

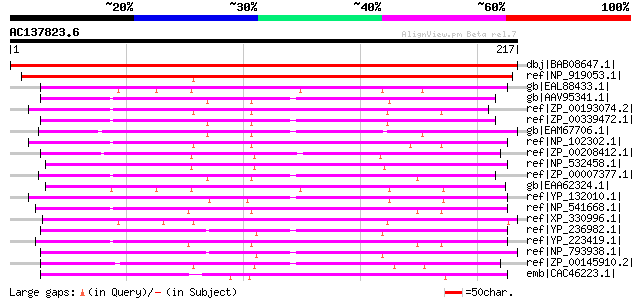
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC137823.6 + phase: 0
(217 letters)
Database: nr
2,540,612 sequences; 863,360,394 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
dbj|BAB08647.1| frnE protein-like [Arabidopsis thaliana] gi|2839... 296 2e-79
ref|NP_919053.1| putative polyketide synthase [Oryza sativa (jap... 268 6e-71
gb|EAL88433.1| DSBA-like thioredoxin domain protein [Aspergillus... 137 3e-31
gb|AAV95341.1| DSBA-like thioredoxin family protein [Silicibacte... 132 5e-30
ref|ZP_00193074.2| COG2761: Predicted dithiol-disulfide isomeras... 129 4e-29
ref|ZP_00339472.1| COG2761: Predicted dithiol-disulfide isomeras... 127 2e-28
gb|EAM67706.1| DSBA oxidoreductase [Jannaschia sp. CCS1] 126 5e-28
ref|NP_102302.1| similar to frnE protein [Mesorhizobium loti MAF... 125 6e-28
ref|ZP_00208412.1| COG2761: Predicted dithiol-disulfide isomeras... 125 6e-28
ref|NP_532458.1| polyketide biosynthesis associated protein [Agr... 123 3e-27
ref|ZP_00007377.1| COG2761: Predicted dithiol-disulfide isomeras... 122 7e-27
gb|EAA62324.1| hypothetical protein AN5143.2 [Aspergillus nidula... 122 7e-27
ref|YP_132010.1| hypothetical 2-hydroxychromene-2-carboxylateiso... 120 3e-26
ref|NP_541668.1| FRNE [Brucella melitensis 16M] gi|23463934|gb|A... 120 3e-26
ref|XP_330996.1| hypothetical protein [Neurospora crassa] gi|289... 120 3e-26
ref|YP_236982.1| DSBA oxidoreductase [Pseudomonas syringae pv. s... 119 8e-26
ref|YP_223419.1| hypothetical FrnE [Brucella abortus biovar 1 st... 117 2e-25
ref|NP_793938.1| 2-hydroxychromene-2-carboxylate isomerase famil... 116 5e-25
ref|ZP_00145910.2| COG2761: Predicted dithiol-disulfide isomeras... 115 9e-25
emb|CAC46223.1| CONSERVED HYPOTHETICAL PROTEIN [Sinorhizobium me... 115 1e-24
>dbj|BAB08647.1| frnE protein-like [Arabidopsis thaliana] gi|28394079|gb|AAO42447.1|
putative frnE protein [Arabidopsis thaliana]
gi|27754699|gb|AAO22793.1| putative frnE protein
[Arabidopsis thaliana] gi|15241578|ref|NP_198706.1| DSBA
oxidoreductase family protein [Arabidopsis thaliana]
Length = 217
Score = 296 bits (759), Expect = 2e-79
Identities = 140/217 (64%), Positives = 176/217 (80%)
Query: 1 MTDTNSAHSEKKLVRIDISSDTVCPWCFVGKKNLDKAIASSKDKYNFEILWHPYQLSPDA 60
M ++ S + KKL++ID+SSD+VCPWCFVGKKNLDKAI +SKD+YNFEI W P+ L P A
Sbjct: 1 MAESASNTASKKLIQIDVSSDSVCPWCFVGKKNLDKAIEASKDQYNFEIRWRPFFLDPSA 60
Query: 61 PKEGIEKREFYRSKFGSRSDQMEARMSEVFRTVGLVYSLSGLTGNTMDSHRLIYFSGQQG 120
PKEG+ K+EFY K+G+R M ARMSEVF+ +GL + +GLTGN++DSHRLI+++G+Q
Sbjct: 61 PKEGVSKKEFYLQKYGNRYQGMFARMSEVFKGLGLEFDTAGLTGNSLDSHRLIHYTGKQA 120
Query: 121 LDKQHDLVEELGLGYFTQEKYIGDQEFLLEAAAKVGIEGAEEFLKNPNNGLKEVEEELKT 180
+KQH LVEEL +GYFTQ K+IGD+EFL+E A KVGIEGAEEFL +PNNG+ EV+EEL
Sbjct: 121 PEKQHTLVEELFIGYFTQGKFIGDREFLVETANKVGIEGAEEFLSDPNNGVTEVKEELAK 180
Query: 181 YSRNITGVPYYVINGSQKLSGGQPPEVFLRAFQAATS 217
YS+NITGVP Y ING KLSG QPPE F AF+AA++
Sbjct: 181 YSKNITGVPNYTINGKVKLSGAQPPETFQSAFKAASA 217
