

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC137821.5 - phase: 0
(589 letters)
Database: nr
2,540,612 sequences; 863,360,394 total letters
Searching..................................................done
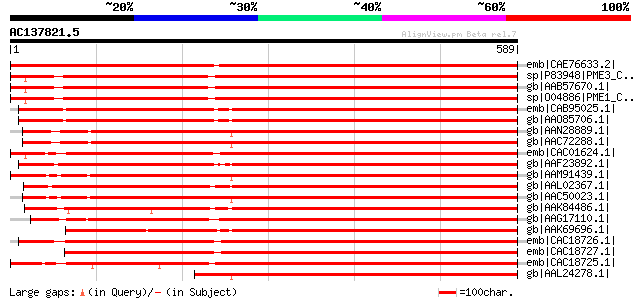
Score E
Sequences producing significant alignments: (bits) Value
emb|CAE76633.2| pectin methylesterase [Cicer arietinum] 1049 0.0
sp|P83948|PME3_CITSI Pectinesterase-3 precursor (Pectin methyles... 838 0.0
gb|AAB57670.1| pectinesterase [Citrus sinensis] 835 0.0
sp|O04886|PME1_CITSI Pectinesterase-1 precursor (Pectin methyles... 833 0.0
emb|CAB95025.1| pectin methylesterase [Nicotiana tabacum] 830 0.0
gb|AAO85706.1| pectin methyl-esterase [Nicotiana benthamiana] 821 0.0
gb|AAN28889.1| At3g14310/MLN21_9 [Arabidopsis thaliana] gi|92795... 818 0.0
gb|AAC72288.1| putative pectin methylesterase [Arabidopsis thali... 814 0.0
emb|CAC01624.1| putative pectin methylesterase [Populus tremula ... 811 0.0
gb|AAF23892.1| pectin methyl esterase [Solanum tuberosum] 809 0.0
gb|AAM91439.1| At1g53830/T18A20_6 [Arabidopsis thaliana] gi|6056... 805 0.0
gb|AAL02367.1| pectin methylesterase [Lycopersicon esculentum] g... 801 0.0
gb|AAC50023.1| ATPME2 precursor [Arabidopsis thaliana] 792 0.0
gb|AAK84486.1| putative thermostable pectinesterase [Citrus sine... 763 0.0
gb|AAG17110.1| putative pectin methylesterase 3 [Linum usitatiss... 694 0.0
gb|AAK69696.1| putative pectin methylesterase LuPME5 [Linum usit... 680 0.0
emb|CAC18726.1| putative pectin methylesterase [Populus tremula ... 620 e-176
emb|CAC18727.1| putative pectin methylesterase [Populus tremula ... 607 e-172
emb|CAC18725.1| putative pectin methylesterase [Populus tremula ... 606 e-172
gb|AAL24278.1| AT3g14310/MLN21_9 [Arabidopsis thaliana] 599 e-170
>emb|CAE76633.2| pectin methylesterase [Cicer arietinum]
Length = 584
Score = 1049 bits (2712), Expect = 0.0
Identities = 518/589 (87%), Positives = 551/589 (92%), Gaps = 5/589 (0%)
Query: 1 MNKIKQSLAGISNSGNKKKLLISLFTTLLIVASIVAIVATTTKNSNKSKNNSIASSSLSL 60
MNKIKQSL ISNS NKKKL SLFT LLI ASI+AI A+ T + N+I SSSLSL
Sbjct: 1 MNKIKQSLTRISNS-NKKKLFFSLFTALLITASIIAISASVTSSQKTKTKNNIVSSSLSL 59
Query: 61 SHHSHAILKSACTTTLYPELCFSAISSEPNITHKITNHKDVISLSLNITTRAVEHNYFTV 120
SHHSH I+K+ACTTTLYP+LCFSAISSEPNITHKI NHKDVISLSLNITTRAVEHN+FTV
Sbjct: 60 SHHSHTIIKTACTTTLYPDLCFSAISSEPNITHKINNHKDVISLSLNITTRAVEHNFFTV 119
Query: 121 EKLLLRKSLTKREKIALHDCLETIDETLDELKEAQNDLVLYPSKKTLYQHADDLKTLISS 180
E LL RK+L++REKIALHDCLETID+TLDELKEAQ DLVLYP+KKTLYQHADDLKTLIS+
Sbjct: 120 ENLLRRKNLSEREKIALHDCLETIDDTLDELKEAQRDLVLYPNKKTLYQHADDLKTLISA 179
Query: 181 AITNQVTCLDGFSHDDADKEVRKVLQEGQIHVEHMCSNALAMTKNMTDKDIAEFEQTNMV 240
AITNQVTCLDGFSHD ADK+VRKVL++GQ+HVEHMCSNALAMTKNMTDKDIA+FE+ N
Sbjct: 180 AITNQVTCLDGFSHDGADKQVRKVLEQGQVHVEHMCSNALAMTKNMTDKDIAKFEENN-- 237
Query: 241 LGSNKNRKLLEEENGVGWPEWISAGDRRLLQGSTVKADVVVAADGSGNFKTVSEAVAAAP 300
+ KNRKLLEEENGV WPEWISAGDRRLLQG+ VKADVVVAADGSGNFKTVSEAVA AP
Sbjct: 238 --NKKNRKLLEEENGVNWPEWISAGDRRLLQGAAVKADVVVAADGSGNFKTVSEAVAGAP 295
Query: 301 LKSSKRYVIKIKAGVYKENVEVPKKKTNIMFLGDGRTNTIITGSRNVVDGSTTFHSATVA 360
LKSSKRYVIKIKAGVYKENVEVPKKK+NIMFLGDG+ NTIIT SRNVVDGSTTFHSATVA
Sbjct: 296 LKSSKRYVIKIKAGVYKENVEVPKKKSNIMFLGDGKKNTIITASRNVVDGSTTFHSATVA 355
Query: 361 IVGGNFLARDITFQNTAGPAKHQAVALRVGADLSAFYNCDIIAYQDTLYVHNNRQFFVNC 420
+VGGNFLARDITFQNTAGP+KHQAVALRVG DLSAFYNCDIIAYQDTLYVHNNRQFFVNC
Sbjct: 356 VVGGNFLARDITFQNTAGPSKHQAVALRVGGDLSAFYNCDIIAYQDTLYVHNNRQFFVNC 415
Query: 421 FISGTVDFIFGNSAVVFQNCDIHARRPNSGQKNMVTAQGRVDPNQNTGIVIQKCRIGATK 480
FISGTVDFIFGNSAVVFQNCDIHAR+P+SGQKNMVTAQGRVDPNQNTGIVIQKCRIGATK
Sbjct: 416 FISGTVDFIFGNSAVVFQNCDIHARKPDSGQKNMVTAQGRVDPNQNTGIVIQKCRIGATK 475
Query: 481 DLEGVKGNFPTYLGRPWKEYSRTVFMQSSISDVIDPVGWHEWNGNFALNTLVYREYQNTG 540
DLEG+KG FPTYLGRPWKEYSRTV MQSSISDVIDP+GWHEWNGNFALNTLVYREYQNTG
Sbjct: 476 DLEGLKGTFPTYLGRPWKEYSRTVIMQSSISDVIDPIGWHEWNGNFALNTLVYREYQNTG 535
Query: 541 PGAGTSKRVTWKGFKVITSAAEAQSFTPGNFIGGSSWLGSTGFPFSLGL 589
PGAGTSKRV WKGFKVITSA+EAQ+FTPGNFIGGS+WLGSTGFPFSLGL
Sbjct: 536 PGAGTSKRVNWKGFKVITSASEAQTFTPGNFIGGSTWLGSTGFPFSLGL 584
>sp|P83948|PME3_CITSI Pectinesterase-3 precursor (Pectin methylesterase 3) (PE 3)
Length = 584
Score = 838 bits (2165), Expect = 0.0
Identities = 419/600 (69%), Positives = 486/600 (80%), Gaps = 27/600 (4%)
Query: 1 MNKIKQSLAGISNSGN----------KKKLLISLFTTLLIVASIVAIVATTTKNSNKSKN 50
M +IK+ +S S KKKL ++LF TLL+VA+++ IVA N N
Sbjct: 1 MTRIKEFFTKLSESSTNQNISNIPKKKKKLFLALFATLLVVAAVIGIVAGVNSRKNSGDN 60
Query: 51 NSIASSSLSLSHHSHAILKSACTTTLYPELCFSAISSEPNITHKITNHKDVISLSLNITT 110
+ HAILKS+C++T YP+LCFSAI++ P + K+T+ KDVI +SLNITT
Sbjct: 61 GN---------EPHHAILKSSCSSTRYPDLCFSAIAAVPEASKKVTSQKDVIEMSLNITT 111
Query: 111 RAVEHNYFTVEKLLLRKSLTKREKIALHDCLETIDETLDELKEAQNDLVLYPSKKTLYQH 170
AVEHNYF ++KLL R +LTKREK+ALHDCLETIDETLDEL +A DL YP+KK+L QH
Sbjct: 112 TAVEHNYFGIQKLLKRTNLTKREKVALHDCLETIDETLDELHKAVEDLEEYPNKKSLSQH 171
Query: 171 ADDLKTLISSAITNQVTCLDGFSHDDADKEVRKVLQEGQIHVEHMCSNALAMTKNMTDKD 230
ADDLKTL+S+A+TNQ TCLDGFSHDDA+K VR L +GQ+HVE MCSNALAM KNMTD D
Sbjct: 172 ADDLKTLMSAAMTNQGTCLDGFSHDDANKHVRDALSDGQVHVEKMCSNALAMIKNMTDTD 231
Query: 231 IAEFEQTNMVLGSNKNRKLLEEENGV-GWPEWISAGDRRLLQGSTVKADVVVAADGSGNF 289
+ M++ ++ NRKL+EE + V GWP W+S GDRRLLQ S+V +VVVAADGSGNF
Sbjct: 232 M-------MIMRTSNNRKLIEETSTVDGWPAWLSTGDRRLLQSSSVTPNVVVAADGSGNF 284
Query: 290 KTVSEAVAAAPLKSSKRYVIKIKAGVYKENVEVPKKKTNIMFLGDGRTNTIITGSRNVVD 349
KTV+ +VAAAP +KRY+I+IKAGVY+ENVEV KK NIMF+GDGRT TIITGSRNVVD
Sbjct: 285 KTVAASVAAAPQGGTKRYIIRIKAGVYRENVEVTKKHKNIMFIGDGRTRTIITGSRNVVD 344
Query: 350 GSTTFHSATVAIVGGNFLARDITFQNTAGPAKHQAVALRVGADLSAFYNCDIIAYQDTLY 409
GSTTF SATVA+VG FLARDITFQNTAGP+KHQAVALRVGADLSAFYNCD++AYQDTLY
Sbjct: 345 GSTTFKSATVAVVGEGFLARDITFQNTAGPSKHQAVALRVGADLSAFYNCDMLAYQDTLY 404
Query: 410 VHNNRQFFVNCFISGTVDFIFGNSAVVFQNCDIHARRPNSGQKNMVTAQGRVDPNQNTGI 469
VH+NRQFFVNC I+GTVDFIFGN+A V QNCDIHAR+PNSGQKNMVTAQGR DPNQNTGI
Sbjct: 405 VHSNRQFFVNCLIAGTVDFIFGNAAAVLQNCDIHARKPNSGQKNMVTAQGRADPNQNTGI 464
