... 587 e-166
gb|AAF23220.1| putative hexose transporter [Arabidopsis thaliana... 585 e-166
emb|CAB80698.1| putative hexose transporter [Arabidopsis thalian... 583 e-165
gb|AAD49995.1| glucose transporter [Arabidopsis thaliana] gi|221... 577 e-163
gb|AAM67326.1| glucose transporter [Arabidopsis thaliana] 577 e-163
emb|CAA39037.1| glucose transporter [Arabidopsis thaliana] 576 e-163
emb|CAA04511.1| hexose transporter [Vitis vinifera] 576 e-163
gb|AAB06594.1| sugar transporter 573 e-162
emb|CAG27609.1| monosaccharide transporter [Populus tremula x Po... 572 e-162
sp|Q41144|STC_RICCO Sugar carrier protein C gi|169718|gb|AAA7976... 571 e-161
gb|AAX47308.1| hexose transporter 7 [Vitis vinifera] 571 e-161
emb|CAD30830.1| monosaccharide-H+ symporter [Datisca glomerata] 567 e-160
emb|CAB07812.1| monosaccharid transport protein [Vicia faba] gi|... 566 e-160
ref|NP_919214.1| putative monosaccharide transport protein MST1 ... 566 e-160
>emb|CAC69074.1| STP13 protein [Arabidopsis thaliana] gi|15240313|ref|NP_198006.1|
hexose transporter, putative [Arabidopsis thaliana]
gi|15450649|gb|AAK96596.1| AT5g26340/F9D12_17
[Arabidopsis thaliana] gi|3319354|gb|AAC26243.1|
contains similarity to sugar transporters (Pfam:
sugar_tr.hmm, score: 395.39) [Arabidopsis thaliana]
gi|9965739|gb|AAG10146.1| putative hexose transporter
MSS1 [Arabidopsis thaliana] gi|7446741|pir||T01853
probable hexose transport protein F9D12.17 - Arabidopsis
thaliana
Length = 526
Score = 736 bits (1899), Expect = 0.0
Identities = 358/499 (71%), Positives = 425/499 (84%)
Query: 1 MAGGGFTTGSSDVVFEARITAAVVISCIMAATGGLMFGYDVGISGGVTSMPSFLQKFFPD 60
M GGGF T ++ V FEA+IT V+ISCIMAATGGLMFGYDVG+SGGVTSMP FL+KFFP
Sbjct: 1 MTGGGFATSANGVEFEAKITPIVIISCIMAATGGLMFGYDVGVSGGVTSMPDFLEKFFPV 60
Query: 61 VYKRTQEHTVLESNYCKYDNQKLQLFTSSLYLAALVASMIASPVTRKLGRKQTMLLAGIL 120
VY++ +SNYCKYDNQ LQLFTSSLYLA L A+ AS TR LGR+ TML+AG+
Sbjct: 61 VYRKVVAGADKDSNYCKYDNQGLQLFTSSLYLAGLTATFFASYTTRTLGRRLTMLIAGVF 120
Query: 121 FIVGTVLSASAGKLILLIFGRILLGCGVGFANQAVPVFLSEIAPTRIRGALNIMFQLNIT 180
FI+G L+A A L +LI GRILLGCGVGFANQAVP+FLSEIAPTRIRG LNI+FQLN+T
Sbjct: 121 FIIGVALNAGAQDLAMLIAGRILLGCGVGFANQAVPLFLSEIAPTRIRGGLNILFQLNVT 180
Query: 181 IGIFIANLVNWFTSKIKGGYGWRVSLAGAIIPAVMLTMGSLIVDDTPNSLIERGFEEKGK 240
IGI ANLVN+ T+KIKGG+GWR+SL A IPA++LT+G+L+V +TPNSL+ERG ++GK
Sbjct: 181 IGILFANLVNYGTAKIKGGWGWRLSLGLAGIPALLLTVGALLVTETPNSLVERGRLDEGK 240
Query: 241 AVLTKIRGVENIEPEFEDILRASKVANEVKSPFKDLVKSHNRPPLIIAICMQVFQQCTGI 300
AVL +IRG +N+EPEF D+L AS++A EVK PF++L++ NRP L+IA+ +Q+FQQCTGI
Sbjct: 241 AVLRRIRGTDNVEPEFADLLEASRLAKEVKHPFRNLLQRRNRPQLVIAVALQIFQQCTGI 300
Query: 301 NAIMFYAPVLFSTLGFHNDASLYSSVITGGVNVLCTLVSVYFVDKAGRRVLLLEACVQMF 360
NAIMFYAPVLFSTLGF +DASLYS+V+TG VNVL TLVS+Y VDK GRRVLLLEA VQMF
Sbjct: 301 NAIMFYAPVLFSTLGFGSDASLYSAVVTGAVNVLSTLVSIYSVDKVGRRVLLLEAGVQMF 360
Query: 361 VSQVVIGIVLGAKLQDHSDSLSKGYAMLVVVMVCTFVASFAWSWGPLGWLIPSETFPLET 420
SQVVI I+LG K+ D S +LSKG+A+LVVVM+CT+VA+FAWSWGPLGWLIPSETFPLET
Sbjct: 361 FSQVVIAIILGVKVTDTSTNLSKGFAILVVVMICTYVAAFAWSWGPLGWLIPSETFPLET 420
Query: 421 RSAGQSVTVFTNMLFTFLIAQAFLSLLCLFKFGIFLFFSAWVFVMGVFTVFLIPETKNIP 480
RSAGQSVTV N+LFTF+IAQAFLS+LC FKFGIF+FFSAWV +M VF +FL+PETKNIP
Sbjct: 421 RSAGQSVTVCVNLLFTFIIAQAFLSMLCHFKFGIFIFFSAWVLIMSVFVMFLLPETKNIP 480
Query: 481 IEDMAETVWKQHWFWRRFM 499
IE+M E VWK+HWFW RFM
Sbjct: 481 IEEMTERVWKKHWFWARFM 499
>emb|CAB52689.1| hexose transporter [Lycopersicon esculentum]
Length = 523
Score = 722 bits (1864), Expect = 0.0
Identities = 356/500 (71%), Positives = 424/500 (84%), Gaps = 2/500 (0%)
Query: 1 MAGGGFTT-GSSDVVFEARITAAVVISCIMAATGGLMFGYDVGISGGVTSMPSFLQKFFP 59
MAGGGFTT G+ FEA+IT V+ISCIMAATGGLMFGYDVG+SGGVTSM FL+KFFP
Sbjct: 1 MAGGGFTTSGNGGTHFEAKITPIVIISCIMAATGGLMFGYDVGVSGGVTSMDPFLKKFFP 60
Query: 60 DVYKRTQEHTVLESNYCKYDNQKLQLFTSSLYLAALVASMIASPVTRKLGRKQTMLLAGI 119
VYKRT+E L+SNYCKYDNQ LQLFTSSLYLA L A+ AS TRKLGR+ TML+AG
Sbjct: 61 TVYKRTKEPG-LDSNYCKYDNQGLQLFTSSLYLAGLTATFFASYTTRKLGRRLTMLIAGC 119
Query: 120 LFIVGTVLSASAGKLILLIFGRILLGCGVGFANQAVPVFLSEIAPTRIRGALNIMFQLNI 179
FI+G VL+A+A L +LI GRILLGCGVGFANQAVP+FLSEIAPTRIRG LNI+FQLN+
Sbjct: 120 FFIIGVVLNAAAQDLAMLIIGRILLGCGVGFANQAVPLFLSEIAPTRIRGGLNILFQLNV 179
Query: 180 TIGIFIANLVNWFTSKIKGGYGWRVSLAGAIIPAVMLTMGSLIVDDTPNSLIERGFEEKG 239
TIGI ANLVN+ T+KI GG+GWR+SL A PAV+LT+G+L V +TPNSLIERG+ E+G
Sbjct: 180 TIGILFANLVNYGTAKISGGWGWRLSLGLAGFPAVLLTLGALFVVETPNSLIERGYLEEG 239
Query: 240 KAVLTKIRGVENIEPEFEDILRASKVANEVKSPFKDLVKSHNRPPLIIAICMQVFQQCTG 299
K VL KIRG +NIEPEF +++ AS+VA +VK PF++L++ NRP LII++ +Q+FQQ TG
Sbjct: 240 KEVLRKIRGTDNIEPEFLELVEASRVAKQVKHPFRNLLQRKNRPQLIISVALQIFQQFTG 299
Query: 300 INAIMFYAPVLFSTLGFHNDASLYSSVITGGVNVLCTLVSVYFVDKAGRRVLLLEACVQM 359
INAIMFYAPVLFSTLGF N A+LYS+VITG VNVL T+VSVY VDK GRRVLLLEA VQM
Sbjct: 300 INAIMFYAPVLFSTLGFGNSAALYSAVITGAVNVLSTVVSVYSVDKLGRRVLLLEAGVQM 359
Query: 360 FVSQVVIGIVLGAKLQDHSDSLSKGYAMLVVVMVCTFVASFAWSWGPLGWLIPSETFPLE 419
+SQ++I I+LG K+ DHSD+LS G+ + VVV++CT+V++FAWSWGPLGWLIPSETFPLE
Sbjct: 360 LLSQIIIAIILGIKVTDHSDNLSHGWGIFVVVLICTYVSAFAWSWGPLGWLIPSETFPLE 419
Query: 420 TRSAGQSVTVFTNMLFTFLIAQAFLSLLCLFKFGIFLFFSAWVFVMGVFTVFLIPETKNI 479
TRSAGQSVTV N+LFTF++AQAFLS+LC FK+GIFLFFS W+FVM +F FL+PETKN+
Sbjct: 420 TRSAGQSVTVCVNLLFTFVMAQAFLSMLCHFKYGIFLFFSGWIFVMSLFVFFLLPETKNV 479
Query: 480 PIEDMAETVWKQHWFWRRFM 499
PIE+M E VWKQHW W+RFM
Sbjct: 480 PIEEMTERVWKQHWLWKRFM 499
>emb|CAA09419.1| hexose transporter protein [Lycopersicon esculentum]