>ref|NP_919053.1| putative polyketide synthase [Oryza sativa (japonica
cultivar-group)] gi|22795241|gb|AAN08213.1| putative
polyketide synthase [Oryza sativa (japonica
cultivar-group)]
Length = 263
Score = 268 bits (686), Expect = 6e-71
Identities = 127/211 (60%), Positives = 166/211 (78%), Gaps = 1/211 (0%)
Query: 6 SAHSEKKLVRIDISSDTVCPWCFVGKKNLDKAIASSKDKYNFEILWHPYQLSPDAPKEGI 65
++++ KKL++ID+SSDTVCPWCFVGKKNL+KA+ +KDK++FE+ WHP+ L+P+APKEGI
Sbjct: 47 ASNAGKKLIQIDVSSDTVCPWCFVGKKNLEKAMEQNKDKFDFEVRWHPFFLNPNAPKEGI 106
Query: 66 EKREFYRSKFGS-RSDQMEARMSEVFRTVGLVYSLSGLTGNTMDSHRLIYFSGQQGLDKQ 124
+K ++YR KFG + + ARM+E+FR +G+ Y +SGLTGNTMDSHRLI +G QG DKQ
Sbjct: 107 KKSDYYRMKFGPIQFEHATARMTEIFRGLGMEYDMSGLTGNTMDSHRLITLAGHQGYDKQ 166
Query: 125 HDLVEELGLGYFTQEKYIGDQEFLLEAAAKVGIEGAEEFLKNPNNGLKEVEEELKTYSRN 184
LVEEL YF K+IGD++ LL+AA KVGIEGAEE L++ N G+ EV+EEL YS
Sbjct: 167 SALVEELFQSYFCHGKFIGDRQVLLDAARKVGIEGAEELLQDSNKGVDEVKEELNKYSSG 226
Query: 185 ITGVPYYVINGSQKLSGGQPPEVFLRAFQAA 215
I+GVP++VING +LSGGQPP F RAF A
Sbjct: 227 ISGVPHFVINGKFQLSGGQPPNAFTRAFDVA 257
>gb|EAL88433.1| DSBA-like thioredoxin domain protein [Aspergillus fumigatus Af293]
Length = 238
Score = 137 bits (344), Expect = 3e-31
Identities = 77/229 (33%), Positives = 125/229 (53%), Gaps = 29/229 (12%)
Query: 14 VRIDISSDTVCPWCFVGKKNLDKAIASSKDKY---NFEILWHPYQLSPDAP-KEGIEKRE 69
+ I I SDTVCPWC+VG + L +AI + K Y F + WH + L+P +P G++KRE
Sbjct: 4 INIQIISDTVCPWCYVGYRRLSRAITAHKAIYPSDTFTLTWHAFYLNPASPGYPGVDKRE 63
Query: 70 FYRSKFG-SRSDQMEARMSEVFRTVGLVYSLSGLTGNTMDSHRLIYFSGQQGLDK----- 123
+Y SKFG R+ + +R++ G+ + G TGNT DSHR+++++G + +
Sbjct: 64 YYASKFGEGRAAAIFSRLAAAEEPDGIAFKFGGRTGNTRDSHRVLWYAGLKEKEAGARGG 123
Query: 124 ----------------QHDLVEELGLGYFTQEKYIGDQEFLLEAAAKVGIE--GAEEFLK 165
Q + E+ YF +EK I D++ L+++AA G++ E+FL+
Sbjct: 124 AAATNGDASEEKVGGLQTRVAEQFFRAYFEEEKNITDRKMLVDSAAAAGLDRGEVEKFLE 183
Query: 166 NPNNGLKEVE-EELKTYSRNITGVPYYVINGSQKLSGGQPPEVFLRAFQ 213
+ + G K+V+ E + R +TGVPY+ + G + G PE FL F+
Sbjct: 184 SGDEGGKDVDLEAERARHRLVTGVPYFTVQGQYAIEGADEPETFLEVFE 232
>gb|AAV95341.1| DSBA-like thioredoxin family protein [Silicibacter pomeroyi DSS-3]
gi|56696938|ref|YP_167300.1| DSBA-like thioredoxin
family protein [Silicibacter pomeroyi DSS-3]
Length = 218
Score = 132 bits (333), Expect = 5e-30
Identities = 71/201 (35%), Positives = 117/201 (57%), Gaps = 9/201 (4%)
Query: 14 VRIDISSDTVCPWCFVGKKNLDKAIASSKDKYNFEILWHPYQLSPDAPKEGIEKREFYRS 73
V++DI SD +CPWC++GK LDKA+A+ D + F I WHP+QL+PD EG+++RE+
Sbjct: 5 VKLDILSDPICPWCYIGKTQLDKALAAIPD-HPFVIEWHPFQLNPDMAPEGMDRREYLER 63
Query: 74 KFGSRSDQME--ARMSEVFRTVGLVYSLSGL--TGNTMDSHRLIYFSGQQGLDKQHDLVE 129
KFG + + A + E GL + T NT+D+HRLI+++G +G KQ + V+
Sbjct: 64 KFGGKEGAVRAYAPVVEHAEKAGLAIDFEAMQRTPNTLDAHRLIHWAGVEG--KQTEAVD 121
Query: 130 ELGLGYFTQEKYIGDQEFLLEAAAKVGIEGAE--EFLKNPNNGLKEVEEELKTYSRNITG 187
L YFT+ + IGD E L + A +G++ A + LK+ + + + + ++
Sbjct: 122 ALFAAYFTEGRDIGDAEVLADIADSIGMDAAVVLKLLKSDADRDDIAKRDSHSREMGVSS 181