Query: 470 VIQKCRIGATKDLEGVKGNFPTYLGRPWKEYSRTVFMQSSISDVIDPVGWHEWNGNFALN 529
VIQK RIGAT DL+ V+G+FPTYLGRPWKEYSRTV MQSSI+DVI P GWHEW+GNFALN
Sbjct: 465 VIQKSRIGATSDLKPVQGSFPTYLGRPWKEYSRTVIMQSSITDVIHPAGWHEWDGNFALN 524
Query: 530 TLVYREYQNTGPGAGTSKRVTWKGFKVITSAAEAQSFTPGNFIGGSSWLGSTGFPFSLGL 589
TL Y E+QN G GAGTS RV WKGF+VITSA EAQ+FTPG+FI GSSWLGSTGFPFSLGL
Sbjct: 525 TLFYGEHQNAGAGAGTSGRVKWKGFRVITSATEAQAFTPGSFIAGSSWLGSTGFPFSLGL 584
>gb|AAB57670.1| pectinesterase [Citrus sinensis]
Length = 584
Score = 835 bits (2156), Expect = 0.0
Identities = 418/600 (69%), Positives = 485/600 (80%), Gaps = 27/600 (4%)
Query: 1 MNKIKQSLAGISNSGN----------KKKLLISLFTTLLIVASIVAIVATTTKNSNKSKN 50
M IK+ +S S + KKKL ++LF TLL+VA+++ IVA N N
Sbjct: 1 MTHIKEFFTKLSESSSNQNISNIPKKKKKLFLALFATLLVVAAVIGIVAGVNSRKNSGDN 60
Query: 51 NSIASSSLSLSHHSHAILKSACTTTLYPELCFSAISSEPNITHKITNHKDVISLSLNITT 110
+ HAILKS+C++T YP+LCFSAI++ P + K+T+ KDVI +SLNITT
Sbjct: 61 GN---------EPHHAILKSSCSSTRYPDLCFSAIAAVPEASKKVTSQKDVIEMSLNITT 111
Query: 111 RAVEHNYFTVEKLLLRKSLTKREKIALHDCLETIDETLDELKEAQNDLVLYPSKKTLYQH 170
AVEHNYF ++KLL R +LTKREK+ALHDCLETIDETLDEL +A DL YP+KK+L QH
Sbjct: 112 TAVEHNYFGIQKLLKRTNLTKREKVALHDCLETIDETLDELHKAVEDLEEYPNKKSLSQH 171
Query: 171 ADDLKTLISSAITNQVTCLDGFSHDDADKEVRKVLQEGQIHVEHMCSNALAMTKNMTDKD 230
ADDLKTL+S+A+TNQ TCLDGFSHDDA+K VR L +GQ+HVE MCSNALAM KNMTD D
Sbjct: 172 ADDLKTLMSAAMTNQGTCLDGFSHDDANKHVRDALSDGQVHVEKMCSNALAMIKNMTDTD 231
Query: 231 IAEFEQTNMVLGSNKNRKLLEEENGV-GWPEWISAGDRRLLQGSTVKADVVVAADGSGNF 289
+ M++ ++ NRKL EE + V GWP W+S GDRRLLQ S+V + VVAADGSGNF
Sbjct: 232 M-------MIMRTSNNRKLTEETSTVDGWPAWLSPGDRRLLQSSSVTPNAVVAADGSGNF 284
Query: 290 KTVSEAVAAAPLKSSKRYVIKIKAGVYKENVEVPKKKTNIMFLGDGRTNTIITGSRNVVD 349
KTV+ AVAAAP +KRY+I+IKAGVY+ENVEV KK NIMF+GDGRT TIITGSRNVVD
Sbjct: 285 KTVAAAVAAAPQGGTKRYIIRIKAGVYRENVEVTKKHKNIMFIGDGRTRTIITGSRNVVD 344
Query: 350 GSTTFHSATVAIVGGNFLARDITFQNTAGPAKHQAVALRVGADLSAFYNCDIIAYQDTLY 409
GSTTF SATVA+VG FLARDITFQNTAGP+KHQAVALRVGADLSAFYNCD++AYQDTLY
Sbjct: 345 GSTTFKSATVAVVGEGFLARDITFQNTAGPSKHQAVALRVGADLSAFYNCDMLAYQDTLY 404
Query: 410 VHNNRQFFVNCFISGTVDFIFGNSAVVFQNCDIHARRPNSGQKNMVTAQGRVDPNQNTGI 469
VH+NRQFFVNC I+GTVDFIFGN+A V QNCDIHAR+PNSGQKNMVTAQGR DPNQNTGI
Sbjct: 405 VHSNRQFFVNCLIAGTVDFIFGNAAAVLQNCDIHARKPNSGQKNMVTAQGRTDPNQNTGI 464
Query: 470 VIQKCRIGATKDLEGVKGNFPTYLGRPWKEYSRTVFMQSSISDVIDPVGWHEWNGNFALN 529
VIQK RIGAT DL+ V+G+FPTYLGRPWKEYSRTV MQSSI+D+I P GWHEW+GNFALN
Sbjct: 465 VIQKSRIGATSDLKPVQGSFPTYLGRPWKEYSRTVIMQSSITDLIHPAGWHEWDGNFALN 524
Query: 530 TLVYREYQNTGPGAGTSKRVTWKGFKVITSAAEAQSFTPGNFIGGSSWLGSTGFPFSLGL 589
TL Y E+QN+G GAGTS RV WKGF+VITSA EAQ+FTPG+FI GSSWLGSTGFPFSLGL
Sbjct: 525 TLFYGEHQNSGAGAGTSGRVKWKGFRVITSATEAQAFTPGSFIAGSSWLGSTGFPFSLGL 584
>sp|O04886|PME1_CITSI Pectinesterase-1 precursor (Pectin methylesterase) (PE)
gi|2098705|gb|AAB57667.1| pectinesterase [Citrus
sinensis]
Length = 584
Score = 833 bits (2152), Expect = 0.0
Identities = 417/600 (69%), Positives = 484/600 (80%), Gaps = 27/600 (4%)
Query: 1 MNKIKQSLAGISNSGN----------KKKLLISLFTTLLIVASIVAIVATTTKNSNKSKN 50
M IK+ +S S + KKKL ++LF TLL+VA+++ IVA N N
Sbjct: 1 MTHIKEFFTKLSESSSNQNISNIPKKKKKLFLALFATLLVVAAVIGIVAGVNSRKNSGDN 60
Query: 51 NSIASSSLSLSHHSHAILKSACTTTLYPELCFSAISSEPNITHKITNHKDVISLSLNITT 110
+ HAILKS+C++T YP+LCFSAI++ P + K+T+ KDVI +SLNITT
Sbjct: 61 GN---------EPHHAILKSSCSSTRYPDLCFSAIAAVPEASKKVTSQKDVIEMSLNITT 111
Query: 111 RAVEHNYFTVEKLLLRKSLTKREKIALHDCLETIDETLDELKEAQNDLVLYPSKKTLYQH 170
AVEHNYF ++KLL R +LTKREK+ALHDCLETIDETLDEL +A DL YP+KK+L QH
Sbjct: 112 TAVEHNYFGIQKLLKRTNLTKREKVALHDCLETIDETLDELHKAVEDLEEYPNKKSLSQH 171
Query: 171 ADDLKTLISSAITNQVTCLDGFSHDDADKEVRKVLQEGQIHVEHMCSNALAMTKNMTDKD 230
ADDLKTL+S+A+TNQ TCLDGFSHDDA+K VR L +GQ+HVE MCSNALAM KNMTD D
Sbjct: 172 ADDLKTLMSAAMTNQGTCLDGFSHDDANKHVRDALSDGQVHVEKMCSNALAMIKNMTDTD 231
Query: 231 IAEFEQTNMVLGSNKNRKLLEEENGV-GWPEWISAGDRRLLQGSTVKADVVVAADGSGNF 289
+ M++ ++ NRKL EE + V GWP W+S GDRRLLQ S+V + VVAADGSGNF
Sbjct: 232 M-------MIMRTSNNRKLTEETSTVDGWPAWLSPGDRRLLQSSSVTPNAVVAADGSGNF 284
Query: 290 KTVSEAVAAAPLKSSKRYVIKIKAGVYKENVEVPKKKTNIMFLGDGRTNTIITGSRNVVD 349
KTV+ AVAAAP +KRY+I+IKAGVY+ENVEV KK NIMF+GDGRT TIITGSRNVVD
Sbjct: 285 KTVAAAVAAAPQGGTKRYIIRIKAGVYRENVEVTKKHKNIMFIGDGRTRTIITGSRNVVD 344
Query: 350 GSTTFHSATVAIVGGNFLARDITFQNTAGPAKHQAVALRVGADLSAFYNCDIIAYQDTLY 409
GSTTF SAT A+VG FLARDITFQNTAGP+KHQAVALRVGADLSAFYNCD++AYQDTLY
Sbjct: 345 GSTTFKSATAAVVGEGFLARDITFQNTAGPSKHQAVALRVGADLSAFYNCDMLAYQDTLY 404
Query: 410 VHNNRQFFVNCFISGTVDFIFGNSAVVFQNCDIHARRPNSGQKNMVTAQGRVDPNQNTGI 469
VH+NRQFFVNC I+GTVDFIFGN+A V QNCDIHAR+PNSGQKNMVTAQGR DPNQNTGI
Sbjct: 405 VHSNRQFFVNCLIAGTVDFIFGNAAAVLQNCDIHARKPNSGQKNMVTAQGRTDPNQNTGI 464
Query: 470 VIQKCRIGATKDLEGVKGNFPTYLGRPWKEYSRTVFMQSSISDVIDPVGWHEWNGNFALN 529
VIQK RIGAT DL+ V+G+FPTYLGRPWKEYSRTV MQSSI+D+I P GWHEW+GNFALN
Sbjct: 465 VIQKSRIGATSDLKPVQGSFPTYLGRPWKEYSRTVIMQSSITDLIHPAGWHEWDGNFALN 524
Query: 530 TLVYREYQNTGPGAGTSKRVTWKGFKVITSAAEAQSFTPGNFIGGSSWLGSTGFPFSLGL 589
TL Y E+QN+G GAGTS RV WKGF+VITSA EAQ+FTPG+FI GSSWLGSTGFPFSLGL
Sbjct: 525 TLFYGEHQNSGAGAGTSGRVKWKGFRVITSATEAQAFTPGSFIAGSSWLGSTGFPFSLGL 584
>emb|CAB95025.1| pectin methylesterase [Nicotiana tabacum]
Length = 579
Score = 830 bits (2143), Expect = 0.0
Identities = 420/580 (72%), Positives = 478/580 (82%), Gaps = 9/580 (1%)
Query: 11 ISNSGNKKKLLISLFTTLLIVASIVAIVATTTKNSNKSKNNSIASSSLSLSHHSHAILKS 70
++ S KKKL +++ ++L+VA+++ +V SN S +++ + S +H AI+KS
Sbjct: 8 VNFSKGKKKLFLTVVASVLLVAAVIGVVVGVKFRSNNSDDHADIQAITSAAH---AIVKS 64
Query: 71 ACTTTLYPELCFSAISSEPNITHKITNHKDVISLSLNITTRAVEHNYFTVEKLL-LRKSL 129
AC TL+PELC+S I+S + + K+T+ KDVI LSLNIT RAV+HN+F VEKL+ RK L
Sbjct: 65 ACENTLHPELCYSTIASVSDFSKKVTSQKDVIELSLNITCRAVQHNFFKVEKLIKTRKGL 124
Query: 130 TKREKIALHDCLETIDETLDELKEAQNDLVLYPSKKTLYQHADDLKTLISSAITNQVTCL 189
REK+ALHDCLETIDETLDEL A DL LYP+KK+L HADDLKTLISSAITNQ TCL
Sbjct: 125 KPREKVALHDCLETIDETLDELHTAIKDLELYPNKKSLKAHADDLKTLISSAITNQETCL 184
Query: 190 DGFSHDDADKEVRKVLQEGQIHVEHMCSNALAMTKNMTDKDIAEFEQTNMVLGSNKNRKL 249
DGFSHDDADK+VRK L +GQ HVE MCSNALAM NMTD DIA ++ G+ NRKL