gi|7446737|pir||T07379 hexose transport protein - tomato
Length = 523
Score = 711 bits (1836), Expect = 0.0
Identities = 351/500 (70%), Positives = 420/500 (83%), Gaps = 2/500 (0%)
Query: 1 MAGGGFTT-GSSDVVFEARITAAVVISCIMAATGGLMFGYDVGISGGVTSMPSFLQKFFP 59
MAGGGFTT G+ + + V+ISCIMAATGGLMFGYDVG+SGGVTSM FL+KFFP
Sbjct: 1 MAGGGFTTSGNGARISRLKSHQLVIISCIMAATGGLMFGYDVGVSGGVTSMDPFLKKFFP 60
Query: 60 DVYKRTQEHTVLESNYCKYDNQKLQLFTSSLYLAALVASMIASPVTRKLGRKQTMLLAGI 119
VYKRT+E L+SNYCKYDNQ LQLFTSSLYLA L A+ AS TRKLGR+ TML+AG
Sbjct: 61 TVYKRTKEPG-LDSNYCKYDNQGLQLFTSSLYLAGLTATFFASYTTRKLGRRLTMLIAGC 119
Query: 120 LFIVGTVLSASAGKLILLIFGRILLGCGVGFANQAVPVFLSEIAPTRIRGALNIMFQLNI 179
FI+G VL+A+A L +LI GRILLGCGVGFANQAVP+FLSEIAPTRIRG LNI+FQLN+
Sbjct: 120 FFIIGVVLNAAAQDLAMLIIGRILLGCGVGFANQAVPLFLSEIAPTRIRGGLNILFQLNV 179
Query: 180 TIGIFIANLVNWFTSKIKGGYGWRVSLAGAIIPAVMLTMGSLIVDDTPNSLIERGFEEKG 239
TIGI ANLVN+ T+KI GG+GWR+SL A PAV+LT+G+L V +TPNSLIERG+ E+G
Sbjct: 180 TIGILFANLVNYGTAKISGGWGWRLSLGLAGFPAVLLTLGALFVVETPNSLIERGYLEEG 239
Query: 240 KAVLTKIRGVENIEPEFEDILRASKVANEVKSPFKDLVKSHNRPPLIIAICMQVFQQCTG 299
K VL KIRG +NIEPEF +++ AS+VA +VK PF++L++ NRP LII++ +Q+FQQ TG
Sbjct: 240 KEVLRKIRGTDNIEPEFLELVEASRVAKQVKHPFRNLLQRKNRPQLIISVALQIFQQFTG 299
Query: 300 INAIMFYAPVLFSTLGFHNDASLYSSVITGGVNVLCTLVSVYFVDKAGRRVLLLEACVQM 359
INAIMFYAPVLFSTLGF N A+LYS+VITG VNVL T+VSVY VDK GRRVLLLEA VQM
Sbjct: 300 INAIMFYAPVLFSTLGFGNSAALYSAVITGAVNVLSTVVSVYSVDKLGRRVLLLEAGVQM 359
Query: 360 FVSQVVIGIVLGAKLQDHSDSLSKGYAMLVVVMVCTFVASFAWSWGPLGWLIPSETFPLE 419
+SQ++I I+LG K+ DHSD+LS G+ + VVV++CT+V++FAWSWGPLGWLIPSETFPLE
Sbjct: 360 LLSQIIIAIILGIKVTDHSDNLSHGWGIFVVVLICTYVSAFAWSWGPLGWLIPSETFPLE 419
Query: 420 TRSAGQSVTVFTNMLFTFLIAQAFLSLLCLFKFGIFLFFSAWVFVMGVFTVFLIPETKNI 479
TRSAGQSVTV N+LFTF++AQAFLS+LC FK+GIFLFFS W+FVM +F FL+PETKN+
Sbjct: 420 TRSAGQSVTVCVNLLFTFVMAQAFLSMLCHFKYGIFLFFSGWIFVMSLFVFFLVPETKNV 479
Query: 480 PIEDMAETVWKQHWFWRRFM 499
PIE+M E VWKQHW W+RFM
Sbjct: 480 PIEEMTERVWKQHWLWKRFM 499
>gb|AAQ24871.1| monosaccharide transporter 4 [Oryza sativa (japonica
cultivar-group)]
Length = 515
Score = 700 bits (1806), Expect = 0.0
Identities = 350/496 (70%), Positives = 412/496 (82%), Gaps = 3/496 (0%)
Query: 4 GGFTTGSSDVVFEARITAAVVISCIMAATGGLMFGYDVGISGGVTSMPSFLQKFFPDVYK 63
GGF+ S V FEA+IT V+ISCIMAATGGLMFGYDVGISGGVTSM FL++FFP V K
Sbjct: 3 GGFSVSGSGVEFEAKITPIVIISCIMAATGGLMFGYDVGISGGVTSMDDFLREFFPTVLK 62
Query: 64 RTQEHTVLESNYCKYDNQKLQLFTSSLYLAALVASMIASPVTRKLGRKQTMLLAGILFIV 123
+ +H ESNYCKYDNQ LQLFTSSLYLA L A+ AS TR+LGR+ TML+AG+ FIV
Sbjct: 63 K--KHEDKESNYCKYDNQGLQLFTSSLYLAGLTATFFASYTTRRLGRRLTMLIAGVFFIV 120
Query: 124 GTVLSASAGKLILLIFGRILLGCGVGFANQAVPVFLSEIAPTRIRGALNIMFQLNITIGI 183
G + + +A L +LI GRILLGCGVGFANQAVP+FLSEIAPTRIRG LNI+FQLN+TIGI
Sbjct: 121 GVIFNGAAQNLAMLIVGRILLGCGVGFANQAVPLFLSEIAPTRIRGGLNILFQLNVTIGI 180
Query: 184 FIANLVNWFTSKIKGGYGWRVSLAGAIIPAVMLTMGSLIVDDTPNSLIERGFEEKGKAVL 243
ANLVN+ T+KI +GWR+SL+ A IPA +LT+G+L V DTPNSLIERG E+GKAVL
Sbjct: 181 LFANLVNYGTAKIHP-WGWRLSLSLAGIPAALLTLGALFVVDTPNSLIERGRLEEGKAVL 239
Query: 244 TKIRGVENIEPEFEDILRASKVANEVKSPFKDLVKSHNRPPLIIAICMQVFQQCTGINAI 303
KIRG +N+EPEF +I+ AS+VA EVK PF++L++ NRP L+IA+ +Q+FQQ TGINAI
Sbjct: 240 RKIRGTDNVEPEFNEIVEASRVAQEVKHPFRNLLQRRNRPQLVIAVLLQIFQQFTGINAI 299
Query: 304 MFYAPVLFSTLGFHNDASLYSSVITGGVNVLCTLVSVYFVDKAGRRVLLLEACVQMFVSQ 363
MFYAPVLF+TLGF DASLYS+VITG VNVL TLVSVY D+ GRR+LLLEA VQMF+SQ
Sbjct: 300 MFYAPVLFNTLGFKTDASLYSAVITGAVNVLSTLVSVYSADRVGRRMLLLEAGVQMFLSQ 359
Query: 364 VVIGIVLGAKLQDHSDSLSKGYAMLVVVMVCTFVASFAWSWGPLGWLIPSETFPLETRSA 423
V I +VLG K+ D SD+L G+A++VVVMVCTFV+SFAWSWGPLGWLIPSETFPLETRSA
Sbjct: 360 VAIAVVLGIKVTDRSDNLGHGWAIMVVVMVCTFVSSFAWSWGPLGWLIPSETFPLETRSA 419
Query: 424 GQSVTVFTNMLFTFLIAQAFLSLLCLFKFGIFLFFSAWVFVMGVFTVFLIPETKNIPIED 483
GQSVTV N+LFTF+IAQAFLS+LC K+ IF FFSAWV VM +F +F +PETKNIPIE+
Sbjct: 420 GQSVTVCVNLLFTFVIAQAFLSMLCHLKYAIFAFFSAWVVVMSLFVLFFLPETKNIPIEE 479
Query: 484 MAETVWKQHWFWRRFM 499
M E VWKQHWFW+RFM
Sbjct: 480 MTERVWKQHWFWKRFM 495
>emb|CAG27608.1| monosaccharide transporter [Populus tremula x Populus tremuloides]
Length = 514
Score = 602 bits (1551), Expect = e-170
Identities = 302/486 (62%), Positives = 380/486 (78%), Gaps = 4/486 (0%)
Query: 15 FEARITAAVVISCIMAATGGLMFGYDVGISGGVTSMPSFLQKFFPDVYKRTQEHTVLESN 74
FE RIT V++ ++AA GGLMFGYD+G+SGGVT+M L+KFF V++R Q+ E+N
Sbjct: 15 FEGRITFNVIVCVVIAACGGLMFGYDIGVSGGVTAMDDVLKKFFYQVWERKQQ--AHENN 72
Query: 75 YCKYDNQKLQLFTSSLYLAALVASMIASPVTRKLGRKQTMLLAGILFIVGTVLSASAGKL 134
YCKYDN+KLQLFTSSLY+AAL+AS +AS K GRK TM LA + FI G L+ A +
Sbjct: 73 YCKYDNKKLQLFTSSLYIAALIASFLASKTCSKFGRKPTMQLASLFFIGGVALTTFAVNI 132
Query: 135 ILLIFGRILLGCGVGFANQAVPVFLSEIAPTRIRGALNIMFQLNITIGIFIANLVNWFTS 194
+LI GR+LLGCGVGFANQAVP+FLSE+AP +IRGALNI FQL ITIGI IAN+VN+
Sbjct: 133 EMLIIGRLLLGCGVGFANQAVPLFLSELAPAKIRGALNISFQLFITIGILIANIVNYVVG 192
Query: 195 KIKGGYGWRVSLAGAIIPAVMLTMGSLIVDDTPNSLIERGFEEKGKAVLTKIRGVENIEP 254
KI YG+R+SL A +PA++L GSL + +TP SLIER E+G+AVL KIRGV+N++
Sbjct: 193 KIHP-YGFRISLGIAGVPALLLCFGSLAIYETPTSLIERKKVEQGRAVLKKIRGVDNVDL 251
Query: 255 EFEDILRASKVANEVKSPFKDLVKSHNRPPLIIAICMQVFQQCTGINAIMFYAPVLFSTL 314
E++ I+ A +VA+++ P+ +L+K +RPPL+IAI MQVFQQ TGINAIMFYAPVLF T+
Sbjct: 252 EYDSIVHACEVASQITQPYHELMKRESRPPLVIAIVMQVFQQFTGINAIMFYAPVLFQTV 311
Query: 315 GFHNDASLYSSVITGGVNVLCTLVSVYFVDKAGRRVLLLEACVQMFVSQVVIGIVLGAKL 374
GF +DA+L SSV+TG VNVL T+VSV VDK GRR LLLEACVQM ++Q +IG VL L
Sbjct: 312 GFGSDAALLSSVVTGLVNVLSTIVSVVLVDKVGRRALLLEACVQMLITQCIIGGVLMKDL 371
