

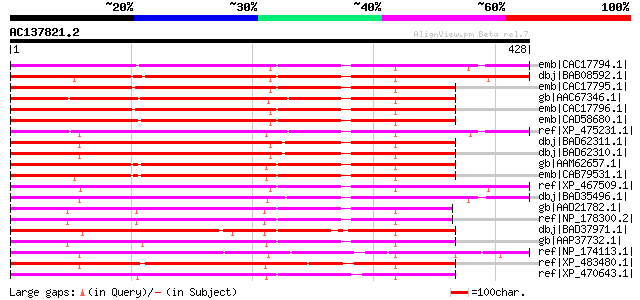
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC137821.2 + phase: 0 /pseudo
(428 letters)
Database: nr
2,540,612 sequences; 863,360,394 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
emb|CAC17794.1| microtubule-associated protein MAP65-1a [Nicotia... 290 8e-77
dbj|BAB08592.1| unnamed protein product [Arabidopsis thaliana] g... 283 6e-75
emb|CAC17795.1| microtubule-associated protein MAP65-1b [Nicotia... 282 1e-74
gb|AAC67346.1| hypothetical protein [Arabidopsis thaliana] gi|25... 281 4e-74
emb|CAC17796.1| microtubule-associated protein MAP65-1c [Nicotia... 279 1e-73
emb|CAD58680.1| 65kD microtubule associated protein [Daucus carota] 275 2e-72
ref|XP_475231.1| putative microtubule-associated protein [Oryza ... 272 1e-71
dbj|BAD62311.1| putative microtubule-associated protein [Oryza s... 271 3e-71
dbj|BAD62310.1| putative microtubule-associated protein [Oryza s... 271 3e-71
gb|AAM62657.1| microtubule-associated protein MAP65-1a [Arabidop... 268 2e-70
emb|CAB79531.1| putative protein [Arabidopsis thaliana] gi|44551... 264 3e-69
ref|XP_467509.1| putative microtubule-associated protein MAP65-1... 253 8e-66
dbj|BAD35496.1| putative microtubule-associated protein MAP65-1a... 247 6e-64
gb|AAD21782.1| unknown protein [Arabidopsis thaliana] gi|2541094... 221 3e-56
ref|NP_178300.2| microtubule associated protein (MAP65/ASE1) fam... 221 3e-56
dbj|BAD37971.1| putative microtubule-associated protein MAP65-1a... 220 6e-56
gb|AAP37732.1| At1g14690 [Arabidopsis thaliana] gi|21539547|gb|A... 219 1e-55
ref|NP_174113.1| microtubule associated protein (MAP65/ASE1) fam... 219 1e-55
ref|XP_483480.1| putative microtubule-associated protein [Oryza ... 211 3e-53
ref|XP_470643.1| Unknown protein [Oryza sativa (japonica cultiva... 209 1e-52
>emb|CAC17794.1| microtubule-associated protein MAP65-1a [Nicotiana tabacum]
Length = 580
Score = 290 bits (741), Expect = 8e-77
Identities = 182/459 (39%), Positives = 277/459 (59%), Gaps = 47/459 (10%)
Query: 1 MLLQIEQEYSEIYHRKVEDTIKHKDYLNKVLDDFQSEIANIASSLGED-YVSF-SRGKGT 58
MLLQI+QE ++Y RKV+ +K + +L + L D + E++ + S+LGE YV + GT
Sbjct: 40 MLLQIDQECLDVYKRKVDQAVKSRAHLLQALADAKVELSRLLSALGEKTYVGIPEKTSGT 99
Query: 59 LKQQLANIIPVVEDLRFKKKERVKEILEIKSQISQIRAEKAGCGQSKGVTDQDVDQCDLT 118
+K+QLA I P +E L +K +R+KE +++SQI +I +E AG + V VD+ DL+
Sbjct: 100 IKEQLAAIAPALEKLWEQKDDRIKEFFDVQSQIQKISSEIAGTREQ--VESLTVDESDLS 157
Query: 119 MEKLGKLKSHLNELQNEKNIRHQKVKSHISTISELSAVMSIDISEILNGIHPSLNDSSNG 178
++KL + ++ L ELQ EK+ R QKV +ST+ +L AV+ +D + +HPSLNDS+
Sbjct: 158 LKKLDEFQAQLQELQKEKSERLQKVLELVSTVHDLCAVLGMDFFSTVTEVHPSLNDSTGV 217
Query: 179 AQQSISDETLARLNESVLLLKQEKQQSLQKVQFLE----ELWDFMGITTDEQKAVCDDVI 234
+SIS++TL+ L ++VL+LK++K+Q L K+Q L +LW+ M T +E++++ D V
Sbjct: 218 QSKSISNDTLSSLAKTVLVLKEDKKQRLHKLQELATQLIDLWNLMD-TPEEERSLFDHVT 276
Query: 235 GLISASVDEVSIHGSLSNDIVKQVDVEVQRLQVLNPSKMKEFVFKRQKELAFKRQNELEE 294
ISASVDEV+I G+L+ D+++Q +VEV+RL L SKM KE+AFKRQ ELE+
Sbjct: 277 CNISASVDEVAIPGALALDLIEQAEVEVERLDQLKASKM--------KEIAFKRQAELED 328
Query: 295 IHRGAHMDMDAKAARQILTDLI--GNIDMSELLQGMDKQIRKAKEQAQSRRDIIDRVAEW 352
I+ AH+++D +AAR+ + LI GN+D +ELL MD QI AKE+A SR++I+D+V +W
Sbjct: 329 IYARAHVEIDTEAAREKIMALIDSGNVDPAELLADMDNQIVNAKEEAHSRKEILDKVEKW 388
Query: 353 KSAAEEEKWLDEYERFI*SVRRKRG-----------------------FLSPRFHLW*RI 389
+A EEE WL++Y R RG L + W
Sbjct: 389 MAACEEESWLEDYNRDDNRYNASRGAHLNLKRAEKARILVNKIPALVDSLVAKTRAW--- 445
Query: 390 *LQK*KHGRQTKKKVSYMKSLHEYNVQRQLREEEKRKSR 428
++ + T V + L EY + R REEEKR+ R
Sbjct: 446 --EQERDTTFTYDGVPLLAMLDEYMMLRHDREEEKRRLR 482
>dbj|BAB08592.1| unnamed protein product [Arabidopsis thaliana]
gi|28416713|gb|AAO42887.1| At5g55230 [Arabidopsis
thaliana] gi|15240485|ref|NP_200334.1| microtubule
associated protein (MAP65/ASE1) family protein
[Arabidopsis thaliana]
Length = 587
Score = 283 bits (725), Expect = 6e-75
Identities = 184/454 (40%), Positives = 277/454 (60%), Gaps = 38/454 (8%)
Query: 1 MLLQIEQEYSEIYHRKVEDTIKHKDYLNKVLDDFQSEIANIASSLGEDYVSF--SRGKGT 58
+LLQIEQE ++Y RKVE K + L + L D +E++++ SLG+ + + GT
Sbjct: 40 LLLQIEQECLDVYKRKVEQAAKSRAELLQTLSDANAELSSLTMSLGDKSLVGIPDKSSGT 99
Query: 59 LKQQLANIIPVVEDLRFKKKERVKEILEIKSQISQIRAEKAGCGQSKGVTDQDVDQCDLT 118
+K+QLA I P +E L +K+ERV+E +++SQI +I + AG G S V VD+ DL+
Sbjct: 100 IKEQLAAIAPALEQLWQQKEERVREFSDVQSQIQKICGDIAG-GLSNEVPI--VDESDLS 156
Query: 119 MEKLGKLKSHLNELQNEKNIRHQKVKSHISTISELSAVMSIDISEILNGIHPSLNDSSNG 178
++KL +S L ELQ EK+ R +KV +ST+ +L AV+ +D + +HPSL++ ++
Sbjct: 157 LKKLDDFQSQLQELQKEKSDRLRKVLEFVSTVHDLCAVLGLDFLSTVTEVHPSLDEDTSV 216
