

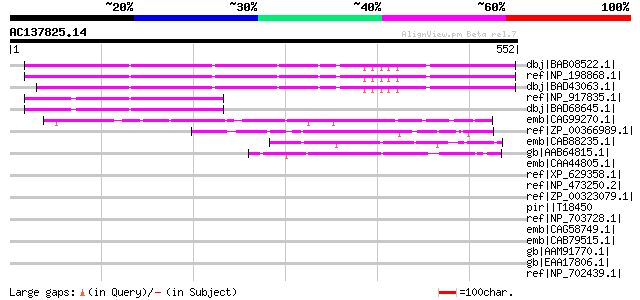
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC137825.14 + phase: 0
(552 letters)
Database: nr
2,540,612 sequences; 863,360,394 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
dbj|BAB08522.1| unnamed protein product [Arabidopsis thaliana] 308 2e-82
ref|NP_198868.1| expressed protein [Arabidopsis thaliana] 308 2e-82
dbj|BAD43063.1| putative protein [Arabidopsis thaliana] 288 2e-76
ref|NP_917835.1| B1158C05.16 [Oryza sativa (japonica cultivar-gr... 68 7e-10
dbj|BAD68645.1| hypothetical protein [Oryza sativa (japonica cul... 68 7e-10
emb|CAG99270.1| unnamed protein product [Kluyveromyces lactis NR... 52 6e-05
ref|ZP_00366989.1| conserved hypothetical protein [Campylobacter... 52 6e-05
emb|CAB88235.1| SPBC1711.05 [Schizosaccharomyces pombe] gi|19112... 49 4e-04
gb|AAB64815.1| Ydr379wp [Saccharomyces cerevisiae] gi|6320587|re... 49 5e-04
emb|CAA44805.1| nucleolin [Xenopus laevis] gi|464252|sp|P20397|N... 48 0.001
ref|XP_629358.1| hypothetical protein DDB0191811 [Dictyostelium ... 47 0.002
ref|NP_473250.2| hypothetical protein [Plasmodium falciparum 3D7... 46 0.004
ref|ZP_00323079.1| hypothetical protein PpenA01001008 [Pediococc... 46 0.004
pir||T18450 hypothetical protein C0570c - malaria parasite (Plas... 46 0.004
ref|NP_703728.1| hypothetical protein, expressed [Plasmodium fal... 45 0.005
emb|CAG58749.1| unnamed protein product [Candida glabrata CBS138... 45 0.005
emb|CAB79515.1| putative protein [Arabidopsis thaliana] gi|72282... 45 0.006
gb|AAM91770.1| unknown protein [Arabidopsis thaliana] gi|1797939... 45 0.006
gb|EAA17806.1| hypothetical protein [Plasmodium yoelii yoelii] 45 0.008
ref|NP_702439.1| hypothetical protein PF14_0550 [Plasmodium falc... 44 0.010
>dbj|BAB08522.1| unnamed protein product [Arabidopsis thaliana]
Length = 654
Score = 308 bits (790), Expect = 2e-82
Identities = 214/591 (36%), Positives = 319/591 (53%), Gaps = 70/591 (11%)
Query: 17 LDNLTTKGLYLLAMIVGGDTVKYERTRSNLKKIIKSSLSSVLSSKRNKDQQLETRKQLFQ 76
LD+LT KGLYL+AMI+ G + +++TR +K+II+ S+ + + E QL Q
Sbjct: 68 LDSLTNKGLYLIAMILTGGSTSFDKTRLKMKEIIRDSVRRDFGKNKEGIGKEEIINQLHQ 127
Query: 77 LLNNPQNYRQRCELLPGSESQFY--HAAVVKVLCGLEKLPSQTLLAMRRKLKGIKAPMPQ 134
+L++P N+R+ C + G + A +KVL L+ L +QTL AM+RKLKG + +PQ
Sbjct: 128 VLSDPANFREDCRMNLGRTPTLHSHRDAAMKVLNELDSLSTQTLRAMKRKLKGSRM-IPQ 186
Query: 135 LQPFKNGWGRNHLIRQVNKISRKMLLELDGGNKLQEPLANAMSVADFSLKLIIGFGSTFL 194
L+ + G R+ LI QV + S KML EL G+KLQE LA A+SV D SLKL G+ +
Sbjct: 187 LKNSRRGQRRSDLINQVRQASEKMLSELSAGDKLQEQLAKALSVVDLSLKLSPGYKTAAA 246
Query: 195 EECYQFSPEVKSLQSDIMQAILSVEKKEVVPLPVLRELQLLIEPKATVVNKSLRKAFVNL 254
+ ++FSPE K+LQ++I++A+ + K V L+ L L ++P+A V N SLR A
Sbjct: 247 TDFFRFSPETKNLQNEIVKAVWLLRK---VRFRELKRLHLCLDPEAEVSNDSLRSAVRKT 303
Query: 255 LTEFLFECSDMNSTPKSLLQILDVINKCSNKSTHDVTLQKEHIEEEVDSILSVSAQTNQI 314
L E+LFECSD+++ PKSL++ L ++N + H V +E IEEE + IL+VSAQ QI
Sbjct: 304 LIEYLFECSDLDTIPKSLMEALSLVNSRTGNVEHKVC-PREAIEEETECILNVSAQVKQI 362
Query: 315 IQDLLPDYEFDQGFTDAYMEEQSEDSDNESDKDEDDSQCSENSLHCSVTPSECLNSDSDK 374
+P+YE DQ F DAYME+ EDSD+ D D+DD EN C++ +E L + +
Sbjct: 363 FCQCIPNYELDQDFGDAYMEDL-EDSDDNDDDDDDDDDDDEN---CNLFDNEKLGNKEES 418
Query: 375 NCDTINLESK--------------------------MRNTPYNNQ--------------C 394
C I LE K +T NQ
Sbjct: 419 RCRNIKLEVKDTQDDAMESSDSDHEESGAECLVLDPTDSTHETNQHDFSSSVNRVVVRDL 478
Query: 395 QEEITEQFS-----TPMSRKNC---DSSAVSLDK-----EPDENIVKRHEFHESYTEAAP 441
E IT + TP S K+ D +++ K E D + ++F +
Sbjct: 479 PESITRVYPRSLYVTPTSNKSIVISDRHDIAMSKTRVKVERDIEMEVDNQFSSRSLFSVE 538
Query: 442 RDKSNFCEEKKPIPTKYSGHKNQYLAAQDACDKTSMLAYNLIGRMLEEFAIAEDLNLDLS 501
KS+ E+KP +KNQYLA Q+ D+TS++A+NLIGR+LE+FA + LNL+
Sbjct: 539 NIKSDDHGEQKP----RRRNKNQYLAIQEISDETSLVAHNLIGRLLEKFADRQRLNLETD 594
Query: 502 KRSYLNCDKQSEDIKETEE--QSSSRKRKGGPPIVRVIEELIPSFPDRYLL 550
+RSYL + + ++ E E Q+SS+ + I+ V++E I S + L+
Sbjct: 595 ERSYLGGESRLQEKVEVSEKKQASSQAKSDEAIILSVVKEQITSLEESVLM 645
>ref|NP_198868.1| expressed protein [Arabidopsis thaliana]
Length = 693
Score = 308 bits (790), Expect = 2e-82
Identities = 214/591 (36%), Positives = 319/591 (53%), Gaps = 70/591 (11%)
Query: 17 LDNLTTKGLYLLAMIVGGDTVKYERTRSNLKKIIKSSLSSVLSSKRNKDQQLETRKQLFQ 76