Query: 375 QDHSDSLSKGYAMLVVVMVCTFVASFAWSWGPLGWLIPSETFPLETRSAGQSVTVFTNML 434
+ + +L G A++VV+MVC FVA FAWSWGPLGWLIPSETFPLETR+AG S V +NML
Sbjct: 372 KT-TGTLPNGDALVVVIMVCVFVAGFAWSWGPLGWLIPSETFPLETRTAGFSFAVSSNML 430
Query: 435 FTFLIAQAFLSLLCLFKFGIFLFFSAWVFVMGVFTVFLIPETKNIPIEDMAETVWKQHWF 494
FTF+IAQAFLS LC K GIF FF+AW+ VMG+F +FL+PETK +P++DM + VWKQHWF
Sbjct: 431 FTFVIAQAFLSTLCHLKAGIFFFFAAWIVVMGLFALFLLPETKGVPVDDMVDRVWKQHWF 490
Query: 495 WRRFMH 500
W+RF +
Sbjct: 491 WKRFFN 496
>gb|AAT90503.1| monosaccharide transport protein 1 [Zea mays]
Length = 523
Score = 590 bits (1522), Expect = e-167
Identities = 289/500 (57%), Positives = 374/500 (74%), Gaps = 4/500 (0%)
Query: 1 MAGGGFTTGSSDVVFEARITAAVVISCIMAATGGLMFGYDVGISGGVTSMPSFLQKFFPD 60
MAGGG + ++T V+++CI+AATGGL+FGYD+GISGGVTSM FL+KFFP+
Sbjct: 1 MAGGGIVNTGGGKDYPGKLTLFVLLTCIVAATGGLIFGYDIGISGGVTSMNPFLEKFFPE 60
Query: 61 VYKRTQEHTVLESNYCKYDNQKLQLFTSSLYLAALVASMIASPVTRKLGRKQTMLLAGIL 120
VY++ QE + YCKYDNQ LQ FTSSLYLAALVAS A+ VTR +GRK +ML+ G+
Sbjct: 61 VYRKKQEAKT--NQYCKYDNQLLQTFTSSLYLAALVASFFAATVTRAVGRKWSMLVGGLT 118
Query: 121 FIVGTVLSASAGKLILLIFGRILLGCGVGFANQAVPVFLSEIAPTRIRGALNIMFQLNIT 180
F+VG L+ +A + +LI GRILLG GVGFANQ+VPV+LSE+AP R+RG LNI FQL IT
Sbjct: 119 FLVGAALNGAAQNIAMLIIGRILLGVGVGFANQSVPVYLSEMAPARLRGMLNIGFQLMIT 178
Query: 181 IGIFIANLVNWFTSKIKGGYGWRVSLAGAIIPAVMLTMGSLIVDDTPNSLIERGFEEKGK 240
IGI A L+N+ T+KIK GYGWRVSLA A +PA ++T+GSL + DTPNSL+ERG E+ +
Sbjct: 179 IGILAAELINYGTNKIKAGYGWRVSLALAAVPAAIITLGSLFLPDTPNSLLERGHPEEAR 238
Query: 241 AVLTKIRGVENIEPEFEDILRASKVANEVKSPFKDLVKSHNRPPLIIAICMQVFQQCTGI 300
+L +IRG ++I E+ D++ AS+ A +V+ P++++++ R L +A+ + FQQ TGI
Sbjct: 239 RMLRRIRGTDDIGEEYADLVAASEEARQVRHPWRNILRRRYRAQLTMAVAIPFFQQLTGI 298
Query: 301 NAIMFYAPVLFSTLGFHNDASLYSSVITGGVNVLCTLVSVYFVDKAGRRVLLLEACVQMF 360
N IMFYAPVLF TLGF NDASL SSVITG VNV T+VS+ VD+ GRR L L+ QM
Sbjct: 299 NVIMFYAPVLFDTLGFKNDASLMSSVITGLVNVFATVVSIVTVDRVGRRKLFLQGGAQMI 358
Query: 361 VSQVVIGIVLGAKL-QDHSDSLSKGYAMLVVVMVCTFVASFAWSWGPLGWLIPSETFPLE 419
V Q+++G ++ AK + ++KGYA +VVV +C +VA FAWSWGPLGWL+PSE FPLE
Sbjct: 359 VCQLIVGTLIAAKFGTSGTGDIAKGYAAVVVVFICAYVAGFAWSWGPLGWLVPSEIFPLE 418
Query: 420 TRSAGQSVTVFTNMLFTFLIAQAFLSLLCLFKFGIFLFFSAWVFVMGVFTVFLIPETKNI 479
R AGQS+ V NM FTF IAQAFL++LC FKFG+F FF+ WV +M VF F +PETKN+
Sbjct: 419 IRPAGQSINVSVNMFFTFCIAQAFLTMLCHFKFGLFYFFAGWVVIMTVFIAFFLPETKNV 478
Query: 480 PIEDMAETVWKQHWFWRRFM 499
PIE+M VWK HWFW+RF+
Sbjct: 479 PIEEMV-LVWKSHWFWKRFI 497
>emb|CAC69068.1| STP8 protein [Arabidopsis thaliana] gi|15240279|ref|NP_197997.1|
sugar transporter, putative [Arabidopsis thaliana]
gi|3319343|gb|AAC26232.1| contains similarity to sugar
transporters (Pfam: sugar_tr.hmm, score: 395.91)
[Arabidopsis thaliana] gi|7446740|pir||T01844 probable
sugar transport protein F9D12.9 - Arabidopsis thaliana
Length = 507
Score = 587 bits (1514), Expect = e-166
Identities = 290/485 (59%), Positives = 371/485 (75%), Gaps = 4/485 (0%)
Query: 15 FEARITAAVVISCIMAATGGLMFGYDVGISGGVTSMPSFLQKFFPDVYKRTQEHTVLESN 74
F+A++T V I I+AA GGL+FGYD+GISGGVT+M FL++FFP VY+R + E+N
Sbjct: 14 FDAKMTVYVFICVIIAAVGGLIFGYDIGISGGVTAMDDFLKEFFPSVYERKKH--AHENN 71
Query: 75 YCKYDNQKLQLFTSSLYLAALVASMIASPVTRKLGRKQTMLLAGILFIVGTVLSASAGKL 134
YCKYDNQ LQLFTSSLYLAALVAS AS KLGR+ TM LA I F++G L+A A +
Sbjct: 72 YCKYDNQFLQLFTSSLYLAALVASFFASATCSKLGRRPTMQLASIFFLIGVGLAAGAVNI 131
Query: 135 ILLIFGRILLGCGVGFANQAVPVFLSEIAPTRIRGALNIMFQLNITIGIFIANLVNWFTS 194
+LI GRILLG GVGF NQAVP+FLSEIAP R+RG LNI+FQL +TIGI IAN+VN+FTS
Sbjct: 132 YMLIIGRILLGFGVGFGNQAVPLFLSEIAPARLRGGLNIVFQLMVTIGILIANIVNYFTS 191
Query: 195 KIKGGYGWRVSLAGAIIPAVMLTMGSLIVDDTPNSLIERGFEEKGKAVLTKIRGVENIEP 254
I YGWR++L GA IPA++L GSL++ +TP SLIER ++GK L KIRGVE+++
Sbjct: 192 SIHP-YGWRIALGGAGIPALILLFGSLLICETPTSLIERNKTKEGKETLKKIRGVEDVDE 250
Query: 255 EFEDILRASKVANEVKSPFKDLVKSHNRPPLIIAICMQVFQQCTGINAIMFYAPVLFSTL 314
E+E I+ A +A +VK P+ L+K +RPP +I + +Q FQQ TGINAIMFYAPVLF T+
Sbjct: 251 EYESIVHACDIARQVKDPYTKLMKPASRPPFVIGMLLQFFQQFTGINAIMFYAPVLFQTV 310
Query: 315 GFHNDASLYSSVITGGVNVLCTLVSVYFVDKAGRRVLLLEACVQMFVSQVVIGIVLGAKL 374
GF NDA+L S+V+TG +NVL T V ++ VDK GRR LLL++ V M + Q+VIGI+L AK
Sbjct: 311 GFGNDAALLSAVVTGTINVLSTFVGIFLVDKTGRRFLLLQSSVHMLICQLVIGIIL-AKD 369
Query: 375 QDHSDSLSKGYAMLVVVMVCTFVASFAWSWGPLGWLIPSETFPLETRSAGQSVTVFTNML 434
D + +L++ A++VV+ VC +V FAWSWGPLGWLIPSETFPLETR+ G ++ V NM
Sbjct: 370 LDVTGTLARPQALVVVIFVCVYVMGFAWSWGPLGWLIPSETFPLETRTEGFALAVSCNMF 429
Query: 435 FTFLIAQAFLSLLCLFKFGIFLFFSAWVFVMGVFTVFLIPETKNIPIEDMAETVWKQHWF 494
FTF+IAQAFLS+LC K GIF FFS W+ VMG+F +F +PETK + I+DM ++VWK HW+
Sbjct: 430 FTFVIAQAFLSMLCAMKSGIFFFFSGWIVVMGLFALFFVPETKGVSIDDMRDSVWKLHWY 489
Query: 495 WRRFM 499
W+RFM
Sbjct: 490 WKRFM 494
>gb|AAF23220.1| putative hexose transporter [Arabidopsis thaliana]
gi|15487256|emb|CAC69073.1| STP6 protein [Arabidopsis
thaliana] gi|15230590|ref|NP_187247.1| sugar
transporter, putative [Arabidopsis thaliana]
Length = 507
Score = 585 bits (1509), Expect = e-166
Identities = 290/485 (59%), Positives = 372/485 (75%), Gaps = 4/485 (0%)
Query: 15 FEARITAAVVISCIMAATGGLMFGYDVGISGGVTSMPSFLQKFFPDVYKRTQEHTVLESN 74
FEA++T V I ++AA GGL+FGYD+GISGGV++M FL++FFP V++R + V E+N
Sbjct: 13 FEAKMTVYVFICVMIAAVGGLIFGYDIGISGGVSAMDDFLKEFFPAVWERKKH--VHENN 70
Query: 75 YCKYDNQKLQLFTSSLYLAALVASMIASPVTRKLGRKQTMLLAGILFIVGTVLSASAGKL 134
YCKYDNQ LQLFTSSLYLAALVAS +AS KLGR+ TM A I F++G L+A A L
Sbjct: 71 YCKYDNQFLQLFTSSLYLAALVASFVASATCSKLGRRPTMQFASIFFLIGVGLTAGAVNL 130