Sbjct: 185 DGFSHDDADKKVRKALLKGQKHVEKMCSNALAMICNMTDTDIANEQKLK---GTTTNRKL 241
Query: 250 LEEENGVGWPEWISAGDRRLLQGSTVKADVVVAADGSGNFKTVSEAVAAAPLKSSKRYVI 309
E+ + WPEW+SAGDRRLLQ STV+ DVVVAADGSGNFKTVSEAVA AP KSSKRYVI
Sbjct: 242 REDNSE--WPEWLSAGDRRLLQSSTVRPDVVVAADGSGNFKTVSEAVAKAPEKSSKRYVI 299
Query: 310 KIKAGVYKENVEVPKKKTNIMFLGDGRTNTIITGSRNVVDGSTTFHSATVAIVGGNFLAR 369
+IKAGVY+ENV+VPKKKTNIMF+GDGR+NTIITGSRNV DGSTTFHSATVA VG FLAR
Sbjct: 300 RIKAGVYRENVDVPKKKTNIMFMGDGRSNTIITGSRNVKDGSTTFHSATVAAVGEKFLAR 359
Query: 370 DITFQNTAGPAKHQAVALRVGADLSAFYNCDIIAYQDTLYVHNNRQFFVNCFISGTVDFI 429
DITFQNTAG AKHQAVALRVG+DLSAFY CDI+AYQD+LYVH+NRQ+FV C I+GTVDFI
Sbjct: 360 DITFQNTAGAAKHQAVALRVGSDLSAFYRCDILAYQDSLYVHSNRQYFVQCLIAGTVDFI 419
Query: 430 FGNSAVVFQNCDIHARRPNSGQKNMVTAQGRVDPNQNTGIVIQKCRIGATKDLEGVKGNF 489
FGN+A V QNCDIHARRP SGQKNMVTAQGR DPNQNTGIVIQKCRIGAT DL V+ +F
Sbjct: 420 FGNAAAVLQNCDIHARRPGSGQKNMVTAQGRSDPNQNTGIVIQKCRIGATSDLRPVQKSF 479
Query: 490 PTYLGRPWKEYSRTVFMQSSISDVIDPVGWHEWNGNFALNTLVYREYQNTGPGAGTSKRV 549
PTYLGRPWKEYSRTV MQSSI+DVI+ GWHEWNGNFALNTL Y EYQNTG GAGTS RV
Sbjct: 480 PTYLGRPWKEYSRTVIMQSSITDVINSAGWHEWNGNFALNTLFYGEYQNTGAGAGTSGRV 539
Query: 550 TWKGFKVITSAAEAQSFTPGNFIGGSSWLGSTGFPFSLGL 589
WKGFKVITSA EAQ++TPG FI G SWL STGFPFSLGL
Sbjct: 540 KWKGFKVITSATEAQAYTPGRFIAGGSWLSSTGFPFSLGL 579
>gb|AAO85706.1| pectin methyl-esterase [Nicotiana benthamiana]
Length = 579
Score = 821 bits (2120), Expect = 0.0
Identities = 416/580 (71%), Positives = 475/580 (81%), Gaps = 9/580 (1%)
Query: 11 ISNSGNKKKLLISLFTTLLIVASIVAIVATTTKNSNKSKNNSIASSSLSLSHHSHAILKS 70
++ S KKKL +++ ++L+VA+++ +V SN S +++ + S +H AI+KS
Sbjct: 8 VNFSKGKKKLFLTVVASVLLVAAVIGVVVGVKFRSNNSDDHADIQAITSAAH---AIVKS 64
Query: 71 ACTTTLYPELCFSAISSEPNITHKITNHKDVISLSLNITTRAVEHNYFTVEKLL-LRKSL 129
AC TL+PELC+S I+S + + K+T+ KDVI LSLNIT RAV+HN+F VEKL+ RK L
Sbjct: 65 ACENTLHPELCYSTIASVSDFSKKVTSQKDVIELSLNITCRAVQHNFFKVEKLIKTRKGL 124
Query: 130 TKREKIALHDCLETIDETLDELKEAQNDLVLYPSKKTLYQHADDLKTLISSAITNQVTCL 189
REK+ALHDCLETIDETLDEL A DL LYP+KK+L HAD LKTLISSAITNQ TCL
Sbjct: 125 KPREKVALHDCLETIDETLDELHTAIKDLELYPNKKSLKAHADGLKTLISSAITNQETCL 184
Query: 190 DGFSHDDADKEVRKVLQEGQIHVEHMCSNALAMTKNMTDKDIAEFEQTNMVLGSNKNRKL 249
DGFSHDDADK+VRK L +GQ HVE MCSNALAM NMTD DIA ++ G+ NRKL
Sbjct: 185 DGFSHDDADKKVRKALLKGQKHVEKMCSNALAMICNMTDTDIANEQKLK---GTTTNRKL 241
Query: 250 LEEENGVGWPEWISAGDRRLLQGSTVKADVVVAADGSGNFKTVSEAVAAAPLKSSKRYVI 309
E+ + WPEW+ AGDRRLLQ STV+ DVVVAADGSGNFKTVSEAVA AP KSSKRYVI
Sbjct: 242 REDNSE--WPEWLPAGDRRLLQSSTVRPDVVVAADGSGNFKTVSEAVAKAPEKSSKRYVI 299
Query: 310 KIKAGVYKENVEVPKKKTNIMFLGDGRTNTIITGSRNVVDGSTTFHSATVAIVGGNFLAR 369
+IKAGVY+ENV+VPKKKTNIMF+GDGR+NTIITGSRNV DGSTTFHSATVA VG FLAR
Sbjct: 300 RIKAGVYRENVDVPKKKTNIMFMGDGRSNTIITGSRNVKDGSTTFHSATVAAVGEKFLAR 359
Query: 370 DITFQNTAGPAKHQAVALRVGADLSAFYNCDIIAYQDTLYVHNNRQFFVNCFISGTVDFI 429
DITFQNTAG AKHQAVALRVG+DLSAFY CDI+AYQD+LYVH+NRQ+FV C I+GTVDFI
Sbjct: 360 DITFQNTAGAAKHQAVALRVGSDLSAFYRCDILAYQDSLYVHSNRQYFVQCLIAGTVDFI 419
Query: 430 FGNSAVVFQNCDIHARRPNSGQKNMVTAQGRVDPNQNTGIVIQKCRIGATKDLEGVKGNF 489
FGN+A V Q+CDIHARRP SGQKNMVTAQGR DPNQNTGIVIQKCRIGAT DL V+ +F
Sbjct: 420 FGNAAAVLQDCDIHARRPGSGQKNMVTAQGRSDPNQNTGIVIQKCRIGATSDLRPVQKSF 479
Query: 490 PTYLGRPWKEYSRTVFMQSSISDVIDPVGWHEWNGNFALNTLVYREYQNTGPGAGTSKRV 549
P YLGRPWKEYSRTV MQSSI+DVI+ GWHEWNGNFALNTL Y EYQNTG GAGTS RV
Sbjct: 480 PMYLGRPWKEYSRTVIMQSSITDVINSAGWHEWNGNFALNTLFYGEYQNTGAGAGTSGRV 539
Query: 550 TWKGFKVITSAAEAQSFTPGNFIGGSSWLGSTGFPFSLGL 589
WKGFKVITSA EAQ++TPG FI G SWL STGFPFSLGL
Sbjct: 540 KWKGFKVITSATEAQAYTPGRFIAGGSWLSSTGFPFSLGL 579
>gb|AAN28889.1| At3g14310/MLN21_9 [Arabidopsis thaliana] gi|9279579|dbj|BAB01037.1|
pectinesterase [Arabidopsis thaliana]
gi|15529256|gb|AAK97722.1| AT3g14310/MLN21_9
[Arabidopsis thaliana] gi|14335010|gb|AAK59769.1|
AT3g14310/MLN21_9 [Arabidopsis thaliana]
gi|15231828|ref|NP_188048.1| pectinesterase family
protein [Arabidopsis thaliana]
Length = 592
Score = 818 bits (2114), Expect = 0.0
Identities = 416/588 (70%), Positives = 467/588 (78%), Gaps = 26/588 (4%)
Query: 16 NKKKLLISLFTTLLIVASIVAIVATTTKNSNKSKNNSIASSSLSLSHHSHAILKSACTTT 75
NKK +L+S LL VA++ I A +K + K +LS SHA+L+S+C++T
Sbjct: 17 NKKLVLLSAAVALLFVAAVAGISAGASKANEKR----------TLSPSSHAVLRSSCSST 66
Query: 76 LYPELCFSAISSEPNITHKITNHKDVISLSLNITTRAVEHNYFTVEKLLL-RKSLTKREK 134
YPELC SA+ + + ++T+ KDVI S+N+T AVEHNYFTV+KL+ RK LT REK
Sbjct: 67 RYPELCISAVVTAGGV--ELTSQKDVIEASVNLTITAVEHNYFTVKKLIKKRKGLTPREK 124
Query: 135 IALHDCLETIDETLDELKEAQNDLVLYPSKKTLYQHADDLKTLISSAITNQVTCLDGFSH 194
ALHDCLETIDETLDEL E DL LYP+KKTL +HA DLKTLISSAITNQ TCLDGFSH
Sbjct: 125 TALHDCLETIDETLDELHETVEDLHLYPTKKTLREHAGDLKTLISSAITNQETCLDGFSH 184
Query: 195 DDADKEVRKVLQEGQIHVEHMCSNALAMTKNMTDKDIAEFEQTNMVLGSNKNRKLLEEEN 254
DDADK+VRK L +GQIHVEHMCSNALAM KNMTD DIA FEQ + +N+ K +E
Sbjct: 185 DDADKQVRKALLKGQIHVEHMCSNALAMIKNMTDTDIANFEQKAKITSNNRKLKEENQET 244
Query: 255 GV-------------GWPEWISAGDRRLLQGSTVKADVVVAADGSGNFKTVSEAVAAAPL 301
V GWP W+SAGDRRLLQGS VKAD VAADGSG FKTV+ AVAAAP
Sbjct: 245 TVAVDIAGAGELDSEGWPTWLSAGDRRLLQGSGVKADATVAADGSGTFKTVAAAVAAAPE 304
Query: 302 KSSKRYVIKIKAGVYKENVEVPKKKTNIMFLGDGRTNTIITGSRNVVDGSTTFHSATVAI 361
S+KRYVI IKAGVY+ENVEV KKK NIMF+GDGRT TIITGSRNVVDGSTTFHSATVA
Sbjct: 305 NSNKRYVIHIKAGVYRENVEVAKKKKNIMFMGDGRTRTIITGSRNVVDGSTTFHSATVAA 364
Query: 362 VGGNFLARDITFQNTAGPAKHQAVALRVGADLSAFYNCDIIAYQDTLYVHNNRQFFVNCF 421
VG FLARDITFQNTAGP+KHQAVALRVG+D SAFYNCD++AYQDTLYVH+NRQFFV C
Sbjct: 365 VGERFLARDITFQNTAGPSKHQAVALRVGSDFSAFYNCDMLAYQDTLYVHSNRQFFVKCL 424
Query: 422 ISGTVDFIFGNSAVVFQNCDIHARRPNSGQKNMVTAQGRVDPNQNTGIVIQKCRIGATKD 481
I+GTVDFIFGN+AVV Q+CDIHARRPNSGQKNMVTAQGR DPNQNTGIVIQKCRIGAT D
Sbjct: 425 IAGTVDFIFGNAAVVLQDCDIHARRPNSGQKNMVTAQGRTDPNQNTGIVIQKCRIGATSD 484
Query: 482 LEGVKGNFPTYLGRPWKEYSRTVFMQSSISDVIDPVGWHEWNGNFALNTLVYREYQNTGP 541
L+ VKG+FPTYLGRPWKEYS+TV MQS+ISDVI P GW EW G FALNTL YREY NTG
Sbjct: 485 LQSVKGSFPTYLGRPWKEYSQTVIMQSAISDVIRPEGWSEWTGTFALNTLTYREYSNTGA 544
Query: 542 GAGTSKRVTWKGFKVITSAAEAQSFTPGNFIGGSSWLGSTGFPFSLGL 589
