

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC137821.7 - phase: 0
(821 letters)
Database: nr
2,540,612 sequences; 863,360,394 total letters
Searching..................................................done
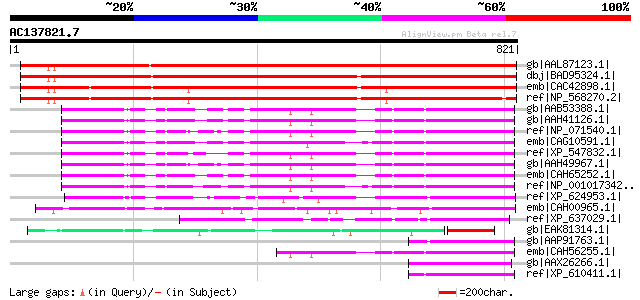
Score E
Sequences producing significant alignments: (bits) Value
gb|AAL87123.1| SEC10 [Arabidopsis thaliana] gi|24418670|sp|Q8RVQ... 1285 0.0
dbj|BAD95324.1| putative protein [Arabidopsis thaliana] 1270 0.0
emb|CAC42898.1| putative protein [Arabidopsis thaliana] 1231 0.0
ref|NP_568270.2| exocyst complex component Sec10-related [Arabid... 1215 0.0
gb|AAB53388.1| brain secretory protein hSec10p [Homo sapiens] gi... 172 4e-41
gb|AAH41126.1| Brain secretory protein SEC10P [Homo sapiens] 172 5e-41
ref|NP_071540.1| component of rsec6/8 secretory complex p71 (71 ... 170 2e-40
emb|CAG10591.1| unnamed protein product [Tetraodon nigroviridis] 170 2e-40
ref|XP_547832.1| PREDICTED: similar to Exocyst complex component... 169 4e-40
gb|AAH49967.1| SEC10-like 1 [Mus musculus] gi|46402177|ref|NP_99... 168 6e-40
emb|CAH65252.1| hypothetical protein [Gallus gallus] gi|60302826... 165 5e-39
ref|NP_001017342.1| hypothetical protein LOC550096 [Xenopus trop... 160 2e-37
ref|XP_624953.1| PREDICTED: similar to ENSANGP00000011291 [Apis ... 159 3e-37
emb|CAH00965.1| unnamed protein product [Kluyveromyces lactis NR... 137 1e-30
ref|XP_637029.1| hypothetical protein DDB0219271 [Dictyostelium ... 134 9e-30
gb|EAK81314.1| hypothetical protein UM00329.1 [Ustilago maydis 5... 133 3e-29
gb|AAP91763.1| brain secretory protein SEC10P-like [Ciona intest... 115 4e-24
emb|CAH56255.1| hypothetical protein [Homo sapiens] 112 5e-23
gb|AAX26266.1| unknown [Schistosoma japonicum] 110 2e-22
ref|XP_610411.1| PREDICTED: similar to Exocyst complex component... 107 1e-21
>gb|AAL87123.1| SEC10 [Arabidopsis thaliana] gi|24418670|sp|Q8RVQ5|SC10_ARATH
Exocyst complex component Sec10
Length = 829
Score = 1285 bits (3324), Expect = 0.0
Identities = 665/816 (81%), Positives = 743/816 (90%), Gaps = 15/816 (1%)
Query: 18 ASSAASFPLILDIDDFKGDFSFDALFGNLVNELLPSFKLEDLEA-------EGADAVQNK 70
+SS S PLILDI+DFKGDFSFDALFGNLVN+LLPSF E+ ++ G D + N
Sbjct: 12 SSSVNSVPLILDIEDFKGDFSFDALFGNLVNDLLPSFLDEEADSGDGHGNIAGVDGLTNG 71
Query: 71 Y-----SQVATSPLFPEVEKLLSLFKDSCKELLELRKQIDGRLHNLKKDVSVQDSKHRRT 125
+ + ++++P FPEV+ LLSLFKD+CKEL++LRKQ+DGRL+ LKK+VS QDSKHR+T
Sbjct: 72 HLRGQSAPLSSAPFFPEVDGLLSLFKDACKELVDLRKQVDGRLNTLKKEVSTQDSKHRKT 131
Query: 126 LAELEKGVDGLFASFARLDSRISSVGQTAAKIGDHLQSADAQRETASQTIELIKYLMEFN 185
L E+EKGVDGLF SFARLD RISSVGQTAAKIGDHLQSADAQRETASQTI+LIKYLMEFN
Sbjct: 132 LTEIEKGVDGLFESFARLDGRISSVGQTAAKIGDHLQSADAQRETASQTIDLIKYLMEFN 191
Query: 186 SSPGDLMELSPLFSDDSRVAEAASIAQKLRSFAEEDIGRHGITAPSAVGNATASRGLEVA 245
SPGDLMELS LFSDDSRVAEAASIAQKLRSFAEEDIGR G A +A GNAT RGLEVA
Sbjct: 192 GSPGDLMELSALFSDDSRVAEAASIAQKLRSFAEEDIGRQG--ASTAAGNATPGRGLEVA 249
Query: 246 VANLQEYCNELENRLLSRFDAASQKRELTTMAECAKILSQFNRGTSAMQHYVATRPMFID 305
VANLQ+YCNELENRLLSRFDAASQ+R+L+TM+ECAKILSQFNRGTSAMQHYVATRPMFID
Sbjct: 250 VANLQDYCNELENRLLSRFDAASQRRDLSTMSECAKILSQFNRGTSAMQHYVATRPMFID 309
Query: 306 VEVMNADTRLVLGDQAAQSSPNNVARGLSSLYKEITDTVRKEAATITAVFPSPNEVMSIL 365
VEVMN+D RLVLGD +Q SP+NVARGLS+L+KEITDTVRKEAATITAVFP+PNEVM+IL
Sbjct: 310 VEVMNSDIRLVLGDHGSQPSPSNVARGLSALFKEITDTVRKEAATITAVFPTPNEVMAIL 369
Query: 366 VQRVLEQRVTALLDKLLVKPSLVNLPSMEEGGLLFYLRMLAVSYEKTQEIARDLRTVGCG 425
VQRVLEQRVT +LDK+L KPSL++ P ++EGGLL YLRMLAV+YE+TQE+A+DLR VGCG
Sbjct: 370 VQRVLEQRVTGILDKILAKPSLMSPPPVQEGGLLLYLRMLAVAYERTQELAKDLRAVGCG 429
Query: 426 DLDVEGLTESLFSSHKDEYPEYEQASLRQLYKVKMEELRAESQ-ISDSSGTIGRSKGATV 484
DLDVE LTESLFSSHKDEYPE+E+ASL+QLY+ KMEELRAESQ +S+SSGTIGRSKGA++
Sbjct: 430 DLDVEDLTESLFSSHKDEYPEHERASLKQLYQAKMEELRAESQQVSESSGTIGRSKGASI 489
Query: 485 ASSQQQISVTVVTEFVRWNEEAITRCNLFSSQPSTLATLVKAVFTCLLDQVSQYIAEGLE 544
+SS QQISVT VTEFVRWNEEAITRC LFSSQP+TLA VKA+FTCLLDQVS YI EGLE
Sbjct: 490 SSSLQQISVTFVTEFVRWNEEAITRCTLFSSQPATLAANVKAIFTCLLDQVSVYITEGLE 549
Query: 545 RARDGLTEAANLRERFVLGTSVSRRVAAAAASAAEAAAAAGESSFRSFMVAVQRSGSSVA 604
RARD L+EAA LRERFVLGTSVSRRVAAAAASAAEAAAAAGESSF+SFMVAVQR GSSVA
Sbjct: 550 RARDSLSEAAALRERFVLGTSVSRRVAAAAASAAEAAAAAGESSFKSFMVAVQRCGSSVA 609
Query: 605 IIQQYFSNSISRLLLPVDGAHAAACEEMATAMSSAEAAAYKGLQQCIETVMAEVERLLSA 664
I+QQYF+NSISRLLLPVDGAHAA+CEEM+TA+S AEAAAYKGLQQCIETVMAEV+RLLS+
Sbjct: 610 IVQQYFANSISRLLLPVDGAHAASCEEMSTALSKAEAAAYKGLQQCIETVMAEVDRLLSS 669
Query: 665 EQKATDYKSPDDGMAPDHRPTNACTRVVAYLSRVLESAFTALEGLNKQAFLSELGNRLHK 724
EQK+TDY+SPDDG+A DHRPTNAC RVVAYLSRVLESAFTALEGLNKQAFL+ELGNRL K
Sbjct: 670 EQKSTDYRSPDDGIASDHRPTNACIRVVAYLSRVLESAFTALEGLNKQAFLTELGNRLEK 729
Query: 725 VLLNHWQKYTFNPSGGLRLKRDITEYGEFVRSFNAPSVDEKFELLGILANVFIVAPESLS 784
+LL HWQK+TFNPSGGLRLKRD+ EY FV+SF APSVDEKFELLGI+ANVFIVAP+SL
Sbjct: 730 LLLTHWQKFTFNPSGGLRLKRDLNEYVGFVKSFGAPSVDEKFELLGIIANVFIVAPDSLP 789
Query: 785 TLFEGTPSIRKDAQRFIQLREDYKSAKLASKLSSLW 820
TLFEG+PSIRKDAQRFIQLREDYKSAKLA+KLSSLW
Sbjct: 790 TLFEGSPSIRKDAQRFIQLREDYKSAKLATKLSSLW 825
>dbj|BAD95324.1| putative protein [Arabidopsis thaliana]
Length = 825
Score = 1270 bits (3286), Expect = 0.0
Identities = 660/816 (80%), Positives = 739/816 (89%), Gaps = 19/816 (2%)
Query: 18 ASSAASFPLILDIDDFKGDFSFDALFGNLVNELLPSFKLEDLEA-------EGADAVQNK 70
+SS S PLILDI+DFKGDFSFDALFGNLVN+LLPSF E+ ++ G D + N
Sbjct: 12 SSSVNSVPLILDIEDFKGDFSFDALFGNLVNDLLPSFLDEEADSGDGHGNIAGVDGLTNG 71
Query: 71 Y-----SQVATSPLFPEVEKLLSLFKDSCKELLELRKQIDGRLHNLKKDVSVQDSKHRRT 125
+ + ++++P FPEV+ LLSLFKD+CKEL++LRKQ+DGRL+ LKK+VS QDSKHR+T
Sbjct: 72 HLRGQSAPLSSAPFFPEVDGLLSLFKDACKELVDLRKQVDGRLNTLKKEVSTQDSKHRKT 131
Query: 126 LAELEKGVDGLFASFARLDSRISSVGQTAAKIGDHLQSADAQRETASQTIELIKYLMEFN 185
L E+EKGVDGLF SFARLD RISSVGQTAAKIGDHLQSADAQRETASQTI+LIKYLMEFN
Sbjct: 132 LTEIEKGVDGLFESFARLDGRISSVGQTAAKIGDHLQSADAQRETASQTIDLIKYLMEFN 191
Query: 186 SSPGDLMELSPLFSDDSRVAEAASIAQKLRSFAEEDIGRHGITAPSAVGNATASRGLEVA 245
SPGDLMELS LFSDDSRVAEAASIAQKLRSFAEEDIGR G +A A GNAT RGLEVA
Sbjct: 192 GSPGDLMELSALFSDDSRVAEAASIAQKLRSFAEEDIGRQGASA--AAGNATPGRGLEVA 249
Query: 246 VANLQEYCNELENRLLSRFDAASQKRELTTMAECAKILSQFNRGTSAMQHYVATRPMFID 305
VANLQ+YCNELENRLLSRFDAASQ+R+L+TM+ECAKILSQFNRGTSAMQHYVATRPMFID
Sbjct: 250 VANLQDYCNELENRLLSRFDAASQRRDLSTMSECAKILSQFNRGTSAMQHYVATRPMFID 309
Query: 306 VEVMNADTRLVLGDQAAQSSPNNVARGLSSLYKEITDTVRKEAATITAVFPSPNEVMSIL 365
VEVMN+D RLVLGD +Q SP+NVARGLS+L+KEITDTVRKEAATITAVFP+PNEVM+IL
Sbjct: 310 VEVMNSDIRLVLGDHGSQPSPSNVARGLSALFKEITDTVRKEAATITAVFPTPNEVMAIL 369
Query: 366 VQRVLEQRVTALLDKLLVKPSLVNLPSMEEGGLLFYLRMLAVSYEKTQEIARDLRTVGCG 425
VQRVLEQRVT +LDK+L KPSL++ P ++EGGLL YLRMLAV+YE+TQE+A+DLR VGCG