Query: 135 ILLIFGRILLGCGVGFANQAVPVFLSEIAPTRIRGALNIMFQLNITIGIFIANLVNWFTS 194
++LI GR+ LG GVGF NQAVP+FLSEIAP ++RG LNI+FQL +TIGI IAN+VN+FT+
Sbjct: 131 VMLIIGRLFLGFGVGFGNQAVPLFLSEIAPAQLRGGLNIVFQLMVTIGILIANIVNYFTA 190
Query: 195 KIKGGYGWRVSLAGAIIPAVMLTMGSLIVDDTPNSLIERGFEEKGKAVLTKIRGVENIEP 254
+ YGWR++L GA IPAV+L GSL++ +TP SLIER E+GK L KIRGV++I
Sbjct: 191 TVHP-YGWRIALGGAGIPAVILLFGSLLIIETPTSLIERNKNEEGKEALRKIRGVDDIND 249
Query: 255 EFEDILRASKVANEVKSPFKDLVKSHNRPPLIIAICMQVFQQCTGINAIMFYAPVLFSTL 314
E+E I+ A +A++VK P++ L+K +RPP II + +Q+FQQ TGINAIMFYAPVLF T+
Sbjct: 250 EYESIVHACDIASQVKDPYRKLLKPASRPPFIIGMLLQLFQQFTGINAIMFYAPVLFQTV 309
Query: 315 GFHNDASLYSSVITGGVNVLCTLVSVYFVDKAGRRVLLLEACVQMFVSQVVIGIVLGAKL 374
GF +DA+L S+VITG +NVL T V +Y VD+ GRR LLL++ V M + Q++IGI+L AK
Sbjct: 310 GFGSDAALLSAVITGSINVLATFVGIYLVDRTGRRFLLLQSSVHMLICQLIIGIIL-AKD 368
Query: 375 QDHSDSLSKGYAMLVVVMVCTFVASFAWSWGPLGWLIPSETFPLETRSAGQSVTVFTNML 434
+ +L + A++VV+ VC +V FAWSWGPLGWLIPSETFPLETRSAG +V V NM
Sbjct: 369 LGVTGTLGRPQALVVVIFVCVYVMGFAWSWGPLGWLIPSETFPLETRSAGFAVAVSCNMF 428
Query: 435 FTFLIAQAFLSLLCLFKFGIFLFFSAWVFVMGVFTVFLIPETKNIPIEDMAETVWKQHWF 494
FTF+IAQAFLS+LC + GIF FFS W+ VMG+F F IPETK I I+DM E+VWK HWF
Sbjct: 429 FTFVIAQAFLSMLCGMRSGIFFFFSGWIIVMGLFAFFFIPETKGIAIDDMRESVWKPHWF 488
Query: 495 WRRFM 499
W+R+M
Sbjct: 489 WKRYM 493
>emb|CAB80698.1| putative hexose transporter [Arabidopsis thaliana]
gi|15487244|emb|CAC69067.1| STP7 protein [Arabidopsis
thaliana] gi|15235215|ref|NP_192114.1| sugar
transporter, putative [Arabidopsis thaliana]
gi|2104529|gb|AAC78697.1| putative hexose transporter
[Arabidopsis thaliana] gi|7446742|pir||T01506 probable
hexose transport protein T10M13.6 - Arabidopsis thaliana
Length = 513
Score = 583 bits (1504), Expect = e-165
Identities = 279/506 (55%), Positives = 384/506 (75%), Gaps = 15/506 (2%)
Query: 1 MAGGGFTTGSSDVV------FEARITAAVVISCIMAATGGLMFGYDVGISGGVTSMPSFL 54
MAGG F G + V ++ ++T+ V+I+C++AA GG +FGYD+GISGGVTSM FL
Sbjct: 1 MAGGSF--GPTGVAKERAEQYQGKVTSYVIIACLVAAIGGSIFGYDIGISGGVTSMDEFL 58
Query: 55 QKFFPDVY-KRTQEHTVLESNYCKYDNQKLQLFTSSLYLAALVASMIASPVTRKLGRKQT 113
++FF VY K+ Q H ESNYCKYDNQ L FTSSLYLA LV++++ASP+TR GR+ +
Sbjct: 59 EEFFHTVYEKKKQAH---ESNYCKYDNQGLAAFTSSLYLAGLVSTLVASPITRNYGRRAS 115
Query: 114 MLLAGILFIVGTVLSASAGKLILLIFGRILLGCGVGFANQAVPVFLSEIAPTRIRGALNI 173
++ GI F++G+ L+A A L +L+ GRI+LG G+GF NQAVP++LSE+APT +RG LN+
Sbjct: 116 IVCGGISFLIGSGLNAGAVNLAMLLAGRIMLGVGIGFGNQAVPLYLSEVAPTHLRGGLNM 175
Query: 174 MFQLNITIGIFIANLVNWFTSKIKGGYGWRVSLAGAIIPAVMLTMGSLIVDDTPNSLIER 233
MFQL TIGIF AN+VN+ T ++K +GWR+SL A PA+++T+G + +TPNSL+ER
Sbjct: 176 MFQLATTIGIFTANMVNYGTQQLKP-WGWRLSLGLAAFPALLMTLGGYFLPETPNSLVER 234
Query: 234 GFEEKGKAVLTKIRGVENIEPEFEDILRASKVANEVKSPFKDLVKSHNRPPLIIAICMQV 293
G E+G+ VL K+RG EN+ E +D++ AS++AN +K PF+++++ +RP L++AICM +
Sbjct: 235 GLTERGRRVLVKLRGTENVNAELQDMVDASELANSIKHPFRNILQKRHRPQLVMAICMPM 294
Query: 294 FQQCTGINAIMFYAPVLFSTLGFHNDASLYSSVITGGVNVLCTLVSVYFVDKAGRRVLLL 353
FQ TGIN+I+FYAPVLF T+GF +ASLYSS +TG V VL T +S+ VD+ GRR LL+
Sbjct: 295 FQILTGINSILFYAPVLFQTMGFGGNASLYSSALTGAVLVLSTFISIGLVDRLGRRALLI 354
Query: 354 EACVQMFVSQVVIGIVLGAKLQDHSDSLSKGYAMLVVVMVCTFVASFAWSWGPLGWLIPS 413
+QM + QV++ ++LG K D+ + LSKGY+++VV+ +C FV +F WSWGPLGW IPS
Sbjct: 355 TGGIQMIICQVIVAVILGVKFGDNQE-LSKGYSVIVVIFICLFVVAFGWSWGPLGWTIPS 413
Query: 414 ETFPLETRSAGQSVTVFTNMLFTFLIAQAFLSLLCLFKFGIFLFFSAWVFVMGVFTVFLI 473
E FPLETRSAGQS+TV N+LFTF+IAQAFL LLC FKFGIFLFF+ WV VM +F FL+
Sbjct: 414 EIFPLETRSAGQSITVAVNLLFTFIIAQAFLGLLCAFKFGIFLFFAGWVTVMTIFVYFLL 473
Query: 474 PETKNIPIEDMAETVWKQHWFWRRFM 499
PETK +PIE+M +W +HWFW++ +
Sbjct: 474 PETKGVPIEEMT-LLWSKHWFWKKVL 498
>gb|AAD49995.1| glucose transporter [Arabidopsis thaliana]
gi|22136870|gb|AAM91779.1| putative glucose transporter
protein [Arabidopsis thaliana]
gi|16604673|gb|AAL24129.1| putative glucose transporter
protein [Arabidopsis thaliana]
gi|15220330|ref|NP_172592.1| glucose transporter (STP1)
[Arabidopsis thaliana] gi|15809962|gb|AAL06908.1|
At1g11260/T28P6_18 [Arabidopsis thaliana]
gi|21542458|sp|P23586|STP1_ARATH Glucose transporter
(Sugar carrier)
Length = 522
Score = 577 bits (1487), Expect = e-163
Identities = 276/501 (55%), Positives = 375/501 (74%), Gaps = 4/501 (0%)
Query: 1 MAGGGFTTGSSDVVFEARITAAVVISCIMAATGGLMFGYDVGISGGVTSMPSFLQKFFPD 60
M GGF G + ++T V+ +C++AA GGL+FGYD+GISGGVTSMPSFL++FFP
Sbjct: 1 MPAGGFVVGDGQKAYPGKLTPFVLFTCVVAAMGGLIFGYDIGISGGVTSMPSFLKRFFPS 60
Query: 61 VYKRTQEHTVLESNYCKYDNQKLQLFTSSLYLAALVASMIASPVTRKLGRKQTMLLAGIL 120
VY++ QE + YC+YD+ L +FTSSLYLAAL++S++AS VTRK GR+ +ML GIL
Sbjct: 61 VYRKQQEDAST-NQYCQYDSPTLTMFTSSLYLAALISSLVASTVTRKFGRRLSMLFGGIL 119
Query: 121 FIVGTVLSASAGKLILLIFGRILLGCGVGFANQAVPVFLSEIAPTRIRGALNIMFQLNIT 180
F G +++ A + +LI GRILLG G+GFANQAVP++LSE+AP + RGALNI FQL+IT
Sbjct: 120 FCAGALINGFAKHVWMLIVGRILLGFGIGFANQAVPLYLSEMAPYKYRGALNIGFQLSIT 179
Query: 181 IGIFIANLVNWFTSKIKGGYGWRVSLAGAIIPAVMLTMGSLIVDDTPNSLIERGFEEKGK 240
IGI +A ++N+F +KIKGG+GWR+SL GA++PA+++T+GSL++ DTPNS+IERG E+ K
Sbjct: 180 IGILVAEVLNYFFAKIKGGWGWRLSLGGAVVPALIITIGSLVLPDTPNSMIERGQHEEAK 239
Query: 241 AVLTKIRGVENIEPEFEDILRASKVANEVKSPFKDLVKSHNRPPLIIAICMQVFQQCTGI 300
L +IRGV+++ EF+D++ ASK + ++ P+++L++ RP L +A+ + FQQ TGI
Sbjct: 240 TKLRRIRGVDDVSQEFDDLVAASKESQSIEHPWRNLLRRKYRPHLTMAVMIPFFQQLTGI 299
Query: 301 NAIMFYAPVLFSTLGFHNDASLYSSVITGGVNVLCTLVSVYFVDKAGRRVLLLEACVQMF 360
N IMFYAPVLF+T+GF DASL S+V+TG VNV TLVS+Y VD+ GRR L LE QM