Query: 179 AQQSISDETLARLNESVLLLKQEKQQSLQKVQFLE----ELWDFMGITTDEQKAVCDDVI 234
+SIS+ETL+RL ++VL LK +K+Q LQK+Q L +LW+ M T DE++ + D V
Sbjct: 217 QSKSISNETLSRLAKTVLTLKDDKKQRLQKLQELATQLIDLWNLMD-TPDEERELFDHVT 275
Query: 235 GLISASVDEVSIHGSLSNDIVKQVDVEVQRLQVLNPSKMKEFVFKRQKELAFKRQNELEE 294
IS+SVDEV++ G+L+ D+++Q +VEV RL L S+M KE+AFK+Q+ELEE
Sbjct: 276 CNISSSVDEVTVPGALARDLIEQAEVEVDRLDQLKASRM--------KEIAFKKQSELEE 327
Query: 295 IHRGAHMDMDAKAARQILTDLI--GNIDMSELLQGMDKQIRKAKEQAQSRRDIIDRVAEW 352
I+ AH++++ ++AR+ + LI GN++ +ELL MD QI KAKE+A SR+DI+DRV +W
Sbjct: 328 IYARAHVEVNPESARERIMSLIDSGNVEPTELLADMDSQISKAKEEAFSRKDILDRVEKW 387
Query: 353 KSAAEEEKWLDEYERFI*SVRRKRG-FLSPRFHLW*RI*LQK----------------*K 395
SA EEE WL++Y R RG L+ + RI + K +
Sbjct: 388 MSACEEESWLEDYNRDQNRYSASRGAHLNLKRAEKARILVSKIPAMVDTLVAKTRAWEEE 447
Query: 396 HGRQ-TKKKVSYMKSLHEYNVQRQLREEEKRKSR 428
H V + L EY + RQ REEEKR+ R
Sbjct: 448 HSMSFAYDGVPLLAMLDEYGMLRQEREEEKRRLR 481
>emb|CAC17795.1| microtubule-associated protein MAP65-1b [Nicotiana tabacum]
Length = 584
Score = 282 bits (722), Expect = 1e-74
Identities = 162/375 (43%), Positives = 251/375 (66%), Gaps = 19/375 (5%)
Query: 1 MLLQIEQEYSEIYHRKVEDTIKHKDYLNKVLDDFQSEIANIASSLGED-YVSF-SRGKGT 58
MLLQI++E ++Y RKV+ +K + +L + L D + E+ + ++LGE Y + G+
Sbjct: 42 MLLQIDRECLDVYKRKVDQAVKSRAHLLEALADAKIELCRLLAALGEKTYAGIPEKASGS 101
Query: 59 LKQQLANIIPVVEDLRFKKKERVKEILEIKSQISQIRAEKAGCGQSKGVTDQDVDQCDLT 118
+K+QLA I P +E+L +K++RVKE +++QI +I +E AG S+ V + VD+ DL+
Sbjct: 102 IKEQLAAIAPALENLWKQKEDRVKEFFNVQAQIEKISSEIAGI--SEHVENPKVDESDLS 159
Query: 119 MEKLGKLKSHLNELQNEKNIRHQKVKSHISTISELSAVMSIDISEILNGIHPSLNDSSNG 178
++KL + ++ L ELQ EK+ R KV +STI +L AV+ +D + +HPSLNDS+
Sbjct: 160 LKKLDEFQAQLQELQKEKSERLHKVLEFVSTIHDLCAVLGLDFFSTITEVHPSLNDSTGV 219
Query: 179 AQQSISDETLARLNESVLLLKQEKQQSLQKVQFLE----ELWDFMGITTDEQKAVCDDVI 234
QSIS++TL+ L+ +VL L ++K+Q LQK+Q L +LW+ M T +E++++ D V
Sbjct: 220 QSQSISNDTLSNLSRTVLALMEDKKQRLQKLQELATQLIDLWNLMD-TPEEERSLFDHVT 278
Query: 235 GLISASVDEVSIHGSLSNDIVKQVDVEVQRLQVLNPSKMKEFVFKRQKELAFKRQNELEE 294
ISASVDEVSI G+L+ D+++Q +VEV+RL L S+M KE+AFK+Q LEE
Sbjct: 279 CNISASVDEVSIPGALALDLIEQAEVEVERLDQLKASRM--------KEIAFKKQAVLEE 330
Query: 295 IHRGAHMDMDAKAARQILTDLI--GNIDMSELLQGMDKQIRKAKEQAQSRRDIIDRVAEW 352
I AH+++D++AARQ + LI GNI+ +ELL MD QI KA E+A SR++I+++V +W
Sbjct: 331 IFAHAHIEIDSEAARQKIMALIESGNIEPAELLTDMDNQIVKANEEAHSRKEILEKVEKW 390
Query: 353 KSAAEEEKWLDEYER 367
+A EEE WL++Y R
Sbjct: 391 MAACEEESWLEDYNR 405
>gb|AAC67346.1| hypothetical protein [Arabidopsis thaliana] gi|25408614|pir||E84808
hypothetical protein At2g38720 [imported] - Arabidopsis
thaliana gi|15224935|ref|NP_181406.1| microtubule
associated protein (MAP65/ASE1) family protein
[Arabidopsis thaliana]
Length = 587
Score = 281 bits (718), Expect = 4e-74
Identities = 167/374 (44%), Positives = 247/374 (65%), Gaps = 18/374 (4%)
Query: 1 MLLQIEQEYSEIYHRKVEDTIKHKDYLNKVLDDFQSEIANIASSLGEDYVSFSRGK-GTL 59
MLL++EQE +IY++KVE T K + L + L ++EIA++ S+LGE VSF++ K G+L
Sbjct: 33 MLLELEQECLDIYNKKVEKTRKFRAELQRSLAQAEAEIASLMSALGEK-VSFAKKKEGSL 91
Query: 60 KQQLANIIPVVEDLRFKKKERVKEILEIKSQISQIRAEKAGCGQSKGVTDQDVDQCDLTM 119
K+Q++++ PV+EDL KK R KE+ E +QI++I + AG + + +VD+ DLT
Sbjct: 92 KEQISSVKPVLEDLLMKKDRRRKELSETLNQIAEITSNIAGNDYTVS-SGSEVDESDLTQ 150
Query: 120 EKLGKLKSHLNELQNEKNIRHQKVKSHISTISELSAVMSIDISEILNGIHPSLNDSSNGA 179
KL +L++ L +L+NEK +R QKV S+IS + ELS ++S D S+ LN +H SL + S
Sbjct: 151 RKLDELRADLQDLRNEKAVRLQKVNSYISAVHELSEILSFDFSKALNSVHSSLTEFSKTH 210
Query: 180 QQSISDETLARLNESVLLLKQEKQQSLQKVQFL----EELWDFMGITTDEQKAVCDDVIG 235
+SIS++TLAR E V LK EK + L K+Q L +ELW+ M DE++ D
Sbjct: 211 SKSISNDTLARFTELVKSLKAEKHERLLKLQGLGRSMQELWNLMETPMDERRRF-DHCSS 269
Query: 236 LISASVDEVSIHGSLSNDIVKQVDVEVQRLQVLNPSKMKEFVFKRQKELAFKRQNELEEI 295
L+S+ D+ G LS DI+++ + EV+RL L SKM KEL FKRQ ELEEI
Sbjct: 270 LLSSLPDDALKKGCLSLDIIREAEDEVRRLNSLKSSKM--------KELVFKRQCELEEI 321
Query: 296 HRGAHMDMDAKAARQILTDLI--GNIDMSELLQGMDKQIRKAKEQAQSRRDIIDRVAEWK 353
RG HMD+++ AAR+ L +LI G+ D+S++L +D QI KA+E+A SR++I+D+V +W+
Sbjct: 322 CRGNHMDINSDAARKSLVELIESGDGDLSDILASIDGQIEKAREEALSRKEILDKVDKWR 381
Query: 354 SAAEEEKWLDEYER 367
A EEE WLD+YE+
Sbjct: 382 HAKEEETWLDDYEK 395
>emb|CAC17796.1| microtubule-associated protein MAP65-1c [Nicotiana tabacum]
Length = 582
Score = 279 bits (713), Expect = 1e-73
Identities = 160/375 (42%), Positives = 251/375 (66%), Gaps = 19/375 (5%)
Query: 1 MLLQIEQEYSEIYHRKVEDTIKHKDYLNKVLDDFQSEIANIASSLGED-YVSF-SRGKGT 58
+LLQ+++E ++Y RKV +K + +L + L D + E+ + ++LGE Y + G+