GAGT+ RV W+GFKVIT+AAEAQ +T G FIGG WL STGFPFSLGL
Sbjct: 545 GAGTANRVKWRGFKVITAAAEAQKYTAGQFIGGGGWLSSTGFPFSLGL 592
>gb|AAC72288.1| putative pectin methylesterase [Arabidopsis thaliana]
Length = 592
Score = 814 bits (2103), Expect = 0.0
Identities = 415/588 (70%), Positives = 465/588 (78%), Gaps = 26/588 (4%)
Query: 16 NKKKLLISLFTTLLIVASIVAIVATTTKNSNKSKNNSIASSSLSLSHHSHAILKSACTTT 75
NKK +L+S LL VA++ I A +K + K +LS SHA+L+S+C++T
Sbjct: 17 NKKLVLLSAAVALLFVAAVAGISAGASKANEKR----------TLSPSSHAVLRSSCSST 66
Query: 76 LYPELCFSAISSEPNITHKITNHKDVISLSLNITTRAVEHNYFTVEKLLL-RKSLTKREK 134
YPELC SA+ + ++T+ KDVI S+N+T AVEHNYFTV+KL+ RK LT REK
Sbjct: 67 RYPELCISAVVTAGAC--ELTSQKDVIEASVNLTITAVEHNYFTVKKLIKKRKGLTPREK 124
Query: 135 IALHDCLETIDETLDELKEAQNDLVLYPSKKTLYQHADDLKTLISSAITNQVTCLDGFSH 194
ALHDCLETIDETLDEL E DL LYP+KKTL +HA DLKTLISSAITNQ TCLDGFSH
Sbjct: 125 TALHDCLETIDETLDELHETVEDLHLYPTKKTLREHAGDLKTLISSAITNQETCLDGFSH 184
Query: 195 DDADKEVRKVLQEGQIHVEHMCSNALAMTKNMTDKDIAEFEQTNMVLGSNKNRKLLEEEN 254
DDADK+VRK L +GQIHVEHMCSNALAM KNMTD DIA FEQ + +N+ K +E
Sbjct: 185 DDADKQVRKALLKGQIHVEHMCSNALAMIKNMTDTDIANFEQKAKITSNNRKLKEENQET 244
Query: 255 GV-------------GWPEWISAGDRRLLQGSTVKADVVVAADGSGNFKTVSEAVAAAPL 301
V GWP W+SAGDRRLLQGS VK D VAADGSG FKTV+ AVAAAP
Sbjct: 245 TVAVDIAGAGELDSEGWPTWLSAGDRRLLQGSGVKRDATVAADGSGTFKTVAAAVAAAPE 304
Query: 302 KSSKRYVIKIKAGVYKENVEVPKKKTNIMFLGDGRTNTIITGSRNVVDGSTTFHSATVAI 361
S+KRYVI IKAGVY+ENVEV KKK NIMF+GDGRT TIITGSRNVVDGSTTFHSATVA
Sbjct: 305 NSNKRYVIHIKAGVYRENVEVAKKKKNIMFMGDGRTRTIITGSRNVVDGSTTFHSATVAA 364
Query: 362 VGGNFLARDITFQNTAGPAKHQAVALRVGADLSAFYNCDIIAYQDTLYVHNNRQFFVNCF 421
VG FLARDITFQNTAGP+KHQAVALRVG+D SAFYNCD++AYQDTLYVH+NRQFFV C
Sbjct: 365 VGERFLARDITFQNTAGPSKHQAVALRVGSDFSAFYNCDMLAYQDTLYVHSNRQFFVKCL 424
Query: 422 ISGTVDFIFGNSAVVFQNCDIHARRPNSGQKNMVTAQGRVDPNQNTGIVIQKCRIGATKD 481
I+GTVDFIFGN+AVV Q+CDIHARRPNSGQKNMVTAQGR DPNQNTGIVIQKCRIGAT D
Sbjct: 425 IAGTVDFIFGNAAVVLQDCDIHARRPNSGQKNMVTAQGRTDPNQNTGIVIQKCRIGATSD 484
Query: 482 LEGVKGNFPTYLGRPWKEYSRTVFMQSSISDVIDPVGWHEWNGNFALNTLVYREYQNTGP 541
L+ VKG+FPTYLGRPWKEYS+TV MQS+ISDVI P GW EW G FALNTL YREY NTG
Sbjct: 485 LQSVKGSFPTYLGRPWKEYSQTVIMQSAISDVIRPEGWSEWTGTFALNTLTYREYSNTGA 544
Query: 542 GAGTSKRVTWKGFKVITSAAEAQSFTPGNFIGGSSWLGSTGFPFSLGL 589
GAGT+ RV W+GFKVIT+AAEAQ +T G FIGG WL STGFPFSLGL
Sbjct: 545 GAGTANRVKWRGFKVITAAAEAQKYTAGQFIGGGGWLSSTGFPFSLGL 592
>emb|CAC01624.1| putative pectin methylesterase [Populus tremula x Populus
tremuloides]
Length = 579
Score = 811 bits (2096), Expect = 0.0
Identities = 415/598 (69%), Positives = 480/598 (79%), Gaps = 28/598 (4%)
Query: 1 MNKIKQSLAGISNSGN-------KKKLLISLFTTLLIVASIVAIVATTTKNSNKSKNNSI 53
M + K L GIS+SGN K+LL++ F L +VA+I A+VA NS+K+ N
Sbjct: 1 MAQDKHPLTGISDSGNHISILKKNKRLLLASFAALFLVATIAAVVAGV--NSHKNGENEG 58
Query: 54 ASSSLSLSHHSHAILKSACTTTLYPELCFSAISSEPNITHKITNHKDVISLSLNITTRAV 113
A HA+LKSAC++TLYPELC+SAI++ P +T + + KDVI LS+N+TT+ V
Sbjct: 59 A----------HAVLKSACSSTLYPELCYSAIATVPGVTGNLASLKDVIELSINLTTKTV 108
Query: 114 EHNYFTVEKLLLRKSLTKREKIALHDCLETIDETLDELKEAQNDLVLYPSKKTLYQHADD 173
+ NYFTVEKL+ + LTKREK ALHDCLETIDETLDEL EAQ D+ YP+KK+L + AD+
Sbjct: 109 QQNYFTVEKLIAKTKLTKREKTALHDCLETIDETLDELHEAQVDISGYPNKKSLKEQADN 168
Query: 174 LKTLISSAITNQVTCLDGFSHDDADKEVRKVLQEGQIHVEHMCSNALAMTKNMTDKDIAE 233
L TL+SSAITNQ TCLDGFSHD ADK+VRK L +GQ HVE MCSNALAM KNMTD DIA
Sbjct: 169 LITLLSSAITNQETCLDGFSHDGADKKVRKALLKGQTHVEKMCSNALAMIKNMTDTDIAN 228
Query: 234 FEQTNMVLGSNKNRKLLEEENGVG--WPEWISAGDRRLLQGSTVKADVVVAADGSGNFKT 291
Q N NRKL EE+ G WPEW+S DRRLLQ S+V +VVVAADGSG++KT
Sbjct: 229 ELQ-------NTNRKLKEEKEGNERVWPEWMSVADRRLLQSSSVTPNVVVAADGSGDYKT 281
Query: 292 VSEAVAAAPLKSSKRYVIKIKAGVYKENVEVPKKKTNIMFLGDGRTNTIITGSRNVVDGS 351
VSEAVAAAP KSSKRY+I+IKAGVY+ENVEVPK K NIMFLGDGR TIIT SRNVVDGS
Sbjct: 282 VSEAVAAAPKKSSKRYIIQIKAGVYRENVEVPKDKHNIMFLGDGRKTTIITASRNVVDGS 341
Query: 352 TTFHSATVAIVGGNFLARDITFQNTAGPAKHQAVALRVGADLSAFYNCDIIAYQDTLYVH 411
TTF SATVA VG FLAR +TF+NTAGP+KHQAVALRVG+DLSAFY CD++AYQDTLYVH
Sbjct: 342 TTFKSATVAAVGQGFLARGVTFENTAGPSKHQAVALRVGSDLSAFYECDMLAYQDTLYVH 401
Query: 412 NNRQFFVNCFISGTVDFIFGNSAVVFQNCDIHARRPNSGQKNMVTAQGRVDPNQNTGIVI 471
+NRQFF+NCF++GTVDFIFGN+A VFQ+CD HARRP+SGQKNMVTAQGR DPNQNTGIVI
Sbjct: 402 SNRQFFINCFVAGTVDFIFGNAAAVFQDCDYHARRPDSGQKNMVTAQGRTDPNQNTGIVI 461
Query: 472 QKCRIGATKDLEGVKGNFPTYLGRPWKEYSRTVFMQSSISDVIDPVGWHEWNGNFALNTL 531
QK RIGAT DL V+ +FPTYLGRPWKEYSRTV MQSSI+DVI P GWHEW+G+FAL+TL
Sbjct: 462 QKSRIGATSDLLPVQSSFPTYLGRPWKEYSRTVIMQSSITDVIQPAGWHEWSGSFALSTL 521
Query: 532 VYREYQNTGPGAGTSKRVTWKGFKVITSAAEAQSFTPGNFIGGSSWLGSTGFPFSLGL 589
Y EYQN+G GAGTS RV W+G+KVITSA EAQ+F PGNFI GSSWLGST FPFSLGL
Sbjct: 522 FYAEYQNSGAGAGTSSRVKWEGYKVITSATEAQAFAPGNFIAGSSWLGSTSFPFSLGL 579
>gb|AAF23892.1| pectin methyl esterase [Solanum tuberosum]
Length = 576
Score = 809 bits (2089), Expect = 0.0
Identities = 409/580 (70%), Positives = 476/580 (81%), Gaps = 12/580 (2%)
Query: 11 ISNSGNKKKLLISLFTTLLIVASIVAIVATTTKNSNKSKNNSIASSSLSLSHHSHAILKS 70
I S KKK+ +++ ++L+VA+++ +VA SN S +++ +++S +HAI+KS
Sbjct: 8 IDFSKRKKKIYLAIVASVLLVAAVIGVVAGVKSRSNNSDDDA---DIMAISSSAHAIVKS 64
Query: 71 ACTTTLYPELCFSAISSEPNITHKITNHKDVISLSLNITTRAVEHNYFTVEKLL-LRKSL 129
AC+ TL+PELC+SAI + + + K+T+ KDVI LSLNIT +AV NY+ V++L+ RK L
Sbjct: 65 ACSNTLHPELCYSAIVNVTDFSKKVTSQKDVIELSLNITVKAVRRNYYAVKELIKTRKGL 124
Query: 130 TKREKIALHDCLETIDETLDELKEAQNDLVLYPSKKTLYQHADDLKTLISSAITNQVTCL 189
T REK+ALHDCLET+DETLDEL A DL LYP+KK+L +HA+DLKTLISSAITNQ TCL
Sbjct: 125 TPREKVALHDCLETMDETLDELHTAVADLELYPNKKSLKEHAEDLKTLISSAITNQETCL 184
Query: 190 DGFSHDDADKEVRKVLQEGQIHVEHMCSNALAMTKNMTDKDIAEFEQTNMVLGSNKNRKL 249
DGFSHD+ADK+VRKVL +GQ HVE MCSNALAM NMT+ DIA + + GS K
Sbjct: 185 DGFSHDEADKKVRKVLLKGQKHVEKMCSNALAMICNMTNTDIANEMKLS---GSRK---- 237
Query: 250 LEEENGVGWPEWISAGDRRLLQGSTVKADVVVAADGSGNFKTVSEAVAAAPLKSSKRYVI 309
L E+NG WPEW+SAGDRRLLQ STV DVVVAADGSG++KTVSEAVA AP KSSKRYVI
Sbjct: 238 LVEDNGE-WPEWLSAGDRRLLQSSTVTPDVVVAADGSGDYKTVSEAVAKAPEKSSKRYVI 296
Query: 310 KIKAGVYKENVEVPKKKTNIMFLGDGRTNTIITGSRNVVDGSTTFHSATVAIVGGNFLAR 369
