

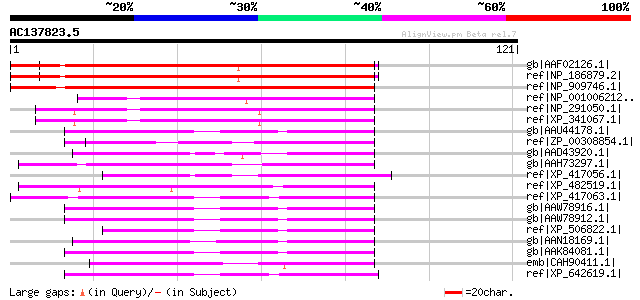
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC137823.5 - phase: 0
(121 letters)
Database: nr
2,540,612 sequences; 863,360,394 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
gb|AAF02126.1| unknown protein [Arabidopsis thaliana] 141 4e-33
ref|NP_186879.2| regulator of chromosome condensation (RCC1) fam... 141 4e-33
ref|NP_909746.1| putative chromosome condensation regulator [Ory... 115 2e-25
ref|NP_001006212.1| similar to Williams-Beuren syndrome chromoso... 49 4e-05
ref|NP_291050.1| Williams-Beuren syndrome chromosome region 16 h... 46 2e-04
ref|XP_341067.1| PREDICTED: similar to Williams-Beuren syndrome ... 46 2e-04
gb|AAU44178.1| ptative chromosome condensation factor [Oryza sat... 46 2e-04
ref|ZP_00308854.1| COG5184: Alpha-tubulin suppressor and related... 46 2e-04
gb|AAD43920.1| UVB-resistance protein UVR8 [Arabidopsis thaliana... 45 3e-04
gb|AAH73297.1| MGC80684 protein [Xenopus laevis] 45 3e-04
ref|XP_417056.1| PREDICTED: similar to Chromosome condensation 1... 45 3e-04
ref|XP_482519.1| putative UVB-resistance protein UVR8 [Oryza sat... 45 4e-04
ref|XP_417063.1| PREDICTED: similar to hypothetical protein [Gal... 45 4e-04
gb|AAW78916.1| putative chromosome condensation factor [Triticum... 45 5e-04
gb|AAW78912.1| putative chromosome condensation factor [Triticum... 45 5e-04
ref|XP_506822.1| PREDICTED P0470G10.15 gene product [Oryza sativ... 45 5e-04
gb|AAN18169.1| At5g63860/MGI19_6 [Arabidopsis thaliana] gi|21928... 45 5e-04
gb|AAK84081.1| putative chromosome condensation factor [Triticum... 45 5e-04
emb|CAH90411.1| hypothetical protein [Pongo pygmaeus] 44 7e-04
ref|XP_642619.1| hypothetical protein DDB0169222 [Dictyostelium ... 44 7e-04
>gb|AAF02126.1| unknown protein [Arabidopsis thaliana]
Length = 456
Score = 141 bits (355), Expect = 4e-33
Identities = 65/87 (74%), Positives = 73/87 (83%), Gaps = 1/87 (1%)
Query: 1 MDIDAILGTTKPVAASIPRKSAIYVWGYNQSGQTGRKEKDDQLRIPKQLPPQLFGCPAGL 60
MDI I+G P + SIP KSAIYVWGYNQSGQTGR E++ LRIPKQLPP+LFGCPAG
Sbjct: 1 MDIGEIIGEVAP-SVSIPTKSAIYVWGYNQSGQTGRNEQEKLLRIPKQLPPELFGCPAGA 59
Query: 61 NTRWLDIACGREHTAAIASDGLLFTWG 87
N+RWLDI+CGREHTAA+ASDG LF WG
Sbjct: 60 NSRWLDISCGREHTAAVASDGSLFAWG 86
Score = 48.5 bits (114), Expect = 4e-05
Identities = 28/82 (34%), Positives = 43/82 (52%), Gaps = 3/82 (3%)
Query: 8 GTTKPVAASIPRKSAIYVWGYNQSGQTGRKEKDDQLRIPKQLPPQL-FGCPAGLNTRWLD 66
GT VA S + + WGYN+ GQ GR + L+ P+ + F A + +
Sbjct: 174 GTAHVVALS--EEGLLQAWGYNEQGQLGRGVTCEGLQAPRVINAYAKFLDEAPELVKIMQ 231
Query: 67 IACGREHTAAIASDGLLFTWGL 88
++CG HTAA++ G ++TWGL
Sbjct: 232 LSCGEYHTAALSDAGEVYTWGL 253
Score = 39.7 bits (91), Expect = 0.017
Identities = 22/76 (28%), Positives = 37/76 (47%), Gaps = 11/76 (14%)
Query: 15 ASIPRKSAIYVWGYNQSGQTGRK--EKDDQLRIPKQLPPQLFGCPAGLN-TRWLDIACGR 71
A++ +Y WG GQ G + D+ IP+++ GL+ ++ACG
Sbjct: 240 AALSDAGEVYTWGLGSMGQLGHVSLQSGDKELIPRRV--------VGLDGVSMKEVACGG 291
Query: 72 EHTAAIASDGLLFTWG 87
HT A++ +G L+ WG
Sbjct: 292 VHTCALSLEGALYAWG 307
Score = 35.4 bits (80), Expect = 0.32
Identities = 19/67 (28%), Positives = 27/67 (39%), Gaps = 16/67 (23%)
Query: 21 SAIYVWGYNQSGQTGRKEKDDQLRIPKQLPPQLFGCPAGLNTRWLDIACGREHTAAIASD 80
S ++VWG NQ P+LF T ++CG H A++ +
Sbjct: 141 SRLWVWGQNQGSNL----------------PRLFSGAFPATTAIRQVSCGTAHVVALSEE 184
Query: 81 GLLFTWG 87
GLL WG
Sbjct: 185 GLLQAWG 191
>ref|NP_186879.2| regulator of chromosome condensation (RCC1) family protein
[Arabidopsis thaliana]
Length = 471
Score = 141 bits (355), Expect = 4e-33
Identities = 65/87 (74%), Positives = 73/87 (83%), Gaps = 1/87 (1%)
Query: 1 MDIDAILGTTKPVAASIPRKSAIYVWGYNQSGQTGRKEKDDQLRIPKQLPPQLFGCPAGL 60
MDI I+G P + SIP KSAIYVWGYNQSGQTGR E++ LRIPKQLPP+LFGCPAG
Sbjct: 1 MDIGEIIGEVAP-SVSIPTKSAIYVWGYNQSGQTGRNEQEKLLRIPKQLPPELFGCPAGA 59
Query: 61 NTRWLDIACGREHTAAIASDGLLFTWG 87
N+RWLDI+CGREHTAA+ASDG LF WG
Sbjct: 60 NSRWLDISCGREHTAAVASDGSLFAWG 86
Score = 48.5 bits (114), Expect = 4e-05
Identities = 28/82 (34%), Positives = 43/82 (52%), Gaps = 3/82 (3%)
Query: 8 GTTKPVAASIPRKSAIYVWGYNQSGQTGRKEKDDQLRIPKQLPPQL-FGCPAGLNTRWLD 66
GT VA S + + WGYN+ GQ GR + L+ P+ + F A + +
Sbjct: 174 GTAHVVALS--EEGLLQAWGYNEQGQLGRGVTCEGLQAPRVINAYAKFLDEAPELVKIMQ 231
Query: 67 IACGREHTAAIASDGLLFTWGL 88
++CG HTAA++ G ++TWGL
Sbjct: 232 LSCGEYHTAALSDAGEVYTWGL 253
