

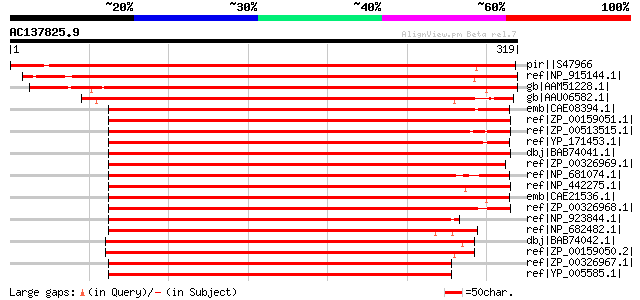
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC137825.9 - phase: 0
(319 letters)
Database: nr
2,540,612 sequences; 863,360,394 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
pir||S47966 probable lipid transfer protein M30 precursor - gard... 530 e-149
ref|NP_915144.1| putative chloroplast membrane-associated 30 kD ... 413 e-114
gb|AAM51228.1| unknown protein [Arabidopsis thaliana] gi|1502797... 411 e-113
gb|AAU06582.1| chloroplast vesicle-inducing protein in plastids ... 248 2e-64
emb|CAE08394.1| similar to chloroplast membrane-associated 30 kD... 225 2e-57
ref|ZP_00159051.1| COG1842: Phage shock protein A (IM30), suppre... 224 3e-57
ref|ZP_00513515.1| PspA/IM30 [Crocosphaera watsonii WH 8501] gi|... 223 4e-57
ref|YP_171453.1| similar to chloroplast membrane-associated 30 k... 223 6e-57
dbj|BAB74041.1| all2342 [Nostoc sp. PCC 7120] gi|17229834|ref|NP... 223 8e-57
ref|ZP_00326969.1| COG1842: Phage shock protein A (IM30), suppre... 213 5e-54
ref|NP_681074.1| lipid transfer protein M30 [Thermosynechococcus... 212 1e-53
ref|NP_442275.1| chloroplast membrane-associated 30 kD protein [... 211 3e-53
emb|CAE21536.1| similar to chloroplast membrane-associated 30 kD... 209 9e-53
ref|ZP_00326968.1| COG1842: Phage shock protein A (IM30), suppre... 209 1e-52
ref|NP_923844.1| hypothetical protein gvip120 [Gloeobacter viola... 209 1e-52
ref|NP_682482.1| lipid transfer protein M30 homolog [Thermosynec... 203 6e-51
dbj|BAB74042.1| all2343 [Nostoc sp. PCC 7120] gi|17229835|ref|NP... 203 6e-51
ref|ZP_00159050.2| COG1842: Phage shock protein A (IM30), suppre... 199 9e-50
ref|ZP_00326967.1| COG1842: Phage shock protein A (IM30), suppre... 198 2e-49
ref|YP_005585.1| phage shock protein A [Thermus thermophilus HB2... 158 2e-37
>pir||S47966 probable lipid transfer protein M30 precursor - garden pea
gi|729842|sp|Q03943|IM30_PEA Membrane-associated 30 kDa
protein, chloroplast precursor (M30)
gi|169107|gb|AAA53545.1| IM30
Length = 323
Score = 530 bits (1366), Expect = e-149
Identities = 283/324 (87%), Positives = 300/324 (92%), Gaps = 8/324 (2%)
Query: 1 MATKLQIFTQLPSAPLQSSSSSSSLRKPLATSFFGTRPANTVKFRGMRIAKPVRGGGAIG 60
M TK QIF+ LPSAPLQ SSS L+KPLAT+ FGTRP +T+KFR MRIAKPVRGGGAIG
Sbjct: 1 MTTKFQIFSGLPSAPLQPSSSL--LKKPLATTLFGTRPVDTLKFRVMRIAKPVRGGGAIG 58
Query: 61 AGMNLFERFTRVVKSYANAIVSSFEDPEKILEQAVLEMNDDLTKMRQATAQVLASQKRME 120
MNLF+RF RVVKSYANA+VS+FEDPEKILEQAVLEMNDDLTKMRQATAQVLASQKR+E
Sbjct: 59 VRMNLFDRFARVVKSYANALVSTFEDPEKILEQAVLEMNDDLTKMRQATAQVLASQKRLE 118
Query: 121 NKYKAATQASEEWYRKAQLALQKGEEDLAREALKRRKSFADNASALKAQLDQQKGVVDSL 180
NKYKAA QASEEWYRKAQLALQKGEEDLAREALKRRKSFADNAS+LKAQLDQQK VVD+L
Sbjct: 119 NKYKAAQQASEEWYRKAQLALQKGEEDLAREALKRRKSFADNASSLKAQLDQQKSVVDNL 178
Query: 181 VSNTRLLESKIQEARSKKDTLKARAQSAKTSTKVSEMLGNVNTSGALSAFEKMEEKVMTM 240
VSNTRLLESKIQEARSKKDTLKARAQSAKT+TKVSEMLGNVNTS ALSAFEKMEEKVMTM
Sbjct: 179 VSNTRLLESKIQEARSKKDTLKARAQSAKTATKVSEMLGNVNTSSALSAFEKMEEKVMTM 238
Query: 241 ESQAEALGQLTSDDLEGKFAMLESSSVDDDLANLKKELSGSSKKGELPPGRS------ST 294
ESQAEALGQLT+DDLEGKFAMLE+SSVDDDLANLKKEL+GSSKKGELPPGRS ST
Sbjct: 239 ESQAEALGQLTTDDLEGKFAMLETSSVDDDLANLKKELAGSSKKGELPPGRSSTTSTTST 298
Query: 295 RTGTPFRDADIETELEQLRQRSKE 318
+TG PFRDADIE ELEQLR+RSKE
Sbjct: 299 KTGNPFRDADIEIELEQLRKRSKE 322
>ref|NP_915144.1| putative chloroplast membrane-associated 30 kD protein [Oryza