Query: 188 VPYYVINGSQKLSGGQPPEVF 208
VP +++ + G QPPE++
Sbjct: 182 VPTFIVANQHAVPGAQPPELW 202
>ref|ZP_00193074.2| COG2761: Predicted dithiol-disulfide isomerase involved in
polyketide biosynthesis [Mesorhizobium sp. BNC1]
Length = 227
Score = 129 bits (325), Expect = 4e-29
Identities = 74/203 (36%), Positives = 115/203 (56%), Gaps = 7/203 (3%)
Query: 9 SEKKLVRIDISSDTVCPWCFVGKKNLDKAIASSKDKYNFEILWHPYQLSPDAPKEGIEKR 68
S+K + +++ SD VCPWCF+G +NL A+A +D + + W PYQL P P EG++++
Sbjct: 2 SDKPALSVNVVSDVVCPWCFLGMRNLQLALADLRD-IDVMVRWRPYQLDPTIPSEGLDRK 60
Query: 69 EFYRSKFGS--RSDQMEARMSEVFRTVGLVYSLSGL--TGNTMDSHRLIYFSGQQGLDKQ 124
+ KFGS R + R+ E+ G+ Y + + NT+D+HR+I ++ G D Q
Sbjct: 61 AYMVQKFGSLERIEAAHDRLREMGTAAGISYHFAAIKVAPNTLDAHRVIRWAATAGEDVQ 120
Query: 125 HDLVEELGLGYFTQEKYIGDQEFLLEAAAKVGIEGA-EEFLKNPNNGLKEVEEELKTYSR 183
+ LVE L YF + + IGD L+EAA + ++ + E L EV+ E++T R
Sbjct: 121 NRLVERLFRLYFEEGQNIGDHAVLIEAARQADMDASLVETLLPTQADRDEVKAEVETAQR 180
Query: 184 -NITGVPYYVINGSQKLSGGQPP 205
ITGVP +++ G L G QPP
Sbjct: 181 MGITGVPCFLLEGRYALMGAQPP 203
>ref|ZP_00339472.1| COG2761: Predicted dithiol-disulfide isomerase involved in
polyketide biosynthesis [Silicibacter sp. TM1040]
Length = 233
Score = 127 bits (319), Expect = 2e-28
Identities = 68/201 (33%), Positives = 119/201 (58%), Gaps = 9/201 (4%)
Query: 14 VRIDISSDTVCPWCFVGKKNLDKAIASSKDKYNFEILWHPYQLSPDAPKEGIEKREFYRS 73
V++DI SD +CPWC++GK +LD+A+A + D + F I WHP+QL+PD P +G+++R + +
Sbjct: 8 VKLDILSDPICPWCYIGKTHLDRALAEAPD-HPFVIEWHPFQLNPDMPGDGMDRRTYLET 66
Query: 74 KFGSRSDQME--ARMSEVFRTVGLVYSLSGL--TGNTMDSHRLIYFSGQQGLDKQHDLVE 129
KFG + + ++ A+++E + G+ + + T NT+++HRLI+++G + KQ D+V+
Sbjct: 67 KFGGKDNAVKVYAQIAEHAQAAGVEINFEAMQRTPNTINAHRLIHWAGLEA--KQSDVVD 124
Query: 130 ELGLGYFTQEKYIGDQEFLLEAAAKVGIEGA--EEFLKNPNNGLKEVEEELKTYSRNITG 187
L YF + K IGD L + A + G+E A E L ++ + + +
Sbjct: 125 ALFKAYFVEGKDIGDPVVLADLAEQAGMERAVVERLLSGDSDAEDIRARDAHSRKMGVNS 184
Query: 188 VPYYVINGSQKLSGGQPPEVF 208
VP +VI + G Q PE++
Sbjct: 185 VPTFVIANQHVVPGAQQPELW 205
>gb|EAM67706.1| DSBA oxidoreductase [Jannaschia sp. CCS1]
Length = 221
Score = 126 bits (316), Expect = 5e-28
Identities = 70/212 (33%), Positives = 120/212 (56%), Gaps = 11/212 (5%)
Query: 13 LVRIDISSDTVCPWCFVGKKNLDKAIASSKDKYNFEILWHPYQLSPDAPKEGIEKREFYR 72
++++DI SD +CPWC++G NL +A+ ++ + F I WHP+QL+PD P++G+++R +
Sbjct: 1 MIKLDILSDPICPWCYIGWSNLARAM-EARPNHPFTIEWHPFQLNPDMPEDGMDRRAYLE 59
Query: 73 SKFGSRSDQME--ARMSEVFRTVGLVYSLSGL--TGNTMDSHRLIYFSGQQGLDKQHDLV 128
KFG + + A ++E GL +L G+ T NT D+HRLI+++G +G +Q LV
Sbjct: 60 GKFGGKDAAVRAYAPVAEKAAEAGLTLNLEGIKRTPNTRDAHRLIHWAGIEG--RQTPLV 117
Query: 129 EELGLGYFTQEKYIGDQEFLLEAAAKVGIEGAEEFLKNPNNGLKEVE---EELKTYSRNI 185
L YF + + IGD++ LLE A VG++ A K ++G + + R +
Sbjct: 118 LALFRAYFVEGRDIGDRDTLLEIAESVGLDRA-MIAKLYDSGADAEDIKARDAHARERGV 176
Query: 186 TGVPYYVINGSQKLSGGQPPEVFLRAFQAATS 217
VP +++ + G QPPE ++ T+
Sbjct: 177 QAVPTFIVANQHAVPGAQPPEQWMAIIDDLTT 208