Score = 39.7 bits (91), Expect = 0.017
Identities = 22/76 (28%), Positives = 37/76 (47%), Gaps = 11/76 (14%)
Query: 15 ASIPRKSAIYVWGYNQSGQTGRK--EKDDQLRIPKQLPPQLFGCPAGLN-TRWLDIACGR 71
A++ +Y WG GQ G + D+ IP+++ GL+ ++ACG
Sbjct: 240 AALSDAGEVYTWGLGSMGQLGHVSLQSGDKELIPRRV--------VGLDGVSMKEVACGG 291
Query: 72 EHTAAIASDGLLFTWG 87
HT A++ +G L+ WG
Sbjct: 292 VHTCALSLEGALYAWG 307
Score = 35.4 bits (80), Expect = 0.32
Identities = 19/67 (28%), Positives = 27/67 (39%), Gaps = 16/67 (23%)
Query: 21 SAIYVWGYNQSGQTGRKEKDDQLRIPKQLPPQLFGCPAGLNTRWLDIACGREHTAAIASD 80
S ++VWG NQ P+LF T ++CG H A++ +
Sbjct: 141 SRLWVWGQNQGSNL----------------PRLFSGAFPATTAIRQVSCGTAHVVALSEE 184
Query: 81 GLLFTWG 87
GLL WG
Sbjct: 185 GLLQAWG 191
>ref|NP_909746.1| putative chromosome condensation regulator [Oryza sativa
(japonica cultivar-group)] gi|22795248|gb|AAN08220.1|
putative chromosome condensation regulator [Oryza
sativa (japonica cultivar-group)]
Length = 505
Score = 115 bits (289), Expect = 2e-25
Identities = 54/87 (62%), Positives = 65/87 (74%), Gaps = 2/87 (2%)
Query: 1 MDIDAILGTTKPVAASIPRKSAIYVWGYNQSGQTGRKEKDDQLRIPKQLPPQLFGCPAGL 60
MD+D +LG + A +P KSAIY+WGYN SGQT RK K+ LRIPK LPP+LF G
Sbjct: 1 MDMDDMLGGLR--VAGVPTKSAIYLWGYNHSGQTARKGKECHLRIPKSLPPKLFKWQDGK 58
Query: 61 NTRWLDIACGREHTAAIASDGLLFTWG 87
N RW+D+ACGR HTAA+ASDG L+TWG
Sbjct: 59 NLRWIDVACGRGHTAAVASDGSLYTWG 85
Score = 43.5 bits (101), Expect = 0.001
Identities = 24/67 (35%), Positives = 37/67 (54%), Gaps = 1/67 (1%)
Query: 23 IYVWGYNQSGQTGRKEKDDQLRIPKQLPPQL-FGCPAGLNTRWLDIACGREHTAAIASDG 81
+ WGYN+ GQ GR + L+ + L F A + + ++CG HTAAI+ +G
Sbjct: 212 LQAWGYNEYGQLGRGCTSEGLQGARVLNAYARFLDEAPELVKIVRVSCGEYHTAAISENG 271
Query: 82 LLFTWGL 88
++TWGL
Sbjct: 272 EVYTWGL 278
Score = 41.6 bits (96), Expect = 0.004
Identities = 23/75 (30%), Positives = 35/75 (46%), Gaps = 9/75 (12%)
Query: 15 ASIPRKSAIYVWGYNQSGQTGR--KEKDDQLRIPKQLPPQLFGCPAGLNTRWLDIACGRE 72
A+I +Y WG GQ G + D+ IP+++ A T D++CG
Sbjct: 265 AAISENGEVYTWGLGSMGQLGHCSLQSGDKELIPRRVV-------ALDRTVIRDVSCGGV 317
Query: 73 HTAAIASDGLLFTWG 87
H+ A+ DG L+ WG
Sbjct: 318 HSCAVTEDGALYAWG 332
>ref|NP_001006212.1| similar to Williams-Beuren syndrome chromosome region 16 homolog;
RCC1-like G exchanging factor-like protein [Gallus
gallus] gi|53133732|emb|CAG32195.1| hypothetical protein
[Gallus gallus]
Length = 454
Score = 48.5 bits (114), Expect = 4e-05
Identities = 24/72 (33%), Positives = 37/72 (51%), Gaps = 4/72 (5%)
Query: 17 IPRKSAIYVWGYNQSGQTGRKEKDDQLRIPKQLPPQLFG-CPAGLNTRWLDIACGREHTA 75
+ + A++VWGY G G+ + +P+ +PP LFG + R + CG H A
Sbjct: 341 LTEEGAVFVWGY---GILGKGPNVTETAVPEMIPPSLFGWSDFSSDVRVARVRCGLSHFA 397
Query: 76 AIASDGLLFTWG 87
A+ + G LF WG
Sbjct: 398 ALTNRGELFVWG 409
Score = 35.8 bits (81), Expect = 0.25
Identities = 20/69 (28%), Positives = 33/69 (46%), Gaps = 6/69 (8%)
Query: 23 IYVWGYNQSGQTGRKEKDDQLRIPKQLPPQLFGCPAGLNTRWLDIACGREHTAAIASDGL 82
++ G N GQ GRK +D++ L QL ++R + + CG++H+ G
Sbjct: 186 VFTMGNNSYGQCGRKVVEDEIYSESHLIHQL----NDFDSRVVQVVCGQDHSLFRTKKGA 241
Query: 83 LFT--WGLD 89
+F WG D
Sbjct: 242 VFACGWGAD 250
Score = 30.8 bits (68), Expect = 7.9
Identities = 28/88 (31%), Positives = 37/88 (41%), Gaps = 7/88 (7%)
Query: 25 VWGY--NQSGQTG--RKEKDDQLRIPKQLPPQLFGCPAGL--NTRWLDIACGREHTAAIA 78
VWG N+ Q G R +D L P P TR L ++CGR H+ +
Sbjct: 122 VWGLGLNKDSQLGFQRSIRDRTKGYEYVLEPSPIPLPLEKPQKTRVLQVSCGRAHSLVLT 181
Query: 79 SDGLLFTWGLDFSYSLLVLLIVHDFIYS 106
+FT G + SY +V D IYS
Sbjct: 182 DSEGVFTMG-NNSYGQCGRKVVEDEIYS 208
>ref|NP_291050.1| Williams-Beuren syndrome chromosome region 16 homolog [Mus
musculus] gi|21552744|gb|AAM62305.1| RCC1-like G
exchanging factor-like protein [Mus musculus]
gi|33244023|gb|AAH55335.1| Williams-Beuren syndrome
chromosome region 16 homolog [Mus musculus]
gi|19353269|gb|AAH24714.1| Williams-Beuren syndrome
chromosome region 16 homolog [Mus musculus]
gi|23822302|sp|Q9CYF5|WBS16_MOUSE Williams-Beuren
syndrome chromosome region 16 protein homolog
gi|12857127|dbj|BAB30902.1| unnamed protein product [Mus
musculus]
Length = 461
Score = 46.2 bits (108), Expect = 2e-04
Identities = 30/88 (34%), Positives = 42/88 (47%), Gaps = 10/88 (11%)
Query: 7 LGTTKPVA------ASIPRKSAIYVWGYNQSGQTGRKEKDDQLRIPKQLPPQLFGCPA-G 59
+G K VA A + + ++VWGY G G+ K + IP+ +PP LFG
Sbjct: 332 VGKVKQVACGGTGCAVLNAEGHVFVWGY---GILGKGPKLLETAIPEMIPPTLFGLTEFN 388