LD+LT KGLYL+AMI+ G + +++TR +K+II+ S+ + + E QL Q
Sbjct: 107 LDSLTNKGLYLIAMILTGGSTSFDKTRLKMKEIIRDSVRRDFGKNKEGIGKEEIINQLHQ 166
Query: 77 LLNNPQNYRQRCELLPGSESQFY--HAAVVKVLCGLEKLPSQTLLAMRRKLKGIKAPMPQ 134
+L++P N+R+ C + G + A +KVL L+ L +QTL AM+RKLKG + +PQ
Sbjct: 167 VLSDPANFREDCRMNLGRTPTLHSHRDAAMKVLNELDSLSTQTLRAMKRKLKGSRM-IPQ 225
Query: 135 LQPFKNGWGRNHLIRQVNKISRKMLLELDGGNKLQEPLANAMSVADFSLKLIIGFGSTFL 194
L+ + G R+ LI QV + S KML EL G+KLQE LA A+SV D SLKL G+ +
Sbjct: 226 LKNSRRGQRRSDLINQVRQASEKMLSELSAGDKLQEQLAKALSVVDLSLKLSPGYKTAAA 285
Query: 195 EECYQFSPEVKSLQSDIMQAILSVEKKEVVPLPVLRELQLLIEPKATVVNKSLRKAFVNL 254
+ ++FSPE K+LQ++I++A+ + K V L+ L L ++P+A V N SLR A
Sbjct: 286 TDFFRFSPETKNLQNEIVKAVWLLRK---VRFRELKRLHLCLDPEAEVSNDSLRSAVRKT 342
Query: 255 LTEFLFECSDMNSTPKSLLQILDVINKCSNKSTHDVTLQKEHIEEEVDSILSVSAQTNQI 314
L E+LFECSD+++ PKSL++ L ++N + H V +E IEEE + IL+VSAQ QI
Sbjct: 343 LIEYLFECSDLDTIPKSLMEALSLVNSRTGNVEHKVC-PREAIEEETECILNVSAQVKQI 401
Query: 315 IQDLLPDYEFDQGFTDAYMEEQSEDSDNESDKDEDDSQCSENSLHCSVTPSECLNSDSDK 374
+P+YE DQ F DAYME+ EDSD+ D D+DD EN C++ +E L + +
Sbjct: 402 FCQCIPNYELDQDFGDAYMEDL-EDSDDNDDDDDDDDDDDEN---CNLFDNEKLGNKEES 457
Query: 375 NCDTINLESK--------------------------MRNTPYNNQ--------------C 394
C I LE K +T NQ
Sbjct: 458 RCRNIKLEVKDTQDDAMESSDSDHEESGAECLVLDPTDSTHETNQHDFSSSVNRVVVRDL 517
Query: 395 QEEITEQFS-----TPMSRKNC---DSSAVSLDK-----EPDENIVKRHEFHESYTEAAP 441
E IT + TP S K+ D +++ K E D + ++F +
Sbjct: 518 PESITRVYPRSLYVTPTSNKSIVISDRHDIAMSKTRVKVERDIEMEVDNQFSSRSLFSVE 577
Query: 442 RDKSNFCEEKKPIPTKYSGHKNQYLAAQDACDKTSMLAYNLIGRMLEEFAIAEDLNLDLS 501
KS+ E+KP +KNQYLA Q+ D+TS++A+NLIGR+LE+FA + LNL+
Sbjct: 578 NIKSDDHGEQKP----RRRNKNQYLAIQEISDETSLVAHNLIGRLLEKFADRQRLNLETD 633
Query: 502 KRSYLNCDKQSEDIKETEE--QSSSRKRKGGPPIVRVIEELIPSFPDRYLL 550
+RSYL + + ++ E E Q+SS+ + I+ V++E I S + L+
Sbjct: 634 ERSYLGGESRLQEKVEVSEKKQASSQAKSDEAIILSVVKEQITSLEESVLM 684
>dbj|BAD43063.1| putative protein [Arabidopsis thaliana]
Length = 574
Score = 288 bits (738), Expect = 2e-76
Identities = 204/578 (35%), Positives = 307/578 (52%), Gaps = 70/578 (12%)
Query: 30 MIVGGDTVKYERTRSNLKKIIKSSLSSVLSSKRNKDQQLETRKQLFQLLNNPQNYRQRCE 89
MI+ G + +++TR +K+II+ S+ + + E QL Q+L++P N+R+ C
Sbjct: 1 MILTGGSTSFDKTRLKMKEIIRDSVRRDFGKNKEGIGKEEIINQLHQVLSDPANFREDCR 60
Query: 90 LLPGSESQFY--HAAVVKVLCGLEKLPSQTLLAMRRKLKGIKAPMPQLQPFKNGWGRNHL 147
+ G + A +KVL L+ L +QTL AM+RKLKG + +PQL+ + G R+ L
Sbjct: 61 MNLGRTPTLHSHRDAAMKVLNELDSLSTQTLRAMKRKLKGSRM-IPQLKNSRRGQRRSDL 119
Query: 148 IRQVNKISRKMLLELDGGNKLQEPLANAMSVADFSLKLIIGFGSTFLEECYQFSPEVKSL 207
I QV + S KML EL G+KLQE LA A+SV D SLKL G+ + + ++FSPE K+L
Sbjct: 120 INQVRQASEKMLSELSAGDKLQEQLAKALSVVDLSLKLSPGYKTAAATDFFRFSPETKNL 179
Query: 208 QSDIMQAILSVEKKEVVPLPVLRELQLLIEPKATVVNKSLRKAFVNLLTEFLFECSDMNS 267
Q++I++A+ + K V L+ L L ++P+A V N SLR A L E+LFECSD+++
Sbjct: 180 QNEIVKAVWLLRK---VRFRELKRLHLCLDPEAEVSNDSLRSAVRKTLIEYLFECSDLDT 236
Query: 268 TPKSLLQILDVINKCSNKSTHDVTLQKEHIEEEVDSILSVSAQTNQIIQDLLPDYEFDQG 327
PKSL++ L ++N + H V +E IEEE + IL+VSAQ QI +P+YE DQ
Sbjct: 237 IPKSLMEALSLVNSRTGNVEHKVC-PREAIEEETECILNVSAQVKQIFCQCIPNYELDQD 295
Query: 328 FTDAYMEEQSEDSDNESDKDEDDSQCSENSLHCSVTPSECLNSDSDKNCDTINLESK--- 384
F DAYME+ EDSD+ D D+DD EN C++ +E L + + C I LE K
Sbjct: 296 FGDAYMEDL-EDSDDNDDDDDDDDDDDEN---CNLFDNEKLGNKEESRCRNIKLEVKDTQ 351
Query: 385 -----------------------MRNTPYNNQ--------------CQEEITEQFS---- 403
+T NQ E IT +
Sbjct: 352 DDAMESSDSDHEESGAECLVLDPTDSTHETNQHDFSSSVNRVVVRDLPESITRVYPRSLY 411
Query: 404 -TPMSRKNC---DSSAVSLDK-----EPDENIVKRHEFHESYTEAAPRDKSNFCEEKKPI 454
TP S K+ D +++ K E D + ++F + KS+ E+KP
Sbjct: 412 VTPTSNKSIVISDRHDIAMSKTRVKVERDIEMEVDNQFSSRSLFSVENIKSDDHGEQKP- 470
Query: 455 PTKYSGHKNQYLAAQDACDKTSMLAYNLIGRMLEEFAIAEDLNLDLSKRSYLNCDKQSED 514
+KNQYLA Q+ D+TS++A+NLIGR+LE+FA + LNL+ +RSYL + + ++
Sbjct: 471 ---RRRNKNQYLAIQEISDETSLVAHNLIGRLLEKFADRQRLNLETDERSYLGGESRLQE 527
Query: 515 IKETEE--QSSSRKRKGGPPIVRVIEELIPSFPDRYLL 550
E E Q+SS+ + I+ V++E I S + L+
Sbjct: 528 KVEVSEKKQASSQAKSDEAIILSVVKEQITSLEESVLM 565
>ref|NP_917835.1| B1158C05.16 [Oryza sativa (japonica cultivar-group)]
Length = 1105
Score = 68.2 bits (165), Expect = 7e-10
Identities = 57/216 (26%), Positives = 104/216 (47%), Gaps = 5/216 (2%)