>ref|NP_102302.1| similar to frnE protein [Mesorhizobium loti MAFF303099]
gi|14021476|dbj|BAB48088.1| mlr0515 [Mesorhizobium loti
MAFF303099]
Length = 226
Score = 125 bits (315), Expect = 6e-28
Identities = 74/211 (35%), Positives = 118/211 (55%), Gaps = 7/211 (3%)
Query: 9 SEKKLVRIDISSDTVCPWCFVGKKNLDKAIASSKDKYNFEILWHPYQLSPDAPKEGIEKR 68
SE V +D+ SD VCPWCF+G+K LDKA+A++ D + I W P+QL P P +G ++R
Sbjct: 2 SEANAVTVDVVSDVVCPWCFIGQKRLDKAVAAAGD-VDVHIRWRPFQLDPTIPPQGKDRR 60
Query: 69 EFYRSKFGS--RSDQMEARMSEVFRTVGLVYSLSGL--TGNTMDSHRLIYFSGQQGLDKQ 124
E+ +KFGS R ++ AR+ + G+ ++ + NT+D+HRLI ++G G Q
Sbjct: 61 EYMLAKFGSDERIREIHARIEPLGEAEGISFAFDAIKVAPNTLDAHRLIRWAGAAGEAVQ 120
Query: 125 HDLVEELGLGYFTQEKYIGDQEFLLEAAAKVGIEGAEEFLKNPNNG-LKEVEEELKTYSR 183
+ LV L F + IGD L+EAA + G++ + P + ++ V E+ T SR
Sbjct: 121 NRLVRRLFQLNFEEGVNIGDHAVLVEAAREAGMDASVVATLLPTDADVEAVRTEIATASR 180
Query: 184 -NITGVPYYVINGSQKLSGGQPPEVFLRAFQ 213
I+GVP +++ G + G Q + A +
Sbjct: 181 MGISGVPCFLLEGKYAVMGAQDVDTLADAIR 211
>ref|ZP_00208412.1| COG2761: Predicted dithiol-disulfide isomerase involved in
polyketide biosynthesis [Magnetospirillum
magnetotacticum MS-1]
Length = 215
Score = 125 bits (315), Expect = 6e-28
Identities = 67/203 (33%), Positives = 117/203 (57%), Gaps = 9/203 (4%)
Query: 14 VRIDISSDTVCPWCFVGKKNLDKAIASSKDKYNFEILWHPYQLSPDAPKEGIEKREFYRS 73
+RI+ DTVCPWC+VGK+ L++A+A + + I+W P+ L+PD P EGI++R +
Sbjct: 1 MRIEYVFDTVCPWCYVGKRRLERALA-QRPETRARIIWRPFLLNPDLPAEGIDRRTYLDR 59
Query: 74 KFG--SRSDQMEARMSEVFRTVGLVYSLSGLT--GNTMDSHRLIYFSGQQGLDKQHDLVE 129
KFG +R ++ A ++ ++ G+ + +T N+++SHR+I ++G G + +LVE
Sbjct: 60 KFGGTARVQRVHAAVAAAGKSEGIDFDFDSITRMPNSLNSHRMIRYAGASGCEA--ELVE 117
Query: 130 ELGLGYFTQEKYIGDQEFLLEAAAKVGI--EGAEEFLKNPNNGLKEVEEELKTYSRNITG 187
L YF Q IGD E L A VG+ + +L + + + + + + + + G
Sbjct: 118 SLYRAYFVQGLDIGDVEVLTAIGASVGLAPDPLRTYLSSDADAVGVLNDNARAHRLGVNG 177
Query: 188 VPYYVINGSQKLSGGQPPEVFLR 210
VP +++GS L+G Q P++ LR
Sbjct: 178 VPCLILDGSYALAGAQEPDILLR 200
>ref|NP_532458.1| polyketide biosynthesis associated protein [Agrobacterium
tumefaciens str. C58] gi|17740217|gb|AAL42774.1|
polyketide biosynthesis associated protein
[Agrobacterium tumefaciens str. C58]
gi|15156878|gb|AAK87544.1| AGR_C_3262p [Agrobacterium
tumefaciens str. C58] gi|15889078|ref|NP_354759.1|
hypothetical protein AGR_C_3262 [Agrobacterium
tumefaciens str. C58] gi|25367092|pir||AH2794 polyketide
biosynthesis associated protein Atu1775 [imported] -
Agrobacterium tumefaciens (strain C58, Dupont)
gi|25367090|pir||G97573 frne protein VCA0178 [imported]
- Agrobacterium tumefaciens (strain C58, Cereon)
Length = 242
Score = 123 bits (309), Expect = 3e-27
Identities = 64/203 (31%), Positives = 110/203 (53%), Gaps = 5/203 (2%)
Query: 16 IDISSDTVCPWCFVGKKNLDKAIASSKDKYNFEILWHPYQLSPDAPKEGIEKREFYRSKF 75
+D+ SD VCPWC++GK LD AIA +D+ + ++ W PY+L+PD P EG++++ K
Sbjct: 26 VDVVSDMVCPWCYLGKARLDLAIAEVQDEISVDVNWRPYRLNPDYPPEGVDQKAALEEKL 85
Query: 76 G-SRSDQMEARMSEVFRTVGLVYSLSGLT--GNTMDSHRLIYFSGQQGLDKQHDLVEELG 132