Query: 60 LNTRWLDIACGREHTAAIASDGLLFTWG 87
+ I CG H AA+ + G LF WG
Sbjct: 389 PEVQVSQIRCGLSHFAALTNKGELFVWG 416
>ref|XP_341067.1| PREDICTED: similar to Williams-Beuren syndrome chromosome region 16
homolog [Rattus norvegicus]
Length = 461
Score = 45.8 bits (107), Expect = 2e-04
Identities = 28/88 (31%), Positives = 41/88 (45%), Gaps = 10/88 (11%)
Query: 7 LGTTKPVA------ASIPRKSAIYVWGYNQSGQTGRKEKDDQLRIPKQLPPQLFGCPA-G 59
+G K VA A + + ++VWGY G G+ + +P+ +PP LFG
Sbjct: 332 VGKVKQVACGGTGCAILNEEGHVFVWGY---GILGKGPNLSETALPEMIPPTLFGLTEFN 388
Query: 60 LNTRWLDIACGREHTAAIASDGLLFTWG 87
+ I CG H AA+ + G LF WG
Sbjct: 389 PEVKVSQIRCGLSHFAALTNKGELFVWG 416
>gb|AAU44178.1| ptative chromosome condensation factor [Oryza sativa (japonica
cultivar-group)]
Length = 917
Score = 45.8 bits (107), Expect = 2e-04
Identities = 23/74 (31%), Positives = 37/74 (49%), Gaps = 8/74 (10%)
Query: 14 AASIPRKSAIYVWGYNQSGQTGRKEKDDQLRIPKQLPPQLFGCPAGLNTRWLDIACGREH 73
AA + R ++ WG + G+ G ++D + P+L A N + +ACG H
Sbjct: 89 AALVTRNGDVFTWGEDSGGRLGHGTREDSVH------PRLVESLAACNVDF--VACGEFH 140
Query: 74 TAAIASDGLLFTWG 87
T A+ + G L+TWG
Sbjct: 141 TCAVTTTGELYTWG 154
Score = 33.5 bits (75), Expect = 1.2
Identities = 21/51 (41%), Positives = 28/51 (54%), Gaps = 7/51 (13%)
Query: 45 IPKQLPPQLFGCPAGLNTRWLDIACGREHTAAIASDGLLFTWGLDFSYSLL 95
IPK++ L G P ++CG HTA I S G LFT+G D S+ +L
Sbjct: 173 IPKRISGALDGLPVAY------VSCGTWHTALITSMGQLFTFG-DGSFGVL 216
Score = 31.6 bits (70), Expect = 4.6
Identities = 21/71 (29%), Positives = 32/71 (44%), Gaps = 7/71 (9%)
Query: 23 IYVWGYNQSGQTGRKEKDDQLRIPKQLPPQLFGCPAGLNTRWL----DIACGREHTAAIA 78
+YVWG + R D +R ++ F P L +R + + CG H A +
Sbjct: 37 VYVWGEVFCENSVRVGSDTIIRSTEKTD---FLLPKPLESRLVLDVYHVDCGVRHAALVT 93
Query: 79 SDGLLFTWGLD 89
+G +FTWG D
Sbjct: 94 RNGDVFTWGED 104
>ref|ZP_00308854.1| COG5184: Alpha-tubulin suppressor and related RCC1
domain-containing proteins [Cytophaga hutchinsonii]
Length = 831
Score = 45.8 bits (107), Expect = 2e-04
Identities = 21/69 (30%), Positives = 38/69 (54%), Gaps = 10/69 (14%)
Query: 19 RKSAIYVWGYNQSGQTGRKEKDDQLRIPKQLPPQLFGCPAGLNTRWLDIACGREHTAAIA 78
R ++ WGYN +GQ G D + ++ P ++ G++ W+ +A G +T A+
Sbjct: 56 RSGELWGWGYNSNGQLG-----DGTTMDRRTPVKI-----GIDINWVAVATGSSYTLALK 105
Query: 79 SDGLLFTWG 87
+DG L++WG
Sbjct: 106 ADGTLWSWG 114
Score = 44.7 bits (104), Expect = 5e-04
Identities = 24/74 (32%), Positives = 34/74 (45%), Gaps = 10/74 (13%)
Query: 14 AASIPRKSAIYVWGYNQSGQTGRKEKDDQLRIPKQLPPQLFGCPAGLNTRWLDIACGREH 73
+ ++ ++ WGYN GQ G D K LP Q+ G W+ I G H
Sbjct: 303 SVALKANGTLWAWGYNVRGQLGYGSGTD-----KHLPVQI-----GTAHTWVAINAGTYH 352
Query: 74 TAAIASDGLLFTWG 87
T + +DG L+TWG
Sbjct: 353 TIGVKADGSLWTWG 366
Score = 42.0 bits (97), Expect = 0.003
Identities = 24/86 (27%), Positives = 41/86 (46%), Gaps = 18/86 (20%)
Query: 5 AILGTTKPVAASIPRKSAIYVWGYNQSGQTGRK---EKDDQLRIPKQLPPQLFGCPAGLN 61
A+ G+ VA + +++ WG N++G+ G ++D +R+ G +
Sbjct: 194 AVAGSNHIVALKVD--GSLWGWGLNEAGEIGNGNTVQQDSPVRV-------------GTD 238
Query: 62 TRWLDIACGREHTAAIASDGLLFTWG 87
W +A G HT AI S+G L+ WG
Sbjct: 239 NDWTTLAAGSNHTLAIKSNGTLWAWG 264
Score = 42.0 bits (97), Expect = 0.003
Identities = 22/73 (30%), Positives = 32/73 (43%), Gaps = 10/73 (13%)
Query: 17 IPRKSAIYVWGYNQSGQTGRKEKDDQLRIPKQLPPQLFGCPAGLNTRWLDIACGREHTAA 76
I ++ WG+N G+ G D+ K P Q+ G + WL G H A
Sbjct: 154 IKANGTLWTWGFNNGGELGIGNNDE-----KHTPVQV-----GADQNWLYAVAGSNHIVA 203
Query: 77 IASDGLLFTWGLD 89
+ DG L+ WGL+
Sbjct: 204 LKVDGSLWGWGLN 216
Score = 40.4 bits (93), Expect = 0.010
Identities = 23/71 (32%), Positives = 34/71 (47%), Gaps = 10/71 (14%)
Query: 17 IPRKSAIYVWGYNQSGQTGRKEKDDQLRIPKQLPPQLFGCPAGLNTRWLDIACGREHTAA 76
I ++ WG N+ GQ G D + K +P QL G + W++IA G H
Sbjct: 555 IKSDGTLWGWGTNRQGQLG-----DGTLVNKYVPVQL-----GTDRDWINIAGGTIHNIC 604
Query: 77 IASDGLLFTWG 87
+ S+G L+ WG
Sbjct: 605 LKSNGTLWAWG 615
Score = 38.9 bits (89), Expect = 0.029
Identities = 19/72 (26%), Positives = 34/72 (46%), Gaps = 8/72 (11%)
Query: 16 SIPRKSAIYVWGYNQSGQTGRKEKDDQLRIPKQLPPQLFGCPAGLNTRWLDIACGREHTA 75
+I ++ WG+N G G P LP Q+ G + W+ ++ G +H+
Sbjct: 253 AIKSNGTLWAWGHNVRGNLGNGTATLD---PATLPVQV-----GTDADWIRVSAGLDHSV 304
Query: 76 AIASDGLLFTWG 87
A+ ++G L+ WG
Sbjct: 305 ALKANGTLWAWG 316
Score = 38.1 bits (87), Expect = 0.050
Identities = 22/71 (30%), Positives = 32/71 (44%), Gaps = 10/71 (14%)