Sbjct: 370 VQRVLEQRVTGILDKILAKPSLMSPPPVQEGGLLLYLRMLAVAYERTQELAKDLRAVGCG 429
Query: 426 DLDVEGLTESLFSSHKDEYPEYEQASLRQLYKVKMEELRAESQ-ISDSSGTIGRSKGATV 484
DLDVE LTESLFSSHKDEYPE+E+ASL+QLY+ KMEELRAESQ +S+SSGTIGRSKGA++
Sbjct: 430 DLDVEDLTESLFSSHKDEYPEHERASLKQLYQAKMEELRAESQQVSESSGTIGRSKGASI 489
Query: 485 ASSQQQISVTVVTEFVRWNEEAITRCNLFSSQPSTLATLVKAVFTCLLDQVSQYIAEGLE 544
+SS QQISVTVVT+FVRWNEEAITRC LFSSQP+TLA VKA+FTCLLDQVS YI EGLE
Sbjct: 490 SSSLQQISVTVVTDFVRWNEEAITRCTLFSSQPATLAANVKAIFTCLLDQVSVYITEGLE 549
Query: 545 RARDGLTEAANLRERFVLGTSVSRRVAAAAASAAEAAAAAGESSFRSFMVAVQRSGSSVA 604
RARD L+EAA LRERFVLG RRVAAAAASAAEAAAAAGESSF+SFMVAVQR GSSVA
Sbjct: 550 RARDSLSEAAALRERFVLG----RRVAAAAASAAEAAAAAGESSFKSFMVAVQRCGSSVA 605
Query: 605 IIQQYFSNSISRLLLPVDGAHAAACEEMATAMSSAEAAAYKGLQQCIETVMAEVERLLSA 664
I+QQYF+NSISRLLLPVDGAHAA+CEEM+TA+S AEAAAYKGLQQCIETVMAEV+RLLS+
Sbjct: 606 IVQQYFANSISRLLLPVDGAHAASCEEMSTALSKAEAAAYKGLQQCIETVMAEVDRLLSS 665
Query: 665 EQKATDYKSPDDGMAPDHRPTNACTRVVAYLSRVLESAFTALEGLNKQAFLSELGNRLHK 724
EQK+TDY+S DDG+A DHRPTNAC RVVAYLSRVLESAFTALEGLNKQAFL+ELGNRL K
Sbjct: 666 EQKSTDYRSTDDGIASDHRPTNACIRVVAYLSRVLESAFTALEGLNKQAFLTELGNRLEK 725
Query: 725 VLLNHWQKYTFNPSGGLRLKRDITEYGEFVRSFNAPSVDEKFELLGILANVFIVAPESLS 784
+LL HWQK+TFNPSGGLRLKRD+ EY FV+SF APSVDEKFELLGI+ANVFIVAP+SL
Sbjct: 726 LLLTHWQKFTFNPSGGLRLKRDLNEYVGFVKSFGAPSVDEKFELLGIIANVFIVAPDSLP 785
Query: 785 TLFEGTPSIRKDAQRFIQLREDYKSAKLASKLSSLW 820
TLFEG+PSIRKDAQRFIQLREDYKSAKLA+KLSSLW
Sbjct: 786 TLFEGSPSIRKDAQRFIQLREDYKSAKLATKLSSLW 821
>emb|CAC42898.1| putative protein [Arabidopsis thaliana]
Length = 863
Score = 1231 bits (3184), Expect = 0.0
Identities = 656/857 (76%), Positives = 734/857 (85%), Gaps = 63/857 (7%)
Query: 18 ASSAASFPLILDIDDFKGDFSFDALFGNLVNELLPSFKLEDLEA-------EGADAVQNK 70
+SS S PLILDI+DFKGDFSFDALFGNLVN+LLPSF E+ ++ G D + N
Sbjct: 12 SSSVNSVPLILDIEDFKGDFSFDALFGNLVNDLLPSFLDEEADSGDGHGNIAGVDGLTNG 71
Query: 71 Y-----SQVATSPLFPEVEKLLSLFKDSCKELLELRKQIDGRLHNLKKDVSVQDSKHRRT 125
+ + ++++P FPEV+ LLSLFKD+CKEL++LRKQ+DGRL+ LKK+VS QDSKHR+T
Sbjct: 72 HLRGQSAPLSSAPFFPEVDGLLSLFKDACKELVDLRKQVDGRLNTLKKEVSTQDSKHRKT 131
Query: 126 LAELEKGVDGLFASFARLDSRISSVGQTAAKIGDHLQSADAQRETASQTIELIKYLMEFN 185
L E GVDGLF SFARLD RISSVGQTAAKIGDHLQSADAQRETASQTI+LIKYLMEFN
Sbjct: 132 LTE---GVDGLFESFARLDGRISSVGQTAAKIGDHLQSADAQRETASQTIDLIKYLMEFN 188
Query: 186 SSPGDLMELSPLFSDDSRVAEAASIAQKLRSFAEEDIGRHGITAPSAVGNATASRGLEVA 245
SPGDLMELS LFSDDSRVAEAASIAQKLRSFAEEDIGR G +A A GNAT RGLEVA
Sbjct: 189 GSPGDLMELSALFSDDSRVAEAASIAQKLRSFAEEDIGRQGASA--AAGNATPGRGLEVA 246
Query: 246 VANLQEYCNELENRLLSRFDAASQKRELTTMAECAKILSQFNR----------------- 288
VANLQ+YCNELENRLLSRFDAASQ+R+L+TM+ECAKILSQ N
Sbjct: 247 VANLQDYCNELENRLLSRFDAASQRRDLSTMSECAKILSQVNALFCSMPLYNSVGKPVLL 306
Query: 289 -----------------GTSAMQHYVATRPMFIDVEVMNADTRLVLGDQAAQSSPNNVAR 331
GTSAMQHYVATRPMFIDVEVMN+D RLVLGD +Q SP+NVAR
Sbjct: 307 SKIRAKAFLTYRVITSWGTSAMQHYVATRPMFIDVEVMNSDIRLVLGDHGSQPSPSNVAR 366
Query: 332 GLSSLYKEITDTVRKEAATITAVFPSPNEVMSILVQRVLEQRVTALLDKLLVKPSLVNLP 391
GLS+L+KEITDTVRKEAATITAVFP+PNEVM+ILVQRVLEQRVT +LDK+L KPSL++ P
Sbjct: 367 GLSALFKEITDTVRKEAATITAVFPTPNEVMAILVQRVLEQRVTGILDKILAKPSLMSPP 426
Query: 392 SMEEGGLLFYLRMLAVSYEKTQEIARDLRTVGCGDLDVEGLTESLFSSHKDEYPEYEQAS 451
++EGGLL YLRMLAV+YE+TQE+A+DLR VGCGDLDVE LTESLFSSHKDEYPE+E+AS
Sbjct: 427 PVQEGGLLLYLRMLAVAYERTQELAKDLRAVGCGDLDVEDLTESLFSSHKDEYPEHERAS 486
Query: 452 LRQLYKVKMEELRAESQ-ISDSSGTIGRSKGATVASSQQQISVTVVTEFVRWNEEAITRC 510
L+QLY+ KMEELRAESQ +S+SSGTIGRSKGA+++SS QQISVTVVT+FVRWNEEAITRC
Sbjct: 487 LKQLYQAKMEELRAESQQVSESSGTIGRSKGASISSSLQQISVTVVTDFVRWNEEAITRC 546
Query: 511 NLFSSQPSTLATLVKAVFTCLLDQVSQYIAEGLERARDGLTEAANLRERFVLGTSVSRRV 570
LFSSQP+TLA VKA+FTCLLDQVS YI EGLERARD L+EAA LRERFVLG RRV
Sbjct: 547 TLFSSQPATLAANVKAIFTCLLDQVSVYITEGLERARDSLSEAAALRERFVLG----RRV 602
Query: 571 AAAAASAAEAAAAAGESSFRSFMVAVQRSGSSVAIIQQ-------YFSNSISRLLLPVDG 623
AAAAASAAEAAAAAGESSF+SFMVAVQR GSSVAI+QQ YF+NSISRLLLPVDG
Sbjct: 603 AAAAASAAEAAAAAGESSFKSFMVAVQRCGSSVAIVQQACWITFMYFANSISRLLLPVDG 662
Query: 624 AHAAACEEMATAMSSAEAAAYKGLQQCIETVMAEVERLLSAEQKATDYKSPDDGMAPDHR 683
AHAA+CEEM+TA+S AEAAAYKGLQQCIETVMAEV+RLLS+EQK+TDY+S DDG+A DHR
Sbjct: 663 AHAASCEEMSTALSKAEAAAYKGLQQCIETVMAEVDRLLSSEQKSTDYRSTDDGIASDHR 722
Query: 684 PTNACTRVVAYLSRVLESAFTALEGLNKQAFLSELGNRLHKVLLNHWQKYTFNPSGGLRL 743
PTNAC RVVAYLSRVLESAFTALEGLNKQAFL+ELGNRL K+LL HWQK+TFNPSGGLRL
Sbjct: 723 PTNACIRVVAYLSRVLESAFTALEGLNKQAFLTELGNRLEKLLLTHWQKFTFNPSGGLRL 782
Query: 744 KRDITEYGEFVRSFNAPSVDEKFELLGILANVFIVAPESLSTLFEGTPSIRKDAQRFIQL 803
KRD+ EY FV+SF APSVDEKFELLGI+ANVFIVAP+SL TLFEG+PSIRKDAQRFIQL
Sbjct: 783 KRDLNEYVGFVKSFGAPSVDEKFELLGIIANVFIVAPDSLPTLFEGSPSIRKDAQRFIQL 842
Query: 804 REDYKSAKLASKLSSLW 820
REDYKSAKLA+KLSSLW
Sbjct: 843 REDYKSAKLATKLSSLW 859
>ref|NP_568270.2| exocyst complex component Sec10-related [Arabidopsis thaliana]
Length = 858
Score = 1215 bits (3144), Expect = 0.0
Identities = 651/857 (75%), Positives = 729/857 (84%), Gaps = 68/857 (7%)
Query: 18 ASSAASFPLILDIDDFKGDFSFDALFGNLVNELLPSFKLEDLEA-------EGADAVQNK 70
+SS S PLILDI+DFKGDFSFDALFGNLVN+LLPSF E+ ++ G D + N
Sbjct: 12 SSSVNSVPLILDIEDFKGDFSFDALFGNLVNDLLPSFLDEEADSGDGHGNIAGVDGLTNG 71
Query: 71 Y-----SQVATSPLFPEVEKLLSLFKDSCKELLELRKQIDGRLHNLKKDVSVQDSKHRRT 125
+ + ++++P FPEV+ LLSLFKD+CKEL++LRKQ+DGRL+ LKK+VS QDSKHR+T
Sbjct: 72 HLRGQSAPLSSAPFFPEVDGLLSLFKDACKELVDLRKQVDGRLNTLKKEVSTQDSKHRKT 131
Query: 126 LAELEKGVDGLFASFARLDSRISSVGQTAAKIGDHLQSADAQRETASQTIELIKYLMEFN 185
L E GVDGLF SFARLD RISSVGQTAAKIGDHLQSADAQRETASQTI+LIKYLMEFN
Sbjct: 132 LTE---GVDGLFESFARLDGRISSVGQTAAKIGDHLQSADAQRETASQTIDLIKYLMEFN 188
Query: 186 SSPGDLMELSPLFSDDSRVAEAASIAQKLRSFAEEDIGRHGITAPSAVGNATASRGLEVA 245
SPGDLMELS LFSDDSRVAEAASIAQKLRSFAEEDIGR G +A A GNAT RGLEVA
Sbjct: 189 GSPGDLMELSALFSDDSRVAEAASIAQKLRSFAEEDIGRQGASA--AAGNATPGRGLEVA 246
Query: 246 VANLQEYCNELENRLLSRFDAASQKRELTTMAECAKILSQFNR----------------- 288
VANLQ+YCNELENRLLSRFDAASQ+R+L+TM+ECAKILSQ N
Sbjct: 247 VANLQDYCNELENRLLSRFDAASQRRDLSTMSECAKILSQVNALFCSMPLYNSVGKPVLL 306
Query: 289 -----------------GTSAMQHYVATRPMFIDVEVMNADTRLVLGDQAAQSSPNNVAR 331
GTSAMQHYVATRPMFIDVEVMN+D RLVLGD +Q SP+NVAR
Sbjct: 307 SKIRAKAFLTYRVITSWGTSAMQHYVATRPMFIDVEVMNSDIRLVLGDHGSQPSPSNVAR 366
Query: 332 GLSSLYKEITDTVRKEAATITAVFPSPNEVMSILVQRVLEQRVTALLDKLLVKPSLVNLP 391
GLS+L+KEITDTVRKEAATITAVFP+PNEVM+ILVQRVLEQRVT +LDK+L KPSL++ P
Sbjct: 367 GLSALFKEITDTVRKEAATITAVFPTPNEVMAILVQRVLEQRVTGILDKILAKPSLMSPP 426
Query: 392 SMEEGGLLFYLRMLAVSYEKTQEIARDLRTVGCGDLDVEGLTESLFSSHKDEYPEYEQAS 451
++EGGLL YLRMLAV+YE+TQE+A+DLR VGCGDLDVE LTESLFSSHKDEYPE+E+AS
Sbjct: 427 PVQEGGLLLYLRMLAVAYERTQELAKDLRAVGCGDLDVEDLTESLFSSHKDEYPEHERAS 486