G R +Q A + E+ + VG+ Y + NT+D+HRL ++ +G D Q +V L
Sbjct: 86 GKDRLEQAHASLVELGKQVGINYDFDAIKIGPNTLDAHRLSLWAHAEGRDVQERIVTALF 145
Query: 133 LGYFTQEKYIGDQEFLLEAAAKVGIEG--AEEFLKNPNNGLKEVEEELKTYSRNITGVPY 190
F + + IGD L + A K G++ L + + + E ++GVP+
Sbjct: 146 KANFEEGRNIGDHAVLTDIAGKAGMDAKVVARLLASDADKDTVIAEIDAAQQMGVSGVPF 205
Query: 191 YVINGSQKLSGGQPPEVFLRAFQ 213
++++ +SG Q P+V + A +
Sbjct: 206 FIVDQKYAISGAQTPDVLIAALR 228
>ref|ZP_00007377.1| COG2761: Predicted dithiol-disulfide isomerase involved in
polyketide biosynthesis [Rhodobacter sphaeroides 2.4.1]
Length = 214
Score = 122 bits (306), Expect = 7e-27
Identities = 68/202 (33%), Positives = 115/202 (56%), Gaps = 9/202 (4%)
Query: 13 LVRIDISSDTVCPWCFVGKKNLDKAIASSKDKYNFEILWHPYQLSPDAPKEGIEKREFYR 72
++R+DI SD VCPWC++GK +LD+A+ + D + F I WHP+QL+PD P EG+++R +
Sbjct: 1 MIRLDIFSDPVCPWCYIGKAHLDRALETRPD-HPFLIEWHPFQLNPDMPPEGMDRRSYLE 59
Query: 73 SKFGSRSDQME--ARMSEVFRTVGLVYSLSGL--TGNTMDSHRLIYFSGQQGLDKQHDLV 128
+KFG R +E A++ GL S + T NT+D+HRLI+++G +G +Q +V
Sbjct: 60 TKFGGRQAAVEVYAQVQAAAEAAGLEIDFSAIERTPNTLDAHRLIHWAGLEG--RQAAVV 117
Query: 129 EELGLGYFTQEKYIGDQEFLLEAAAKVGIEGAEEFLKNPNNGLKE--VEEELKTYSRNIT 186
L GYF + IG E L + A + G++ A ++ +E + ++ +
Sbjct: 118 SALFRGYFREGLDIGVPEVLADIAGRCGMDRALTLRLLSSDADREDLAARDADARAKGVR 177
Query: 187 GVPYYVINGSQKLSGGQPPEVF 208
VP +++ + G QP E++
Sbjct: 178 AVPTFLVARRHVVPGAQPVELW 199
>gb|EAA62324.1| hypothetical protein AN5143.2 [Aspergillus nidulans FGSC A4]
gi|67537946|ref|XP_662747.1| hypothetical protein
AN5143_2 [Aspergillus nidulans FGSC A4]
gi|49095638|ref|XP_409280.1| hypothetical protein
AN5143.2 [Aspergillus nidulans FGSC A4]
Length = 971
Score = 122 bits (306), Expect = 7e-27
Identities = 76/223 (34%), Positives = 119/223 (53%), Gaps = 26/223 (11%)
Query: 16 IDISSDTVCPWCFVGKKNLDKAIASSK---DKYNFEILWHPYQLSPDAP-KEGIEKREFY 71
I I SD+VCPWC+VG + L +AI + K F I W P+ L+ +P G+ KR+FY
Sbjct: 741 IQIISDSVCPWCYVGYRRLSRAITTHKLTNPLDTFTITWSPFYLNASSPGYPGVNKRQFY 800
Query: 72 RSKFG-SRSDQMEARMSEVFRTVGLVYSLSGLTGNTMDSHRLIYFSGQQGLDK------- 123
+KFG +R+ + R++ V G+ +S G TG T DSHR+I+ +G++ ++
Sbjct: 801 ENKFGAARTGAIFERLAAVGEGEGIKFSFGGQTGKTRDSHRVIWLAGKKEREQREKGEGV 860
Query: 124 ------------QHDLVEELGLGYFTQEKYIGDQEFLLEAAAKVGIEGAE-EFLKNPNNG 170
Q +VE L YF +EK I ++ L+EAA G++ +E E + + G
Sbjct: 861 AGKIENGVIGGLQTRVVERLFRAYFEEEKNITERAVLVEAAVGGGLDKSEVEGFLDSDVG 920
Query: 171 LKEVEEELKTYSRN-ITGVPYYVINGSQKLSGGQPPEVFLRAF 212
EV+ + + R +TGVPY+++ G + G PE FL F
Sbjct: 921 GVEVDRDAEGARRQFVTGVPYFMVQGQYAIEGADEPETFLEVF 963
>ref|YP_132010.1| hypothetical 2-hydroxychromene-2-carboxylateisomerase family
protein [Photobacterium profundum SS9]
gi|46915438|emb|CAG22210.1| hypothetical
2-hydroxychromene-2-carboxylateisomerase family protein
[Photobacterium profundum SS9]
Length = 218
Score = 120 bits (301), Expect = 3e-26
Identities = 68/212 (32%), Positives = 120/212 (56%), Gaps = 9/212 (4%)