Query: 17 LDNLTTKGLYLLAMIVGGDTVKYERTRSNLKKIIKSSLSSVLSSKRNKDQQLETRKQLFQ 76
++ L+ + L+ +A IV + V +E+TR + KII+ L L++ R+++ QL
Sbjct: 317 INKLSNEALHSVANIVTHNRVSFEKTRPAMNKIIEDHLPQYLANLRDENDM----SQLSH 372
Query: 77 LLNNPQNYRQRCELLPGSESQFYHAAVVKVLCGLEKLPSQTLLAMRRKLKGIKAPMPQLQ 136
+L N + R + S +++ + L L L Q+L+AM+RKL I P+
Sbjct: 373 ILTNSFSCRSNSLNIATPISPKMLSSINQALNVLGTLTIQSLVAMKRKLDEISF-TPKFS 431
Query: 137 PFKNGWGRNHLIRQVNKISRKMLLELDGGNKLQEPLANAMSVADFSLKLIIGFGSTFLEE 196
+ H++ + K KM+ + L + LA A+SV + K + E
Sbjct: 432 FVPRISRKAHMVTVIRKQCNKMISRVGESGDLPKNLAKALSVVNLYRKQELKCMDISQAE 491
Query: 197 CYQFSPEVKSLQSDIMQAILSVEKKEVVPLPVLREL 232
+ FS + LQ+D++ AI S++K + L +LR +
Sbjct: 492 FFPFSKKAIFLQNDVLNAIWSIQKLKKGDLKLLRPI 527
>dbj|BAD68645.1| hypothetical protein [Oryza sativa (japonica cultivar-group)]
gi|55295997|dbj|BAD68888.1| hypothetical protein [Oryza
sativa (japonica cultivar-group)]
Length = 475
Score = 68.2 bits (165), Expect = 7e-10
Identities = 57/216 (26%), Positives = 104/216 (47%), Gaps = 5/216 (2%)
Query: 17 LDNLTTKGLYLLAMIVGGDTVKYERTRSNLKKIIKSSLSSVLSSKRNKDQQLETRKQLFQ 76
++ L+ + L+ +A IV + V +E+TR + KII+ L L++ R+++ QL
Sbjct: 158 INKLSNEALHSVANIVTHNRVSFEKTRPAMNKIIEDHLPQYLANLRDENDM----SQLSH 213
Query: 77 LLNNPQNYRQRCELLPGSESQFYHAAVVKVLCGLEKLPSQTLLAMRRKLKGIKAPMPQLQ 136
+L N + R + S +++ + L L L Q+L+AM+RKL I P+
Sbjct: 214 ILTNSFSCRSNSLNIATPISPKMLSSINQALNVLGTLTIQSLVAMKRKLDEISF-TPKFS 272
Query: 137 PFKNGWGRNHLIRQVNKISRKMLLELDGGNKLQEPLANAMSVADFSLKLIIGFGSTFLEE 196
+ H++ + K KM+ + L + LA A+SV + K + E
Sbjct: 273 FVPRISRKAHMVTVIRKQCNKMISRVGESGDLPKNLAKALSVVNLYRKQELKCMDISQAE 332
Query: 197 CYQFSPEVKSLQSDIMQAILSVEKKEVVPLPVLREL 232
+ FS + LQ+D++ AI S++K + L +LR +
Sbjct: 333 FFPFSKKAIFLQNDVLNAIWSIQKLKKGDLKLLRPI 368
>emb|CAG99270.1| unnamed protein product [Kluyveromyces lactis NRRL Y-1140]
gi|50308363|ref|XP_454183.1| unnamed protein product
[Kluyveromyces lactis]
Length = 1755
Score = 51.6 bits (122), Expect = 6e-05
Identities = 108/518 (20%), Positives = 213/518 (40%), Gaps = 59/518 (11%)
Query: 37 VKYERTRSNLKKI------IKSSLSSVLSSKRNKDQQLETRKQLFQLLNNPQNYRQRCEL 90
VK+ +TR++L+K + +SL+S L + LET K + N N R
Sbjct: 701 VKFNKTRADLEKSTIENENVINSLNSKLEKLSKSLEHLETEKVNAETELNQLNERHSALE 760
Query: 91 LPGSESQFYHAAVVKVLCGLEKLPSQTLLAMRRKLKGIKAPMPQLQPFKNGWGRNHLIRQ 150
+ ++++ +V LE+L ++ K I Q+ +K N L ++
Sbjct: 761 VELNDTKIALESVNNKSQKLEELK-------QKNTKAIAVNQEQIDKYKEKI--NGLQQK 811
Query: 151 VNKISRKMLLELDGGNKLQEPLANAMSVADFSLKLIIGFGSTFLE-ECYQFSPEVKSLQS 209
V +IS L DG NK+ L ++ + LK + S LE + ++S E +L+
Sbjct: 812 VKEISESKLKAEDGINKMSRELFT-LTKENGKLKEDLKSHSKKLEIQEKKYSSETANLEK 870
Query: 210 DIMQAILSVEKKEVVPLPVLRELQLLIEPKATVVNKSLRKAFVNLLTEFLFECSDMNST- 268
+ + V++ ++ + L E T+++ N E + S+ S
Sbjct: 871 QLKERGTEVQELRERISEDIKRIDTL-EKNVTILS--------NQKIELETKLSNQTSLI 921
Query: 269 PKSLLQILDVINKCSNKSTHDVTLQKEHIEEEVDSILSVSAQTNQIIQDLLPDYE---FD 325
PK ++ + N + TL K+ +E+E ++ ++ N I+ L + E D
Sbjct: 922 PKLTEKLKGLANNYKDLENERDTLAKKILEKE-EANKTIMQNLNSEIESLNKEREEMRLD 980
Query: 326 QGFTDAYMEEQSEDSDNESDK--DEDDS-------------QCSENSLHCSVTPSECLNS 370
+ Y +++ E+ D + K E++S +C T E NS
Sbjct: 981 LQYAAEYHQKEKENFDAHTQKLTSENNSKSESIISLQTKLDECERQIKEYKTTNEELKNS 1040
Query: 371 DSDKNCDTINLESKMRNTPYNNQCQEEITEQFSTPMSRKNCDSSAVSLDKEPDENIVKRH 430
N I LES + + + +E E+ + + N + +V +K DE + + +
Sbjct: 1041 LHALNVKCIELESSLESAKQSTDNSDETIEELNDSVIAINDELQSVLAEK--DELLKQNN 1098
Query: 431 EFHESYT--EAAPRDKSNFCEEKKPIPTKYSGHKNQYL-AAQDACDKTSMLAYNLIGRML 487
+ +E + ++K++ C+ + K S N+ + ++++ DK +L
Sbjct: 1099 KINEELCNYQQELQEKADSCQ---GLQDKISSLNNEIMQISEESNDKIKLLE----ASNE 1151
Query: 488 EEFAIAEDLNLDLSKRSYLNCDKQSEDIKETEEQSSSR 525
E+ A +DL ++S N D ++E +K + SR
Sbjct: 1152 EKVAEIKDLKSEISNIKQ-NADTKAEKLKSEIDALKSR 1188
>ref|ZP_00366989.1| conserved hypothetical protein [Campylobacter coli RM2228]
gi|57020971|gb|EAL57635.1| conserved hypothetical
protein [Campylobacter coli RM2228]
Length = 769
Score = 51.6 bits (122), Expect = 6e-05
Identities = 73/338 (21%), Positives = 141/338 (41%), Gaps = 38/338 (11%)
Query: 199 QFSPEVKSLQSDIMQAILSVEKKEVVPLPVLRELQLLIEPKATVVNKSLRKAFVNLLTEF 258
+ + E+ ++++D +A+ E + +L++ E L N+L +