Sbjct: 40 ILLQMDRECLDVYKRKVGQAVKSRAHLLEALADAKIELCRLLAALGEKTYAGIPEKASGS 99
Query: 59 LKQQLANIIPVVEDLRFKKKERVKEILEIKSQISQIRAEKAGCGQSKGVTDQDVDQCDLT 118
+K+QLA I P +E+L +K++RVKE ++++QI +I +E AG S+ V + VD+ DL+
Sbjct: 100 IKEQLAAIAPALENLWKQKEDRVKEFFDVQAQIEKISSEIAGI--SEHVENPKVDESDLS 157
Query: 119 MEKLGKLKSHLNELQNEKNIRHQKVKSHISTISELSAVMSIDISEILNGIHPSLNDSSNG 178
++KL + ++ L ELQ EK+ R KV +STI +L AV+ +D + +HPSLNDS+
Sbjct: 158 LKKLDEFQAQLQELQKEKSERLHKVLEFVSTIHDLCAVLGLDFFGTITEVHPSLNDSTGV 217
Query: 179 AQQSISDETLARLNESVLLLKQEKQQSLQKVQFLE----ELWDFMGITTDEQKAVCDDVI 234
+SISD+TL+ L+ +VL L ++K+Q LQK+Q L +LW+ M T +E++++ D V
Sbjct: 218 QSKSISDDTLSNLSRTVLALTEDKKQRLQKLQELTTQLIDLWNLMD-TPEEERSLFDHVC 276
Query: 235 GLISASVDEVSIHGSLSNDIVKQVDVEVQRLQVLNPSKMKEFVFKRQKELAFKRQNELEE 294
ISASVDEVSI G+L+ D+++Q +VEV+RL L S+M KE+AFK+Q LEE
Sbjct: 277 CNISASVDEVSIPGALALDLIEQAEVEVERLDQLKASRM--------KEIAFKKQAVLEE 328
Query: 295 IHRGAHMDMDAKAARQILTDLI--GNIDMSELLQGMDKQIRKAKEQAQSRRDIIDRVAEW 352
I AH+++D++AARQ + LI GNI+ +ELL MD QI KA E+A SR++I+++V +W
Sbjct: 329 IFAHAHVEIDSEAARQKIMTLIESGNIEPAELLTDMDNQIVKANEEAHSRKEILEKVEKW 388
Query: 353 KSAAEEEKWLDEYER 367
+A EEE WL++Y R
Sbjct: 389 MAACEEESWLEDYNR 403
>emb|CAD58680.1| 65kD microtubule associated protein [Daucus carota]
Length = 576
Score = 275 bits (704), Expect = 2e-72
Identities = 160/375 (42%), Positives = 249/375 (65%), Gaps = 19/375 (5%)
Query: 1 MLLQIEQEYSEIYHRKVEDTIKHKDYLNKVLDDFQSEIANIASSLGED-YVSF-SRGKGT 58
MLLQ+EQE ++Y RKV+ +K + +L + L D Q E+ ++ S+LGE YV GT
Sbjct: 40 MLLQLEQECLDVYKRKVDHAVKSRAHLLQSLADGQVELHSLMSALGEKTYVGIPDNTSGT 99
Query: 59 LKQQLANIIPVVEDLRFKKKERVKEILEIKSQISQIRAEKAGCGQSKGVTDQDVDQCDLT 118
+K+QL+ I P +E L +K ER+KE +++SQI + G + G VD+ DL+
Sbjct: 100 IKEQLSAIAPTLEQLWKQKDERIKEFSDVQSQIQNDMCQITGTIEKVG--SPVVDESDLS 157
Query: 119 MEKLGKLKSHLNELQNEKNIRHQKVKSHISTISELSAVMSIDISEILNGIHPSLNDSSNG 178
++KL + ++ L ELQ EK+ R KV +ST+ +L AV+ +D + +HPSL+DS
Sbjct: 158 LKKLDEFQAKLQELQKEKSDRLHKVLEFVSTVHDLCAVLRMDFFSTVTEVHPSLDDSIGV 217
Query: 179 AQQSISDETLARLNESVLLLKQEKQQSLQKVQFLE----ELWDFMGITTDEQKAVCDDVI 234
+SIS++TL++L ++VL L+++K+Q L K+Q L +LW+ M T++E++++ D V
Sbjct: 218 QSKSISNDTLSQLAKTVLALEEDKEQRLHKLQELAKQLIDLWNLMD-TSEEERSLFDHVT 276
Query: 235 GLISASVDEVSIHGSLSNDIVKQVDVEVQRLQVLNPSKMKEFVFKRQKELAFKRQNELEE 294
+SASVDEV+I G+L+ D+++Q +VEV+RL L S+M KE+AFKRQ ELEE
Sbjct: 277 CNVSASVDEVTIPGALALDLIEQAEVEVERLDQLKASRM--------KEIAFKRQAELEE 328
Query: 295 IHRGAHMDMDAKAARQILTDLI--GNIDMSELLQGMDKQIRKAKEQAQSRRDIIDRVAEW 352
I+ AH+++D++AAR+ + LI G+++ +ELL MD QI KAKE+A SR+DI+D+V +W
Sbjct: 329 IYVHAHVEIDSEAARERIMALIDSGDVEPAELLDDMDNQIVKAKEEALSRKDILDKVEKW 388
Query: 353 KSAAEEEKWLDEYER 367
SA EEE WL++Y R
Sbjct: 389 MSACEEESWLEDYNR 403
>ref|XP_475231.1| putative microtubule-associated protein [Oryza sativa (japonica
cultivar-group)] gi|49328159|gb|AAT58855.1| putative
microtubule-associated protein [Oryza sativa (japonica
cultivar-group)]
Length = 570
Score = 272 bits (696), Expect = 1e-71
Identities = 179/460 (38%), Positives = 261/460 (55%), Gaps = 47/460 (10%)
Query: 1 MLLQIEQEYSEIYHRKVEDTIKHKDYLNKVLDDFQSEIANIASSLGEDYVSFSRGK---G 57
M+LQ+E++ +Y +KVE T K K+ L + L + +I I S+LGE SFSR + G
Sbjct: 34 MILQLEEDCLNVYRKKVEQTRKQKEDLIEELSFGELDIEKILSALGERE-SFSRVEKLGG 92
Query: 58 TLKQQLANIIPVVEDLRFKKKERVKEILEIKSQISQIRAEKAGCGQSKGVTDQDVDQCDL 117
TL +QLA + PV+EDLR ++ ERV+E + +++QI ++ AE +G ++ VD+ +L
Sbjct: 93 TLLEQLAKVEPVLEDLRRRRDERVEEFMVVQAQIVRLHAEISGTIENGDPVPPLVDETNL 152
Query: 118 TMEKLGKLKSHLNELQNEKNIRHQKVKSHISTISELSAVMSIDISEILNGIHPSLNDSSN 177
++ +L + KS L ELQ EKN+R QK+ I+ I E+ +MS+D+ + L +HPS +
Sbjct: 153 SLRRLEEFKSQLKELQTEKNLRLQKIDVQINCIHEICNMMSLDLKKELYDVHPSFVELGR 212
Query: 178 GAQQSISDETLARLNESVLLLKQEKQQSLQKVQ----FLEELWDFMGITTDEQKAVCDDV 233
SISD TL RL V L QEK+Q L+K+Q L ELW+ M T EQK D V
Sbjct: 213 TTSMSISDSTLERLAGKVHSLNQEKKQRLRKLQDLGSTLIELWNLMDTPTAEQKCF-DHV 271
Query: 234 IGLISASVDEVSIHGSLSNDIVKQVDVEVQRLQVLNPSKMKEFVFKRQKELAFKRQNELE 293
LIS S G L+ +++++V+VEV+RL L SKM KEL K+ ELE
Sbjct: 272 TSLISVSPSTKMPQGCLARELIEKVEVEVKRLNCLKASKM--------KELVLKKMIELE 323
Query: 294 EIHRGAHMDMDAKAARQILTDLI--GNIDMSELLQGMDKQIRKAKEQAQSRRDIIDRVAE 351
EI++ HMD+D+ R+IL DLI G D+S+LL GMD +I KA+E A SR++I+++V +
Sbjct: 324 EIYKSVHMDIDSDYERRILNDLIDSGKADLSDLLTGMDGRITKAREHALSRKEILEKVEK 383
Query: 352 WKSAAEEEKWLDEYERFI*SVRRKRGF-----------------------LSPRFHLW*R 388
W A+EEE WLDEYE+ RG L+ + W
Sbjct: 384 WTLASEEESWLDEYEKDQNRYNAGRGAHKNLKHAEKARMLVSKIPSLLENLTAKIKAW-- 441
Query: 389 I*LQK*KHGRQTKKKVSYMKSLHEYNVQRQLREEEKRKSR 428
+K K+ + SL EY +RQ ++EEKR+SR