Query: 452 LRQLYKVKMEELRAESQ-ISDSSGTIGRSKGATVASSQQQISVTVVTEFVRWNEEAITRC 510
L+QLY+ KMEELRAESQ +S+SSGTIGRSKGA+++SS QQISVTVVT+FVRWNEEAITRC
Sbjct: 487 LKQLYQAKMEELRAESQQVSESSGTIGRSKGASISSSLQQISVTVVTDFVRWNEEAITRC 546
Query: 511 NLFSSQPSTLATLVKAVFTCLLDQVSQYIAEGLERARDGLTEAANLRERFVLGTSVSRRV 570
LFSSQP+TLA VKA+FTCLLDQVS YI EGLERARD L+EAA LRERFVLG RRV
Sbjct: 547 TLFSSQPATLAANVKAIFTCLLDQVSVYITEGLERARDSLSEAAALRERFVLG----RRV 602
Query: 571 AAAAASAAEAAAAAGESSFRSFMVAVQRSGSSVAIIQQ-------YFSNSISRLLLPVDG 623
AAAAASAAEAAAAAGESSF+SFMVAVQR GSSVAI+QQ YF+NSISRLLLPVDG
Sbjct: 603 AAAAASAAEAAAAAGESSFKSFMVAVQRCGSSVAIVQQACWITFMYFANSISRLLLPVDG 662
Query: 624 AHAAACEEMATAMSSAEAAAYKGLQQCIETVMAEVERLLSAEQKATDYKSPDDGMAPDHR 683
AHAA+CEEM+TA+S AEAAAYKGLQQCIETVMAEV+RLLS+EQK+TDY+S DDG+A DHR
Sbjct: 663 AHAASCEEMSTALSKAEAAAYKGLQQCIETVMAEVDRLLSSEQKSTDYRSTDDGIASDHR 722
Query: 684 PTNACTRVVAYLSRVLESAFTALEGLNKQAFLSELGNRLHKVLLNHWQKYTFNPSGGLRL 743
PTNAC RVVAYLSRVLESAFTALEGLNKQAFL+ELGNRL K+LL HWQK+TFNPSGGLRL
Sbjct: 723 PTNACIRVVAYLSRVLESAFTALEGLNKQAFLTELGNRLEKLLLTHWQKFTFNPSGGLRL 782
Query: 744 KRDITEYGEFVRSFNAPSVDEKFELLGILANVFIVAPESLSTLFEGTPSIRKDAQRFIQL 803
KRD+ EY FV+SF APSVDEKFELLGI+ANVFIVAP+SL TLFEG+PSIRKDAQ
Sbjct: 783 KRDLNEYVGFVKSFGAPSVDEKFELLGIIANVFIVAPDSLPTLFEGSPSIRKDAQ----- 837
Query: 804 REDYKSAKLASKLSSLW 820
REDYKSAKLA+KLSSLW
Sbjct: 838 REDYKSAKLATKLSSLW 854
>gb|AAB53388.1| brain secretory protein hSec10p [Homo sapiens]
gi|5730037|ref|NP_006535.1| brain secretory protein
SEC10P [Homo sapiens] gi|24418661|sp|O00471|SC10_HUMAN
Exocyst complex component Sec10 (hSec10)
Length = 708
Score = 172 bits (436), Expect = 4e-41
Identities = 173/757 (22%), Positives = 317/757 (41%), Gaps = 111/757 (14%)
Query: 84 EKLLSLFKDSCKELLELRKQIDGRLHNLKKDVSVQDSKHRRTLAELEKGVDGLFASFARL 143
++LL F + +EL + ++I ++ L++ + + + + EL+K F F L
Sbjct: 40 KRLLEEFVNHIQELQIMDERIQRKVEKLEQQCQKEAKEFAKKVQELQKSNQVAFQHFQEL 99
Query: 144 DSRISSVGQTAAKIGDHLQSADAQRETASQTIELIKYLMEFNSSPGDLMELSPLFSDDSR 203
D IS V +GD L+ + R+ A + +L+KY EF G+L S +F++ +
Sbjct: 100 DEHISYVATKVCHLGDQLEGVNTPRQRAVEAQKLMKYFNEFLD--GELK--SDVFTNSEK 155
Query: 204 VAEAASIAQKLRSFAEEDIGRHGITAPSAVGNATASRGLEVAVANLQEYCNELENRLLSR 263
+ EAA I QKL A+E R EV +Y ++LE +L+
Sbjct: 156 IKEAADIIQKLHLIAQE---------------LPFDRFSEVKSKIASKY-HDLECQLIQE 199
Query: 264 FDAASQKRELTTMAECAKILSQFNRGTSAMQHYVATRPMFIDVEVMNADTRLVLGDQAAQ 323
F +A ++ E++ M E A +L F + + Y+ Q
Sbjct: 200 FTSAQRRGEISRMREVAAVLLHFKGYSHCVDVYIKQC-------------------QEGA 240
Query: 324 SSPNNVARGLSSLYKEITDTVRKEAATITAVFPSPNEVMSILVQRVLEQRVTALLDKLLV 383
N++ L + + V +F +P V++ L+Q V E ++ + + +
Sbjct: 241 YLRNDIFEDAGILCQRVNKQVGD-------IFSNPETVLAKLIQNVFEIKLQSFVKE--- 290
Query: 384 KPSLVNLPSMEEGGLLFYLRMLAVSYEKTQEIARDLRTVGCG---DLDVEGLTESLFSSH 440
L + YL+ L Y +T ++ L G + L +S+F S+
Sbjct: 291 -----QLEECRKSDAEQYLKNLYDLYTRTTNLSSKLMEFNLGTDKQTFLSKLIKSIFISY 345
Query: 441 KDEYPEYEQASLR--------QLYKVKMEELRA--ESQISDSSGTIGRSKGATVASS--- 487
+ Y E E L+ + Y K + R+ I D I + + S
Sbjct: 346 LENYIEVETGYLKSRSAMILQRYYDSKNHQKRSIGTGGIQDLKERIRQRTNLPLGPSIDT 405
Query: 488 --QQQISVTVVTEFVRWNEEAITRCNLFSSQPSTLATLVKAVFTCLLDQVS-QYIAEGLE 544
+ +S VV ++ ++A RC+ S PS L +FT L++ + ++I LE
Sbjct: 406 HGETFLSQEVVVNLLQETKQAFERCHRLSD-PSDLPRNAFRIFTILVEFLCIEHIDYALE 464
Query: 545 RARDGLTEAANLRERFVLGTSVSRRVAAAAASAAEAAAAAGESSFRSFMVAVQRSGSSVA 604
G+ + + F+ VQ++ +
Sbjct: 465 TGLAGIPSSDSRNANLY------------------------------FLDVVQQANTIFH 494
Query: 605 IIQQYFSNSISRLLLPVDGAHAAAC-EEMATAMSSAEAAAYKGLQQCIETVMAEVERLLS 663
+ + F++ + L+ + C ++ + E G+ + + ++ +++ +L+
Sbjct: 495 LFDKQFNDHLMPLIS--SSPKLSECLQKKKEIIEQMEMKLDTGIDRTLNCMIGQMKHILA 552
Query: 664 AEQKATDYKSPDDGMAPDHRPTNACTRVVAYLSRVLESAFTALEGLNKQAFLSELGNRLH 723
AEQK TD+K P+D + TNAC +V AY+ + +E +++G N L ELG R H
Sbjct: 553 AEQKKTDFK-PEDENNVLIQYTNACVKVCAYVRKQVEKIKNSMDGKNVDTVLMELGVRFH 611
Query: 724 KVLLNHWQKYTFNPSGGLRLKRDITEYGEFVRSFNAPSVDEKFELLGILANVFIVAPESL 783
+++ H Q+Y+++ GG+ D+ EY + + F P V F+ L L N+ +VAP++L
Sbjct: 612 RLIYEHLQQYSYSCMGGMLAICDVAEYRKCAKDFKIPMVLHLFDTLHALCNLLVVAPDNL 671
Query: 784 STLFEGTPSIRKD---AQRFIQLREDYKSAKLASKLS 817
+ G D F+QLR DY+SA+LA S
Sbjct: 672 KQVCSGEQLANLDKNILHSFVQLRADYRSARLARHFS 708
>gb|AAH41126.1| Brain secretory protein SEC10P [Homo sapiens]
Length = 708
Score = 172 bits (435), Expect = 5e-41
Identities = 173/757 (22%), Positives = 317/757 (41%), Gaps = 111/757 (14%)
Query: 84 EKLLSLFKDSCKELLELRKQIDGRLHNLKKDVSVQDSKHRRTLAELEKGVDGLFASFARL 143
++LL F + +EL + ++I ++ L++ + + + + EL+K F F L
Sbjct: 40 KRLLEEFVNHIQELQIMDERIQRKVEKLEQQCQKEAKEFAKKVQELQKSNQVAFQHFQEL 99
Query: 144 DSRISSVGQTAAKIGDHLQSADAQRETASQTIELIKYLMEFNSSPGDLMELSPLFSDDSR 203
D IS V +GD L+ + R+ A + +L+KY EF G+L S +F++ +
Sbjct: 100 DEHISYVATKVCHLGDQLEGVNTPRQRAVEAQKLMKYFNEFLD--GELK--SDVFTNSEK 155
Query: 204 VAEAASIAQKLRSFAEEDIGRHGITAPSAVGNATASRGLEVAVANLQEYCNELENRLLSR 263
+ EAA I QKL A+E R EV +Y ++LE +L+
Sbjct: 156 IKEAADIIQKLHLIAQE---------------LPFDRFSEVKSKIASKY-HDLECQLIQE 199
Query: 264 FDAASQKRELTTMAECAKILSQFNRGTSAMQHYVATRPMFIDVEVMNADTRLVLGDQAAQ 323
F +A ++ E++ M E A +L F + + Y+ Q
Sbjct: 200 FTSAQRRGEISRMREVAAVLLHFKGYSHCVDVYIKQC-------------------QEGA 240
Query: 324 SSPNNVARGLSSLYKEITDTVRKEAATITAVFPSPNEVMSILVQRVLEQRVTALLDKLLV 383
N++ L + + V +F +P V++ L+Q V E ++ + + +
Sbjct: 241 YLRNDIFEDAGILCQRVNKQVGD-------IFSNPETVLAKLIQNVFEIKLQSFVKE--- 290
Query: 384 KPSLVNLPSMEEGGLLFYLRMLAVSYEKTQEIARDLRTVGCG---DLDVEGLTESLFSSH 440
L + YL+ L Y +T ++ L G + L +S+F S+
Sbjct: 291 -----QLEECRKSDAEQYLKNLYDLYTRTTNLSSKLMEFNLGTDKQTFLSKLIKSIFISY 345
Query: 441 KDEYPEYEQASLR--------QLYKVKMEELRA--ESQISDSSGTIGRSKGATVASS--- 487
+ Y E E L+ + Y K + R+ I D I + + S
Sbjct: 346 LENYIEVETGYLKSRSAMILQRYYDSKNHQKRSIGTGGIQDLKERIRQRTNLPLGPSIDT 405
Query: 488 --QQQISVTVVTEFVRWNEEAITRCNLFSSQPSTLATLVKAVFTCLLDQVS-QYIAEGLE 544
+ +S VV ++ ++A RC+ S PS L +FT L++ + ++I LE
Sbjct: 406 HGETFLSQEVVVNLLQETKQAFERCHRLSD-PSDLPRNAFRIFTILVEFLCIEHIDYALE 464
Query: 545 RARDGLTEAANLRERFVLGTSVSRRVAAAAASAAEAAAAAGESSFRSFMVAVQRSGSSVA 604
G+ + + F+ VQ++ +
Sbjct: 465 TGLAGIPSSDSRNANLY------------------------------FLDVVQQANTIFH 494
Query: 605 IIQQYFSNSISRLLLPVDGAHAAAC-EEMATAMSSAEAAAYKGLQQCIETVMAEVERLLS 663
+ + F++ + L+ + C ++ + E G+ + + ++ +++ +L+
Sbjct: 495 LFDKQFNDHLMPLIS--SSPKLSECLQKKKEIIEQMEMKLDTGIDRTLNCMIRQMKHILA 552
Query: 664 AEQKATDYKSPDDGMAPDHRPTNACTRVVAYLSRVLESAFTALEGLNKQAFLSELGNRLH 723
AEQK TD+K P+D + TNAC +V AY+ + +E +++G N L ELG R H
Sbjct: 553 AEQKKTDFK-PEDENNVLIQYTNACVKVCAYVRKQVEKIKNSMDGKNVDTVLMELGVRFH 611
Query: 724 KVLLNHWQKYTFNPSGGLRLKRDITEYGEFVRSFNAPSVDEKFELLGILANVFIVAPESL 783
+++ H Q+Y+++ GG+ D+ EY + + F P V F+ L L N+ +VAP++L