Sbjct: 262 KIAKELNAIKNDFFKALNRPENLMIKDTELLKQSAKAFE--------KLENTLKNILGDH 313
Query: 259 LFECSDMNSTPKSLLQILDVINKCSNKSTHDVTLQKEHIEEEVDSILSVSAQTNQIIQDL 318
+ S +SLL + I N++ + ++ E+I+E++ AQ I ++
Sbjct: 314 FPKTPQKESILESLLNHKENIK---NENIKNELIKNENIKEDI------KAQNEDINKN- 363
Query: 319 LPDYEFDQGFTDAYMEEQSEDSDNESDKDEDDSQCSENSLHCSVTPSECLNSDSDKNC-D 377
+Y D+ + +E+SE+ + K E+ ++ + + E L S D + +
Sbjct: 364 -DEYTKDESPDSSIKQEESEEKSPQVPKKENSLDDNDADIKQDIKKEENLESQEDFHPKE 422
Query: 378 TINLESKMRNTPYNNQCQEEITEQFSTPMSRKNCDSSAVSLDKEPD----ENIVKRH-EF 432
NL+SK N N Q + TE+ + KN S+ + E ENI K + +F
Sbjct: 423 DQNLDSKTENRKDNTQDSKNKTEENQAENNTKNSTSNQDKIKDEKQEKIKENITKENPKF 482
Query: 433 HESYTEAAPRDKSNFCEEKKPIPTKYSGHKNQYLAAQDACDKTSMLAYNLIGRMLEEFAI 492
+E+ TE P T+ + N AQ+ + N++ + F++
Sbjct: 483 YETKTE-----NKTSANTHNPNNTQNTQQLNNQ-TAQNIIKNQEFIKQNIVKNL--AFSV 534
Query: 493 AEDLNL----DLSKRSYLNCDKQSEDIKETEEQSSSRK 526
E+L+L DLSK K +E +KE E + + K
Sbjct: 535 -ENLDLEQVQDLSKSLSNLSRKVNESLKELEPHTQNAK 571
>emb|CAB88235.1| SPBC1711.05 [Schizosaccharomyces pombe]
gi|19112670|ref|NP_595878.1| hypothetical protein
SPBC1711.05 [Schizosaccharomyces pombe 972h-]
Length = 451
Score = 48.9 bits (115), Expect = 4e-04
Identities = 63/264 (23%), Positives = 110/264 (40%), Gaps = 32/264 (12%)
Query: 283 SNKSTHDVTLQKEHIEEEVDSILSVSAQTNQIIQDLLPDYEFDQGFTDAYMEEQSEDSDN 342
S+ S+ + E E DS S S+ ++ + D + +D+ E SEDSD+
Sbjct: 144 SDSSSSSSDSESESSSEGSDSSSSSSSSESESSSE---DNDSSSSSSDSESESSSEDSDS 200
Query: 343 ESDKDEDDSQCS----ENSLHCSVTPSECLNSDSDKNCDTINLESKMRNTPYNNQCQEEI 398
S + +S+ S ++S S + SE + D+D + + + ES+ + ++
Sbjct: 201 SSSSSDSESESSSEGSDSSSSSSSSESESSSEDNDSSSSSSDSESESSSEDSDSSSSSSD 260
Query: 399 TEQFSTPMSRKNCDSSAVSLDKEPDENIVKRHEFHESYTEAAPRDKSNFCEEKKPIPTKY 458
+E S S K+ DSS+ S D E D + ES +E + D ++ + +
Sbjct: 261 SESES---SSKDSDSSSNSSDSEDDSSSDSSDSESESSSEDS--DSTSSSSDSDSSSSSE 315
Query: 459 SGHKNQ------YLAAQDACDKTSMLAYNLIGRMLEEFAIAEDLNLDLSKRSYLNCDKQS 512
G+ N ++AQ + + TS EE +D D SK + ++
Sbjct: 316 DGNSNTDTTTSGEVSAQSSTNSTSS----------EESTSVKD--EDSSKIHDKSLKRKH 363
Query: 513 EDIKETEEQSSSRKRKGGPPIVRV 536
ED + + SSR K P RV
Sbjct: 364 EDDESSTSTKSSRTTK--TPFTRV 385
Score = 42.7 bits (99), Expect = 0.030
Identities = 53/278 (19%), Positives = 109/278 (39%), Gaps = 8/278 (2%)
Query: 251 FVNLLTEFLFECSDMN----STPKSLLQILDVINKCSNKSTHDVTLQKEHIEEEVDSILS 306
+V FL E D + K+LL +L +T DV K+ E S+
Sbjct: 23 YVKTAQTFLKETGDKDLAKGKVKKNLLDLLSQKEFLPYLTTEDVGKHKKTKE----SLEK 78
Query: 307 VSAQTNQIIQDLLPDYEFDQGFTDAYMEEQSEDSDNESDKDEDDSQCSENSLHCSVTPSE 366
+ + +I + P + + S++SD+ S + E S+ +++S S + SE
Sbjct: 79 SNDDSQKISKKGAPPEKAHSSSEASGSGSSSDESDSSSSESESSSEDNDSSSSSSDSESE 138
Query: 367 CLNSDSDKNCDTINLESKMRNTPYNNQCQEEITEQFSTPMSRKNCDSSAVSLDKEPDENI 426
+ DSD + + + ES+ + ++ +E S+ + SS+ S + E+
Sbjct: 139 SSSEDSDSSSSSSDSESESSSEGSDSSSSSSSSESESSSEDNDSSSSSSDSESESSSEDS 198
Query: 427 VKRHEFHESYTEAAPRDKSNFCEEKKPIPTKYSGHKNQYLAAQDACDKTSMLAYNLIGRM 486
+S +E++ + S + ++ D+ ++S +
Sbjct: 199 DSSSSSSDSESESSSEGSDSSSSSSSSESESSSEDNDSSSSSSDSESESSSEDSDSSSSS 258
Query: 487 LEEFAIAEDLNLDLSKRSYLNCDKQSEDIKETEEQSSS 524
+ + + + D S S + D S D ++E +SSS
Sbjct: 259 SDSESESSSKDSDSSSNSSDSEDDSSSDSSDSESESSS 296
Score = 40.4 bits (93), Expect = 0.15
Identities = 45/177 (25%), Positives = 76/177 (42%), Gaps = 20/177 (11%)
Query: 261 ECSDMNSTPKSLLQILDVINKCSNKSTHDVTLQKEHIEEEVDSILSVSAQTNQIIQDLLP 320
E SD +S+ S + S+ S+ D + E E+ DS S S ++
Sbjct: 214 EGSDSSSSSSSSESESSSEDNDSSSSSSDS--ESESSSEDSDSSSSSSDSESESSSK--- 268
Query: 321 DYEFDQGFTDAYMEEQSEDSDNESDKDEDDSQCSENSLHCSVTPSECLNSDSDKNCDTIN 380
D + +D+ + S+ SD+ES+ +DS + +S S + S + D + N DT
Sbjct: 269 DSDSSSNSSDSEDDSSSDSSDSESESSSEDSDSTSSS---SDSDSSSSSEDGNSNTDTTT 325
Query: 381 LESKMRNTPYNNQCQEEITEQFSTPMSRKNCDSSAVSLDKEPDENIVKRHEFHESYT 437
+ N+ EE T S K+ DSS + D+++ ++HE ES T
Sbjct: 326 SGEVSAQSSTNSTSSEEST-------SVKDEDSSKIH-----DKSLKRKHEDDESST 370
>gb|AAB64815.1| Ydr379wp [Saccharomyces cerevisiae] gi|6320587|ref|NP_010667.1|
GTPase-activating protein for the polarity-establishment
protein Cdc42p; implicated in control of septin
organization, pheromone response, and haploid invasive
growth; Rga2p [Saccharomyces cerevisiae]
gi|31076883|sp|Q06407|RGA2_YEAST Rho-type
GTPase-activating protein 2 gi|2131456|pir||S61174