Sbjct: 442 ---EKENGVPFMYDKIRLLDSLEEYTSRRQQKDEEKRRSR 478
>dbj|BAD62311.1| putative microtubule-associated protein [Oryza sativa (japonica
cultivar-group)] gi|54290620|dbj|BAD62191.1| putative
microtubule-associated protein [Oryza sativa (japonica
cultivar-group)]
Length = 576
Score = 271 bits (693), Expect = 3e-71
Identities = 159/376 (42%), Positives = 240/376 (63%), Gaps = 18/376 (4%)
Query: 1 MLLQIEQEYSEIYHRKVEDTIKHKDYLNKVLDDFQSEIANIASSLGEDYVSFSRGK--GT 58
+L Q++QE ++Y RKV+ +D L + LDD + E+A + S+LGE ++ + K GT
Sbjct: 31 VLYQLDQECLDVYKRKVDQATDSRDLLIQALDDSKIELARLLSALGEKAIARTPEKTSGT 90
Query: 59 LKQQLANIIPVVEDLRFKKKERVKEILEIKSQISQIRAEKAGCGQ-SKGVTDQDVDQCDL 117
+KQQLA I P +E L +K ERV+E + ++SQI QI E AG + + V V++ DL
Sbjct: 91 IKQQLAAIAPTLEKLNKQKNERVREFVNVQSQIDQICGEIAGTTEVGEKVATPQVNEDDL 150
Query: 118 TMEKLGKLKSHLNELQNEKNIRHQKVKSHISTISELSAVMSIDISEILNGIHPSLNDSSN 177
T+E+L + +S L EL+ EK+ R +KV ++S I L V+ +D + +HPSL+DS
Sbjct: 151 TLERLEEFRSQLQELEKEKSNRLEKVLDYVSMIHNLCTVLGMDFLSTVTEVHPSLDDSIG 210
Query: 178 GAQQSISDETLARLNESVLLLKQEKQQSLQKVQFLE----ELWDFMGITTDEQKAVCDDV 233
+SIS++TL++L+++V L ++K+ L K+Q L +LWD M E +++ D V
Sbjct: 211 DNCKSISNDTLSKLDKTVATLNEDKKSRLSKLQELAGQLYDLWDLMDAPMQE-RSMFDHV 269
Query: 234 IGLISASVDEVSIHGSLSNDIVKQVDVEVQRLQVLNPSKMKEFVFKRQKELAFKRQNELE 293
SASVD+V+ G+L+ D+++Q +VEVQRL L SKM KE+AFK+Q ELE
Sbjct: 270 TCNRSASVDKVTAPGALALDLIEQAEVEVQRLDQLKYSKM--------KEIAFKKQTELE 321
Query: 294 EIHRGAHMDMDAKAARQILTDLI--GNIDMSELLQGMDKQIRKAKEQAQSRRDIIDRVAE 351
+I+ GAHM +D AA + + LI GNI+ SEL+ M+ QI KAKE+A SR++I+D+V
Sbjct: 322 DIYAGAHMVIDTAAAHEKILALIEAGNIEPSELIADMESQISKAKEEALSRKEILDKVER 381
Query: 352 WKSAAEEEKWLDEYER 367
W SA EEE WL++Y R
Sbjct: 382 WMSACEEESWLEDYNR 397
>dbj|BAD62310.1| putative microtubule-associated protein [Oryza sativa (japonica
cultivar-group)] gi|54290619|dbj|BAD62190.1| putative
microtubule-associated protein [Oryza sativa (japonica
cultivar-group)]
Length = 460
Score = 271 bits (693), Expect = 3e-71
Identities = 159/376 (42%), Positives = 240/376 (63%), Gaps = 18/376 (4%)
Query: 1 MLLQIEQEYSEIYHRKVEDTIKHKDYLNKVLDDFQSEIANIASSLGEDYVSFSRGK--GT 58
+L Q++QE ++Y RKV+ +D L + LDD + E+A + S+LGE ++ + K GT
Sbjct: 31 VLYQLDQECLDVYKRKVDQATDSRDLLIQALDDSKIELARLLSALGEKAIARTPEKTSGT 90
Query: 59 LKQQLANIIPVVEDLRFKKKERVKEILEIKSQISQIRAEKAGCGQ-SKGVTDQDVDQCDL 117
+KQQLA I P +E L +K ERV+E + ++SQI QI E AG + + V V++ DL
Sbjct: 91 IKQQLAAIAPTLEKLNKQKNERVREFVNVQSQIDQICGEIAGTTEVGEKVATPQVNEDDL 150
Query: 118 TMEKLGKLKSHLNELQNEKNIRHQKVKSHISTISELSAVMSIDISEILNGIHPSLNDSSN 177
T+E+L + +S L EL+ EK+ R +KV ++S I L V+ +D + +HPSL+DS
Sbjct: 151 TLERLEEFRSQLQELEKEKSNRLEKVLDYVSMIHNLCTVLGMDFLSTVTEVHPSLDDSIG 210
Query: 178 GAQQSISDETLARLNESVLLLKQEKQQSLQKVQFLE----ELWDFMGITTDEQKAVCDDV 233
+SIS++TL++L+++V L ++K+ L K+Q L +LWD M E +++ D V
Sbjct: 211 DNCKSISNDTLSKLDKTVATLNEDKKSRLSKLQELAGQLYDLWDLMDAPMQE-RSMFDHV 269
Query: 234 IGLISASVDEVSIHGSLSNDIVKQVDVEVQRLQVLNPSKMKEFVFKRQKELAFKRQNELE 293
SASVD+V+ G+L+ D+++Q +VEVQRL L SKM KE+AFK+Q ELE
Sbjct: 270 TCNRSASVDKVTAPGALALDLIEQAEVEVQRLDQLKYSKM--------KEIAFKKQTELE 321
Query: 294 EIHRGAHMDMDAKAARQILTDLI--GNIDMSELLQGMDKQIRKAKEQAQSRRDIIDRVAE 351
+I+ GAHM +D AA + + LI GNI+ SEL+ M+ QI KAKE+A SR++I+D+V
Sbjct: 322 DIYAGAHMVIDTAAAHEKILALIEAGNIEPSELIADMESQISKAKEEALSRKEILDKVER 381
Query: 352 WKSAAEEEKWLDEYER 367
W SA EEE WL++Y R
Sbjct: 382 WMSACEEESWLEDYNR 397
>gb|AAM62657.1| microtubule-associated protein MAP65-1a [Arabidopsis thaliana]
gi|18416818|ref|NP_567756.1| microtubule associated
protein (MAP65/ASE1) family protein [Arabidopsis
thaliana]
Length = 578
Score = 268 bits (685), Expect = 2e-70
Identities = 159/375 (42%), Positives = 243/375 (64%), Gaps = 20/375 (5%)
Query: 1 MLLQIEQEYSEIYHRKVEDTIKHKDYLNKVLDDFQSEIANIASSLGE-DYVSF-SRGKGT 58
+LLQIE+E +Y +KVE K + L + L D E++N+ ++LGE Y+ + GT
Sbjct: 40 LLLQIEEECLNVYKKKVELAAKSRAELLQTLSDATVELSNLTTALGEKSYIDIPDKTSGT 99
Query: 59 LKQQLANIIPVVEDLRFKKKERVKEILEIKSQISQIRAEKAGCGQSKGVTDQDVDQCDLT 118
+K+QL+ I P +E L +K+ERV+ +++SQI +I E AG G + G VD+ DL+
Sbjct: 100 IKEQLSAIAPALEQLWQQKEERVRAFSDVQSQIQKICEEIAG-GLNNG--PHVVDETDLS 156
Query: 119 MEKLGKLKSHLNELQNEKNIRHQKVKSHISTISELSAVMSIDISEILNGIHPSLNDSSNG 178
+++L + L ELQ EK+ R QKV +ST+ +L AV+ +D + +HPSL++++
Sbjct: 157 LKRLDDFQRKLQELQKEKSDRLQKVLEFVSTVHDLCAVLRLDFLSTVTEVHPSLDEANGV 216
Query: 179 AQQSISDETLARLNESVLLLKQEKQQSLQKVQ----FLEELWDFMGITTDEQKAVCDDVI 234
+SIS+ETLARL ++VL LK++K Q L+K+Q L +LW+ M T+DE++ + D V
Sbjct: 217 QTKSISNETLARLAKTVLTLKEDKMQRLKKLQELATQLTDLWNLMD-TSDEERELFDHVT 275
Query: 235 GLISASVDEVSIHGSLSNDIVKQVDVEVQRLQVLNPSKMKEFVFKRQKELAFKRQNELEE 294