Query: 9 SEKKLVRIDISSDTVCPWCFVGKKNLDKAIASSKDKYNFEILWHPYQLSPDAPKEGIEKR 68
S+ + +RIDI SD VCPWC +G + L++A+ + + K N E+ W P+Q++PD P EG
Sbjct: 2 SKLRNIRIDIVSDVVCPWCIIGYRRLEEALGAFEGKINVELYWQPFQINPDMPSEGENLG 61
Query: 69 EFYRSKFGSRSDQMEA---RMSEVFRTVGLVYSLS--GLTGNTMDSHRLIYFSGQQGLDK 123
E K+G S+Q +A + ++ ++ ++ + NT +H+L++++ +QG +
Sbjct: 62 EHLTKKYGLTSEQSQANRENLIQIGESLDFTFNFTPEXKIYNTFKAHQLLHWAAKQG--R 119
Query: 124 QHDLVEELGLGYFTQEKYIGDQEFLLEAAAKVGIEGAE-EFLKNPNNGLKEVEEELKTYS 182
QH L L YFT++K D E L+ AA +VG++G E + +V+ +T++
Sbjct: 120 QHALKLALFDAYFTEQKDPSDIELLVTAAMQVGLDGEEARAVLTDERFADDVKMNQQTWT 179
Query: 183 RN-ITGVPYYVINGSQKLSGGQPPEVFLRAFQ 213
+ I VP V++ +SG Q PE F+++ Q
Sbjct: 180 NSGIQSVPSIVLDQKYLISGAQDPETFIQSIQ 211
>ref|NP_541668.1| FRNE [Brucella melitensis 16M] gi|23463934|gb|AAN33766.1| frnE
protein, putative [Brucella suis 1330]
gi|17984876|gb|AAL53932.1| FRNE [Brucella melitensis
16M] gi|25367097|pir||AI3595 frnE protein [imported] -
Brucella melitensis (strain 16M)
gi|23500321|ref|NP_699761.1| frnE protein, putative
[Brucella suis 1330]
Length = 224
Score = 120 bits (301), Expect = 3e-26
Identities = 70/208 (33%), Positives = 114/208 (54%), Gaps = 7/208 (3%)
Query: 12 KLVRIDISSDTVCPWCFVGKKNLDKAIASSKDKYNFEILWHPYQLSPDAPKEGIEKREFY 71
K + ID+ SD VCPWCF+G+K L+ A+AS D EI W P+QL P P G +++ +
Sbjct: 4 KKITIDVVSDVVCPWCFLGRKRLEAALASLPD-VEAEIRWRPFQLDPTLPPHGKDRQTYL 62
Query: 72 RSKF--GSRSDQMEARMSEVFRTVGLVYSLSGL--TGNTMDSHRLIYFSGQQGLDKQHDL 127
R KF GS+ D ++ + G+V+ + NT+D+HR+I+++ Q D Q +
Sbjct: 63 REKFGTGSKIDDSHRQLRALGEEYGIVFDFDAIIRAPNTLDAHRVIHWAAQAAPDTQDRM 122
Query: 128 VEELGLGYFTQEKYIGDQEFLLEAAAKVGIEGAEEFLKNPNNGLKE-VEEELKTYSR-NI 185
V L YF Q + IG+ E L++AAA V ++ + K+ + +E+ T +R +
Sbjct: 123 VGMLFSLYFEQGQDIGNHEVLVDAAASVSMDAEVVARLLQSEADKDTIRDEIATANRIGV 182
Query: 186 TGVPYYVINGSQKLSGGQPPEVFLRAFQ 213
GVP ++I+ + G Q P+V A +
Sbjct: 183 RGVPCFIIDQKYAVMGAQTPDVLADAIR 210
>ref|XP_330996.1| hypothetical protein [Neurospora crassa] gi|28921079|gb|EAA30397.1|
hypothetical protein [Neurospora crassa]
Length = 233
Score = 120 bits (300), Expect = 3e-26
Identities = 71/217 (32%), Positives = 113/217 (51%), Gaps = 14/217 (6%)
Query: 15 RIDISSDTVCPWCFVGKKNLDKAIASSKDKY------NFEILWHPYQLSPDAPKEG-IEK 67
+I + SD +CPWC+VGK L++AI KD F + WHP+ L P PK G I+
Sbjct: 5 QIKVVSDIICPWCYVGKARLERAIKLYKDVIPGGANDTFTVTWHPFYLDPSLPKTGGIDP 64
Query: 68 REFYRSKFGS--RSDQMEARMSEVFRTVGLVYSLSGLTGNTMDSHRLIYFSGQQGLDKQH 125
+ + K GS R + AR+ + G+ +SL+G GNT ++HRLI + + + ++
Sbjct: 65 KAYLGKKLGSPERLAMVHARLKAIGEGEGIKFSLNGRIGNTRNAHRLIQLAKTKSNEIEN 124
Query: 126 DLVEELGLGYFTQEKYIGDQEFLLEAAAKVGIEGA--EEFLKNPNNGLKEVEEELKTYSR 183
L + ++ + + L+ A + G++GA E +L + G E + +
Sbjct: 125 KTAAALFQLHHEEDGDVSSNDMLIAAGKRAGLDGAEVESWLASDRGGEDVDREVAEAQRK 184
Query: 184 NITGVPYYVINGSQKLSGGQPPEVFLRAF---QAATS 217
I GVP + ING +LSG Q PE F++ F +AATS
Sbjct: 185 GIHGVPNFTINGQSELSGAQDPETFVQEFLRVKAATS 221
>ref|YP_236982.1| DSBA oxidoreductase [Pseudomonas syringae pv. syringae B728a]