sativa (japonica cultivar-group)]
gi|20161290|dbj|BAB90216.1| putative membrane-associated
30 kDa protein, chloroplast precursor [Oryza sativa
(japonica cultivar-group)] gi|21952843|dbj|BAC06258.1|
putative chloroplast membrane-associated 30 kD protein
[Oryza sativa (japonica cultivar-group)]
gi|44888076|sp|Q8S0J7|IM30_ORYSA Probable
membrane-associated 30 kDa protein, chloroplast
precursor
Length = 317
Score = 413 bits (1062), Expect = e-114
Identities = 226/315 (71%), Positives = 262/315 (82%), Gaps = 9/315 (2%)
Query: 9 TQLPSAPLQSSSSSSSLRKPLATSFFGTRPANTVKFRGMRIAKP-VRGGGAIGAGMNLFE 67
T L AP +S+S R L TSF +V R +++ + V G NL +
Sbjct: 8 TSLRLAP-PPPASASFRRTALRTSFLN----GSVSLRLIQVRQSNVNRFKCNGIRSNLLD 62
Query: 68 RFTRVVKSYANAIVSSFEDPEKILEQAVLEMNDDLTKMRQATAQVLASQKRMENKYKAAT 127
RF+RVVKSYANA++SSFEDPEKIL+QAVLEMNDDLTKMRQATAQVLASQKR+ENKYKAA
Sbjct: 63 RFSRVVKSYANAVLSSFEDPEKILDQAVLEMNDDLTKMRQATAQVLASQKRLENKYKAAE 122
Query: 128 QASEEWYRKAQLALQKGEEDLAREALKRRKSFADNASALKAQLDQQKGVVDSLVSNTRLL 187
QAS++WYR+AQLALQKG+EDLAREALKRRKS+ADNAS+LKAQLDQQKGVV++LVSNTR+L
Sbjct: 123 QASDDWYRRAQLALQKGDEDLAREALKRRKSYADNASSLKAQLDQQKGVVENLVSNTRVL 182
Query: 188 ESKIQEARSKKDTLKARAQSAKTSTKVSEMLGNVNTSGALSAFEKMEEKVMTMESQAEAL 247
ESKI EA+ KKDTLKARAQSAKTSTKVSEMLGNVNTSGALSAFEKMEEKVM MESQAEAL
Sbjct: 183 ESKIAEAKQKKDTLKARAQSAKTSTKVSEMLGNVNTSGALSAFEKMEEKVMAMESQAEAL 242
Query: 248 GQLTSDDLEGKFAMLESSSVDDDLANLKKELSGSSKKGELPPGR---SSTRTGTPFRDAD 304
GQL +DDLEGKFA+LE+SSVDDDLA +KKE+SGSS KGELPPGR S++ PFRD +
Sbjct: 243 GQLATDDLEGKFALLETSSVDDDLAQMKKEISGSSSKGELPPGRTAVSNSGAARPFRDIE 302
Query: 305 IETELEQLRQRSKEF 319
IE EL +LR+++ E+
Sbjct: 303 IENELNELRKKANEY 317
>gb|AAM51228.1| unknown protein [Arabidopsis thaliana] gi|15027977|gb|AAK76519.1|
unknown protein [Arabidopsis thaliana]
gi|18408237|ref|NP_564846.1| PspA/IM30 family protein
[Arabidopsis thaliana] gi|3335333|gb|AAC27135.1| Similar
to chloroplast membrane-associated 30KD protein
precursor (IM30) gb|M73744 from Pisum sativum. ESTs
gb|N37557, gb|W43887 and gb|AA042479 come from this
gene. [Arabidopsis thaliana]
gi|44888073|sp|O80796|IM30_ARATH Probable
membrane-associated 30 kDa protein, chloroplast
precursor gi|7447853|pir||T02347 probable lipid transfer
protein T8F5.2 - Arabidopsis thaliana
Length = 330
Score = 411 bits (1057), Expect = e-113
Identities = 221/312 (70%), Positives = 266/312 (84%), Gaps = 7/312 (2%)
Query: 13 SAPLQSSSSSSSLR-KPLATSFFGTRPANTVKFRGMRIA--KPVRGGGAIGAGMNLFERF 69
S+P SS+ SLR PL TSFFG + ++ +R+A +R G GA MNLFERF
Sbjct: 21 SSPSTSSNRPCSLRILPLRTSFFGNS-SGALRVNVLRLACDNRLRCNGH-GATMNLFERF 78
Query: 70 TRVVKSYANAIVSSFEDPEKILEQAVLEMNDDLTKMRQATAQVLASQKRMENKYKAATQA 129
+RVVKSYANA++SSFEDPEKILEQ V+EMN DLTKMRQATAQVLASQK+++NKYKAA Q+
Sbjct: 79 SRVVKSYANALISSFEDPEKILEQTVIEMNSDLTKMRQATAQVLASQKQLQNKYKAAQQS 138
Query: 130 SEEWYRKAQLALQKGEEDLAREALKRRKSFADNASALKAQLDQQKGVVDSLVSNTRLLES 189
S++WY++AQLAL KG+EDLAREALKRRKSFADNA+ALK QLDQQKGVVD+LVSNTRLLES
Sbjct: 139 SDDWYKRAQLALAKGDEDLAREALKRRKSFADNATALKTQLDQQKGVVDNLVSNTRLLES 198
Query: 190 KIQEARSKKDTLKARAQSAKTSTKVSEMLGNVNTSGALSAFEKMEEKVMTMESQAEALGQ 249
KIQEA++KKDTL ARA++AKT+TKV EM+G VNTSGALSAFEKMEEKVM MES+A+AL Q
Sbjct: 199 KIQEAKAKKDTLLARARTAKTATKVQEMIGTVNTSGALSAFEKMEEKVMAMESEADALTQ 258
Query: 250 LTSDDLEGKFAMLESSSVDDDLANLKKELSGSSKKGELPPGRSSTRTGT--PFRDADIET 307