Sbjct: 300 NVIMFYAPVLFNTIGFTTDASLMSAVVTGSVNVAATLVSIYGVDRWGRRFLFLEGGTQML 359
Query: 361 VSQVVIGIVLGAK--LQDHSDSLSKGYAMLVVVMVCTFVASFAWSWGPLGWLIPSETFPL 418
+ Q V+ +GAK + L K YA++VV +C +VA FAWSWGPLGWL+PSE FPL
Sbjct: 360 ICQAVVAACIGAKFGVDGTPGELPKWYAIVVVTFICIYVAGFAWSWGPLGWLVPSEIFPL 419
Query: 419 ETRSAGQSVTVFTNMLFTFLIAQAFLSLLCLFKFGIFLFFSAWVFVMGVFTVFLIPETKN 478
E RSA QS+TV NM+FTF+IAQ FL++LC KFG+FL F+ +V VM +F +PETK
Sbjct: 420 EIRSAAQSITVSVNMIFTFIIAQIFLTMLCHLKFGLFLVFAFFVVVMSIFVYIFLPETKG 479
Query: 479 IPIEDMAETVWKQHWFWRRFM 499
IPIE+M + VW+ HW+W RF+
Sbjct: 480 IPIEEMGQ-VWRSHWYWSRFV 499
>gb|AAM67326.1| glucose transporter [Arabidopsis thaliana]
Length = 522
Score = 577 bits (1486), Expect = e-163
Identities = 276/501 (55%), Positives = 375/501 (74%), Gaps = 4/501 (0%)
Query: 1 MAGGGFTTGSSDVVFEARITAAVVISCIMAATGGLMFGYDVGISGGVTSMPSFLQKFFPD 60
M GGF G + ++T V+ +C++AA GGL+FGYD+GISGGVTSMPSFL++FFP
Sbjct: 1 MPAGGFVVGDGQKAYPGKLTPFVLFTCVVAAMGGLIFGYDIGISGGVTSMPSFLKRFFPS 60
Query: 61 VYKRTQEHTVLESNYCKYDNQKLQLFTSSLYLAALVASMIASPVTRKLGRKQTMLLAGIL 120
VY++ QE + YC+YD+ L +FTSSLYLAAL++S++AS VTRK GR+ +ML GIL
Sbjct: 61 VYRKQQEDAST-NQYCQYDSPTLTMFTSSLYLAALISSLVASTVTRKFGRRLSMLFGGIL 119
Query: 121 FIVGTVLSASAGKLILLIFGRILLGCGVGFANQAVPVFLSEIAPTRIRGALNIMFQLNIT 180
F G +++ A + +LI GRILLG G+GFANQAVP++LSE+AP + RGALNI FQL+IT
Sbjct: 120 FCAGALINGFAKHVWMLIVGRILLGFGIGFANQAVPLYLSEMAPYKYRGALNIGFQLSIT 179
Query: 181 IGIFIANLVNWFTSKIKGGYGWRVSLAGAIIPAVMLTMGSLIVDDTPNSLIERGFEEKGK 240
IGI +A ++N+F +KIKGG+GWR+SL GA++PA+++T+GSL++ DTPNS+IERG E+ K
Sbjct: 180 IGILVAEVLNYFFAKIKGGWGWRLSLGGAVVPALIITIGSLVLPDTPNSMIERGQHEEAK 239
Query: 241 AVLTKIRGVENIEPEFEDILRASKVANEVKSPFKDLVKSHNRPPLIIAICMQVFQQCTGI 300
L +IRGV+++ EF+D++ ASK + ++ P+++L++ RP L +A+ + FQQ TGI
Sbjct: 240 TKLRRIRGVDDVSQEFDDLVAASKESQSIEHPWRNLLRRKYRPHLTMAVMIPFFQQLTGI 299
Query: 301 NAIMFYAPVLFSTLGFHNDASLYSSVITGGVNVLCTLVSVYFVDKAGRRVLLLEACVQMF 360
N IMFYAPVLF+T+GF DASL S+V+TG VNV TLVS+Y VD+ GRR L LE QM
Sbjct: 300 NVIMFYAPVLFNTIGFTTDASLMSAVVTGSVNVAATLVSIYGVDRWGRRFLFLEGGTQML 359
Query: 361 VSQVVIGIVLGAK--LQDHSDSLSKGYAMLVVVMVCTFVASFAWSWGPLGWLIPSETFPL 418
+ Q V+ +GAK + L K YA++VV +C +VA FAWSWGPLGWL+PSE FPL
Sbjct: 360 ICQAVVAACIGAKFGVDGTPGELPKWYAIVVVTFICIYVAGFAWSWGPLGWLVPSEIFPL 419
Query: 419 ETRSAGQSVTVFTNMLFTFLIAQAFLSLLCLFKFGIFLFFSAWVFVMGVFTVFLIPETKN 478
E RSA QS+TV NM+FTF+IAQ FL++LC KFG+FL F+ +V VM +F +PETK
Sbjct: 420 EIRSAAQSITVSVNMIFTFIIAQIFLTMLCHLKFGLFLVFAFFVVVMSIFEYIFLPETKG 479
Query: 479 IPIEDMAETVWKQHWFWRRFM 499
IPIE+M + VW+ HW+W RF+
Sbjct: 480 IPIEEMGQ-VWRSHWYWSRFV 499
>emb|CAA39037.1| glucose transporter [Arabidopsis thaliana]
Length = 522
Score = 576 bits (1484), Expect = e-163
Identities = 276/501 (55%), Positives = 375/501 (74%), Gaps = 4/501 (0%)
Query: 1 MAGGGFTTGSSDVVFEARITAAVVISCIMAATGGLMFGYDVGISGGVTSMPSFLQKFFPD 60
M GGF G + ++T V+ +C++AA GGL+FGYD+GISGGVTSMPSFL++FFP
Sbjct: 1 MPAGGFVVGDGQKAYPGKLTPFVLFTCVVAAMGGLIFGYDIGISGGVTSMPSFLKRFFPS 60
Query: 61 VYKRTQEHTVLESNYCKYDNQKLQLFTSSLYLAALVASMIASPVTRKLGRKQTMLLAGIL 120
VY++ QE + YC+YD+ L +FTSSLYLAAL++S++AS VTRK GR+ +ML GIL
Sbjct: 61 VYRKQQEDAST-NQYCQYDSPTLTMFTSSLYLAALISSLVASTVTRKFGRRLSMLFGGIL 119
Query: 121 FIVGTVLSASAGKLILLIFGRILLGCGVGFANQAVPVFLSEIAPTRIRGALNIMFQLNIT 180
F G +++ A + +LI GRILLG G+GFANQAVP++LSE+AP + RGALNI FQL+IT
Sbjct: 120 FCAGALINGFAKHVWMLIVGRILLGFGIGFANQAVPLYLSEMAPYKYRGALNIGFQLSIT 179
Query: 181 IGIFIANLVNWFTSKIKGGYGWRVSLAGAIIPAVMLTMGSLIVDDTPNSLIERGFEEKGK 240
IGI +A ++N+F +KIKGG+GWR+SL GA++PA+++T+GSL++ DTPNS+IERG E+ K
Sbjct: 180 IGILVAEVLNYFFAKIKGGWGWRLSLGGAVVPALIITIGSLVLPDTPNSMIERGQHEEAK 239
Query: 241 AVLTKIRGVENIEPEFEDILRASKVANEVKSPFKDLVKSHNRPPLIIAICMQVFQQCTGI 300
L +IRGV+++ EF+D++ ASK + ++ P+++L++ RP L +A+ + FQQ TGI
Sbjct: 240 TKLRRIRGVDDVSQEFDDLVAASKESQSIEHPWRNLLRRKYRPHLTMAVMIPFFQQLTGI 299
Query: 301 NAIMFYAPVLFSTLGFHNDASLYSSVITGGVNVLCTLVSVYFVDKAGRRVLLLEACVQMF 360
N IMFYAPVLF+T+GF DASL S+V+TG VNV TLVS+Y VD+ GRR L LE QM
Sbjct: 300 NVIMFYAPVLFNTIGFTTDASLMSAVVTGSVNVGATLVSIYGVDRWGRRFLFLEGGTQML 359
Query: 361 VSQVVIGIVLGAK--LQDHSDSLSKGYAMLVVVMVCTFVASFAWSWGPLGWLIPSETFPL 418
+ Q V+ +GAK + L K YA++VV +C +VA FAWSWGPLGWL+PSE FPL
Sbjct: 360 ICQAVVAACIGAKFGVDGTPGELPKWYAIVVVTFICIYVAGFAWSWGPLGWLVPSEIFPL 419
Query: 419 ETRSAGQSVTVFTNMLFTFLIAQAFLSLLCLFKFGIFLFFSAWVFVMGVFTVFLIPETKN 478
E RSA QS+TV NM+FTF+IAQ FL++LC KFG+FL F+ +V VM +F +PETK
Sbjct: 420 EIRSAAQSITVSVNMIFTFIIAQIFLTMLCHLKFGLFLVFAFFVVVMSIFVYIFLPETKG 479
Query: 479 IPIEDMAETVWKQHWFWRRFM 499
IPIE+M + VW+ HW+W RF+
Sbjct: 480 IPIEEMGQ-VWRSHWYWSRFV 499
>emb|CAA04511.1| hexose transporter [Vitis vinifera]
Length = 519
Score = 576 bits (1484), Expect = e-163
Identities = 282/501 (56%), Positives = 379/501 (75%), Gaps = 5/501 (0%)
Query: 2 AGGGFTTGSSDVVFEARITAAVVISCIMAATGGLMFGYDVGISGGVTSMPSFLQKFFPDV 61
A GGF G+ + +T V ++C++AA GGL+FGYD+GISGGVTSM FLQKFFP V
Sbjct: 3 AVGGFDKGTGKA-YPGNLTPYVTVTCVVAAMGGLIFGYDIGISGGVTSMAPFLQKFFPSV 61
Query: 62 YKRTQEHTVLESNYCKYDNQKLQLFTSSLYLAALVASMIASPVTRKLGRKQTMLLAGILF 121
Y++ + + YCK+D++ L LFTSSLYLAAL++S++A+ VTRK GRK +ML G+LF
Sbjct: 62 YRK-EALDKSTNQYCKFDSETLTLFTSSLYLAALLSSLVAATVTRKFGRKLSMLFGGLLF 120
Query: 122 IVGTVLSASAGKLILLIFGRILLGCGVGFANQAVPVFLSEIAPTRIRGALNIMFQLNITI 181
G +++ +A + +LI GRILLG G+GFANQ+VP++LSE+AP + RGALNI FQL+ITI
Sbjct: 121 CAGAIINGAAKAVWMLIVGRILLGFGIGFANQSVPLYLSEMAPYKYRGALNIGFQLSITI 180
Query: 182 GIFIANLVNWFTSKIKGGYGWRVSLAGAIIPAVMLTMGSLIVDDTPNSLIERGFEEKGKA 241