ISASV EV+ G+L+ D+++Q +VEV RL L S+M KE+AFK+Q+ELEE
Sbjct: 276 SNISASVHEVTASGALALDLIEQAEVEVDRLDQLKSSRM--------KEIAFKKQSELEE 327
Query: 295 IHRGAHMDMDAKAARQILTDLI--GNIDMSELLQGMDKQIRKAKEQAQSRRDIIDRVAEW 352
I+ AH+++ + R+ + LI GN + +ELL MD QI KAKE+A SR++I+DRV +W
Sbjct: 328 IYARAHIEIKPEVVRERIMSLIDAGNTEPTELLADMDSQIAKAKEEAFSRKEILDRVEKW 387
Query: 353 KSAAEEEKWLDEYER 367
SA EEE WL++Y R
Sbjct: 388 MSACEEESWLEDYNR 402
>emb|CAB79531.1| putative protein [Arabidopsis thaliana] gi|4455199|emb|CAB36522.1|
putative protein [Arabidopsis thaliana]
gi|7485328|pir||T04799 hypothetical protein F10M23.100 -
Arabidopsis thaliana
Length = 473
Score = 264 bits (675), Expect = 3e-69
Identities = 159/385 (41%), Positives = 243/385 (62%), Gaps = 30/385 (7%)
Query: 1 MLLQIEQEYSEIYHRKVEDTIKHKDYLNKVLDDFQSEIANIASSLGE-DYVSF------- 52
+LLQIE+E +Y +KVE K + L + L D E++N+ ++LGE Y+
Sbjct: 40 LLLQIEEECLNVYKKKVELAAKSRAELLQTLSDATVELSNLTTALGEKSYIDICDSMSLF 99
Query: 53 ----SRGKGTLKQQLANIIPVVEDLRFKKKERVKEILEIKSQISQIRAEKAGCGQSKGVT 108
+ GT+K+QL+ I P +E L +K+ERV+ +++SQI +I E AG G + G
Sbjct: 100 PLQPDKTSGTIKEQLSAIAPALEQLWQQKEERVRAFSDVQSQIQKICEEIAG-GLNNG-- 156
Query: 109 DQDVDQCDLTMEKLGKLKSHLNELQNEKNIRHQKVKSHISTISELSAVMSIDISEILNGI 168
VD+ DL++++L + L ELQ EK+ R QKV +ST+ +L AV+ +D + +
Sbjct: 157 PHVVDETDLSLKRLDDFQRKLQELQKEKSDRLQKVLEFVSTVHDLCAVLRLDFLSTVTEV 216
Query: 169 HPSLNDSSNGAQQSISDETLARLNESVLLLKQEKQQSLQKVQ----FLEELWDFMGITTD 224
HPSL++++ +SIS+ETLARL ++VL LK++K Q L+K+Q L +LW+ M T+D
Sbjct: 217 HPSLDEANGVQTKSISNETLARLAKTVLTLKEDKMQRLKKLQELATQLTDLWNLMD-TSD 275
Query: 225 EQKAVCDDVIGLISASVDEVSIHGSLSNDIVKQVDVEVQRLQVLNPSKMKEFVFKRQKEL 284
E++ + D V ISASV EV+ G+L+ D+++Q +VEV RL L S+M KE+
Sbjct: 276 EERELFDHVTSNISASVHEVTASGALALDLIEQAEVEVDRLDQLKSSRM--------KEI 327
Query: 285 AFKRQNELEEIHRGAHMDMDAKAARQILTDLI--GNIDMSELLQGMDKQIRKAKEQAQSR 342
AFK+Q+ELEEI+ AH+++ + R+ + LI GN + +ELL MD QI KAKE+A SR
Sbjct: 328 AFKKQSELEEIYARAHIEIKPEVVRERIMSLIDAGNTEPTELLADMDSQIAKAKEEAFSR 387
Query: 343 RDIIDRVAEWKSAAEEEKWLDEYER 367
++I+DRV +W SA EEE WL++Y R
Sbjct: 388 KEILDRVEKWMSACEEESWLEDYNR 412
>ref|XP_467509.1| putative microtubule-associated protein MAP65-1a [Oryza sativa
(japonica cultivar-group)] gi|51964314|ref|XP_506942.1|
PREDICTED OJ1008_D06.15 gene product [Oryza sativa
(japonica cultivar-group)] gi|45735837|dbj|BAD12872.1|
putative microtubule-associated protein MAP65-1a [Oryza
sativa (japonica cultivar-group)]
Length = 589
Score = 253 bits (646), Expect = 8e-66
Identities = 168/455 (36%), Positives = 261/455 (56%), Gaps = 36/455 (7%)
Query: 1 MLLQIEQEYSEIYHRKVEDTIKHKDYLNKVLDDFQSEIANIASSLGEDYVSFSRGKG--T 58
+L Q++QE ++Y RKV+ K +D L + LD ++E+A +AS+LGE + S K T
Sbjct: 34 VLFQLDQECLDVYKRKVDQATKSRDLLLQALDYSKTELARLASALGEKSIDISPEKTART 93
Query: 59 LKQQLANIIPVVEDLRFKKKERVKEILEIKSQISQIRAEKAGCGQ-SKGVTDQDVDQCDL 117
+K+QL I P +E L KKKER+KE+ I+S+I QIR E AG + + V +++ DL
Sbjct: 94 IKEQLTAIAPTLEQLGKKKKERIKELANIQSRIEQIRGEIAGTLEMGQQVALPQINEDDL 153
Query: 118 TMEKLGKLKSHLNELQNEKNIRHQKVKSHISTISELSAVMSIDISEILNGIHPSLNDSSN 177
T+ KL + + L EL+ EK+ R +KV H+ + +L V+ +D + +H SL+DS
Sbjct: 154 TVRKLREFQLQLQELEKEKSRRLEKVLEHVGMVHDLCNVLGMDFFRTITQVHSSLDDSIG 213
Query: 178 GAQQSISDETLARLNESVLLLKQEKQQSLQKVQFLE----ELWDFMGITTDEQKAVCDDV 233
++IS+ETL++L+ ++ L ++K+ L+K+Q L +LWD M T E++++ D V
Sbjct: 214 NEHKNISNETLSKLDRTIGTLNEDKRLRLEKLQELATQLYDLWDLMD-TPVEERSLFDHV 272
Query: 234 IGLISASVDEVSIHGSLSNDIVKQVDVEVQRLQVLNPSKMKEFVFKRQKELAFKRQNELE 293
+A+V+EV + G+L+ D++ Q EV+RL L SKM KE+AFK+Q LE
Sbjct: 273 SCNRTATVEEVMVPGALAVDVIDQAQTEVERLDQLKYSKM--------KEIAFKKQAILE 324
Query: 294 EIHRGAHMDMDAKAARQILTDLI--GNIDMSELLQGMDKQIRKAKEQAQSRRDIIDRVAE 351
+I+ H+ +D A + + LI GN++ SEL+ MD QI KAKE+A SR++I+D+V
Sbjct: 325 DIYASTHVVLDTAVAHEKIQALIESGNMEPSELIADMDSQILKAKEEALSRKEILDKVER 384
Query: 352 WKSAAEEEKWLDEYERFI*SVRRKRG-FLSPRFHLW*RI*LQK----------------* 394
W S+ EEE WL++Y R RG L+ + RI + K
Sbjct: 385 WISSCEEESWLEDYSRDDNRYNSGRGAHLNLKRAEKARILVSKIPALVETLVAKTRAWEE 444
Query: 395 KHGRQ-TKKKVSYMKSLHEYNVQRQLREEEKRKSR 428
HG VS + L EY + RQ REEEK++ R
Sbjct: 445 NHGLPFMYDGVSLLAMLDEYVILRQEREEEKKRMR 479
>dbj|BAD35496.1| putative microtubule-associated protein MAP65-1a [Oryza sativa
(japonica cultivar-group)]
Length = 581
Score = 247 bits (630), Expect = 6e-64
Identities = 168/461 (36%), Positives = 261/461 (56%), Gaps = 48/461 (10%)
Query: 1 MLLQIEQEYSEIYHRKVEDTIKHKDYLNKVLDDFQSEIANIASSLGEDYVSFSRGK--GT 58
MLLQ+EQE ++Y RKV+ + L + L + +SE++ + +LGE +S K GT
Sbjct: 40 MLLQLEQECLDVYRRKVDQASNSRARLLQQLANAKSELSRLLCALGELSISGIPDKTTGT 99