+ +D+LEGKF MLE+SSVDDDLA+LKKELSGSSKKGELPPGRS+ T PF+D++IE
Sbjct: 259 IGTDELEGKFQMLETSSVDDDLADLKKELSGSSKKGELPPGRSTVAASTRYPFKDSEIEN 318
Query: 308 ELEQLRQRSKEF 319
EL +LR+++ +F
Sbjct: 319 ELNELRRKANDF 330
>gb|AAU06582.1| chloroplast vesicle-inducing protein in plastids 1 [Chlamydomonas
reinhardtii]
Length = 288
Score = 248 bits (633), Expect = 2e-64
Identities = 139/277 (50%), Positives = 193/277 (69%), Gaps = 14/277 (5%)
Query: 46 GMRIAKPV--RGGGAIGAGMNLFERFTRVVKSYANAIVSSFEDPEKILEQAVLEMNDDLT 103
G+R A P R A+ NLF R R+V S+A +VS+ EDPEK+L+Q V EM DL
Sbjct: 19 GVRSAVPAVRRSRKAVVVQANLFSRAARIVNSWATNVVSNAEDPEKLLDQVVEEMQGDLI 78
Query: 104 KMRQATAQVLASQKRMENKYKAATQASEEWYRKAQLALQKGEEDLAREALKRRKSFADNA 163
KMRQA A +LA QK++E KYK A +++W R+A+LA+QKGE+DLA+EALKRRK++ + A
Sbjct: 79 KMRQAAATILAQQKQIETKYKQAQTTADDWLRRAELAVQKGEDDLAKEALKRRKTYQEQA 138
Query: 164 SALKAQLDQQKGVVDSLVSNTRLLESKIQEARSKKDTLKARAQSAKTSTKVSEMLGNVNT 223
LK Q+DQ G +++NTR LE+K+QEARSKK+TLKARA SAKTS ++ EM+ +NT
Sbjct: 139 DQLKVQVDQLSGASGDVLNNTRALEAKLQEARSKKETLKARAASAKTSQQIQEMMSGLNT 198
Query: 224 SGALSAFEKMEEKVMTMESQAEALGQLT-SDDLEGKFAMLESSSVDDDLANLKKEL--SG 280
S A+ AF+KME+KV++ME+QAE+ L SD ++ KF LES +VDD+LA LK+ + S
Sbjct: 199 SNAVVAFDKMEQKVLSMEAQAESTKMLVGSDTIDNKFKQLESGTVDDELAALKRGMLPST 258
Query: 281 SSKKGELPPGRSSTRTGTPFRDADIETELEQLRQRSK 317
+S G LP R+ DA ++ ELE LR++++
Sbjct: 259 TSAVGSLPEPRAV--------DA-LDLELEALRKKAR 286
>emb|CAE08394.1| similar to chloroplast membrane-associated 30 kD protein
[Synechococcus sp. WH 8102] gi|33866411|ref|NP_897970.1|
similar to chloroplast membrane-associated 30 kD protein
[Synechococcus sp. WH 8102]
Length = 263
Score = 225 bits (573), Expect = 2e-57
Identities = 118/253 (46%), Positives = 172/253 (67%), Gaps = 2/253 (0%)
Query: 63 MNLFERFTRVVKSYANAIVSSFEDPEKILEQAVLEMNDDLTKMRQATAQVLASQKRMENK 122
M F+R R+V++ ANA VSS EDP KIL+Q+V +M DL K+RQA A +ASQKR+ ++
Sbjct: 7 MGFFDRLGRLVRANANAAVSSMEDPSKILDQSVADMQSDLVKLRQAVALAIASQKRLRSQ 66
Query: 123 YKAATQASEEWYRKAQLALQKGEEDLAREALKRRKSFADNASALKAQLDQQKGVVDSLVS 182
+ A+ + WY +A+LAL+KGEEDLAREAL RRK+F + A++L AQ+ Q G V++L
Sbjct: 67 AEQASSQATTWYERAELALKKGEEDLAREALTRRKTFQETATSLNAQVQSQDGQVETLKK 126
Query: 183 NTRLLESKIQEARSKKDTLKARAQSAKTSTKVSEMLGNVNTSGALSAFEKMEEKVMTMES 242
+ LE KI EA++KKD LKARAQ+A+ ++ +GN+ T+ A++AFE+MEEKV ME+
Sbjct: 127 SLVALEGKIAEAKTKKDMLKARAQAAQAQQQLQSAVGNIGTNSAMAAFERMEEKVEAMEA 186
Query: 243 QAEALGQLTSDDLEGKFAMLE-SSSVDDDLANLKKELSGSSKKGELPPGRSSTRTGTPFR 301
+A +L DLE +FA LE S VDD+LA L+ +L G + LP S + TP +
Sbjct: 187 TGQAAAELAGADLESQFAALEGGSDVDDELAALRNQLKGGPESVALPASEQS-QAVTPVK 245
Query: 302 DADIETELEQLRQ 314
+++ +LE L++
Sbjct: 246 VEEVDADLEDLKR 258
>ref|ZP_00159051.1| COG1842: Phage shock protein A (IM30), suppresses sigma54-dependent
transcription [Anabaena variabilis ATCC 29413]
Length = 258
Score = 224 bits (571), Expect = 3e-57
Identities = 118/254 (46%), Positives = 176/254 (68%), Gaps = 1/254 (0%)
Query: 63 MNLFERFTRVVKSYANAIVSSFEDPEKILEQAVLEMNDDLTKMRQATAQVLASQKRMENK 122
M LF+R RVV + N +V+ EDPEK+LEQA+LEM +DL ++RQ AQ +A+QKR E +
Sbjct: 1 MGLFDRIKRVVSANLNDLVNKAEDPEKMLEQAILEMQEDLVQLRQGVAQAIAAQKRTEKQ 60
Query: 123 YKAATQASEEWYRKAQLALQKGEEDLAREALKRRKSFADNASALKAQLDQQKGVVDSLVS 182
Y A +W R AQLALQKG+E+LAR+AL+R+K++ + ++ALK LDQQ V++L
Sbjct: 61 YTDAQNEINKWQRNAQLALQKGDENLARQALERKKTYTETSTALKTSLDQQSTQVETLKR 120
Query: 183 NTRLLESKIQEARSKKDTLKARAQSAKTSTKVSEMLGNVNTSGALSAFEKMEEKVMTMES 242