+IKAGVY+ENV+VPKKKTNIMF+GDGR+NTIIT SRNV DGSTTFHSATVA VG FLAR
Sbjct: 297 RIKAGVYRENVDVPKKKTNIMFMGDGRSNTIITASRNVQDGSTTFHSATVAAVGEKFLAR 356
Query: 370 DITFQNTAGPAKHQAVALRVGADLSAFYNCDIIAYQDTLYVHNNRQFFVNCFISGTVDFI 429
DITFQNTAG +KHQAVALRVG+DLSAFY CDI+AYQDTLYVH+NRQFFV C ++GTVDFI
Sbjct: 357 DITFQNTAGASKHQAVALRVGSDLSAFYKCDILAYQDTLYVHSNRQFFVQCLVAGTVDFI 416
Query: 430 FGNSAVVFQNCDIHARRPNSGQKNMVTAQGRVDPNQNTGIVIQKCRIGATKDLEGVKGNF 489
FGN A V Q+CDIHARRP SGQKNMVTAQGR DPNQNTGIVIQKCRIGAT DL V+ +F
Sbjct: 417 FGNGAAVLQDCDIHARRPGSGQKNMVTAQGRTDPNQNTGIVIQKCRIGATSDLRPVQKSF 476
Query: 490 PTYLGRPWKEYSRTVFMQSSISDVIDPVGWHEWNGNFALNTLVYREYQNTGPGAGTSKRV 549
PTYLGRPWKEYSRTV MQSSI+DVI P GWHEWNGNFALNTL Y EY NTG GA TS RV
Sbjct: 477 PTYLGRPWKEYSRTVIMQSSITDVIQPAGWHEWNGNFALNTLFYGEYANTGAGAATSGRV 536
Query: 550 TWKGFKVITSAAEAQSFTPGNFIGGSSWLGSTGFPFSLGL 589
WKG KVITS+ EAQ++TPG+FI G SWL STGFPFSLGL
Sbjct: 537 KWKGHKVITSSTEAQAYTPGSFIAGGSWLSSTGFPFSLGL 576
>gb|AAM91439.1| At1g53830/T18A20_6 [Arabidopsis thaliana] gi|6056392|gb|AAF02856.1|
pectinesterase 2 [Arabidopsis thaliana]
gi|13605623|gb|AAK32805.1| At1g53830/T18A20_6
[Arabidopsis thaliana] gi|15220955|ref|NP_175786.1|
pectinesterase family protein [Arabidopsis thaliana]
gi|17865767|sp|Q42534|PME2_ARATH Pectinesterase-2
precursor (Pectin methylesterase 2) (PE 2)
Length = 587
Score = 805 bits (2078), Expect = 0.0
Identities = 416/596 (69%), Positives = 474/596 (78%), Gaps = 16/596 (2%)
Query: 1 MNKIKQSLAGISNSGNKKKLLISLFT-TLLIVASIVAIVATTTKNSNKSKNNSIASSSLS 59
M IK+ ++ S+ N KKL++S LL++ASIV I ATTT N++KN I +
Sbjct: 1 MAPIKEFISKFSDFKNNKKLILSSAAIALLLLASIVGIAATTT---NQNKNQKITT---- 53
Query: 60 LSHHSHAILKSACTTTLYPELCFSAISSEPNITHKITNHKDVISLSLNITTRAVEHNYFT 119
LS SHAILKS C++TLYPELCFSA+++ ++T+ K+VI SLN+TT+AV+HNYF
Sbjct: 54 LSSTSHAILKSVCSSTLYPELCFSAVAATGG--KELTSQKEVIEASLNLTTKAVKHNYFA 111
Query: 120 VEKLLL-RKSLTKREKIALHDCLETIDETLDELKEAQNDLVLYPSKKTLYQHADDLKTLI 178
V+KL+ RK LT RE ALHDCLETIDETLDEL A DL YP +K+L +HADDLKTLI
Sbjct: 112 VKKLIAKRKGLTPREVTALHDCLETIDETLDELHVAVEDLHQYPKQKSLRKHADDLKTLI 171
Query: 179 SSAITNQVTCLDGFSHDDADKEVRKVLQEGQIHVEHMCSNALAMTKNMTDKDIAEFE--Q 236
SSAITNQ TCLDGFS+DDAD++VRK L +GQ+HVEHMCSNALAM KNMT+ DIA FE
Sbjct: 172 SSAITNQGTCLDGFSYDDADRKVRKALLKGQVHVEHMCSNALAMIKNMTETDIANFELRD 231
Query: 237 TNMVLGSNKNRKLLEEENGV---GWPEWISAGDRRLLQGSTVKADVVVAADGSGNFKTVS 293
+ +N NRKL E + GWP+W+S GDRRLLQGST+KAD VA DGSG+F TV+
Sbjct: 232 KSSTFTNNNNRKLKEVTGDLDSDGWPKWLSVGDRRLLQGSTIKADATVADDGSGDFTTVA 291
Query: 294 EAVAAAPLKSSKRYVIKIKAGVYKENVEVPKKKTNIMFLGDGRTNTIITGSRNVVDGSTT 353
AVAAAP KS+KR+VI IKAGVY+ENVEV KKKTNIMFLGDGR TIITGSRNVVDGSTT
Sbjct: 292 AAVAAAPEKSNKRFVIHIKAGVYRENVEVTKKKTNIMFLGDGRGKTIITGSRNVVDGSTT 351
Query: 354 FHSATVAIVGGNFLARDITFQNTAGPAKHQAVALRVGADLSAFYNCDIIAYQDTLYVHNN 413
FHSATVA VG FLARDITFQNTAGP+KHQAVALRVG+D SAFY CD+ AYQDTLYVH+N
Sbjct: 352 FHSATVAAVGERFLARDITFQNTAGPSKHQAVALRVGSDFSAFYQCDMFAYQDTLYVHSN 411
Query: 414 RQFFVNCFISGTVDFIFGNSAVVFQNCDIHARRPNSGQKNMVTAQGRVDPNQNTGIVIQK 473
RQFFV C I+GTVDFIFGN+A V Q+CDI+ARRPNSGQKNMVTAQGR DPNQNTGIVIQ
Sbjct: 412 RQFFVKCHITGTVDFIFGNAAAVLQDCDINARRPNSGQKNMVTAQGRSDPNQNTGIVIQN 471
Query: 474 CRIGATKDLEGVKGNFPTYLGRPWKEYSRTVFMQSSISDVIDPVGWHEWNGNFALNTLVY 533
CRIG T DL VKG FPTYLGRPWKEYSRTV MQS ISDVI P GWHEW+G+FAL+TL Y
Sbjct: 472 CRIGGTSDLLAVKGTFPTYLGRPWKEYSRTVIMQSDISDVIRPEGWHEWSGSFALDTLTY 531
Query: 534 REYQNTGPGAGTSKRVTWKGFKVITSAAEAQSFTPGNFIGGSSWLGSTGFPFSLGL 589
REY N G GAGT+ RV WKG+KVITS EAQ FT G FIGG WL STGFPFSL L
Sbjct: 532 REYLNRGGGAGTANRVKWKGYKVITSDTEAQPFTAGQFIGGGGWLASTGFPFSLSL 587
>gb|AAL02367.1| pectin methylesterase [Lycopersicon esculentum]
gi|1222552|gb|AAD09283.1| pectin methylesterase
[Lycopersicon esculentum] gi|7447374|pir||T07848
pectinesterase (EC 3.1.1.11) - tomato
gi|6093740|sp|Q43143|PMEU_LYCES Pectinesterase U1
precursor (Pectin methylesterase) (PE)
Length = 583
Score = 801 bits (2068), Expect = 0.0
Identities = 401/574 (69%), Positives = 469/574 (80%), Gaps = 9/574 (1%)
Query: 17 KKKLLISLFTTLLIVASIVAIVATTTKNSNKSKNNSIASSSLSLSHHSHAILKSACTTTL 76
KKK+ +++ ++L+VA+++ +VA +S KN+ + +++S +HAI+KSAC+ TL
Sbjct: 18 KKKIYLAIVASVLLVAAVIGVVAGVKSHS---KNSDDHADIMAISSSAHAIVKSACSNTL 74
Query: 77 YPELCFSAISSEPNITHKITNHKDVISLSLNITTRAVEHNYFTVEKLL-LRKSLTKREKI 135
+PELC+SAI + + + K+T+ KDVI LSLNIT +AV NY+ V++L+ RK LT REK+
Sbjct: 75 HPELCYSAIVNVSDFSKKVTSQKDVIELSLNITVKAVRRNYYAVKELIKTRKGLTPREKV 134
Query: 136 ALHDCLETIDETLDELKEAQNDLVLYPSKKTLYQHADDLKTLISSAITNQVTCLDGFSHD 195
ALHDCLET+DETLDEL A DL LYP+KK+L +H +DLKTLISSAITNQ TCLDGFSHD
Sbjct: 135 ALHDCLETMDETLDELHTAVEDLELYPNKKSLKEHVEDLKTLISSAITNQETCLDGFSHD 194
Query: 196 DADKEVRKVLQEGQIHVEHMCSNALAMTKNMTDKDIAEFEQTNMVLGSNKNRKLLEEENG 255
+ADK+VRKVL +GQ HVE MCSNALAM NMTD DIA M L + N + L E+NG
Sbjct: 195 EADKKVRKVLLKGQKHVEKMCSNALAMICNMTDTDIAN----EMKLSAPANNRKLVEDNG 250
Query: 256 VGWPEWISAGDRRLLQGSTVKADVVVAADGSGNFKTVSEAVAAAPLKSSKRYVIKIKAGV 315
WPEW+SAGDRRLLQ STV DVVVAADGSG++KTVSEAV AP KSSKRYVI+IKAGV
Sbjct: 251 E-WPEWLSAGDRRLLQSSTVTPDVVVAADGSGDYKTVSEAVRKAPEKSSKRYVIRIKAGV 309
Query: 316 YKENVEVPKKKTNIMFLGDGRTNTIITGSRNVVDGSTTFHSATVAIVGGNFLARDITFQN 375
Y+ENV+VPKKKTNIMF+GDG++NTIIT SRNV DGSTTFHSATV V G LARDITFQN
Sbjct: 310 YRENVDVPKKKTNIMFMGDGKSNTIITASRNVQDGSTTFHSATVVRVAGKVLARDITFQN 369
Query: 376 TAGPAKHQAVALRVGADLSAFYNCDIIAYQDTLYVHNNRQFFVNCFISGTVDFIFGNSAV 435
TAG +KHQAVAL VG+DLSAFY CD++AYQDTLYVH+NRQFFV C ++GTVDFIFGN A
Sbjct: 370 TAGASKHQAVALCVGSDLSAFYRCDMLAYQDTLYVHSNRQFFVQCLVAGTVDFIFGNGAA 429
Query: 436 VFQNCDIHARRPNSGQKNMVTAQGRVDPNQNTGIVIQKCRIGATKDLEGVKGNFPTYLGR 495
VFQ+CDIHARRP SGQKNMVTAQGR DPNQNTGIVIQKCRIGAT DL V+ +FPTYLGR
Sbjct: 430 VFQDCDIHARRPGSGQKNMVTAQGRTDPNQNTGIVIQKCRIGATSDLRPVQKSFPTYLGR 489
Query: 496 PWKEYSRTVFMQSSISDVIDPVGWHEWNGNFALNTLVYREYQNTGPGAGTSKRVTWKGFK 555
PWKEYSRTV MQSSI+DVI P GWHEWNGNFAL+TL Y EY NTG GA TS RV WKG K
Sbjct: 490 PWKEYSRTVIMQSSITDVIQPAGWHEWNGNFALDTLFYGEYANTGAGAPTSGRVKWKGHK 549
Query: 556 VITSAAEAQSFTPGNFIGGSSWLGSTGFPFSLGL 589
VITS+ EAQ++TPG FI G SWL STGFPFSLGL
Sbjct: 550 VITSSTEAQAYTPGRFIAGGSWLSSTGFPFSLGL 583