Sbjct: 612 RLIYEHLQQYSYSCMGGMLAICDVAEYRKCAKDFKIPMVLHLFDTLHALCNLLVVAPDNL 671
Query: 784 STLFEGTPSIRKD---AQRFIQLREDYKSAKLASKLS 817
+ G D F+QLR DY+SA+LA S
Sbjct: 672 KQVCSGEQLANLDKNILHSFVQLRADYRSARLARHFS 708
>ref|NP_071540.1| component of rsec6/8 secretory complex p71 (71 kDa) [Rattus
norvegicus] gi|1903420|gb|AAC53096.1| p71 [Rattus
norvegicus] gi|7514120|pir||JC6329 yeast secretory
protein homolog rsec6/8 complex - rat
gi|24418658|sp|P97878|SC10_RAT Exocyst complex component
Sec10 (71 kDa component of rsec6/8 secretory complex)
(p71)
Length = 708
Score = 170 bits (430), Expect = 2e-40
Identities = 176/757 (23%), Positives = 320/757 (42%), Gaps = 111/757 (14%)
Query: 84 EKLLSLFKDSCKELLELRKQIDGRLHNLKKDVSVQDSKHRRTLAELEKGVDGLFASFARL 143
++LL F + +EL + ++I ++ L++ + + + + EL+K F F L
Sbjct: 40 KRLLEEFVNHIQELQIMDERIQRKVEKLEQQCQKEAKEFAKKVQELQKSNQVAFQHFQEL 99
Query: 144 DSRISSVGQTAAKIGDHLQSADAQRETASQTIELIKYLMEFNSSPGDLMELSPLFSDDSR 203
D IS V +GD L+ + R+ A + +L+KY EF G+L S +F++ +
Sbjct: 100 DEHISYVATKVCHLGDQLEGVNTPRQRAVEAQKLMKYFNEFLD--GELK--SDVFTNPEK 155
Query: 204 VAEAASIAQKLRSFAEEDIGRHGITAPSAVGNATASRGLEVAVANLQEYCNELENRLLSR 263
+ EAA + QKL A+E R EV +Y ++LE +L+
Sbjct: 156 IKEAADVIQKLHLIAQE---------------LPFDRFSEVKSKIASKY-HDLECQLIQE 199
Query: 264 FDAASQKRELTTMAECAKILSQFNRGTSAMQHYVATRPMFIDVEVMNADTRLVLGDQAAQ 323
F +A ++ E++ M E A +L F +G S IDV + L + +
Sbjct: 200 FTSAQRRGEVSRMREVAAVLLHF-KGYSHC----------IDVYIKQCQEGAYLRNDIFE 248
Query: 324 SSPNNVARGLSSLYKEITDTVRKEAATITAVFPSPNEVMSILVQRVLEQRVTALLDKLLV 383
+ R + K++ D +F +P V++ L+Q V E ++ + +
Sbjct: 249 DAAILCQR----VNKQVGD-----------IFSNPEAVLAKLIQNVFEVKLQSFVKD--- 290
Query: 384 KPSLVNLPSMEEGGLLFYLRMLAVSYEKTQEIARDLRTVGCG---DLDVEGLTESLFSSH 440
L + YL+ L Y +T ++ L G + L +S+F S+
Sbjct: 291 -----QLEECRKSDAEQYLKSLYDLYTRTTSLSSKLMEFNLGTDKQTFLSKLIKSIFVSY 345
Query: 441 KDEYPEYEQASLR--------QLYKVKMEELRA--ESQISDSSGTIGRSKGATVASS--- 487
+ Y E E L+ + Y K + R+ I D I + + S
Sbjct: 346 LENYIEVEIGYLKSRSAMILQRYYDSKNHQKRSIGTGGIQDLKERIRQRTNLPLGPSIDT 405
Query: 488 --QQQISVTVVTEFVRWNEEAITRCNLFSSQPSTLATLVKAVFTCLLDQVS-QYIAEGLE 544
+ +S VV ++ ++A RC+ S PS L +FT L++ + ++I LE
Sbjct: 406 HGETFLSQEVVVNLLQETKQAFERCHRLSD-PSDLPRNAFRIFTILVEFLCIEHIDYALE 464
Query: 545 RARDGLTEAANLRERFVLGTSVSRRVAAAAASAAEAAAAAGESSFRSFMVAVQRSGSSVA 604
G+ + + F+ VQ++ +
Sbjct: 465 TGLAGIPSSDSRNANLY------------------------------FLDVVQQANTIFH 494
Query: 605 IIQQYFSNSISRLLLPVDGAHAAAC-EEMATAMSSAEAAAYKGLQQCIETVMAEVERLLS 663
+ + F++ + L+ + C ++ + E G+ + + ++ +++ +L+
Sbjct: 495 LFDKQFNDHLMPLIS--SSPKLSECLQKKKEIIEQMEMKLDTGIDRTLNCMIGQMKHILA 552
Query: 664 AEQKATDYKSPDDGMAPDHRPTNACTRVVAYLSRVLESAFTALEGLNKQAFLSELGNRLH 723
AEQK TD+K P+D + TNAC +V AY+ + +E +++G N L ELG R H
Sbjct: 553 AEQKKTDFK-PEDENNVLIQYTNACVKVCAYVRKQVEKIKNSMDGKNVDTVLMELGVRFH 611
Query: 724 KVLLNHWQKYTFNPSGGLRLKRDITEYGEFVRSFNAPSVDEKFELLGILANVFIVAPESL 783
++ H Q+Y+++ GG+ D+ EY + + F P V F+ L L N+ +VAP++L
Sbjct: 612 RLTYEHLQQYSYSCMGGMLAICDVAEYRKCAKDFKIPMVLHLFDTLHALCNLLVVAPDNL 671
Query: 784 STLFEGTPSIRKD---AQRFIQLREDYKSAKLASKLS 817
+ G D F+QLR DY+SA+LA S
Sbjct: 672 KQVCSGEQLANLDKNILHSFVQLRADYRSARLARHFS 708
>emb|CAG10591.1| unnamed protein product [Tetraodon nigroviridis]
Length = 699
Score = 170 bits (430), Expect = 2e-40
Identities = 175/757 (23%), Positives = 320/757 (42%), Gaps = 111/757 (14%)
Query: 84 EKLLSLFKDSCKELLELRKQIDGRLHNLKKDVSVQDSKHRRTLAELEKGVDGLFASFARL 143
++LL F++ +EL +L ++I ++ L+ + + + +L + F F L
Sbjct: 31 KRLLEEFENHIEELKQLDEKIQRKVEKLEHQCQREAKEFAHKVQDLYRSNQVAFQHFQEL 90
Query: 144 DSRISSVGQTAAKIGDHLQSADAQRETASQTIELIKYLMEFNSSPGDLMELSPLFSDDSR 203
D IS V +GD L+ + R+ A + L+ Y EF GDL S +F++ +
Sbjct: 91 DEHISYVATKVCHLGDQLEGVNTPRQRAVEAQRLMTYFNEFLD--GDLR--SDVFNNPDK 146
Query: 204 VAEAASIAQKLRSFAEEDIGRHGITAPSAVGNATASRGLEVAVANLQEYCNELENRLLSR 263
+ EAA I QKL A+E R +V +Y ++LE +L+
Sbjct: 147 IKEAADIIQKLHLIAQE---------------LPFDRFPDVKAKIASKY-HDLERQLIQE 190
Query: 264 FDAASQKRELTTMAECAKILSQFNRGTSAMQHYVATRPMFIDVEVMNADTRLVLGDQAAQ 323
F AA ++ E+ M E A +L F + Y+ Q
Sbjct: 191 FTAAQRRGEIGRMREVAAVLLHFKGYAHCVDVYIKQC-------------------QEGA 231
Query: 324 SSPNNVARGLSSLYKEITDTVRKEAATITAVFPSPNEVMSILVQRVLEQRVTALLDKLLV 383
N+V + L + + V + VF SP VM+ L+Q + E ++ + +
Sbjct: 232 YLRNDVFEDTAILCQRVNKQVGE-------VFSSPETVMAKLIQNIFENKLQSHVKD--- 281
Query: 384 KPSLVNLPSMEEGGLLFYLRMLAVSYEKTQEIARDLRTVGCGD---LDVEGLTESLFSSH 440
L + YL+ L Y +T +A L G + L +S+FS++
Sbjct: 282 -----RLDGTRHSDVEQYLKNLYDLYTRTTALATKLTEFNLGSDKHTFLSKLIKSIFSTY 336
Query: 441 KDEYPEYEQASLRQLYKVKME---ELRAESQISDSSGTIGRSK-----------GATVAS 486
+ Y + E+ LR + ++ + + + S +G+I K G + +
Sbjct: 337 LESYIDMEKEYLRNRGAMILQRYYDSKNHQKRSIGTGSIQELKERIRQRTNLSLGPVIDT 396
Query: 487 SQQQ-ISVTVVTEFVRWNEEAITRCNLFSSQPSTLATLVKAVFTCLLDQVS-QYIAEGLE 544
+ +S +V ++ A RC+ S PS L ++F L++ + ++I LE
Sbjct: 397 HGETFLSPELVVNLLQETRHAFERCHRLSD-PSDLPKNAFSIFLLLVEHLCVEHIDYALE 455
Query: 545 RARDGLTEAANLRERFVLGTSVSRRVAAAAASAAEAAAAAGESSFRSFMVAVQRSGSSVA 604
+ +A +++A +++ F+ VQ++ S
Sbjct: 456 -------------------------IGLSAIPSSDA-----KNANLYFLDVVQQANSIFH 485
Query: 605 IIQQYFSNSISRLLLPVDGAHAAAC-EEMATAMSSAEAAAYKGLQQCIETVMAEVERLLS 663
+ + F + + L+ A C + + E G+ + I ++ +++ +L+
Sbjct: 486 LFDKQFHDQLMPLIS--SSPKLAECLHKKKEVIEQMEVKLDTGIDRTINCMVGQMKYILA 543
Query: 664 AEQKATDYKSPDDGMAPDHRPTNACTRVVAYLSRVLESAFTALEGLNKQAFLSELGNRLH 723
EQK TD+K P+D + T AC++V Y+SR +E +++G N L+ELG R H
Sbjct: 544 TEQKKTDFK-PEDENNVMIQYTTACSKVCVYVSRQVEHVRKSMDGKNVDTVLTELGVRFH 602
Query: 724 KVLLNHWQKYTFNPSGGLRLKRDITEYGEFVRSFNAPSVDEKFELLGILANVFIVAPESL 783
+++ H Q+Y+++ GG+ D+ EY + F P V + F+ L L N+ +VAP++L
Sbjct: 603 RLIHEHLQQYSYSSMGGMLAICDVAEYRRCAKDFRVPLVLQLFDTLHALCNLLVVAPDNL 662
Query: 784 STLFEG---TPSIRKDAQRFIQLREDYKSAKLASKLS 817
+ G T R F+QLR DY+SA+L S
Sbjct: 663 KQVCSGEQLTNLDRNLLHAFVQLRVDYRSARLGRHFS 699
>ref|XP_547832.1| PREDICTED: similar to Exocyst complex component Sec10 (hSec10)
[Canis familiaris]
Length = 734
Score = 169 bits (427), Expect = 4e-40
Identities = 172/757 (22%), Positives = 313/757 (40%), Gaps = 126/757 (16%)
Query: 84 EKLLSLFKDSCKELLELRKQIDGRLHNLKKDVSVQDSKHRRTLAELEKGVDGLFASFARL 143
++LL F + +EL + ++I ++ L++ + + + + EL+K F F L
Sbjct: 81 KRLLEEFVNHIQELQIMDERIQRKVEKLEQQCQKEAKEFAKKVQELQKSNQVAFQHFQEL 140
Query: 144 DSRISSVGQTAAKIGDHLQSADAQRETASQTIELIKYLMEFNSSPGDLMELSPLFSDDSR 203
D IS V +GD L+ + R+ A + +L+KY EF G+L S +F++ +
Sbjct: 141 DEHISYVATKVCHLGDQLEGVNTPRQRAVEAQKLMKYFNEFLD--GELK--SDVFTNSEK 196
Query: 204 VAEAASIAQKLRSFAEEDIGRHGITAPSAVGNATASRGLEVAVANLQEYCNELENRLLSR 263
+ EAA I QKL A+E R EV +Y ++LE +L+
Sbjct: 197 IKEAADIIQKLHLIAQE---------------LPFDRFSEVKSKIASKY-HDLECQLIQE 240