N LESKI EA++KK+ LKAR +AK ++ M+ +N+S A++AFE+MEEKV+ E+
Sbjct: 121 NLIQLESKISEAKTKKEMLKARITTAKAQEQLEGMVRGMNSSSAMAAFERMEEKVLMQEA 180
Query: 243 QAEALGQLTSDDLEGKFAMLESSS-VDDDLANLKKELSGSSKKGELPPGRSSTRTGTPFR 301
+A++ +L DLE +FA LESSS VDD+LA LK L+ + ++ LPP + ++ P
Sbjct: 181 RAQSAAELAGADLETQFARLESSSDVDDELAALKASLAPAPEQNLLPPQQQEQQSTAPKS 240
Query: 302 DADIETELEQLRQR 315
+++EL+ LR++
Sbjct: 241 KEVVDSELDALRKQ 254
>ref|ZP_00513515.1| PspA/IM30 [Crocosphaera watsonii WH 8501]
gi|67857479|gb|EAM52718.1| PspA/IM30 [Crocosphaera
watsonii WH 8501]
Length = 257
Score = 223 bits (569), Expect = 4e-57
Identities = 120/255 (47%), Positives = 179/255 (70%), Gaps = 4/255 (1%)
Query: 63 MNLFERFTRVVKSYANAIVSSFEDPEKILEQAVLEMNDDLTKMRQATAQVLASQKRMENK 122
M LF+R RVV++ N +VS EDPEKILEQ + +M DL +++QA AQ + SQKR E +
Sbjct: 1 MGLFDRLGRVVRANLNDLVSKAEDPEKILEQTIEDMGKDLIELKQAFAQTMGSQKRTEGQ 60
Query: 123 YKAATQASEEWYRKAQLALQKGEEDLAREALKRRKSFADNASALKAQLDQQKGVVDSLVS 182
Y A Q + W ++A+LAL KGEE LAREAL R+KSF D A+ LK QLDQQ ++++
Sbjct: 61 YTKALQEANSWEQRAKLALSKGEETLAREALVRKKSFTDTATTLKEQLDQQNAQIETMRR 120
Query: 183 NTRLLESKIQEARSKKDTLKARAQSAKTSTKVSEMLGNVNTSGALSAFEKMEEKVMTMES 242
N LESKI EA++KKD L +RA++AK + + + +NTSG+++AFE+ME KV+ ME+
Sbjct: 121 NLTALESKIAEAKTKKDMLVSRAKAAKANEALQSTMSGLNTSGSMAAFERMENKVLDMEA 180
Query: 243 QAEALGQLTSDDLEGKFAMLES-SSVDDDLANLKKELSGSSK-KGELPPGRSSTRTGTPF 300
+++A+G++ LE +FA LE+ S V+D+LA LK +++GS++ K LP G S+ TP
Sbjct: 181 RSQAVGEIGGSSLENQFAELEAGSGVEDELAMLKAQMTGSAEPKAALPEG-STEDKATP- 238
Query: 301 RDADIETELEQLRQR 315
+D+ ++ EL+ LR++
Sbjct: 239 KDSAVDAELDDLRRQ 253
>ref|YP_171453.1| similar to chloroplast membrane-associated 30 kD protein
[Synechococcus elongatus PCC 6301]
gi|56685711|dbj|BAD78933.1| similar to chloroplast
membrane-associated 30 kD protein [Synechococcus
elongatus PCC 6301] gi|46129498|ref|ZP_00163945.2|
COG1842: Phage shock protein A (IM30), suppresses
sigma54-dependent transcription [Synechococcus elongatus
PCC 7942]
Length = 257
Score = 223 bits (568), Expect = 6e-57
Identities = 119/254 (46%), Positives = 180/254 (70%), Gaps = 4/254 (1%)
Query: 63 MNLFERFTRVVKSYANAIVSSFEDPEKILEQAVLEMNDDLTKMRQATAQVLASQKRMENK 122
M LF+R +RVV++ N +VS EDPEKILEQA+++M +DL ++RQA A +A QKR+E +
Sbjct: 1 MGLFDRISRVVRANMNDMVSKAEDPEKILEQALMDMQEDLIQLRQAVATAIAGQKRIEQQ 60
Query: 123 YKAATQASEEWYRKAQLALQKGEEDLAREALKRRKSFADNASALKAQLDQQKGVVDSLVS 182
A + W ++AQLALQKG+EDLAR+AL R+K+F + A ALK QLDQQ G VD+L
Sbjct: 61 MNQAQTQAASWQQRAQLALQKGQEDLARQALSRKKTFNETAVALKTQLDQQAGQVDTLKR 120
Query: 183 NTRLLESKIQEARSKKDTLKARAQSAKTSTKVSEMLGNVNTSGALSAFEKMEEKVMTMES 242
N LE KI EA++KKD LKARAQ+AK ++ +G ++T+ +++AFE+MEEKV++ME+
Sbjct: 121 NLVALEGKIAEAKTKKDMLKARAQAAKAQEQLQSAVGRLSTNTSMAAFERMEEKVLSMEA 180
Query: 243 QAEALGQLTSDDLEGKFAMLES-SSVDDDLANLKKELSGS-SKKGELPPGRSSTRTGTPF 300
+A+A +L DLE +F LE+ V+D+LA +K +L+G+ + LPPG ++ + +
Sbjct: 181 KAQASAELAGTDLESQFVSLEAGGDVEDELAAMKAQLAGAPTATTALPPGAATPVSVS-- 238
Query: 301 RDADIETELEQLRQ 314
+ + ++ ELE L++
Sbjct: 239 QPSTVDAELEALKR 252
>dbj|BAB74041.1| all2342 [Nostoc sp. PCC 7120] gi|17229834|ref|NP_486382.1|
hypothetical protein all2342 [Nostoc sp. PCC 7120]
gi|25315831|pir||AG2098 hypothetical protein all2342
[imported] - Nostoc sp. (strain PCC 7120)
Length = 258
Score = 223 bits (567), Expect = 8e-57
Identities = 117/254 (46%), Positives = 174/254 (68%), Gaps = 1/254 (0%)