hypothetical protein YDR379w - yeast (Saccharomyces
cerevisiae)
Length = 1009
Score = 48.5 bits (114), Expect = 5e-04
Identities = 69/284 (24%), Positives = 123/284 (43%), Gaps = 29/284 (10%)
Query: 261 ECSDMNSTPKSLLQILDVINKCSNKSTHDVTLQKEHIEE-----EVDSILSVSAQTNQII 315
+C + K LL L+ K S+K + L + ++ ++ VS I
Sbjct: 121 DCHEKLLRKKQLL--LENQTKNSSKEDFPIKLPERSVKRPLSPTRINGKSDVSTNNTAIS 178
Query: 316 QDLLPDYEFDQGFTDAYM--EEQSEDSDNESDKDEDDSQCSENSLHC-SVTPSECLNSDS 372
++L+ E DQ T + +E+ E S N+++ +++ E S H +V+ + LNS
Sbjct: 179 KNLVSSNE-DQQLTPQVLVSQERDESSLNDNNDNDNSKDREETSSHARTVSIDDILNSTL 237
Query: 373 DKNCDTINLESKMRNTPYNNQCQEEITEQFSTPMSRKNCDSSAVSLDKEPDENIVKRHEF 432
+ + ++I +S + N Y N+ E++T + P R N DS V + P+ N F
Sbjct: 238 EHDSNSIEEQSLVDNEDYINKMGEDVTYRLLKP-QRANRDSIVVKDPRIPNSNSNANRFF 296
Query: 433 HESYTEAAPRDKSNFCEEKKPIPTKYSGHKNQYLAAQDACDK-TSMLAYNLIGRMLEEFA 491
E +D ++ E + + T +++ DK TS L + +M EE
Sbjct: 297 SIYDKEETDKDDTDNKENEIIVNT-----------PRNSTDKITSPLNSPMAVQMNEEVE 345
Query: 492 IAEDLNLDLSKRSYLNCDKQSEDIKETEEQSSSRKRKGGPPIVR 535
L L LS+ + N +K S+ I + S+S+ PI R
Sbjct: 346 PPHGLALTLSEATKEN-NKSSQGI----QTSTSKSMNHVSPITR 384
>emb|CAA44805.1| nucleolin [Xenopus laevis] gi|464252|sp|P20397|NUCL_XENLA Nucleolin
(Protein C23)
Length = 651
Score = 47.8 bits (112), Expect = 0.001
Identities = 54/204 (26%), Positives = 80/204 (38%), Gaps = 23/204 (11%)
Query: 338 EDSDNESDKDEDDSQCSENSLHCSVTPSECLNSDSDKNCDTINLESKMRNTPYNNQCQEE 397
EDS+ E D +EDDS E + TP++ + + K TP N Q +
Sbjct: 26 EDSEEEEDMEEDDSSDEEVEVPVKKTPAKKTATPAKATPGKAATPGKKGATPAKNGKQAK 85
Query: 398 ITEQFSTPMSRKNCDSSAVSLDKEPDEN--IVKRHEFHESYTEAAPRDKSNFCEE----K 451
E + DS + D++P +N + K+ + +E D+ EE K
Sbjct: 86 KQES-----EEEEDDSDEEAEDQKPIKNKPVAKKAVAKKEESEEDDDDEDESEEEKAVAK 140
Query: 452 KPIPTKYSGHKNQYLAAQD---ACDKTSMLAYNLIGRMLEEFAIAEDLNLDLSKRSYLNC 508
KP P K K Q +D + D+ +A L G+ + A +D D
Sbjct: 141 KPTPAKKPAGKKQESEEEDDEESEDEPMEVAPALKGKKTAQAAEEDDEEED--------- 191
Query: 509 DKQSEDIKETEEQSSSRKRKGGPP 532
D ED + EEQ S KRK P
Sbjct: 192 DDDEEDDDDEEEQQGSAKRKKEMP 215
>ref|XP_629358.1| hypothetical protein DDB0191811 [Dictyostelium discoideum]
gi|60462719|gb|EAL60921.1| hypothetical protein
DDB0191811 [Dictyostelium discoideum]
Length = 942
Score = 46.6 bits (109), Expect = 0.002
Identities = 65/330 (19%), Positives = 133/330 (39%), Gaps = 50/330 (15%)
Query: 237 EPKATVVNKSLRKAFVNLLTEFLFECSDMNSTPKSLLQILDVINKCSNKS---------- 286
+P + ++ SL+K FL S S+ S+L D+I K KS
Sbjct: 43 DPSSFSLSSSLKKRPTENTASFL---SSSLSSSSSILSADDLIKKNRRKSFGESKEKEEK 99
Query: 287 --THDVTLQKEHIEEEVDSILSVSAQTNQIIQDLLPDYEFDQG---------FTDAYMEE 335
+ +KE +E+ S S + T L P +++ +
Sbjct: 100 EKEKEKEKEKEKEKEKEKSSSSTTTTTTTTTSKLKPKSTLTSSSSSSSHIATYSEPLKDH 159
Query: 336 QSEDSDNESDKDEDDSQCSENSLHCSVTPSECLNSDSDKNCDTINL-----ESKMRNTPY 390
SE + E +K+++ + +++ + ++TP+ + K+ ++ N+ + +T
Sbjct: 160 SSEPKEKEKEKEKEKEKENKDKSNSTLTPTNVVKKLKRKSIESGNIFQFDQAKESLSTSS 219
Query: 391 NNQCQEEITEQFSTPMSRKNCDSSAVSLDKEPDENIVKR-------------HEFHESYT 437
++ T +T S SS++S DKEP E+ K+ +E+
Sbjct: 220 SSSSGTTATSTTTTTTS----SSSSISKDKEPKESNKKKLKVSVQSSQTEENENENENQL 275
Query: 438 EAAPRDKSNFCEEKKPIPTKYSGHKNQYLAAQDACDKTSMLAYNLIGRMLEEFAIAEDLN 497
E P +KS+ + K + +G K + ++ K S ++ I +L++ ++ +
Sbjct: 276 EEQPEEKSSKKKVSKTSSLESTGEKKKKRSSTSGTSKKSS-TFDSIKSILDD---DDEND 331
Query: 498 LDLSKRSYLNCDKQSEDIKETEEQSSSRKR 527
D K N +K E K+ +++ SS K+
Sbjct: 332 FDYEKDDNGNGEKVKEKEKKKKKKKSSSKK 361
>ref|NP_473250.2| hypothetical protein [Plasmodium falciparum 3D7]
gi|7672224|emb|CAA15592.2| hypothetical protein
[Plasmodium falciparum 3D7]
Length = 594
Score = 45.8 bits (107), Expect = 0.004
Identities = 48/220 (21%), Positives = 88/220 (39%), Gaps = 32/220 (14%)
Query: 279 INKCSNKSTHDVTLQKE-----HIEEEVDS------------ILSVSAQTNQIIQDLLPD 321
IN +K+ ++ + ++ HIEE +++ + V+ + Q I +L +
Sbjct: 366 INNEDHKNVNNTQINEDIQIIKHIEENIENEHDKLEETQEKKMEDVAQKDEQQIDNLENE 425
Query: 322 YEFDQ----GFTDAYMEEQSEDSDNESDKDEDDSQCSENSLHCSVTPSE-CLNSDSD-KN 375
+ ++ G D E + E+ E DK+E+D + + C E C + + D K
Sbjct: 426 KDTEKKEADGGEDKEKEGKEEEDKEEEDKEEEDKKEEDKKEECKEEYKEDCKDKEEDKKE 485
Query: 376 CDTINLESKMRNTPYNNQCQEEITEQFSTPMSRKNCDSSAVSLDKEPD-ENIVKRHEFHE 434
C K +C+EE E+ +K C DKE D E ++ + E