Query: 59 LKQQLANIIPVVEDLRFKKKERVKEILEIKSQISQIRAEKAGCGQ-SKGVTDQDVDQCDL 117
+K+QL I P +E L +K +RV+E ++ QI IR E AG Q + V++ DL
Sbjct: 100 IKEQLEAISPFLEKLCREKDKRVREFAGVQLQIQTIRGEIAGSLQVGDHMETPRVNEDDL 159
Query: 118 TMEKLGKLKSHLNELQNEKNIRHQKVKSHISTISELSAVMSIDISEILNGIHPSLNDSSN 177
+ +KL + S L LQ EK+ R K+ +S++ +L +V+ +D + +HPSLNDS
Sbjct: 160 STKKLNEFLSELQALQKEKSNRLHKILDFVSSVHDLCSVLGMDFLSTVTEVHPSLNDSVG 219
Query: 178 GAQQSISDETLARLNESVLLLKQEKQQSLQKVQF----LEELWDFMGITTDEQKAVCDDV 233
+SISD TL++L++ V+ LK+EK + L+++Q L +LW+ M + DE++ + D V
Sbjct: 220 AEFKSISDATLSKLSKMVIQLKEEKSKRLERIQALASQLTDLWNLMDTSADERQ-LFDHV 278
Query: 234 IGLISASVDEVSIHGSLSNDIVKQVDVEVQRLQVLNPSKMKEFVFKRQKELAFKRQNELE 293
IS+++DEV+ G+L D+++Q ++EV+RL L S+M K++AFKRQ ELE
Sbjct: 279 TCNISSTLDEVTAPGALDIDLIEQAELEVERLDQLKASRM--------KDIAFKRQTELE 330
Query: 294 EIHRGAHMDMDAKAAR-QILTDLIGNI-DMSELLQGMDKQIRKAKEQAQSRRDIIDRVAE 351
+I+ AH+ +D AAR +ILT + +I + SELL M+ QI KAKE+A SR+DI+++V
Sbjct: 331 DIYAQAHITIDTSAARDRILTVIDSSIFEPSELLADMENQILKAKEEALSRKDILEKVER 390
Query: 352 WKSAAEEEKWLDEYERFI*SVRRKRG-----------------------FLSPRFHLW*R 388
W SA EEE WL++Y + RG L + W
Sbjct: 391 WMSACEEESWLEDYSQDDNRYSATRGAHLNLKRAEKARLLVSKIPVIVDTLMAKTRAW-- 448
Query: 389 I*LQK*KHGRQ-TKKKVSYMKSLHEYNVQRQLREEEKRKSR 428
+ +HG + V + L EY V RQ +EEEKR+ R
Sbjct: 449 ----EQEHGMPFSYDGVHLLAMLDEYKVLRQQKEEEKRRMR 485
>gb|AAD21782.1| unknown protein [Arabidopsis thaliana] gi|25410943|pir||F84430
hypothetical protein At2g01910 [imported] - Arabidopsis
thaliana
Length = 608
Score = 221 bits (564), Expect = 3e-56
Identities = 132/378 (34%), Positives = 218/378 (56%), Gaps = 22/378 (5%)
Query: 1 MLLQIEQEYSEIYHRKVEDTIKHKDYLNKVLDDFQSEIANIASSLG----EDYVSFSRGK 56
ML+++E+E +IY RKV++ K L++ + ++E+A++ ++LG + +G
Sbjct: 42 MLMELERECLQIYQRKVDEAANSKAKLHQSVASIEAEVASLMAALGVLNINSPIKLDKGS 101
Query: 57 GTLKQQLANIIPVVEDLRFKKKERVKEILEIKSQISQIRAEKAGCGQ--SKGVT-DQDVD 113
+LK++LA + P+VE+LR +K+ER+K+ +IK+QI +I E +G +K + ++
Sbjct: 102 KSLKEKLAAVTPLVEELRIQKEERMKQFSDIKAQIEKISGEISGYSDHLNKAMNISLTLE 161
Query: 114 QCDLTMEKLGKLKSHLNELQNEKNIRHQKVKSHISTISELSAVMSIDISEILNGIHPSLN 173
+ DLT+ L + ++HL LQ EK+ R KV +++ + L V+ +D S+ ++ +HPSL+
Sbjct: 162 EQDLTLRNLNEYQTHLRTLQKEKSDRLNKVLGYVNEVHALCGVLGVDFSQTVSAVHPSLH 221
Query: 174 DSSNGAQQSISDETLARLNESVLLLKQEKQQSLQK----VQFLEELWDFMGITTDEQKAV 229
+ +ISD TL L + LK E++ QK V L ELW+ M T E +
Sbjct: 222 RTDQEQSTNISDSTLEGLEHMIQKLKTERKSRFQKLKDVVASLFELWNLMD-TPQEDRTK 280
Query: 230 CDDVIGLISASVDEVSIHGSLSNDIVKQVDVEVQRLQVLNPSKMKEFVFKRQKELAFKRQ 289
V ++ +S ++ G LS + ++QV EV L L S+M KEL KR+
Sbjct: 281 FGKVTYVVRSSEANITEPGILSTETIEQVSTEVDSLSKLKASRM--------KELVMKRR 332
Query: 290 NELEEIHRGAHMDMDAKAARQILTDLI--GNIDMSELLQGMDKQIRKAKEQAQSRRDIID 347
+ELE++ R H+ D + + T LI G +D SELL ++ QI K K++AQSR+DI+D
Sbjct: 333 SELEDLCRLTHIQPDTSTSAEKSTALIDSGLVDPSELLANIEMQINKIKDEAQSRKDIMD 392
Query: 348 RVAEWKSAAEEEKWLDEY 365
R+ W SA EEE WL+EY
Sbjct: 393 RIDRWLSACEEENWLEEY 410
>ref|NP_178300.2| microtubule associated protein (MAP65/ASE1) family protein
[Arabidopsis thaliana]
Length = 567
Score = 221 bits (564), Expect = 3e-56
Identities = 132/378 (34%), Positives = 218/378 (56%), Gaps = 22/378 (5%)
Query: 1 MLLQIEQEYSEIYHRKVEDTIKHKDYLNKVLDDFQSEIANIASSLG----EDYVSFSRGK 56
ML+++E+E +IY RKV++ K L++ + ++E+A++ ++LG + +G
Sbjct: 1 MLMELERECLQIYQRKVDEAANSKAKLHQSVASIEAEVASLMAALGVLNINSPIKLDKGS 60
Query: 57 GTLKQQLANIIPVVEDLRFKKKERVKEILEIKSQISQIRAEKAGCGQ--SKGVT-DQDVD 113
+LK++LA + P+VE+LR +K+ER+K+ +IK+QI +I E +G +K + ++
Sbjct: 61 KSLKEKLAAVTPLVEELRIQKEERMKQFSDIKAQIEKISGEISGYSDHLNKAMNISLTLE 120
Query: 114 QCDLTMEKLGKLKSHLNELQNEKNIRHQKVKSHISTISELSAVMSIDISEILNGIHPSLN 173
+ DLT+ L + ++HL LQ EK+ R KV +++ + L V+ +D S+ ++ +HPSL+
Sbjct: 121 EQDLTLRNLNEYQTHLRTLQKEKSDRLNKVLGYVNEVHALCGVLGVDFSQTVSAVHPSLH 180
Query: 174 DSSNGAQQSISDETLARLNESVLLLKQEKQQSLQK----VQFLEELWDFMGITTDEQKAV 229
+ +ISD TL L + LK E++ QK V L ELW+ M T E +
Sbjct: 181 RTDQEQSTNISDSTLEGLEHMIQKLKTERKSRFQKLKDVVASLFELWNLMD-TPQEDRTK 239
Query: 230 CDDVIGLISASVDEVSIHGSLSNDIVKQVDVEVQRLQVLNPSKMKEFVFKRQKELAFKRQ 289
V ++ +S ++ G LS + ++QV EV L L S+M KEL KR+
Sbjct: 240 FGKVTYVVRSSEANITEPGILSTETIEQVSTEVDSLSKLKASRM--------KELVMKRR 291
Query: 290 NELEEIHRGAHMDMDAKAARQILTDLI--GNIDMSELLQGMDKQIRKAKEQAQSRRDIID 347
+ELE++ R H+ D + + T LI G +D SELL ++ QI K K++AQSR+DI+D
Sbjct: 292 SELEDLCRLTHIQPDTSTSAEKSTALIDSGLVDPSELLANIEMQINKIKDEAQSRKDIMD 351
Query: 348 RVAEWKSAAEEEKWLDEY 365
R+ W SA EEE WL+EY
Sbjct: 352 RIDRWLSACEEENWLEEY 369
>dbj|BAD37971.1| putative microtubule-associated protein MAP65-1a [Oryza sativa
(japonica cultivar-group)]