Query: 63 MNLFERFTRVVKSYANAIVSSFEDPEKILEQAVLEMNDDLTKMRQATAQVLASQKRMENK 122
M LF+R RVV + N +V+ EDPEK+LEQA+LEM +DL ++RQ AQ +A+QKR E +
Sbjct: 1 MGLFDRIKRVVSANLNDLVNKAEDPEKMLEQAILEMQEDLVQLRQGVAQAIAAQKRTEKQ 60
Query: 123 YKAATQASEEWYRKAQLALQKGEEDLAREALKRRKSFADNASALKAQLDQQKGVVDSLVS 182
Y A +W R AQLALQKG+E+LAR+AL+R+K++ + +ALK LDQQ V++L
Sbjct: 61 YSDAQNEINKWQRNAQLALQKGDENLARQALERKKTYTETGTALKTSLDQQSTQVETLKR 120
Query: 183 NTRLLESKIQEARSKKDTLKARAQSAKTSTKVSEMLGNVNTSGALSAFEKMEEKVMTMES 242
N LESKI EA++KK+ LKAR +AK ++ M+ +N+S A++AFE+MEEKV+ E+
Sbjct: 121 NLIQLESKISEAKTKKEMLKARITTAKAQEQLEGMVRGMNSSSAMAAFERMEEKVLMQEA 180
Query: 243 QAEALGQLTSDDLEGKFAMLES-SSVDDDLANLKKELSGSSKKGELPPGRSSTRTGTPFR 301
+A++ +L DLE +FA LES S VDD+LA LK L+ + ++ LPP + ++ P
Sbjct: 181 RAQSAAELAGADLETQFARLESGSDVDDELAALKASLAPAPEQNLLPPQQQEQQSTAPKS 240
Query: 302 DADIETELEQLRQR 315
+++EL+ LR++
Sbjct: 241 KEVVDSELDALRKQ 254
>ref|ZP_00326969.1| COG1842: Phage shock protein A (IM30), suppresses sigma54-dependent
transcription [Trichodesmium erythraeum IMS101]
Length = 254
Score = 213 bits (543), Expect = 5e-54
Identities = 106/251 (42%), Positives = 172/251 (68%), Gaps = 1/251 (0%)
Query: 63 MNLFERFTRVVKSYANAIVSSFEDPEKILEQAVLEMNDDLTKMRQATAQVLASQKRMENK 122
M LF+R +R+ ++ N IV+ EDPEK+LEQ++++M +DL ++RQA A +A+QKR + +
Sbjct: 1 MGLFDRISRIARANVNDIVNKAEDPEKVLEQSIIDMQEDLVQLRQAVASAIATQKRTQQQ 60
Query: 123 YKAATQASEEWYRKAQLALQKGEEDLAREALKRRKSFADNASALKAQLDQQKGVVDSLVS 182
+K A +++WY +A+LAL+KG+++LA+EAL+R+ +++D AS +K L+ Q V++L
Sbjct: 61 HKQAESQAQQWYSRAELALKKGDDNLAKEALQRKTTYSDTASTIKVSLESQTSQVETLKR 120
Query: 183 NTRLLESKIQEARSKKDTLKARAQSAKTSTKVSEMLGNVNTSGALSAFEKMEEKVMTMES 242
N LE KI EA++KKD L AR ++AKT +++ +G++NTSGA++AF++MEEKV+ ME+
Sbjct: 121 NLTALEGKISEAKAKKDILNARLKAAKTQEQLNNTIGSLNTSGAMAAFDRMEEKVLQMEA 180
Query: 243 QAEALGQLTSDDLEGKFAMLESSS-VDDDLANLKKELSGSSKKGELPPGRSSTRTGTPFR 301
+++A +L LE F LE++S VDD+LA +K +L G S P S P
Sbjct: 181 RSQASAELAGAGLEEDFRQLEANSDVDDELAAMKAKLVGGSVNQSQLPASSQGSVEKPVV 240
Query: 302 DADIETELEQL 312
D ++E +QL
Sbjct: 241 DDELEDLRKQL 251
>ref|NP_681074.1| lipid transfer protein M30 [Thermosynechococcus elongatus BP-1]
gi|22294004|dbj|BAC07836.1| tlr0283 [Thermosynechococcus
elongatus BP-1]
Length = 243
Score = 212 bits (540), Expect = 1e-53
Identities = 114/252 (45%), Positives = 171/252 (67%), Gaps = 10/252 (3%)
Query: 63 MNLFERFTRVVKSYANAIVSSFEDPEKILEQAVLEMNDDLTKMRQATAQVLASQKRMENK 122
M LF+R +RVV+S+ NA+V + EDPEKILEQ ++EM D+L +RQA AQ +ASQKR+E +
Sbjct: 1 MGLFDRVSRVVRSWLNALVGAAEDPEKILEQTLIEMQDNLVTLRQAVAQAIASQKRLEQQ 60
Query: 123 YKAATQASEEWYRKAQLALQKGEEDLAREALKRRKSFADNASALKAQLDQQKGVVDSLVS 182
+ Q + EW R+A++ALQ G+E LA EAL R+K+ + A ALK QL+Q V++L
Sbjct: 61 FNQNQQQAAEWERRAKIALQHGDEQLALEALSRKKTALNAAMALKGQLEQSVTQVEALKK 120
Query: 183 NTRLLESKIQEARSKKDTLKARAQSAKTSTKVSEMLGNVNTSGALSAFEKMEEKVMTMES 242
+ LESKI EAR++K+ L ARA++AK S ++ + ++N + ++AFE+MEE+V +E+
Sbjct: 121 QMQQLESKIAEARTRKEMLVARARAAKASEQLQQTFSSLNINAPMAAFERMEERVQELEA 180
Query: 243 QAEALGQLTSDDLEGKFAMLESSSVDDDLANLKKELSGSSKKGELPPGRSSTRTGTPFRD 302
+++A+ +L SD LE KFA LE+ SV++DLA LK EL+ +L P + TP D
Sbjct: 181 RSQAVAELNSDTLEAKFAALEAGSVEEDLARLKAELANP----QLSP------SSTPAYD 230
Query: 303 ADIETELEQLRQ 314
++E QL +
Sbjct: 231 PELEALRRQLEK 242
>ref|NP_442275.1| chloroplast membrane-associated 30 kD protein [Synechocystis sp.