Sbjct: 486 CKDKEEGKKECKEEGKKECKEECKEE-----GKKECKEEGKEEDKEKDKEENNEKEDKEE 540
Query: 435 SYTEAAPRDKSNFCEEKKPIPTKYSGHKNQYLAAQDACDK 474
++N+ EE+ +P K H+ +DA K
Sbjct: 541 EINSHLNIHETNYDEEQLGLPKKMESHQED---KEDAVQK 577
>ref|ZP_00323079.1| hypothetical protein PpenA01001008 [Pediococcus pentosaceus ATCC
25745]
Length = 2334
Score = 45.8 bits (107), Expect = 0.004
Identities = 47/242 (19%), Positives = 93/242 (38%), Gaps = 23/242 (9%)
Query: 263 SDMNSTPKSLLQILDVINKCSNKSTHDVTLQKEHIEEEVDSILSVSAQTNQIIQDLLPDY 322
SD ST S + + I+ ++KST D + + S S T+ + D
Sbjct: 1449 SDSRSTSTS---VSNSISDSNSKSTSDSRSTSTSVSDSTSDSASTSHSTSDSVSTSNSDS 1505
Query: 323 EF----DQGFTDAYMEEQSEDSDNESDKDEDDSQCSENSLHCSVTPSECLNSDSDKNCDT 378
+ D + + DS+++SD + NS+ S++ S NSDS D+
Sbjct: 1506 DSKSTSDSRSASTSVSDSKSDSESKSDSTSKSDSITSNSISESISTS---NSDSSSKSDS 1562
Query: 379 INL-ESKMRNTPYNNQCQEEITEQFST--PMSRKNCD----------SSAVSLDKEPDEN 425
++ +S+ +T ++ + + ST +S N D S++ S+ ++
Sbjct: 1563 KSISDSRSTSTSVSDSTSDSASTSHSTSDSVSTSNSDSDSKSMSESRSTSTSVSDSTSDS 1622
Query: 426 IVKRHEFHESYTEAAPRDKSNFCEEKKPIPTKYSGHKNQYLAAQDACDKTSMLAYNLIGR 485
H +S + + S + + T S K+ + D+ K+ + N I
Sbjct: 1623 ASTSHSTSDSVSTSKSDSSSKSTSDSRSTSTSISDSKSDSASKSDSISKSDSITSNSISE 1682
Query: 486 ML 487
+
Sbjct: 1683 SI 1684
Score = 40.8 bits (94), Expect = 0.11
Identities = 56/290 (19%), Positives = 106/290 (36%), Gaps = 39/290 (13%)
Query: 263 SDMNSTPKSLLQILDVINKCSNKSTHDVTLQKEHIEEEVDSIL------------SVSAQ 310
SD ST S+ + S ++ V+ K E + DSI S+S
Sbjct: 1058 SDSRSTSTSVSDSISKSMSDSRSTSTSVSDSKSDSESKSDSISKSDSITSNSISESISTS 1117
Query: 311 TNQIIQDLLPDYEFDQGFTDAYMEEQSEDSDNESDKDEDDSQCSENSLHCSVTPSECLNS 370
+ I D D T + + DS ++SD + +S+ S++ S +S
Sbjct: 1118 NSDSISDSNSKSTSDSRSTSTSISDSKSDSASKSDSVSKSDSITSDSISESISTSNS-DS 1176
Query: 371 DSDKNCDTINLESKMRNTPYNNQCQEEITEQFSTP--MSRKNCDSS-------------- 414
SD N + + +S+ +T ++ + + ST +S N DSS
Sbjct: 1177 ISDSNSKSTS-DSRSTSTSVSDSKSDSASTSHSTSDSVSTSNSDSSSKSDSVSTSDSRST 1235
Query: 415 AVSLDKEPDENIVKRHEFHESYTEAAPRDKSNFCEEKKPIPTKYSGHKNQYLAAQDACDK 474
+ S+ ++ H +S + + S + + T S K+ + D+ K
Sbjct: 1236 STSISDSTSDSASTSHSTSDSVSTSNSDSDSKSTSDSRSASTSVSDSKSDSASKSDSTSK 1295
Query: 475 TSMLAYNLIGRMLEEFAIAEDLNLDLSKRSYLNCDKQSEDIKETEEQSSS 524
+ + N +I+E ++ S S + K + D + T S+
Sbjct: 1296 SDSITSN---------SISESISTSNSDSSSKSDSKSTSDSRSTSTSVSN 1336
Score = 39.3 bits (90), Expect = 0.33
Identities = 50/214 (23%), Positives = 80/214 (37%), Gaps = 14/214 (6%)
Query: 263 SDMNSTPKSLLQILDVINKCSNKSTHDV-TLQKEHIEEEVDSILSVSAQTNQIIQDLLPD 321
SD ST S + N S+ + V T V S SA T+ D +
Sbjct: 1989 SDSTSTSHSTSDSVSTSNSDSSSKSDSVSTSDSRSASTSVSDSTSDSASTSHSTSDSVST 2048
Query: 322 YEFDQGFTDAYMEEQSEDSDNESDKDEDDSQCSENSLHCSVTPSECLNSDSDKNCDTINL 381
D + + DS + S D + S ++ H + NSDS D+I+
Sbjct: 2049 SNSDSSSKSD--SKSASDSRSTSTSVSDSTSNSTSASHSTSDSVSTSNSDSSSKSDSIST 2106
Query: 382 -ESKMRNTPYNNQCQEEITEQFSTPMSRKNCDSSAVSLDKEPDENIVKRHEFH-----ES 435
ES+ +T ++ ++ ST S + S+++S D N H+ H ES
Sbjct: 2107 SESRSASTSVSDSTSVSTSDSRSTSTSISDSTSNSIS-----DGNGSTSHDNHHGSGSES 2161
Query: 436 YTEAAPRDKSNFCEEKKPIPTKYSGHKNQYLAAQ 469
+E+ + S+ I S KN A+Q
Sbjct: 2162 NSESGSQSTSDSQSISTSISDSTSDSKNSGSASQ 2195
Score = 37.4 bits (85), Expect = 1.3
Identities = 41/171 (23%), Positives = 68/171 (38%), Gaps = 11/171 (6%)
Query: 263 SDMNSTPKSLLQILDVINKCSN-KSTHDVTLQKEHIEEEVDSILSVSAQTNQIIQDLLPD 321
SD ST S + N S+ KS + + + S S T+ + D
Sbjct: 1580 SDSASTSHSTSDSVSTSNSDSDSKSMSESRSTSTSVSDSTSDSASTSHSTSDSVSTSKSD 1639
Query: 322 YEF----DQGFTDAYMEEQSEDSDNESDKDEDDSQCSENSLHCSVTPSECLNSDSDKNCD 377
D T + + DS ++SD + NS+ S++ S+ SDS D
Sbjct: 1640 SSSKSTSDSRSTSTSISDSKSDSASKSDSISKSDSITSNSISESISTSK---SDSSSKSD 1696
Query: 378 TINL-ESKMRNTPYNNQCQEEITEQFSTP--MSRKNCDSSAVSLDKEPDEN 425
+ + ES+ +T ++ + I+ ST +S N DSS+ S K E+
Sbjct: 1697 SKSTSESRSASTSVSDSTSDSISTSHSTSDSVSTSNSDSSSKSDSKSTSES 1747
Score = 35.4 bits (80), Expect = 4.8
Identities = 39/174 (22%), Positives = 68/174 (38%), Gaps = 16/174 (9%)
Query: 263 SDMNSTPKSLLQILDVINKCSNKSTHDVTLQKEHIEEEVDSILSVSAQTNQIIQDLLPDY 322
SD ST S D +K ++ S T + I + S S + + D D
Sbjct: 1769 SDSVSTSNS-----DSSSKSASDSRSTSTSVSDSISDSNSKSTSDSRSASTSVSDSTSDS 1823
Query: 323 EFDQGFTDAYMEEQSEDSDNESDKD--------EDDSQCSENSLHCSVTPSECLNSDSDK 374