gi|63257848|gb|AAY38944.1| DSBA oxidoreductase
[Pseudomonas syringae pv. syringae B728a]
Length = 215
Score = 119 bits (297), Expect = 8e-26
Identities = 74/207 (35%), Positives = 111/207 (52%), Gaps = 10/207 (4%)
Query: 14 VRIDISSDTVCPWCFVGKKNLDKAIASSKDKYNFEILWHPYQLSPDAPKEGIEKREFYRS 73
++ID SD CPWC +G + LD+A+ + D+ I + P++L+P+ P EG + +E
Sbjct: 5 LKIDFISDVSCPWCVIGLRALDQALEALGDEVQAHIHFQPFELNPNMPAEGQDIKEHIAE 64
Query: 74 KFGSRSDQMEARMSEVFRTVGLVYSLSGLTG-----NTMDSHRLIYFSGQQGLDKQHDLV 128
K+GS +Q+E + E R G + +G NT D+HRL++++ QQG KQH L
Sbjct: 65 KYGSTPEQIEG-IHETIRERGAELGFTFASGERRIYNTFDAHRLLHWAEQQG--KQHALK 121
Query: 129 EELGLGYFTQEKYIGDQEFLLEAAAKVGIE--GAEEFLKNPNNGLKEVEEELKTYSRNIT 186
+ L YF+ K D + L A KVG++ A+ L + + E E SR IT
Sbjct: 122 QALFEAYFSDLKDPSDHQTLAAVAQKVGLDRLRAQAILGSDEYTAEVREAEQLWTSRGIT 181
Query: 187 GVPYYVINGSQKLSGGQPPEVFLRAFQ 213
VP V N +SGGQP EVF+ A +
Sbjct: 182 SVPTMVFNDQYAVSGGQPVEVFVSAIR 208
>ref|YP_223419.1| hypothetical FrnE [Brucella abortus biovar 1 str. 9-941]
gi|62197759|gb|AAX76058.1| hypothetical FrnE [Brucella
abortus biovar 1 str. 9-941]
Length = 224
Score = 117 bits (294), Expect = 2e-25
Identities = 69/208 (33%), Positives = 113/208 (54%), Gaps = 7/208 (3%)
Query: 12 KLVRIDISSDTVCPWCFVGKKNLDKAIASSKDKYNFEILWHPYQLSPDAPKEGIEKREFY 71
K + ID+ SD VCPWCF+G+K L+ A+AS D EI W P+QL P P G +++ +
Sbjct: 4 KKITIDVVSDVVCPWCFLGRKRLEAALASLPD-VEAEIRWRPFQLDPTLPPHGKDRQTYL 62
Query: 72 RSKF--GSRSDQMEARMSEVFRTVGLVYSLSGL--TGNTMDSHRLIYFSGQQGLDKQHDL 127
R KF GS+ D ++ + G+V+ + NT+D+HR+I+++ Q D Q +
Sbjct: 63 REKFGTGSKIDDSHRQLRALGEEYGIVFDFDAIIRAPNTLDAHRVIHWAAQAAPDTQDRM 122
Query: 128 VEELGLGYFTQEKYIGDQEFLLEAAAKVGIEGAEEFLKNPNNGLKE-VEEELKTYSR-NI 185
V L YF Q + IG+ E L++AAA V ++ + K+ + +E+ T + +
Sbjct: 123 VGMLFSLYFEQGQDIGNHEVLVDAAASVSMDAEVVARLLQSEADKDTIRDEIATANPIGV 182
Query: 186 TGVPYYVINGSQKLSGGQPPEVFLRAFQ 213
GVP ++I+ + G Q P+V A +
Sbjct: 183 RGVPCFIIDQKYAVMGAQTPDVLADAIR 210
>ref|NP_793938.1| 2-hydroxychromene-2-carboxylate isomerase family protein
[Pseudomonas syringae pv. tomato str. DC3000]
gi|28854570|gb|AAO57633.1|
2-hydroxychromene-2-carboxylate isomerase family protein
[Pseudomonas syringae pv. tomato str. DC3000]
Length = 215
Score = 116 bits (290), Expect = 5e-25
Identities = 73/211 (34%), Positives = 114/211 (53%), Gaps = 10/211 (4%)
Query: 14 VRIDISSDTVCPWCFVGKKNLDKAIASSKDKYNFEILWHPYQLSPDAPKEGIEKREFYRS 73
++ID SD CPWC +G + LD+A+ + D+ +I + P++L+P+ P EG + +E
Sbjct: 5 LKIDFISDVSCPWCVIGLRALDQALEALGDEVQAQIHFQPFELNPNMPAEGQDIKEHIAE 64
Query: 74 KFGSRSDQMEARMSEVFRTVGLVYSLSGLTG-----NTMDSHRLIYFSGQQGLDKQHDLV 128
K+GS +Q+EA + E R G + G NT D+HRL++++ Q+G KQ L
Sbjct: 65 KYGSTPEQIEA-IHETIRERGAELGFTFGKGERRIYNTFDAHRLLHWAEQEG--KQPALK 121
Query: 129 EELGLGYFTQEKYIGDQEFLLEAAAKVGIE--GAEEFLKNPNNGLKEVEEELKTYSRNIT 186
+ L + YF++ K + L + A KVG++ A+ L + + E E SR IT
Sbjct: 122 QALFVAYFSELKDPSSHQTLADVAQKVGLDRLRAQAILDSDEFASEVREAEQLWTSRGIT 181
Query: 187 GVPYYVINGSQKLSGGQPPEVFLRAFQAATS 217
VP V N +SGGQP EVF+ A + S