PCC 6803] gi|1001614|dbj|BAA10345.1| chloroplast
membrane-associated 30 kD protein [Synechocystis sp. PCC
6803] gi|3123076|sp|Q55707|Y617_SYNY3 Hypothetical
protein sll0617
Length = 267
Score = 211 bits (536), Expect = 3e-53
Identities = 118/263 (44%), Positives = 175/263 (65%), Gaps = 10/263 (3%)
Query: 63 MNLFERFTRVVKSYANAIVSSFEDPEKILEQAVLEMNDDLTKMRQATAQVLASQKRMENK 122
M LF+R RVV++ N +VS EDPEK+LEQAV++M +DL ++RQA A+ +A +KR E +
Sbjct: 1 MGLFDRLGRVVRANLNDLVSKAEDPEKVLEQAVIDMQEDLVQLRQAVARTIAEEKRTEQR 60
Query: 123 YKAATQASEEWYRKAQLALQKGEEDLAREALKRRKSFADNASALKAQLDQQKGVVDSLVS 182
TQ +++W +A+LAL GEE+LAREAL R+KS D A+A + QL QQ+ + ++L
Sbjct: 61 LNQDTQEAKKWEDRAKLALTNGEENLAREALARKKSLTDTAAAYQTQLAQQRTMSENLRR 120
Query: 183 NTRLLESKIQEARSKKDTLKARAQSAKTSTKVSEMLGNVNTSGALSAFEKMEEKVMTMES 242
N LE+KI EA++KK+ L+ARA++AK + ++ + LG + TS A SAFE+ME KV+ ME+
Sbjct: 121 NLAALEAKISEAKTKKNMLQARAKAAKANAELQQTLGGLGTSSATSAFERMENKVLDMEA 180
Query: 243 QAEALGQLTSDDLEGKFAMLESSS-VDDDLANLKKELSGSSKKG--------ELPPGRSS 293
++A G+L +E +FA LE+SS V+D+LA LK ++G + G E P SS
Sbjct: 181 TSQAAGELAGFGIENQFAQLEASSGVEDELAALKASMAGGALPGTSAATPQLEAAPVDSS 240
Query: 294 TRTGTPFR-DADIETELEQLRQR 315
+ DA I+ EL+ LR+R
Sbjct: 241 VPANNASQDDAVIDQELDDLRRR 263
>emb|CAE21536.1| similar to chloroplast membrane-associated 30 kD protein
[Prochlorococcus marinus str. MIT 9313]
gi|33863628|ref|NP_895188.1| similar to chloroplast
membrane-associated 30 kD protein [Prochlorococcus
marinus str. MIT 9313]
Length = 267
Score = 209 bits (532), Expect = 9e-53
Identities = 112/262 (42%), Positives = 172/262 (64%), Gaps = 10/262 (3%)
Query: 63 MNLFERFTRVVKSYANAIVSSFEDPEKILEQAVLEMNDDLTKMRQATAQVLASQKRMENK 122
M F+R +R++++ N +VS EDP KIL+Q+V +M DL K+RQA A +ASQKR+ N+
Sbjct: 1 MGFFDRLSRLLRANLNDLVSKAEDPVKILDQSVADMQADLVKLRQAVAMAIASQKRLRNQ 60
Query: 123 YKAATQASEEWYRKAQLALQKGEEDLAREALKRRKSFADNASALKAQLDQQKGVVDSLVS 182
A WY +A+LALQKGEEDLA+EAL RRK F ++++AL QL Q+G V++L
Sbjct: 61 ADQAEGQVRTWYERAELALQKGEEDLAKEALTRRKGFQESSTALTNQLKGQEGQVETLKR 120
Query: 183 NTRLLESKIQEARSKKDTLKARAQSAKTSTKVSEMLGNVNTSGALSAFEKMEEKVMTMES 242
+ LE KI EAR+KKD LKARAQ+AK ++ +GN+ T+ A++AF++ME+KV +E+
Sbjct: 121 SLVALEGKIAEARTKKDMLKARAQAAKAQQQLQSAVGNLGTNSAMAAFDRMEDKVQALEA 180
Query: 243 QAEALGQLTSDDLEGKFAMLE-SSSVDDDLANLKKELSGSSKKGELPPGRSSTRTGT--- 298
++A +L DLE +FA LE + VDD+L+ L++ L G ++ LP +S+ +
Sbjct: 181 SSQAAAELAGADLESQFAALEGGNDVDDELSALRQRLEGGAEAVALPAAETSSLEESKDA 240
Query: 299 ------PFRDADIETELEQLRQ 314
+ A+++ ELE+L++
Sbjct: 241 NGPEVEAVKVAEVDAELEELKR 262
>ref|ZP_00326968.1| COG1842: Phage shock protein A (IM30), suppresses sigma54-dependent
transcription [Trichodesmium erythraeum IMS101]
Length = 254
Score = 209 bits (531), Expect = 1e-52
Identities = 108/255 (42%), Positives = 174/255 (67%), Gaps = 7/255 (2%)
Query: 63 MNLFERFTRVVKSYANAIVSSFEDPEKILEQAVLEMNDDLTKMRQATAQVLASQKRMENK 122
M LF+R +R+ ++ N +V++ EDP+KILEQ++L+M +DL ++RQA A +A+QKR + +
Sbjct: 1 MGLFDRISRIFRANVNEMVNNAEDPQKILEQSILDMQEDLVQLRQAVASGIATQKRTQQQ 60
Query: 123 YKAATQASEEWYRKAQLALQKGEEDLAREALKRRKSFADNASALKAQLDQQKGVVDSLVS 182
Y A +++WY +AQLALQKG+E+LA+EAL+R+ +++ A+ +K L+ Q V++L
Sbjct: 61 YNQAESQAKQWYSRAQLALQKGDENLAKEALQRKNTYSQTANTIKVSLESQTSQVETLKR 120
Query: 183 NTRLLESKIQEARSKKDTLKARAQSAKTSTKVSEMLGNVNTSGALSAFEKMEEKVMTMES 242
N LE KI EA++KKD L AR ++AKT +++ +G++N SGA++AFE+MEEKV ME+
Sbjct: 121 NLTALEGKISEAKAKKDILNARLKAAKTQEQLNNTIGSLNNSGAMAAFERMEEKVFQMEA 180
Query: 243 QAEALGQLTSDDLEGKFAMLESSS-VDDDLANLKKEL-SGSSKKGELPPGRSSTRTGTPF 300
+++A +L LE F LE+SS VDD+LA +K +L GS + +LP +
Sbjct: 181 RSQASAELAGVGLEEDFRKLEASSDVDDELAAMKAKLVGGSVNQSQLPASSQGSA----- 235
Query: 301 RDADIETELEQLRQR 315
+ ++ ELE LR++
Sbjct: 236 EKSVVDDELEDLRKQ 250
>ref|NP_923844.1| hypothetical protein gvip120 [Gloeobacter violaceus PCC 7421]