Query: 17 IPRKSAIYVWGYNQSGQTGRKEKDDQLRIPKQLPPQLFGCPAGLNTRWLDIACGREHTAA 76
I +++ WG+N G+ G L +P ++ G T W+ I G HT
Sbjct: 505 IQADGSLWSWGHNYDGELGLGTNLKTL-VPTRV---------GTGTDWISIKTGVGHTLG 554
Query: 77 IASDGLLFTWG 87
I SDG L+ WG
Sbjct: 555 IKSDGTLWGWG 565
Score = 37.4 bits (85), Expect = 0.085
Identities = 24/73 (32%), Positives = 35/73 (47%), Gaps = 11/73 (15%)
Query: 23 IYVWGYNQSGQTGRKEKDDQLRIPKQLPPQLFGCPAGLNTRWLDIACGR-EHTAAIASDG 81
++ WG N SGQ G D P+ LP Q+ G T W+ IA + I +DG
Sbjct: 460 LWAWGDNFSGQLG-----DGTEQPRMLPKQI-----GTATTWVSIAAASGVQSFGIQADG 509
Query: 82 LLFTWGLDFSYSL 94
L++WG ++ L
Sbjct: 510 SLWSWGHNYDGEL 522
Score = 34.3 bits (77), Expect = 0.72
Identities = 19/88 (21%), Positives = 34/88 (38%), Gaps = 13/88 (14%)
Query: 7 LGTTKPVAASIPRKSAIYVWGYNQSGQTGRKEKDDQLRIPKQLPPQLFGCPAGLNTRWLD 66
+ + + + I+ WG N G+ + ++ G + W
Sbjct: 397 IAASNAFSMGLKANGTIWTWGLNPFETDGQYKNSSPVQ-------------TGSDQNWKS 443
Query: 67 IACGREHTAAIASDGLLFTWGLDFSYSL 94
IA G + A+ +DG L+ WG +FS L
Sbjct: 444 IATGSNYILALKADGTLWAWGDNFSGQL 471
Score = 31.2 bits (69), Expect = 6.1
Identities = 17/68 (25%), Positives = 31/68 (45%), Gaps = 9/68 (13%)
Query: 22 AIYVWGYNQSGQTGRKEKDDQLRIPKQLPPQLFGCPAGLNTRWLDIACGREHTAAIASDG 81
+++ WG N + Q G DQ +P + P W+ IA + + ++G
Sbjct: 361 SLWTWGDNTNAQLGDGGTADQ-----PVPHSIRTAPDD----WISIAASNAFSMGLKANG 411
Query: 82 LLFTWGLD 89
++TWGL+
Sbjct: 412 TIWTWGLN 419
Score = 30.8 bits (68), Expect = 7.9
Identities = 20/72 (27%), Positives = 31/72 (42%), Gaps = 10/72 (13%)
Query: 23 IYVWGYNQSGQTGRKEKDDQLRIPKQLPPQLFGCPAGLNTRWLDIACGREHTAAIASDGL 82
++ WG N GQ G Q R+ + Q W+D+ + + AI +DG
Sbjct: 611 LWAWGRNNYGQVGTGNLITQDRMVQISAEQ----------NWIDVYASLDQSFAIRADGS 660
Query: 83 LFTWGLDFSYSL 94
L+ GL+ S L
Sbjct: 661 LWACGLNSSGQL 672
>gb|AAD43920.1| UVB-resistance protein UVR8 [Arabidopsis thaliana]
gi|11358913|pir||T50662 UVB-resistance protein UVR8
[imported] - Arabidopsis thaliana
Length = 440
Score = 45.4 bits (106), Expect = 3e-04
Identities = 26/73 (35%), Positives = 41/73 (55%), Gaps = 10/73 (13%)
Query: 16 SIPRKSAIYVWGYNQSGQTGRKEKDDQLRIPKQLPPQLF-GCPAGLNTRWLDIACGREHT 74
++ + + WG NQ+GQ G + +D L +P+++ Q F G P + +A G EHT
Sbjct: 134 AVTMEGEVQSWGRNQNGQLGLGDTEDSL-VPQKI--QAFEGIPIKM------VAAGAEHT 184
Query: 75 AAIASDGLLFTWG 87
AA+ DG L+ WG
Sbjct: 185 AAVTEDGDLYGWG 197
Score = 40.4 bits (93), Expect = 0.010
Identities = 25/72 (34%), Positives = 38/72 (52%), Gaps = 8/72 (11%)
Query: 16 SIPRKSAIYVWGYNQSGQTGRKEKDDQLRIPKQLPPQLFGCPAGLNTRWLDIACGREHTA 75
S+ A+Y +G+++ GQ G + +D L IP +L A N+ I+ G HT
Sbjct: 238 SVSYSGALYTYGWSKYGQLGHGDLEDHL-IPHKLE-------ALSNSFISQISGGWRHTM 289
Query: 76 AIASDGLLFTWG 87
A+ SDG L+ WG
Sbjct: 290 ALTSDGKLYGWG 301
Score = 35.4 bits (80), Expect = 0.32
Identities = 18/73 (24%), Positives = 38/73 (51%), Gaps = 8/73 (10%)
Query: 15 ASIPRKSAIYVWGYNQSGQTGRKEKDDQLRIPKQLPPQLFGCPAGLNTRWLDIACGREHT 74
A++ +Y WG+ + G G ++ D+L +P+++ + + +ACG HT
Sbjct: 185 AAVTEDGDLYGWGWGRYGNLGLGDRTDRL-VPERVT-------STGGEKMSMVACGWRHT 236
Query: 75 AAIASDGLLFTWG 87
+++ G L+T+G
Sbjct: 237 ISVSYSGALYTYG 249
Score = 34.3 bits (77), Expect = 0.72
Identities = 18/65 (27%), Positives = 29/65 (43%), Gaps = 8/65 (12%)
Query: 23 IYVWGYNQSGQTGRKEKDDQLRIPKQLPPQLFGCPAGLNTRWLDIACGREHTAAIASDGL 82
+Y WG+N+ GQ G DQ + P + + + ++CG HT A+
Sbjct: 297 LYGWGWNKFGQVGVGNNLDQCSPVQVRFPD--------DQKVVQVSCGWRHTLAVTERNN 348
Query: 83 LFTWG 87
+F WG
Sbjct: 349 VFAWG 353
Score = 32.0 bits (71), Expect = 3.6
Identities = 17/63 (26%), Positives = 30/63 (46%), Gaps = 9/63 (14%)
Query: 26 WGYNQSGQTGRKEKDDQLRIPKQLPPQLFGCPAGLNTRWLDIACGREHTAAIASDGL-LF 84
WG + GQ G + +D + P QL + + + CG +HT A + G+ ++
Sbjct: 39 WGRGEDGQLGHGDAED-----RPSPTQLSALDGH---QIVSVTCGADHTVAYSQSGMEVY 90
Query: 85 TWG 87
+WG
Sbjct: 91 SWG 93
>gb|AAH73297.1| MGC80684 protein [Xenopus laevis]
Length = 1028
Score = 45.4 bits (106), Expect = 3e-04
Identities = 27/85 (31%), Positives = 40/85 (46%), Gaps = 9/85 (10%)
Query: 3 IDAILGTTKPVAASIPRKSAIYVWGYNQSGQTGRKEKDDQLRIPKQLPPQLFGCPAGLNT 62
+D GT VA + IY WG GQ G + + PK++ L N
Sbjct: 87 VDVSCGTNHSVAMC--DEGNIYSWGDGSEGQLGTGKFSSKNFTPKRITGLL-------NR 137
Query: 63 RWLDIACGREHTAAIASDGLLFTWG 87
+ + I+CG H+ A+A DG +F+WG
Sbjct: 138 KIIQISCGNFHSVALAEDGRVFSWG 162
Score = 38.5 bits (88), Expect = 0.038
Identities = 21/74 (28%), Positives = 34/74 (45%), Gaps = 6/74 (8%)