GI +AN++N+F +KIKGG+GWR+SL GA++PA+++T+GSL++ DTPNS+IERG E K
Sbjct: 181 GILVANILNYFFAKIKGGWGWRLSLGGAVVPALIITVGSLVLPDTPNSMIERGQHEGAKT 240
Query: 242 VLTKIRGVENIEPEFEDILRASKVANEVKSPFKDLVKSHNRPPLIIAICMQVFQQCTGIN 301
L +IRGV+++E EF D++ AS+ + V+ P+++L++ RP L +AI + FQQ TGIN
Sbjct: 241 KLRRIRGVDDVEEEFNDLVVASEASKLVEHPWRNLLQRKYRPHLTMAILIPFFQQLTGIN 300
Query: 302 AIMFYAPVLFSTLGFHNDASLYSSVITGGVNVLCTLVSVYFVDKAGRRVLLLEACVQMFV 361
IMFYAPVLF T+GF +DASL S+VITGGVNVL T+VS+Y VDK GRR L LE QM +
Sbjct: 301 VIMFYAPVLFKTIGFADDASLMSAVITGGVNVLATIVSIYGVDKWGRRFLFLEGGTQMLI 360
Query: 362 SQVVIGIVLGAK--LQDHSDSLSKGYAMLVVVMVCTFVASFAWSWGPLGWLIPSETFPLE 419
QV++ +G K + +L K YA++VV+ +C +V+ FAWSWGPLGWL+PSE FPLE
Sbjct: 361 CQVIVATCIGVKFGVDGEPGALPKWYAIVVVLFICVYVSGFAWSWGPLGWLVPSEIFPLE 420
Query: 420 TRSAGQSVTVFTNMLFTFLIAQAFLSLLCLFKFGIFLFFSAWVFVMGVFTVFLIPETKNI 479
RSA QSV V NM FTF+IAQ FL++LC KFG+FLFF+ +V VM F F +PETK I
Sbjct: 421 IRSAAQSVNVSVNMFFTFIIAQIFLNMLCHMKFGLFLFFAFFVVVMSFFIYFFLPETKGI 480
Query: 480 PIEDMAETVWKQHWFWRRFMH 500
PIE+MAE VWK HWFW R+++
Sbjct: 481 PIEEMAE-VWKSHWFWSRYVN 500
>gb|AAB06594.1| sugar transporter
Length = 518
Score = 573 bits (1478), Expect = e-162
Identities = 279/501 (55%), Positives = 381/501 (75%), Gaps = 4/501 (0%)
Query: 1 MAGGGFTTGSSDVVFEARITAAVVISCIMAATGGLMFGYDVGISGGVTSMPSFLQKFFPD 60
MAGGG G + + +T V I+CI+AA GGL+FGYD+GISGGVTSM FL+KFFP
Sbjct: 1 MAGGGIPIGGGNKEYPGNLTPFVTITCIVAAMGGLIFGYDIGISGGVTSMDPFLKKFFPA 60
Query: 61 VYKRTQEHTVLESNYCKYDNQKLQLFTSSLYLAALVASMIASPVTRKLGRKQTMLLAGIL 120
VY++ + + YC+YD+Q L +FTSSLYLAAL++S++AS +TR+ GRK +ML G+L
Sbjct: 61 VYRKKNKDKST-NQYCQYDSQTLTMFTSSLYLAALLSSLVASTITRRFGRKLSMLFGGLL 119
Query: 121 FIVGTVLSASAGKLILLIFGRILLGCGVGFANQAVPVFLSEIAPTRIRGALNIMFQLNIT 180
F+VG +++ A + +LI GRILLG G+GFANQ VP++LSE+AP + RGALNI FQL+IT
Sbjct: 120 FLVGALINGFANHVWMLIVGRILLGFGIGFANQPVPLYLSEMAPYKYRGALNIGFQLSIT 179
Query: 181 IGIFIANLVNWFTSKIKGGYGWRVSLAGAIIPAVMLTMGSLIVDDTPNSLIERGFEEKGK 240
IGI +AN++N+F +KIKGG+GWR+SL GA++PA+++T+GSL++ DTPNS+IERG + K
Sbjct: 180 IGILVANVLNYFFAKIKGGWGWRLSLGGAMVPALIITIGSLVLPDTPNSMIERGDRDGAK 239
Query: 241 AVLTKIRGVENIEPEFEDILRASKVANEVKSPFKDLVKSHNRPPLIIAICMQVFQQCTGI 300
A L +IRG+E+++ EF D++ AS+ + +V++P+++L++ RP L +A+ + FQQ TGI
Sbjct: 240 AQLKRIRGIEDVDEEFNDLVAASEASMQVENPWRNLLQRKYRPQLTMAVLIPFFQQFTGI 299
Query: 301 NAIMFYAPVLFSTLGFHNDASLYSSVITGGVNVLCTLVSVYFVDKAGRRVLLLEACVQMF 360
N IMFYAPVLF+++GF +DASL S+VITG VNV+ T VS+Y VDK GRR L LE QM
Sbjct: 300 NVIMFYAPVLFNSIGFKDDASLMSAVITGVVNVVATCVSIYGVDKWGRRALFLEGGAQML 359
Query: 361 VSQVVIGIVLGAKL--QDHSDSLSKGYAMLVVVMVCTFVASFAWSWGPLGWLIPSETFPL 418
+ QV + +GAK + +L + YA++VV+ +C +VA FAWSWGPLGWL+PSE FPL
Sbjct: 360 ICQVAVAAAIGAKFGTSGNPGNLPEWYAIVVVLFICIYVAGFAWSWGPLGWLVPSEIFPL 419
Query: 419 ETRSAGQSVTVFTNMLFTFLIAQAFLSLLCLFKFGIFLFFSAWVFVMGVFTVFLIPETKN 478
E RSA QSV V NMLFTFL+AQ FL +LC KFG+FLFF+ +V VM ++ FL+PETK
Sbjct: 420 EIRSAAQSVNVSVNMLFTFLVAQVFLIMLCHMKFGLFLFFAFFVLVMSIYVFFLLPETKG 479
Query: 479 IPIEDMAETVWKQHWFWRRFM 499
IPIE+M + VWK H FW RF+
Sbjct: 480 IPIEEM-DRVWKSHPFWSRFV 499
>emb|CAG27609.1| monosaccharide transporter [Populus tremula x Populus tremuloides]
Length = 522
Score = 572 bits (1475), Expect = e-162
Identities = 273/501 (54%), Positives = 381/501 (75%), Gaps = 4/501 (0%)
Query: 1 MAGGGFTTGSSDVVFEARITAAVVISCIMAATGGLMFGYDVGISGGVTSMPSFLQKFFPD 60
M G G + + +T V ++CI+AA GGL+FGYD+GISGGVTSMPSFL+KFFP
Sbjct: 1 MPAVGIAVGDNKKEYPGNLTPFVTVTCIVAAMGGLIFGYDIGISGGVTSMPSFLKKFFPS 60
Query: 61 VYKRTQEHTVLESNYCKYDNQKLQLFTSSLYLAALVASMIASPVTRKLGRKQTMLLAGIL 120
VY++ QE + YC+YD+Q L +FTSSLYLAAL+AS++AS VTRK GRK +ML G+L
Sbjct: 61 VYRKQQEDAT-SNQYCQYDSQTLTMFTSSLYLAALLASLVASIVTRKFGRKLSMLFGGVL 119
Query: 121 FIVGTVLSASAGKLILLIFGRILLGCGVGFANQAVPVFLSEIAPTRIRGALNIMFQLNIT 180
F G +++ A + +LI GRILLG G+GFANQ+VP++LSE+AP + RGALNI FQL+IT
Sbjct: 120 FCAGAIINGFAQAVWMLILGRILLGFGIGFANQSVPLYLSEMAPYKFRGALNIGFQLSIT 179
Query: 181 IGIFIANLVNWFTSKIKGGYGWRVSLAGAIIPAVMLTMGSLIVDDTPNSLIERGFEEKGK 240
IGI +AN++N+F +KI GG+GWR+SL GA++PA+++T+GSL++ DTPNS+IERG ++ +
Sbjct: 180 IGILVANVLNYFFAKIHGGWGWRLSLGGAMVPALIITVGSLVLPDTPNSMIERGQHDEAR 239
Query: 241 AVLTKIRGVENIEPEFEDILRASKVANEVKSPFKDLVKSHNRPPLIIAICMQVFQQCTGI 300
L ++RGV++++ EF D++ AS+ + +V+ P+++L++ RP + +A+ + FQQ TGI
Sbjct: 240 EKLRRVRGVDDVDEEFNDLVAASEASMKVEHPWRNLLQRKYRPHITMAVMIPFFQQLTGI 299
Query: 301 NAIMFYAPVLFSTLGFHNDASLYSSVITGGVNVLCTLVSVYFVDKAGRRVLLLEACVQMF 360
N IMFYAPVLF+T+GF ++ASL S+VITG VNV+ T+VS+Y VDK GRR L LE QM
Sbjct: 300 NVIMFYAPVLFNTIGFGSNASLMSAVITGVVNVVATMVSIYGVDKWGRRFLFLEGGFQML 359
Query: 361 VSQVVIGIVLGAK--LQDHSDSLSKGYAMLVVVMVCTFVASFAWSWGPLGWLIPSETFPL 418
+ Q V+ +GAK + + L K YA++VV+ +C +VA FAWSWGPLGWL+PSE FPL
Sbjct: 360 ICQAVVAACIGAKFGVNGNPGELPKWYAIVVVLFICIYVAGFAWSWGPLGWLVPSEFFPL 419
Query: 419 ETRSAGQSVTVFTNMLFTFLIAQAFLSLLCLFKFGIFLFFSAWVFVMGVFTVFLIPETKN 478
E RSA QS++V NMLFTF++AQ FL++LC KFG+FLFF+ +V +M +F + +PETK
Sbjct: 420 EIRSAAQSISVSVNMLFTFIVAQIFLTMLCHLKFGLFLFFAFFVVLMSIFVYYFLPETKG 479
Query: 479 IPIEDMAETVWKQHWFWRRFM 499
IPIE+M + VWK HWFW R++
Sbjct: 480 IPIEEMGQ-VWKTHWFWSRYV 499
>sp|Q41144|STC_RICCO Sugar carrier protein C gi|169718|gb|AAA79761.1| sugar carrier
protein
Length = 523
Score = 571 bits (1472), Expect = e-161
Identities = 273/499 (54%), Positives = 380/499 (75%), Gaps = 4/499 (0%)
Query: 3 GGGFTTGSSDVVFEARITAAVVISCIMAATGGLMFGYDVGISGGVTSMPSFLQKFFPDVY 62
GG +G + V+ +T V ++C++AA GGL+FGYD+GISGGVTSM SFL+KFFP VY
Sbjct: 5 GGIPPSGGNRKVYPGNLTLYVTVTCVVAAMGGLIFGYDIGISGGVTSMDSFLKKFFPSVY 64