gi|35211461|dbj|BAC88839.1| pspA [Gloeobacter violaceus
PCC 7421]
Length = 237
Score = 209 bits (531), Expect = 1e-52
Identities = 109/222 (49%), Positives = 160/222 (71%), Gaps = 2/222 (0%)
Query: 63 MNLFERFTRVVKSYANAIVSSFEDPEKILEQAVLEMNDDLTKMRQATAQVLASQKRMENK 122
M LF+R V+KS N++V+ EDPEK+L Q V +M +DL ++RQA AQ +AS+KR+E +
Sbjct: 1 MGLFDRIAAVLKSNLNSVVTKAEDPEKMLNQTVNDMQEDLVQLRQAVAQAIASEKRIEQQ 60
Query: 123 YKAATQASEEWYRKAQLALQKGEEDLAREALKRRKSFADNASALKAQLDQQKGVVDSLVS 182
Y A + EW R+A LA+ K ++LAREAL RRKSF+++A+ LK QL+QQ+ V +L +
Sbjct: 61 YLQADAQANEWQRRAALAVSKDNDELAREALTRRKSFSESATGLKTQLEQQRKTVATLKT 120
Query: 183 NTRLLESKIQEARSKKDTLKARAQSAKTSTKVSEMLGNVNTSGALSAFEKMEEKVMTMES 242
N LE KI EA++KKD L ARA+SAK + ++++ LG VNT G+ S FE+MEEKV +E+
Sbjct: 121 NLTGLEGKISEAKAKKDLLVARARSAKATEQINQTLGKVNTGGSFSTFERMEEKVNELEA 180
Query: 243 QAEALGQLTSDDLEGKFAMLES-SSVDDDLANLKKELSGSSK 283
+++A+ +L +D+LE +F +LES V+D+L LK +L GS K
Sbjct: 181 RSQAVAELATDNLEDQFKVLESGGGVEDELLALKSQL-GSQK 221
>ref|NP_682482.1| lipid transfer protein M30 homolog [Thermosynechococcus elongatus
BP-1] gi|22295417|dbj|BAC09244.1| tll1692
[Thermosynechococcus elongatus BP-1]
Length = 281
Score = 203 bits (516), Expect = 6e-51
Identities = 107/239 (44%), Positives = 164/239 (67%), Gaps = 7/239 (2%)
Query: 63 MNLFERFTRVVKSYANAIVSSFEDPEKILEQAVLEMNDDLTKMRQATAQVLASQKRMENK 122
M L +R V++S NA+VS+ EDPEKILEQ +++MN DL K+RQA AQ +A+QKR+E +
Sbjct: 1 MGLLDRVGMVIRSNLNALVSAAEDPEKILEQTIIDMNADLVKLRQAVAQAIAAQKRLEQQ 60
Query: 123 YKAATQASEEWYRKAQLALQKGEEDLAREALKRRKSFADNASALKAQLDQQKGVVDSLVS 182
Y + +++W +A+LAL +G+E LA EAL R+K+ AD A+ALKAQ+DQ V +L
Sbjct: 61 YAQNLKDAQQWEERARLALSRGDEALALEALNRKKTAADTAAALKAQVDQMAAQVKTLKD 120
Query: 183 NTRLLESKIQEARSKKDTLKARAQSAKTSTKVSEMLGNVNTSGALSAFEKMEEKVMTMES 242
N LESKI EA++KK+ LKAR ++AK S ++ + + V TS A++AFE+ME+KV+ ME+
Sbjct: 121 NLTALESKISEAKTKKEMLKARVRAAKASEQIHQAVSKVGTSNAMAAFERMEDKVLQMEA 180
Query: 243 QAEALGQLTSDDLEGKFAMLESSS----VDDDLANLKKE---LSGSSKKGELPPGRSST 294
+++A+ +L+ D LE +F L+SS+ V+ +L +K++ L + G LP S +
Sbjct: 181 RSQAIAELSGDTLESQFEALQSSANRQDVEFELLEMKRQMGLLPDAPSAGTLPQTSSGS 239
>dbj|BAB74042.1| all2343 [Nostoc sp. PCC 7120] gi|17229835|ref|NP_486383.1|
hypothetical protein all2343 [Nostoc sp. PCC 7120]
gi|25315833|pir||AH2098 hypothetical protein all2343
[imported] - Nostoc sp. (strain PCC 7120)
Length = 274
Score = 203 bits (516), Expect = 6e-51
Identities = 110/236 (46%), Positives = 163/236 (68%), Gaps = 4/236 (1%)
Query: 61 AGMNLFERFTRVVKSYANAIVSSFEDPEKILEQAVLEMNDDLTKMRQATAQVLASQKRME 120
A M L +R RV+ + N++V+S EDPEKILEQAV+EM D+L ++RQ AQ +A+QKR E
Sbjct: 38 AEMELIKRIQRVISANFNSVVASKEDPEKILEQAVMEMQDNLLQLRQGVAQAIATQKRTE 97
Query: 121 NKYKAATQASEEWYRKAQLALQKGEEDLAREALKRRKSFADNASALKAQLDQQKGVVDSL 180
+ +A +EEWYR+AQLALQ+G E LAREAL +R+S+ D A +Q+ +Q +VD L
Sbjct: 98 RQAASAQSTAEEWYRRAQLALQQGNEPLAREALTKRQSYVDAAKTFSSQIGEQIEIVDRL 157
Query: 181 VSNTRLLESKIQEARSKKDTLKARAQSAKTSTKVSEMLGNVNTSGALSAFEKMEEKVMTM 240
+ R LE KI EA++KKD ARA+SA+ S K+ EML + + +LSAFE+MEEKV+ +
Sbjct: 158 KKDMRSLELKIAEAKTKKDMYIARARSAEASYKLQEMLSGGSATSSLSAFERMEEKVLQI 217
Query: 241 ESQAEALGQLTSDDLEGKFAMLESSS-VDDDLANLKKELSGSSK---KGELPPGRS 292
E+++E + Q+ SDDL+ +F LE+++ VD LA +K + + + + +LP G +
Sbjct: 218 EAKSELITQMGSDDLQKQFDSLEAANDVDSQLAAMKTQALSAGENTPQQQLPQGEN 273
>ref|ZP_00159050.2| COG1842: Phage shock protein A (IM30), suppresses sigma54-dependent
transcription [Anabaena variabilis ATCC 29413]
Length = 272
Score = 199 bits (506), Expect = 9e-50
Identities = 108/236 (45%), Positives = 163/236 (68%), Gaps = 4/236 (1%)
Query: 61 AGMNLFERFTRVVKSYANAIVSSFEDPEKILEQAVLEMNDDLTKMRQATAQVLASQKRME 120
A M L +R RV+ + N++V+S EDPEKILEQAV+EM ++L ++RQ AQ +A+QKR E