Query: 264 FDAASQKRELTTMAECAKILSQFNRGTSAMQHYVATRPMFIDVEVMNADTRLVLGDQAAQ 323
F +A ++ E++ M E A +L F +F D ++ +GD
Sbjct: 241 FTSAQRRGEISRMREVAAVLLHFKGA-------YLRNDIFEDAAILCQRVNKQVGD---- 289
Query: 324 SSPNNVARGLSSLYKEITDTVRKEAATITAVFPSPNEVMSILVQRVLEQRVTALLDKLLV 383
+F +P V++ L+Q V E ++ + +
Sbjct: 290 ------------------------------IFSNPETVLAKLIQNVFEIKLQSFVKD--- 316
Query: 384 KPSLVNLPSMEEGGLLFYLRMLAVSYEKTQEIARDLRTVGCG---DLDVEGLTESLFSSH 440
L + YL+ L Y +T ++ L G + L +S+F S+
Sbjct: 317 -----QLEECRKSDAEQYLKNLYDLYTRTTNLSSKLMEFNLGTDKQTFLSKLIKSIFISY 371
Query: 441 KDEYPEYEQASLR--------QLYKVKMEELRA--ESQISDSSGTIGRSKGATVASS--- 487
+ Y E E L+ + Y K + R+ I D I + + S
Sbjct: 372 LENYIEVETGYLKSRSAMILQRYYDSKNHQKRSIGTGGIQDLKERIRQRTNLPLGPSIDT 431
Query: 488 --QQQISVTVVTEFVRWNEEAITRCNLFSSQPSTLATLVKAVFTCLLDQVS-QYIAEGLE 544
+ +S VV ++ ++A RC+ S PS L +FT L++ + ++I LE
Sbjct: 432 HGETFLSQEVVVNLLQETKQAFERCHRLSD-PSDLPRNAFRIFTILVEFLCIEHIDYALE 490
Query: 545 RARDGLTEAANLRERFVLGTSVSRRVAAAAASAAEAAAAAGESSFRSFMVAVQRSGSSVA 604
G+ + + F+ VQ++ +
Sbjct: 491 TGLAGIPSSDSRNANLY------------------------------FLDVVQQANTIFH 520
Query: 605 IIQQYFSNSISRLLLPVDGAHAAAC-EEMATAMSSAEAAAYKGLQQCIETVMAEVERLLS 663
+ + F++ + L+ + C ++ + E G+ + + ++ +++ +L+
Sbjct: 521 LFDKQFNDHLMPLIS--SSPKLSECLQKKKEIIEQMEMKLDTGIDRTLNCMIGQMKHILA 578
Query: 664 AEQKATDYKSPDDGMAPDHRPTNACTRVVAYLSRVLESAFTALEGLNKQAFLSELGNRLH 723
AEQK TD+K P+D + TNAC +V AY+ + +E +++G N L ELG R H
Sbjct: 579 AEQKKTDFK-PEDENNVLIQYTNACVKVCAYVRKQVEKIKNSMDGKNVDTVLMELGVRFH 637
Query: 724 KVLLNHWQKYTFNPSGGLRLKRDITEYGEFVRSFNAPSVDEKFELLGILANVFIVAPESL 783
+++ H Q+Y+++ GG+ D+ EY + + F P V + F+ L L N+ +VAP++L
Sbjct: 638 RLIYEHLQQYSYSCMGGMLAICDVAEYRKCAKDFKIPLVLQLFDTLHALCNLLVVAPDNL 697
Query: 784 STLFEGTPSIRKD---AQRFIQLREDYKSAKLASKLS 817
+ G D F+QLR DY+SA+LA S
Sbjct: 698 KQVCSGEQLANLDKNILHSFVQLRADYRSARLARHFS 734
>gb|AAH49967.1| SEC10-like 1 [Mus musculus] gi|46402177|ref|NP_997097.1| SEC10-like
1 [Mus musculus]
Length = 708
Score = 168 bits (426), Expect = 6e-40
Identities = 175/757 (23%), Positives = 320/757 (42%), Gaps = 111/757 (14%)
Query: 84 EKLLSLFKDSCKELLELRKQIDGRLHNLKKDVSVQDSKHRRTLAELEKGVDGLFASFARL 143
++LL F + +EL + ++I ++ L++ + + + + EL+K F F L
Sbjct: 40 KRLLEEFVNHIQELQIMDERIQRKVEKLEQQCQKEAKEFAKKVQELQKSNQVAFQHFQEL 99
Query: 144 DSRISSVGQTAAKIGDHLQSADAQRETASQTIELIKYLMEFNSSPGDLMELSPLFSDDSR 203
D IS V +GD L+ + R+ A + +L+KY EF G+L S +F++ +
Sbjct: 100 DEHISYVATKVCHLGDQLEGVNTPRQRAVEAQKLMKYFNEFLD--GELK--SDVFTNSEK 155
Query: 204 VAEAASIAQKLRSFAEEDIGRHGITAPSAVGNATASRGLEVAVANLQEYCNELENRLLSR 263
+ EAA + QKL A+E R EV +Y ++LE +L+
Sbjct: 156 IKEAADVIQKLHLIAQE---------------LPFDRFSEVKSKIASKY-HDLECQLIQE 199
Query: 264 FDAASQKRELTTMAECAKILSQFNRGTSAMQHYVATRPMFIDVEVMNADTRLVLGDQAAQ 323
F +A ++ E++ M E A +L F +G S IDV + L + +
Sbjct: 200 FTSAQRRGEVSRMREVAAVLLHF-KGYSHC----------IDVYIKQCQEGAYLRNDIFE 248
Query: 324 SSPNNVARGLSSLYKEITDTVRKEAATITAVFPSPNEVMSILVQRVLEQRVTALLDKLLV 383
+ R + K++ D +F +P V++ L+Q V E ++ + +
Sbjct: 249 DAAILCQR----VNKQVGD-----------IFSNPEAVLAKLIQSVFEIKLQSFVKD--- 290
Query: 384 KPSLVNLPSMEEGGLLFYLRMLAVSYEKTQEIARDLRTVGCG---DLDVEGLTESLFSSH 440
L + YL+ L Y +T ++ L G + L +S+F S+
Sbjct: 291 -----QLEECRKSDAEQYLKSLYDLYTRTTGLSSKLMEFNLGTDKQTFLSKLIKSIFISY 345
Query: 441 KDEYPEYEQASLR--------QLYKVKMEELRA--ESQISDSSGTIGRSKGATVASS--- 487
+ Y E E L+ + Y K + R+ I D I + + S
Sbjct: 346 LENYIEVEIGYLKSRSAMILQRYYDSKNHQKRSIGTGGIQDLKERIRQRTNLPLGPSIDT 405
Query: 488 --QQQISVTVVTEFVRWNEEAITRCNLFSSQPSTLATLVKAVFTCLLDQVS-QYIAEGLE 544
+ +S VV ++ ++A RC+ S PS L +FT L++ + ++I LE
Sbjct: 406 HGETFLSQEVVVNLLQETKQAFERCHRLSD-PSDLPRNAFRIFTILVEFLCIEHIDYALE 464
Query: 545 RARDGLTEAANLRERFVLGTSVSRRVAAAAASAAEAAAAAGESSFRSFMVAVQRSGSSVA 604
G+ + + F+ VQ++ +
Sbjct: 465 TGLAGIPSSDSRNANLY------------------------------FLDVVQQANTIFH 494
Query: 605 IIQQYFSNSISRLLLPVDGAHAAAC-EEMATAMSSAEAAAYKGLQQCIETVMAEVERLLS 663
+ + F++ + L+ + C ++ + E G+ + + ++ +++ +L+
Sbjct: 495 LFDKQFNDHLMPLIS--SSPKLSECLQKKKEIIEQMEMKLDTGIDRTLNCMIGQMKHILA 552
Query: 664 AEQKATDYKSPDDGMAPDHRPTNACTRVVAYLSRVLESAFTALEGLNKQAFLSELGNRLH 723
AEQK TD+K P+D + TNAC +V Y+ + +E +++G N L ELG R H
Sbjct: 553 AEQKKTDFK-PEDENNVLIQYTNACVKVCVYVRKQVEKIKNSMDGKNVDTVLMELGVRFH 611
Query: 724 KVLLNHWQKYTFNPSGGLRLKRDITEYGEFVRSFNAPSVDEKFELLGILANVFIVAPESL 783
+++ H Q+Y+++ GG+ D+ EY + + F P V F+ L L N+ +VAP++L
Sbjct: 612 RLIYEHLQQYSYSCMGGMLAICDVAEYRKCAKDFKIPMVLHLFDTLHALCNLLVVAPDNL 671
Query: 784 STLFEGTPSIRKD---AQRFIQLREDYKSAKLASKLS 817
+ G D F+QLR DY+SA+LA S
Sbjct: 672 KQVCSGEQLANLDKNILHSFVQLRADYRSARLARHFS 708
>emb|CAH65252.1| hypothetical protein [Gallus gallus]
gi|60302826|ref|NP_001012607.1| similar to Exocyst
complex component Sec10 (hSec10) [Gallus gallus]
Length = 707
Score = 165 bits (418), Expect = 5e-39
Identities = 170/757 (22%), Positives = 317/757 (41%), Gaps = 111/757 (14%)
Query: 84 EKLLSLFKDSCKELLELRKQIDGRLHNLKKDVSVQDSKHRRTLAELEKGVDGLFASFARL 143
++LL F + +EL + ++I ++ L++ + + + + EL+K F F L
Sbjct: 39 KRLLEEFVNHIQELQVMDERIQRKVEKLEQQCQKEAKEFAKKVQELQKSNQVAFQHFQEL 98
Query: 144 DSRISSVGQTAAKIGDHLQSADAQRETASQTIELIKYLMEFNSSPGDLMELSPLFSDDSR 203
D IS V +GD L+ + R+ A + +L+KY EF G+L S +F++ +
Sbjct: 99 DEHISYVATKVCHLGDQLEGVNTPRQRAVEAQKLMKYFNEFLD--GELK--SDVFTNSEK 154
Query: 204 VAEAASIAQKLRSFAEEDIGRHGITAPSAVGNATASRGLEVAVANLQEYCNELENRLLSR 263
+ EAA I QKL A+E R EV +Y ++LE +L+
Sbjct: 155 IKEAADIIQKLHLIAQE---------------LPFDRFSEVKSKIASKY-HDLECQLIQE 198
Query: 264 FDAASQKRELTTMAECAKILSQFNRGTSAMQHYVATRPMFIDVEVMNADTRLVLGDQAAQ 323
F +A ++ E++ M E A +L F + + Y+ Q
Sbjct: 199 FTSAQRRGEISRMREVAAVLLHFKGYSHCVDVYIKQC-------------------QEGA 239
Query: 324 SSPNNVARGLSSLYKEITDTVRKEAATITAVFPSPNEVMSILVQRVLEQRVTALLDKLLV 383
N+V + L + + V + VF +P V++ L+Q + E ++ + +
Sbjct: 240 FLRNDVFEDAAILCQRVNKQVGE-------VFSNPETVLAKLIQNIFEVKLQSYVKD--- 289
Query: 384 KPSLVNLPSMEEGGLLFYLRMLAVSYEKTQEIARDLRTVGCG---DLDVEGLTESLFSSH 440
L + YL+ L Y +T ++ L G + L +S+F S+
Sbjct: 290 -----QLEEHRKSDAEQYLKNLYDLYTRTTNLSSKLMEFNLGTDKQTFLSKLIKSIFISY 344
Query: 441 KDEYPEYEQASLR--------QLYKVKMEELRA--ESQISDSSGTIGRSKGATVASS--- 487
+ Y E E L+ + Y K + R+ I D I + + S
Sbjct: 345 LENYIEVEIGYLKSRSAMILQRYYDSKNHQKRSIGTGGIQDLKERIRQRTNLPLGPSIDT 404
Query: 488 --QQQISVTVVTEFVRWNEEAITRCNLFSSQPSTLATLVKAVFTCLLDQV-SQYIAEGLE 544
+ +S VV ++ ++A RC+ S PS L +F+ L+D + +++I LE
Sbjct: 405 HGETFLSQEVVVNLLQETKQAFERCHRLSD-PSDLPKNAFRIFSMLVDFLCTEHIDYALE 463
Query: 545 RARDGLTEAANLRERFVLGTSVSRRVAAAAASAAEAAAAAGESSFRSFMVAVQRSGSSVA 604
G+ + + F+ V ++ +
Sbjct: 464 TGLAGIPSSDSKNANLY------------------------------FLDVVHQANTIFH 493