T + + DSD++S D D + S ++ H + NSDS
Sbjct: 1824 TSTSHSTSDSVSTSNSDSDSKSMSDSRSTSTSISDSTSDSTSTSHSTSDSVSTSNSDSSS 1883
Query: 375 NCDTINL-ESKMRNTPYNNQCQEEITEQFST--PMSRKNCDSSAVSLDKEPDEN 425
D+++ +S+ +T ++ + + ST +S N DSS+ S K ++
Sbjct: 1884 KSDSVSTSDSRSASTSVSDSTSDSTSTSHSTSDSVSTSNSDSSSKSDSKSASDS 1937
>pir||T18450 hypothetical protein C0570c - malaria parasite (Plasmodium
falciparum)
Length = 419
Score = 45.8 bits (107), Expect = 0.004
Identities = 48/220 (21%), Positives = 88/220 (39%), Gaps = 32/220 (14%)
Query: 279 INKCSNKSTHDVTLQKE-----HIEEEVDS------------ILSVSAQTNQIIQDLLPD 321
IN +K+ ++ + ++ HIEE +++ + V+ + Q I +L +
Sbjct: 191 INNEDHKNVNNTQINEDIQIIKHIEENIENEHDKLEETQEKKMEDVAQKDEQQIDNLENE 250
Query: 322 YEFDQ----GFTDAYMEEQSEDSDNESDKDEDDSQCSENSLHCSVTPSE-CLNSDSD-KN 375
+ ++ G D E + E+ E DK+E+D + + C E C + + D K
Sbjct: 251 KDTEKKEADGGEDKEKEGKEEEDKEEEDKEEEDKKEEDKKEECKEEYKEDCKDKEEDKKE 310
Query: 376 CDTINLESKMRNTPYNNQCQEEITEQFSTPMSRKNCDSSAVSLDKEPD-ENIVKRHEFHE 434
C K +C+EE E+ +K C DKE D E ++ + E
Sbjct: 311 CKDKEEGKKECKEEGKKECKEECKEE-----GKKECKEEGKEEDKEKDKEENNEKEDKEE 365
Query: 435 SYTEAAPRDKSNFCEEKKPIPTKYSGHKNQYLAAQDACDK 474
++N+ EE+ +P K H+ +DA K
Sbjct: 366 EINSHLNIHETNYDEEQLGLPKKMESHQED---KEDAVQK 402
>ref|NP_703728.1| hypothetical protein, expressed [Plasmodium falciparum 3D7]
gi|46362298|emb|CAG25236.1| hypothetical protein,
expressed [Plasmodium falciparum 3D7]
Length = 2402
Score = 45.4 bits (106), Expect = 0.005
Identities = 58/263 (22%), Positives = 108/263 (41%), Gaps = 27/263 (10%)
Query: 205 KSLQSDIMQAILSVEKKEVVPLPVLRELQLLIEPKAT-VVNKSLRKAFVNLLTEFLFEC- 262
K L I + K ++PL +R LQ+ K T V+N S+ + +N++ F+F
Sbjct: 531 KRLDFKIHPKFFHIPNKSILPLCSIRLLQINDAMKFTKVLNVSVSSSSLNIINLFMFSII 590
Query: 263 ---SDMNSTPKSLLQILDVI--NKCSNKSTHDVTLQKEHIEEEVDSILSVSAQTNQIIQD 317
S N + +S + ++ + C + D + E ++ ++ N
Sbjct: 591 FNSSKNNVSVQSFSAPIPLLMGDVCGTEKDGDEMTHGNVLHAENNNDVNGDNGDNG---- 646
Query: 318 LLPDYEFDQGFTDAYMEEQSEDSDNESDKDEDDSQCSENSLHCSVTPSECLNSDSDKNCD 377
D + D G D ++ + D DN D +++D+ +N+ + E N+D + N D
Sbjct: 647 ---DNDDDNGDNDDNNDDNN-DEDNNDDNNDEDNNNDDNNDEDN-NNDEDNNNDDNNNND 701
Query: 378 TINLESKMRNTPYNNQCQEEITEQFSTPMSRKNCDSSAVSLDKEPDE-----NIVKRHEF 432
N + +T + + C + ST + K S++KE D+ N VK++
Sbjct: 702 DNNNDGNNVSTYHKDHCAK------STLENEKYTFDECTSINKEIDDINHHYNFVKKNSL 755
Query: 433 HESYTEAAPRDKSNFCEEKKPIP 455
E Y K E+ + IP
Sbjct: 756 SEPYENIVNILKYKQSEDMEKIP 778
>emb|CAG58749.1| unnamed protein product [Candida glabrata CBS138]
gi|50286801|ref|XP_445830.1| unnamed protein product
[Candida glabrata]
Length = 425
Score = 45.4 bits (106), Expect = 0.005
Identities = 38/172 (22%), Positives = 73/172 (42%), Gaps = 13/172 (7%)
Query: 286 STHDVTLQKEHIEEEVDSILSVSAQTNQIIQDLLPDYEFDQGFTDAYMEEQSEDSDNESD 345
S D+ L K+ +E DS S S+ + + +D+ E S+D + +
Sbjct: 10 SKKDLKLAKKEVESSSDSSSSSSSSS-------------ESSSSDSSSSESSDDEEPKKT 56
Query: 346 KDEDDSQCSENSLHCSVTPSECLNSDSDKNCDTINLESKMRNTPYNNQCQEEITEQFSTP 405
K+ DS S++S S + SE + +S ++ + S ++ + +E ++ +
Sbjct: 57 KESSDSSSSDSSSSESDSDSESDSEESKEDKKESSSSSSSSSSDSESDSEESKEDKKESS 116
Query: 406 MSRKNCDSSAVSLDKEPDENIVKRHEFHESYTEAAPRDKSNFCEEKKPIPTK 457
S + SS+ S D E D + E E+ E + + EE++ P K
Sbjct: 117 DSESSSSSSSSSSDSESDSDSSSSDEEEEAKEEKDSKKRKAEDEEEESSPKK 168
Score = 37.7 bits (86), Expect = 0.96
Identities = 34/170 (20%), Positives = 79/170 (46%), Gaps = 1/170 (0%)
Query: 240 ATVVNKSLRKAFVNLLTEFLFECSDMNSTPKSLLQILDVINKCSNKSTHDVTLQKEHIEE 299
A K + K + L + + SD +S+ S + + S++S+ D +K
Sbjct: 2 AKSTTKKVSKKDLKLAKKEVESSSDSSSSSSSSSESSSS-DSSSSESSDDEEPKKTKESS 60
Query: 300 EVDSILSVSAQTNQIIQDLLPDYEFDQGFTDAYMEEQSEDSDNESDKDEDDSQCSENSLH 359
+ S S S++++ + + + D+ + + S DS+++S++ ++D + S +S
Sbjct: 61 DSSSSDSSSSESDSDSESDSEESKEDKKESSSSSSSSSSDSESDSEESKEDKKESSDSES 120
Query: 360 CSVTPSECLNSDSDKNCDTINLESKMRNTPYNNQCQEEITEQFSTPMSRK 409
S + S +S+SD + + + E + + + + + E E+ S+P K
Sbjct: 121 SSSSSSSSSDSESDSDSSSSDEEEEAKEEKDSKKRKAEDEEEESSPKKAK 170
>emb|CAB79515.1| putative protein [Arabidopsis thaliana] gi|7228247|emb|CAB77061.1|
putative protein [Arabidopsis thaliana]
gi|7487043|pir||T08926 hypothetical protein T15N24.50 -
Arabidopsis thaliana
Length = 626
Score = 45.1 bits (105), Expect = 0.006