>gb|AAC50023.1| ATPME2 precursor [Arabidopsis thaliana]
Length = 582
Score = 792 bits (2045), Expect = 0.0
Identities = 409/580 (70%), Positives = 461/580 (78%), Gaps = 15/580 (2%)
Query: 16 NKKKLLISLFTTLLIVASIVAIVATTTKNSNKSKNNSIASSSLSLSHHSHAILKSACTTT 75
NKK +L S LL++ASIV I ATTT N++KN I + L SHAILKS C++T
Sbjct: 12 NKKLILSSAAIALLLLASIVGIAATTT---NQNKNQKITT----LFSTSHAILKSVCSST 64
Query: 76 LYPELCFSAISSEPNITHKITNHKDVISLSLNITTRAVEHNYFTVEKLLL-RKSLTKREK 134
LYPELCFSA+++ ++T+ K+VI SLN+TT+AV+HNYF V+KL+ RK LT RE
Sbjct: 65 LYPELCFSAVAATGG--KELTSQKEVIEASLNLTTKAVKHNYFAVKKLIAKRKGLTPREV 122
Query: 135 IALHDCLETIDETLDELKEAQNDLVLYPSKKTLYQHADDLKTLISSAITNQVTCLDGFSH 194
ALHDCLETIDETLDEL A DL YP +K+L +HADDLKTLISSAITNQ TCLDGFS+
Sbjct: 123 TALHDCLETIDETLDELHVAVEDLHQYPKQKSLRKHADDLKTLISSAITNQGTCLDGFSY 182
Query: 195 DDADKEVRKVLQEGQIHVEHMCSNALAMTKNMTDKDIAEFEQTNMV--LGSNKNRKLLEE 252
DDAD++VRK L +GQ+HVEHMCSNALAM KNMT+ DIA FE + L + RKL E
Sbjct: 183 DDADRKVRKALLKGQVHVEHMCSNALAMIKNMTETDIANFELRDKFFNLHQQQQRKLKEV 242
Query: 253 ENGV---GWPEWISAGDRRLLQGSTVKADVVVAADGSGNFKTVSEAVAAAPLKSSKRYVI 309
+ GWP+W+S GDRRLLQGST+KAD VA DGSG+F S AVAAAP KS+KR+VI
Sbjct: 243 TGDLDSDGWPKWLSVGDRRLLQGSTIKADATVADDGSGDFDNGSAAVAAAPEKSNKRFVI 302
Query: 310 KIKAGVYKENVEVPKKKTNIMFLGDGRTNTIITGSRNVVDGSTTFHSATVAIVGGNFLAR 369
IKAGVY+ENVEV KKKTNIMFLGDGR TIITGSRNVVDGSTTFHSATVA VG FLAR
Sbjct: 303 HIKAGVYRENVEVTKKKTNIMFLGDGRGKTIITGSRNVVDGSTTFHSATVAAVGERFLAR 362
Query: 370 DITFQNTAGPAKHQAVALRVGADLSAFYNCDIIAYQDTLYVHNNRQFFVNCFISGTVDFI 429
DITFQNTAGP+KHQAVALRVG+D SAFY CD+ AYQDTLYVH+NRQFFV C I+GTVDFI
Sbjct: 363 DITFQNTAGPSKHQAVALRVGSDFSAFYQCDMFAYQDTLYVHSNRQFFVKCHITGTVDFI 422
Query: 430 FGNSAVVFQNCDIHARRPNSGQKNMVTAQGRVDPNQNTGIVIQKCRIGATKDLEGVKGNF 489
FGN+A V Q+CDI+ARRPNSGQKNMVTAQGR DPNQNTGIVIQ CRIG T DL VKG F
Sbjct: 423 FGNAAAVLQDCDINARRPNSGQKNMVTAQGRSDPNQNTGIVIQNCRIGGTSDLLAVKGTF 482
Query: 490 PTYLGRPWKEYSRTVFMQSSISDVIDPVGWHEWNGNFALNTLVYREYQNTGPGAGTSKRV 549
PTYLGRPWKEYSRTV MQS ISDVI P GWHEW+G+FAL+TL YREY N G GAGT+ RV
Sbjct: 483 PTYLGRPWKEYSRTVIMQSDISDVIRPEGWHEWSGSFALDTLTYREYLNRGGGAGTANRV 542
Query: 550 TWKGFKVITSAAEAQSFTPGNFIGGSSWLGSTGFPFSLGL 589
WKG+KVITS EAQ FT G FIGG WL STGFPFSL L
Sbjct: 543 KWKGYKVITSDTEAQPFTAGQFIGGGGWLASTGFPFSLSL 582
>gb|AAK84486.1| putative thermostable pectinesterase [Citrus sinensis]
gi|24250746|gb|AAK84485.1| putative thermostable
pectinesterase [Citrus sinensis]
Length = 631
Score = 763 bits (1969), Expect = 0.0
Identities = 383/582 (65%), Positives = 467/582 (79%), Gaps = 24/582 (4%)
Query: 18 KKLLISLFTTLLIVASIVAIVATTTKNSNKSKNNSIASSSLSLSHHSHA----ILKSACT 73
+KL ++LF +L+V ++V I+AT NS+ + + IA+ H+HA +++S+C+
Sbjct: 64 RKLFLALFAVVLVVTAVVTIIATRKNNSSNTNDEHIAA-------HAHAYAQPVIRSSCS 116
Query: 74 TTLYPELCFSAISSEPNIT-HKITNHKDVISLSLNITTRAVEHNYFTVEKLLL--RKSLT 130
TLYPELCFSA+S+ P K+ + KDVI LSLN+T AV+HNYF ++KL+ + +LT
Sbjct: 117 ATLYPELCFSALSAAPAAAVSKVNSPKDVIRLSLNLTITAVQHNYFAIKKLITTRKSTLT 176
Query: 131 KREKIALHDCLETIDETLDELKEAQNDLVLYPS---KKTLYQHADDLKTLISSAITNQVT 187
KREK +LHDCLE +DETLDEL + +++L YP+ K++ + AD+LK L+S+A+TNQ T
Sbjct: 177 KREKTSLHDCLEMVDETLDELYKTEHELQGYPAAANNKSIAEQADELKILVSAAMTNQET 236
Query: 188 CLDGFSHDDADKEVRKVLQEGQIHVEHMCSNALAMTKNMTDKDIAEFEQTNMVLGSNKNR 247
CLDGFSH+ ADK++R+ L EGQ+HV HMCSNALAM KNMTD DI + ++V +K R
Sbjct: 237 CLDGFSHERADKKIREELMEGQMHVFHMCSNALAMIKNMTDGDIGK----DIVDHYSKAR 292
Query: 248 KLLEEENGVGWPEWISAGDRRLLQGSTVKADVVVAADGSGNFKTVSEAVAAAPLKSSKRY 307
+L +E WPEW+SAGDRRLLQ +TV DV VAADGSGN+ TV+ AVAAAP SS+RY
Sbjct: 293 RLDDETK---WPEWLSAGDRRLLQATTVVPDVTVAADGSGNYLTVAAAVAAAPEGSSRRY 349
Query: 308 VIKIKAGVYKENVEVPKKKTNIMFLGDGRTNTIITGSRNVVDGSTTFHSATVAIVGGNFL 367
+I+IKAG Y+ENVEVPKKK N+MF+GDGRT TIITGSRNVVDGSTTF+SATVA+VG FL
Sbjct: 350 IIRIKAGEYRENVEVPKKKINLMFIGDGRTTTIITGSRNVVDGSTTFNSATVAVVGDGFL 409
Query: 368 ARDITFQNTAGPAKHQAVALRVGADLSAFYNCDIIAYQDTLYVHNNRQFFVNCFISGTVD 427
ARDITFQNTAGP+KHQAVALRVG+DLSAFY CD++AYQDTLYVH+ RQF+ +C I+GTVD
Sbjct: 410 ARDITFQNTAGPSKHQAVALRVGSDLSAFYRCDMLAYQDTLYVHSLRQFYTSCIIAGTVD 469
Query: 428 FIFGNSAVVFQNCDIHARRPNSGQKNMVTAQGRVDPNQNTGIVIQKCRIGATKDLEGVKG 487
FIFGN+A VFQNCDIHARRPN Q+NMVTAQGR DPNQNTGIVIQKCRIGAT DL VKG
Sbjct: 470 FIFGNAAAVFQNCDIHARRPNPNQRNMVTAQGRDDPNQNTGIVIQKCRIGATSDLLAVKG 529
Query: 488 NFPTYLGRPWKEYSRTVFMQSSISDVIDPVGWHEWNGNFALNTLVYREYQNTGPGAGTSK 547
+F TYLGRPWK YSRTV MQS ISDVI+P GW+EW+GNFAL+TL Y EYQNTG GA TS
Sbjct: 530 SFETYLGRPWKRYSRTVVMQSDISDVINPAGWYEWSGNFALDTLFYAEYQNTGAGADTSN 589
Query: 548 RVTWKGFKVITSAAEAQSFTPGNFIGGSSWLGSTGFPFSLGL 589
RV W FKVITSAAEAQ++T NFI GS+WLGSTGFPFSLGL
Sbjct: 590 RVKWSTFKVITSAAEAQTYTAANFIAGSTWLGSTGFPFSLGL 631
>gb|AAG17110.1| putative pectin methylesterase 3 [Linum usitatissimum]
Length = 555
Score = 694 bits (1790), Expect = 0.0
Identities = 354/567 (62%), Positives = 429/567 (75%), Gaps = 20/567 (3%)
Query: 25 FTTLLIVASIVAIVATTTKNSNKSKNNSIASSSLSLSHHSHAILKSACTTTLYPELCFSA 84
F TLL + I+ +T+T ++ +S+++S AIL S+C+ T +P+LCFS+
Sbjct: 7 FLTLLNLLIIILASSTSTFVASTRTYSSLSASRC-------AILTSSCSNTRHPDLCFSS 59
Query: 85 ISSEPNITHKITNHKDVISLSLNITTRAVEHNYFTVEKLL-LRKSLTKREKIALHDCLET 143
++S P + + DVI S+N+T +V N V K L R LT R + AL DC+ET
Sbjct: 60 LASAP-VHVSLNTQMDVIKASINVTCTSVLRNIAAVNKALSTRTDLTPRSRSALKDCVET 118
Query: 144 IDETLDELKEAQNDLVLYPSKKTLYQHADDLKTLISSAITNQVTCLDGFSHDDADKEVRK 203
+ +LDEL A +L YP+KK++ +HADDLKTL+S+A TNQ TCLDGFSHDD++K+VRK
Sbjct: 119 MSTSLDELHVALAELDEYPNKKSITRHADDLKTLLSAATTNQETCLDGFSHDDSEKKVRK 178
Query: 204 VLQEGQIHVEHMCSNALAMTKNMTDKDIAEFEQTNMVLGSNKNRKLLEEENGVGWPEWIS 263
L+ G + VE MC NAL M NMT+ D+A S N E + WP W+
Sbjct: 179 TLETGPVRVEKMCGNALGMIVNMTETDMA----------SATNAVNTEGGSSGSWPIWMK 228
Query: 264 AGDRRLLQ-GSTVKADVVVAADGSGNFKTVSEAVAAAPLKSSKRYVIKIKAGVYKENVEV 322
GDRRLLQ G+TV +VVVAADGSG ++ VSEAVAAAP KSSKRYVI+IKAG+Y+ENVEV
Sbjct: 229 GGDRRLLQAGTTVTPNVVVAADGSGKYRRVSEAVAAAPSKSSKRYVIRIKAGIYRENVEV 288
Query: 323 PKKKTNIMFLGDGRTNTIITGSRNVVDGSTTFHSATVAIVGGNFLARDITFQNTAGPAKH 382
PK KTNIMF+GDGR+NTIITG++NVVDGSTTF+SATVA+VG FLARDITFQNTAGP+KH
Sbjct: 289 PKDKTNIMFVGDGRSNTIITGNKNVVDGSTTFNSATVAVVGQGFLARDITFQNTAGPSKH 348