Length = 594
Score = 220 bits (561), Expect = 6e-56
Identities = 137/378 (36%), Positives = 234/378 (61%), Gaps = 22/378 (5%)
Query: 1 MLLQIEQEYSEIYHRKVEDTIKHKDYLNKVLDDFQSEIANIASSLGEDYVSFSRGKGT-- 58
+L +IE+E E+Y RKV+D + + L++ + ++E+A++ ++LGE + + K
Sbjct: 38 VLSEIERECLEVYRRKVDDANRTRVQLHQSVATKEAEVASLVATLGEHKLYLKKDKSVVP 97
Query: 59 LKQQLANIIPVVEDLRFKKKERVKEILEIKSQISQIRAEKAGCGQSKGVTDQ-DVDQCDL 117
LK+QLA ++PV+E+L+ KK+ER+K+ +I+SQI +IR+E + + VD+ DL
Sbjct: 98 LKEQLAAVVPVLENLKGKKEERLKQFSDIQSQIEKIRSELSEYSDGDDKANSLIVDENDL 157
Query: 118 TMEKLGKLKSHLNELQNEKNIRHQKVKSHISTISELSAVMSIDISEILNGIHPSLNDSSN 177
+ KL ++ L+ LQ EK+ R KV +++ + L V+ ID + +NGIHPSL+ N
Sbjct: 158 STRKLNNYQAQLHALQKEKSDRLHKVLEYVNEVHCLCGVLGIDFGKTVNGIHPSLH--QN 215
Query: 178 GAQQS--ISDETLARLNESVLLLKQEKQQSLQK----VQFLEELWDFMGITTDEQKAVCD 231
G +QS IS+ TL L ++ LK E++ + K ++ L +LW M + E++ +
Sbjct: 216 GLEQSTNISNSTLEGLANTISNLKAEQRSRIDKMRETMESLCKLWKLMD-SPQEERRQFN 274
Query: 232 DVIGLISASVDEVSIHGSLSNDIVKQVDVEVQRLQVLNPSKMKEFVFKRQKELAFKRQNE 291
V+ ++ +S +E+ G LS + ++++ EV+RL +K+K +R KE+ K+++E
Sbjct: 275 RVLSVLISSEEEILSPGVLSQETIEKMGAEVERL-----TKLKA---RRLKEIFMKKRSE 326
Query: 292 LEEIHRGAHMDMDAKAARQILTDLI--GNIDMSELLQGMDKQIRKAKEQAQSRRDIIDRV 349
LEEI R AH++ DA A + ++I G ID SELL ++ QI KAKE++ SR+DI+DR+
Sbjct: 327 LEEICRSAHIEPDASTAPEQTNEMIDSGMIDPSELLAKLESQILKAKEESLSRKDIMDRI 386
Query: 350 AEWKSAAEEEKWLDEYER 367
+W SA +EE WL+EY +
Sbjct: 387 NKWISACDEEAWLEEYNQ 404
>gb|AAP37732.1| At1g14690 [Arabidopsis thaliana] gi|21539547|gb|AAM53326.1| unknown
protein [Arabidopsis thaliana]
Length = 603
Score = 219 bits (559), Expect = 1e-55
Identities = 132/380 (34%), Positives = 219/380 (56%), Gaps = 22/380 (5%)
Query: 1 MLLQIEQEYSEIYHRKVEDTIKHKDYLNKVLDDFQSEIANIASSLG----EDYVSFSRGK 56
ML+++E+E EIY RKV++ K L++ L ++EIA++ ++LG + G
Sbjct: 42 MLMELEKECLEIYRRKVDEAANSKAQLHQSLVSIEAEIASLLAALGVFNSHSPMKAKEGS 101
Query: 57 GTLKQQLANIIPVVEDLRFKKKERVKEILEIKSQISQIRAEKAGCGQSKGVT---DQDVD 113
+LK++LA + P++EDLR +K ER+K+ ++IK+QI ++ E +G T +D
Sbjct: 102 KSLKEKLAAVRPMLEDLRLQKDERMKQFVDIKAQIEKMSGEISGYSDQLNKTMVGSLALD 161
Query: 114 QCDLTMEKLGKLKSHLNELQNEKNIRHQKVKSHISTISELSAVMSIDISEILNGIHPSLN 173
+ DLT+ KL + ++HL LQ EK+ R KV +++ + L V+ +D + ++ +HPSL+
Sbjct: 162 EQDLTLRKLNEYQTHLRSLQKEKSDRLNKVLDYVNEVHTLCGVLGVDFGQTVSEVHPSLH 221
Query: 174 DSSNGAQQSISDETLARLNESVLLLKQEKQQSLQKVQ----FLEELWDFMGITTDEQKAV 229
+ + +ISD+TL L+ + LK E+ QK++ L ELW+ M T+ E++
Sbjct: 222 RTDHEQSTNISDDTLDGLHHMIHKLKTERSVRFQKLKDVAGSLFELWNLMD-TSQEERTK 280
Query: 230 CDDVIGLISASVDEVSIHGSLSNDIVKQVDVEVQRLQVLNPSKMKEFVFKRQKELAFKRQ 289
V ++ +S +++ LS++ ++QV EV L S+M KEL KR+
Sbjct: 281 FASVSYVVRSSESDITEPNILSSETIEQVSAEVDCFNKLKASRM--------KELVMKRR 332
Query: 290 NELEEIHRGAHMDMDAKAARQILTDLI--GNIDMSELLQGMDKQIRKAKEQAQSRRDIID 347
ELE + R AH++ D + + T LI G +D SELL ++ I K KE+A SR++IID
Sbjct: 333 TELENLCRLAHIEADTSTSLEKSTALIDSGLVDPSELLTNIELHINKIKEEAHSRKEIID 392
Query: 348 RVAEWKSAAEEEKWLDEYER 367
R+ W SA EEE WL+EY +
Sbjct: 393 RIDRWLSACEEENWLEEYNQ 412
>ref|NP_174113.1| microtubule associated protein (MAP65/ASE1) family protein
[Arabidopsis thaliana] gi|25518581|pir||E86404
hypothetical protein F13K9.3 - Arabidopsis thaliana
gi|12322987|gb|AAG51477.1| hypothetical protein
[Arabidopsis thaliana]
Length = 592
Score = 219 bits (558), Expect = 1e-55
Identities = 159/456 (34%), Positives = 253/456 (54%), Gaps = 40/456 (8%)
Query: 1 MLLQIEQEYSEIYHRKVEDTIKHKDYLNKVLDDFQSEIANIASSLGEDYVSFSRGK--GT 58
+LL IEQE E Y RKV+ + L++ L + ++E+ + LGE V K GT
Sbjct: 74 VLLDIEQECVEAYRRKVDHANVSRSRLHQELAESEAELTHFLLCLGERSVPGRPEKKGGT 133
Query: 59 LKQQLANIIPVVEDLRFKKKERVKEILEIKSQISQIRAEKAGCGQSKGVTDQ-DVDQCDL 117
L++QL +I P + ++R +K ERVK+ +K +I +I AE AG + T + +D DL
Sbjct: 134 LREQLDSIAPALREMRLRKDERVKQFRSVKGEIQKISAEIAGRSTYEDSTRKITIDDNDL 193
Query: 118 TMEKLGKLKSHLNELQNEKNIRHQKVKSHISTISELSAVMSIDISEILNGIHPSLNDSSN 177
+ +KL + ++ L+ L +EKN R QKV +I I +LSA + + S I+ IHPSLND
Sbjct: 194 SNKKLEEYQNELHRLHDEKNERLQKVDIYICAIRDLSATLGTEASMIITKIHPSLNDLY- 252
Query: 178 GAQQSISDETLARLNESVLLLKQEKQQSLQKVQFL----EELWDFMGITTDEQKAVCDDV 233
G ++ISD+ L +LN +V+ L++EK + L+K+ L LW+ M + ++++ V
Sbjct: 253 GISKNISDDILKKLNGTVVSLEEEKHKRLEKLHHLGRALSNLWNLMDASYEDRQKFFH-V 311
Query: 234 IGLISASVDEVSIHGSLSNDIVKQVDVEVQRLQVLNPSKMKEFVFKRQKELAFKRQNELE 293
I L+S++ +V GS++ DI++Q + EV+RL L S++KE K+QKEL E
Sbjct: 312 IDLLSSAPSDVCAPGSITLDIIQQAEAEVKRLDQLKASRIKELFIKKQKEL--------E 363
Query: 294 EIHRGAHMDMDAKAARQILTDLI--GNIDMSELLQGMDKQIRKAKEQAQSRRDIIDRVAE 351
+ +HM+ + I T+L+ G +D +LL MD++I +AKE+A SR+ II++V