Identities = 52/215 (24%), Positives = 91/215 (42%), Gaps = 17/215 (7%)
Query: 289 DVTLQKEHIEEEVDSILSVSAQTNQIIQDLLPDYEFDQGFTDAYMEEQSEDSDNESDKDE 348
+V+ +E EEE + S + ++ DL D + ++G D+ ++ +D D++ D DE
Sbjct: 56 EVSTDEEEEEEENEQ----SDEGSESGSDLFSDGD-EEGNNDS--DDDDDDDDDDDDDDE 108
Query: 349 DDSQCSENSLHCSVTPSECLNSDSDKNCDTINLESKMRNTPYNNQCQ-EEITEQFSTPMS 407
D +E+ L S + SD D + LE K R + + ++ ++F +
Sbjct: 109 DAEPLAEDFLDGSDNEEVTMGSDLDSDSGGSKLERKSRAIDRKRKKEVQDADDEFKMNIK 168
Query: 408 RKNCDSSAVSLDKEPDENIVKRHEFH--ESYTEAAPRDKSNFCEEKKPIPTKYSGHKNQY 465
K D + KE +E + + + R SNF K + K H+
Sbjct: 169 EKP-DEFQLPTQKELEEEARRPPDLPSLQMRIREIVRILSNF----KDLKPKGDKHERND 223
Query: 466 LAAQDACDKTSMLAYN--LIGRMLEEFAIAEDLNL 498
Q D +S YN LIG ++E F + E + L
Sbjct: 224 YVGQLKADLSSYYGYNEFLIGTLIEMFPVVELMEL 258
>gb|AAM91770.1| unknown protein [Arabidopsis thaliana] gi|17979390|gb|AAL49920.1|
unknown protein [Arabidopsis thaliana]
gi|22328952|ref|NP_194390.2| nucleolar protein, putative
[Arabidopsis thaliana]
Length = 671
Score = 45.1 bits (105), Expect = 0.006
Identities = 52/215 (24%), Positives = 91/215 (42%), Gaps = 17/215 (7%)
Query: 289 DVTLQKEHIEEEVDSILSVSAQTNQIIQDLLPDYEFDQGFTDAYMEEQSEDSDNESDKDE 348
+V+ +E EEE + S + ++ DL D + ++G D+ ++ +D D++ D DE
Sbjct: 56 EVSTDEEEEEEENEQ----SDEGSESGSDLFSDGD-EEGNNDS--DDDDDDDDDDDDDDE 108
Query: 349 DDSQCSENSLHCSVTPSECLNSDSDKNCDTINLESKMRNTPYNNQCQ-EEITEQFSTPMS 407
D +E+ L S + SD D + LE K R + + ++ ++F +
Sbjct: 109 DAEPLAEDFLDGSDNEEVTMGSDLDSDSGGSKLERKSRAIDRKRKKEVQDADDEFKMNIK 168
Query: 408 RKNCDSSAVSLDKEPDENIVKRHEFH--ESYTEAAPRDKSNFCEEKKPIPTKYSGHKNQY 465
K D + KE +E + + + R SNF K + K H+
Sbjct: 169 EKP-DEFQLPTQKELEEEARRPPDLPSLQMRIREIVRILSNF----KDLKPKGDKHERND 223
Query: 466 LAAQDACDKTSMLAYN--LIGRMLEEFAIAEDLNL 498
Q D +S YN LIG ++E F + E + L
Sbjct: 224 YVGQLKADLSSYYGYNEFLIGTLIEMFPVVELMEL 258
>gb|EAA17806.1| hypothetical protein [Plasmodium yoelii yoelii]
Length = 650
Score = 44.7 bits (104), Expect = 0.008
Identities = 36/156 (23%), Positives = 68/156 (43%), Gaps = 7/156 (4%)
Query: 277 DVINKCSNKSTHDVTLQKEHIEEEV-----DSILSVSAQTNQIIQDLLPDYEFDQGFTDA 331
D+I+ + T +KE I+ +V + L + +++ D+ D D D
Sbjct: 154 DIIDVVHTQREPLQTAEKEEIKNDVKLKENSTNLKKNKDADEMDDDM-DDENLDTSVEDE 212
Query: 332 YMEEQSEDSDNESDKDEDDSQCSENSLHCSVTPSECLNSDSDKNCDTINLESKMRNTPYN 391
Y + + ED D ESD D++D + EN + S++N + +++ +N N
Sbjct: 213 YNDNEDEDDDFESDDDKEDDE-KENQKQNQKEKEKHSEEQSEQNEEQNEEQNEEQNEEQN 271
Query: 392 NQCQEEITEQFSTPMSRKNCDSSAVSLDKEPDENIV 427
+ EE E+ + N + +S +K ENI+
Sbjct: 272 EEQNEEQNEKLYKQIIIDNDKETEISGNKNTKENIL 307
>ref|NP_702439.1| hypothetical protein PF14_0550 [Plasmodium falciparum 3D7]
gi|23497623|gb|AAN37163.1| hypothetical protein
[Plasmodium falciparum 3D7]
Length = 1478
Score = 44.3 bits (103), Expect = 0.010
Identities = 32/120 (26%), Positives = 60/120 (49%), Gaps = 15/120 (12%)
Query: 334 EEQSEDS-DNESDKDEDDSQCSENSLHCSVTPSECLNSDSDKNCDTINLESKMRN----- 387
E+Q+ED+ DN +KD D S + + S N++++ N + N + N
Sbjct: 1128 EKQNEDNKDNMDNKDNKDFINSHENETKNELQSNSKNNNNNNNNNNNNNNNNNNNNYDDD 1187
Query: 388 -------TPYNNQCQEEITEQFSTPMSRKNCDSSAVSLDKEPDE--NIVKRHEFHESYTE 438
T YNN+ +++I F+ ++ + +SAVS+D+E E NI +HE ++ + +
Sbjct: 1188 DLVINIKTKYNNKNEKDIKSDFTLNLNSDDVTTSAVSIDEEKKEENNIYNKHEKNQEHNK 1247
Database: nr
Posted date: Jul 5, 2005 12:34 AM
Number of letters in database: 863,360,394
Number of sequences in database: 2,540,612
Lambda K H
0.314 0.131 0.366
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 902,328,385
Number of Sequences: 2540612
Number of extensions: 38069385
Number of successful extensions: 168400
Number of sequences better than 10.0: 740
Number of HSP's better than 10.0 without gapping: 47
Number of HSP's successfully gapped in prelim test: 759
Number of HSP's that attempted gapping in prelim test: 161246
Number of HSP's gapped (non-prelim): 3281
length of query: 552
length of database: 863,360,394
effective HSP length: 133
effective length of query: 419
effective length of database: 525,458,998
effective search space: 220167320162
effective search space used: 220167320162
T: 11
A: 40
X1: 16 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (22.0 bits)
S2: 78 (34.7 bits)
Medicago: description of AC137825.14