Query: 16 SIPRKSAIYVWGYNQSGQTGRKEKDDQLRIPKQLPPQLFGCPAGLNTRWLDIACGREHTA 75
++ ++ WG N +GQ G K + P L + I+CG EHTA
Sbjct: 204 ALSMSGTVFAWGRNNAGQLGFKNDAKK----GTFKPYAVDSLRDLGVAY--ISCGEEHTA 257
Query: 76 AIASDGLLFTWGLD 89
+ DG+++T+G D
Sbjct: 258 VLNKDGIVYTFGDD 271
Score = 33.1 bits (74), Expect = 1.6
Identities = 19/74 (25%), Positives = 32/74 (42%), Gaps = 7/74 (9%)
Query: 14 AASIPRKSAIYVWGYNQSGQTGRKEKDDQLRIPKQLPPQLFGCPAGLNTRWLDIACGREH 73
+ ++ ++ WG N+ GQ G +I Q PQL G+ + + G
Sbjct: 149 SVALAEDGRVFSWGQNKCGQLGLGS-----QIINQATPQLVKSLKGIPL--VQVTAGGSQ 201
Query: 74 TAAIASDGLLFTWG 87
T A++ G +F WG
Sbjct: 202 TFALSMSGTVFAWG 215
>ref|XP_417056.1| PREDICTED: similar to Chromosome condensation 1-like [Gallus
gallus]
Length = 463
Score = 45.4 bits (106), Expect = 3e-04
Identities = 22/69 (31%), Positives = 37/69 (52%), Gaps = 7/69 (10%)
Query: 23 IYVWGYNQSGQTGRKEKDDQLRIPKQLPPQLFGCPAGLNTRWLDIACGREHTAAIASDGL 82
+Y WG+N Q G + L +P Q+ L +N + +++ACG H+ + SDG
Sbjct: 274 VYTWGHNAYSQLGNGTTNHGL-VPCQVSTNL------VNKKVIEVACGSHHSMVLTSDGE 326
Query: 83 LFTWGLDFS 91
++TWG + S
Sbjct: 327 VYTWGYNNS 335
Score = 41.2 bits (95), Expect = 0.006
Identities = 24/80 (30%), Positives = 35/80 (43%), Gaps = 9/80 (11%)
Query: 16 SIPRKSAIYVWGYNQSGQTGRKEKDDQLRIPKQLPPQLFGCPAGLNTRWLDIACGREHTA 75
++ +YVWGYN +GQ G +Q P A R +ACG HT
Sbjct: 373 AVVENGEVYVWGYNGNGQLGLGSSGNQ--------PTPCRIAALQGIRVQRVACGCAHTL 424
Query: 76 AIASDGLLFTWGLDFSYSLL 95
+ +G ++ WG + SY L
Sbjct: 425 VLTDEGQIYAWGAN-SYGQL 443
Score = 41.2 bits (95), Expect = 0.006
Identities = 21/65 (32%), Positives = 34/65 (52%), Gaps = 7/65 (10%)
Query: 23 IYVWGYNQSGQTGRKEKDDQLRIPKQLPPQLFGCPAGLNTRWLDIACGREHTAAIASDGL 82
+Y WGYN SGQ G +Q +P ++ C N ++IACG+ + A+ +G
Sbjct: 327 VYTWGYNNSGQVGSGSTANQ-----PIPRRVTSCLQ--NKVVVNIACGQMCSMAVVENGE 379
Query: 83 LFTWG 87
++ WG
Sbjct: 380 VYVWG 384
>ref|XP_482519.1| putative UVB-resistance protein UVR8 [Oryza sativa (japonica
cultivar-group)] gi|51964906|ref|XP_507237.1| PREDICTED
OJ1124_B05.9 gene product [Oryza sativa (japonica
cultivar-group)] gi|38175475|dbj|BAD01172.1| putative
UVB-resistance protein UVR8 [Oryza sativa (japonica
cultivar-group)]
Length = 395
Score = 45.1 bits (105), Expect = 4e-04
Identities = 29/94 (30%), Positives = 44/94 (45%), Gaps = 11/94 (11%)
Query: 3 IDAILGTTKPVAA-------SIPRKSAIYVWGYNQSGQTGRK--EKDDQLRIPKQLPPQL 53
+DA+ G + AA ++ K Y WG N+ GQ G + K+D R ++ P
Sbjct: 91 VDALAGVSIVQAAIGGWHCLAVDDKGRAYAWGGNEYGQCGEEAERKEDGTRALRRDIPIP 150
Query: 54 FGCPAGLNTRWLDIACGREHTAAIASDGLLFTWG 87
GC L R +A G H+ + DG ++TWG
Sbjct: 151 QGCAPKLKVR--QVAAGGTHSVVLTQDGHVWTWG 182
Score = 35.0 bits (79), Expect = 0.42
Identities = 24/78 (30%), Positives = 35/78 (44%), Gaps = 14/78 (17%)
Query: 12 PVAASIPRKSAIYVWGYNQSGQTGRK--EKDDQLRIPKQLPPQLFGCPAGLNTRWLDIAC 69
P A+S+P +A+ WG + GQ G E++D R L +
Sbjct: 4 PPASSLP--TAVLAWGSGEDGQLGMGGYEEEDWARGVAALDALAVSA----------VVA 51
Query: 70 GREHTAAIASDGLLFTWG 87
G ++ AI +DG LFTWG
Sbjct: 52 GSRNSLAICADGRLFTWG 69
Score = 35.0 bits (79), Expect = 0.42
Identities = 16/74 (21%), Positives = 38/74 (50%), Gaps = 7/74 (9%)
Query: 14 AASIPRKSAIYVWGYNQSGQTGRKEKDDQLRIPKQLPPQLFGCPAGLNTRWLDIACGREH 73
+ ++ ++ +Y WG + G+ G + +P+++ AG + + ++CG H
Sbjct: 267 STALTKEGEVYAWGRGEHGRLGFGDDKSSHMVPQKVE-----LLAGEDI--IQVSCGGTH 319
Query: 74 TAAIASDGLLFTWG 87
+ A+ DG +F++G
Sbjct: 320 SVALTRDGRMFSYG 333
Score = 34.3 bits (77), Expect = 0.72
Identities = 18/65 (27%), Positives = 32/65 (48%), Gaps = 8/65 (12%)
Query: 23 IYVWGYNQSGQTGRKEKDDQLRIPKQLPPQLFGCPAGLNTRWLDIACGREHTAAIASDGL 82
++ WG N+ GQ G + P+ P ++ G + +DIA G H+ A+ +G
Sbjct: 224 LWAWGNNEYGQLGTGDTQ-----PRSQPIRVEGLS---DLSLVDIAAGGWHSTALTKEGE 275
Query: 83 LFTWG 87
++ WG
Sbjct: 276 VYAWG 280
>ref|XP_417063.1| PREDICTED: similar to hypothetical protein [Gallus gallus]
Length = 550
Score = 45.1 bits (105), Expect = 4e-04
Identities = 29/87 (33%), Positives = 39/87 (44%), Gaps = 10/87 (11%)
Query: 1 MDIDAILGTTKPVAASIPRKSAIYVWGYNQSGQTGRKEKDDQLRIPKQLPPQLFGCPAGL 60
M + G T +A + +Y WGYN +GQ G +QL P G+
Sbjct: 201 MVVGIACGQTSSMA--VVNNGEVYGWGYNGNGQLGLGNNGNQLT------PCRVAALHGV 252
Query: 61 NTRWLDIACGREHTAAIASDGLLFTWG 87
L IACG HT A+ +GLL+ WG
Sbjct: 253 CI--LQIACGYAHTLALTDEGLLYAWG 277
Score = 38.9 bits (89), Expect = 0.029
Identities = 20/65 (30%), Positives = 33/65 (50%), Gaps = 7/65 (10%)