Query: 383 QAVALRVGADLSAFYNCDIIAYQDTLYVHNNRQFFVNCFISGTVDFIFGNSAVVFQNCDI 442
QAVALRVGADL+AFY CD +AYQDTLYVH+NRQFF+NC + GTVDFIFGNSA VFQNCDI
Sbjct: 349 QAVALRVGADLAAFYRCDFLAYQDTLYVHSNRQFFINCLVVGTVDFIFGNSAAVFQNCDI 408
Query: 443 HARRPNSGQKNMVTAQGRVDPNQNTGIVIQKCRIGATKDLEGVKGNFPTYLGRPWKEYSR 502
HARRPN GQKNM+TA GR DPNQNTGIVIQK RI AT DL+ VKG+F TYLGRPWK Y+R
Sbjct: 409 HARRPNPGQKNMLTAHGRTDPNQNTGIVIQKSRIAATSDLQSVKGSFGTYLGRPWKAYAR 468
Query: 503 TVFMQSSISDVIDPVGWHEWNGNFALNTLVYREYQNTGPGAGTSKRVTWKGFKVITSAAE 562
TV MQS+ISDV+ P GWHEW+GNFALNTL Y E++N+G G+G + RV WKG KVI+S AE
Sbjct: 469 TVIMQSTISDVVHPAGWHEWDGNFALNTLFYGEHKNSGAGSGVNGRVKWKGHKVISSDAE 528
Query: 563 AQSFTPGNFIGGSSWLGSTGFPFSLGL 589
A FTPG FI G SWLGST FPF+LGL
Sbjct: 529 AAGFTPGRFIAGGSWLGSTTFPFTLGL 555
>gb|AAK69696.1| putative pectin methylesterase LuPME5 [Linum usitatissimum]
Length = 553
Score = 680 bits (1755), Expect = 0.0
Identities = 342/527 (64%), Positives = 419/527 (78%), Gaps = 8/527 (1%)
Query: 65 HAILKSACTTTLYPELCFSAISSEPNITHKITNHKDVISLSLNITTRAVEHNYFTVEKLL 124
H ++KS+C+TTLYPELC SA S+ + I DV+ LSLN T AV+ N ++K++
Sbjct: 33 HPVIKSSCSTTLYPELCHSAASASAAVLSDIKTTTDVVDLSLNATIAAVQANNQAIKKII 92
Query: 125 LRKSL--TKREKIALHDCLETIDETLDELKEAQNDLVLYPSKKTLYQHADDLKTLISSAI 182
+SL TKREK AL DC+E ET+DE + +L + KK+ + +DLKTL+S+A+
Sbjct: 93 SSRSLSLTKREKAALADCIELCGETMDEPVKTIEEL--HGKKKSAAERGEDLKTLLSAAM 150
Query: 183 TNQVTCLDGFSHDDADKEVRKVLQEGQIHVEHMCSNALAMTKNMTDKDIAEFEQTNMVLG 242
TNQ TCLDGFSHD DK+VR++L GQ +V MCSN+LAM +N+T++++ +T L
Sbjct: 151 TNQETCLDGFSHDKGDKKVRELLAAGQTNVGRMCSNSLAMVENITEEEVFREGKTASFL- 209
Query: 243 SNKNRKLLEEENGVGWPEWISAGDRRLLQGSTVKADVVVAADGSGNFKTVSEAVAAAPLK 302
++ RK+ EEE+G WP WISAGDRRLLQ TV +VVVAADGSGNF+TVS+AVAAAP
Sbjct: 210 -SEGRKMGEEEDG--WPRWISAGDRRLLQAGTVTPNVVVAADGSGNFRTVSQAVAAAPEG 266
Query: 303 SSKRYVIKIKAGVYKENVEVPKKKTNIMFLGDGRTNTIITGSRNVVDGSTTFHSATVAIV 362
S+ RYVI+IKAGVY+E + VPKKKTN+MF+GDGRT+TIITGS NVVDGSTTF+SATVA+V
Sbjct: 267 STSRYVIRIKAGVYRETLVVPKKKTNLMFVGDGRTSTIITGSMNVVDGSTTFNSATVAVV 326
Query: 363 GGNFLARDITFQNTAGPAKHQAVALRVGADLSAFYNCDIIAYQDTLYVHNNRQFFVNCFI 422
G F+ARD+TFQNTAGP+KHQAVALRV AD +AFY CD++AYQDTLYVH+ RQF+V+CFI
Sbjct: 327 GDRFMARDLTFQNTAGPSKHQAVALRVNADFTAFYRCDMLAYQDTLYVHSLRQFYVSCFI 386
Query: 423 SGTVDFIFGNSAVVFQNCDIHARRPNSGQKNMVTAQGRVDPNQNTGIVIQKCRIGATKDL 482
+GTVDFIFGN+AVV QNCDIHARRPNSGQ+NMVTAQGR DPNQNTGIVIQKCRIGAT+DL
Sbjct: 387 AGTVDFIFGNAAVVLQNCDIHARRPNSGQRNMVTAQGRDDPNQNTGIVIQKCRIGATQDL 446
Query: 483 EGVKGNFPTYLGRPWKEYSRTVFMQSSISDVIDPVGWHEWNGNFALNTLVYREYQNTGPG 542
V+ + +YLGRPWK YSRTV MQ+ IS+VI P GW W+GNFAL TL YREY NTG G
Sbjct: 447 LQVQSSVESYLGRPWKMYSRTVIMQTDISNVIRPAGWFMWDGNFALATLTYREYANTGAG 506
Query: 543 AGTSKRVTWKGFKVITSAAEAQSFTPGNFIGGSSWLGSTGFPFSLGL 589
+GTS RV W G+KVITSA+EAQ F P +FIGG+SWL +TGFPFSL L
Sbjct: 507 SGTSGRVRWGGYKVITSASEAQPFAPRSFIGGASWLPATGFPFSLDL 553
>emb|CAC18726.1| putative pectin methylesterase [Populus tremula x Populus
tremuloides]
Length = 574
Score = 620 bits (1600), Expect = e-176
Identities = 324/585 (55%), Positives = 413/585 (70%), Gaps = 24/585 (4%)
Query: 11 ISNSGNKKKLLISLFTTLLIVASIVAIVATTTKNSNKSKNNSIASSSLSLSHHSHAILKS 70
IS S K+L++++F + L+VA+I+AI + N +KN++ +HA+L +
Sbjct: 8 ISMSKKNKRLVLAIFASFLLVATIIAISIGVNSHKNSTKNDA-----------AHALLMA 56
Query: 71 ACTTTLYPELCFSAISSEPNITHKITNHKDVISLSLNITTRAVEHNYFTVEKLLLRKSLT 130
+C +T YP+LC+SA + P+ + + K VI ++N T A+ K+L ++ T
Sbjct: 57 SCNSTRYPDLCYSAATCFPDDSGNSGDPKAVILKNINATIDAINSKKIEANKILSTENPT 116
Query: 131 KREKIALHDCLETIDETLDELKEAQNDLVLYPSKKTLYQ--HADDLKTLISSAITNQVTC 188
K++K AL DC + D +L +L + +L P+ K L Q +AD+L T +S+ +N+ +C
Sbjct: 117 KKQKTALEDCTKNYDSSLADLDKVWGELNRNPNNKKLQQQSYADELATKVSACKSNEDSC 176
Query: 189 LDGFSHDDADKEVRKV-LQEGQIHVEHMCSNALAMTKNMTD--KDIAEFEQTNMVLGSNK 245
DGFSH +E R + L + + MCSN LA+ K +T+ K IA +T
Sbjct: 177 FDGFSHSSFLREFRDIFLGSSEDNAGKMCSNTLALIKTLTEGTKAIANRLKTT------- 229
Query: 246 NRKLLEEENG-VGWPEWISAGDRRLLQGSTVKADVVVAADGSGNFKTVSEAVAAAPLKSS 304
+RKL EE++ GWPEW+S DRRL Q S + DVVV+ADGSG ++TVS AVAAAP S+
Sbjct: 230 SRKLKEEDDSDEGWPEWLSVTDRRLFQSSLLTPDVVVSADGSGKYRTVSAAVAAAPKHSA 289
Query: 305 KRYVIKIKAGVYKENVEVPKKKTNIMFLGDGRTNTIITGSRNVVDGSTTFHSATVAIVGG 364
KRY+IKIKAGVY+ENVEVP +KTNIMFLGDGR TIIT SRNVVDG TT+HSATVA+VG
Sbjct: 290 KRYIIKIKAGVYRENVEVPSEKTNIMFLGDGRKRTIITASRNVVDGGTTYHSATVAVVGK 349
Query: 365 NFLARDITFQNTAGPAKHQAVALRVGADLSAFYNCDIIAYQDTLYVHNNRQFFVNCFISG 424
FLARDITFQNTAG +K+QAVALRV +D +AFY C ++AYQ+TL+VH+NRQFF NC+I+G
Sbjct: 350 GFLARDITFQNTAGASKYQAVALRVESDFAAFYKCGMVAYQNTLHVHSNRQFFTNCYIAG 409
Query: 425 TVDFIFGNSAVVFQNCDIHARRPNSGQKNMVTAQGRVDPNQNTGIVIQKCRIGATKDLEG 484
TVDFIFGNSA VFQ+CDI ARR N GQ +TAQGR DPNQNTGIVIQK RIG T DL+
Sbjct: 410 TVDFIFGNSAAVFQDCDIRARRANPGQTITITAQGRSDPNQNTGIVIQKSRIGGTPDLQH 469
Query: 485 VKGNFPTYLGRPWKEYSRTVFMQSSISDVIDPVGWHEWNGNFALNTLVYREYQNTGPGAG 544
+ NF +LGRPWKEYSRTV MQSSISDVI P GW EW G FAL+TL + EY+N+G GAG
Sbjct: 470 ARSNFSAFLGRPWKEYSRTVIMQSSISDVISPAGWREWKGRFALDTLHFAEYENSGAGAG 529
Query: 545 TSKRVTWKGFKVITSAAEAQSFTPGNFIGGSSWLGSTGFPFSLGL 589
TS RV WKG+KVIT A EAQ+FT NFI GSSWL ST FPFSLGL
Sbjct: 530 TSGRVPWKGYKVITDATEAQAFTARNFITGSSWLKSTTFPFSLGL 574
>emb|CAC18727.1| putative pectin methylesterase [Populus tremula x Populus
tremuloides]
Length = 536
Score = 607 bits (1566), Expect = e-172
Identities = 315/532 (59%), Positives = 387/532 (72%), Gaps = 13/532 (2%)
Query: 64 SHAILKSACTTTLYPELCFSAISSEPNITHKITNHKDVISLSLNITTRAVEHNYFTVEKL 123
+HA+L ++C +T YP+LC+SA SS P+ + K + K VI ++N T A+ K+
Sbjct: 12 AHALLMASCNSTRYPDLCYSAASSFPDDSGKSGDPKAVILKNINATIDAINSKKIEANKI 71
Query: 124 LLRKSLTKREKIALHDCLETIDETLDELKEAQNDLVLYPSKKTLYQ--HADDLKTLISSA 181
L ++ TK++K AL DC + D +L +L + +L P+ K L Q +AD+L T +S+
Sbjct: 72 LSTENPTKKQKTALEDCTKNYDSSLADLDKVWGELNRNPNNKKLQQQSYADELTTKVSAC 131
Query: 182 ITNQVTCLDGFSHDDADKEVRKV-LQEGQIHVEHMCSNALAMTKNMTD--KDIAEFEQTN 238
+N+ +C DGFSH + R + L + + MCSN LA+ K +T+ K IA +T
Sbjct: 132 KSNEDSCFDGFSHSSFLRGFRDIFLGSSEDNAGKMCSNTLALIKTLTEGTKAIANRLKTT 191
Query: 239 MVLGSNKNRKLLEEENG-VGWPEWISAGDRRLLQGSTVKADVVVAADGSGNFKTVSEAVA 297