Query: 605 IIQQYFSNSISRLLLPVDGAHAAAC-EEMATAMSSAEAAAYKGLQQCIETVMAEVERLLS 663
+ + F++ + L+ + C ++ + E G+ + + ++ +++ +L+
Sbjct: 494 LFDKQFNDHLMPLIS--SSPKLSECLQKKKDIIEQMEVKLDMGIDRTLNCMIGQMKHILA 551
Query: 664 AEQKATDYKSPDDGMAPDHRPTNACTRVVAYLSRVLESAFTALEGLNKQAFLSELGNRLH 723
AEQK TD+K P+D + TNAC +V +Y+ + +E +++G N L E G R H
Sbjct: 552 AEQKKTDFK-PEDENNVLIQYTNACVKVCSYVRKQVEKIRKSMDGKNVDTVLMEFGVRFH 610
Query: 724 KVLLNHWQKYTFNPSGGLRLKRDITEYGEFVRSFNAPSVDEKFELLGILANVFIVAPESL 783
+++ H Q+Y+++ GG+ D+ EY + + F V + F+ L L N+ +VAP++L
Sbjct: 611 RLIYEHLQQYSYSCMGGMLAICDVAEYRKCAKDFKIALVLQLFDTLHALCNLLVVAPDNL 670
Query: 784 STLFEGTPSIRKD---AQRFIQLREDYKSAKLASKLS 817
+ G D F+QLR DY+SA+LA S
Sbjct: 671 KQVCSGEQLANLDKNILHSFVQLRVDYRSARLARHFS 707
>ref|NP_001017342.1| hypothetical protein LOC550096 [Xenopus tropicalis]
Length = 708
Score = 160 bits (404), Expect = 2e-37
Identities = 169/757 (22%), Positives = 315/757 (41%), Gaps = 111/757 (14%)
Query: 84 EKLLSLFKDSCKELLELRKQIDGRLHNLKKDVSVQDSKHRRTLAELEKGVDGLFASFARL 143
++LL F + EL + ++I ++ L++ + + + + EL+K F F L
Sbjct: 40 KRLLEQFVNHIDELKLMDERIQRKVEKLEQQCQKEAKEFAKKVQELQKSNQVAFQHFQEL 99
Query: 144 DSRISSVGQTAAKIGDHLQSADAQRETASQTIELIKYLMEFNSSPGDLMELSPLFSDDSR 203
D IS V +GD L+ + R+ A + +L+ Y EF G+L S +F++ +
Sbjct: 100 DEHISYVATKVCHLGDQLEGVNTPRQRAVEAQKLMTYFNEFLD--GELR--SDVFNNPEK 155
Query: 204 VAEAASIAQKLRSFAEEDIGRHGITAPSAVGNATASRGLEVAVANLQEYCNELENRLLSR 263
+ EAA I QKL A+E R EV +Y ++LE++L+
Sbjct: 156 IKEAADIIQKLHLIAQE---------------LPFDRFAEVKSKIASKY-HDLEHQLIQE 199
Query: 264 FDAASQKRELTTMAECAKILSQFNRGTSAMQHYVATRPMFIDVEVMNADTRLVLGDQAAQ 323
F +A ++ E++ M E A +L F + + Y+ + G
Sbjct: 200 FTSAQRRGEISRMREVAAVLLHFKGYSHCVDVYI---------------NQCQEGAYLKY 244
Query: 324 SSPNNVARGLSSLYKEITDTVRKEAATITAVFPSPNEVMSILVQRVLEQRVTALLDKLLV 383
N A + +E+ D +F +P V++ L+Q + E ++ + + +
Sbjct: 245 DIFENSAVLCQRVNREVGD-----------IFSNPEIVLAKLIQNIFEVKIQSCVKE--- 290
Query: 384 KPSLVNLPSMEEGGLLFYLRMLAVSYEKTQEIARDLRTVGCG---DLDVEGLTESLFSSH 440
L + YL+ L Y +T ++ L G + L +S+F S+
Sbjct: 291 -----QLDECRKSDAEQYLKSLYDLYTRTTGLSSKLMEFNLGTDKQTFLSKLIKSIFISY 345
Query: 441 KDEYPEYEQASLR--------QLYKVKMEELR--AESQISDSSGTIGRSKGATVASS--- 487
+ Y + E A LR + Y K + R I D I + + S
Sbjct: 346 LENYIDVETAYLRSRSSVILQRYYDSKNHQKRPIGGGGIQDLKERIRQRTNLPLGPSIDT 405
Query: 488 --QQQISVTVVTEFVRWNEEAITRCNLFSSQPSTLATLVKAVFTCLLDQVS-QYIAEGLE 544
+ +S VV ++ + A RC+ S PS L +F+ L+D + ++I LE
Sbjct: 406 HGETFLSQEVVVNLLQETKHAFERCHKLSD-PSDLPKNAFRIFSILVDYLCIEHIDYALE 464
Query: 545 RARDGLTEAANLRERFVLGTSVSRRVAAAAASAAEAAAAAGESSFRSFMVAVQRSGSSVA 604
+ A + ++ +++ F V ++ +
Sbjct: 465 -------------------------IGLVAIPSPDS-----KNANLYFFDVVHQANTIFH 494
Query: 605 IIQQYFSNSISRLLLPVDGAHAAAC-EEMATAMSSAEAAAYKGLQQCIETVMAEVERLLS 663
+ + F++ + L+ C ++ + E G+ + I ++ +++++L+
Sbjct: 495 LFDKQFNDHLMPLIS--SSPKLTECLQKKKEIIEQMEVKLDTGIDRTINCMVGQMKQILA 552
Query: 664 AEQKATDYKSPDDGMAPDHRPTNACTRVVAYLSRVLESAFTALEGLNKQAFLSELGNRLH 723
AEQK TD+K P+D + T AC +V Y+ + +E +++G N L ELG R H
Sbjct: 553 AEQKKTDFK-PEDENNVLIQYTTACVKVCLYVRKQVEKIRNSMDGKNVDTVLMELGVRFH 611
Query: 724 KVLLNHWQKYTFNPSGGLRLKRDITEYGEFVRSFNAPSVDEKFELLGILANVFIVAPESL 783
+++ H Q+++++ GG+ D+ EY + F P V + F+ L L N+ +VAP++L
Sbjct: 612 RLIYEHLQQFSYSSMGGMLAICDVAEYRRCAKDFKIPLVLQLFDTLHSLCNLLVVAPDNL 671
Query: 784 STLFEGTPSIRKD---AQRFIQLREDYKSAKLASKLS 817
+ G D F+QLR DY+ A+LA S
Sbjct: 672 KQVCSGEQLANLDKNILHSFVQLRADYRPARLARHFS 708
>ref|XP_624953.1| PREDICTED: similar to ENSANGP00000011291 [Apis mellifera]
Length = 716
Score = 159 bits (403), Expect = 3e-37
Identities = 168/759 (22%), Positives = 307/759 (40%), Gaps = 122/759 (16%)
Query: 90 FKDSCKELLELRKQIDGRLHNLKKDVSVQDSKHRRTLAELEKGVDGLFASFARLDSRISS 149
F + K+L L+++ + L+ + +++KH + EL++ F +LD RI+
Sbjct: 49 FLQAIKDLQILQERQQKKCDKLETALKDEEAKHILEILELQERNKHSIDLFHQLDERINL 108
Query: 150 VGQTAAKIGDHLQSADAQRETASQTIELIKYLMEFNSSPGDLMELSPLFSDDSRVAEAAS 209
V +GD L+S + R A + +L+++ +F SPG L + P+F+D S + EAA
Sbjct: 109 VATKVLHLGDQLESVNTPRARAVEAQKLMRHFSDF-LSPGPLTD--PIFTDKSSLYEAAD 165
Query: 210 IAQKLRSFAEEDIGRHGITAPSAVGNATASRGLEVAVANLQEYCNELENRLLSRFDAASQ 269
+ QKL ++E S E A + +E+E L+ F A
Sbjct: 166 VIQKLHLISQE----------------LPSEKFEHAKKKITAKYDEIERNLIEEFVRAHN 209
Query: 270 KRELTTMAECAKILSQFNRGTSAMQHYVATRPMFIDVEVMNADTRLVLGDQAAQSSPNNV 329
+ + M E A +L+ F + + ++ +Q+ S
Sbjct: 210 REDANRMRELASVLTHFKGYSQCIDAFI---------------------EQSQMGS---- 244
Query: 330 ARGLSSLYKEITDTVRKEAATITAVFPSPNEVMSILVQRVLEQRVTALLDKLLVKPSLVN 389
G +++++ K + VF +P +VM+ V + R L K ++
Sbjct: 245 -FGGKDVFQDVIPMCTKYHKLMQQVFTNPEQVMAKFVLNIYHLR--------LQKYAVAK 295
Query: 390 LPSMEEGGLLFYLRMLAVSYEKTQEIARDLRTVG-CGD-LDVEGLTESLFSSHKD----- 442
L + YL+ L Y +T +++ +L+ C D + + LT ++F + D
Sbjct: 296 LADKTDSDK--YLKNLYDLYTRTVKLSTELKMFNICTDEMYLTKLTRNIFQKYLDTYIII 353
Query: 443 --------------EYPEYEQASLRQLYKVKMEELRAESQISDSSGTIGRSKGATVASSQ 488
EY E + +QL +ELR + Q +G +A +
Sbjct: 354 EIKALREKSAALLIEYYESKNHQKKQLQSGGFQELRRDLQ-----AVLGARTNINIAQIE 408
Query: 489 QQISVTVVTE-----FVRWNEEAITRCNLFSSQPSTLATLVKAVFTCLLDQVSQYIAEGL 543
T ++E ++ ++ A RC L S ++ L +S+++ L
Sbjct: 409 NYGGETFLSEELAIALLQRSKVAFQRCQLLSKSDEIPMNALQIFEILLQYLISEHVDYAL 468
Query: 544 ERARDGLTEAANLRERFVLGTSVSRRVAAAAASAAEAAAAAGESSFRSFMVAVQRSGSSV 603
E + + + + F V++ + V
Sbjct: 469 ELGLQSVPIPESRTQPEI-----------------------------HFFNIVRQCNAIV 499
Query: 604 AIIQQYFSNSISRLLLPVDGAHAAACEEMATAMSSAEAAAYKGLQQCIETVMAEVERLLS 663
++++ F++S+ L++ H + + + GL + I ++ V+ L
Sbjct: 500 RLLEEQFNDSVIPLIVSTP-KHGDCMLKKKIILDQIDMKLETGLDRSINAIIGWVKVYLQ 558
Query: 664 AEQKATDYKSPDDGMAPDHRPTNACTRVVAYLSRVLESAFTALEGLNKQAFLSELGNRLH 723
EQ+ TD+K D D T+AC VV Y++ ++ + L+G N L+ELG R H
Sbjct: 559 NEQRKTDFKPETD---VDTLSTSACLTVVQYVNGMIRHIRSTLDGKNLNNVLTELGVRFH 615
Query: 724 KVLLNHWQKYTFNPSGGLRLKRDITEYGEFVRSFNAPSVDEKFELLGILANVFIVAPESL 783
KV+ +H Q++ FN +G + D+ EY + V+ V F+ L L N+ +V PE+L
Sbjct: 616 KVIYDHLQQFQFNSAGAMCAICDMNEYRKCVKELEIDLVTSLFDTLYALCNLLLVKPENL 675
Query: 784 STLFEGTPSIRKDAQ---RFIQLREDYKSAKLASKLSSL 819
+ G D FIQLR DYK+ KLA+ L L
Sbjct: 676 KQVCTGDQLATLDRNVLINFIQLRTDYKTQKLANCLKGL 714
>emb|CAH00965.1| unnamed protein product [Kluyveromyces lactis NRRL Y-1140]
gi|50307751|ref|XP_453869.1| unnamed protein product
[Kluyveromyces lactis]
Length = 817
Score = 137 bits (346), Expect = 1e-30
Identities = 177/831 (21%), Positives = 348/831 (41%), Gaps = 92/831 (11%)
Query: 42 LFGNLVNELLPSFKLEDLEAEGADAVQN----KYSQVATSPLFPEVEKLLSLFKDSCKEL 97
L G VNE + + + A A N + Q+ P E +L KEL
Sbjct: 21 LSGLTVNEFVTELSKDQASKQNAPAAVNASIPENKQLDAKPFIRTFESVL-------KEL 73
Query: 98 LELRKQIDGRLHNLKKDVSVQDSKHRRTLAELEKGVDGLFASFARLDSRISSVGQTAAKI 157