Query: 63 KRTQEHTVLESNYCKYDNQKLQLFTSSLYLAALVASMIASPVTRKLGRKQTMLLAGILFI 122
++ + + YC+YD+Q L +FTSSLYLAAL+AS++AS +TRK GRK +ML G+LF
Sbjct: 65 RKKKADES-SNQYCQYDSQTLTMFTSSLYLAALIASLVASTITRKFGRKLSMLFGGVLFC 123
Query: 123 VGTVLSASAGKLILLIFGRILLGCGVGFANQAVPVFLSEIAPTRIRGALNIMFQLNITIG 182
G +++ +A + +LI GRILLG G+GFANQ+VP++LSE+AP + RGALNI FQL+ITIG
Sbjct: 124 AGAIINGAAKAVWMLILGRILLGFGIGFANQSVPLYLSEMAPYKYRGALNIGFQLSITIG 183
Query: 183 IFIANLVNWFTSKIKGGYGWRVSLAGAIIPAVMLTMGSLIVDDTPNSLIERGFEEKGKAV 242
I +AN++N+F +KIKGG+GWR+SL GA++PA+++T+GSL++ DTPNS+IERG E+ +A
Sbjct: 184 ILVANVLNYFFAKIKGGWGWRLSLGGAMVPALIITVGSLVLPDTPNSMIERGQHEEARAH 243
Query: 243 LTKIRGVENIEPEFEDILRASKVANEVKSPFKDLVKSHNRPPLIIAICMQVFQQCTGINA 302
L ++RGVE+++ EF D++ AS+ + +V+ P+++L++ RP L +AI + FQQ TGIN
Sbjct: 244 LKRVRGVEDVDEEFTDLVHASEDSKKVEHPWRNLLQRKYRPHLSMAIAIPFFQQLTGINV 303
Query: 303 IMFYAPVLFSTLGFHNDASLYSSVITGGVNVLCTLVSVYFVDKAGRRVLLLEACVQMFVS 362
IMFYAPVLF T+GF +DA+L S+VITG VNV T+VS+Y VDK GRR L LE VQM +
Sbjct: 304 IMFYAPVLFDTIGFGSDAALMSAVITGLVNVFATMVSIYGVDKWGRRFLFLEGGVQMLIC 363
Query: 363 QVVIGIVLGAK--LQDHSDSLSKGYAMLVVVMVCTFVASFAWSWGPLGWLIPSETFPLET 420
Q ++ +GAK + L + YA++VV+ +C +V+ FAWSWGPLGWL+PSE FPLE
Sbjct: 364 QAIVAACIGAKFGVDGAPGDLPQWYAVVVVLFICIYVSGFAWSWGPLGWLVPSEIFPLEI 423
Query: 421 RSAGQSVTVFTNMLFTFLIAQAFLSLLCLFKFGIFLFFSAWVFVMGVFTVFLIPETKNIP 480
RSA QSV V NM FTF++AQ FL +LC KFG+F+FFS +V +M +F + +PETK IP
Sbjct: 424 RSAAQSVNVSVNMFFTFVVAQVFLIMLCHLKFGLFIFFSFFVLIMSIFVYYFLPETKGIP 483
Query: 481 IEDMAETVWKQHWFWRRFM 499
IE+M + VWKQHW+W R++
Sbjct: 484 IEEMGQ-VWKQHWYWSRYV 501
>gb|AAX47308.1| hexose transporter 7 [Vitis vinifera]
Length = 526
Score = 571 bits (1472), Expect = e-161
Identities = 274/487 (56%), Positives = 374/487 (76%), Gaps = 10/487 (2%)
Query: 15 FEARITAAVVISCIMAATGGLMFGYDVGISGGVTSMPSFLQKFFPDVY--KRTQEHTVLE 72
++ R+T VV++C++AA GG +FGYD+G+SGGVTSM +FL+KFF VY KR E E
Sbjct: 21 YKGRLTTYVVVACLVAAVGGAIFGYDIGVSGGVTSMDTFLEKFFHTVYLKKRRAE----E 76
Query: 73 SNYCKYDNQKLQLFTSSLYLAALVASMIASPVTRKLGRKQTMLLAGILFIVGTVLSASAG 132
+YCKY++Q L FTSSLYLA LVAS++ASP+TRK GR+ +++ GI F++G L+A+A
Sbjct: 77 DHYCKYNDQGLAAFTSSLYLAGLVASIVASPITRKYGRRASIVCGGISFLIGAALNAAAV 136
Query: 133 KLILLIFGRILLGCGVGFANQAVPVFLSEIAPTRIRGALNIMFQLNITIGIFIANLVNWF 192
L +L+ GRI+LG G+GF +QAVP++LSE+AP +RGALN+MFQL T GIF AN++N+
Sbjct: 137 NLAMLLSGRIMLGIGIGFGDQAVPLYLSEMAPAHLRGALNMMFQLATTTGIFTANMINYG 196
Query: 193 TSKIKGGYGWRVSLAGAIIPAVMLTMGSLIVDDTPNSLIERGFEEKGKAVLTKIRGVENI 252
T+K+ +GWR+SL A +PA+++T+G L + +TPNSLIERG EKG+ VL +IRG +
Sbjct: 197 TAKLPS-WGWRLSLGLAALPAILMTVGGLFLPETPNSLIERGSREKGRRVLERIRGTNEV 255
Query: 253 EPEFEDILRASKVANEVKSPFKDLVKSHNRPPLIIAICMQVFQQCTGINAIMFYAPVLFS 312
+ EFEDI+ AS++AN +K PF+++++ NRP L++AICM FQ GIN+I+FYAPVLF
Sbjct: 256 DAEFEDIVDASELANSIKHPFRNILERRNRPQLVMAICMPAFQILNGINSILFYAPVLFQ 315
Query: 313 TLGFHNDASLYSSVITGGVNVLCTLVSVYFVDKAGRRVLLLEACVQMFVSQVVIGIVLGA 372
T+GF N A+LYSS +TG V VL T+VS+ VD+ GRRVLL+ +QM + QV + I+LG
Sbjct: 316 TMGFGN-ATLYSSALTGAVLVLSTVVSIGLVDRLGRRVLLISGGIQMVLCQVTVAIILGV 374
Query: 373 KLQDHSDSLSKGYAMLVVVMVCTFVASFAWSWGPLGWLIPSETFPLETRSAGQSVTVFTN 432
K + D LSKGY++LVV+++C FV +F WSWGPLGW +PSE FPLETRSAGQS+TV N
Sbjct: 375 KFGSN-DGLSKGYSVLVVIVICLFVIAFGWSWGPLGWTVPSEIFPLETRSAGQSITVVVN 433
Query: 433 MLFTFLIAQAFLSLLCLFKFGIFLFFSAWVFVMGVFTVFLIPETKNIPIEDMAETVWKQH 492
+LFTF+IAQ FLS+LC FK GIFLFF+ W+ +M +F F +PETK +PIE+M VWK+H
Sbjct: 434 LLFTFIIAQCFLSMLCSFKHGIFLFFAGWIVIMTLFVYFFLPETKGVPIEEMI-FVWKKH 492
Query: 493 WFWRRFM 499
WFW+R +
Sbjct: 493 WFWKRMV 499
>emb|CAD30830.1| monosaccharide-H+ symporter [Datisca glomerata]
Length = 523
Score = 567 bits (1461), Expect = e-160
Identities = 275/500 (55%), Positives = 378/500 (75%), Gaps = 4/500 (0%)
Query: 2 AGGGFTTGSSDVVFEARITAAVVISCIMAATGGLMFGYDVGISGGVTSMPSFLQKFFPDV 61
A GG G S + +T V I+CI+AA GGL+FGYD+GISGGVTSM SFL+KFFP V
Sbjct: 3 AVGGIVVGGSKKEYPGNLTPYVTITCIVAAMGGLIFGYDIGISGGVTSMDSFLKKFFPAV 62
Query: 62 YKRTQEHTVLESNYCKYDNQKLQLFTSSLYLAALVASMIASPVTRKLGRKQTMLLAGILF 121
Y++ +E + YC+YD+Q L +FTSSLYLAAL+AS++AS +TRK GR+ +ML GILF
Sbjct: 63 YRK-KELDSTTNQYCQYDSQTLTMFTSSLYLAALLASIVASTITRKFGRRLSMLFGGILF 121
Query: 122 IVGTVLSASAGKLILLIFGRILLGCGVGFANQAVPVFLSEIAPTRIRGALNIMFQLNITI 181
G +++ A + +LI GR+ LG G+GF+NQ+VP++LSE+AP + RGALNI FQL+ITI
Sbjct: 122 CAGAIINGFAQAVWMLILGRMFLGFGIGFSNQSVPLYLSEMAPYKYRGALNIGFQLSITI 181
Query: 182 GIFIANLVNWFTSKIKGGYGWRVSLAGAIIPAVMLTMGSLIVDDTPNSLIERGFEEKGKA 241
GI +AN++N+F +KI+GG+GWR+SL GA++PA+++T+GSL++ DTPNSLIERG ++ ++
Sbjct: 182 GILVANVLNYFFAKIRGGWGWRLSLGGAMVPALIITVGSLLLPDTPNSLIERGNRDEARS 241
Query: 242 VLTKIRGVENIEPEFEDILRASKVANEVKSPFKDLVKSHNRPPLIIAICMQVFQQCTGIN 301
L ++RGV++++ EF D++ AS+ + +V+ P+ +L++ RP L +AI + FQQ TGIN
Sbjct: 242 KLQRVRGVDDVDEEFNDLVAASEESKQVEHPWTNLLRRKYRPHLAMAILIPFFQQLTGIN 301
Query: 302 AIMFYAPVLFSTLGFHNDASLYSSVITGGVNVLCTLVSVYFVDKAGRRVLLLEACVQMFV 361
IMFYAPVLF+T+GF +DASL S+VITG VNV TLVS+Y VDK GRR L LE QM +
Sbjct: 302 VIMFYAPVLFNTIGFGSDASLMSAVITGCVNVAGTLVSIYGVDKWGRRFLFLEGGFQMLI 361
Query: 362 SQVVIGIVLGAK--LQDHSDSLSKGYAMLVVVMVCTFVASFAWSWGPLGWLIPSETFPLE 419
Q V+ +GAK + + L K YA++VV+ +C +VA F+WSWGPLGWL+PSE+FPLE
Sbjct: 362 CQAVVAAAIGAKFGVNGNPGELPKWYAIVVVLFICIYVAGFSWSWGPLGWLVPSESFPLE 421
Query: 420 TRSAGQSVTVFTNMLFTFLIAQAFLSLLCLFKFGIFLFFSAWVFVMGVFTVFLIPETKNI 479
RSA QS+ V NM+FTF IAQ FL++LC KFG+F+FF+ +V VM +F F +PETK I
Sbjct: 422 IRSAAQSINVSVNMIFTFAIAQIFLTMLCHLKFGLFIFFAFFVVVMSIFVYFFLPETKGI 481
Query: 480 PIEDMAETVWKQHWFWRRFM 499