Query: 23 IYVWGYNQSGQTGRKEKDDQLRIPKQLPPQLFGCPAGLNTRWLDIACGREHTAAIASDGL 82
+Y WG+N Q G + + P Q+ C L + +++ACG H+ A++ DG
Sbjct: 115 VYAWGHNGYSQLGNGATNQGVT-----PVQV--CTNLLLKKVIEVACGSHHSMALSFDGD 167
Query: 83 LFTWG 87
L+ WG
Sbjct: 168 LYAWG 172
Score = 36.6 bits (83), Expect = 0.14
Identities = 19/65 (29%), Positives = 32/65 (49%), Gaps = 7/65 (10%)
Query: 23 IYVWGYNQSGQTGRKEKDDQLRIPKQLPPQLFGCPAGLNTRWLDIACGREHTAAIASDGL 82
+Y WGYN GQ G +Q P ++ C + + IACG+ + A+ ++G
Sbjct: 168 LYAWGYNNCGQVGSGSTANQ-----PTPRRVSNCLQ--DKMVVGIACGQTSSMAVVNNGE 220
Query: 83 LFTWG 87
++ WG
Sbjct: 221 VYGWG 225
>gb|AAW78916.1| putative chromosome condensation factor [Triticum aestivum]
Length = 907
Score = 44.7 bits (104), Expect = 5e-04
Identities = 22/74 (29%), Positives = 37/74 (49%), Gaps = 8/74 (10%)
Query: 14 AASIPRKSAIYVWGYNQSGQTGRKEKDDQLRIPKQLPPQLFGCPAGLNTRWLDIACGREH 73
A+ + R ++ WG + G+ G ++D + P+L A N + +ACG H
Sbjct: 89 ASLVTRNGEVFTWGEDSGGRLGHGTREDSVH------PRLVESLAACNIDF--VACGEFH 140
Query: 74 TAAIASDGLLFTWG 87
T A+ + G L+TWG
Sbjct: 141 TCAVTTTGELYTWG 154
Score = 32.7 bits (73), Expect = 2.1
Identities = 22/75 (29%), Positives = 32/75 (42%), Gaps = 15/75 (20%)
Query: 23 IYVWGYNQSGQTGRKEKDDQLR--------IPKQLPPQLFGCPAGLNTRWLDIACGREHT 74
+YVWG + R D +R +PK L L L+ +D CG H
Sbjct: 37 VYVWGEVVCDNSARTSSDTVIRSTGKTDFLLPKPLESNLV-----LDVYHVD--CGVRHA 89
Query: 75 AAIASDGLLFTWGLD 89
+ + +G +FTWG D
Sbjct: 90 SLVTRNGEVFTWGED 104
Score = 31.2 bits (69), Expect = 6.1
Identities = 19/47 (40%), Positives = 28/47 (59%), Gaps = 3/47 (6%)
Query: 49 LPPQLFGCPAGLNTRWLDIACGREHTAAIASDGLLFTWGLDFSYSLL 95
+P ++ G GL + ++CG HTA I S G LFT+G D S+ +L
Sbjct: 173 IPRRISGPLEGLQIAY--VSCGTWHTALITSMGQLFTFG-DGSFGVL 216
>gb|AAW78912.1| putative chromosome condensation factor [Triticum turgidum]
Length = 882
Score = 44.7 bits (104), Expect = 5e-04
Identities = 22/74 (29%), Positives = 37/74 (49%), Gaps = 8/74 (10%)
Query: 14 AASIPRKSAIYVWGYNQSGQTGRKEKDDQLRIPKQLPPQLFGCPAGLNTRWLDIACGREH 73
A+ + R ++ WG + G+ G ++D + P+L A N + +ACG H
Sbjct: 89 ASLVTRNGEVFTWGEDSGGRLGHGTREDSVH------PRLVESLAACNIDF--VACGEFH 140
Query: 74 TAAIASDGLLFTWG 87
T A+ + G L+TWG
Sbjct: 141 TCAVTTTGELYTWG 154
Score = 32.7 bits (73), Expect = 2.1
Identities = 22/75 (29%), Positives = 32/75 (42%), Gaps = 15/75 (20%)
Query: 23 IYVWGYNQSGQTGRKEKDDQLR--------IPKQLPPQLFGCPAGLNTRWLDIACGREHT 74
+YVWG + R D +R +PK L L L+ +D CG H
Sbjct: 37 VYVWGEVVCDNSARTSSDTVIRSTGKTDFLLPKPLESNLV-----LDVYHVD--CGVRHA 89
Query: 75 AAIASDGLLFTWGLD 89
+ + +G +FTWG D
Sbjct: 90 SLVTRNGEVFTWGED 104
Score = 31.2 bits (69), Expect = 6.1
Identities = 19/47 (40%), Positives = 28/47 (59%), Gaps = 3/47 (6%)
Query: 49 LPPQLFGCPAGLNTRWLDIACGREHTAAIASDGLLFTWGLDFSYSLL 95
+P ++ G GL + ++CG HTA I S G LFT+G D S+ +L
Sbjct: 173 IPRRISGPLEGLQIAY--VSCGTWHTALITSMGQLFTFG-DGSFGVL 216
>ref|XP_506822.1| PREDICTED P0470G10.15 gene product [Oryza sativa (japonica
cultivar-group)] gi|50909401|ref|XP_466189.1| putative
UVB-resistance protein (UVR8) [Oryza sativa (japonica
cultivar-group)] gi|50725773|dbj|BAD33304.1| putative
UVB-resistance protein (UVR8) [Oryza sativa (japonica
cultivar-group)]
Length = 453
Score = 44.7 bits (104), Expect = 5e-04
Identities = 25/65 (38%), Positives = 33/65 (50%), Gaps = 8/65 (12%)
Query: 23 IYVWGYNQSGQTGRKEKDDQLRIPKQLPPQLFGCPAGLNTRWLDIACGREHTAAIASDGL 82
++ WG NQ+GQ G +D L PQ G+ + IA G EHTAA+ DG
Sbjct: 155 VHSWGRNQNGQLGLGNTEDSLL------PQKIQAFEGVRVKM--IAAGAEHTAAVTEDGD 206
Query: 83 LFTWG 87
L+ WG
Sbjct: 207 LYGWG 211
Score = 40.4 bits (93), Expect = 0.010
Identities = 21/74 (28%), Positives = 42/74 (56%), Gaps = 10/74 (13%)
Query: 15 ASIPRKSAIYVWGYNQSGQTGRKEKDDQLRIPKQLPPQLFGCPAGLN-TRWLDIACGREH 73
A++ +Y WG+ + G G ++DD+L IP+++ + +N + + +ACG H
Sbjct: 199 AAVTEDGDLYGWGWGRYGNLGLGDRDDRL-IPEKV--------SSVNGQKMVLVACGWRH 249
Query: 74 TAAIASDGLLFTWG 87
T ++S G ++T+G
Sbjct: 250 TITVSSSGSIYTYG 263
Score = 39.3 bits (90), Expect = 0.022
Identities = 23/72 (31%), Positives = 39/72 (53%), Gaps = 8/72 (11%)
Query: 16 SIPRKSAIYVWGYNQSGQTGRKEKDDQLRIPKQLPPQLFGCPAGLNTRWLDIACGREHTA 75
++ +IY +G+++ GQ G + +D L +P +L A +T I+ G HT
Sbjct: 252 TVSSSGSIYTYGWSKYGQLGHGDFEDHL-VPHKLE-------ALKDTTISQISGGWRHTM 303
Query: 76 AIASDGLLFTWG 87
A+A+DG L+ WG
Sbjct: 304 ALAADGKLYGWG 315
Score = 35.8 bits (81), Expect = 0.25