+L+K+ R L + VS + H +++ + + ++ +LD ++S+V Q + +
Sbjct: 74 KKLQKETLSRSGQLTQQVSQMEISHAKSIMVSRGQLKDIVQNYDQLDHKLSAVTQVVSPL 133
Query: 158 GDHLQSADAQRETASQTIELIKYLMEFNSSPGDLMELSPLF--SDDSRVAEAASIAQKLR 215
G L+ + Q+ +++ELI + F GD EL+ L SD + + A I + L
Sbjct: 134 GTKLEKSIKQKNAYIKSVELISFYASFLEE-GDCPELNKLLESSDKRDICQGALILKSLL 192
Query: 216 SFAEEDIGRHGITAPSAVGNATASRGLEVAVANLQEYCNELENRLLSRFDAASQKRELTT 275
++ + + + EV +++ + E R+L F+AA ++ + +
Sbjct: 193 ILTKK------------LDTKSVPKTQEVTQI-IEQLATDFEVRILQGFNAAYRENDYSR 239
Query: 276 MAECAKILSQFNRGTSAMQHYVATRPMFIDVEVMNADTRLVLGDQAAQSSPNNVARGLSS 335
+ + A IL FN G + ++++ F + +A + D ++ N +
Sbjct: 240 LHQLAWILDTFNNGVNIIKNFTDKHQFFEESSNFSAKESALFTDDVFKAELMNPDSHILK 299
Query: 336 LYKEITD-------TVRKEAATITAVFPS--PNEVMSILVQRVLEQR----VTALLDKLL 382
+++ + + E+ + VF + PN VM + +++V E + V LL+ L
Sbjct: 300 YDEKVVEYIQSVFNAIENESKVVATVFENKAPN-VMHLFIEKVFELKLRPLVVFLLNSAL 358
Query: 383 VKPSLVNLPSMEEGGLLFYLRMLAVSYEKTQEIARDLRTVGCGDLD--VEGLTESLFSSH 440
SL + S++ Y + S ++ E +L+ G + +E ++LF
Sbjct: 359 SLSSLAYVRSLQS-----YFSITNNSVKQLAEFLSNLKMDSDGKVSACLEKCWKNLFIDI 413
Query: 441 KDEYPEYEQASLRQLYKVKMEELRAESQISDSSGTIGRSKG-----ATVASSQQQISVTV 495
+ +Y + R L V +++ ++ IS S RS T S ++ +
Sbjct: 414 LFDRSKYFEVEKRTLENVLVQKA-SDFNISFDSDIRPRSLANKLNEVTDFKSSPELKILT 472
Query: 496 VTEFVRWNEEAITRCNLFSSQ-----PSTLATLVKAV----FTC-LLDQVSQYIAEGLER 545
R+++ + N F + P+ L K + FT +D + + E L R
Sbjct: 473 SRSGGRFSQLNLFLKNKFEKRFEHENPAQLNITSKKIDRDYFTLSYIDLMIKCSVESLAR 532
Query: 546 ARDGLTEAANLRERFVLGTSVSRRVAAAAASAAEAAAAA-------GESSFR-SFMVAVQ 597
+ + A+ ++ TS+ + + SA E A A E+S F+ +
Sbjct: 533 VTELVPAIADDFTYELVETSLIGVIDSYIESALEVAHAQLNSIDIHRETSIELKFLKYIS 592
Query: 598 RSGSSVAIIQQYFSNSISRLLLPVDGAHAAACEEMATAMSSAEAAAYKGLQQCIETVMAE 657
+ ++I+ Y + + LL C + + + A +K + + ++ E
Sbjct: 593 TATEMISIVSAYIKSIVLPLL--------QNCPSIKRKVIALSNAYFKKCELLMNLLIVE 644
Query: 658 VERLLS-------AEQKATDYKSPDDGMAPDHRPTNACTRVVAYLSRVLESAFTALEGLN 710
+LL+ ++QK D+ S + T T +V LS V + L+ N
Sbjct: 645 TSKLLNQKFTLILSKQKRKDFLSKSQDFLD--QDTETTTELVNVLSSVYSQSALYLKNGN 702
Query: 711 KQAFLSELGNRLHKVLLNHWQKYTFNPSGGLRLKRDITEYGEFVRSFNAPSVDEKFELLG 770
+AFL++LG+ L + LLNH++K+ + +GG+ + +DI + V ++ ++EKF L
Sbjct: 703 LKAFLTKLGDNLFEQLLNHYKKFQVSSTGGVIVTKDIIAFQTAVEDWDVEELNEKFATLR 762
Query: 771 ILANVFIVAPESLSTLFEGTPSIRKDAQ---RFIQLREDYKSAKLASKLSS 818
L+N+F V PE L++L + + D + +I REDY K +
Sbjct: 763 ELSNLFTVQPELLTSLTKEGRLVNVDREIIDEYISKREDYNQDGFIGKFKA 813
>ref|XP_637029.1| hypothetical protein DDB0219271 [Dictyostelium discoideum]
gi|60465502|gb|EAL63587.1| hypothetical protein
DDB0219271 [Dictyostelium discoideum]
Length = 520
Score = 134 bits (338), Expect = 9e-30
Identities = 126/542 (23%), Positives = 228/542 (41%), Gaps = 55/542 (10%)
Query: 276 MAECAKILSQFNRGTSAMQHYVATRPMFIDVEVMNADTRLVLGDQAAQSSPNNVA-RGLS 334
M +CA L FN G Y+ MF D++ D L NN+
Sbjct: 1 MKQCATTLHGFNGGERCRSRYIQKLKMFFDIDSFRKDENLANNITKRLIRGNNIVDTRFE 60
Query: 335 SLYKEITDTVRKEAATITAVFPSPNEVMSILVQRVLEQRVTALLDKLLVKPSLVNLPSME 394
Y +I V E I VF + M++L+ R+ EQRV ++ +L S+E
Sbjct: 61 IFYTDILKDVSHEQMVIQNVFVNQTSAMAMLIIRLFEQRVRLFIENVL---------SLE 111
Query: 395 EGGLLFYLRMLAVSYEKTQEIARD-LRTVGCGDLDVEGLTESLFSSHKDEYPEYEQASLR 453
+ +L+ + ++ T+++ D L++ G +D+ L S+F +++ Y + E L
Sbjct: 112 SNNVSMFLQTVHYAFNSTKKLLVDPLQSYGIVGVDLNQLLNSIFYQYQEGYIQKETTYLV 171
Query: 454 QLYKVKMEELRAESQISDSSGTIGRSKGATVASSQQQISVTVVTEFVRWNEEAITRCNLF 513
L++ + E Q D + ++ + FV+ E A+TR
Sbjct: 172 SLFQSNVVEECERLQTLDRYSMY----------LEDGLNPEITQMFVQQTENALTRSYTL 221
Query: 514 SSQPSTLATLVKAVFTCLLDQVSQYIAEGLERARDGLTEAANLRERFVLGTSVSRRVAAA 573
S + LA +K +F +L+ + + + FVL + + +
Sbjct: 222 SLD-NILADNIKTIFFLMLEYLFEKYS------------------MFVLNKYIELPMIPS 262
Query: 574 AASAAEAAAAAGESS--FRSFMVAVQRSGSSVAIIQQYFSNSISRLLLPVDGAHAAACEE 631
++S + + S FR + Q G ++ Q + I ++ + ++
Sbjct: 263 SSSNVDTKSIQDSISQLFRVVLSINQIVGQIQSMFQVFVLPHIQTSMI----VQSQCSDQ 318
Query: 632 MATAMSSAEAAAYKGLQQCIETVMAEVERLLSAEQKATDYKSPDDGMAPDHRPTNACTRV 691
+ +SS E GL+ + T++ +E+ L Q DY D D+ T+ C V
Sbjct: 319 LYFNISSLENTINTGLENSLTTMIQLIEKTL-LPQGRNDYLIDDY----DNSVTDTCASV 373
Query: 692 VAYLSRVLESAFTALEGLNKQAFLSELGNRLHKVLLNHWQKYTFNPS-GGLRLKRDITEY 750
+ + + A L+G N ++ ELG + V +NH++K+ G L+L RD+TEY
Sbjct: 374 IKLIQSFYDMAKINLQGKNFHIYVEELGLKSQFVFINHFKKFKIGQGIGTLKLMRDLTEY 433
Query: 751 GEFVRSFNAPSVDEKFELLGILANVFIVAPESLSTLFEG---TPSIRKDAQRFIQLREDY 807
+ F + VD+ FELL ++ + +V PE+ + EG T ++D FI+ R D+
Sbjct: 434 RNLSKQFKSQKVDDAFELLFEISKLHLVNPENFKLVIEGGALTRMSKQDLIIFIKQRSDF 493
Query: 808 KS 809
KS
Sbjct: 494 KS 495
>gb|EAK81314.1| hypothetical protein UM00329.1 [Ustilago maydis 521]
gi|49067308|ref|XP_397944.1| hypothetical protein
UM00329.1 [Ustilago maydis 521]
Length = 1133
Score = 133 bits (334), Expect = 3e-29
Identities = 167/715 (23%), Positives = 279/715 (38%), Gaps = 112/715 (15%)
Query: 30 IDDFKGDFSFDALFGNLVNELLPSFKLEDLEAEGADAVQNKYSQVATSPLFPEVEKLLSL 89
+D F+ DF+ + L ++L+ K + P P +
Sbjct: 51 LDTFQNDFNTNDFIAQLTSKLVQRSKADP------------------GPFNPR--PFIRT 90
Query: 90 FKDSCKELLELRKQIDGRLHNLKKDVSVQDSKHRRTLAELEKGVDGLFASFARLDSRISS 149
F+ + +L ++R Q+D + L V V +S + + L EL K + SF L+ RIS
Sbjct: 91 FEAAMDQLAQIRAQVDAQSQQLSSAVHVAESAYTKKLDELSKNFVAVGNSFNSLEDRISE 150
Query: 150 VGQTAAKIGDHLQSADAQRETASQTIELIKYLMEFNSSPGDLMELSPLFSDDSRVA--EA 207
VG+TA +IG+ L++ D QR AS+ +LI++ F + GD +L L + R +
Sbjct: 151 VGRTAIRIGEQLETIDKQRNRASEAHDLIEFYYMF--ARGDTTKLEHLRKEGGREGRIKT 208
Query: 208 ASIAQKLRSFAEEDIGRHGITAPSAVGNATASRGLEVAVANLQEYCNELENRLLSRFDAA 267
A IA++L +A+ G E ++ YC E +L FD
Sbjct: 209 AVIARRL----------------AAISREVDVAGAEQTRDSIDRYCERFERDMLKLFDKF 252
Query: 268 SQKRELTTMAECAKILSQFNRGTSAMQHYVATRPMFID---------VEVMNADTRLVLG 318
++ + M+ AK+L FN G S +Q YV FI +E+ T +
Sbjct: 253 YRRSDPKMMSHIAKVLQGFNGGASCVQIYVNQHDFFISKDRVGEANGIELSQIWTHMADP 312
Query: 319 DQAAQSSPNNVARGLSSLYKEITDTVRKEAATITAVFPSPNEVMSILVQRVLEQRVTALL 378
D A P L++L+ EI TV EA I+AVFP+P VM +QRV Q V +
Sbjct: 313 DHA----PPKSEPSLTALFNEIRQTVEIEAQIISAVFPNPLVVMQTFLQRVFAQSVQGFV 368
Query: 379 DKLLVKPSLVNLPSMEEGGLLFYLRMLAVSYEKTQEIARDLRTVGCGDLDVEGLTESLFS 438
+ ++ K + +N PS G G++D + L
Sbjct: 369 EVIMDKAADINAPSFVGAG---------------------------GNIDTKTAALELNP 401
Query: 439 SHKDEYPEYEQASLRQLYKVKMEELRAESQISDSSGTIGRSKGATVASSQQQISVTVVTE 498
+ A LR L+ + L + + G AS+ VV++
Sbjct: 402 PSNSQPTTSHLAFLRALHMARSSALSLVNDLKLYDYRGAGIIGPAYASAHTANGDGVVSD 461
Query: 499 FVR--WNEEAITRCNLFSSQPSTLATL----VKAVFTCLLDQVSQYI---AEGLERARDG 549
+R N+ A L + S LAT+ V+ +F ++ + +YI + L G
Sbjct: 462 SIRLLGNDFAAGASQLAGASASPLATMLDQSVEELFVPYMEGI-RYIDRESRSLADLYAG 520
Query: 550 L---------TEAANLRERFVLGTSVSRRVAAAAASAAEAAAAAGESSFRSFMVAVQRSG 600
L T AA R++ G ++ R+ + ++A+++ S +