Sbjct: 364 DTCNMSHMETPSTEMGNI-TNLVDSGEVDHVDLLAAMDEKIARAKEEAASRKGIIEKVDR 422
Query: 352 WKSAAEEEKWLDEYE-------------RFI*SVRRKRGFLSPRFHLW*RI*LQK*KHGR 398
W A++EE+WL+EY+ R + R R +S L I L K K
Sbjct: 423 WMLASDEERWLEEYDQDENRYSVSRNAHRNLRRAERARITVSKISGLVESI-LVKAKSWE 481
Query: 399 QTKKK------VSYMKSLHEYNVQRQLREEEKRKSR 428
++K V + L EYN RQ +E EK++ R
Sbjct: 482 VERQKVFLYNEVPLVAMLQEYNKLRQEKEMEKQRLR 517
>ref|XP_483480.1| putative microtubule-associated protein [Oryza sativa (japonica
cultivar-group)] gi|42407887|dbj|BAD09028.1| putative
microtubule-associated protein [Oryza sativa (japonica
cultivar-group)]
Length = 488
Score = 211 bits (537), Expect = 3e-53
Identities = 130/376 (34%), Positives = 228/376 (60%), Gaps = 21/376 (5%)
Query: 1 MLLQIEQEYSEIYHRKVEDTIKHKDYLNKVLDDFQSEIANIASSLGEDYVSFSRGK--GT 58
+L +IEQE E+Y RKV + L++ L + ++E N+ SLGE K GT
Sbjct: 44 ILQEIEQECQEVYRRKVNSANMSRIQLHQALAESEAEFTNLLLSLGERSFPGRPEKMTGT 103
Query: 59 LKQQLANIIPVVEDLRFKKKERVKEILEIKSQISQIRAEKAGCGQSKGVTDQDVDQCDLT 118
LK+QL +I P +++++ +K+ R+K+ E++++I +I +E AG +++ +T V+Q DL+
Sbjct: 104 LKEQLNSITPALQEMQMRKEARLKQFREVQTEIQRIASEIAGRPENEAIT---VNQEDLS 160
Query: 119 MEKLGKLKSHLNELQNEKNIRHQKVKSHISTISELSAVMSIDISEILNGIHPSLNDSSNG 178
++KL + +S L L+ EK+ R KV+ + I + +M +D S+IL+ +H SL D +N
Sbjct: 161 LKKLEEHQSELQRLKREKSDRLCKVEEYKVLIHNYAKIMGMDPSKILSNVHTSLLDGAND 220
Query: 179 AQ-QSISDETLARLNESVLLLKQEKQQSLQKV----QFLEELWDFMGITTDEQKAVCDDV 233
Q ++ISD+ L +LN V LK+EK Q + K+ + L +LW+ + +E++ +
Sbjct: 221 QQTKNISDDILNKLNTMVQQLKEEKNQRMDKLHSLGKALTKLWNILDTNMEERRPYGEIK 280
Query: 234 IGLISASVDEVSIHGSLSNDIVKQVDVEVQRLQVLNPSKMKEFVFKRQKELAFKRQNELE 293
I +++ + GSL+ + +++++ EVQRL L SKMKE L +Q E++
Sbjct: 281 IYSMTSGSSMLG-PGSLTLETIQKIESEVQRLDHLKASKMKE--------LFMIKQTEIK 331
Query: 294 EIHRGAHMDMDAKAARQILTDLI--GNIDMSELLQGMDKQIRKAKEQAQSRRDIIDRVAE 351
EI + +HMDM + + D+I G++D +LL+ MD+ I K KE+A SR++I+D+V +
Sbjct: 332 EICKKSHMDMPYQTEMNKIMDVIMSGDVDHDDLLKTMDEYIYKVKEEATSRKEIMDKVEK 391
Query: 352 WKSAAEEEKWLDEYER 367
W ++ +EE+WL+EY R
Sbjct: 392 WMASCDEERWLEEYSR 407
>ref|XP_470643.1| Unknown protein [Oryza sativa (japonica cultivar-group)]
gi|27357984|gb|AAO06976.1| Unknown protein [Oryza sativa
(japonica cultivar-group)]
Length = 607
Score = 209 bits (532), Expect = 1e-52
Identities = 126/376 (33%), Positives = 219/376 (57%), Gaps = 18/376 (4%)
Query: 1 MLLQIEQEYSEIYHRKVEDTIKHKDYLNKVLDDFQSEIANIASSLGEDYVSFSRG-KGTL 59
M L++E E +Y RKV+ + L + L ++E+ + +S+GE F K +L
Sbjct: 46 MFLELETECMRVYRRKVDSANAERSQLRQSLMAKEAELKVLVASIGEITPKFKVDEKQSL 105
Query: 60 KQQLANIIPVVEDLRFKKKERVKEILEIKSQISQIRAEKAGCGQS--KGVTDQDVDQCDL 117
K+QLA + P++EDLR KK+ER+K+ ++SQI +I+A+ + G + D DL
Sbjct: 106 KEQLAKVTPLLEDLRSKKEERIKQFSLVQSQIEKIKAQISDHNNQHDNGPVNHSKDNHDL 165
Query: 118 TMEKLGKLKSHLNELQNEKNIRHQKVKSHISTISELSAVMSIDISEILNGIHPSLNDSSN 177
+ +L L++ L LQ EK+ R QKV ++ + L +V+ +D ++ + +HPSL+ +++
Sbjct: 166 STRRLSDLQAELRNLQKEKSDRLQKVFIYVDEVHCLCSVLGMDFAKTVKDVHPSLHGANS 225
Query: 178 GAQQSISDETLARLNESVLLLKQEKQQSLQKVQ----FLEELWDFMGITTDEQKAVCDDV 233
+ISD TL L E++L LK EK+ + K+Q L +LW+ M +T++++ V
Sbjct: 226 ENSTNISDSTLEGLTETILKLKAEKRTRVSKLQEIVGKLHKLWNLME-STEQERRHFTRV 284
Query: 234 IGLISASVDEVSIHGSLSNDIVKQVDVEVQRLQVLNPSKMKEFVFKRQKELAFKRQNELE 293
++ ++ +E++ LS + +++ + EV+RL S+MKE V K++ ELE
Sbjct: 285 AAVLGSTEEEITSSSVLSLETIQETEEEVERLTKQKASRMKELVLKKRL--------ELE 336
Query: 294 EIHRGAHMDMDAKAARQILTDLI--GNIDMSELLQGMDKQIRKAKEQAQSRRDIIDRVAE 351
+I AHM+ D A + +T LI G +D ELL ++ QI KA+E++ +R+DI+++V
Sbjct: 337 DICSNAHMEPDMSTAPEKITALIDSGLVDPCELLSSIETQIAKAREESLTRKDIMEKVDR 396
Query: 352 WKSAAEEEKWLDEYER 367
W SA +EE WL+EY +
Sbjct: 397 WLSACDEETWLEEYNQ 412
Database: nr
Posted date: Jul 5, 2005 12:34 AM
Number of letters in database: 863,360,394
Number of sequences in database: 2,540,612
Lambda K H
0.321 0.135 0.371
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 613,466,761
Number of Sequences: 2540612
Number of extensions: 23263276
Number of successful extensions: 111791
Number of sequences better than 10.0: 2013
Number of HSP's better than 10.0 without gapping: 78
Number of HSP's successfully gapped in prelim test: 1988
Number of HSP's that attempted gapping in prelim test: 107771
Number of HSP's gapped (non-prelim): 4982
length of query: 428
length of database: 863,360,394
effective HSP length: 131
effective length of query: 297
effective length of database: 530,540,222
effective search space: 157570445934
effective search space used: 157570445934
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 76 (33.9 bits)
Medicago: description of AC137821.2