Identities = 19/70 (27%), Positives = 32/70 (45%), Gaps = 18/70 (25%)
Query: 23 IYVWGYNQSGQTGRKEKDD-----QLRIPKQLPPQLFGCPAGLNTRWLDIACGREHTAAI 77
+Y WG+N+ GQ G + +D Q+ P + + + +ACG HT A+
Sbjct: 311 LYGWGWNKFGQVGVGDNEDHCSPVQVNFPNE-------------QKVVQVACGWRHTLAL 357
Query: 78 ASDGLLFTWG 87
+F+WG
Sbjct: 358 TEAKNVFSWG 367
Score = 35.0 bits (79), Expect = 0.42
Identities = 19/63 (30%), Positives = 32/63 (50%), Gaps = 5/63 (7%)
Query: 26 WGYNQSGQTGRKEKDDQLRIPKQLPPQLFGCPAGLNTRWLDIACGREHTAAIASDGL-LF 84
WG + GQ G + +D+ +P L P G+ + + CG +HT A + + L L+
Sbjct: 49 WGRGEDGQLGHGDAEDR-PVPTVLTAAFDDAPGGVAS---VVICGADHTTAYSDEELQLY 104
Query: 85 TWG 87
+WG
Sbjct: 105 SWG 107
Score = 31.6 bits (70), Expect = 4.6
Identities = 19/65 (29%), Positives = 26/65 (39%), Gaps = 8/65 (12%)
Query: 23 IYVWGYNQSGQTGRKEKDDQLRIPKQLPPQLFGCPAGLNTRWLDIACGREHTAAIASDGL 82
+Y WG+ G+ G D PQ G+ R IACG H A+ G
Sbjct: 103 LYSWGWGDFGRLGHGNSSDVFN------PQPIQALQGV--RITQIACGDSHCLAVTVAGH 154
Query: 83 LFTWG 87
+ +WG
Sbjct: 155 VHSWG 159
>gb|AAN18169.1| At5g63860/MGI19_6 [Arabidopsis thaliana] gi|21928123|gb|AAM78089.1|
AT5g63860/MGI19_6 [Arabidopsis thaliana]
gi|10177674|dbj|BAB11034.1| UVB-resistance protein UVR8
[Arabidopsis thaliana] gi|15237540|ref|NP_201191.1|
UVB-resistance protein (UVR8) [Arabidopsis thaliana]
Length = 440
Score = 44.7 bits (104), Expect = 5e-04
Identities = 24/72 (33%), Positives = 36/72 (49%), Gaps = 8/72 (11%)
Query: 16 SIPRKSAIYVWGYNQSGQTGRKEKDDQLRIPKQLPPQLFGCPAGLNTRWLDIACGREHTA 75
++ + + WG NQ+GQ G + +D L PQ G+ + +A G EHTA
Sbjct: 134 AVTMEGEVQSWGRNQNGQLGLGDTEDSL------VPQKIQAFEGIRIKM--VAAGAEHTA 185
Query: 76 AIASDGLLFTWG 87
A+ DG L+ WG
Sbjct: 186 AVTEDGDLYGWG 197
Score = 40.4 bits (93), Expect = 0.010
Identities = 25/72 (34%), Positives = 38/72 (52%), Gaps = 8/72 (11%)
Query: 16 SIPRKSAIYVWGYNQSGQTGRKEKDDQLRIPKQLPPQLFGCPAGLNTRWLDIACGREHTA 75
S+ A+Y +G+++ GQ G + +D L IP +L A N+ I+ G HT
Sbjct: 238 SVSYSGALYTYGWSKYGQLGHGDLEDHL-IPHKLE-------ALSNSFISQISGGWRHTM 289
Query: 76 AIASDGLLFTWG 87
A+ SDG L+ WG
Sbjct: 290 ALTSDGKLYGWG 301
Score = 35.4 bits (80), Expect = 0.32
Identities = 18/73 (24%), Positives = 38/73 (51%), Gaps = 8/73 (10%)
Query: 15 ASIPRKSAIYVWGYNQSGQTGRKEKDDQLRIPKQLPPQLFGCPAGLNTRWLDIACGREHT 74
A++ +Y WG+ + G G ++ D+L +P+++ + + +ACG HT
Sbjct: 185 AAVTEDGDLYGWGWGRYGNLGLGDRTDRL-VPERVT-------STGGEKMSMVACGWRHT 236
Query: 75 AAIASDGLLFTWG 87
+++ G L+T+G
Sbjct: 237 ISVSYSGALYTYG 249
Score = 34.3 bits (77), Expect = 0.72
Identities = 18/65 (27%), Positives = 29/65 (43%), Gaps = 8/65 (12%)
Query: 23 IYVWGYNQSGQTGRKEKDDQLRIPKQLPPQLFGCPAGLNTRWLDIACGREHTAAIASDGL 82
+Y WG+N+ GQ G DQ + P + + + ++CG HT A+
Sbjct: 297 LYGWGWNKFGQVGVGNNLDQCSPVQVRFPD--------DQKVVQVSCGWRHTLAVTERNN 348
Query: 83 LFTWG 87
+F WG
Sbjct: 349 VFAWG 353
Score = 32.0 bits (71), Expect = 3.6
Identities = 17/63 (26%), Positives = 30/63 (46%), Gaps = 9/63 (14%)
Query: 26 WGYNQSGQTGRKEKDDQLRIPKQLPPQLFGCPAGLNTRWLDIACGREHTAAIASDGL-LF 84
WG + GQ G + +D + P QL + + + CG +HT A + G+ ++
Sbjct: 39 WGRGEDGQLGHGDAED-----RPSPTQLSALDGH---QIVSVTCGADHTVAYSQSGMEVY 90
Query: 85 TWG 87
+WG
Sbjct: 91 SWG 93
>gb|AAK84081.1| putative chromosome condensation factor [Triticum monococcum]
Length = 907
Score = 44.7 bits (104), Expect = 5e-04
Identities = 22/74 (29%), Positives = 37/74 (49%), Gaps = 8/74 (10%)
Query: 14 AASIPRKSAIYVWGYNQSGQTGRKEKDDQLRIPKQLPPQLFGCPAGLNTRWLDIACGREH 73
A+ + R ++ WG + G+ G ++D + P+L A N + +ACG H
Sbjct: 89 ASLVTRNGEVFTWGEDSGGRLGHGTREDSVH------PRLVESLAACNIDF--VACGEFH 140
Query: 74 TAAIASDGLLFTWG 87
T A+ + G L+TWG
Sbjct: 141 TCAVTTTGELYTWG 154
Score = 32.7 bits (73), Expect = 2.1
Identities = 22/75 (29%), Positives = 32/75 (42%), Gaps = 15/75 (20%)
Query: 23 IYVWGYNQSGQTGRKEKDDQLR--------IPKQLPPQLFGCPAGLNTRWLDIACGREHT 74
+YVWG + R D +R +PK L L L+ +D CG H
Sbjct: 37 VYVWGEVVCDNSARTSSDTVIRSTGKTDFLLPKPLESNLV-----LDVYHVD--CGVRHA 89
Query: 75 AAIASDGLLFTWGLD 89
+ + +G +FTWG D
Sbjct: 90 SLVTRNGEVFTWGED 104
Score = 31.2 bits (69), Expect = 6.1
Identities = 19/47 (40%), Positives = 28/47 (59%), Gaps = 3/47 (6%)
Query: 49 LPPQLFGCPAGLNTRWLDIACGREHTAAIASDGLLFTWGLDFSYSLL 95
+P ++ G GL + ++CG HTA I S G LFT+G D S+ +L
Sbjct: 173 IPRRISGPLEGLQIAY--VSCGTWHTALITSMGQLFTFG-DGSFGVL 216
>emb|CAH90411.1| hypothetical protein [Pongo pygmaeus]
Length = 975
Score = 44.3 bits (103), Expect = 7e-04