Sbjct: 521 LLFKFISFHRTSAAAKRDK-ASGNTIFNRMRNQITATSDASSSTSSLGGSSNTAQTSTAA 579
Query: 601 SSVAIIQQYFSNSISRLLLPVDGAHAAACEE---MATAMSSAEAAAYKGLQ-------QC 650
SS + +S L V G H A TA S+A A+A +
Sbjct: 580 SSTVSTSKTSFFKLSNLADRVRGTHNTAATSSTLSVTAGSTATASAESSVDGQHVAGGAH 639
Query: 651 IETVMAEVERLLSAE--QKATDYKSPDDGMAPDHRPTNACTRVVAYLSRVLESAF 703
+E + + + LS + +K + + G D + + L RVL SA+
Sbjct: 640 VEDELDDTDGDLSLDIAEKMLRWHAESIGRCVDLSSPSEVPKATFALLRVLTSAY 694
Score = 62.8 bits (151), Expect = 4e-08
Identities = 28/76 (36%), Positives = 48/76 (62%)
Query: 710 NKQAFLSELGNRLHKVLLNHWQKYTFNPSGGLRLKRDITEYGEFVRSFNAPSVDEKFELL 769
N ++FL+E+G H +LL H +KYT + +GG+ L +D+ Y + + +F P + ++FE+L
Sbjct: 917 NAESFLTEIGVAFHGLLLEHLRKYTVSATGGIMLTKDLAVYQDAIGTFGIPVLSDRFEML 976
Query: 770 GILANVFIVAPESLST 785
L N+FIV L +
Sbjct: 977 RQLGNLFIVQASVLKS 992
Score = 37.7 bits (86), Expect = 1.5
Identities = 55/242 (22%), Positives = 93/242 (37%), Gaps = 35/242 (14%)
Query: 465 AESQISDSSGTIGRSKGATVASSQQQISVTVVTEFVRWNEEAITRCNLFSSQPSTLATLV 524
AES + G + + +S+ + + +RW+ E+I RC SS PS +
Sbjct: 625 AESSVDGQHVAGGAHVEDELDDTDGDLSLDIAEKMLRWHAESIGRCVDLSS-PSEVP--- 680
Query: 525 KAVFTCLLDQVSQYIAEGLERARDGLTEAANLRERFVLGTSVSRRVAAAAASAAEAAAAA 584
KA F L S YI +E A D A AA +A + A
Sbjct: 681 KATFALLRVLTSAYIKTYVETALDS---------------------ALAAVTAQDIRGAV 719
Query: 585 GESSFRSFMVAVQRSGSSVAIIQQYFSNSISRLLLPVDGAHAAACEEMAT----AMSSAE 640
S ++V RS + + Q++ N+ LLP+ + EM + E
Sbjct: 720 MPDP--SAAMSVVRSVELLISLWQHYVNT---ALLPLASTSVTSRREMMIFNNHNLLRVE 774
Query: 641 AAAYKGLQQCIETVMAEVERLLSAEQKATDYKSPDDGMAPDHRPTNACTRVVAYLSRVLE 700
+Q+ + V+ + LS ++K D+ +D +A + T C L +V +
Sbjct: 775 GKCDALMQKIADNVVGFLSNRLSTQRK-NDFTPKNDDLAFARQNTEPCVACSEALEKVQQ 833
Query: 701 SA 702
+A
Sbjct: 834 AA 835
>gb|AAP91763.1| brain secretory protein SEC10P-like [Ciona intestinalis]
Length = 194
Score = 115 bits (289), Expect = 4e-24
Identities = 59/175 (33%), Positives = 98/175 (55%), Gaps = 6/175 (3%)
Query: 646 GLQQCIETVMAEVERLLSAEQKATDYKSPDDGMAPDHRPTNACTRVVAYLSRVLESAFTA 705
G+ +C+ +V+ + ++ EQK +D+ + P + TNAC + Y+ +V+++ T+
Sbjct: 23 GIDKCLSSVIGWMRHIMRTEQKKSDFSTDQP---PQQQYTNACQMLCKYVKKVIDTMRTS 79
Query: 706 LEGLNKQAFLSELGNRLHKVLLNHWQKYTFNPSGGLRLKRDITEYGEFVRSFNAPSVDEK 765
L+G N A L E G R H+++ +H Q+Y+F+ GG+ D+ EYG+ + P V+
Sbjct: 80 LDGNNVDAVLKEFGTRYHRLVYDHLQQYSFSSMGGMMAICDVNEYGKCAQQLQVPFVNSL 139
Query: 766 FELLGILANVFIVAPESLSTLFEGTPSIRKD---AQRFIQLREDYKSAKLASKLS 817
FE L L N+ +VAPE+L + G D F+QLR DY+SA+LA S
Sbjct: 140 FESLHSLCNLLVVAPENLRQVCSGDQFANLDRAVLHTFVQLRSDYRSARLAKHFS 194
>emb|CAH56255.1| hypothetical protein [Homo sapiens]
Length = 396
Score = 112 bits (280), Expect = 5e-23
Identities = 98/406 (24%), Positives = 178/406 (43%), Gaps = 54/406 (13%)
Query: 432 LTESLFSSHKDEYPEYEQASLR--------QLYKVKMEELRA--ESQISDSSGTIGRSKG 481
L +S+F S+ + Y E E L+ + Y K + R+ I D I +
Sbjct: 25 LIKSIFISYLENYIEVETGYLKSRSAMILQRYYDSKNHQKRSIGTGGIQDLKERIRQRTN 84
Query: 482 ATVASS-----QQQISVTVVTEFVRWNEEAITRCNLFSSQPSTLATLVKAVFTCLLDQVS 536
+ S + +S VV ++ ++A RC+ S PS L +FT L++ +
Sbjct: 85 LPLGPSIDTHGETFLSQEVVVNLLQETKQAFERCHRLSD-PSDLPRNAFRIFTILVEFLC 143
Query: 537 -QYIAEGLERARDGLTEAANLRERFVLGTSVSRRVAAAAASAAEAAAAAGESSFRSFMVA 595
++I LE G+ + + F+
Sbjct: 144 IEHIDYALETGLAGIPSSDSRNANLY------------------------------FLDV 173
Query: 596 VQRSGSSVAIIQQYFSNSISRLLLPVDGAHAAAC-EEMATAMSSAEAAAYKGLQQCIETV 654
VQ++ + + + F++ + L+ + C ++ + E G+ + + +
Sbjct: 174 VQQANTIFHLFDKQFNDHLMPLIS--SSPKLSECLQKKKEIIEQMEMKLDTGIDRTLNCM 231
Query: 655 MAEVERLLSAEQKATDYKSPDDGMAPDHRPTNACTRVVAYLSRVLESAFTALEGLNKQAF 714
+ +++ +L+AEQK TD+K P+D + TNAC +V AY+ + +E +++G N
Sbjct: 232 IGQMKHILAAEQKKTDFK-PEDENNVLIQYTNACVKVCAYVRKQVEKIKNSMDGKNVDTV 290
Query: 715 LSELGNRLHKVLLNHWQKYTFNPSGGLRLKRDITEYGEFVRSFNAPSVDEKFELLGILAN 774
L ELG R H+++ H Q+Y+++ GG+ D+ EY + + F P V F+ L L N
Sbjct: 291 LMELGVRFHRLIYEHLQQYSYSCMGGMLAICDVAEYRKCAKDFKIPMVLHLFDTLHALCN 350
Query: 775 VFIVAPESLSTLFEGTPSIRKD---AQRFIQLREDYKSAKLASKLS 817
+ +VAP++L + G D F+QLR DY+SA+LA S
Sbjct: 351 LLVVAPDNLKQVCSGEQLANLDKNILHSFVQLRADYRSARLARHFS 396
>gb|AAX26266.1| unknown [Schistosoma japonicum]
Length = 212
Score = 110 bits (274), Expect = 2e-22
Identities = 58/173 (33%), Positives = 103/173 (59%), Gaps = 6/173 (3%)
Query: 646 GLQQCIETVMAEVERLLSAEQKATDYK---SPDDGMAPDHRPTNACTRVVAYLSRVLESA 702
GL++C+ ++ V+ LLS EQ+ TD++ + + G+ + P+ AC VV +L+ +
Sbjct: 30 GLEKCLNLAISRVQYLLSNEQRKTDFRPDTNANSGLITSNPPSLACQHVVGFLTHLNRET 89
Query: 703 FTALEGLNKQAFLSELGNRLHKVLLNHWQKYTFNPSGGLRLKRDITEYGEFVRSFNAPSV 762
L+G N + FL E G +L++ L++H+ ++F+ +GG +D+T Y E + F +P+V
Sbjct: 90 HQHLDGQNLKTFLHEFGMKLNRTLVDHFYNFSFSDTGGFVAMQDVTAYREIAKHFESPTV 149
Query: 763 DEKFELLGILANVFIVAPESLSTLFEG--TPSIRKD-AQRFIQLREDYKSAKL 812
+ F++L L N+ ++ PE++ + + I KD FIQLR DYK+AKL
Sbjct: 150 NLVFDILLKLMNLMLIKPENVQQVVQDYLQSGIPKDLLMNFIQLRTDYKTAKL 202
>ref|XP_610411.1| PREDICTED: similar to Exocyst complex component Sec10 (hSec10),
partial [Bos taurus]
Length = 276
Score = 107 bits (268), Expect = 1e-21
Identities = 58/175 (33%), Positives = 99/175 (56%), Gaps = 4/175 (2%)
Query: 646 GLQQCIETVMAEVERLLSAEQKATDYKSPDDGMAPDHRPTNACTRVVAYLSRVLESAFTA 705
G+ + + ++ +++ +L+AEQK TD+K P+D + TNAC +V AY+ + +E +
Sbjct: 103 GIDRTLNCMIGQMKHILAAEQKKTDFK-PEDENNVLIQYTNACVKVCAYVRKQVEKIKNS 161
Query: 706 LEGLNKQAFLSELGNRLHKVLLNHWQKYTFNPSGGLRLKRDITEYGEFVRSFNAPSVDEK 765
++G N L ELG R H+++ H Q+Y+++ GG+ D+ EY + + F P V +
Sbjct: 162 MDGKNVDTVLMELGVRFHRLIYEHLQQYSYSCMGGMLAICDVAEYRKCAKDFKIPLVLQL 221
Query: 766 FELLGILANVFIVAPESLSTLFEGTPSIRKD---AQRFIQLREDYKSAKLASKLS 817
F+ L L N+ +V P++L + G D F+QLR DY+SA+LA S
Sbjct: 222 FDTLHALCNLLVVNPDNLKQVCSGEQLANLDKNILHSFVQLRADYRSARLARHFS 276
Database: nr
Posted date: Jul 5, 2005 12:34 AM
Number of letters in database: 863,360,394
Number of sequences in database: 2,540,612
Lambda K H
0.315 0.129 0.348
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 1,161,401,404
Number of Sequences: 2540612
Number of extensions: 44263588
Number of successful extensions: 163059
Number of sequences better than 10.0: 306
Number of HSP's better than 10.0 without gapping: 74
Number of HSP's successfully gapped in prelim test: 238
Number of HSP's that attempted gapping in prelim test: 162234
Number of HSP's gapped (non-prelim): 900
length of query: 821
length of database: 863,360,394
effective HSP length: 137
effective length of query: 684
effective length of database: 515,296,550
effective search space: 352462840200
effective search space used: 352462840200
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.6 bits)
S2: 79 (35.0 bits)
Medicago: description of AC137821.7