+RKL EE++ GWPEW+S DRRL Q S + DVVVAADGSG ++TVS AVA
Sbjct: 192 -------SRKLKEEDDSDEGWPEWLSVTDRRLFQSSLLTPDVVVAADGSGKYRTVSAAVA 244
Query: 298 AAPLKSSKRYVIKIKAGVYKENVEVPKKKTNIMFLGDGRTNTIITGSRNVVDGSTTFHSA 357
AAP S+KRY+IKIKAGVY+ENVEVP +KTNIMFLGDGR TIIT SRNVVDG TT+HSA
Sbjct: 245 AAPKHSAKRYIIKIKAGVYRENVEVPSEKTNIMFLGDGRKKTIITASRNVVDGGTTYHSA 304
Query: 358 TVAIVGGNFLARDITFQNTAGPAKHQAVALRVGADLSAFYNCDIIAYQDTLYVHNNRQFF 417
TVA+VG FLARDITFQNTAG +K+QAVALRV +D +AFY C ++AYQ+TL+VH+NRQFF
Sbjct: 305 TVAVVGKGFLARDITFQNTAGASKYQAVALRVESDFAAFYKCGVVAYQNTLHVHSNRQFF 364
Query: 418 VNCFISGTVDFIFGNSAVVFQNCDIHARRPNSGQKNMVTAQGRVDPNQNTGIVIQKCRIG 477
N +I+GTVDFIFGNSA VFQ+CDI ARRPN GQ +TAQGR DPNQNTGIVIQK RIG
Sbjct: 365 TNSYIAGTVDFIFGNSAAVFQDCDIRARRPNPGQTITITAQGRSDPNQNTGIVIQKSRIG 424
Query: 478 ATKDLEGVKGNFPTYLGRPWKEYSRTVFMQSSISDVIDPVGWHEWNGNFALNTLVYREYQ 537
AT DL+ + NF YLGRPWKEYSRTV MQSSISDVI P GW EW G FALNTL + EY+
Sbjct: 425 ATPDLQHARSNFSVYLGRPWKEYSRTVIMQSSISDVISPAGWREWKGRFALNTLHFAEYE 484
Query: 538 NTGPGAGTSKRVTWKGFKVITSAAEAQSFTPGNFIGGSSWLGSTGFPFSLGL 589
N+G GAGTS RV WKG+KVIT A EAQ+FT NFI GSSWL ST FPFSLGL
Sbjct: 485 NSGAGAGTSGRVPWKGYKVITDATEAQAFTARNFITGSSWLKSTTFPFSLGL 536
>emb|CAC18725.1| putative pectin methylesterase [Populus tremula x Populus
tremuloides]
Length = 588
Score = 606 bits (1563), Expect = e-172
Identities = 326/597 (54%), Positives = 409/597 (67%), Gaps = 25/597 (4%)
Query: 1 MNKIKQSLAGISNSGNKKKLLISLFTTLLIVASIVAIVATTTKNSNKSKNNSIASSSLSL 60
+ +I S IS S K L ++LF +LL+VA++ A+V T NS S N A
Sbjct: 9 LTEISDSGKHISFSHKNKSLSLALFVSLLLVATLAAVV--TPVNSQNSNKNGAA------ 60
Query: 61 SHHSHAILKSACTTTLYPELCFSAISSEPNITHK---ITNHKDVISLSLNITTRAVEHNY 117
H+I+K +C++T YPELC+SAI++ P I + DV+ S+ T +A++ N
Sbjct: 61 ----HSIIKMSCSSTRYPELCYSAIANGPGAAASLAAINDENDVLIESIKATQQAIDTNT 116
Query: 118 FTVE--KLLLRKSLTKREKIALHDCLETIDETLDELKEAQNDLVLYPSKKTLYQHAD--- 172
+E K + LT ++ AL + + + +L+ A+ L+ Y ++ L D
Sbjct: 117 AGIESYKTTNKMKLTDQQNDALDASTDNNELSQSDLQNAEQSLLYYTNEIPLSDDQDAGP 176
Query: 173 DLKTLISSAITNQVTCLDGFSHDDADKEVRKVLQEGQIHVEHMCSNALAMTKNMTDKDIA 232
D+ T +SS IT Q T +DGFSH ADKEVRK + +G +V MC N LAM+ NMT A
Sbjct: 177 DINTPLSSCITYQDTIMDGFSHTAADKEVRKDISDGVDNVRKMCMNTLAMSMNMTATRTA 236
Query: 233 EFEQTNMVLGSNKNRKLLEEENGVGWPEWISAGDRRLLQGSTVKADVVVAADGSGNFKTV 292
N + + +N K N GWP+W+S +RRLLQ S++ DVVVAADGSGN+ TV
Sbjct: 237 -----NELKTTKRNLKEENSRNEGGWPKWLSVANRRLLQSSSLTPDVVVAADGSGNYSTV 291
Query: 293 SEAVAAAPLKSSKRYVIKIKAGVYKENVEVPKKKTNIMFLGDGRTNTIITGSRNVVDGST 352
S AVAAAP +SSKRY+I+IKAGVY+E V+VP KT++MFLGDGR TIIT SR+VVDG T
Sbjct: 292 SAAVAAAPTRSSKRYIIRIKAGVYRETVQVPINKTSLMFLGDGRRKTIITASRSVVDGIT 351
Query: 353 TFHSATVAIVGGNFLARDITFQNTAGPAKHQAVALRVGADLSAFYNCDIIAYQDTLYVHN 412
F SATVA +G FLARDI F+NTAGP+ QAVALRV +D +AFY C+++ YQDTL+VH
Sbjct: 352 AFRSATVAAMGEGFLARDIAFENTAGPSNRQAVALRVSSDRAAFYKCNVLGYQDTLHVHA 411
Query: 413 NRQFFVNCFISGTVDFIFGNSAVVFQNCDIHARRPNSGQKNMVTAQGRVDPNQNTGIVIQ 472
NRQFF+NC I+GTVDFIFGNSAVVFQ+CDIHARRPN GQ +TAQGR DPNQ TGIVIQ
Sbjct: 412 NRQFFINCLIAGTVDFIFGNSAVVFQDCDIHARRPNPGQTITITAQGRSDPNQKTGIVIQ 471
Query: 473 KCRIGATKDLEGVKGNFPTYLGRPWKEYSRTVFMQSSISDVIDPVGWHEWNGNFALNTLV 532
K RI AT DL V+ NF YLGRPWKE+SRTV MQSSISDVI+ GW EW G +ALNTL
Sbjct: 472 KSRIHATSDLLPVRSNFSAYLGRPWKEHSRTVVMQSSISDVINRAGWLEWRGKYALNTLY 531
Query: 533 YREYQNTGPGAGTSKRVTWKGFKVITSAAEAQSFTPGNFIGGSSWLGSTGFPFSLGL 589
Y EY N+G GA TS+RVTWKG+KVIT+ AEA+SFTP NFI GS+WL ST FPFSL L
Sbjct: 532 YGEYNNSGAGAATSERVTWKGYKVITATAEAKSFTPRNFIAGSTWLKSTTFPFSLDL 588
>gb|AAL24278.1| AT3g14310/MLN21_9 [Arabidopsis thaliana]
Length = 388
Score = 599 bits (1545), Expect = e-170
Identities = 296/388 (76%), Positives = 321/388 (82%), Gaps = 13/388 (3%)
Query: 215 MCSNALAMTKNMTDKDIAEFEQTNMVLGSNKNRKLLEEENGV-------------GWPEW 261
MCSNALAM KNMTD DIA FEQ + +N+ K +E V GWP W
Sbjct: 1 MCSNALAMIKNMTDTDIANFEQKAKITSNNRKLKEENQETTVAVDIAGAGELDSEGWPTW 60
Query: 262 ISAGDRRLLQGSTVKADVVVAADGSGNFKTVSEAVAAAPLKSSKRYVIKIKAGVYKENVE 321
+SAGDRRLLQGS VKAD VAADGSG FKTV+ AVAAAP S+KRYVI IKAGVY+ENVE
Sbjct: 61 LSAGDRRLLQGSGVKADATVAADGSGTFKTVAAAVAAAPENSNKRYVIHIKAGVYRENVE 120
Query: 322 VPKKKTNIMFLGDGRTNTIITGSRNVVDGSTTFHSATVAIVGGNFLARDITFQNTAGPAK 381
V KKK NIMF+GDGRT TIITGSRNVVDGSTTFHSATVA VG FLARDITFQNTAGP+K
Sbjct: 121 VAKKKKNIMFMGDGRTRTIITGSRNVVDGSTTFHSATVAAVGERFLARDITFQNTAGPSK 180
Query: 382 HQAVALRVGADLSAFYNCDIIAYQDTLYVHNNRQFFVNCFISGTVDFIFGNSAVVFQNCD 441
HQAVALRVG+D SAFYNCD++AYQDTLYVH+NRQFFV C I+GTVDFIFGN+AVV Q+CD
Sbjct: 181 HQAVALRVGSDFSAFYNCDMLAYQDTLYVHSNRQFFVKCLIAGTVDFIFGNAAVVLQDCD 240
Query: 442 IHARRPNSGQKNMVTAQGRVDPNQNTGIVIQKCRIGATKDLEGVKGNFPTYLGRPWKEYS 501
IHARRPNSGQKNMVTAQGR DPNQNTGIVIQKCRIGAT DL+ VKG+FPTYLGRPWKEYS
Sbjct: 241 IHARRPNSGQKNMVTAQGRTDPNQNTGIVIQKCRIGATSDLQSVKGSFPTYLGRPWKEYS 300
Query: 502 RTVFMQSSISDVIDPVGWHEWNGNFALNTLVYREYQNTGPGAGTSKRVTWKGFKVITSAA 561
+TV MQS+ISDVI P GW EW G FALNTL YREY NTG GAGT+ RV W+GFKVIT+AA
Sbjct: 301 QTVIMQSAISDVIRPEGWSEWTGTFALNTLTYREYSNTGAGAGTANRVKWRGFKVITAAA 360
Query: 562 EAQSFTPGNFIGGSSWLGSTGFPFSLGL 589
EAQ +T G FIGG WL STGFPFSLGL
Sbjct: 361 EAQKYTAGQFIGGGGWLSSTGFPFSLGL 388
Database: nr
Posted date: Jul 5, 2005 12:34 AM
Number of letters in database: 863,360,394
Number of sequences in database: 2,540,612
Lambda K H
0.316 0.131 0.378
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 954,712,836
Number of Sequences: 2540612
Number of extensions: 40305046
Number of successful extensions: 115929
Number of sequences better than 10.0: 459
Number of HSP's better than 10.0 without gapping: 344
Number of HSP's successfully gapped in prelim test: 115
Number of HSP's that attempted gapping in prelim test: 114476
Number of HSP's gapped (non-prelim): 579
length of query: 589
length of database: 863,360,394
effective HSP length: 134
effective length of query: 455
effective length of database: 522,918,386
effective search space: 237927865630
effective search space used: 237927865630
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.6 bits)
S2: 78 (34.7 bits)
Medicago: description of AC137821.5