Sbjct: 182 SVPTMVFNDQYAVSGGQPVEVFVSAIRQMLS 212
>ref|ZP_00145910.2| COG2761: Predicted dithiol-disulfide isomerase involved in
polyketide biosynthesis [Psychrobacter sp. 273-4]
Length = 217
Score = 115 bits (288), Expect = 9e-25
Identities = 71/204 (34%), Positives = 110/204 (53%), Gaps = 11/204 (5%)
Query: 14 VRIDISSDTVCPWCFVGKKNLDKAIASSKDKYNFEILWHPYQLSPDAPKEGIEKREFYRS 73
++IDI SD VCPWC +G + L +A+ + + EI WHP++L+P+ P EG RE
Sbjct: 8 LQIDIVSDVVCPWCIIGYRQLAEALKDTNTAH--EIHWHPFELNPNMPAEGQNLREHIIE 65
Query: 74 KFGS---RSDQMEARMSEVFRTVGLVYSLSGLT--GNTMDSHRLIYFSGQQGLDKQHDLV 128
K+GS S++ RM+ VG ++ + T NT + H+L++++ QQG HDL
Sbjct: 66 KYGSSVQESNESRTRMAAAGADVGFEFNFTDDTRMHNTFNLHQLLHWADQQG--HMHDLK 123
Query: 129 EELGLGYFTQEKYIGDQEFLLEAAAKVGIEGAEEF-LKNPNNGLKEVEE-ELKTYSRNIT 186
+ L +FT + I D L + AA++G++ AE + K+V E E + + I
Sbjct: 124 QALFTAHFTNNRNISDNAVLADIAAEIGLDRAEALAVVEDQRFAKDVREAEQHSAQQGIQ 183
Query: 187 GVPYYVINGSQKLSGGQPPEVFLR 210
GVP + NG +SG Q E F R
Sbjct: 184 GVPAVIFNGRHLVSGAQGIENFTR 207
>emb|CAC46223.1| CONSERVED HYPOTHETICAL PROTEIN [Sinorhizobium meliloti]
gi|15965397|ref|NP_385750.1| hypothetical protein
SMc00952 [Sinorhizobium meliloti 1021]
Length = 221
Score = 115 bits (287), Expect = 1e-24
Identities = 72/211 (34%), Positives = 108/211 (51%), Gaps = 16/211 (7%)
Query: 14 VRIDISSDTVCPWCFVGKKNLDKAIASSKDKYNFEILWHPYQLSPDAPKEGIEKREFYRS 73
V IDI SD VCPWC++GK LD+AIA+ + N I W PYQL+PD P EGI+ +E +
Sbjct: 4 VNIDIVSDVVCPWCYLGKNRLDQAIANVAGEINVAIQWRPYQLNPDLPPEGIDHKEHLAA 63
Query: 74 KFGSRSDQMEARMSEVFRTV-------GLVYSLSG--LTGNTMDSHRLIYFSGQQGLDKQ 124
K G +A + E RT+ G+ + ++ NT+D+HRLI ++ G Q
Sbjct: 64 KLGG-----QAAVDEAHRTLEGLGVEDGIAFDFDAVKVSPNTLDAHRLIRWAATGGEAAQ 118
Query: 125 HDLVEELGLGYFTQEKYIGDQEFLLEAAAKVGIEGAEEFLKNPNNGLKEVEEELKTYSRN 184
+V L F + + +GDQ LL+ A + G+E ++ K+ + +R
Sbjct: 119 DAVVSLLFKANFEEGRNLGDQAVLLDIAEQAGLERPVIAALLASDADKDAVRQEIDMARE 178
Query: 185 I--TGVPYYVINGSQKLSGGQPPEVFLRAFQ 213
I TGVP ++I + G Q EV A +
Sbjct: 179 IGVTGVPCFIIEQQYAVMGAQSVEVLTSALR 209
Database: nr
Posted date: Jul 5, 2005 12:34 AM
Number of letters in database: 863,360,394
Number of sequences in database: 2,540,612
Lambda K H
0.314 0.134 0.384
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 368,695,460
Number of Sequences: 2540612
Number of extensions: 15138936
Number of successful extensions: 29711
Number of sequences better than 10.0: 154
Number of HSP's better than 10.0 without gapping: 107
Number of HSP's successfully gapped in prelim test: 47
Number of HSP's that attempted gapping in prelim test: 29339
Number of HSP's gapped (non-prelim): 159
length of query: 217
length of database: 863,360,394
effective HSP length: 123
effective length of query: 94
effective length of database: 550,865,118
effective search space: 51781321092
effective search space used: 51781321092
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (22.0 bits)
S2: 72 (32.3 bits)
Medicago: description of AC137823.6