PIE+M VWK HW+W RF+
Sbjct: 482 PIEEMGR-VWKSHWYWSRFV 500
>emb|CAB07812.1| monosaccharid transport protein [Vicia faba] gi|7446727|pir||T12199
monosaccharid transport protein - fava bean
Length = 516
Score = 566 bits (1459), Expect = e-160
Identities = 277/502 (55%), Positives = 383/502 (76%), Gaps = 6/502 (1%)
Query: 1 MAGGGFTTGSSDVVFEARITAAVVISCIMAATGGLMFGYDVGISGGVTSMPSFLQKFFPD 60
M G G+ + + +T V I+C++AA GGL+FGYD+GISGGVTSM FL+KFFP
Sbjct: 1 MPAAGIPIGAGNKEYPGNLTPFVTITCVVAAMGGLIFGYDIGISGGVTSMNPFLEKFFPA 60
Query: 61 VY-KRTQEHTVLESNYCKYDNQKLQLFTSSLYLAALVASMIASPVTRKLGRKQTMLLAGI 119
VY K+ +H+ ++ YC+YD++ L LFTSSLYLAAL++S++AS +TR+ GRK +ML G+
Sbjct: 61 VYRKKNAQHS--KNQYCQYDSETLTLFTSSLYLAALLSSVVASTITRRFGRKLSMLFGGL 118
Query: 120 LFIVGTVLSASAGKLILLIFGRILLGCGVGFANQAVPVFLSEIAPTRIRGALNIMFQLNI 179
LF+VG +++ A + +LI GRILLG G+GFANQ+VP++LSE+AP + RGALNI FQL+I
Sbjct: 119 LFLVGALINGLAQNVAMLIVGRILLGFGIGFANQSVPLYLSEMAPYKYRGALNIGFQLSI 178
Query: 180 TIGIFIANLVNWFTSKIKGGYGWRVSLAGAIIPAVMLTMGSLIVDDTPNSLIERGFEEKG 239
TIGI +AN++N+F +KIKGG+GWR+SL GA++PA+++T+GSLI+ DTPNS+IERG +
Sbjct: 179 TIGILVANILNYFFAKIKGGWGWRLSLGGAMVPALIITIGSLILPDTPNSMIERGDRDGA 238
Query: 240 KAVLTKIRGVENIEPEFEDILRASKVANEVKSPFKDLVKSHNRPPLIIAICMQVFQQCTG 299
KA L +IRGVE+++ EF D++ AS+ + +V++P+++L++ RP L +A+ + FQQ TG
Sbjct: 239 KAQLKRIRGVEDVDEEFNDLVAASETSMQVENPWRNLLQRKYRPQLTMAVLIPFFQQFTG 298
Query: 300 INAIMFYAPVLFSTLGFHNDASLYSSVITGGVNVLCTLVSVYFVDKAGRRVLLLEACVQM 359
IN IMFYAPVLF+++GF +DASL S+VITG VNV+ T VS+Y VDK GRR L LE VQM
Sbjct: 299 INVIMFYAPVLFNSIGFKDDASLMSAVITGVVNVVATCVSIYGVDKWGRRALFLEGGVQM 358
Query: 360 FVSQVVIGIVLGAKLQDHSD--SLSKGYAMLVVVMVCTFVASFAWSWGPLGWLIPSETFP 417
+ QV + + + AK + L K YA++VV+ +C +VA FAWSWGPLGWL+PSE FP
Sbjct: 359 LICQVAVAVSIAAKFGTSGEPGDLPKWYAIVVVLFICIYVAGFAWSWGPLGWLVPSEIFP 418
Query: 418 LETRSAGQSVTVFTNMLFTFLIAQAFLSLLCLFKFGIFLFFSAWVFVMGVFTVFLIPETK 477
LE RSA QSV V NMLFTFL+AQ FL++LC KFG+FLFF+ +V VM ++ ++PETK
Sbjct: 419 LEIRSAAQSVNVSVNMLFTFLVAQIFLTMLCHMKFGLFLFFAFFVVVMTIYIYTMLPETK 478
Query: 478 NIPIEDMAETVWKQHWFWRRFM 499
IPIE+M + VWK H +W RF+
Sbjct: 479 GIPIEEM-DRVWKSHPYWSRFV 499
>ref|NP_919214.1| putative monosaccharide transport protein MST1 [Oryza sativa
(japonica cultivar-group)] gi|50508995|dbj|BAD31944.1|
putative monosaccharide transport protein MST1 [Oryza
sativa (japonica cultivar-group)]
gi|22324466|dbj|BAC10381.1| putative monosaccharide
transport protein MST1 [Oryza sativa (japonica
cultivar-group)] gi|11991114|dbj|BAB19864.1|
monosaccharide transporter 3 [Oryza sativa]
Length = 518
Score = 566 bits (1458), Expect = e-160
Identities = 282/501 (56%), Positives = 367/501 (72%), Gaps = 4/501 (0%)
Query: 1 MAGGGFTTGSSDVVFEARITAAVVISCIMAATGGLMFGYDVGISGGVTSMPSFLQKFFPD 60
MAGG + + + ++T V +C++AATGGL+FGYD+GISGGVTSM FL+KFFP+
Sbjct: 1 MAGGAVVSTGAGKDYPGKLTLFVFFTCVVAATGGLIFGYDIGISGGVTSMDPFLRKFFPE 60
Query: 61 VYKRTQEHTVLESNYCKYDNQKLQLFTSSLYLAALVASMIASPVTRKLGRKQTMLLAGIL 120
VY++ Q + YCKYDNQ LQ FTSSLYLAALV+S A+ VTR LGRK +M G+
Sbjct: 61 VYRKKQMADK-NNQYCKYDNQLLQTFTSSLYLAALVSSFFAATVTRVLGRKWSMFAGGLT 119
Query: 121 FIVGTVLSASAGKLILLIFGRILLGCGVGFANQAVPVFLSEIAPTRIRGALNIMFQLNIT 180
F++G L+ +A + +LI GRILLG GVGFANQ+VPV+LSE+AP R+RG LNI FQL IT
Sbjct: 120 FLIGAALNGAAENVAMLIVGRILLGVGVGFANQSVPVYLSEMAPARLRGMLNIGFQLMIT 179
Query: 181 IGIFIANLVNWFTSKIKGGYGWRVSLAGAIIPAVMLTMGSLIVDDTPNSLIERGFEEKGK 240
IGI A L+N+ T+KIK G+GWRVSLA A +PA ++T+GSL + DTPNSLI+RG E +
Sbjct: 180 IGILAAELINYGTAKIKAGWGWRVSLALAAVPAAIITLGSLFLPDTPNSLIDRGHPEAAE 239
Query: 241 AVLTKIRGVE-NIEPEFEDILRASKVANEVKSPFKDLVKSHNRPPLIIAICMQVFQQCTG 299
+L +IRG + ++ E+ D++ AS+ + V+ P++++++ R L +AIC+ FQQ TG
Sbjct: 240 RMLRRIRGSDVDVSEEYADLVAASEESKLVQHPWRNILRRKYRAQLTMAICIPFFQQLTG 299
Query: 300 INAIMFYAPVLFSTLGFHNDASLYSSVITGGVNVLCTLVSVYFVDKAGRRVLLLEACVQM 359
IN IMFYAPVLF TLGF +DASL S+VITG VNV TLVS++ VD+ GRR L L+ QM
Sbjct: 300 INVIMFYAPVLFDTLGFKSDASLMSAVITGLVNVFATLVSIFTVDRLGRRKLFLQGGAQM 359
Query: 360 FVSQVVIGIVLGAKLQDHS-DSLSKGYAMLVVVMVCTFVASFAWSWGPLGWLIPSETFPL 418
V QVV+G ++ K + KGYA +VV+ +C +VA FAWSWGPLGWL+PSE FPL
Sbjct: 360 VVCQVVVGTLIAVKFGTSGIGDIPKGYAAVVVLFICMYVAGFAWSWGPLGWLVPSEIFPL 419
Query: 419 ETRSAGQSVTVFTNMLFTFLIAQAFLSLLCLFKFGIFLFFSAWVFVMGVFTVFLIPETKN 478
E R AGQS+ V NMLFTF+IAQAFL++LC KFG+F FF+ WV +M VF +PETKN
Sbjct: 420 EIRPAGQSINVSVNMLFTFVIAQAFLTMLCHMKFGLFYFFAGWVVIMTVFIALFLPETKN 479
Query: 479 IPIEDMAETVWKQHWFWRRFM 499
+PIE+M VWK HWFWRRF+
Sbjct: 480 VPIEEMV-LVWKSHWFWRRFI 499
Database: nr
Posted date: Jul 5, 2005 12:34 AM
Number of letters in database: 863,360,394
Number of sequences in database: 2,540,612
Lambda K H
0.327 0.141 0.426
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 790,992,470
Number of Sequences: 2540612
Number of extensions: 31757368
Number of successful extensions: 124832
Number of sequences better than 10.0: 4340
Number of HSP's better than 10.0 without gapping: 2419
Number of HSP's successfully gapped in prelim test: 1922
Number of HSP's that attempted gapping in prelim test: 114653
Number of HSP's gapped (non-prelim): 5908
length of query: 500
length of database: 863,360,394
effective HSP length: 132
effective length of query: 368
effective length of database: 527,999,610
effective search space: 194303856480
effective search space used: 194303856480
T: 11
A: 40
X1: 15 ( 7.1 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 40 (21.7 bits)
S2: 77 (34.3 bits)
Medicago: description of AC137838.10