Identities = 22/69 (31%), Positives = 34/69 (48%), Gaps = 9/69 (13%)
Query: 20 KSAIYVWGYNQSGQTGRKEKDDQLRIPKQLPPQLFGCPAGLNTRW-LDIACGREHTAAIA 78
+ ++ WG GQ G +D + +P+ + LN + L ++CG H A+A
Sbjct: 102 RGQLFSWGAGSDGQLGLMTTEDSVAVPRLIQK--------LNQQTILQVSCGNWHCLALA 153
Query: 79 SDGLLFTWG 87
SDG FTWG
Sbjct: 154 SDGQFFTWG 162
Score = 38.1 bits (87), Expect = 0.050
Identities = 32/97 (32%), Positives = 46/97 (46%), Gaps = 17/97 (17%)
Query: 22 AIYVWGYNQSGQTGRKEKDDQLRIPKQLPPQLFGCPAGL--NTRWLDIACGREHTAAIAS 79
A++ WG N +GQ G ++ D ++ P C L + + I+CG EHTA +
Sbjct: 210 AVFGWGMNNAGQLGLSDEKD-----RESP-----CHVKLLRTQKVVYISCGEEHTAVLTK 259
Query: 80 DGLLFTWGLDFSYSLLVLLIVHDFIYSFKNCFIVLEL 116
G +FT+G SY L HD + N VLEL
Sbjct: 260 SGGVFTFGAG-SYGQL----GHDSMNDEVNPRRVLEL 291
Score = 35.8 bits (81), Expect = 0.25
Identities = 19/65 (29%), Positives = 31/65 (47%), Gaps = 10/65 (15%)
Query: 23 IYVWGYNQSGQTGRKEKDDQLRIPKQLPPQLFGCPAGLNTRWLDIACGREHTAAIASDGL 82
+Y G N GQ G + ++ P+ G A + + +ACG H+ A++ G
Sbjct: 55 VYTCGLNTKGQLGHVREGNK--------PEQIGALADQHI--IHVACGESHSLALSDRGQ 104
Query: 83 LFTWG 87
LF+WG
Sbjct: 105 LFSWG 109
Score = 34.7 bits (78), Expect = 0.55
Identities = 21/74 (28%), Positives = 36/74 (48%), Gaps = 9/74 (12%)
Query: 15 ASIPRKSAIYVWGYNQSGQTGRKEKDDQLRIPKQLPPQLFGCPAGLNTRWLDIACGREHT 74
A + + ++ +G GQ G +D++ + L +L G IACGR+HT
Sbjct: 255 AVLTKSGGVFTFGAGSYGQLGHDSMNDEVNPRRVL--ELMGSEV------TQIACGRQHT 306
Query: 75 AA-IASDGLLFTWG 87
A + S GL++ +G
Sbjct: 307 LAFVPSSGLIYAFG 320
>ref|XP_642619.1| hypothetical protein DDB0169222 [Dictyostelium discoideum]
gi|21281379|gb|AAM45266.1| similar to Homo sapiens
(Human). HERC2 protein [Dictyostelium discoideum]
gi|60470703|gb|EAL68677.1| hypothetical protein
DDB0169222 [Dictyostelium discoideum]
Length = 849
Score = 44.3 bits (103), Expect = 7e-04
Identities = 22/75 (29%), Positives = 37/75 (49%), Gaps = 8/75 (10%)
Query: 14 AASIPRKSAIYVWGYNQSGQTGRKEKDDQLRIPKQLPPQLFGCPAGLNTRWLDIACGREH 73
+A++ + +Y WG+N+ GQ G + PQL +N + + CGR+H
Sbjct: 224 SAAVTKSGRLYTWGFNEKGQLGLGNRWFHST------PQLVKTLIDVNI--VSVVCGRQH 275
Query: 74 TAAIASDGLLFTWGL 88
AI G +++WGL
Sbjct: 276 ICAITDQGEVYSWGL 290
Score = 32.7 bits (73), Expect = 2.1
Identities = 23/72 (31%), Positives = 32/72 (43%), Gaps = 6/72 (8%)
Query: 16 SIPRKSAIYVWGYNQSGQTGRKEKDDQLRIPKQLPPQLFGCPAGLNTRWLDIACGREHTA 75
+I + +Y WG GQ G L PK++ Q F LN R +ACG T
Sbjct: 278 AITDQGEVYSWGLGVFGQLGHGNVKSYLH-PKKI--QQF---VELNERIAQVACGSNFTM 331
Query: 76 AIASDGLLFTWG 87
+ GLL+ +G
Sbjct: 332 VRSVQGLLYAFG 343
Score = 32.0 bits (71), Expect = 3.6
Identities = 29/101 (28%), Positives = 45/101 (43%), Gaps = 15/101 (14%)
Query: 3 IDAILGTTKPVAASIPRKSAIYVWGYNQSGQTGRKE----KDDQLRIPKQLPPQLFGCPA 58
I + G +A S R +Y WG ++ GQ G + D + + K P L +
Sbjct: 37 IQTVSGEAHSLAVS--RFGDVYSWGRSKEGQLGIGQGYGGSDKVMFVAK---PTLI--KS 89
Query: 59 GLNTRWLDIACGREHTAAIASDGLLFTWG----LDFSYSLL 95
+ R + +ACG H+ A+ G ++ WG LD S S L
Sbjct: 90 LQHERIIKVACGNFHSLALTDMGKVYEWGQLHRLDESQSSL 130
Score = 31.6 bits (70), Expect = 4.6
Identities = 19/76 (25%), Positives = 35/76 (46%), Gaps = 4/76 (5%)
Query: 23 IYVWGYNQSGQTGRKEKDDQLRIPKQLPPQLFGCPA---GLNTRWL-DIACGREHTAAIA 78
+Y +G+ + GQ G E+ L + + P L T+ + ++ACG HT +
Sbjct: 339 LYAFGHGEYGQLGSTEETQHLDFGGRDNHFKYSIPIVVKSLETKKIKNVACGHLHTIVVT 398
Query: 79 SDGLLFTWGLDFSYSL 94
+ ++ WG S +L
Sbjct: 399 DENEVYQWGWGSSGAL 414
Database: nr
Posted date: Jul 5, 2005 12:34 AM
Number of letters in database: 863,360,394
Number of sequences in database: 2,540,612
Lambda K H
0.326 0.143 0.458
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 215,095,907
Number of Sequences: 2540612
Number of extensions: 8555797
Number of successful extensions: 25350
Number of sequences better than 10.0: 512
Number of HSP's better than 10.0 without gapping: 217
Number of HSP's successfully gapped in prelim test: 295
Number of HSP's that attempted gapping in prelim test: 22940
Number of HSP's gapped (non-prelim): 2289
length of query: 121
length of database: 863,360,394
effective HSP length: 97
effective length of query: 24
effective length of database: 616,921,030
effective search space: 14806104720
effective search space used: 14806104720
T: 11
A: 40
X1: 15 ( 7.0 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 40 (21.6 bits)
S2: 68 (30.8 bits)
Medicago: description of AC137823.5