Sbjct: 36 AEMELIKRIQRVISANFNSVVASKEDPEKILEQAVMEMQENLLRLRQGVAQAIATQKRTE 95
Query: 121 NKYKAATQASEEWYRKAQLALQKGEEDLAREALKRRKSFADNASALKAQLDQQKGVVDSL 180
+ +A +EEWYR+AQLALQ+G E LAREAL +R+S+ D A +Q+ +Q +VD L
Sbjct: 96 RQAASAQSTAEEWYRRAQLALQQGNELLAREALTKRQSYVDAAKTFSSQIGEQIEIVDRL 155
Query: 181 VSNTRLLESKIQEARSKKDTLKARAQSAKTSTKVSEMLGNVNTSGALSAFEKMEEKVMTM 240
+ R LE KI EA++KKD ARA+SA+ S K+ EML + + +L+AFE+MEEKV+ +
Sbjct: 156 KKDMRSLELKIAEAKTKKDMYIARARSAEASYKLQEMLSGGSATSSLNAFERMEEKVLQI 215
Query: 241 ESQAEALGQLTSDDLEGKFAMLESSS-VDDDLANLKKEL---SGSSKKGELPPGRS 292
E+++E + Q+ SDDL+ +F LE+++ VD LA +K + ++ + +LP G +
Sbjct: 216 EAKSELITQMGSDDLQKQFDSLEAANDVDSQLAAMKTQALSEGENTPQQQLPQGEN 271
>ref|ZP_00326967.1| COG1842: Phage shock protein A (IM30), suppresses sigma54-dependent
transcription [Trichodesmium erythraeum IMS101]
Length = 221
Score = 198 bits (504), Expect = 2e-49
Identities = 99/217 (45%), Positives = 151/217 (68%), Gaps = 1/217 (0%)
Query: 63 MNLFERFTRVVKSYANAIVSSFEDPEKILEQAVLEMNDDLTKMRQATAQVLASQKRMENK 122
M +F+ F RV+++ N+++S+ EDPEKILEQ + M +DL K+RQ A+ +A+QK+ E +
Sbjct: 1 MGIFDHFWRVIRANVNSLISNAEDPEKILEQTIKNMQNDLVKLRQVVAEAIATQKQTERQ 60
Query: 123 YKAATQASEEWYRKAQLALQKGEEDLAREALKRRKSFADNASALKAQLDQQKGVVDSLVS 182
A ++EWYR+A LALQKG+E+LAREAL RR S+ + A+ +K Q++QQK VV+ L
Sbjct: 61 SYQAKSTADEWYRRAHLALQKGQENLAREALTRRNSYQETATVMKVQMEQQKQVVEKLKQ 120
Query: 183 NTRLLESKIQEARSKKDTLKARAQSAKTSTKVSEMLGNVNTSGALSAFEKMEEKVMTMES 242
N + LE K+ EA+ KK+ ARA+SA+ S K+++ML VNT LS FE+MEEK+ +ES
Sbjct: 121 NMKQLEHKVSEAKLKKNMYIARARSARASEKLNKMLDLVNTGNTLSTFEEMEEKITQLES 180
Query: 243 QAEALGQLTSDDLEGKFAMLESSS-VDDDLANLKKEL 278
Q++ + +L D LE KF LE + ++ +L +K ++
Sbjct: 181 QSQMIAELGIDSLEKKFNTLEGDNHIEAELKAMKAKI 217
>ref|YP_005585.1| phage shock protein A [Thermus thermophilus HB27]
gi|46197545|gb|AAS81958.1| phage shock protein A
[Thermus thermophilus HB27]
Length = 220
Score = 158 bits (400), Expect = 2e-37
Identities = 86/217 (39%), Positives = 135/217 (61%), Gaps = 1/217 (0%)
Query: 63 MNLFERFTRVVKSYANAIVSSFEDPEKILEQAVLEMNDDLTKMRQATAQVLASQKRMENK 122
M L++R +R+V++ N ++ EDPEKI+EQA+L+M L + R+ A+ LA QKR+E +
Sbjct: 1 MTLWDRLSRLVRANLNDLLRRAEDPEKIIEQALLDMKQALREAREQVAEALAEQKRLERE 60
Query: 123 YKAATQASEEWYRKAQLALQKGEEDLAREALKRRKSFADNASALKAQLDQQKGVVDSLVS 182
++ + + W KA+ AL+ G EDLAREAL+R+K D A KAQL + K + + L++
Sbjct: 61 VESHEREAALWEEKAREALKAGREDLAREALRRKKRALDLAEGFKAQLSEHKALTERLMT 120
Query: 183 NTRLLESKIQEARSKKDTLKARAQSAKTSTKVSEMLGNVNTSGALSAFEKMEEKVMTMES 242
+ LE+KI EA ++K L AR + + + V M ++ AL AFE+ME +++ ME
Sbjct: 121 QLKALEAKIDEAEARKKLLLARKKGVEAAETVRRMESRLDQHPALEAFEEMEARILAMED 180
Query: 243 QAEALGQLTSDDLEGKF-AMLESSSVDDDLANLKKEL 278
+ +AL L DLE +F A+ V++DL LK+EL
Sbjct: 181 RHQALEALDQADLEKEFQALSAEKEVEEDLLRLKREL 217
Database: nr
Posted date: Jul 5, 2005 12:34 AM
Number of letters in database: 863,360,394
Number of sequences in database: 2,540,612
Lambda K H
0.309 0.123 0.316
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 428,188,603
Number of Sequences: 2540612
Number of extensions: 15960973
Number of successful extensions: 70413
Number of sequences better than 10.0: 2695
Number of HSP's better than 10.0 without gapping: 194
Number of HSP's successfully gapped in prelim test: 2551
Number of HSP's that attempted gapping in prelim test: 67550
Number of HSP's gapped (non-prelim): 4966
length of query: 319
length of database: 863,360,394
effective HSP length: 128
effective length of query: 191
effective length of database: 538,162,058
effective search space: 102788953078
effective search space used: 102788953078
T: 11
A: 40
X1: 16 ( 7.1 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (21.7 bits)
S2: 75 (33.5 bits)
Medicago: description of AC137825.9


